Abstract
The superfamily of tripartite motif-containing (TRIM) proteins is conserved throughout the metazoan kingdom and has expanded rapidly during vertebrate evolution; there are now more than 60 TRIM proteins known in humans and mice. Many TRIM proteins are induced by type I and type II interferons, which are crucial for many aspects of resistance to pathogens, and several are known to be required for the restriction of infection by lentiviruses. In this Review, we describe recent data that reveal broader antiviral and antimicrobial activities of TRIM proteins and discuss their involvement in the regulation of pathogen-recognition and transcriptional pathways in host defence.
Innate immune responses are triggered by the engagement of Toll-like receptors (TLRs) or other pattern-recognition receptors (PRRs) by bacterial, viral or fungal components1,2. Ligation of PRRs initiates various immune responses, including the production of cytokines and the initiation of pro-inflammatory and adaptive immune responses. Interferons (IFNs) are produced in response to these stimuli and are often crucial for the induction of effective immunity3–6.
Recent studies have shown that many members of the tripartite motif-containing (TRIM) superfamily are expressed in response to IFNs and are involved in a broad range of biological processes that are associated with innate immunity7,8. The TRIM or RBCC motif, which defines this superfamily, comprises a RING domain, one or two B-box domains and an associated coiled-coil domain in the amino-terminal region7,9. The presence of a RING domain, which can mediate the conjugation of proteins with ubiquitin, with small ubiquitin-like modifier (SUMO) or with the ubiquitin-like molecule IFN-stimulated protein of 15 kDa (ISG15), contributes to the biological flexibility of TRIM proteins. In addition, TRIM proteins often self-associate through their coiled-coil domains to form large protein complexes that occupy discrete cytoplasmic or nuclear subcompartments7. In addition to their role in innate immunity, TRIM proteins are involved in a broad range of biological processes, including some that underlie genetic disorders, neurological disorders and cancers10. TRIM proteins are present in all metazoans, but the number of family members varies between species; there are more members in humans (65 members; TABLE 1, see Supplementary information S1 (figure)) and mice (64 members) than in worms (~20 members) and flies (<10 members), which suggests that the family has evolved extensively. It is of note, however, that the precise number of TRIM family members in each species is currently uncertain owing to the divergent domain composition of the family members. Although it is clear that they arise from a common ancestral gene, TRIM genes have evolved independently, as shown by their generally scattered presence throughout the genome and by the acquisition of species-specific functions.
Table 1.
Schematic structures of tripartite motif-containing protein families*
| Family‡ | N-terminal region (RBCC motif)|| | C-terminal region | Family members |
|---|---|---|---|
| C-I |
|
|
MID1, MID2, TRIM9, TRIM36, TRIM46, TRIM67 |
| C-II |
|
|
TRIM54, TRIM55, TRIM63 |
| C-III |
|
|
TRIM42 |
| C-IV |
|
|
TRIML1, TRIM4, TRIM5α, TRIM6, TRIM7, TRIM10, TRIM11, TRIM15, TRIM17, TRIM21, TRIM22, TRIM25, TRIM26, TRIM27, TRIM34, TRIM35, TRIM38, TRIM39, TRIM41, TRIM43, TRIM47, TRIM48, TRIM49, TRIM50, TRIM53, TRIM58, TRIM60, TRIM62, TRIM64, TRIM65, TRIM68, TRIM69, TRIM72, TRIM75 |
| C-V |
|
|
PML, TRIM8, TRIM31, TRIM40, TRIM52, TRIM56, TRIM61, TRIM73, TRIM74 |
| C-VI |
|
|
TRIM24, TRIM28, TRIM33 |
| C-VII |
|
|
TRIM2, TRIM3, TRIM32, TRIM71 |
| C-VIII |
|
|
TRIM37 |
| C-IX |
|
|
TRIM23 |
| C-X§ |
|
|
TRIM45 |
| C-XI§ |
|
|
TRIM13, TRIM59 |
A more detailed structure for each human tripartite motif-containing (TRIM) family member is presented in Supplementary information S1 (figure).
The classification of nine human TRIM protein families denoted C-I to C-IX is based on differences in carboxy-terminal-domain composition as defined by Short and Cox36.
Two new families have been added to reclassify TRIM proteins based on new sequence information.
Dotted outlines indicate domains that are not present in all TRIM family members.
ARF, ADP ribosylation factor-like; B, B-box; BR, bromodomain; CC, coiled-coil; COS, C-terminal subgroup one signature; FN3, fibronectin type 3; FIL, filamin-type immunoglobulin; MATH, meprin and tumour-necrosis factor receptor-associated factor homology; MID, midline; N-terminal, amino-terminal; PHD, plant homeodomain; PML, promyelocytic leukaemia; R, RING finger; TM, transmembrane.
In this Review, we discuss recent findings that illustrate the involvement of TRIM proteins in innate immunity and, in some cases, in genetic diseases and autoimmunity. We focus on the recent explosion of information on the effects of TRIM proteins on the life cycle of HIV and their involvement in the signalling pathways that are triggered by TLRs and other PRRs.
TRIM proteins
The most striking feature of TRIM superfamily proteins is the highly conserved order of domains in the RBCC motif10–12 (TABLE 1). The conservation of this domain ordering in TRIM proteins from various species indicates that the RBCC motif is the defining characteristic of this superfamily. In family members that lack one of the domains of the RBCC motif, the other domains are conserved in order and spacing. Structural analyses of these domains in several family members have greatly increased our understanding of the contribution of each domain to TRIM protein function.
The RING domain
The RING domain is a zinc-binding motif that is located within 10–20 amino acids of the first methionine in the N-terminal portion of almost all TRIM proteins7,9. General insights into the function of RING domains came primarily from studies of the RING-domain-containing protein CBL (Casitas B-lineage lymphoma), which showed that RING domains mediate ubiquitylation events13–15. Crystal and solution structures for RING domains of TRIM family members have recently been described (see Supplementary information S2 (figure)). The RING domains of many TRIM family members, including TRIM5α, TRIM8, TRIM11, TRIM21 (also known as RO52), TRIM22 and TRIM25, have now been shown to confer E3 ubiquitin ligase activity, which allows them to mediate ubiquitylation events16–22. The E3 ubiquitin ligase activity of the RING domain is now known to be crucial for the anti-HIV functions of some TRIM proteins and for mediating their effects on signalling pathways following innate immune receptor triggering (TABLE 2; see later). Interestingly, TRIM25 can also modify itself and other proteins by conjugating ISG15 in a RING-domain-independent manner23,24. Furthermore, the RING domains of both promyelocytic leukaemia (PML; also known as TRIM19) and TRIM63 are known to associate with the SUMO-conjugating enzyme UBE2I (ubiquitin-conjugating enzyme E2I), which suggests that they might be involved in sumoylation10.
Table 2.
Tripartite motif-containing proteins
| Gene | Alias | Cytoband | Isoforms | Cellular location* | Expression in response to IFNγ and LPS‡ | E3 ubiquitin ligase activity | Disease association | Refs |
|---|---|---|---|---|---|---|---|---|
| MID2 | TRIM1 | Xq22 | 2 | Cytoplasm, microtubules | ↔ | ND | Candidate for X-linked FG syndrome | 85,122 |
| TRIM5 | NA | 11p15 | 3 | Cytoplasm | ↑ | Yes | ND | 44,85 |
| TRIM8 | NA | 10q24.3 | 1 | ND | ↑ | Yes | ND | 21,85 |
| TRIM11 | NA | 1q42.13 | 1 | Cytoplasm, nucleus | ND | Yes | Contributes to Alzheimer’s- disease-like neuronal insults | 18,72 |
| TRIM15 | NA | 6p21.3 | 1 | ND | ↔ | ND | ND | 85 |
| MID1 | TRIM18 | Xp22 | 1 | Cytoplasm, microtubules | ↑ | Yes | Mutated in X-linked Opitz G/BBB syndrome | 29,85,123 |
| PML | TRIM19 | 15q22 | 9 | Nucleus | ↑ | ND | PML–RAR fusion oncogene in AML | 85,124 |
| MEFV | TRIM20 | 16p13.3 | 1 | Cytoplasm, microtubules, nucleus | ↔ | ND | Mutated in FMF | 47,48, 85,125 |
| TRIM21 | RO52 | 11p15.5 | 1 | Cytoplasm, nucleus | ↑ | Yes | Encodes RO52 autoantigen in SLE and Sjögren’s syndrome | 85, 104,105 |
| TRIM22 | NA | 11p15 | 1 | Nucleus | ↑ | Yes | ND | 22,85 |
| TRIM25 | NA | 17q23.2 | 1 | ND | ↑ | Yes | ND | 85,96 |
| TRIM26 | NA | 6p21.3 | 1 | ND | ↑ | ND | ND | 85 |
| TRIM27 | NA | 6p22 | 1 | Cytoplasm, nucleus | ↓ | ND | TRIM27–RET fusion oncogene in thyroid cancer | 85,126 |
| TRIM28 | TIF1B | 19q13.4 | 1 | Nucleus | ↔ | Yes | ND | 55,85 |
| TRIM31 | NA | 6p21.3 | 1 | ND | ↑ | ND | ND | 85 |
| TRIM32 | NA | 9q33.1 | 1 | Nucleus | ↓ | Yes | Mutated in limb-girdle muscular dystrophy | 85, 120,127 |
| TRIM34 | NA | 11p15 | 3 | ND | ↑ | ND | ND | 85 |
| TRIM38 | NA | 6p21.3 | 1 | ND | ↑ | ND | ND | 85 |
| TRIM62 | NA | 1p35.1 | 1 | ND | ↑ | ND | ND | 85 |
| TRIM63 | MURF1 | 1p34–p33 | 1 | Cytoplasm, nucleus | ND | Yes | ND | 121 |
| TRIM68 | SS-56 | 11p15.4 | 1 | Cytoplasm | ↑ | ND | Encodes SS-56 autoantigen in SLE and Sjögren’s syndrome | 85,117 |
Based on UniProt Consortium database.
↑ indicates expression is induced by IFNγ and LPS; ↓ indicates expression is downregulated; ↔ indicates expression is unaffected.
AML, acute myeloid leukaemia; FMF, familial Mediterranean fever; IFNγ, interferon-γ; LPS, lipopolysaccharide; MEFV, Mediterranean fever; MID, midline; MURF1, muscle ring-finger protein 1; NA, not applicable; ND, not determined; PML, promyelocytic leukaemia; SLE, systemic lupus erythematosus; TIF1B, translation-initiation factor 1B; TRIM, tripartite motif-containing.
The B-box domains
The B-box domains are also zinc- binding motifs, and B-box 1 and B-box 2 have different consensus sequences between members of the TRIM superfamily. TRIM proteins can contain one or both of these elements, although B-box 1 is never present without B-box 2 (TABLE 1). Structural studies of several human TRIM B-box 1 and B-box 2 domains have shown that they have ternary structures that are similar to those of RING domains, which suggests that all three domains have evolved from a common ancestral domain (see Supplementary information S2 (figure)).
B-box domains have been shown to contribute to innate resistance to HIV and to the development of a genetic disorder that shows a mendelian pattern of inheritance. More specifically, studies of the HIV restriction factor TRIM5α showed that B-box 2 influences the recognition of the viral capsid by the carboxy-terminal PRYSPRY domain26 and the ability of TRIM15 to mediate HIV restriction27. In addition, mutations in both B-box domains of midline 1 (MID1; also known as TRIM18) have been identified in patients with X-linked Opitz G/ BBB syndrome28,29 (TABLE 2). This syndrome is characterized by malformations of the ventral midline, including increased distance between the eyes and abnormally placed opening of the male urethra. Most MID1 mutations in patients with this syndrome are found in the C-terminal fibronectin type 3 and PRYSPRY domains (see later), with fewer mutations found in the B-box 1 or coiled-coil domains30.
MID1 is generally present in the cytoplasm and associated with microtubules. The RING domain of activated MID1 has E3 ubiquitin ligase activity, which mediates the ubiquitylation of the microtubule-associated phosphatase PP2AC (protein phosphatase 2, catalytic subunit, α isoform), which leads to PP2AC proteasomal degradation and displacement from microtubules29. The mutations in MID1 result in its cytosolic accumulation and in the aberrant localization of PP2AC, thereby altering the normal regulatory functions of PP2AC in epithelial–mesenchymal-cell transition, programmed cell death and cell migration, all of which are crucial for the proper development of midline structures.
The coiled-coil domain
The B-box domains are almost always followed by a coiled-coil domain, which is a motif found in many proteins families. This domain mediates homomeric and heteromeric interactions among TRIM family members and other proteins, in particular self-association. Sequence and structural studies have shown that these domains are diverse and that they vary from structures with a single continuous coil to ones with three contiguous but non-continuous coiled subdomains (see Supplementary information S1, S2 (figures)). Protein–protein interactions involving coil-coiled domains promote the generation of high-molecular-mass complexes31 that, among other functions, define specific subcellular structures7. Perhaps the best known example is the assembly of macromolecular nuclear structures that are termed nuclear bodies, which are composed of the sumoylated scaffolding protein PML and numerous other proteins that have been implicated in various cellular processes (recently reviewed in REFS 32,33). In addition, it has been established that the coiled-coil domain of TRIM5α is required for trimerization and has an important role in the restriction of viral infectivity by this TRIM protein34,35.
Classification and C-terminal domains
The classification of human TRIM family members, which was devised by Short and Cox36, is based on the composition of sequences in the C-terminal region in relation to the RBCC motif. The C-terminal region can contain any of 10 distinct motifs alone or in combination, which results in the designation of 9 families, C-I to C-IX36 (TABLE 1). This classification system highlights the association of particular domains with: specific subcellular localization (for example, families with members that contain the COS (C-terminal subgroup one signature) domain localize with microtubules); cell-specific expression (C-II family members are expressed by muscle cells); and function (C-VI family members are associated with transcriptional regulation). We have provisionally added two new families to this classification system: the C-X family and the C-XI family. More specifically, the C-X family includes TRIM45, which was previously included in the C-VII family but is no longer thought to have NHL repeats. In addition, the C-XI family includes TRIM13 and TRIM59, which were previously included in the C-IV family (members of which do not have a recognized C-terminal domain) but were recently shown to have C-terminal transmembrane domains. If other TRIM proteins are found to have transmembrane domains, we propose that they should fall into this category. At least nine other proteins appear as TRIM family members in searches of various databases, such as those of the HUGO Gene Nomenclature Committee or the UniProt Consortium. However, these proteins are not considered to be true TRIM proteins as they all lack RING domains and other elements of the canonical RBCC motif.
The COS box
The COS box is localized immediately downstream of the coiled-coil sequences in a subset of TRIM family members, but also in several non-TRIM proteins36. Studies have shown that this sequence is responsible for mediating binding to microtubules33. MID2 (also known as TRIM1), which has a COS box, has been shown to have antiretroviral activity37,38.
The PRY and SPRY domains
The C-terminal sequences that are most commonly found in TRIM family members are the ~61-amino-acid PRY and ~140-amino-acid SPRY domains. These domains are present in 11 different human protein families, but most prominently in human and mouse TRIM family members39,40. The SPRY domain is found alone in 39 human TRIM family members, and is fused to the PRY domain (to form the PRYSPRY domain; also referred to as B30.2) in 24 family members (see Supplementary information S1 (figure)). The SPRY domain is evolutionarily ancient and can be found in animals, plants and fungi. By comparison, the PRYSPRY domain is found only in vertebrate species, including humans, mice, chickens and frogs41. The recent evolution of SPRY to PRYSPRY and the expansion in the number of genes that contain this domain has interesting parallels to the evolution and expansion of immune receptors that occurred after the appearance of adaptive immune mechanisms42,43. The possibility that this expansion is indicative of selective pressures for immune defence is suggested by the contribution of the PRYSPRY domain TRIM5 orthologues to the innate immune restriction of retroviruses in various species19,34,38,44–46.
Analysis of the crystal structure of the PRYSPRY domain reveals that it is a dimer in which a donor sequence from one molecule binds to an acceptor sequence from the other molecule to form a putative interaction site, which is similar to the interaction seen in antigen–antibody complexes40. This suggests that the PRYSPRY domain can bind to target proteins that present donor strands with different sequences, and might imply that sequence polymorphisms in the donor–acceptor sequences of the 24 PRYSPRY-domain-containing TRIM proteins may specify discrete sets of target proteins that uniquely contribute to the function of each member.
Strikingly, mutations in the non-conforming (that is, does not contain a RING domain) TRIM protein MEFV (Mediterranean fever; also known as TRIM20) that are responsible for the hereditary disease familial Mediterranean fever (FMF)47,48 (TABLE 2) map precisely to this interaction site, which suggests that these mutations interfere with the ability of the PRYSPRY domain to interact with its targets. How the FMF-associated mutations lead to increased inflammation is not yet clear, but it is possible that they alter the activity of pyrin, which is encoded by MEFV, thereby leading to the dysregulated activation of interleukin-1β (IL-1β) and nuclear factor-κB (NF-κB)48.
Our understanding of how PRY and SPRY sequences interact to exert their combined biological effects is still limited. However, recent structural studies of the PRYSPRY-containing autoantigen TRIM21 showed that it is a unique Fc receptor for IgG, and that binding of TRIM21 to the immunoglobulin heavy chain sequences of IgG is mediated through two binding pockets, one in the PRY domain and the other in the SPRY domain12,49 (see Supplementary information S2 (figure)). This binding mechanism is unlike that of known mammalian Fc receptors for IgG and might allow TRIM21 to crosslink the B-cell receptor, thereby resulting in T-cell-independent B-cell activation as a trigger for autoimmunity.
Other C-terminal domains
The C terminus of TRIM family members can also contain various other domains (TABLE 1). More specifically, fibronectin type 3 (FN3) domains, which can contain binding sites for DNA and heparin, are found in some TRIM proteins. The C-VI TRIM family proteins have plant homeodomains (PHDs), which are found in nuclear proteins and are thought to be involved in chromatin-mediated transcriptional regulation. In TRIM proteins, PHDs are always paired with bromodomains, which recognize acetylated lysine residues such as those on the N-terminal tails of histones. This pairing mediates transcriptional repression50–53. Moreover, domains with multiple NHL repeats of 40 residues can be involved in protein–protein interactions. In addition, filamin-type immunoglobulin domains might be involved in dimerization and actin crosslinking. Finally, ADP ribosylation factor-like (ARF) domains are involved in intracellular vesicular trafficking, and a MATH (meprin and tumour-necrosis factor receptor (TNFR)-associated factor (TRAF) homology) domain is necessary and sufficient for self-association and receptor interactions by some TRAF proteins. The contribution of many of these domains to the regulation of innate immune responses has yet to be established, but the contribution of other domains is well known.
Expression patterns of TRIM proteins
The tissue distribution of mRNAs and proteins that are encoded by various TRIM genes is extremely heterogeneous7,36 (see Tigem and GeneCards websites). Nearly half the genes are ubiquitously expressed in adult tissues, whereas the expression patterns for the other genes can vary from a single tissue to multiple organs. Mouse Trim30 is the only TRIM gene that is known to have restricted expression in adult haematopoietic tissue and is only expressed in the spleen7.
By using enhanced green fluorescent protein (EGFP)–TRIM fusion proteins, several distinct expression patterns of TRIM proteins in the cytoplasm or nucleus of transfected cells have been observed7,36. Co-localization studies using these fusion proteins and specific markers for well-defined cytoplasmic and nuclear structures (such as mitochondria, Golgi apparatus and spliceosomes) showed that all the TRIM proteins, with the exception of MID1 and MID2, occupy previously unrecognized subcellular compartments7,36. Mutations that altered the coiled-coil domains resulted in a diffuse redistribution of these proteins, which supports a role for the coiled-coil domain in TRIM protein multimerization. Furthermore, proteins with mutations in the RING or B-box domains either showed altered localization or formed aberrant structures. These observations indicate that individual TRIM proteins may function in unique subcellular compartments.
It should be noted that many TRIM genes have differentially spliced isoforms, which can exhibit different subcellular distributions and protein–protein interactions7. For example, there are approximately nine or more isoforms of PML that can localize to either the nucleus or the cytoplasm, and this differential localization is responsible for achieving different functions33,54. In addition, individual isoforms might be differentially susceptible to post-translational modifications, including ubiquitylation, sumoylation, ISGylation and phosphorylation, which can markedly affect protein expression levels55 and subcellular localization56.
TRIM proteins and retroviral infections
A recent study showed that HIV requires more than 250 host factors for successful infection27. In addition to host factors that are hijacked by viruses, many elements of the innate immune system are directed at containing viral spread before the adaptive immune system becomes involved. Some of the factors that are used by the innate immune system to confer resistance to retro-viruses include the mouse retroviral restriction factor Fv1 (Friend-virus-susceptibility factor 1)57,58 and a family of cytidine deaminases that is exemplified by human APOBEC3G (apolipoprotein-B mRNA-editing enzyme, catalytic polypeptide-like 3G)59,60. Interestingly, rhesus monkey TRIM5α (rhTRIM5α) was the first prominent example of species-specific retroviral restriction by a component of the innate immune response; rhTRIM5α functions as the main inhibitor of HIV-1 replication in Old World monkey cells44. Although emerging roles for additional TRIM proteins in restricting infection by HIV and other retroviruses have been previously described11,61, a series of studies that was published this year has greatly expanded our understanding of the function of several TRIM proteins with antiretroviral activity22,27,38. Although much is known about the relationship between mouse TRIM proteins and murine leukaemia viruses (MLV), in this Review we mainly discuss HIV.
Restricting HIV infection and release
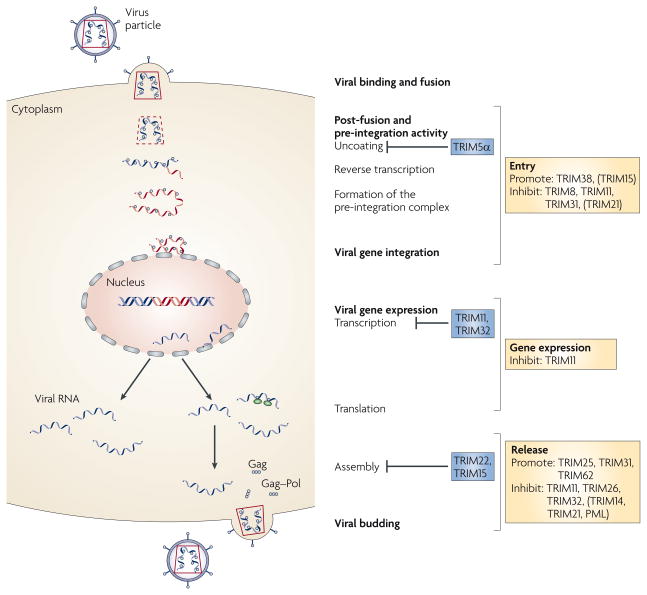
The most extensive analyses of the effects of TRIM proteins on retro-viral infections have come from a study of the effects of human and mouse TRIM proteins on cell binding, fusion and budding of HIV, MLV and avian leukosis viruses38. The results of this and other studies have identified numerous TRIM family members that interfere with several stages of the HIV life cycle. This life cycle can be summarized as viral binding and fusion, post-fusion and pre-integration activity, viral gene integration, viral gene expression and viral budding (FIG. 1).
Figure 1. Effects of human and mouse TRIM proteins on infection with HIV.
Stages in the life cycle of HIV, from cell binding and fusion to release from the cell11,22,27,118, are depicted on the left. Human tripartite motif-containing (TRIM) family members that are known to inhibit specific stages of the cycle are shown alongside. TRIM family members that are known to affect viral entry, viral gene expression or viral release, but for which the exact stage affected is not known, are shown on the right. Here, the definition of viral entry includes events from binding and fusion of the virus to its cellular receptors to the expression of viral proteins. Viral release relates to the presence of infectious virus or virus-encoded proteins in the supernatant of cultured infected cells22,38. The TRIM proteins that affect gene expression include those that might affect viral transcription or translation from the integrated provirus. Proteins with moderate inhibitory or promoting effects are listed in parentheses. PML, promyelocytic leukaemia.
For most TRIM proteins, the exact point of intersection with the viral life cycle and the precise mechanisms of action are not known. It should be noted that almost all the current information on the effect of TRIM proteins on HIV infection comes from studies of a limited number of cell lines in which the TRIM proteins were overexpressed or knocked down. Therefore, the current information may not relate to events in different cell lineages, states of differentiation or activation, functions of alternative transcripts or varying post-translational modifications. With this caveat in mind, several generalizations can be made. First, distinct TRIM proteins function at specific stages of the HIV life cycle. This finding is in keeping with the idea that mammalian species have been subject to selection pressures by similar pathogenic retroviruses, even though no mouse lentiviruses have been identified. Second, although the effects of most TRIM proteins are inhibitory, a minority can function to promote HIV infection38. Third, individual TRIM proteins, such as TRIM11 and TRIM22, can inhibit different stages of the viral life cycle. Fourth, evolutionary sequence conservation between humans, mice and other species, such as rhesus monkeys, cannot necessarily be equated with conservation of function41.
The effects of individual TRIM family members on specific steps of the HIV life cycle include: interference with uncoating of the viral pre-integration complex by human TRIM5α and rhTRIM5α but not by the mouse homologue19,34,38,44; repression of viral gene expression by TRIM11 and TRIM32 (and possibly by TRIM22, although these findings are debated)38,62,63; and inhibition of viral assembly by TRIM22 and TRIM15 (REFS 22,38).
TRIM5α-mediated restriction of uncoating of the viral pre-integration complex involves the recognition of sequences that are present on the viral capsid in the context of an intact mature viral core by the PRYSPRY domain64,65. Although human TRIM5α is much less effective than rhTRIM5α in restricting HIV infection, it was found that a single amino-acid substitution in the PRY domain of the human sequence conferred potent HIV restriction ability to human TRIM5α (REFS 66,67), even though the 52-residue region of the PRY domains of the monkey and human proteins still differed by 14 amino acids67.
The gene that encodes human TRIM5α is located on chromosome 11, in a small cluster of closely related TRIM genes (TRIM22, TRIM6 and TRIM34) (TABLE 2). Interestingly, evolutionary genetic analyses showed that TRIM5 and TRIM22 have evolved under positive selection, whereas the linked TRIM6 and TRIM34 genes have not. This selection has been directed to the portion of the PRYSPRY domain that is involved in the recognition of capsid proteins68, such that TRIM22 specifically binds to the HIV capsid and induces the inhibition of HIV-particle production22. By contrast, mutations in the common human TRIM5α variants that are structurally remote from those that were found to be evolutionarily selected had no effect on HIV disease progression69 and therefore are not indicative of selective pressures.
TRIM5α is normally located in distinct, dynamic cytoplasmic clusters that are termed cytoplasmic bodies, and its expression is rapidly turned over. Following infection with HIV, these cytoplasmic bodies form around the HIV pre-integration complexes, and this is a requisite step in retroviral restriction70. In the cytoplasmic bodies, TRIM5α, which has E3 ubiquitin ligase activity, becomes autoubiquitylated and is rapidly degraded by the proteasome, whereas the pre-integration complexes seem to remain ubiquitin-free. The insertion of TRIM5α into the capsid lattice of the viral cores, and the subsequent displacement of the capsid lattice during proteasomal degradation of TRIM5α, could destabilize it, thereby causing it to dissociate into individual capsid proteins in the absence of direct capsid protein degradation70,71. TRIM5α-mediated loss of the capsid protein is thought to be proteasome dependent, as the HIV–TRIM5α complexes remain intact in the presence of a proteasome inhibitor65. Unexpectedly, the levels of TRIM5α expression in cultured cells are negatively affected by TRIM11 and TRIM34, which have been found to co-localize with TRIM5α in cytoplasmic bodies38. It has been suggested that TRIM11 might achieve this effect by promoting the degradation and repressing the transcription of TRIM5α, similar to its effects on other cellular proteins18,72. It is not known whether repression of TRIM5α expression by TRIM11 and TRIM34 facilitates HIV replication in vivo.
Recently, an active TRIM5 protein was identified in rabbits45. Rabbit TRIM5 was found to be closely related to the TRIM5 proteins of primates and cattle, and similarly restricts the replication of multiple unrelated retroviruses, which suggests that they evolved from a common ancestral gene. The finding that the genome of rabbits contains an endogenous type K lentivirus73 indicates that lentiviruses might have been the driving force that was responsible for the conserved antiviral activities of TRIM5 orthologues.
Domain-dependent effects of TRIM proteins on HIV
Although many TRIM proteins contain E3 ubiquitin ligase activity (TABLE 2), the importance of this function in restricting HIV replication has only recently been assessed38. The antiviral activities of TRIM11 (REF. 38) and TRIM22 (REF. 22) were found to be crucially dependent on a functional E3 ubiquitin ligase (RING) domain. TRIM62 was shown to promote the release of HIV from infected cells, and a TRIM62 protein that has a mutation in its RING domain was more active than the native protein in promoting HIV release38. However, the inhibitory activity of TRIM15 on viral release was unaffected by the loss of E3 ubiquitin ligase activity38.
Intermolecular and intramolecular interactions have also been shown to be important for the functions of individual TRIM family members. The antiviral function of TRIM15 was found to require the interaction of its B-box domain with the HIV Gag precursor protein40. In addition, HIV capsid recognition by the TRIM5α SPRY domain was significantly affected by alterations in B-box 2 sequences26. By contrast, TRIM5α trimerization, which is required for the formation of cytoplasmic bodies, is dependent on the coiled-coil domain34.
PML, one of the best studied TRIM family members, also inhibits the growth of numerous RNA and DNA viruses32,33. PML is required for the formation of nuclear bodies that contain a large number of proteins, some of which are present constitutively, whereas others are present only transiently. PML is expressed as a family of alternatively spliced isoforms that are associated with specific activities, which include the cellular processes of oncogenesis, DNA-damage and stress responses, apoptosis, senescence, and resistance to viral infections. PML expression is induced by IFNs in many cells74. Induction of PML expression by macrophages and dendritic cells (DCs) requires IFN-regulatory factor 8 (IRF8)75, as well as several other proteins from the nuclear bodies, including SP100 and p53. Conjugation of SUMO to PML is required for its localization to the nuclear bodies76.
The contributions of PML and nuclear bodies to defence against various RNA viruses is conferred through p53-dependent and p53-independent pathways. More specifically, resistance to infection by the human foamy retrovirus, vesicular stomatitis virus, rabies virus, lymphocytic choriomeningitis virus and influenza virus is p53 dependent, whereas resistance to infection by polio virus is p53 independent32. A role for PML in the defence against RNA viruses is supported by the observation that some of the viruses have evolved distinct mechanisms to counteract the repressive effects of PML and nuclear bodies77,78.
The relationship between DNA viruses and PML and nuclear bodies is more complex. This is because nuclear bodies are closely associated with ‘replication compartments’ for viral DNA, and regulatory proteins of large DNA viruses (such as adenoviruses and herpes viruses) sometimes localize to and disrupt the structure of the nuclear bodies32,79. In addition, the genomes of smaller DNA viruses that do not disrupt nuclear bodies are sensitive to cell-mediated repression. Interestingly, herpes simplex virus encodes a regulatory protein, ICP0 (also known as RL2), that has a RING domain with E3 ubiquitin ligase activity. ICP0 localizes to nuclear bodies, degrades PML and blocks PML-mediated inhibition of viral gene expression and virus production80.
The relationship between PML, nuclear bodies and RNA or DNA viruses must be interpreted with caution as it is often based on the study of only some PML isoforms and at expression levels that may not be physiological, such as those based on overexpression studies or on studies using PML- or p53-deficient mice32. It is, however, clear that PML and its involvement in the structure and function of nuclear bodies influence a wide range of cellular pathways that are involved in the defence against many RNA and DNA viruses.
TRIM proteins and IFN
Type I and type II IFNs have potent antiviral and antimicrobial activities3,4,81. Type I IFNs (IFNα and IFNβ) are produced in large amounts by plasmacytoid DCs and other types of DC82–84, as well as by other cell types after viral infection. Type II IFN (IFNγ) is synthesized by natural killer (NK) cells and T cells in response to IL-12 and type I IFNs. Both types of IFN act through JAK (Janus kinase)–STAT (signal transducer and activator of transcription) signalling pathways and induce the transcription of a large, overlapping set of target genes that collectively establish innate immunity3–6. IFNs have a particularly important role in activating many of the innate immune functions of macrophages and DCs, and in promoting the initiation of adaptive immunity. In addition, IFNs induce the expression of many TRIM family members (FIG. 2, TABLE 2).
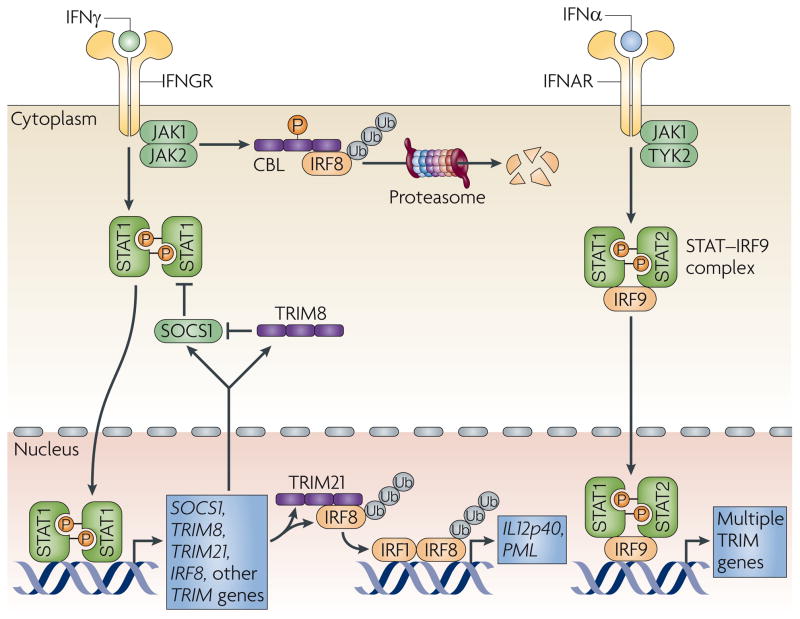
Figure 2. TRIM proteins in IFN signalling.
Interferon-γ (IFNγ) and type I IFNs (IFNα and IFNβ) activate STAT1 (signal transducer and activator of transcription 1) and the STAT–IRF9 (IFN-regulatory factor 9) complex, respectively, which leads to the induction of transcription of many tripartite motif-containing (TRIM) genes8,85 (see also TABLE 2). TRIM8 activation, which is induced by IFNγ and STAT1, blocks suppressor of cytokine signalling 1 (SOCS1), a negative regulator of IFNγ signalling21. TRIM21, which is activated by IFNs, interacts with IFN-induced IRF8, which leads to its ubiquitylation105. Ubiquitylated IRF8 then increases transcription from the promoter of IL12p40 (interleukin-12, p40 subunit) and presumably increases the transcription of other target genes, including PML (promyelocytic leukaemia). IRF8 is also ubiquitylated by Casitas B-lineage lymphoma (CBL), which leads to proteasome-mediated degradation119. IFNAR, IFNα receptor; IFNGR, IFNγ receptor; JAK, Janus kinase; TYK2, tyrosine kinase 2; Ub, ubiquitin.
Induction of TRIM proteins by IFNs
Studies of human monocytes and macrophages stimulated with IFNγ and the TLR4 ligand lipopolysaccharide showed that the expression of 26 of the 44 TRIM genes that were examined was upregulated85. An analysis of mouse primary macrophages, DCs and T cells showed that 17 of 28 TRIM genes tested were upregulated following infection with influenza virus or induction of TLR signalling in a type-I-IFN-dependent manner8. In support of a direct responsiveness to IFNs, the promoters of some of the TRIM genes, including TRIM5, have an IFN-stimulated response element (ISRE) that drives transcription86. These data highlight the close relationship between some TRIM proteins and host defence, and suggest that responses that are directed against pathogens reflect the selective forces that could drive the evolution of the TRIM family.
Given that IFN responses vary between different cell types and are modulated by various signalling pathways87, it is likely that the expression of additional TRIM genes is IFN responsive and is modulated by related signalling mechanisms. Importantly, of the many TRIM proteins that have been reported to have antiretroviral activities22,38, IFN responsiveness has been directly shown only for TRIM5α, TRIM22 and PML22,74,88. These observations are consistent with early studies showing that type I IFNs inhibited HIV replication89,90. Although the mechanisms by which IFNs exert antiretroviral activities remain elusive, it is becoming clear that TRIM family members are prominent mediators of these antiretroviral activities.
Some TRIM genes that are stimulated by IFNs regulate signalling pathways that affect innate immunity. Although this line of research is still in its infancy, the available literature indicates that TRIM proteins are broadly involved in various steps of these signalling pathways.
Regulation of IFN signalling by TRIM8
Once activated, IFN-induced signalling pathways are promptly attenuated by multiple mechanisms that act to prevent uncontrolled inflammatory responses. For example, suppressor of cytokine signalling 1 (SOCS1) interferes with the activity of JAK proteins to downregulate IFN-activated gene expression. TRIM8, a protein that is induced by IFNγ in many cells, was shown to interact with SOCS1 (REF. 21) (FIG. 2) and to inhibit SOCS1-mediated downregulation of IFNγ signalling in transfection experiments. Although E3 ubiquitin ligase activity has not been shown for TRIM8, co-expression of TRIM8 with SOCS1 promoted SOCS1 degradation, which suggests that TRIM8 destabilizes SOCS1, thereby resulting in the downregulation of SOCS1 activity following IFNγ stimulation.
TRIM proteins in PRR signalling pathways
More than 13 TLRs sense various pathogen components and trigger signalling pathways that activate NF-κB, IRF3 and IRF7 (REFS 1,2). NF-κB stimulates the expression of pro-inflammatory cytokines, whereas IRF3 and IRF7 activate IFN transcription. In addition, viral RNAs are recognized by RNA helicases of the retinoic-acid-inducible gene I (RIG-I) signalling pathway91,92. Recent literature has provided the first evidence that TRIM proteins are involved in regulating these pathways (FIG. 3).
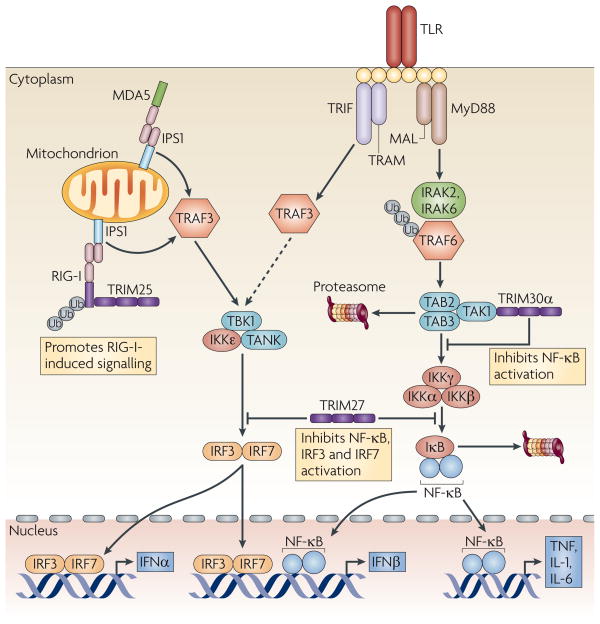
Figure 3. TRIM proteins in TLR, RIG-I and MDA5 signalling pathways.
Following engagement by ligand, Toll-like receptors (TLRs) that are expressed on the cell surface (TLR2, TLR4 and TLR5) and on endosomes (TLR3, TLR7, TLR8 and TLR9) activate downstream signalling pathways that lead to the activation of nuclear factor-κB (NF-κB), interferon (IFN)-regulatory factor 3 (IRF3) and IRF7. This induces the expression of type I IFNs and pro-inflammatory cytokines, such as tumour-necrosis factor (TNF), interleukin-1 (IL-1) and IL-6. Binding of double-stranded RNA or 5′ phosphorylated single-stranded RNA by the RNA helicases retinoic-acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), respectively, also activates the two pathways91. Tripartite motif-containing protein 30α (TRIM30α) binds to TAK1 (transforming growth factor-β (TGFβ)-activated kinase 1) and destabilizes TAK1-binding protein 2 (TAB2) and TAB3 to inhibit NF-κB activation96. TRIM27 binds to IκB (inhibitor of NF-κB) kinases (IKKs) and blocks the activation of NF-κB and of IRF3 and IRF7 (REF. 100). TRIM25 binds to RIG-I to conjugate ubiquitin, thereby promoting the activation of downstream pathways to increase cytokine induction17. IPS1, IFNB-promoter stimulator 1; IRAK, IL-1-receptor-associated kinase; MAL, myeloid differentiation primary-response gene 88 (MyD88)-adaptor-like protein; TBK1, TNF-receptor-associated factor (TRAF)-family-member-associated NF-κB activator (TANK)-binding kinase 1; TRAM, Toll/IL-1-receptor-domain-containing adaptor protein inducing IFNβ (TRIF)-related adaptor molecule; Ub, ubiquitin.
Ubiquitylation of RIG-I
TRIM25 is an IFN-inducible member of the C-IV family of TRIM proteins (TABLE 1). TRIM25 ubiquitylates the cell-cycle regulator 14-3-3σ (also known as stratifin), which results in negative regulation of cell-cycle progression93,94. In addition, TRIM25 can conjugate ISG15 to 14-3-3σ (REF. 23). A recent study shows an important role for TRIM25 in the RIG-I pathway17. TRIM25 interacts with the cas-pase-recruitment domain (CARD) of RIG-I through its PRYSPRY domain, which results in the ubiquitylation of RIG-I and, consequently, the induction of type I IFN production and NF-κB activity17. Interestingly, a recent paper described the ISGylation of RIG-I95, which suggests that TRIM25 may be involved in both the ubiquitylation and the ISGylation of RIG-I and therefore might have a role in both positive and negative regulation of type I IFN signalling. As other proteins in the PRR pathways contain CARDs, TRIM25 may also regulate the activity of other proteins that are involved in mediating innate immune responses. Indeed, it has been reported that embryonic fibroblasts from Trim25−/− mice are less effective in controlling the replication of Sendai virus than those of wild-type mice. Finally, as noted above, TRIM25 inhibits late-stage replication of HIV and MLV38.
Negative regulation of TLR signalling
The TLR-induced activation of NF-κB that is downstream of TRAF6 initially involves the activation of three protein kinases: TAB1 (transforming growth factor-β (TGFβ-activated kinase 1 (TAK1)-binding protein 1), TAB2 and TAB3. This in turn activates TAK1, which is required for the activation of IκB (inhibitor of NF-κB) kinase-α (IKKα) and IKKβ. IKKα and IKKβ trigger the degradation of IκB. A recent study showed that mouse TRIM30α interacts with TAK1 and promotes the degradation of TAB2 and TAB3, resulting in the inhibition of TRAF6 ubiquitylation and consequently the inhibition of NF-κB activity96 (FIG. 3), which ultimately reduced the induction of pro-inflammatory cytokine expression. As TRIM30α expression is upregulated by viruses and TLRs in an IFN-dependent manner8, this inhibition may represent a post-activation state of negative regulation of NF-κB activity. Because TAK1 has a crucial role in activating NF-κB through other signalling pathways, such as those that are initiated by TNFR1 and IL-1R97, mouse TRIM30α might have a role in regulating inflammation in addition to regulating TLR signalling. TRIM30α is a mouse-specific TRIM with no orthologue in humans, so the mechanism of TAK1 regulation probably differs between rodents and humans. However, it remains possible that TRIM proteins in humans have a similar role in regulating NF-κB activity.
TRIM27, also known as the RET finger protein, was shown previously to interact with the tumour-suppressor protein retinoblastoma 1 and with the Mi-2β-containing histone deacetylase complex to repress transcription98,99. More recently, it was reported that TRIM27 interacts with the IKK family of kinases and regulates both NF-κB-mediated and IRF-mediated gene expression100. This study also showed that TRIM27 interacts with three IKK proteins — IKKα, IKKβ and IKKε — and with TBK1 (TRAF-family-member-associated NF-κB activator (TANK)-binding kinase 1). These interactions with IKKs seem to take place in the cytoplasm and prevent IKK-mediated nuclear translocation of IRF3 without inhibiting its dimerization100. Overexpression of TRIM27 inhibits NF-κB-driven and IRF3-driven reporter gene expression, whereas this transcription was increased by small interfering RNA (siRNA)-mediated knockdown of TRIM27 expression. This suggests that TRIM27 negatively regulates the function of both NF-κB and IRF3. The mechanism by which TRIM27 regulates the activity of IKKs is currently unknown; however, because inhibition was observed in a TRIM27 mutant that lacked the RING domain, this function may not involve its E3 ubiquitin ligase activity100.
In addition, TRIM27 was shown to interact with SUMO-conjugating E3 ubiquitin ligases of the PIAS (protein inhibitor of activated STAT) family and to become sumoylated, which changed its distribution and activity101. Given that PIAS1 and PIAS3 are involved in STAT1- and STAT3-mediated cytokine signalling102, the function of some TRIM proteins, including TRIM27, may be closely linked to sumoylation and ISGylation. It was recently shown that nitric oxide promotes the degradation of PIAS3 through ubiquitylation by TRIM32 (REF. 103). Given that IFNs stimulate the production of nitric oxide, it is possible that the TRIM family has a role in regulating SUMO-dependent pathways that contribute to innate immunity.
Regulation of cytokine gene transcription by TRIM21
TRIM21, an IFN-inducible TRIM family member, has long been known as an autoantigen (RO52 or SS-A) that is recognized by antibodies in the sera of patients with Sjögren’s syndrome and of patients with systemic lupus erythematosus (SLE)104. TRIM21 mostly resides in the cytoplasm, although low levels are expressed in the nucleus, and it binds to the Fc region of IgG12, which could contribute to the pathogenesis of these diseases. Recently, TRIM21 was shown to interact with the transcription factor IRF8 following stimulation by IFN and TLR ligation, which suggests a role for this protein in innate immunity105 (FIG. 2). This interaction, which occurs in the nucleus, stimulated the ubiquitylation of IRF8 and increased the ability of IRF8 to drive Il12p40 gene transcription in mice. Supporting the hypothesis that TRIM21 mediates IRF8 ubiquitylation, TRIM21 was shown to act as an E3 ubiquitin ligase for itself and other proteins106,107. Consistent with the observation that TRIM21 interacts with IRF8 in the nucleus, TRIM21 was shown to accumulate in the nucleus of various human cells that had been stimulated with type I IFNs108. The ability of ubiquitylated IRF8 to increase cytokine expression is consistent with earlier studies that showed that ubiquitylation can increase the activity of some transcription factors109. As IRF8 regulates the expression of several other genes that are important for innate immunity, including those encoding type I IFNs, MHC class II molecules, inducible nitric-oxide synthase and natural-resistance-associated macrophage protein 1 (REFS 110–112), TRIM21 may have a broader role in host defence than is currently appreciated. IRF8 may also link TRIM21 to PML, as the expression of IRF8 is required for PML expression by macrophages and DCs75. A recent study reports that TRIM21 ubiquitylates IRF3 following activation, which leads to its degradation113.
Furthermore, IRF8, alone or together with IRF4, modulates B-cell development and function at multiple stages of differentiation114,115. As TRIM21 is expressed by lymphocytes, myeloid cells and DCs, it might also regulate IRF8 activity in B cells114. It is therefore possible that a role for TRIM21 as an autoantigen might be, in part, due to its regulation of the function of IRF8. In addition, TRIM21 may regulate T-cell activation or proliferation, as overexpression of TRIM21 was shown to increase IL-2 production by CD28-stimulated Jurkat T cells116. Interestingly, TRIM68, which is encoded by a gene that is adjacent to TRIM21 and has a structure that is similar to TRIM21, is another autoantigen (SS-56) that is found in patients with Sjögren’s syndrome and SLE117. This raises the possibility that TRIM21 and TRIM68 may have similar functions.
Concluding remarks
The versatility of the TRIM proteins is due to their diversity, splicing variants, differences in tissue expression and subcellular localization, but also to the increasingly well understood interactions of their N-terminal and C-terminal domains with other proteins. Their biological effects are also reflected in their ability to interact directly with viral components, either alone or in combination with other cellular proteins, and to modulate signalling pathways that are triggered by the engagement of PRRs. This downstream regulation affects the expression of both type I and type II IFNs, and of cytokines that are involved in pro-inflammatory responses and in promoting different aspects of the adaptive immune response.
At present, our knowledge of the mechanisms by which the TRIM family proteins restrict viral growth and influence signalling pathways is limited. This is due, in part, to our imperfect ability to piece together the functions of the different domains that mediate ubiquitylation, ISGylation and sumoylation, the nature of the ubiquitin-conjugating enzyme (E2) and its target proteins, the modes of modification that inhibit or increase activity, and the counterbalancing activities of the large families of deubiquitylating enzymes. Structural studies will have an increasingly important role in providing a detailed picture of the protein–protein interactions between members of this family that are important in innate and adaptive immunity. Finally, progress in this field may open new horizons for developing strategies to prevent or combat infectious diseases, inflammatory conditions and autoimmune disorders.
Supplementary Material
Acknowledgments
We thank J. Weddle for his assistance in the preparation of this manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Child Health and Human Development, and National Institute of Allergy and Infectious Diseases.
Glossary
- Ubiquitylation
The covalent conjugation of ubiquitin, a 76 amino-acid protein, to other proteins. In addition to targeting proteins for proteasome-mediated degradation, ubiquitylation affects protein stability, trafficking and gene expression, among other functions
- E3 ubiquitin ligase
An enzyme that covalently attaches ubiquitin to specific target proteins at defined lysine residues, resulting in monoubiquitylation, multiubiquitylation or polyubiquitylation of the target protein. E3 ubiquitin ligases act together with a ubiquitin- activating enzyme (E1) and a ubiquitin-conjugating enzyme (E2)
- Sumoylation
The post-translational modification of proteins that involves the covalent attachment of small ubiquitin-related modifier 1 (SUMO1)–SUMO4 and that regulates the interactions of those proteins with other macromolecules
- Nuclear bodies
Punctuated, dot-like structures in the nucleus that are visible through a light microscope. The interferon-inducible proteins promyelocytic leukaemia (PML), SP100 and ISG20, as well as SUMO1, are found in nuclear bodies
- Familial Mediterranean fever
(FMF). The most common familial inflammatory disease that is characterized by self-limited attacks of fever and serositis. FMF is transmitted in an autosomal recessive pattern and is caused by mutations in the PRYSPRY domain of the MEFV gene, which encodes the protein pyrin
- Positive selection
A process of natural selection that increases the frequency of beneficial mutations in a population
- p53
A major transcription factor that is activated by numerous genotoxic insults and subsequently induces cell-cycle arrest, cellular senescence or apoptosis. p53 is frequently mutated or functionally inactivated in cancer
- Plasmacytoid DC
A dendritic cell (DC) that is morphologically similar to the antibody-producing plasma cell and produces very high levels of type I interferons (IFNs) in response to viral infection and Toll-like receptor (TLR) stimulation. Plasmacytoid DCs express TLR7 and TLR9, but low levels of TLR3, and the induction of type I IFNs by plasmacytoid DCs does not seem to rely on the retinoic-acid-inducible gene I (RIG-I) signalling pathway
- 14-3-3σ
A member of a family of conserved proteins that are present in all eukaryotic organisms and are involved in diverse cellular processes, such as apoptosis, intracellular signalling and cell-cycle regulation. 14-3-3 proteins function as adaptors in protein interactions and can regulate protein localization and enzymatic activity
- Small interfering RNA
Short double-stranded RNAs of 19–23 nucleotides that induce RNA interference, which is a post-transcriptional process that leads to gene silencing in a sequence-specific manner
- Systemic lupus erythematosus
(SLE). SLE is a multigenic, chronic and potentially fatal autoimmune disorder that is characterized by a broad range of clinical abnormalities. SLE can affect nearly all components of the immune system
Footnotes
DATABASES
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
CBL | IRF3 | IRF7 | ISG15 | MEFV | MID1 | PIAS1 | PIAS3 | PML | SOCS1 | TRIM5α | TRIM6 | TRIM8 | TRIM11 | TRIM21 | TRIM22 | TRIM25 | TRIM27 | TRIM34 | UBE2I
OMIM: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM
Sjögren’s syndrome | X-linked Opitz G/BBB syndrome
FURTHER INFORMATION
GeneCards: http://www.genecards.org
HUGO Gene Nomenclature Committee: http://www.genenames.org
Tigem website: http://www.tigem.it
UniProt Consortium: http://www.uniprot.org
See online article: S1 (figure) | S2 (figure)
References
- 1.Kawai T, Akira S. TLR signaling. Semin Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 2.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nature Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 3.Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-γ. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 4.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 5.Beutler B, et al. Genetic analysis of resistance to viral infection. Nature Rev Immunol. 2007;7:753–766. doi: 10.1038/nri2174. [DOI] [PubMed] [Google Scholar]
- 6.Trinchieri G, Sher A. Cooperation of Toll-like receptor signals in innate immune defence. Nature Rev Immunol. 2007;7:179–190. doi: 10.1038/nri2038. [DOI] [PubMed] [Google Scholar]
- 7.Reymond A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. This study shows that many of the 37 human TRIM family members examined self-associate to form aggregates that occupy several, previously unrecognized, subcellular compartments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajsbaum R, Stoye JP, O’Garra A. Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur J Immunol. 2008;38:619–630. doi: 10.1002/eji.200737916. The authors screened for the expression of 29 mouse TRIM genes in T cells, macrophages and DCs, and showed that many TRIM genes are induced following infection with influenza virus and TLR stimulation, and that this induction is, for the most part, dependent on type I IFNs. [DOI] [PubMed] [Google Scholar]
- 9.Torok M, Etkin LD. Two B or not two B? Overview of the rapidly expanding B-box family of proteins. Differentiation. 2001;67:63–71. doi: 10.1046/j.1432-0436.2001.067003063.x. [DOI] [PubMed] [Google Scholar]
- 10.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 11.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nature Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 12.James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY- mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci USA. 2007;104:6200–6205. doi: 10.1073/pnas.0609174104. This study shows that the PRYSPRY domain of TRIM21 comprises a β-sandwich of antiparallel β-sheets and identifies binding pockets for IgG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joazeiro CA, et al. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 14.Lorick KL, et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waterman H, Levkowitz G, Alroy I, Yarden Y. The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. J Biol Chem. 1999;274:22151–22154. doi: 10.1074/jbc.274.32.22151. [DOI] [PubMed] [Google Scholar]
- 16.Sabile A, et al. Regulation of p27 degradation and S-phase progression by Ro52 RING finger protein. Mol Cell Biol. 2006;26:5994–6004. doi: 10.1128/MCB.01630-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. This paper shows that TRIM25 binds to RIG-I and conjugates ubiquitin to its CARD domain. This ubiquitylation increases downstream signalling to elicit host antiviral responses. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa H, Tachikawa H, Miura Y, Takahashi N. TRIM11 binds to and destabilizes a key component of the activator-mediated cofactor complex (ARC105) through the ubiquitin-proteasome system. FEBS Lett. 2006;580:4784–4792. doi: 10.1016/j.febslet.2006.07.066. [DOI] [PubMed] [Google Scholar]
- 19.Diaz-Griffero F, et al. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006;349:300–315. doi: 10.1016/j.virol.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 20.Yamauchi K, Wada K, Tanji K, Tanaka M, Kamitani T. Ubiquitination of E3 ubiquitin ligase TRIM5α and its potential role. FEBS J. 2008;275:1540–1555. doi: 10.1111/j.1742-4658.2008.06313.x. [DOI] [PubMed] [Google Scholar]
- 21.Toniato E, et al. TRIM8/GERP RING finger protein interacts with SOCS-1. J Biol Chem. 2002;277:37315–37322. doi: 10.1074/jbc.M205900200. In this study, TRIM8, which is induced by IFNγ, is identified as a factor that interacts with SOCS1 and this interaction leads to its destabilization and reduced IFNγ-dependent reporter activity. [DOI] [PubMed] [Google Scholar]
- 22.Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 2008;4:e1000007. doi: 10.1371/journal.ppat.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou W, Zhang DE. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J Biol Chem. 2006;281:3989–3994. doi: 10.1074/jbc.M510787200. This study shows that the RING domain of TRIM25 can mediate ISGylation and ubiquitylation. [DOI] [PubMed] [Google Scholar]
- 24.Zou W, Wang J, Zhang DE. Negative regulation of ISG15 E3 ligase EFP through its autoISGylation. Biochem Biophys Res Commun. 2007;354:321–327. doi: 10.1016/j.bbrc.2006.12.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tao H, et al. Structure of the MID1 tandem B-boxes reveals an interaction reminiscent of intermolecular ring heterodimers. Biochemistry. 2008;47:2450–2457. doi: 10.1021/bi7018496. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Song B, Xiang SH, Sodroski J. Functional interplay between the B-BOX 2 and the B30.2(SPRY) domains of TRIM5α. Virology. 2007;366:234–244. doi: 10.1016/j.virol.2007.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 28.Ferrentino R, Bassi MT, Chitayat D, Tabolacci E, Meroni G. MID1 mutation screening in a large cohort of Opitz G/BBB syndrome patients: twenty-nine novel mutations identified. Hum Mutat. 2007;28:206–207. doi: 10.1002/humu.9480. [DOI] [PubMed] [Google Scholar]
- 29.Trockenbacher A, et al. MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nature Genet. 2001;29:287–294. doi: 10.1038/ng762. [DOI] [PubMed] [Google Scholar]
- 30.Schweiger S, Schneider R. The MID1/PP2A complex: A key to the pathogenesis of Opitz BBB/G syndrome. Bioessays. 2003;25:356–366. doi: 10.1002/bies.10256. [DOI] [PubMed] [Google Scholar]
- 31.Minucci S, et al. Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol Cell. 2000;5:811–820. doi: 10.1016/s1097-2765(00)80321-4. [DOI] [PubMed] [Google Scholar]
- 32.Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nature Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 34.Javanbakht H, et al. Characterization of TRIM5α trimerization and its contribution to human immunodeficiency virus capsid binding. Virology. 2006;353:234–246. doi: 10.1016/j.virol.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 35.Mische CC, et al. Retroviral restriction factor TRIM5α is a trimer. J Virol. 2005;79:14446–14450. doi: 10.1128/JVI.79.22.14446-14450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Short KM, Cox TC. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J Biol Chem. 2006;281:8970–8980. doi: 10.1074/jbc.M512755200. In this study, the authors identify a new COS domain that is present in the C-terminal region of subsets of the TRIM superfamily and is required for the interaction with microtubules. This observation provides the basis for a new classification for the TRIM superfamily based on domains that are located in the C terminus. [DOI] [PubMed] [Google Scholar]
- 37.Yap MW, Nisole S, Lynch C, Stoye JP. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci USA. 2004;101:10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uchil PD, Quinlan BD, Chan WT, Luna JM, Mothes W. Trim E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 2008;4:e16. doi: 10.1371/journal.ppat.0040016. In this study, the authors screen 36 human and 19 mouse TRIM proteins for their effects on the life cycle of HIV-1, MLV and avian leukosis virus in cultured cells and show that many TRIM proteins inhibit early and late stages of retrovirus infections. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woo JS, Suh HY, Park SY, Oh BH. Structural basis for protein recognition by B30.2/SPRY domains. Mol Cell. 2006;24:967–976. doi: 10.1016/j.molcel.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 40.Grutter C, et al. Structure of the PRYSPRY-domain: implications for autoinflammatory diseases. FEBS Lett. 2006;580:99–106. doi: 10.1016/j.febslet.2005.11.076. The authors identify PRYSPRY dimers that form an interaction site for binding target proteins. This site is specifically mutated in MEFV in patients with familial Mediterranean fever. [DOI] [PubMed] [Google Scholar]
- 41.Rhodes DA, de Bono B, Trowsdale J. Relationship between SPRY and B30.2 protein domains. Evolution of a component of immune defence? Immunology. 2005;116:411–417. doi: 10.1111/j.1365-2567.2005.02248.x. The authors show that the acquisition of PRY domains by evolutionarily ancient SPRY sequences to generate the PRYSPRY domain is a modern vertebrate adaptation that is associated with the rapid expansion of PRYSPRY-containing proteins with broad roles in innate immunity, particularly to retroviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flajnik MF, Du Pasquier L. Evolution of innate and adaptive immunity: can we draw a line? Trends Immunol. 2004;25:640–644. doi: 10.1016/j.it.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Pancer Z, Cooper MD. The evolution of adaptive immunity. Annu Rev Immunol. 2006;24:497–518. doi: 10.1146/annurev.immunol.24.021605.090542. [DOI] [PubMed] [Google Scholar]
- 44.Stremlau M, et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. This is the first demonstration that TRIM5α is a host-cell component that interrupts the life cycle of HIV at the stage of capsid uncoating. The authors show that rhTRIM5α is more efficient than human TRIM5α at blocking this stage and that TRIM5α also restricts simian immunodeficiency virus in a species-specific manner. [DOI] [PubMed] [Google Scholar]
- 45.Schaller T, Hue S, Towers GJ. An active TRIM5 protein in rabbits indicates a common antiviral ancestor for mammalian TRIM5 proteins. J Virol. 2007;81:11713–11721. doi: 10.1128/JVI.01468-07. The identification of TRIM5 and an endogenous lentivirus in rabbits shows that TRIM5-related proteins in many vertebrates are derived from a common ancestor with antiretroviral properties. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Si Z, et al. Evolution of a cytoplasmic tripartite motif (TRIM) protein in cows that restricts retroviral infection. Proc Natl Acad Sci USA. 2006;103:7454–7459. doi: 10.1073/pnas.0600771103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.TIF Consortium 1997. Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranen fever. Cell. 1997;90:797–807. doi: 10.1016/s0092-8674(00)80539-5. [DOI] [PubMed] [Google Scholar]
- 48.Stojanov S, Kastner DL. Familial autoinflammatory diseases: genetics, pathogenesis and treatment. Curr Opin Rheum. 2005;17:586–599. doi: 10.1097/bor.0000174210.78449.6b. [DOI] [PubMed] [Google Scholar]
- 49.Keeble AH, Khan Z, Forster A, James LC. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc Natl Acad Sci USA. 2008;105:6045–6050. doi: 10.1073/pnas.0800159105. These authors show that TRIM21 is a novel IgG receptor that is functionally distinct from previously described Fc receptors and is highly conserved among mammals. Its unique structure could promote the formation of highly crosslinked immune complexes that contribute to the pathogenesis of Sjögren’s syndrome and SLE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Douarin B, Nielsen AL, You J, Chambon P, Losson R. TIF1α: a chromatin-specific mediator for the ligand-dependent activation function AF-2 of nuclear receptors? Biochem Soc Trans. 1997;25:605–612. doi: 10.1042/bst0250605. [DOI] [PubMed] [Google Scholar]
- 51.Friedman JR, et al. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 52.Klugbauer S, Rabes HM. The transcription coactivator HTIF1 and a related protein are fused to the RET receptor tyrosine kinase in childhood papillary thyroid carcinomas. Oncogene. 1999;18:4388–4393. doi: 10.1038/sj.onc.1202824. [DOI] [PubMed] [Google Scholar]
- 53.Ivanov AV, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20:7223–7233. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- 55.Scaglioni PP, et al. A CK2-dependent mechanism for degradation of the PML tumor suppressor. Cell. 2006;126:269–283. doi: 10.1016/j.cell.2006.05.041. [DOI] [PubMed] [Google Scholar]
- 56.Zhong S, et al. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
- 57.Lilly F, Pincus T, George Kleln SW, Alexander H. Advances in Cancer Research. Academic; New York: 1973. pp. 231–277. [Google Scholar]
- 58.Best S, Tissier PL, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene. Fvl Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 59.Goff SP. Genetic control of retrovirus susceptibility in mammalian cells. Annu Rev Genet. 2004;38:61–85. doi: 10.1146/annurev.genet.38.072902.094136. [DOI] [PubMed] [Google Scholar]
- 60.Rosenberg BR, Papavasiliou FN. Beyond SHM and CSR: AID and related cytidine deaminases in the host response to viral infection. Adv Immunol. 2007;94:215–244. doi: 10.1016/S0065-2776(06)94007-3. [DOI] [PubMed] [Google Scholar]
- 61.Towers GJ. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology. 2007;4:40. doi: 10.1186/1742-4690-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tissot C, Mechti N. Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J Biol Chem. 1995;270:14891–14898. doi: 10.1074/jbc.270.25.14891. [DOI] [PubMed] [Google Scholar]
- 63.Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. This paper shows that TRIM28 silences the transcription of MLV in embryonic carcinoma and embryonic stem cells, thereby identifying a host-cell factor that limits retroviral replication in embryonic cells. [DOI] [PubMed] [Google Scholar]
- 64.Sebastian S, Luban J. TRIM5α selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5α restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci USA. 2006;103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li X, et al. Functional replacement of the RING, B-BOX 2, and coiled-coil domains of tripartite motif 5α (TRIM5α) by heterologous TRIM domains. J Virol. 2006;80:6198–6206. doi: 10.1128/JVI.00283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 68.Sawyer SL, Emerman M, Malik HS. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 2007;3:e197. doi: 10.1371/journal.ppat.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goldschmidt V, et al. Role of common human TRIM5α variants in HIV-1 disease progression. Retrovirology. 2006;3:54. doi: 10.1186/1742-4690-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Campbell EM, Perez O, Anderson JL, Hope TJ. Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5α. J Cell Biol. 2008;180:549–561. doi: 10.1083/jcb.200706154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rold CJ, Aiken C. Proteasomal degradation of TRIM5α during retrovirus restriction. PLoS Pathog. 2008;4:e1000074. doi: 10.1371/journal.ppat.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Niikura T, et al. A tripartite motif protein TRIM11 binds and destabilizes Humanin, a neuroprotective peptide against Alzheimer’s disease-relevant insults. Eur J Neurosci. 2003;17:1150–1158. doi: 10.1046/j.1460-9568.2003.02553.x. [DOI] [PubMed] [Google Scholar]
- 73.Katzourakis A, Tristem M, Pybus OG, Gifford RJ. Discovery and analysis of the first endogenous lentivirus. Proc Natl Acad Sci USA. 2007;104:6261–6265. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Regad T, et al. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J. 2001;20:3495–3505. doi: 10.1093/emboj/20.13.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dror N, et al. Interferon regulatory factor-8 is indispensable for the expression of promyelocytic leukemia and the formation of nuclear bodies in myeloid cells. J Biol Chem. 2007;282:5633–5640. doi: 10.1074/jbc.M607825200. [DOI] [PubMed] [Google Scholar]
- 76.Lallemand-Breitenbach V, et al. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor α degradation. J Exp Med. 2001;193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pampin M, Simonin Y, Blondel B, Percherancier Y, Chelbi-Alix MK. Cross talk between PML and p53 during poliovirus infection: implications for antiviral defense. J Virol. 2006;80:8582–8592. doi: 10.1128/JVI.00031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iki S, et al. Serum-dependent expression of promyelocytic leukemia protein suppresses propagation of influenza virus. Virology. 2005;343:106–115. doi: 10.1016/j.virol.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 79.Rosa-Calatrava MR, et al. Adenovirus protein IX sequesters host-cell promyelocytic leukaemia protein and contributes to efficient viral proliferation. EMBO Rep. 2003;4:969–975. doi: 10.1038/sj.embor.embor943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Everett RD, et al. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol. 2006;80:7995–8005. doi: 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pestka S. The interferons: 50 years after their discovery, there is much more to learn. J Biol Chem. 2007;282:20047–20051. doi: 10.1074/jbc.R700004200. [DOI] [PubMed] [Google Scholar]
- 82.Banchereau J, et al. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 83.Asselin-Paturel C, Trinchieri G. Production of type I interferons: plasmacytoid dendritic cells and beyond. J Exp Med. 2005;202:461–465. doi: 10.1084/jem.20051395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gabriele L, Ozato K. The role of the interferon regulatory factor (IRF) family in dendritic cell development and function. Cytokine Growth Factor Rev. 2007;18:503–510. doi: 10.1016/j.cytogfr.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 85.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte- to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 86.Asaoka K, et al. A retrovirus restriction factor TRIM5α is transcriptionally regulated by interferons. Biochem Biophys Res Commun. 2005;338:1950–1956. doi: 10.1016/j.bbrc.2005.10.173. [DOI] [PubMed] [Google Scholar]
- 87.van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 88.Sakuma R, Noser JA, Ohmine S, Ikeda Y. Rhesus monkey TRIM5α restricts HIV-1 production through rapid degradation of viral Gag polyproteins. Nature Med. 2007;13:631–635. doi: 10.1038/nm1562. [DOI] [PubMed] [Google Scholar]
- 89.Agy MB, Acker RL, Sherbert CH, Katze MG. Interferon treatment inhibits virus replication in HIV-1- and SIV-infected CD4+ T-cell lines by distinct mechanisms: evidence for decreased stability and aberrant processing of HIV-1 proteins. Virology. 1995;214:379–386. doi: 10.1006/viro.1995.0047. [DOI] [PubMed] [Google Scholar]
- 90.Fernie BF, Poli G, Fauci AS. α interferon suppresses virion but not soluble human immunodeficiency virus antigen production in chronically infected T-lymphocytic cells. J Virol. 1991;65:3968–3971. doi: 10.1128/jvi.65.7.3968-3971.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoneyama M, Fujita T. RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev. 2007;18:545–551. doi: 10.1016/j.cytogfr.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 92.Takeuchi O, Akira S. MDA5/RIG-I and virus recognition. Curr Opin Immunol. 2008;20:17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 93.Nakasato N, et al. A ubiquitin E3 ligase Efp is up-regulated by interferons and conjugated with ISG15. Biochem Biophys Res Commun. 2006;351:540–546. doi: 10.1016/j.bbrc.2006.10.061. [DOI] [PubMed] [Google Scholar]
- 94.Urano T, et al. Efp targets 14-3-3 σ for proteolysis and promotes breast tumour growth. Nature. 2002;417:871–875. doi: 10.1038/nature00826. [DOI] [PubMed] [Google Scholar]
- 95.Kim MJ, Hwang SY, Imaizumi T, Yoo JY. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J Virol. 2008;82:1474–1483. doi: 10.1128/JVI.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shi M, et al. TRIM30α negatively regulates TLR-mediated NF-κB activation by targeting TAB2 and TAB3 for degradation. Nature Immunol. 2008;9:369–377. doi: 10.1038/ni1577. TRIM30α, a mouse-only TRIM protein, interacts with the TAB2–TAB3–TAK1 complex. TRIM30α destabilizes TAB2–TAB3 interactions to prevent TRAF6 ubiquitylation, thereby inhibiting TLR-mediated NF-κB activation. [DOI] [PubMed] [Google Scholar]
- 97.Shim JH, et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathwaysin vivo. Genes Dev. 2005;19:2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Krutzfeldt M, et al. Selective ablation of retinoblastoma protein function by the RET finger protein. Mol Cell. 2005;18:213–224. doi: 10.1016/j.molcel.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 99.Shimono Y, et al. Mi-2β associates with BRG1 and RET finger protein at the distinct regions with transcriptional activating and repressing abilities. J Biol Chem. 2003;278:51638–51645. doi: 10.1074/jbc.M309198200. [DOI] [PubMed] [Google Scholar]
- 100.Zha J, et al. The Ret finger protein inhibits signaling mediated by the noncanonical and canonical IκB kinase family members. J Immunol. 2006;176:1072–1080. doi: 10.4049/jimmunol.176.2.1072. In this paper, TRIM27 is found to bind to IKKs that activate NF-κB and IRF3–IRF7. Knockdown of TRIM27 expression leads to increased NF-κB and IRF3–IRF7 activation, which indicates that this TRIM protein is a negative regulator of TLR and other signalling pathways. [DOI] [PubMed] [Google Scholar]
- 101.Matsuura T, et al. PIAS proteins are involved in the SUMO-1 modification, intracellular translocation and transcriptional repressive activity of RET finger protein. Exp Cell Res. 2005;308:65–77. doi: 10.1016/j.yexcr.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 102.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nature Rev Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 103.Qu J, et al. Nitric oxide destabilizes pias3 and regulates sumoylation. PLoS ONE. 2007;2:e1085. doi: 10.1371/journal.pone.0001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ben-Chetrit E, Chan EK, Sullivan KF, Tan EM. A 52-kD protein is a novel component of the SS-A/Ro antigenic particle. J Exp Med. 1988;167:1560–1571. doi: 10.1084/jem.167.5.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kong HJ, et al. Cutting Edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J Immunol. 2007;179:26–30. doi: 10.4049/jimmunol.179.1.26. This study shows that TRIM21 binds to IRF8 in macrophages following IFN or TLR stimulation. TRIM21 ubiquitylated IRF8 and increased the transcriptional activity of IRF8 for IL12p40 expression. [DOI] [PubMed] [Google Scholar]
- 106.Espinosa A, et al. The Sjögren’s syndrome-associated autoantigen Ro52 is an E3 ligase that regulates proliferation and cell death. J Immunol. 2006;176:6277–6285. doi: 10.4049/jimmunol.176.10.6277. [DOI] [PubMed] [Google Scholar]
- 107.Wada K, Kamitani T. UnpEL/Usp4 is ubiquitinated by Ro52 and deubiquitinated by itself. Biochem Biophys Res Commun. 2006;342:253–258. doi: 10.1016/j.bbrc.2006.01.144. [DOI] [PubMed] [Google Scholar]
- 108.Strandberg L, et al. Interferon-α induces up-regulation and nuclear translocation of the Ro52 autoantigen as detected by a panel of novel Ro52-specific monoclonal antibodies. J Clin Immunol. 2008;28:220–231. doi: 10.1007/s10875-007-9157-0. [DOI] [PubMed] [Google Scholar]
- 109.Ben-Neriah Y. Regulatory function of ubiquitination in the immmune system. Nature Immunol. 2002;3:20–26. doi: 10.1038/ni0102-20. [DOI] [PubMed] [Google Scholar]
- 110.Alter-Koltunoff M, et al. Innate immunity to intraphagosomal pathogens is mediated by interferon regulatory factor 8 (IRF-8) that stimulates the expression of macrophage-specific nramp1 through antagonizing repression by c-Myc. J Biol Chem. 2008;283:2724–2733. doi: 10.1074/jbc.M707704200. [DOI] [PubMed] [Google Scholar]
- 111.Tamura T, et al. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- 112.Tailor P, et al. The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity. 2007;27:228–239. doi: 10.1016/j.immuni.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Higgs R, et al. The E3 ubiquitin ligase Ro52 negatively regulates IFN-β production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol. 2008;181:1780–1786. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee CH, et al. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/ IFN consensus sequence-binding protein. J Exp Med. 2006;203:63–72. doi: 10.1084/jem.20051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ma S, Turetsky A, Trinh L, Lu R. IFN regulatory factor 4 and 8 promote Ig light chain κ locus activation in pre-B cell development. J Immunol. 2006;177:7898–7904. doi: 10.4049/jimmunol.177.11.7898. [DOI] [PubMed] [Google Scholar]
- 116.Ishii T, et al. SS-A/Ro52, an autoantigen involved in CD28-mediated IL-2 production. J Immunol. 2003;170:3653–3661. doi: 10.4049/jimmunol.170.7.3653. [DOI] [PubMed] [Google Scholar]
- 117.Billaut-Mulot O, et al. SS-56, a novel cellular target of autoantibody responses in Sjögren syndrome and systemic lupus erythematosus. J Clin Invest. 2001;108:861–869. doi: 10.1172/JCI13469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fridell RA, Harding LS, Bogerd HP, Cullen BR. Identification of a novel human zinc finger protein that specifically interacts with the activation domain of lentiviral Tat proteins. Virology. 1995;209:347–357. doi: 10.1006/viro.1995.1266. [DOI] [PubMed] [Google Scholar]
- 119.Xiong H, et al. Ubiquitin-dependent degradation of interferon regulatory factor-8 mediated by Cbl down-regulates interleukin-12 expression. J Biol Chem. 2005;280:23531–23539. doi: 10.1074/jbc.M414296200. [DOI] [PubMed] [Google Scholar]
- 120.Horn EJ, et al. RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis. 2004;25:157–167. doi: 10.1093/carcin/bgh003. [DOI] [PubMed] [Google Scholar]
- 121.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 122.Jehee FS, et al. An Xq22.3 duplication detected by comparative genomic hybridization microarray (Array-CGH) defines a new locus (FGS5) for FG syndrome. Am J Med Genet A. 2005;139:221–226. doi: 10.1002/ajmg.a.30991. [DOI] [PubMed] [Google Scholar]
- 123.Dal Zotto L, et al. The mouse Mid1 gene: implications for the pathogenesis of Opitz syndrome and the evolution of the mammalian pseudoautosomal region. Hum Mol Genet. 1998;7:489–499. doi: 10.1093/hmg/7.3.489. [DOI] [PubMed] [Google Scholar]
- 124.Reiter A, Lengfelder E, Grimwade D. Pathogenesis, diagnosis and monitoring of residual disease in acute promyelocytic leukaemia. Acta Haematol. 2004;112:55–67. doi: 10.1159/000077560. [DOI] [PubMed] [Google Scholar]
- 125.El-Shanti H, Majeed HA, El-Khateeb M. Familial mediterranean fever in Arabs. Lancet. 2006;367:1016–1024. doi: 10.1016/S0140-6736(06)68430-4. [DOI] [PubMed] [Google Scholar]
- 126.Saenko V, et al. Novel tumorigenic rearrangement, Δrfp/ret, in a papillary thyroid carcinoma from externally irradiated patient. Mutat Res. 2003;527:81–90. doi: 10.1016/s0027-5107(03)00056-3. [DOI] [PubMed] [Google Scholar]
- 127.Guglieri M, Magri F, Comi GP. Molecular etiopathogenesis of limb girdle muscular and congenital muscular dystrophies: boundaries and contiguities. Clin Chim Acta. 2005;361:54–79. doi: 10.1016/j.cccn.2005.05.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.