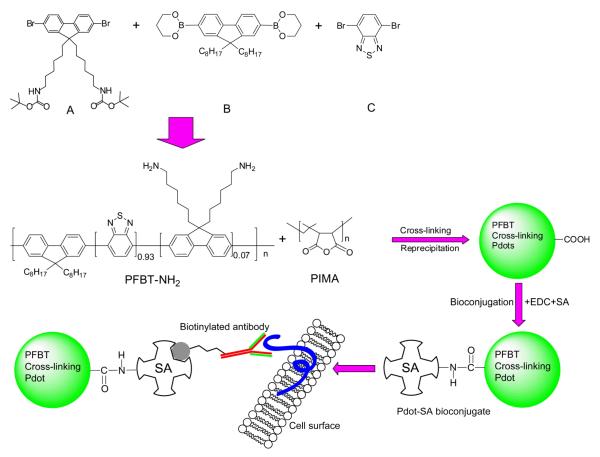
Semiconducting polymer dots (Pdots) as fluorescent probes exhibit superior characteristics such as high brightness, fast emission rates, non-blinking, and non-toxic features. [1-11] These properties make them well-suited for biological imaging and biosensing applications. [12-17] For biological applications, however, the surface of Pdots needs to be functionalized for subsequent bioconjugation and cellular targeting. [18, 19] To this end, several methods have been employed to modify the surface of Pdots, such as employing phospholipid encapsulation, [20] and surfactant miniemulsion. [21] Recently, we have developed a co-condensation method to functionalize the Pdot surface, where an amphiphilic polymer bearing functional groups was physically blended with semiconducting polymer to create surface reactive groups for bioconjugation. [4, 5] In these studies, we have also made quantitative comparison between Pdots, Qdots, and organic dyes, and have shown Pdots offer excellent optical performance. [4] The cytotoxicity of Pdots has also been studied in previous published reports using cultured cells, and these studies have generally suggested that Pdots have good biocompatibility. [13, 22-24] Therefore, successful bioconjugation via these schemes opens up a new and practical way to employ the highly fluorescent Pdotbioconjugates for a wide variety of biological applications.
For further improving the probe performance, however, several limitations in the current functionalization methods need to be overcome. Because Pdot formation is driven by hydrophobic interactions, polymer molecules in the Pdots are physically associated with each other. In many cases, the functional molecules may fall off from the nanoparticles due to the relatively weak non-covalent interactions. In the co-condensation scheme, for example, selection of amphiphilic functional polymer encounters an obvious paradox. Large molecular weight is preferable for more stable functionalization, because they are less likely to dissociate from the formed Pdots; however, high blending ratio is required to uniformly distribute large amphiphilic molecules into each particle, a requirement that both sacrifices the per-particle fluorescence brightness and yield large particle size for accommodating large amphiphilic molecules. Selection of small amphiphilic molecule can lower the blending ratio, enhance per-particle brightness, and overcome the above issue, but the use of low molecular weight amphiphilic polymer can result in unstable functionalization. To translate Pdots for use in biological studies, small size is an important feature, because it offers better tissue and subcellular penetration while minimizing non-specific interactions. In our previous methods [4, 5, 25], it was difficult in general to achieve Pdots with sizes smaller than 10 nm.
To overcome these drawbacks, we developed a cross-linking strategy to covalently bond the functional polymer molecules to the Pdots. In this study, a semiconducting polymer with side-chain amine groups was first synthesized and a functional polymer, Poly(isobutylene-alt-maleic anhydride) (PIMA) or Poly(styrene-co-maleic anhydride) (PSMA), was used as a cross-linker. Each PIMA or PSMA molecule comprises multiple reactive units that spontaneously react with side-chain amine groups of the polymer, thereby forming covalent cross links with the semiconducting polymer while simultaneously providing carboxyl groups for bioconjugation. In addition to providing stable functionalized Pdots, one important finding is that this approach can reliably form Pdots that are very small (10 nm or less) in size, characteristics that are difficult to achieve with most other methods for preparing Pdots.
To determine the stability enhancement offered by this cross-linking approach, we employed a dye-leaching method to compare the stability of the cross-linked Pdots with Pdots functionalized by the co-condensation method described previously. We applied the cross-linked Pdots to label the surface of breast cancer cells and the microtubules of cervical cancer cells to demonstrate their performance in biological imaging as well as flow cytometry experiments. Stable functionalization together with the small sizes provided by cross-linked Pdots is significant for improving labeling efficiency and sensitivity in biological assays.
Highly bright Pdots can be functionalized by the co-condensation strategy where amphiphilic polymer molecules bearing functional groups are co-condensed with semiconducting polymer to modify the Pdot surface. To characterize the stability of cross-linked Pdots versus co-condensed Pdots, we employed a dye-leaching method to investigate the stability of the two respective types of Pdots. First, Pdots were functionalized by the co-condensation method, where a small amphiphilic polymer poly(styrene-co-maleic anhydride) (PSMA) with a molecular weight of 1600 was physically blended with a semiconducting polymer poly[(9,9-dioctylfluorenyl-2,7-diyl)-co-(1,4-benzo-{2,1’,3}-thiadazole)] (PFBT) to form carboxyl-functionalized PFBT dots. We then conjugated a Rhodamine-amine dye to the PFBT dots by formation of amide bonds catalyzed by 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) (Supporting Scheme S1). The resulting Pdot-Rhodamine nanoparticles were purified from free Rhodamine molecules by a desalting column (Bio-Rad Econo-Pac® 10DG columns). A small amount of surfactant such as Triton X-100 was added to the aqueous solution of Pdot-Rhodamine to mimic the swelling conditions usually encountered in biological systems. A centrifugal filtration device (Amicon® Ultra-4 Centrifugal Filter with a molecular weight cutoff of 100,000) was used to separate Pdots from the filtrate containing Rhodamine molecules that dissociated from the Pdots.
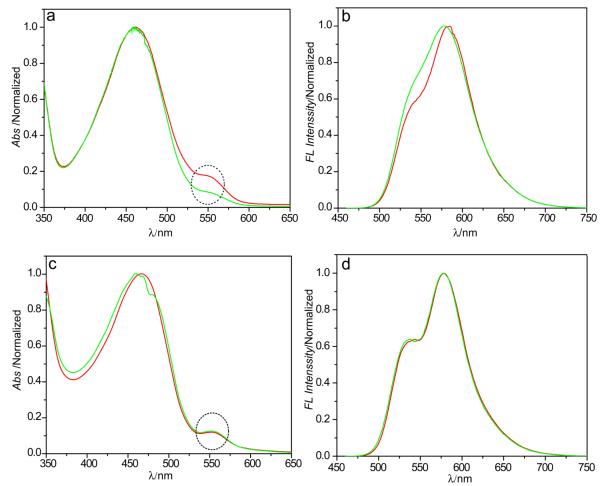
The absorption and fluorescence spectra of Pdot-Rhodamine solutions were measured before and after centrifugal filtration. For the Pdot-Rhodamine sample after column purification, a clear absorption peak from Rhodamine dye was observed, while this peak was absent in the control sample without using EDC. This indicates that the Rhodamine dyes were covalently linked to PSMA molecules that are physically associated with Pdots. After centrifugal filtration, a clear decrease in the Rhdoamine absorption was observed for the Pdot-Rhodamine nanoparticles (Figure 1a), indicating part of the functional PSMA molecules were dissociated from the Pdots. The fluorescence spectra (Figure 1b) of the Pdot-Rhodamine also exhibited some spectral changes due to the reduced energy transfer from PFBT donor to Rhodamine acceptor, consistent with the loss of PSMA-Rhodamine molecules from the Pdots.
Figure 1.
Stability of functionalized Pdots. The absorption (left) and fluorescence (right) spectra of Pdots doped with Rhodamine. (a, b) PFBT/PSMA-Rhodamine Pdots functionalized using the physical-blending method. (c, d) PFBT-PSMA-Rhodamine Pdots formed by the covalent cross-linking approach. The circled regions indicate the absorption peaks of Rhodamine, which clearly show significant dye leaching from PSMA/PSMA-Rhodamine Pdots prepared by physical blending, but no leaching in PFBT-PSMA-Rhodamine cross-linked Pdots.
In a separate experiment, Rhodamine-NH2 was covalently linked to PSMA first, and then the PSMA-NH-Rhodamine was physically blended with PFBT polymer to form Pdots (Supporting Scheme S2). The dye leaching experiment was further performed on this type of Pdot-Rhodamine particles, which show similar level of dye leaching as the EDC-catalyzed Pdot-Rhodamine particles (Supporting Figure S1). Therefore, these leaching results indicate the PSMA functionalization by the co-condensation method is not stable, and the targeting molecules detached from the particles can significantly reduce labeling efficiency in many cellular labeling and biological assays.
To overcome this stability problem, we developed a cross-linking strategy that covalently links the functional molecules to the Pdots. PFBT polymers with side-chain amine groups (PFBT-NH2) were synthesized based on Suzuki coupling. The cross-linker PSMA molecules comprise multiple amine-reactive units, which can covalently bond together one or several PFBT molecules in the Pdots and simultaneously create surface carboxyl groups (Supporting Scheme S2). PFBT-NH2 and PSMA was mixed in tetrahydrofuran (THF) and the reaction was allowed to proceed for 24 hours, after which a solution of Rhodamine-NH2 in DMSO was added and the reaction lasted for another 24 hours. Finally, the cross-linked Pdot-Rhodamine particles were prepared from the above mixture by the reprecipitation method.
We then used the same dye-leaching method to characterize the stability of the cross-linked Pdots. As shown in the spectra of the Pdot-Rhodamine before and after centrifugal filtration, both absorption (Figure 1c) and fluorescence spectra (Figure 1d) remain unchanged, indicating the cross-linking functionalization is highly stable and permanent. It is worth noting that the Pdots prepared from the pure PFBT-NH2 alone are positively charged, and are sticky, unstable, and prone to aggregation. Cross-linking with PSMA modified the Pdot surface to be negatively charged (Zeta potential was -57 mV), and resulted in stable, non-sticky, and functionalized Pdots. The cross-linking approach can be applied to other polymer species. For example, we have also cross-linked with another semiconducting polymer polyfluorene with side-chain amine groups (PFO-NH2), which also yielded stably functionalized Pdots, indicating the method is general for other fluorescent polymers.
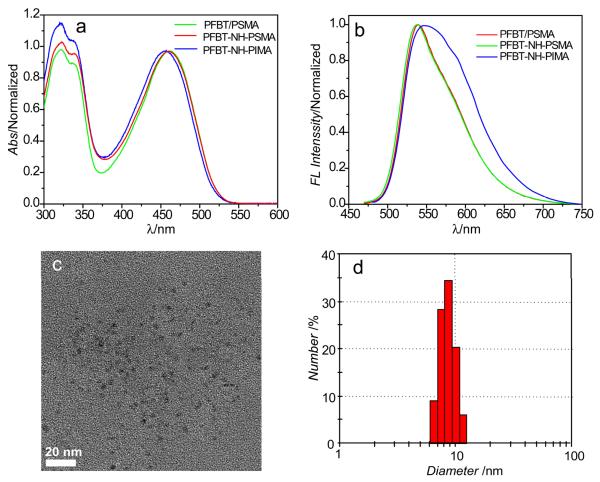
The method is also flexible and alternative functional molecules can be used as cross-linkers. We used another polymer, Poly(isobutylene-alt-maleic anhydride) (PIMA), to cross link with PFBT-NH2 to form Pdots (PFBT-NH-PIMA). After reacting PFBT-NH2 and PIMA for 24 hours, Pdots were prepared by rapid injection of a solution of the polymer mixture in THF into water under ultrasonication. We then investigated the optical properties of the cross-linked Pdots to determine any effect the cross-linking may have on the formed Pdots. Figure 2 shows the absorption and fluorescence spectra of PSMA and PIMA cross-linked Pdots as well as those physically blended with PSMA. As indicated in the absorption spectra (Figure 2a), the cross-linked PFBT-NH-PIMA Pdots showed a slight blue-shift in absorption, while the PFBT-NH-PSMA showed similar absorption profile as compared with Pdots that were physically blended with PSMA. Correspondingly, the cross-linked PFBT-NH-PIMA Pdots exhibited red-shifted and broadened fluorescence spectrum, but no obvious change was observed for the PFBT-NH-PSMA cross-linked Pdots. These results indicate the cross-linking with the polymer side chains may modify the optical properties and that spectral shift and broadening are strongly dependent on the cross-linking molecules. In this regard, the PIMA cross-linked side chains were more hydrophilic than the PSMA cross-linked ones after they were completely hydrolyzed in water, thus the resultant PFBT-NH-PIMA Pdots showed obvious red-shift and broadening in the fluorescence spectra. This finding also indicates that the cross-linking molecule needs to be carefully selected. Nevertheless, the two types of cross-linked Pdots showed relatively high quantum yields, for example, 21% for PFBT-NH-PIMA and 23% for PFBT-NH-PSMA, respectively. These values are comparable to that of the pure PFBT dots. In addition to quantum yield, photostability is another important photophysical property to consider when employing fluorescent probes for bioimaging. In previous studies, we have established that our PFBT/PS-PEG Pdots have exellent photostability. [4] Therefore, here, we wanted to compare the photostability between our cross-linked PFBT-NH-PIMA Pdots and PFBT/PS-PEG Pdots to see whether cross linking would have any effect on photostability. We found the obtained intensity trajectories of these two types of Pdots were similar (Figure S2), thus indicating they had comparable photostability and that cross linking did not have any negative effects on the photostability of Pdots.
Figure 2.
Optical and physical properties of Pdots. (a) Absorption and (b) fluorescence spectra of physically blended PFBT-PSMA Pdots, cross-linked PFBT-NH-PSMA Pdots, and cross-linked PFBT-NH-PIMA Pdots in water. (c) Typical TEM image of cross-linked PFBT-NH-PIMA Pdots. Scale bar: 20 nm. (d) Histogram showing the hydrodynamic particle size of PFBT-NH-PIMA Pdots measured by DLS (average diameter = 8.7 nm).
The particle size was investigated by transmission electron microscopy (TEM, Figure 2c) and dynamic light scattering (DLS, Figure 2d). Importantly, the PFBT-NH-PIMA cross-linked Pdots exhibit extremely small particle size (~9 nm, average hydrodynamic diameter) with a narrow size distribution. The average size of the Pdots as measured by TEM was even smaller (~ 5 nm in diameter). This discrepancy was likely caused by the fact that TEM measured the diameter of the Pdots after they had been dried on the surface, while DLS determined the diameter of Pdots in solution where they were hydrated and thus in a more “swollen” state. [26, 27] The PFBT-NH-PSMA Pdots also show small particle size (~11 nm), as compared with the PFBT dots (~20 nm) physically blended with PSMA under the same preparation conditions (Supporting Figure S3). Similarly, another semiconducting polymer PFO-NH2 was cross-linked with PIMA, and the resulting PFO-NH-PIMA Pdots also showed small particle size (~10 nm) and narrow distribution, as determined by TEM and DLS measurements (Supporting Figure S4). These results indicate that the cross-linking method provides a general approach to prepare Pdots with small particle size (~10 nm), a feat that is difficult to accomplish using nanoprecipitation of non cross-linked Pdots. Because Pdots are formed by hydrophobic interactions, covalent cross-linking with different molecules can change the hydrophobic index and molecular weight of the polymer precursor, therefore affecting the inter- or intra-molecular interactions during particle formation. These results also indicate that the side-chain modification can be useful for finely tuning the particle size for preferable applications. Furthermore, the small particle size and narrow size distribution are desirable in many biological studies.
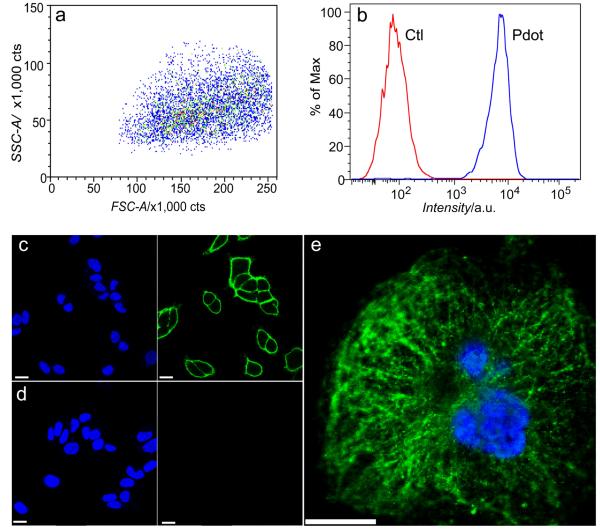
Finally, to apply Pdots for cellular imaging, we successfully performed bioconjugation of streptadvidin to the cross-linked functionalized Pdots, and demonstrated specific cell surface labeling using the Pdot-bioconjugates (Scheme 1). The final streptavidin conjugated Pdots was used to label a cell-surface receptor EpCAM, which is widely used for detection of circulating tumor cells. MCF-7 breast cancer cells were sequentially incubated with biotinylated primary anti-EpCAM antibody and Pdot-streptavidin conjugates. As shown by the flow cytometry results (Figure 3a and 3b), the Pdot-streptavidin probes effectively labeled EpCAM receptors on the cell surface, while no labeling was observed in the control sample (identical conditions but without primary antibody). Fluorescence imaging further confirmed the specific cellular labeling with Pdot probes (Figure 3 and Figure S5). As shown in Figure 3c, the Pdot-streptavidin effectively labeled EpCAM receptors on the cell surface. In the control experiments, where the biotinylated primary antibody was absent, no fluorescence on the cell surface was detected (Figure 3d), confirming specific cellular labeling without non-specific binding. Furthermore, we carried out subcellular microtubule labeling in HeLa cells (Figure 3e and Figure S5). From the images, it is noted that individual tubular structures can be clearly seen and well-resolved. This result further proves that cross-linked Pdot conjugates have compact and small size as well as ultralow nonspecific binding in the subcellular environment, because such high quality images of subcellular structures could not be observed using our larger PFBT-streptavidin probes (prepared by physical blending) under the same labeling conditions as in our previous report. [4] Therefore, in the current Pdot-bioconjugates, the biomolecules, functional molecules, and semiconducting polymer molecules are all covalently linked together, providing a small, robust, and bright fluorescent probe for extensive biological applications.
Scheme 1.
Schematic outlining the preparation of small cross-linked PFBT-NH-PIMA Pdots conjugated to streptadvidin for cellular imaging.
Figure 3.
Cellular labeling using cross-linked Pdots. (a, b) Flow cytometry measurements made using cross-linked PFBT-NH-PIMA Pdot-streptavidin. Blue curve in (b) shows the positive labeling and red curve shows the negative labeling in the control, where identical experimental conditions were followed but in the absence of primary biotinylated antibody. (c, d) Confocal fluorescence microscopy images of MCF-7 breast-cancer cells labeled with cross-linked PFBT- NH-PIMA Pdot-streptavidin. Panel c shows positive labeling and panel d shows negative labeling performed under the same condition but in the absence of biotinylated primary antibody. Images from left to right: blue fluorescence from the nucleus stain Hoechst 34580; green fluorescence images from Pdot. (e) Confocal fluorescence microscopy image of microtubules in HeLa cells labeled with cross-linked Pdot-streptavidin. The blue channel was from the nucleus stain, while the green channel showed emission from PFBT-NH-PIMA Pdots. The HeLa cells were incubated with both biotinylated anti-α-tubulin and PFBT-NH-PIMA Pdot-streptavidin. All the scale bars represent 20 μm for (c), (d) and (e).
In summary, we developed a cross-linking strategy that covalently binds functional molecules to the polymer dots while simultaneously providing functional groups for biomolecular conjugation. Semiconducting polymer with side-chain amine groups was synthesized and a functional molecule comprising multiple amine-reactive units, such as PIMA and PSMA, was used as a cross-linker. We used a dye-leaching experiment to show the stability of the cross-linked Pdots was much superior over Pdots formed using the previously described method of co-condensation of semiconducting polymers with an amphilic polymer. In addition to exhibiting high stability as anticipated, importanly cross-linked Pdots are small in size. Small Pdots may be expecially useful in live cell or in vivo studies, where size plays an important role in the targeting (e.g. with cell penetrating peptides) and biodistribution properties of the nanoparticle. Future research will focus on further controlling the size of cross-linked Pdots by adjusting the amount of cross-linker molecules and by selection of different cross-linker molecules. We conjugated streptavidin to the cross-linked Pdots and applied them to label cell surface receptors and subcellular microtubules for fluorescence imaging as well as flow cytometry experiments. This study presents a method to form covalently and stably functionalized Pdots that are significant for improving labeling efficiency and sensitivity in cellular assays.
Experimental
Materials and methods
All of the chemicals and solvents were purchased from Sigma-Aldrich unless indicated elsewhere. 1H-NMR and 13C-NMR spectra were recorded on a Bruker AV500 spectrometer. The MASS spectrum was measured by the Bruker Autoflex II Compass 1.0. The molecular weight of polymers was measured by the GPC method (Viscotek TDA305 GPC), and polystyrene was used as the standard (THF as eluent). The particle size and zeta-potentials were characterized by dynamic light scattering (Malvern Zetasizer NanoZS). For the TEM measurements, one drop of the Pdot dispersion was placed on a formvar/carbon-coated copper grid. After evaporation of the water, the nanoparticles were imaged with a transmission electron microscope (FEI Tecnai F20). UV-Vis absorption spectra were recorded in a 1 cm quartz cuvette with a DU 720 scanning spectrophotometer (Beckman Coulter, Inc., CA USA). Fluorescence spectra were obtained using a Fluorolog-3 fluorometer (HORIBA Jobin Yvon, NJ USA). Fluorescence quantum yields were measured using a Hamamatsu photonic multichannel analyzer C10027 equipped with a CCD integrating sphere. Flow cytometry was operated on a BD FACS Canto flow cytometer (BD Biosciences, San Jose, CA, USA). A 488 nm laser was used for excitation and emission was collected through Alex 488 channel equipped with a 500 nm long-pass filter and a 530/30 nm band-pass filter.
Preparation of PFBT/PSMA Pdots by co-condensation and Rhodamine conjugation
First, PFBT/PSMA Pdots were prepared by using the reprecipitation method. PFBT in THF solution (250 μL, 1 mg/mL) and PSMA in THF solution (50 μL, 1 mg/mL) were added to anhydrous THF (4.7 mL). The polymer mixture was injected quickly to MilliQ water (10 mL) under sonication. THF was removed by nitrogen stripping, and the solution was concentrated to 5 mL (50 μg/mL) by continuous nitrogen stripping on a 90°C hotplate, followed by filtration through a 0.2 micron filter. The above Pdot solution (4 mL) was used to react with Rhodamine-NH2 by the EDC catalyzed reaction. Rhodamine-NH2 in DMSO solution (2 μL, 5 mg/mL) was added to Pdot solution (4 mL, 50 μg/mL) under magnetic stirring, and then fresh EDC in water (80 μL, 5 mg/mL) was added to the mixture. The solution was stirred for 4 hours at room temperature. The Pdot-Rhodamine particles were then purified from free Rhodamine molecules by a Bio-Rad Econo-Pac 10DG column (Hercules, CA, USA) to remove the free Rhodamine-NH2 molecules.
Covalent cross-linking PSMA with PFBT-NH2 and Rhodamine-NH 2
PFBT-3.5%NH2 (10 mg) and PSMA (2 mg) were dissolved in anhydrous THF (50 mL). The mixture was kept stirring for 24 h under reflux and N2 flow protection. Then, Rhodamine-NH2 in anhydrous DMSO solution (70 μL, 5 mg/mL) was added into the above reacted solution, and the reaction was allowed to proceed for another 24 h. The resultant solution was evaporated to remove solvent and was washed with ethanol 3 times (10 mL/time); furthermore the product was dried under vacuum. Finally, the powder product was dissolved in THF and filtered through a 0.2 μm PVDF filter. The obtained solution was adjusted to have the concentration of 1 mg/mL for subsequent Pdot preparation.
Covalent cross-linking of PSMA with PFBT-NH2
PFBT-3.5%NH2 (10 mg) and PSMA (2 mg) were dissolved in anhydrous THF (50 mL). Then, the mixture was kept stirring under reflux and N2 flow for 48 h. The resultant solution was evaporated to remove solvent and then dried under vacuum. Finally, the product was dissolved in THF and filtered through a 0.2 μm PVDF filter. The obtained solution was adjusted to have the concentration of 1 mg/mL for subsequent Pdot preparation.
Covalent cross-linking of PIMA with PFBT-NH 2 or PFO-NH 2
PFBT-3.5%NH2 (12 mg) or PFO-5%NH2 and PIMA (3 mg) were dissolved in THF (50 mL). Then the mixture was kept stirring under reflux and N2 flow protection for 48 h. The resultant solution was evaporated to remove solvent and then dried under vacuum. Finally, the product was dissolved in THF and filtered through a 0.2 μm PVDF filter. The obtained solution was adjusted to have the concentration of 1 mg/mL for subsequent Pdot preparation.
Preparation of cross-linked Pdots
The cross-linked Pdots were prepared from the cross-linked polymer precursor obtained above by the reprecipitation method. Briefly, the cross-linked polymer (PFBT-NH-PIMA, or PFBT-NH-PSMA, or PFO-NH-PIMA) was diluted in THF solution to a concentration of 0.05 mg/mL. The diluted polymer in THF solution (5 mL) was injected quickly into MilliQ water (10 mL) under sonication. THF was removed by nitrogen stripping, and the solution was concentrated by continuous nitrogen stripping to 5 mL on a 90 °C hotplate, followed by filtration through a 0.2 micron filter.
Supplementary Material
Acknowledgment
We gratefully acknowledge support from the National Institutes of Health (CA147831 and NS062725).
Footnotes
Supporting Information Available Synthesis of polymers, cellular experiments (cell culture, flow cytometry, cellurlar imaging set up and protocols), absorption and fluorescence spectra of PFBT/PSMA-Rhodamine Pdots, DLS data of PFBT-NH-PSMA Pdots, DLS and TEM data of PFO-NH-PIMA Pdots, fluorescence images of Pdot labeled cells. This material is available free of charge via the internet at the Wiley Online Library.
Contributor Information
Dr. Jiangbo Yu, Department of Chemistry, University of Washington Seattle, Washington 98195, United States.
Prof. Changfeng Wu, State Key Laboratory on Integrated Optoelectronics, College of Electronic Science and Engineering, Jilin University Changchun 130012, China.
Dr. Xuanjun Zhang, Department of Chemistry, University of Washington Seattle, Washington 98195, United States
Dr. Fangmao Ye, Department of Chemistry, University of Washington Seattle, Washington 98195, United States
Dr. Maria Elena Gallina, Department of Chemistry, University of Washington Seattle, Washington 98195, United States
Dr. Yu Rong, Department of Chemistry, University of Washington Seattle, Washington 98195, United States
Dr. Yizhe Wu, Department of Chemistry, University of Washington Seattle, Washington 98195, United States
Dr. Wei Sun, Department of Chemistry, University of Washington Seattle, Washington 98195, United States
Dr. Yang-Hsiang Chan, Department of Chemistry, University of Washington Seattle, Washington 98195, United States
Prof. Daniel T. Chiu, Department of Chemistry, University of Washington Seattle, Washington 98195, United States.
References
- [1].Wu C, Bull B, Szymanski C, Christensen K, McNeill J. ACS Nano. 2008;2:2415. doi: 10.1021/nn800590n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yu J, Wu C, Sahu S, Fernando L, Szymanski C, McNeill J. J. Am. Chem. Soc. 2009;131:18410. doi: 10.1021/ja907228q. [DOI] [PubMed] [Google Scholar]
- [3].Yu J, Wu C, Tian Z, McNeill J. Nano Lett. 2012;12(3):1300. doi: 10.1021/nl203784m. [DOI] [PubMed] [Google Scholar]
- [4].Wu C, Schneider T, Zeigler M, Yu J, Schiro P, Burnham D, McNeill JD, Chiu DT. J. Am. Chem. Soc. 2010;132:15410. doi: 10.1021/ja107196s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wu C, Jin Y, Schneider T, Burnham DR, Smith PB, Chiu DT. Angew. Chem. Int. Ed. 2010;49:9436. doi: 10.1002/anie.201004260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kaeser A, Schenning APHJ. Adv. Mater. 2010;22:2985. doi: 10.1002/adma.201000427. [DOI] [PubMed] [Google Scholar]
- [7].Pecher J, Mecking S. Chem. Rev. 2010;110:6260. doi: 10.1021/cr100132y. [DOI] [PubMed] [Google Scholar]
- [8].Petkau K, Kaeser A, Fischer I, Brunsveld L, Schenning APHJ. J. Am. Chem. Soc. 2011;133:17063. doi: 10.1021/ja2075345. [DOI] [PubMed] [Google Scholar]
- [9].Ko Y-J, Mendez E, Moon JH. Macromolecules. 2011;44:5527. doi: 10.1021/ma200661h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schutze F, Stempfle B, Jungst C, Woll D, Zumbusch A, Mecking S. Chem. Comm. 2012;48 doi: 10.1039/c2cc17066c. [DOI] [PubMed] [Google Scholar]
- [11].Childress ES, Roberts CA, Sherwood DY, LeGuyader CLM, Harbron EJ. Anal. Chem. 2012;84:1235. doi: 10.1021/ac300022y. [DOI] [PubMed] [Google Scholar]
- [12].Rahim NAA, McDaniel W, Bardon K, Srinivasan S, Vickerman V, So PTC, Moon JH. Adv. Mater. 2009;21:3492. [Google Scholar]
- [13].Feng X, Lv F, Liu L, Tang H, Xing C, Yang Q, Wang S. ACS Appl. Mater. Interfaces. 2010;2:2429. doi: 10.1021/am100435k. [DOI] [PubMed] [Google Scholar]
- [14].Pu K-Y, Li K, Zhang X, Liu B. Adv. Mater. 2010;22:4186. doi: 10.1002/adma.201001544. [DOI] [PubMed] [Google Scholar]
- [15].Li K, Liu Y, Pu K-Y, Feng S-S, Zhan R, Liu B. Adv. Funct. Mater. 2011;21:287. [Google Scholar]
- [16].Ye FM, Wu CF, Jin YH, Chan YH, Zhang XJ, Chiu DT. J. Am. Chem. Soc. 2011;133:8146. doi: 10.1021/ja202945g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chan YH, Wu CF, Ye FM, Jin YH, Smith PB, Chiu DT. Anal. Chem. 2011;83:1448. doi: 10.1021/ac103140x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ye F, Wu C, Jin Y, Wang M, Chan Y-H, Yu J, Sun W, Hayden S, Chiu DT. Chem. Comm. 2012;48:1778. doi: 10.1039/c2cc16486h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jin Y, Ye F, Wu C, Chan Y-H, Chiu DT. Chem. Comm. 2012;48 doi: 10.1039/c2cc17703j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Howes P, Green M, Levitt J, Suhling K, Hughes M. J. Am. Chem. Soc. 2010;132:3989. doi: 10.1021/ja1002179. [DOI] [PubMed] [Google Scholar]
- [21].Landfester K. Annu. Rev. Mater. Res. 2006;36:231. [Google Scholar]
- [22].Frazer RQ, Byron RT, Osborne PB, West KP. J Long Term Eff. Med. Implants. 2005;15:11. doi: 10.1615/jlongtermeffmedimplants.v15.i6.60. [DOI] [PubMed] [Google Scholar]
- [23].Fernando LP, Kandel PK, Yu J, McNeill J, Ackroyd PC, Christensen KA. Biomacromolecules. 2010;11:2675. doi: 10.1021/bm1007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sun B, Sun MJ, Gu Z, Shen QD, Jiang SJ, Xu Y, Wang Y. Macromolecules. 2010;43:10348. [Google Scholar]
- [25].Wu C, Hansen S, Hou Q, Yu J, Zeigler M, Jin Y, Burnham D, McNeill J, Olson J, Chiu DT. Angew. Chem. Int. Ed. 2011;50:3430. doi: 10.1002/anie.201007461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cheng F, Zhang K, Chen D, Zhu L, Jiang M. Macromolecules. 2009;42:7108. [Google Scholar]
- [27].Ogawa M, Kataoka H, Nitahara S, Fujimoto H, Aoki H, Ito S, Narazaki M, Matsuda T. Bull. Chem. Soc. Jap. 2012;85:79. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.