Abstract
Mammalian CST (CTC1-STN1-TEN1) associates with telomeres and depletion of CTC1 or STN1 causes telomere defects. However, the function of mammalian CST remains poorly understood. We show here that depletion of CST subunits leads to both telomeric and non-telomeric phenotypes associated with DNA replication defects. Stable knockdown of CTC1 or STN1 increases the incidence of anaphase bridges and multi-telomeric signals, indicating genomic and telomeric instability. STN1 knockdown also delays replication through the telomere indicating a role in replication fork passage through this natural barrier. Furthermore, we find that STN1 plays a novel role in genome-wide replication restart after hydroxyurea (HU)-induced replication fork stalling. STN1 depletion leads to reduced EdU incorporation after HU release. However, most forks rapidly resume replication, indicating replisome integrity is largely intact and STN1 depletion has little effect on fork restart. Instead, STN1 depletion leads to a decrease in new origin firing. Our findings suggest that CST rescues stalled replication forks during conditions of replication stress, such as those found at natural replication barriers, likely by facilitating dormant origin firing.
Keywords: CTC1, DNA replication, replication origin, STN1, telomeres
Introduction
Mammalian telomeres consist of kilobases of duplex T2AG3/C3TA2 repeats bound by a six-protein complex called shelterin (Palm and de Lange, 2008; Stewart et al, 2012). The G-rich strand ends in a 12–400 nt ssDNA 3′ overhang which is proposed to invade the duplex region to create a telomeric loop (t-loop) that caps the chromosome terminus. Telomeres pose a unique problem for the replication machinery due to their repetitive nature and unusual terminal structure (Gilson and Geli, 2007; Stewart et al, 2012). The duplex region is replicated by the conventional DNA replication machinery. However, the heterochromatic nature of this region and its potential to form secondary structures appear to impede passage of the replication fork (Paeschke et al, 2010). The t-loop may provide an additional barrier to the fork. In human cells that express telomerase, the 3′ overhang is elongated by telomerase soon after passage of the replication fork (Zhao et al, 2009). The extended overhang is then partially filled at the end of S-phase. This C-strand fill-in is a stepwise process and is thought to occur through the recruitment of DNA polymerase α-primase (pol α) (Zhao et al, 2009).
Recent findings indicate that a number of extra proteins are needed in addition to the standard replication machinery to properly replicate the telomeric duplex. These include TRF1, FEN1, BLM, RTEL, RECQL4, BRCA2 and RAD51 (Sfeir et al, 2009; Badie et al, 2010; Saharia et al, 2010; Ghosh et al, 2011). Depletion of these proteins causes the appearance of multi-telomeric signals (MTS) during telomere FISH analysis on metaphase chromosomes. MTS have also been called fragile telomeres because they share features of common fragile sites which are observed cytogenetically as gaps or breaks in chromosomes (Durkin and Glover, 2007). While the actual nature of the MTS is poorly understood, like fragile sites, they can form under conditions of replication stress and replication fork stalling (Durkin and Glover, 2007; Chan et al, 2009; Sfeir et al, 2009).
Fork stalling can be induced by a number of factors, such as repetitive or complex DNA sequences, depletion of nucleotide pools and DNA damage (Durkin and Glover, 2007; Branzei and Foiani, 2010; Petermann and Helleday, 2010). Once stalled, replication must be rapidly restarted to maintain genome stability. In the absence of restart, replication forks collapse leading to regions of ssDNA, DNA double-strand breaks and unwanted recombination events. The mechanisms underlying replication restart at telomeres, fragile sites and other sites of difficult-to-replicate DNA are not fully understood. The work described here indicates that the recently identified mammalian CTC1-STN1-TEN1 (CST) complex promotes DNA replication restart at both telomeric and non-telomeric sites.
Mammalian CST resembles the Cdc13-Stn1-Ten1 complex (ScCST) from Saccharomyces cerevisiae in that the STN1 and TEN1 subunits are conserved, both complexes share a structural similarity to Replication Protein A (RPA) and both bind tightly to ssDNA (Giraud-Panis et al, 2010; Price et al, 2010). ScCST is responsible for protecting yeast telomeres through sequence-specific binding to the G-strand overhang (Pennock et al, 2001; Shore and Bianchi, 2009) and for coordinating G- and C-strand synthesis during telomere replication via interactions with telomerase and pol α (Qi and Zakian, 2000; Chandra et al, 2001; Puglisi et al, 2008). Mammalian CST also localizes to telomeres and its depletion causes changes in telomere structure (see below) indicating a role in telomere maintenance (Miyake et al, 2009; Surovtseva et al, 2009). Unlike its yeast counterpart, mammalian CST does not appear to be required for telomere protection but it may play a similar role in telomere replication.
The proposed role for mammalian CST is in the C-strand fill-in that occurs following extension of the 3′ overhang by telomerase (Price et al, 2010). Recent studies show that this fill-in occurs hours after replication of the telomere duplex (Zhao et al, 2009). Since a replisome is unlikely to be present hours after telomeres have replicated and telomerase has acted, CST is proposed to recruit pol α to initiate the fill-in reaction (Zhao et al, 2009). However, several lines of evidence suggested that CST might also have non-telomeric functions. First, CST binding to ssDNA is sequence independent and only ∼20% of STN1 foci localize to telomeres (Miyake et al, 2009). Second, depletion of CTC1 results in increased γH2AX foci, which do not colocalize with telomeres (Miyake et al, 2009; Surovtseva et al, 2009). Finally, CTC1 and STN1 were initially isolated as subunits of a pol α accessory factor (AAF) that increases pol α processivity and affinity for an ssDNA template (Goulian et al, 1990; Casteel et al, 2009). More recent work indicates that Xenopus CST shares many of these same properties (Nakaoka et al, 2012).
Interestingly, a new series of clinical studies has identified mutations in the CTC1 subunit of CST as the cause of human disease (Anderson et al, 2012; Keller et al, 2012; Polvi et al, 2012). Initially, CTC1 mutations were shown to underlie Coats plus, a severe pleiotropic disorder with many clinical manifestations ranging from retinal telangiectasia, intracranial calcification with leukodystrophy and brain cysts, to predisposition to fractures and gastrointestinal bleeding. A subset of the clinical features of Coats plus overlap those found in disorders caused by deficiencies in telomere maintenance, the so-called telomere syndromes. Moreover, analysis of telomere length revealed that cells from some, but not all, patients had shortened telomeres (Anderson et al, 2012; Polvi et al, 2012). Most recently, a CTC1 mutation has been found to manifest as dyskeratosis congenita with the classical features of a defect in telomere maintenance (Keller et al, 2012). Thus, the clinical manifestations and cellular phenotypes of CTC1 mutations support both telomeric and non-telomeric roles for CST in human biology.
Here, we provide evidence that CST has both telomeric and non-telomeric roles in resolving problems associated with DNA replication. At the telomere, stable knockdown of either CTC1 or STN1 caused MTS and delayed telomere replication. STN1 depletion also leads to a general decrease in both new origin firing and the resumption of DNA replication following treatment with hydroxyurea (HU) to induce genome-wide fork stalling. Together, our findings suggest that human CST plays a key role in replication restart as a specialized replication factor, which promotes DNA replication under conditions of replication stress or at natural replication barriers.
Results
Knockdown of CTC1 or STN1 promotes genome instability and MTS
We previously showed that acute depletion of human CTC1 with siRNA leads to an increase in γH2AX staining, chromatin bridges and a variety of telomere defects including increased telomere loss and G-overhang elongation (Surovtseva et al, 2009). To determine whether STN1 knockdown causes similar defects, we created cell lines with stable knockdown of either STN1 or CTC1 by infecting HeLa 1.2.11 cells with shRNA-encoding lentivirus. Single cell clones were established and the level of knockdown was assessed by immunoblotting and RT–qPCR for STN1 and RT-qPCR alone for CTC1 (due to the lack of a suitable antibody). STN1 mRNA levels were decreased by 60–80% and the protein was barely detectable (Figure 1A and B). CTC1 mRNA levels were decreased by ∼70% (Figure 1B). One of the shSTN1 clones (shSTN1-7) was subsequently transfected with a construct expressing sh-resistant Flag-tagged STN1 (shSTN1-7 Res; Figure 1A) and the resulting cells were used to verify that phenotypes were specific to STN1 depletion and not off-target effects.
Figure 1.
Depletion of CTC1 or STN1 causes genomic instability in HeLa 1.2.11 cells. (A) Western blot showing knockdown of STN1 (42 kDa) in shSTN1 clones and re-expression of an sh-resistant Flag-STN1 (shSTN1-7 Res). Loading control is α-Actinin (100 kDa). Lanes 1–3 contain 25 μg of protein and lane 4 contains 10 μg. Numbers below gel indicate the level of STN1 relative to non-target control (shNT) after normalization to α-Actinin. (B) RT–qPCR of STN1 and CTC1 mRNA in different clones. Levels are relative to shNT with normalization to GAPDH (mean±s.e.m., n=3 independent experiments). (C, D) Anaphase bridges observed after release of control, shSTN1 or shCTC1 clones from nocadozole block (mean±s.e.m., n⩾3 independent experiments). NT, non-target; WT, wild type.
The shSTN1 clones divided at a normal rate and showed no significant growth defects (Supplementary Figure 1A and see below). These results contrast to previous findings with HeLa cells in which acute siRNA knockdown of STN1 caused cell death (Dai et al, 2010). The robust growth of the shRNA clones probably reflects the lower level of knockdown.
Our previous studies showed that complete loss of CTC1 causes anaphase bridge formation in Arabidopsis (Surovtseva et al, 2009). Whether lack of CTC1 has a similar effect in human cells was not established because very few cells entered metaphase after the acute CTC1 depletion (Surovtseva et al, 2009). When we examined the HeLa 1.2.11 stable CTC1 and STN1 knockdown cell lines we found they had normal levels of metaphase and anaphase cells and we observed that depletion of either protein caused a significant increase in the frequency of anaphase bridges (Figure 1C and D). The increase due to STN1 depletion was largely prevented by expression of the sh-resistant Flag-STN1. The STN1 knockdown clones also exhibited other phenotypes previously observed after siRNA knockdown of CTC1 including an increase in the number of micronuclei and an increase in the average length of the telomeric G-strand overhang (Supplementary Figure 1B and C), suggesting that, like CTC1, STN1 prevents genome instability and helps maintain G-overhang length.
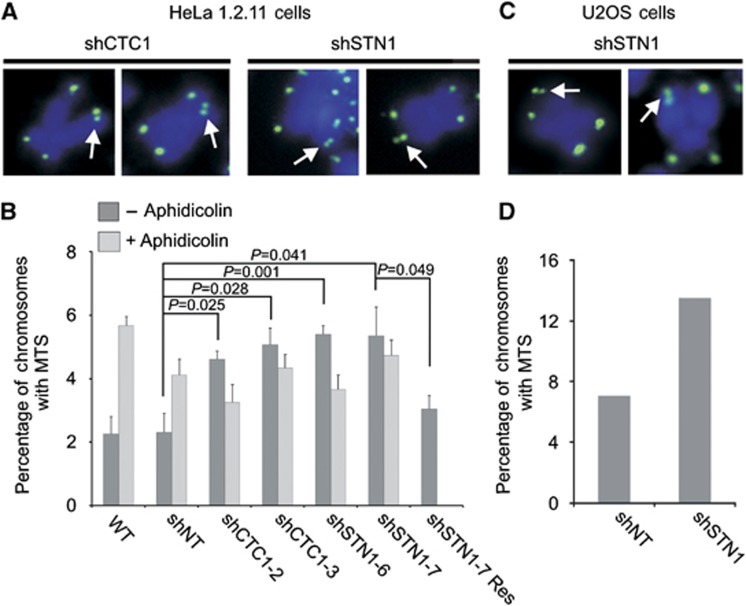
One difference in the phenotype seen after acute CTC1 knockdown versus stable CTC1 or STN1 depletion was the nature of the telomere defects visible by fluorescence in-situ hybridization (FISH). While acute knockdown of CTC1 in HeLa S3 cells (a strain with telomeres of 3–7 kb) yielded an increase in chromosomes lacking telomeric FISH signals (Surovtseva et al, 2009), a similar increase in signal-free ends was not observed in the shCTC1 and shSTN1 HeLa 1.2.11 clones (telomeres of 10–20 kb). Instead, and as previously reported, we observed an increase in the number of chromosome ends with MTS (Figure 2A and B; Supplementary Figure 2A; Price et al, 2010). To further explore this finding, we examined MTS occurrence in two separate shSTN1 and shCTC1 clones and after rescue of the STN1 knockdown with the sh-resistant Flag-STN1. With each knockdown clone, we observed an approximately two-fold increase in MTS that was rescued in the sh-resistant Flag-STN1 cell line (Figure 2B; Supplementary Table 1).
Figure 2.
CTC1 or STN1 depletion cause multi-telomeric signals (MTS). (A, C) Telomere FISH of HeLa 1.2.11 shCTC1 or shSTN1 clones (A) or U2OS shSTN1 clone (C) showing examples of MTS (white arrows). Green, FITC-telomere probe; blue, DAPI. (B) Quantification of MTS in HeLa 1.2.11 cell lines. Metaphase spreads were made from cells grown±0.25 μg/ml aphidicolin for 16 h prior to the addition of colchicine or colcemid (mean±s.e.m., n⩾3 independent experiments). (D) Quantification of MTS from a single experiment with a U2OS STN1 knockdown clone. NT, non-target; WT, wild type.
To determine whether our ability to detect signal-free ends in HeLa S3 after CTC1 siRNA knockdown but not in HeLa 1.2.11 after stable CTC1 knockdown reflected use of different HeLa strains or the different knockdown approach, we used siRNA to deplete CTC1 in HeLa 1.2.11 cells (Supplementary Figure 2B; Supplementary Table 1). The siRNA depletion caused a large increase in signal-free ends and a smaller, but consistent, increase in MTS (Supplementary Figure 2C). Thus, acute CTC1 knockdown appears to favour telomere loss over MTS formation while stable knockdown of CTC1 or STN1 causes MTS alone. It is likely that acute knockdown of CTC1 in the HeLa S3 cells also caused some MTS but the short telomeres made them difficult to detect.
We next examined whether stable STN1 knockdown caused MTS or loss of telomere signal in U2OS cells, an ALT cell line with long heterogeneous telomeres (>20 kb). As with the HeLa 1.2.11 cells, we observed an increase in MTS but not in signal-free ends (Figure 2C and D; Supplementary Figure 3C and D; Supplementary Table 1). Interestingly, STN1 knockdown caused a significant growth defect in U2OS cells. Although the level of knockdown was similar to that observed in the HeLa 1.2.11 cells, the STN1-depleted U2OS cells grew more slowly than control U2OS cells (Supplementary Figure 3A and B) and single cell clones only survived for a few weeks, causing us to examine a knockdown pool to verify the MTS phenotype (Supplementary Figure 3C and D). Overall, we conclude that both STN1 and CTC1 are needed for genome stability and telomere maintenance.
CST depletion slows replication through the telomeric tract
Given that the appearance of MTS can reflect problems associated with replication through the duplex region of the telomere (Sfeir et al, 2009; Saharia et al, 2010), we suspected that the MTS caused by STN1 or CTC1 depletion might stem from a similar cause. To explore this possibility, we examined the effect of combining STN1 or CTC1 knockdown with aphidicolin treatment. Aphidicolin causes replication stress by inhibiting DNA polymerase α, δ and ε (Cheng and Kuchta, 1993) and is known to induce MTS (Sfeir et al, 2009). As expected, aphidicolin treatment caused an increase in MTS levels in both the WT and non-target (shNT) control cell lines (Figure 2B; Supplementary Table 1). However, treatment with aphidicolin in the context of STN1 or CTC1 depletion resulted in an epistatic-like interaction in which the levels of MTS remained similar or slightly decreased relative to those observed without aphidicolin treatment. These findings suggest that CST and DNA polymerase α, δ and/or ε act within a common pathway to prevent MTS formation. While the experiment does not address the exact step at which CST and aphidicolin interface, it supports a role for CST in replication of the telomeric duplex.
To test more directly for the role of CST in telomere replication, we next asked whether STN1 depletion delays the overall rate of replication through either the telomere duplex or the bulk of the genome. For this experiment, the HeLa 1.2.11 shSTN1-7, shSTN1-7Res and shNT clones were synchronized at G1/S with a double-thymidine block, released into fresh media and allowed to enter S-phase (Supplementary Figures 4E and 5E). Cells were then pulsed labelled with either BrdU or EdU for consecutive 1.5 h intervals (Figure 3A) and harvested at the end of each time point. First, we examined the overall rate of genome replication. The EdU-labelled cells were fixed, the EdU was reacted with fluorophore and the relative amount of EdU uptake measured by FACS. As shown in Figure 3B and Supplementary Figures 4A, 4B, 5A and 5B, the rate of EdU uptake in the shSTN1, shSTN1-7-Res and control shNT cells was essentially identical throughout S-phase indicating that, at this level of knockdown, STN1 depletion does not affect the rate of whole genome replication.
Figure 3.
STN1 depletion delays telomere replication but does not affect the rate of bulk genomic DNA replication. (A) Experimental timeline. HeLa 1.2.11 cells were released from a double-thymidine block into S-phase and incubated with BrdU or EdU for consecutive 1.5 h intervals. (B) Rates of bulk genomic DNA replication were determined by EdU uptake. Graph shows EdU incorporated at consecutive time periods (Mean EdU staining X % EdU-positive cells). (C–E) Rates of telomere replication throughout S-phase. (C) BrdU-labelled DNA from 4.5 and 6 h time points was subject to CsCl sedimentation to separate leading and lagging strand telomeres. Telomeric DNA from each gradient fraction was quantified by slot blot hybridization. (D) Per cent of newly replicated leading strand telomere signals relative to the total telomere signal for each time period throughout S-phase. (E) Examples of slot blot used to obtain data in (C) and (D). Data are representative of three independent experiments. NT, non-target.
Next, we quantified the amount of telomeric DNA replicated at each time point throughout S-phase. DNA from the BrdU-labelled cells was isolated, restriction digested and subjected to CsCl density gradient centrifugation to separate unreplicated and replicated telomeres (Chai et al, 2006). The gradient was fractionated and the relative amount of telomeric DNA in each fraction was quantified by slot blot hybridization using a telomeric DNA probe (Figure 3C–E; Supplementary Figures 4C, 4D, 5C and 5D). Telomeres replicated by leading strand synthesis incorporate multiple BrdU molecules per telomeric repeat (TTAGGG) and hence sediment at a higher density than telomeres replicated by lagging strand synthesis (CCCTAA) and both are separated from any unreplicated telomeric DNA (Chai et al, 2006; Zhao et al, 2011).
Quantification of replicated telomeric DNA from the shNT, shSTN1-7 and shSTN1-7 Res cells revealed a considerable difference in the timing of telomere replication in the control versus the STN1-depleted cells (Figure 3C–E; Supplementary Figures 4C, 4D, 5C and 5D). Although all three cell types initiated telomere replication in a similar manner, the STN1 knockdown cells completed replication more slowly such that telomere replication reached a maximum and then declined 1.5–3 h earlier in the control cells (Figure 3D; Supplementary Figures 4D and 5D). While there was some experiment-to-experiment variation in the timing of maximal telomere replication, the delay in the STN1 knockdown cells was very consistent and was readily apparent regardless of whether we quantified the total amount of replicated telomere DNA (Supplementary Figure 4D) or the amount of the leading strand peak (which was more visible at early time points) (Figure 3D; Supplementary Figure 5D). Thus, our results indicate that STN1 depletion slows replication of the telomere duplex without affecting the rate of bulk genomic DNA replication. We therefore conclude that components of the CST complex play a specific role in promoting efficient replication of the telomeric tract. The above findings also provide strong support for our proposal that the MTS observed after STN1 or CTC1 depletion result from problems associated with telomere replication.
STN1 and TRF1 promote telomere replication via different pathways
Since CST does not appear to be a general replication factor, possible functions for STN1 in replication of the telomere duplex DNA could include helping prevent replication fork stalling or promoting subsequent replication restart. To learn more about the role of CST in these processes, we next examined the effect of STN1 and TRF1 co-depletion on MTS frequency. TRF1 helps prevent fork stalling during telomere replication and the increased stalling caused by TRF1 depletion results in elevated MTS (Martinez et al, 2009; Sfeir et al, 2009). Thus, analysis of MTS levels after co-depletion of STN1 and TRF1 should indicate whether STN1 affects fork stalling via the same or different pathways.
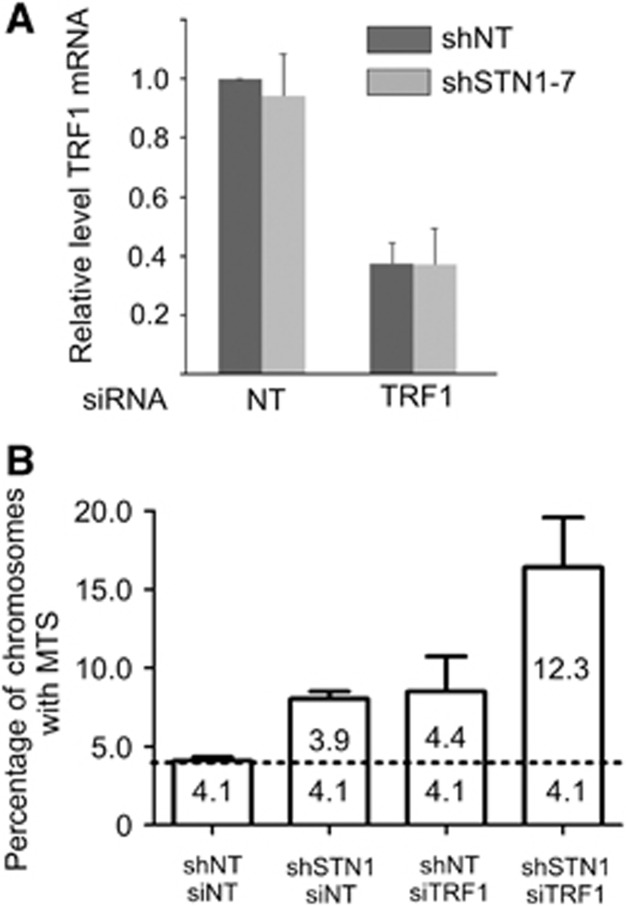
TRF1 was depleted in the HeLa 1.2.11 shSTN1-7 clone by transfecting cells twice, 24 h apart, with a previously characterized siRNA (Ohishi et al, 2010). Forty-eight hours after the second transfection, RNA was extracted and the level of knockdown measured by RT–qPCR (Figure 4A). Telomere FISH was performed 48 h after the second transfection and the number of MTS determined. As expected, MTS levels were increased approximately two-fold above background with either TRF1 or STN1 single knockdown (Figure 4B). The overall background levels of MTS were higher than observed in previous experiments with HeLa 1.2.11 (compare Figure 2B and Figure 4B). This was most likely due to the siRNA transfection as the fold increase with STN1 knockdown was similar in both experiments. When we compared MTS levels caused by co-depletion of STN1 and TRF1 relative to those observed after STN1 or TRF1 single knockdown, we consistently observed a greater than additive increase in MTS (Figure 4B; Supplementary Table 1). This result indicates that STN1 and TRF1 affect different processes during telomere replication. Given that CTC1 and STN1 appear to function in conjunction with DNA polymerase to prevent MTS formation (Figure 2B) and CST acts as a DNA pol α affinity factor (Goulian and Heard, 1990; Goulian et al, 1990), the result also suggests that the increase in MTS after STN1 depletion might reflect a role for STN1 in replication restart rather than in the prevention of fork stalling.
Figure 4.
Co-depletion of TRF1 and STN1 causes an additive increase in MTS. (A) Relative level of TRF1 mRNA 48 h after siRNA transfection as measured by RT–qPCR with normalization to GAPDH (mean±s.e.m., n=3 independent experiments). (B) Quantification of MTS (mean±s.e.m., n=3 independent experiments). Dashed line indicates the background level of MTS. NT, non-target.
STN1 promotes genome-wide replication restart after HU-induced fork stalling
Since CST appears to have both telomeric and non-telomeric functions, we hypothesized that CST might promote recovery from replication fork stalling at non-telomeric locations. To test this possibility, we examined whether STN1 promotes DNA replication restart after fork stalling across the genome. Replication fork stalling was induced by treatment with HU, a ribonucleotide reductase inhibitor, which stalls DNA polymerization by depleting nucleotide pools (Koc et al, 2004). Cells were treated with moderately high levels of HU (2 mM) over a short time frame (2 h) to avoid inducing fork collapse (Petermann and Helleday, 2010; Petermann et al, 2010).
After HU treatment, cells were released into media containing EdU for 30 min to label cells that resumed replication. Cells were then fixed, the EdU was reacted with fluorophore and actively replicating cells identified by immunofluorescence (Figure 5A). Following image capture, the mean fluorescence intensity of the EdU signal was quantified. The results are presented as both the per cent of nuclei at different Arbitrary Fluorescence Units (AFU) (Figure 5B) and the average fluorescence intensity of all nuclei counted (Figure 5C). HU-induced fork stalling was verified by the lack of EdU incorporation during HU treatment.
Figure 5.
Replication restart after HU treatment is inhibited by STN1 depletion. (A–C) Cells were treated for 2 h with 2 mM HU and released into medium containing EdU for 30 min. (A) EdU incorporation by HeLa 1.2.11 clones after release from HU. Blue, DAPI; green, EdU. (B) Quantification of the levels of EdU uptake after release from HU, as measured by mean fluorescence intensity (mean±s.e.m., n⩾3 independent experiments). Each bar indicates the total number of nuclei above or below the AFU given below. AFU, arbitrary fluorescence units. Nuclei below 10 AFU are considered as EdU negative, those above 10 AFU are EdU positive. (C) Average AFU values of all nuclei following HU removal for both HeLa1.2.11 knockdown clones and pools of U2OS knockdown cells (mean±s.e.m., n⩾3 independent experiments). NT, non-target.
As anticipated, recovery from HU treatment resulted in a significant decrease in EdU uptake relative to untreated cells, reflecting gradual recovery from fork stalling. Interestingly, the HU caused a slower recovery (less EdU uptake) in the shSTN1 HeLa 1.2.11 clones than in the shNT control cells (Figure 5C). Furthermore, STN1 depletion caused the per cent of EdU-negative nuclei (mean AFU ⩽10) to increase and the per cent of nuclei with higher levels of EdU incorporation (AFU >20) to decrease (Figure 5B). These effects were largely rescued by expression of the sh-resistant Flag-STN1 allele (Figure 5B and C). Importantly, without HU treatment, the per cent of EdU-positive cells and the levels of EdU incorporation were similar in the shSTN1 and control HeLa 1.2.11 cells, indicating that the decrease in EdU incorporation after HU treatment was not due to inherent differences in the number of cells in S-phase or rates of replication (Figure 5C). We therefore conclude that STN1 depletion delays replication restart after fork stalling in HeLa 1.2.11 cells.
To examine whether the deficiency in replication restart was a general phenomenon, we also examined EdU incorporation in STN1-depleted U2OS cells (Figure 5C; Supplementary Figure 6). As observed with HeLa 1.2.11 cells, U2OS cells showed a greater decrease in EdU incorporation after release from HU than the shNT control cells. For the U2OS shSTN1 cells, the decrease in EdU incorporation was greater than could be accounted for by their slower growth (Supplementary Figure 6B). Overall, our results indicate that STN1 not only facilitates replication through telomeres but also genome-wide replication restart after fork stalling.
Checkpoint activation through CHK1 is unaffected by depletion of STN1
ATR signalling appears to stabilize stalled replication forks, thus preventing fork collapse and allowing resumption of DNA replication through the restart of stalled forks (Paulsen and Cimprich, 2007; Chanoux et al, 2009). Since a variety of factors found at the replication fork (e.g., Tim/Tipin and Claspin) enhance ATR signalling and hence the stability of stalled forks (Smith et al, 2009; Leman et al, 2010; Kemp et al, 2010), we questioned whether STN1 or the CST complex might also promote replication restart in this manner. The question seemed particularly pertinent given the ability of CTC1 and STN1 to act as a pol α affinity factor (Goulian et al, 1990). In Xenopus, fork stalling causes hyperloading of pol α and synthesis of short DNA primers which are required for loading of the RAD9-RAD1-HUS1 checkpoint clamp and subsequent ATR activation (Yan and Michael, 2009; Van et al, 2010). Thus, a possible function of CST would be to load pol α for primer synthesis and ATR signalling.
To assess this possibility, we chose to examine phosphorylation of the downstream target CHK1 (Paulsen and Cimprich, 2007) after HU treatment. HeLa 1.2.11 shSTN1 and shNT clones were treated with HU for 1 h and then allowed to recover for 10 or 30 min. Whole cell lysates were analysed by western blot using antibody to phospho-CHK1. Following HU treatment, we observed robust CHK1 phosphorylation in multiple shSTN1 clones. While the actual levels of total CHK1 and phospho-CHK1 varied in a clone-specific manner, there was no consistent correlation between the level of STN1 expression and either the magnitude of the response to HU (Supplementary Figure 7) or the rate at which the response decayed (unpublished observation). We therefore conclude that STN1 is not required for ATR activation after replication fork stalling. Thus, CST is unlikely to promote replication restart by preventing fork collapse through activation of the ATR-signalling pathway.
STN1 depletion causes a decrease in new origin firing after HU-induced fork stalling
To further explore whether CST plays a role in the restart of stalled replication forks, we used the disappearance of RPA foci after release from HU as a way to assess whether STN1 depletion causes a delay in the repair of stalled forks (Zou and Elledge, 2003; Robison et al, 2004). shSTN1-7 or control (shNT and shSTN1-7-Res) clones were grown in EdU for 20 min to label cells in S-phase and then treated with HU for 2 h. After release into fresh media, the cells were fixed at various time points and incubated with antibody to RPA34 to visualize RPA foci. S-phase cells were identified by EdU staining. As expected, confocal microscopy revealed large numbers of RPA foci in the S-phase cells immediately after HU treatment (Figure 6). These foci rapidly disappeared and were almost gone 12 min after release in both the STN1 depleted and control cells (Figure 6, compare No HU with 12 min release). Quantification of the overall RPA signal intensity within the nuclei revealed that STN1 depletion caused no significant change in the rate at which the foci disappeared (Supplementary Figure 8). The observed rapid loss of RPA indicates that the replisome remains largely intact after short-term HU treatment and suggests that, in this situation, STN1 is not required at the majority of forks to restart replication.
Figure 6.
Rapid disappearance of RPA foci after release from HU. shNT (top row of each panel) or shSTN1-7 (bottom row of each panel) cells were incubated with EdU to label S-phase cells, then treated with 2 mM HU for 2 h and fixed without release (top left), released for 8 min prior to fixation (top right) or 12 minutes prior to fixation (bottom left). No HU treatment is shown (bottom right) to represent RPA foci that occur naturally during replication. Images are shown from a single experiment and are representative of three independent experiments. Red, RPA; green, EdU; blue, DAPI. NT, non-target.
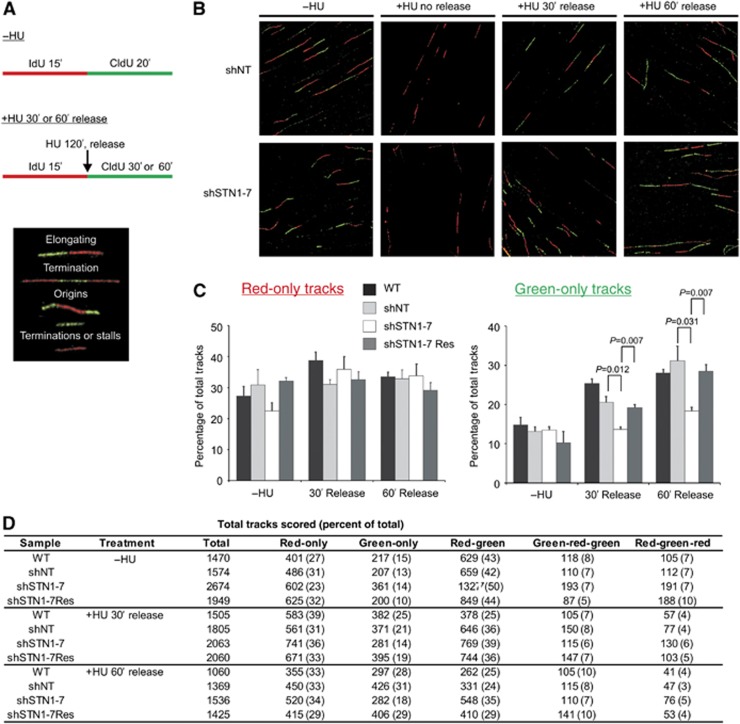
An alternative mechanism through which CST might promote replication restart is by facilitating new origin firing. To distinguish between possible effects of STN1 depletion on origin firing versus fork restart, we turned to DNA fibre analysis to directly examine DNA replication events at the molecular level. HeLa 1.2.11 cells were labelled with IdU for 15 min. Replication was then stalled by the addition of HU for 2 h. The cells were released into media containing CldU for either 30 or 60 min, harvested, lysed on microscope slides and the DNA fibres spread as previously described (Chastain et al, 2006; Figure 7A). The IdU and CldU labelled DNA tracks were stained with antibodies to IdU and CldU, visualized by confocal microscopy and replication events were quantified (Figure 7). Untreated cells were used to determine the background level of replication events (−HU). Control cells incubated with CldU during the HU treatment showed only red (IdU) tracks and demonstrated that the HU caused efficient fork stalling (Figure 7B, +HU No Release).
Figure 7.
STN1 depletion leads to decreased new origin firing following release from HU-induced fork stalling. (A) Schematic of experimental approach and types of DNA fibres scored. HeLa 1.2.11 cells were pulse labelled with IdU and CldU, as indicated, to label individual replication forks. DNA fibre spreading was then performed (see Materials and methods) and IdU/CldU visualized by immunofluorescent labelling. Images indicate the different replication events observed. (B) Representative images of DNA fibres. Red, IdU; green, CldU. (C) Graphical representation of the percentage of red-only (stalls or terminations) or green-only (new origins fired during CldU pulse) tracks (mean±s.e.m., n=3 independent experiments). (D) Total number of tracks scored. In parenthesis is the percentage of the total number of tracks scored. NT, non-target; WT, wild type.
When we determined the fraction of forks that remained stalled after HU release (red-only tracks) we found no significant difference between the STN1-depleted and control cells (WT, shNT and STN1-7-Res; Figure 7C and D). In each case, the cells displayed rapid recovery from fork stalling with the majority of forks resuming replication (red-green tracks) within the 30-min time frame (Figure 7D). In contrast, we found that STN1 depletion caused a striking decrease in the level of new origin firing (green-only tracks). This was apparent at both the 30- and 60-min time points, indicating a continued inhibition of origin firing. Thus, the DNA fibre analysis not only corroborated the RPA study (Figure 6) by demonstrating rapid, STN1-independent, recovery of stalled forks after HU treatment but also revealed an unexpected role for STN1 in origin firing. Moreover, the reduction in origin firing when STN1-depleted cells are released from HU can explain the reduction in EdU incorporation observed under these same conditions (Figure 5).
Discussion
Here, we show that the STN1 subunit of the human CST complex is required for efficient replication of the duplex region of the telomere. We also show that STN1 promotes genome-wide replication restart following HU-induced replication fork stalling and this effect is exerted at the level of new origin firing. Each of these findings were quite unexpected because the budding yeast ScCST complex has only been shown to function in telomerase- and pol α-mediated extension of the extreme chromosome terminus and not in replication of duplex DNA (Shore and Bianchi, 2009; Giraud-Panis et al, 2010). However, our discoveries help explain the anaphase bridges and MTS observed after CTC1 or STN1 depletion. Studies suggest that anaphase bridges can arise after failure to rescue stalled forks (Chan et al, 2009; Naim and Rosselli, 2009; Kawabata et al, 2011). Likewise, MTS arise from problems during replication of the telomere duplex that lead to fork stalling or a deficiency in fork rescue (Sfeir et al, 2009; Saharia et al, 2010).
Our analysis of MTS levels following STN1 and TRF1 co-depletion indicates that STN1 and TRF1 promote telomere replication in different ways. Given that TRF1 depletion causes fork stalling and STN1 facilitates replication restart after HU treatment, our results suggest that the mechanism by which STN1 aids replication through the telomere, and thus prevents formation of MTS, is by promoting the restart of replication after fork stalling. At present the defect that underlies MTS formation is unknown, but as MTS do not appear to reflect actual breaks in the chromosome, one possibility is that replication fork stalling results in the loading of incorrect histone marks and this affects subsequent metaphase chromosome condensation in the region of the stall (Jasencakova et al, 2010).
Roles of CST in DNA replication
Overall our results indicate that STN1, and possibly the whole CST complex, functions as a specialized replication factor that is needed under conditions of replication stress. Thus, STN1/CST may play a key role in replicating regions of the genome that pose a natural barrier to the replication fork (e.g., telomeres, common fragile sites or tri-nucleotide repeats) or certain lesions needing repair. Our finding that STN1 functions in replication restart is particularly interesting given the original work demonstrating that mammalian CTC1 and STN1 (AAF) stimulate pol α affinity for ssDNA templates (Goulian et al, 1990) and more recent work indicating that Xenopus CST also promotes priming of replication on ssDNA (Nakaoka et al, 2012). These observations raise the possibility that STN1/CST may be responsible for loading pol α during replication restart.
Rescue of stalled forks is thought to involve various forms of fork remodelling and Holiday junction formation (Petermann and Helleday, 2010). One common feature of proposed models for fork restart is the need to re-prime DNA synthesis and reload a replisome at the re-modelled fork in an origin-independent manner. Since fork stalling and remodelling can lead to extended regions of ssDNA (Sogo et al, 2002; Byun et al, 2005; Petermann and Helleday, 2010), one obvious function for STN1/CST would be to recruit pol α to facilitate fill-in of these regions in much the same way that it is thought to promote C-strand fill-in during telomere replication (Zhao et al, 2009; Price et al, 2010). Although we did not find convincing evidence for this activity during recovery from HU treatment, it is possible that CST functions in such a manner if the replisome is damaged and pol α needs to be re-attached before a fork can resume replication. In our DNA fibre analysis experiments, the initial (IdU) labelling period was sufficiently long to allow some forks to naturally terminate replication. Consequently, the termination reactions in STN1-depleted cells could have obscured a small increase in red-only tracks caused by forks with a damaged replisome failing to resume replication. Given the significant delay in telomere replication observed after STN1 depletion during an unperturbed cell cycle (Figure 3) and the low abundance of replication origins within the telomeric tract (Sfeir et al, 2009; Drosopoulos et al, 2012), it is also possible that CST mediated re-priming at stalled forks is important at telomeres and other natural replication barriers. Future studies will be required to fully understand whether CST plays such a role at these sites.
The discovery that STN1/CST functions in replication restart by promoting origin firing was both unexpected and intriguing because CST does not localize to replication foci (Miyake et al, 2009) and STN1 depletion does not affect the rate of bulk genomic DNA replication in an unperturbed cell cycle (Figure 3). Thus, there was no reason to suppose that CST normally localizes to origins as part of the replication initiation complex. However, our experiments do not directly address whether STN1/CST functions in the firing of primary origins when HU-treated cells first enter S-phase or in the firing of dormant origins in a replicon where a stalled fork has failed to restart. Both the shSTN1 and control cells become partially synchronized at the G1/S boundary during HU treatment, resulting in a subsequent burst in primary origin firing when this subset of cells enter S-phase. The remainder of the increase is expected to come from firing of dormant origins near stalled replication forks (Blow et al, 2011). Given that STN1 depletion does not cause an S-phase delay in an unperturbed cell cycle, we speculate that CST is involved in the firing of dormant origins, due to replication stress, rather than the firing of primary origins.
Rescue of replication through dormant origin firing was previously shown to predominate over fork restart after prolonged HU treatment (24 h), which leads to fork collapse (Petermann et al, 2010). However, a recent study found that dormant origin firing is a common event during HU-induced stalling presumably as an attempt to rescue stalled replication forks and resume DNA replication (Karnani and Dutta, 2011). Although the dormant origins were able to fire in the presence of HU, they rapidly stalled within ∼2 kb of initiation. The above studies indicate that dormant origin firing is an important response to HU-induced replication stress. We suspect that this same phenomenon is responsible for the increase in new origin firing that we observed after release of control cells from short-term HU treatment (Figure 7C). Given the accompanying rapid loss of RPA foci and the minor increase in stalled forks (Figures 6 and 7), we suspect that DNA replication resumes by concomitant fork restart and dormant origin firing. Since STN1 depletion leads to decreased new origin firing, our results suggest that CST plays an important role in the firing of dormant origins under conditions of replication stress.
Dormant origins are licensed prior to the onset of S-phase through loading of the MCM2-7 helicase by CDC6 and CDT1 (Blow et al, 2011). How they are regulated in mammalian cells is poorly understood. One possibility is that all origins within a cluster are qualitatively similar so which origins fire and which remain dormant is purely stochastic. However, in budding yeast, firing of dormant origins and the timing of primary origin firing seems to be controlled through competition for limiting factors needed to assemble the replication initiation complex (Mantiero et al, 2011). If the same situation exists in mammalian cells, then specialized factors may be needed for the unscheduled assembly of replication initiation complexes at dormant origins in response to replication stress. CST could serve as such a factor by facilitating loading of pol α.
CST and shelterin
The co-existence of shelterin and CST at the telomere suggests a division of labour among the complexes responsible for telomere replication and end protection. While shelterin stands out as the dedicated telomere complex that is responsible for protecting mammalian telomeres (Palm and de Lange, 2008; Stewart et al, 2012), CST is needed for replication through the telomere duplex DNA (this publication) and for C-strand fill-in synthesis following telomerase action (Wang, F et al, manuscript in preparation). Exactly how shelterin and CST cooperate in telomere replication remains to be determined. However, STN1 has been shown to interact with the shelterin protein TPP1 in mouse embryonic stem cells and disruption of this interaction leads to telomere lengthening (Wan et al, 2009). Thus, perhaps TPP1 helps recruit CST to regions associated with replication fork stalling within the telomeric duplex. Since TPP1 also recruits telomerase and enhances telomerase processivity (Xin et al, 2007; Abreu et al, 2010; Latrick and Cech, 2010), it is possible that an additional role of TPP1 is to coordinate telomeric G-strand synthesis by first recruiting telomerase and then CST to the DNA terminus. CST would then recruit DNA pol α for C-strand fill-in, as appears to occur in budding yeast.
Thus far, ScCST has not been shown to function in a manner equivalent to human STN1/CST in replication of duplex DNA. However, the sequence specificity of Cdc13 for telomeric DNA is not conserved in other species of budding yeast, suggesting alternative roles for CST even within this phylum (Mandell et al, 2011). Moreover, overexpression of ScStn1 results in phenotypes suggestive of a general role in the response to replication stress including decreased fork progression, DNA pol α-dependent interference with the S-phase checkpoint and ScStn1 mislocalization to sites across the chromosome (Gasparyan et al, 2009). Thus, it seems likely that future studies will reveal non-telomeric replicative functions for CST-like complexes or CST components regardless of whether the telomere is packaged by CST or a shelterin-type complex.
Materials and methods
Cell culture
HeLa 1.2.11 cells were cultured in RPMI-1640 and U2OS cells in DMEM supplemented with 10% FBS, antibiotics and glutamine.
Stable shRNA knockdown clones and siRNA knockdown
Antibodies
The following antibodies were used for immunoblotting: α-Actinin from Santa Cruz (sc-17829), OBFC1 (STN1) from Abcam (ab89250), phospho-Chk1(Ser345) from Cell Signaling (2341), Chk1 from Santa Cruz (sc-8408), Goat α-Mouse-HRP (Thermo Scientific) and Goat α-Rabbit-HRP (Thermo Scientific). See Supplementary data for immunoblotting protocols.
Anaphase bridge analysis
HeLa 1.2.11 cells plated on coverslips were treated with nocodazole at 50 ng/ml for ∼4 h. Cells were washed three times with PBS and incubated in medium without nocodazole for 30–90 min. Cells were fixed with 3% formaldehyde in PBS for 10 min at room temperature, rinsed twice with PBS and mounted with mounting medium containing 0.2 μg/ml DAPI.
Telomere FISH
HeLa 1.2.11 or U2OS cells were plated and grown overnight to 30–40% confluency. Colcemid (0.5 μg/ml) or colchicine (0.1 μg/ml) was then added for ∼1.5 h. Metaphase spreads were made and telomere FISH performed, as described (Dimitrova et al, 2008), with an FITC-(TTAGGG)3 probe (Biosynthesis) and the following steps to amplify the FITC signal. After the final hybridization wash and dehydration, the slides were incubated with 1 × PBG (1 × PBS with 0.5% BSA and 0.2% cold water fish gelatin) for 20 min followed by incubation with 6 μg/ml biotinylated anti-fluorescein (Vector Laboratories) for 1–2 h at room temperature. The slides were then washed three times with 1 × PBG and incubated with 16 μg/ml fluorescein-avidin (Vector Laboratories) for 30–60 min at 37°C in a humidified chamber. The slides were washed three times with 1 × PBG, dehydrated with an EtOH series and mounted with mounting medium containing 0.5 μg/ml DAPI. For aphidicolin treatment, 0.25 μg/ml was added for ∼16 h and the cells washed three times prior to the addition of colcemid or colchicine. For the U2OS experiments, after 2 h in colcemid (0.05 μg/ml), cells were collected every 30 min for 2.5 h by mitotic shake-off.
Replication restart assay
HeLa 1.2.11 or U2OS cells were plated onto coverslips and grown overnight to ∼30% confluency. HU (2 mM, Sigma) was then added for 2 h. The cells were then washed three times with pre-warmed serum-free media. Normal growth media with 50 μM EdU (Invitrogen) was added and cells were incubated at 37°C for 30 min. After EdU incorporation, the coverslips were fixed with MeOH at −20°C for 10 min, processed using the Click-iT EdU AlexaFluor 488 Imaging Kit (Invitrogen), as instructed, and mounted with mounting medium containing 0.2 μg/ml DAPI.
Cell-cycle synchronization and analysis of bulk genomic and telomeric DNA replication rates
HeLa 1.2.11 cells were synchronized at the G1/S boundary, by treatment with 2 mM thymidine (Sigma) for 14 h. Cells were then washed three times with PBS and released into pre-warmed culture medium. A second thymidine block was initiated 9 h later and after an additional 14 h, cells were washed and released into pre-warmed medium. Cells were then pulsed labelled with BrdU (100 μM) or EdU (50 μM) for consecutive 1.5 h intervals as indicated in Figure 3A and genomic DNA was isolated at the end of each labelling period. The rate of bulk genomic DNA replication was determined by quantifying EdU uptake. Cells were fixed with MeOH, processed using the Click-It EdU AlexaFluor 488 kit (Invitrogen) and stained with propidium iodide. They were then analysed by FACS to determine the percentage of cells that were in S-phase and EdU positive (gate shown in Supplementary Figures 4A and 5A) and the mean intensity determined. The total amount of EdU uptake was calculated for each labelling period by multiplying the percentage of EdU-positive cells with their mean value. The rate of telomeric DNA replication was determined by quantifying the amount of telomeric DNA newly replicated during each time period. Separation of leading and lagging telomeres was performed as described (Chai et al, 2006). Briefly, genomic DNA from BrdU-labelled cells was digested with Hinf I and Msp I then mixed with a CsCl solution and subjected to ultracentrifugation at 55 000 r.p.m. for 20 h at 25°C. Fractions were collected and the density for each fraction measured with a refractometer. The amount of telomeric DNA in each fraction was determined by slot blot analysis and hybridization with a telomeric oligonucleotide probe. The signal was normalized by max-min normalization, which transformed the maximum numbers to 100, minimum number to 0 and fits the other data points in between. The percentage of newly synthesized leading telomere was calculated by dividing the replicated leading strand peak by the sum of all peaks (corresponding to unreplicated and replicated leading and lagging strand telomeric DNA).
Quantification of RPA after recovery from fork stalling
HeLa 1.2.11 shNT, shSTN1-7 and shSTN1-7 Res cells were plated at ∼5 × 104 in 24-well plates on glass coverslips and grown overnight. Cells were subsequently pulsed for 20 min with 50 μm EdU, followed by stalling of replication forks with 2 mM HU for 2 h or no HU, as a control. Following three quick PBS washes, the cells were released into normal growth medium for the times indicated. Prior to fixation, cells were incubated in CSK buffer (10 mM HEPES, 300 mM sucrose, 100 mM NaCl and 3 mM MgCl2) twice for 2 min each followed by extraction in 0.5% Triton/CSK for 4 min to visualize repair foci. Cells were subsequently fixed in 4% formaldehyde for 20 min, followed by permeabilization with 0.5% NP-40 for 5 min. Coverslips were then blocked with 2% BSA for 30 min, incubated with anti-RPA antibody (1:100) for 2 h (RPA34-19, Calbiochem), and RPA visualized with AlexaFluor594 (1:1000) (Invitrogen). Staining of EdU was performed using Click-It AlexaFluor488 according to manufacturer’s instructions. Finally, DNA was stained with DAPI and the coverslips mounted on a slide with FluoroGel (EMS).
DNA fibre analysis
HeLa 1.2.11 cell lines were plated at ∼5 × 105 cell/6 cm plate and grown overnight. The cells were pulse labelled with 50 μM IdU (Sigma) for 15 min. The control cells (−HU) were then pulsed labelled with 100 μM CldU (Sigma) for 20 min. To the rest of the plates, 2 mM HU was added for 2 h. After HU treatment, the cells were washed three times with pre-warmed serum-free media. The cells were then released into pre-warmed normal growth media with 100 μM CldU and incubated at 37°C for either 30 or 60 min (+HU 30′ or 60′ Release). As another control to ensure forks were stalled with HU treatment, one set of plates was not released from HU but instead 100 μM CldU was added 1 h into the HU treatment and the cells incubated for another hour (+HU No Release). The cells were then collected and DNA fibres prepared as previously described (Chastain et al, 2006). The slides were fixed with 3:1 methanol:acetic acid for 2 min, allowed to dry overnight and stored at −20°C for at least 24 h. The slides were then immunostained as described (Chastain et al, 2006), with minor modifications. Briefly, the slides were treated with 2.5 N HCl for 30 min at room temperature, washed once with 1 × PBS+0.1% Tween-20 and twice with 1 × PBS. In all, 5% BSA in 1 × PBS was then used to block the slides. Slides were then incubated for 1 h at room temperature with mouse α-BrdU (1:500, Becton Dickson) and rat α-BrdU (1:500, Accurate Chemical) to detect IdU and CldU, respectively. Following incubation, the slides were washed for 13–14 min with a stringency buffer (10 mM Tris pH 7.4, 300 mM NaCl, 0.2% Tween-20, 0.2% Igepal CA-630) to reduce non-specific binding. Slides were then washed twice with 1 × PBS, blocked with 5% BSA in 1 × PBS for 30 min at room temperature and incubated for 30 min with AlexaFluor 594 rabbit α-mouse (1:1000) and AlexaFluor 488 chicken α-rat (1:750) (Invitrogen). Next, the slides were washed once with 1 × PBS+0.1% Tween-20, twice with 1 × PBS and blocked with 5% BSA. Tertiary antibodies, AlexaFluor 594 goat α-rabbit (1:1000) and AlexaFluor 488 (1:750), were added and the slides washed, as described for secondary antibodies. The slides were then dehydrated with an EtOH series and mounted with Pro-long Gold Anti-fade (Invitrogen). All antibodies were diluted in 5% BSA in 1 × PBS+0.1% Tween-20.
Data acquisition and analysis
Telomere FISH images were taken at × 1000 and replication restart and anaphase cells images at × 200 with a Nikon Eclipse E400 fluorescent microscope, equipped with a Spot 2 digital camera (Diagnostic Instruments Inc). MTS and signal-free ends were scored blindly by trained individuals. At least 200 chromosomes were analysed per independent telomere FISH experiment, unless otherwise indicated, and at least 200 anaphase cells were scored per independent anaphase bridge experiment. Average AFU measurements of nuclei, for the replication restart assay, were obtained using ImageJ software. Briefly, nuclei were defined by particle analysis, with watershed. These defined regions of interest (ROI) were then overlaid onto the image with the EdU signal. The mean AFU was then acquired for each ROI. These numbers were used to determine the average AFU of all nuclei and create bins for the nuclei above or below a given AFU. At least 700 or 400 nuclei were scored for each independent HeLa 1.2.11 or U2OS replication restart experiment, respectively.
Confocal images shown in Figure 6 were acquired at × 630 magnification using a Zeiss LSM710 microscope. Images for quantification were acquired at × 400 using a Zeiss Axioplan 2 microscope equipped with a Zeiss Axiocam MRM camera and analysed using ImageJ. Binning of AFU’s in Supplementary Figure 8 was performed as described for replication restart assays, a minimum of 500 nuclei (∼150 EdU+ cells) were analysed for each condition in three independent experiments.
DNA fibres were visualized with a Zeiss LSM710 confocal microscope under × 630. At least 200 fibres and 5 images were scored for each independent experiment. Scoring of fibres was performed using software described previously (Wang et al, 2011). The software recognizes the DNA fibres and creates an excel spreadsheet with the raw data. Parameters are then adjusted within the excel file. To set the parameters, an entire independent experiment was scored and parameters were then set to reflect what was observed. These exact parameters were then applied to each independent experiment. The parameters include a min size of any red or green segment (2 pixels), minimum track length (5 pixels), per cent of discontinuity within the track (<10%), a signal to noise ratio threshold (3) and a maximum track thickness (<10 pixels) to avoid scoring bundled DNA fibre. These parameters greatly reduced the number of DNA fibre bundles, background staining and stretched fibres included in the analysis.
Statistical analysis
The Student’s two-tailed unpaired t-test was used to determine statistical significance and the P-values obtained are indicated in the figures.
Supplementary Material
Acknowledgments
We thank Vidjaya L Premkumar, Anuradha de Silva and Alex Barnhill for help with MTS scoring, replication restart scoring and cell culture work; Andi Griesinger for assisting in selection of the CTC1 shRNA clones and development of the telomere FISH protocol; and Birgit Ehmer for assistance with the confocal microscope. This work was supported by NIH grants GM041803 to CMP and AGO1228 to WEW, and USPHS grant ES018918 and NIEHS grant P30ES010126 to PDC. JAS was supported by F32GM097833 and T32CA117846 and CK by T32CA117846.
Author contributions: JAS designed and performed the MTS, replication restart analyses and DNA fibre analysis, and contributed to other experiments. He also took a lead role in manuscript preparation. FW designed and executed the TRF1 knockdown and replication rate experiments. He also contributed to other experiments. CK performed the RPA analysis and contributed to other experiments. MFC performed the CHK1 analysis and the CTC1 siRNA knockdown. She also contributed other experiments and prepared many of the reagents. CMP contributed to project design and manuscript preparation. JAS, FW, MFC, CK and CMP discussed and contributed to project planning on a regular basis. PDC taught JAS how to perform the DNA fibre analysis. He also supervised and helped perform the data analysis. WEW suggested concepts and contributed to design of the replication rate analysis and to manuscript writing.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP (2010) TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol 30: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BH, Kasher PR, Mayer J, Szynkiewicz M, Jenkinson EM, Bhaskar SS, Urquhart JE, Daly SB, Dickerson JE, O'Sullivan J, Leibundgut EO, Muter J, Abdel-Salem GM, Babul-Hirji R, Baxter P, Berger A, Bonafe L, Brunstom-Hernandez JE, Buckard JA, Chitayat D et al. (2012) Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat Genet 44: 338–342 [DOI] [PubMed] [Google Scholar]
- Badie S, Escandell JM, Bouwman P, Carlos AR, Thanasoula M, Gallardo MM, Suram A, Jaco I, Benitez J, Herbig U, Blasco MA, Jonkers J, Tarsounas M (2010) BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat Struct Mol Biol 17: 1461–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ, Jackson DA (2011) How dormant origins promote complete genome replication. Trends Biochem Sci 36: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2010) Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 19: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB (2009) A DNA polymerase-{alpha}{middle dot}primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem 284: 5807–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Du Q, Shay JW, Wright WE (2006) Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell 21: 427–435 [DOI] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID (2009) Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11: 753–760 [DOI] [PubMed] [Google Scholar]
- Chandra A, Hughes TR, Nugent CI, Lundblad V (2001) Cdc13 both positively and negatively regulates telomere replication. Genes Dev 15: 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ (2009) ATR and H2AX cooperate in maintaining genome stability under replication stress. J Biol Chem 284: 5994–6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain PD 2nd, Heffernan TP, Nevis KR, Lin L, Kaufmann WK, Kaufman DG, Cordeiro-Stone M (2006) Checkpoint regulation of replication dynamics in UV-irradiated human cells. Cell Cycle 5: 2160–2167 [DOI] [PubMed] [Google Scholar]
- Cheng CH, Kuchta RD (1993) DNA polymerase epsilon: aphidicolin inhibition and the relationship between polymerase and exonuclease activity. Biochemistry 32: 8568–8574 [DOI] [PubMed] [Google Scholar]
- Dai X, Huang C, Bhusari A, Sampathi S, Schubert K, Chai W (2010) Molecular steps of G-overhang generation at human telomeres and its function in chromosome end protection. EMBO J 29: 2788–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T (2008) 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosopoulos WC, Kosiyatrakul ST, Yan Z, Calderano SG, Schildkraut CL (2012) Human telomeres replicate using chromosome-specific, rather than universal, replication programs. J Cell Biol 197: 253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SG, Glover TW (2007) Chromosome fragile sites. Annu Rev Genet 41: 169–192 [DOI] [PubMed] [Google Scholar]
- Gasparyan HJ, Xu L, Petreaca RC, Rex AE, Small VY, Bhogal NS, Julius JA, Warsi TH, Bachant J, Aparicio OM, Nugent CI (2009) Yeast telomere capping protein Stn1 overrides DNA replication control through the S phase checkpoint. Proc Natl Acad Sci USA 106: 2206–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AK, Rossi ML, Singh DK, Dunn C, Ramamoorthy M, Croteau DL, Liu Y, Bohr VA (2011) RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J Biol Chem 287: 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Geli V (2007) How telomeres are replicated. Nat Rev Mol Cell Biol 8: 825–838 [DOI] [PubMed] [Google Scholar]
- Giraud-Panis MJ, Teixeira MT, Geli V, Gilson E (2010) CST meets shelterin to keep telomeres in check. Mol Cell 39: 665–676 [DOI] [PubMed] [Google Scholar]
- Goulian M, Heard CJ (1990) The mechanism of action of an accessory protein for DNA polymerase alpha/primase. J Biol Chem 265: 13231–13239 [PubMed] [Google Scholar]
- Goulian M, Heard CJ, Grimm SL (1990) Purification and properties of an accessory protein for DNA polymerase alpha/primase. J Biol Chem 265: 13221–13230 [PubMed] [Google Scholar]
- Jasencakova Z, Scharf AN, Ask K, Corpet A, Imhof A, Almouzni G, Groth A (2010) Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell 37: 736–743 [DOI] [PubMed] [Google Scholar]
- Karnani N, Dutta A (2011) The effect of the intra-S-phase checkpoint on origins of replication in human cells. Genes Dev 25: 621–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata T, Luebben SW, Yamaguchi S, Ilves I, Matise I, Buske T, Botchan MR, Shima N (2011) Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell 41: 543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller RB, Gagne KE, Usmani GN, Asdourian GK, Williams DA, Hofmann I, Agarwal S (2012) CTC1 mutations in a patient with dyskeratosis congenita. Pediatr Blood Cancer 59: 311–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Akan Z, Yilmaz S, Grillo M, Smith-Roe SL, Kang TH, Cordeiro-Stone M, Kaufmann WK, Abraham RT, Sancar A, Unsal-Kacmaz K (2010) Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J Biol Chem 285: 16562–16571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc A, Wheeler LJ, Mathews CK, Merrill GF (2004) Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J Biol Chem 279: 223–230 [DOI] [PubMed] [Google Scholar]
- Latrick CM, Cech TR (2010) POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J 29: 924–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman AR, Noguchi C, Lee CY, Noguchi E (2010) Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J Cell Sci 123: 660–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell EK, Gelinas AD, Wuttke DS, Lundblad V (2011) Sequence-specific binding to telomeric DNA is not a conserved property of the cdc13 DNA binding domain. Biochemistry 50: 6289–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D, Mackenzie A, Donaldson A, Zegerman P (2011) Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J 30: 4805–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernandez-Capetillo O, Tarsounas M, Blasco MA (2009) Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev 23: 2060–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F (2009) RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell 36: 193–206 [DOI] [PubMed] [Google Scholar]
- Naim V, Rosselli F (2009) The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol 11: 761–768 [DOI] [PubMed] [Google Scholar]
- Nakaoka H, Nishiyama A, Saito M, Ishikawa F (2012) Xenopus laevis Ctc1-Stn1-Ten1 (xCST) protein complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J Biol Chem 287: 619–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi T, Hirota T, Tsuruo T, Seimiya H (2010) TRF1 mediates mitotic abnormalities induced by Aurora-A overexpression. Cancer Res 70: 2041–2052 [DOI] [PubMed] [Google Scholar]
- Paeschke K, McDonald KR, Zakian VA (2010) Telomeres: structures in need of unwinding. FEBS Lett 584: 3760–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Cimprich KA (2007) The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 6: 953–966 [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V (2001) Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387–396 [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T (2010) Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 11: 683–687 [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T (2010) Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37: 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polvi A, Linnankivi T, Kivela T, Herva R, Keating JP, Makitie O, Pareyson D, Vainionpaa L, Lahtinen J, Hovatta I, Pihko H, Lehesjoki AE (2012) Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am J Hum Genet 90: 540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE (2010) Evolution of CST function in telomere maintenance. Cell Cycle 9: 3157–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D (2008) Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J 27: 2328–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA (2000) The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev 14: 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- Robison JG, Elliott J, Dixon K, Oakley GG (2004) Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J Biol Chem 279: 34802–34810 [DOI] [PubMed] [Google Scholar]
- Saharia A, Teasley DC, Duxin JP, Dao B, Chiappinelli KB, Stewart SA (2010) FEN1 ensures telomere stability by facilitating replication fork re-initiation. J Biol Chem 285: 27057–27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D, Bianchi A (2009) Telomere length regulation: coupling DNA end processing to feedback regulation of telomerase. EMBO J 28: 2309–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Fu MA, Brown EJ (2009) Tim-Tipin dysfunction creates an indispensible reliance on the ATR-Chk1 pathway for continued DNA synthesis. J Cell Biol 187: 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- Stewart JA, Chaiken MF, Wang F, Price CM (2012) Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat Res 730: 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE (2009) Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell 36: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van C, Yan S, Michael WM, Waga S, Cimprich KA (2010) Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J Cell Biol 189: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, Qin J, Songyang Z, Liu D (2009) OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem 284: 26725–26731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chastain P, Yap PT, Cheng JZ, Kaufman D, Guo L, Shen D (2011) Automated DNA fiber tracking and measurement. Proceedings of the 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 30 March–2 April 2011, pp1349–1352 [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z (2007) TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 445: 559–562 [DOI] [PubMed] [Google Scholar]
- Yan S, Michael WM (2009) TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: implications for a replication restart-based mechanism for ATR checkpoint activation. Cell Cycle 8: 2877–2884 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE (2009) Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138: 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Shay JW, Wright WE (2011) Telomere terminal G/C strand synthesis: measuring telomerase action and C-rich fill-in. Methods Mol Biol 735: 63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.