Abstract
The Rpd3S histone deacetylase complex represses cryptic transcription initiation within coding regions by maintaining the hypo-acetylated state of transcribed chromatin. Rpd3S recognizes methylation of histone H3 at lysine 36 (H3K36me), which is required for its deacetylation activity. Rpd3S is able to function over a wide range of H3K36me levels, making this a unique system to examine how chromatin regulators tolerate the reduction of their recognition signal. Here, we demonstrated that Rpd3S makes histone modification-independent contacts with nucleosomes, and that Rpd3S prefers di-nucleosome templates since two binding surfaces can be readily accessed simultaneously. Importantly, this multivalent mode of interaction across two linked nucleosomes allows Rpd3S to tolerate a two-fold intramolecular reduction of H3K36me. Our data suggest that chromatin regulators utilize an intrinsic di-nucleosome-recognition mechanism to prevent compromised function when their primary recognition modifications are diluted.
Keywords: chromatin recognition, di-nucleosome, epigenetics, HDAC
Introduction
Histone posttranslational modifications (HPTMs) regulate most, if not all, nuclear processes that involve chromatin (Kouzarides, 2007). HPTMs are also considered to be epigenetic marks that establish transcription programs in daughter cells to resemble those in mother cells (Hammoud et al, 2009; Probst et al, 2009; Kaufman and Rando, 2010; Moazed, 2011). Some HPTMs can directly influence chromatin structure (Shogren-Knaak et al, 2006), whereas most HPTMs serve as a signalling platform to recruit effector proteins, which in turn determine the ultimate states of chromatin (Strahl and Allis, 2000).
A typical protein modification generates a binary signal, such as phosphorylation at one residue representing ON whereas de-phosphorylation at the same residue indicating OFF. Since there are many symmetric features within the nucleosome structure, such as two copies of each histone and the symmetric shape of nucleosome core particle, it remains unclear whether HPTM signals follow the usual binary rule, or if HPTM readers can interpret a gradient of signals. This is an important issue particularly considering the highly debated epigenetic aspects of histone modifications. Newly synthesized histones are not believed to carry HPTMs specific to given genomic loci. Therefore, replicated chromatin faces a potential two-fold dilution of these HPTMs when new histones are deposited on the daughter strands after replication (Henikoff and Shilatifard, 2011). Recent quantitative mass-spectrometry analyses demonstrated that many transcription-related HPTMs are indeed diluted upon replication (Zee et al, 2010; Xu et al, 2011). How diluted HPTMs are transmitted and eventually restored to the parental pattern remains largely unknown (Bonasio et al, 2010). So how do cells cope with this transient dilution before HPTMs are restored or re-established?
The Set2–Rpd3S pathway is responsible for maintaining hypo-acetylated states of transcribed chromatin, thereby suppressing cryptic transcription initiation (Joshi and Struhl, 2005; Keogh et al, 2005; Carrozza et al, 2005b). The Rpd3S histone deacetylase (HDAC) complex is initially recruited to coding regions through the phosphorylated C-terminal domain of RNA polymerase II (Pol II) (Drouin et al, 2010; Govind et al, 2010). Locally enriched Rpd3S recognizes nucleosomes that are methylated at H3K36 by Set2 through combined actions of two reading modules: the chromodomain of Eaf3 (CHDEaf3), which binds to methylated H3K36, and the PHD domain of Rco1 (PHDRco1) (Li et al, 2007b). This multivalent interaction leads to the engagement of Rpd3S with chromatin substrates and subsequent deacetylation. In this pathway, the H3K36me signal is required, but not sufficient to influence the chromatin state; instead, it is histone acetylation regulated by Rpd3S that controls the accessibility of cryptic promoters to the transcription machinery (Li et al, 2007a). Therefore, Rpd3S function must be maintained to ensure correct function of this pathway. Rpd3S can discriminate different states of H3K36me, however, H3K36me2 seems to be sufficient to suppress cryptic transcription (Li et al, 2009). Interestingly, it was found that in a PAF1 deletion yeast strain, H3K36me3 is eradicated and the amount of K36me2 is also significantly lowered at coding regions. However, Rpd3S retains its normal function, as typical Set2–Rpd3S phenotypes including histone hyper-acetylation and cryptic transcription were not detected in this mutant (Chu et al, 2007; Youdell et al, 2008; Li et al, 2009). These results suggest that Rpd3S functions over a wide range of H3K36me levels in a normal cellular context, which provides us with a suitable system to examine how chromatin regulators tolerate the reduction of their recognition signal.
Here, using this well-studied HPTM-signalling pathway, we set out to explore the molecular mechanism by which chromatin regulators sense HPTM gradients. Our biochemical analysis suggested that chromatin effectors recognize the di-nucleosome as a unit through a multivalent mode of interaction, which includes HPTM-dependent and HPTM-independent binding. This intrinsic ability makes these chromatin complexes capable of tolerating minor fluctuation of their primary HPTMs recognition signals.
Results
We have shown previously that Rpd3S can distinguish different states of H3K36me. Even with significantly reduced levels of H3K36me2, Rpd3S is still able to suppress cryptic transcription (Li et al, 2009). To understand the molecular mechanism underlying this apparent reading tolerance phenomenon, we took a biochemical approach to examine how Rpd3S recognizes cognate substrates and carries out its enzymatic activity. Two critical domains are required for Rpd3S function: CHDEaf3 contributes to overall affinity and provides specificity towards H3K36me, and PHDRco1 is required for Rpd3S function both in vitro and in vivo (Li et al, 2007b, 2007c). However, how these two domains achieve synergistic binding remains unresolved.
Rpd3S makes K36me3-independent contact with nucleosomes
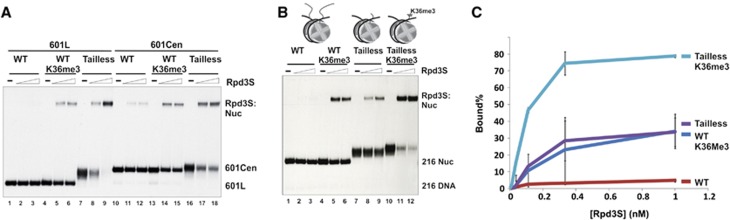
We first investigated the roles of histone tails in Rpd3S binding. Recombinant Xenopus core histone octamers were formed by mixing different combinations of wild-type (WT) and tail-truncated histones as described previously (Dorigo et al, 2003) (Supplementary Figure 1A–C). These histone octamers were then reconstituted into mono-nucleosomes with a radio-labelled DNA probe containing the ‘601’ nucleosome positioning sequence (Thastrom et al, 2004) (Supplementary Figure 1D). Using these chromatin templates in a gel-mobility shift assay, we found that Rpd3S preferentially recognizes tailless nucleosomes (Figure 1A). This enhanced binding does not depend on nucleosome positioning, since it can be detected using both central and lateral positioned nucleosomes (Figure 1A). Preferential binding of Rpd3S requires a linker DNA (Supplementary Figure 2, lanes 10–12), and is not sequence specific since a similar result was obtained when another template was used (Li et al, 2007b) (Supplementary Figure 2, lanes 13–18). In our tailless construct of histone H3, only the first 27 amino acids of histone H3 were truncated; thus, the peptide sequence that is involved in H3K36me binding is still intact. Therefore, we asked whether this enhanced binding of Rpd3S to tailless nucleosomes still depends on H3K36me. To this end, we took advantage of the methyl-lysine analogue technology to prepare chemically methylated histone H3 at K36 (Simon et al, 2007). As shown in Figure 1B (lanes 10–12) and 1C, H3K36me3 further stimulates the binding of Rpd3S to tailless nucleosomes, suggesting that Rpd3S binds to tailless nucleosomes in a physiologically relevant configuration.
Figure 1.
Rpd3S prefers tailless nucleosomes. (A) Rpd3S preferentially binds to tailless nucleosomes independent of translational positioning as demonstrated by gel-mobility shift assays. In 601CEN and 601L fragments, the 601 sequence is placed at the centre and lateral positions of a 216-bp DNA, respectively. (B) The binding of Rpd3S to tailless nucleosomes is stimulated by H3K36me3. The compositions of tail-truncated histone octamers are listed below (‘g’ stands for the globular domain of histones; underlined subunits are full-length histones). Tailless (or TL): gH3/gH4/gH2A/gH2B; Tailless K36me3: gH3Kc36me3/gH4/gH2A/gH2B. (C) Quantification of EMSA results over a wide range of concentration (0.01 nM, 0.033 nM, 0.1 nM, 0.33 nM and 1 nM of Rpd3S) based on three independent experiments.
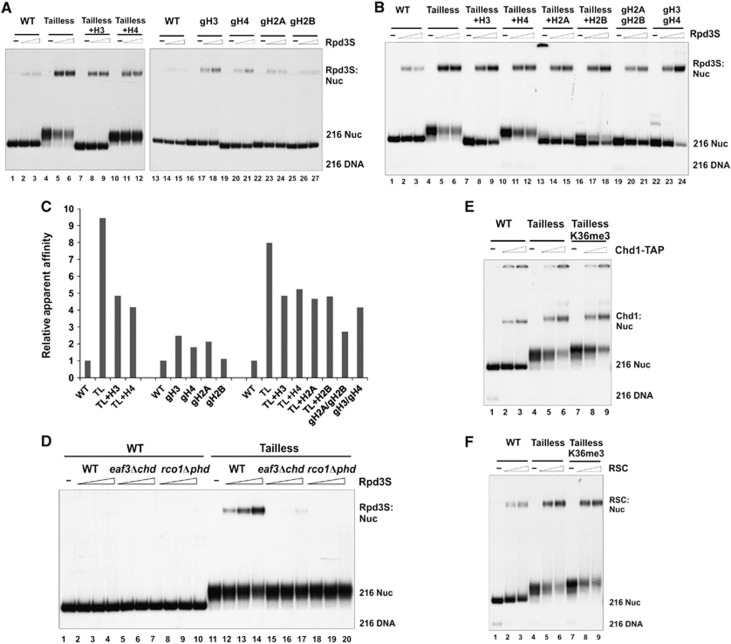
Intriguingly, we found that the binding of Rpd3S to tailless nucleosomes is proportional to the number of tails that have been removed (Figure 2A–C). In other words, histone tails seem to exert additive interference on Rpd3S binding, and there is no clear preference for any histone tail (Figure 2C). To rule out the possibility that tailless nucleosomes form aggregates that in turn increase non-specific binding of Rpd3S, we tested these nucleosomes in chromatin-remodelling assays. Indeed, the tailless nucleosomes could be efficiently mobilized by remodelling enzymes, suggesting that these tailless nucleosomes are normal, highly organized structural entities (Supplementary Figure 2B). Consistent with a previous publication (Clapier et al, 2001), we also found that histone H4 tails are required for the full remodelling activity of ACF (Supplementary Figure 2B, lanes 7–9 and 19–24). Therefore, we demonstrated that progressively increased affinity of Rpd3S upon tail removal is not due to aggregation of nucleosomes. Finally, we showed that even with much stronger affinity, the binding of Rpd3S to tailless nucleosomes still depends on CHDEaf3 and PHDRco1 (Figure 2D), indicating that all critical contact points for Rpd3S are within tailless nucleosomes. Indeed, preferential binding to tailless nucleosomes is a common feature among chromatin regulators, as we also demonstrated that two chromatin remodelers, Chd1 and RSC both prefer tailless nucleosomes (Figure 2E and F).
Figure 2.
Rpd3S can contact nucleosomes in a K36me-independent manner. (A–C) The binding of Rpd3S is inversely proportional to the number of histone tails within nucleosomes. (A, B) Gel-mobility shift assays using various forms of WT or tailless nucleosomes. The nomenclature of tail-truncated nucleosomes follows the general rules listed in Figure 1. Tailless (or TL): gH3/gH4/gH2A/gH2B; Tailless+H3: H3/gH4/gH2A/gH2B; Tailless+H4: gH3/H4/gH2A/gH2B; Tailless+H2A: gH3/gH4/H2A/gH2B; Tailless+H2B: gH3/gH4/gH2A/H2B; gH2A/gH2B: H3/H4/gH2A/gH2B; gH3/gH4: gH3/gH4/H2A/H2B; gH3: gH3/H4/H2A/H2B; gH4: H3/gH4/H2A/H2B; gH2A: H3/H4/gH2A/H2B; gH2B: H3/H4/H2A/gH2B. (C) Relative apparent affinities of Rpd3S to each tailless nucleosome. The intensity of gel-shift bands from each nucleosome were quantified using the USI imaging processing software. The ratio between mutant and WT nucleosomes (on the same gel) was defined as relative apparent affinity. (D) The binding of Rpd3S to tailless nucleosome requires PHDRco1 and CHDEaf3. (E, F) Preferential binding to tailless nucleosomes is common among chromatin-associated factors. Two chromatin remodelers (Chd1-TAP (E) and RSC (F)) were tested for their ability to bind to tailless nucleosomes. Both complexes showed increased affinity to tailless nucleosomes, although neither could distinguish between H3K36 methylated and unmodified nucleosomes.
Two not mutually exclusive models can explain why Rpd3S favours tailless nucleosomes: (1) histone N-terminal tails can directly contact nucleosomal DNA (Mutskov et al, 1998), thereby competing for the contact points with Rpd3S; (2) flexible histone tails can interfere with the ability of Rpd3S to bind the globular domains of nucleosomes. We favour the second model, since interaction with the core histone globular domain has emerged as a rather prevalent phenomenon among chromatin-associated complexes, such as the Snf5 subunit of Swi/Snf, the BAH domain of Sir3, and the Rcc1 protein (Dechassa et al, 2008; Makde et al, 2010; Armache et al, 2011). Most importantly, our data strongly suggest that Rpd3S binding involves multiple nucleosomal contacts including methylated H3K36 and a modification-independent binding surface on tailless nucleosomes.
Rpd3S preferentially binds di-nucleosomes
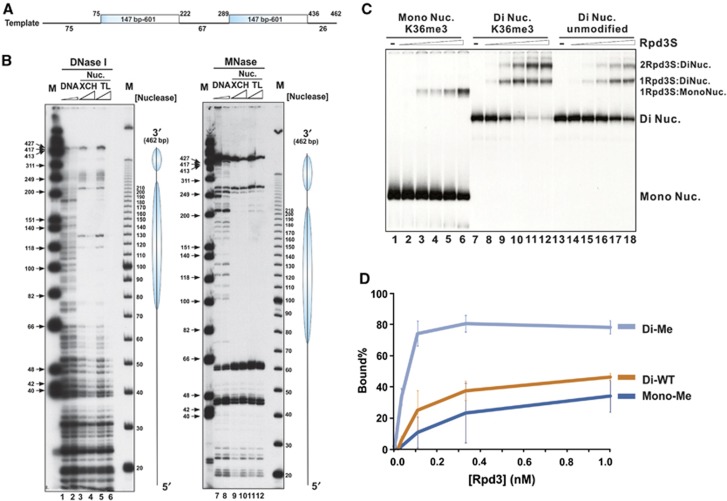
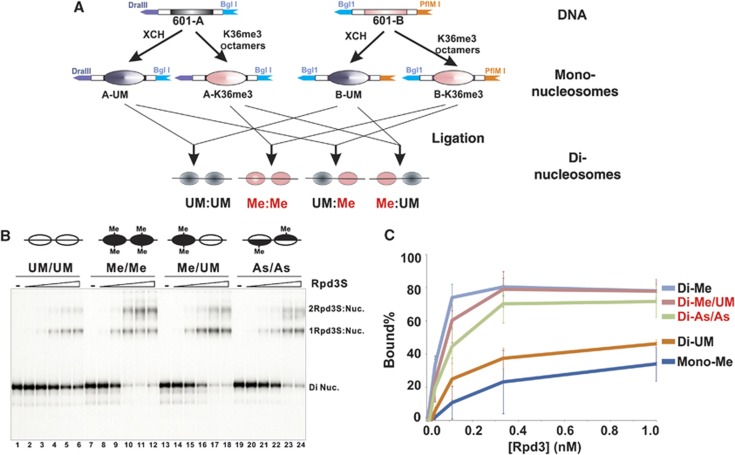
Given that Rpd3S recognizes at least two distinct regions within a nucleosome, we speculated that flexible histone tails may block CHDEaf3 and PHDRco1 from simultaneously contacting different nucleosome interacting surfaces. Therefore, we hypothesized that Rpd3S prefers di-nucleosome templates because two interacting interfaces can be presented from two separate nucleosomes within one di-nucleosome molecule, thereby minimizing the potential spatial constraints of mono-nucleosomes. To test this possibility, we designed a DNA template containing two 601 positioning sequences (Figure 3A). The integrity of reconstituted di-nucleosomes and the position of each nucleosome were confirmed by nuclease accessibility assays using both Micrococcal Nuclease (MNase) and DNaseI (Figure 3B). The digestion pattern of the template DNA indicated that the probe DNA was properly protected by two well-positioned nucleosomes.
Figure 3.
Rpd3S prefers di-nucleosomes. (A, B) Nucleosomes are properly positioned on di-nucleosomal templates. (A) A diagram of the di-nucleosome DNA template. (B) Nucleosome positioning of reconstituted di-nucleosomal templates was determined by DNaseI and MNase accessibility assays. XCH-Xenopus core histones; TL-tailless histones. (C) Rpd3S prefers di-nucleosomes over mono-nucleosomes as revealed by EMSA assays. Equal amounts of nucleosomes were used between mono- and di-nucleosomal templates. In all, 0.01 nM, 0.033 nM, 0.1 nM, 0.33 nM and 1 nM of Rpd3S were used. (D) Quantification of EMSA results shown in (C) based on three independent experiments.
When we mixed Rpd3S with these di-nucleosomes in an EMSA assay, two band shifts were observed as shown in Figure 3C (lanes 8–12 and lanes 14–18). We assigned the faster-migrating retarded species as one Rpd3S binding to one di-nucleosome (1Rpd3S:DiNuc) and the slower retarded species as two Rpd3Ss binding to one di-nucleosome (2Rpd3S:DiNuc). This band assignment was based on the following three considerations: (i) Rpd3S exists as a monomer in solution. Using size-exclusion chromatography, both recombinant and native Rpd3S have been estimated to be a 670-kDa complex, which is significantly bigger than the molar mass total of all five subunits (∼400 kDa) (Carrozza et al, 2005b; Govind et al, 2010). Because the elution volume of size-exclusion chromatography also depends on the shape, charge, and matrix interactions of the complex, we decided to use a solution-based method to determine the subunit stoichiometry/molar mass of the complex. Analytical ultracentrifugation sedimentation velocity (AUC-SV) analysis was chosen because of our previous success on similar large complexes (Padrick et al, 2011). The recombinant Rpd3 complex used in this analysis was prepared from a baculovirus expression system (Govind et al, 2010) (Supplementary Figure 3A). Although we detected some aggregation and degradation products in this sample, a large part of the signal (47%) belonged to a species that has a sedimentation coefficient under standard conditions (s20,w) of about 14.6S (Supplementary Figure 3B). The frictional ratio (fr) of this material is about 1.4, indicating a molar mass of about 410 000 g/mol, which corresponds well to a monomer of the Rpd3S complex (398 734 g/mol-calculated mass total). Moreover, this value is consistent with a size estimation based on our ongoing electron-microscopic structural study of the same complex (in collaboration with the Asturias lab, Scripps). Therefore, we concluded that Rpd3S is a monomeric complex and does not form homodimers under our solution conditions, although we cannot exclude the possibility that Rpd3S contains more than one copy of small subunits. (ii) The 1Rpd3S:DiNuc and 1Rpd3S:monoNuc species were the only gel-shift bands visible when low concentrations of Rpd3S were used and nucleosomal substrates were in molar excess. These conditions make it very improbable for two already less abundant Rpd3S monomers to bind to one molecule of di-nucleosome. (iii) When increasing amounts of mono-nucleosomes were added to a fixed amount of Rpd3S that is at a much lower concentration than the nucleosomes, we did not observe any additional shift (Supplementary Figure 3C). This finding essentially rules out the possibility that one Rpd3S complex can bind to two mono-nucleosomes simultaneously.
After establishing the stoichiometry of Rpd3S di-nucleosome binding, we next performed a side-by-side comparison of Rpd3S bound to mono-nucleosomes or di-nucleosomes. Since equimolar quantities of di-nucleosomes contain twice the number of nucleosomes as mono-nucleosomes, we decided to normalize this local concentration difference by the following approach. A common primer (5′) shared between mono- and di-nucleosome templates was end-labelled using 32P, then split and paired with corresponding unlabelled primers (3′) to amplify the two DNA probes. Because the two PCR products shared the same labelled primer, we could measure the radioactivity incorporation of the final gel-purified nucleosomes to determine the relative amount of each nucleosome. As shown in Figure 3C, the counts for mono-nucleosomes are twice as much as that for di-nucleosomes, so that the molar amounts of nucleosomes in mono- and di-nucleosome templates are equal. Indeed, we found that Rpd3S binding to di-nucleosomes is greater than its binding to mono-nucleosomes, and that Rpd3S:di-nucleosome binding is further stimulated by H3K36me (Figure 3C and D).
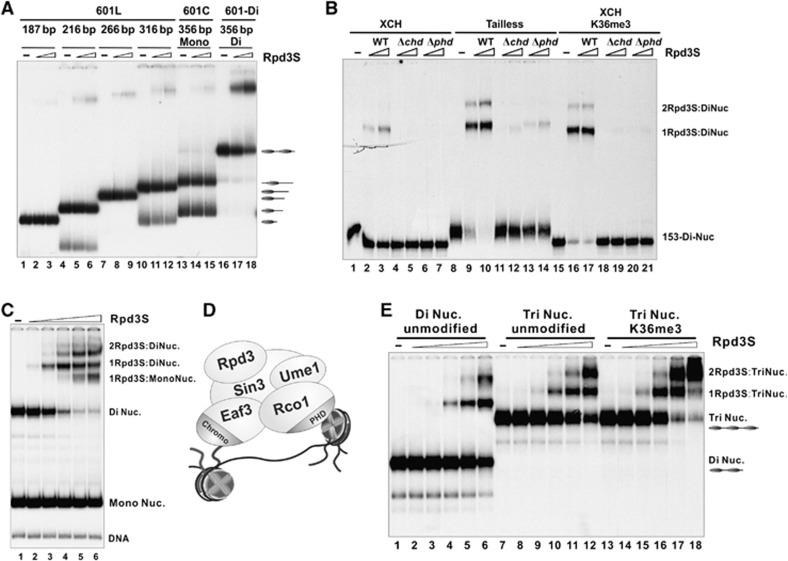
We next tested whether the increased binding of Rpd3S to di-nucleosomes is a result of the additional DNA present in this template. Rpd3S was incubated with a serials of mono-nucleosomes containing linker DNA ranging from 40 to 180 bp (Figure 4A). Rpd3S displayed a similar affinity toward all of the mono-nucleosomes tested. More importantly, when mono- and di-nucleosomal templates containing the same DNA sequence were compared (Figure 4A, lanes 13–18), di-nucleosomes showed stronger binding than mono-nucleosomes (whose composition was confirmed by MNase/DNaseI mapping (Supplementary Figure 4).
Figure 4.
Rpd3S complex bridges two nucleosomes within a di-nucleosome template. (A) Mono-nucleosomes bearing various lengths of linker DNA were compared with di-nucleosomes for Rpd3S binding. The position of each nucleosome within the DNA template is labelled on the right. (B) The binding of Rpd3S to tailless or H3K36-methylated di-nucleosomes depends on PHDRco1 and CHDEaf3. (C) A competition experiment for Rpd3S. Mono-nucleosomes and di-nucleosomes were mixed at 2:1 molar ratio before an increasing amount of Rpd3S was added into the reaction. (D) A bridging model in which Rpd3S simultaneously contacts two nucleosomes within a di-nucleosome template. (E) Rpd3S binds to tri-nucleosomes and di-nucleosomes in a similar manner. The same titrations of Rpd3S were used in all binding reactions in each panel.
Thus far, we observed three additive factors that can enhance Rpd3S chromatin recognition: increased H3K36me, removal of histone tails and di-nucleosome templates. Even for the substrates in which two of above factors were combined (Figure 4B, lanes 8–14 and 15–21), the binding of Rpd3S still required CHDEaf3 and PHDRco1 (Figure 4B, lanes 11–14 and 18–21), suggesting that Rpd3S di-nucleosome binding is mediated by a similar multivalency as previously described (Li et al, 2007b). We also confirmed that even a single mutation at the H3K36me-binding pocket of CHDEaf3 can disrupt the binding of Rpd3S to di-nucleosome templates (Supplementary Figure 5).
To demonstrate the preference of Rpd3S to di-nucleosomes in a more direct manner, Rpd3S was added to mono-nucleosomes and di-nucleosomes in a competition binding reaction. Equal concentrations (on a mono-nucleosome basis) of mono- and di-nucleosomes were mixed before Rpd3S was added to the reaction. As expected, Rpd3S favours di-nucleosomes (Figure 4C). Only when free di-nucleosomes are depleted does Rpd3S start to interact with mono-nucleosomes (Figure 4C, lanes 5 and 6). We noted that at a low concentration of Rpd3S (Figure 4C, lane3), 1Rpd3S:DiNuc was the predominant species. The fact that the single Rpd3S complex prefers di-nucleosomes strongly suggests that more free energy for binding is available from this template. Di-nucleosomes may adopt a conformation that is more compatible with Rpd3S binding, or the tethering of a second nucleosome to the first may enhance binding at a low-affinity site on Rpd3S. More importantly, since binding of only one Rpd3S molecule was observed under this condition, this result effectively eliminates the possibility that a synergistic interaction between two Rpd3S molecules is required for di-nucleosome interaction. The result is consistent with the AUC described earlier, since the Rpd3S complex also existed as a monomer in that analysis.
In summary, we conclude that Rpd3S prefers di-nucleosomes by contacting two nucleosomes simultaneously as illustrated in Figure 4D. Furthermore, we found that the binding of Rpd3S to tri-nucleosomes is similar to its binding to di-nucleosomes (Figure 4E), which is consistent with the notion that Rpd3S contains two reading modules, and can only bind to two nucleosomes at a time.
Rpd3S can tolerate an intramolecular two-fold reduction of H3K36me via di-nucleosome recognition
Given that Rpd3S bridges two linked nucleosomes (Figure 4D) and this binding requires H3K36me-dependent and -independent chromatin contacts (presumably with the core histone globular domain), we speculated that Rpd3S only requires one of the di-nucleosomes to be methylated for optimal binding to occur. To test this hypothesis, we prepared a hybrid di-nucleosomal template that includes one H3K36 tri-methylated and one unmodified nucleosome (Me/UM). This species was created by ligating separately reconstituted mono-nucleosomes through a non-palindromic site (Figure 5A; Supplementary Figure 6A). As shown in Figure 5B and C, in which the amount of unbound nucleosomes were quantified and considered as a reflection of the binding strength, hybrid di-nucleosomes display a similar binding pattern to fully methylated di-nucleosomes. The orientation of hybrid di-nucleosome is not important, as a similar result was observed when UM/Me di-nucleosomes were used.
Figure 5.
The binding of Rpd3S to di-nucleosomes is not sensitive to an intramolecular two-fold reduction in H3K36 methylation. (A) A schematic illustration of the strategy to generate di-nucleosomes that consist of two nucleosomes with different modification patterns. Ligating two individually reconstituted mono-nucleosomes through non-palindromic restriction sites formed these nucleosomes. (B) EMSA results. UM-unmodified; Me-K36me3; As-asymmetrically tri-methylated at K36 (each histone octamer only contains one methylated histone H3). (C) Quantification of EMSA results shown in (B) based on three independent experiments.
We next examined another scenario of intramolecular reduction of H3K36me, in which each nucleosome contains one methylated histone H3 and one unmodified one. These asymmetrically methylated (or hemi-methylated) nucleosomes were prepared as described previously (Li and Shogren-Knaak, 2008) (Supplementary Figure 7). Di-nucleosomes bearing these hemi-methylated histone octamers appear to bind to Rpd3S better than unmodified di-nucleosomes (Figure 5B, lanes 19–24 and 5C). Collectively, these results indicate that binding of Rpd3S to chromatin can tolerate an intramolecular two-fold reduction of H3K36me within di-nucleosomes through HPTM-independent and H3K36me-dependent contacts.
Methylated di-nucleosomes are preferentially deacetylated by Rpd3S
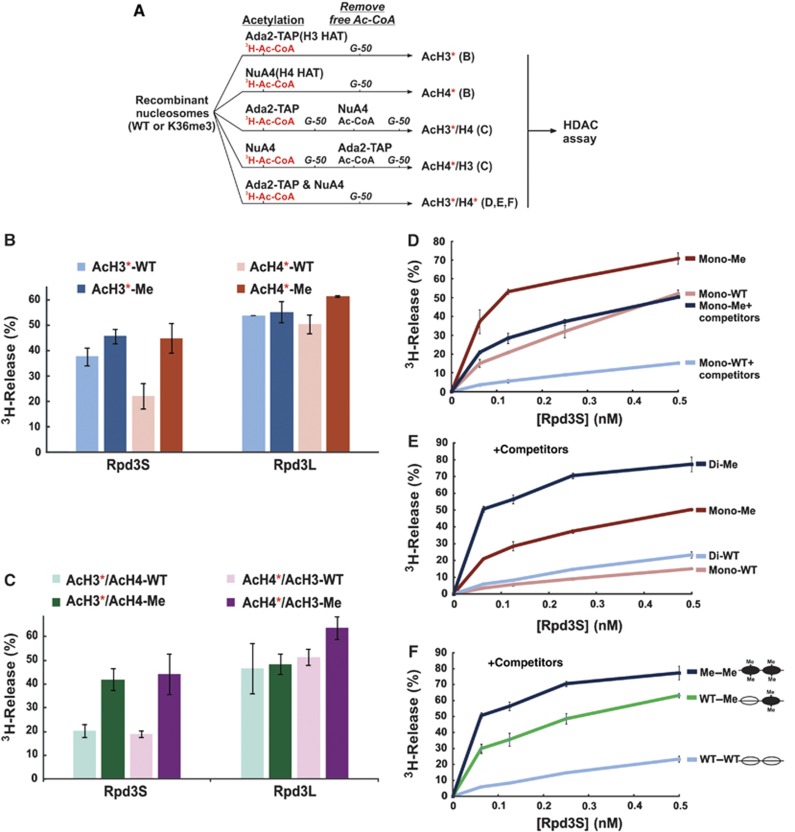
We next asked whether the binding properties of Rpd3S described above correlate with its HDAC activity in vitro. Currently, most studies on HDAC activity rely on 3H-labelled core histones or histone peptides, which are often not the physiological substrates for many HDACs. To overcome this potential shortcoming, we developed a robust nucleosome-based HDAC assay to examine HPTM cross-talk. The strategy (illustrated in Figure 6A) is to pre-acetylate various specifically modified nucleosomes using known histone acetyltransferase (HAT) complexes, thereby incorporating 3H-labelled acetyl-group to these nucleosomes as HDAC substrates.
Figure 6.
Methylated di-nucleosomes are preferentially deacetylated by Rpd3S. (A) A scheme for making specifically modified nucleosomal substrates for HDAC assays. * represents 3H-labelled histones. The letters in parentheses represent the panel number of the HDAC assays for which the substrates prepared from each branch were used. (B, C) Rpd3S preferentially deacetylates histone H3 and H4 of K36 methylated nucleosomes. (B) WT and methylated mono-nucleosomes were acetylated by either NuA4 or Ada2-TAP in the presence of 3H-Acetyl-CoA. Unincorporated 3H-Acetyl-CoAwas removed by passing through a G50 gel-filtration column. The resulting nucleosomes (acetylated H3 or acetylated H4) were used in HDAC assay. (C) Sequential acetylation with either 3H-Acetyl-CoA or non-radioactive Acetyl-CoA was carried out as illustrated in (A), and nucleosomes was purified after each step using a gel-filtration column. (D) In the presence of competitors, Rpd3S HDAC activity displays more dependence on H3K36me. In all, 100 ng of indicated nucleosomal substrates was used in each reaction, in which a five-fold molar excess of HeLa oligo-nucleosomes in comparison to each indicated di-nucleosome substrates were added. The ratios of HDAC on methylated and unmodified nucleosomes were defined as MSF. The average of MSF at different concentrations of Rpd3S in the absence of competitors is 2.0; while the average of MSF with competitors is 4.5. (E) Rpd3S prefers di-nucleosome substrates. HDAC assays were performed under the same condition as (D) with indicated nucleosomal substrates. (F) In the presence of competitor nucleosomes, Rpd3S differentially deacetylates di-nucleosome substrates based on their H3K36me status.
We first wanted to determine the substrate specificity of Rpd3S. Recombinant mono-nucleosomes were labelled using either a mixture of three HATs, SAGA, ADA and SLIK, which primarily target histone H3 (Lee et al, 2004; Lee and Workman, 2007) or NuA4, which mainly acetylates histone H4 (Grant et al, 1997) in the presence of 3H-acetyl-CoA (Figure 6A, the top two branches). As shown in Figure 6B, Rpd3S deacetylates H4 in a H3K36me-dependent manner; however, its activity on H3 is not as sensitive to H3K36me. To further validate the H3K36me specificity detected here, we also examined the HDAC activity of another Rpd3-containing complex, Rpd3L, which turned out to be H3K36me independent (Figure 6B), in agreement with its previously characterized in-vivo activity (Carrozza et al, 2005a).
Since these two types of nucleosomes are not only labelled differently, but also carried distinct acetylation patterns, we adopted a sequential modification (or hot-and-cold) approach to minimize any differences. As illustrated in Figure 6A (third and fourth branch), the H3 HAT and the H4 HAT were reciprocally used to incorporate 3H-acetyl-CoA and to acetylate the rest of the nucleosomes. In this way, we generated two comparable nucleosomes with the only difference being which histone (H3 or H4) is labelled with 3H. Using these sets of substrates, we demonstrated that Rpd3S preferentially deacetylates histone H3 and H4 in H3K36 methylated nucleosomes (Figure 6C), suggesting that Rpd3S is a nucleosomal deacetylase for both histone H3 and H4. This result is consistent with in-vivo observations that the levels of acetylated H3 and H4 both increased in a ΔSET2 mutant or in various Rpd3S mutants (Keogh et al, 2005; Carrozza et al, 2005b). More importantly, this H3K36me-stimulated Rpd3S HDAC activity also provides an explanation for an earlier finding that H3K36me is required for full activity of Rpd3S in vivo after its initial recruitment to coding regions (Drouin et al, 2010; Govind et al, 2010). Because we did not observe any noticeable activity differences of Rpd3S on histone H3 or H4, all nucleosomes used in the following experiments were labelled by incubating with an H3 HAT and an H4 HAT at the same time (Figure 6; last branch).
To further optimize the HDAC reactions, we decided to mimic physiological conditions by adding oligo-nucleosome competitors into the HDAC reaction. Rpd3S was titrated over a wide range of concentrations to maximize the ability to detect minor differences between substrates. As expected, adding competitor nucleosomes reduced the overall activity of Rpd3S on both substrates (Figure 6D). However, Rpd3S HDAC activity appeared to be more dependent on H3K36me under this condition, as the methylation stimulating factors (MSFs) increased by more than two-fold (see figure legend).
We next sought to compare Rpd3S HDAC activity on mono-nucleosomes and di-nucleosomes substrates (Figure 6E) in the presence of competitors. When mono-nucleosomes and di-nucleosomes in the same modification state were compared, Rpd3S displays strong preference for di-nucleosomal templates over mono-nucleosomes (Figure 6E). Consistent with the EMSA result (Figure 3C), Rpd3S favours methylated substrates even in the context of di-nucleosomes (comparing the two blue lines in Figure 6E).
Finally, we asked if Rpd3S HDAC activity could tolerate a two-fold intramolecular dilution of H3K36me. Ligating one unmodified mono-nucleosome to one K36me3 mono-nucleosome via a non-palindromic site generated a hetero-nucleosome template (Supplementary Figure 6B). These di-nucleosomes were purified through preparative native electrophoresis and subjected to acetylation labelling as illustrated in Figure 6A (last branch). In the presence of competitor nucleosomes, the extent of Rpd3S activity on hetero-di-nucleosome (UM–Me) is more comparable to that on fully methylated di-nucleosome than that on unmodified ones. Therefore, we concluded that a two-fold dilution of H3K36me is tolerated by Rpd3S.
Discussion
It is generally accepted that the functional readouts of most HPTMs are dictated by the particular effectors/readers that they recruit to specific chromosome locations (Yun et al, 2011). Especially for those factors that are involved in maintaining genome integrity, it would be advantageous to have an intrinsic capacity to remain bound to chromatin even in the event of minor fluctuations in the primary recruitment signal. Here, our biochemical analysis of Rpd3S binding to its cognate substrates has revealed a novel molecular mechanism by which this chromatin regulator tolerates the reduction of HPTM signals. The principle of this safeguard mechanism is to use one reading module to recognize targeting HPTMs and another domain to interact with neighbouring nucleosomes independent of their HPTM states, thereby providing the potential to tolerate the dilution of targeting HPTMs. This mechanism should have general implications, since many chromatin regulators possess this multivalent recognition potential (Yun et al, 2011). Heterochromatin protein 1 (HP1) has been shown to interact with the histone-fold domain of H3 (Nielsen et al, 2002) and recognize H3K9 methylation. PRC1 binds tailless nucleosomes (Breiling et al, 1999; Shao et al, 1999) and recognizes H3K27 methylated tails (Francis et al, 2004). Moreover, chromatin factors commonly display preference for oligo-nucleosomes. For instance, Bdf1 forms a di-nucleosome complex with the +1 and +2 nucleosomes (Koerber et al, 2009); HP1 prefers oligo-nucleosomes (Nielsen et al, 2002; Canzio et al, 2011); PRC2 shows strong histone methyltransferase activity on oligo-nucleosomes (Cao et al, 2002), PRC1 contacts about 3–4 nucleosomes (Francis et al, 2004); and tri-nucleosomes are the minimal unit to detect the binding of the yeast SIR complex (Martino et al, 2009). More importantly, this molecular mechanism also provides a potential solution for chromatin-remodelling factors to function immediately after DNA replication, when the parental HPTM pattern has not been fully restored on both daughter strands.
We propose that a di-nucleosome-recognition mechanism enables chromatin effectors to endure an at least two-fold dilution of their primary recruiting HPTMs. However, under physiological conditions, the threshold for reading tolerance may not be limited to two-fold. For example, deletion of PAF1 eradicates H3K36me3 and lowers the amount of H3K36me2 more than two-fold. However, Rpd3S retains its normal function, as we did not observe any histone acetylation defects or a cryptic transcription phenotype in PAF1 mutant cells (Chu et al, 2007; Youdell et al, 2008; Li et al, 2009). This result suggests that Rpd3S can tolerate a significant reduction of H3K36me in the normal cellular context. We found that when competitors were included in the histone deacetylation reaction, the increased stringency accentuated the ability of Rpd3S to distinguish between methylated and unmodified substrates (Figure 6D). As shown in Figure 5, the binding affinity of Rpd3S toward hetero-di-nucleosomes or hemi-methylated nucleosomes is much closer to that of fully methylated nucleosomes than to that of unmodified nucleosomes. However, we can consistently detect modest binding differences among three methylated nucleosomes. Importantly, the slightly better binding of hetero-di-nucleosomes over hemi-methylated nucleosomes cannot be easily explained using a statistical argument, since these two nucleosomes contain exactly the same number of methyl groups. There seems to be a tolerance threshold for Rpd3S binding. In light of histone modification in the di-nucleosome context, for instance, H3K36me3 on di-nucleosomes could generate at least five different signals simply based on the number of methyl groups (0–4). This signal gradient may provide a certain level of plasticity for cells to maintain genome integrity while dealing with challenges, such as replication errors and damage repairs.
Here, we show that Rpd3S binds to di-nucleosomes and tri-nucleosomes similarly (Figure 4E), which may reflect the fact that this particular complex contains two reading modules (CHDEaf3 and PHDRco1). Many complexes have multiple chromatin-recognition modules. Even one subunit can contain multiple domains that can read different HPTM signals (Ruthenburg et al, 2011). Therefore, it will be interesting to see in future experiments whether a complex with more reading modules can display stronger reading tolerance, e.g., during DNA replication when combinations of PTM are changed simultaneously.
Materials and methods
Protein purification
The Rpd3S complexes (WT, the rco1Δphd, eaf3Δchd and two Eaf3 chromo domain point mutants) were purified through tandem affinity purification (Rco1-TAP) as described (Li et al, 2007b) using corresponding yeast strains listed in Supplementary Table 4. Chromatin-remodelling complexes RSC (Rsc2-TAP), Swi/Snf (Snf6-TAP), ISW1 (Isw1-TAP), CHD1 (Chd1-TAP), NuA4 (Epl1-TAP) and Ada2-TAP (containing a mixture of the SAGA, SLIK and ADA HAT complexes that primarily modify histone H3 (Lee et al, 2004)) were also purified using the tandem-purification approach. Drosophila ACF (ATP-utilizing chromatin assembly and remodelling factor) was prepared by coexpression of the Acf1 and ISWI subunits using a baculovirus system in Sf21 insect cells and purified through single-step FLAG purification. Recombinant Rpd3S was reconstituted by coexpression of five different baculovirus that correspond to each subunit and purified through Rco1–Flag (Supplementary Figure 3). The concentrations of all proteins/protein complexes were determined by either protein standard curves or common protein marks. For all direct comparisons, each component involved was quantified side-by-side on the same gel to minimize the estimation errors.
Nucleosome preparation and chromatin-based biochemical assays
Detailed procedures of nucleosome preparation and other chromatin assays are described in the Supplementary data. Recombinant Xenopus histones were individually purified and reconstituted into histone octamers using a previous protocol (Li et al, 2007b). All histone H3K36 methylated histones were prepared using the Methyl-Lysine Analog (MLA) approach (Simon et al, 2007; Li et al, 2009). The chemical reactions result in almost all lysines being converted to methylation mimics, as was confirmed by mass-spectrometry analysis and/or western blotting.
EMSA assay
EMSA reactions were carried out in a 15-μl system containing 10 mM HEPES pH 7.8, 50 mM KCl, 4 mM MgCl2, 5 mM DTT, 0.25 mg/ml BSA, 5% Glycerol and 0.1 mM PMSF (1 × EMSA buffer). The procedures for sliding assays were described previously (Li et al, 2005).
In principle, with the same concentration of DNA, di-nucleosomes templates contain twice as many nucleosomes as mono-nucleosome templates. This difference was taken into consideration when mono-nucleosomes and di-nucleosomes are directly compared (as shown in Figure 2A and B). Twenty pmol of Primer P1111 was 32P end-labelled and then split in half. Half of labelled P1111 was paired with cold p772 using pBL630 as templates to generate the mono-nucleosome probe (ChrT03). The other half was mixed with cold P1112 using pBL634 as templates to create di-nucleosomal DNA (ChrT04). Given that these two probes share a primer that has the same radioactivity incorporation efficiency, we normalized the amount of each nucleosomes used in EMSA based on the radioactivity of final gel-purified nucleosomes.
HDAC assay
To prepare specifically acetylated nucleosomal templates for HDAC assays, 20 μg of reconstituted nucleosomes was mixed with 30 μl of 5 × HAT buffer (250 mM Tris–HCl pH 8.0, 5 mM DTT, 25% Glycerol, 0.03% NP40 and 1 mM PMSF), 6 μl of 20 μM 3H-acetyl-CoA (or cold acetyl-CoA), and the HATs (as illustrated in Figure 6A) in a 150-μl reaction. After 20 h incubation at 30°C, the reaction mixtures were passed through a G-50 mini column (GE) prewashed with 1 × HAT buffer to remove unincorporated acetyl-CoA. The overall acetylation level of resulting nucleosomes was determined by standard filter-binding assay using a scintillation counter or western blots (Yun et al, 2012).
In all, 5 μl of acetylated nucleosomes (0.10 μg) were mixed with a proper amount of HDAC and the final volume was adjusted to 15 μl using CEB Buffer (10 mM Tris–HCl pH=8.0; 150 mM NaCl; 1 mM magnesium acetate; 1 mM Imidazole; 2 mM EGTA pH8.0; 10 mM βME; 0.1% NP40; 10% Glycerol). After a 1-h incubation at 30°C, 20 μl of H2O and 36 μl of 1 M HCl/0.4 M acetic acid was added to stop the reactions along with 800 μl of ethyl acetate. The mixtures were vigorously vortexed for 5 s and centrifuged at 12 000 g for 10 min at 4°C. In all, 720 μl of supernatant was then transferred to a scintillation vial containing 4 ml of scintillation fluid for liquid counting. In Figure 6D–F, 0.5 μg of HeLa oligonucleosomes were added to each reaction as competitors.
Plasmids and yeast strains
All plasmids and yeast strains used in this study are listed in Supplementary tables 2 and 4 respectively.
Supplementary Material
Acknowledgments
We thank Dr Karolin Luger for plasmids. We are grateful to Drs Lee Kraus, Michael Carey, Samantha Pattenden and Jerry Workman for critical comments on the manuscript. JH was supported by the National Research Foundation of Korea Grant (MEST) (NRF-2010-R13-0029523). BL was a WA ‘Tex’ Moncrief, Jr Scholar in Medical Research and was supported by grants from the National Institutes of Health (R01GM090077), the Welch Foundation (I-1713), the March of Dimes Foundation, and the American Heart Association.
Author contributions: JH, JW, CHL, MY, CB and BL conceived the project and designed the experiments. JH, JW, CHL, MY, DG CB performed the experiments. BL wrote the manuscript and CB edited it.
Footnotes
The authors declare that they have no conflict of interest.
References
- Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE (2011) Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0A resolution. Science 334: 977–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D (2010) Molecular signals of epigenetic states. Science 330: 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiling A, Bonte E, Ferrari S, Becker PB, Paro R (1999) The Drosophila polycomb protein interacts with nucleosomal core particles in vitro via its repression domain. Mol Cell Biol 19: 8451–8460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, Al-Sady B (2011) Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell 41: 67–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Florens L, Swanson SK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL (2005a) Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim Biophys Acta 1731: 77–87discussion 75–6 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL (2005b) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Chu Y, Simic R, Warner MH, Arndt KM, Prelich G (2007) Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. Embo J 26: 4646–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Längst G, Corona DF, Becker PB, Nightingale KP (2001) Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol 21: 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B (2008) Architecture of the SWI/SNF-nucleosome complex. Mol Cell Biol 28: 6010–6021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Bystricky K, Richmond TJ (2003) Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol 327: 85–96 [DOI] [PubMed] [Google Scholar]
- Drouin S, Laramee L, Jacques PE, Forest A, Bergeron M, Robert F (2010) DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet 6: e1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL (2004) Chromatin compaction by a polycomb group protein complex. Science 306: 1574–1577 [DOI] [PubMed] [Google Scholar]
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG (2010) Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell 39: 234–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11: 1640–1650 [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR (2009) Distinctive chromatin in human sperm packages genes for embryo development. Nature 460: 473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A (2011) Histone modification: cause or cog? Trends Genet 27: 389–396 [DOI] [PubMed] [Google Scholar]
- Joshi AA, Struhl K (2005) Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20: 971–978 [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Rando OJ (2010) Chromatin as a potential carrier of heritable information. Curr Opin Cell Biol 22: 284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605 [DOI] [PubMed] [Google Scholar]
- Koerber RT, Rhee HS, Jiang C, Pugh BF (2009) Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol Cell 35: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Lee KK, Prochasson P, Florens L, Swanson SK, Washburn MP, Workman JL (2004) Proteomic analysis of chromatin-modifying complexes in Saccharomyces cerevisiae identifies novel subunits. Biochem Soc Trans 32: 899–903 [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL (2007) Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol 8: 284–295 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007a) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL (2007b) Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL (2007c) Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev 21: 1422–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Jackson J, Simon MD, Fleharty B, Gogol M, Seidel C, Workman JL, Shilatifard A (2009) Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J Biol Chem 284: 7970–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL (2005) Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci USA 102: 18385–18390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Shogren-Knaak MA (2008) Cross-talk between histone H3 tails produces cooperative nucleosome acetylation. Proc Natl Acad Sci USA 105: 18243–18248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makde RD, England JR, Yennawar HP, Tan S (2010) Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467: 562–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM (2009) Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell 33: 323–334 [DOI] [PubMed] [Google Scholar]
- Moazed D (2011) Mechanisms for the inheritance of chromatin states. Cell 146: 510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutskov V, Gerber D, Angelov D, Ausio J, Workman J, Dimitrov S (1998) Persistent interactions of core histone tails with nucleosomal DNA following acetylation and transcription factor binding. Mol Cell Biol 18: 6293–6304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED (2002) Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416: 103–107 [DOI] [PubMed] [Google Scholar]
- Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK (2011) Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA 108: E472–E479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G (2009) Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10: 192–206 [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, Allis CD (2011) Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145: 692–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE (1999) Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98: 37–46 [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM (2007) The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128: 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Thastrom A, Bingham LM, Widom J (2004) Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J Mol Biol 338: 695–709 [DOI] [PubMed] [Google Scholar]
- Xu M, Wang W, Chen S, Zhu B (2011) A model for mitotic inheritance of histone lysine methylation. EMBO Rep 13: 60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youdell ML, Kizer KO, Kisseleva-Romanova E, Fuchs SM, Duro E, Strahl BD, Mellor J (2008) Roles for Ctk1 and Spt6 in regulating the different methylation states of Histone H3 lysine 36. Mol Cell Biol 28: 4915–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M, Ruan C, Huh JW, Li B (2012) Reconstitution of modified chromatin templates for in vitro functional assays. Methods Mol Biol 833: 237–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M, Wu J, Workman JL, Li B (2011) Readers of histone modifications. Cell Res 21: 564–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA (2010) In vivo residue-specific histone methylation dynamics. J Biol Chem 285: 3341–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.