Abstract
RNA-binding E3 ubiquitin ligases were recently identified, though their function remains unclear. While studying the regulation of the MHC class I (MHC-I) pathway, we here characterize a novel role for ubiquitin in mRNA degradation. MHC-I molecules provide ligands for both cytotoxic T-lymphocytes as well as natural killer (NK) cells, and play a central role in innate and adaptive immunity. MHC-I cell-surface expression is closely monitored by NK cells, whose killer immunoglobulin-like receptors encode MHC-I-specific activatory and inhibitory receptors, implying that MHC-I expression needs to be tightly regulated. In a functional siRNA ubiquitome screen we identified MEX-3C, a novel RNA-binding ubiquitin E3 ligase, as responsible for the post-transcriptional, allotype-specific regulation of MHC-I. MEX-3C binds the 3′UTR of HLA-A2 mRNA, inducing its RING-dependent degradation. The RING domain of MEX-3C is not required for HLA-A2 cell-surface downregulation, but regulates the degradation of HLA-A2 mRNA. We have therefore uncovered a novel post-transcriptional pathway for regulation of HLA-A allotypes and provide a link between ubiquitination and mRNA degradation.
Keywords: 3′UTR, antigen presentation, HLA-A, MHC class I, mRNA decay, NK cells, ubiquitin E3 ligase
Introduction
Cell-surface MHC-I provides key ligands for the different receptor families on CTL and NK cells. Signals received by the killer cell immunoglobulin-like receptors (KIRs) balance the NK-cell response between tolerance of healthy cells and killing of unhealthy cells (Parham, 2005). Since small changes in MHC-I levels affect susceptibility to NK killing (Holmes et al, 2011), surface expression of MHC-I must be finely regulated at each stage of the antigen-presentation pathway (Wearsch and Cresswell, 2008), and precise regulation of MHC-I expression is essential for cell survival. Post-translational modification of MHC-I molecules by ubiquitin provides a potent mechanism for regulating MHC-I turnover and several viral as well as cellular ubiquitin E3 ligases (Randow and Lehner, 2009) regulate MHC-I assembly in the endoplasmic reticulum and at the cell surface. Although ubiquitin is best recognized for its role in post-translational protein regulation, a potential role in the regulation of RNA stability has emerged from the finding that at least 15 E3 ubiquitin ligases encode predicted RNA-binding domains (RBDs) (Cano et al, 2010). While there is some understanding of how these proteins regulate mRNA, their requirement for E3 ligase activity is unclear. Turnover of AU-rich cytokine mRNAs is dependent on ubiquitination (Laroia et al, 2002), though the link between mRNA turnover and ubiquitination is not defined. Roquin, an RNA-binding E3 ligase regulates Icos mRNA stability through the repressive action of a particular miRNA (miR-101) (Vinuesa et al, 2005; Yu et al, 2007), though again, the requirement for its E3 ligase activity remains unclear.
MHC-I ubiquitination is involved in the post-translational regulation of MHC-I assembly, but ubiquitin is not known to play a role in the transcriptional or post-transcriptional regulation of MHC-I. Our recent experiments showed that ubiquitin is involved in the regulation of cell-surface MHC-I, since overexpression of a ubiquitin mutant markedly increased cell-surface HLA-A2 (Duncan et al, 2010). We now extend these findings, and in a functional siRNA screen identify MEX-3C, a novel RNA-binding E3 ubiquitin ligase, as responsible for the post-transcriptional regulation of common HLA-A allotypes. MEX-3C is highly expressed in cells of the innate immune system, in particular activated NK cells, and affects the threshold of killing by these cells. We show that MEX-3C binds and induces the RING-dependent degradation of HLA-A2 mRNA through its 3′UTR. The RING domain of MEX-3C is not required for HLA-A2 surface downregulation, but is required for HLA-A2 mRNA degradation. These results reveal a novel mechanism for MHC-I allotype-specific regulation and provide, for the first time, a link between ubiquitination and mRNA decay.
Results
The Ubiquitin-I44A mutant identifies a role for post-transcriptional regulation of HLA-A2 in HEK293 cells
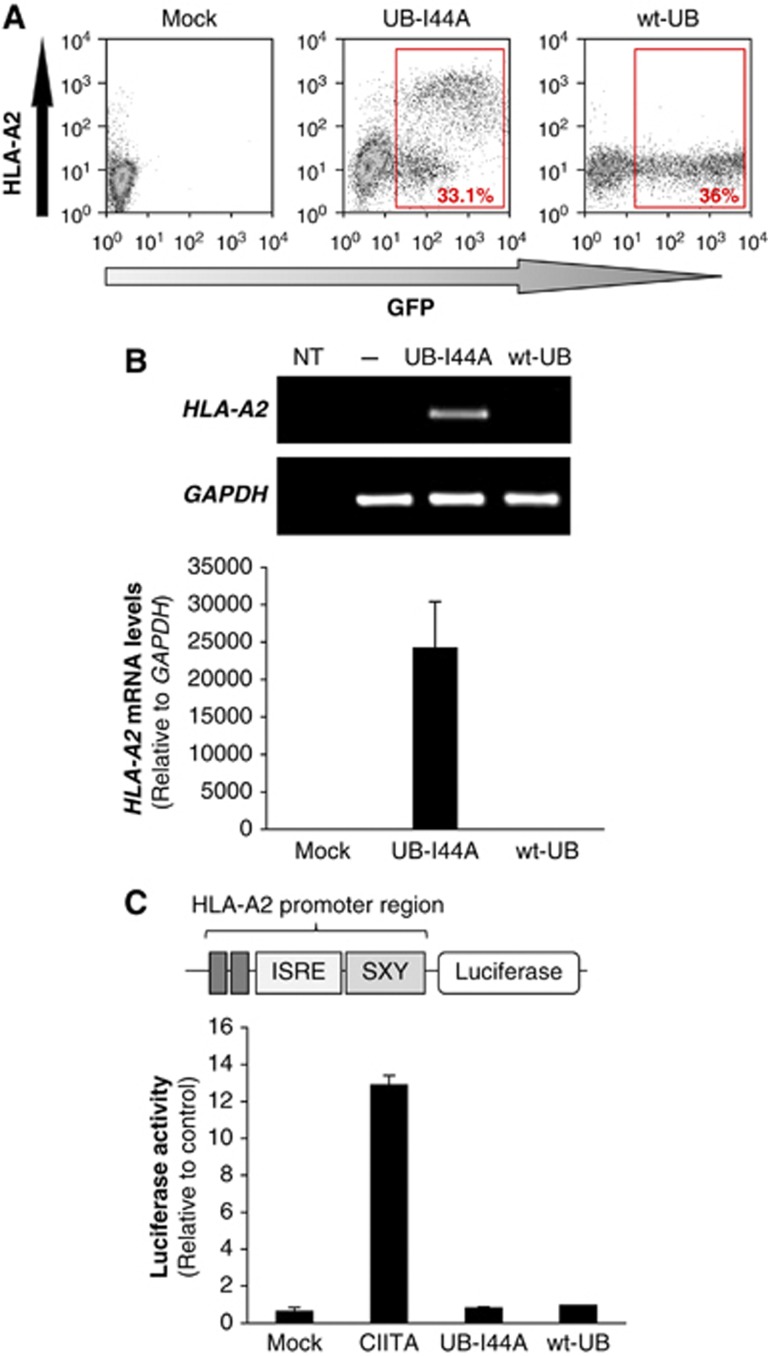
We recently showed that the I44A ubiquitin mutant (UB-I44A), which has a mutation in its hydrophobic binding surface, causes a dramatic increase in cell-surface HLA-A2 (Figure 1A and Duncan et al, 2010) in HEK293 cells, with no effect on HLA-B/C or -E (Duncan et al, 2010). Whether ubiquitin is required for the transcriptional or post-transcriptional regulation of MHC-I is unknown. To investigate this further, we used quantitative RT–PCR to show that UB-I44A transfection markedly increased HLA-A2 mRNA levels in these cells (Figure 1B). To our surprise, this increase in mRNA did not reflect de-novo initiation of HLA-A2 transcription, as neither wild-type ubiquitin nor the UB-I44A mutant increased reporter luciferase activity in cells expressing luciferase driven by the HLA-A*0201 promoter (pGL3-A230; Gobin and van den Elsen, 2001) (Figure 1C). Transfection of the positive control MHC class II transactivator (CIITA) significantly increased luciferase activity (Figure 1C). The increase in HLA-A2-specific mRNA was therefore most likely to be post-transcriptional, rather than due to increased HLA-A2 mRNA transcription. Since these findings suggested a role for ubiquitin in the regulation of HLA-A2 mRNA stability, we screened HEK293 cells with a custom siRNA library, targeting the putative E3 ubiquitin ligase family (Stagg et al, 2009) to identify the ubiquitin E3 ligase responsible.
Figure 1.
The Ubiquitin-I44A mutant induces a post-transcriptional increase in HLA-A2 mRNA. (A) Cytofluorometric analysis of cell-surface HLA-A2 expression in HEK293 cells expressing GFP-tagged UB-I44A or wt-UB. Cells were labelled with mAb BB7.2 for HLA-A2 expression. Red boxes show % of Ubiquitin–GFP expressing cells. (B) UB-I44A expression increases HLA-A2 mRNA levels. HEK293 cells were sorted by GFP levels, a surrogate marker for ubiquitin expression. HLA-A2 transcripts were measured by qRT–PCR and normalized against GAPDH levels, using the ddCt method. Relative transcript levels are visualized in a 1.8% agarose gel (top panel) and expressed as mean±s.d. (bottom panel). Data are representative of three experiments. NT: no template control. (C) UB-I44A does not affect HLA-A2 promoter activity. HEK293 cells were co-transfected with pGL3-A230, a luciferase reporter driven by the HLA-A*0201 promoter, together with CIITA (positive control), empty vector (mock control) or the wild-type (wt) and UB-I44A ubiquitin constructs. Luciferase activity was determined 48 h post-transfection. Results are relative to mock control levels (set as 1) and expressed as mean±s.d. of three independent experiments (N=3).
A targeted siRNA screen identifies MEX-3C, a novel RNA-binding E3 ubiquitin ligase, as involved in HLA-A2 regulation
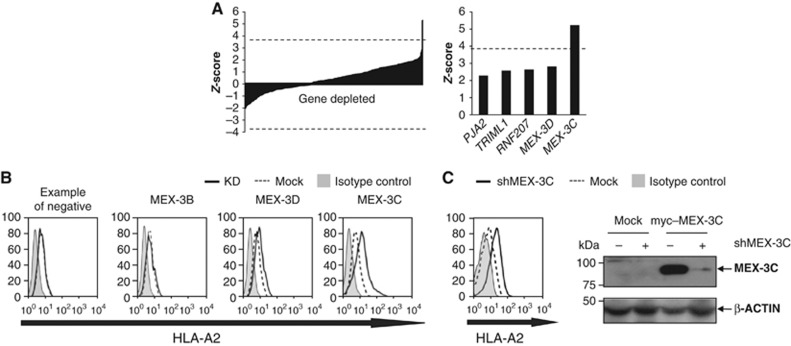
The rationale of the screen was our prediction that depletion of the E3 ligase required for HLA-A2 regulation would increase cell-surface HLA-A2 levels, similar to the effect observed with the UB-I44A mutant (Figure 1A). We screened a ubiquitome library, specific for RING and Hect E3 ligases. Of the 375 E3 ligases screened, depletion of only one, MEX-3C, significantly increased HLA-A2 surface levels (Figure 2A and B), with a Z-score (5.23) significantly above the Bonferroni-corrected P-value threshold (P=0.05) of 3.82. The increased cell-surface HLA-A2 expression was achieved with four independent MEX-3C siRNAs (Supplementary Figure S1A). Effective on-target depletion of endogenous MEX-3C mRNA was detected by qRT–PCR; with no effect on its homologues MEX-3A, MEX-3B and MEX-3D (Supplementary Figure S1B). Although the MEX-3C-specific antibody was insufficiently sensitive to detect endogenous MEX-3C in HEK293s, effective depletion of exogenous myc–MEX-3C was seen by immunoblot, confirming specificity of the siRNA (Figure 2C), which had no activity against the related myc–MEX-3D (data not shown).
Figure 2.
A targeted siRNA screen identifies a role for MEX-3C, a novel RNA-binding E3 ubiquitin ligase, in HLA-A2 regulation. (A) siRNA-mediated depletion of MEX-3C significantly increases cell-surface HLA-A2 expression. The left bar chart shows the Z-scores for the 375 E3 ligases screened in three independent experiments. Of the 375 E3 ligases screened, only MEX-3C depletion gave a significant increase in HLA-A2 expression. The graph on the right shows the top five Z-scores. MEX-3C’s Z-score (5.23) was significantly above the Bonferroni-corrected P-value threshold (P=0.05) of 3.82 (dashed line). (B) Flow cytometric analysis of cell-surface HLA-A2 levels in mock (dashed line) versus siRNA-treated cells (solid line). Isotype control (solid grey). (C) Stable shRNA-mediated depletion of MEX-3C increases cell-surface HLA-A2. (Left) Cytofluorometric analysis of HLA-A2 surface levels in parental (mock shRNA depleted, dashed line) HEK293 and cells stably transduced with a shRNAmir against MEX-3C (shMEX-3C; solid line). (Right) Immunoblot analysis of MEX-3C in HEK293 cells expressing myc-tagged MEX-3C or control cells (mock), both stably transduced with shMEX-3C. The shMEX-3C sequence is identical to the targeting oligonucleotide #2 of the original screening pool. β-Actin was used as loading reference levels.
The MEX-3 protein family of evolutionarily conserved RNA-binding E3 ligases are recruited to P-bodies and involved in post-transcriptional regulation (Buchet-Poyau et al, 2007). There are four human MEX-3 proteins, each containing two tandem repeat mRNA-binding KH domains and a C-terminal E3 ubiquitin ligase RING domain (Figure 3A; Buchet-Poyau et al, 2007). MEX-3C directly binds RNA via the KH domains (Buchet-Poyau et al, 2007), though the sequence specificity and affinity is undefined. MEX-3C also interacts directly with the argonaute AGO1 and AGO2 proteins, co-immunoprecipitates poly(A)-binding protein (PABP) and shuttles from the nucleus to the cytoplasm via the CRM-1 complex (Buchet-Poyau et al, 2007).
Figure 3.
MEX-3C’s ubiquitin ligase activity is required for HLA-A2 mRNA degradation. (A) Schematic representation of MEX-3C protein. Asterisks indicate the residues mutated in the mutKH MEX-3C (G249D; G343D), while the arrowhead indicates the position of the premature stop codon in RINGless MEX-3C (1–636). (B) Over-expression of wild-type and RINGless MEX-3C reduce HLA-A2 levels in HEK293T cells. Cytofluorometric analysis of cell-surface HLA-A2 in HEK293T cells expressing the indicated MEX-3C cDNAs, co-transfected with GFP cDNA (10%) as a surrogate marker for MEX-3C expression (see also Supplementary Figure S1C for MEX-3C immunoblots). TfR levels were measured as control. HLA-A2 geometric fluorescent means (GFM) in transfected GFPhigh cells for the empty vector control and wtMEX-3C were 90.5 and 29.4, respectively, resulting in a three-fold cell-surface downregulation of HLA-A2. A similar effect was seen in cells transfected with RINGless MEX-3C (GFM=43.2); while treatment with MEX-3C KH mutant caused only a minor decrease in cell-surface HLA-A2 expression (GFM=74.8). (C) The RING domain of MEX-3C is required for HLA-A2 mRNA degradation. Endogenous HLA-A2 mRNA levels were analysed by qRT–PCR from sorted GFPhigh (MEX-3C (includes wild-type or RINGless MEX-3C) expressing) and GFP− (control) populations 48 h post-transfection (see also Supplementary Figure S1D for MEX-3C protein levels). A significant reduction in HLA-A2 transcript levels (0.44±0.10; N=5; P-value=0.0001) was seen in wtMEX-3C expressing cells when compared with control cells (set at 1±0.25) all normalized to GAPDH. In contrast, RINGless MEX-3C shows a modest increase in HLA-A2 transcripts (1.23±0.14; N=5) compared with control cells. Results are expressed as the mean±s.d. of five independent experiments (***P-value=0.0001).
Overexpression of MEX-3C reduces HLA-A2 levels in HEK293T cells
Since depletion of endogenous MEX-3C increased HLA-A2 expression, we examined how MEX-3C overexpression affects HLA-A2 in HEK293T cells, which have higher resting HLA-A2 levels than HEK293s. As predicted from the MEX-3C RNAi experiments, exogenous expression of wtMEX-3C (together with pMax–GFP at a 10:1 ratio so that GFP expression provides a surrogate marker for exogenous MEX-3C) decreased both HLA-A2 mRNA and surface HLA-A2 expression (three-fold) (Figure 3B and C). A similar decrease in surface HLA-A2 expression was seen in cells transfected with RINGless MEX-3C (aa 1–636), which has no ubiquitination activity. In contrast, the MEX-3C KH mutant (G249D; G343D), with single-point mutations in each of the two RBDs, caused only a minor decrease in cell-surface HLA-A2 (Figure 3B; Supplementary Figure S1C).
The RING domain of MEX-3C is required for HLA-A2 mRNA degradation
MEX-3C’s E3 ubiquitin ligase activity was readily confirmed by immunoblotting (Supplementary Figure S2A). However, since wt and RINGless MEX-3C both decreased cell-surface HLA-A2 expression, the role of MEX-3C’s C-terminal RING domain is unclear (Figure 3B), raising the question whether the E3 ligase activity is required for HLA-A2 mRNA regulation?
MEX-3C originates from an evolutionarily conserved family of RNA-binding proteins. The founder member, MEX-3 from Caenorhabditis elegans is a key effector of cell polarity, controlling the spatial patterning of PAL-1 in the nematode embryo. Disruption of the mex-3 locus is embryonic lethal and MEX-3 is thought to prevent translation of the PAL-1 mRNA through binding the PAL-1 3′UTR (Draper et al, 1996; Hunter and Kenyon, 1996). The two KH domains required for RNA binding retain ∼80% sequence identity with their four human MEX-3 (hMEX-3A–D) paralogues. Evolutionary diversification from C. elegans lead to the acquisition of a C-terminal RING finger, evident in the Drosophila orthologue, with subsequent expansion from a single dMex-3 (RNA-binding+RING finger domain) to four genes on different chromosomes in higher eukaryotes, including humans. To try and identify a role for the MEX-3C RING, we measured endogenous HLA-A2 mRNA in cells expressing high levels of MEX-3C (sorted for GFPhigh) and GFP− (control) populations following MEX-3C transfections (Figure 3C). A significant reduction in HLA-A2 mRNA is seen in wtMEX-3C cells compared with control cells (Figure 3C). In contrast, cells expressing RINGless MEX-3C showed no decrease in HLA-A2 mRNA, despite, like wtMEX-3C showing reduced cell-surface HLA-A2 levels (Figure 3B; Supplementary Figure S1D). Indeed, these cells show a modest increase in HLA-A2 mRNA, confirming an absolute requirement of MEX-3C’s RING in mRNA degradation. Therefore, degradation of HLA-A2 mRNA by MEX-3C is dependent on a functional RING domain and implies a critical role for ubiquitin ligase activity in mRNA decay.
MEX-3C binds HLA-A2 mRNA in vivo
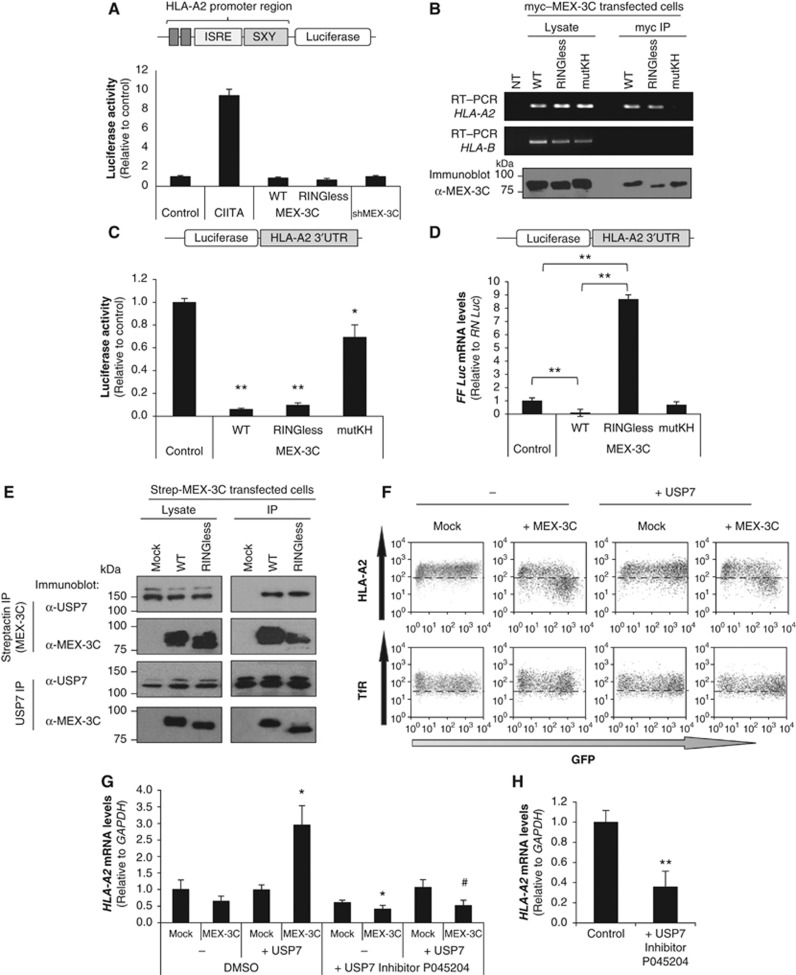
To confirm that the decreased HLA-A2 mRNA levels seen with wtMEX-3C was due to mRNA decay, and not decreased transcription, we used the previously described reporter assay (Figure 1C) to show that neither wt nor RINGless MEX-3C affected HLA-A*0201 promoter activity (Figure 4A). MEX-3C binds mRNA through its KH domains (Buchet-Poyau et al, 2007), and we confirmed a direct interaction between both wt and RINGless MEX-3C and HLA-A2 mRNA by RT–PCR (Buchet-Poyau et al, 2007), whereas no HLA-A2-specific mRNA was immunoprecipitated with the MEX-3C KH mutant (Figure 4B). Furthermore, the closely related HLA-B mRNA was not detected in MEX-3C immunoprecipitates (Figure 4B).
Figure 4.
MEX-3C regulates HLA-A2 levels through its mRNA 3′UTR. (A) MEX-3C does not affect HLA-A2 promoter activity. HEK293Ts were co-transfected with a luciferase reporter driven by the HLA-A*0201 promoter, together with controls or the indicated MEX-3C constructs. (B) MEX-3C binds HLA-A2 mRNA in vivo. MEX-3C was immunoprecipitated (IP) from HEK293Ts expressing the indicated myc-tagged–MEX-3C cDNAs. RT–PCR was performed on either total RNA from lysates, or following IP with anti-myc antibody (top panel). MEX-3C immunoblots were performed on lysates or anti-myc IPs (bottom panel). NT: No template. (C) MEX-3C regulates HLA-A2 levels by its mRNA 3′UTR. The HLA-A*0201-3′UTR was cloned downstream of firefly luciferase and co-transfected into HEK293Ts with the indicated MEX-3C constructs. Luciferase activity in wtMEX-3C expressing cells (0.06±0.001; N=4; P-value =0.0005) showed a 16-fold decrease compared with control cells (set as 1). RINGless-MEX-3C (0.10±0.002; N=4; P-value=0.006) was also markedly decreased, an effect that was partially abrogated by the mutKH-MEX-3C (0.69±0.01; N=4). **P-value=0.0005 versus Control; *P-value=0.001 versus wtMEX-3C. Results are expressed as the mean±s.d. of four independent experiments. (D) The RING domain of MEX-3C is required for mRNA degradation. FF-Luc-HLA-A2-3′UTR mRNA levels were analysed by qRT–PCR from cells expressing the different MEX-3C mutants as indicated. Results are expressed as the mean±s.d. of three independent experiments. **P-value <0.001. (E) MEX-3C associates with the DUB USP7. HEK293Ts were co-transfected with Strep-His-wt or RINGless MEX-3C. Cells were lysed in 1% NP-40 and immunoprecipitated on Streptactin beads (MEX-3C IPs) or for endogenous USP7, and immunoblotted for USP7 and MEX-3C. (F) Cytofluorometric analysis of cell-surface HLA-A2 in HEK293Ts expressing wtMEX-3C and/or USP7 cDNAs. GFP expression is a surrogate marker for MEX-3C and USP7 expression. TfR levels were measured as control. The HLA-A2 geometric fluorescent means in transfected (GFP+) cells are: Mock=177; MEX-3C only=48; USP7 only=164 and MEX-3C+USP7=45.7. (G) The Ubiquitin ligase activity of MEX-3C is required for HLA-A2 mRNA degradation. Endogenous HLA-A2 mRNA levels were analysed by qRT–PCR from unsorted cells expressing MEX-3C and/or exogenous USP7. A significant accumulation of HLA-A2 transcripts (3.1±0.2; compared with control) was detected when MEX-3C and USP7 were co-expressed. This effect was abrogated by inhibiting USP7 activity. Results are expressed as the mean±s.d. of three independent experiments. *P-value<0.05 versus control; #P-value<0.05 versus USP7+MEX-3C cells. (H) USP7 inhibition results in a rapid decrease in HLA-A2 mRNA levels (0.36±0.1; versus control=1). NKL cells (see later) were incubated with USP7 inhibitor P045204 (10 μM for 4 h). Endogenous HLA-A2 mRNA levels were analysed by qRT–PCR. Results are expressed as the mean±s.d. of three independent experiments. **P-value<0.001.
MEX-3C regulates HLA-A2 levels through its mRNA 3′UTR
Since MEX-3C binds poly-A-binding protein, potentially directing it to the 3′UTR of mRNAs (Buchet-Poyau et al, 2007), we wanted to determine whether MEX-3C could regulate firefly luciferase protein expression (measured as luciferase activity) through the HLA-A2 3′UTR. The HLA-A*0201 3′UTR was cloned downstream of the 3′region of the firefly (FF) luciferase gene in the pGL3 control vector, which utilizes FF luciferase as the primary reporter to monitor mRNA regulation. Renilla luciferase was used to normalize transfection levels. Both wt and RINGless MEX-3C markedly downregulate FF luciferase activity (Figure 4C), an effect almost completely abrogated by the mutKH MEX-3C (0.69±0.01), in accordance with its decreased mRNA binding. These results are consistent with our previous data (Figure 3B) showing that wt and RINGless MEX-3C reduce cell-surface HLA-A2 through the HLA-A2 3′UTR. While expression of both wt and RINGless MEX-3C had decreased surface HLA-A2 protein expression (Figure 3C), only wtMEX-3C had decreased HLA-A2 transcripts. This important finding was further examined by measuring luciferase mRNA levels in the FF-Luc-HLA-A2 3′UTR constructs, following expression of wt and RINGless MEX-3C, which both decrease FF-luciferase protein levels (Figure 4C). As seen with endogenous HLA-A2 transcripts (Figure 3C), wt MEX-3C expression markedly decreased FF-Luc-HLA-A2 3′UTR mRNA (Figure 4D). In contrast, following RINGless MEX-3C expression, there was no decrease in FF-Luc-HLA-A2 3′UTR mRNA, but a marked elevation in FF-Luc-HLA-A2 3′UTR mRNA levels is evident (Figure 4D), despite almost undetectable FF-luciferase protein levels (Figure 4C). Indeed, expression of RINGless MEX-3C caused an eight-fold increase in FF-Luc-HLA-A2 3′UTR mRNA compared with control (Figure 4D), confirming the requirement of MEX-3C’s RING, and by implication E3 ligase activity, in mRNA degradation. Taken together (Figures 3C and 4D), our results emphasize that ubiquitin ligase activity is required for mRNA degradation. In the absence of the RING, RNA binding by MEX-3C is sufficient to prevent translation or ‘sequester’ the bound mRNA. Under these circumstances, the mRNA is neither translated nor degraded, causing a decrease in HLA-A2 protein expression. This situation appears analogous to MEX-3’s activity in C. elegans, prior to MEX-3’s evolutionary acquisition of a RING, where translation of its substrate PAL-1 mRNA is repressed but PAL-1 mRNA is not degraded, as seen with RINGless MEX-3C on HLA-A2 mRNA. We infer that the MEX-3 gene products were initially RNA-binding and later acquired ubiquitin E3 ligase activity, which not only prevents mRNA translation, but now also promotes degradation.
USP7 is the DUB for MEX-3C, and confirms the requirement of E3 ubiquitin ligase activity in HLA-A2 mRNA decay
To identify additional MEX-3C-binding partners, we performed a MEX-3C (Streptactin) pull-down followed by mass spectrometry analysis (Supplementary Table S1). We found that both wt and RINGless MEX-3C bind the deubiquitinating (DUB) enzyme USP7, a finding readily confirmed by immunoprecipitation (Strep-MEX-3C or endogenous USP7) and immunoblotting in both directions (Figure 4E). The interaction was further confirmed in NK cells where endogenous MEX-3C associates with endogenous USP7 (see later and Supplementary Figure S7A). Neither overexpression nor depletion of USP7 affected MEX-3C protein levels (Supplementary Figure S7B and C). To determine how this DUB might regulate MEX-3C’s activity, we overexpressed USP7 together with wtMEX-3C. A phenotype similar to that observed with RINGless MEX-3C is seen, in that HLA-A2 cell-surface expression was decreased (Figure 4F), but no HLA-A2 mRNA degradation was seen (Figure 4G). Indeed, expression of USP7, when co-expressed with wtMEX-3C, caused a significant three-fold increase in endogenous HLA-A2 mRNA compared with the control in unsorted HEK293T cells (Figure 4G). Conversely, HLA-A2 mRNA levels decreased when USP7 activity was inhibited in HEK293T cells and NKL cells (see later and Figure 4G and H). In the presence of the USP7 DUB, more HLA-A2 is bound to wtMEX-3C, as overexpression of USP7 increases the amount of HLA-A2 mRNA co-immunopreciptated with MEX-3C (as measured by qRT–PCR) (Supplementary Figure S7D).
Ubiquitin ligase activity is therefore required for mRNA degradation and can be prevented either by removal of the RING (RINGless MEX-3C) or by overexpression of the USP7 DUB. These results also imply that USP7 has DUB activity against either MEX-3C itself, or other substrate(s) of MEX-3C.
MEX-3C is induced in activated human NK cells
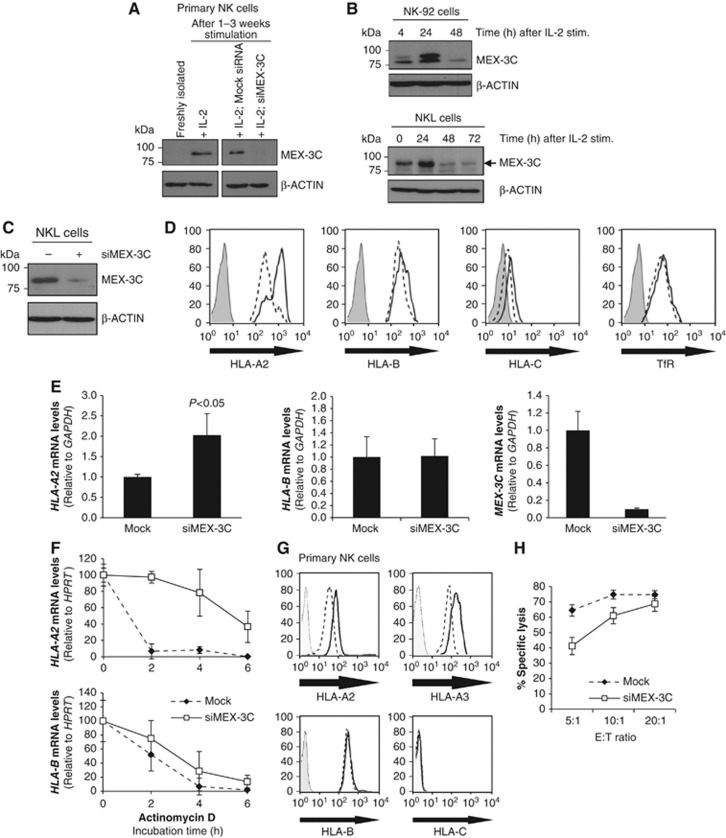
Mex-3C mRNA is reported to be preferentially expressed in cells of the immune system, particularly NK cells (http://biogps.gnf.org). Immunoblot analysis of MEX-3C in primary human NK cells and two NK cell lines (NK-92; NKL) was consistent with this transcriptional analysis (Figure 5). Despite not being detected in freshly isolated primary NK cells, MEX-3C was markedly upregulated following NK cell activation, with MEX-3C identity confirmed by siRNA-mediated depletion (Figure 5A). This IL-2 responsive expression of MEX-3C was also seen in NK-92 and NKL cells, peaking 24 h post-stimulation, and downregulated by 48 h (Figure 5B). These results are specific to the NK lineage, as MEX-3C expression was not detected in IL-2-activated primary CD4+ T-cells (Supplementary Figure S3).
Figure 5.
MEX-3C specifically regulates HLA-A but not HLA-B or HLA-C expression in primary human NK cells and modulates NK cell killing. (A) Immunoblot analysis of MEX-3C from freshly isolated and activated primary human NK cells. NK cells were purified from healthy blood donors and activated for 1–3 weeks. MEX-3C siRNA nucleofection results in efficient depletion in NK cells at 72 h post-nucleofection. (B) Immunoblot analysis of MEX-3C in the NK-92 (upper panel) and NKL cell lines (lower panel), following stimulation with IL-2 (350 U/ml) at the different time points shown. (C) Immunoblot analysis of MEX-3C downregulation in NKL cells after 72 h post-nucleofection. (D) Depletion of MEX-3C in NKL cells results in an allotype-specific increase of surface HLA-A2. Cytofluorometric analysis of HLA-A2, -B, -C, and TfR levels in mock (dashed line) and siMEX-3C cells (solid line). Isotype control (solid grey). (E) MEX-3C depletion increases HLA-A2 but not HLA-B mRNA levels in NKL cells. HLA-A2, pan-HLA-B, and MEX-3C mRNA levels were measured by qRT–PCR in mock and siMEX-3C nucleofected cells. Quantitative RT–PCR analysis of MEX-3C siRNA knockdown cells showed a significant 2.03±0.56-fold increase in HLA-A2 mRNA levels over mock cells (P-value=0.010, n=3). Transcript levels are relative to GAPDH and normalized to mock levels, set as 1 and expressed as the mean±s.d. of three independent experiments (P-value=0.01). (F) Depletion of MEX-3C increases HLA-A2 mRNA half-life. Mock (dashed line) or siMEX-3C (solid line) treated NKL cells were incubated with Actinomycin D (5 μg/ml). HLA-A2 and HLA-B mRNA levels were measured by qRT–PCR and normalized against HPRT. Results are the mean±s.d. of three independent experiments. (G) Depletion of MEX-3C in primary human NK cells results in an allotype-specific increase of surface HLA-A (HLA-A2 and -A3) but not HLA-B or -C. Cytofluorometric analysis of HLA-A2, -A3, -B, and -C levels in mock (dashed line) and siMEX-3C nucleofected cells (solid line). Isotype control (solid grey). (H) Reduced target cell killing in MEX-3C depleted primary NK cells. 51Cr release assay (percent specific lysis) using mock siRNA (dashed line) or siMEX-3C (solid line) nucleofected NK cells against K562 target cells. Error bars show the standard error of the six replicates at each point. The results are representative of three independent experiments using polyclonal NK cells from unrelated donors.
MEX-3C specifically regulates HLA-A but not HLA-B/C expression in NK cells and modulates NK cell killing
As initially observed in HEK293 cells, siRNA-mediated depletion of MEX-3C from the more physiologically relevant NKL cells also increased surface HLA-A2 expression (Figure 5C and D), and HLA-A2 mRNA levels (Figure 5E). Depletion of MEX-3C caused a four-fold increase in HLA-A2 mRNA half-life, from 1.48 h in mock cells to 5.68 h in siMEX-3C cells while having no significant effect on the corresponding HLA-B mRNA half-life (Figure 5F). The 3′UTR of the common HLA-A/B/C alleles have a high degree of identity (Supplementary Figure S5A), and a role for post-transcriptional regulation of MHC-I mRNA decay was described for HLA-C (McCutcheon et al, 1995), where HLA-Cw*0702 mRNA stability is post-transcriptionally regulated through binding miR-148 (Kulkarni et al, 2011). This prompted us to test whether MEX-3C also regulates HLA-C mRNA. MEX-3C depletion in human primary NK cells as well as NKL cells showed no effect on HLA-B/C expression (Figure 5D–G). Consistent with these results and in contrast to HLA-A2, a luciferase-3′UTR reporter assay for the HLA-Cw*0602 and Cw*0702 alleles showed no effect of MEX-3C on HLA-C mRNA stability (Supplementary Figure S5B). To see whether other HLA-A allotypes were also affected, MEX-3C was depleted from activated primary human NK cells and showed an increase in both HLA-A2 and HLA-A3 surface expression (Figure 5G). These two alleles (HLA-A2 and HLA-A3) along with HLA-A01, A11 and A24 are the most common HLA-A alleles and share identical 3′UTR sequences (Supplementary Figure S4), implying that all HLA-A alleles are regulated by MEX-3C.
To correlate a functional phenotype with the observed increase in HLA-A expression following MEX-3C depletion, we compared NK cell killing in the presence or absence of MEX-3C. siRNA-mediated depletion of MEX-3C from primary activated human NK cells increased HLA-A expression and was associated with reduced killing of target cells (Figure 5H), suggesting that altered HLA-A expression affects the threshold for killing by these cells.
Discussion
The identification of MEX-3C as an RNA-binding ubiquitin E3 ligase required for the post-transcriptional regulation of HLA-A allotypes suggests a novel role for ubiquitin in the control of RNA degradation. MEX-3C regulates expression of HLA-A2 by binding its 3′UTR and subsequently degrading HLA-A2 transcripts in an ubiquitin-dependent manner. Indeed, the HLA-A2 mRNA half-life is extended 3.8-fold following MEX-3C depletion. The link between ubiquitination and mRNA decay, although previously suggested for the turnover of some AU-rich mRNAs (Laroia et al, 2002), is not well defined. Here, we show that the RING domain of MEX-3C, and hence its ubiquitin ligase activity, is absolutely required for the degradation of HLA-A2 mRNA, providing a link between ubiquitination activity and mRNA decay.
MEX-3C originates from an evolutionarily conserved family of RNA-binding proteins. The founder member, MEX-3 from C. elegans contains two KH domains that bind and prevent translation of the PAL-1 mRNA (Draper et al, 1996; Hunter and Kenyon, 1996) with no effect in PAL-1 mRNA stability (Draper et al, 1996). The subsequent evolutionary diversification from C. elegans is associated with the acquisition of a C-terminal RING finger (e.g., as seen in Drosophila) with serial duplication events from a single dMex-3 (RNA-binding+RING finger domain) gene, to four genes in higher eukaryotes. Hence, MEX-3 gene products were initially RNA-binding translational repressors and later acquired ubiquitin E3 ligase activity which, as described above, is required for mRNA degradation.
The finding that the USP7 deubiquitinating enzyme antagonizes MEX-3C’s ability to degrade mRNA provides additional evidence for the requirement of ubiquitin in mRNA decay. USP7’s deubiquitinating activity could either be directed against MEX-3C itself, which readily auto-ubiquitinates (Supplementary Figure S2B), or against other undefined MEX-3C protein substrates. We therefore suggest a preliminary model for MEX-3C-mediated mRNA degradation (Supplementary Figure S6). MEX-3C recognizes and binds the 3′UTR of target mRNA (HLA-A2) through its KH domains, and shuttles its mRNA cargo from the nucleus to the cytosol via the CRM-1 pathway (Buchet-Poyau et al, 2007). In the cytosol, MEX-3C interacts with Argonaute proteins (Buchet-Poyau et al, 2007), linking HLA-A2 mRNA with the RISC-mediated mRNA decay pathway (Supplementary Figure S6). The simplest model would suggest that when MEX-3C is released from USP7 DUB activity it is itself autoubiquitinated and degraded, allowing the specific mRNA to be transferred to the RNA degradation machinery (Supplementary Figure S6). In the case of RINGless MEX-3C, the HLA-A2 mRNA is sequestered and not translated, causing equivalent HLA-A2 protein downregulation as wild-type MEX-3C, but in this situation the mRNA is not degraded. RINGless MEX-3C therefore represses HLA-A2 mRNA translation, and the presence of the RING confers mRNA degradation activity.
How and exactly where the mRNA is sequestered in the absence of ubiquitin ligase activity, and which proteins are ubiquitinated to promote mRNA degradation remains to be determined. Eukaryotic mRNAs are protected from decay by the 5′ N7-methylguanosine cap and the 3′ poly(A) tail. Initiation of mRNA decay requires an event that exposes the mRNA ends, by decapping or deadenylation, to 5′-to-3′ or 3′-to-5′ exonucleases (Mühlemann and Lykke-Andersen, 2010). The disassembly of mRNPs is essential to allow access of the degradation machinery to the targeted mRNA. Since the RING domain of MEX-3C is clearly required for mRNA degradation, MEX-3C may ubiquitinate components of the mRNP complex, promoting disassembly of the complex and therefore allowing access to the exonucleases.
MEX-3C is not the only RNA-binding protein with ubiquitin ligase activity. Within the >600 E3 ubiquitin ligases, we identified a group of 15 genes, whose products encode RNA-binding motifs as well as RING finger domains and are therefore predicted to bind RNA and have ubiquitin E3 ligase activity (Cano et al, 2010). The best characterized is Roquin which, like MEX-3C, contains an E3 ligase RING finger domain, but has a CCCH RBD, rather than the tandem repeat KH domains seen in MEX-3C. Roquin was first identified in a screen for autoimmune regulators in mice, in which a mutation in Roquin resulted in a lupus-like pathology. Roquin deficiency increased Icos mRNA and cell-surface Icos expression on T cells, causing the accumulation of lymphocytes associated with a lupus-like autoimmune syndrome (Vinuesa et al, 2005). Like MEX-3C, Roquin localizes to cytosolic RNA granules implicated in mRNA stability, though they differ in the mechanism for mRNA decay. While Roquin participates in the decapping pathway (Glasmacher et al, 2010), MEX-3C is implicated in RISC-mediated decay.
The relatively restricted expression of MEX-3C to NK cells, together with its increased expression upon cell activation, suggests a role for MEX-3C in fine-tuning HLA-A expression. We find that depletion of MEX-3C from activated primary NK cells leads to an increase in HLA-A allotype expression with a concomitant decrease in target cell killing. The HLA-A restricted specificity of MEX-3C was unanticipated, and the high 3′UTR sequence conservation (Supplementary Figure S4) of the common HLA-A alleles implies that they are all regulated by MEX-3C, suggesting an unexpected benefit of allotype-specific regulation.
Selective loss of expression of alleles of the HLA-A locus (Kageshita et al, 1993; Ferrone and Marincola, 1995), particularly HLA-A2, the commonest Caucasian HLA allele, is a common feature of tumours and may facilitate CTL escape. A plausible explanation as to why NK cells might specifically decrease cell-surface HLA-A expression upon activation is to reset the threshold of inhibitory receptors (e.g., KIRs, LILRs) for NK cell killing, as reported between murine Ly49 and MHC-I where cis H2Dd sequesters Ly49A, relieving NK cells from the suppressive effect of unengaged Ly49A (Held and Mariuzza, 2008; Chalifour et al, 2009). The increase in MEX-3C that occurs upon NK cell activation will decrease surface HLA-A, freeing the potential inhibitory receptor from cis binding, and therefore decreasing the threshold for NK-mediated killing. It is likely that MEX-3C binds and regulates other mRNA species. Future work will focus on identifying these additional mRNA targets and should give further insight into MEX-3C’s broader physiological relevance and role in NK cell function.
In conclusion, we have identified a post-transcriptional pathway for the specific regulation of HLA-A allotypes in activated NK cells that may be particularly important in setting the threshold of NK cell killing. MEX-3C, a novel RNA-binding ubiquitin E3 ligase regulates expression of HLA-A2 by binding its 3′UTR and subsequently degrading HLA-A2 transcripts. The RING domain of MEX-3C is absolutely required for HLA-A2 mRNA degradation, providing a direct link between ubiquitination and mRNA decay.
Materials and methods
Cells, plasmids, and transfections
HEK293 FlpIn cells were grown in RPMI 1640 medium supplemented with 10% FCS. Cells were transfected using 293-Transit Reagent (Mirus Bio) and analysed by flow cytometry or immunoblotting at 48 or 72 h following transfection.
Primary polyclonal NK cells were purified and cultured as previously described (Wills et al, 2005). NK killing assays (51Cr release assays) were performed with K562 target cells as described (Wills et al, 2005). NKL and NK-92 cells were grown in RPMI 1640 medium supplemented with 10% FCS, 10% Human AB serum (Sigma) and 350 U/ml of IL-2.
MEX-3C siRNA-mediated depletion in NK cells was performed using Amaxa Nucleofector kit V, program X-01 and 800 pmoles of siRNA oligonucleotides. Mock knockdowns were performed using RISC-free siGLO control (Thermo Fisher Scientific).
The pCMV-Tag3B-MEX-3C constructs are previously described (Buchet-Poyau et al, 2007). The RINGless and mutKH forms of MEX-3C were achieved by site-directed mutagenesis as described in Buchet-Poyau et al (2007), and using the following primers: For RINGless MEX-3C: M3C-FOR: 5′-CTAGGGATCCGATGCCCAGCGGCAGCTC-3′ and RingKO-M3C-REV: 5-CTAGGGCGCGCCATCACTAGTCGTGCTTTCGTCTTGATT-3′; and for the double KH mutant: KH1For: 5′-CGAGATCGTCGATCGCCAGGGTTG-3′ and KH1Rev: 5′-CAACCCTGGCGATCGACGATCTCG-3′ using the previously described single KH mutant as template (Buchet-Poyau et al, 2007).
The USP7 inhibitor P045204 (Weinstock et al, 2012) was a kind gift from Dr Benjamin Nicholson (Progenra, Inc.). Cells were incubated with inhibitor at 10 μM for 4 h.
Luciferase assays
The HLA-A*0201 promoter firefly luciferase reporter construct (pGL3-A230) was a gift from PJ van den Elsen (Kurnick et al, 2001). The firefly luciferase HLA-Cw*0602 and Cw*0702 3′UTR reporter constructs (pGL3 based) were kindly provided by M Carrington (Kulkarni et al, 2011). A TK Renilla luciferase reporter (pRL-TK) gene was co-transfected at a 1:10 ratio to provide an internal control. For the FF-Luc-HLA-A2 3′UTR reporter construct, the 3′UTR of HLA-A*0201 was PCR-amplified using HEK293T cDNA as template and the following primers: A2-3UTR-FOR 5′-ATCTGCTAGCCTATAGTGTGAGA and A2-3UTR-REV 5′-ATCTTCTAGATTTAATAGGGAAGGAAGAAGTTACAGC. The 3′UTR was subsequently cloned downstream of the firefly luciferase expression cassette of the pGL3 control vector (Promega). The correct sequence and orientation was verified by sequencing. Luciferase activity was measured at 48 h post-transfection using the Dual-Luciferase Reporter Assay System, as described by the manufacturer (Promega). All assays were performed in triplicate, with the Renilla-luciferase control used to standardize transfection efficiency. Results are relative to control levels (set as 1), and expressed as the mean±s.d. of at least three independent experiments.
siRNA screening
Cells were reverse transfected with a custom E3 ubiquitin ligase siRNA library (Thermo Fisher Scientific) (Stagg et al, 2009) using Oligofectamine (Invitrogen) at 80 nM final concentration. Each gene was targeted with an siRNA pool of four individual oligonucleotides.
The hMEX-3C siRNA pool was subsequently deconvoluted into four individual oligonucleotides (#1 [J-006989-05]: 5′-GGAGUGAUCCUUCUGGUAA-3′; #2 [J-006989-06]: 5′-GAAACUAUAUAGAGCUCAA-3′; #3 [J-006989-07]: 5′-CAAAUGGUACCAAUAGUUA-3′; and #4 [J-006989-08]: 5′-GCGCAAGAAUGAUAUCCAA-3′; Thermo Fisher Scientific). Cells were cultured for 60 h and then assayed by FACSCalibur (BD). Data were analysed with FlowJo. Z-scores were used to standardize our data, which had a normal distribution (99% of data had Z-scores between −2 and 2). Plates were screened n=2 or n=4 when it appeared a hit might be present. Data were normalized to mock-transfected controls (five on each plate), and the mean was taken of repeats where present. Z-scores were calculated from the mean and standard deviation of the entire data set. Using a Bonferroni-corrected P-value (0.05), a Z-score of >3.82 is statistically significant.
The MEX-3C stable knockdown cell line was generated by transducing HEK293 cells with pGIPZ lentiviral shRNAmir vector against MEX-3C (shMEX-3C)[172-0044-H8] (Open Biosystems) followed by puromycin (1 μg/ml) selection.
Flow cytometry
Cells were stained with primary antibodies in PBS+5% FCS and visualized with goat anti-mouse Cy5-conjugated secondary antibody (Jackson ImmunoResearch Laboratories); except when directly conjugated primary antibodies were used. Cells were fixed in PBS with 1% PFA, read on a FACSCalibur (BD), and analysed in FlowJo. For flow cytometry analysis, the following Abs were used: AF488-conjugated mouse anti-HLA-A2 (Serotec), mAb BB7.2 (anti-HLA-A2), mAb 4E (anti-HLA-B), mAb DT9 (anti-HLA-C) (a kind gift from Ashley Moffett (University of Cambridge)), anti-IgG2b isotype-control (eBioscience), and mAb anti-human CD71(TfR (transferrin receptor)) (Santa Cruz Biotechnology). Fluorescent secondary Abs were from Molecular Probes (Eugene, OR) and HRP-conjugated secondary Abs from Jackson ImmunoResearch Laboratories (West Grove, PA).
Immunoprecipitations and immunoblotting
Cells were lysed at 48 or 72 h post-transfection for 30 min in 2 × Laemli buffer (+ DTT+Protease inhibitors (Roche)) plus benzonase nuclease (Sigma) and heated at 55°C for 10 min prior to 8% SDS–PAGE. Samples were transferred to PVDF (Millipore), the membranes were blocked for 1 h at room temperature, and incubated with primary antibody overnight at 4°C. Primary antibodies used were rabbit polyclonal anti-RKHD2 (MEX-3C) antibody (Pro-Sci Incorporated) used at 1:1500 dilution and incubated overnight in PBST containing 5% milk. Mouse monoclonal anti-ACTIN (Sigma) or anti-myc (9E10) (Roche) were used at 1:10 000 and 1:5000 dilutions, respectively. Membranes were developed in West Pico Extended Chemiluminescent substrate (Thermo Fisher Scientific, Waltham, MA).
For immunoprecipitations, cells were lysed 48 h post-transfection lysed in 1% NP-40 in TBS with 1 μM ZnCl2, 0.5 mM PMSF, 10 mM iodoacetamide, 10 U/ml RNAsin (Promega), Roche complete protease inhibitor for 20 min on ice. Strep-His-tagged proteins were immunoprecipitated with Streptactin sepharose beads (IBA GmbH). After three washes in lysis buffer containing RNAsin, samples were eluted in SDS sample buffer (10 min at 98°C). Immunoprecipitated proteins were then separated by SDS–PAGE, and immunoblotted as described above. For ubiquitinated species, PVDF membranes were denatured in 6 M Guanidinium Chloride before probing with P4D1 antibody.
USP7 immunoprecipitations were performed as described (Nathan et al, 2008), using USP7-specific antisera (Bethyl Laboratories) and Protein A Sepharose beads. The USP7 vector, pCI-USP7, was donated by Roger Everett.
For mass spectrometry analysis of Strep-wtMEX-3C pull-down, HEK293T cells were transfected with pQE empty vector (Mock) or wt Strep-His–MEX-3C, lysed in 1% NP-40 and immunoprecipitated (IP) on Streptactin beads as described above. Co-immunoprecipitated proteins were digested with trypsin using the filter-aided sample preparation (FASP) protocol (Wisniewski et al, 2009; Weekes et al, 2010) and analysed by LC–MSMS. Raw spectra were processed using Proteome Discoverer 1.2 and searched against a Uniprot Human database (downloaded 190811, 54523 sequences) using Mascot Daemon 2.3.2. A false-discovery rate (FDR) for peptides of 0.05 was applied and reported proteins required a minimum of two peptides and a score higher than 35.
RNA extraction and qRT–PCR analysis
Total RNA was extracted using the RNeasy Plus kit (Qiagen). Total RNA (2 μg) was reverse transcribed into cDNA using a poly(d)T primer and Super RT reverse transcriptase (HT Biotechnology Ltd) following the manufacturer’s instructions.
Real-time qRT–PCR was performed using the ABI Prism 7700HT Sequence Detector Systems (Applied Biosystems) and SYBR Green Master mix kit (Applied Biosystems). Briefly, all reactions were performed with 120 ng of cDNA, 12.5 μl of SYBR GREEN PCR master mix and 0.2 μM forward and reverse primers in a final reaction volume of 25 μl. Cycling parameters were 95°C for 10 min, followed by 40 cycles of 94°C for 30 s, 58°C for 1 min. Primer sequences for HLA-A2-specific RT–PCR were described in Wang et al (1999) and are as follows: A2For(5pE1A2): 5′-TCCTGCTACTCTCGGGGGCT, A2REV(AP2): 5′-TCACTTTCCGTGCTCCCC. MEX-3C RT–PCR primers were: hM3CF:5′-TGAACGGGGAGCAGGCG-3′, hM3CR:5′-TGACTTGGACGGTGGTTTGA-3′ as previously described (Buchet-Poyau et al, 2007). GAPDH was used as an internal control to normalize the difference in the amount of input cDNA. GAPDH primers used: GAPDH-FOR: 5′-ATGGGGAAGGTGAAGGTCG and GAPDH-REV: 5′-CTCCACGACGTACTCAGCG. MEX-3A, MEX-3B and MEX-3C qRT–PCR primers are described in Buchet-Poyau et al (2007). Firefly and Renilla luciferase primers and PCR conditions were as described in Zamorano et al (2006).
Supplementary Material
Acknowledgments
We thank all the members of the Lehner laboratory, as well as Dr Jernej Ule for helpful discussions. This work was supported by the Medical Research Council and the Wellcome Trust.
Author contributions: FC, HB, LMD, MRW, and PJL performed the experiments and wrote the manuscript. KB-P and MB provided the reagents.
Footnotes
The authors declare that they have no conflict of interest.
References
- Buchet-Poyau K, Courchet J, Le Hir H, Séraphin B, Scoazec JY, Duret L, Domon-Dell C, Freund JN, Billaud M (2007) Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res 35: 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano F, Miranda-Saavedra D, Lehner PJ (2010) RNA-binding E3 ubiquitin ligases: novel players in nucleic acid regulation. Biochem Soc Trans 38: 1621–1626 [DOI] [PubMed] [Google Scholar]
- Chalifour A, Scarpellino L, Back J, Brodin P, Devèvre E, Gros F, Lévy F, Leclercq G, Höglund P, Beermann F, Held W (2009) A Role for cis interaction between the inhibitory Ly49A receptor and MHC class I for natural killer cell education. Immunity 30: 337–347 [DOI] [PubMed] [Google Scholar]
- Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR (1996) MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell 87: 205–216 [DOI] [PubMed] [Google Scholar]
- Duncan LM, Nathan JA, Lehner PJ (2010) Stabilization of an E3 ligase-E2-ubiquitin complex increases cell surface MHC class I expression. J Immunol 184: 6978–6985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrone S, Marincola FM (1995) Loss of HLA class I antigens by melanoma cells: molecular mechanisms, functional significance and clinical relevance. Immunol Today 16: 487–494 [DOI] [PubMed] [Google Scholar]
- Glasmacher E, Hoefig KP, Vogel KU, Rath N, Du L, Wolf C, Kremmer E, Wang X, Heissmeyer V (2010) Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat Immunol 11: 725–733 [DOI] [PubMed] [Google Scholar]
- Gobin SJ, van den Elsen PJ (2001) Locus-specific regulation of HLA-A and HLA-B expression is not determined by nucleotide variation in the X2 box promoter element. Blood 97: 1518–1521 [DOI] [PubMed] [Google Scholar]
- Held W, Mariuzza RA (2008) Cis interactions of immunoreceptors with MHC and non-MHC ligands. Nat Rev Immunol 8: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes TD, El-Sherbiny YM, Davison A, Clough SL, Blair GE, Cook GP (2011) A human NK cell activation/inhibition threshold allows small changes in the target cell surface phenotype to dramatically alter susceptibility to NK cells. J Immunol 186: 1538–1545 [DOI] [PubMed] [Google Scholar]
- Hunter CP, Kenyon C (1996) Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell 87: 217–226 [DOI] [PubMed] [Google Scholar]
- Kageshita T, Wang Z, Calorini L, Yoshii A, Kimura T, Ono T, Gattoni-Celli S, Ferrone S (1993) Selective loss of human leukocyte class I allospecificities and staining of melanoma cells by monoclonal antibodies recognizing monomorphic determinants of class I human leukocyte antigens. Cancer Res 53: 3349–3354 [PubMed] [Google Scholar]
- Kulkarni S, Savan R, Qi Y, Gao X, Yuki Y, Bass SE, Martin MP, Hunt P, Deeks SG, Telenti A, Pereyra F, Goldstein D, Wolinsky S, Walker B, Young HA, Carrington M (2011) Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature 472: 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurnick JT, Ramirez-Montagut T, Boyle LA, Andrews DM, Pandolfi F, Durda PJ, Butera D, Dunn IS, Benson EM, Gobin SJ, van den Elsen PJ (2001) A novel autocrine pathway of tumor escape from immune recognition: melanoma cell lines produce a soluble protein that diminishes expression of the gene encoding the melanocyte lineage melan-A/MART-1 antigen through down-modulation of its promoter. J Immunol 167: 1204–1211 [DOI] [PubMed] [Google Scholar]
- Laroia G, Sarkar B, Schneider RJ (2002) Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc Nat Acad Sci USA 99: 1842–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JA, Gumperz J, Smith KD, Lutz CT, Parham P (1995) Low HLA-C expression at cell surfaces correlates with increased turnover of heavy chain mRNA. J Exp Med 181: 2085–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlemann O, Lykke-Andersen J (2010) How and where are nonsense mRNAs degraded in mammalian cells? RNA Biol 7: 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan JA, Sengupta S, Wood SA, Admon A, Markson G, Sanderson C, Lehner PJ (2008) The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic 9: 1130–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P (2005) MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol 5: 201–214 [DOI] [PubMed] [Google Scholar]
- Randow F, Lehner PJ (2009) Viral avoidance and exploitation of the ubiquitin system. Nat Cell Biol 11: 527–534 [DOI] [PubMed] [Google Scholar]
- Stagg HR, Thomas M, van den Boomen D, Wiertz EJ, Drabkin HA, Gemmill RM, Lehner PJ (2009) The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J Cell Biol 186: 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, Yu D, Domaschenz H, Whittle B, Lambe T, Roberts IS, Copley RR, Bell JI, Cornall RJ, Goodnow CC (2005) A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature 435: 452–458 [DOI] [PubMed] [Google Scholar]
- Wang Z, Marincola FM, Rivoltini L, Parmiani G, Ferrone S (1999) Selective histocompatibility leukocyte antigen (HLA)-A2 loss caused by aberrant pre-mRNA splicing in 624MEL28 melanoma cells. J Exp Med 190: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearsch PA, Cresswell P (2008) The quality control of MHC class I peptide loading. Curr Opin Cell Biol 20: 624–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekes MP, Antrobus R, Lill JR, Duncan LM, Hör S, Lehner PJ (2010) Comparative analysis of techniques to purify plasma membrane proteins. J Biomol Tech 21: 108–115 [PMC free article] [PubMed] [Google Scholar]
- Weinstock J, Wu J, Cao P, Kingsbury WD, McDermott JL, Kodrasov MP, McKelvey DM, Kumar KGS, Goldenberg SJ, Mattern MR, Nicholson B (2012) Selective dual inhibitors of the cancer-related deubiquitylating proteases USP7 and USP47. Med Chem Lett (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills MR, Ashiru O, Reeves MB, Okecha G, Trowsdale J, Tomasec P, Wilkinson GW, Sinclair J, Sissons JG (2005) Human cytomegalovirus encodes an MHC class I-like molecule (UL142) that functions to inhibit NK cell lysis. J Immunol 175: 7457–7465 [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
- Yu D, Tan AH, Hu X, Athanasopoulos V, Simpson N, Silva DG, Hutloff A, Giles KM, Leedman PJ, Lam KP, Goodnow CC, Vinuesa CG (2007) Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature 450: 299–303 [DOI] [PubMed] [Google Scholar]
- Zamorano R, Suchindran S, Gainer JV (2006) 3′-Untranslated region of the type 2 bradykinin receptor is a potent regulator of gene expression. Am J Physiol Renal Physiol 290: F456–F464 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.