Abstract
Stomatin proteins oligomerize at membranes and have been implicated in ion channel regulation and membrane trafficking. To obtain mechanistic insights into their function, we determined three crystal structures of the conserved stomatin domain of mouse stomatin that assembles into a banana-shaped dimer. We show that dimerization is crucial for the repression of acid-sensing ion channel 3 (ASIC3) activity. A hydrophobic pocket at the inside of the concave surface is open in the presence of an internal peptide ligand and closes in the absence of this ligand, and we demonstrate a function of this pocket in the inhibition of ASIC3 activity. In one crystal form, stomatin assembles via two conserved surfaces into a cylindrical oligomer, and these oligomerization surfaces are also essential for the inhibition of ASIC3-mediated currents. The assembly mode of stomatin uncovered in this study might serve as a model to understand oligomerization processes of related membrane-remodelling proteins, such as flotillin and prohibitin.
Keywords: ASIC, ion channel regulation, membrane trafficking, protein structure, stomatin family
Introduction
The 32-kDa peripheral membrane protein stomatin is the founding member of the stomatin family, which in mammals includes stomatin-like proteins (STOMLs) 1–3 and the kidney-specific podocin (reviewed in Lapatsina et al, 2012a). These proteins are characterized by a conserved domain of ∼120 residues, the stomatin domain, which is a member of the SPFH (Stomatin, Prohibitin, Flotillin, HflK/C) domain family found in many proteins throughout all kingdoms of life. Membrane targeting of stomatins is mediated by a 20 amino-acid amino (N)-terminal membrane insertion domain featuring a conserved proline (Kadurin et al, 2009). An exception is STOML-2, which lacks this hairpin anchor. The N- and carboxy (C)-terminal regions of stomatins are cytoplasmic and unique for each family member (Salzer qet al, 1993; Seidel and Prohaska, 1998; Boute et al, 2000; Owczarek et al, 2001). Stomatin has been found to be phosphorylated (Salzer et al, 1993), palmitoylated (Snyers et al, 1999b) and to associate with lipid rafts (Snyers et al, 1999a) where it forms higher-order oligomers (Snyers et al, 1998). Furthermore, STOML-1 was shown to form hetero-oligomers with stomatin at endosomal membranes (Mairhofer et al, 2009). Stomatin can also form hetero-oligomers with STOML-3 in sensory neurons and this interaction requires the presence of the N-terminal membrane anchoring domain (Lapatsina et al, 2012b). Stomatin is absent in the erythrocyte membrane of patients suffering from a specific form of haemolytic anaemia (Stewart et al, 1992). Since the erythrocyte membrane of affected individuals is leaky to monovalent cations, it was proposed that stomatin regulates the activity of ion channels (Gallagher and Forget, 1995; Unfried et al, 1995). Subsequent studies, however, showed that loss of stomatin is not directly responsible for the observed membrane leakiness (Zhu et al, 1999; Fricke et al, 2003). A central function for the Caenorhabditis elegans stomatin homologue MEC-2 in touch sensation has been convincingly demonstrated (Huang et al, 1995). MEC-2 is present in a complex with the acid-sensing ion channel (ASIC)-related, Na+ selective, MEC-4/MEC-10 ion channel complex (Goodman et al, 2002; Cueva et al, 2007), and all three subunits of this complex are required to transduce mechanical signals in C. elegans (O’Hagan et al, 2005). When co-expressed with MEC-4/MEC-10, MEC-2 increases the amplitude of amiloride-sensitive currents 40-fold, suggesting a positive regulation (Goodman et al, 2002). Strikingly, mouse models lacking STOML-3 also show complete loss of mechanosensitivity in 40% of cutaneous mechanoreceptors (Wetzel et al, 2007), indicating a conserved function of stomatin family members in mammalian mechanosensation. In contrast, over-expression of mouse stomatin inhibits the amplitude of proton-gated currents in cells overexpressing mouse ASIC3 and increases the speed at which ASIC2a channels inactivate (Price et al, 2004).
No high-resolution structural data are available for the mammalian stomatin family, but the crystal structure of a homologue from Pyrococcus horikoshii (ph) has recently been determined (Yokoyama et al, 2008). The analysis of ph stomatin has revealed a crucial role for the stomatin domain in the formation of trimers and possibly higher-order oligomers. Interestingly, ASICs were also shown to have a trimeric channel architecture (Jasti et al, 2007; Gonzales et al, 2009), suggesting a symmetric interaction with a putative stomatin trimer. To understand the role of oligomerization in ion channel regulation, we have used X-ray crystallography to characterize mammalian stomatin and performed a structure-based analysis of its modulation of ASIC family members. Here, we demonstrate that, unlike in ph stomatin, mammalian stomatin forms a banana-shaped dimer and higher-order oligomers that are crucial for its regulation of ion channel function.
Results
Structure of the stomatin dimer
A construct comprising the mouse stomatin domain (amino acids 86–213, stomatin86–213; Figure 1A) was expressed in bacteria and purified to homogeneity. Crystals of this construct diffracted up to 2.4 Å. The phase problem was solved by molecular replacement using the ph stomatin domain as a search model, and the model was refined to an Rwork/Rfree of 21.7/26.8% (Table I).
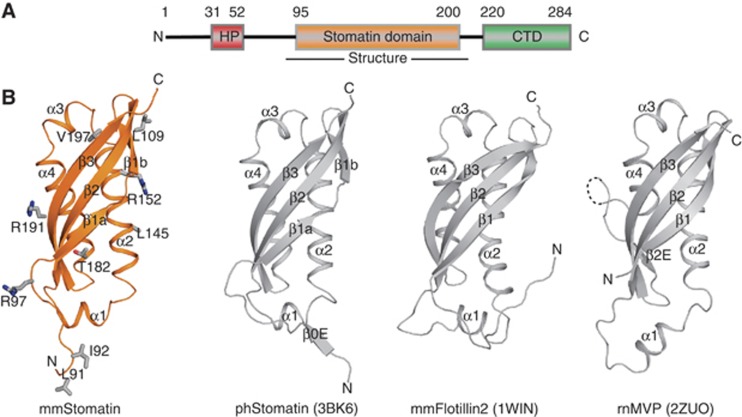
Figure 1.
Structure of the mouse stomatin domain. (A) Structure-based domain architecture of mammalian stomatin. HP, hydrophobic hairpin; CTD, C-terminal domain. (B) Structure of the mouse stomatin domain (left) with residues mutated in this study shown in stick representation. The crystal structure of the ph stomatin domain (pdb 3BK6), the NMR structure of mouse flotillin-2 (pdb 1WIN) and the crystal structure of the SPFH domain (amino acids 519–646) of the rat major vault protein (pdb 2ZUO) are shown for comparison. All structures contain a central antiparallel β-sheet composed of three β-strands and a similar arrangement of their α-helices. ph stomatin features an additional β-strand, while flotillin has an elongated loop connecting β2 and α1. The SPFH domain of the major vault protein has an extra β-sheet β2E interspersed shortly before β3 and an unresolved loop of unknown function.
Table 1. Data collection and refinement statistics.
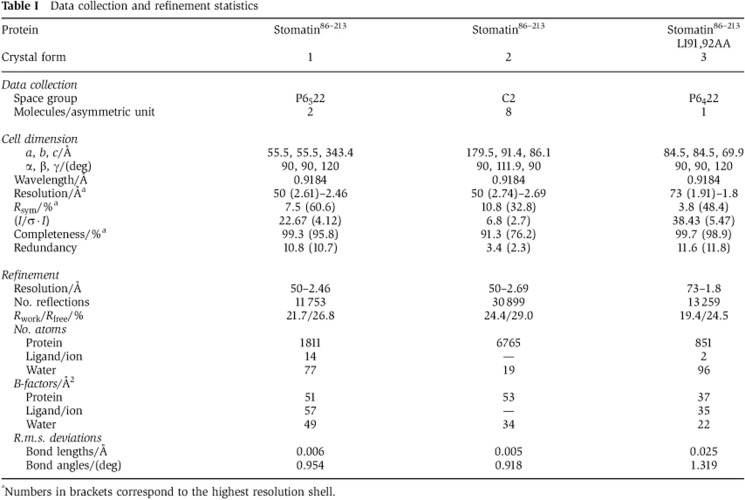
aNumbers in brackets correspond to the highest resolution shell.
Mouse stomatin86–213 has a mixed α/β-fold (Figure 1B; Supplementary Figure 1A). The N-terminal β-strand 1 is subdivided into β1a and β1b by a short loop and together with β2 and β3 forms an anti-parallel, curved β-sheet. Helices α2 and α4 extend in parallel and occupy the groove of the sheet, whereas the short α1 and α3 helices are oriented perpendicularly at both ends of the sheet. N- and C-termini are located at opposing sides of the molecule. The core of the mouse stomatin domain is very similar to that of ph stomatin (root mean square deviation (r.m.s.d.) of 1.5 Å along 107 aligned residues), with only minor deviations in α1, α2 and β1a (Figure 1B). Ph stomatin harbours an additional unique β-strand (β0) at the N-terminus (Figure 1B). The corresponding region in stomatin86–213 forms a loop. β0 mediates trimerization of ph stomatin by forming an interstrand contact to the neighbouring molecule (Supplementary Figure 1B) (Yokoyama et al, 2008). Also, flotillin2 (pdb 1WIN) and the SPFH domain of the major vault protein (Tanaka et al, 2009) share a similar fold to the mouse stomatin domain (r.m.s.d. of 2 Å along 106 amino acids and of 2.6 Å along 109 aligned residues, respectively, Figure 1B, Supplementary Figure 1C).
The mouse stomatin domain dimerized in the crystal via the C-terminal four residues of β3 (amino acids 196–199), which form a symmetric intermolecular β-sheet with the opposing molecule burying 600 Å2 surface area per molecule (Figure 2A and B). The interaction between the monomers features only main chain, no side chain interactions. The resulting stomatin dimer has a banana-shape, where α2–α4 form the outer and the β-sheet the inner surface of the banana. The N-termini are located at opposite ends of the dimerization interface.
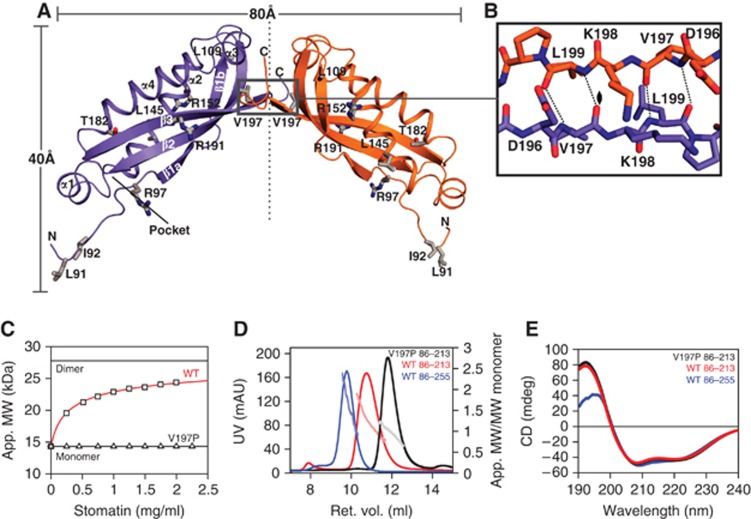
Figure 2.
The stomatin dimer. (A) Cartoon representation of the stomatin dimer, with the N- and C-termini indicated. Residues mutated in this study are shown in stick representation. (B) Details of the dimerization site at the C-terminus of the stomatin domain featuring an intermolecular β-sheet. (C) Sedimentation equilibrium experiments to determine apparent molecular masses at different protein concentrations. Data for stomatin86–213 were fitted to a monomer–dimer equilibrium resulting in a KD of 37±1 μM, where the error refers to the standard error of the fit. The V197P mutation prevented dimerization. (D) Analytical gel filtration of stomatin86–213, stomatin86–255 and stomatin86–213 V197P. The absolute molecular masses as determined by right-angle laser scattering analysis are indicated in light colours and refer to the right y axis. (E) CD spectra of stomatin86–213 (red), stomatin86–255 (blue) and stomatin86–213 V197P (black).
To analyse the relevance of the stomatin dimer for assembly in solution, analytical ultracentrifugation (AUC) experiments were carried out (Figure 2C). In these experiments, stomatin86–213 was observed in a monomer–dimer equilibrium with a dissociation constant (KD) of 37 μM. Also in analytical gel filtration experiments, coupled to a right-angle light scattering device, stomatin86–213 showed a monomer–dimer equilibrium (Figure 2D). To exclude the possibility that the C-terminal truncation of the crystallized construct caused artificial dimer formation, we analysed a C-terminal extended construct, stomatin86–255, which corresponds in length to the crystallized trimeric ph stomatin construct. Stomatin86–255 also eluted in a monomer–dimer equilibrium in analytical gel filtration (Figure 2D), indicating that residues 214–255 of mouse stomatin do not influence oligomerization.
To test whether the dimer interface in the crystal corresponds to the observed dimer in solution, a mutagenesis approach was applied. Since the intermolecular β-sheet interface employed only main chain contacts, we initially sought to disrupt the interface by electrostatic repulsion, using the V197D mutation, where the two negatively charged aspartate side chains from different monomers would directly oppose each other. However, this variant was insoluble. To disrupt β3 locally, we then introduced the V197P mutation into the stomatin86–213 construct. In circular dichroism (CD) measurements, this mutant showed a very similar spectrum as stomatin86–213, indicating that the mutation did not grossly disturb the fold of the stomatin domain (Figure 2E). In analytical ultracentrifugation, however, the V197P mutation completely prevented dimerization (Figure 2C). Also in analytical gel filtration experiments, stomatin86–213 V197P was mostly monomeric (Figure 2D).
To analyse whether the V197P mutation can disrupt self-association of full-length stomatin (from here on described as stomatin) in living cells, we used Bimolecular Fluorescence Complementation (BiFC) (Hu et al, 2002). In this assay, protein–protein interactions are detected by monitoring the development of a fluorescent complex comprising the N- and C-terminal fragments of YFP (each fused to the C-terminal end of stomatin or a stomatin variant; Supplementary Figure 2). For stomatin, we previously showed a robust fluorescence signal development at the plasma membrane and in intracellular structures (Lapatsina et al, 2012b). BiFC stomatin constructs carrying the V197P mutation showed a substantially reduced fluorescence signal consistent with the idea that the protein does not efficiently oligomerize in cells (Supplementary Figure 2).
To determine the function of the stomatin dimer in the modulation of ASICs, electrophysiological recordings were performed. For this purpose, Chinese hamster ovary (CHO) cells were used because they are devoid of endogenous proton-gated ASIC-like currents (Garcia-Anoveros et al, 2001; Cadiou et al, 2007; Smith et al, 2007) and are easier to handle than other cells such as primary mouse fibroblasts (Supplementary Figure 3). In CHO cells overexpressing ASIC3, application of a low pH solution immediately evokes an inward current, which rapidly inactivates and is followed by a smaller, sustained current throughout the period of low pH stimulation (Figure 3A). In agreement with previous data (Price et al, 2004), co-expression of stomatin with ASIC3 was associated with a large inhibition of ASIC3 peak current amplitude. Accordingly, we observed that at both pH 6 and pH 4, CHO cells co-expressing ASIC3 and stomatin had significantly smaller currents compared with ASIC3 alone (Figure 3A). In contrast, stomatin V197P showed no inhibitory action on ASIC3 proton-gated currents. This effect was not caused by different expression levels, since stomatin and stomatin V197P were expressed at similar levels (Supplementary Figure 4A).
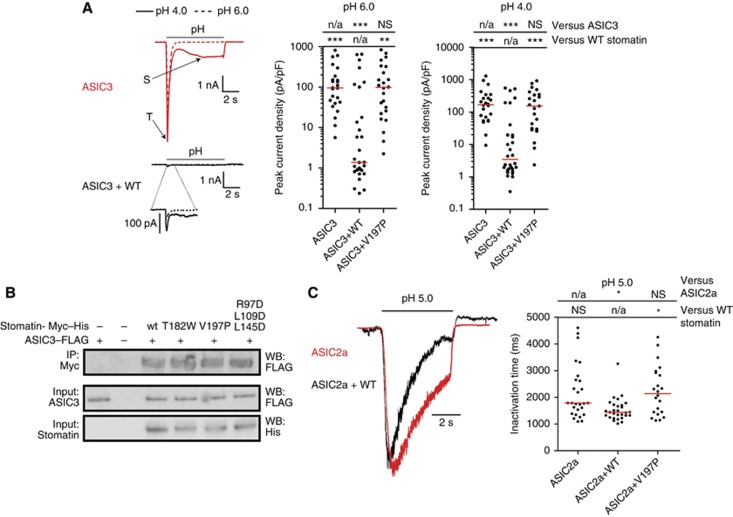
Figure 3.
Stomatin dimerization is crucial for the inhibition of ASIC3 current amplitudes. (A) ASIC3-mediated currents in CHO cells (upper, red traces) have both transient (T) and sustained (S) phases; the S-phase is larger at pH 4.0 (solid line) than at pH 6.0 (dotted line). Co-transfection with stomatin results in inhibition of proton-gated currents (lower, black traces). In dot plots of peak current density, each dot represents one cell and the red lines show the median for each data set. The Kruskal–Wallis test followed by Dunn’s post test and statistical comparisons of peak current density compared with ASIC3 expressed alone and ASIC3 co-expressed with stomatin are shown. (B) Co-immunoprecipitations of FLAG-tagged ASIC3 from CHO cell lysates using Myc-tagged stomatin or the indicated mutants. (C) ASIC2a-mediated currents in CHO cells, normalized for current amplitude, showing that co-transfection of stomatin (black) decreases the inactivation time of pH 5.0 activated ASIC2a transient currents (red). In dot plots of inactivation time, each dot represents one cell and the red lines show the median for each data set. The Kruskal–Wallis test followed by Dunn’s post test and statistical comparisons of inactivation time compared with ASIC2a expressed alone and ASIC2a co-expressed with stomatin are shown. n/a, not applicable, NS, not significant, *P<0.05; **P<0.01 and ***P<0.001. Figure source data can be found with the Supplementary data.
Using mCherry-tagged stomatin and eGFP-tagged ASIC3 constructs, stomatin appeared to co-localize with ASIC3 in intracellular structures and, to a lesser extent, at the plasma membrane (Supplementary Figure 4B) (Price et al, 2004). The V197P mutant showed a similar distribution (Supplementary Figure 4B). Also in BiFC assays, stomatin and the V197P mutant co-localized with ASIC3 at intracellular structures and at the plasma membrane (Supplementary Figure 5).
To unequivocally show a physical interaction between ASIC3 and stomatin, pull-down studies were performed (Figure 3B). In agreement with previous data (Price et al, 2004; Lapatsina et al, 2012b), ASIC3 was co-immunoprecipitated with Myc-tagged stomatin and, to a similar extent, with the V197P mutant. These results indicate that dimerization of stomatin is not a prerequisite for a physical interaction with ASICs.
Stomatin is also known to accelerate the inactivation of ASIC2a proton-gated currents (Price et al, 2004), a phenomenon that we also observed using a shift to pH 5 (Figure 3C), but not to pH 4 (Supplementary Figure 6). Unlike current amplitude, which is dependent on the amount of functional ion channels present in the plasma membrane, the inactivation time of ASIC2a-mediated currents is a mechanistic feature of ion channel gating. Therefore, faster ASIC2a inactivation times in the presence of stomatin are likely representative of a direct interaction between ASIC2a and stomatin or a modulation of the local membrane environment.
Similarly to the loss of inhibitory action upon ASIC3, stomatin V197P had no significant effect on the inactivation time of ASIC2a proton-gated currents (Figure 3C; Supplementary Figure 6). Taken together, these experiments indicate that the functional minimal building block of eukaryotic stomatin involved in the modulation of ASIC2a and ASIC3 currents is a banana-shaped dimer.
A hydrophobic pocket is important for stomatin function
Analysis of the crystal packing revealed that Leu91 and Ile92 in the N-terminal loop preceding the stomatin domain protrude into a hydrophobic pocket at the concave face of an opposing stomatin86–213 dimer (Figure 4A). The hydrophobic pocket had a volume of 350 Å3 and its inner surface was formed by the partially conserved residues Phe101, Tyr124, Val126, Ile136, Leu178 and Thr182 (Figure 4B). This assembly resulted in a linear oligomeric structure, which was further intertwined with an opposing oligomer resulting in a double helical architecture (Supplementary Figure 7A). In a second crystal form, a similar double helical structure containing a stomatin86–213 dimer was observed (Table I; Supplementary Figure 7B), and the hydrophobic pocket was also occupied by Leu91 and Ile92 of a neighbouring molecule.
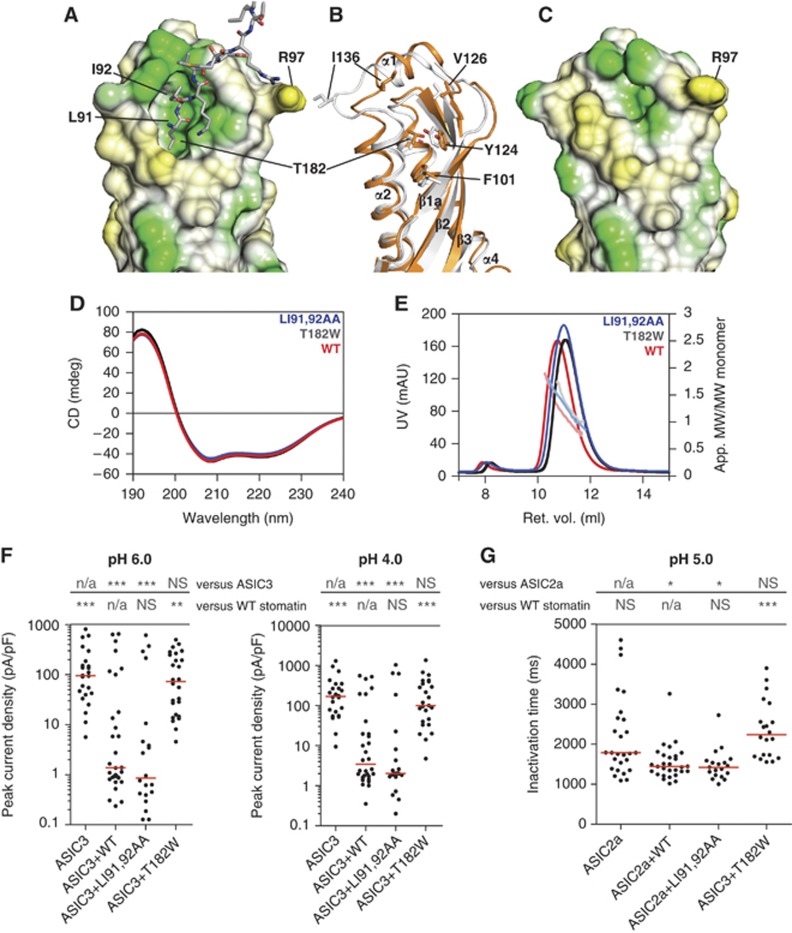
Figure 4.
A dynamic hydrophobic pocket is essential for stomatin function. (A) Lipophilic surface potential of stomatin86–213 where hydrophobic residues are indicated in green and polar residues in yellow. A hydrophobic pocket is filled by residues Leu91 and Ile92 of an opposing monomer. (B) Structural rearrangements in α1, α2 and β2 induce pocket closure, as seen by a superposition of the open (orange) and closed (white) pocket structures. (C) In stomatin86–213 LI91,92AA, the pocket is partially closed. (D) CD measurements for stomatin86–213 (red) and the LI91,92AA (blue) and T182W mutants (black). (E) Analytical gel filtration experiments for the indicated stomatin86–213 mutants. (F) Electrophysiological recordings of CHO cells co-transfected with ASIC3 and the indicated stomatin constructs at pH 4.0 and pH 6.0, as described in Figure 3A. (G) Inactivation times for CHO cells transfected with ASIC2a and the indicated stomatin constructs as described in Figure 3C.
To analyse the architecture of the pocket in the absence of Leu91 and Ile92, we mutated the two residues to alanine. According to the structure, this mutation completely removes the interaction site between the two dimers. Stomatin86–213 LI91,92AA was crystallized and the structure solved by molecular replacement (Table I). Also in this crystal form 3, a banana-shaped dimer built via β3 was found, but the hydrophobic pocket was empty and greatly reduced in size (volume of 150 Å3; Figure 4C). In particular, the binding site for Leu91 was completely occupied by Tyr124 and Asp140, whereas the cleft occupied by Ile92 in crystal form 1 and 2 was still present and even widened by a movement of Ile136 (Figure 4B and C). Pocket closure of the Leu91-binding site was caused by rearrangements in α1, α2 and β2, which moved towards the pocket (Figure 4B).
To test the importance of this pocket for the inhibition of ASICs, we introduced a mutation expected to close the pocket (T182W). In density gradient centrifugation experiments, a similar mutation, T182A, was shown not to alter oligomerization of human stomatin (Umlauf et al, 2006). Stomatin86–213 T182W showed no apparent folding defect in CD measurements (Figure 4D) and eluted as monomer/dimer in analytical gel filtration experiments (Figure 4E), indicating that dimer formation is not affected by this mutation. When overexpressed in CHO cells, stomatin T182W showed similar expression levels as stomatin and displayed a similar cellular distribution when it was fluorescently tagged (Supplementary Figures 4 and 5). Furthermore, stomatin T182W still interacted with ASIC3 in pull-down experiments (Figure 3B). However, in contrast to stomatin, stomatin T182W lost the ability to modulate proton-gated ASIC3 as shown in whole-cell recordings (Figure 4F). Furthermore, this T182W mutant no longer caused a significant decrease in the inactivation time of ASIC2a proton-gated currents at pH 5.0 (Figure 4G). At pH 4.0, it even prolonged inactivation times (Supplementary Figure 6).
As expected from the crystal structure, the fold and dimer formation of stomatin86–213 LI91,92AA was not affected (Figure 4D and E). Also the cellular localization of fluorescently tagged stomatin LI91,92AA was comparable to stomatin (Supplementary Figures 4 and 5). Furthermore, we still observed the typical modulation of ASIC3 and ASIC2a currents for stomatin LI91,92AA (Figure 4F and G; Supplementary Figure 6). These results indicate that the hydrophobic pocket is crucial for the function of stomatin in inhibiting ASIC2 and ASIC3. However, Leu91 and Ile92 in the loop preceding the stomatin domain do not appear to be the physiological targets of this pocket for inhibition of ASIC function. Accordingly, the helical filaments observed in crystal form 1 and 2 have no apparent functional relevance.
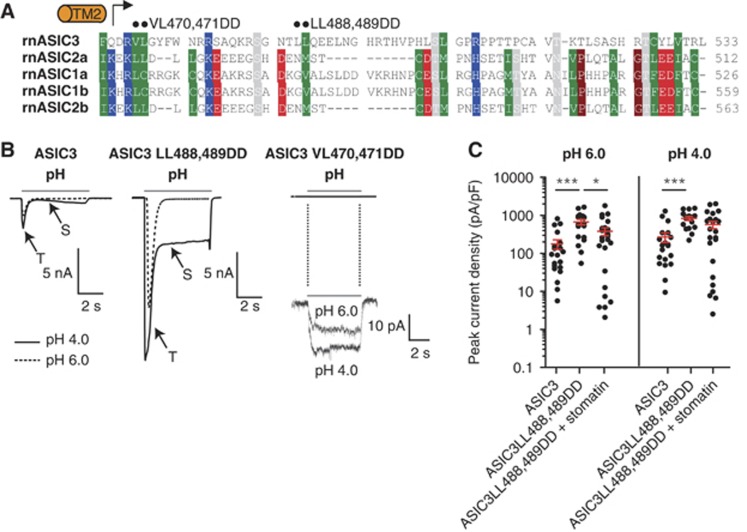
The C-terminus of ASIC3 contains a Leu488, Leu489 motif resembling the N-terminal Leu91, Ile92 in stomatin (Figure 5A). Assuming that these residues might interact with the stomatin hydrophobic pocket, we mutated both to aspartic acid (LL488,489DD) to disrupt such a putative interaction. The ASIC3 Leu488, Leu489 mutant still co-localized with stomatin at internal membranes and the plasma membrane (Supplementary Figures 4 and 5). It also interacted with stomatin in pull-down assays (Supplementary Figure 8A), indicating that these two amino acids are not crucial for the interaction with stomatin. Interestingly, the LL488,489DD mutant showed a substantial increase in proton-gated transient and sustained current amplitudes (Figure 5B). The regulation of this mutant by stomatin was partially lost: Transient amplitudes of this mutant at pH 6.0 were still affected by the presence of stomatin (Figure 5C), but stomatin did not cause an effect on sustained current amplitudes at pH 6.0 or transient and sustained current amplitudes at pH 4.0 (Figure 5C; Supplementary Figure 8). These data suggest that the C-terminus of ASIC3 is important for modulation by stomatin. Mutations in a second hydrophobic motif in the C-terminus of ASIC3 (VL470,471DD) resulted in a non-functional ASIC3 channel (Figure 5B).
Figure 5.
A mutation in the C-terminus of ASIC3 affects the modulation by stomatin. (A) Sequence alignment of the C-termini of the ASIC family following transmembrane helix 2 (TM2). The positions of the mutations analysed in this study are indicated by black dots. Type conserved residues are labelled as in Supplementary Figure 1A. (B) Example traces of proton-evoked currents in cells overexpressing either ASIC3, ASIC3 LL488,489DD or ASIC3 VL470,471DD. The ASIC3 LL488, 489DD mutant showed increased transient (T) and sustained (S) current amplitudes compared with ASIC3. In contrast, the ASIC3 VL470,471DD mutant is non-functional; a 250-fold magnification of the example trace is shown below. (C) Quantification of transient currents mediated by ASIC3, ASIC3 LL488,489DD and ASIC3 LL488,489DD together with stomatin. Each dot corresponds to one cell. ***P<0.001; *P<0.05.
Higher-order oligomerization of stomatin
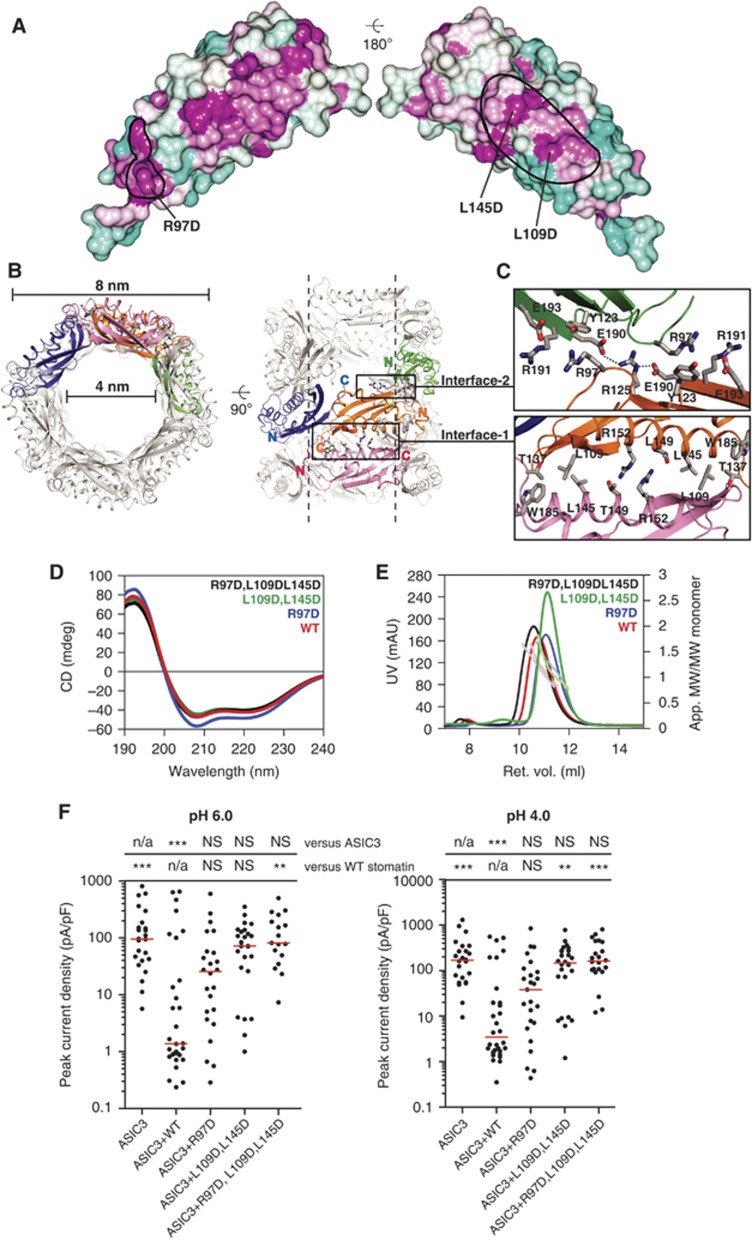
When analysing the crystal packing of crystal form 3 (Table I), we noticed that the stomatin86–213 LI91,92AA dimers assembled in tubular structures via two conserved symmetric interfaces (Figure 6A and B; Supplementary Figure 7C). Interface-1 has a buried surface area per molecule of 700 Å2 (Figure 6C). In particular, Trp185 and Leu145 form a hydrophobic surface at the periphery of interface-1, to which Leu109 of the opposing molecule binds. In the centre of interface-1, Arg152 forms a hydrogen bond with Thr149 of the opposing molecule. Interestingly, Trp184 in human stomatin corresponding to Trp185 in mouse stomatin has previously been shown to be important for oligomerization of stomatin (Umlauf et al, 2006).
Figure 6.
Higher-order oligomerization of stomatin. (A) Solvent-accessible residues in the stomatin domain are coloured according to sequence conservation within the mammalian stomatin family using a gradient from violet (highly conserved) to cyan (not conserved). The oligomerization interfaces in crystal form 3 are indicated by black lines. (B) Two views on the tubular structures formed by stomatin dimers in crystal form 3. (C) Detailed views on the conserved interfaces. (D) CD measurements for stomatin86–213 (red), and the R97D (blue), L109D, L145D (green) and R97D, L109D, L145D (black) mutants. (E) Analytical gel filtration analysis for the indicated stomatin86–213 mutants. (F) Electrophysiological recordings of CHO cells transfected with ASIC3 and the indicated stomatin constructs at pH 4.0 and pH 6.0, as described in Figure 3A.
Interface-2 has a buried surface area of 300 Å2 and features an interaction of the invariant Arg97 with a conserved surface-exposed amino acid triplet formed by Tyr123, Glu193 and Arg191 (Figure 6C).
To test the relevance of these interfaces for the function of stomatin, mutagenesis studies were performed. To disrupt interface-1, we generated a stomatin86–213 L109D,L145D mutant, which behaved like stomatin86–213 in CD spectroscopy (Figure 6D) and eluted as stomatin86–213 in analytical gel filtration experiments (Figure 6E). In CHO cells, stomatin L109D,L145D also showed similar expression levels compared with stomatin and localized similarly (Supplementary Figures 4 and 5). However, in whole-cell recordings, stomatin L109D,L145D failed to inhibit ASIC3 at pH 4.0 and peak current density was significantly greater than in the presence of stomatin (Figure 6F). At pH 6.0, stomatin L109D,L145D had an intermediate inhibitory action; although peak current density was not significantly lower than the peak current density of ASIC3 expressed alone, it was also not significantly greater than the peak current density of ASIC3 co-expressed with stomatin (Figure 6F). Disruption of interface-2 with stomatin R97D led to an intermediate phenotype so that ASIC3 proton-gated current amplitudes at both pH 4.0 and pH 6.0 were not significantly different from wild-type ASIC3 alone or ASIC3 co-expressed with stomatin (Figure 6F). Interestingly, the R184C mutation in MEC-2 corresponding to Arg97 in mouse stomatin leads to reduced touch sensitivity (Chalfie and Sulston, 1981).
A combined triple mutant in both interfaces, stomatin86–213 R97D, L109D, L145D, showed a similar CD spectrum compared with stomatin86–213 (Figure 6D) and a similar gel filtration profile (Figure 6E). Stomatin R97D, L109D, L145D also co-localized with ASIC3 in CHO cells (Supplementary Figures 4 and 5) and could be co-immunoprecipitated with ASIC3 (Figure 3B). However, this triple mutant failed to inhibit ASIC3 currents at both pH 4.0 and pH 6.0, suggesting a synergistic action of these two interfaces in the inhibition of ASIC currents. These data are consistent with a model whereby both interface-1 and interface-2 participate in higher-order stomatin oligomer formation in cells that can regulate ion channel activity.
Discussion
Stomatin-domain proteins exert powerful physiological effects on ion channels (Goodman et al, 2002; Price et al, 2004; Martinez-Salgado et al, 2007; Wetzel et al, 2007; Lapatsina et al, 2012a, 2012b). Proton-gated ASIC3-mediated current amplitude is suppressed and ASIC2a current inactivation time is accelerated in the presence of stomatin (Price et al, 2004), and we have used this as an assay of stomatin function. By solving the core structure of the eukaryotic stomatin domain and performing a structure–function analysis, we were able to define three key regions of the stomatin molecule necessary for its ability to modulate ASIC2a and 3. First, we show that the basic building block of stomatin is a banana-shaped dimer, which is formed by an intermolecular β-sheet at the C-terminus of the stomatin domain and is necessary for the modulation of ASICs. Second, we have identified a dynamic hydrophobic pocket in the stomatin domain, the closure of which prevents modulation of ASICs without affecting dimerization. This pocket can accommodate a dipeptide of two amino acids with branched aliphatic side chains (Leu, Ile). We have also provided evidence that a leucine–leucine dipeptide in the C-terminus of ASIC3 could contribute to channel modulation by stomatin. Finally, we found two conserved surfaces in the stomatin domain, which could be involved in the formation of ring-like structures. Disruption of the potential interaction sites in this oligomer rendered the stomatin molecule ineffective in modulating ASICs. This finding raises the interesting possibility that these two interfaces are used in plasma membrane-associated stomatin oligomers.
There is plentiful evidence that stomatin and other stomatin domain containing proteins can form higher-order oligomers (Snyers et al, 1998; Umlauf et al, 2006). These oligomers are thought to create microenvironments at membranes required for specific cellular functions, for example, the control of ion channels (Huber et al, 2006; Lapatsina et al, 2012b). In our crystal form 3, we observed ring-like stomatin oligomers with an outer diameter of 8 nm. Slight alterations in the assembly interfaces might lead to oligomers of different diameter in the cell. Furthermore, the C-terminal region of stomatin was shown to contribute to oligomerization (Umlauf et al, 2006). For cyanobacterial stomatin (Boehm et al, 2009) and yeast prohibitin (Tatsuta et al, 2005), ring-like oligomers were identified by single-particle electron microscopy analysis, supporting the idea that stomatin-domain proteins may assemble at the membrane in ring-like structures with a banana-shaped dimer as the building block. Also most of the other SPFH-domain proteins, such as prohibitin, erlins and podocin localize to distinct membrane microdomains in the cell (Browman et al, 2007). Thus, our model of stomatin oligomerization might be relevant for many other SPFH domain containing proteins, although the SPFH domain can also be engaged in different oligomerization modes (Supplementary Figure 1B and C).
Striking parallels to the well-known membrane scaffolds of the BAR (Bin/Amphiphysin/Rvs) family are apparent. BAR-domain also assemble into banana-shaped dimers with similar μM affinities to that shown for the stomatin dimer here (Peter et al, 2004; Frost et al, 2009). Furthermore, BAR-domain dimers assemble via low affinity interactions into helical structures involving interactions at the tip and the lateral side of the dimer and may thus exert changes in membrane curvature, which often is supported by the insertion of N-terminal amphipathic helices (Shimada et al, 2007; Frost et al, 2008). The membrane insertion domain of stomatins might exert a similar function in targeting and concentrating stomatin at cellular membranes and creating membrane curvature. In the absence of this hydrophobic hairpin, the affinity for membranes and lipid droplets appears to be low and stomatin is cytoplasmic (Umlauf et al, 2004; Lapatsina et al, 2012b). Accordingly, the stomatin construct used in this study did not contain the membrane anchor and also did not bind to liposomes in vitro.
The activity of ASICs can be modulated by phosphorylation, and an interaction of their C-termini with the PDZ (PSD95, Dlg1 and zo-1)- and BAR-domain containing protein PICK1 (protein interacting with C-kinase1) is required for this modulation (Baron et al, 2002; Duggan et al, 2002; Leonard et al, 2003; Deval et al, 2004). Other PDZ-domain containing proteins, like PSD95 and Lin-7b, have also been shown to interact with the C-terminus of ASICs and can exert either positive or negative modulation of pH-gated currents (Hruska-Hageman et al, 2004). In these cases, it is thought that channel activity is primarily modulated by regulating the number of channels on the plasma membrane. In the present study, we have identified a C-terminal di-leucine motif in ASIC3, which appears to be involved in stomatin modulation without affecting ASIC3 membrane targeting. Mutation of the di-leucine motif to aspartic acid greatly increased current amplitudes without affecting the physical interaction with stomatin (Figure 5). This suggests that stomatin primarily regulates functional properties of the ASIC3 channel via this di-leucine motif. Conversely, closure of the hydrophobic pocket in stomatin with the T182W mutation abolished stomatin’s ability to negatively regulate ASIC3 (Figure 4).
MEC-2 is a positive regulator of mechanosensitive ion channels such as MEC-4 and MEC-10 (O’Hagan et al, 2005; Lapatsina et al, 2012a). The mechanosensitive ion channel targets of mammalian stomatin-domain containing proteins are, however, still unknown (Smith and Lewin, 2009; Lumpkin et al, 2010; Poole et al, 2011; Lapatsina et al, 2012a). The functional domains of the stomatin molecule identified here as essential for the negative regulation of ASICs may be equally important for the regulation of mechanotransduction. Interestingly, the conserved surface-exposed Arg97 at interface-2, is at an identical position to a mutant allele u64 of mec-2 that produces some touch insensitivity in worms (Chalfie and Sulston, 1981). Finally, hetero-dimerization of flotillins, and, possibly, higher-order oligomerization, induces membrane curvature and vesicle budding at the plasma membrane during clathrin-independent endocytosis (Frick et al, 2007). Thus, it is conceivable that the architecture of the ring-like stomatin oligomers is fundamental for an understanding of a wide range of different cellular functions involving SPFH-domain proteins.
Materials and methods
Protein expression and purification
Constructs of mouse stomatin (amino acids 86–213, stomatin86–213, and amino acids 86–255, stomatin86–255) and the indicated point mutants were expressed as N-terminal GST–fusion proteins in Escherichia coli BL21 DE3 phage resistant Rosetta (Novagen), including a PreScission protease cleavage site between the GST and stomatin constructs. In all constructs, Cys87 was mutated to serine to prevent oxidative cross-linking. Bacteria were grown to an OD600 of 0.4 in TB medium when protein expression was induced with 80 μM IPTG, followed by overnight expression at 18°C. Cells were resuspended in ice-cold lysis buffer (50 mM HEPES/NaOH, pH 7.5, 500 mM NaCl, 0.1 mM Pefabloc SC (Roth), 1 μM DNAse (Roth)) and lyzed using a microfluidizer (Microfluidics, Newton, USA). After centrifugation at 100 000 g for 45 min at 4°C, the supernatant was applied on a GSH-column pre-equilibrated with lysis buffer and extensively washed with 50 mM HEPES/NaOH, pH 7.5, 500 mM NaCl. In the same buffer, overnight cleavage of the GST tag was performed at 4°C in the presence of 250 μg PreScission protease which was directly added to the GSH beads. The protein was eluted, concentrated and further purified by size exclusion chromatography on a Superdex75 26/60 column pre-equilibrated with 10 mM HEPES/NaOH, pH 7.5, 150 mM NaCl. Peak fractions of the protein were pooled and concentrated to ∼30 mg/ml. The protein was flash-frozen in liquid nitrogen and stored at −80°C. For all constructs, the yield of purified protein was ∼1.5 mg/l bacteria culture.
Crystallization and structure determination
Before crystallization, proteins were diluted with 10 mM HEPES/NaOH, pH 7.5, 150 mM NaCl. All crystallization trials were performed at 20°C.
The sitting-drop vapour diffusion method in 96-well plate format was used to crystallize stomatin86–213. In all, 300 nl protein at 6 mg/ml was mixed with an equal volume of reservoir solution containing 50 mM cadmium sulphate, 1 M sodium acetate and 100 mM HEPES/NaOH, pH 7.5. After 6 days, hexagonal crystals appeared. Crystals were transferred in a cryo-solution containing 100 mM HEPES/NaOH pH 7.5, 150 mM NaCl, 50 mM cadmium sulphate, 1 M sodium acetate and 25% glycerol. All crystals were cryo-cooled by plunging them in liquid nitrogen.
For crystal form 2 of stomatin86–213, the hanging-drop vapour diffusion method was used in 24-well plates containing 0.7 ml reservoir solution. Crystallization drops were composed of 1 μl protein and 1 μl reservoir solution containing 100 mM HEPES/NaOH, pH 7.5, 7% ethanol, 5% glycerol, 800 mM sodium acetate. Rhombohedral crystals appeared after 6 days. The cryo-solution contained 200 mM HEPES/NaOH, pH 7.5, 100 mM NaCl, 800 mM sodium acetate, 25% glycerol.
Stomatin86–213 LI91,92AA was crystallized using the hanging-drop vapour diffusion method in 24-well plates. Crystallization drops were composed of 1 μl protein and 1 μl reservoir solution containing 100 mM HEPES/NaOH, pH 7.5, 5% ethanol, 20 mM cadmium sulphate. Hexagonal crystals appeared after 7 days. The cryo-solution contained 100 mM HEPES/NaOH, pH 7.5, 150 mM NaCl, 20 mM cadmium sulphate and 25% glycerol.
Data sets of single crystals were recorded at beamline 14.1, BESSY II, Berlin, Germany with a Rayonics MX-225 CCD detector or at the microfocus setup at Swiss Light Source PX06SA beamline, equipped with a PILATUS M6 detector. Initial indexing and determination of an optimal data collection strategy was done using Mosflm (Leslie, 2006). Recorded intensities were integrated with the programme XDS (Kabsch, 2010) or HKL-2000 (Otwinowski and Minor, 1997). Molecular replacement was carried out using the programs MOLREP (Vagin and Teplyakov, 1997) via the CCP4 graphical interface version 6.0.2 (COLLABORATIVE COMPUTATIONAL PROJECT, 1994), using the ph stomatin domain as a search model. Figures were prepared using PyMOL (Schrödinger LLC, 2003). The plugin VASCo (Steinkellner et al, 2009) was used to illustrate the hydrophobic surfaces. The cavity was analysed using the CASTp server (Dundas et al, 2006). The size of the interfaces was calculated with the help of the PISA server (Krissinel and Henrick, 2007). The 3D figure was prepared with pymol and Adobe Acrobat 9 Pro Extended (Kumar et al, 2010). Use of Adobe Acrobat Reader 9.0 (or a higher version) is required to activate the 3D figure.
Circular dichroism
Protein samples were diluted to 0.2 g/l in a buffer containing 150 mM NaF and 10 mM Na2HPO4, pH7.4. CD measurements with three replicates were performed using a Chirascan spectrometer (Applied Photophysics) in the wavelength spectrum of 190240, nm. For data analysis, the web DICHROWEB server (Whitmore and Wallace, 2004) with the CDSSTR algorithm was used.
Right-angle light scattering
A coupled RALS-refractive index detector (Malvern) was connected in line to an analytical gel filtration column Superdex75 10/300 to determine absolute molecular masses of the applied proteins. Data were analysed with the provided OmniSec software. The running buffer contained 100 mM HEPES/NaOH, pH 7.5, 150 mM NaCl. For each protein sample, 100 μl of a 1.5 mg/ml protein solution was applied.
Analytical ultracentrifugation
Molecular mass studies of stomatin variants in 10 mM HEPES/NaOH, pH 7.5, 150 mM NaCl were performed in an XL-A type analytical ultracentrifuge (Beckman) equipped with UV absorbance optics. Sedimentation equilibrium experiments were carried out using six-channel cells with 12 mm optical path length and the capacity to handle three solvent–solution pairs of about 70 μl liquid. Sedimentation equilibrium was reached after an equilibrium speed of 24 000 r.p.m. for about 30 h at 10°C. The radial absorbance in each compartment was recorded at three different wavelengths between 270 and 290 nm, depending on the concentration used in the experiments. Molecular mass determinations employed the global fit of the three radial distributions using the programs POLYMOLE or POLYMOLA (Behlke et al, 1997). When proteins adopt a monomer–dimer equilibrium, the molecular mass, M, can be treated approximately as a weight average parameter (Mw). This value is a composite of the monomer molecular mass (Mm) and that of the dimer (Md) and the partial concentrations of monomers, cm, and dimers, cd.
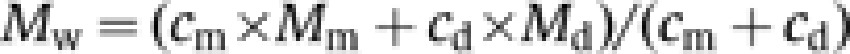 |
Therefore, the equilibrium constant, Kd, can be derived with
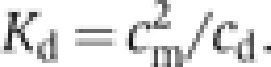 |
Cellular localization
CHO cells were grown to a density of 60% in Dulbecco’s Modified Eagle’s Medium supplemented with 10% bovine serum albumin, 100 units/ml penicillin, 100 μg/ml streptomycin (Roth) and 4.5 mM glucose as supplement. Transfections with plasmids encoding mouse stomatin–mCherry and rat eGFP–ASIC3 were performed using Fugene (Roche). At 24 h post-transfection, cells were fixed with 5% paraformaldehyde at room temperature, washed with PBS, water and mounted on cover slides using Mowiol (Roth). Images were acquired using Leica SP5 confocal microscope and analysed using the ImageJ software.
Immunoprecipitation and immunoblotting
CHO cells were co-transfected with plasmids encoding Myc-His-tagged stomatin or of the indicated stomatin variants and plasmids encoding FLAG-tagged ASIC3. After 24 h, cells were solubilized with lysis buffer containing 1% TritonX-100, 0.1% SDS, 10 mM Tris–HCl, pH 7.6, 150 mM NaCl, 100 μM 0.1 mM Pefabloc SC (Roth). Following ultracentrifugation at 100 000 g for 20 min at 4°C, 500 μg of detergent-soluble protein was subjected to immunoprecipitation using anti-c-Myc (Millipore catalogue # 06-340) and protein A-Sepharose and then washed three times in lysis buffer. Proteins were eluted from the beads with SDS sample buffer (4% SDS, 0.4% Bromphenol blue, 40% glycerol, 200 mM Tris–HCl, pH 6.8) by incubation for 5 min at 60°C and separated on SDS–PAGE. The proteins were transferred to nitrocellulose membrane and blocked by incubation in 5% milk powder in TBST (50 mM TRIS, pH 7.4, 150 mM NaCl, 0.1% Tween 20) for 1 h. Immunoblots were incubated with anti-polyhistidin-peroxidase conjugate (Sigma Aldrich Catalogue # A7058) or anti-FLAG (Sigma Aldrich Catalogue # F3165) antibodies o/n at 4°C. The blots were washed three times with TBST in 5% milk powder in TBST and incubated with horseradish-peroxidase conjugate anti-mouse antibody and washed three times with TBST. Bound antibodies were detected using the enhanced chemiluminescence kit (Amersham). For expression tests, a monoclonal mouse antibody directed against the c-Myc tag (sc-40 9E10 Santa Cruz, California), or α-Tubulin (T 6793, Sigma-Aldrich, Munich) was used.
Bimolecular fluorescence complementation assays
BiFC was used to determine the localization of interaction between ASIC3 and stomatin variants in CHO cells. A construct of ASIC3 fused at its C-terminus to a C-terminal fragment of YFP was co-expressed with a construct of stomatin fused at its C-terminus with an N-terminal fragment of YFP according to Hu et al (2002). A fluorescent signal develops if the N- and C-terminal fragments of YFP come within 5 nm of each other. The BiFC signal was visualized 24 h after transfection in living cells using epifluorescent microscopy (Olympus IX71, × 100 oil-immersion objective).
Self-association of stomatin was detected by monitoring BiFC signal development over time. Human embryonic kidney (HEK) cells were transfected with BiFC constructs of stomatin or stomatin V197P fused at their C-termini with N- and C-terminal fragments of YFP. Eight hours after transfection, cells were transferred into a 384-well plate and fluorescence development was monitored every 30 min for 16 h at 37°C using a TECAN M1000 plate reader (imaging conditions: excitation: 515 nm, bandwidth 8 nm; emission: 535 nm, bandwidth 8 nm). The slope was determined by a linear fit to the increase in relative fluorescence intensity over time (excitation: 515 nm, bandwidth 8 nm; emission: 535 nm, bandwidth 8 nm).
Electrophysiological measurement and analysis
CHO cell transfections were conducted with Lipofectamine LTX (Invitrogen) according to the manufacturer’s protocol. Briefly, plasmids were diluted into Opti-MEM (Gibco) at a ratio of 4:1 (stomatin:ASICx), with a DNA concentration of 2 μg/40 mm dish and Lipofectamine LTX was subsequently added. The ASIC cDNAs encoded for rat ASIC2a and rat ASIC3. During the 30-min incubation period, the culture medium was replaced with Opti-MEM followed by adding the transfection mixture by careful dropping. After 4 h, the transfection medium was replaced and cells were used 24 h later for whole-cell electrophysiology. The following solutions were used: extracellular (in mM) – NaCl (140), KCl (4), CaCl2 (2), MgCl2 (1), glucose (4), HEPES (10), adjusted to pH 7.4 with NaOH (for solutions below pH 6.0, MES was used in place of HEPES) and intracellular – KCl (110), NaCl (10), MgCl2 (1), EGTA (1) and HEPES (10), adjusted to pH 7.3 with KOH. Patch pipettes were pulled (Flaming-Brown puller, Sutter Instruments) from borosilicate glass capillaries (Hilgenberg) and had a resistance of 3–6 MΩ. Recordings were made using an EPC-9 amplifier (HEKA) and Patchmaster© software (HEKA). Whole-cell currents were recorded at 20 kHz, pipette and membrane capacitance were compensated using Patchmaster macros and series resistance was compensated by 70%. Cells were stimulated with a 5-s pulse of an acidic solution, pH 4 and 6 for ASIC3 transfected cells (randomly applied, 2 min wash between stimulations) and pH 4.0 and 5.0 for ASIC2a transfected cells. Analysis was carried out using Fitmaster (HEKA) and GraphPad Prizm (GraphPad Software, Inc.), current amplitudes were normalized to cell capacitance and values expressed as pA/pF. Stomatin displays dose-related effects upon ASICs (Price et al, 2004) and upon transfection of stomatin with ASIC3, we also observed proton-gated currents of varying amplitudes. Differences between peak current densities (ASIC3 experiments) and inactivation times (ASIC2a experiments) were assessed using the Kruskal–Wallis test followed by Dunn’s post test.
Tail-derived mouse adult fibroblast (MAF) cultures were prepared from adult stomatin-/- mice and wild-type BL6 littermates. Tail tips, 0.5 cm in length, were cut into small pieces under sterile conditions and incubated for 2 h in DPBS containing 0.2% collagenase and 2 U/ml dispase (Roche). Sedimented cells were cultured in DMEM medium (Gibco) containing 10% FCS, 1% Pen/Strep and L-glutamine. In all, 35 000 cells were plated on a 3.5-cm dish and transfected on next day (30–40% confluence). In all, 29 μl of FuGENE 6 (Promega) were added to 230 μl of OPTI-MEM (Gibco) and incubated for 5 min. In all, 9 μg of plasmid DNA (ASIC3:GFP=4:1) were added to the mixture and incubated for additional 30 min before adding drop wise to the cells. Cells were assayed 24–48 h after transfection. Electrophysiological recordings were performed as described above.
Accession numbers
PDB coordinates of the three stomatin crystal forms have been submitted to the PDB database (pdb codes 4FVF, 4FVG, 4FVJ).
Supplementary Material
Acknowledgments
We acknowledge O Ristau for advice in analytical ultracentrifugation analysis; S Werner, M Papst, H Thränhardt for technical assistance; CS Hee for help with the preparation of the 3D pdf figure and the BESSY II and SLS staff for assistance during data collection. This project was supported by grants of the German Research Council (SFB449/B18 to OD and GRL, SFB958/A09 to GRL and KP, SFB958/A12 to OD), by a Career Development Award of the International Human Frontier Science Program Organization and an EMBO YIP award (to OD).
Author contributions: JB, EJS, GRL and OD designed the research; JB, EJS, KP, DO, LL, AK and JB performed the research; JB, EJS, DS, KP, DO, LL, JB, GRL and OD analysed the data; JB, EJS, GRL and OD wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Baron A, Deval E, Salinas M, Lingueglia E, Voilley N, Lazdunski M (2002) Protein kinase C stimulates the acid-sensing ion channel ASIC2a via the PDZ domain-containing protein PICK1. J Biol Chem 277: 50463–50468 [DOI] [PubMed] [Google Scholar]
- Behlke J, Ristau O, Schonfeld HJ (1997) Nucleotide-dependent complex formation between the Escherichia coli chaperonins GroEL and GroES studied under equilibrium conditions. Biochemistry 36: 5149–5156 [DOI] [PubMed] [Google Scholar]
- Boehm M, Nield J, Zhang P, Aro EM, Komenda J, Nixon PJ (2009) Structural and mutational analysis of band 7 proteins in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 191: 6425–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boute N, Gribouval O, Roselli S, Benessy F, Lee H, Fuchshuber A, Dahan K, Gubler MC, Niaudet P, Antignac C (2000) NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet 24: 349–354 [DOI] [PubMed] [Google Scholar]
- Browman DT, Hoegg MB, Robbins SM (2007) The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol 17: 394–402 [DOI] [PubMed] [Google Scholar]
- Cadiou H, Studer M, Jones NG, Smith ES, Ballard A, McMahon SB, McNaughton PA (2007) Modulation of acid-sensing ion channel activity by nitric oxide. J Neurosci 27: 13251–13260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Sulston J (1981) Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol 82: 358–370 [DOI] [PubMed] [Google Scholar]
- COLLABORATIVE COMPUTATIONAL PROJECT. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cueva JG, Mulholland A, Goodman MB (2007) Nanoscale organization of the MEC-4 DEG/ENaC sensory mechanotransduction channel in Caenorhabditis elegans touch receptor neurons. J Neurosci 27: 14089–14098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval E, Salinas M, Baron A, Lingueglia E, Lazdunski M (2004) ASIC2b-dependent regulation of ASIC3, an essential acid-sensing ion channel subunit in sensory neurons via the partner protein PICK-1. J Biol Chem 279: 19531–19539 [DOI] [PubMed] [Google Scholar]
- Duggan A, Garcia-Anoveros J, Corey DP (2002) The PDZ domain protein PICK1 and the sodium channel BNaC1 interact and localize at mechanosensory terminals of dorsal root ganglion neurons and dendrites of central neurons. J Biol Chem 277: 5203–5208 [DOI] [PubMed] [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res 34: W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick M, Bright NA, Riento K, Bray A, Merrified C, Nichols BJ (2007) Coassembly of flotillins induces formation of membrane microdomains, membrane curvature, and vesicle budding. Curr Biol 17: 1151–1156 [DOI] [PubMed] [Google Scholar]
- Fricke B, Argent AC, Chetty MC, Pizzey AR, Turner EJ, Ho MM, Iolascon A, von DM, Stewart GW (2003) The ‘stomatin’ gene and protein in overhydrated hereditary stomatocytosis. Blood 102: 2268–2277 [DOI] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De CP, Unger VM (2008) Structural basis of membrane invagination by F-BAR domains. Cell 132: 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Unger VM, De CP (2009) The BAR domain superfamily: membrane-molding macromolecules. Cell 137: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher PG, Forget BG (1995) Structure, organization, and expression of the human band 7.2b gene, a candidate gene for hereditary hydrocytosis. J Biol Chem 270: 26358–26363 [DOI] [PubMed] [Google Scholar]
- Garcia-Anoveros J, Samad TA, Zuvela-Jelaska L, Woolf CJ, Corey DP (2001) Transport and localization of the DEG/ENaC ion channel BNaC1alpha to peripheral mechanosensory terminals of dorsal root ganglia neurons. J Neurosci 21: 2678–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales EB, Kawate T, Gouaux E (2009) Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature 460: 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MB, Ernstrom GG, Chelur DS, O’Hagan R, Yao CA, Chalfie M (2002) MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature 415: 1039–1042 [DOI] [PubMed] [Google Scholar]
- Hruska-Hageman AM, Benson CJ, Leonard AS, Price MP, Welsh MJ (2004) PSD-95 and Lin-7b interact with acid-sensing ion channel-3 and have opposite effects on H+-gated current. J Biol Chem 279: 46962–46968 [DOI] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Huang M, Gu G, Ferguson EL, Chalfie M (1995) A stomatin-like protein necessary for mechanosensation in C. elegans. Nature 378: 292–295 [DOI] [PubMed] [Google Scholar]
- Huber TB, Schermer B, Müller RU, Höhne M, Bartram M, Calixto A, Hagmann H, Reinhardt C, Koos F, Kunzelmann K, Shirokova E, Krautwurst D, Harteneck C, Simons M, Pavenstädt H, Kerjaschki D, Thiele C, Walz G, Chalfie M, Benzing T (2006) Podocin and MEC-2 bind cholesterol to regulate the activity of associated ion channels. Proc Natl Acad Sci USA 103: 17079–17086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasti J, Furukawa H, Gonzales EB, Gouaux E (2007) Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature 449: 316–323 [DOI] [PubMed] [Google Scholar]
- Kabsch W (2010) XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurin I, Huber S, Grunder S (2009) A single conserved proline residue determines the membrane topology of stomatin. Biochem J 418: 587–594 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Kumar P, Ziegler A, Grahn A, Hee CS, Ziegler A (2010) Leaving the structural ivory tower, assisted by interactive 3D PDF. Trends Biochem Sci 35: 419–422 [DOI] [PubMed] [Google Scholar]
- Lapatsina L, Brand J, Poole K, Daumke O, Lewin GR (2012a) Stomatin-domain proteins. Eur J Cell Biol 91: 240–245 [DOI] [PubMed] [Google Scholar]
- Lapatsina L, Jira J, Smith ES, Poole K, Kozlenkov A, Bilbao D, Lewin GR, Heppenstall PA (2012b) Regulation of ASIC channels by a stomatin/STOML3 complex located in a mobile vesicle pool in sensory neurons. Open Biol 2: 120096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AS, Yermolaieva O, Hruska-Hageman A, Askwith CC, Price MP, Wemmie JA, Welsh MJ (2003) cAMP-dependent protein kinase phosphorylation of the acid-sensing ion channel-1 regulates its binding to the protein interacting with C-kinase-1. Proc Natl Acad Sci USA 100: 2029–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AG (2006) The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr 62: 48–57 [DOI] [PubMed] [Google Scholar]
- Lumpkin EA, Marshall KL, Nelson AM (2010) The cell biology of touch. J Cell Biol 191: 237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairhofer M, Steiner M, Salzer U, Prohaska R (2009) Stomatin-like protein-1 interacts with stomatin and is targeted to late endosomes. J Biol Chem 284: 29218–29229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salgado C, Benckendorff AG, Chiang LY, Wang R, Milenkovic N, Wetzel C, Hu J, Stucky CL, Parra MG, Mohandas N, Lewin GR (2007) Stomatin and sensory neuron mechanotransduction. J Neurophysiol 98: 3802–3808 [DOI] [PubMed] [Google Scholar]
- O’Hagan R, Chalfie M, Goodman MB (2005) The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat Neurosci 8: 43–50 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Owczarek CM, Treutlein HR, Portbury KJ, Gulluyan LM, Kola I, Hertzog PJ (2001) A novel member of the STOMATIN/EPB72/mec-2 family, stomatin-like 2 (STOML2), is ubiquitously expressed and localizes to HSA chromosome 9p13.1. Cytogenet Cell Genet 92: 196–203 [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303: 495–499 [DOI] [PubMed] [Google Scholar]
- Poole K, Lechner SG, Lewin GR (2011) The molecular and genetic basis of touch. InThe Handbook of Touch Hertenstein MJ, Weiss SJ (eds) pp59–84New York, USA: Springer Publications [Google Scholar]
- Price MP, Thompson RJ, Eshcol JO, Wemmie JA, Benson CJ (2004) Stomatin modulates gating of acid-sensing ion channels. J Biol Chem 279: 53886–53891 [DOI] [PubMed] [Google Scholar]
- Salzer U, Ahorn H, Prohaska R (1993) Identification of the phosphorylation site on human erythrocyte band 7 integral membrane protein: implications for a monotopic protein structure. Biochim Biophys Acta 1151: 149–152 [DOI] [PubMed] [Google Scholar]
- Schrödinger LLC. (2003) The PyMOL Molecular Graphics System. Version 1.4.1 [Google Scholar]
- Seidel G, Prohaska R (1998) Molecular cloning of hSLP-1, a novel human brain-specific member of the band 7/MEC-2 family similar to Caenorhabditis elegans UNC-24. Gene 225: 23–29 [DOI] [PubMed] [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, Liu ZJ, Wang BC, Yamamoto M, Terada T, Miyazawa A, Tanaka A, Sugano S, Shirouzu M, Nagayama K, Takenawa T et al. (2007) Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell 129: 761–772 [DOI] [PubMed] [Google Scholar]
- Smith ES, Lewin GR (2009) Nociceptors: a phylogenetic view. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 195: 1089–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ES, Zhang X, Cadiou H, McNaughton PA (2007) Proton binding sites involved in the activation of acid-sensing ion channel ASIC2a. Neurosci Lett 426: 12–17 [DOI] [PubMed] [Google Scholar]
- Snyers L, Umlauf E, Prohaska R (1998) Oligomeric nature of the integral membrane protein stomatin. J Biol Chem 273: 17221–17226 [DOI] [PubMed] [Google Scholar]
- Snyers L, Umlauf E, Prohaska R (1999a) Association of stomatin with lipid-protein complexes in the plasma membrane and the endocytic compartment. Eur J Cell Biol 78: 802–812 [DOI] [PubMed] [Google Scholar]
- Snyers L, Umlauf E, Prohaska R (1999b) Cysteine 29 is the major palmitoylation site on stomatin. FEBS Lett 449: 101–104 [DOI] [PubMed] [Google Scholar]
- Steinkellner G, Rader R, Thallinger GG, Kratky C, Gruber K (2009) VASCo: computation and visualization of annotated protein surface contacts. BMC Bioinformatics 10: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GW, Hepworth-Jones BE, Keen JN, Dash BC, Argent AC, Casimir CM (1992) Isolation of cDNA coding for an ubiquitous membrane protein deficient in high Na+, low K+ stomatocytic erythrocytes. Blood 79: 1593–1601 [PubMed] [Google Scholar]
- Tanaka H, Kato K, Yamashita E, Sumizawa T, Zhou Y, Yao M, Iwasaki K, Yoshimura M, Tsukihara T (2009) The structure of rat liver vault at 3.5 angstrom resolution. Science 323: 384–388 [DOI] [PubMed] [Google Scholar]
- Tatsuta T, Model K, Langer T (2005) Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol Biol Cell 16: 248–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf E, Csaszar E, Moertelmaier M, Schuetz GJ, Parton RG, Prohaska R (2004) Association of stomatin with lipid bodies. J Biol Chem 279: 23699–23709 [DOI] [PubMed] [Google Scholar]
- Umlauf E, Mairhofer M, Prohaska R (2006) Characterization of the stomatin domain involved in homo-oligomerization and lipid raft association. J Biol Chem 281: 23349–23356 [DOI] [PubMed] [Google Scholar]
- Unfried I, Entler B, Prohaska R (1995) The organization of the gene (EPB72) encoding the human erythrocyte band 7 integral membrane protein (protein 7.2b). Genomics 30: 521–528 [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Cryst 30: 1022–1025 [Google Scholar]
- Wetzel C, Hu J, Riethmacher D, Benckendorff A, Harder L, Eilers A, Moshourab R, Kozlenkov A, Labuz D, Caspani O, Erdmann B, Machelska H, Heppenstall PA, Lewin GR (2007) A stomatin-domain protein essential for touch sensation in the mouse. Nature 445: 206–209 [DOI] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res 32: W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H, Fujii S, Matsui I (2008) Crystal structure of a core domain of stomatin from Pyrococcus horikoshii Illustrates a novel trimeric and coiled-coil fold. J Mol Biol 376: 868–878 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Paszty C, Turetsky T, Tsai S, Kuypers FA, Lee G, Cooper P, Gallagher PG, Stevens ME, Rubin E, Mohandas N, Mentzer WC (1999) Stomatocytosis is absent in ‘stomatin’-deficient murine red blood cells. Blood 93: 2404–2410 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.