Abstract
In a cell-based assay for novel inhibitors, we have discovered that two glycosides of 5-thiomannose, each containing an interglycosidic nitrogen atom, prevented the correct zymogen processing of the prohormone proopiomelanocortinin (POMC) and the transcription factor sterol-regulatory element-binding protein-2 (SREBP-2) in mouse pituitary cells and Chinese hamster ovary (CHO) cells, respectively. In the case of SREBP-2, these effects were correlated with the altered N-linked glycosylation of subtilisin / kexin-like isozyme-1 (SKI-1), the protease responsible for SREBP-2 processing under sterol-limiting conditions. Further examination of the effects of these compounds in CHO cells showed that they caused extensive protein hypoglycosylation in a manner similar to type I congenital disorders of glycosylation (CDGs) since the remaining N-glycans in treated cells were complete (normal) structures. The underglycosylation of glycoproteins in 5-thiomannoside-treated cells is now shown to be caused by the compromised biosynthesis of the dolichol-linked oligosaccharide (DLO) N-glycosylation donor, although the nucleotide sugars required for the synthesis of DLOs were not reduced under these conditions, nor are these effects reversed upon the addition of exogenous mannose. Analysis of DLO intermediates by fluorophore-assisted carbohydrate electrophoresis demonstrates that 5-thiomannose-containing glycosides block DLO biosynthesis most likely at a stage prior to the GlcNAc2Man3 intermediate, on the cytosolic face of the endoplasmic reticulum.
Keywords: 5-thiomannosides, glycoproteins, dolichol-linked oligosaccharides, mannosyltransferases, nucleotide sugars
Introduction
In order to probe the relationships between a glycan’s composition and / or structure and its biological function, the ability to manipulate glycosylation patterns is required. One powerful approach for elucidating glycan function is the genetic manipulation of cell lines or organisms in which specific glycosyltransferases or glycosidases have either been deleted or overexpressed.[1] The use of genetic techniques is often quite difficult, however, since the biosynthesis of glycans is not linear nor is it template-driven. Furthermore, the mutation of genes involved in glycan assembly, especially in animal models, often produces lethal[2] or pleiomorphic phenotypes—in human patients, these are collectively diagnosed as congenital disorders of glycosylation (CDGs). In addition, many glycan processing enzymes are only absolutely essential only for embryonic development.[3] Alternatively, many glycan – protein relationships have been elucidated through the use of chemical agents that affect glycan biosynthesis or metabolism. These small molecule inhibitors permit the facile investigation of glycosylation in many different cell lines, tissues or organisms and represent a crucial complement to genetic methods of glycan analysis. Given the key role glycans play in human diseases[4, 5] some of these inhibitors are the focus of drug development efforts, and the search for inhibitors or modulators of glycosylation continues to be an area of active research.[6]
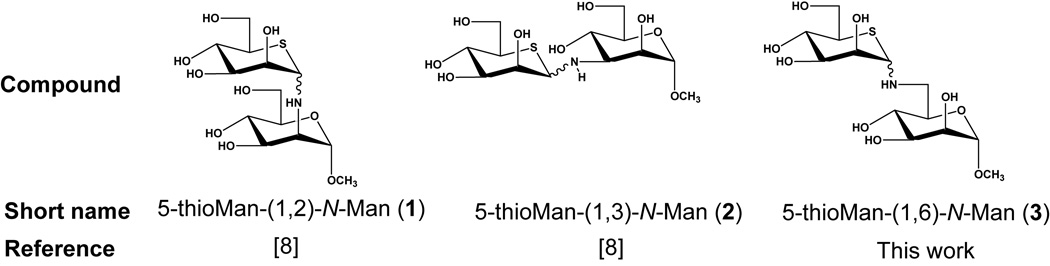
We have recently screened a small library of synthetic carbohydrate-like molecules for effects on the proteolytic processing and / or intracellular targeting of the glycosylated prohormone proopiomelanocortinin (POMC) by proprotein convertase 1/3 (PC1/3).[7] The asparagine-(or N-) linked glycans were shown to play key roles in the processing of POMC into β-lipotroic hormone and β-endorphin. We also showed that two heteroatom-containing disaccharide analogues (Figure 1), each containing a non-reducing 5-thiomannose moiety [8] (these compounds are collectively referred to as 5-thiomannosides), could be taken up by cells in which they not only inhibited PC1 activity but also greatly reduced the activity of a related proprotein convertase known as subtilisin / kexin isozyme-1 (SKI-1), a potential therapeutic target, given its role in several viral infections[9] and in sterol metabolism.[10] Significantly, in Chinese hamster ovary (CHO) cells grown in the presence of 5-thioMan-(1–2)-N-Man (1) and 5-thioMan-(1–3)-N-Man (2), both SKI-1 autocatalytic activation and its substrate, sterol-regulatory element binding protein-2 (SREBP-2), processing were inhibited in a fashion which correlated with altered N-glycosylation. Specifically, both of these compounds were found to reduce the extent of SKI-1 glycosylation, causing a reduction of the apparent molecular weight of all SKI-1 isoforms on SDS-PAGE gels. We now report studies intended to probe the mechanism by which 5-thiomannosides affect protein glycosylation.
Figure 1.
The structures of the heteroatom-containing 5-thiomannosides used in this study. These compounds were synthesized as inseparable mixtures of α/β anomers and tested as such since these S/N disaccharide analogues slowly mutarotate in aqueous solutions.
Glycosides composed of thiosugars in which the ring oxygen atom has been replaced with a sulfur atom have unique structural and chemical properties and intriguing bioactivities.[11] For example, although 5-thioxylopyranosides hydrolyze faster than their oxygen congeners[12] and 5-thioglucopyranosyl carbenium ions, the intermediates of glycoside hydrolysis, are formed 800 times faster than the parent oxygen-containing glycosyl fluorides,[13] it has nevertheless been demonstrated that some thioglycosides are very resistant to enzyme-catalyzed hydrolysis. Thus, both 5-thioGlc-(1–4)-N-Glc[14] and 5-thioGlc-(1–6)-N-Glc[15] were enzymatically hydrolyzed at retarded rates. Similarly, 5-thioGlc-α1–4-Glc containing oxygen, sulfur or selenium in the interglycosidic linkage are also inhibitors of glucoamylase G2, albeit weaker than the nitrogen-congener.[16] In addition to their altered reactivity, some thiosugars, especially those in their reducing forms, are known to bind to monosaccharide-binding proteins with higher affinity than their oxygen analogues.[17–19] Similarly, some[20] thiosugar-containing oligosaccharides bind more tightly to their cognate proteins than their oxygen congeners. These properties have led us,[8, 14, 16, 21, 22] and others, [15, 23–26] to synthesize many derivatives of 5-thioglycosides as potential inhibitors of glycosidase enzymes.
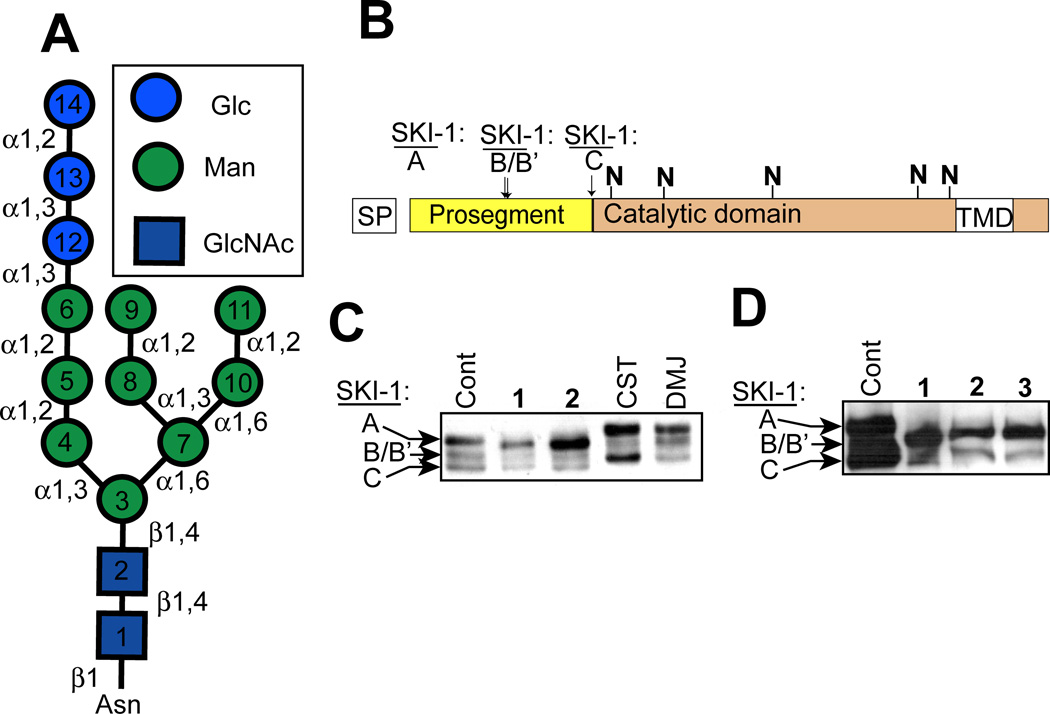
Although designed as glycosidase inhibitors it, is possible that 1 and 2 affect SKI-1 glycosylation via other mechanisms. For example, close mimics of glycan substrates (i.e. acceptor analogues) have been used to investigate the structural requirements of specific glycosyltransferases[27–29] and some, in particular those with modifications to sugar residues which prevent them from being acceptors, are inhibitors of their target enzymes in vitro[30–32] and in cultured cells.[33] Alternatively, rather than acting as inhibitors, the 5-thiomannosides may act as decoy substrates, thereby providing glycosyltransferases with an excess of alternative substrates—effectively preventing the transfer of a monosaccharide to the intended glycoprotein—a phenomenon known as oligosaccharide priming.[34–36] In the context of studies in cultured cells, the hydrolase-resistance properties of thio-sugar-containing disaccharides would be predicted to increase their effective cellular lifetimes. It is conceivable, therefore, that 1 and 2 affect SKI-1 glycosylation either by preventing correct N-glycan processing by α-mannosidases or by altering N-glycan biosynthesis by acting as glycosyltransferase inhibitors or decoy substrates. For example, 2 could mimic the free Man-α-(1–3)-Man linkage found in Man-α-(1–6)-[Man-α-(1–3)-]Man-β-(1–4)-GlcNAc-β-(1–4)-GlcNAc-β-1-Asn, the substrate for GlcNAc T1, a key step in the conversion of high-mannose N-glycans into hybrid- or complex-type structures. Indeed, substrate-like trisaccharides are inhibitors of this enzyme in in vitro assays.[32] Alternatively, multiple points of inhibition could be envisioned which involve the mannosyltransferases responsible for the assembly of dolichol-linked oligosaccharides (DLOs), specifically of the construction of dolichol-PP-GlcNAc2Man9Glc3—the donor substrate for N-glycosylation—since this structure contains two Man-α-(1–3)-Man and four Man-α-(1–2)-Man moieties (Figure 2A). Thus, the 5-thiomannoside inhibitors tested could have affected SKI-1 glycosylation (Figure 2B) in a fashion resembling either class I or II CDGs (reviewed in [37]). In the former, sites of N-glycosylation are skipped by the oligosaccharyltransferase (OST) complex due to its decreased affinity for truncated dolichol-linked oligosaccharides,[38] while in class II CDGs, truncated glycans are observed on glycoproteins due to defects in the subsequent processing of N-glycans by both glycosidases and glycosyltransferases.
Figure 2.
5-Thiomannosides affect the glycosylation of SKI-1 in a way that does not resemble known glycosidase inhibitors. (A) Structure of the 14-residue oligosaccharide which is attached to a specific asparagine residue(s) of a protein to create an N-linked glycoprotein. This oligosaccharides is transferred from a dolichol-pyrophosphate-linked donor which itself is assembled stepwise on both sides of the ER membrane. Monosaccharide units are added to this dolichol-linked oligosaccharide (DLO) in the order in which they are numbered. The DLO precursors themselves are nucleotide-sugar-(residues 1–7) or dolichol-phosphate- (residues 8–14) activated monosaccharides. (B) SKI-1, which is predicted to contain up to five N-glycans (denoted with the “N”), is initially biosynthesized as an inactive proprotein (SKI-1A) and its zymogen activation occurs upon the sequential processing of the inhibitory prosegment at sites B/B’ and C. SKI-1C is the only substrate-active isoform and is only produced within the Golgi; A and B/B’ isoforms are ER-localized. (C) SKI-1 zymogen activation and changes in N-glycosylation in CHO cells grown in the presence of the 5-thiomannosides (1 and 2) and the confirmed glycosidase inhibitors castanospermine (CST) and deoxymannojrimycin (DMJ) was examined by immunoblot analysis. (D) Identical analysis was performed in cells treated with 3 which was predicted to be active since N-glycans contain two α-(1–6)-linked dimannose moieties.
CHO cells have proven to be a valuable model system for examining N-glycosylation defects since they are easy to culture and transfect and since many glycosylation mutants are well characterized in these cells.[39, 40] Furthermore, CHO cells are a preferred mammalian cell line for the production of therapeutic glycoproteins since the glycan structures they produce are very similar to those biosynthesized by human cells.[41] In many studies, model proteins are utilized to elucidate the biochemical basis for N-glycosylation defects—for example serum transferin from human CDG-patients,[42] and interferon-gamma[43] or erythropoietin[44] in CHO cells. For these reasons, we chose to use SKI-1 expressed in CHO cells as a model to further probe the mechanism by which 5-thiomannosides affect protein glycosylation.
Results
5-Thiomannosides do not act like well known ER glycosidase inhibitors
Mechanistic studies on the mode of action of 5-thiomannosides were initially carried out by comparing these compounds with others which are known to affect N-glycosylation. Compounds 1 and 2 did not affect SKI-1 N-glycosylation in a similar fashion as the ER glycosidase inhibitor castanospermine (CST) and the mannosidase inhibitor deoxymannojirimicin (DMJ) (Figure 2C), consistent with our previous report on the negligible impact of glycosidase inhibition on SREBP-2 processing by SKI-1.[7] In contrast to 1 and 2, neither CST nor DMJ greatly decreased SKI-1 autocatalytic zymogen activation (to SKI-1C). Nor did CST or DMJ increase the electrophoretic mobility of all three SKI-1 isoforms as observed for 1 and 2. Note that SKI-1 produced within cells treated with 1 and 2 was still glycosylated, since all isoforms were sensitive to PNGaseF under these conditions.[7] Since N-glycans contain Man-α-(1–2)-, (1–3)-, and (1–6)-Man linkages (Figure 2A), we predicted that 5-thioMan-(1–6)-N-Man (3) would have a similar effect on SKI-1 as the (1–2)- and (1–3)-linked analogues. This compound was synthesized (see Supporting Information) and tested in SRD-12B cells (Figure 2D). As predicted, 3 also affected SKI-1 zymogen activation and decreased its apparent molecular weight. These results suggest that all three 5-thiomannosides likely affect N-glycosylation through a similar mechanism, an effect that cannot be replicated by other known glycosidase inhibitors.
Reduced N-glycosylation in CHO K1 cells: Lectin blotting
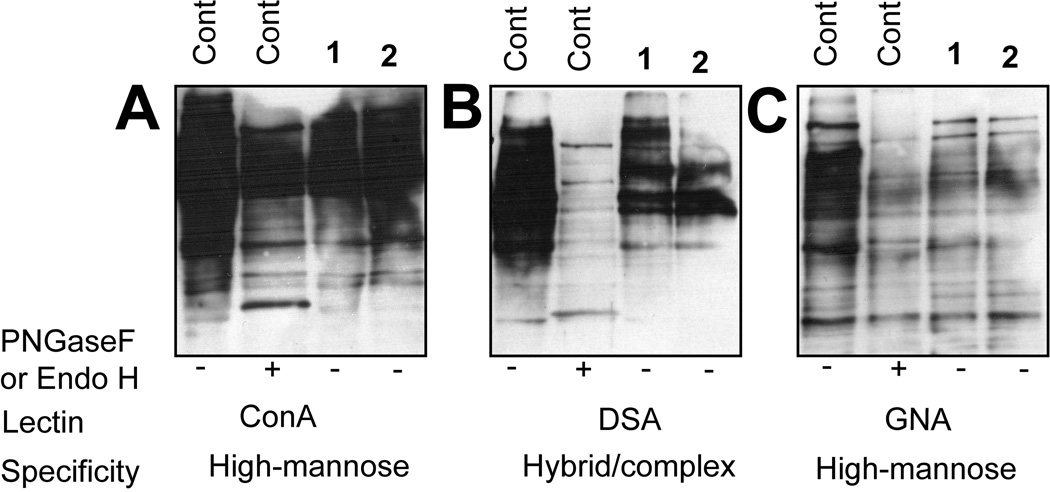
To rule out the possibility that these compounds only affect SKI-1 glycosylation in SRD-12B cells, lectin blotting experiments in CHO K1 cells were performed (Figure 3). For each lectin tested, identical amounts of protein were resolved and blotted; the amount of protein loaded per lane was also normalized. Treatment with either 1 or 2 reduced the extent of protein N-linked glycosylation relative to control cells, an effect that was especially pronounced when blots were probed with the high mannose-specific lectin GNA (Figure 3C). Blotting with ConA (Figure 3A) also demonstrated a decrease in high mannose N-glycans although this was less pronounced than data obtained with GNA. Similarly, the β-(1–4)-GlcNAc-specific lectin DSA (Figure 3B) showed decreased reactivity with proteins isolated from treated cells, correlating well with the reduced binding observed for the mannose-specific lectins. These data are consistent with the inhibitors either affecting N-glycan structure in the ER (since high-mannose structures are affected) or the efficiency of protein glycosylation (i.e. fewer N-glycans per protein molecule) by inhibiting the biosynthesis of DLO donors. Nevertheless, it is clear that the glycosylation defect observed for SKI-1 is not limited to this protein nor the SRD-12B cells in which it was expressed.
Figure 3.
5-Thiomannosides decrease the extent of N-glycosylation observed in CHO K1 cells. Western blots of cell lysates isolated from treated cells were probed with the lectins (A) ConA, (B) DSA, and (C) GNA. Protein concentrations were measured and equal amounts of protein were added to each lane. The high-mannose-specific glycosidase EndoH was used only of GNA.
Analysis of N-glycans in cells treated with 5- thiomannosides
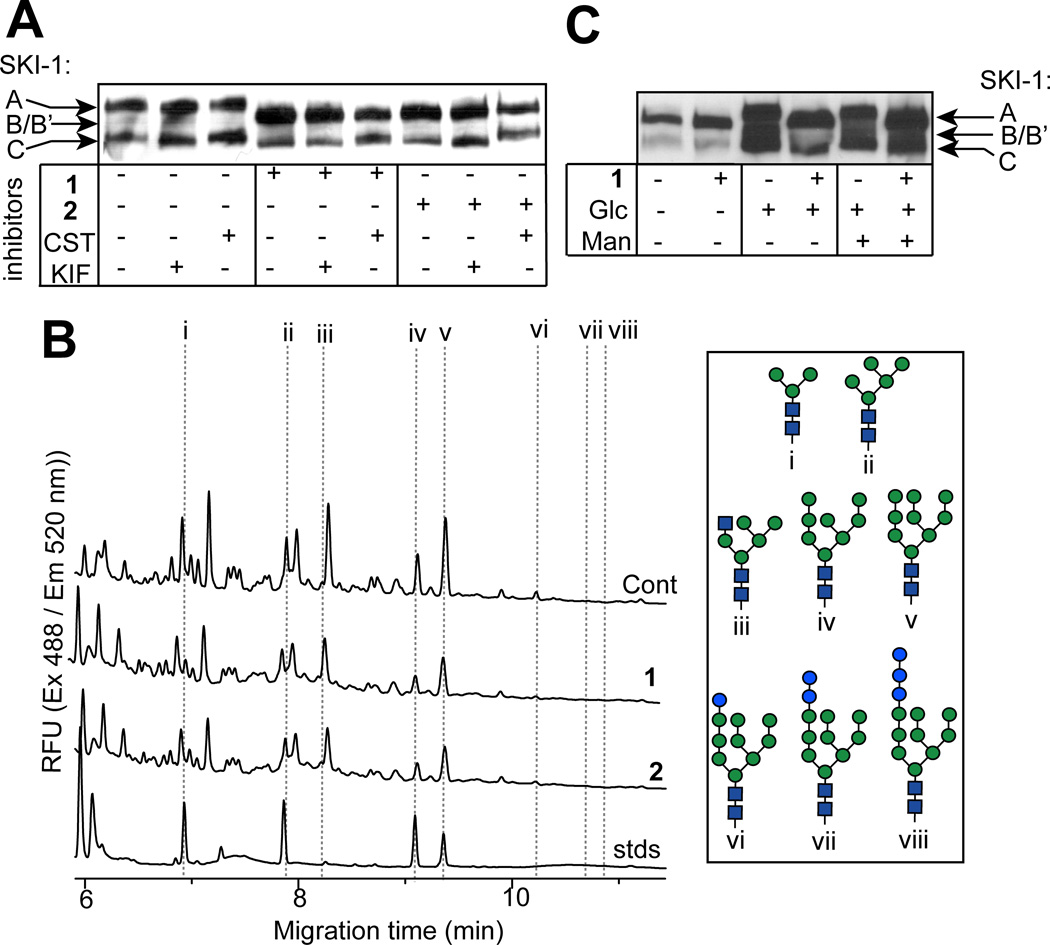
Although CDGs in which truncated DLOs are produced often display an absence of N-glycans at acceptor sites—due to the substrate specificity of the oligosaccharyltransferase (OST) for complete (mature) DLOs—CHO cell mutants have been isolated in which truncated N-glycans are transferred to proteins from incomplete DLO donors.[39] To investigate whether this occurs in CHO cells treated with 5-thiomannosides, we analyzed SKI-1 in cells treated simultaneously with these inhibitors and inhibitors of ER glycosidases (Figure 4A), which would effectively trap N-glycans transferred from untruncated DLOs in a state unprocessed by ER glycosidases or Golgi glycosyltransferases. However, if N-glycans in 5-thiomannoside-treated cells are significantly truncated prior to their transfer onto SKI-1, essentially missing the monosaccharide units normally removed by processing enzymes, blocking the activity of ER glycosidases would have little effect on later processing by Golgi transferases. The changes in SKI-1C (the only Golgi-localized isoform) electrophoretic mobility in cells co-incubated with 1 or 2 and either CST or the α-mannosidase inhibitor kifunensine (KIF) were indistinguishable from those observed in cells that were not exposed to the 5-thiomannosides. This suggests that SKI-1 in treated cells does not contain aberrant N-glycans and that the apparent decrease in the molecular weight of SKI-1 in treated cells is the result of transferring fewer numbers of otherwise complete DLO-associated oligosaccharides (Figure 2A) onto SKI-1 in treated cells.
Figure 4.
5-Thiomannosides do not cause truncated N-glycans to be transferred onto SKI-1. (A) Co-incubation of cells with either 1 or 2 and ER-glucosidase inhibitors does not decrease the electrophoretic mobility of SKI-1 nor does (B) capillary electrophoresis (CE) analysis of CHO-derived N-glycans demonstrate any qualitative difference between untreated or 5-thiomannoside-treated cells. The structures of i, ii, iv and v are inferred from their co-migration with pure standards, while peaks iii, and vi – viii can be identified due to their increased abundance in swainsonine- or canstanospermine-treated cells, respectively. (C) SKI-1 hypoglycosylation in the presence of 1 cannot be corrected by growing cells under high-nutrient conditions.
To unambiguously determine if altered N-glycan structures account for the altered mass of SKI-1, N-glycans isolated from PNGaseF-digested glycoproteins derived from both CHO K1 and SRD-12B cells, were purified and analyzed by capillary electrophoresis (CE) (Figure 4B). The most prominent intracellular N-linked oligosaccharide which could be identified in both cells lines was GlcNAc2Man9 (peak v). The identity of this glycan was established based on its co-migration with a standard of the same composition and based on the fact that this peak specifically increased when samples were obtained from cells that were treated with KIF (Supporting Information). No unusual glycans were detected upon analysis of N-glycans obtained from 5-thiomannose-treated cells, a result which cannot be attributed to the rapid N-glycan processing by glycosidases since GlcNAc2Man9 was the major high mannose oligosaccharide observed in both 1- and 2-treated cells. Similarly, no aberrant N-glycan structures were observed upon CE analysis of N-glycans derived from cells co-treated with 5-thiomannosides and CST or KIF, or in N-glycans derived from secreted glycoproteins (not shown). These data indicate that the reason for reduced glycosylation of SKI-1 is not due to altered structures of N-glycans but rather transfer of fewer numbers of otherwise normal structures, probably as a result of partially inhibited DLO biosynthesis—a common occurrence in many type I CDGs. Consequently, we saw no evidence of significant numbers of truncated N-glycans transferred to SKI-1.
In certain situations, N-glycosylation can be reduced under glucose-limiting conditions due to impaired DLO biosynthesis,[45] and furthermore, the effects of some CDGs in fibroblasts isolated from human patients could be ameliorated upon the addition of exogenous mannose.[46] Therefore, in order to investigate whether 5-thiomannosides affect DLO biosynthesis, and hence protein N-glycosylation, by altering sugar metabolism we investigated the effects of these compounds on SKI-1 derived from cells grown under nutrient-limiting conditions (Figure 4C). Glucose- deprivation (1 mM rather than the 18 mM glucose normally included in the cell-culture media) reduced SKI-1 glycosylation (lane i), an effect that was not made worse by 1 (lane ii). Although growing cells under high glucose conditions (lane iii) rescued SKI-1 N-glycosylation, nevertheless 1 still caused a reduction in SKI-1’s molecular weight (lane iv). Similarly, the addition of 5 mM mannose to the cell-culture media also failed to correct the glycosylation defect induced by the 5-thiomannoside (lanes v and vi). In summary, these results demonstrate that 1 and 2 induced protein hypoglycosylation by reducing, but not completely blocking, the extent of GlcNAc2Man9Glc3 transfer from DLOs to nascent polypeptides, effects that could not be reversed upon the addition of excess monosaccharide precursors of DLO biosynthesis.
Mannose-incorporation into DLOs is inhibited in cells treated with 5-thiomannosides
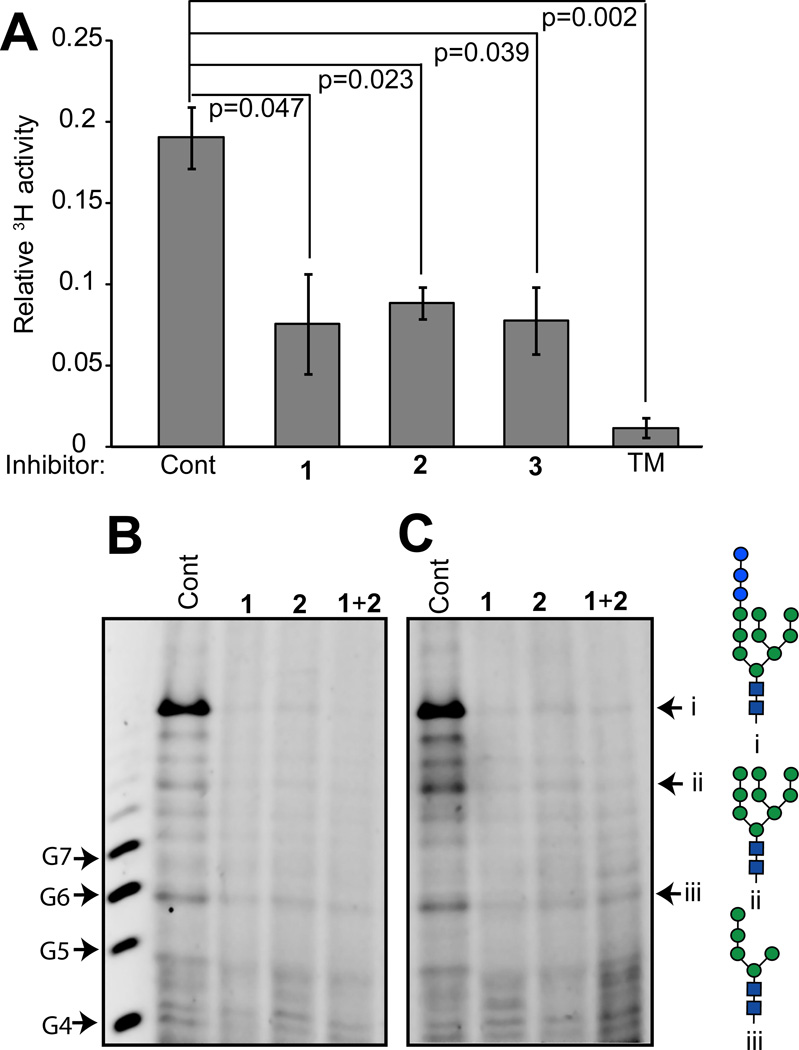
Given that the monosaccharide units of the 5-thiomannosides tested were of the mannose configuration (Figure 1A) and that inhibitors containing the α-mannose linkages common to N-glycans (Figure 2A) affected SKI-1 glycosylation equally (Figure 2C), we hypothesized that if these compounds affected DLO biosynthesis they might specifically block mannose incorporation. This hypothesis was tested by pulsing cells, pre-incubated with inhibitors, with 2-[3H]-mannose and then partitioning the various glycoconjugates into different solvents. The incorporation of [3H]-mannose into DLOs, which are soluble in a mixture of chloroform : methanol : water (10:10:3), was measured by liquid scintillation counting, and the fraction in DLOs relative to the total intracellular [3H] pool between treated and control groups of cells was compared (Figure 5A). All three 5-thiomannosides were observed to significantly decrease the amount of 3H-mannose incorporated into DLOs. Tunicamycin, a known specific inhibitor of DLO assembly, was used to validate this assay and confirm that the 5-thiomannosides blocked assembly of DLOs.
Figure 5.
5-Thiomannosides significantly inhibit the biosynthesis of dolichol-linked oligosaccharides. (A) Solvent partitioning of cell extracts into organic or aqueous solvents demonstrate that reduced amounts of [3H]-mannose are incorporated into cells treated with the 1 – 3. The [3H] activity of the DLO pools are reported as a fraction of the total amount of intracellular activity. N = 3 for each condition and the data are reported ± standard deviation. p–values were calculated using the unpaired, two-tailed student’s t-test. Fluorophore-assisted carbohydrate electrophoresis (FACE) analysis of DLOs extracted from (B) CHO K1 and (C) SRD-12B cells demonstrates that in both cell lines 1 and 2 reduced DLO biosynthesis relative to non-treated cells. The positions of standards i – iii are established through the use of purified standards. G4 – G7 refer to dextran oligomers of four to seven glucose units.
A common feature of many CDG models in which a particular DLO pathway glycosyltransferase is presumed to be defective (for example some alg mutants in yeast, Lec mutants in CHO cells, or fibroblasts isolated from human patients or animals) is the accumulation of appropriate truncated DLO intermediates. Therefore, we reasoned that if different 5-thiomannoside analogues blocked [3H]-mannose incorporation at different points in DLO biosynthesis, a variety of structures should accumulate, and furthermore, that the simultaneous addition of two different 5-thiomannosides should have an identical effect to the inhibitor which blocks the earliest acting glycosyltransferase. DLOs from 5-thiomannoside-treated cells were extracted into organic solvents, subjected to weak acid hydrolysis (to remove the dolichol-pyrophosphate moiety), permethylated, and characterized by mass spectrometry; however, no useful data were obtained. When similarly prepared DLOs from CHO K1 (Figure 5B) and SRD-12B (Figure 5C) cells treated with 1 or 2 were analyzed by fluorophore-assisted carbohydrate electrophoresis (FACE), it was observed that DLO biosynthesis was severely compromised compared to non-treated cells, although no specific intermediates were found to accumulate. The FACE system shown in Figure 5 was designed to resolve DLO glycans with structures GlcNAc2Man3 and higher. The results therefore suggest that the 5-thiomannosides either specifically inhibit glycosyltransferase steps in DLO biosynthesis earlier than GlcNAc2Man3, or prevent the synthesis of saccharide donors for those transferases.
Effects of 5-thiomannose on SKI-1 glycosylation
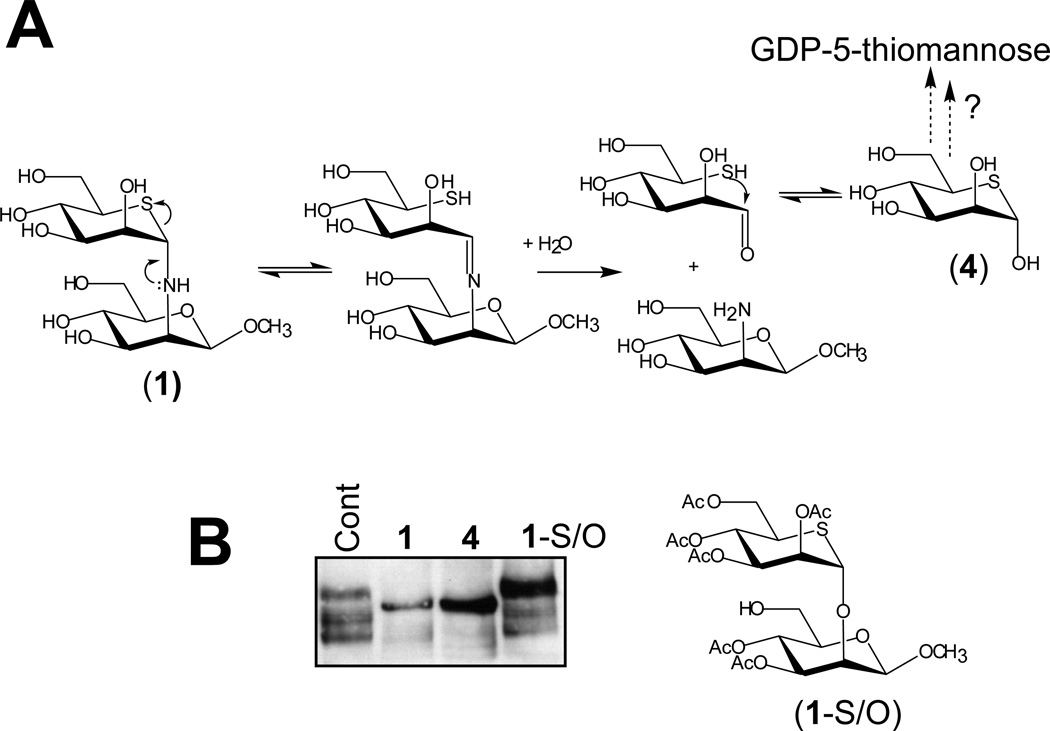
As shown in Figure 4C, the 5-thiomannosides inhibited the DLO pathway in a fashion which appeared to be unaffected by addition of exogenous mannose, suggesting that the compounds themselves or metabolites of these compounds acted in a manner that was resistant to competition by mannose. Further, we reasoned that the three different 5- thiomannosides could generate identical effects in cells if they were metabolized to the same compound. Common to 1 – 3 is the 5-thiomannose (4) moiety which, if produced within cells, may be activated as GDP-5-thiomannose (5) (Figure 6A), the 5-thio-analogue of the natural mannosyl donor GDP-mannose, which is directly or indirectly involved in mannose incorporation into DLOs (Figure 2A). GDP-5-thiomannose is a poor substrate for yeast α-(1–2)-mannosyltransferases [47]. Similarly, the 5-thio-analogues of GDP-fucose, UDP-galactose, and UDP-GlcNAc are also poor substrates for their respective glycosyltransferases.[48] Thus, we hypothesized that if 1 – 3 were hydrolyzed into 4—which itself would become activated as GDP-5-thiomannose—then 4 alone should replicate the effects of 1 – 3 on SKI-1 glycosylation. This proved to be the case as 4 alone induced SKI-1 hypoglycosylation in treated cells (Figure 6B). In contrast, 5-thioMan-α-(1–2)-O-Man (1-S/O),[22] containing an interglycosidic oxygen atom, which is resistant to the type of hydrolysis described for the S/N analogue, did not appear to affect SKI-1 glycosylation (Figure 6B). Therefore, the most likely mode of action of compounds 1–3 appears to be conversion to 4, perhaps in concert with further conversion to 5, with these compounds exerting their effects even in the presence of exogenously added mannose.
Figure 6.
The affects of the 5-thiomannosides (1–3) can be replicated by their common intermediate—5-thiomannose (4). (A) A proposed hydrolysis reaction for 1 proceeds through an iminium intermediate to yield 5-thiomannose which may possibly become activated as the poor mannosyltransferase substrate GDP-5-thiomannose. (B) 5-thiomannose replicates the affects of 1 on SKI-1 N-glycosylation and zymogen activation while a more hydrolase resistant analogue does not.
Nucleotide sugar analysis of cells treated with 1, 2 and 4
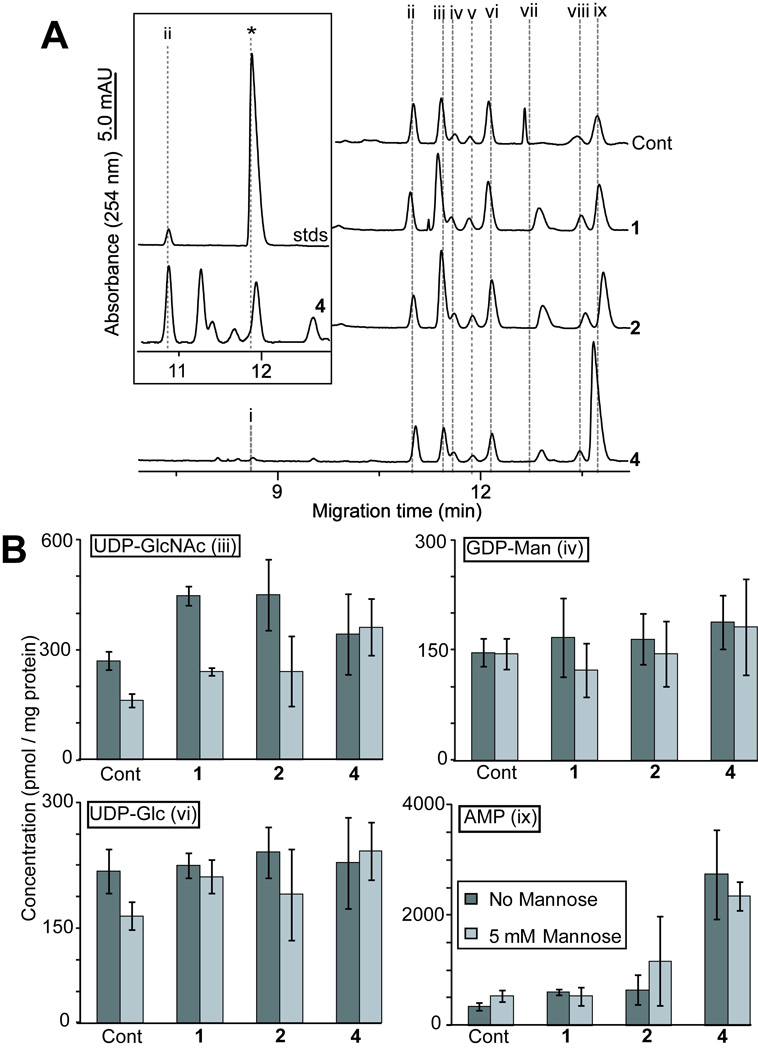
To further assess the plausibility of the 5-thiomannosides inhibiting DLO biosynthesis upon their conversion to 5-thiomannose and subsequent metabolism into GDP-5-thiomannose, we sought to detect this latter compound within treated cells. The analysis of total cellular nucleotide sugar levels would also permit the monitoring for the biosynthesis of all nucleotide-sugar precursors for the first seven reactions in the DLO pathway. Nucleotide sugars were extracted from 5-thiomannoside (1 and 2)- or 4-treated cells grown under both high- and low-mannose conditions (Figure 7A) since it was hypothesized that exogenous mannose would decrease the rate at which GDP-5-thiomannose (5) would be formed within treated cells by direct competition with 4. GDP-5-thiomannose was synthetically prepared to serve as a standard for capillary electrophoresis (CE) analysis of nucleotide sugars. This compound migrates just ahead of the naturally occurring UDP-Glc (Figure 7A, inset), and these two nucleotide sugars could be distinguished upon spiking synthetic GDP-5-thiomannose into samples of nucleotide sugars extracted from cells (not shown). Nucleotide sugar extracts from either 1-, 2- or 4-treated cells did not appear to contain any detectable GDP-5-thiomannose upon their CE analysis. Furthermore, none of the inhibitors prevented the biosynthesis of the DLO precursors UDP-GlcNAc, UDP-Glc or, importantly, GDP-Man (Figure 7B). Indeed, compounds 1 and 2 appeared to increase UDP-GlcNAc concentrations within treated cells, although the addition of 5 mM mannose to the cell culture medium prevented this effect. Significantly, only 4 caused a large increase in the AMP levels of treated cells; this increase could not be prevented in samples that had been supplemented with mannose. The inability of the 5-thiomannosides to affect AMP levels suggests that these compounds affect N-glycosylation by a different mechanism than 5-thiomannose itself.
Figure 7.
The 5-Thiomannosides and 5-thiomannose are not converted into GDP-5-thiomannose nor do they prevent the metabolism of nucleotide sugars used in DLO biosynthesis. (A) Representative CE electropherograms obtained from nucleotide sugars extracted from CHO K1 cells grown in the presence of 1, 2 and 4. The identity of peaks was established on the basis of their co-migration with the following standards: CMP-Neu5Ac (i), GDP-Glc, internal standard (ii), UDP-GlcNAc (iii), GDP-Man (iv), GDP-Fuc (v), UDP-Glc (vi), UDP-GalNAc (vii), UDP-Gal (viii) and AMP (ix). Inset is the electropherogram region of 4-treated cells predicted to contain GDP-5-thiomannose (* denotes the position of the synthetically prepared standard). GDP-Glc was added to cell lysates before extraction and the area of this peak was normalized to allow for the (B) calculation of the relative changes in nucleotide sugar levels upon treatment with inhibitors under normal (dark bars) and mannose-supplemented (to 5 mM, light bars) conditions. All data are reported ± standard deviation (n = 3).
Discussion
It is well known that conditions that interfere with the correct biosynthesis of DLOs results in N-glycosylation macroheterogeneity by causing N-glycosylation sequons to be entirely skipped by the oligosaccharyl transferase complex;[49] indeed, this is the basis of many class I congenital disorders of glycosylation. The evidence presented above demonstrates that SKI-1 hypoglycosylation in the presence of three different 5-thiomannosides ultimately results from their inhibition of DLO biosynthesis. Although these compounds profoundly inhibited DLO biosynthesis, they did not completely block this process, as demonstrated by the binding of lectins to lysates obtained from treated cells (Figure 3B) and by the normal structures of N-glycans observed through CE analysis (Figure 4B). The inhibition of DLO biosynthesis at a specific point should result in the accumulation of an oligosaccharide intermediate at the step immediately preceding the point of inhibition. It is on this basis, in fact, that many CDGs are diagnosed upon the analysis of DLO biosynthesis in fibroblasts derived from human patients [45]. Although this biosynthetic pathway was still partially functional in 5-thiomannoside-treated cells, analysis of DLO intermediates demonstrated that no specific structure accumulated (Figure 5B,C). Instead, all intermediates with GlcNAc2Man3 and above were found at greatly reduced levels, suggesting that each compound tested (due to compound availability only 1 and 2 were tested) inhibited DLO biosynthesis either at an earlier step or at multiple points; these results led to the hypothesis that the three different 5-thiomannosides inhibited DLO assembly upon their metabolism to a common intermediate, namely 5-thiomannose (4).
5-Thiomannose did induce SKI-1 hypoglycosylation (Figure 6B). One mechanism by which this compound could cause this effect is upon its activation to a poor mannosyltransferase donnor—GDP-5-thioMan. CE analysis of CHO-derived nucleotide sugars, however, did not provide any evidence that 5-thioMan itself was activated as a nucleotide sugar (Figure 7A, inset). While this observation does not rule out the possibility that the 5-thiomannosides affect DLO biosynthesis after their metabolism to 4, mass spectrometric analysis of conditioned extracts of cells that had been treated for 14 h with 1 and 2 indicated that these compounds could be recovered intact from cells (Supporting Information). Furthermore, nucleotide sugar analysis shows that the 5-thiomannosides differ from 5-thioMan in their effects on cells. The most apparent difference is that 5-thioMan causes a significant increase in the amount of AMP observed in treated cells. This effect is very similar to a phenomenon known as the “honeybee syndrome” [50]. Honeybees have low amounts of phosphomannose isomerase, the enzyme that converts Man-6-PO4 into the glycolysis intermediate Fruc-6-PO4. When honeybees are fed Man as their sole energy source it is rapidly phosphorylated to Man-6-PO4 which is very inefficiently converted to Fruc-6- PO4. The accumulating Man-6-PO4 in these insects is then degraded by a phosphatase and again phosphorylated by a rapidly decreasing supply of ATP, and so on in a futile cycle, ultimately resulting in death. 5-Thiomannose appears to have a similar effect in cells. Although sample hydrolysis during analysis and / or nucleotide extraction prevented the reliable measurement of ATP (samples were stored at 10 °C in pure H2O during CE analysis), it was nevertheless clear that in most 5-thioMan-treated samples (under both high and normal Man conditions), ATP was greatly diminished (not shown). The reduction in ATP levels in 5-thioMan-treated cells, therefore, likely induces hypoglycosylation by a mechanism similar to that observed in Glc-starved cells.[51] In further support of this hypothesis, it should be noted that the glycosylation macroheterogeneity of recombinant interferon-γ expressed in anchorage-independent CHO cells has been linked to the adenylate energy charge of these cells.[43]
In contrast to 4, the negligible effect of 1 and 2 on AMP levels indicates that the latter affect DLO levels in a different fashion. There is no apparent lack of nucleotide sugars which are DLO precursors in 5-thiomannoside-treated cells (Figure 7B), suggesting that these compounds do not inhibit DLO biosynthesis by interfering with the cell’s primary metabolism. It is tempting to speculate that if the 5-thiomannosides cause hypoglycosylation (lectin blotting demonstrates a global lack of high-Man N-glycans (Figures3A,C), then there will be less of a demand for UDP-GlcNAc, a sugar donor required for N-glycan branching and elongation into complex- and hybrid-type structures. Indeed, in cells grown in media without added mannose a slight increase in UDP-GlcNAc levels was observed in 5-thiomannoside-treated cells. Cells grown under high-mannose conditions did not cause an increase in UDP-GlcNAc levels (Figure 7B), although in the case of 1 they did not relieve the hypoglycosylation of SKI-1 (Figure 4C). Thus it appears that mannose only partially rescues cells from the effects of the 5-thiomannosides. These results suggest that the 5-thiomannosides directly inhibit early (presumably cytosolic) transferases responsible for DLO biosynthesis. Interestingly, two of the cytosolic GDP-Man-dependent transferases required to make DLOs are bifunctional enzymes. ALG 2 catalyzes the formation of both α-(1–3) and α-(1–6) bonds, forming Man-α-(1–3)-Man-α1-R, where R is Dol-PP-GlcNAc2Man1,[52] (Figure 2A, residues 4 and 5), while ALG 11 catalyzes the successive transfer of two α-(1–2)-linked Man residues to form Man-α-(1–2)-Man-α1-R[53] (Figure 2A, residues 6 and 7). 5-ThioMan-(1–3/6)-N-Man (2 and 3) may therefore both inhibit one, or both, ALG2-catalyzed reactions while 5-thioMan-(1–2)-N-Man (1) might inhibit the two ALG11-catalyzed reactions. Each of these scenarios presumes that the 5-thiomannosides act as substrate mimics and each would account for the inability to observe a single discrete DLO intermediate in treated cells. Alternatively, these compounds may act as oligosaccharide primers, accepting a mannose residue from mannosyltransferases in place of the natural DLO substrate. This hypothesis is questioned by several observations. First, unmodified 5-thiomannosides were recovered from cells and detected by mass spectrometry (Supporting Information) but no evidence was obtained for compounds consisting of 5-thiomannosides that had been elongated by a single hexose residue. Second, the addition of exogenous mannose should allow for cells to minimize the impact of these non-productive transfer reactions. Nevertheless, further experiments are necessary to distinguish between these two possibilities.
Conclusion
In summary, 5-thiomannosides containing an interglycosidic nitrogen atom specifically inhibit DLO biosynthesis in CHO K1 cells and as a result induce the hypoglycosylation of N-linked glycoproteins. These compounds have also previously been demonstrated to be active in AtT-20 cells, a mouse pituitary cell line.[7] Although the exact mechanism of their bioactivities remains to be elucidated, it is clear that this class of compounds affects an early step in DLO biosynthesis. Since the 5-thiomannosides only partially reduce protein N-glycosylation, they may be valuable tools by which the cell- and / or tissue-specific effects of class I CDGs can be chemically induced and may therefore be useful in the further characterization of the biochemical basis of the pathological effects of these disorders.
Experimental Section
Chemicals and test inhibitors
The glycosidase inhibitors castanospermine (CST), swainsonine (SW) and 1-deoxymannojirimicin (DMJ) were obtained from Toronto Research Chemicals, Inc. (North York, ON), while kifunensine (KIF) was purchased from LC Scientific Inc. (Concord, ON). Tunicamycin (TM) was purchased from Sigma (St. Louis, MO). The 5-thiomannoside inhibitors 5-thioMan-(1–2)-N-Man and 5-thioMan-(1–3)-N-Man were synthesized as previously described.[8] An acetylated, O-linked disaccharide, 5-thioMan-α-(1–2)-O-Man-OAc, was also synthesized according to reported procedures.[22] In addition, a 1,6-linked analogue, 5-thioMan-(1–6)-N-Man, was prepared by the condensation of 5-thio-D-mannose and methyl-6-amino-6-deoxy-α-D-mannopyranoside[54] under identical conditions to those reported for the 1,2- and 1,3-linked congeners (the NMR data for this compound are given in the Supplementary Material). All inhibitors were stored at −20 °C as stock solutions in H2O, with the exception of 5-thioMan-(1–2)-O-Man-OAc which was soluble in DMSO. Due to their presumed poor membrane permeability, all disaccharide analogues were used at 5 mM except for the O-acetylated derivative which was tested at 150 µM. The known glycosidase inhibitors were typically tested at concentrations of 1.2, 0.4 and 5 mM for CST, KIF, and DMJ, respectively. TM was used at a concentration of 2 µg / mL. GDP-5-thio-D-mannose was synthesized as previously reported [47] and purified by ion-paired HPLC.[55] All other nucleotide sugars were purchased from Sigma and stock solutions in H2O were stored at −20 °C.
Cell lines and culture
Cells were maintained in monolayer culture on 10 cm tissue culture plates at 5 % CO2 and 37 °C. CHO K1 and CHO SRD-12B cells[56] stably expressing V5-epitope tagged human SKI-1[57] were both grown in a 1:1 mixture of DMEM and Ham’s F12 medium (Invitrogen / Gibco, Carlsbad, CA) containing 5 % (v/v) FBS (Gibco), unless otherwise indicated. Gentamicin (Sigma) was always included in the medium of SRD-12B cells at a concentration of 400 µg / mL but was excluded when inhibitors were tested. Cells were routinely passed using trypsin-EDTA (Sigma) every three to four days. For experiments performed under nutrient limiting conditions, cells were grown in glucose- and serum-free DMEM (Gibco) supplemented with 1 or 10 mM glucose or 10 mM glucose + 5 mM mannose and 0.3 mM DL-proline.
Immunoblot analysis of SKI-1 zymogen activation
Confluent monolayers of SRD-12B cells grown on 3.5 cm culture plates were incubated in the presence or absence of inhibitors for 14 h after which the media was removed and cells were washed with ice cold PBS. Cells were lysed, on ice, by adding 1× radioimmunoprecipitation assay (RIPA) (100 µL) buffer containing a 1x protease inhibitor cocktail (Roche, Basel, SW). Cells were scraped off the plates, and the resulting suspension was vigorously vortexed and centrifuged (10,000 rpm, 10 min, 4 °C). The supernatants were resolved by SDS-PAGE on 6 % gels run at 4 °C and transferred to BioRad (Hercules, CA) Trans-Blot nitrocellulose membranes. SKI-1 was detected by probing the blots with HRP-conjugated anti-V5 antibodies (Bethyl Laboratories, Montgomery, TX) (1:8,000 dilution) in PBS + 0.1% Tween-20 (PBS-T) containing 5 % skim milk powder, over night at 4 °C. Immunoreactive material was visualized using the GE Healthcare (Little Chalfont, UK) ECL Plus western blot detection kit according to the manufacturer’s instructions.
Lectin blot analysis of CHO K1 cell lysates
CHO K1 cells were treated with inhibitors, and lysates were prepared as described above. Protein concentrations were measured by the BioRad DC Protein Assay and normalized amounts were resolved on 10 % polyacrylamide gels by SDS-PAGE. Identical amounts of each sample were analyzed in each lectin blot. Controls were prepared by treating cell lysates, obtained from non-treated cells, with PNGaseF or, for GNA only, ENDO H (New England Biolabs, Inc., Ipswich, MA) according to the manufacturer’s instructions. ConA, (EY Laboratories, Inc., San Mateo, CA), DSA and GNA (Vector Laboratories, Inc., Burlingame, CA) were purchased as their biotin-conjugates and used at dilutions of 1:8000 (ConA) and 1:500 (DSA and GNA) in PBS-T. Blots were incubated with lectins at 4 °C overnight, washed for 1 h with 4 changes of PBS-T, then blocked with 1% BSA for 0.5 h, and subsequently probed with Thermo / Pierce (Rockford, IL) HRP-conjugated streptavidin. After washing blots for an additional 1 h, bound lectins were visualized with ECL Plus.
Isolation and purification of N-glycans
Confluent monolayers of CHO K1 or SRD-12B cells (~2×106 cells / plate) grown in the presence of inhibitors for 14 h were washed with ice cold PBS (1×10 mL) before harvesting by gently scraping them off the plates. Cells were pelleted by centrifugation (10 min, 1000 rpm, 4 °C), and N-glycans were obtained according to methods adopted by the Consortium for Functional Glycomics [58], with slight modifications. Briefly, cell pellets were suspended in 50 mM Tris, pH 7.4 + 0.1 % (w/v) SDS, sonicated (2 blasts×30 s, 10 % power) with a W-375 ultrasonic processor (Heat Systems Ultrasonics, Inc.), dialyzed across a 3.5 kDa membrane (Spectrum Laboratories, Inc.) against 50 mM NH4HCO3, pH 7.4 (2×24 h), and lyophilized. Proteins were reduced in 2 mg / mL dithiothreitol (Sigma) in 0.6 M Tris, pH 8.5 (1 h, 37 °C), alkylated upon the addition of iodoacetamide (Sigma) to 6 mg / mL (2 h, 20 °C, dark), exchanged into 50 mM NH4HCO3, pH 8.5 using 10 kDa NMWCO centrifugal filter units (Millipore), and lyophilized. The reduced and alkylated samples were digested with 1 mg / mL porcine trypsin (Sigma) in 50 mM NH4HCO3, pH 8.5 and the resulting peptides were purified by solid-phase extractrion (SPE) on C18 cartridges (Waters). Material eluting with 20 – 60 % (v / v) 1-propanol containing 5 % (v / v) AcOH was pooled, lyophilized, and the N-glycans on the resulting glycopeptides were released upon treatment with 40 U / µL PNGaseF (24 h, 37 °C). Buffer salts and peptides were removed from the N-glycans using 200 mg porous graphite-containing Hypercarb SPE cartridges (Thermo Scientific) essentially according to the method of Redmond and Packer.[59] Appropriate SPE fractions were passed through 0.22 µm filters and lyophilized.
Capillary electrophoresis analysis of N-glycans
Dried N-glycans were labelled by reductive amination with 4 µL 100 mM 8-aminopyrene-1,3,6-trisulfonate (APTS) (Beckman-Coulter, Mississauga, ON) and 500 mM NaBH3CN (Fluka) in 50 % (v / v) THF containing 7.5 % (v / v) acetic acid, as described by Guttman et al.[60] The labelling was terminated by the addition of 18 MΩ H2O to 100 µL final volume. Samples were typically further diluted with H2O before CE analysis. CE separations were carried out using a ProteomeLab PA800 (Beckman-Coulter) equipped with a laser-induced fluorescence (LIF) detector and a 488 nm argon-ion laser. All separations were carried out in reverse polarity using a 25 mM NaOAc buffer, pH 4.74, containing polymeric additives (Beckman-Coulter), a constant voltage of 30 kV and coated N-CHO capillaries (Beckman-Coulter) of 50 µm internal diameter×50 cm effective length. All injections were made from the cathode side of the capillary by applying a pressure of 0.5 psi for 10 s. The identity of key N-glycans was established through the use of purified standards (Prozyme / Glyko, Hayward, CA) labelled with APTS as described above or by comparison with N-glycans obtained from cells treated with CST or SW.
3H-mannose incorporation into lipid-linked oligosaccharide (DLOs) pools
Confluent monolayers (~107 cells) of CHO K1 cells were pre-incubated for 5 h with inhibitors before they were harvested with 37 °C PBS containing 10 mM EDTA, transferred to a glass tube, and washed with DMEM containing 1 mM glucose. Cells were re-suspended in media containing inhibitors and 1 mCi/mL 2-[3H]-mannose (American Radiolabeled Chemicals, Inc., St. Louis, MO), and incubated at 37 °C with gentle agitation for 15 min. Samples were chased for an additional 5 min with media containing 10 mM mannose and immediately pelleted by centrifugation (3000×g, 4 °C, 10 min) in a pre-chilled centrifuge. Cell pellets were sequentially extracted with CM (chloroform : methanol = 2 : 1,v / v), H2O, and CMW (chloroform : methanol : H2O = 10 : 10 : 3, v / v / v) [61] and the 3H activity incorporated into each solvent(s) was measured by scintillation counting. The extent of 2-[3H]-mannose incorporation into DLO pools was determined by calculating the ratio of 3H activity in the CMW fraction relative to the total intracellular activity.
Fluorophore-assisted electrophoresis (FACE) analysis of DLOs
DLOs were extracted and analyzed by FACE exactly as previously described.[61] Each sample was derived from the pooled contents of two 10 cm plates of cells that had been exposed to inhibitors for 24 h. Briefly, cell monolayers were washed twice with ice cold phosphate-bufered saline, then harvested by scraping into methanol followed by disruption in a bath sonicator. The cellular residue was dried by centrifugal evaporation under vacuum, and then sequentially extracted with CM (2:1), H2O, and CMW (10:10:3). The CMW extract was applied to a DEAE-cellulose (acetate form) column. Contaminants were removed by elution with 3 mM NH4OAc (in CMW), and DLOs were recovered by elution with 300 mM NH4OAc (in CMW). The DLO fraction was desalted by phase separation and the glycan portion was released from dolichol upon weak acid treatment. Charged contaminants were eliminated by passing the released glycans over an ion exchange resin. The glycans were then labeled with ANDS and resolved on a FACE oligosaccharide profiling gel.
Extraction and purification of nucleotide sugars
CHO K1 cells were extracted into 70 % ethanol according to established procedures.[62, 63] The crude extracts were spiked with GDP-Glc (200 pmol) as an internal standard, dissolved in 18 MΩ H2O (0.5 mL), and extracted using Supleco (Bellefonte, PA) EnviCarb SPE cartridges (200 mg).[55] Sugar nucleotides were stored at −20 °C until analysis. Extracts were prepared from cells grown in normal media or under high mannose (5 mM) conditions.
Analysis of sugar nucleotide pools by CE
Extracted sugar nucleotides were dissolved in H2O (200 µL) and an aliquot was diluted 1 : 4 prior to characterization by CE on a ProteomeLab PA800 according to the method of Feng et al.[64] which had been adapted with an electrokinetic sample injection method as described by Chien and Burgi.[65] Electrophoresis was carried out at a constant voltage of 26 kV (which produced a current of roughly 80 – 90 µA) rather than 30 kV as previously used by Gloster et al.[48] as this allowed for better resolution between UDP-GlcNAc and GDP-Man. Electropherograms were derived by measuring the absorbance at 254 (± 10) nm at a rate of 4 Hz. Peaks were integrated using 32 Karat 5.0 software (Beckman-Coulter) and all data were normalized to the area of the GDP-Glc internal standard. The concentrations of sugar nucleotides and nucleotide phosphates within samples were determined from their respective calibration curves (Supplementary Material). Care was taken to ensure that standards and samples contained equal amounts of internal standard (GDP-Glc) and were injected for an equal length of time. All extractions were performed in triplicate, and in each case the nucleotide sugar concentration is expressed relative to the amount of protein (as a proxy of cell number) in each sample.
Supplementary Material
Acknowledgements
We thank B. D. Johnston and G. Gu for synthesizing the 5-thiomannosides (1 and 2) and 5-thiomannose (4). BMP is grateful to the Natural Sciences and Engineering Research Council of Canada for financial support. MAL thanks the National Institutes of Health (GM38545) and the Welch Foundation (I-1168). Part of this work was supported by grants from the Canadian Institutes for Health Research (MOP 44363) and a Canada Research Chair (216684) to NGS.
Abbreviations
- APTS
8-aminopyrene-1,3,6-trisulfonate
- CDG
congential disorder of glycosylation
- CE
capillary electrophoresis
- CHO
Chinese hamster ovary
- ConA
Concanavalin A
- CST
castanospermine
- DLO
dolichol-linked oligosaccharide
- DMEM
Dulbeco’s modified Eagle medium
- DMJ
deoxymannojirimycin
- DSA
Dantura stramonium lectin
- ER
endoplasmic reticulum
- FACE
fluorophore-assisted carbohydrate electrophoresis
- Fruc
fructose
- Fuc
fucose
- Gal
galactose
- GDP
guanosine diphosphate
- GNA
galanthus nivalis lectin
- GalNAc
N-acetylgalactosamine
- Glc
glucose
- GlcNAc
N-acetylglucosamine
- Man
mannose
- Neu5Ac
N-acetylneuraminic acid
- SREBP-2
sterol-regulatory-element-binding protein-2
- SKI-1
subtilisin/kexin like isozyme-1
- SRD-12B
sterol regulatory defective-12B cells
- 5-thioMan
5-thiomannose
- TM
tunicamycin
- UDP
uridine diphosphate
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org.proxy.lib.sfu.ca/10.1002/chem.2010xxxxx.
References
- 1.Burda P, Jakob CA, Beinhauer J, Hegemann JH, Aebi M. Glycobiology. 1999;9:617–625. doi: 10.1093/glycob/9.6.617. [DOI] [PubMed] [Google Scholar]
- 2.Tremblay LO, Kovács EN, Daniels E, Wong NK, Sutton-Smith M, Morris HR, Dell A, Marcinkiewicz E, Seidah NG, McKerlie C, Herscovics A. J. Biol. Chem. 2007;282:2558–2566. doi: 10.1074/jbc.M608661200. [DOI] [PubMed] [Google Scholar]
- 3.Haltiwanger RS, Lowe JB. Annu. Rev. Biochem. 2004;73:491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- 4.Dennis JW, Nabi IR, Demetriou M. Cell. 2009;139:1229–1241. doi: 10.1016/j.cell.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alavi A, Axford JS. Rheumatology. 2008;47:760–770. doi: 10.1093/rheumatology/ken081. [DOI] [PubMed] [Google Scholar]
- 6.Brown JR, Crawford BE, Esko JD. Crit. Rev. Biochem. Mol. Biol. 2008;42:481–515. doi: 10.1080/10409230701751611. [DOI] [PubMed] [Google Scholar]
- 7.Zandberg WF, Benjannet S, Hamelin J, Pinto BM, Seidah NG. Glycobiology. 2011;21:1290–1300. doi: 10.1093/glycob/cwr060. [DOI] [PubMed] [Google Scholar]
- 8.Johnston BD, Pinto BM. J. Org. Chem. 1998;63:5797–5800. doi: 10.1021/jo980168r. [DOI] [PubMed] [Google Scholar]
- 9.Lenz O, ter Meulen J, Klenk H-D, Seidah NG, Garten W. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12701–12705. doi: 10.1073/pnas.221447598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seidah NG, Khatib AM, Prat A. Biol. Chem. 2006;387:871–877. doi: 10.1515/BC.2006.110. [DOI] [PubMed] [Google Scholar]
- 11.Yuasa H, Hashimoto H. Rev. Heteroatom Chem. 1999;19:35–65. [Google Scholar]
- 12.Whistler RL, Es TV. J. Org. Chem. 1963;28:2303–2304. [Google Scholar]
- 13.Johnston BD, Indurugalla D, Pinto BM, Bennet AJ. J. Am. Chem. Soc. 2001;123:12698–12699. doi: 10.1021/ja017118f. [DOI] [PubMed] [Google Scholar]
- 14.Andrews JS, Weimar T, Frandsen TP, Svensson B, Pinto BM. J. Am. Chem. Soc. 1995;117:10799–10804. [Google Scholar]
- 15.Yuasa H, Hindsgaul O, Palcic MM. J. Am. Chem. Soc. 1992;114:5891–5892. [Google Scholar]
- 16.Mehta S, Andrews JS, Svensson B, Pinto BM. J. Am. Chem. Soc. 1995;117:9783–9790. [Google Scholar]
- 17.Rangarajan M, Hartley BS. Biochem. J. 1992;283:223–233. doi: 10.1042/bj2830223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machado de Domenech EE, Sols A. FEBS Lett. 1980;119:174–176. doi: 10.1016/0014-5793(80)81024-6. [DOI] [PubMed] [Google Scholar]
- 19.Kajimoto T, Liu KKC, Pederson RL, Zhong Z, Ichikawa Y, Porco JA, Wong CH. J. Am. Chem. Soc. 1991;113:6187–6196. [Google Scholar]
- 20.Izumi M, Tsuruta O, Hashimoto H, Yazawa S. Tetrahedron. Lett. 1996;37:1809–1812. [Google Scholar]
- 21.Randell KD, Johnston BD, Pinto BM. Carbohydr. Res. 2000;326:145–150. doi: 10.1016/s0008-6215(00)00044-6. [DOI] [PubMed] [Google Scholar]
- 22.Johnston BD, Pinto BM. Carbohydr. Res. 1998;310:17–25. [Google Scholar]
- 23.Hironobu H, Masashi K, Hideya Y. Chem. Eur. J. 1996;2:556–560. [Google Scholar]
- 24.Ding Y, Hindsgaul O. Bioorg. Med. Chem. Lett. 1998;8:1215–1220. doi: 10.1016/s0960-894x(98)00198-x. [DOI] [PubMed] [Google Scholar]
- 25.Izumi M, Suhara Y, Ichikawa Y. J. Org. Chem. 1998;63:4811–4816. [Google Scholar]
- 26.Yuasa H, Jouyabu M, Mitsuhashi N, Hashimoto H. Res. Lett. Org. Chem. 2008 4 pages. [Google Scholar]
- 27.Chung SJ, Takayama S, Wong C-H. Bioorg. Med. Chem. Lett. 1998;8:3359–3364. doi: 10.1016/s0960-894x(98)00618-0. [DOI] [PubMed] [Google Scholar]
- 28.Möller G, Reck F, Paulsen H, Kaur K, Sarkar M, Schachter H, Brockhausen I. Glycoconjugate J. 1992;9:180–190. doi: 10.1007/BF00731163. [DOI] [PubMed] [Google Scholar]
- 29.Subramaniam V, Gurcha SS, Besra GS, Lowary TL. Bioorg. Med. Chem. 2005;13:1083–1094. doi: 10.1016/j.bmc.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 30.Helland A-C, Hindsgaul O, Palcic MM, Stults CLM, Macher BA. Carbohydr. Res. 1995;276:91–98. doi: 10.1016/0008-6215(95)00165-p. [DOI] [PubMed] [Google Scholar]
- 31.Lu P-P, Hindsgaul O, Li H, Palcic MM. Carbohydr. Res. 1997;303:283–291. doi: 10.1016/s0008-6215(97)00174-2. [DOI] [PubMed] [Google Scholar]
- 32.Reck F, Springer M, Meinjohanns E, Paulsen H, Brockhausen I, Schachter H. Glycoconjugate J. 1995;12:747–754. doi: 10.1007/BF00731234. [DOI] [PubMed] [Google Scholar]
- 33.Laferté S, Chan NWC, Sujino K, Lowary TL, Palcic MM. Eur. J. Biochem. 2000;267:4840–4849. doi: 10.1046/j.1432-1327.2000.01544.x. [DOI] [PubMed] [Google Scholar]
- 34.Kuan SF, Byrd JC, Basbaum C, Kim YS. J. Biol. Chem. 1989;264:19271–19277. [PubMed] [Google Scholar]
- 35.Sarkar AK, Brown JR, Esko JD. Carbohydr. Res. 2000;329:287–300. doi: 10.1016/s0008-6215(00)00200-7. [DOI] [PubMed] [Google Scholar]
- 36.Sarkar AK, Fritz TA, Taylor WH, Esko JD. Proc. Natl. Acad. Sci.USA. 1995;92:3323–3327. doi: 10.1073/pnas.92.8.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freeze HH, Aebi M. Cur. Opin. Struct. Biol. 2005;15:490–498. doi: 10.1016/j.sbi.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 38.Silberstein S, Gilmore R. The FASEB J. 1996;10:849–858. [PubMed] [Google Scholar]
- 39.Patnaik SK, Stanley P, Minoru F. Meth. Enzymol. 2006;416:159–182. doi: 10.1016/S0076-6879(06)16011-5. [DOI] [PubMed] [Google Scholar]
- 40.Hong Y, Sundaram S, Shin D-J, Stanley P. J. Biol. Chem. 2004;279:49894–49901. doi: 10.1074/jbc.M410121200. [DOI] [PubMed] [Google Scholar]
- 41.Butler M. App. Microbiol. Biotech. 2005;68:283–291. doi: 10.1007/s00253-005-1980-8. [DOI] [PubMed] [Google Scholar]
- 42.Jaeken J, van Eijkb HG, van der Heulb C, Corbeela L, Eeckelsa R, Eggermonta E. Clin. Chim. Acta. 1984;144:245–247. doi: 10.1016/0009-8981(84)90059-7. [DOI] [PubMed] [Google Scholar]
- 43.Kochanowski N, Blanchard F, Cacan R, Chirat F, Guedon E, Marc A, Goergen JL. Biotech. Bioeng. 2008;100:721–733. doi: 10.1002/bit.21816. [DOI] [PubMed] [Google Scholar]
- 44.Yang M, Butler M. Biotech. Bioeng. 2000;68:370–380. doi: 10.1002/(sici)1097-0290(20000520)68:4<370::aid-bit2>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 45.Gao N, Shang J, Lehrman MA. J. Biol. Chem. 2005;280:17901–17909. doi: 10.1074/jbc.M500510200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rush JS, Panneerselvam K, Waechter CJ, Freeze HH. Glycobiol. 2000;10:829–835. doi: 10.1093/glycob/10.8.829. [DOI] [PubMed] [Google Scholar]
- 47.Tsuruta O, Yuasa H, Hashimoto H, Sujino K, Otter A, Li H, Palcic MM. J. Org. Chem. 2003;68:6400–6406. doi: 10.1021/jo0300035. [DOI] [PubMed] [Google Scholar]
- 48.Gloster TM, Zandberg WF, Heinonen JE, Shen DL, Deng L, Vocadlo DJ. Nat. Chem. Biol. 2011;7:174–181. doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karaoglu D, Kelleher DJ, Gilmore R. Biochem. 2001;40:12193–12206. doi: 10.1021/bi0111911. [DOI] [PubMed] [Google Scholar]
- 50.Sols A, Cadenas E, Alvarado F. Science. 1960;131:297–298. doi: 10.1126/science.131.3396.297. [DOI] [PubMed] [Google Scholar]
- 51.Rearick JI, Chapman A, Kornfeld S. J. Biol. Chem. 1981;256:6255–6261. [PubMed] [Google Scholar]
- 52.Kämpf M, Absmanner B, Schwarz M, Lehle L. J. Biol. Chem. 2009;284:11900–11912. doi: 10.1074/jbc.M806416200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Reilly MK, Zhang G, Imperiali B. Biochem. 2006;45:9593–9603. doi: 10.1021/bi060878o. [DOI] [PubMed] [Google Scholar]
- 54.Reina JJ, Maldonado OS, Tabarani G, Fieschi F, Rojo J. Bioconj. Chem. 2007;18:963–969. doi: 10.1021/bc060369z. [DOI] [PubMed] [Google Scholar]
- 55.Räbinä J, Mäki M, Savilahti EM, Järvinen N, Penttilä L, Renkonen R. Glycoconjugate J. 2001;18:799–805. doi: 10.1023/a:1021107602535. [DOI] [PubMed] [Google Scholar]
- 56.Rawson RB, Cheng D, Brown MS, Goldstein JL. J. Biol. Chem. 1998;273:28261–28269. doi: 10.1074/jbc.273.43.28261. [DOI] [PubMed] [Google Scholar]
- 57.Elagoz A, Benjannet S, Mammarbassi A, Wickham L, Seidah NG. J. Biol. Chem. 2002;277:11265–11275. doi: 10.1074/jbc.M109011200. [DOI] [PubMed] [Google Scholar]
- 58.JangLee J, North SJ, SuttonSmith M, Goldberg D, Panico M, Morris H, Haslam S, Dell A, Minoru F. Meth. Enzym. 2006;415:59–86. doi: 10.1016/S0076-6879(06)15005-3. [DOI] [PubMed] [Google Scholar]
- 59.Redmond JW, Packer NH. Carbohydr. Res. 1999;319:74–79. [Google Scholar]
- 60.Guttman A, Chen FTA, Evangelista RA, Cooke N. Anal. Biochem. 1996;233:234–242. doi: 10.1006/abio.1996.0034. [DOI] [PubMed] [Google Scholar]
- 61.Gao N, Lehrman MA. Meth. Enzym. 2006;415:3–20. doi: 10.1016/S0076-6879(06)15001-6. [DOI] [PubMed] [Google Scholar]
- 62.Tomiya N, Ailor E, Lawrence SM, Betenbaugh MJ, Lee YC. Anal. Biochem. 2001;293:129–137. doi: 10.1006/abio.2001.5091. [DOI] [PubMed] [Google Scholar]
- 63.Ferguson MAJ, Turnock DC. Eukaryotic Cell. 2007;6:1450–1463. doi: 10.1128/EC.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feng H-T, Wong N, Wee S, Lee MM. J. Chromatogr. B. 2008;870:131–134. doi: 10.1016/j.jchromb.2008.05.038. [DOI] [PubMed] [Google Scholar]
- 65.Chien R-L, Burgi DS. J. Chromatogr. A. 1991;559:141–152. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.