SUMMARY
Nephronophthisis-related ciliopathies (NPHP-RC) are degenerative recessive diseases that affect kidney, retina and brain. Genetic defects in NPHP gene products that localize to cilia and centrosomes defined them as ‘ciliopathies’. However, disease mechanisms remain poorly understood. Here we identify by whole exome resequencing, mutations of MRE11, ZNF423, and CEP164 as causing NPHP-RC. All three genes function within the DNA damage response (DDR) pathway, hitherto not implicated in ciliopathies. We demonstrate that, upon induced DNA damage, the NPHP-RC proteins ZNF423, CEP164 and NPHP10 colocalize to nuclear foci positive for TIP60, known to activate ATM at sites of DNA damage. We show that knockdown of CEP164 or ZNF423 causes sensitivity to DNA damaging agents, and that cep164 knockdown in zebrafish results in dysregulated DDR and an NPHP-RC phenotype. We identify TTBK2, CCDC92, NPHP3 and DVL3 as novel CEP164 interaction partners. Our findings link degenerative diseases of kidney and retina, disorders of increasing prevalence, to mechanisms of DDR.
INTRODUCTION
Nephronophthisis (NPHP) is a recessive cystic kidney disease that represents the most frequent genetic cause of end-stage kidney disease in the first three decades of life. NPHP-related ciliopathies (NPHP-RC) are single-gene recessive disorders that affect kidney, retina, brain and liver by prenatal-onset dysplasia or by organ degeneration and fibrosis in early adulthood. Identification of recessive mutations in more than 10 different genes (NPHP1-NPHP10) (Attanasio et al., 2007; Delous et al., 2007; Hildebrandt et al., 1997; Mollet et al., 2002; Olbrich et al., 2003; Otto et al., 2002; Otto, 2005; Otto et al., 2010a; Otto et al., 2003; Otto et al., 2008; Sayer et al., 2006; Valente et al., 2006) revealed that their gene products share localization at the primary cilia-centrosomes complex and mitotic spindle poles in a cell cycle dependent manner, characterizing them as retinal-renal “ciliopathies” (Ansley et al., 2003; Hildebrandt et al., 2011). Multiple signaling pathways downstream of cilia have been implicated in the disease mechanisms of NPHP-RC, including Wnt signaling (Germino, 2005; Simons et al., 2005) and Shh signaling (Huangfu and Anderson, 2005; Huangfu et al., 2003). However, despite convergence of ciliopathy pathogenesis at cilia and centrosomes it remains largely unknown what signaling pathways downstream of cilia and centrosome function operate in the disease mechanisms that generate the NPHP-RC phenotypes.
Centrosomal proteins have been recently implicated in DNA damage response (DDR). Both pericentrin (PCNT), a core centrosomal protein (Doxsey et al., 1994), as well as CEP152, encoding a centrosomal protein required for centriolar duplication (Blachon et al., 2008), are defective in Seckel syndrome, an autosomal recessive disorder characterized by dwarfism, microcephaly and mental retardation (Griffith et al., 2008; Kalay et al., 2011; Rauch et al., 2008). Consistent with the first mutation identified in Seckel syndrome being in ataxia-telangiectasia mutated and RAD3-related (ATR), a key phosphoinositide 3-kinase-related protein kinase involved in DDR signaling (O’Driscoll et al., 2003), PCNT- and CEP152-mutant cells are also defective in ATR-dependent DDR signaling, although the mechanism of the signaling defect is not fully understood. It has been proposed that PCNT signals G2/M arrest by affecting centrosomal localization of CHK1 (Tibelius et al., 2009). CEP152 interacts with CINP, a cell-cycle checkpoint protein, which itself binds to ATR-interacting protein (ATRIP) and regulates ATR-dependent signaling, resistance to replication stress, and G2 checkpoint integrity (Lovejoy et al., 2009).
Recessive null mutations in certain NPHP genes cause severe, congenital-onset phenotypes (Meckel syndrome) of dysplasia and malformation in kidney (polycystic dysplastic kidneys), eye (coloboma/microphthalmia), cerebellum (vermis hypoplasia in Joubert syndrome), and liver (cysts, ductal plate malformation), whereas hypomorphic mutations in the same gene cause late-onset, degenerative phenotypes such as renal tubular degeneration with fibrosis (nephronophthisis), retinal degeneration (Senior-Loken syndrome; SLSN), and liver fibrosis (Chaki et al., 2011; Hildebrandt et al., 2011). However, disease mechanisms of neither the dysplastic nor the degenerative phenotypes of NPHP-RC are understood. This situation is partially due to the fact that the more than 10 known NPHP genes explain less than 50% of all cases with NPHP-RC, and that many of the single-gene causes of NPHP-RC are still unknown (Otto et al., 2010b). The finding that some of the recently identified genetic causes of NPHP-RC are exceedingly rare (Attanasio et al., 2007) necessitates the ability to identify novel single-gene causes of NPHP-RC in single affected families. To achieve this goal we developed a strategy that combines homozygosity mapping with whole exome resequencing (WER) (Otto et al., 2010a). Because this approach allows identification of multiple different causes of NPHP-RC within a short time frame it has the potential of delineating pathogenic pathways.
Using this approach, we identify here mutations in 3 new NPHP-RC genes, MRE11, ZNF423, and CEP164, which together suggest involvement of a DNA damage response (DDR) signaling pathway in NPHP-RC pathogenesis.
RESULTS
Whole exome resequencing accelerates discovery of NPHP-RC genes
Identification of monogenic causes of ciliopathies is limited by their rarity (Attanasio et al., 2007), necessitating methods to identify ciliopathy-causing genes in single families, which include whole exome resequencing (WER). However, WER typically yields hundreds of variants from normal reference sequence as “candidate mutations” (Ng et al., 2009), whereas only a single-gene mutation will represent the disease cause. To overcome this limitation, we here combined WER with homozygosity mapping (Hildebrandt et al., 2009c) in sib pairs affected with NPHP-RC and performed functional analysis of the identified genes (Otto et al., 2010a).
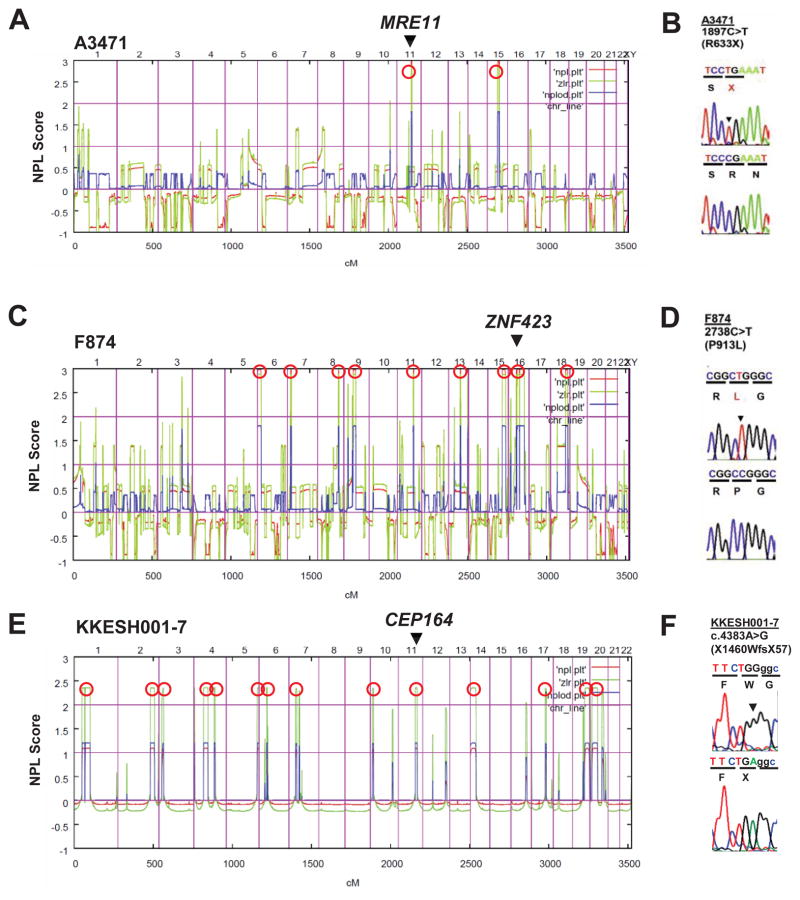
Homozygosity mapping yielded positional candidate regions of homozygosity by descent (Hildebrandt et al., 2009c) in families A3471 (2 regions), F874 (9 regions), and KKESH001-7 (14 regions) (Figure 1), who had one or more features of NPHP-RC, including NPHP, retinal degeneration, liver fibrosis, or cerebellar degeneration/hypoplasia (Table 1). We then performed WER in one affected individual of each of the three NPHP-RC families (Ng et al., 2009; Otto et al., 2010a). Remarkably, each of three NPHP-RC genes consecutively identified by this approach, MRE11, ZNF423 and CEP164, suggested a functional connection to the DDR pathway (Figure 1, Table 1).
Figure 1. Identification of recessive mutations in MRE11, ZNF423 and CEP164 in NPHP-RC using homozygosity mapping and WER.
Data regarding homozygosity mapping and mutations are shown for family A3471 with MRE11 mutation (A–B), family F874 with ZNF423 mutation (C–D), and family KKESH001 with CEP164 mutation (E–F). (A, C, E) Non-parametric lod scores (NPL) are plotted across the human genome in 3 families (A3471, F874 and KKESH001) with NPHP-RC (see also Table 1). The x-axis shows SNP positions on human chromosomes concatenated from p-ter (left) to q-ter (right). Genetic distance is given in cM. Maximum NPL peaks (Hildebrandt et al., 2009b) (red circles) indicate candidate regions of homozygosity by descent. The genes MRE11, ZNF423 and CEP164 are positioned (arrow heads) within one of the maximum NPL peaks.
(B, D, F) Homozygous mutations detected in families with NPHP-RC. Family number (underlined), mutation (arrowheads) and predicted translational changes (in parenthesis) are indicated (see also Table 1). Sequence traces are shown for mutations above normal controls. (For additional mutations in other families see also Table 1 and Figure S2).
Table 1.
Mutations of MRE11A, ZNF423 and CEP164 in families with NPHP-RC.
| Family-Individual | Ethnic origin | Nucleotide alterationa,b (Hg19 position) | Deduced protein change | Exon/Intron (state) | Continuous amino acid sequence conservation | Parental consanguinity | Kidney (age at ESKF) | Eye (age at RD) | Other (at age) |
|---|---|---|---|---|---|---|---|---|---|
| MRE11 | |||||||||
| F3471 -21 -22 |
Pakistani | c.1897C>T (Chr11: 94,170,372) | p.R633X | 16 (hom) | N/A | Yes | No renal failure | normal | -21: CVA (MRI), ataxia, dysarthria, myoclonus -22: CVA (MRI), ataxia |
| ZNF423 | |||||||||
| F874 -21 -22 |
Turkey | c.2738C>T (Chr16: 49,670,325) | p.P913L | 5 (hom) | (Danio rerio) | Yes | NPHP | ND | -21 and -22: CVH Infantile NPHP Situs inversus |
| A106 -21 -22 |
Iceland | c.1518delC (Chr16: 49,671,545) | p.P506fsX43 | 5 (het) | (Xenopus trop) | No | PKD | LCA | CVH (Joubert) |
| A111 -21 |
? | c.3829C>T (Chr16: 49,525,212) | p.H1277Y | 9 (het) | (Danio rerio) | ? | PKD | RD | CVH, NPHP, perinatal breathing abnormality, tongue tumor |
| CEP164 | |||||||||
| F319 -21 -22 |
Turkey | c.32A>C (Chr11: 117,209,334) | p.Q11P | 3 (hom) | Ch. R.c | Yes | NPHP, no Bx -21: (8yr) -22: (8yr) |
-21: RD (8/11yr, not yet blind) -22: no RP at 8 yrs |
-21: obesity? no LF -22: obesity? LF? |
| F59 -21 -22 -23 |
USA (Europe) | c.277C>T, (Chr11: 117,222,588) c.1573C>T (Chr11: 117,252,580) |
p.R93W, p.Q525X | 5 (het) 13 (het) |
Ch. R.c N/A |
No | NPHP, no Bx -21: (9 yr) -22: (8 yr) -23: normal |
-21: RD (6 yr) -22: LCA (legally blind at 5 mo) -23: (2 yr) |
-22: NY (birth), mild AI -23: seizuresd, substantial DD, mild ID |
| NPH505 | ND | c.1726C>T (Chr11: 117,257,920) | p.R576X | 15 (hom) | N/A | Yes | NPHP, Bx (8 yr) | RD and flat ERG (not blind) | CVH, FD, bilateral PD, bronchiectasis (1 mo), abnormal LFT, obesity |
| KKESH001-7 | Saudi | c.4383A>G (Chr11: 117,282,884) | p.X1460W fsX57 | 33 (hom) | N/A | Yes | normal | (RD) LCA, flat ERG (blind <2 yr) | N/A |
All mutations were absent from >270 healthy control individuals and from the ESP Exome Variant Sever data base, except the CEP164 variant p.R576X (allele frequency in European Americans 1/7,019).
cDNA mutation numbering is based on human reference sequences NM_014956.4 for MRE11, NM_015069.2 for ZNF423, and NP_055771 for CEP164, where +1 corresponds to the A of ATG start translation codon.
Ch.R, Chlamydomonas Reinhardtii;
Seizures were intractable, generalized and/or partial complex.
AI, aortic insufficiency; Bx, Kidney biopsy; CVH, cerebeller vermis hypoplasia; DD, developmental delay; ERG, electroretinogram; ESKF, end-stage kidney failure; FD, facial dysmorphism; het, heterozygous; hom, homozygous; ID, intellectual disability; LCA, Leber congenital amaurosis; LF, liver fibrosis; LFT, liver function tests; MRI, magnetic resonance imaging; N/A, not applicable; ND, no data; NPHP, nephronophthisis; NPHP-RC, nephronophthisis-related ciliopathies; NY, nystagmus; PD, polydactyly; RD, retinal degeneration; SS, short stature; yr, year/years.
A mutation of MRE11 causes progressive cerebellar degeneration
In family F3471 two siblings had cerebellar vermis hypoplasia (CVH), a central feature of NPHP-RC (Table 1). Mapping regions of homozygosity by descent yielded 2 candidate loci (Figure 1A). WER detected a homozygous truncation mutation (p.R633X) of MRE11 (Figure 1B; Table 1) previously described for CVH in a Pakistani family (Stewart et al., 1999). Family F3471 is also from Pakistan, suggesting a founder effect for this allele. MRE11 is an essential component of the ATM-Chk2 pathway of DDR (Figure S1), where it recruits ATM (ataxia telangiectasia-mutated) to sites of DNA double-strand breaks (Figure S1A). Rediscovery of this MRE11 mutation in family F3471 thus generated an unexpected link between NPHP-RC phenotype and the ATM pathway of DDR signaling (Figure S1A).
Patients with the NPHP-RC Joubert syndrome have defects in ZNF423
Another link of NPHP-RC to the ATM pathway of DDR signaling emerged from homozygosity mapping and WER in two siblings (F874) with infantile onset NPHP, CVH, and situs inversus (Table 1). SNP mapping yielded nine candidate regions of homozygosity by descent (Figure 1C). We identified in both affected individuals a homozygous missense mutation (p.P913L; conserved in vertebrates) of ZNF423 (Figure 1D). In addition, when examining 96 additional Joubert syndrome subjects, we detected two heterozygous-only mutations of ZNF423: p.P506fsX43 in family A106 and p.H1277Y in individual A111-21 (Table 1). Mutations of the mouse ortholog Zfp423 cause reduced proliferation and abnormal development of midline neural progenitors resulting in a loss of the cerebellar vermis (Alcaraz et al., 2006; Cheng et al., 2007) similar to that seen in Joubert syndrome patients with CVH.
ZNF423 encodes a protein with 30 zinc fingers (Figure 2A). Interestingly, a mouse mutation disrupting only the last zinc finger, similar to p.H1277Y, is sufficient to cause a severe phenotype (Cheng et al., 2007). Mouse models also display phenotypic variability that is subject to modifier genes, environment, and stochastic effects (Alcaraz et al., 2011; Alcaraz et al., 2006), consistent with the variable presentations of NPHP-RC patients. The homozygous mutation p.P913L, located between zinc fingers 21 and 22 (Figure 2A), most likely exerts recessive loss-of-function, analogous to the Zfp423 mouse models. Of the heterozygous-only mutations, one (p.N506fsX43) truncates the protein before the 11th zinc finger, whereas the other mutation (p.H1277Y) abrogates a zinc-coordinating histidine that is part of the knuckle in the last zinc finger, which is required for interactions with EBF family transcription factors (Tsai and Reed, 1998), and is likely critical for ZNF423 function (Figure 2A).
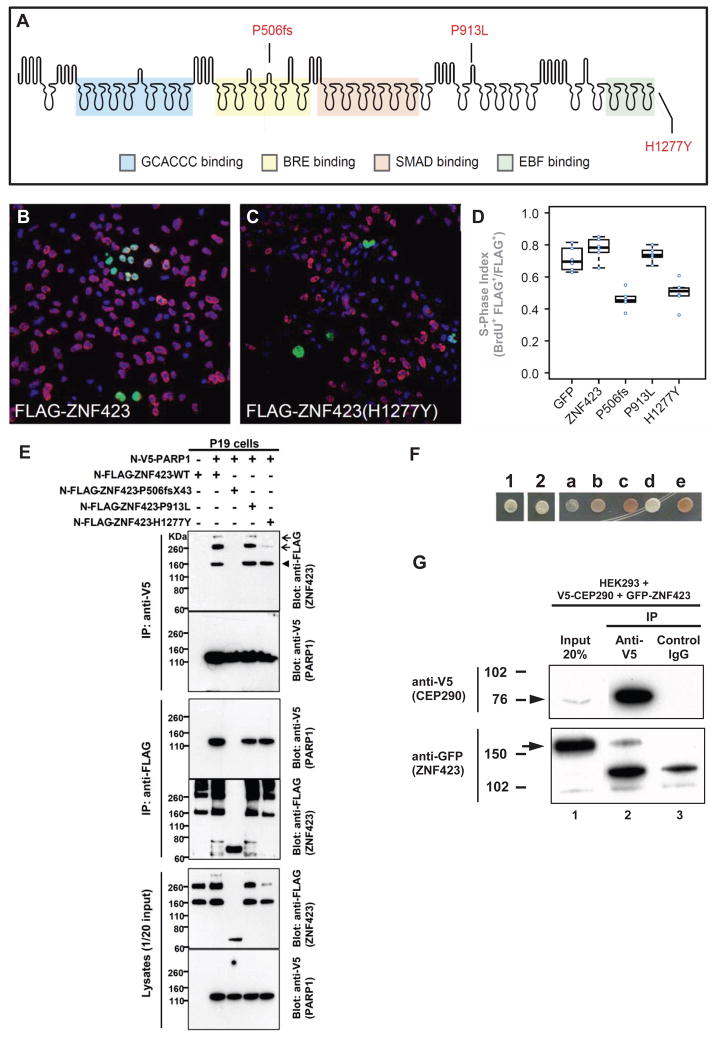
Figure 2. Two ZNF423 mutations have dominant negative characteristics (A–D), ZNF423 mutation abrogates interaction with PARP1 (E), and ZNF423 directly interacts with the NPHP-RC protein CEP290/NPHP6 (F–G).
(A) Amino acid residues altered by NPHP-RC mutations in ZNF423 are drawn in relation to functional annotation of its 30 Zn-fingers.
(B–D) S-phase index assay (fraction of transfected cells incorporating BrdU) for P19 cells transfected with either wildtype or mutant ZNF423. (B) Representative field of cells transfected with wildtype ZNF423 shows high frequency of BrdU+ FLAG+ double-positive cells. (C) ZNF423–H1277Y transfected cells exhibits fewer FLAG–positive cells and a lower proportion that are double positive. (D) S-phase index measured in duplicate transfections for each of three DNA preparations per construct. A GFP construct was used as a nonspecific control. Constructs with P506fsX43 and H1277Y mutations detected in NPHP-RC show significantly reduced S-phase index (p>10−5, ANOVA with post-hoc TukeyHSD).
(E) ZNF423 interacts with PARP1. P19 cells were co-transfected with expression constructs for N-terminally V5-tagged human full-length ZNF423 and FLAG-tagged human full length PARP1. Comparable amounts of both proteins were present in all lysates (lower panels). Proteins were precipitated, using anti-V5 (upper panels) and anti-Flag antibodies (middle panels), respectively. Reciprocal coIP demonstrates interaction between ZNF423 and PARP1. Note that the ZFN423 mutation P506fsX43 abrogates this interaction (arrow head) and that mutation H1277Y diminishes ZNF423 multimerization (arrow).
(F–G) ZNF423 directly interacts with CEP290/NPHP6.
(F) A human fetal brain yeast two-hybrid library screened with human CEP290/NPHP6 (JAS2; aa 1917–2479) fused to the DNA-binding domain of GAL4 (pDEST32) identified ZNF423 as a direct interaction partner of CEP290/NPHP6. The interaction was confirmed using direct yeast two-hybrid assay where 1 and 2 represent colony growth of CEP290 bait with ZNF423 prey. a–e are controls for colony growth on medium deficient in histidine, leucine and tryptophan.
(G) HEK293T were cotransfected with human V5-tagged partial human V5-CEP290 clone and GFP-tagged full-length human ZNF423 clone. Immunoprecipitation with anti-V5 (lane 2), but not control IgG (lane 3) precipitated both the V5-tagged CEP290 (arrowhead) as well as gfp-tagged ZNF423 (arrow).
We next examined whether the heterozygous-only mutations (Table 1) lead to loss of function via a dominant mechanism, using a proliferation assay in P19 cells (Figure 2B–D). Mutations were engineered into a FLAG-tagged ZNF423 cDNA and assayed by a S-phase index, defined as the proportion of transfected cells that incorporate BrdU in 1 h, 48 h after transfection. Simple loss of function alleles should not interfere with endogenous Zfp423 activity in this assay. Indeed, overexpression of either wild-type or the homozygous p.P913L allele had no effect (Figure 2D). However, transfection with either the p.P506fsX43 frame-shifting allele, which removes the zinc fingers required for SMAD and EBF interactions, and the H1277Y substitution allele, which destroys the terminal zinc finger required for EBF interaction, reduced the mitotic index to little more than half that of cells transfected with GFP control vector or other alleles of ZNF423 (Figure 2B–D). A dominant mechanism is plausible for the two heterozygous mutations, as each is predicted to interfere selectively with a subset of interaction domains (Figure 2A). Neither subject had siblings, and DNA from parents was not available to determine whether the mutations occurred de novo.
We detected five additional putative mutations in highly conserved (including histidine knuckle) residues of ZNF423 among Joubert syndrome families (Table S1). While these mutations have not been confirmed functionally, the high incidence of predicted deleterious mutations found in patients but absent from 270 healthy control individuals, dbSNP, and 1,000 Genomes Project data further support identification of ZNF423 as a causal gene in NPHP-RC and JBS.
ZNF423/OAZ was recently shown to interact with the DNA ds-damage sensor PARP1 (poly-ADP ribosyl polymerase 1) (Ku et al., 2003), which recruits MRE11 and ATM to sites of DNA damage (Figure S1A). This indirectly linked ZNF423 to the ATM pathway of DNA damage signaling (Figure S1A). We therefore tested whether ZNF423 mutations affect interaction between ZNF423 and PARP1. Coimmunoprecipitation verified the association of ZNF423 and PARP1 in reciprocal assays (Figure 2E). More importantly, the truncating mutation P506fsX43, which we detected in a JBTS patient (Table 1), abrogates this interaction (Figure 2E), while H1277Y inhibits multimerization of ZNF423 (Figure 2E). In addition, depletion of ZNF423 mRNA caused sensitivity to DNA damaging agents (see below).
Furthermore, we identified ZNF423 as a direct interaction partner of CEP290/NPHP6, which is mutated in NPHP-RC (Sayer et al., 2006; Valente et al., 2006). In a yeast two-hybrid screen of human fetal brain library with a CEP290 (JAS2; amino acids 1917–2479) ‘bait’ we found 3 in-frame “prey” sequences corresponding to ZNF423 (amino acids 178–406). This interaction was confirmed in a direct yeast two-hybrid assay (Figure 2F) and by co-immunoprecipitation of CEP290/NPHP6 with ZNF423 expressed in HEK293T cells (Figure 2G). CEP290/NPHP6 is known to interact with the NPHP-RC protein NPHP5 (Schafer et al., 2008) and localizes to the ciliary transition zone (Sang et al., 2011). A nuclear function of CEP290/NPHP6 is likely: it contains a nuclear localization sequence (NLS), binds the transcription factor ATF4, and localizes to the nucleus by cell fractionation (Sayer et al., 2006). Direct binding between ZNF423 and CEP290/NPHP6, whose mutation also causes a CVH phenotype (Sayer et al., 2006; Valente et al., 2006), is consonant with other NPHP-RC protein interactions in dynamic complexes (Sang et al., 2011).
Mutations of CEP164 cause NPHP-RC
Leber congenital amaurosis (LCA) is an early-onset form of isolated retinal degeneration (RD) that can be allelic with NPHP-RC. For example, null mutations of CEP290/NPHP6 cause severe multiorgan NPHP-RC variants of JBTS and MKS syndromes (Helou et al., 2007), whereas hypomorphic mutations cause LCA only (Chang et al., 2006; den Hollander et al., 2006). By homozygosity mapping in a Saudi family (KKESH001) of first-cousin parents with a child who had LCA with nystagmus, hyperopic discs, vascular attenuation, diffuse retinal pigment epithelium atrophy, and non-recordable ERG (Table 1), we obtained 14 candidate regions (Figure 1E). By whole exome resequencing we detected a homozygous point mutation in CEP164 (centrosomal protein 164 kDa) that abolished the termination codon, adding 57 amino acid residues to the open reading frame (p.X1460WfsX57) (Figure 1F, Table 1). The mutation was absent from 96 Saudi healthy controls and from 224 North American LCA patients who lack mutations in other known LCA genes.
We also had considered CEP164 a candidate gene for NPHP-RC, because it is part of the human centrosomal proteome (Andersen et al., 2003). We performed exon-PCR and Sanger sequencing of all 31 coding exons for one affected individual in each of 856 different NPHP-RC families (see Experimental Methods Online). We detected both mutated CEP164 alleles in each of 3 additional families with NPHP-RC (Table 1; Figure S2). Specifically, we found i) a homozygous missense mutation (p.Q11P; conserved to Chlamydomonas) in two siblings of family F319 with Senior-Loken syndrome (NPHP with retinal degeneration) (Table 1), ii) compound heterozygosity for a missense mutation (p.R93W; conserved to Chlamydomonas) and a truncating mutation (p.Q525X) in three siblings of family F59 with Senior-Loken syndrome, one of whom also had seizures and intellectual disability and, iii) a homozygous nonsense mutation (p.R576X) in individual NPH-505 with Senior-Loken syndrome, partial vermis aplasia and bronchiectasis (Table 1). In individuals with LCA we also detected heterozygous mutations, for which we were unable to detect a second recessive allele. These were p.R93W, also present in F59, and p.Q1410X, which was detected in two families with phenotypic features of Joubert syndrome and oral-facial-digital syndrome (OFG6), and heterozygously in two out of 96 healthy control individuals (data not shown). All other mutations were absent from >96 ethnically matched healthy control individuals and from healthy controls of the 1000 genomes project (Table 1). We thereby identified recessive mutations of CEP164 as a new cause of NPHP-RC. Because of the significant overlap of phenotypic features with other forms of NPHP-RC we introduce the alias NPHP14 for the CEP164 protein (http://www.ncbi.nlm.nih.gov/omim/?term=nphp*).
Although the number of families with CEP164 mutation is small, our findings suggest a genotype-phenotype correlation (Table 1). First, the patient with the mildest phenotype had isolated LCA only and a homozygous mutation (p.X1460WfsX57) that removes the termination codon, adding 57 new amino acids to the C-terminus. This may reduce protein function, because overexpression of full-length CEP164 with a C-terminal GFP tag strongly reduced centrosomal labeling compared to an N-terminal GFP tag (Figure 3A–D). Second, the homozygous missense allele (p.Q11P) caused a multi-organ phenotype of NPHP with retinal degeneration (Senior-Loken syndrome; SLSN). Third, compound heterozygosity for one missense and one truncating allele (p.R93W; p.Q525X) caused SLSN with additional central nervous system involvement. Fourth, a homozygous truncating mutation (p.R576X) caused a severe phenotype that combined Joubert syndrome (NPHP, retinal degeneration and vermis hypoplasia) with bronchiectasis, a classic feature of primary ciliary dyskinesia (PCD), a disease of motile cilia. These data further support the gradient of genotype-phenotype correlations characteristic of NPHP-RC, in which null mutations cause the severe dysplastic phenotypes of Meckel syndrome and Joubert syndrome, whereas hypomorphic alleles cause the milder degenerative phenotypes of NPHP and SLSN (Hildebrandt et al., 2011).
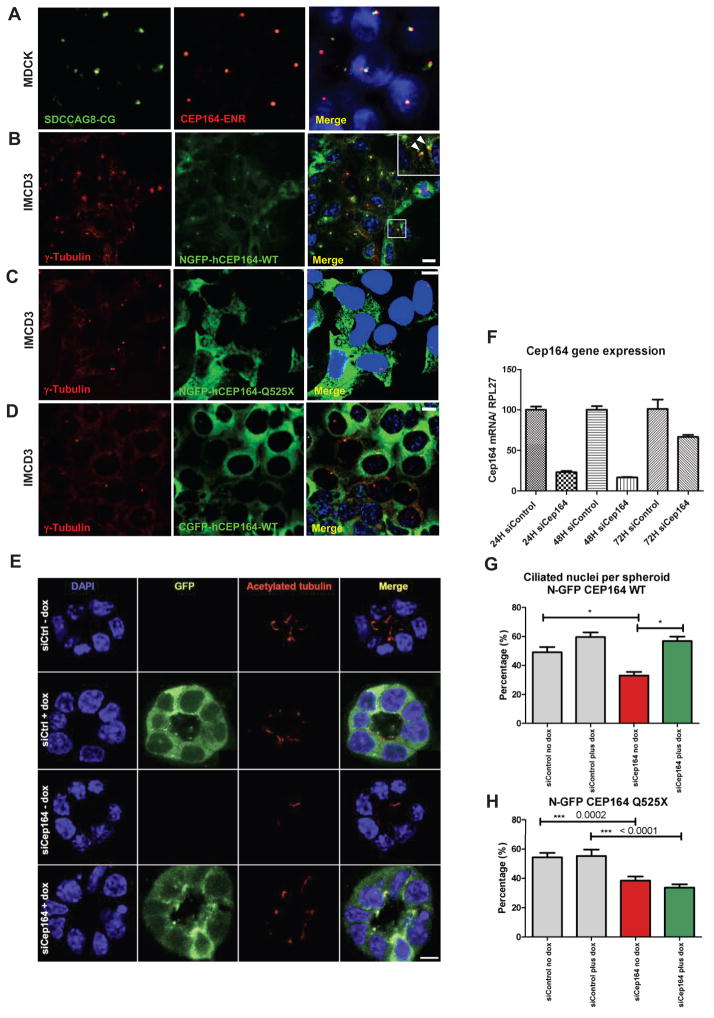
Figure 3. (A–D) Expression of mutant CEP164 in renal epithelial cells abrogates localization to centrosomes.
(A) Immunofluorescence using α-SDCCAG8/NPHP10-CG antibody, labels both centrioles, whereas α-CEP164-ENR antibody demonstrates CEP164 staining at the mother centriole only.
(B) Inducible overexpression of N-terminally GFP-tagged human full-length CEP164 isoform 1 (NGFP-CEP164-WT) in IMCD3 cells demonstrates, in addition to a cytoplasmic expression pattern, localization at one of the two centrioles (inset, arrow heads) consistent with selective localization to the mother centriole (Graser et al., 2007). Both centrioles are stained with an anti-γ-tubulin antibody.
(C) In contrast, the centrosomal signal is abrogated upon overexpression of an N-terminally GFP-tagged truncated CEP164 construct representing the mutation p.Q525X.
(D) The number of centrosomes positive for CEP164 is reduced upon overexpression of C-terminally GFP-tagged human full-length CEP164 isoform 1 (CGFP-hCEP164-WT), which mimics the mutation p.X1460fs57 that causes a read-through of the stop-codon X1460, adding 57 aberrant amino acid residues to the C-terminus of CEP164. Similar data was obtained upon CEP164 expression in hTERT-RPE cells (see also Figure S3B–D).
IMCD3 cells were stably transfected with the respective CEP164 constructs in a retroviral vector for doxycyclin-inducible expression (pRetroX-Tight-Pur). Scale bars represent 10 μm.
(E–H) Knockdown of Cep164 disrupts ciliary frequency.
(E) Depletion of Cep164 by siRNA (F) causes a ciliary defect in 3D spheroid growth assays. IMCD3 cells transfected with either siCtrl or siCep164 were grown to spheroids in 72 hours and immunostained for acetylated tubulin (red). DAPI stains nuclei (blue). Doxycycline induced stably transfected NGFP-hCEP164-WT (green). Space bar represents 5 μm.
(G) Nuclei and cilia were scored within a single spheroid to generate ciliary frequencies. siCep164 transfected cells manifest lower cilia frequencies (33%) compared to control transfected IMCD3 cells (49%). Induction of NGFP-hCEP164-WT in siCep164 transfected cells rescues this ciliary defect (57%). 50 spheroids per condition were analysed in three independent experiments. Error bars represent SEM, n=3, *p-value <0.0002.
(H) Ciliary frequency is not rescued by mutant CEP164. Ciliary frequencies are reduced in siCep164 transfected IMCD3 cells (39%) compared to control siCtrl transfected IMCD3 cells (54%). Induction of NGFP-hCEP164-Q525X does not rescue this ciliary defect (34%). 50 spheroids per condition were analyzed. Error bars represent SEM, ***p-value <0.0002.
See also Figure S3.
CEP164 is transcribed into 3 common isoforms (Figure S2A–C). In addition to the centrosome, CEP164 is part of the photoreceptor sensory cilium proteome (Liu et al., 2007). Homozygous truncating mutations found in exons common to isoforms 1 and 3, but not in isoform 2 support the idea that isoforms 1 and/or 3 are relevant for the NPHP-RC phenotype (Figure S2B). The deduced CEP164 amino acid sequence has a WW domain (aa 57–89), which interacts with the DDR protein ATRIP (Figure S2D). CEP164 also contains a segment of 6 coiled-coil domains (Figure S1C), which are frequently found in NPHP-RC genes (Hildebrandt et al., 2009a). CEP164 is conserved across species including the green alga Chlamydomonas reinhardtii, suggesting a conserved function with strong sequence constraints. To study expression and subcellular localization of the CEP164 protein we utilized antibodies against human CEP164 and characterized them by immunoblotting and by immunofluorescence (Figure S3).
Mutation of CEP164 abrogates mother centriole localization
By confocal microscopy of GFP-labeled CEP164 protein with other labels, we show that CEP164 colocalizes in hTERT-RPE cells with the mother centriole, with the mitotic spindle poles, and with the abscission structure in a cell cycle-dependent way (Figure S4), a feature characteristic of proteins involved in single-gene ciliopathies (Otto et al., 2010a) (Graser et al., 2007).
We then examined subcellular localization of CEP164 in mouse retina (Figure S3C) as well as in MDCK2 (Madin Darby canine kidney) cell lines, IMCD3 (mouse kidney inner medullary collecting duct) cell lines (Figure 3), and hTERT-RPE (human retinal pigment epithelium) cell lines (Figure S3D–F, Figure S5A) that stably express GFP-tagged human full length CEP164 wild type isoform 1 from a doxycyclin-inducible stable expression construct (NGFP-hCEP164-WT). We found that upon IF using the α-CEP164-NR antibody in MDCK cells as well as upon induction with doxycyclin (10 ng/ml) of N-terminally GFP tagged NGFP-hCEP164-WT in IMCD3 cells, CEP164 localizes to mother centrioles of cells (Figure 3A–B). In contrast, the signal at centrosomes was abrogated upon overexpression of an N-terminally GFP-tagged truncated CEP164 construct representing the mutation p.Q525X (Figure 3C). Furthermore, the signal occurred at a reduced number of centrosomes upon overexpression of C-terminally GFP-tagged human full-length construct (Figure 3D), which mimics the mutation p.X1460WfsX57 occurring in NPHP-RC family KKESH001 that causes a read-through of the stop-codon X1460, adding 57 aberrant amino acid residues to the C-terminus of CEP164 (Table 1). Corresponding data were obtained upon CEP164 expression in hTERT_RPE cells (Figure S3D–E). We thus demonstrated lack of centrosomal localization for the truncating mutation p.Q525X and for an equivalent of the p.Q1460WextX57 mutation.
Endogenous Cep164 levels regulate cilia in 3D epithelial spheroid structures
Loss of function of several genes that cause nephronophthisis in NPHP-RC cause disruption of 3D architecture of renal epithelial cell culture (Otto et al., 2010a; Sang et al., 2011). To evaluate CEP164 by this criterion, we transfected murine kidney IMCD3 cells with siRNA oligonucleotides against murine Cep164, or random sequences (Ctrl) in 3D spheroid growth assays. After 3 days, siCtrl transfected cells formed spheroid structures with a clear lumen, apical cilia, defined tight junctions and clear basolateral structure. Cells transfected with siCep164 developed spheroids with overall normal architecture and size, but with markedly reduced frequency of cilia (Figure 3E–H). While 49% of the spheroid cells transfected with siCtrl generated detectable cilia, only 33% of the siCep164 transfected cells grown in 3D cultures were ciliated (p<0.0001). We conclude that Cep164 affects ciliogenesis or maintenance, but that the overall architecture of renal 3D growths is not as grossly affected as we have previously seen for knockdown of other NPHP-RC genes (Sang et al., 2011).
To exclude the possibility that the effect observed was due to off-target effects, we used two IMCD3 clones stably transfected with inducible full-length human CEP164 (IMCD3-NGFP-CEP164-WT clones 2 and 8). Both lines form spheroids in 3D cultures. We then repeated siRNA of the endogenous murine Cep164 and observed no irregularities except for reduced cilia frequency. However, after doxycycline induction of siCep164-resistant human NGFP-CEP164, ciliary frequencies were restored, with 57% of the siCep164 transfected cells ciliated (p=<0.0001) (Figure 3G). Importantly, rescue was not observed for IMCD3 cells expressing a patient mutation (NGFP-CEP164-Q525X, Figure 3H). We conclude that reducing cellular levels of Cep164 affects renal ciliation frequencies without gross disturbances to tubular architecture, a phenomenon that can be rescued with WT but not mutant CEP164.
NPHP-RC proteins colocalize with the DDR protein TIP60 to nuclear foci
A non-centrosomal localization for CEP164 was recently described by demonstrating its translocation to nuclear foci in response to DNA damage (Pan and Lee, 2009; Sivasubramaniam et al., 2008). In this context CEP164 is thought to play a role in DNA damage-response (DDR) signaling where it interacts with the DDR protein ATRIP (Figure S2D), is activated by the DDR proteins ATM and ATR, and is necessary for checkpoint-1 (Chk1) activation. Abrogation of CEP164 function leads to loss of G2/M cell cycle checkpoint and aberrant nuclear divisions (Sivasubramaniam et al., 2008). Because the NPHP-RC gene products MRE11 and CEP164 are essential components of DDR signaling, and because ZNF423 interacts with the bona fide DDR protein PARP1 (Ku et al., 2003), our identification of mutations in these genes strongly suggested a role of DDR signaling in the pathogenesis of NPHP-RC (Figure S1). We therefore examined nuclear foci localization for the newly identified NPHP-RC causing gene products ZNF423 and CEP164 and for previously identified NPHP-RC gene products.
Localization of SDCCAG8 (alias NPHP10), in which we had previously identified NPHP-RC mutations (Otto et al., 2010a), shows nuclear foci in hTERT-RPE cells in addition to its centrosomal localization (Figure 4B–D). Transient shRNA knockdown confirmed specificity of the signal (Figure S4B–D). SDCCAG8/NPHP10 did not colocalize with markers for PLM bodies (Janderova-Rossmeislova et al., 2007) or CENP-C (marking chromosomal centromeres) (Figure S5A). In contrast SDCCAG8/NPHP10 fully colocalized with SC35 in hTERT-RPE cells (Figure 4A–C). SC35, also known as serine/arginine-rich splicing factor 2 (SRSF2), is a splicing factor that plays a role in DDR by controlling cell fate decisions in response to DNA damaging agents (Edmond et al., 2010; Reinhardt et al., 2011). SC35 marks hubs of enhanced gene expression (Szczerbal and Bridger, 2010), is phosphorylated by topoisomerase I (Elias et al., 2003), and is required for genomic stability during mammalian organogenesis (Xiao et al., 2007). Moreover, ZNF423 also fully colocalizes (Figure 4D), and CEP164 partially colocalizes (Figure 4E) with SC35 in nuclear foci. Consequently, ZNF423 and CEP164 also colocalize with SDCCAG8/NPHP10 in SC35-positive nuclear foci (Figure 4F,G).
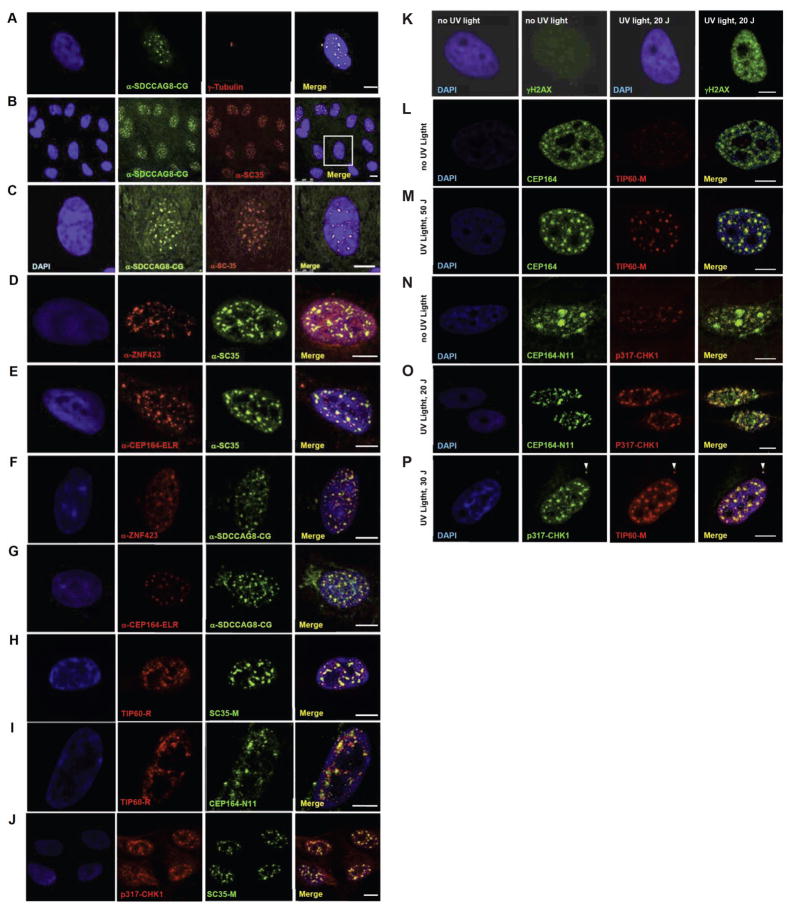
Figure 4. (A–P) Colocalization upon immunofluorescence of the NPHP-RC proteins SDCCAG8/NPHP10, ZNF423 and CEP164 to nuclear foci that are positive for the DDR signaling proteins SC35, TIP60 and Chk1 in hTERT-RPE cells.
(A–G) Colocalization of NPHP-RC proteins with SC35 in nuclear foci. SDCCAG8/NPHP10 (A–C) and ZNF423 (D) fully colocalize to nuclear foci with SC35, and (E) CEP164 partially colocalizes with SC35. SDCCAG8/NPHP10 also colocalizes with the newly identified NPHP-RC proteins ZNF423 (F) and CEP164 (G).
(H–J) Colocalization of NPHP-RC proteins with the DDR protein TIP60 I nuclear foci.
(H) TIP60 fully colocalizes with SC35.
(I) TIP60 partially colocalizes with CEP164.
(J) Chk1 fully colocalizes with SC35/SRSF2. DNA is stained in blue with DAPI. Scale bars represent 5μm.
(K–P) Colocalization of DDR and NPHP proteins upon induction of DDR by UV radiation in HeLa cells.
(K) Following irradiation of HeLa cells with UV light at 20 J/m2 a strong immunofluorescence signal of an anti-γH2AX antibody indicates activation of DDR.
(L–M) Upon irradiation with UV light, CEP164-positive nuclear foci condense and colocalize with newly appearing TIP60 foci of similar size.
(N–O) In untreated cells (N) a pattern of broad CEP164 speckles, which are CHK1-negative and locate to DAPI-negative domains, changes to a pattern of multiple smaller foci (O) that are double-positive for both, CEP164-N11 and CHK1.
(P) p317-CHK1 fully colocalizes with TIP60 to nuclear foci and to the centrosome (arrow head).
See also Figures S4, S5.
SC35 functions within a TIP60 complex, in which TIP60 acetylates SC35 on lysine 52 (Figure S1B), modifying the role of SC35 in the promotion of apoptosis and inhibition of G2/M arrest (Edmond et al., 2010), which is regulated by the checkpoint proteins Chk1 and Chk2 (Figure S1D). Interestingly, the TIP60 protein, together with the heterotrimeric MRN complex (of which MRE11 is a component) constitutes the major activator of ATM within the ATM pathway of DDR signaling (Ciccia and Elledge, 2010) (Figure S1A). In hTERT-RPE cells the ATM activator TIP60 colocalizes to nuclear foci with SC5/SRSF2 (Figure 4H) and partially with the newly identified NPHP-RC protein CEP164 (Figure 4I). We thus identify a new group of NPHP-RC proteins and demonstrate that they colocalize to nuclear foci with the DDR proteins TIP60 and SC35. These gene products include the newly identified NPHP-RC proteins in ZNF423 and CEP164 as well as SDCCAG8/NPHP10. Interestingly, the protein OFD1, which is mutated in the ciliopathy oral-facial-digital syndrome, is part of the TIP60 complex, which is a major ATM activator within DDR signaling; we recently identified OFD1 as a direct interaction partner of SDCCAG8/NPHP10 (Figure S1B) (Otto et al., 2010a).
CEP164CEP164 colocalizes with the DDR proteins TIP60 and CHK1 in nuclear foci upon DNA damage
Because one of the central mechanisms controlled by DDR signaling is cell cycle regulation through phosphorylation of checkpoint-1 (Chk1) and checkpoint-2 (Chk2) proteins (Figure S1D), we also tested whether checkpoint proteins are recruited to SC25/SRSF2-positive nuclear foci. SC35 and p317-Chk1 colocalize to nuclear foci in hTERT-RPE cells (Figure 4J). As colocalization conveys only a static image of DDR components, we then tested whether localization of CEP164 to nuclear foci was inducible by DNA damage as suggested (Sivasubramaniam et al., 2008). Following irradiation with 20–50 J/m2 of UV light, CEP164-positive nuclear foci condensed to larger size and colocalized with newly appearing TIP60 foci of similar size (Figure 4K–M). Similarly, a pattern of broad CEP164 speckles, which were CHK1-negative and locate to DAPI-negative domains in untreated cells (Figure 4N), changed to a pattern of multiple smaller foci that were positive for both CEP164 and CHK1 (Figure 4O). Colocalization of TIP60 and p317-CHK1 in these foci (Figure 4P) demonstrated that the UV-inducible foci observed in HeLa cells are likely equivalent to the foci positive for TIP60 and SC35 that we observed as positive for NPHP-RC proteins in hTERT-RPE cells (Figure 4A–J). We thus demonstrate that CEP164 translocates in response to DNA damage to nuclear foci that contain the DDR proteins TIP60 and CHK1.
Reducing endogenous levels of Cep164 causes anaphase lagging chromosomes
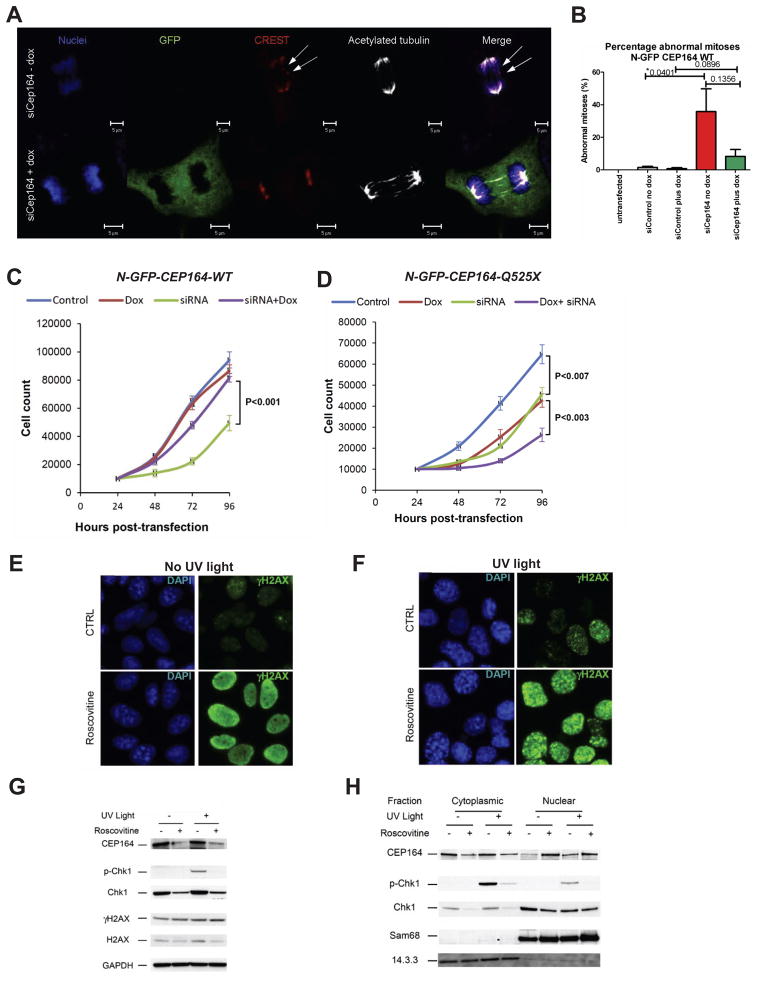
Lagging chromosomes on anaphase spindles (”anaphase lag”) are a hallmark of many mutations that affect mitotic checkpoint integrity. Acute reduction of Cep164 by siCep164 knockdown in IMCD3 cells exacerbated the incidence of anaphase lag from 1% in siCtrl controls to 21% in siCep164-treated cells, in over 250 anaphases scored from unsynchronized cells in a total of five independent experiments (Figure 5A–B, p=0.04). CREST antiserum and DAPI confirmed the presence of incomplete mitotic congression and unattached kinetochores during late anaphase. This phenomenon was specific, since doxycycline-inducible expression of WT-CEP164 during Cep164 siRNA knockdown reduced the incidence of anaphase lag to just 4% (Figure 5B). Untransfected (non-siRNA treatment) IMCD3 cells had no detectable anaphase lag (0%). These data indicate a requirement for Cep164 at the G2/M checkpoint.
Figure 5. Knockdown of Cep164 causes anaphase lag and retarded cell growth.
(A–B) Knockdown of CEP164 causes anaphase lag. siCep164 knockdown in IMCD3 cells increased anaphase lag incidence from 1% after siCtrl to 21% after siCep164-treated cells (n >250 anaphases, five independent experiments). CREST antiserum (red) and DAPI (blue) confirmed the presence of incomplete mitotic congression and unattached kinetochores during late anaphase (white arrows). Doxycycline-inducible expression of WT-CEP164 during Cep164 siRNA knockdown reduced the incidence of anaphase lag to 4%, whereas untransfected IMCD3 cells had no detectable anaphase lag (0%) (B). Bars represent SEM, p values (student T-test) are indicated above the bar graph.
(C–D) Transient knockdown of Cep164 inhibits proliferation, which is rescued by wild type but not mutant CEP164. In clonally selected and doxycycline (Dox)-inducible mouse IMCD3 cells siRNA knockdown was performed.
(C) IMCD3 cells depleted of murine Cep164 grew more slowly (siRNA, green line) than non-depleted cells (control, blue line) or the non-depleted cells induced to express human wild type CEP164 (Dox, red line). Expression of WT Cep164 in siRNA-depleted cells rescued the slow growth phenotype of Cep164 depletion (siRNA+dox, purple line).
(D) As in C, except mutant Cep164 cDNA (CEP164-Q525X) was expressed under doxycyclin control. Expression of this allele itself had a negative impact on cell growth (green line), suggesting a dominant negative effect. An even greater negative effect was seen when the endogenous Cep164 was depleted in cells expressing CEP164-Q525X (siRNA+dox, purple line). The average counts are plotted with standard deviations. Asterisks indicates significant differences by unpaired Student’s t-test (P< 0.05).
(E–H) The effect of roscovitine on UV-induced DDR. Cells were UV-irradiated with 30 J/m2 and analyzed 1 h post UV-irradiation. Where indicated, cells were pre-incubated for 24 h with the CDK inhibitor roscovitine (80 μM). (E–F) Immunofluorescence analysis showed that roscovitine triggered uniform nuclear distribution of γH2AX (activated H2AX phosphorylated at Ser139) in non UV-irradiated cells suggesting partial DDR activation (E). UV radiation caused enhanced γH2Ax staining with a prominent nuclear foci pattern, characteristic of strong DDR activation (F).
(G–H) The effect of roscovitine on UV-triggered subcellular localization of CEP164 and Chk1. CEP164 and Chk1 proteins, along with nuclear marker Sam68 and cytoplasmic marker 14.3.3 were analyzed by western blot. Roscovitine decreased the amount of CEP164 present in control and UV-irradiated cells (G). This was most likely due to translocation of CEP164 into the nucleus upon roscovitine treatment as shown by subcellular fractionation (H). As expected, UV radiation increased phosphorylation of Chk1 at Ser317 (p-Chk1) (G), and roscovitine decreased Chk1 protein expression and abrogated UV-induced p-Chk1 in both cytoplasm and nucleus (G–H). Proteins 14.3.3 and Sam68 serve as controls for cytoplasmic vs. nuclear fraction, respectively. See also Figure S6.
Human wild type CEP164 but not its NPHP-RC truncation rescues IMCD3 cell proliferation
In clonally selected IMCD3 cells expressing wild type human CEP164 cDNA construct N-GFP-CEP164-WT under doxycycline (Dox) control, depletion of endogenous mouse Cep164 retarded proliferation in comparison to either undepleted control cells or undepleted cells that were Dox-induced to overexpress N-GFP-CEP164-WT alone (Figure 5C). Cep164-depleted growth was rescued by Dox-induced expression of human N-GFP-CEP164-WT (Figure 5C). Cells expressing truncated cDNA construct N-GFP-CEP164-Q525X, modeling the NPHP-RC mutation in family F59, exhibited retarded growth, even when the endogenous Cep164 was present (Figure 5D), consistent with a dominant negative effect. Further depletion of the endogenous Cep164 in N-GFP-CEP164-Q525X expressing cells showed an additive effect on growth retardation, confirming the dominant negative effect of N-GFP-CEP164-Q525X in this experimental system (Figure 5D).
CDK inhibition translocates Cep164 to the nucleus
CEP164 is required for DNA damage-induced phosphorylation of Chk1, and down regulation of CEP164 significantly reduces DDR (Maude and Enders, 2005; Sivasubramaniam et al., 2008). On the other hand, the DDR pathway can also be activated by the small molecule CDK inhibitor roscovitine, which in addition reduces Chk1 expression (Maude and Enders, 2005). Roscovitine also reduces the development of kidney cysts in the Nphp9 mouse model, Jck (Bukanov et al., 2006). We therefore tested the influence of roscovitine (targeting CDK2, 5, 7 and 9) on DDR activation in IMCD3 cells. Immunofluorescence shows increased uniform distribution of γH2AX (activated H2AX phosphorylated at Ser139) in the nucleus of IMCD3 cells upon roscovitine treatment in irradiated cells, indicating partial DDR activation (Figure 5E). Second, in cells treated with roscovitine, UV irradiation caused enhanced γH2AX staining with a prominent nuclear foci pattern, characteristic of strong DDR activation (Figure 5F). Immunoblotting showed that roscovitine decreased the amount of CEP164 present in both control and UV-irradiated cells (Figure 5G–H). This was most likely due to translocation of CEP164 into the nucleus upon roscovitine treatment, as shown by subcellular fractionation (Figure 5H). As expected, UV radiation increased phosphorylation of Chk1 at Ser317 (p-Chk1) (Figure 5G), and roscovitine decreased Chk1 protein expression and abrogated UV-induced p-Chk1 in both cytoplasm and nucleus (Figure 5G–H). Together, these data indicate that CDK inhibition by roscovitine causes nuclear translocation of CEP164 and inhibits Chk1 activation. γH2AX activation by roscovitine may restore cell cycle control by Chk2 activation instead (Maude and Enders, 2005).
CEP164 directly interacts with CCDC92 and TTBK2
NPHP-RC proteins are known to interact with other NPHP-RC proteins in dynamic complexes that have been termed the “NPHP-JBTS-MKS interaction network” (Sang et al., 2011). In order to identify novel direct interaction partners of CEP164 we performed yeast two-hybrid screening of both a random-primed bovine retina and an oligo-dT primed human retina cDNA library. Full length and partial cDNA clones of CEP164 were used as baits: CEP164fl (encoding the full length protein), CEP1641-550 (encoding amino acids 1-550), CEP164551-1100 (encoding amino acids 551-1100) and CEP1641101-1460 (encoding amino acids 1101-1460). We identified CCDC92 (coiled coil domain containing protein 92) and TTBK2 (tau tubulin kinase 2) as direct interactors of CEP164 (Figure S5C–J). CCDC92 was the protein with the most positive clones in the screen with the human oligo-dT retinal cDNA library, while TTBK2 appeared to be the main CEP164 interactor in the bovine library. Two overlapping CCDC92 clones were found with baits CEP164fl, CEP1641-550 and CEP1641101-1460 in the human library, however, no CCDC92 clones were identified with the bovine library (Figure S5C). TTBK2 was identified in both screens, with 12 hits (4 different clones) in the bovine, and a single clone in the human retina cDNA library (Figure S5C). Interactions between CEP164 and each of these new partners were validated by GST pulldown (Figure S5D) and co-immunoprecipitation (Figure S5E–H). Immunofluorescence showed that CCDC92 fully colocalizes with CEP164 at the mother centriole (Figure S5I). TTBK2 weakly colocalizes with CEP164 at one of the centrioles, but yields a strong signal at the mid body in dividing hTERT-RPE cells (Figure S5J). Tau tubulin kinase 2 (TTBK2) is a member of the casein kinase family encoded by the gene mutated in cerebellar ataxia type 11 (Houlden et al., 2007). TTBK2 can phosphorylate tubulin, and its kinase activity is required for the phosphorylation of tau by GSK-3β. Tau is a microtubule associated protein that is also found in the nucleus, where it is a key player in the early stress response/DNA damage protection of neurons (Sultan et al., 2011).
CEP164 interacts with NPHP3 and DVL3
To determine whether CEP164 interacts with known NPHP-RC proteins, HEK 293T cells were co-transfected with N-terminally V5-tagged human full-length CEP164 and the seven different FLAG-tagged human full length proteins NPHP1-NPHP5, NPHP8, NPHP9 or the control protein CD2AP. NPHP proteins were precipitated, using anti-Flag M2 beads. We detected interaction of CEP164 with NPHP3 and weakly with NPHP4 (Figure S6A–B), demonstrating that CEP164 is in a complex with other known NPHP-RC proteins (Figure S1A–B). The DDR protein DDB1 interacted with NPHP2 (Figure S6C–D).
The dishevelled protein (Dvl) is a central component of the Wnt pathway and it has been shown that NPHP2/inversin interacts with Dvl targeting it for proteasomal degradation, thereby liberating the β-catenin destruction complex and triggering a switch from canonical to non-canonical Wnt signaling (Germino, 2005; Simons et al., 2005). In loss of function of the NPHP-RC protein NPHP2/INVS, which causes NPHP type 2 in humans (Otto et al., 2003), this switch is lacking (Simons et al., 2005). By immunoprecipitation coupled to mass spectrometry using endogenous Dvl3 as bait we here identify interaction between Dvl3 and CEP164 (Figure S6E–H). Immunocytochemistry reveals that endogenous Dvl3 and CEP164 share centrosomal localization in most cells analyzed (Figure S6A). To further study CEP164-Dvl3 interaction we performed GST-pull down assay in HEK293T cells as well as domain mapping, using a set of Dvl3 deletion constructs (Angers et al., 2006). We demonstrate that GST-CEP164 (aa2-195), which was shown to interact with the DDR protein ATRIP (Sivasubramaniam et al., 2008), is sufficient to pull down endogenous Dvl3 from the cellular lysate (Figure S6B). Domain mapping for Dvl3 suggests that CEP164 interacts with the proline-rich region of Dvl3, because only mutants containing this sequence efficiently co-immunoprecipitate with CEP164-GFP (Figure S6C). Interestingly, only wild type CEP164-mCherryRFP but not the NPHP-RC causing mutant CEP164-Q525X detected in family F59 (Table 1) can be efficiently immunoprecipitated with Dvl3 (Figure S6D), further supporting its pathogenic role.
cep164 loss of function causes NPHP-RC and DDR activation in zebrafish
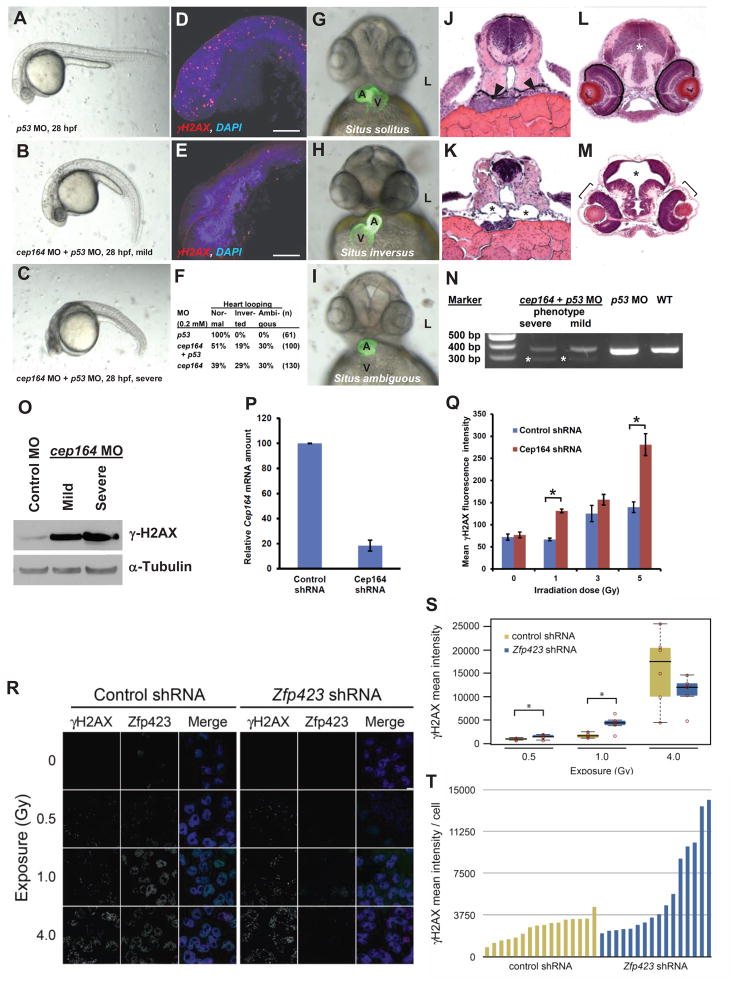
To test in a vertebrate animal model whether loss of cep164 function results in both, an NPHP-RC phenotype as well as DDR activation, we performed cep164 knockdown in zebrafish embryos using a morpholino-oligonucleotide (MO) that targets the exon 7 splice donor site of cep164 (Fig. 6). A p53 MO was injected to reduce off-target MO effects (Robu et al., 2007). At 28 hours post fertilization (hpf) we observed the ciliopathy phenotypes of ventral body axis curvature and cell death (Fig. 6A–C). Embryos showed increased expression of phosphorylated γH2AX (Fig. 6D–E). At 48 hpf, cep164 morphants displayed the typical ciliopathy phenotype of abnormal heart looping as a laterality defect (Fig. 6F–I). Furthermore, at 72 hpf, embryos developed further NPHP-RC phenotypes, including pronephric tubule cysts (Fig. 6J–K), as well as hydrocephalus and retinal dysplasia (Fig. 6L–O). To test in a vertebrate animal model whether loss of cep164 function results in both, an NPHP-RC phenotype as well as DDR activation, we performed cep164 knockdown in zebrafish embryos using a morpholino-oligonucleotide (MO) that targets the exon 7 splice donor site of cep164 (Fig. 6). A p53 MO was injected to reduce off-target MO effects (Robu et al., 2007). At 28 hours post fertilization (hpf) we observed the ciliopathy phenotypes of ventral body axis curvature and cell death (Fig. 6A–C). Embryos showed increased expression of phosphorylated γH2AX (Fig. 6D–E). At 48 hpf, cep164 morphants displayed the typical ciliopathy phenotype of abnormal heart looping as a laterality defect (Fig. 6F–I). Furthermore, at 72 hpf, embryos developed further NPHP-RC phenotypes, including pronephric tubule cysts (Fig. 6J–K), as well as hydrocephalus and retinal dysplasia (Fig. 6L–O).
Figure 6. Knockdown of cep164 in zebrafish embryos results in ciliopathy phenotypes, and knockdown of Cep164 or Zfp423(Znf423) causes sensitivity to DNA damage.
A morpholino-oligonucleotide (cep164 MO) targeting the exon 7 splice donor site of zebrafish cep164 was injected into fertilized eggs at the one to four-cells stage together with p53 MO (0.2 mM) to minimize non-specific MO effects.
(A–E) Whereas p53 MO injection (n=67) did not produce any phenotype (A), coinjection of cep164 MO at 28 hpf caused the mild ciliopathy phenotype of ventral body axis curvature in 48% of embryos (60/125) (B). 50% of embryos (62/125) showed severe cell death throughout the body as judged by grey-appearing cells in the head region (C). Embryos with severe cell death also showed increased expression of phosphorylated γH2AX (D) compared to p53 MO control (E). Most embryos with massive cell death did not survive beyond 48 hpf.
(F–I) At 48 hpf, surviving cep164 morphants displayed the ciliopathy phenotype of laterality defects. Whereas p53 MO did not cause any abnormal heart looping (F,G), cep164 MO caused inverted heart looping (H) or ambiguous heart looping (I). (A, atrium; L, left; V, ventricle)
(J–M) At 72 hpf, embryos developed further ciliopathy phenotypes. When compared to p53 MO controls (J), pronephric tubules (arrow heads) exhibited cystic dilation (K, asterisks) in 25% (7/28) of embryos. In addition, when compared to p53 MO controls (L), 0% (0/67) of embryos showed kidney cysts, hydrocephalus (asterisk), or retinal dysplasia (brackets) (M).
(N) At 0.2 mM, cep164 MO knockdown effectively altered mRNA processing as revealed by RT-PCR. The wild type (WT) mRNA product is 339 bp. A shorter aberrantly spliced mRNA product appeared in cep164 morphants (asterisks), and the normal mRNA product was significantly reduced. p53 MO alone did not affect cep164 mRNA processing. See also Figure S7.
(O) Quantification of γ-H2AX levels in cep164 MO morphants. Whole fish lysates were prepared from morphants injected with control MO (p53 0.2 mM) or cep164 MO (p53 0.2 mM, cep164 0.2 mM). Injection of cep164-targeting MO causes upregulation of γ-H2AX in cep164 kd embryos signifying perturbed DDR. γ-H2AX levels correlate with the phenotypic severity of the cep164 morphants (see panels A–C). Anti-α-tubulin antibody was used to show equal loading.
(P–Q) Cep164-deficient IMCD3 cells exhibit radiation sensitivity. In IMCD3 cells transduced with shRNA retrovirus, Cep164 expression was suppressed by shRNA knockdown to about 20% of control as judged by qPCR (P). Cep164 knockdown resulted in a dose-dependent increase of γH2AX positive cells in a FACS assay, signifying increased radiation sensitivity to IR and perturbed DDR. See also Figure S8. In (Q) the level of significance of two-tailed t-test (p<0.001) is indicated by an asterisk.
(R–T) Zfp423(Znf423)-deficient P19 cells exhibit radiation sensitivity. P19 cells transduced with shRNA lentivirus were exposed to the indicated level X-irradiation. Zfp423 and γH2AX immunofluorescence was quantified in matched replicate cultures for each virus 2h after irradiation.
(R) Representative images illustrate dose-responsiveness of γH2AX and effective knockdown of Zfp423 expression.
(S) γH2AX intensity normalized to DAPI+ nuclei is increased following IR at 0.5 and 1.0 Gy, signifying increased IR sensitivity and perturbed DDR (2 fields from each of 6 replicate cultures per condition). Asterisks, uncorrected pair-wise p<0.05, Mann-Whitney U test, 2 tails.
(T) Histogram shows average γH2AX intensity per cell in 16 additional replicate cultures for each shRNA at 1.0 Gy exposure. P=0.018, Mann-Whitney U test, 2 tails. See also Figure S8.
Depletion of CEP164 or ZNF423(Zfp423) causes sensitivity to DNA damaging agents
To assess whether depletion of CEP164 causes sensitivity to DNA damage, Cep164 expression was stably suppressed in the mouse renal cell line IMCD3 (Fig. 6P–Q). Cep164 knockdown resulted in a dose-dependent increase of γH2AX intensity levels in a FACS analysis, signifying increased radiation sensitivity to IR and perturbed DDR. Cellular sensitivity to IR was also seen in cells depleted of CEP164 using a multicolor competition assay (MCA) (Smogorzewska et al., 2007) (Figure S7A–B).
To test whether ZNF423(Zfp423) affects DDR, we examined P19 cells, which express high levels of endogenous Zfp423 (Fig. 6R–T). Replicate cultures infected with lentivirus expressing either scrambled control or Zfp423-targeted shRNA were exposed to 0–10 Gy of X-irradiation and imaged for Zfp423 and nuclear γH2AX foci (Fig. 6R). Quantification showed significantly increased γH2AX intensities in Zfp423-depleted cells at lower (0.5 and 1.0 Gy) exposures (Fig. 6S), but the effect was non-significant when corrected for number of exposures. To determine whether sensitivity to lower dose is reproducible, we exposed 32 additional cultures at 1.0 Gy (Fig. 6T). Normalized γH2AX fluorescence in Zfp423 knockdown had both higher mean (9.6 vs. 4.7) and median (6.6 vs. 5.2) values than control (Fig. 6T). These data replicate the radiation sensitivity with high significance (p=0.018, Mann-Whitney U test, 2 tails), indicating that P19 cells require Zfp423 for quantitatively normal DDR. These results are further confirmed by multicolor competition assays Figure S7C.
DISCUSSION
Disease gene identification implicates NPHP-RC proteins in DDR
We here provide evidence that NPHP-RC proteins may play a role in DDR signaling by demonstrating, i) mutation of MRE11, ZNF423 and CEP164 in NPHP-RC individuals; ii) abrogation by an NPHP-RC mutation of the ZNF423 interaction with the DDR protein PARP1; iii) colocalization of the centrosomal proteins ZNF423, CEP164, and SDCCAG8/NPHP10 to nuclear foci with the DDR protein TIP60; iv) cellular sensitivity to DNA damaging agents upon knockdown of ZNF423 or CEP164, v) occurrence of dysregulated DDR and an NPHP-RC phenotype in zebrafish upon knockdown of cep164.
Relations of DDR to the NPHP-RC phenotype of ataxia
Many DDR signaling proteins have been shown to localize to nuclear foci as well as to centrosomes. These include among others p53, MRE11, TIP60, ATR, CHK1, CHK2, BRCA1, Rad51 (Kramer et al., 2004). In addition, dual localization of proteins at centrosomes as well as at nuclear foci has been demonstrated for multiple known DDR proteins related to ataxia or CVH. This includes MRE11 as well as the DDR proteins ATR (ataxia telangiectasia-related), CEP152, and pericentrin, all three of which are mutated in the Seckel syndrome (primordial dwarfism), a progeria syndrome with CVH (Griffith et al., 2008; Kalay et al., 2011; Murga et al., 2009). Individuals with mutations in the three NPHP-RC-causing genes that we identify here, MRE11, ZNF423 and CEP164, share the NPHP-RC phenotype of CVH phenotype and ataxia. The first protein that strongly linked DDR signaling to the ataxia phenotype was the protein ATM (ataxia telangiectasia mutated) (Savitsky et al., 1995). Shortly thereafter, mutation of MRE11, which is a direct interaction partner and activator of ATM within the MRN complex (Figure S1A), was demonstrated to cause ataxia (Stewart et al., 1999), a finding that we confirm here. The other interaction partner necessary for ATM activation within DDR signaling is the TIP60 complex, a histone acetyltransferase (HAT) multisubunit complex (Ciccia and Elledge, 2010) (Figure S1A). The TIP60 protein interacts with SC35 (Figure S1C) and, upon genotoxic stress, ceases to inhibit SC35, which causes to G2/M arrest (Edmond et al., 2010). Interestingly, we identify here in individuals with NPHP-RC, CVH and ataxia, mutations in proteins that colocalize to nuclear foci with TIP60 and/or its interaction partner SC35. These proteins are ZNF423, CEP164 and the previously identified NPHP-RC protein SDCCAG8/NPHP10 (Otto et al., 2010a).
As a further link between ataxia and mutation of NPHP-RC proteins, we had previously identified in individuals with NPHP-RC and ataxia mutation of ATXN10 (encoding ataxin-10), and demonstrated interaction of ATXN10 with NPHP5 (Sang et al., 2011) (Figure S1C). We also identify here as a direct interaction partner of CEP164 the protein TTBK2 (Figure S8), which if defective causes spinocerebellar ataxia 11 (SCA11) or pure ataxia (Houlden et al., 2007). Finally, we demonstrate that NPHP3, another protein that is deficient in NPHP-RC with CVH and ataxia (Bergmann et al., 2008), interacts with CEP164 (Figure S9). Our findings support the notion that many products of genes, which if mutated cause ataxia or NPHP-RC with ataxia, play a role in DDR and are part of a dynamic protein complex.
A DDR-based pathogenic hypothesis of dysplasia and degeneration in NPHP-RC
We here generate evidence that proteins, which if defective cause NPHP-RC, exhibit dual localization at centrosomes as well as in nuclear foci, and that they play a role in DDR. We also demonstrate the parallel occurrence of DDR defects with an NPHP-RC phenotype upon cep164 knockdown in zebrafish. We therefore propose that defects in DDR may participate in the pathogenesis of NPHP-RC. Whereas, multiple signaling pathways have been implicated in the pathogenesis of NPHP-RC (Hildebrandt et al., 2011), including non-canonical (Simons et al., 2005) and canonical Wnt signaling (Yu et al., 2009), Shh signaling (Huangfu et al., 2003), and mitotic spindle orientation (Fischer et al., 2006), none of them consistently explains the phenotypes observed. Particularly, none of these mechanisms provides a model for the dichotomy of dysplasia phenotypes resulting from null-alleles of NPHP-RC genes versus degenerative phenotypes resulting from hypomorphic alleles of the same NPHP-RC genes. Based on the findings described in this work, we propose a pathogenic hypothesis for NPHP-RC that implicates DDR signaling as a relevant disease mechanism. Within this hypothesis loss of function of NPHP-RC proteins with a dual role in DDR and centrosomal signaling, would cause disturbance of cell cycle checkpoint control, which is particularly detrimental for embryonic and adult progenitor cell survival. This notion is in accordance with the orthologous mouse model for ZNF423 loss of function, the Zfp423−/− mouse, in which CVH with ataxia is caused by defective adult progenitor cell survival in the cerebellum (Alcaraz et al., 2006).
Within this pathogenic hypothesis for NPHP-RC, a DDR signaling defect would lead to impairment of cell cycle checkpoint control, which in turn would cause lack of progenitor cells. This hypothesis could lend a possible explanation to the following persisting conundrum of NPHP-RC pathogenesis: In certain NPHP genes (e.g., NPHP3, 6, or 8) null mutations cause severe, congenital-onset phenotypes of dysplasia and malformation in kidney, eye, CVH, and liver, whereas hypomorphic mutations in the same gene cause only late-onset degenerative phenotypes such as renal tubular degeneration and fibrosis (nephronophthisis), retinal degeneration (Senior-Loken syndrome), and liver fibrosis. However, the disease mechanisms of neither the degenerative nor the dysplastic phenotypes are understood. These findings suggest that null mutations act during morphogenesis in embryonic development causing dysplasia and malformation, whereas hypomorphic mutations act during the “chronic” processes of tissue maintenance and repair, which are spread out over months or years of the life of an organism. Because DDR signaling is in high demand under conditions of high proliferation during development (morphogenesis), causing high “replication stress” to progenitor cells, tissue dysplasia would be an expected pathogenic outcome. Conversely, during tissue maintenance, low replication stress would be expected, but persistent DDR impairment would allow slow accumulation of DNA damage with a phenotype of tissue degeneration.
At least two related findings support this model: i) In a mouse conditional knockout model of the cystic kidney disease gene Pkhd1, knockout of the gene before two weeks of postnatal life, up to which a high proliferation state prevailed, caused (dysplastic) kidney cysts, whereas knockdown after two weeks of postnatal life, when proliferation rate was shown to be dramatically reduced, only caused occasional cysts, the number of which increased when tissue injury was induced (Piontek et al., 2007). This phenomenon could be explained by different degrees of replication stress, and thereby DDR activation, under different proliferation rates. ii) In Seckel syndrome (primordial dwarfism), a progeria syndrome with CVH caused by mutation of the centrosomal and DDR proteins ATR, CEP152, or pericentrin, the degree of cerebellar impairment is dependent on cell proliferation state (Kalay et al., 2011; Murga et al., 2009; Rauch et al., 2008).
Thus a DDR-related pathomechanism could explain how recessive mutation of the same NPHP-RC gene may cause both, dysplasia and malformation (in Meckel syndrome) versus slow tissue degeneration (in NPHP) due to high versus low replication stress, respectively. In this model null mutations would act during the high proliferation states of embryonic development/morphogenesis with high replication stress, causing organ dysplasia and malformation, whereas hypomorphic mutations of the same NPHP-RC gene would act during the low-proliferation state of tissue maintenance with low proliferation stress, causing organ degeneration and fibrosis by accumulation of DNA damage over the course of years. Both phenotypes could be mediated through rapid loss of embryonic progenitor cells versus slow loss of adult progenitor cells. Under this hypothesis the degenerative phenotypes of NPHP-RC would represent a disease mechanism of organ-specific premature aging mediated by DDR defects.
A DDR-based pathogenic hypothesis might explain specific organ involvement in NPHP-RC
Another conundrum of NPHP-RC pathogenesis is the question why organ degeneration occurs in specific organs and at characteristic sites. Most prominently there is involvement of kidney, eye, liver and cerebellum. In the kidney, fibrosis and cysts are most prominent at the corticomedully junction of the kidneys. In the eye, there is retinal degeneration. In the liver, fibrosis is strictly periportal, i.e. surrounding the bile ducts. Along the lines of a DRR-related pathogenic hypothesis of NPHP-RC (see above), it is tempting to speculate that the specific tissue regions or cell types affected in NPHP-RC are more strongly exposed to genotoxins. In the kidney, the distal convoluted tubule segment, for which we here show increased γH2AX staining as a reflection of pronounced DDR (Figure 6D), and around which most fibrotic changes occur, could be more strongly exposed to genotoxins such as hydroxyurea. Retinal degeneration could theoretically be caused by slow postnatal accumulation of UV light-induced DNA damage. Most strikingly, bile duct-surrounding cholangiocytes in the liver are the one mammalian cell type that is most strongly exposed to genotoxins that are generated as metabolic end-products by the liver for excretion in bile.
In summary, a testable pathogenic hypothesis of NPHP-RC that implicates DDR signaling, impaired cell cycle checkpoint control with lack of progenitor cells might potentially explain some of the ill understood features of ciliopathies:
It might provide a mechanism for the dual phenotypes of degeneration/dysplasia seen in NPHP-RC in kidney, eye, cerebellum and liver.
It would implicate in the NPHP-RC pathogenesis, lack of response to replication stress-sensing as a functional basis for understanding the dualism of dysplasia that occurs in high-proliferation states during development/morphogenesis or repair versus degeneration, which occurs during the low proliferation state of tissue maintenance.
It would characterize the degenerative phenotypes as diseases of organ-specific premature aging, thereby pointing in new directions for identification of small compounds for therapy including cyclin inhibitors.
EXPERIMENTAL PROCEDURES
Research subjects
We obtained blood samples and pedigrees following informed consent from individuals with NPHP-RC and/or their parents. Approval for human subjects research was obtained from the University of Michigan Institutional Review Board, McGill University Health Centre and the other institutions involved. The diagnosis of NPHP-RC was based on published clinical criteria (Chaki et al., 2011). Rat studies were performed according to Dutch Animal Welfare laws and were locally reviewed by the University of Utrecht Animal Ethical Committee (DEC). Human subjects provided informed consent to the use of their tissue for research purposes.
Linkage analysis
For genome-wide homozygosity mapping the GeneChip® Human Mapping 250k StyI Array from Affymetrix was used. Non-parametric LOD scores were calculated using a modified version of the program GENEHUNTER 2.1 (Kruglyak et al., 1996; Strauch et al., 2000) through stepwise use of a sliding window with sets of 110 SNPs and the program ALLEGRO (Gudbjartsson et al., 2000) in order to identify regions of homozygosity as described (Hildebrandt et al., 2009c; Sayer et al., 2006) using a disease allele frequency of 0.0001 and Caucasian marker allele frequencies.
Bioinformatics
Genetic location is according to the February 2009 Human Genome Browser data (http://www.genome.ucsc.edu).
Statistical analysis
Student’s two-tailed non-paired t-tests and normal distribution two-tailed z-tests were carried out using pooled standard error and s.d. values to determine the statistical significance of different cohorts.
Supplementary Material
ARTICLE HIGHLIGHTS.
Mutations of ZNF423 or CEP164 are novel causes of retinal-renal ciliopathies.
The gene products colocalize with TIP60 to both, centrosomes and nuclear foci.
Knockdown of ZNF423 or CEP164 impairs DNA damage response (DDR) signaling.
Knockdown of cep164 in zebrafish causes a ciliopathy phenotype and dysregulated DDR.
Acknowledgments
We are grateful to the study individuals for their contributions.
This research was supported by grants from the National Institutes of Health to F.H. (DK068306, DK090917), to B.A.H (NS054871, NS060109), to N.K. (HD042601, DK075972, DK072301), from the March of Dimes Foundation to F.H. (FY11-241), from the Center for Organogenesis of the University of Michigan to F.H., by grants from the Netherlands Organization for Scientific Research to R.R. (NWO Vidi-91786396) and to R.H.G. (NWO Vidi-917.66.354), by the Avenir-INSERM program, the Agence Nationale pour la Recherche, the Union Nationale pour les Aveugles et Déficients Visuels, RETINA France, Programme Hospitalier de Recherche National 2007, and the Association Bardet-Biedl, France to H.D. and C.S.
This research was supported by grants from the European Community’s Seventh Framework Programme FP7/2009 under grant agreement no: 241955, SYSCILIA (to R.H.G., G.G.S., N.K., R.R., H.O., and G.W.).
This research was supported by grants from the Dutch Kidney Foundation (KJPB09.009 and IP11.58 to H.H.A).
R.C. is supported by grants from the Retina Research Foundation and the National Eye Institute (R01EY018571). H.W. was supported by postdoctoral fellowship F32EY19430 of the National Institutes of Health.
R.K.K. is supported by FFB-Canada, CIHR, FRSQ, and Reseau Vision.
V.B. is supported by MSM0021622430 (Ministry of Education, Youth and Sports of the Czech Republic), AVOZ50040507, AVOZ50040702 (Academy of Sciences of the Czech Republic), 204/09/H058, and 204/09/0498 (Czech Science Foundation) and EMBO Installation Grant; I.C. is supported by the programme Brno PhD Talent“ of South Moravian Center for International Mobility.
S.S. is a laureate of the Equipe FRM (DEQ20071210558) and the Agence National de la Recherche (R09087KS, R11012KK).
This study was supported by the DFG (SCHE1562 and SFB832 to B.S.; SFB829 to T.B.; SFB592 to H.O.).
T.B. was supported by SFB829 of the German Research Foundation. B.S. was supported by SFB832 of the German Research Foundation.
J.A.S. is a GlaxoSmithKline clinician scientist.
J.S.A is supported by the Lundbeck Foundation.
F.H. is an Investigator of the Howard Hughes Medical Institute, a Frederick G. L. Huetwell Professor, and a Doris Duke Distinguished Clinical Scientist.
A.S. is supported by the Burroughs Wellcome Fund Career Award for Medical Scientists and the Doris Duke Charitable Foundation Clinical Scientist Development Awards, and is a Rita Allen Foundation and an Irma T. Hirschl scholar.
We thank Emad B. Abboud, Ali A. Al-Rajhi, and Richard Alan Lewis for sample collection, Randy Moon (University of Washington) for providing plasmids and to Peter Konik (University of South Bohemia) for help with the mass-spectrometry, Carsten Janke for providing GT335 antibody, Jeff Salisbury for providing centrin-2 antibody, Eric Nigg for providing Cep164 antibody, Bo Chang for Cep290 antibody, and Frits Meeuwsen and Nathalie Prince for technical assistance, and Patricia Furman for contribution of clinical data.
This work utilized two Cores of the Michigan Diabetes Research and Training Center funded by DK020572 from the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
ACCESSION NUMBERS
CEP164, NP_055771; MRE11, NM_014956.4 ; ZNF423, NM_015069.2; CCDC92, NP_079416; TTBK2; NP_775771 and NP_001193000.
Competing interests statement
The authors declare that they have no competing financial interests.
AUTHOR CONTRIBUTIONS
M.C., R.C., H.W., A.C., G.R., S.J., S.J.A., S.N., J.O.T., M.A., G.N., P.N., S.L., and E.A.O. generated data on total genome linkage, whole exome resequencing, and gene identification.
R.A., performed high-resolution confocal microscopy and UV sensitivity studies.
Spheroid assay, anaphase lag, tissue γH2AX, and cell cycle studies were contributed by R.H.G, G.G.S., J.A.J. and A.K.G., who generated clonal inducible cell lines. J.A.J contributed to all studies in rats. R.G. supplied human samples and all blinded pathological analysis.
R.C. independently mapped and identified SDCCAG8 in family KKSEH001-7.
W.Z., S.W., C.A., and I.D. generated phenotypic data in zebrafish.
T.W.H., R.A., M.C., H.Y.G., A.K.G., L.E., and J.A.S. performed immunofluorescence, subcellular localization and coIP studies.
C.J.H and B.A.H. performed genetic and functional studies on ZNF423.
Protein interaction data was generated by J.S.A., K.V., L.E. and J.A.S. for ZNF423 and CEP290, by H.H.A., J.V.R., M.M.O., S.J.F.L., and R.R. for TTKB2 and CCDC92, by A.G. and G.W. for NPHP3, by I.C., R.S.G. and V.B. for Dvl3, by B.S., M.L. and T.B. and by J.S.
J.A.J. contributed to control studies in rats.
Roscovitine effect was studied by H.H. and O.I.-B.
S.S., C.A., R.K.K., H.R., I.L., A.N., C.S., H.D., R.M., J.G.G., P.F., S.A., D.G.D., A.L., C.G., N.K., L.P., E.B.A, A.A.A-R., R.A.L. J.R.L., H.O., and C.A.J. recruited patients, gathered detailed clinical information for the study, and/or performed exon sequencing.
A.S., E.G., J.M.S., and R.S. performed DDR studies on CEP164 and ZNF423.
F.H. conceived of and directed the entire project, and wrote the paper with additional input from collaborators.
Supplemental information includes Extended Experimental procedures, one table, 7 figures and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcaraz WA, Chen E, Valdes P, Kim E, Lo YH, Vo J, Hamilton BA. Modifier genes and non-genetic factors reshape anatomical deficits in Zfp423-deficient mice. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaraz WA, Gold DA, Raponi E, Gent PM, Concepcion D, Hamilton BA. Zfp423 controls proliferation and differentiation of neural precursors in cerebellar vermis formation. Proc Natl Acad Sci U S A. 2006;103:19424–19429. doi: 10.1073/pnas.0609184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, MacCoss MJ, Moon RT. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol. 2006;8:348–357. doi: 10.1038/ncb1381. [DOI] [PubMed] [Google Scholar]
- Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, Kim JC, Ross AJ, Eichers ER, Teslovich TM, et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- Attanasio M, Uhlenhaut NH, Sousa VH, O’Toole JF, Otto E, Anlag K, Klugmann C, Treier AC, Helou J, Sayer JA, et al. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nat Genet. 2007;39:1018–1024. doi: 10.1038/ng2072. [DOI] [PubMed] [Google Scholar]
- Bergmann C, Fliegauf M, Bruchle NO, Frank V, Olbrich H, Kirschner J, Schermer B, Schmedding I, Kispert A, Kranzlin B, et al. Loss of nephrocystin-3 function can cause embryonic lethality, Meckel-Gruber-like syndrome, situs inversus, and renal-hepatic-pancreatic dysplasia. Am J Hum Genet. 2008;82:959–970. doi: 10.1016/j.ajhg.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachon S, Gopalakrishnan J, Omori Y, Polyanovsky A, Church A, Nicastro D, Malicki J, Avidor-Reiss T. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics. 2008;180:2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukanov NO, Smith LA, Klinger KW, Ledbetter SR, Ibraghimov-Beskrovnaya O. Long-lasting arrest of murine polycystic kidney disease with CDK inhibitor roscovitine. Nature. 2006;444:949–952. doi: 10.1038/nature05348. [DOI] [PubMed] [Google Scholar]
- Chaki M, Hoefele J, Allen SJ, Ramaswami G, Janssen S, Bergmann C, Heckenlively JR, Otto EA, Hildebrandt F. Genotype-phenotype correlation in 440 patients with NPHP-related ciliopathies. Kidney Int. 2011 doi: 10.1038/ki.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Khanna H, Hawes N, Jimeno D, He S, Lillo C, Parapuram SK, Cheng H, Scott A, Hurd RE, et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum Mol Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LE, Zhang J, Reed RR. The transcription factor Zfp423/OAZ is required for cerebellar development and CNS midline patterning. Dev Biol. 2007;307:43–52. doi: 10.1016/j.ydbio.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delous M, Baala L, Salomon R, Laclef C, Vierkotten J, Tory K, Golzio C, Lacoste T, Besse L, Ozilou C, et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat Genet. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- den Hollander AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KE, Zonneveld MN, Strom TM, Meitinger T, Brunner HG, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- Edmond V, Moysan E, Khochbin S, Matthias P, Brambilla C, Brambilla E, Gazzeri S, Eymin B. Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J. 2010 doi: 10.1038/emboj.2010.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias E, Lalun N, Lorenzato M, Blache L, Chelidze P, O’Donohue MF, Ploton D, Bobichon H. Cell-cycle-dependent three-dimensional redistribution of nuclear proteins, P 120, pKi-67, and SC 35 splicing factor, in the presence of the topoisomerase I inhibitor camptothecin. Exp Cell Res. 2003;291:176–188. doi: 10.1016/s0014-4827(03)00377-x. [DOI] [PubMed] [Google Scholar]
- Fischer E, Legue E, Doyen A, Nato F, Nicolas JF, Torres V, Yaniv M, Pontoglio M. Defective planar cell polarity in polycystic kidney disease. Nat Genet. 2006;38:21–23. doi: 10.1038/ng1701. [DOI] [PubMed] [Google Scholar]
- Germino GG. Linking cilia to Wnts. Nat Genet. 2005;37:455–457. doi: 10.1038/ng0505-455. [DOI] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith E, Walker S, Martin CA, Vagnarelli P, Stiff T, Vernay B, Al Sanna N, Saggar A, Hamel B, Earnshaw WC, et al. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet. 2008;40:232–236. doi: 10.1038/ng.2007.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- Helou J, Otto EA, Attanasio M, Allen SJ, Parisi MA, Glass I, Utsch B, Hashmi S, Fazzi E, Omran H, et al. Mutation analysis of NPHP6/CEP290 in patients with Joubert syndrome and Senior-Loken syndrome. J Med Genet. 2007;44:657–663. doi: 10.1136/jmg.2007.052027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Attanasio M, Otto E. Nephronophthisis: disease mechanisms of a ciliopathy. J Am Soc Nephrol. 2009a;20:23–35. doi: 10.1681/ASN.2008050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Heeringa SF, Ruschendorf F, Attanasio M, Nurnberg G, Becker C, Seelow D, Huebner N, Chernin G, Vlangos CN, et al. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 2009b;5:e1000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Heeringa SF, Rüschendorf F, Attanasio M, Nürnberg G, Becker C, Seelow D, Huebner N, Chernin G, Vlangos CN, et al. A Systematic Approach to Mapping Recessive Disease Genes in Individuals from Outbred Populations. PloS Genetics. 2009c;5:31000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Otto E, Rensing C, Nothwang HG, Vollmer M, Adolphs J, Hanusch H, Brandis M. A novel gene encoding an SH3 domain protein is mutated in nephronophthisis type 1. Nat Genet. 1997;17:149–153. doi: 10.1038/ng1097-149. [DOI] [PubMed] [Google Scholar]
- Houlden H, Johnson J, Gardner-Thorpe C, Lashley T, Hernandez D, Worth P, Singleton AB, Hilton DA, Holton J, Revesz T, et al. Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11. Nat Genet. 2007;39:1434–1436. doi: 10.1038/ng.2007.43. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Janderova-Rossmeislova L, Novakova Z, Vlasakova J, Philimonenko V, Hozak P, Hodny Z. PML protein association with specific nucleolar structures differs in normal, tumor and senescent human cells. J Struct Biol. 2007;159:56–70. doi: 10.1016/j.jsb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, Bicknell LS, Kayserili H, Li Y, Tuysuz B, Nurnberg G, et al. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet. 2011;43:23–26. doi: 10.1038/ng.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Lukas J, Bartek J. Checking out the centrosome. Cell Cycle. 2004;3:1390–1393. doi: 10.4161/cc.3.11.1252. [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- Ku MC, Stewart S, Hata A. Poly(ADP-ribose) polymerase 1 interacts with OAZ and regulates BMP-target genes. Biochem Biophys Res Commun. 2003;311:702–707. doi: 10.1016/j.bbrc.2003.10.053. [DOI] [PubMed] [Google Scholar]
- Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Jr, Rux JJ, Speicher DW, Pierce EA. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy CA, Xu X, Bansbach CE, Glick GG, Zhao R, Ye F, Sirbu BM, Titus LC, Shyr Y, Cortez D. Functional genomic screens identify CINP as a genome maintenance protein. Proc Natl Acad Sci U S A. 2009;106:19304–19309. doi: 10.1073/pnas.0909345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Enders GH. Cdk inhibition in human cells compromises chk1 function and activates a DNA damage response. Cancer Res. 2005;65:780–786. [PubMed] [Google Scholar]
- Mollet G, Salomon R, Gribouval O, Silbermann F, Bacq D, Landthaler G, Milford D, Nayir A, Rizzoni G, Antignac C, et al. The gene mutated in juvenile nephronophthisis type 4 encodes a novel protein that interacts with nephrocystin. Nat Genet. 2002;32:300–305. doi: 10.1038/ng996. [DOI] [PubMed] [Google Scholar]
- Murga M, Bunting S, Montana MF, Soria R, Mulero F, Canamero M, Lee Y, McKinnon PJ, Nussenzweig A, Fernandez-Capetillo O. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet. 2009;41:891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- Olbrich H, Fliegauf M, Hoefele J, Kispert A, Otto E, Volz A, Wolf MT, Sasmaz G, Trauer U, Reinhardt R, et al. Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat Genet. 2003;34:455–459. doi: 10.1038/ng1216. [DOI] [PubMed] [Google Scholar]
- Otto E, Hoefele J, Ruf R, Mueller AM, Hiller KS, Wolf MT, Schuermann MJ, Becker A, Birkenhager R, Sudbrak R, et al. A gene mutated in nephronophthisis and retinitis pigmentosa encodes a novel protein, nephroretinin, conserved in evolution. Am J Hum Genet. 2002;71:1167–1171. doi: 10.1086/344395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto E, Loeys B, Khanna H, Hellemans J, Sudbrak R, Fan S, Muerb U, O’Toole JF, Helou J, Attanasio M, Utsch B, Sayer JA, Lillo C, Jimeno D, Coucke P, De Paepe A, Reinhardt R, Klages S, Tsuda M, Kawakami I, Kusakabe T, Omran H, Imm A, Tippens M, Raymond PA, Hill J, Beales P, He S, Kispert A, Margolis B, Williams DS, Swaroop A, Hildebrandt F. A novel ciliary IQ domain protein, NPHP5, is mutated in Senior-Loken syndrome (nephronophthisis with retinitis pigmentosa), and interacts with RPGR and calmodulin. Nat Genet. 2005;37:282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- Otto EA, Hurd TW, Airik R, Chaki M, Zhou W, Stoetzel C, Patil SB, Levy S, Ghosh AK, Murga-Zamalloa CA, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010a;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto EA, Ramaswami G, Janssen S, Chaki M, Allen SJ, Zhou W, Airik R, Hurd TW, Ghosh AK, Wolf MT, et al. Mutation analysis of 18 nephronophthisis associated ciliopathy disease genes using a DNA pooling and next generation sequencing strategy. J Med Genet. 2010b doi: 10.1136/jmg.2010.082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto EA, Schermer B, Obara T, O’Toole JF, Hiller KS, Mueller AM, Ruf RG, Hoefele J, Beekmann F, Landau D, et al. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat Genet. 2003;34:413–420. doi: 10.1038/ng1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto EA, Trapp ML, Schultheiss UT, Helou J, Quarmby LM, Hildebrandt F. NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. J Am Soc Nephrol. 2008;19:587–592. doi: 10.1681/ASN.2007040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YR, Lee EY. UV-dependent interaction between Cep164 and XPA mediates localization of Cep164 at sites of DNA damage and UV sensitivity. Cell Cycle. 2009;8:655–664. doi: 10.4161/cc.8.4.7844. [DOI] [PubMed] [Google Scholar]
- Piontek K, Menezes LF, Garcia-Gonzalez MA, Huso DL, Germino GG. A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat Med. 2007;13:1490–1495. doi: 10.1038/nm1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Thiel CT, Schindler D, Wick U, Crow YJ, Ekici AB, van Essen AJ, Goecke TO, Al-Gazali L, Chrzanowska KH, et al. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science. 2008;319:816–819. doi: 10.1126/science.1151174. [DOI] [PubMed] [Google Scholar]
- Reinhardt HC, Cannell IG, Morandell S, Yaffe MB. Is post-transcriptional stabilization, splicing and translation of selective mRNAs a key to the DNA damage response? Cell Cycle. 2011;10:23–27. doi: 10.4161/cc.10.1.14351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, Baye LM, Wen X, Scales SJ, Kwong M, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Sayer JA, Otto EA, O’Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV, et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- Schafer T, Putz M, Lienkamp S, Ganner A, Bergbreiter A, Ramachandran H, Gieloff V, Gerner M, Mattonet C, Czarnecki PG, et al. Genetic and physical interaction between the NPHP5 and NPHP6 gene products. Hum Mol Genet. 2008;17:3655–3662. doi: 10.1093/hmg/ddn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Gloy J, Ganner A, Bullerkotte A, Bashkurov M, Kronig C, Schermer B, Benzing T, Cabello OA, Jenny A, et al. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat Genet. 2005;37:537–543. doi: 10.1038/ng1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubramaniam S, Sun X, Pan YR, Wang S, Lee EY. Cep164 is a mediator protein required for the maintenance of genomic stability through modulation of MDC1, RPA, and CHK1. Genes Dev. 2008;22:587–600. doi: 10.1101/gad.1627708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Strauch K, Fimmers R, Kurz T, Deichmann KA, Wienker TF, Baur MP. Parametric and nonparametric multipoint linkage analysis with imprinting and two-locus-trait models: application to mite sensitization. Am J Hum Genet. 2000;66:1945–1957. doi: 10.1086/302911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan A, Nesslany F, Violet M, Begard S, Loyens A, Talahari S, Mansuroglu Z, Marzin D, Sergeant N, Humez S, et al. Nuclear tau, a key player in neuronal DNA protection. J Biol Chem. 2011;286:4566–4575. doi: 10.1074/jbc.M110.199976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczerbal I, Bridger JM. Association of adipogenic genes with SC-35 domains during porcine adipogenesis. Chromosome Res. 2010;18:887–895. doi: 10.1007/s10577-010-9176-1. [DOI] [PubMed] [Google Scholar]
- Tibelius A, Marhold J, Zentgraf H, Heilig CE, Neitzel H, Ducommun B, Rauch A, Ho AD, Bartek J, Kramer A. Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. J Cell Biol. 2009;185:1149–1157. doi: 10.1083/jcb.200810159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, Reed RR. Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz. Mol Cell Biol. 1998;18:6447–6456. doi: 10.1128/mcb.18.11.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Silhavy JL, Brancati F, Barrano G, Krishnaswami SR, Castori M, Lancaster MA, Boltshauser E, Boccone L, Al-Gazali L, et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- Xiao R, Sun Y, Ding JH, Lin S, Rose DW, Rosenfeld MG, Fu XD, Li X. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol Cell Biol. 2007;27:5393–5402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Carroll TJ, Rajagopal J, Kobayashi A, Ren Q, McMahon AP. A Wnt7b-dependent pathway regulates the orientation of epithelial cell division and establishes the cortico-medullary axis of the mammalian kidney. Development. 2009;136:161–171. doi: 10.1242/dev.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.