Abstract
To discover novel biomarkers for early detection of human lung squamous cell cancer (LSCC) and explore possible mechanisms of LSCC carcinogenesis, iTRAQ-tagging combined with two dimensional liquid chromatography tandem MS analysis was used to identify differentially expressed proteins in human bronchial epithelial carcinogenic process using laser capture microdissection-purified normal bronchial epithelium (NBE), squamous metaplasia (SM), atypical hyperplasia (AH), carcinoma in situ (CIS) and invasive LSCC. As a result, 102 differentially expressed proteins were identified, and three differential proteins (GSTP1, HSPB1 and CKB) showing progressively expressional changes in the carcinogenic process were selectively validated by Western blotting. Immunohistochemistry was performed to detect the expression of the three proteins in an independent set of paraffin-embedded archival specimens including various stage tissues of bronchial epithelial carcinogenesis, and their ability for early detection of LSCC was evaluated by receiver operating characteristic analysis. The results showed that the combination of the three proteins could perfectly discriminate NBE from preneoplastic lesions (SM, AH and CIS) from invasive LSCC, achieving a sensitivity of 96% and a specificity of 92% in discriminating NBE from preneoplatic lesions, a sensitivity of 100% and a specificity of 98% in discriminating NBE from invasive LSCC, and a sensitivity of 92% and a specificity of 91% in discriminating preneoplatic lesions from invasive LSCC, respectively. Furthermore, we knocked down GSTP1 in immortalized human bronchial epithelial cell line 16HBE cells, and then measured their susceptibility to carcinogen benzo(a)pyrene-induced cell transformation. The results showed that GSTP1 knockdown significantly increased the efficiency of benzo(a)pyrene-induced 16HBE cell transformation. The present data first time show that GSTP1, HSPB1 and CKB are novel potential biomarkers for early detection of LSCC, and GSTP1 down-regulation is involved in human bronchial epithelial carcinogenesis.
Lung cancer is the most frequently occurring malignancy with increasing incidence and is the leading cause of mortality in cancer-related deaths in China and worldwide (1, 2). Although great improvement has been made in diagnosis and treatment of lung cancer, the overall patients' survival is still very low and does not exceed 15% (3). The poor prognosis of this cancer is mainly explained by the fact that the diagnosis is generally made only at advanced stages because of the lack of reliable, early diagnostic biomarkers and the limited understanding of its carcinogenic mechanisms. Therefore, identification of biomarkers for early detection of lung cancer is mandatory, in turn leading to more effective treatment and reduction of mortality.
Lung squamous cell carcinoma (LSCC)1 originated from the bronchial epithelial cells is the most common histological type of lung cancer. It is known that carcinogenesis of LSCC is a multistage process and the result of multistep accumulation of genetic and epigenetic alterations (4). With exposure to environmental carcinogens, bronchial epithelial carcinogenesis often progresses in the following manner: hyperplasia, squamous metaplasia (SM), atypical hyperplasia (AH), cancer in situ (CIS) and invasive cancer (5). LSCC is the end-point of a whole range of morphological abnormalities that are displayed in the bronchial epithelia of the patients with LSCC and/or smokers (5), and that could be used to identify key proteins associated with the ongoing carcinogenic process.
Analysis of differentially expressed proteins in LSCC using proteomics revealed that expression and modified levels of proteins have some predictive power for clinical outcome and personalized risk assessment (6–9). Our previous studies using proteomics based on 2-DE and MS identified the differential tissue and serum proteins in LSCC leading to discovery of potential biomarkers for diagnosis or prognosis of LSCC (10–13). Although a number of proteomic studies on lung cancer have been reported (6–17), little is known about the changes of protein expressional profiles in the human bronchial epithelial carcinogenic process (18), and there are no clinically established biomarkers available for early detection of LSCC. Comparative proteomics analysis of successive stages of human bronchial epithelial carcinogenesis is the most direct and persuasive way to find biomarkers for early diagnosis of LSCC. A major obstacle, however, to the analysis of tissue specimens is tissue heterogeneity, which is particularly relevant to bronchial preneoplastic lesions as these tissues only include a little of target cells. Several approaches have been employed to obtain homogeneous cell populations from a heterogeneous tissue, such as short-term cell culture and laser capture microdissection (LCM). Since 1996, LCM has emerged as a good choice for purifying target cells from tissues (19).
Isobaric tags for relative and absolute quantitation (iTRAQ) in combination with two dimensional liquid chromatography tandem MS (2D LC-MS/MS) analysis is emerging as one of the more powerful quantitative proteomics methodologies in the search for tumor biomarkers (20–23). In the iTRAQ technology, tagging is on primary amines, thus potentially allowing the tagging of most tryptic peptides. The multiplexing ability afforded by the iTRAQ reagents, which are available in four to eight different tags, is ideally suited for our study because it provides us with a means to simultaneously compare proteomes in successive stages of human bronchial epithelial carcinogenesis.
To search biomarkers for early detection of LSCC and explore the possible mechanisms of bronchial epithelial carcinogenesis, in this study iTRAQ tagging followed by 2D LC-MS/MS was performed to identify differential proteins among LCM-purified bronchial epithelial carcinogenic tissues, and some differentially expressed proteins identified by proteomics were selectively validated. Furthermore, values of the three differential proteins (GSTP1, HSPB1, and CKB) with progressively expressional alterations in the bronchial epithelial carcinogenic process for early detection of LSCC were assessed by immunohistochemistry and receiver operating characteristic (ROC) curve analysis, and the roles of GSTP1 in human bronchial epithelial carcinogenic process were analyzed. We first time show that GSTP1, HSPB1, and CKB are potential biomarkers for early detection of LSCC, and demonstrate that GSTP1 is involved in human bronchial epithelial carcinogenesis.
EXPERIMENTAL PROCEDURES
Sample Collection, Laser Capture Microdissection and Protein Extraction
All fresh tissues from the LSCC patients undergoing curative surgery and receiving neither chemotherapy nor radiotherapy were obtained from Department of Cardiothoracic Surgery, The Second Xiangya Hospital of Central South University, China, and used for a proteomics analysis. The patients signed an informed consent form for the study which was approved by the local ethical committee. After surgery, tumor tissues and bronchi were removed from the resected pulmonary lobes, and stored at −80 °C. Normal bronchial epithelium (NBE), squamous metaplasia (SM), atypical hyperplasia (AH), carcinoma in situ (CIS) and invasive LSCC were obtained from the bronchi or tumor tissues, and diagnosed by pathological examination of a H&E-stained frozen tissue sections according to the 1999 World Health Organization/International Association for the Study of Lung Cancer classification (24). LCM was performed with a Leica AS LMD system to purify the cells of interest from each type of tissue as previously described by us (25). Each cell population was determined to be 95% homogeneous by microscopic visualization of the captured cells (supplementary Fig. S1).
The microdissected cells were dissolved in lysis buffer (7 m urea, 2 m thiourea, 65 mm dithiothreitol, 0.1 mm phenylmethylsulfonyl fluoride) at 4 °C for 1 h, and then centrifuged at 12,000 rpm for 30 min at 4 °C. The supernatant was collected, and the protein concentration was determined by 2D Quantification kit (Amersham Biosciences). To diminish the effect of sample biological variation on the results of a proteomics analysis, equal amounts of protein from the microdissected cells of 10 different individuals were pooled to generate one common sample for each type of tissue (NBE, SM, AH/CIS, and invasive LSCC), in turn obtaining the four pooled protein samples used for iTRAQ labeling.
An independent set of formalin-fixed and paraffin-embedded archival tissue specimens including 66 cases of NBE, 64 cases of SM, 60 cases of AH, 13 cases of CIS, 66 cases of invasive LSCC was obtained from bronchoscopic or surgical procedures at the People Hospital of Hunan Province, Changsha, China, and used for immunohistochemical staining. The patients recruited in this study received neither chemotherapy nor radiotherapy. The parameters of patients and tissue specimens are shown in supplementary Table S1.
Protein Digestion and Labeling with iTRAQ Reagents
Trypsin digestion and iTRAQ labeling were performed according to the manufacturer's protocol (Applied Biosystems, Foster City, CA). Briefly, 100 μg protein of each pooled sample was reduced and alkylated, and then digested overnight at 37 °C with trypsin (mass spectrometry grade; Promega, Madison, WI) and labeled with iTRAQ™ reagents (Applied Biosystems) as follows: NBE, iTRAQ reagent 117; SM, iTRAQ reagents 114, AH/CIS, iTRAQ reagents 116; and invasive LSCC, iTRAQ reagent 115. Four labeled digests were then mixed and dried.
Off-line 2D LC-MS/MS
The mixed peptides were fractionated by strong cation exchange chromatography on a 20AD HPLC system (Shimadzu) using a polysulfoethyl column (2.1 × 100 mm, 5 μm, 300 Å; The Nest Group Inc.) as previously described by us (26). Briefly, the mixed peptides were desalted with Sep-Pak Cartridge (Waters, Milford, MA), diluted with the loading buffer (10 mm KH2PO4 in 25% acetonitrile, pH 2.8) and loaded onto the column. Buffer A was identical in composition to the loading buffer, and buffer B was same as buffer A except containing 350 mm KCl. Separation was performed using a linear binary gradient of 0–80% buffer B in buffer A at a flow rate of 200 μl/min for 60 min. The absorbance at 214 nm and 280 nm was monitored, and a total of 30 strong cation exchange fractions were collected along the gradient.
Each strong cation exchange fraction was dried down, dissolved in buffer C (5% acetonitrile, 0.1% formic acid), and analyzed on Qstar XL (Applied Biosystems) as previously described by us (26). Briefly, peptides were separated on a reverse-phase (RB) column (ZORBAX 300SB-C18 column, 5 μm, 300Å, 0.1 × 15 mm; Micromass) using a 20AD HPLC system (Shimadzu). The HPLC gradient was 5–35% buffer D (95% acetonitrile, 0.1% formic acid) in buffer C at a flow rate of 0.2 μl/min for 65 min. Survey scans were acquired from 400–1800 with up to four precursors selected for MS/MS from m/z 100–2000 using a dynamic exclusion of 30S. The iTRAQ labeled peptides fragmented under collision-induced dissociation conditions to give reporter ions at 114.1, 115.1, 116.1, and 117.1 Th. The ratios of peak areas of the iTRAQ reporter ions reflect the relative abundances of the peptides and, consequently, the proteins in the samples. Larger, sequence-information-rich fragment ions were also produced under these MS/MS conditions and gave the identity of the protein from which the peptide originated. iTRAQ labeling followed by 2D LC-MS/MS analysis was repeated in triplicate to diminish the effect of experimental variation on the results of a proteomics analysis.
Data Analysis
The software used for data acquisition was Analyst QS 1.1 (Applied Biosystems). The software used for protein identification and quantitation was ProteinPilot™ 3.0 software (Software Revision Number: 114732; Applied Biosystems) with the integrated ParagonTM search algorithm and Pro GroupTM algorithm (Revision Number: 3.0.0.0, 113442; Applied Biosystems). The precursor tolerance was set to 0.2 Da, and the iTRAQ fragment tolerance was set to 0.2 Da. The data analysis parameters were set as follows: Sample type: iTRAQ (peptide labeled); Cys alkylation: MMTS; Digestion: Trypsin; Instrument: QSTAR ESI; Species: Homo sapiens; ID Focus: Biological modifications; Database: International Protein Index human database (version: 3.45; 143958 entries); Search Effort: Thorough; Max missed cleavages: 2; FDR Analysis: Yes; User Modified Parameter Files: No; Bias Correction: Auto; Background Correction: Yes. Identified proteins were grouped by the software to minimize redundancy. All peptides used for the calculation of protein ratios were unique to the given protein or proteins within the group, and peptides that were common to other isoforms or proteins of the same family were ignored. The protein confidence threshold cutoff is 1.3 (unused ProtScore) with at least one peptide with 95% confidence. The average iTRAQ ratios from the triplicate experiments were calculated for each protein. In addition, false discovery rate (FDR) for the protein identification was calculated by searching against a concatenated reversed database. The FDR was calculated based on the following formula: FDR = 2 × nrev/(ntar+ nrev)(27). Nrev is the number of peptide hits matched to the “reverse” protein, and ntar is the number of peptide hits matched to the target protein.
Western Blotting
Western blotting was performed in one independent set of microdissected tissues (NBE, SM, AH, CIS, and invasive LSCC, each 10 cases) as previously described by us (25). Briefly, 30 μg of lysates were separated by 8% SDS-PAGE, and transferred to polyvinylidene difluoride membranes. Blots were incubated with primary anti-GSTP1 antibody (1:500; Abcam, Cambridge, MA), anti-HSPB1 antibody (1:1000; Abcam) or anti-CKB antibody (1:400; Sigma) overnight at 4 °C, followed by incubation with a horseradish peroxidase-conjugated secondary antibody (1:3000; Amersham Biosciences) for 1 h at room temperature. The signal was visualized with ECL detection reagent (Amersham Biosciences) and quantitated by densitometry using ImageQuant image analysis system (Storm Optical Scanner, Molecular Dynamics, Sunnyvale, CA). β-actin was simultaneously detected using mouse anti-β-actin antibody (1:3000; Sigma) as a loading control.
Immunohistochemistry and Evaluation of Staining
Immunohistochemistry was performed on formalin-fixed and paraffin-embedded tissue sections using a standard. Briefly, 4 μm of tissue sections were deparaffinized, rehydrated, and treated with an antigen retrieval solution (10 mmol/L sodium citrate buffer, pH 6.0). The sections were incubated with anti-GSTP1(1:200; Abcam), anti-HSPB1 (1:100; Abcam) or anti-CKB(1:250, Sigma) antibody overnight at 4 °C, and then were incubated with 1:1000 dilution of biotinylated secondary antibody followed by avidin-biotin peroxidase complex (DAKO) according to the manufacturer's instructions. Finally, tissue sections were incubated with 3′, 3′-diaminobenzidine (Sigma) until a brown color developed, and counterstained with Harris' modified hematoxylin. In negative controls, primary antibodies were omitted.
Immunostaining was blindly evaluated by two investigators in an effort to provide a consensus on staining patterns by light microscopy. A quantitative score was performed by adding the score of staining area and the score of staining intensity for each case to assess the expression levels of the proteins as previously described by us (26). First, a quantitative score was performed by estimating the percentage of immunopositive cells: 0, no staining of cells in any microscopic fields; 1+, <30% of tissue stained positive; 2+, between 30 and 60% stained positive; and 3+, >60% stained positive. Second, the intensity of staining was scored by evaluating the average staining intensity of the positive cells (0, no staining; 1+, mild staining; 2+, moderate staining; 3+, intense staining). Finally a total score (ranging from 0–6) was obtained by adding the area score and the intensity score for each case. A combined staining score of ≤2 was considered to be negative staining (no expression); a score between 3 and 4 was considered to be moderate staining (expression); and a score between 5 and 6 was considered to be strong staining (high expression).
Statistical Analysis of Immunohistochemical Data
Statistical analysis was performed using SPSS 15.0. Difference of GSTP1, HSPB1, and CKB protein expression between the two stages of bronchial epithelial carcinogenesis (NBE versus SM, AH/CIS or LSCC; SM versus AH/CIS or invasive LSCC; AH/CIS versus invasive LSCC) was analyzed using Mann-Whitney U test. Because of the small number of CIS, it was combined with AH into one group. Moreover, the three proteins were individually, and as a panel, assessed for its ability to discriminate NBE from preneoplastic lesions (SM, AH, and CIS) from invasive LSCC by evaluating its ROC curve based on the immunohistochemistry scores as previously described by us (28). Sensitivity, specificity, positive predictive value, and negative predictive value of the three proteins were calculated individually and as a panel. A two-sided p < 0.05 was considered significant.
Stable Transfection of GSTP1 siRNA Plasmid into Human Bronchial Epithelial Cells
GSTP1 siRNA plasmid pLKO.1-GSTP1-shRNAs and empty vector pLKO.1 were purchased from Openbiosystems. For stable transfection, 1 × 107 immortalized human bronchial epithelial cell line 16HBE cells were transfected with pLKO.1-GSTP1-shRNA and pLKO.1 using Lipofectamine 2000 Reagent (Invitrogen), respectively, according to the manufacturer's instructions. After 14 days of selection in Dulbecco's modified Eagle's medium (DMEM) medium containing 1.0 μg/ml puromycin (Invitrogen), individual puromycin-resistant colonies were isolated and expanded. The expression of GSTP1 in these clones was determined by Western blotting.
Cell Culture and Carcinogen Exposures
Stably transfected 16HBE cells with pLKO.1-GSTP1-shRNAs and empty vector, and untransfected cells were cultured to 30–40% confluence in DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA). The cells were exposed to 1 μm B[a]P(Sigma) or vehicle (DMSO; Sigma) for 1 day and then recovered in fresh medium without B[a]P for 6 days. After repeated treatment with B[a]P or DMSO for 16 weeks, the cells were harvested, and subjected to analyses of cell transformation characteristics including cell proliferation, anchorage dependent and independent colony formation, cell cycle and apoptosis.
Analysis of Cell Growth in Low Serum Medium by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) assay
The cells in DMEM medium containing 0.5% fetal calf serum were plated at 1 × 104 cells per well in 96-well tissue culture plates, and grew for 7 days. Every 24 h, 20 μl of MTT (5 mg/ml; Sigma) was added to wells, and the medium was removed after 4 h of incubation. 150 μl DMSO was added to each well for 10 min at room temperature. The absorbance of each well was read with a Bio-Tek Instruments EL310 Microplate Autoreader at 490 nm. MTT assay was performed three times in triplicate.
Anchorage Dependent and Independent Colony Formation Assays
Plate colony formation and soft agar colony formation assays were done as previously described by us (29). For plate colony formation assay, the cells in DMEM medium containing 10% fetal calf serum (FCS) were seeded at 1 × 103 cells per well in six-well tissue culture plates. After growth for 10 days at 37 °C, the dishes were stained with crystal violet (Sigma) and colonies of >50 cells were counted under microscope. For soft agar colony formation assay, the cells suspended in 0.3% agar (Sigma) containing DMEM medium and 10% FCS at a density of 5 × 103 cells/ml. Next, 1 ml of the cell suspension was placed over 1 ml of 0.5% agar containing DMEM medium and 10% FCS in 6-well tissue culture plates. After plating, 1 ml of DMEM medium containing 10% FCS was added to the soft agar cultures and replenished every 3 days. Cells were allowed to grow for 12 days and colonies consisting of >50 cells were counted under microscope. All assays were performed three times in triplicate.
Flow Cytometry Analysis
The cells (1 × 106 cells) were harvested, washed twice with cold PBS buffer and fixed with 70% cold ethanol at 4 °C overnight. The fixed cells was then centrifuged, suspended in a buffer (100 mm sodium citrate and 0.1% Triton X-100), and incubated for 15 min at room temperature. The cells were incubated with 10 μg/ml RNase A (Sigma) for 10 min at room temperature, and DNA was stained with 40 μg/ml propidium iodide at 37 °C for 30 min. Samples were immediately analyzed by a FACScan flow cytometry (Becton Dickinson). For cell cycle analysis, the cells were grown in DMEM medium containing 10% FCS for 24 h, and for cell apoptosis analysis, the cells were grown in serum free DMEM medium for 24 h. Three independent experiments were done.
Hoechst 33258 Staining of Apoptotic Cells
The cells were cultured with DMEM medium containing 10% FCS in six-well tissue culture plates for 24 h and then cultured in serum free DMEM medium for another 24 h, washed with PBS, fixed with 4% paraformaldehyde for 30 min at 4 °C, and stained with 5 μg/ml cell-permeable DNA dye Hoechst 33258 (Sigma) dissolved in Hanks' buffer in the dark for 10 min. Apoptotic cells were identified on the basis of the presence of highly condensed or fragmented nuclei. To calculate the percentage of apoptotic cells, at least 200 cells from three randomized microscopic fields were counted.
Bioinformatics Analysis
To identify coregulated proteins, Hierarchical clustering was performed on the differentially expressed proteins during bronchial epithelial carcinogenesis using Cluster 3.0 and Java TreeView-1.1.6-win. Co-regulated proteins were annotated by GO using DAVID software (30). GO terms with computed p values less than 0.05 were considered as significantly enriched. KEGG pathway analysis was performed with the protein-protein interaction network of the differential proteins in each group using Cytoscape (V2.8.2) with the ClueGO v1.4 plugin. A protein-protein interaction network was constructed using VisAnt toolkit (31, 32), in which the differential proteins in each group served as a bait, and the proteins have a direct experimental interaction with the bait proteins in the databases. KEGG pathway was considered statistically significant when the corrected p value was less than 0.01.
RESULTS
Identification of Differentially Expressed Proteins during Human Bronchial Epithelial Carcinogenesis Using iTRAQ Labeling and 2D LC-MS/MS
A total of 387 nonredundant proteins were repeatedly identified by triplicate iTRAQ labeling and 2D LC-MS/MS analyses, 87.1% of which were identified with ≥2 peptide matches. The FDR for proteins identification based on searching against a reversed database was 0.0063, 0.0138, and 0.0162 in the triplicate experiments, respectively. The detailed information including information of peptide sequences, protein quantification date, average iTRAQ ratio, and distinct and common peptides with a group of proteins for these identified proteins is shown in supplementary Table S2, and the CID spectra of 50 proteins based on the single peptide identification are shown in supplementary Fig. S2.
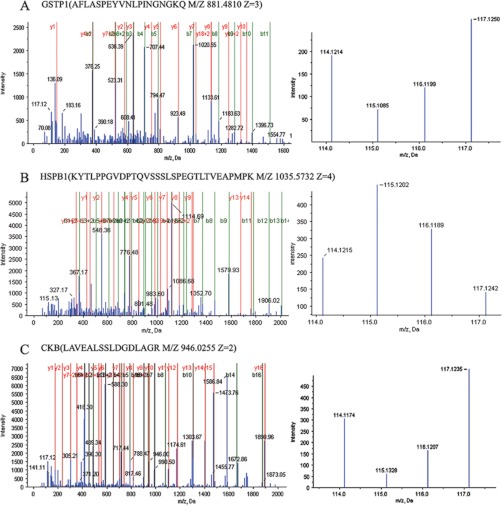
To identify the differentially expressed proteins in the bronchial epithelial carcinogenic process, protein expressional profiles between the two stages of this process (NBE versus SM, AH/CIS or LSCC; SM versus AH/CIS or LSCC; AH/CIS versus LSCC) were compared. A total of six comparisons were performed. The proteins met the following criteria were confidently considered as differentially expressed proteins: (1) proteins were repeatedly identified by the triplicate experiments; (2) proteins were identified based on ≥2 peptides; (3) proteins showed an averaged ratio-fold change ≥1.5 or ≤0.667 in the triplicate experiments between the two stages (t test, p < 0.05), and (4) proteins should differentially expressed in NBE and LSCC. As a result, 102 proteins were found to be differentially expressed in at least one of the six comparisons except differentially expressed in NBE and LSCC. The names of these 102 proteins and the stages at which their expression is significantly changed are shown in Table I. The detailed information for these differential proteins is reported in supplementary Table S2 (shown in bold). Among these differential proteins, six proteins (HSPB1, GSTP1, CKB, S100A9, SELENBP1, isoform 1 of guanylate-binding protein 6) showed progressively expressional changes during the carcinogenic process. MS/MS spectra used for the identification and quantitation of GSTP1, HSPB1, and CKB with progressively expressional changes are shown in Fig. 1.
Fig. 1.
MS/MS spectra used for the identification and quantitation of GSTP1, HSPB1 and CKB. A, (left) the sequence AFLASPEYVNLPINGNGKQ allows the identification of GSTP1; (right) the released iTRAQ reporter ions provide the relative quantitation of GSTP1 from the four tissues evaluated. B, (left) the sequence KYTLPPGVDPTQVSSSLSPEGTLTVEAPMPK allows the identification of HSPB1; (right) the released iTRAQ reporter ions provide the relative quantitation of HSPB1 from the four tissues evaluated. C, (left) the sequence LAVEALSSLDGDLAGR allows the identification of CKB; (right) the released iTRAQ reporter ions provide the relative quantitation of CKB from the four tissues evaluated. NBE, labeled with iTRAQ reagent 117; SM, labeled with iTRAQ reagents 114, AH/CIS, labeled with iTRAQ reagent 116; and invasive LSCC, labeled with iTRAQ reagent 115.
Table I. Differentially expressed proteins during human bronchial epithelial carcinogenesis.
| No | Accession # | Protein name | NBE vs. SM | NBE vs. AH/CIS | NBE vs. LSCC | SM vs. AH/CIS | SM vs. LSCC | AH/CIS vs. LSCC |
|---|---|---|---|---|---|---|---|---|
| 1 | IPI00029733.1 | Aldo-keto reductase family 1 member C1 | ↓0.028 | ↓0.625 | ↓0.017 | ↓0.017 | ||
| 2 | IPI00025512.2 | Heat shock protein beta-1 | ↓0.275 | ↓0.186 | ↓0.050 | ↓0.574 | ↓0.127 | ↓0.205 |
| 3 | IPI00221222.7 | Activated RNA polymerase II transcriptional coactivator p15 | ↓0.276 | ↓0.215 | ↓0.050 | ↓0.142 | ↓0.197 | |
| 4 | IPI00027462.1 | Protein S100-A9 | ↓0.148 | ↓0.067 | ↓0.064 | ↓0.380 | ↓0.281 | ↓0.640 |
| 5 | IPI00411765.3 | Isoform 2 of 14–3-3 protein sigma | ↓0.078 | ↓0.055 | ↓0.106 | ↓0.608 | ↑2.067 | |
| 6 | IPI00010214.1 | Protein S100-A14 | ↓0.273 | ↓0.242 | ↓0.126 | ↓0.432 | ↓0.501 | |
| 7 | IPI00217963.3 | type I cytoskeletal 16 | ↑2.112 | ↓0.067 | ↓0.140 | ↓0.037 | ↓0.088 | ↑2.530 |
| 8 | IPI00375746.4 | Isoform 1 of Guanylate-binding protein 6 | ↓0.538 | ↓0.330 | ↓0.181 | ↓0.621 | ↓0.333 | ↓0.526 |
| 9 | IPI00017334.1 | Prohibitin | ↓0.188 | ↓0.226 | ↓0.240 | |||
| 10 | IPI00453473.6 | Histone H4 | ↓0.371 | ↓0.064 | ↓0.206 | ↓0.118 | ||
| 11 | IPI00218988.4 | Isoform 2 of Adenylate kinase isoenzyme 2, mitochondrial | ↓0.402 | ↓0.611 | ↓0.220 | ↓0.360 | ↓0.237 | |
| 12 | IPI00738499.2 | Ferritin light chain | ↓0.225 | ↓0.250 | ↓0.283 | |||
| 13 | IPI00032140.4 | Serpin H1 precursor | ↓0.231 | ↓0.233 | ↓0.298 | |||
| 14 | IPI00549725.6 | Phosphoglycerate mutase 1 | ↓0.239 | ↑2.391 | ↓0.380 | ↓0.171 | ||
| 15 | IPI00013933.2 | Isoform DPI of Desmoplakin | ↓0.328 | ↓0.164 | ↓0.269 | ↓0.465 | ||
| 16 | IPI00887678.1 | LOC654188 similar to peptidylprolyl isomerase A-like | ↓0.434 | ↓0.447 | ↓0.270 | ↓0.570 | ||
| 17 | IPI00021926.2 | 26S protease regulatory subunit S10B | ↓0.446 | ↓0.584 | ↓0.281 | ↓0.427 | ↓0.322 | |
| 18 | IPI00018873.1 | Isoform 1 of Nicotinamide phosphoribosyltransferase | ↓0.102 | ↓0.285 | ↓0.170 | ↓0.373 | ||
| 19 | IPI00021347.1 | Ubiquitin-conjugating enzyme E2 L3 | ↑1.963 | ↓0.625 | ↓0.290 | ↓0.192 | ↓0.096 | ↓0.313 |
| 20 | IPI00300725.7 | type II cytoskeletal 6A | ↓0.032 | ↓0.019 | ↓0.293 | ↓0.268 | ↑19.069 | |
| 21 | IPI00024933.3 | 60S ribosomal protein L12 | ↓0.612 | ↓0.298 | ↓0.300 | ↓0.453 | ||
| 22 | IPI00219018.7 | Glyceraldehyde-3-phosphate dehydrogenase | ↓0.371 | ↓0.327 | ↓0.307 | |||
| 23 | IPI00009866.6 | type I cytoskeletal 13 | ↓0.033 | ↓0.047 | ↓0.314 | ↑1.609 | ||
| 24 | IPI00642971.3 | eukaryotic translation elongation factor 1 delta isoform 1 | ↓0.350 | ↓0.469 | ↓0.519 | |||
| 25 | IPI00007047.1 | Protein S100-A8 | ↓0.481 | ↓0.268 | ↓0.350 | ↓0.508 | ||
| 26 | IPI00646656.2 | Asparaginyl-tRNA synthetase variant (Fragment) | ↓0.359 | ↓0.292 | ↓0.330 | |||
| 27 | IPI00644989.2 | Isoform 1 of Protein disulfide-isomerase A6 precursor | ↓0.364 | ↓0.389 | ↓0.382 | |||
| 28 | IPI00015842.1 | Reticulocalbin-1 precursor | ↓0.374 | ↓0.367 | ||||
| 29 | IPI00789551.1 | Uncharacterized protein MATR3 | ↓0.393 | ↓0.463 | ↓0.264 | |||
| 30 | IPI00294834.6 | Aspartyl/asparaginyl beta-hydroxylase | ↓0.412 | ↓0.409 | ||||
| 31 | IPI00746438.2 | Isoform 2 of 60S ribosomal protein L11 | ↓0.458 | ↓0.543 | ||||
| 32 | IPI00295400.1 | Tryptophanyl-tRNA synthetase | ↑2.413 | ↓0.459 | ↓0.187 | ↓0.227 | ||
| 33 | IPI00003362.2 | HSPA5 protein | ↓0.460 | ↓0.446 | ↓0.333 | |||
| 34 | IPI00026230.1 | Heterogeneous nuclear ribonucleoprotein H2 | ↓0.552 | ↓0.466 | ↓0.572 | |||
| 35 | IPI00465315.6 | Cytochrome c | ↓0.481 | ↓0.639 | ↓0.598 | |||
| 36 | IPI00011654.2 | Tubulin beta chain | ↓0.544 | ↓0.405 | ↓0.491 | |||
| 37 | IPI00747533.2 | PGD 56 kDa protein | ↓0.514 | ↓0.436 | ||||
| 38 | IPI00024284.4 | Basement membrane-specific heparan sulfate proteoglycan core protein precursor | ↓0.518 | ↓0.664 | ↓0.468 | ↓0.466 | ||
| 39 | IPI00748905.1 | NAPB protein | ↓0.528 | |||||
| 40 | IPI00796333.1 | ALDOA 45 kDa protein | ↓0.534 | ↓0.527 | ↓0.409 | |||
| 41 | IPI00027497.5 | Glucose-6-phosphate isomerase | ↑1.593 | ↓0.550 | ↓0.472 | ↓0.348 | ||
| 42 | IPI00291006.1 | Malate dehydrogenase | ↓0.628 | ↓0.514 | ↓0.551 | |||
| 43 | IPI00848342.1 | Lactotransferrin precursor | ↓0.559 | ↓0.424 | ||||
| 44 | IPI00658109.1 | LOC100133623 Creatine kinase | ↓0.585 | |||||
| 45 | IPI00003519.1 | 116 kDa U5 small nuclear ribonucleoprotein component | ↓0.588 | ↓0.560 | ||||
| 46 | IPI00218319.3 | Isoform 2 of Tropomyosin alpha-3 chain | ↓0.637 | |||||
| 47 | IPI00296534.1 | Isoform D of Fibulin-1 precursor | ↑1.561 | ↓0.659 | ↓0.425 | ↓0.527 | ||
| 48 | IPI00009032.1 | Lupus La protein | ↑1.558 | ↑1.526 | ||||
| 49 | IPI00375145.1 | Isoform Short of Ubiquitin carboxyl-terminal hydrolase 5 | ↑1.590 | ↑2.116 | ||||
| 50 | IPI00450768.7 | type I cytoskeletal 17 | ↓0.308 | ↓0.054 | ↑1.658 | ↓0.120 | ↑4.042 | ↑18.003 |
| 51 | IPI00220766.5 | Lactoylglutathione lyase | ↑1.767 | ↑1.681 | ||||
| 52 | IPI00014898.2 | Isoform 1 of Plectin-1 | ↑2.008 | ↑2.818 | ↑2.325 | |||
| 53 | IPI00472724.1 | Elongation factor 1-alpha-like 3 | ↓0.575 | ↑2.163 | ↑3.112 | ↑3.911 | ||
| 54 | IPI00020599.1 | Calreticulin precursor | ↑2.211 | ↑2.493 | ||||
| 55 | IPI00303476.1 | ATP synthase subunit beta, mitochondrial precursor | ↑2.273 | ↑2.510 | ↑2.102 | |||
| 56 | IPI00027107.5 | Tu translation elongation factor, mitochondrial precursor | ↑2.314 | ↑2.168 | ||||
| 57 | IPI00411704.9 | Isoform 1 of Eukaryotic translation initiation factor 5A-1 | ↑2.480 | ↑2.484 | ||||
| 58 | IPI00440493.2 | ATP synthase subunit alpha | ↑2.483 | ↑2.485 | ↑2.017 | |||
| 59 | IPI00853547.1 | glucose-6-phosphate dehydrogenase isoform a | ↓0.389 | ↑2.717 | ↓0.486 | ↑3.546 | ↑7.099 | |
| 60 | IPI00183695.9 | Protein S100-A10 | ↓0.488 | ↓0.318 | ↑2.844 | ↓0.430 | ↑3.682 | ↑5.345 |
| 61 | IPI00027444.1 | Leukocyte elastase inhibitor | ↓0.578 | ↑2.870 | ↑3.251 | ↑1.659 | ||
| 62 | IPI00465084.6 | Desmin | ↑2.945 | ↑2.889 | ↑2.987 | |||
| 63 | IPI00017704.3 | Coactosin-like protein | ↑3.002 | ↑3.058 | ||||
| 64 | IPI00465431.7 | Galectin-3 | ↑1.935 | ↑3.092 | ↑1.614 | ↑2.623 | ↑1.665 | |
| 65 | IPI00218918.5 | Annexin A1 | ↓0.601 | ↑3.121 | ↑3.689 | ↑4.724 | ||
| 66 | IPI00003865.1 | Isoform 1 of Heat shock cognate 71 kDa protein | ↑3.308 | ↑2.422 | ||||
| 67 | IPI00022793.5 | Trifunctional enzyme subunit beta | ↑3.464 | |||||
| 68 | IPI00291136.4 | Collagen alpha-1(VI) chain precursor | ↑3.531 | |||||
| 69 | IPI00010471.5 | Plastin-2 | ↑3.799 | ↑2.276 | ↑2.728 | |||
| 70 | IPI00216691.5 | Profilin-1 | ↑3.993 | ↑6.700 | ||||
| 71 | IPI00644087.1 | LMNA Progerin | ↑4.163 | ↑4.348 | ↑4.717 | |||
| 72 | IPI00018219.1 | Transforming growth factor-beta-induced protein ig-h3 precursor | ↓0.539 | ↑4.280 | ||||
| 73 | IPI00180675.4 | Tubulin alpha-1A chain | ↑2.233 | ↑3.115 | ↑4.472 | |||
| 74 | IPI00745729.2 | SELENBP1 54 kDa protein | ↑1.555 | ↑2.421 | ↑4.488 | ↑1.770 | ↑3.342 | ↑1.906 |
| 75 | IPI00792011.1 | Calcyphosin | ↑4.478 | ↑6.808 | ↑4.708 | |||
| 76 | IPI00022200.2 | alpha 3 type VI collagen isoform 1 precursor | ↑1.732 | ↑4.922 | ↑1.959 | ↑5.435 | ||
| 77 | IPI00329801.12 | Annexin A5 | ↑2.365 | ↑1.524 | ↑5.297 | ↓0.639 | ↑2.465 | ↑3.831 |
| 78 | IPI00455383.4 | Isoform 2 of Clathrin heavy chain 1 | ↑5.639 | ↑4.592 | ↑5.243 | |||
| 79 | IPI00000105.4 | Major vault protein | ↑5.967 | ↑1.595 | ↑7.013 | ↑4.683 | ||
| 80 | IPI00414320.1 | Annexin A11 | ↑2.011 | ↑6.030 | ↑2.286 | |||
| 81 | IPI00291005.8 | Malate dehydrogenase | ↑2.891 | ↑6.101 | ↑4.535 | |||
| 82 | IPI00022204.2 | Isoform 1 of Serpin B3 | ↑3.041 | ↑7.684 | ↑2.968 | ↑7.552 | ||
| 83 | IPI00219757.13 | Glutathione S-transferase P | ↑2.451 | ↑4.819 | ↑8.054 | ↑1.954 | ↑3.761 | ↑1.971 |
| 84 | IPI00654709.1 | ALDH3A1 protein (Fragment) | ↑8.193 | ↑6.242 | ||||
| 85 | IPI00745872.2 | Isoform 1 of Serum albumin precursor | ↑2.648 | ↑8.322 | ↑2.029 | ↑6.751 | ↑4.002 | |
| 86 | IPI00872684.1 | EZR 69 kDa protein | ↑1.826 | ↑2.245 | ↑9.195 | ↑5.606 | ↑4.613 | |
| 87 | IPI00009123.1 | Nucleobindin-2 precursor | ↑6.538 | ↑9.796 | ↑4.263 | ↑7.419 | ||
| 88 | IPI00027463.1 | Protein S100-A6 | ↑1.566 | ↑9.869 | ↑6.702 | ↑4.461 | ||
| 89 | IPI00887291.1 | hypothetical LOC729659 | ↓0.649 | ↑10.526 | ↑11.910 | ↑13.996 | ||
| 90 | IPI00553177.1 | Isoform 1 of Alpha-1-antitrypsin precursor | ↑12.862 | ↑13.994 | ↑15.337 | |||
| 91 | IPI00152295.1 | Spermatogenesis-associated protein 18 | ↑3.627 | ↑12.957 | ↑1.804 | |||
| 92 | IPI00847342.1 | keratin 7 | ↑13.664 | ↑11.683 | ↑11.397 | |||
| 93 | IPI00024915.2 | Isoform Mitochondrial of Peroxiredoxin-5 | ↑2.128 | ↑8.200 | ↑13.800 | ↑4.058 | ↑8.016 | |
| 94 | IPI00007427.2 | AGR2 | ↑3.253 | ↑14.088 | ↑4.492 | ↑17.958 | ↑6.054 | |
| 95 | IPI00554788.5 | type I cytoskeletal 18 | ↑1.528 | ↑14.101 | ↑11.035 | ↑10.365 | ||
| 96 | IPI00000816.1 | 14–3-3 protein epsilon | ↑1.729 | ↑16.565 | ↑14.496 | ↑13.096 | ||
| 97 | IPI00294739.1 | SAM domain and HD domain-containing protein 1 | ↑1.558 | ↑19.613 | ↑13.165 | |||
| 98 | IPI00554648.3 | type II cytoskeletal 8 | ↑2.225 | ↑21.377 | ↑1.866 | ↑19.352 | ↑15.184 | |
| 99 | IPI00807545.1 | Isoform 3 of Heterogeneous nuclear ribonucleoprotein K | ↑21.953 | ↑21.770 | ↑22.728 | |||
| 100 | IPI00022977.1 | Creatine kinase B-type | ↑2.576 | ↑6.028 | ↑22.071 | ↑2.372 | ↑12.224 | ↑5.840 |
| 101 | IPI00795633.1 | CLU | ↑22.850 | ↑23.117 | ↑21.293 | |||
| 102 | IPI00640817.1 | AK1 Adenylate kinase 1 | ↑2.325 | ↑25.104 | ↑17.557 | ↑13.796 |
Validation of Differentially Expressed Proteins Indentified by Proteomics
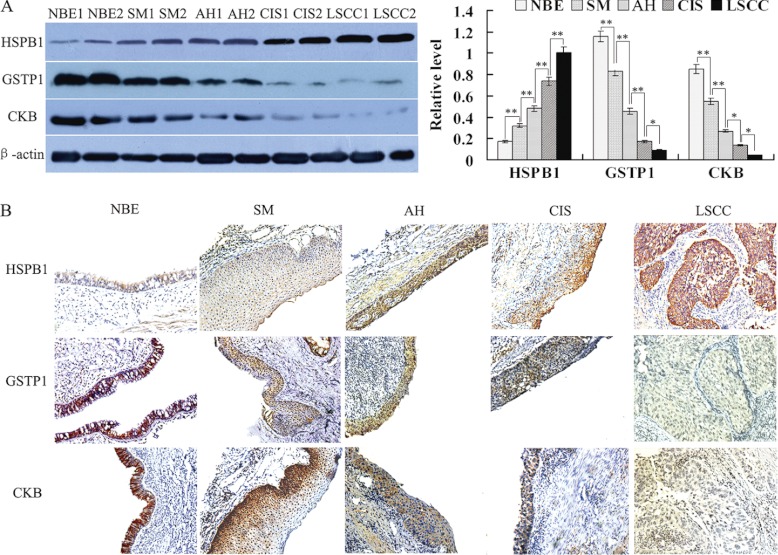
Three proteins (HSPB1, GSTP1, and CKB) with progressively expressional changes during the bronchial epithelial carcinogenesis identified by MS analysis were chosen for verification. Western blotting was performed to detect the expressional levels of the three proteins in one independent set of LCM-purified tissues including NBE, SM, AH, CIS and invasive LSCC, 10 cases for each tissue. As shown in Fig. 2A, HSPB1 expression was progressively increased, whereas expressions of GSTP1 and CKB was progressively decreased along with evolution of bronchial epithelial carcinogenesis, which is consistent with the findings in MS analysis.
Fig. 2.
Expressional changes of HSPB1, GSTP1, and CKB in the human bronchial epithelial carcinogenic process. A, (left) a representative result of Western blotting shows the expressions of HSPB1, GSTP1, and CKB in the microdissected NBE, SM, AH, CIS, and invasive LSCC; (right) histogram shows the expression levels of the three proteins in these tissues as determined by densitometric analysis. β-actin is used as the internal loading control. Columns, mean from 10 cases of tissues; bars, S.D. (*, p < 0.05; **, p < 0.01 by One-way ANOVA). B, a representative result of immunohistochemistry shows the expression of HSPB1, GSTP1, and CKB in the NBE, SM, AH, CIS, and invasive LSCC. Original magnification, ×200.
Values of HSPB1, GSTP1, and CKB as Biomarkers for Early Detection of LSCC
Immunohistochemistry was performed to detect the expressional levels of the three proteins (HSPB1, GSTP1, and CKB) in an independent set of archival tissue specimens including NBE, SM, AH, CIS, and invasive LSCC. As shown in Fig. 2B and Table II, HSPB1 expression was progressively increased, whereas expressions of GSTP1 and CKB was progressively decreased along with evolution of bronchial epithelial carcinogenesis, which also supports the above MS findings. Moreover, there was significantly different in the expressional levels of HSPB1, GSTP1, and CKB in the two stages of bronchial epithelial carcinogenic process (Table II).
Table II. The difference of HSPB1, GSTP1, CKB expression in bronchial epithelial carcinogenic process. p < 0.01 or 0.05 by Mann-Whitney U test, NBE vs. SM, AH/CIS, or LSCC. SM vs. AH/CIS or LSCC; AH/CIS vs. LSSC.
| n | Score |
|||
|---|---|---|---|---|
| Low (0–2) | Moderate (3–4) | High (5–6) | ||
| HSPB1 | ||||
| NBE | 66 | 34 | 25 | 7 |
| SM | 64 | 18 | 32 | 14 |
| AH/CIS | 68 | 8 | 32 | 28 |
| HLSC | 66 | 3 | 15 | 48 |
| GSTP1 | ||||
| NBE | 66 | 7 | 23 | 36 |
| SM | 64 | 12 | 28 | 24 |
| AH/CIS | 71 | 27 | 29 | 15 |
| HLSC | 66 | 41 | 18 | 7 |
| CKB | ||||
| NBE | 66 | 8 | 23 | 35 |
| SM | 64 | 16 | 27 | 21 |
| AH/CIS | 73 | 28 | 32 | 13 |
| HLSC | 66 | 47 | 15 | 4 |
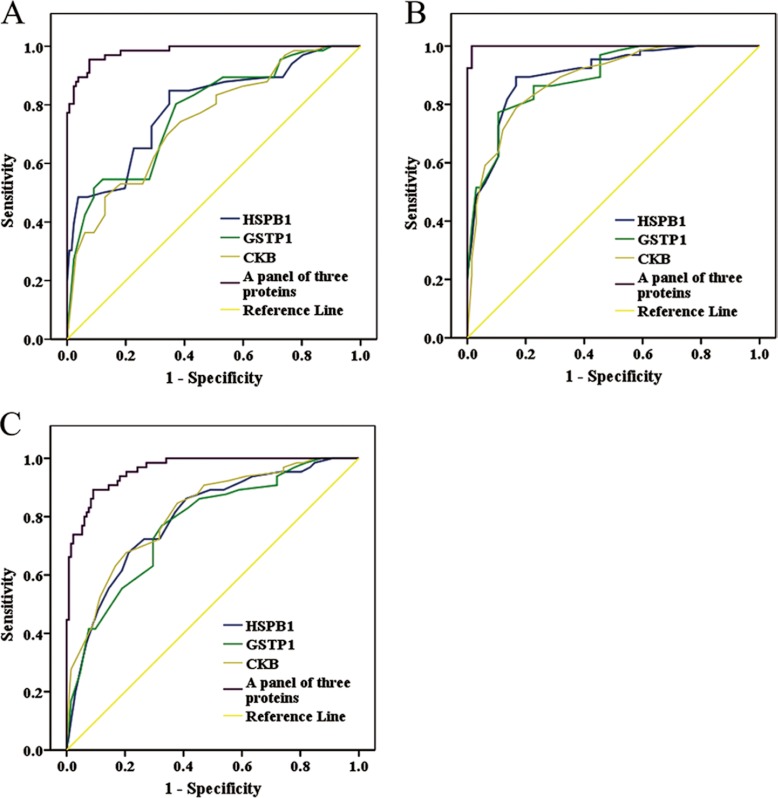
The ability of the three proteins in distinguishing NBE from preneoplastic lesions (SM, AH, and CIS) from invasive LSCC was analyzed by determining the ROC curves of the three proteins individually and as a panel. The area under the curve (AUC) of the three proteins is listed in Table III–V together with their individual and collective values of merit. When individual protein serves as a biomarker, their sensitivity and specificity are 74–85% and 61–65% in discriminating NBE from preneoplastic lesions, 77–89% and 83–89% in discriminating NBE from invasive LSCC, and 68–77% and 67–80% in discriminating preneoplastic lesions from invasive LSCC, respectively (Fig. 3; Table III–V). As a panel, the three proteins achieved a sensitivity of 96% and a specificity of 92% in discriminating NBE from preneoplastic lesions, a sensitivity of 100% and a specificity of 98% in discriminating NBE from invasive LSCC, and a sensitivity of 92% and a specificity of 91% in discriminating preneoplastic lesions from invasive LSCC (Fig. 3 and Table III–V).
Table III. Receiver operating characteristics from IHC scores of the three proteins in distinguishing NBE from preneoplastic lesions (SM, AH, and CIS) individually and as a panel. PPV, positive predict value; NPV, negative predict value; AUC, area under the curve.
| Proteins | Sensitivity | Specificity | PPV | NPV | AUC |
|---|---|---|---|---|---|
| HSPB1 | 0.85 | 0.65 | 0.88 | 0.50 | 0.79 |
| GSTP1 | 0.80 | 0.63 | 0.87 | 0.55 | 0.78 |
| CKB | 0.74 | 0.61 | 0.88 | 0.43 | 0.75 |
| A panel of three proteins | 0.96 | 0.92 | 0.97 | 0.86 | 0.98 |
Table IV. Receiver operating characteristics from IHC scores of the three proteins in distinguishing NBE from invasive LSCC individually and as a panel.
| Proteins | Sensitivity | Specificity | PPV | NPV | AUC |
|---|---|---|---|---|---|
| HSPB1 | 0.89 | 0.83 | 0.83 | 0.86 | 0.90 |
| GSTP1 | 0.77 | 0.89 | 0.77 | 0.82 | 0.89 |
| CKB | 0.79 | 0.83 | 0.77 | 0.83 | 0.89 |
| A panel of three proteins | 1.00 | 0.98 | 0.99 | 0.99 | 1.00 |
Table V. Receiver operating characteristics from IHC scores of the three proteins in distinguishing preneoplastic lesions (SM, AH and CIS) from invasive LSCC individually and as a panel.
| Proteins | Sensitivity | Specificity | PPV | NPV | AUC |
|---|---|---|---|---|---|
| HSPB1 | 0.68 | 0.79 | 0.86 | 0.55 | 0.80 |
| GSTP1 | 0.77 | 0.67 | 0.84 | 0.51 | 0.77 |
| CKB | 0.68 | 0.80 | 0.89 | 0.52 | 0.81 |
| A panel of three proteins | 0.92 | 0.91 | 0.92 | 0.83 | 0.96 |
Fig. 3.
Efficacy of HSPB1, GSTP1, and CKB in discriminating NBE from preneoplastic lesions(SM, AH, and CIS) from invasive LSCC. Receiver operating characteristic (ROC) curves of HSPB1, GSTP1, and CKB in discriminating NBE from preneoplastic lesions (A), NBE from invasive LSCC (B), and preneoplastic lesions from invasive LSCC (C), individually and as a panel.
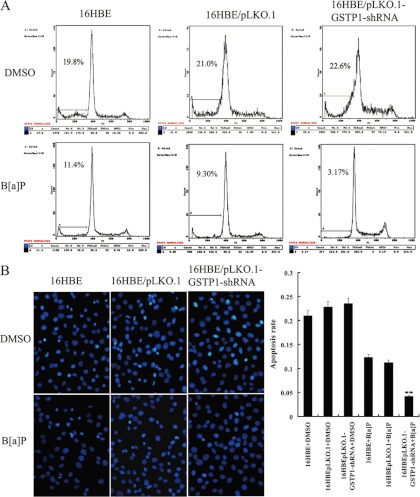
Knockdown of GSTP1 Increased the Susceptibility of Human Bronchial Epithelial Cell Transformation Induced by B[a]P
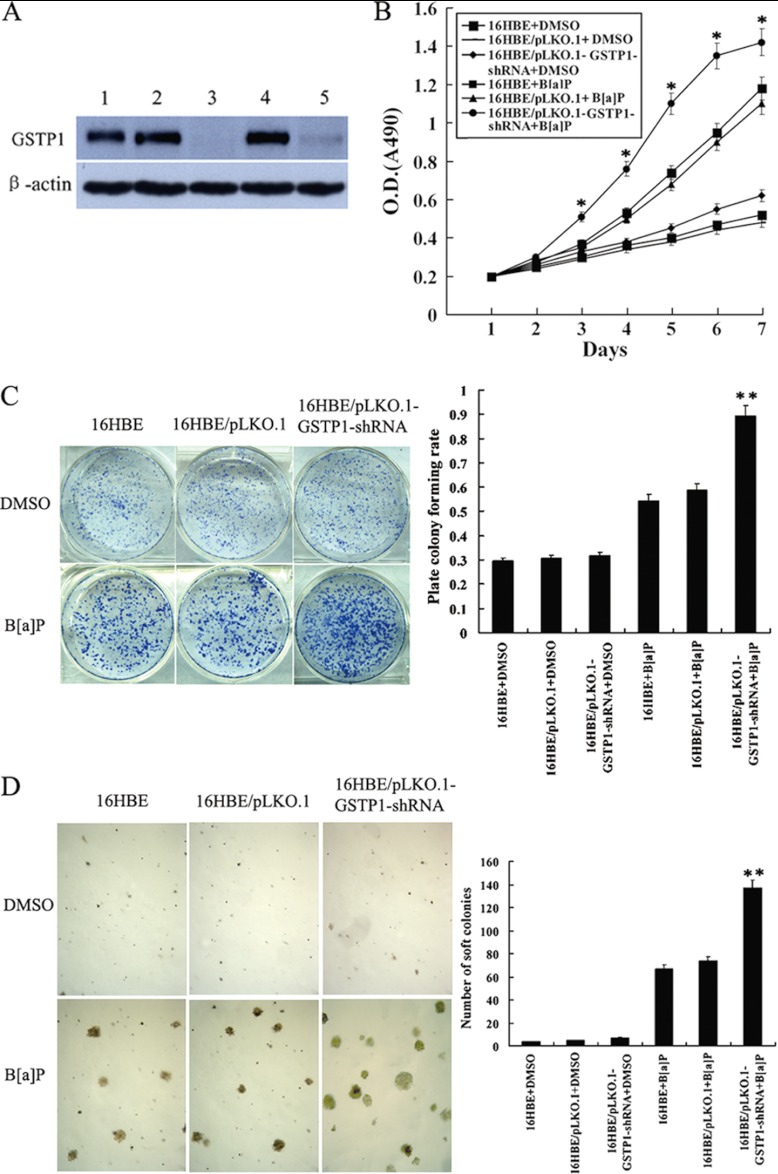
To know whether down-regulation of GSTP1 is involved in bronchial epithelial carcinogenesis, we generated stably transfected human bronchial epithelial cell line 16HBE cells with knockdown of GSTP1 (Fig. 4A), and measured the susceptibility of the transfected 16HBE cell transformation induced by B[a]P. After repeated treatment with 1 μm B[a]P for 16 weeks, significant differences in transformation efficiency between 16HBE cells with knockdown of GSTP1 and control cells (empty vector-transfected 16HBE cells and untransfected cells) were seen: (1) MTT assay showed that cell growth rate in low serum medium is significantly higher in 16HBE cells with knockdown of GSTP1 than in control cells (Fig. 4B); (2) anchorage dependent and independent colony formation assays showed that about 1.7-fold more plate colonies and about two-fold more soft agar colonies developed from 16HBE cells with knockdown of GSTP1 compared with control cells(Figs. 4C and 4D); (3) flow cytometric analysis revealed that a significant increase of S phase populations with a corresponding decrease of G0/G1 phase in 16HBE cells with knockdown of GSTP1 compared with the control cells (Table VI); and (4) both Hoechst 33258 staining and flow cytometric analysis of apoptotic cells showed less apoptotic cells are detected in 6HBE cells with knockdown of GSTP1 than in the control cells (Fig. 5). Taken together, these results demonstrated that knockdown of GSTP1 increased the susceptibility of human bronchial epithelial cell transformation induced by B[a]P, supporting that GSTP1 down-regulation is involved in human bronchial epithelial carcinogenesis.
Fig. 4.
The effects of GSTP1 gene knockout on the B[a]P-induced human bronchial epithelial cell transformation. A, Western blotting shows GSTP1 expression in the untransfected (1), empty vector pLKO.1-transfected (2, 4), and pLKO.1-GSTP1-shRNA-tansfected 16HBE cells (3, 5). β-actin is used as an internal control for loading. B, Cell growth in low serum medium after exposed to B[a]P for 16 weeks. Cells were subjected to MTT assay as described in “Experimental Procedures.” Three experiments were done; points, mean; bars, S.D. (*, p < 0.05 versus untransfected or empty vector-transfected 16HBE cells after exposed to B[a]P by Student's t test). C, anchorage dependent colony growth after cells exposed to B[a]P for 16 weeks. (left) cells were subjected to plate colony formation assay as described in “Experimental Procedures,” and colonies were stained with crystal violet and photographed under microscope; (right) the histogram showed plate colony formation rates. Three experiments were done; columns, mean; bars, S.D. (**, p < 0.01 versus untransfected or empty vector -transfected 16HBE cells after exposed to B[a]P by One-way ANOVA). D, Anchorage independent colony growth after cells exposed to B[a]P for 16 weeks. (left) cells were subjected to soft agar colony formation assay as described in “Experimental procedures,” and colonies were photographed under microscope; (right) the histogram showed number of soft agar colonies in 10 randomly chosen microscopic fields using a 5× objective. Three experiments were done; columns, mean; bars, S.D. (**, p < 0.01 versus untransfected or empty vector -transfected 16HBE cells after exposed to B[a]P by One-way ANOVA). Cell proliferation, and plate and soft agar colony growth of the cells exposed to vehicle (DMSO) for 16 weeks are also shown and used as controls. 16HBE, untransfected cells; 16HBE/pLKO.1, empty vector pLKO.1-transfected cells; 16HBE/pLKO.1-GSTP1-shRNA, pLKO.1-GSTP1-shRNA -tansfected cells.
Table VI. Cell cycle distribution of 16HBE with knockdown of GSTP1 and controls after repeated treatment with 1 μm B[a]P for 16 weeks. * p < 0.05 vs. 16HBE/pLKO.1+ B[a]P or 16HBE+ B[a]P by t-test. Cell cycle distribution of the cells exposed to vehicle (DMSO) for 16 weeks is also shown and used as controls.
| Cell lines | Number of cell (%) |
||
|---|---|---|---|
| G0/G1 | S | G2/M | |
| 16HBE+DMSO | 76.10 ± 6.29 | 12.18 ± 3.15 | 11.72 ± 3.33 |
| 16HBE/pLKO.1+DMSO | 75.56 ± 3.72 | 12.47 ± 2.17 | 11.97 ± 2.13 |
| 16HBE/pLKO.1-GSTP1-shRNA+ DMSO | 73.74 ± 2.08 | 13.46 ± 2.08 | 12.79 ± 1.87 |
| 16HBE+ B[a]P | 71.88 ± 2.99 | 15.10 ± 3.38 | 13.01 ± 2.76 |
| 16HBE/pLKO.1+ B[a]P | 71.38 ± 5.77 | 15.44 ± 3.63 | 13.18 ± 2.20 |
| 16HBE/pLKO.1-GSTP1-shRNA+ B[a]P | 54.67 ± 5.24* | 29.75 ± 5.25* | 15.58 ± 1.35 |
Fig. 5.
The effects of GSTP1 knockdown on the apoptosis of B[a]P-transformed human bronchial epithelial cells. A, A representative result of flow cytometry analysis of cell apoptosis cultured in serum free medium after exposed to B[a]P for 16 weeks. Cells were grown in serum free DMEM medium for 24h, and then assessed for apoptosis by flow cytometry as described in “Experimental procedures.” B, (right) Hoechst 33258 staining of cell apoptosis cultured in serum free medium after exposed to B[a]P for 16 weeks. Cells were grown in serum free DMEM medium for 24h, and then assessed for apoptosis using the cell-permeable DNA dye Hoechst 33258. Apoptotic nuclei showing intense fluorescence corresponding to chromatin condensation; (left) a histogram showed the cell apoptotic rates. Three experiments were done; columns, mean; bars, S.D.(**, p < 0.05 versus untransfected or empty vector -transfected 16HBE cells exposed to B[a]P). Apoptosis of the cells exposed to vehicle (DMSO) for 16 weeks is also shown and used as controls. 16HBE, untransfected cells; 16HBE/pLKO.1, empty vector pLKO.1-transfected cells; 16HBE/pLKO.1-GSTP1-shRNA, pLKO.1- GSTP1-shRNA -tansfected cells.
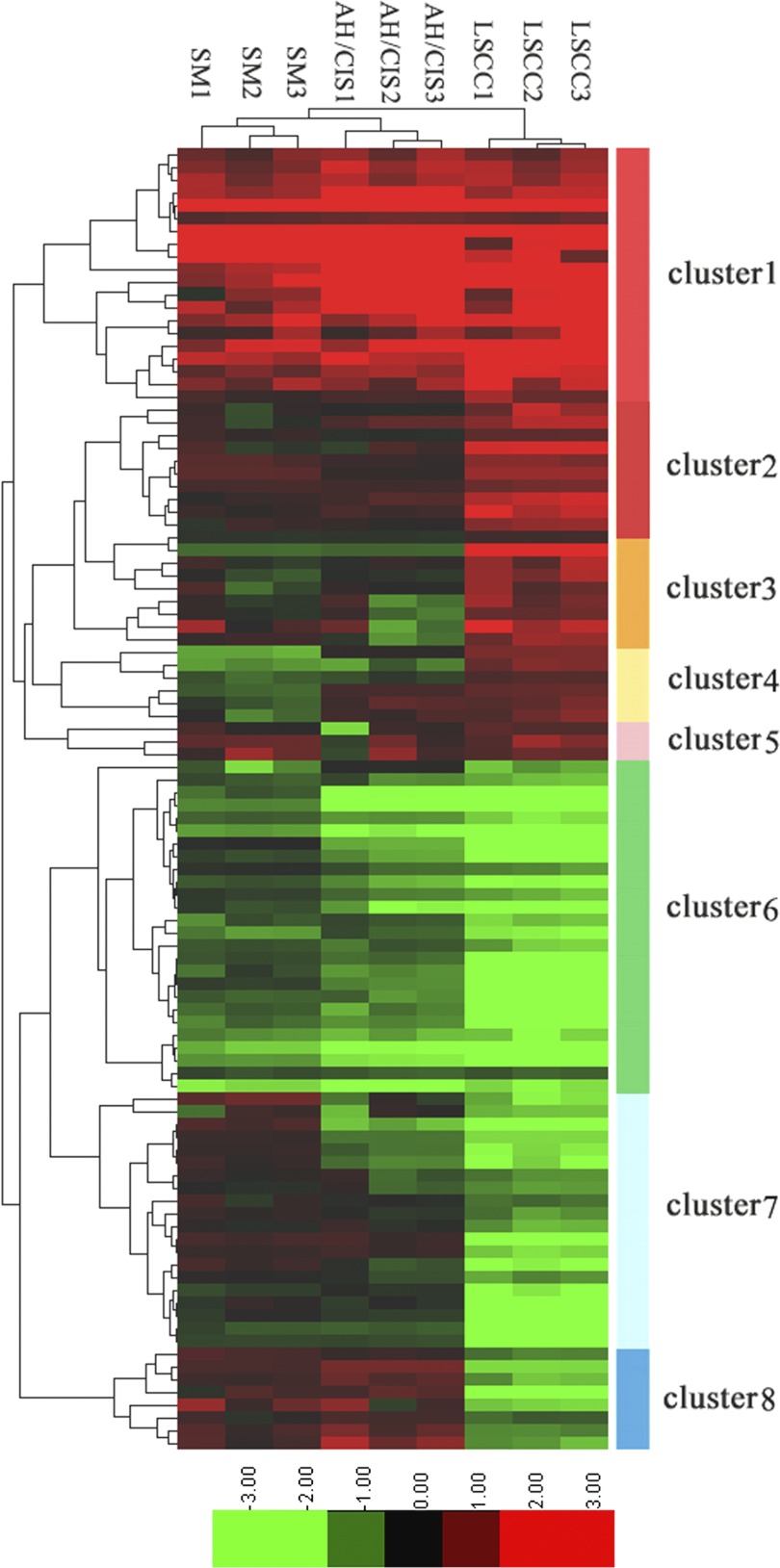
Hierarchical Clustering, Gene-ontology and KEGG Pathways Analysis of the differential proteins
To get more insight on the biological significance of the differentially expressed proteins in bronchial epithelial carcinogenic process, hierarchical clustering was performed on 102 differentially expressed proteins. All differentially expressed proteins were hierarchically grouped into eight clusters and three groups (Fig. 6). Group 1 consists of clusters 1 and 2, the proteins of which were up-regulated in preneoplastic lesions (SM, AH/CIS), and invasive LSCC versus NBE, and exhibited the highest expression in invasive LSCC. Group 2 (cluster 6) includes the proteins that was down-regulated in preneoplastic lesions (SM, AH/CIS), and invasive LSCC versus NBE, and exhibited the lowest expression in invasive LSCC. Group 3 consists of clusters 3, 4, 5, 7, and 8, the proteins of which were up-regulated or down-regulated in a certain stage of bronchial epithelial carcinogenic process. The proteins within the same cluster are coregulated proteins, and may have similar biological functions during bronchial epithelial carcinogenesis. GO analysis showed that each group is enriched with the proteins of different functions, and may play a distinctive role during bronchial epithelial carcinogenesis (supplementary Table S3). KEGG pathway analysis revealed that the proteins in three groups are involved in cancer-associated signaling pathways such as MAPK signaling pathway, apoptosis, cell cycle and p53 signaling pathway, and ErbB signaling pathway (supplementary Figs. S3, S4, and S5). The differentially expressed proteins may play a role in bronchial epithelial carcinogenesis by these signaling pathways.
Fig. 6.
Hierarchical clustering of 102 differentially expressed proteins. Log2 of normalized counts of differential proteins were clustered. Red clusters denote high levels of expression whereas light green clusters denote low levels of expression. The x axis represents the samples whereas the y axis represents the proteins. The panel on the right shows the 8 different clusters. SM, AH/CIS, and LSCC represent SM, AH/CIS, or LSCC versus NBE, respectively.
DISCUSSION
LSCC carcinogenesis is a multistage process from normal to preneoplastic lesions and then on to carcinoma (4). Identification of proteins with altered expression as a manifestation of human bronchial epithelial carcinogenesis is important in discovery of biomarkers for early detection of LSCC. In this study, iTRAQ labeling combined with 2D LC-MS/MS was used to identify differentially expressed proteins during bronchial epithelial carcinogenesis. As a result, 102 differentially expressed proteins were identified, and three differential proteins (GSTP1, HSPB1, and CKB) showing progressively expressional changes during the carcinogenic process were selectively validated. Next, we evaluated the ability of three candidate biomarkers (GSTP1, HSPB1, and CKB) for early detection of LSCC, finding that panel of the three proteins can perfectly distinguish NBE from preneoplastic lesions from invasive LSCC with high sensitivity and specificity. The results suggest that the three proteins are potentials biomarkers for early detection of LSCC.
Glutathione S-transferase P1 (GSTP1), a major detoxification enzyme and stress response signaling protein, is an important part of cellular defense against endogenous and exogenous chemicals such as chemical carcinogens and chemotherapeutic drugs (33). Deregulation of GSTP1 has been reported in lung cancer, and is related to the chemosensitivity and prognosis of the patients (34–36). A lot of studies have focused on the polymorphism and promoter methylation of GSTP1 gene and their biological significances in lung cancer (37, 38), and showed that GSTP1 polymorphism decreased GSTP1 activity, and was associated with risk of lung cancer (39, 40).
To know whether down-regulation of GSTP1 is involved in bronchial epithelial carcinogenesis, we knocked down GSTP1 in immortalized human bronchial epithelial line 16HBE cells, and then detected whether GSTP1 knockdown increased the susceptibility of cell transformation induced by carcinogen B[a]P. After weekly exposed to 1 μm B[a]P for 16 weeks, transformation efficiency of 16HBE cells with GSTP1 knockdown was significantly higher than that of control cells, and GSTP1 knockdown increased the susceptibility of bronchial epithelial cell transformation induced by B[a]P, demonstrating that GSTP1 plays an important role in human bronchial epithelial carcinogenesis. To our knowledge, this is the first report to establish a correlation between GSTP1 down-regulation and carcinogenesis of human bronchial epithelium, and GSTP1 as a potential biomarker for early detection of LSCC.
Polycyclic aromatic hydrocarbons such as B[a]P are main lung carcinogens within tobacco smoke (41), and the source of DNA adducts (42). B[a]P is activated by the cytochrome P450 system and epoxide hydrolase to electrophilic reactive metabolite B[a]P-diolepoxide(BPDE), which is highly mutagenic and carcinogenic (43, 44). GSTP1 catalyzes the detoxification of ultimate carcinogen BPDE by the conjugation of reduced glutathione (GSH) with BPDE (45). Therefore, it is conceivable that GSTP1 down-regulation enhances the level of B[a]P DNA adducts and the frequency of gene mutation, then increasing the risk of bronchial epithelial carcinogenesis. In fact, epidemiologic evidence has indicated that GSTP1 is an important factor in individual susceptibility to smoking-induced lung cancer, and activity-altering polymorphisms in GSTP1 were found to be potential risk modifiers in lung cancer development (46).
HSPB1 (heat shock protein beta-1, also called HSP27), a key member of the small heat-shock protein family, is ubiquitously expressed at low levels and can be induced by various physiological or environmental stresses (47). HSPB1 plays crucial roles in carcinogenesis and progression through inhibition of cell apoptosis and senescence, two essential traits of cancer cells (48–50). Various human cancers have been reported to exhibit overexpression of HSPB1, and the tumorigenic potentials of HSPB1 have been demonstrated in both experimental animal models and cell biologic studies (51–53). HSPB1 has been considered as an independent prognosis marker for cancers because its overexpression was involved in chemoradiotherapeutic resistance of tumor cells (54). Our previous study have found that HSPB1 was overexpressed in human LSCC, especially in those with lymph node metastasis and higher clinical stages, indicating the roles of HSPB1 in the progression and metastasis of LSCC(55).
In the present study, we found that the expression of HSPB1 is progressively increased during human bronchial epithelial carcinogenesis, and can serve as a biomarker for early detection of LSCC. To our knowledge, this is the first study to evaluate HSPB1 expression in human bronchial epithelium carcinogenic process and its early diagnostic significance in LSCC. Interestingly, previous studies have showed that HSPB1 was overexpressed in preneoplastic lesions in the uterine cervix and gastric cancer, and has an early diagnostic significance for these two tumors (56, 57), which also supports HSPB1 as biomarker for early detection of LSCC. It has been reported that HSPB1 overexpression was involved in liver carcinogenesis induced by chemical carcinogens (58), and HSPB1 expression in benign proliferating breast lesions increased risk of cell malignant progression (59). Therefore, it is conceivable that HSPB1 up-regulation may confer a higher susceptibility of human bronchial epithelial cells to carcinogenic stimuli, and contribute to cell survival and growth advantage, leading to evolution of bronchial epithelial carcinogenesis.
Creatine kinase is an enzyme involved in energy transduction pathways, and exists in tissues and serum as a dimer of its two isoenzymes (CKB and CKM). Creatine kinase brain-type (CKB) is predominantly expressed in normal lung, colon and liver tissues (60). CKB is overexpressed in a wide variety of cancers, and can serve as a penitential biomarker for various human tumors (61–63). In this study, we found that the expression of CKB was progressively decreased during human bronchial epithelial carcinogenesis, and can serve as a biomarker for early detection of LSCC. To our knowledge, this is the first study to evaluate CKB expression in human bronchial epithelium carcinogenic process and its early diagnostic significance in LSCC. Interestingly, previous studies have showed that CKB down-regulation is involved in oral carcinogenesis (64), CKB expression was decreased in colon adenocarcinoma and LSCC compared with their originated normal tissues (65, 66), and transfection of a dominant-negative CKB into colon cancer cells caused remarkable changes in cell shape, adhesion, and invasion, and resulted in an epithelial-to-mesenchymal transition (EMT) in these cells (67). Our results, together with these reports, suggest that CKB down-regulation is involved in human bronchial epithelial carcinogenesis.
To get more insight on the biological significance of the differentially expressed proteins in bronchial epithelial carcinogenic process, Hierarchical clustering, gene-ontology and KEGG pathways analysis were performed on 102 differential proteins. Hierarchical clustering analysis of differentially expressed proteins showed stage-specific and coregulated expression profiles. GO analysis showed that each functional group may play a distinctive role during bronchial epithelial carcinogenesis. KEGG pathway analysis revealed that the differentially expressed proteins are involved in cancer-associated signaling pathways. The data provide valuable information for further study of molecular mechanisms that govern the normal to malignant conversion of human bronchial epithelium.
CONCLUSION
The use of iTRAQ-labeling combined with 2D LC-MS/MS identified 102 differentially expressed proteins in human bronchial epithelial carcinogenic process, and three differential proteins (GSTP1, HSPB1, and CKB) with progressively expressional changes were selectively verified. We found that panel of the three proteins can serve as novel potential biomarkers for early detection of LSCC. We further showed that GSTP1 knockdown increased the susceptibility of bronchial epithelial cell transformation induced by B[a]P, first time demonstrating that GSTP1 plays an important role in human bronchial epithelial carcinogenesis. The findings reported here could have potential clinical value in early diagnosis of LSCC, and provide valuable information for further study of molecular mechanisms that govern the normal to malignant conversion of human bronchial epithelium.
Acknowledgments
We thank the Proteome Center of Fudan University, China, for technical assistance.
Footnotes
* This work was supported by National Nature Science Foundation of China (30973290, 81172559, 81172302), Lotus Scholars Program of Hunan Province, China (2007–362), Key Research Program from Science and Technology Department of Hunan Province, China (2010FJ2009), Nature Science Foundation of Hunan Province, China (11JJ2045), Scientific Research Fund of Hunan Provincial Education Department, China (09C837), and Scientific Research Fund of Hunan Provincial Health Department, China (B2010-037).
 This article contains supplemental Figs. S1 to S5 and Tables S1 to S3.
This article contains supplemental Figs. S1 to S5 and Tables S1 to S3.
1 The abbreviations used are:
- LSCC
- lung squamous cell cancer
- NBE
- normal bronchial epithelium
- SM
- squamous metaplasia
- AH
- atypical hyperplasia
- CIS
- carcinoma in situ
- iTRAQ
- isobaric tags for relative and absolute quantitation
- RP
- reverse phase
- LCM
- laser capture microdissection
- GSTP1
- glutathione S-transferase P1
- HSPB1
- heat shock protein beta-1
- CKB
- creatine kinase brain-type
- 2D LC-MS/MS
- two-dimensional liquid chromatography-mass spectrometry
- ROC
- receiver operating characteristic
- AUC
- area under the curve
- IHC
- immunohistochemistry
- B[a]P
- benzo(a)pyrene.
REFERENCES
- 1. Parkin D. M. (2001) Global cancer statistics in the year 2000. Lancet Oncol. 2, 533–543 [DOI] [PubMed] [Google Scholar]
- 2. Yang L., Parkin D. M., Li L. D., Chen Y. D., Bray F. (2004) Estimation and projection of the national profile of cancer mortality in China: 1991–2005. Br. J. Cancer 90, 2157–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Howe H. L., Wingo P. A., Thun M. J., Ries L. A., Rosenberg H. M., Feigal E. G., Edwards B. K. (2001) Annual report to the nation on the status of cancer (1973 through 1998), featuring cancers with recent increasing trends. J. Natl. Cancer Inst. 93, 824–842 [DOI] [PubMed] [Google Scholar]
- 4. Greenberg A. K., Yee H., Rom W. N. (2002) Preneoplastic lesions of the lung. Respir Res. 3, 20–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Auerbach O., Hammond E. C., Garfinkel L. (1979) Changes in bronchial epithelium in relation to cigarette smoking 1955–1966 VS 1970–1977. N. Engl. J. Med. 300, 381–385 [DOI] [PubMed] [Google Scholar]
- 6. Deng B., Ye N., Luo G., Chen X., Wang Y. (2005) Proteomics analysis of stage-specific proteins expressed in human squamous cell lung carcinoma tissues. Cancer Biomark. 1, 279–286 [DOI] [PubMed] [Google Scholar]
- 7. Planque C., Kulasingam V., Smith C. R., Reckamp K., Goodglick L., Diamandis E.P. (2009) Identification of five candidate lung cancer biomarkers by proteomics analysis of conditioned media of four lung cancer cell lines. Mol. Cell. Proteomics 8, 2746–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Patz E. F., Jr., Campa M. J., Gottlin E. B., Kusmartseva I., Guan X. R., Herndon J. E., 2nd2 (2007) Panel of serum biomarkers for the diagnosis of lung cancer. J. Clin. Oncol. 25, 5578–5583 [DOI] [PubMed] [Google Scholar]
- 9. Zeng X., Hood B. L., Sun M., Conrads T. P., Day R. S., Weissfeld J. L., Siegfried J. M., Bigbee W. L. (2010) Lung cancer serum biomarker discovery using glycoprotein capture and liquid chromatography mass spectrometry. J. Proteome Res. 9, 6440–6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li C., Xiao Z., Chen Z., Zhang X., Li J., Wu X., Li X., Yi H., Li M., Zhu G., Liang S. (2006) Proteome analysis of human lung squamous carcinoma. Proteomics 6, 547–558 [DOI] [PubMed] [Google Scholar]
- 11. Yang F., Xiao Z. Q., Zhang X. Z., Li C., Zhang P. F., Li M. Y., Chen Y., Zhu G. Q., Sun Y., Liu Y. F., Chen Z. C. (2007) Identification of tumor antigens in human lung squamous carcinoma by serological proteome analysis. J. Proteome Res. 6, 751–758 [DOI] [PubMed] [Google Scholar]
- 12. Li D. J., Deng G., Xiao Z. Q., Yao H. X., Li C., Peng F., Li M. Y., Zhang P. F., Chen Y. H., Chen Z. C. (2009) Identificating 14–3-3 sigma as a lymph node metastasis-related protein in human lung squamous carcinoma. Cancer Lett. 279, 65–73 [DOI] [PubMed] [Google Scholar]
- 13. Zhang X. Z., Xiao Z. F., Li C., Xiao Z. Q., Yang F., Li D. J., Li M. Y., Li F., Chen Z. C. (2009) Triosephosphate isomerase and peroxiredoxin 6, two novel serum markers for human lung squamous cell carcinoma. Cancer Sci. 100, 2396–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brichory F., Beer D., Le Naour F., Giordano T., Hanash S. (2001) Proteomics-based identification of protein gene product 9.5 as a tumor antigen that induces a humoral immune response in lung cancer. Cancer Res. 61, 7908–7912 [PubMed] [Google Scholar]
- 15. Ocak S., Chaurand P., Massion P. P. (2009) Mass spectrometry-based proteomic profiling of lung cancer. Proc. Am. Thorac. Soc. 6, 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hassanein M., Rahman J. S., Chaurand P., Massion P. P. (2011) Advances in proteomic strategies toward the early detection of lung cancer. Proc. Am. Thorac. Soc. 8, 183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bharti A., Ma P. C., Salgia R. (2007) Biomarker discovery in lung cancer–promises and challenges of clinical proteomics. Mass. Spectrom. Rev. 26, 451–466 [DOI] [PubMed] [Google Scholar]
- 18. Rahman S. M., Shyr Y., Yildiz P. B., Gonzalez A. L., Li H., Zhang X., Chaurand P., Yanagisawa K., Slovis B. S., Miller R. F., Ninan M., Miller Y. E., Franklin W. A., Caprioli R. M., Carbone D. P., Massion P.P. (2005) Proteomic patterns of preinvasive bronchial lesions. Am. J. Respir. Crit. Care Med. 172, 1556–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emmert-Buck M. R., Bonner R. F., Smith P. D., Chuaqui R. F., Zhuang Z., Goldstein S. R., Weiss R. A., Liotta L.A. (1996) Laser capture microdissection. Science 274, 998–1001 [DOI] [PubMed] [Google Scholar]
- 20. Zieske L.R. (2006) A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J. Exp. Bot. 57, 1501–1508 [DOI] [PubMed] [Google Scholar]
- 21. Ralhan R., Desouza L. V., Matta A., Chandra, Tripathy S., Ghanny S., Datta, Gupta S., Bahadur S., Siu K. W. (2008) Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Mol. Cell. Proteomics 7, 1162–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. DeSouza L. V., Grigull J., Ghanny S., Dubé V., Romaschin A. D., Colgan T. J., Siu K.W. (2007) Endometrial carcinoma biomarker discovery and verification using differentially tagged clinical samples with multidimensional liquid chromatography and tandem mass spectrometry. Mol. Cell. Proteomics 6, 1170–1182 [DOI] [PubMed] [Google Scholar]
- 23. Pierce A., Unwin R. D., Evans C. A., Griffiths S., Carney L., Zhang L., Jaworska E., Lee C. F., Blinco D., Okoniewski M. J., Miller C. J., Bitton D. A., Spooncer E., Whetton A. D. (2008) Eight-channel iTRAQ enables comparison of the activity of six leukemogenic tyrosine kinases. Mol. Cell. Proteomics 7, 853–863 [DOI] [PubMed] [Google Scholar]
- 24. Travis W. D., Colby T. V., Corrin B., Shimosato Y., Brambilla E. (1999) Histological typing of lung and pleural tumours, 3rd Edn., Springer-Verlag, Berlin [Google Scholar]
- 25. Cheng A. L., Huang W. G., Chen Z. C., Peng F., Zhang P. F., Li M. Y., Li F., Li J. L., Li C., Yi H., Yi B., Xiao Z. Q. (2008) Identification of novel nasopharyngeal carcinoma biomarkers by laser capture microdissection and proteomic analysis. Clin. Cancer Res. 14, 435–445 [DOI] [PubMed] [Google Scholar]
- 26. Xiao Z., Li G., Chen Y., Li M., Peng F., Li C., Li F., Yu Y., Ouyang Y., Xiao Z., Chen Z. (2010) Quantitative proteomic analysis of formalin-fixed and paraffin-embedded nasopharyngeal carcinoma using iTRAQ labeling, two-dimensional liquid chromatography, and tandem mass spectrometry. J. Histochem. Cytochem. 58, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 28. Feng X. P., Yi H., Li M. Y., Li X. H., Yi B., Zhang P. F., Li C., Peng F., Tang C. E., Li J. L., Chen Z. C., Xiao Z. Q. (2010) Identification of biomarkers for predicting nasopharyngeal carcinoma response to radiotherapy by proteomics. Cancer Res. 70, 3450–3462 [DOI] [PubMed] [Google Scholar]
- 29. Sun Y., Yi H., Yang Y., Yu Y., Ouyang Y., Yang F., Xiao Z., Chen Z. (2009) Functional characterization of p53 in nasopharyngeal carcinoma by stable shRNA expression. Int. J. Oncol. 34, 1017–1027 [DOI] [PubMed] [Google Scholar]
- 30. Dennis G, Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 31. Hu Z., Mellor J., Wu J., DeLisi C. (2004) VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu Z., Mellor J., Wu J., Yamada T., Holloway D., Delisi C. (2005) VisANT: data-integrating visual framework for biological networks and modules. Nucleic Acids Res. 33, W352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hayes J. D., Pulford D. J. (1995) (1995) The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 30, 445–600 [DOI] [PubMed] [Google Scholar]
- 34. Sekine I., Minna J. D., Nishio K., Tamura T., Saijo N. (2006) A literature review of molecular markers predictive of clinical response to cytotoxic chemotherapy in patients with lung cancer. J. Thorac. Oncol. 1, 31–37 [PubMed] [Google Scholar]
- 35. Vlachogeorgos G, S, Manali E. D., Blana E., Legaki S., Karagiannidis N., Polychronopoulos V. S., Roussos C. (2008) Placental isoform glutathione S-transferase and P-glycoprotein expression in advanced non-small cell lung cancer: association with response to treatment and survival. Cancer 114, 519–526 [DOI] [PubMed] [Google Scholar]
- 36. Hirano T., Kato H., Maeda M., Gong Y., Shou Y., Nakamura M., Maeda J., Yashima K., Kato Y., Akimoto S., Ohira T., Tsuboi M., Ikeda N. (2005) Identification of postoperative adjuvant chemotherapy responders in non-small cell lung cancer by novel biomarker. Int. J. Cancer 117, 460–468 [DOI] [PubMed] [Google Scholar]
- 37. Sun N., Sun X., Chen B., Cheng H., Feng J., Cheng L., Lu Z. (2010) MRP2 and GSTP1 polymorphisms and chemotherapy response in advanced non-small cell lung cancer. Cancer Chemother. Pharmacol. 65, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gao P., Yang X., Xue Y. W., Zhang X. F., Wang Y., Liu W. J., Wu X. J. (2009) Promoter methylation of glutathione S-transferase pi1 and multidrug resistance gene 1 in bronchioloalveolar carcinoma and its correlation with DNA methyltransferase 1 expression. Cancer 115, 3222–3232 [DOI] [PubMed] [Google Scholar]
- 39. Cote M. L., Chen W., Smith D. W., Benhamou S., Bouchardy C., Butkiewicz D., Fong K. M., Gené M., Hirvonen A., Kiyohara C., Larsen J. E., Lin P., Raaschou-Nielsen O., Povey A. C., Reszka E., Risch A., Schneider J., Schwartz A. G., Sorensen M., To-Figueras J., Tokudome S., Pu Y., Yang P., Wenzlaff A. S., Wikman H., Taioli E. (2009) Meta- and pooled analysis of GSTP1 polymorphism and lung cancer: a HuGE-GSEC review. Am. J. Epidemiol. 169, 802–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zienolddiny S., Campa D., Lind H., Ryberg D., Skaug V., Stangeland L. B., Canzian F., Haugen A. (2008) A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of non-small cell lung cancer in smokers. Carcinogenesis 29, 1164–1169 [DOI] [PubMed] [Google Scholar]
- 41. Hoffmann D., Djordjevic M. V., Hoffmann I. (1997) The changing cigarette. Prev. Med. 26, 427–434 [DOI] [PubMed] [Google Scholar]
- 42. Osada H., Takahashi T. (2002) Genetic alterations of multiple tumor suppressors and oncogenes in the carcinogenesis and progression of lung cancer. Oncogene 21, 7421–7434 [DOI] [PubMed] [Google Scholar]
- 43. Sims P., Grover P. L. (1974) Epoxides in polycyclic aromatic hydrocarbon metabolism and carcinogenesis. Adv. Cancer Res. 20, 165–274 [DOI] [PubMed] [Google Scholar]
- 44. Baird W. M., Hooven L. A., Mahadevan B. (2005) Carcinogenic polycyclic aromatic hydrocarbon-DNA adducts and mechanism of action. Environ. Mol. Mutagen. 45, 106–114 [DOI] [PubMed] [Google Scholar]
- 45. Hayes J. D., Flanagan J. U., Jowsey I. R. (2005) Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 45, 51–88 [DOI] [PubMed] [Google Scholar]
- 46. Reszka E., Wasowicz W. (2001) Significance of genetic polymorphisms in glutathione S-transferase multigene family and lung cancer risk. Int. J. Occup. Med. Environ. Health 14, 99–113 [PubMed] [Google Scholar]
- 47. Ciocca D. R., Oesterreich S., Chamness G. C., McGuire W. L., Fuqua S. A. (1993) Biological and clinical implications of heat shock protein 27,000 (Hsp27): a review. J. Natl. Cancer Inst. 85, 1558–1570 [DOI] [PubMed] [Google Scholar]
- 48. Paul C., Manero F., Gonin S., Kretz-Remy C., Virot S., Arrigo A. P. (2002) Hsp27 as a negative regulator of cytochrome C release. Mol. Cell. Biol. 22, 816–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. O'Callaghan-Sunol C., Gabai V. L., Sherman M. Y. (2007) Hsp27 modulates p53 signaling and suppresses cellular senescence. Cancer Res. 67, 11779–11788 [DOI] [PubMed] [Google Scholar]
- 50. Calderwood S. K., Khaleque M. A., Sawyer D. B., Ciocca D. R. (2006) Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem. Sci. 31, 164–172 [DOI] [PubMed] [Google Scholar]
- 51. Kim E. H., Lee H. J., Lee D. H., Bae S., Soh J. W., Jeoung D., Kim J., Cho C. K., Lee Y. J., Lee Y. S. (2007) Inhibition of heat shock protein 27-mediated resistance to DNA damaging agents by a novel PKC delta-V5 heptapeptide. Cancer Res. 67, 6333–6341 [DOI] [PubMed] [Google Scholar]
- 52. Garrido C., Fromentin A., Bonnotte B., Favre N., Moutet M., Arrigo A. P., Mehlen P., Solary E. (1998) Heat shock protein 27 enhances the tumorigenicity of immunogenic rat colon carcinoma cell clones. Cancer Res. 58, 5495–5499 [PubMed] [Google Scholar]
- 53. Kamada M., So A., Muramaki M., Rocchi P., Beraldi E., Gleave M. (2007) Hsp27 knockdown using nucleotide-based therapies inhibit tumor growth and enhance chemotherapy in human bladder cancer cells. Mol. Cancer Ther. 6, 299–308 [DOI] [PubMed] [Google Scholar]
- 54. Ciocca D. R., Calderwood S. K. (2005) Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 10, 86–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yao H., Zhang Z., Xiao Z., Chen Y., Li C., Zhang P., Li M., Liu Y., Guan Y., Yu Y., Chen Z. (2009) Identification of metastasis associated proteins in human lung squamous carcinoma using two-dimensional difference gel electrophoresis and laser capture microdissection. Lung Cancer 65, 41–48 [DOI] [PubMed] [Google Scholar]
- 56. Lomnytska M. I., Becker S., Bodin I., Olsson A., Hellman K., Hellström A. C., Mints M., Hellman U., Auer G., Andersson S. (2011) Differential expression of ANXA6, HSP27, PRDX2, NCF2, and TPM4 during uterine cervix carcinogenesis: diagnostic and prognostic value. Br. J. Cancer 104, 110–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kapranos N., Kominea A., Konstantinopoulos P. A., Savva S., Artelaris S., Vandoros G., Sotiropoulou-Bonikou G., Papavassiliou A. G. (2002) Expression of the 27-kDa heat shock protein (HSP27) in gastric carcinomas and adjacent normal, metaplastic, and dysplastic gastric mucosa, and its prognostic significance. J. Cancer Res. Clin. Oncol. 128, 426–432 [DOI] [PubMed] [Google Scholar]
- 58. Lindeman B., Skarpen E., Huitfeldt H. S. (1998) Stress protein expression in rat liver during tumour promotion: induction of heat-shock protein 27 in hepatocytes exposed to 2-acetylaminofluorene. Carcinogenesis 19, 1559–1563 [DOI] [PubMed] [Google Scholar]
- 59. O'Neill P. A., Shaaban A. M., West C. R., Dodson A., Jarvis C., Moore P., Davies M. P., Sibson D. R., Foster C. S. (2004) Increased risk of malignant progression in benign proliferating breast lesions defined by expression of heat shock protein 27. Br. J. Cancer 90, 182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Joseph J., Cardesa A., Carreras J. (1997) Creatine kinase activity and isoenzymes in lung, colon and liver carcinomas. Br. J. Cancer 76, 600–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zarghami N., Giai M., Yu H., Roagna R., Ponzone R., Katsaros D., Sismondi P., Diamandis E. P. (1996) Creatine kinase BB isoenzyme levels in tumour cytosols and survival of breast cancer patients. Br. J. Cancer 73, 386–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liippo K. K., Terho T. (1991) Concomitant monitoring of serum neuron-specific enolase and creatine kinase BB in small cell lung cancer. Acta Oncol. 30, 321–324 [DOI] [PubMed] [Google Scholar]
- 63. Huddleston H. G., Wong K. K., Welch W. R., Berkowitz R. S., Mok S. C. (2005) Clinical applications of microarray technology: creatine kinase B is an up-regulated gene in epithelial ovarian cancer and shows promise as a serum marker. Gynecol. Oncol. 96, 77–83 [DOI] [PubMed] [Google Scholar]
- 64. Lin L. M., Chen Y. K. (1991) Creatine kinase isoenzymes activity in serum and buccal pouch tissue of hamsters during DMBA-induced squamous cell carcinogenesis. J. Oral Pathol. Med. 20, 479–485 [DOI] [PubMed] [Google Scholar]
- 65. Balasubramani M., Day B. W., Schoen R. E., Getzenberg R. H. (2006) Altered expression and localization of creatine kinase B, heterogeneous nuclear ribonucleoprotein F, and high mobility group box 1 protein in the nuclear matrix associated with colon cancer. Cancer Res. 66, 763–769 [DOI] [PubMed] [Google Scholar]
- 66. Joseph J., Cardesa A., Carreras J. (1997) Creatine kinase activity and isoenzymes in lung, colon and liver carcinomas. Br. J. Cancer 76, 600–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mooney S. M., Rajagopalan K., Williams B. H., Zeng Y., Christudass C. S., Li Y., Yin B., Kulkarni P., Getzenberg R. H. (2011) Creatine kinase brain overexpression protects colorectal cells from various metabolic and non-metabolic stresses. J. Cell. Biochem. 112, 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]