Abstract
The molecular pathways involved in neovascularization of regenerating tissues and tumor angiogenesis resemble each other. However, the regulatory mechanisms of neovascularization under neoplastic circumstances are unbalanced leading to abnormal protein expression patterns resulting in the formation of defective and often abortive tumor vessels. Because gliomas are among the most vascularized tumors, we compared the protein expression profiles of proliferating vessels in glioblastoma with those in tissues in which physiological angiogenesis takes place. By using a combination of laser microdissection and LTQ Orbitrap mass spectrometry comparisons of protein profiles were made. The approach yielded 29 and 12 differentially expressed proteins for glioblastoma and endometrium blood vessels, respectively. The aberrant expression of five proteins, i.e. periostin, tenascin-C, TGF-beta induced protein, integrin alpha-V, and laminin subunit beta-2 were validated by immunohistochemistry. In addition, pathway analysis of the differentially expressed proteins was performed and significant differences in the usage of angiogenic pathways were found. We conclude that there are essential differences in protein expression profiles between tumor and normal physiological angiogenesis.
Neovascularization is a complex process taking place under physiological and pathological circumstances. There is large overlap in the cellular components, regulatory factors, and signaling mechanisms acting in the angiogenic process of regeneration, embryonic development, and tumor vascularization (1). In both physiological and pathological neovascularization signaling mechanisms, growth factors and their receptors, cell adhesion molecules and their specific extracellular matrix ligands take part (2). Angiocrine molecules like basic fibroblast growth factor and vascular endothelial growth factor and their receptors have been identified in the context of physiological and tumor angiogenesis (3). It is believed that the angiogenic switch is always triggered by hypoxia, starting with the up-regulation of vascular endothelial growth factor. The process of neovascularization consists not only of sprouting angiogenesis, but also of activation of endothelial precursor cells with the capacity to form de novo blood vessels (vasculogenesis). Vasculogenesis is also part of tumor vascularization but there is dispute about its relative importance (4, 5). Despite the similarities there are obvious differences between physiological vascularization and angiogenesis in tumors. Under physiological conditions, the regulatory mechanisms are well-coordinated and balanced and endothelial cell functions are tightly orchestrated by both pro- and anti-angiogenic factors. In tumor angiogenesis however, there is an excess of pro-angiogenic factors leading to uncoordinated proliferation and tubulogenesis of endothelial cells and migration of mural cells like pericytes (6). It is likely that the molecular differences between physiological angiogenesis and neovascularization in tumors are mainly at the level of regulation of pathways and overexpression of particular proteins.
In order to identify proteins that are specifically expressed in tumor angiogenesis, comparisons with protein profiles of blood vessels in which active normal angiogenesis take place are necessary. Physiological angiogenesis occurs in adults during the menstrual cycle and in repair or regeneration of tissue during wound healing (7). Therefore, we included blood vessels from proliferating endometrium (representing physiological angiogenesis) in the present analysis. A better model for tumor angiogenesis than that taking place in glial neoplasms is hardly imaginable and therefore, we implicated the blood vessels of glioblastomas in this study. Of all tumors, gliomas are among the most vascularized ones. Most glial tumors develop from low-grade, relatively benign neoplasms into high-grade tumors. Glioblastomas (or glioblastoma multiforme; GBM1) are gliomas of the highest malignancy grade. These tumors are the most frequently encountered primary brain tumors in humans. GBMs are highly infiltrative tumors that show rapid clinical progression. Most patients succumb in less than a year after the diagnosis is made. In contrast to their low-grade counterparts, GBMs show high proliferation and cell density, and notorious microvascular proliferation and necrosis (as a sequel of the bad quality of the blood vessels) (8). It is believed that angiogenesis, the formation of new blood vessels from pre-existing vasculature, not de novo vasculogenesis from individual cells, is the dominant mechanism in the development of tumor vasculature (9, 10).
The aim of the present investigation was to elucidate differences between regenerative (physiological) angiogenesis and angiogenesis in neoplasms at the protein level. The identification of differences in protein expression patterns are of paramount clinical importance: in some situations the formation of new blood vessels should be stimulated, whereas in others the main goal is to repress neovascularization. To this end, we microdissected the blood vessels from GBM and proliferating endometrium by using laser capture microdissection. The microdissected blood vessel subsets were analyzed by nano liquid chromatography LTQ Orbitrap mass spectrometry. Differentially expressed proteins detected in either group were characterized and linked to molecular pathways. In addition, a selection of differentially expressed proteins was validated by immunohistochemistry.
MATERIALS AND METHODS
Patient Samples
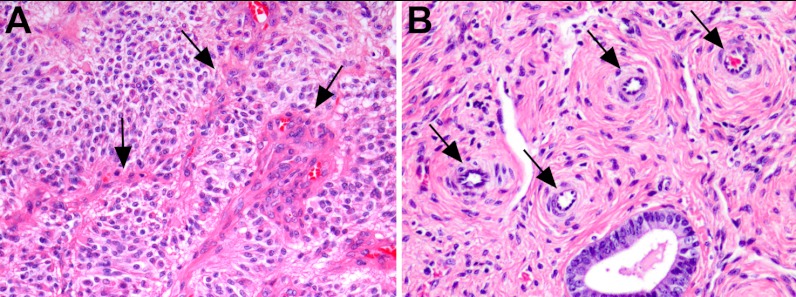
Ten fresh-frozen surgical samples of GBM located in the cerebral hemispheres were taken from the files of the Department of Pathology, Erasmus MC, Rotterdam, The Netherlands. Five patients were male. The ages ranged between 41 and 86 years (median 67.5 years). In addition, 10 fresh-frozen endometrium samples were collected at the Department of Gynecology, Erasmus Medical Center, Rotterdam, The Netherlands. The samples were collected from premenopausal women who had undergone hysterectomies for diseases not involving the endometrium. Their ages ranged between 39 and 46 years (median of 43 years). Sections of 5 μm from each sample were counterstained and examined by a pathologist (JMK) to verify the presence of blood vessels (Fig. 1).
Fig. 1.
Proliferating blood vessels in a glioblastoma (A) and endometrium (B) sample (H&E, magnification 400×).
The use of all samples was approved by the Medical Ethical Committee of the Erasmus Medical Center Rotterdam, The Netherlands.
Laser Capture Microdissection
The procedure was as previously described (12). Briefly, cryosections of 10 μm were made from each sample and mounted on polyethylene naphthalate covered glass slides (P.A.L.M. Microlaser Technologies AG, Bernried, Germany). The slides were fixed in 70% ice-cold ethanol for a maximum of 2 h. Before laser microdissection, the slides were stained with hematoxylin and dehydrated in a series of ethanol solutions and left to air-dry for 5 min. The P.A.L.M. laser microdissector and pressure catapulting device, type P-MB, was used with PalmRobo version 2.2 software at 40 × magnification. An area of 200,000 μm2 was microdissected from each sample, resulting in ∼2000 cells/sample (estimated cell volume: 10 × 10 × 10 μm). Altogether, four groups of samples were laser microdissected: glioma blood vessels, glioma tissue surrounding the blood vessels, endometrium blood vessels, and endometrium tissue surrounding the blood vessels. In addition, a corresponding sized area of the polyethylene naphthalate membrane was laser microdissected and used as a negative control. As internal quality control, three sections of a glioma sample without microdissection were collected. The preparation and analysis of the internal quality controls was similar to that of the test samples.
Sample Preparation
The procedure for sample preparation was as previously published (12). For laser microdissection, 7 μl of 0.1% RapiGest (Waters, Milford, MA) in 50 mm NH4HCO3 pH 8.0 was used to collect the laser microdissected tissue areas. Protein LoBind, 0.5-ml Eppendorf tubes (Eppendorf, Hamburg, Germany) containing the samples were stored at −80 °C until the time of preparation. After thawing the samples, 15 μl of RapiGest solution were added to each tube to bring the final volume of each sample to 22 μl. The cells were disrupted by external sonication for 1 min at 70% amplitude at a maximum temperature of 25 °C (Branson Ultrasonics, Danbury, CT). For protein solubilization and denaturation, the samples were incubated at 37 and 100 °C for 5 and 15 min, respectively. To each sample, 1.5 μl of 100 ng/μl gold grade trypsin (Promega, Madison, WI) in 3 mm Tris-HCl diluted 1:10 in 50 mm NH4HCO3 was added and incubated overnight at 37 °C. To inactivate trypsin and to degrade the RapiGest, 3 μl of 25% trifluoroacetic acid were added and samples were incubated for 30 min at 37 °C. Samples were centrifuged at maximum speed for 15 min at 4 °C and the supernatant was transferred to LC-vials (Waters, Milford, MA) and measured by nanoLC Orbitrap mass spectrometry. The internal quality control sample was diluted at 1:400 using LC-MS grade water in order to obtain a comparable peptide concentration to that of the laser microdissected blood vessel samples. The internal control sample was prepared with the other samples and measured at regular intervals (every five samples) over the measurement period of about 30 days.
LTQ Orbitrap Measurements
Nano LC-MS measurements were carried out on an Ultimate 3000 nano LC system (Dionex, Germering, Germany) online coupled to a hybrid linear ion trap/Orbitrap MS (LTQ Orbitrap XL; Thermo Fisher Scientific, Bremen, Germany). The total volumes of the digested samples (∼20 μl) were loaded onto a C18 trap column (C18 PepMap, 300 μm ID × 5 mm, 5 μm particle size, 100 Å pore size; Dionex, Amsterdam, The Netherlands) and washed for 10 min using a flow rate of 25 μl/min of 0.1% trifluoroacetic acid. The trap column was switched in-line with the analytical column (PepMap C18, 75 μm ID × 150 mm, 3 μm particle and 100 Å pore size; Dionex, Amsterdam, The Netherlands) and peptides were eluted with the following binary (A and B) gradient: 0–25% solvent B in 120 min; 25–50% solvent B from 120–180 min; solvent A consists of 2% acetonitrile and 0.1% formic in water and solvent B consists of 80% acetonitrile and 0.08% formic acid in water. The column flow rate was set to 300 nL/min. For MS detection a data dependent acquisition method was used: a high resolution survey scan from 400–1800 Da was obtained in the Orbitrap (value of target of automatic gain control 106, resolution 30,000 at 400 m/z; lock mass was set to 445.120025 u (protonated (Si(CH3)2O)6)1. Based on this survey scan the five most intensive ions were consecutively isolated (automatic gain control target set to 104 ions) and fragmented by collision activated dissociation applying 35% normalized collision energy in the linear ion trap. After precursors were selected for MS/MS, they were excluded for 3 min from fragmentation. Samples were prepared and measured in a randomized way. We measured the internal quality control sample once in every five measurements.
Protein Identification
From raw mass spectrometer data files, tandem MS (MS/MS) spectra were extracted by Mascot Deamon (Matrix Science, London, UK) version 2.2.2 using the Xcalibur extract msn tool (version 2.07) into mgf files. All mgf files were analyzed using Mascot. The set-up was designed to search the UniProt (release 15.6) database. The Mascot search engine was used with fragment ion mass tolerance of 0.5 Da and a parent ion tolerance of 10 ppm. Oxidation of methionine was specified in Mascot as a variable modification. A minimum ion score of 25 was required for identification. Scaffold software (Version, 2_05_01), [www.proteomesoftware.com] (Portland, OR), was used to summarize and filter MS/MS based peptides and protein identifications. Peptide identifications were accepted if they exceeded a peptide probability of 95.0%. Protein identifications were accepted if protein probability exceeded 99.0% and at least two peptides were identified. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped.
Label-Free Quantitation
Scaffold was used to generate a data file of the identified proteins including the number of sequenced peptides (spectral counts) that were found in each sample. On this data file we performed a Significance Analysis of Microarrays (SAM, Stanford University) version 3.1 and performed a multiclass comparison with false discovery rate of 5%. In the SAM analysis the relative frequency of occurrence of each protein in the four groups of samples was calculated. The relative frequency of occurrence indicates the mean differences between the protein measurements in each class, versus the mean of all measurements for the particular protein. Therefore, the positive values mean that the protein was measured in that class more than the average measurements of that protein in all the other classes. The negative values mean that the protein was measured in that class less than the average measurement of the same protein in other classes. The proteins that were significantly expressed in either one of the blood vessel groups as compared with the other three groups were taken as differentially expressed.
In addition the data was analyzed using the Progenesis LC-MS Software package (Version, 2.5), Non Linear Dynamics, New Castle UK. We aligned all blood vessel samples of glioma and endometrium using one of the glioma blood vessel samples as reference. Identification was added to the Progenesis result file by performing a Mascot database search with above described parameters. The statistical analyses of the data was performed using Progenesis and Partek® Genomics Suite™ version 6.09.0129 software [http://www.partek.com/]. The zero values in the data matrix obtained by Progenesis were removed and the distribution of the normalized abundance values was log2-transformed and subsequently an ANOVA analysis was performed in Partek® Genomics Suite™.
Pathway Analysis
The lists of differentially expressed proteins in glioma blood vessels derived from the data analysis were combined and uploaded into the Ingenuity IPA system version, 7.1 [www.ingenuity.com] to generate biological networks relating to glioma angiogenesis. In addition, we uploaded the lists of differentially expressed proteins from the endometrium blood vessels into the Ingenuity IPA system to generate biological networks relating to physiological angiogenesis. Subsequently, we compared the biological networks of the two groups in order to search for differences in the regulation of proteins in the pathways.
Validation by Immunohistochemistry
Eight proteins were validated by immunohistochemistry on formalin-fixed, paraffin-embedded tissue sections; basement membrane-specific heparan sulfate proteoglycan core protein (P98160), integrin alpha-V (P06756), laminin subunit beta-2 (Q6PCB0), collagen alpha-1 (XVIII) chain (P39060), integrin-linked protein kinase (Q13418), tenascin C (P24821), transforming growth factor-beta-induced protein ig-h3 (Q15582), and periostin (Q15063) (Table I). To investigate expressional variation between the two investigated vessel groups in independent tissue samples, ten additional samples from each group of glioma, endometrium, and normal brain were immunostained. Further, in order to determine the specificity of the above mentioned proteins to glioma angiogenesis, a series of other gliomas, carcinomas, vascular malformations, reactive conditions in which angiogenesis takes place, normal brain samples and placentas were tested for the presence of these proteins. More details about these samples are available in Table II. Immunohistochemical staining was performed following the manufacturer's procedures (alkaline phosphate technique) on 5-μm paraffin sections. We followed the same procedure as previously described (15).
Table I. Antibodies used for immunohistochemical validation.
| Antibody | Company | Treatment | dilution (Ab:buffer) |
|---|---|---|---|
| Basement membrane-specific heparan sulfate proteoglycan core protein | Abcam, Cambridge, UK | Microwave | 50 |
| Collagen alpha-1 (XVIII) chain | Atlas antibodies, Stockholm, Sweden | Microwave | 100 |
| Laminin subunit beta-2 | Dako, Glostrup, Denmark | Pronase incubation (10 minutes) | 10 |
| Periostin | Atlas antibodies, Stockholm, Sweden | Microwave | 100 |
| Tenascin C | Abcam, Cambridge, UK | Microwave | 100 |
| Transforming growth factor-beta-induced protein ig-h3 | Sigma (Atlas antibodies), Stockholm, Sweden | Microwave | 400 |
| Integrin alpha-V | Santa Cruz Biotechnology, Heidelberg, Germany | Microwave | 50 |
| Integrin-linked protein kinase | Bioworld Technology, inc, Suffolk, UK | Microwave | 50 |
Table II. Overview of patients samples used for validation by immunohistochemistry.
| Tissue type | # of females | Age range of females (years) | # of males | Age range of males (years) |
|---|---|---|---|---|
| Glioblastoma | 1 | 30 | 9 | 32–57 |
| Normal brain samples | 5 | 39–76 | 5 | 28–62 |
| Pilocytic astrocytoma | 3 | 17–56 | 3 | 7–15 |
| Ependymoma | 0 | – | 3 | 7–21 |
| Anaplastic oligodendroglioma | 0 | – | 4 | 44–61 |
| Renal cell carcinoma | 2 | 57–74 | 5 | 24–67 |
| Arteriovenous malformation (AVM) | 1 | 18 | 5 | 35–59 |
| Cavernous hemangioma | 4 | 4–50 | 4 | 6–75 |
| Ischemic infarction of brain | 0 | – | 2 | 54–57 |
| Placenta | 3 | 28–44 | 0 | – |
| Endometrium | 13 | 23–81 | 0 | – |
RESULTS
The internal quality control was measured seven times at equal intervals during the measurement period of the complete sample set. The average CV of the spectral counts of the identified proteins in the sample was 20.8% and the average number of the identified proteins was 206 with a standard deviation of 16. No significant changes in the number of identified proteins were observed over the period of time in which measurements were repeated for internal quality control samples indicating that both the experimental set-up and the protein digests were stable over the measurement period of 30 days. The number of identified proteins did not change significantly during the measurement period of 30 days.
In total 694 proteins were identified in the four sample groups. The spectral counts of all identified proteins as observed in the Scaffold software were used in SAM analysis with a false discovery rate of 5% resulting in 152 differentially expressed proteins. We categorized these 152 differentially expressed proteins based on their relative frequency of occurrence (SAM analysis) in the sample groups. We considered a protein to be specific for a particular sample group if its occurrence was at least twice the occurrence encountered in any of the other groups. Out of the 152 differentially expressed proteins 29 were found in the GBM blood vessel group (Table IIIA) whereas 12 were found in the endometrium blood vessel group (Table IIIB).
Table III. A: Differentially expressed proteins in glioma angiogenesis based on the spectral counts of each protein after performing SAM analyses. p values = ANOVA of peptide abundances in Progenesis of GV and EV, ¥ indicates the proteins that were selected for IHC validation, *** represents a significant p value (p < 0.05), ND = Not detected in the Progenesis analyses. GV = glioma blood vessels, GT = glial tumor tissue, EV = endometrium blood vessels, ET = endometrial glands and stroma, RFO = the relative frequency of occurrence, the values rank each protein in each class. The RFO indicates the mean differences between the protein measurements in each class, versus the mean of all measurements for the particular protein. Therefore, the positive values = the protein was measured in that class more than the average measurements of that protein in all the other classes. The negative values = the protein was measured in that class less than the average measurement of the same protein in other classes. The color grade represents the frequency of occurrence (SAM analysis), dark green is the highest, dark red is the lowest and the different grade of yellow/orange indicates all the in between values.
| Protein | p value | Gene | Accession # | GV RFO | EV RFO | GT RFO | ET RFO |
|---|---|---|---|---|---|---|---|
| Laminin subunit gamma-1 | *** | LAMC1 | P11047 | 2.33 | −0.07 | −1.46 | −0.65 |
| Basement membrane-specific heparan sulfate proteoglycan core protein ¥ | *** | HSPG2 | P98160 | 2.27 | −0.36 | −1.15 | −0.64 |
| Agrin | *** | AGRN | O00468 | 2.02 | −0.51 | −0.66 | −0.78 |
| Plastin-3 | PLS3 | P13797 | 1.98 | −0.46 | −0.73 | −0.73 | |
| Integrin alpha-V ¥ | ITGAV | P06756 | 1.95 | −0.89 | −0.15 | −0.89 | |
| Laminin subunit alpha-5 | *** | LAMA5 | O15230 | 1.90 | −0.08 | −0.93 | −0.79 |
| von Willebrand factor A domain-containing protein 1 | VWA1 | Q6PCB0 | 1.81 | −0.72 | −0.34 | −0.72 | |
| Laminin subunit beta-2 ¥ | *** | LAMB2 | P55268 | 1.78 | −0.76 | −0.44 | −0.53 |
| Collagen alpha-1(XVIII) chain ¥ | *** | COL18A1 | P39060 | 1.77 | −0.38 | −0.63 | −0.69 |
| 40S ribosomal protein S16 | RPS16 | P62249 | 1.58 | −0.83 | 0.08 | −0.83 | |
| Band 3 anion transport protein | *** | SLC4A1 | P02730 | 1.56 | −0.44 | −0.46 | −0.61 |
| Integrin-linked protein kinase ¥ | ND | ILK | Q13418 | 1.54 | −0.37 | −1.01 | −0.05 |
| Adenylyl cyclase-associated protein 1 | CAP1 | Q01518 | 1.52 | −0.95 | −0.05 | −0.51 | |
| Nestin | NES | P48681 | 1.46 | −1.06 | 0.64 | −1.11 | |
| Fibronectin | *** | FN1 | P02751 | 1.46 | −0.43 | −0.47 | −0.51 |
| T-complex protein 1 subunit beta | CCT2 | P78371 | 1.44 | −0.44 | −0.04 | −0.96 | |
| Laminin subunit beta-1 | *** | LAMB1 | P07942 | 1.43 | −0.31 | −0.53 | −0.53 |
| Glutamate dehydrogenase 1, mitochondrial | GLUD1 | P00367 | 1.43 | −0.74 | −0.12 | −0.56 | |
| T-complex protein 1 subunit epsilon | CCT5 | P48643 | 1.43 | −0.51 | −0.37 | −0.51 | |
| Tenascin-C ¥ | *** | TNC | P24821 | 1.43 | −0.68 | −0.10 | −0.64 |
| Chloride intracellular channel protein 1 | ND | CLIC1 | O00299 | 1.41 | −0.44 | −0.14 | −0.81 |
| Nidogen-1 | *** | NID1 | P14543 | 1.30 | −0.15 | −0.68 | −0.40 |
| Nidogen-2 | *** | NID2 | Q14112 | 1.28 | −0.48 | −0.49 | −0.26 |
| Fermitin family homolog 3 | FERMT3 | Q86UX7 | 1.28 | −0.66 | −0.31 | −0.27 | |
| Transforming growth factor-beta-induced protein ig-h3 ¥ | *** | TGFBI | Q15582 | 1.20 | −0.40 | −0.68 | −0.05 |
| Elongation factor 2 | *** | EEF2 | P13639 | 1.15 | −0.62 | −0.22 | −0.29 |
| Coactosin-like protein | COTL1 | Q14019 | 1.09 | −0.57 | 0.04 | −0.57 | |
| CD99 antigen | ND | CD99 | P14209 | 1.07 | −0.49 | −0.32 | −0.23 |
| Periostin ¥ | *** | POSTN | Q15063 | 1.06 | −0.48 | −0.10 | −0.48 |
B: Differentially expressed proteins in endometrium angiogenesis on the spectral counts of each protein after performing SAM analyses. p values = ANOVA of peptide abundances in Progenesis of GV and EV, *** represents a significant p value (p < 0.05), ND = Not detected in the Progenesis analyses. GV = glioma blood vessels, GT = glial tumor tissue, EV = endometrium blood vessels, ET = endometrial glands and stroma, RFO = the relative frequency of occurrence, the values are the means to rank each protein in each class. The RFO indicates the mean differences between the protein measurements in each class, versus the mean of all measurements for the particular protein. Therefore, the positive values = the protein was measured in that class more than the average measurements of that protein in all the other classes. The negative values = the protein was measured in that class less than the average measurement of the same protein in other classes. The color grade represents the frequency of occurrence (SAM analysis), dark green is the highest, dark red is the lowest and the different grade of yellow/orange indicates all the in between values.
| Protein | p value | Gene | Accession # | EV RFO | GV RFO | GT RFO | ET RFO |
|---|---|---|---|---|---|---|---|
| Myosin-Ic | *** | MYO1C | O00159 | 2.39 | −1.16 | −1.78 | 0.72 |
| Prolargin | ND | PRELP | P51888 | 1.93 | −0.20 | −1.61 | 0.04 |
| Polymerase I and transcript release factor | *** | PTRF | Q6NZI2 | 1.75 | −0.72 | −1.17 | 0.26 |
| Caveolin-1 | CAV1 | Q03135 | 1.53 | −0.36 | −1.31 | 0.27 | |
| Poly(rC)-binding protein 1 | PCBP1 | Q15365 | 1.51 | −0.71 | −0.89 | 0.18 | |
| Cadherin-13 | *** | CDH13 | P55290 | 1.46 | −1.05 | −0.72 | 0.38 |
| Protein kinase C delta-binding protein | PRKCDBP | Q969G5 | 1.39 | −0.67 | −0.67 | 0.02 | |
| Keratin, type II cytoskeletal 1b | KRT77 | Q7Z794 | 1.27 | −0.59 | −0.11 | −0.56 | |
| Actin, aortic smooth muscle | ACTA2 | P62736 | 1.24 | −0.24 | −1.56 | 0.72 | |
| Complement factor B | CFB | P00751 | 1.24 | −0.72 | −0.42 | −0.06 | |
| Proliferation-associated protein 2G4 | ND | PA2G4 | Q9UQ80 | 1.09 | −0.35 | −0.35 | −0.35 |
| Myoferlin | ND | FER1L3 | Q9NZM1 | 0.98 | −0.30 | −0.35 | −0.30 |
The raw spectral counts as obtained via Scaffold were also compared with Progenesis. Successful alignment of the data derived from the blood vessels from the endometrium and GBM was achieved. By using the Progenesis results, 18 out of the 41 proteins appeared to be differentially expressed (ANOVA; p < 0.05) between the sample groups. (3-B = Table IIIB), The significant proteins in the ANOVA analyses are indicated in 3-B = Table IIIB.
Pathway Analysis
The list of 29 proteins, which were significantly up-regulated in GBM blood vessels, was uploaded in IPA and mapped against the database. IPA could map all proteins in three different networks. The first matched network designated as “tissue development and cell-to-cell signaling” had a score of 50 and contained 20 of the identified proteins. Ten of the 20 proteins were associated with a function designated as “cardiovascular system development and function” and five proteins, namely: collagen alpha-1(XVIII) chain, laminin subunit alpha-5, laminin subunit gamma-1, fibronectin, and integrin alpha-V appeared to be related to angiogenesis. In addition, the 12 differentially expressed proteins identified in the endometrium blood vessels were uploaded in IPA and were mapped to four different networks. The first matched network called “tissue development, embryonic development” contained 11 proteins. Nine proteins showed a direct relation with the function called “Cardiovascular System Development and function” and three proteins (caveolin-1, myosin-Ic, and protein kinase C delta-binding protein) appeared to be related with proliferation of endothelial cells. At the level of molecular and cellular functions, the differentially expressed proteins that were identified in the glioma blood vessels were associated with cell-to-cell signaling, cellular movements, and cell morphology. In contrast, the differentially expressed proteins in the endometrium blood vessels were associated with molecular transport and excretion of proteins.
Immunohistochemistry
Eight proteins were selected for immunohistochemical validation based on the availability of antibodies (Table 3A). Six of these proteins had been significantly emerged from both the SAM and the ANOVA analysis. Integrin-alpha V and integrin-linked protein kinase were selected based on their relation to angiogenesis (12, 13) (Supplemental file S1 and S2).
The overexpression of five out of eight proteins in glioma blood vessels compared with endometrium vessels was confirmed by immunohistochemistry. The proliferated blood vessels of GBM samples were immunopositive for periostin, TGF-β induced protein ig-h3, integrin-alpha V, tenascin C, and laminin (Fig. 2). The other three proteins, e.g. basement membrane-specific heparan sulfate proteoglycan core protein, proteins collagen alpha-1 (XVIII) chain, and integrin-linked protein kinase, were not present in the GBM blood vessels only, but also found in the endometrium blood vessels by immunohistochemistry.
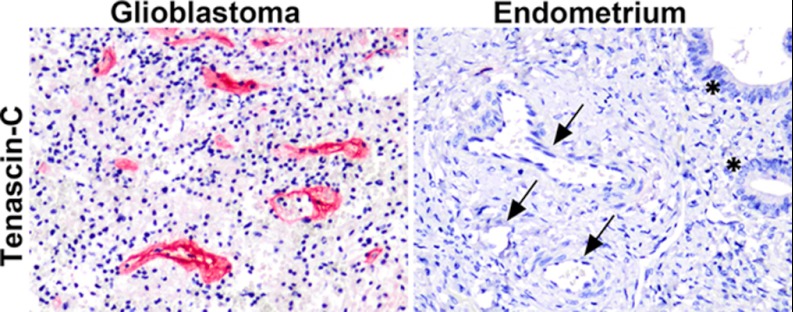
Fig. 2.
Expression pattern of tenascin C in glioblastoma and endometrium blood vessels. Overexpression of tenascin-C in blood vessels of GBM, whereas no immunostaining in the endometrial blood vessels is observed (arrows). The stars represent the endometrial glands (magnification 200×).
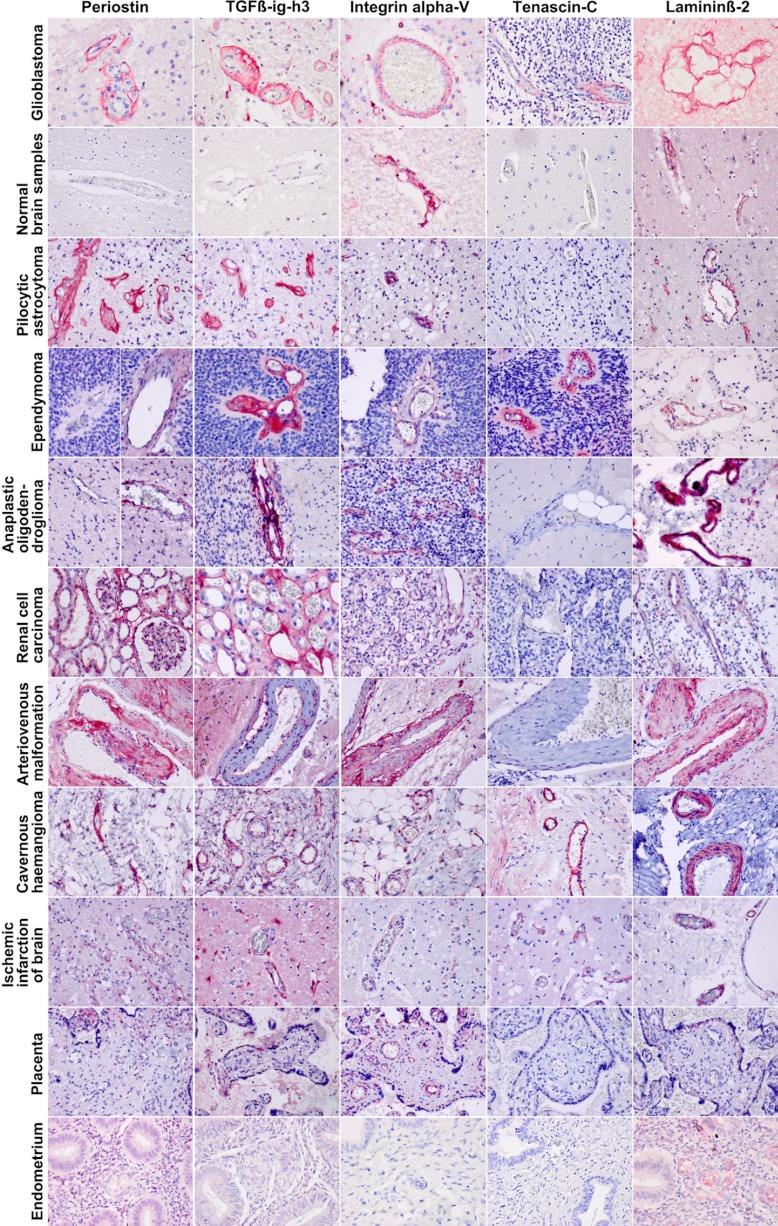
The results of the immunostaining of various glioma types showed some variation in expression of the above mentioned proteins in the blood vessels, depending on the stage of proliferation; high expression in young sprouts, low in sclerotic, abortive vessels. The expression of each protein was constant among the ten different samples that were tested in each group. For comparison of the results of immunohistochemistry, we included other nonglial tumors and reactive conditions. The results of immunostaining of carcinomas, vascular malformations, reactive conditions, angiogenesis in placentas, and normal blood vessels in normal brains varied and are summarized in Table IV and Fig. 3.
Table IV. Samples used for extra validation by immunohistochemistry. From each tissue type different numbers of sample were used, the information is indicated in Table II. Positive: the protein was expressed in all samples we tested from a tissue type and in all blood vessels but not in the surrounding tissue. Negative: the protein was not expressed in any of the blood vessels tested in a certain tissue type. Variably positive: the protein was expressed in some of the blood vessels and all samples from a tissue type showed comparable expression.
| Tissue Type | Periostin (Q15063) | TGFβ-ig-h3 (Q15582) | Integrin alpha-V (P06756) | Tenascin-C (P24821) | Lamininβ-2 (P55268) |
|---|---|---|---|---|---|
| Glioma | |||||
| Glioblastoma | Positive | Positive | Positive | Positive | Positive |
| Normal brain samples | Negative | Negative | Positive | Negative | Positive |
| Pilocytic astrocytoma | Positive | Positive | Positive | Negative | Slightly Positive |
| Ependymoma | Variably + | Positive | Positive | Positive | Positive |
| Anaplastic oligodendroglioma | Variably + | Positive | Positive | Negative | Positive |
| Renal cell carcinoma | Positive | Negative | Positive | Negative | Positive |
| Vascular malformation | |||||
| Arteriovenous malformation (AVM) | Positive | Variably + | Positive | Negative | Positive |
| Cavernous hemangioma | Positive | Positive | Positive | Variably + | Arteries + |
| Veins – | |||||
| Reactive condition | |||||
| Ischemic infarction of brain | Positive | Positive | Positive | Positive | Positive |
| Tissues with physiological angiogenesis | |||||
| Placenta | Vessels – Stroma + | Vessels – Stroma + | Positive | Negative | Vessels – Basal lamina + |
| Endometrium | Negative | Negative | Negative | Negative | Positive |
Fig. 3.
Immunohistochemical validation of part of the differentially expressed proteins in various tissue sections (see also Table VI). The overexpression of periostatin, TGF-beta, integrin alpha V, tenascin C, and laminin in the vasculature of GBM and also in pilocytic astrocytoma, ependymoma, and anaplastic oligodendroglioma row 1, 3–5. The sample set was extended with a nonglial tumor (renal cell carcinoma) of which the vasculature was variably immunopositive for the various proteins (sixth row). The nonneoplastic arteriovenous malformation (AVM) and cavernous hemangioma (CH) stained positive for all proteins tested, except for immunonegativity for laminin of the AVM. Remarkably, the blood vessels in ischemic brain tissue were immunopositive for all proteins found in the glioma vessels. These results were constant among all the samples that were used in each type of tissue.
DISCUSSION
Although significant progress has been made in the field of proteomics over the last two decades, the identification and quantification of the entire proteome of tissue samples is not possible. The analysis of complex protein mixtures is currently one of the most challenging subjects in the area of proteomics. The normal biological variation in protein expression among cells of identical lineage is around 15% to 30% (15). Under situations of stress or in tumors, an even higher variation of protein expression is expected (15, 16). A major source of this expressional variation is caused by differences in the structure and function of different cell populations present in individual samples. The complexity of human biopsy samples is still a considerable obstacle in proteomics analyses (17). Reduction of the complexity of protein mixtures can be reached at various levels. First, at the level of the tissue, methods of sample purification applied prior to analysis may well improve the accuracy of detection and increase the chance to identify low abundant proteins (18). By laser capture microdissection particular microscopic structures (like blood vessels) can be targeted and isolated and therefore reducing the chance of averaging out proteins of interest. Further, there are methods of enhancing the detection of proteins in the proteomics technology using very low numbers of cells. In several studies, it was proven that the use of advanced methods of fractionation prior to measuring the peptide digests in any of the mass spectrometers increases the number of identified proteins (12, 18, 19–20, 21). In order to reduce the complexity of the samples in the present study and enrich for blood vessels in our glioblastoma and endometrium blood vessel samples, laser microdissection was applied to separate the blood vessels from the surrounding tissue. Additionally, the samples were fractionated in a solvent gradient using nano liquid chromatographic separation on-line coupled with a mass spectrometer. The rapid scan rate, high mass accuracy and sensitivity of the LTQ Orbitrap assisted in the identification of relatively large numbers of proteins from small numbers of cells of no more than 2000 cells (∼270 ng total protein, estimating that one cell has ∼135 pg total protein). Because of the relatively large differences between the tissue types used in this study, alignment of all data was not possible and label-free comparison for Progenesis analysis was only possible for the groups of microdissected blood vessels. The spectral counts of all 694 identified proteins were taken into consideration for SAM analysis in which comparisons of the four groups were made. Proteins that were relatively up-regulated in either glioblastoma blood vessels or endometrium blood vessels were identified. The combination of both approaches resulted in a list of reliably differentially expressed proteins. A portion of the proteins that emerged from both analytic strategies was successfully validated by immunohistochemistry. The results of immunohistochemical validation proved that the combination of laser capture microdissection and subsequent nano-LC Orbitrap mass spectrometry is a powerful approach to find tissue specific proteins. Nevertheless, some identified proteins were immunohistochemically detected in glioblastoma as well as endometrium blood vessels. It may well be that the differences detected by the LTQ Orbitrap mass spectrometry did not always match the discriminative power of immunohistochemistry. Further, there may be issues of the specificity of particular antibodies.
The blood vessels in GBM display phenotypical changes ranging from incipient proliferation of endothelium to sarcomatous vascular structures and these changes can all be encountered in the vasculature of the same tumor. The endothelial, pericytic and smooth muscle cells take various spatial positions within the vascular walls depending on the size of the proliferated vessel (22). The endothelial cells show increased numbers of caveolae and fenestrations, prominent pinocytotic vesicles, diminished numbers of tight junctions, leading to leakage and disruption of the blood-brain barrier functions (4). In a previous study, we identified several proteins that were specifically up-regulated in glioma vasculature although not in resting blood vessels of normal brain (12, 23). In the present study we did not compare the glioma blood vessels with the resting vessels of normal brain, but with vessels taken from tissue in which active angiogenesis takes place, i.e., endometrium vessels instead. We found specific up-regulation of 29 proteins in the GBM. All proteins are known to take part in angiogenic pathways. Because these proteins are expressed at significantly lower levels in the endometrium samples, we consider them as characteristic of angiogenesis in neoplasia. Fourteen of the 29 proteins appear to be structural proteins; two are integrins and five are enzymes. Periostin is an extracellular matrix protein that is involved in cell adhesion (24). It regulates cell function and cell-matrix interaction but is not a component of the basal lamina itself. Periostin was found in the blood vessels of nonsmall cell lung cancers and plays a critical role in cardiac remodeling (25, 26). Recently, Kii et al. proved that periostin promotes the incorporation of tenascin-C into the ECM and mediates the formation of the meshwork architecture of the ECM (27). Tenacin-C was also found overexpressed in the GBM blood vessels in the present study. Tenascin-C is an extracellular matrix protein that participates in normal fetal development and wound healing (28). In GBM its presence correlates well with microvascular density (28). Tenascin-C mediates vascular endothelial growth factor expression (VEGF) (29), which is up-regulated and induced by hypoxia inducible factor (HIF-1) under hypoxic conditions (30). The finding of the specific expression of integrin alpha V in glioma angiogenesis in this study is also of interest because this protein serves as a receptor for the extracellular matrix. The relation of integrin alpha V and angiogenesis has been described in several types of cancer among which cervical cancers (31), ovarian carcinoma (32), breast cancer (33), and melanoma (34). We also found overexpression of transforming growth factor-beta-induced protein ig-h3 in the GBM blood vessels (35). TGFβ regulates the expression of various proteins TGFβ induced protein ig-h3, periostin, integrin alpha V, tenascin-C, fibronectin, colligin 2, caldesmon, acidic calponin, and basement membrane-specific heparan sulfate proteoglycan core protein (35, 36). Further investigations of the interrelationships between these proteins may reveal their relative importance and whether they should be considered potential targets for therapeutic anti-angiogenic interventions.
Footnotes
1 The abbreviations used are:
- GBM
- glioblastoma multiforme
- SAM
- significance analysis of microarrays.
REFERENCES
- 1. Ribatti D. (2010) Erythropoietin and tumor angiogenesis. Stem Cells Dev. 19, 1–4 [DOI] [PubMed] [Google Scholar]
- 2. Somanath P. R., Ciocea A., Byzova T. V. (2009) Integrin and growth factor receptor alliance in angiogenesis. Cell Biochem. Biophys. 53, 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ribatti D. (2010) The seminal work of Werner Risau in the study of the development of the vascular system. Int. J. Dev. Biol. 54, 567–572 [DOI] [PubMed] [Google Scholar]
- 4. Li Calzi S., Neu M. B., Shaw L. C., Kielczewski J. L., Moldovan N. I., Grant M. B. (2010) EPCs and pathological angiogenesis: when good cells go bad. Microvasc. Res. 79, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wickersheim A., Kerber M., de Miguel L. S., Plate K. H., Machein M. R. (2009) Endothelial progenitor cells do not contribute to tumor endothelium in primary and metastatic tumors. Int. J. Cancer 125, 1771–1777 [DOI] [PubMed] [Google Scholar]
- 6. Pozzi A., Zent R. (2009) Regulation of endothelial cell functions by basement membrane- and arachidonic acid-derived products. Wiley Interdiscip. Rev. Syst. Biol. Med. 1, 254–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van Langendonckt A., Donnez J., Defrère S., Dunselman G. A., Groothuis P. G. (2008) Antiangiogenic and vascular-disrupting agents in endometriosis: pitfalls and promises. Mol. Hum. Reprod. 14, 259–268 [DOI] [PubMed] [Google Scholar]
- 8. Holland E. C. (2000) Glioblastoma multiforme: the terminator. Proc. Natl. Acad. Sci. U.S.A. 97, 6242–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Argyriou A. A., Giannopoulou E., Kalofonos H. P. (2009) Angiogenesis and anti-angiogenic molecularly targeted therapies in malignant gliomas. Oncology 77, 1–11 [DOI] [PubMed] [Google Scholar]
- 10. Hanahan D., Folkman J. (1996) Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell 86, 353–364 [DOI] [PubMed] [Google Scholar]
- 11. Mustafa D. A., Burgers P. C., Dekker L. J., Charif H., Titulaer M. K., Smitt P. A., Luider T. M., Kros J. M. (2007) Identification of glioma neovascularization-related proteins by using MALDI-FTMS and nano-LC fractionation to microdissected tumor vessels. Mol. Cell Proteomics 6, 1147–1157 [DOI] [PubMed] [Google Scholar]
- 12. Gladson C. L. (1996) Expression of integrin alpha v beta 3 in small blood vessels of glioblastoma tumors. J. Neuropathol. Exp. Neurol. 55, 1143–1149 [DOI] [PubMed] [Google Scholar]
- 13. Koul D., Shen R., Bergh S., Lu Y., de Groot J. F., Liu T. J., Mills G. B., Yung W. K. (2005) Targeting integrin-linked kinase inhibits Akt signaling pathways and decreases tumor progression of human glioblastoma. Mol. Cancer Ther. 4, 1681–1688 [DOI] [PubMed] [Google Scholar]
- 14. Sigal A., Milo R., Cohen A., Geva-Zatorsky N., Klein Y., Liron Y., Rosenfeld N., Danon T., Perzov N., Alon U. (2006) Variability and memory of protein levels in human cells. Nature 444, 643–646 [DOI] [PubMed] [Google Scholar]
- 15. Stingl C., van Vilsteren F. G., Guzel C., Ten Kate F. J., Visser M., Krishnadath K. K., Bergman J. J., Luider T. M. (2011) Reproducibility of Protein Identification of Selected Cell Types in Barrett's Esophagus Analyzed by Combining Laser-Capture Microdissection and Mass Spectrometry. J. Proteome Res. 10, 288–298 [DOI] [PubMed] [Google Scholar]
- 16. Martinez-Pinna R., Martin-Ventura J. L., Mas S., Blanco-Colio L. M., Tuñon J., Egido J. (2008) Proteomics in atherosclerosis. Curr. Atheroscler. Rep. 10, 209–215 [DOI] [PubMed] [Google Scholar]
- 17. Johann D. J., Rodriguez-Canales J., Mukherjee S., Prieto D. A., Hanson J. C., Emmert-Buck M., Blonder J. (2009) Approaching solid tumor heterogeneity on a cellular basis by tissue proteomics using laser capture microdissection and biological mass spectrometry. J. Proteome Res. 8, 2310–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shen Y., Tolić N., Masselon C., Pasa-Tolić L., Camp D. G., 2nd, Lipton M. S., Anderson G. A., Smith R. D. (2004) Nanoscale proteomics. Anal. Bioanal. Chem. 378, 1037–1045 [DOI] [PubMed] [Google Scholar]
- 19. de Groot C. J., Steegers-Theunissen R. P., Güzel C., Steegers E. A., Luider T. M. (2005) Peptide patterns of laser dissected human trophoblasts analyzed by matrix-assisted laser desorption/ionisation-time of flight mass spectrometry. Proteomics 5, 597–607 [DOI] [PubMed] [Google Scholar]
- 20. Umar A., Dalebout J. C., Timmermans A. M., Foekens J. A., Luider T. M. (2005) Method optimisation for peptide profiling of microdissected breast carcinoma tissue by matrix-assisted laser desorption/ionisation-time of flight and matrix-assisted laser desorption/ionisation-time of flight/time of flight-mass spectrometry. Proteomics 5, 2680–2688 [DOI] [PubMed] [Google Scholar]
- 21. Umar A., Luider T. M., Foekens J. A., Pasa-Tolic L. (2007) NanoLC-FT-ICR MS improves proteome coverage attainable for approximately 3000 laser-microdissected breast carcinoma cells. Proteomics 7, 323–329 [DOI] [PubMed] [Google Scholar]
- 22. Mustafa D., van der Weiden M., Zheng P., Nigg A., Luider T. M., Kros J. M. (2008) Expression Sites of Colligin 2 in Glioma Blood Vessels. Brain Pathol. 20, 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng P. P., Luider T. M., Pieters R., Avezaat C. J., van den Bent M. J., Sillevis Smitt P. A., Kros J. M. (2003) Identification of tumor-related proteins by proteomic analysis of cerebrospinal fluid from patients with primary brain tumors. J. Neuropathol. Exp. Neurol. 62, 855–862 [DOI] [PubMed] [Google Scholar]
- 24. Takayama G., Arima K., Kanaji T., Toda S., Tanaka H., Shoji S., McKenzie A. N., Nagai H., Hotokebuchi T., Izuhara K. (2006) Periostin: a novel component of subepithelial fibrosis of bronchial asthma downstream of IL-4 and IL-13 signals. J. Allergy Clin. Immunol. 118, 98–104 [DOI] [PubMed] [Google Scholar]
- 25. Soltermann A., Tischler V., Arbogast S., Braun J., Probst-Hensch N., Weder W., Moch H., Kristiansen G. (2008) Prognostic significance of epithelial-mesenchymal and mesenchymal-epithelial transition protein expression in non-small cell lung cancer. Clin. Cancer Res. 14, 7430–7437 [DOI] [PubMed] [Google Scholar]
- 26. Kühn B., del Monte F., Hajjar R. J., Chang Y. S., Lebeche D., Arab S., Keating M. T. (2007) Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat. Med. 13, 962–969 [DOI] [PubMed] [Google Scholar]
- 27. Kii I., Nishiyama T., Li M., Matsumoto K., Saito M., Amizuka N., Kudo A. (2010) Incorporation of tenascin-C into the extracellular matrix by periostin underlies an extracellular meshwork architecture. J. Biol. Chem. 285, 2028–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Behrem S., Zarković K., Eskinja N., Jonjić N. (2005) Distribution pattern of tenascin-C in glioblastoma: correlation with angiogenesis and tumor cell proliferation. Pathol. Oncol. Res. 11, 229–235 [DOI] [PubMed] [Google Scholar]
- 29. Tanaka K., Hiraiwa N., Hashimoto H., Yamazaki Y., Kusakabe M. (2004) Tenascin-C regulates angiogenesis in tumor through the regulation of vascular endothelial growth factor expression. Int. J. Cancer 108, 31–40 [DOI] [PubMed] [Google Scholar]
- 30. Rong Y., Durden D. L., Van Meir E. G., Brat D. J. (2006) ‘Pseudopalisading’ necrosis in glioblastoma: a familiar morphologic feature that links vascular pathology, hypoxia, and angiogenesis. J. Neuropathol. Exp. Neurol. 65, 529–539 [DOI] [PubMed] [Google Scholar]
- 31. Chattopadhyay N., Mitra A., Frei E., Chatterjee A. (2001) Human cervical tumor cell (SiHa) surface alphavbeta3 integrin receptor has associated matrix metalloproteinase (MMP-2) activity. J. Cancer Res. Clin. Oncol. 127, 653–658 [DOI] [PubMed] [Google Scholar]
- 32. Ahmed N., Riley C., Rice G., Quinn M. (2005) Role of integrin receptors for fibronectin, collagen and laminin in the regulation of ovarian carcinoma functions in response to a matrix microenvironment. Clin. Exp. Metastasis 22, 391–402 [DOI] [PubMed] [Google Scholar]
- 33. Lin H. Y., Lansing L., Merillon J. M., Davis F. B., Tang H. Y., Shih A., Vitrac X., Krisa S., Keating T., Cao H. J., Bergh J., Quackenbush S., Davis P. J. (2006) Integrin alphaVbeta3 contains a receptor site for resveratrol. FASEB J. 20, 1742–1744 [DOI] [PubMed] [Google Scholar]
- 34. Koistinen P., Ahonen M., Kähäri V. M., Heino J. (2004) alphaV integrin promotes in vitro and in vivo survival of cells in metastatic melanoma. Int. J. Cancer 112, 61–70 [DOI] [PubMed] [Google Scholar]
- 35. Ha S. W., Bae J. S., Yeo H. J., Lee S. H., Choi J. Y., Sohn Y. K., Kim J. G., Kim I. S., Kim B. W. (2003) TGF-beta-induced protein beta ig-h3 is upregulated by high glucose in vascular smooth muscle cells. J. Cell Biochem. 88, 774–782 [DOI] [PubMed] [Google Scholar]
- 36. Dobaczewski M., Gonzalez-Quesada C., Frangogiannis N. G. (2010) The extracellular matrix as a modulator of the inflammatory and reparative response following myocardial infarction. J. Mol. Cell Cardiol. 48, 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]