Abstract
Cerebrospinal fluid is routinely collected for the diagnosis and monitoring of patients with neurological malignancies. However, little is known as to how its constituents may change in a patient when presented with a malignant glioma. Here, we used a targeted mass-spectrometry based metabolomics platform using selected reaction monitoring with positive/negative switching and profiled the relative levels of over 124 polar metabolites present in patient cerebrospinal fluid. We analyzed the metabolic profiles from 10 patients presenting malignant gliomas and seven control patients that did not present malignancy to test whether a small sample size could provide statistically significant signatures. We carried out multiple unbiased forms of classification using a series of unsupervised techniques and identified metabolic signatures that distinguish malignant glioma patients from the control patients. One subtype identified contained metabolites enriched in citric acid cycle components. Newly diagnosed patients segregated into a different subtype and exhibited low levels of metabolites involved in tryptophan metabolism, which may indicate the absence of an inflammatory signature. Together our results provide the first global assessment of the polar metabolic composition in cerebrospinal fluid that accompanies malignancy, and demonstrate that data obtained from high throughput mass spectrometry technology may have suitable predictive capabilities for the identification of biomarkers and classification of neurological diseases.
Patients with malignant gliomas (MG)1 have poor prognosis. Those with glioblastomas have a median survival of 14.6 months and 5-year survival is only 9.8% despite aggressive treatment with temozolomide chemo-irradiation (1). Patients with anaplastic gliomas have a slightly better prognosis with a median survival of 3 to 10 years depending on the molecular genetics of the tumor (2, 3). Therefore, having a means of inquiring the biological state of the malignant glioma would guide the application of proper treatment. Despite the high resolution of magnetic resonance imaging (MRI), only macroscopic structural information is obtained and this measurement does not reveal the underlying biology of the malignant glioma. Although tumor tissue analysis performed serially over time is possible, brain biopsies have inherent sampling errors because of the heterogeneous nature of the tumor and resections may lead to neurological deficits (4, 5).
These limitations led us to search for an improved and/or complementary method of evaluating malignant gliomas using biomarkers in the cerebrospinal fluid (CSF). Because the CSF bathes the tissues of the central nervous system (CNS), it provides an attractive source of material for clinical diagnostics. CSF is readily accessible either by lumbar puncture or reservoir sampling, which is less invasive and can be performed serially and may potentially yield a more integrated view of the tumor's activity (6, 7). As a result, CSF-derived biomarkers may provide an earlier diagnosis than MRI, obviate invasive procedures, offer information on the tumor's biological state, prognosticate patient survival, and/or predict treatment responses.
CSF derived from patients presenting malignant gliomas may contain signatures of altered metabolism known to occur in tumor cells and may further harbor signatures of secondary, noncell-autonomous effects that arise from the tumor microenvironment (8–11). Such physiological processes include inflammation, endocrine pathophysiology, and cellular debris resulting from necrotic cells that accumulates as a byproduct of disrupted tissue architecture (12).
Metabolic profiling has also been used as a diagnostic tool in the setting of human cancer. Studies of metabolites from patient urine that were discovered using metabolomics technology have recently come into focus as possible biomarkers for metastatic prostate cancer and renal cell carcinoma (13, 14). For example, one study used mass spectrometry to probe the metabolic composition of urine in patients with prostate cancer at different stages of the disease. The study identified a collection of metabolites that correlated with advanced metastatic prostate cancer including glycine and N-methylglycine (sarcosine) (13).
There is additional motivation for the study of metabolism in the biological fluid of cancer patients, given extensive evidence of altered cellular metabolism during tumorigenesis. In particular there is increasing evidence that many of the recurrent genetic alterations that contribute to the pathogenesis of gliomas induce changes in cellular metabolism (15). Recurrent genetic events in glioma such as MYC amplification, PTEN deletion or protein loss and EFGR amplification have multiple downstream metabolic targets (16). Also the metabolic genes IDH1 and IDH2 are mutated in ∼12% of primary gliomas through a gain-of-function mutation that alters the enzymatic activity of the protein product that results in the production of 2-hydroxyglutarate (17). It is therefore likely that the metabolic alterations observed in malignant glioma cells may propagate to global changes in the composition of CSF.
Recent metabolic profiling of CSF has revealed a rich composition of diverse metabolites present in high concentrations in normal patient CSF (18–23). Further clinical studies have found alterations in the composition of CSF under pathological conditions. One study identified gross alterations in metabolic profiles in patients diagnosed with schizophrenia (24). Another study showed that macaques infected with simian immunodeficiency virus undergo detectable changes in the composition of their CSF (25). Together, these studies suggested that current metabolomics technology may allow for the possibility of detecting alterations in the CSF of patients with malignant gliomas that could serve as useful biomarkers and/or assisting with diagnosis.
Using a liquid chromatography/tandem mass spectrometry (LC-MS/MS) based platform employing selected reaction monitoring (SRM) with positive and negative ion switching using a 5500 QTRAP hybrid dual quadrupole linear ion trap mass spectrometer (Qq-IT) (26–30), we investigated whether we could identify unique molecular features in the CSF of patients with malignant gliomas using a limited sample set. The platform utilizes hydrophilic interaction chromatography (HILIC) at pH = 9.0 (31, 32). 254 unique polar metabolites from 285 SRM scans were targeted from a single 16-min experiment without chromatographic scheduling. The platform was designed to include as many polar metabolites that we could that cover major metabolic pathways including glycolysis, TCA cycle, the pentose phosphate pathway, amino acid metabolism, nucleotide metabolism, etc. to study cancer cell metabolism. We robustly quantified the relative levels of 124 water-soluble metabolites using microliter quantities (250 μl) of patient CSF in a cohort of patients with malignant gliomas and a control cohort without any malignancy. Many of these metabolites overlap with those detected in previous CSF metabolomics studies referenced above. Using multiple computational algorithms to classify the data in an unbiased manner, we identified significant differences in the metabolite composition between patients with the disease and controls, as well as those with newly diagnosed and recurrent malignant gliomas. Together, our findings demonstrate that the metabolite composition of the CSF may provide clinically relevant biomarkers and may provide insights into the mechanisms underlying the pathogenesis of malignant gliomas. Ultimately, these findings would provide a basis for additional clinical studies that can determine the sensitivity and specificity, as well as the predictive value, of the metabolite biomarkers identified in the CSF.
EXPERIMENTAL PROCEDURES
Patients and CSF
Aliquots of 5–10 ml of CSF were obtained from male and female patients aged 27 through 67 and of varying disease state in the Brain Tumor Clinic at Beth Israel Deaconess Medical Center (BIDMC) by ETW and staff. CSF samples were collected at the time of neurological evaluation when there was an indication for lumbar puncture or sampling from a ventricular reservoir. All samples were collected from lumbar puncture except one (patient 3 in Table I) whose CSF was taken from a ventricular drain. Patient consents were obtained for CSF storage and CSF biomarker analysis under institutional review board (IRB)-approved protocols at BIDMC. Samples were stored at −80 °C until the time of the experiment. Other clinical CSF parameters, such as white blood cell (WBC) count, total protein level, glucose level, LDH level, and cytology were also tabulated. New versus recurrent diagnosis, survival time, tumor size estimated through MRI, and survival status at end of study, age, and gender were also recorded.
Table I. (A) Clinical profile of patients with malignant gliomas and (B) their CSF profiles. GBM, glioblastoma; AOA, anaplastic oligoastrocytoma; and AA, anaplastic astrocytoma; XRT, radiation therapy; TMZ, temozolomide; and CPT-11, irinotecan. Newly Diagnosed (N), Recurrent (R). Δ(Initial–Sample Date) (time from diagnosis to sample collection). WBC–White Blood Cell Count (number/μl), Protein (mg/dL), Glucose (mg/dL), LDH (International Units/L). T1 GAD–2D tumor size estimate from Gadolinium MRI. FLAIR–2D tumor size estimate from FLAIR MRI.
| A | ||||||
|---|---|---|---|---|---|---|
| Patient | Age | Gender | DIAGNOSIS | Newly Diagnosed or Recurrent | Δ (Initial - Sample Date) (months) | Survival (months) |
| 1 | 61 | M | GBM | N | 0 | 8 |
| 2 | 59 | M | GBM | N | 0 | 9 |
| 3 | 45 | F | AOA | R | 34 | 34 |
| 4 | 67 | M | GBM | R | 9 | 17 |
| 5 | 57 | M | AOA | N | 0 | 21 |
| 6 | 27 | F | AA | R | 3 | 27 |
| 7 | 55 | M | GBM | N | 0 | 37 |
| 8 | 41 | F | AA | R | 8 | 27 |
| 9 | 52 | M | GBM | R | 7 | 18 |
| 10 | 46 | F | GBM | R | 12 | 14 |
| B | ||||||||
|---|---|---|---|---|---|---|---|---|
| Patient | WBC | Protein | Glucose | LDH | T1 Gad (cm2) | FLAIR (cm2) | Δ (FLAIR - T1 Gad) (cm2) | Treatment |
| 1 | 2 | 23 | 63 | 11 | 1 | 23.8 | 22.8 | |
| 2 | 2 | 47 | 70 | 9 | 5.7 | 37.6 | 31.9 | |
| 3 | 3 | 209 | 87 | N/A | 9 | 39.1 | 30.1 | XRT, TMZ, Avastin, CPT-11 |
| 4 | 1 | 96 | 67 | 56 | 23.9 | 43.6 | 19.7 | XRT, TMZ, ZD6474, NovoTTF |
| 5 | 2 | 63 | 63 | 19 | 0.7 | 11.6 | 10.9 | |
| 6 | 3 | 61 | 93 | N/A | 2.6 | 3.7 | 1.1 | XRT, TMZ |
| 7 | 1 | 51 | 73 | 26 | 1.2 | 36.5 | 35.3 | |
| 8 | 5 | 57 | 77 | 22 | 0.6 | 13.2 | 12.6 | XRT, TMZ, Avastin, CPT-11 |
| 9 | 1 | 41 | 63 | N/A | 17.6 | 28.5 | 10.9 | XRT, TMZ, NovoTTF |
| 10 | 4 | 158 | 76 | 56 | 10.2 | 37.6 | 27.4 | XRT, TMZ, CPT-11 |
Sample Preparation
Two hundred and fifty microliters of CSF were subjected to overnight precipitation in 80% methanol at −20 °C followed by centrifugation at 13,000 rpm for 10 min at 4 °C. Supernatants were collected and dried under vacuum and reconstituted in 50 μl of 95:5 LC/MS grade water/HPLC grade acetonitrile and then cleared by centrifugation at 13,000 rpm for 5 min. Supernatants were then dried to a pellet using a SpeedVac (Thermo Fisher Scientific) and stored at −80 °C for analysis.
Targeted Mass Spectrometry
Samples were resuspended using 20 μl LC/MS grade water for mass spectrometry. Ten microliters were injected and analyzed using a 5500 QTRAP triple quadrupole mass spectrometer (AB/SCIEX) coupled to a Prominence UFLC HPLC system (Shimadzu) via SRM of a total of 285 SRM transitions using positive and negative polarity switching corresponding to 254 unique endogenous water soluble metabolites. Some metabolites were targeted in both positive and negative ion mode. Electrospray ionization voltage was +4900V in positive ion mode and −4500V in negative ion mode with a source temperature of 475 °C. The dwell time was 4 ms per SRM transition and the total duty cycle time for all metabolites was 1.89 s resulting in ∼9–12 data points acquired per detected metabolite. Samples were delivered to the 5500 QTRAP using a 2.0 mm i.d × 15 cm Luna NH2 hydrophilic interaction chromatography (HILIC) column (Phenomenex, Torrance, CA) at 300 μl/min. Gradients were run starting from 85% buffer B (HPLC grade acetonitrile) to 40% B from 0–5 min; 40% B to 0% B from 5–16 min; 0% B was held from 16–24 min; 0% B to 85% B from 24–25 min; 85% B was held for 7 min to re-equilibrate the column. Buffer A was comprised of 20 mm ammonium hydroxide/20 mm ammonium acetate (pH = 9.0) in 95:5 water/acetonitrile. Peak areas from the total ion current for each metabolite SRM transition were integrated using MultiQuant version 1.1 software (AB/SCIEX) via the MQ4 peak integration algorithm using a minimum of eight data points with a 30 s retention time window.
Computational Analysis
All calculations were carried out using the R programming suite (http://www.r-project.org/). The resulting integrations from the MultiQuant software provided a starting point for subsequent analyses. All metabolites undetected in at least one sample were excluded from the analysis. We chose this more conservative convention because we found it to be more robust than considering imputations for undetected metabolites, which can lead to statistical artifacts. This pruning step reduced the number of metabolites to 124. Principal components analysis (PCA) on the peak integrations was carried out using the prcomp() method in the R statistical computing language (http://www.r-project.org/). As is standard, data were normalized to the mean and data transformed to standard units. To assess the statistical significance, we used a Monte Carlo analysis. We considered a set of n = 100,000 random matrices with normally distributed matrix elements of mean and variance identical to that of the original data. We computed the eigenvalue spectrum of each matrix and a histogram for each principal component. p values were obtained by comparing the eigenvalues for each principal component in the actual data to the histogram obtained from random data. p values were found to be 7.7 × 10−15, 2.6 × 10−5, and 1.8 × 10−9 respectively for the three largest components. Pathway analysis using the KEGG (www.genome.jp/kegg/) pathway database was carried out using metaboanalyst (www.metaboanalyst.ca) (33). The absolute value of the coefficients (loadings) for each component was obtained and then ranked from highest to lowest. The top 40 values were used for the pathway analysis. The overlay of the k-means clustering was carried out using the kmeans() method in R. k = 3 clusters were obtained starting with three partitions using 20 iterations to ensure convergence. Hierarchical clustering was performed using Ward's method in the hclust() method that minimizes the variance in each cluster (34). All reported R2 values were obtained from Pearson correlations. p values were obtained using two-tailed t test statistics except for the principal components analysis for which a one-tailed statistic was used.
Ethics Approval
The research study described here was performed under IRB-approved protocol #: 2007-P-000381/2 at BIDMC (Federal Wide Assurance # 00003245). This certifies that the research study referenced was authorized by the IRB for research involving human subjects. All administrative requirements for the above referenced protocol have been met.
RESULTS
Study Design and Metabolomics Platform
CSF was collected from patients (Table I) as part of their routine clinical care under an institutionally approved IRB protocol. Among the 10 patients with malignant gliomas, four had newly diagnosed and six had recurrent disease (Table I). There were six glioblastomas, two anaplastic astrocytomas, and two anaplastic oligoastrocytomas (Table I). CSF samples from seven control subjects without any malignancy were also analyzed. All samples were collected from lumbar puncture except one (patient 3 in Table I) whose CSF was taken from a ventricular drain.
In the Qq-IT triple quadrupole system that employs SRM, the first quadrupole isolates a precursor ion and transfers it into the second quadrupole for fragmentation via collision induced dissociation whereby the third analyzer (linear ion trap) then isolates a selected fragment ion for quantitative analysis. Positive and negative ion switching was possible because of the 50 millisecond (msec) switch time and 4 msec dwell time in the 5500 QTRAP. The entire duty cycle for 285 SRM scans were performed in ∼1.89 s. Approximately nine to 12 data points were collected per peak with ∼8–9 s peak-width at half height.
Metabolomics of Patient-derived CSF
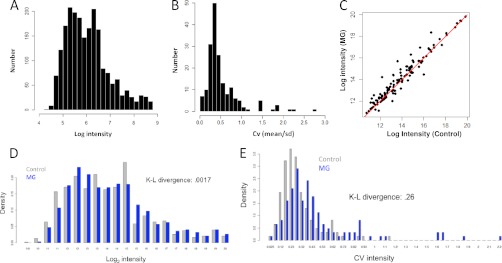
Using our metabolomics platform, we robustly (observed recorded peak area intensity across each patient sample having a signal-to-noise > 2:1) measured over 124 polar metabolites from 17 samples (10 malignant gliomas and seven controls) in total from patient CSF (supplemental Table S1). An analysis of the distribution of intensities of the recorded concentrations showed approximately a log-normal distribution (Fig. 1A). An inspection of the width of the distribution showed a dynamic range of ∼4 orders of magnitude (Fig. 1A). To understand the extent of variation in the data, the coefficient of variation (standard deviation/mean) or CV was computed for each metabolite. An analysis of the histogram revealed that metabolite CV's are concentrated from 0.25–0.75 (Fig. 1B). An analysis of the average intensities of the malignant glioma samples plotted against control samples revealed substantial differences in the average metabolite intensities (Fig. 1C). Eighty-six metabolites were on average higher in the malignant glioma samples and 38 metabolites were higher on average in the control samples. These differences can be better visualized when plotting the histogram of intensities in control samples (blue) and compared with MG samples (gray) (Fig. 1D). From the histograms, it is apparent that gross similarities (Kullback-Leibler (K-L) divergence = 0.0017) in the metabolite intensities of malignant gliomas and controls are obtained suggesting that metabolites levels are typically on the same order of magnitude in control versus MG subjects. These differences in intensities could further be observed by analyzing the differences in the coefficient of variation (CV), as expressed by histogram (Fig. 1E), of controls and MG patients. Interestingly, the histograms of the CV exhibited a larger divergence (K-L divergence = 0.26) demonstrating that the MG patients showed more variability in their metabolite composition. This finding suggested that better-distinguished subtypes of metabolite composition might be obtained in MG samples than in control samples. These observations also indicate that a diverse set of metabolites can be identified in a small volume (less than or equal to 250 μl) of CSF. Thus, a mass-spectrometry based metabolomics analysis of CSF could potentially distinguish malignant glioma patients from those without any malignancy as well as fine structure within the MG population.
Fig. 1.
Data collection and analysis. A, Histogram of the log-intensities of polar metabolites measured by targeted LC-MS/MS across 17 samples. B, Histogram of coefficient of variation (CV) of measured metabolites across 17 samples. C, Scatterplot of average metabolite intensities of control samples (x axis) against malignant glioma (MG) samples. One hundred and eighteen metabolites lie above the line and 71 below the line showing a statistically significant difference. D, Comparison of histogram densities of the log-intensities of measured metabolites of MG samples (blue) and control samples (gray). Each histogram was scaled by dividing values by the total count for each to account for differences in number of data points. Kullback-Leibler (K-L) divergence between the distributions was measured to be 0.0017. E, Comparison of histogram densitry of the CV (standard deviation/mean) of measured metabolites of MG samples (blue) and control samples (gray). K-L divergence between the distributions was observed to be 0.29.
Metabolic Signatures Unique to Malignant Glioma CSF Samples
An analysis of the metabolite intensities and CVs revealed that gross differences exist in the small molecule composition of the CSF from these two patient-cohorts. This observation led us to analyze global differences in the small molecule composition. One commonly used method to identify patterned, global differences in multivariate data sets involves using hierarchical clustering (35–37). Such clustering methods have been employed to study the organization of mRNA profiles in different samples in microarray collections of primary tumors (38). These methods are becoming increasingly appreciated in the oncology clinic for diagnosis and prognosis (39).
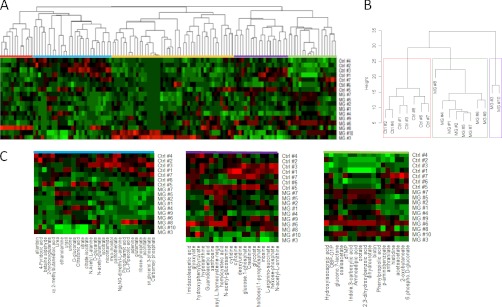
An unsupervised hierarchical clustering algorithm using Ward's method (34) that minimizes the variance within clusters was carried out. Analysis of the resulting classification revealed distinct patterns of metabolites both enhanced and decreased in malignant glioma CSF samples relative to the control samples (Fig. 2A). The tree structure (Fig. 2B) revealed three separate branches corresponding to two classes of cancer patients and one additional branch corresponding to the control samples. Interestingly, the CSF samples from patients that have newly diagnosed malignant gliomas (Table I) (Patients 1, 2, 5, and 7) form a single cluster that is separated in distance from the other samples. Fig. 2C shows metabolite signatures that contribute to the clusters distinguishing MG from control subjects. Therefore, with no a priori information, this hierarchical classification not only segregated malignant glioma samples from controls, but also patients who have recurrent disease from those with newly diagnosed malignant gliomas (Figs. 2A, B). Importantly, these changes cannot be distinguished from MRI measurements because there was no correlation between cluster group and MRI measurements.
Fig. 2.
Hierarchical clustering of CSF metabolite composition. A, Unsupervised hierarchical clustering of CSF metabolite profiles. B, Dendrogam obtained from unsupervised hierarchical clustering of CSF metabolite profiles for malignant glioma (MG) and control (Ctrl) samples. The height of the dendrogram represents Euclidean distance. Boxes are drawn around the three clusters. C, Profiles of three signatures that distinguish MG from control samples.
To gain additional insights into our ability to classify malignant glioma patients based on the metabolic composition of CSF, additional unsupervised learning methods were examined. Although overall, different classification methods should give consistent results, different methods may reveal different details about the fine structure of the data. Thus we also carried out a principal components analysis (PCA). Using PCA, correlations in the metabolite levels are computed and the matrix of these correlations is rotated into directions (eigenvectors) that account for the largest sample variance (40). An analysis of these directions provides an unbiased means of clustering the samples by accounting for the relative contribution of each direction to each sample. Metabolites that comprise the coefficients of these eigenvectors can give a biological interpretation to the computation by identifying groups of metabolites that covary.
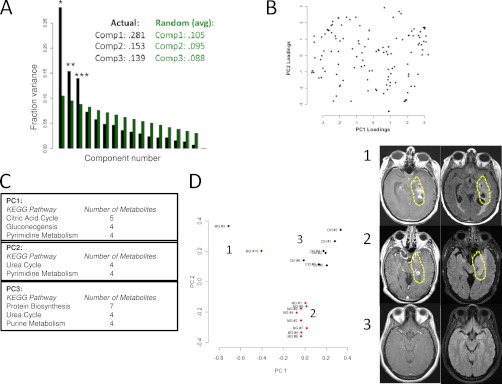
We first assessed the statistical significance of a PCA analysis on the raw data. A Monte Carlo algorithm (methods) was used to compute a distribution of eigenvalues obtained from random data of equal mean and variance to that of the measured data. A one-tailed p value for each eigenvalue was obtained from this distribution. It was found that the first, second, and third components were highly statistically significant (*, p = 8.3 × 10−15; **, p = 2.6 × 10−5; and ***, p = 1.8 × 10−9 respectively). Components one, two, and three captured 28.1%, 15.3%, and 13.9% percent of the variance respectively (Fig. 3A).
Fig. 3.
Principal component analysis (PCA) of CSF metabolite composition. A, Fraction of variance explained (Fraction variance) plotted across principal component number. Black bars indicate values obtained from measured data. Green bars indicate average values obtained from a PCA analysis of n = 100,000 random, normally distributed data of dimension, mean, and variance equivalent to the measured data. Numerical values of the first three components are shown in the caption for each case. *denotes p = 8.3e-15, ** denotes p = 2.6e-5, *** denotes p = 1.8e-9. p values were obtained from Monte Carlo simulations. B, Loadings plot obtained from the PCA analysis. Coefficients of eigenvectors for the first and second principal components are shown. C, Kegg pathway analysis of the first three significant princpal components. The first column lists the pathway and the second column lists the number of metabolites identified within the pathway for the top 40 coefficients in magnitude for each principal component. Abbreviations: Phe, Phenylalanine; Tyr, Tyrosine; Ala, Alanine. D, (left) Projection of individual samples onto the first two principal components. Colors correspond to cluster membership as assigned by k-means clustering with k = 3. Samples from malignant glioma patients segregate into two groups and the control samples segregate into a separate group. (right) Representative MRI images of the Tumors from patients in group 1 and group 2.
A loadings plot which contains coefficients of the first two eigenvectors is shown in Fig. 3B. From inspection of Fig. 3B, it is apparent that there is overlap in the largest contributing coefficients but some metabolite contributions are unique to each component. To further interpret the components, we used the KEGG database to interpret the coefficients by overlaying the top 40 coefficients onto the KEGG metabolic pathway map (Fig. 3C). From an analysis of the pathways involved in each principal component, it is apparent that metabolites from pyrimidine metabolism contribute to both components (Fig. 3C). Metabolites from the citric acid cycle (n = 5), gluconeogenesis (n = 4), and pyrimidine metabolism (n = 4) appear in the first component. Metabolites from the urea cycle (n = 4) and pyrimidine metabolism (n = 4) emerge in the second component. The third component comprises metabolites from protein biosynthesis (n = 7), the urea cycle (n = 4), and purine metabolism. Together, these findings suggest that distinct, interpretable biological process are defining the glioma samples and accounting for its separation from control samples.
A scatter plot (scores plot) of the projections of each sample onto the first two principal components is shown in Fig. 3D. From the scatter plot, it is apparent that two subsets of malignant glioma patients were identified and they are distinguished from the control samples. When a k-means clustering was overlaid onto each sample by denoting each cluster with a different color (Fig. 3D), a similar pattern is observed. Thus, while both the hierarchical clustering and PCA give similar overall results, each method identified different details within the data.
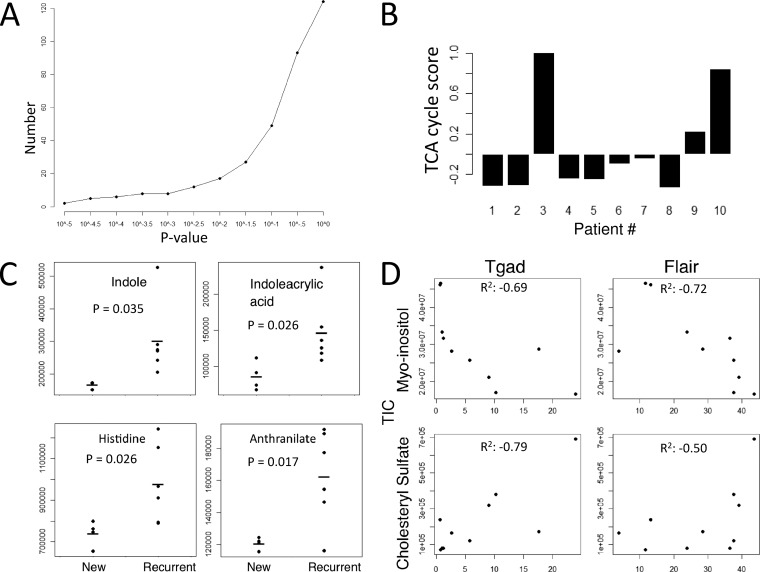
We next considered the metabolites that differ overall from controls to glioma patients. Although the classification algorithms were able to define finer structure in the data which points to the existence of subtypes, these subtypes (with the exception of the separation of recurrent and newly diagnosed) are not yet defined clinically. We therefore chose an overall comparison since such an analysis could have the most immediate clinical application for diagnostic biomarker discovery. Thirty-nine metabolites significantly changed in the CSF of the malignant gliomas relative to the control samples using a more stringent two-tailed t test statistic (p < 0.05) (Fig. 4A, Table II). These metabolites originate from several metabolic pathways such as amino acid, lipid, pyrimidine, and central carbon metabolism. One recently identified metabolite biomarker in glioma patients is 2-hydroxyglutarate (2-HG). Interestingly, the level of 2-HG is also several folds higher in patient 3 which could indicate the presence of an IDH1 mutation in the tumor (17).
Fig. 4.
Metabolite signatures of GBM subtypes and correlations with clinical parameters. A, Number of metabolites that change in the CSF of malignant glioma samples relative to the control patients as a function of p value cutoff using a two-tailed t test statistic. B, TCA cycle signature associated with patients 3 and 10 and its relative contribution across patient samples. Signature score is defined as the normalized sum of metabolite intensity of the citric acid cycle components (succinate, fumarate, alpha-ketoglutarate, malate, oxaloacetate, isocitrate, and citrate). C, Tryptophan metabolism intermediates associated with recurrent disease (p value < 0.05). D, Example metabolites cholesteryl-sulfate and myo-inositol that positively and negatively correlate (R2) with tumor size as measured with MRI using Tgad and FLAIR (x axis) and correlated with the integrated total ion current (TIC) peak areas (y axis) obtained from targeted LC-MS/MS measurements.
Table II. Metabolite changes in malignant glioma vs. control patients.
| Metabolite | ID | Fold change (Malignant glioma vs. control) | p value |
|---|---|---|---|
| biotin | C00120 | 2.96 | 1.03E-06 |
| glucono.d-lactone | C00198 | 2.14 | 6.57E-06 |
| dihydroorotate | C00337 | 2.19 | 1.65E-05 |
| orotate | C00295 | 2.15 | 1.80E-05 |
| 2,3-dihydroxybenzoic acid | C00196 | 2.13 | 2.96E-05 |
| Indole.3-carboxylic acid | HMDB03320 | 1.88 | 7.93E-05 |
| Acetylcarnitine DL | C02571 | 5.23 | 1.30E-04 |
| Aminoadipic acid | C00956 | 1.90 | 1.82E-04 |
| proline | C00148 | 2.83 | 1.23E-03 |
| Phenyllactic acid | C01479 | 1.56 | 1.30E-03 |
| 2.Hydroxy.2-methylbutanedioic acid | C02612 | 0.22 | 1.59E-03 |
| Phenylpropiolic acid | HMDB00563 | 1.26 | 1.78E-03 |
| dTMP | C00364 | 1.53 | 4.93E-03 |
| N6-Acetyl-l-lysine | C02727 | 1.46 | 5.27E-03 |
| oxaloacetate | C00036 | 1.77 | 6.80E-03 |
| Acetyllysine | C02727 | 1.45 | 8.51E-03 |
| shikimate | C04236 | 3.69 | 8.61E-03 |
| Atrolactic acid | HMDB00475 | 1.66 | 1.26E-02 |
| methionine | C00073 | 2.27 | 1.27E-02 |
| taurine | C00245 | 1.50 | 1.29E-02 |
| purine | C00465 | 1.47 | 1.48E-02 |
| N-acetyl-glutamine | HMDB06029 | 1.43 | 1.80E-02 |
| 2-ketohaxanoic acid | HMDB01864 | 3.16 | 1.93E-02 |
| ribose-phosphate | C00117 | 0.73 | 2.15E-02 |
| myo-inositol | C00137 | 0.65 | 2.27E-02 |
| lysine | C00047 | 1.54 | 2.86E-02 |
| glucosamine | C00329 | 1.38 | 3.02E-02 |
| adenine | C00147 | 0.49 | 3.23E-02 |
| nicotinamide | C00153 | 0.34 | 3.40E-02 |
| thiamine | C01081 | 7.99 | 3.51E-02 |
| phenylalanine | C00079 | 1.34 | 3.53E-02 |
| S-methyl-5-thioadenosine | C00170 | 1.50 | 4.07E-02 |
| hypoxanthine | C00262 | 0.72 | 4.14E-02 |
| isocitrate | C00311 | 0.72 | 4.36E-02 |
| serine | C00065 | 1.58 | 4.74E-02 |
| 7-methylguanosine | HMDB01107 | 1.59 | 4.85E-02 |
| glutamine | C00064 | 1.36 | 4.85E-02 |
| glucose.1-phosphate | C00103 | 0.91 | 4.86E-02 |
The different subtypes of metabolic profiles observed in the CSF of malignant glioma patients included a group characterized by new diagnosis (Patients 1, 2, 5, and 7) and a set of two patients (Patients 3 and 10) with recurrent disease that exhibit a similar profile but have distinct profiles from the other CSF samples (Fig. 3B). An inspection of the signature of metabolites that delineates patients 3 and 10 revealed a specific enrichment in metabolites derived from the citric acid (TCA) cycle (Fig. 4B). This increase in TCA cycle metabolites was not observed in other patients suggesting that this signature may identify a subset of malignant glioma patients at disease recurrence.
An inspection of metabolites specific to newly-diagnosed patients revealed seven metabolites that change (p < 0.05) in this subset relative to the patients with recurrent disease (Fig. 4C). Interestingly, many of these metabolites are involved in tryptophan and histidine metabolism. These metabolites include indoleacrylic acid (p = 0.026), indole (p = 0.035), histidine (p = 0.03), and anthranilate (p = 0.017). An enzyme in this pathway indolamine 2,3 dioxygenase is commonly used as a biomarker for immune activity (41). Aberrant activity of this enzyme is also causally implicated in immune-mediated neurological disorders (42). It is thus likely the alterations in the concentrations of these metabolites are indicative of immune activity in these patients.
Correlations with Clinical Parameters
Potential clinical applications of the metabolomics analysis require that the measurements be compared with known methods used in clinical settings. Because clinical parameters were available for the patients in this study, we therefore assessed whether the relative levels of individual metabolites correlated with measures of tumor size. Two parameters that are used to estimate tumor size in glioma patients are MRI-based measurements of T1 relaxation of gadolinium (Tgad) and the T1 relaxation using fluid attenuation inversion recovery (FLAIR) (43). Although higher-order, multivariate analyses such as partial least squares regression are important for describing complex responses, such analyses to give helpful results would require larger sample sizes. Therefore we considered univariate correlations.
We correlated these measurements of tumor-size with metabolites and found that eight metabolites correlated with Tgad and ten correlated with FLAIR (R2 > 0.50) (supplemental Table S1, Fig. 4D). Of these metabolites, the levels of six of these correlated with both measurements. The levels of four metabolites are correlated positively and two are negatively correlated with tumor size. These metabolites are myo-inositol (Tgad R2 = –0.69, FLAIR R2 = −0.72), acetylcarnitine (Tgad R2 = 0.54, FLAIR R2 = 0.67), cytidine (Tgad R2 = –0.56, FLAIR R2 = –0.62), acetoacetate (Tgad R2 = 0.57, FLAIR R2 = 0.60), phenylpropiolic acid (Tgad R2 = 0.72, FLAIR R2 = 0.57), and cholesteryl sulfate (Tgad R2 = 0.79, FLAIR R2 = 0.50).
Another parameter of clinical interest is patient survival. We therefore assessed metabolites that correlate with patient survival. Fourteen metabolites were found to correlate with patient survival (R2 > 0.50) (supplemental Table S1). Three of these metabolites (panthothenate, biotin, and taurine) are common dietary supplements suggesting that the consumption of these compounds can be detected in the CSF. The other metabolites involve intermediates in nucleotide and amino acid metabolism (supplemental Table S1).
DISCUSSION
We present a global polar metabolome analysis of human CSF from a small cohort of patients with malignant gliomas. In this study, we tested and show that statistically significant signatures of glioma and normal patients can be obtained from a small sample size. The study explored (1) an advanced positive and negative switching LC-MS/MS platform for the fragmentation and detection of metabolites, (2) unsupervised classification algorithms to detect aggregate differences of metabolites between patient samples, and (3) the inherent physiology of systemic cancer-associated metabolism. Our approach achieved success for several reasons. The LC-MS/MS SRM based platform consists of a highly sensitive hybrid triple quadrupole MS and allows for chromatographic separation with selective and ultrafast detection with positive and negative switching. This is important because clinically relevant metabolites may exist transiently and in low-abundance due to inherent instability in biological fluids. Also, we used multiple classification algorithms, including hierarchical clustering and principle component analysis, which provides independent validation of signatures that distinguish malignant gliomas from control samples as well as patients with recurrent and newly diagnosed disease. Lastly, an additional explanation for the success of our approach might be that metabolites resulting from abnormal metabolic chemical reactions can accumulate in the CSF in an exponential fashion with respect to the noise. The signal-to-noise ratio of clinically relevant biomarkers from the metabolomics platform is different from that in genomics and proteomics where the rise of metabolites is exponential, while genetic and protein products give linear signals, with respect to the amount of abnormal tissue. Therefore, despite the small sample size in our cohort, the robust differences in certain metabolite levels suggests that there is a great potential for metabolite biomarker discovery in CSF and, when taken in aggregate, metabolite signatures may also have the potential to be used as diagnostic biomarkers.
Because diffusion rates are much higher for small molecules than for cells and proteins, the site of CSF collection, lumbar versus ventricular, is likely less relevant than the steady-state levels of other biological materials. Gerber et al. investigated CSF biomarkers for bacterial meningitis and found that lactate from either lumbar or ventricular source had good linear correlation (r = 0.79, p < 0.001) as opposed to leukocyte (r = 0.39, p = 0.02) or protein levels (r = 0.42, p = 0.006) (44). Each of our patient samples was taken from the lumbar thecal sac except for subject 3 whose CSF was collected from a ventricular drain. Therefore, our collection method is unlikely to confound our analysis of the metabolome in the CSF. However, a rigorous demonstration of whether or not the absolute level of a given metabolite would be different between lumbar and ventricular sites would require the simultaneous collection and subsequent metabolomic analysis of CSF from both sites and such a collection is ethically unsound in routine clinical care. Furthermore, the cluster dendrogram demonstrated that subjects 3 and 10 are different from the rest of the malignant cohort. Indeed, these two subjects have larger tumors and later stage disease. However, it remains to be determined whether the differences found in the metabolome from these two subjects (or the differences in the recurrent versus newly-diagnosed patients) are a result of tumor progression or treatment effect on the brain. A future study to lend more insight might exploit the prospective and periodic sampling of CSF while correlating the changes of metabolite composition with the histology of recurrent tumor or treatment effect.
There are a number of sources giving rise to the detected metabolites in the CSF, including the tumor, stroma, and inflammatory cells that have migrated into the tumor microenvironment. First, malignant gliomas have altered metabolism directly induced by somatic mutations such as MYC amplification, PTEN deletion or protein loss, EGFR amplification, and IDH1/IDH2 mutations. The metabolic enzymes therefore catalyze reactions that result in the accumulation of abnormal metabolites in the CSF even though the tumor volume is at most less than one-tenth of the total volume of the brain and spinal cord. For example, IDH1 mutant cells can secrete enough 2-hydroxyglutarate into the cell culture medium to achieve a two molar concentration within 24 h (17, 45). Indeed, we were able to detect high levels of 2-hydroxyglutarate in patient 3 suggesting that this individual may have an IDH1-mutated malignant glioma. We found that the CSF myo-inositol levels decrease, whereas cholesteryl sulfate level increases linearly respect to tumor size as measured by Tgad and FLAIR according to the Macdonald's criteria. Myo-inositol is a cerebral osmolyte found to be significantly lowered in glioblastomas and anaplastic astrocytomas than in low-grade gliomas and normal brain tissue as detected by magnetic resonance spectroscopy (MRS) (46, 47). Consistent with the previous MRS findings that decreasing myo-inositol level was associated with increasing aggressiveness of the glioma phenotype, our data also showed that the tumor size in our cohort negatively correlated with myo-inositol levels in the CSF. In contrast, cholesteryl sulfate is an important component of the cell membrane and it is distributed into the extracellular fluid such as the plasma (48). The CSF of our cohort had cholesteryl sulfate that positively correlated with the tumor size. Furthermore, accumulation of cholesteryl sulfate was seen in human bronchial epithelial cells undergoing squamous metaplasia (49) and this observation suggests that this metabolite may arise from the oncogenic transformation of normal tissue. Lastly, tryptophan metabolism is important for the activation and suppression of inflammation (41, 42, 50, 51). In our cohort with recurrent malignant gliomas, we found significantly elevated indole, indoleacrylic acid, and anthranilic acid, which are metabolites involved in the tryptophan metabolism, as compared with newly diagnosed patients. In addition, histidine, an essential amino acid and a precursor for the pro-inflammatory histamine, was also found to be markedly elevated in our patients with recurrent disease. These inflammation-associated metabolites may be byproducts of the innate immune system.
The accumulation of CSF metabolites from the TCA cycle may indicate advanced disease. Patients 3 and 10 both had recurrent malignant gliomas, with CSF samples taken 34 and 12 months from initial diagnosis, and their tumor sizes were large as measured by gadolinium-enhanced T1 and FLAIR images on head MRI. MRI is insufficient to delineate the entire tumor due to the infiltrative nature of the malignant glioma because tumor size is estimated based on the amount of contrast leakage from tumor vasculature. Because there are glioma cells outside of the area of active tumor angiogenesis, gadolinium enhancement as shown on T1 MRI underestimates the extent of the tumor. In contrast, FLAIR signal abnormality overestimates tumor size because FLAIR hyperintensity not only represents infiltrative tumor but also cerebral edema and radiation changes in the brain (52).
Together our study provides the first demonstration of metabolite profiling in the CSF of malignant Glioma patients. Our findings conclude that mass spectrometry-based metabolomics methods offer a promising technology for the discovery of biomarkers of malignant Glioma from sampling biological fluid. Several biologically interpretable candidate biomarkers from both individual metabolites as well as from collective metabolite signatures were obtained. It is our hope that this study extends the suitability of metabolomics technology and motivates the metabolic analysis of CSF for further research purposes in oncology such as a clinical trial with a larger patient cohort.
Acknowledgments
We thank Matt Vander Heiden and Joshua Rabinowitz for helpful discussions.
Footnotes
* This work was supported in part by National Institutes Of Health (NIH) 3P30CA006516–45S7 and NIH P01 CA120964 to J.M.A. J.W.L. is supported by a fellowship from the American Cancer Society. L.C.C. acknowledges funding from NIH R01-GM41890. S.S., K.D.S., and E.T.W. are supported by the A Reason To Ride research fund.
 This article contains supplemental Table S1.
This article contains supplemental Table S1.
1 The abbreviations used are:
- MG
- malignant gliomas
- CNS
- central nervous systems
- SRM
- selected reaction monitoring
- CSF
- cerebrospinal fluid
- HILIC
- hydrophilic interaction chromatography
- TCA
- the citric acid cycle
- LC-MS/MS
- tandem mass spectrometry
- MRI
- magnetic resonance imaging.
REFERENCES
- 1. Stupp R., Hegi M. E., Mason W. P., van den Bent M. J., Taphoorn M. J. B., Janzer R. C., Ludwin S. K., Allgeier A., Fisher B., Belanger K., Hau P., Brandes A. A., Gijtenbeek J., Marosi C., Vecht C. J., Mokhtari K., Wesseling P., Villa S., Eisenhauer E., Gorlia T., Weller M., Lacombe D., Cairncross J. G., Mirimanoff R. O., European Org R., Treatment; Canc Brain Tumour Grp; Radiat Oncol G., and Natl Canc Inst Canada Clin T (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 10, 459–466 [DOI] [PubMed] [Google Scholar]
- 2. Cairncross G., Berkey B., Shaw E., Jenkins R., Scheithauer B., Brachman D., Buckner J., Fink K., Souhami L., Laperierre N., Mehta M., Curran W. (2006) Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma: Intergroup Radiation Therapy Oncology Group Trial 9402. J. Clin. Oncol. 24, 2707–2714 [DOI] [PubMed] [Google Scholar]
- 3. Louis D. N., Holland E. C., Cairncross J. G. (2001) Glioma classification - A molecular reappraisal. Am. J. Pathol. 159, 779–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chandrasoma P. T., Smith M. M., Apuzzo M. L. J. (1989) Stereotactic biopsy in the diagnosis of brain masses - Comparison of results of biopsy and resected surgical specimen. Neurosurgery 24, 160–165 [DOI] [PubMed] [Google Scholar]
- 5. Glantz M. J., Burger P. C., Herndon J. E., 2nd, Friedman A. H., Cairncross J. G., Vick N. A., Schold S. C., Jr. (1991) Influence of the type of surgery on the histologic diagnosis in patients with anaplastic gliomas. Neurology 41, 1741–1744 [DOI] [PubMed] [Google Scholar]
- 6. Swanson K., Lok E., Song S., Dessain J., Wong E. T. (2009) Cerebrospinal fluid (CSF) Biomarkers for breast cancer (BC) brain metastases. Ann. Neurol. 66, S58 [Google Scholar]
- 7. Lehtinen M. K., Zappaterra M. W., Chen X., Yang Y. J., Hill A. D., Lun M. L., Maynard T., Gonzalez D., Kim S., Ye P., D'Ercole A. J., Wong E. T., LaMantia A. S., Walsh C. A. (2010) The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron 69, 893–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Locasale J. W., Cantley L. C., Vander Heiden M. G. (2009) Cancer's insatiable appetite. Nat. Biotechnol. 27, 916–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luo J., Solimini N. L., Elledge S. J. (2009) Principles of Cancer Therapy: Oncogene and Non-oncogene Addiction. Cell 136, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Michelakis ED S. G., Dromparis P, Webster L, Haromy A, Niven E, Maguire C, Gammer TL, Mackey JR, Fulton D, Abdulkarim B, McMurtry MS, Petruk KC. (2010) Metabolic modulation of glioblastoma with dichloroacetate. Sci. Translation Med. 2, 31–34 [DOI] [PubMed] [Google Scholar]
- 12. Tlsty T. D., Coussens L. M. (2006) Tumor stroma and regulation of cancer development. Annu. Rev. Pathol.-Mech. Dis. 1, 119–150 [DOI] [PubMed] [Google Scholar]
- 13. Sreekumar A., Poisson L. M., Rajendiran T. M., Khan A. P., Cao Q., Yu J., Laxman B., Mehra R., Lonigro R. J., Li Y., Nyati M. K., Ahsan A., Kalyana-Sundaram S., Han B., Cao X. H., Byun J., Omenn G. S., Ghosh D., Pennathur S., Alexander D. C., Berger A., Shuster J. R., Wei J. T., Varambally S., Beecher C., Chinnaiyan A. M. (2009) Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457, 910–914 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 14. Lin L., Huang Z. Z., Gao Y., Yan X. M., Xing J. C., Hang W. (2011) LC-MS based serum metabonomic analysis for renal cell carcinoma diagnosis, staging, and biomarker discovery. J. Proteome Res. 10, 1396–1405 [DOI] [PubMed] [Google Scholar]
- 15. Parsons D. W., Jones S., Zhang X., Lin J. C., Leary R. J., Angenendt P., Mankoo P., Carter H., Siu I. M., Gallia G. L., Olivi A., McLendon R., Rasheed B. A., Keir S., Nikolskaya T., Nikolsky Y., Busam D. A., Tekleab H., Diaz L. A., Hartigan J., Smith D. R., Strausberg R. L., Marie S. K. N., Shinjo S. M., Yan H., Riggins G. J., Bigner D. D., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V. E., Kinzler K. W. (2008) An integrated genomic analysis of human glioblastoma Multiforme. Science 321, 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deberardinis R. J., Lum J. J., Hatzivassiliou G., Thompson C. B. (2008) The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7, 11–20 [DOI] [PubMed] [Google Scholar]
- 17. Dang L., White D. W., Gross S., Bennett B. D., Bittinger M. A., Driggers E. M., Fantin V. R., Jang H. G., Jin S., Keenan M. C., Marks K. M., Prins R. M., Ward P. S., Yen K. E., Liau L. M., Rabinowitz J. D., Cantley L. C., Thompson C. B., Heiden M. G. V., Su S. M. (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wuolikainen A., Moritz T., Marklund S. L., Antti H., Andersen P. M. Disease-related changes in the cerebrospinal fluid metabolome in amyotrophic lateral sclerosis detected by GC/TOFMS. PLoS One 6, e17947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rosenling T., Slim C. L., Christin C., Coulier L., Shi S., Stoop M. P., Bosman J., Suits F., Horvatovich P. L., Stockhofe-Zurwieden N., Vreeken R., Hankemeier T., van Gool A. J., Luider T. M., Bischoff R. (2009) The effect of preanalytical factors on stability of the proteome and selected metabolites in cerebrospinal fluid (CSF). J. Proteome Res. 8, 5511–5522 [DOI] [PubMed] [Google Scholar]
- 20. Wishart D. S., Lewis M. J., Morrissey J. A., Flegel M. D., Jeroncic K., Xiong Y. P., Cheng D., Eisner R., Gautam B., Tzur D., Sawhney S., Bamforth F., Greiner R., Li L. (2008) The human cerebrospinal fluid metabolome. J. Chromatogr. B 871, 164–173 [DOI] [PubMed] [Google Scholar]
- 21. Crews B., Wikoff W. R., Patti G. J., Woo H. K., Kalisiak E., Heideker J., Siuzdak G. (2009) Variability analysis of human plasma and cerebral spinal fluid reveals statistical significance of changes in mass spectrometry-based metabolomics data. Anal. Chem. 81, 8538–8544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stoop M. P., Coulier L., Rosenling T., Shi S., Smolinska A. M., Buydens L., Ampt K., Stingl C., Dane A., Muilwijk B., Luitwieler R. L., Sillevis Smitt P. A., Hintzen R. Q., Bischoff R., Wijmenga S. S., Hankemeier T., van Gool A. J., Luider T. M. (2010) Quantitative proteomics and metabolomics analysis of normal human cerebrospinal fluid samples. Mol. Cell. Proteomics 9, 2063–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rosenling T., Stoop M. P., Smolinska A., Muilwijk B., Coulier L., Shi S., Dane A., Christin C., Suits F., Horvatovich P. L., Wijmenga S. S., Buydens L. M., Vreeken R., Hankemeier T., van Gool A. J., Luider T. M., Bischoff R. The impact of delayed storage on the measured proteome and metabolome of human cerebrospinal fluid. Clin. Chem. 57, 1703–1711 [DOI] [PubMed] [Google Scholar]
- 24. Holmes E., Tsang T. M., Huang J. T. J., Leweke F. M., Koethe D., Gerth C. W., Nolden B. M., Gross S., Schreiber D., Nicholson J. K., Bahn S. (2006) Metabolic profiling of CSF: Evidence that early intervention may impact on disease progression and outcome in schizophrenia. PLos Med. 3, 1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wikoff W. R., Pendyala G., Siuzdak G., Fox H. S. (2008) Metabolomic analysis of the cerebrospinal fluid reveals changes in phospholipase expression in the CNS of SIV-infected macaques. J. Clin. Invest. 2661–2669 Page [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yuan M., Breitkopf S. B., Yang X., Asara J. M. (2012) A Positive/Negative Switching Targeted Mass Spectrometry Based Metabolomics Platform for Bodily Fluids, Cells, Fresh and Fixed Tissue. Nature Protocols In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kelly A. D., Breitkopf S. B., Yuan M., Goldsmith J., Spentzos D., Asara J. M. Metabolomic Profiling from Formalin-Fixed, Paraffin-Embedded Tumor Tissue Using Targeted LC/MS/MS: Application in Sarcoma. PLoS One 6, e25357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Locasale J. W., Grassian A. R., Melman T., Lyssiotis C. A., Mattaini K. R., Bass A. J., Heffron G., Metallo C. M., Muranen T., Sharfi H., Sasaki A. T., Anastasiou D., Mullarky E., Vokes N. I., Sasaki M., Beroukhim R., Stephanopoulos G., Ligon A. H., Meyerson M., Richardson A. L., Chin L., Wagner G., Asara J. M., Brugge J. S., Cantley L. C., Vander Heiden M. G. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 43, 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anastasiou D., Poulogiannis G., Asara J. M., Boxer M. B., Jiang J. K., Shen M., Bellinger G., Sasaki A. T., Locasale J. W., Auld D. S., Thomas C. J., Vander Heiden M. G., Cantley L. C. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 334, 1278–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yi C. H., Pan H., Seebacher J., Jang I. H., Hyberts S. G., Heffron G. J., Vander Heiden M. G., Yang R., Li F., Locasale J. W., Sharfi H., Zhai B., Rodriguez-Mias R., Luithardt H., Cantley L. C., Daley G. Q., Asara J. M., Gygi S. P., Wagner G., Liu C. F., Yuan J. Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell 146, 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bajad S. U., Lu W. Y., Kimball E. H., Yuan J., Peterson C., Rabinowitz J. D. (2006) Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A 1125, 76–88 [DOI] [PubMed] [Google Scholar]
- 32. Lu W., Bennett B. D., Rabinowitz J. D. (2008) Analytical strategies for LC-MS-based targeted metabolomics. J. Chromatogr. B 871, 236–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xia J., Wishart D. S. Metabolomic data processing, analysis, and interpretation using MetaboAnalyst. Curr. Protoc. Bioinformatics Chapter 14, Unit 14 10 [DOI] [PubMed] [Google Scholar]
- 34. Ward J. H. (1963) Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 58, 236 [Google Scholar]
- 35. Quackenbush J. (2001) Computational analysis of microarray data. Nat. Rev. Genet. 2, 418–427 [DOI] [PubMed] [Google Scholar]
- 36. Sorlie T., Perou C. M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M. B., van de Rijn M., Jeffrey S. S., Thorsen T., Quist H., Matese J. C., Brown P. O., Botstein D., Lonning P. E., Borresen-Dale A. L. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A. 98, 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sorlie T., Tibshirani R., Parker J., Hastie T., Marron J. S., Nobel A., Deng S., Johnsen H., Pesich R., Geisler S., Demeter J., Perou C. M., Lonning P. E., Brown P. O., Borresen-Dale A. L., Botstein D. (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. U. S. A. 100, 8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Collisson E. A., Sadanandam A., Olson P., Gibb W. J., Truitt M., Gu S. D., Cooc J., Weinkle J., Kim G. E., Jakkula L., Feiler H. S., Ko A. H., Olshen A. B., Danenberg K. L., Tempero M. A., Spellman P. T., Hanahan D., Gray J. W. (2011) Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 17, 500–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhu C. Q., Ding K. Y., Strumpf D., Weir B. A., Meyerson M., Pennell N., Thomas R. K., Naoki K., Ladd-Acosta C., Liu N., Pintilie M., Der S., Seymour L., Jurisica I., Shepherd F. A., Tsao M. S. (2010) Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J. Clin. Oncol. 28, 4417–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ringnér M. (2008) What is principal component analysis? Nat. Biotechnol. 26, 303–304 [DOI] [PubMed] [Google Scholar]
- 41. Munn D. H., Zhou M., Attwood J. T., Bondarev I., Conway S. J., Marshall B., Brown C., Mellor A. L. (1998) Prevention of allogeneic fetal rejection by tryptophan catabolism. Science 281, 1191–1193 [DOI] [PubMed] [Google Scholar]
- 42. Dantzer R., O'Connor J. C., Freund G. G., Johnson R. W., Kelley K. W. (2008) From inflammation to sickness and depression: when the immune system subjugates the brain. Nat. Rev. Neurosci. 9, 46–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lacroix M., Abi-Said D., Fourney D. R., Gokaslan Z. L., Shi W. M., DeMonte F., Lang F. F., McCutcheon I. E., Hassenbusch S. J., Holland E., Hess K., Michael C., Miller D., Sawaya R. (2001) A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J. Neurosurg. 95, 190–198 [DOI] [PubMed] [Google Scholar]
- 44. Gerber J., Tumani H., Kolenda H., Nau R. (1998) Lumbar and ventricular CSF protein, leukocytes, and lactate in suspected bacterial CNS infections. Neurology 51, 1710–1714 [DOI] [PubMed] [Google Scholar]
- 45. Bralten L. B. C., Kloosterhof N. K., Balvers R., Sacchetti A., Lapre L., Lamfers M., Leenstra S., de Jonge H., Kros J. M., Jansen E. E. W., Struys E. A., Jakobs C., Salomons G. S., Diks S. H., Peppelenbosch M., Kremer A., Hoogenraad C. C., Smitt P., French P. J. (2011) IDH1 R132H decreases proliferation of glioma cell lines in vitro and in vivo. Ann. Neurol. 69, 455–463 [DOI] [PubMed] [Google Scholar]
- 46. Kallenberg K., Bock H. C., Helms G., Jung K., Wrede A., Buhk J. H., Giese A., Frahm J., Strik H., Dechent P., Knauth M. (2009) Untreated glioblastoma multiforme: increased myo-inositol and glutamine levels in the contralateral cerebral hemisphere at proton MR spectroscopy. Radiology 253, 805–812 [DOI] [PubMed] [Google Scholar]
- 47. Castillo M., Smith J. K., Kwock L. (2000) Correlation of myo-inositol levels and grading of cerebral astrocytomas. Am. J. Neuroradiol. 21, 1645–1649 [PMC free article] [PubMed] [Google Scholar]
- 48. Strott C. A., Higashi Y. (2003) Cholesterol sulfate in human physiology: what's it all about? J. Lipid Res. 44, 1268–1278 [DOI] [PubMed] [Google Scholar]
- 49. Rearick J. I., Stoner G. D., George M. A., Jetten A. M. (1988) Cholesterol sulfate accumulation in tumorigenic and nontumorigenic rat esophageal epithelial cells: evidence for defective differentiation control in tumorigenic cells. Cancer Res. 48, 5289–5295 [PubMed] [Google Scholar]
- 50. Romani L., Fallarino F., De Luca A., Montagnoli C., D'Angelo C., Zelante T., Vacca C., Bistoni F., Fioretti M. C., Grohmann U., Segal B. H., Puccetti P. (2008) Defective tryptophan catabolism underlies inflammation in mouse chronic granulomatous disease. Nature 451, 211–215 [DOI] [PubMed] [Google Scholar]
- 51. Grivennikov S. I., Greten F. R., Karin M. (2010) Immunity, Inflammation, and Cancer. Cell 140, 883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wong E. T. (2006) Tumor growth, invasion, and angiogenesis in malignant gliomas. J. Neuro-Oncol. 77, 295–296 [DOI] [PubMed] [Google Scholar]