Abstract
Monkeypox virus (MPXV) is comprised of two clades: Congo Basin MPXV, with an associated case fatality rate of 10%, and Western African MPXV, which is associated with less severe infection and minimal lethality. We thus postulated that Congo Basin and West African MPXV would differentially modulate host cell responses and, as many host responses are regulated through phosphorylation independent of transcription or translation, we employed systems kinomics with peptide arrays to investigate these functional host responses. Using this approach we have demonstrated that Congo Basin MPXV infection selectively down-regulates host responses as compared with West African MPXV, including growth factor- and apoptosis-related responses. These results were confirmed using fluorescence-activated cell sorting analysis demonstrating that West African MPXV infection resulted in a significant increase in apoptosis in human monocytes as compared with Congo Basin MPXV. Further, differentially phosphorylated kinases were identified through comparison of our MPXV data sets and validated as potential targets for pharmacological inhibition of Congo Basin MPXV infection, including increased Akt S473 phosphorylation and decreased p53 S15 phosphorylation. Inhibition of Akt S473 phosphorylation resulted in a significant decrease in Congo Basin MPXV virus yield (261-fold) but did not affect West African MPXV. In addition, treatment with staurosporine, an apoptosis activator resulted in a 49-fold greater decrease in Congo Basin MPXV yields as compared with West African MPXV. Thus, using a systems kinomics approach, our investigation demonstrates that West African and Congo Basin MPXV differentially modulate host cell responses and has identified potential host targets of therapeutic interest.
Monkeypox virus (MPXV)1 is a member of the genus Orthopoxvirus, which also includes vaccinia virus (VACV), ectromelia virus, cowpox virus (CPXV), and variola virus (VARV), the causative agent of smallpox. MPXV causes zoonotic disease that manifests similarly to smallpox with associated case fatality rates of ∼10% (1). Although MPXV was first isolated from cynomolgus macaques in 1958 in Denmark (2) there was limited scientific interest in the virus until it was demonstrated in the 1970s that MPXV could cause lethal infection in humans (3). Following the cessation of smallpox vaccination there has been a dramatic increase in MPXV incidence in the Democratic Republic of the Congo over the past 30 years (4) and it is estimated that ∼50% of the general population is not protected against MPXV or VARV (4). These concerns have been compounded by the first reported incidence of MPXV outside of the African continent following the accidental introduction of MPXV in the Midwestern United States in 2003 (5). Further concerns have also been raised regarding the potential use of MPXV as a bioterrorism agent thus resulting in its classification as a Class C Select Agent (1, 6).
Monkeypox virus is comprised of two distinct clades that are genetically, clinically, and geographically distinct. Congo Basin MPXV, also known as Central African MPXV, has associated case fatality rates of ∼10% in non-vaccinated individuals as compared with the minimal lethality associated with the less-virulent West African MPXV clade (7). Lending further support, comparative infection models in non-human primates (8), mice (9, 10), prairie dogs (11, 12) and ground squirrels (13) have all demonstrated greater lethality or morbidity associated with Congo Basin MPXV infection when compared with West African MPXV. Additionally, an outbreak of West African MPXV in the U.S. in 2003 following importation of MPXV-infected rodents from Ghana resulted in 69 diagnosed MPXV cases; however, disease severity was relatively mild with no fatalities (5). Although these reports have demonstrated a definitive difference in virulence between the two MPXV clades, there is a paucity of information regarding the virus or host factors that mediate the divergent pathogenesis. Recently, investigations of the global gene expression programs of both the host and virus during Congo Basin MPXV infection have provided insight into the underlying mechanisms of MPXV disease pathogenesis. In particular, independent investigations of host responses to Congo Basin MPXV infection by Rubins et al. (14) and Alkhalil et al. (15) have demonstrated global suppression of host gene expression programs following viral infection. Interestingly, these included the modulation of such diverse host responses as the regulation of histone expression, cytoskeletal rearrangement, cell cycle progression, and interferon-associated gene expression (14, 15). This is perhaps unsurprising as it was demonstrated that the host response modifier genes of Congo Basin MPXV are transcribed at steady-state levels throughout the course of infection (16). Corresponding investigations for West African MPXV have not been reported.
Although studies of global gene expression have been informative, it is increasingly appreciated that many cellular processes are regulated independently of changes in transcription or translation through post-translational modifications of host proteins. For example, phosphorylation is one of the most pivotal biological mechanisms for regulation of cellular processes with 518 annotated human kinase genes and ∼100,000 human phosphorylation sites identified to date (17, 18). As virtually all cell signaling processes are regulated by phosphotransfer reactions, and aberrant kinase activity has been implicated in a variety of diseases, kinases are an attractive target for therapeutic intervention (19, 20). The priority that has been placed on the development of kinase inhibitors for the treatment of a variety of human diseases such as cancer has resulted in the development of tremendous libraries of potential inhibitors that may have other applications for treatment of infectious diseases.
Kinome profiling through global analysis of kinase abundance, activity, phosphorylation status, and substrate specificity provides a novel mechanism for investigating disease pathogenesis through the activation or repression of host cell signal transduction pathways (20). For example, a recent investigation by Bowick et al. utilized kinome peptide arrays to identify host cell signaling nodes of interest that were differentially modulated by two variants of Pichinde virus producing either lethal or self-limiting disease (21). In addition, numerous pathogens, including poxviruses, have been shown to target host cellular processes as a part of their pathogenic mechanism through host protein mimicry (22, 23), including through production of eukaryotic-like kinases (24–26). Such pathogen-encoded effectors may be equally attractive therapeutic targets as their host-encoded counterparts. Thus, systems kinomics with kinome peptide arrays represents a novel methodology for investigating host responses to clinically relevant infectious diseases and identification of potential therapeutic targets.
As direct comparison of the genomes of West African and Congo Basin MPXV demonstrate significant variability in the regions coding for host response modifier proteins, we postulated that the differential virulence of the two MPXV clades is related to the differential modulation of host cell signaling pathways following infection (27, 28). Thus, we sought to investigate host signaling pathway responses to West African and Congo Basin MPXV insult with peptide arrays comprised of human kinase targets for cell growth and differentiation, stress responses, and innate immunity. Host kinome responses to CPXV and VACV were also included for comparison of host response conservation across the orthopoxvirus genus. In the hierarchical clustering analysis Congo Basin MPXV demonstrated similar target phosphorylation patterns to CPXV and moderately to VACV; however, there was limited similarity between West African and Congo Basin MPXV-induced phosphorylation patterns. Congo Basin MPXV infection resulted in a significant down-regulation of host cell responses as compared with infection with West African MPXV as demonstrated through pathway over-representation analysis (ORA) with InnateDB. The down-regulated pathways were related primarily to growth and proliferation, apoptosis, and immune surveillance. The biological relevance of the differential pathways identified was demonstrated through flow cytometry and cell proliferation assays as West African MPXV-infected monocytes had significantly increased apoptotic and cell proliferative responses. Further, pharmacological inhibition of selected host targets differentially phosphorylated following infection by the two MPXV clades validated our systems kinomics results. Thus, we have employed systems kinomics for the investigation of host responses to MPXV infection, and demonstrate for the first time that West African and Congo Basin MPXV induce significantly different host cell signaling pathway activities following viral infection.
MATERIALS AND METHODS
Cell and Virus Conditions
MPXV Zaire 79 and MPXV Sierra Leone 70 strains were propagated in BSC-1 cells at a MOI of 0.1 for 4 days. BSC-1 cells were maintained in MEM supplemented with 10% fetal calf serum and 1% penicillin and streptomycin. Virus stocks were prepared by disruption of BSC-1 cells by successive freeze thaw followed by purification using sucrose gradients. Virus was quantified in a standard plaque assay on either Vero E6 or BSC-1 cells as described previously (19). VACV Western Reserve and CPXV Brighton Red were generated in a similar fashion. Human THP-1 monocytes (ATCC TIB-202R) were maintained in RPMI 1640 medium supplemented with 10% (v/v) heat inactivated fetal bovine serum, 2 mm l-glutamine and 1 mm sodium pyruvate. All cultures were maintained at 37 °C in a humidified 5% (v/v) CO2 incubator.
Viral Infection
Cells were plated in six-well plates and rested for 24 h prior to infection. Cells were infected with either Congo Basin MPXV or West African MPXV at a MOI of 3 or mock infected with an equivalent fraction of culture medium free of any virus. All viral infections were performed at the National Institutes of Health at Biosafety Level 3 in accordance with NIH/CDC Biosafety in Microbiological and Biomedical Laboratories guidelines, as well as in accordance with CDC Select Agent regulations. Virus was incubated with host cells for 1 h at 37 °C with periodic rocking. Following incubation, monocytes were washed twice with phosphate-buffered saline (PBS), resuspended with fresh RPMI 1640 media with 2% (v/v) fetal bovine serum and incubated for 24 h.
Kinome Analysis
Design, construction and application of the peptide arrays were based upon a previously reported protocol with the following modifications (56). Virus-infected and mock-infected THP-1 monocytes were pelleted following incubation and cell lysate was prepared and incubated with human kinome arrays (JPT Technologies, Berlin, Germany). Briefly, cell pellets were lysed with 100 μl of lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm ethylenediamine tetraacetic acid, 1 mm ethylene glycol tetraacetic acid, 1% Triton-X100, 2.5 mm sodium pyrophosphate, 1 mm Na3VO4, 1 mm NaF, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 mm phenylmethylsulfonyl fluoride) and incubated on ice for 10 min. Following incubation, cell lysates were filtered through Amicon 100K filters (Millipore) for 15 min at 4 °C to remove intact viral particles. Subsequent peptide array processing was performed under BSL2 conditions. A 70 μl aliquot of the supernatant was mixed with 10 μl of the activation mix (50% glycerol, 50 μm ATP, 60 mm MgCl2, 0.05% Brij-35 and 0.25 mg/ml bovine serum albumin) and incubated on a peptide array for 2 h at 37 °C. Arrays were subsequently washed once with PBS containing 1% Triton X-100 and submerged in PRO-Q Diamond Phosphoprotein Stain (Invitrogen, Carlsbad, CA) with gentle agitation in the dark for 1 h. Following staining, arrays were washed in destain [20% acetonitrile, 50 mm sodium acetate, pH 4.0 (Sigma)] 3X for 10 min/wash with the addition of fresh destain each time. A final wash was performed with dH2O and placed in 50 ml conical tubes and air-dried for 20 min. Remaining moisture on the arrays was removed by centrifugation of the arrays at 300 × g for 3 min. Array images were acquired using an Axon 4000B microarray scanner at 532–560 nm with a 580 nm filter to detect dye fluorescence. Images were collected using the GenePix 6.0 software (MDS, Foster City, CA). Signal intensity values were collected using the GenePix 6.0 Software (MDS) with the following settings: scanner saturation level 65535, background calculation done using local feature background, signal mean and background mean intensity values used for analysis, local background features excludes 2 pixels, and width of background set to 3 feature diameters. Intensity values for the spots and background were collected for each array.
Kinome Data Preprocessing
The specific responses of each peptide were calculated by subtracting background intensity from foreground intensity. The resulting data were transformed using the variance stabilization model (57), previously trained by a larger MAP kinome data set, to bring all the transformed data onto the same scale while alleviating variance-mean-dependence. In addition, for each of the 300 peptides in a single treatment, the intensities induced by the treatments were subtracted by the intensities from the biological control (i.e. THP-1 + control media) and test statistics calculated. Average intensities were then taken over the three transformed replicate intensities and these values subjected to hierarchical clustering analysis. The R package variance stabilization was used for the transformation (58).
Treatment-Treatment Variability Analysis
Peptide phosphorylations were subjected to paired t-tests to compare their signal intensities under a treatment condition with those under the control condition. Four tests were done for each peptide. Specifically, the tests were Congo Basin MPXV versus THP-1, West African MPXV versus THP-1, CPXV BR versus THP-1, and VACV WR versus THP-1. The p value cutoff was chosen to be 0.20. Formally, the test statistic (TS) was calculated as:
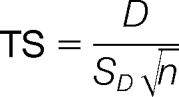 |
where D is the mean of the differences between responses for the same peptides induced by two different treatments, SD the standard deviation of the differences, and n is the number of replicates for that peptide in each treatment (i.e. 3 in our data set). Finally, the p values for the phosphorylation and dephosphorylation events were calculated as P[TS > t(n−1)] and P[TS < t(n−1)], respectively (i.e. one-sided t test). Peptides with significant (p < 0.20) changes in phosphorylation were selected using PERL and BASH scripts. The paired t test was done using R built-in function t.test with paired = True.
Hierarchical Clustering Analysis
The preprocessed data was subjected to hierarchical clustering and principle component analysis (PCA) to cluster treatments based on their kinome profiles. Specifically, for hierarchical clustering, McQuitty + (1 - Pearson Correlation) was used. Briefly, each treatment vector was considered as a singleton (i.e. a cluster with a single element) at the initial stage of the clustering. This method uses (1 - Pearson correlation) to calculate the distances between any two vectors of treatment, say X and Y. Formally, the Pearson correlation is computed as:
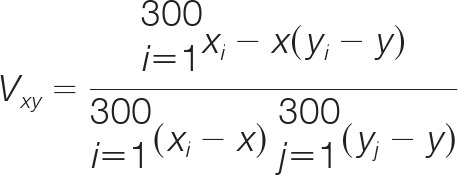 |
and,
In addition, the McQuitty method updates the distance between the two clusters in such a way that upon merging cluster CX and cluster CY into a new cluster CXY, the distance between CXY and each of the remaining clusters, say CR, is calculated in concern with the sizes of CX and CY. Mathematically, let the size of CX be nX and size of CY be nY, then:
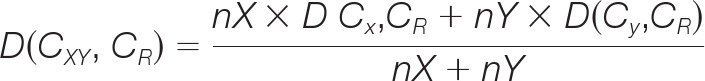 |
The hierarchical clustering was augmented by a heatmap which is also generated using the R function heatmap.2. The function converts the intensity values to statistical z-scores, and then the z-scores are encoded as color (green/red) intensities. Green usually means a value lower than the mean; red a value higher.
Pathway Analysis of Differentially Phosphorylated Peptides
InnateDb (www.innatedb.com) is a publically available resource which, based on levels of either differential expression or phosphorylation, predicts biological pathways based on experiment fold change datasets. Pathways are assigned a probability value (p) based on the number of proteins present for a particular pathway as well as the degree to which they are differentially expressed or modified relative to a control condition. For our investigation input data was limited to peptides which showed consistent responses across the biological replicates (p < 0.05) as well as statistically significant changes from the control condition (p < 0.10).
WST-1 Cell Proliferation Assay
Cell proliferation in the MPXV-infected and mock-infected THP-1 monocytes was performed using the WST-1 proliferation assay (Millipore). Briefly, THP-1s (2 × 104 cells) were plated and rested overnight followed by 24 h MPXV infection as described above. WST-1/ECS solution was then added at a ratio of 1:10 and incubated at 37 °C for 2 h. Absorbance readings were acquired on a plate reader at a wavelength of 450 nm. Independent experiments were done in duplicate or triplicate and were repeated at least three times.
Apoptosis Assays
Induction of apoptosis in MPXV-infected THP-1 monocytes was determined using the ApoAlert Annexin V assay (Clontech) according to the manufacturer's instructions. Briefly, THP-1 monocytes were infected as described above. Following 24 h infection, cells were rinsed and resuspended with 1X binding buffer followed by the addition of 500 ng of Annexin V-FITC and Propidium iodide and incubated at room temperature in the dark for 10 min. Cells were subsequently washed with 1X binding buffer, incubated in fixative solution for 1 h at room temperature (PBS, 4% formaldehyde, 1% pluronic acid), and finally washed and resuspended in 1X binding buffer. Percentage of apoptotic cells was determined by BD® FACS analysis. Independent experiments were done in duplicate or triplicate and were repeated at least three times.
Caspase-3 activity was assessed using the EnzChek Caspase-3 Assay Kit (Invitrogen) according to the manufacturer's instructions. Briefly, MPXV-infected and mock-infected THP-1 monocytes were harvested by centrifugation 24 h postinfection and washed with PBS. Cell pellets were resuspended in 1X cell lysis buffer for 30 min on ice and centrifuged to remove cellular debris. Z-DEVD-AMC substrate was subsequently added and samples were incubated at room temperature for 30 min and fluorescence was measured at 441 nm on a standard plate reader.
Met PhosphoELISAS
THP-1 monocytes were infected or mock-infected as described above. At 24 h post-infection cells were harvested by centrifugation, washed with PBS, and lysed on ice for 10 min with 1X lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm ethylenediamine tetraacetic acid, 1 mm ethylene glycol tetraacetic acid, 1% Triton-X100, 2.5 mm sodium pyrophosphate, 1 mm Na3VO4, 1 mm NaF, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 mm phenylmethylsulfonyl fluoride). Phosphorylated c-Met was measured using the PathScan Phospho-Met (Tyr1234/1235) enzyme-linked immunosorbent assay (ELISA) Assay Kit (Cell Signaling) following the manufacturer's instructions. Experiments were done in duplicate were repeated independently at least three times.
Pathway Inhibitor Assays
For the inhibitor studies THP-1 monocytes were pretreated for 30 min with LY29002 (20 μm; Sigma Aldrich), Akt-X (15 μm; EMD Biosciences, San Diego, CA) nutlin 3 (10 μm; Sigma Aldrich), Met Kinase Inhibitor (1 μm or 10 μm; Santa Cruz Biotechnology, Santa Cruz, CA), staurosporine (10 μm; Enzo Lifesciences, Farmingdale, NY), SB-202190 (10 μm; Enzo Lifesciences) or BML-257 (10 μm; Enzo Lifesciences). Control cells were also employed in the absence of inhibitors. Cells were subsequently infected as described above in the continued presence or absence of inhibitor and harvested by centrifugation at 24 h post-infection followed by washing with cold PBS. Cells were subsequently disrupted by freezing and thawing and virus was collected from the supernatant of centrifuged cells and assayed for infectivity as described previously (19). Each experiment was run in duplicate and the results are reported as average values. The data was confirmed by at least three independent experiments with identical results.
RESULTS
West African and Congo Basin MPXV Induce Differential Host Target Phosphorylation
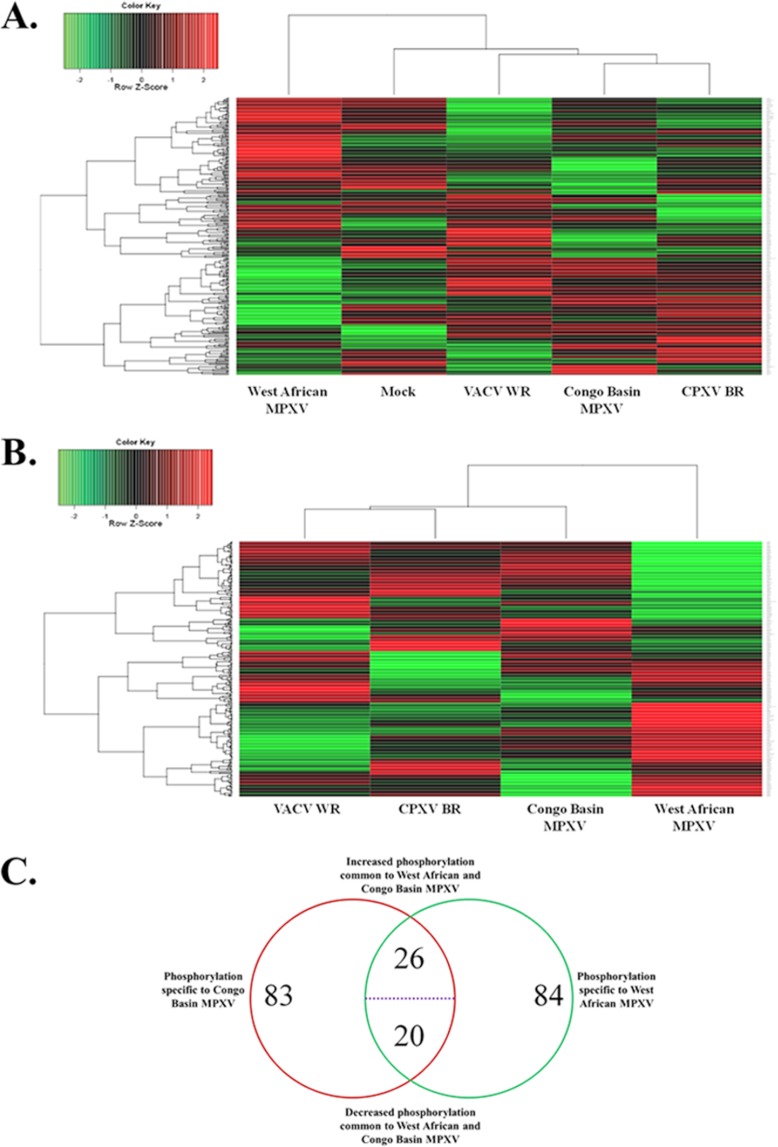
We postulated that the virulence differences associated with West African and Congo Basin MPXV may be attributed to the differential modulation of host cell signaling pathways following infection. Thus, we employed systems kinomics (20, 29, 30) with high-throughput human kinome peptide arrays to study the global activation state of host kinases, the kinome, following West African or Congo Basin MPXV infection. The resultant kinome data sets were subjected to hierarchical cluster analysis for comparative analysis and visualization of changes to host cell phosphorylation following infection. Conserved patterns of host kinome responses across the orthopoxvirus genus were also compared by the incorporation of CPXV BR and VACV WR. Interestingly, West African and Congo Basin MPXV demonstrated weak patterns of clustering independent of subtraction of the mock infected control (Figs. 1A, 1B). Further, Congo Basin MPXV-induced host phosphorylations clustered strongly with those induced by CPXV BR and VACV WR. Pairwise comparison of the MPXV data sets using Student's t test revealed a number of differential phosphorylations between either of the MPXV clades and the mock-infected condition or between the two MPXV clades directly. The commonality of peptide phosphorylations between West African and Congo Basin MPXV is presented in Fig. 1C. Of the peptide phosphorylations that displayed high technical reproducibility (as described in the materials and methods), 129 and 130 peptides were differentially phosphorylated relative to the mock-infected control for Congo Basin and West African MPXV, respectively. Approximately one-third of the differentially phosphorylated peptides were conserved in both identity and direction of phosphorylation changes (Fig. 1C). This is not unanticipated as the limited amino acid sequence variability between the MPXV clades (27) would suggest some degree of conserved interaction with the host. The remaining peptide phosphorylation differences represented phosphorylation events specific to a particular MPXV clade and represent differential trends in responses (increased versus decreased phosphorylation).
Fig. 1.
Analysis of orthopoxvirus-infected kinome data sets. Peptide phosphorylation was assessed by densitometry, scaled and normalized using GeneSpring 6.0 software. For hierarchical clustering, McQuitty + (1 - Pearson Correlation) was used. A, hierarchical clustering of the orthopoxvirus infected kinome data sets alongside the mock-infected control data set. B, cluster analysis of the orthopoxvirus infected kinome data sets following background subtraction of the mock-infected control data set. C, commonality between West African MPXV and Congo Basin MPXV spot phosphorylations represented as a Venn diagram. Spots which demonstrated a significant (p < 0.20) differential phosphorylation between the MPXV-infected and mock-infected control were compiled into a data set for comparative analysis.
Congo Basin MPXV Selectively Down-regulates Host Signaling Pathways As Compared With West African MPXV
To gain insight into the relation between our kinome data and differential mechanisms of host response modulation employed by either West African or Congo Basin MPXV, we employed pathway ORA with the online software InnateDB (31). Input data for the analysis was limited to significant differences in peptide target phosphorylations (p < 0.10) relative to the mock-infected controls to ensure that identified pathways represented conserved biological responses.
For our initial pathway analysis we focused on protein phosphorylations that were common only to Congo Basin or West African MPXV infection (Fig. 1C). Pathway ORA of protein phosphorylations common to the Congo Basin MPXV infection data set resulted in a limited subset of host signaling pathways (Table I). Notably, FoxO family signaling, a pathway related to the regulation of cellular processes (32)), was the only pathway found to be up-regulated in activity. Interestingly, the activities of pathways related to growth factor responses [Growth hormone signaling pathways, Melanoma] were predicted to be down-regulated. Down-regulation of the Cytokine-Cytokine Receptor Interaction Pathway in our analysis lent further support for the ability of Congo Basin MPXV to subvert immune defenses (Table I). In contrast, protein phosphorylations common to West African MPXV infection resulted in a broader subset of pathways identified by our ORA (Table II). Up-regulated pathways were primarily related to immune activation, including interleukin 2 (IL-2) and IL-6 signaling pathways (33), and growth factor responses, including fibroblast growth factor (FGF) (34), epidermal growth factor (EGFR) (35, 36), and platelet derived growth factor beta (PDGFβ). In contrast, down-regulated pathways were found to be primarily related to transforming growth factor beta (TGFβ) signaling responses (Table II).
Table I. Pathway over representation analysis of uncommon proteins differentially phosphorylated in response to Congo basin MPXV infection.
| Pathway Name | Proteins in Pathway | Increased Phosphorylations | Up-Regulated Pathway P Value | Decreased Phosphorylations | Down-Regulated Pathway P Value |
|---|---|---|---|---|---|
| FoxO family signaling | 4 | 4 | 0.074 | 0 | 1 |
| Cytokine-cytokine receptor interaction | 3 | 0 | 1 | 3 | 1 |
| Growth hormone signaling pathway | 3 | 0 | 1 | 3 | 1 |
| Melanoma | 7 | 2 | 0.98 | 5 | 1 |
Table II. Pathway over representation analysis of uncommon proteins differentially phosphorylated in response to West African MPXV infection.
| Pathway Name | Proteins in Pathway | Increased Phosphorylations | Up-Regulated Pathway P Value | Decreased Phosphorylations | Down-Regulated Pathway P Value |
|---|---|---|---|---|---|
| FGF signaling pathway | 6 | 6 | 0.027 | 0 | 1 |
| IL2 | 6 | 6 | 0.027 | 0 | 1 |
| ErbB1 downstream signaling | 9 | 8 | 0.034 | 1 | |
| Signaling events mediated by TCPTP | 5 | 5 | 0.052 | 0 | 1 |
| IL6 | 8 | 7 | 0.062 | 0 | 1 |
| KitReceptor | 4 | 4 | 0.099 | 0 | 1 |
| LPA receptor mediated events | 4 | 4 | 0.099 | 0 | 1 |
| PDGFR-beta signaling pathway | 4 | 4 | 0.099 | 0 | 1 |
| Regulation of nuclear SMAD2/3 signaling | 4 | 4 | 0.099 | 0 | 1 |
| TGF-beta receptor signaling | 6 | 1 | 1 | 5 | 0.027 |
| ALK1 signaling events | 3 | 0 | 1 | 3 | 0.054 |
| Chronic myeloid leukemia | 7 | 2 | 0.99 | 5 | 0.070 |
| Colorectal cancer | 7 | 2 | 0.99 | 5 | 0.070 |
Although these comparisons suggested that Congo Basin and West African MPXV differentially modulate host cell response following infection, the limited information from our initial pathway ORA compelled us to investigate this phenomenon further through direct comparison of the complete Congo Basin and West African MPXV kinome array data sets. We hypothesized that this comparison would provide a more detailed comparison by including protein phosphorylations that were common to both MPXV infection groups as well as those that were uncommon. As predicted, the direct comparison of the complete data sets yielded a much broader range of pathways differentially modulated by the two MPXV clades (Table III). Interestingly, Congo Basin MPXV infection resulted in the global down-regulation of host cellular responses as compared with West African MPXV following direct comparison of the two kinome data sets. As found in our initial comparative the down-regulated pathways were primarily related to growth factor signaling: including fibroblast growth factor (FGF) signaling (34), BCR-ABL-mediated signaling (17), and growth hormone signaling pathway (18)). However, pathways related to subversion of apoptotic responses were also identified, including the FAS signaling pathway (32), direct p53 effectors (37), regulation of bad phosphorylation (38), and phosphatase and tensin homolog (PTEN) dependent cell cycle arrest and apoptosis (16). Thus, our pathway ORA suggest that the Congo Basin MPXV selectively down-regulated host cell responses primarily related to growth factor signaling and apoptotic responses as compared with the less virulent West African MPXV clade.
Table III. Differential host cell signaling responses to Congo Basin MPXV-infected monocytes as compared to West African MPXV-infected monocytes. InnateDB is a publically available pathway analysis tool. Based on levels of differential expression or phosphorylation InnateDB is able to predict pathways that are consistent with the experimental data. Pathways are assigned a probability value (p) based on the number of proteins present for a particular pathway. It also provides the number of uploaded pathways associated with a particular pathway as well as the subset of individual proteins that are differentially phosphorylated. For this investigation fold change cutoffs were set at 80% confidence of the difference between the infected and mock-infected treatment.
| Pathway Name | Proteins in Pathway | Increased Phosphorylations | Up-Regulated Pathway P Value | Decreased Phosphorylations | Down-Regulated Pathway P Value |
|---|---|---|---|---|---|
| Agrin in postsynaptic differentiation | 3 | 3 | 0.068 | 0 | 1 |
| FGF signaling pathway | 8 | 0 | 1 | 8 | 0.0018 |
| Inhibition of cellular proliferation by gleevec | 8 | 0 | 1 | 7 | 0.021 |
| Ctcf: first multivalent nuclear factor | 5 | 0 | 1 | 5 | 0.022 |
| Signaling events activated by Hepatocyte Growth Factor Receptor (c-Met) | 12 | 2 | 0.99 | 9 | 0.042 |
| Signaling events mediated by Stem cell factor receptor (c-Kit) | 12 | 2 | 0.99 | 9 | 0.042 |
| Tpo signaling pathway | 7 | 0 | 1 | 6 | 0.043 |
| BMP2 signaling pathway(through Smad) | 4 | 0 | 1 | 4 | 0.049 |
| C-MYB transcription factor network | 4 | 0 | 1 | 4 | 0.049 |
| Calcineurin-regulated NFAT-dependent transcription in lymphocytes | 4 | 0 | 1 | 4 | 0.049 |
| Calcium signaling in the CD4+ TCR pathway | 4 | 0 | 1 | 4 | 0.049 |
| Class I PI3K signaling events mediated by Akt | 4 | 0 | 1 | 4 | 0.049 |
| Pten dependent cell cycle arrest and apoptosis | 4 | 0 | 1 | 4 | 0.049 |
| Regulation of bad phosphorylation | 4 | 0 | 1 | 4 | 0.049 |
| Direct p53 effectors | 6 | 1 | 0.97 | 5 | 0.083 |
| ErbB2/ErbB3 signaling events | 6 | 0 | 1 | 5 | 0.083 |
| FAS signaling pathway (CD95) | 6 | 1 | 0.97 | 5 | 0.083 |
| Osteopontin-mediated events | 6 | 1 | 0.97 | 5 | 0.083 |
| Role of Calcineurin-dependent NFAT signaling in lymphocytes | 6 | 0 | 1 | 5 | 0.083 |
Biological Validation of Signaling Responses Identified Through Kinome Analysis
Following the demonstration that Congo Basin MPXV infection down-regulated pathways related to growth factor signaling/cell proliferation and apoptosis as compared with West African MPXV, we sought to biologically validate these results. Importantly, we first demonstrated that the growth kinetics and level of virus replication of the two MPXV clades were similar throughout the course of our infection studies (supplemental Fig. S1).
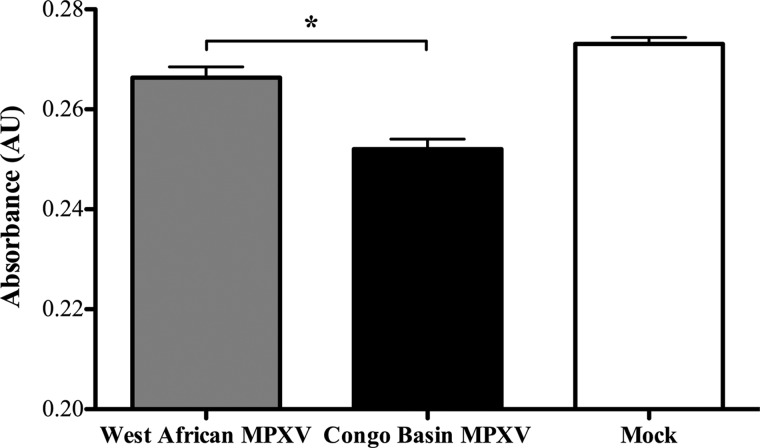
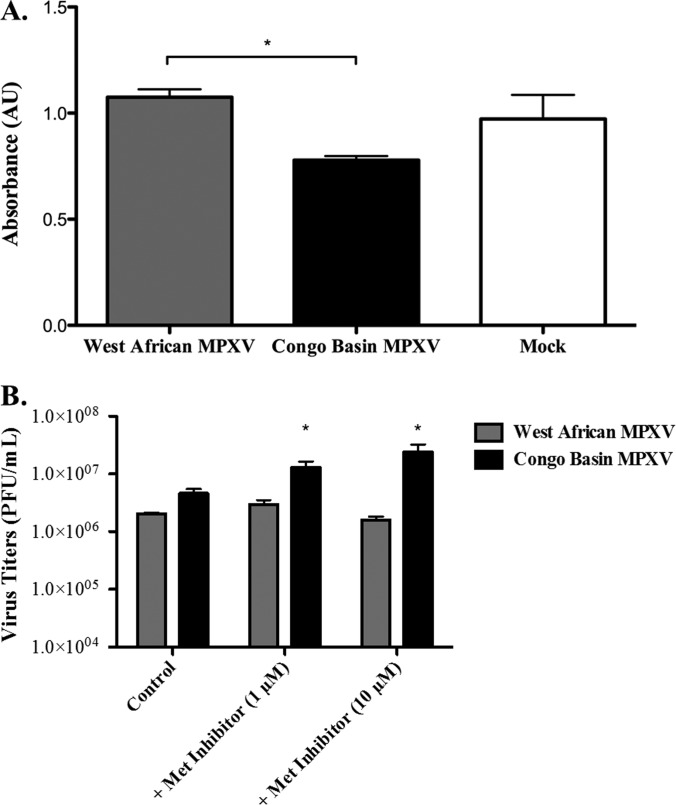
First, our pathway ORA data for the direct comparison of Congo Basin and West African MPXV suggested that Congo Basin MPXV down-regulated cell proliferative responses as compared with West African MPXV (Table III). Thus, we monitored cell proliferation responses to the two MPXV viruses through a WST-1 assay as this assay is routinely used to investigate cell proliferation, cell viability and cytotoxicity. Our data demonstrate that Congo Basin MPXV infection resulted in a moderate but significant reduction in THP-1 cell proliferation as compared with West African MPXV (Fig. 2; data presented relative to mock-infected cells). This is consistent with the differential host responses identified through our pathway ORA.
Fig. 2.
Comparison of the effect of West African MPXV or Congo Basin MPXV on cell proliferation. Human THP-1 monocytes were seeded in a 96-well plate at 1 × 105 cell/well with 100 μl of culture medium and infected at an MOI of 3 with West African or Congo Basin MPXV for 24 h. Cell proliferation was assessed through the addition of WST-1 reagent 30 min at the 24 h time point and absorbance was measured at 450 nm in a multiwall plate reader. Data is presented as the mean of three independent experiments ± S.D. *, p < 0.1. A Student's t test was used for the comparison.
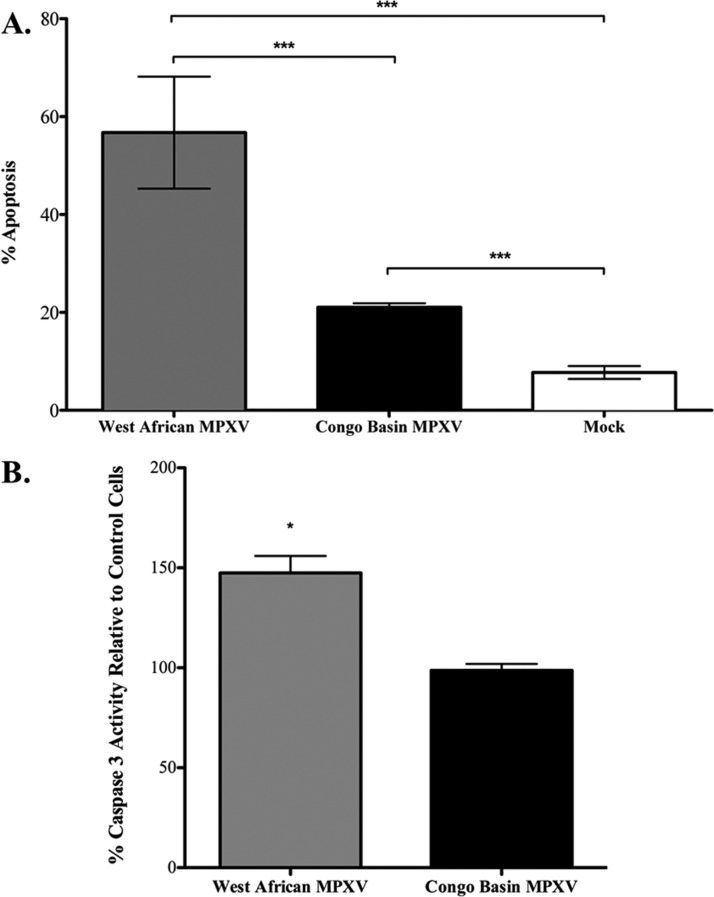
The direct comparison of the kinome array data sets for Congo Basin and West African MPXV also suggested that West African MPXV selectively down-regulated apoptotic responses in host cells (Table I). Using fluorescence-activated cell sorting (FACS) analysis of AnnexinV staining we demonstrated a significant increase in host cell apoptosis following West African MPXV infection as compared with Congo Basin MPXV (Fig. 3A). This was further corroborated through a caspase 3 activity assay demonstrating a significant increase in caspase 3 activity in West African MPXV-infected cells relative to Congo Basin MPXV-infected cells (Fig. 3B; data presented relative to mock-infected control cells).
Fig. 3.
West African MPXV infection results in increased apoptosis as compared with Congo Basin MPXV. Human THP-1 monocytes (1 × 106 cells) were infected at an MOI of 3 and seeded in a six-well plate with 3 ml of culture medium and incubated for 24 h. A, FACS analysis of THP-1 monocyte apoptosis 24 h postinfection by the two MPXV clades. Data is presented as the mean of three independent experiments ± S.D. B, Caspase 3 activity of West African or Congo Basin MPXV-infected THP-1 monocyte lysates relative to the mock-infected THP-1 cells. Data is presented as the mean of three independent experiments with identical results ± S.D. *, p < 0.1; ***, p < 0.001. A Student's t test was used for the comparison.
To further validate our kinome array data we selected a target that was common in all iterations of the pathway ORA, c-Met. c-Met-related signaling pathways were found to be repressed by Congo Basin MPXV in both Table I and Table III. We investigated the phosphorylation status of c-Met in response to both Congo Basin and West African MPXV directly through phosphospecific ELISA. Our array data demonstrated that c-Met Y1234 phosphorylation was significantly repressed (–1.4-fold) in Congo Basin MPXV infected monocytes as compared with West African MPXV. Using a c-Met Y1234 phosphospecific ELISA we demonstrated that Congo Basin MPXV-infected cell lysates had a significant reduction (–1.4-fold) in c-Met Y1234 phosphorylation as compared with lysates from West African MPXV-infected monocytes (Fig. 4A). Further, pharmacological inhibition of c-Met phosphorylation with 1 μm or 10 μm of Met Kinase Inhibitor 30 min prior to infection with the two MPXV clades (multiplicity of infection (MOI) = 3) resulted in a concentration-dependent increase in Congo Basin MPXV viral titers as compared with untreated Congo Basin MPXV-infected cells and no significant effect on West African MPXV infection (Fig. 4B). Taken together, the results of our biological validation functionally confirmed our kinome array data demonstrating the reliability and power of this approach.
Fig. 4.
West African MPXV infection increases Met phosphorylation as compared with Congo Basin MPXV. Human THP-1 monocytes (1 × 106 cells) were infected at an MOI of 3 and seeded in a six-well plate with 3 ml of culture medium and incubated for 24 h. A, Met PhosphoELISA analysis of West African or Congo Basin MPXV-infected THP-1 monocytes relative to the mock-infected THP-1 cells. Phosphorylated Met (Tyr1234/1235) was determined by phosphoELISA assay as described in Methods. B, Effect Met phosphorylation inhibition on West African and Congo Basin MPXV virus yields. Human THP-1 monocytes were either left untreated or treated for 30 min with Met Kinase Inhibitor (1 μm or 10 μm) prior to virus infection. Cells were subsequently infected with West African MPXV or Congo Basin MPXV at an MOI of 3 in the absence or continued presence of inhibitor for 24 h. Viruses were then collected and viral titers were determined. Data is presented as the means of three independent experiments ± S.D. *, p < 0.1. A Student's t test was used for the comparison and p values were based on comparison to the control infected conditions.
Disruption of Akt S473 and p38α Phosphorylation, or Activation of Apoptosis, Significantly Inhibits Congo Basin MPXV Replication as Compared with West African MPXV
In addition to increasing our understanding of mechanisms of viral pathogenesis, our kinome data and pathway ORA provided the identities of specific host pathways and kinases that were differentially modulated by Congo Basin and West African MPXV. Previously, it has been demonstrated that poxviruses can selectively modulate host responses mediated by the PBK/Akt pathway for regulation of viral replication (39, 40). Soares et al. demonstrated that the selective pharmacological inhibition of Akt S473 phosphorylation with Akt-X or T308 phosphorylation with LY294002 reduced CPXV and VACV virus yields by 80–90% (40). Our kinome array data suggested that phosphorylation of Akt S473 was significantly increased (1.8 fold) following Congo Basin MPXV but was unaffected following West African MPXV infection. Akt T308 phosphorylation was comparably increased in both West African and Congo Basin MPXV infected cells as compared with the mock-infected controls (supplemental Table S5). Thus, we sought to investigate the effects of pharmacological inhibition of Akt phosphorylation on West African and Congo Basin MPXV viral replication. We performed one-step viral growth curves for both MPXV clades in the presence or absence of LY294002 (20 μm) or Akt-X (15 μm). This concentration of inhibitors had no significant effect on cell viability (data not shown). Cells were left untreated or were pretreated with inhibitors for 30 min prior to infection at an MOI of 3 in the continued presence of inhibitors. At 24 h post-infection virus was collected and assayed for infectivity. Inhibition of Akt S308 by LY294002 had no significant inhibitory effect on virus production in monocytes infected with either MPXV clade (Fig. 5). However, inhibition of Akt S473 phosphorylation with Akt-X selectively inhibited Congo Basin MPXV virus replication (261-fold reduction in virus yield), whereas there was no significant inhibition West African MPXV infection as compared with the untreated infected controls (Fig. 5). Interestingly, pharmacological inhibition of Akt translocation with BML-257 demonstrated a similar inhibitory effect (∼3.5-fold difference) on the two MPXV clades suggesting a central role for Akt in the life cycles of both MPXV clades.
Fig. 5.
Inhibitors of Akt or p38 alpha phosphorylation, or activation of apoptosis, decrease Congo Basin MPXV viral replication as compared with West African MPXV. Human THP-1 monocytes were either left untreated or treated for 30 min with LY294002 (20 μm), Akt-X (15 μm), nutlin 3 (10 μm), staurosporine (10 μm), SB-202190 (10 μm), or BML-257 (10 μm) prior to virus infection. Cells were subsequently infected with West African MPXV or Congo Basin MPXV at an MOI of 3 in the absence or continued presence of the pharmacological agents for 24 h. Viruses were then collected and viral titers were determined. Data are representative of three independent experiments ± S.D. *, p < 0.1; **, p < 0.01; ***, p < 0.001. A Student's t test was used for the comparison and p values were based on comparison to the control infected conditions.
Recently, Zachos et al. have demonstrated that herpes simplex virus type 1 (HSV-1)-enhanced infection through stimulation of p38 MAPK signaling (41). Analysis of our kinome data sets also suggested that Congo Basin MPXV infection resulted in increased p38 phosphorylation (p38α and p38δ) as compared with West African MPXV (1.5-fold and 2.4 fold, respectively). We had also found that pre-treatment of HEK293 cells with SB-202190, a p38α inhibitor, resulted in a significant inhibition of a GFP expressing Congo Basin MPXV (Data not shown) suggesting a central role for p38 in Congo Basin MPXV infection. Pretreatment of cells with SB-202190 resulted in decreased West African and Congo Basin MPXV virus yields; however, addition of the inhibitor had a 9.5-fold greater inhibitory effect on Congo Basin MPXV virus production. SB-202190 was not cytotoxic to cells at the concentration tested.
As Congo Basin MPXV selectively repressed apoptosis in infected monocytes as compared with West African MPXV, and this correlated with reduced phosphorylation of p53 in our kinome array comparison (–1.6-fold), we investigated the effect of chemical antagonism of apoptosis on MPXV virus yields. We postulated that increased apoptosis in Congo Basin MPXV-infected monocytes would result in reduced virus yields as compared with West African MPXV. Cells were pretreated with staurosporine (10 μm) and were subsequently infected with MPXV as above. Cells treated with staurosporine were >94% viable after 24 h. Activation of apoptosis resulted in significantly reduced viral yields in both Congo Basin MPXV-infected cells (495-fold decrease) and West African MPXV-infected cells (10-fold decrease) (Fig. 5). However, the ∼50-fold greater inhibition of Congo Basin MPXV suggests a much stronger effect of apoptosis activation on Congo Basin MPXV virus replication. These data indicate the rapid translation of kinomic data into rational strategies for antiviral target discovery.
DISCUSSION
Although global eradication of smallpox was announced in 1980 there is a paucity of information regarding the molecular mechanisms underlying VARV disease pathogenesis. The critical importance of understanding the molecular mechanisms of poxvirus disease is further underscored by the increasing incidence of MPXV, a surrogate for VARV studies, and the accidental introduction of MPXV into the United States in 2003. Through a combination of systems kinomics and biological and functional validation assays we have demonstrated that West African and Congo Basin MPXV differentially modulate host cell responses and in particular those related to cell proliferation, cell survival and apoptosis.
Our investigation demonstrated that Congo Basin MPXV selectively downregulated pathways largely related to apoptotic events, including p53- and Fas-mediated cell signaling events, and cell proliferation, and enhanced cell survival as compared with West African MPXV. Indeed, the ability of poxviruses to subvert pro-apoptotic host cell responses, in particular through gene-encoded apoptosis inhibitors, is well documented (40, 42–45). Analysis of our kinome data sets suggested that the anti-apoptotic activities associated with Congo Basin MPXV were largely related to p53-mediated signaling apoptosis pathways and supported by FACS analysis and caspase 3 activity assays. Proteomic analysis of West African and Congo Basin MPXV has demonstrated >99% amino acid sequence conservation between the two MPXV clades; however, differences within the two sequences were sequestered to host response modifier proteins at the terminal ends of the genome (28). Interestingly, BR-203, an ortholog of myxoma virus M-T4 believed to have a role in subversion of host apoptotic responses (32, 46), is predicted to be severely truncated in West African MPXV though the full-length gene appears in Congo Basin MPXV (42). Thus, it is enticing to speculate that BR-203 plays a central role in the virulence associated with Congo Basin MPXV and may be a novel target for therapeutic investigations. Further, there is an increasing appreciation for subversion of host apoptotic responses as a means of facilitating viral replication (47). As apoptosis is an important function of innate immunity for limiting pathogen dissemination it is unsurprising that viruses have evolved diverse mechanisms for subverting host apoptotic responses.
To gain further insight into the mechanisms of MPXV-mediated host response modulation we directly compared the kinome data sets from the two MPXV clades. Pathway ORA suggested that Congo Basin MPXV-infection represses host signaling pathways largely related to growth factor signaling and cell proliferative responses as compared with the less virulent West African MPXV clade. Although it may seem counter-intuitive for a more virulent virus to reduce cell proliferative responses, investigations of vFGF, a baculovirus FGF mimic, have demonstrated that vFGF increases virus dissemination and reduces the time to death for the host (48). The ability of vFGF to bind and activate the host FGF receptor results in virus-mediated host cell proliferation. Signaling events mediated by the hepatocyte growth factor receptor c-Met were identified through pathway ORA in both the phosphorylation events common to Congo Basin MPXV and following direct comparison to the West African MPXV kinome data sets. This coupled with our phosphoELISA data and Met Kinase Inhibitor data suggested that c-Met occupied a central role in Congo Basin MPXV infection of monocytes. This suggests that Congo Basin MPXV selectively modulates c-Met-mediated signaling pathways, and perhaps other growth factor-mediated signaling events, during infection. Indeed, the repression of monocyte cell proliferation by Congo Basin MPXV may highlight a secondary mechanism for the virus to subvert potential host cell apoptosis following enhanced, and potentially uncontrolled, cell proliferation. Recent evidence from Ryan and colleagues suggests that chronic hepatitis C infection may impede host immune responses by disabling dendritic cells as a mechanism to reduce pathogen recognition and initiation of the innate immune response (49). Moreover, pathway ORA of phosphorylation events common to Congo Basin MPXV infection suggested that the FoxO signaling pathways were distinct to infection by this MPXV clade. FoxO signaling pathways have been implicated in the delicate balance between cell survival and cell death (50). As FoxO activation can block cell proliferation and promote quiescence in many cell types it is enticing to speculate that this may reflect a vital role for the activation of FoxO signaling pathways in subversion of host immunity by Congo Basin MPXV. Thus, our data suggests that the increased virulence associated with Congo Basin MPXV as compared with West African MPXV may be a consequence of both the precise modulation of programmed cell death, cell cycle progression and promotion of cell survival. Further investigations focusing on the selective modulation of the pathways identified in our comparison of the two MPXV clade data sets may identify previously unknown roles of these pathways in viral infection.
A recent investigation by Soares et al. highlighted the central role of Akt phosphorylation in orthopoxvirus infection (40). Our kinome analysis demonstrated that West African and Congo Basin MPXV differentially modulate Akt S473 phosphorylation. Importantly, pharmacological inhibition of S473 phosphorylation with Akt-X significantly inhibited viral replication in Congo Basin MPXV-infected cells; however, no reduction was demonstrated for West African MPXV-infected cells. Thus, our data suggest that the two MPXV clades may utilize different host cell signaling pathways for viral replication and the precise mechanisms underlying these differences require further exploration. Interestingly, pretreatment with BML-257, an Akt translocation inhibitor, had similar inhibitory effects on West African and Congo Basin MPXV. This indicates that the modulation of Akt-mediated signaling by Congo Basin MPXV is likely related to the phosphorylation state of Akt rather than its cellular localization. Our investigation also suggests a central role for p38α, and potentially p38-MAPK signaling, in Congo Basin MPXV infections. Our kinome data demonstrated that Congo Basin MPXV infection results in increased phosphorylation of the p38α and treatment with the p38α inhibitor SB-202190 resulted in a 9.5-fold greater decrease in Congo Basin MPXV virus yields as compared with West African MPXV. Although the activation of p38 MAPK signaling is classically related to stress responses and activation of cell death (51), there is increasing evidence that under certain circumstances p38 MAPK can enhance cell survival. Indeed, p38 MAPK activation has been demonstrated to enhance cell survival responses to DNA damage (52, 53). Further, Zachos et al. have demonstrated that herpes simplex virus type 1 (HSV-1)-mediated stimulation of p38 MAPK resulted in enhanced viral transcription and increased virus yields (41).
Although systems kinomics has been employed extensively in cancer research (20) there has been limited application to the investigation of viral infections. Thus, molecular investigations of poxvirus disease pathogenesis have relied on gene expression studies. Rubins et al. demonstrated that MPXV genes related to immunomodulation were expressed throughout the course of infection (54). Alkhalil et al. and Rubins et al. recently demonstrated that Congo Basin MPXV infection down-regulated a broad range of host responses (15). More recently, Brown et al. demonstrated that MPXV insult resulted in reductions to host protein expression levels, most notably those participating in structural or metabolic processes (55). Although these investigations have provided important information regarding modulation of host immunity by Congo Basin MPXV, the integration of systems kinomics has provided pertinent functional information regarding the differential host response to Congo Basin MPXV and the less-virulent West African MPXV clade and identified potential host targets for therapeutic intervention.
Footnotes
* This study was supported, in part, by the National Institute of Allergy and Infectious Diseases, Division of Intramural Research.
 This article contains supplemental Fig. S1 and Tables S1 to S5.
This article contains supplemental Fig. S1 and Tables S1 to S5.
1 The abbreviations used are:
- MPXV
- Monkeypox virus
- CPXV
- Cowpox virus
- FBS
- Fetal bovine serum
- FGF
- Fibroblast growth factor
- HSV
- Herpes simplex virus
- IGF1
- Insulin-like growth factor 1
- MAPK
- Mitogen activated protein kinase
- ORA
- Over-representation analysis
- PTEN
- Phosphatase and tensin homolog
- TLR
- Toll-like receptor
- VACV
- Vaccinia virus
- VARV
- Variola virus.
REFERENCES
- 1. Parker S., Nuara A., Buller R. M., Schultz D. A. (2007) Human monkeypox: an emerging zoonotic disease. Future Microbiol. 2, 17–34 [DOI] [PubMed] [Google Scholar]
- 2. Von Magnus P., Anderson E. K., Petersen K. B., Birch-Andersen A. (1959) A pox-like disease in cynomolgus monkeys. Acta Path. Microbiol. Scand. 46, 156–176 [Google Scholar]
- 3. Ladnyj I. D., Ziegler P., Kima E. (1972) A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bull World Health Organ. 46, 593–597 [PMC free article] [PubMed] [Google Scholar]
- 4. Rimoin A. W., Mulembakani P. M., Johnston S. C., Lloyd Smith J. O., Kisalu N. K., Kinkela T. L., Blumberg S., Thomassen H. A., Pike B. L., Fair J. N., Wolfe N. D., Shongo R. L., Graham B. S., Formenty P., Okitolonda E., Hensley L. E., Meyer H., Wright L. L., Muyembe J. J. (2010) Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc. Natl. Acad. Sci. U.S.A. 107, 16262–16267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reed K. D., Melski J. W., Graham M. B., Regnery R. L., Sotir M. J., Wegner M. V., Kazmierczak J. J., Stratman E. J., Li Y., Fairley J. A., Swain G. R., Olson V. A., Sargent E. K., Kehl S. C., Frace M. A., Kline R., Foldy S. L., Davis J. P., Damon I. K. (2004) The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 350, 342–350 [DOI] [PubMed] [Google Scholar]
- 6. Jahrling P. B., Fritz E. A., Hensley L. E. (2005) Countermeasures to the bioterrorist threat of smallpox. Curr. Mol. Med. 5, 817–826 [DOI] [PubMed] [Google Scholar]
- 7. Jezek Z., Grab B., Paluku K. M., Szczeniowski M. V. (1988) Human monkeypox: disease pattern, incidence and attack rates in a rural area of northern Zaire. Trop. Geogr. Med. 40, 73–83 [PubMed] [Google Scholar]
- 8. Saijo M., Ami Y., Suzaki Y., Nagata N., Iwata N., Hasegawa H., Iizuka I., Shiota T., Sakai K., Ogata M., Fukushi S., Mizutani T., Sata T., Kurata T., Kurane I., Morikawa S. (2009) Virulence and pathophysiology of the Congo Basin and West African strains of monkeypox virus in non-human primates. J. Gen. Virol. 90, 2266–2271 [DOI] [PubMed] [Google Scholar]
- 9. Osorio J. E., Iams K. P., Meteyer C. U., Rocke T. E. (2009) Comparison of monkeypox viruses pathogenesis in mice by in vivo imaging. PLoS One 4, e6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hutson C. L., Abel J. A., Carroll D. S., Olson V. A., Braden Z. H., Hughes C. M., Dillon M., Hopkins C., Karem K. L., Damon I. K., Osorio J. E. Comparison of West African and Congo Basin monkeypox viruses in BALB/c and C57BL/6 mice. PLoS One 5, e8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hutson C. L., Carroll D. S., Self J., Weiss S., Hughes C. M., Braden Z., Olson V. A., Smith S. K., Karem K. L., Regnery R. L., Damon I. K. (2010) Dosage comparison of Congo Basin and West African strains of monkeypox virus using a prairie dog animal model of systemic orthopoxvirus disease. Virology 402, 72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hutson C. L., Olson V. A., Carroll D. S., Abel J. A., Hughes C. M., Braden Z. H., Weiss S., Self J., Osorio J. E., Hudson P. N., Dillon M., Karem K. L., Damon I. K., Regnery R. L. (2009) A prairie dog animal model of systemic orthopoxvirus disease using West African and Congo Basin strains of monkeypox virus. J. Gen. Virol. 90, 323–333 [DOI] [PubMed] [Google Scholar]
- 13. Sbrana E., Xiao S. Y., Newman P. C., Tesh R. B. (2007) Comparative pathology of North American and central African strains of monkeypox virus in a ground squirrel model of the disease. Am. J. Trop. Med. Hyg. 76, 155–164 [PubMed] [Google Scholar]
- 14. Rubins K. H., Hensley L. E., Relman D. A., Brown P. O. Stunned silence: gene expression programs in human cells infected with monkeypox or vaccinia virus. PLoS One 6, e15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alkhalil A., Hammamieh R., Hardick J., Ichou M. A., Jett M., Ibrahim S. (2010) Gene expression profiling of monkeypox virus-infected cells reveals novel interfaces for host-virus interactions. Virol. J. 7, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rubins K. H., Hensley L. E., Bell G. W., Wang C., Lefkowitz E. J., Brown P. O., Relman D. A. (2008) Comparative analysis of viral gene expression programs during poxvirus infection: a transcriptional map of the vaccinia and monkeypox genomes. PLoS One 3, e2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shenolikar S. (2007) Analysis of protein phosphatases: toolbox for unraveling cell signaling networks. Methods Mol. Biol. 365, 1–8 [DOI] [PubMed] [Google Scholar]
- 18. Zhang H., Zha X., Tan Y., Hornbeck P. V., Mastrangelo A. J., Alessi D. R., Polakiewicz R. D., Comb M. J. (2002) Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J. Biol. Chem. 277, 39379–39387 [DOI] [PubMed] [Google Scholar]
- 19. Johnson R. F., Dyall J., Ragland D. R., Huzella L., Byrum R., Jett C., St Claire M., Smith A. L., Paragas J., Blaney J. E., Jahrling P. B. (2011) Comparative analysis of monkeypox virus infection of cynomolgus macaques by the intravenous or intrabronchial inoculation route. J. Virol. 85, 2112–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Piersma S. R., Labots M., Verheul H. M., Jiménez C. R. (2010) Strategies for kinome profiling in cancer and potential clinical applications: chemical proteomics and array-based methods. Anal. Bioanal. Chem. 397, 3163–3171 [DOI] [PubMed] [Google Scholar]
- 21. Bowick G. C., Fennewald S. M., Scott E. P., Zhang L., Elsom B. L., Aronson J. F., Spratt H. M., Luxon B. A., Gorenstein D. G., Herzog N. K. (2007) Identification of differentially activated cell-signaling networks associated with pichinde virus pathogenesis by using systems kinomics. J. Virol. 81, 1923–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bahar M. W., Graham S. C., Chen R. A., Cooray S., Smith G. L., Stuart D. I., Grimes J. M. (2011) How vaccinia virus has evolved to subvert the host immune response. J. Structural Biol. 175, 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seet B. T., Johnston J. B., Brunetti C. R., Barrett J. W., Everett H., Cameron C., Sypula J., Nazarian S. H., Lucas A., McFadden G. (2003) Poxviruses and immune evasion. Annu. Rev. Immunol. 21, 377–423 [DOI] [PubMed] [Google Scholar]
- 24. Jung M., Finnen R. L., Neron C. E., Banfield B. W. (2011) The alphaherpesvirus serine/threonine kinase us3 disrupts promyelocytic leukemia protein nuclear bodies. J. Virol. 85, 5301–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klerkx E. P., Lazo P. A., Askjaer P. (2009) Emerging biological functions of the vaccinia-related kinase (VRK) family. Histol. Histopathol. 24, 749–759 [DOI] [PubMed] [Google Scholar]
- 26. Valbuena A., Sanz-Garcia M., Lopez-Sanchez I., Vega F. M., Lazo P. A. (2011) Roles of VRK1 as a new player in the control of biological processes required for cell division. Cell. Signal. 23, 1267–1272 [DOI] [PubMed] [Google Scholar]
- 27. Chen N., Li G., Liszewski M. K., Atkinson J. P., Jahrling P. B., Feng Z., Schriewer J., Buck C., Wang C., Lefkowitz E. J., Esposito J. J., Harms T., Damon I. K., Roper R. L., Upton C., Buller R. M. (2005) Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology 340, 46–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Likos A. M., Sammons S. A., Olson V. A., Frace A. M., Li Y., Olsen-Rasmussen M., Davidson W., Galloway R., Khristova M. L., Reynolds M. G., Zhao H., Carroll D. S., Curns A., Formenty P., Esposito J. J., Regnery R. L., Damon I. K. (2005) A tale of two clades: monkeypox viruses. J. Gen. Virol. 86, 2661–2672 [DOI] [PubMed] [Google Scholar]
- 29. Jalal S., Kindrachuk J., Napper S. (2007) Phosphoproteome and kinome analysis: unique perspectives on the same problem. Curr. Anal. Chem. 3, 1–15 [Google Scholar]
- 30. Sopko R., Andrews B. J. (2008) Linking the kinome and phosphorylome–a comprehensive review of approaches to find kinase targets. Mol. Biosyst. 4, 920–933 [DOI] [PubMed] [Google Scholar]
- 31. Goff A. J., Chapman J., Foster C., Wlazlowski C., Shamblin J., Lin K., Kreiselmeier N., Mucker E., Paragas J., Lawler J., Hensley L. (2011) A novel respiratory model of infection with monkeypox virus in cynomolgus macaques. J. Virol. 85, 4898–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barry M., Hnatiuk S., Mossman K., Lee S. F., Boshkov L., McFadden G. (1997) The myxoma virus M-T4 gene encodes a novel RDEL-containing protein that is retained within the endoplasmic reticulum and is important for the productive infection of lymphocytes. Virology 239, 360–377 [DOI] [PubMed] [Google Scholar]
- 33. Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 34. Reeves P. M., Bommarius B., Lebeis S., McNulty S., Christensen J., Swimm A., Chahroudi A., Chavan R., Feinberg M. B., Veach D., Bornmann W., Sherman M., Kalman D. (2005) Disabling poxvirus pathogenesis by inhibition of Abl-family tyrosine kinases. Nat. Med. 11, 731–739 [DOI] [PubMed] [Google Scholar]
- 35. Secchiero P., Bosco R., Celeghini C., Zauli G. (2011) Recent advances in the therapeutic perspectives of nutlin-3. Curr. Pharm. Des. 17, 569–577 [DOI] [PubMed] [Google Scholar]
- 36. Secchiero P., di Iasio M. G., Gonelli A., Zauli G. (2008) The MDM2 inhibitor Nutlins as an innovative therapeutic tool for the treatment of haematological malignancies. Curr. Pharm. Des. 14, 2100–2110 [DOI] [PubMed] [Google Scholar]
- 37. Bálint E. E., Vousden K. H. (2001) Activation and activities of the p53 tumour suppressor protein. Br. J. Cancer 85, 1813–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chang F., Steelman L. S., Shelton J. G., Lee J. T., Navolanic P. M., Blalock W. L., Franklin R., McCubrey J. A. (2003) Regulation of cell cycle progression and apoptosis by the Ras/Raf/MEK/ERK pathway (Review). Int. J. Oncol. 22, 469–480 [PubMed] [Google Scholar]
- 39. Wang G., Barrett J. W., Stanford M., Werden S. J., Johnston J. B., Gao X., Sun M., Cheng J. Q., McFadden G. (2006) Infection of human cancer cells with myxoma virus requires Akt activation via interaction with a viral ankyrin-repeat host range factor. Proc. Natl. Acad. Sci. U.S.A. 103, 4640–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Soares J. A., Leite F. G., Andrade L. G., Torres A. A., De Sousa L. P., Barcelos L. S., Teixeira M. M., Ferreira P. C., Kroon E. G., Souto-Padrón T., Bonjardim C. A. (2009) Activation of the PI3K/Akt pathway early during vaccinia and cowpox virus infections is required for both host survival and viral replication. J. Virol. 83, 6883–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zachos G., Koffa M., Preston C. M., Clements J. B., Conner J. (2001) Herpes simplex virus type 1 blocks the apoptotic host cell defense mechanisms that target Bcl-2 and manipulates activation of p38 mitogen-activated protein kinase to improve viral replication. J. Virol. 75, 2710–2728 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42. Weaver J. R., Isaacs S. N. (2008) Monkeypox virus and insights into its immunomodulatory proteins. Immunol. Rev. 225, 96–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sedger L. M., Osvath S. R., Xu X. M., Li G., Chan F. K., Barrett J. W., McFadden G. (2006) Poxvirus tumor necrosis factor receptor (TNFR)-like T2 proteins contain a conserved preligand assembly domain that inhibits cellular TNFR1-induced cell death. J. Virol. 80, 9300–9309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wasilenko S. T., Banadyga L., Bond D., Barry M. (2005) The vaccinia virus F1L protein interacts with the proapoptotic protein Bak and inhibits Bak activation. J. Virol. 79, 14031–14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wasilenko S. T., Stewart T. L., Meyers A. F., Barry M. (2003) Vaccinia virus encodes a previously uncharacterized mitochondrial-associated inhibitor of apoptosis. Proc. Natl. Acad. Sci. U.S.A. 100, 14345–14350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hnatiuk S., Barry M., Zeng W., Liu L., Lucas A., Percy D., McFadden G. (1999) Role of the C-terminal RDEL motif of the myxoma virus M-T4 protein in terms of apoptosis regulation and viral pathogenesis. Virology 263, 290–306 [DOI] [PubMed] [Google Scholar]
- 47. Best S. M. (2008) Viral subversion of apoptotic enzymes: escape from death row. Annu. Rev. Microbiol. 62, 171–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Passarelli A. L. (2011) Barriers to success: how baculoviruses establish efficient systemic infections. Virology 411, 383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ryan E. J., Stevenson N. J., Hegarty J. E., O'Farrelly C. (2011) Chronic hepatitis C infection blocks the ability of dendritic cells to secrete IFN-alpha and stimulate T-cell proliferation. J. Viral. Hepat. 18, 840–851 [DOI] [PubMed] [Google Scholar]
- 50. Burgering B. M., Medema R. H. (2003) Decisions on life and death: FOXO Forkhead transcription factors are in command when PKB/Akt is off duty. J. Leukoc. Biol. 73, 689–701 [DOI] [PubMed] [Google Scholar]
- 51. Thornton T. M., Rincon M. (2009) Non-classical p38 map kinase functions: cell cycle checkpoints and survival. Int. J. Biol. Sci. 5, 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cappellini A., Tazzari P. L., Mantovani I., Billi A. M., Tassi C., Ricci F., Conte R., Martelli A. M. (2005) Antiapoptotic role of p38 mitogen activated protein kinase in Jurkat T cells and normal human T lymphocytes treated with 8-methoxypsoralen and ultraviolet-A radiation. Apoptosis 10, 141–152 [DOI] [PubMed] [Google Scholar]
- 53. Kurosu T., Takahashi Y., Fukuda T., Koyama T., Miki T., Miura O. (2005) p38 MAP kinase plays a role in G2 checkpoint activation and inhibits apoptosis of human B cell lymphoma cells treated with etoposide. Apoptosis 10, 1111–1120 [DOI] [PubMed] [Google Scholar]
- 54. Breman J. G., Kalisa-Ruti, Steniowski M. V., Zanotto E., Gromyko A. I., Arita I. (1980) Human monkeypox, 1970–79. Bull. World Health Organ. 58, 165–182 [PMC free article] [PubMed] [Google Scholar]
- 55. Brown J. N., Estep R. D., Lopez-Ferrer D., Brewer H. M., Clauss T. R., Manes N. P., O'Connor M., Li H., Adkins J. N., Wong S. W., Smith R. D. (2010) Characterization of macaque pulmonary fluid proteome during monkeypox infection: dynamics of host response. Mol. Cell. Proteomics 9, 2760–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jalal S., Arsenault R., Potter A. A., Babiuk L. A., Griebel P. J., Napper S. (2009) Genome to kinome: species-specific peptide arrays for kinome analysis. Sci. Signal. 2, pl1. [DOI] [PubMed] [Google Scholar]
- 57. Huber W., von Heydebreck A., Sultmann H., Poustka A., Vingron M. (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 Suppl 1, S96–104 [DOI] [PubMed] [Google Scholar]
- 58. Bloomfield V. (2009) Computer Simulation and Data Analysis in Molecular Biology and Biophysics: An Introduction Using R, Springer, Dodrecht Heidelberg London New York [Google Scholar]