Abstract
Membrane proteins play key roles in the development and progression of cancer. We have studied differentially expressed membrane proteins in glioblastoma multiforme (GBM), the most common and aggressive type of primary brain tumor, by high resolution LC-MS/MS mass spectrometry and quantitation by iTRAQ. A total of 1834 membrane proteins were identified with high confidence, of which 356 proteins were found to be altered by 2-fold change or more (198 up- and 158 down-regulated); 56% of them are known membrane proteins associated with major cellular processes. Mass spectrometry results were confirmed for representative proteins on individual specimens by immunohistochemistry. On mapping of the differentially expressed proteins to cellular pathways and functional networks, we notably observed many calcium-binding proteins to be altered, implicating deregulation of calcium signaling and homeostasis in GBM, a pathway also found to be enriched in the report (Dong, H., Luo, L., Hong, S., Siu, H., Xiao, Y., Jin, L., Chen, R., and Xiong, M. (2010) Integrated analysis of mutations, miRNA and mRNA expression in glioblastoma. BMC Syst. Biol. 4, 163) based on The Cancer Genome Atlas analysis of GBMs. Annotations of the 356 proteins identified by us with The Cancer Genome Atlas transcriptome data set indicated overlap with 295 corresponding transcripts, which included 49 potential miRNA targets; many transcripts correlated with proteins in their expression status. Nearly 50% of the differentially expressed proteins could be classified as transmembrane domain or signal sequence-containing proteins (159 of 356) with potential of appearance in cerebrospinal fluid or plasma. Interestingly, 75 of them have been already reported in normal cerebrospinal fluid or plasma along with other proteins. This first, in-depth analysis of the differentially expressed membrane proteome of GBM confirms genes/proteins that have been implicated in earlier studies, as well as reveals novel candidates that are being reported for the first time in GBM or any other cancer that could be investigated further for clinical applications.
Gliomas are primary tumors of the central nervous system with astrocytomas constituting 40% of them. Glioblastoma multiforme (GBM)1 is the most aggressive of the malignant astrocytomas. They occur mainly in adults with a mean survival period of 8–12 months. These aggressive tumors exhibit local metastasis and are resistant to current modalities of treatment, generally resulting in tumor recurrence (1).
Recent years have witnessed many genomics and transcriptomics efforts to understand molecular processes associated with GBM tumorigenesis (2). They have indicated gene signatures that may be associated with the treatment response, aggressive tumor groups, or tumor subtypes representing cellular heterogeneity. Applying next generation sequencing technology, massive parallel sequencing signature, Lin et al. (3) observed 4535 differentially expressed genes associated with GBM. Integration of these with independently published data sets suggested a 38-gene survival set and a 9-gene set associated with poor outcome. They also showed a positive association with markers of stem-like cells, including CD133 and nestin (4). In a large scale analysis of GBM by The Cancer Genome Atlas (TCGA) Network, a total of 601 genes was sequenced for detection of somatic mutations, and expression of over 12,000 genes was measured in more than 200 GBM cases to understand altered genes and pathways on the basis of copy number variations, mutation frequency analyses or differentially expressed transcripts including miRNAs (5, 6). Analysis based on TCGA transcriptome data revealed that GBMs could be subtyped into four different groups, each defined by a signature of 210 transcripts (7). In another effort, a differentially expressed gene signature of 214 genes characterized CD133-positive cancer stem cells studied against the negative cells from GBM. The gene signature was also identified for tumor aggressiveness and excessive mutations associated with the younger patient group from TCGA study (8).
Several proteomic studies have also been reported providing important insights in the pathophysiology of these tumors (1, 9). In our previous study, we compared whole tissue extracts from various grades of glioma with control specimens employing a two-dimensional electrophoresis-MS approach and identified 72 differentially expressed proteins, of which 27 were present in multiple tumor specimens (10). Iwadate et al. (11) analyzed various grades of gliomas by a two-dimensional electrophoresis-MS approach and identified possible markers that may correlate with survival of patients. Khalil and James (12) identified 97 differentially expressed proteins from whole tumor tissue extracts employing a more sensitive two-dimensional electrophoresis DIGE-MALDI-TOF/TOF approach. However, the two-dimensional gel-based quantitation approach is biased to identify relatively high abundant proteins and may miss low abundance regulatory proteins including membrane proteins. Gel-free approaches (LC-MS/MS) are being widely used for clinical tissue or cell line analysis. In a recent study, Melchior et al. (13) carried out a proteomic analysis of GBM tumor tissues by employing two-dimensional LC-MS/MS with both bottom-up and semi top-down approaches and identified 2660 unique proteins representing the GBM proteome. iTRAQ is a technology integrated with LC-MS analysis for measuring protein expression levels in complex samples (14). iTRAQ reagents contain isobaric tags designed to bind specifically to amine groups of peptides and enabling quantitation through measurement of relative intensities of the reporter ions generated upon MS/MS fragmentation.
Membrane proteins and their interactions with the surrounding microenvironment have direct implications in tumor proliferation and metastasis (15). Some earlier studies have focused on membrane proteome but using GBM cell lines and xenograft models (16, 17). The aim of the current study was to build a high confidence, tumor-associated, differentially expressed membrane protein panel from clinical specimens of GBM, as a first step toward their targeted validation in a clinical setting by us and others in the community. For this, we analyzed differentially expressed membrane proteins from the microsomal fraction of the clinical specimens of GBM using high resolution LC-MS/MS mass spectrometry and quantitation by iTRAQ. Proteins identified include several key members implicated in GBMs in earlier reports, as well as many functionally important novel proteins. The data set along with its extensive annotation with the published transcriptomic data sets reveals a number of differentially expressed membrane proteins of GBM that are functionally significant and potentially associated with GBM pathogenesis, which could be further investigated for clinical applications.
EXPERIMENTAL PROCEDURES
Sample Collection
Specimens were collected at the time of surgery and snap frozen in liquid nitrogen at Nizam's Institute of Medical Sciences, Hyderabad, India. All of the samples were collected with informed consent from the patients and approval of the institutional ethics committee. One part of the tissue was used for histopathology studies. Of over 100 surgical biopsies collected, 45 were astrocytomas, with 22 of them being grouped as glioblastomas based on clinical and histopathological evaluation as per World Health Organization guidelines. Of these, six samples were from subjects (three males and three females) of 50–60 years of age (the most susceptible group) and were selected for proteomic analysis. All were supratentorial glioblastomas mostly involving frontal lobe. Morphologically, all showed classic histological features of GBM including brisk mitosis, pleomorphism, necrosis, and microvascular proliferation. Immunohistochemistry for glial fibrallary acidic protein, vimentin and S100-B were positive in all cases with high Ki-67 labeling index. The frozen tissue was stored at −80 °C until used for proteomic analysis. Gliomas do not generally have defined surgical margins. For this reason, brain tissue obtained from epilepsy surgeries is generally used as experimental controls in research studies. Three samples used as controls in this study were from temporal lobe epilepsy, which occurs in young adult individuals (20–30 years). Multiple sections from the temporal neocortex were studied, both morphologically and immunohistochemically. The temporal cortex that was used as control did not show any abnormalities by light microscopy. Further, immunohistochemistry (IHC) with antibodies directed against phosphorylated neurofilament and synaptophysin proteins did not reveal any abnormal neurons in the cortex.
Subcellular Fractionation for Enrichment of Microsomal Proteins
For enrichment of membrane proteins, tissues from GBM patients (n = 6; three males and three females) or control subjects (n = 3; two males and one female) were pooled separately, and subcellular fractionation was carried out as described by Cox and Emili (18). This method yields a microsomal fraction that consists of endoplasmic reticulum, Golgi, intracellular vesicles, and plasma membrane proteins. Briefly, pooled tissue samples (1 g) were washed with phosphate-buffered saline, excised into small pieces, and then homogenized with a Dounce homogenizer (PISCO, Kolkata, India) in 3 ml of 250 STMDPS buffer (250 mm sucrose, 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 1 mm DTT, 25 μg ml−1 spermine, and 1 mm PMSF; Sigma-Aldrich). The homogenate was centrifuged at 4 °C, for 15 min at 800 × g, and supernatant (S1) was collected. The pellet was washed with 250 STMDPS buffer and centrifuged, and the supernatant (S2) was pooled with S1. The pooled supernatants were precentrifuged in the cold for 15 min at 6,000 × g followed by ultracentrifugation for 1 h at 1,00,000 × g. The pellet was resuspended in ME buffer (20 mm Tris-HCl, pH 7.8, 0.4 m NaCl, 15% glycerol, 1 mm DTT, 1 mm PMSF, and 1.5% Triton X-100; Sigma-Aldrich) and incubated for 60 min with gentle rocking at 4 °C followed by centrifugation for 30 min at 4 °C at 9,000 × g. The supernatant containing membrane proteins was stored at −80 °C. Protein amount was estimated using Bradford method and used for proteomic analysis to identify differentially expressed proteins.
iTRAQ Labeling
Membrane proteins from the pooled control or tumor samples were digested with trypsin, and the peptides were labeled with iTRAQ reagents according to the manufacturer's instructions (iTRAQ Reagents Multiplex kit; Applied Biosystems/MDS Sciex, Foster City, CA). Briefly, 80 μg of membrane proteins were vacuum-dried and resuspended in 20 μl of dissolution buffer and 1 μl of denaturant at room temperature. The samples were reduced, alkylated, and trypsinized (with 4 μg of modified sequencing grade trypsin; Promega, Madison, WI) for 16 h, at 37 °C. Trypsin-digested samples were labeled with four different iTRAQ reagents dissolved in 70 μl of ethanol at room temperature for 1 h. The reactions were quenched with glycine (10 mm). Sample labeling was as follows: tumor tissue samples with 114 and 116 tags and control samples with 115 and 117 tags. All of the four labeled samples were pooled, vacuum-dried, and subjected to fractionation through a strong cation exchange (SCX) column.
SCX Fractionation
The pooled sample after iTRAQ labeling was resuspended in 1 ml of buffer A (10 mm KH2PO4, 25% (v/v) ACN, pH 2.9) and separated on a SCX column (Zorbax 300-SCX; 5 μm, 2.1-mm inner diameter × 50 mm; Agilent Technologies, Santa Clara, CA) at a flow rate of 700 μl/min with a 40 min gradient (5 min, 0–5% buffer B (buffer A + 1 m KCl); 5 min, 5–10%; 5 min, 10–23%; 5 min, 23–50%; 10 min, 50–100%; 10 min, 100% B). One minute fractions were collected, vacuum-dried, and desalted using C18 cartridge (Pierce) as per the manufacturer's instructions. After desalting, consecutive fractions were pooled to obtain a total of eight fractions for LC-MS analysis. Comparable peptide quantities approximated from SCX chromatograms were taken for LC-MS analysis.
LC-MS/MS Analysis
Nanoflow electrospray ionization tandem mass spectrometric analysis of peptide samples was carried out using linear trap quadrupole OrbitrapVelos (Thermo Scientific, Bremen, Germany) interfaced with the Agilent 1200 Series nanoflow LC system. The chromatographic capillary columns used were packed with Magic C18 AQ (particle size, 5 μm; pore size, 100 Å; Michrom Bioresources, Auburn, CA) reversed phase material in 100% ACN at a pressure of 1000 p.s.i. The peptide sample from each SCX fraction was enriched using a trap column (75 μm × 2 cm) at a flow rate of 3 μl/min and separated on an analytical column (75 μm × 10 cm) at a flow rate of 350 nl/min. The peptides were eluted using a linear gradient of 7–30% ACN over 65 min. Mass spectrometric analysis was carried out in a data-dependent manner with full scans acquired using the Orbitrap mass analyzer at a mass resolution of 60,000 at 400 m/z. For each MS cycle, the 20 most intense precursor ions from a survey scan were selected for MS/MS, and fragmentation was detected at a mass resolution of 15,000 at m/z 400. The fragmentation was carried out using higher energy collision dissociation as the activation method with 40% normalized collision energy. The ions selected for fragmentation were excluded for 30 s. The automatic gain control for full FT MS was set to 1 million ions and for FT MS/MS was set to 0.1 million ions with a maximum time of accumulation of 500 ms, respectively. For accurate mass measurements, the lock mass option was enabled.
Protein Identification and Quantitation
The MS data were analyzed using Proteome Discoverer (Thermo Fisher Scientific; Beta Version 1.2.0.208). The workflow consisted of a spectrum selector and a reporter ion quantifier. MS/MS search was carried out using Sequest search algorithm against the National Center for Biotechnology Information RefSeq database (release 40) containing 31,811 proteins. Search parameters included trypsin as the enzyme with one missed cleavage allowed; oxidation of methionine was set as a dynamic modification, whereas alkylation at cysteine and iTRAQ modification at the N terminus of the peptide and lysine were set as static modifications. Precursor and fragment mass tolerance were set to 20 ppm and 0.1 Da, respectively. The peptide and protein data were extracted using high peptide confidence and top one peptide rank filters. Unique peptide(s) for each protein identified were used to determine relative protein content in the two samples. The false discovery rate was calculated by enabling the peptide sequence analysis using a decoy database. High confidence peptide identifications were obtained by setting a target false discovery rate threshold of 1% at the peptide level. Relative quantitation of proteins was carried out based on the relative intensities of reporter ions released during MS/MS fragmentation of peptides. Whenever a protein was identified with multiple peptides, relative intensities of the two reporter ions for each of the peptide identifiers for a protein were used for averaging and assessing the percentage of variability to determine relative quantity of a protein.
Bioinformatic Analysis
Bioinformatic analysis and annotations of the proteins identified were carried out based on their biological functions and cellular localizations as per Human Protein Reference Database (http://www.hprd.org) (19), which is in compliance with Gene Ontology standards. Pathway grouping was done using the Ingenuity Pathway Knowledge Base (Ingenuity Systems, Redwood City, CA). Deeper annotations in the context of cancer and GBMs were done by accessing specific published information and is provided as supplemental information.
Immunohistochemistry
Formalin-fixed paraffin-embedded individual GBM tissue sections or commercially available tissue microarrays (Folio Biosciences, Columbus, OH) with 35 GBM tissue cores and five cases of normal brain in duplicate were used for the IHC studies. The following primary antibodies were used: anti-ANXA2 mouse monoclonal, 1:300; anti-XRCC6 mouse monoclonal, 1:300; anti-SCARB2 mouse monoclonal, 1:200; anti-CAMKIIA mouse monoclonal, 1:200; anti-GOLIM4 rabbit polyclonal, 1:150; anti-RAB3A rabbit polyclonal, 1:50; and anti-SV2A rabbit polyclonal, 1:150. All of the monoclonal antibodies were obtained from Santa Cruz Biotechnologies and mono-specific polyclonal antibodies were from Sigma-Aldrich. Dilutions for primary and secondary antibody were optimized with the positive control tissues. After deparaffinization and rehydration, antigen retrieval was performed by immersing the slide in antigen retrieval buffer (10 mm sodium citrate, 0.05% Tween 20, pH 6.0) at 95 °C for 5 min using pressure cooker. Endogenous peroxidases were blocked with 0.03% hydrogen peroxide, and nonspecific binding was blocked with 2% fetal calf serum in Tris-buffered saline with 0.1% Triton X-100 (TBST, pH 7.6). The sections were then incubated for 1 h at room temperature with primary antibodies followed by peroxidase-labeled polymer conjugate to anti-mouse or anti-rabbit immunoglobulins for 1 h and developed with diaminobenzidine system. The sections were counter stained with the Mayer's hematoxylin and dehydrated, and the image was taken under microscope.
Western Blot Analysis
Membrane proteins extracted from pooled GBM or control tissues were resolved by SDS-PAGE (4–20% gradient precast gel; Invitrogen). The protein bands were electro transferred to a PVDF membrane (Millipore, Bedford, MA), blocked with 2% (v/v) BSA in TBST (150 mm NaCl, 20 mm Tris, 0.1% Tween 20, pH 7.4) for 2 h at room temperature, followed by incubation with primary antibody (anti-ATL3, 1:1000 (polyclonal rabbit, Abcam); anti-NEGR1, 1:1000 (polyclonal rabbit, Abcam); anti-PALM2, 1:200 (monoclonal mouse, Santa Cruz Biotechnologies); anti-calreticulin, 1:200 (polyclonal rabbit, Santa Cruz Biotechnologies); anti-β-actin, 1:1000, (polyclonal rabbit, Thermo-Scientific)) diluted with 1% BSA in TBST at room temperature for 2 h. After extensive wash with TBST, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody (anti-mouse or anti-rabbit; Bangalore Genei, India) diluted with 1% BSA in TBST for 90 min at room temperature. The membranes were developed using Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore). Calreticulin was used as a marker to observe membrane enrichment, whereas some of the selected proteins PALM2, NEGR1, and ATL3 were validated by Western blot analysis with actin as loading control.
RESULTS
We have analyzed the microsomal protein fraction isolated from pooled clinical specimens of GBM by LC-MS/MS using iTRAQ to generate data of differentially expressed membrane proteins. The overall workflow for the analysis is shown in Fig. 1A. This resource of the differentially expressed membrane proteins can then be extended in clinical experimental designs with individual specimens, in a targeted manner. To enrich the membrane proteins, subcellular fractionation was carried out, and microsomal fractions of the pooled tissues were prepared as described under “Experimental Procedures” (18). The microsomal fraction includes endoplasmic reticulum, Golgi, intracellular vesicles, and plasma membranes. Enrichment of membrane proteins was assessed by comparing the relative levels of the membrane protein calreticulin (55 kDa) in the microsomal and cytoplasmic fractions by Western blot analysis (Fig. 1B).
Fig. 1.
A, workflow for sample preparation and analysis of differentially expressed membrane proteins from GBM. Details of subcellular preparation of the membrane fraction, iTRAQ labeling, and SCX and nanoLC chromatography are provided under “Experimental Procedures.” B, Western blot of calreticulin showing ascertain membrane protein enrichment. The microsomal membrane protein preparation and the cytoplasmic proteins were immunoblotted using commercial calreticulin antibodies as described under “Experimental Procedures.”
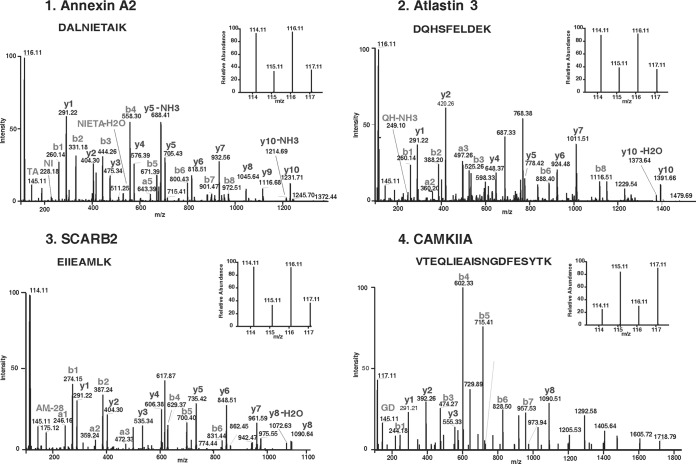
Equal amounts of protein from membrane fractions of the tumor and control specimens were digested with trypsin, and the tryptic peptides were labeled with iTRAQ reagents (114 and 116 for tumor specimens and 115 and 117 for the controls). This strategy provided internal technical replicates for the two types of samples. The iTRAQ-labeled peptide samples were pooled and fractionated using SCX column chromatography, and the eluted fractions (n = 8) were analyzed by LC-MS/MS. The data was searched against National Center for Biotechnology Information RefSeq database (version 40) using Protein Discoverer (version 1.2) using SEQUEST. A total of 14,637 iTRAQ-labeled peptides were identified that mapped to 1834 proteins, of which 1199 were based on ≥2 peptides and were included to assess differential expression. Fold changes were determined based on the ratios of the peak areas of iTRAQ reporter ions for the corresponding peptides from the tumor and control samples. In this manner, 643 proteins were identified with a threshold of 1.5-fold change (supplemental Table 1A). From this list, a subset of proteins with ≥2-fold changes was extracted and was found to comprise of 356 proteins (198 up-regulated and 158 down-regulated; supplemental Table 1B). Unless mentioned otherwise, we used this subset of 356 proteins for all subsequent bioinformatics analysis, biological annotations, and interpretation. The protein identifications were based on a minimum of two peptides for each protein. 73 proteins were identified with two peptides, 59 were identified with three peptides, and the remaining 224 proteins were identified with four or more peptides. We observed a high concordance between the two reporter ion intensities for a given peptide, for virtually all of the peptides identified. More than 90% of the peptides in the data set yielded at least 2-fold difference each. For obtaining an average fold change for a protein, we used all of the peptides identifying that protein. When we compared our data set with the differentially expressed GBM transcriptome recently published by TCGA group (7), we observed matches for more than 80% of the candidates (see below). Thus, taken together, we believe our protein identifications and quantifications are of high confidence and are consistent with the results of other transcriptome studies. MS/MS spectra along with reporter ions of peptides belonging to representative proteins are shown in Fig. 2.
Fig. 2.
MS/MS spectra of peptides with their reporter ions for representative differentially expressed proteins. Panel 1–3 show relative intensities of reporter ions for up regulated proteins annexin A2 (AnxA2), atlastin 3 (ATL3) and scavenger receptor B2 (SCARB2) respectively, and panel 4 shows the same for down regulated protein calmodulin-dependent protein kinase II alpha (CAMKIIA). MS and MS/MS spectra acquisition are described under “Experimental Procedures.”
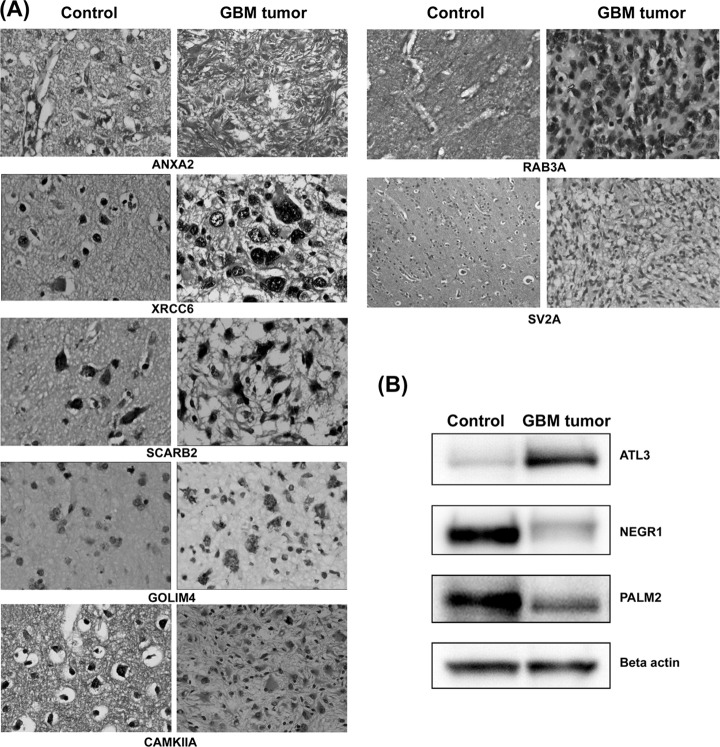
To confirm the differential expression observed by iTRAQ analysis, the expression levels of select proteins were further examined using IHC or Western blotting (if IHC-compatible antibodies were not available). We selected candidates on the basis of 1) the extent of differential expression, 2) functional significance of the protein, or 3) novelty as a differentially expressed protein (see “Discussion”). Seven proteins (ANXA2, XRCC6, SCARB2, GOLIM4, CAMKIIA, RAB3A, and SV2A) were thus selected, and IHC was performed with six individual cases. Comparison with the primary transcriptome data published by TCGA Network supports altered expression of these proteins (6). Other differentially expressed proteins such as atlastin 3 (ATL3), paralemmin 2 (PALM2), and neuronal growth regulator 1 (NEGR1), which have important biological functions, were examined by Western blots because of nonavailability of the IHC-compatible antibodies. IHC images and Western blot results are shown in Fig. 3 (A and B), and details of the IHC results from six individual patients are provided in supplemental Table 2. All of the proteins showed a positive correlation with the MS results.
Fig. 3.
A, immunohistochemistry of individual GBM tissue sections. Up-regulation of AnxA2, XRCC6, SCARB2, and GOLIM4 and down-regulation of CAMKIIA, RAB3A, and SV2A were confirmed in GBM specimens. IHC images of control brain sections and GBM tumor sections are shown. IHC protocol is described under “Experimental Procedures.” The staining and scoring details are shown in supplemental Table 3. B, Western blot analysis. Differential protein expression of atlastin 3 (ATL3), neuronal growth regulator 1 (NEGR1), and paralemmin 2 (PALM2) in GBM tissue is shown. β-Actin was used as protein loading control.
We further examined ANXA2, XRCC6, SCARB2, GOLIM4, and CAMKIIA proteins on commercial tissue microarrays (n = 35 in duplicates), for their expression across a larger set of GBM samples compared with normal samples. Except GOLIM4, four proteins: ANXA2, CAMKIIA, SCARB2, and XRCC6, showed differential expression in GBM tumor sections compared with normal samples. Across 35 GBM cases, moderate to strong staining was seen for ANXA2 in 26 cases, XRCC6 in 29 cases, SCARB2 in 23 cases, and GOLIM4 in 8 cases, whereas down-regulated protein CAMKIIA showed negative to mild staining in 33 cases. The staining intensity and distribution across the section was scored as negative (0), mild (1+), moderate (2+), or strong (3+). The distribution of staining was generally above 70% in most of the cases. The IHC details are provided in Table I.
Table I. Tissue microarray analysis of select differentially expressed proteins.
Commercial tissue microarrays with 35 GBM cases and five cases of normal brain in duplicate were used for the analysis. The details of scoring are given under “Results.”
| Protein | Proteomic data | Samples | No. of cases showing expression | Staining intensity |
|||
|---|---|---|---|---|---|---|---|
| 0 | 1+ | 2+ | 3+ | ||||
| ANXA2 | Up-regulated | Case | 26/35 | 9 | 0 | 7 | 19 |
| Control | 3 | 1 | 1 | 0 | |||
| XRCC6 | Up-regulated | Case | 34/35 | 1 | 5 | 11 | 18 |
| Control | 1 | 2 | 2 | 0 | |||
| SCARB2 | Up-regulated | Case | 26/35 | 9 | 3 | 12 | 11 |
| Control | 1 | 3 | 1 | 0 | |||
| GOLIM4 | Up-regulated | Case | 15/35 | 24 | 3 | 7 | 1 |
| Control | 4 | 1 | 0 | 0 | |||
| CAMKIIA | Down-regulated | Case | 5/35 | 32 | 1 | 2 | 0 |
| Control | 1 | 1 | 2 | 1 | |||
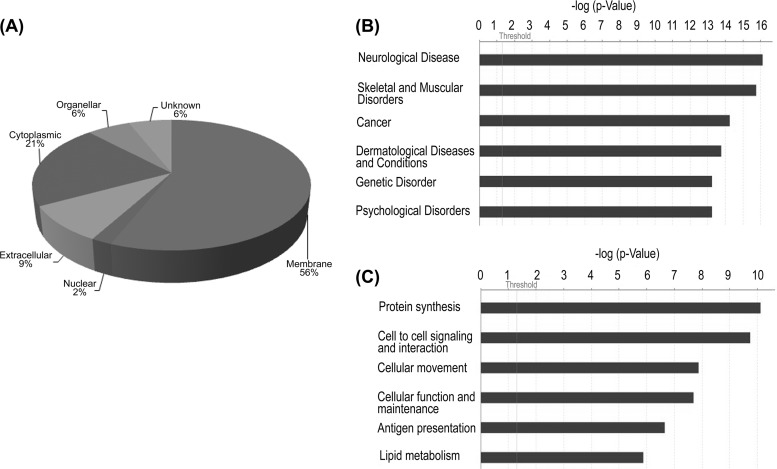
Subcellular classification of the 356 differentially expressed proteins using Human Protein Reference Database revealed that majority (56%) of them could be classified as membrane proteins with the rest representing other classes (Fig. 4A). To understand the biological significance of these molecules, we also analyzed these proteins using the Ingenuity Pathway Knowledge Base and identified networks, molecular and cellular processes, canonical pathways, and diseases and disorders that are most relevant to the data set. The top networks identified include cell to cell signaling and interaction, cell morphology, cellular function, and maintenance. The top three canonical pathways identified with this data set were acute phase response signaling, caveolar-mediated endocytosis signaling, and calcium signaling. Neurological diseases, cytoskelal and muscular disorders, and cancer were the major diseases identified, with nearly 50% of the proteins mapping on them (Fig. 4B). Protein synthesis, cell-to-cell signaling and interaction, cellular movement, and antigen presentation were the major molecular and cellular processes identified by Ingenuity Pathway Analysis (Fig. 4C). The protein IDs and p values associated with networks and canonical pathways are shown in Tables II and III. Those for molecular and cellular processes and diseases are shown in supplemental Table 3. Among the networks observed, many calcium-regulatory proteins were identified and include several known isoforms of S100 proteins (A4, 8, 9, 10, and 11) annexins (A1, 2, 4, and 5), and others such as integrins (ITGB1, ITGB2, ITGAM, and ITGA5) and receptors like MRC2. All of these proteins were overexpressed in our study.
Fig. 4.
A, subcellular classification of proteins identified from microsomal fraction based on annotation with reference to Human Protein Reference Database (www.hprd.org). B, disease association of altered proteins. C, major cellular processes found altered in GBM as assessed by the Ingenuity Pathway Knowledge base. Proteins listed in supplemental Table 1B were used for both of the analyses.
Table II. Ingenuity Pathway Analysis of the differentially expressed membrane proteins associated with major networks and processes (A).
Differentially expressed proteins from supplemental Table IA were used for the analysis and are shown in bold type. Only the top three networks are shown.
| Top functions | Associated molecules | Score | Focus molecules |
|---|---|---|---|
| Cell morphology, cellular development, cell-to-cell signaling and interaction | A2M, Akt, ANXA5, Ap1, ATP1A2, calmodulin, CAMP, CD44, CD63, CD74, CD163, CRMP1, EGFR, ERK1/2, F2, focal adhesion kinase, HLA-DR, HP, Hsp27, HSPA5, HSPB1, IQGAP1, KCNQ2, MAPT, p38 MAPK, PLG, PPIB, PRKAR2B, Rac, SNCA, SNCB, SNCG, SPARC, TNC, VTN | 33 | 26 |
| Cell morphology, cellular development, cancer | Actin, α-tubulin, APOC3, ATP1B1, CHI3L1, CRIP2, DPYSL2, eotaxin, ERK, FCER1G, FCGR3A, FLNA, hCG, histone h3, IgG, ITGA5, ITGB1, Jnk, LGALS3, MGST1, MSN, MVP, NAMPT, PDGF BB, PODXL, PTPRC, PTX3, RPL10A, S100A11, SLC4A4, TUBB2A, TUBB2C, VCL, VIM, YBX1 | 33 | 26 |
| Cellular function and maintenance, cell-to-cell signaling and interaction, hematological system development and function | 14-3-3, α-actinin, AMPH, ANXA1, APOH, B2M, BCAP31, C3, CALR, CAMK2D, CAMK2G, CANX, CLU, DNM1, Fibrinogen, HLA-A, HLA-B27, HLA-C, ICAM1, IL12 (complex), Immunoglobulin, ITGAM, ITGB2, LDL, LTF, MHC Class I (complex), MPO, NF-κB (complex), PACSIN1, PDIA3, RHOB, SLC2A5, Tap, TCR, TLN1 | 29 | 24 |
Table III. Ingenuity Pathway Analysis of the differentially expressed membrane proteins associated with canonical pathways.
Differentially expressed proteins from supplemental Table IA were used for the analysis and are shown in bold type. Only the top three pathways are shown.
| Canonical pathways | Associated molecules | p value | Ratio |
|---|---|---|---|
| Acute phase response signaling | HPX, FTL, ORM1/ORM2, C3, APOH, APOA1, APOA2, C9, CP, F2, FGG, PLG, ALB, HP, ITIH2, TF, C4B (includes others), FGB, A2M | 4.27 E-09 | 19/172 |
| Caveolar-mediated endocytosis signaling | ITGB1, B2M, ITGB2, ALB, ITGAM, HLA-A, ITSN1, FLNA, FLNC, ITGA5, EGFR, HLA-C | 4.57 E-08 | 12/81 |
| Calcium signaling | RAP2A, CALR, MYL6, ITPR2, TPM2, SLC8A2, ATP2B2, CAMK2A, CAMK2D, PRKAR2B, PPP3R1, MYH9, ASPH, TPM4, PPP3CA, CAMK2G, CAMK2B | 1.47 E-07 | 17/185 |
We also carried out a detailed comparison of our data with the published transcriptomic and protein data sets that are relevant to GBM pathogenesis, as well as their clinical and cellular heterogeneity. In a pilot project, TCGA Network group carried out a large scale study of gene expression, copy number variations, and mutation frequency to identify altered genes and pathways in GBMs (5). The transcriptome master list derived from TCGA data consisted of 11,861 genes with relative gene estimates (7). We compared our 356 differentially expressed protein set for the presence of the corresponding transcripts in this master list and found 295 matching transcripts (supplemental Table 4), including those for seven proteins highlighted and tested by IHC (see above). For each of the 295 matching transcripts, there are over 200 (corresponding to the number of specimens used) fold change data points, representing sample-wise differential expression (up, down, or no change) that varied widely across the specimens. Independently, using primary TCGA data, Dong et al. (6) generated a subset of 1697 differentially expressed transcripts with unified fold change values through statistical analysis. We also compared our differentially expressed protein data set with the corresponding gene expressions at the transcript level and found 105 of them to be positively correlated (Fig. 5 and supplemental Table 4); nine of them have been reported as cancer candidate genes (CHI3L1, MSN, RPN1, RTN1, SNAP25, SV2B, SYN2, SYT1, and VSNL1). Apparently, the remaining transcript matches, which were present in the primary data of TCGA (see above), did not pass the statistical evaluations because of their low frequency of occurrence across multiple samples. The analysis of Dong et al. (6) also identified calcium signaling pathway as the most enriched pathway with p value 4.45 × 10−5, a pathway also implicated from our proteomic data, as described above. Forty-nine of 356 proteins identified by us were found to correspond to targets of top 19 miRNAs reported in the study (supplemental Table 4). These included 15 targets for GBM-associated miRNAs and the rest for other cancer associated miRNAs: mir-155, mir-16, and mir-21.
Fig. 5.

Comparison of differential proteins identified in our study with TCGA transcriptome data for which unified differential expression estimates were available (6). A total of 105 of 106 differentially expressed proteins and the corresponding transcripts showed positive correlation, of which 40 were up-regulated (A) and 65 were down-regulated (B). Fold changes observed for proteomic and transcriptomic data were transformed to log 2 values and represented in the bar diagram.
Further, bioinformatic analysis to identify signal peptide or transmembrane (TM) domain containing proteins, using Signal P 3.0 and TMHMM 2.0 software, yielded a total of 159 proteins in this category (supplemental Table 5). Among the TM domain-containing proteins, 18 proteins are known to have cell receptor activity (Table IV) and include complement receptors, scavenger receptors, and integrin receptors. Macrophage scavenger receptor 1 (MSR1), scavenger receptor B2 (SCARB2), and CD163 were shown to be specifically expressed by tumor-associated macrophages indicating their presence in GBM tumor microenvironment (20). Further, signal/TM domain containing proteins have strong potential to be secreted or released into proximal fluids such as cerebrospinal fluid or blood plasma. When we examined our list of 356 proteins for their presence among the proteins reported in normal cerebrospinal fluid or plasma (21, 22), we found 77 detected in the cerebrospinal fluid and 163 in the plasma (supplemental Table 5), including 75 of 159 TM or signal sequence containing proteins. Some of the important ones include protease inhibitor, SERPINA3; brain-specific molecule tenascin-R (TNR) involved in cell adhesion; and tripeptidyl-peptidase I (TPP1), a serine protease, and tumor-associated proteins like SPARC and S100A9.
Table IV. Integral membrane proteins with receptor activity and plasma membrane localization identified in the analysis with reference to Human Protein Reference Database (www.hprd.org).
| Symbol | Protein name | Peptides | Fold change |
|---|---|---|---|
| MRC2 | Mannose receptor, C type 2 | 2 | 7.6 |
| FCER1G | Fc fragment of IgE, high affinity I, receptor for γ polypeptide precursor | 2 | 6.4 |
| FCGR3A | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) isoform d precursor | 2 | 5.0 |
| HLA-DRA | Major histocompatibility complex, class II, DR α precursor | 6 | 4.4 |
| CD44 | CD44 antigen isoform 4 precursor | 6 | 4.4 |
| CD74 | CD74 antigen isoform b | 3 | 4.4 |
| MSR1 | Macrophage scavenger receptor 1 isoform type 1 | 3 | 4.0 |
| HLA-C | Major histocompatibility complex, class I, C precursor | 6 | 3.9 |
| CD163 | CD163 antigen isoform b | 5 | 3.8 |
| ICAM1 | Intercellular adhesion molecule 1 precursor | 4 | 3.4 |
| EGFR | Epidermal growth factor receptor isoform b precursor | 3 | 3.3 |
| ITGB2 | Integrin, β2 precursor | 9 | 3.1 |
| HLA-A | Major histocompatibility complex, class I, A precursor | 5 | 3.0 |
| SCARB2 | Scavenger receptor class B, member 2 | 6 | 2.7 |
| PTPRC | Protein-tyrosine phosphatase, receptor type, C isoform 2 precursor | 12 | 2.6 |
| ITGB1 | Integrin β1 isoform 1A precursor | 9 | 2.6 |
| ITGA5 | Integrin α5 precursor | 2 | 2.2 |
| BCAP31 | B-cell receptor-associated protein 31 isoform b | 7 | 2.0 |
DISCUSSION
This analysis represents the first differentially expressed membrane proteome study of human glioblastoma specimens. When compared with the large scale transcriptome study by TCGA Network using more than 200 clinical GBM specimens (7), we observed corresponding transcript matches with more than 80% of the proteins identified (see “Results”). Extensive comparison of these proteins with literature and publically available large transcript or protein data sets reveal genes of regulatory importance, miRNA targets, and proteins with signal sequences and TM domains (supplemental Table 4). These comparisons provide support for some of the earlier observations, as well as reveal new molecules in the context of GBM; some of them are new even in the context of cancer, in general (supplemental Table 6). We believe that this high confidence data set has the merit for further clinical investigations of GBM.
The differentially expressed proteins include some of the major candidates already documented for GBM, for example, overexpression of epidermal growth factor receptor and chitinase 3 like 1 (CHI3L1), also known as YKL-40 (23, 24). Erythrocyte membrane protein (EPB41L3) implicated as a tumor suppressor (25); paralammin 2 (PALM2), a membrane protein involved in cell motility and morphogenesis (26); and neuronal growth regulator (NEGR1), a cell adhesion molecule preferentially expressed in brain and reported to be associated with ovarian and other cancers (27), were also found to be down-regulated in our study and are novel in the context of GBM. Other proteins such as ATP-dependent DNA helicase II (XRCC6), Golgi integral membrane protein (GOLIM4), scavenger receptor B2 (SCARB2), and atlastin 3 (ATL3) were found up-regulated and are entirely new in the context of any cancer. XRCC6, also known as KU-70, is a DNA repair protein with helicase activity localized in the nucleus (28) and also recently found to be present on cell surface of GBM cell lines and possibly involved in cell-to-cell interaction and migration (29). Atlastins (ATL3) belong to a family of GTPases localized on endoplasmic reticulum membranes and are considered to be membrane tethers and implicated in the maintenance of normal membrane structures (30, 31). The list also includes the well verified miRNA target proteins such as SCAMP1, RTN4, and BASP1 (which are down-regulated) for mir-155, -16, and -21, all known to be overexpressed in multiple cancers.
A linear comparison with the transcriptome-based altered pathways in GBM implied by the study of TCGA or other groups were not feasible because of the relative sizes and nature of the data sets. However, independent Ingenuity Pathway Analysis of the differentially expressed proteins revealed involvement of key molecules such as epidermal growth factor receptor, annexins, and integrins in cell to cell signaling and interaction networks, suggesting a complex tumor microenvironment. All of these proteins were found to be up-regulated in our study. Epidermal growth factor receptor overexpression promoting cell proliferation is well documented for GBM and many other cancers (22). It is linked to many cascading regulatory pathways including the RTK pathway identified in TCGA study (5). Annexins in general, have various intra- and extracellular roles in a range of cellular processes such as ANXA2 in cell signaling and ion transport and ANXA1 in cell division and apoptosis (32–34). Integrins are a large family of heterodimeric transmembrane glycoproteins and function as cell-matrix and cell-cell adhesion receptor molecules and regulate complex ion and molecular environment around the cells (35). We observed overexpression of integrins (ITGB1, ITGB2, ITGAM, and ITGA5) in the present study.
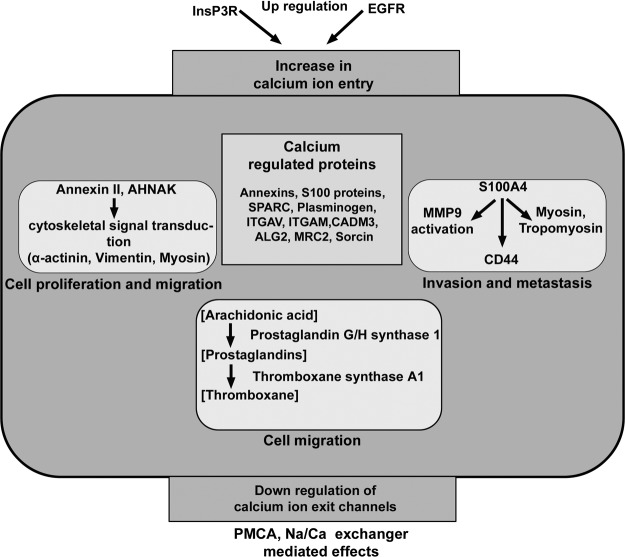
Three top canonical pathways revealed are acute phase response signaling, caveolar signaling, and calcium signaling (Table III). Acute phase response is a set of inflammatory response initiated by immune cells in the tumor microenvironment. Association of caveolar-mediated endocytosis and integrin recycling in cancer development and progression is known (36). Calcium ions act as secondary messengers and regulate various cellular processes through calcium-modulated proteins. It is interesting to note that calcium signaling, a major regulatory pathway mapped with our data set, was also found to be enriched in the differentially expressed transcriptome derived from TCGA data (6). Calcium channels and pumps localized on plasma membrane and endoplasmic reticulum regulate calcium ion levels in the cells important for many functional networks. We observed altered expression of several calcium-binding proteins in the present study. These proteins include calcium transporters, sensors, regulatory, and effector proteins. Malignant gliomas are also shown to have an overall increased formation of prostaglandins and thromboxanes, when compared with meningiomas and normal brain and may influence calcium ion levels (37). Up-regulation of the two enzymes thromboxane A synthase 1 (TBXAS1) and prostaglandin G/H synthase 1 (PTGS1) is consistent to speculate higher level of prostaglandin synthesis from arachidonic acid. Changes in prostaglandin synthesis are also associated in some other neurological disorders (38). Fig. 6 shows interplay of these molecules, network, and pathways, the overall result of which suggests elevation of calcium ions in the tumor cells. If so, it may be linked to cytotoxic effects like high necrosis observed in these tumors or linked to apoptosis process but limited by the other spatio-temporal parameters required to bring up the final effect. Nonetheless, consistent with the possible elevation of calcium ion levels as speculated, the analysis reveals deregulation in the family of calcium-binding proteins (Table V). They include several known isoforms of S100 proteins, annexins, and others like integrins (see “Results”). Among them, S100A4 is known to play role in tumor metastasis (39), and we find it up-regulated in our study. We also observe several known interacting partners of S100A4 and other S100 proteins to be differentially expressed (Table VI). For example, S100A4 is known to interact with tropomyosin (TPM4) and myosin (MYH9) (up-regulated) and suggested to affect the cytoskeleton of the tumor cells (39). A recent study reported that the ANXA2/S100A10 and AHNAK complex regulates actin cytoskeleton and cell membrane architecture (40). ANXA2 and AHNAK have also been shown to be downstream effectors for calcium-mediated signaling involved in cell proliferation and migration (41). The changes in the calcium-binding S100 proteins and their interactors imply cytoskeletal changes and processes favoring tumor cell proliferation and metastasis, in a calcium-dependent manner. Together, the above observations are indicative of deregulation of calcium homeostasis and calcium signaling, rendering it a hallmark of these tumors, analogous with other aggressive cancers (42). The protein subsets from altered pathways or interacting networks could be further investigated first for technical validation and then for clinical validations. Interestingly, calcium regulatory proteins ANXA2 and CAMKIIA tested in the tissue microarray analysis (Table I) have already shown encouraging results with 35 individual cases and also indicate the same trend at the transcript level in TCGA data (7, 6).
Fig. 6.
Scheme showing differentially expressed proteins identified and their known functional roles in calcium homeostasis and cancer-related events. Several calcium-binding proteins such as calcium transporters, sensors, and regulatory proteins found to be differentially expressed along with their association with tumor progression and metastasis is also shown. The overall effect suggested calcium ion elevation.
Table V. Differentially expressed calcium-binding proteins identified in the study.
The list was extracted from supplemental Table IA and assignment of calcium binding property was made with reference to Human Protein Reference Database (www.hprd.org).
| Symbol | Protein name | Peptides | Fold change |
|---|---|---|---|
| PLG | Plasminogen | 3 | 10.2 |
| MRC2 | Mannose receptor, C type 2 | 2 | 7.6 |
| S100A9 | S100 calcium-binding protein A9 | 7 | 6.2 |
| S100A10 | S100 calcium binding protein A10 | 2 | 6.0 |
| S100A8 | S100 calcium-binding protein A8 | 5 | 5.6 |
| ANXA5 | Annexin 5 | 14 | 5.3 |
| ANXA1 | Annexin I | 13 | 4.8 |
| ANXA2 | Annexin A2 isoform 2 | 17 | 3.8 |
| ITGAM | Integrin α M isoform 2 precursor | 7 | 3.5 |
| SPARC | Secreted protein, acidic, cysteine-rich precursor | 2 | 3.5 |
| MPO | Myeloperoxidase | 13 | 3.3 |
| CANX | Calnexin precursor | 16 | 3.2 |
| ITGB2 | Integrin, β2 precursor | 9 | 3.1 |
| PON2 | Paraoxonase 2 isoform 2 | 6 | 3.0 |
| CALU | Calumenin isoform a precursor | 9 | 2.8 |
| FGG | Fibrinogen, γ chain isoform γ-A precursor | 3 | 2.8 |
| ANXA4 | Annexin IV | 5 | 2.7 |
| ITGB1 | Integrin β1 isoform 1A precursor | 9 | 2.6 |
| TPM4 | Tropomyosin 4 isoform 2 | 9 | 2.5 |
| ITPR2 | Inositol 1,4,5-triphosphate receptor, type 2 | 4 | 2.4 |
| SSR1 | Signal sequence receptor, α precursor | 2 | 2.4 |
| RCN1 | Reticulocalbin 1 precursor | 6 | 2.4 |
| HSP90B1 | Heat shock protein 90-kDa β, member 1 precursor | 29 | 2.4 |
| F2 | Coagulation factor II preproprotein | 5 | 2.3 |
| ITGA5 | Integrin α5 precursor | 2 | 2.2 |
| LPCAT1 | Lysophosphatidylcholineacyltransferase 1 | 3 | 2.2 |
| ASPH | Aspartate β-hydroxylase isoform f | 4 | 2.2 |
| S100A4 | S100 calcium-binding protein A4 | 2 | 2.2 |
| CCDC47 | Coiled-coil domain containing 47 precursor | 5 | 2.2 |
| MYL6 | Myosin, light chain 6, alkali, smooth muscle and nonmuscle isoform 2 | 8 | 2.2 |
| LMAN2 | Lectin, mannose-binding 2 precursor | 6 | 2.1 |
| SRI | Sorcin isoform b | 4 | 2.1 |
| CAPN5 | Calpain 5 | 3 | 2.1 |
| CALR | Calreticulin precursor | 13 | 2.0 |
| ATP2B2 | Plasma membrane calcium ATPase 2 isoform 2 | 20 | 0.5 |
| SYT5 | Synaptotagmin V | 4 | 0.4 |
| CAMK2G | Calcium/calmodulin-dependent protein kinase II γ isoform 4 | 6 | 0.4 |
| ITSN1 | Intersectin 1 isoform ITSN-s | 2 | 0.4 |
| PPP3CA | Protein phosphatase 3, catalytic subunit, α isoform isoform 3 | 4 | 0.4 |
| SYP | Synaptophysin | 3 | 0.4 |
| SCG2 | Secretogranin II precursor | 4 | 0.4 |
| CADM3 | Cell adhesion molecule 3 isoform 2 | 5 | 0.4 |
| SLC8A2 | Solute carrier family 8 member 2 precursor | 12 | 0.4 |
| VSNL1 | Visinin-like 1 | 10 | 0.4 |
| PPP3R1 | Protein phosphatase 3, regulatory subunit B, α isoform 1 | 2 | 0.3 |
| SYT1 | Synaptotagmin I | 17 | 0.3 |
Table VI. Differentially expressed S100 proteins and their interactors (as per Human Protein Reference Database) identified in the analysis.
The S100 proteins marked in bold type, and target proteins are identified in our study.
| S100 proteins | Interacting proteins | Peptides | Fold change |
|---|---|---|---|
| S100A10 | Plasminogen | 3 | 10.1 |
| S100A8/A9 | Cytochrome B | 3 | 6.0 |
| S100B, S100A6, S100A1 | Annexin V | 14 | 5.3 |
| S100A11 | Annexin I | 13 | 4.8 |
| S100A10, S100A6, S100B | Annexin II | 17 | 3.8 |
| S100A8/A9, S100B | Vimentin | 22 | 3.2 |
| S100A1, S100B | GFAP | 21 | 2.8 |
| S100B | IQGAP1 | 8 | 2.5 |
| S100A4 | Myosin heavy chain | 70 | 2.5 |
| S100B | AHNAK | 32 | 2.4 |
| S100B | Calponin | 2 | 2.3 |
| S100A4, S100A6 | Tropomyosin | 5 | 2.0 |
| S100A4 | Septin6 | 8 | 0.5 |
| S100A1, S100B | Aldolase C | 11 | 0.4 |
| S100B | Neuromodulin | 10 | 0.4 |
| S100A4 | Septin7 | 11 | 0.4 |
| S100A13 | Synaptotagmin | 17 | 0.3 |
| S100A1 | Synapsin | 17 | 0.2 |
| S100A1 | MAPT | 7 | 0.1 |
Further, our analysis also reveals many TM domain- or signal sequence-containing proteins that have strong likelihood of being released in the cerebrospinal fluid and plasma (Table VII). YKL-40 included in this list is already shown by others as a serum marker for GBM (24). Other proteins included in the list are SPARC, S100A9, and SERPINA3, which are already implicated in GBM or other cancers (43–45). We are investigating many of these potential candidates for their differential status in these body fluids of GBM patients (work in progress). As discovery phase biomarker candidates go through the process of clinical validation, their number shrinks drastically on account of the clinical heterogeneity encountered. Generating a large portfolio of differentially expressed proteins of high confidence with functional relevance to tumor condition is important to meet this challenge, and we believe our analysis provides such leads in terms of functional protein groups and proteins with secretory potential with many of them being novel. They are thus important for further investigations in therapeutic or diagnostic perspectives.
Table VII. Differentially expressed proteins with signal sequence or transmembrane (TM) domains and their detectability in normal cerebrospinal fluid (CSF) or plasma.
The protein list was extracted from supplemental Table IA after comparison with data from Refs. 21 and 22.
| Gene symbol | Protein name | Peptides | Fold | Signal/TM | CSF | Plasma |
|---|---|---|---|---|---|---|
| A1BG | α1B-Glycoprotein precursor | 3 | 3.2 | Signal | + | + |
| A2M | α2-Macroglobulin precursor | 17 | 2.2 | Signal | + | + |
| ALB | Albumin preproprotein | 36 | 3.7 | Signal | + | + |
| APOA1 | Apolipoprotein A-I preproprotein | 11 | 3.1 | Signal | + | + |
| APOA2 | Apolipoprotein A-II preproprotein | 2 | 2.3 | Signal | + | + |
| APOC1 | Apolipoprotein C-I precursor | 3 | 2.2 | Signal | + | + |
| APOC3 | Apolipoprotein C-III precursor | 2 | 2.3 | Signal | + | + |
| ARHGDIB | Rho GDP dissociation inhibitor β | 2 | 3.5 | TM | + | |
| ATP1A3 | Na+/K+-ATPase α3 subunit | 42 | 0.4 | TM | + | |
| B2M | β2-microglobulin precursor | 2 | 5.8 | Signal | + | + |
| BCAN | Brevican isoform 1 | 5 | 0.4 | TM | + | + |
| C3 | Complement component 3 precursor | 19 | 2.4 | Signal | + | + |
| C4B | Complement component 4B preproprotein | 10 | 2.3 | Signal | + | |
| C9 | Complement component 9 precursor | 3 | 2.2 | TM | + | + |
| CALR | Calreticulin precursor | 13 | 2.0 | Signal | + | + |
| CALU | Calumenin isoform a precursor | 9 | 2.8 | Signal | + | |
| CANX | Calnexin precursor | 16 | 3.2 | Signal | + | |
| CD163 | CD163 antigen isoform b | 5 | 3.8 | Signal | + | + |
| CD44 | CD44 antigen isoform 4 precursor | 6 | 4.4 | Signal | + | + |
| CHI3L1 | Chitinase 3-like 1 precursor | 3 | 14.7 | Signal | + | + |
| CLU | Clusterin isoform 2 | 9 | 2.2 | Signal | + | + |
| CNTNAP2 | Cell recognition molecule Caspr2 precursor | 3 | 0.5 | TM | + | |
| CP | Ceruloplasmin precursor | 12 | 4.1 | Signal | + | + |
| EGFR | Epidermal growth factor receptor isoform b precursor | 3 | 3.3 | Signal | + | + |
| F2 | Coagulation factor II preproprotein | 5 | 2.3 | Signal | + | + |
| FCGR3A | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) isoform d precursor | 2 | 5.0 | TM | + | |
| FGB | Fibrinogen, β chain preproprotein | 3 | 2.0 | TM | + | + |
| FLNA | Filamin A, α isoform 1 | 31 | 2.9 | TM | + | + |
| GALNT2 | Polypeptide N-acetylgalactosaminyltransferase 2 | 2 | 3.1 | TM | + | + |
| GANAB | Neutral α-glucosidase AB isoform 2 | 15 | 2.2 | Signal | + | |
| GLUD1 | Glutamate dehydrogenase 1 precursor | 14 | 0.5 | Signal | + | |
| GOLIM4 | Golgi integral membrane protein 4 | 4 | 5.6 | Signal | + | |
| GOT2 | Aspartate aminotransferase 2 precursor | 5 | 0.5 | Signal | + | + |
| HLA-A | Major histocompatibility complex, class I, A precursor | 5 | 3.0 | Signal | + | + |
| HLA-C | Major histocompatibility complex, class I, C precursor | 6 | 3.9 | Signal | + | |
| HLA-DRA | Major histocompatibility complex, class II, DR α precursor | 6 | 4.4 | TM | + | |
| HP | Haptoglobin isoform 1 preproprotein | 9 | 3.4 | Signal | + | |
| HSP90B1 | Heat shock protein 90-kDa β, member 1 precursor | 29 | 2.4 | TM | + | + |
| ICAM1 | Intercellular adhesion molecule 1 precursor | 4 | 3.4 | Signal | + | |
| ITGA5 | Integrin α5 precursor | 2 | 2.2 | TM | + | |
| ITGB1 | Integrin β1 isoform 1A precursor | 9 | 2.6 | TM | + | + |
| L1CAM | L1 cell adhesion molecule isoform 3 precursor | 16 | 0.5 | TM | + | |
| LAMP2 | Lysosomal-associated membrane protein 2 isoform A precursor | 3 | 2.2 | TM | + | + |
| LMAN2 | Lectin, mannose-binding 2 precursor | 6 | 2.1 | TM | + | + |
| MLEC | Malectin precursor | 5 | 2.4 | TM | + | |
| MPO | Myeloperoxidase | 13 | 3.3 | Signal | + | |
| MRC2 | Mannose receptor, C type 2 | 2 | 7.6 | TM | + | + |
| MSN | Moesin | 19 | 3.9 | TM | + | + |
| MYH9 | Myosin, heavy polypeptide 9, nonmuscle | 70 | 2.5 | TM | + | |
| NCAM1 | Neural cell adhesion molecule 1 isoform 2 | 20 | 0.4 | TM | + | + |
| NEGR1 | Neuronal growth regulator 1 precursor | 5 | 0.5 | Signal | + | + |
| OPCML | Opioid binding protein/cell adhesion molecule-like isoform b preproprotein | 3 | 0.5 | Signal | + | + |
| ORM1 | Orosomucoid 1 precursor | 6 | 4.9 | Signal | + | + |
| PDIA3 | Protein-disulfide isomerase A3 precursor | 19 | 2.3 | Signal | + | |
| PDIA6 | Protein-disulfide isomerase A6 precursor | 9 | 2.6 | Signal | + | |
| PLG | Plasminogen | 3 | 10.2 | Signal | + | + |
| PODXL | Podocalyxin-like isoform 2 precursor | 3 | 3.3 | Signal | + | |
| PTPRC | Protein-tyrosine phosphatase, receptor type, C isoform 2 precursor | 12 | 2.6 | TM | + | |
| PTPRZ1 | Protein-tyrosine phosphatase, receptor-type, ζ1 precursor | 8 | 0.5 | TM | + | |
| RTN4 | Reticulon 4 isoform C | 4 | 0.2 | Signal | + | |
| RTN4 | Reticulon 4 isoform B | 6 | 3.2 | TM | + | |
| S100A8 | S100 calcium-binding protein A8 | 5 | 5.6 | TM | + | + |
| SERPINA3 | Serpin peptidase inhibitor, clade A, member 3 precursor | 2 | 3.0 | Signal | + | + |
| SLC2A1 | Solute carrier family 2 (facilitated glucose transporter), member 1 | 5 | 2.2 | TM | + | |
| SLC4A1 | Solute carrier family 4, anion exchanger, member 1 | 11 | 2.4 | TM | + | |
| SNCA | α-Synuclein isoform NACP112 | 5 | 0.3 | TM | + | |
| SPARC | Secreted protein, acidic, cysteine-rich precursor | 2 | 3.5 | Signal | + | + |
| TF | Transferrin precursor | 22 | 2.1 | Signal | + | + |
| TLN1 | Talin 1 | 7 | 2.0 | TM | + | + |
| TNC | Tenascin C precursor | 5 | 2.8 | TM | + | |
| TNR | Tenascin R precursor | 19 | 0.4 | TM | + | |
| TPP1 | Tripeptidyl-peptidase I preproprotein | 4 | 2.5 | Signal | + | + |
| VCL | Vinculin isoform VCL | 4 | 2.2 | TM | + | |
| VIM | Vimentin | 22 | 3.2 | TM | + | |
| VTN | Vitronectin precursor | 3 | 4.7 | Signal | + | + |
Acknowledgments
The Mass Spectrometry Facility at the Institute of Bioinformatics in Bangalore, India used for the analysis was established under support from the Department of Biotechnology, Govt. of India in a collaborative project with National Institute of Mental Health and Neurosciences, Bangalore, India. We gratefully acknowledge technical support in IHC studies from T. Avinash Raj from Centre for Cellular and Molecular Biology, Hyderabad, India and P. Madhavan from Nizam's Institute of Medical Sciences, Hyderabad, India. SRISTEK Hyderabad was involved in specimen collections, coordination, and clinical documentation.
Footnotes
* This work was supported by the Council of Scientific and Industrial Research, Government of India, under Network Project IAP 001 at the Centre for Cellular and Molecular Biology, Hyderabad, India. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- GBM
- glioblastoma multiforme
- iTRAQ
- isobaric tags for relative and absolute quantitation
- SCX
- strong cation exchange
- IHC
- immunohistochemistry
- TM
- transmembrane
- TCGA
- The Cancer Genome Atlas
- miRNA
- microRNA.
REFERENCES
- 1. Niclou S. P., Fack F., Rajcevic U. (2010) Glioma proteomics: Status and perspectives. J. Proteomics 73, 1823–1838 [DOI] [PubMed] [Google Scholar]
- 2. Sirdeshmukh R., Santosh V., Srikanth A. (2006) Differential protein expression, protein profiles of human gliomas and clinical implications. Bio Arrays: From Basics to Diagnostics (Appasani K., ed) 1st Ed., pp. 145–169, Humana Press, Totowa, NJ [Google Scholar]
- 3. Lin B., Madan A., Yoon J. G., Fang X., Yan X., Kim T. K., Hwang D., Hood L., Foltz G. (2010) Massively parallel signature sequencing and bioinformatics analysis identifies up-regulation of TGFBI and SOX4 in human glioblastoma. PLoS One 5, e10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Colman H., Zhang L., Sulman E. P., McDonald J. M., Shooshtari N. L., Rivera A., Popoff S., Nutt C. L., Louis D. N., Cairncross J. G., Gilbert M. R., Phillips H. S., Mehta M. P., Chakravarti A., Pelloski C. E., Bhat K., Feuerstein B. G., Jenkins R. B., Aldape K. (2010) A multigene predictor of outcome in glioblastoma. Neuro. Oncol. 12, 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. The Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dong H., Luo L., Hong S., Siu H., Xiao Y., Jin L., Chen R., Xiong M. (2010) Integrated analysis of mutations, miRNA and mRNA expression in glioblastoma. BMC Syst. Biol. 4, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Verhaak R. G., Hoadley K. A., Purdom E., Wang V., Qi Y., Wilkerson M. D., Miller C. R., Ding L., Golub T., Mesirov J. P., Alexe G., Lawrence M., O'Kelly M., Tamayo P., Weir B. A., Gabriel S., Winckler W., Gupta S., Jakkula L., Feiler H. S., Hodgson J. G., James C. D., Sarkaria J. N., Brennan C., Kahn A., Spellman P. T., Wilson R. K., Speed T. P., Gray J. W., Meyerson M., Getz G., Perou C. M., Hayes D. N.; Cancer Genome Atlas Research Network (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yan X., Ma L., Yi D., Yoon J. G., Diercks A., Foltz G., Price N. D., Hood L. E., Tian Q. (2011) A CD133-related gene expression signature identifies an aggressive glioblastoma subtype with excessive mutations. Proc. Natl. Acad. Sci. U.S.A. 108, 1591–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deighton R. F., McGregor R., Kemp J., McCulloch J., Whittle I. R. (2010) Glioma pathophysiology: Insights emerging from proteomics. Brain Pathol. 20, 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chumbalkar V. C., Subhashini C., Dhople V. M., Sundaram C. S., Jagannadham M. V., Kumar K. N., Srinivas P. N., Mythili R., Rao M. K., Kulkarni M. J., Hegde S., Hegde A. S., Samual C., Santosh V., Singh L., Sirdeshmukh R. (2005) Differential protein expression in human gliomas and molecular insights. Proteomics 5, 1167–1177 [DOI] [PubMed] [Google Scholar]
- 11. Iwadate Y., Sakaida T., Hiwasa T., Nagai Y., Ishikura H., Takiguchi M., Yamaura A. (2004) Molecular classification and survival prediction in human gliomas based on proteome analysis. Cancer Res. 64, 2496–2501 [DOI] [PubMed] [Google Scholar]
- 12. Khalil A. A., James P. (2007) Biomarker discovery: A proteomic approach for brain cancer profiling. Cancer Sci. 98, 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Melchior K., Tholey A., Heisel S., Keller A., Lenhof H. P., Meese E., Huber C. G. (2009) Proteomic study of human glioblastomamultiforme tissue employing complementary two-dimensional liquid chromatography- and mass spectrometry-based approaches. J. Proteome Res. 8, 4604–4614 [DOI] [PubMed] [Google Scholar]
- 14. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 15. Leth-Larsen R., Lund R. R., Ditzel H. J. (2010) Plasma membrane proteomics and its application in clinical cancer biomarker discovery. Mol. Cell. Proteomics 9, 1369–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seyfried N. T., Huysentruyt L. C., Atwood J. A., 3rd, Xia Q., Seyfried T. N., Orlando R. (2008) Up-regulation of NG2 proteoglycan and interferon-induced transmembrane proteins 1 and 3 in mouse astrocytoma: A membrane proteomics approach. Cancer Lett. 263, 243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rajcevic U., Petersen K., Knol J. C., Loos M., Bougnaud S., Klychnikov O., Li K. W., Pham T. V., Wang J., Miletic H., Peng Z., Bjerkvig R., Jimenez C. R., Niclou S. P. (2009) iTRAQ-based proteomics profiling reveals increased metabolic activity and cellular cross-talk in angiogenic compared with invasive glioblastoma phenotype. Mol. Cell. Proteomics 8, 2595–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cox B., Emili A. (2006) Tissue subcellular fractionation and protein extraction for use in mass-spectrometry-based proteomics. Nat. Protoc. 1, 1872–1878 [DOI] [PubMed] [Google Scholar]
- 19. Keshava Prasad T. S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A., Balakrishnan L., Marimuthu A., Banerjee S., Somanathan D. S., Sebastian A., Rani S., Ray S., Harrys Kishore C. J., Kanth S., Ahmed M., Kashyap M. K., Mohmood R., Ramachandra Y. L., Krishna V., Rahiman B. A., Mohan S., Ranganathan P., Ramabadran S., Chaerkady R., Pandey A. (2009) Human Protein Reference Database: 2009 update. Nucleic Acids Res. 37, D767–D772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mantovani A., Sozzani S., Locati M., Allavena P., Sica A. (2002) Macrophage polarization: Tumour-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23, 549–555 [DOI] [PubMed] [Google Scholar]
- 21. Zougman A., Pilch B., Podtelejnikov A., Kiehntopf M., Schnabel C., Kumar C., Mann M. (2008) Integrated analysis of the cerebrospinal fluid peptidome and proteome. J. Proteome Res. 7, 386–399 [DOI] [PubMed] [Google Scholar]
- 22. Farrah T., Deutsch E. W., Omenn G. S., Campbell D. S., Sun Z., Bletz J. A., Mallick P., Katz J. E., Malmström J., Ossola R., Watts J. D., Lin B., Zhang H., Moritz R. L., Aebersold R. (2011) A high-confidence human plasma proteome reference set with estimated concentrations in Peptide Atlas. Mol. Cell. Proteomics 10, M110.006353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bryant J. A., Finn R. S., Slamon D. J., Cloughesy T. F., Charles A. C. (2004) EGF activates intracellular and intercellular calcium signaling by distinct pathways in tumour cells. Cancer Biol. Ther. 3, 1243–1249 [DOI] [PubMed] [Google Scholar]
- 24. Tanwar M. K., Gilbert M. R., Holland E. C. (2002) Gene expression microarray analysis reveals YKL-40 to be a potential serum marker for malignant character in human glioma. Cancer Res. 62, 4364–4368 [PubMed] [Google Scholar]
- 25. Charboneau A. L., Singh V., Yu T., Newsham I. F. (2002) Suppression of growth and increased cellular attachment after expression of DAL-1 in MCF-7 breast cancer cells. Int. J. Cancer 100, 181–188 [DOI] [PubMed] [Google Scholar]
- 26. Kutzleb C., Sanders G., Yamamoto R., Wang X., Lichte B., Petrasch-Parwez E., Kilimann M. W. (1998) Paralemmin, a prenyl-palmitoyl-anchored phosphoprotein abundant in neurons and implicated in plasma membrane dynamics and cell process formation. J. Cell Biol. 143, 795–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ntougkos E., Rush R., Scott D., Frankenberg T., Gabra H., Smyth J. F., Sellar G. C. (2005) The IgLON family in epithelial ovarian cancer: Expression profiles and clinicopathologic correlates. Clin. Cancer Res. 11, 5764–5768 [DOI] [PubMed] [Google Scholar]
- 28. Jackson S. P. (2002) Sensing and repairing DNA double-strand breaks. Carcinogenesis 23, 687–696 [DOI] [PubMed] [Google Scholar]
- 29. Monferran S., Paupert J., Dauvillier S., Salles B., Muller C. (2004) The membrane form of the DNA repair protein Ku interacts at the cell surface with metalloproteinase 9. EMBO J. 23, 3758–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Orso G., Pendin D., Liu S., Tosetto J., Moss T. J., Faust J. E., Micaroni M., Egorova A., Martinuzzi A., McNew J. A., Daga A. (2009) Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460, 978–983 [DOI] [PubMed] [Google Scholar]
- 31. Rismanchi N., Soderblom C., Stadler J., Zhu P. P., Blackstone C. (2008) Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum. Mol. Genet. 17, 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gerke V., Moss S. E. (2002) Annexins: From structure to function. Physiol. Rev. 82, 331–371 [DOI] [PubMed] [Google Scholar]
- 33. Mussunoor S., Murray G. I. (2008) The role of annexins in tumour development and progression. J. Pathol. 216, 131–140 [DOI] [PubMed] [Google Scholar]
- 34. Gerke V., Creutz C. E., Moss S. E. (2005) Annexins: Linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 6, 449–461 [DOI] [PubMed] [Google Scholar]
- 35. Desgrosellier J. S., Cheresh D. A. (2010) Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 10, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shin S., Wolgamott L., Yoon S. (2012) Integrin trafficking and tumour progression. Int. J. Cell Biol. doi:10.1155/2012/516789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gaetani P., Butti G., Chiabrando C., Danova M., Castelli M. G., Riccardi A., Assietti R., Paoletti P. (1991) A study on the biological behavior of human brain tumours: Part I. Arachidonic acid metabolism and DNA content. J. Neurooncol. 10, 233–240 [DOI] [PubMed] [Google Scholar]
- 38. Hwang D., Lee I. Y., Yoo H., Gehlenborg N., Cho J. H., Petritis B., Baxter D., Pitstick R., Young R., Spicer D., Price N. D., Hohmann J. G., Dearmond S. J., Carlson G. A., Hood L. E. (2009) A systems approach to prion disease. Mol. Syst. Biol. 5, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mazzucchelli L. (2002) Protein S100A4: Too long overlooked by pathologists? Am. J. Pathol. 160, 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Benaud C., Gentil B. J., Assard N., Court M., Garin J., Delphin C., Baudier J. (2004) AHNAK interaction with the annexin 2/S100A10 complex regulates cell membrane cytoarchitecture. J. Cell Biol. 164, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang G., Kernan K. A., Thomas A., Collins S., Song Y., Li L., Zhu W., Leboeuf R. C., Eddy A. A. (2009) A novel signaling pathway: Fibroblast nicotinic receptor α1 binds urokinase and promotes renal fibrosis. J. Biol. Chem. 284, 29050–29064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Monteith G. R., McAndrew D., Faddy H. M., Roberts-Thomson S. J. (2007) Calcium and cancer: Targeting Ca2+ transport. Nat. Rev. Cancer 7, 519–530 [DOI] [PubMed] [Google Scholar]
- 43. Seno T., Harada H., Kohno S., Teraoka M., Inoue A., Ohnishi T. (2009) Downregulation of SPARC expression inhibits cell migration and invasion in malignant gliomas. Int. J. Oncol. 34, 707–715 [DOI] [PubMed] [Google Scholar]
- 44. Németh J., Angel P., Hess J. (2009) Dual role of S100A8 and S100A9 in inflammation-associated cancer. Anti-Inflamm. Anti-Allergy Agents Med. Chem. 8, 329–336 [Google Scholar]
- 45. Kloth J. N., Gorter A., Fleuren G. J., Oosting J., Uljee S., ter Haar N., Dreef E. J., Kenter G. G., Jordanova E. S. (2008) Elevated expression of SerpinA1 and SerpinA3 in HLA-positive cervical carcinoma. J. Pathol. 215, 222–230 [DOI] [PubMed] [Google Scholar]