Abstract
Thermococcus onnurineus NA1, a sulfur-reducing hyperthermophilic archaeon, is capable of H2-producing growth, considered to be hydrogenogenic carboxydotrophy. Utilization of formate as a sole energy source has been well studied in T. onnurineus NA1. However, whether formate can be used as its carbon source remains unknown. To obtain a global view of the metabolic characteristics of H2-producing growth, a quantitative proteome analysis of T. onnurineus NA1 grown on formate, CO, and starch was performed by combining one-dimensional SDS-PAGE with nano UPLC-MSE. A total of 587 proteins corresponding to 29.7% of the encoding genes were identified, and the major metabolic pathways (especially energy metabolism) were characterized at the protein level. Expression of glycolytic enzymes was common but more highly induced in starch-grown cells. In contrast, enzymes involved in key steps of the gluconeogenesis and pentose phosphate pathways were strongly up-regulated in formate-grown cells, suggesting that formate could be utilized as a carbon source by T. onnurineus NA1. In accordance with the genomic analysis, comprehensive proteomic analysis also revealed a number of hydrogenase clusters apparently associated with formate metabolism. On the other hand, CODH and CO-induced hydrogenases belonging to the Hyg4-II cluster, as well as sulfhydrogenase-I and Mbx, were prominently expressed during CO culture. Our data suggest that CO can be utilized as a sole energy source for H2 production via an electron transport mechanism and that CO2 produced from catabolism or CO oxidation by CODH and CO-induced hydrogenases may subsequently be assimilated into the organic carbon. Overall, proteomic comparison of formate- and CO-grown cells with starch-grown cells revealed that a single carbon compound, such as formate and CO, can be utilized as an efficient substrate to provide cellular carbon and/or energy by T. onnurineus NA1.
Hyperthermophilic archaea can use a wide variety of carbon and energy sources. Hyperthermophiles are widely distributed in extreme habitats such as deep-sea thermal vents, hot springs, and deep oil reservoirs (1–3). So far, the most frequently studied hyperthermophiles are from the genera Thermococcus and Pyrococcus, which belong to the order Thermococcales (4). These are ecologically important hyperthermophilic archaea for understanding the physiology and metabolic activity of microbial consortia within marine hot-water ecosystems. Members of the order Thermococcales are anaerobic heterotrophs that utilize various complex substrates with elemental sulfur (S0) or protons as electron acceptors (4–6). Unlike other Thermococcales, Thermococcus strain AM4 (7) and Thermococcus onnurineus NA1 (8) are capable of lithotrophic carbon monoxide-dependent hydrogenogenic growth. These Thermococcus strains use CO as a carbon and energy source by converting it into carbon dioxide (CO2). In addition, several hyperthermophilic archaea of the genus Thermococcus can grow with formate as an electron donor, producing hydrogen gas (9). T. onnurineus NA1 is a sulfur-reducing hyperthermophilic archaeon isolated from a deep sea hydrothermal vent area in the Eastern Manus Basin of Papua New Guinea (10). It is one of the more metabolically versatile hyperthermophiles in that it can use one-carbon (C1) substrates such as formate and CO, as well as multi-carbon substrates such as starch as the sole source of carbon and energy. Despite new insights into the mechanisms involved in the coupling of formate oxidation to energy conservation in T. onnurineus NA1 (9), the metabolic processes unique to growth on formate remain to be characterized. Typically, many hyperthermophiles are heterotrophs or grow on acetate. Most enzymes in the catabolic pathway of hyperthermophilic archaea have been identified and characterized. However, the anabolic pathway used by T. onnurineus NA1 to anaerobically convert these one-carbon substrates into cellular carbon is poorly understood. Therefore, study of one-carbon metabolism involved in H2-producing growth in T. onnurineus NA1 is of considerable interest because one-carbon substrates (i.e. methanol, carbon dioxide, and formate) are important for energy metabolism and carbon fixation pathways in some archaea (e.g. methanogenic archaea) and thus are expected to have many potential applications to industrial processes.
Recently, complete genome sequences have been determined for a number of Thermococcales, including three representative Pyrococcus species (11–13) and four Thermococcus strains: Thermococcus sibiricus (6), T. onnurineus NA1 (8), Thermococcus kodakaraensis KOD1 (14), and Thermococcus gammatolerans (15). Comparative genomic studies on these Thermococcales have paved the way for exploring the features and functions of genes involved in major metabolic pathways. Using a whole genome shotgun approach, the structure of the T. onnurineus NA1 genome was accurately determined, indicating that it contains 1,847,607 base pairs and 1976 predicted open reading frames (8). Moreover, genetic predictions based on these genomic data have provided clues for identifying genes correlated with biological functions (6, 8, 14). In particular, genomic analysis of T. onnurineus NA1 revealed several distinct hydrogenase gene clusters involved in H2 metabolism and carboxydotrophic pathways (16). Compared with genome analysis, only a handful of proteomic studies have been performed on Thermococcus strains. In T. gammatolerans, the first archaeal genome-wide proteome investigation was performed using an LC-MS/MS shotgun approach at the primary genome annotation stage (15). This study demonstrated that T. gammatolerans utilizes various metabolic pathways during growth in nutrient-rich medium (15). However, few studies have examined changes in the protein expression profile of T. onnurineus NA1 (17, 18). A previous 2-DE/MS-MS proteome analysis provided the first global view of the metabolic pathways of T. onnurineus NA1 during heterotrophic growth (17). Using an SDS-PAGE/LC-MS/MS shotgun proteomic approach, we recently made significant progress in identifying the metabolic enzymes specific for hydrogen production under carboxydotrophic culture conditions (18). Although recent genome analyses and proteomic characterizations have revealed general genomic features and metabolic pathways, a comprehensive understanding of the overall metabolism of T. onnurineus NA1 is lacking. Furthermore, a quantitative comparative analysis of all enzymes needed for central carbon flow, electron transfer, and/or energy conservation under different growth conditions has not been published.
Here we describe a quantitative proteome analysis of T. onnurineus NA1 cells using one-dimensional SDS-PAGE coupled with nano-UPLC-MSE.1 We examined the metabolism of T. onnurineus NA1 during H2-producing growth on different substrates. The major metabolic pathways proposed from a genomic analysis were characterized by comparing the protein expression profiles of cultures grown in the presence of formate, CO, and starch as sole carbon and/or energy sources. For the first time, we can draw a more complete metabolic picture of T. onnurineus NA1 at the protein level, which may facilitate a greater understanding of the metabolic adaptation of hyperthermophiles to extreme environments.
EXPERIMENTAL PROCEDURES
Strain and Culture Conditions
T. onnurineus NA1 (10) was cultured in yeast extract/peptone/sulfur medium that contained 35.0 g/liter NaCl, 3.3 g/liter Na2SO4, 0.05 g/liter KCl, 0.02 g/liter H3BO3, 8.8 g/liter MgCl2·6H2O, 0.05 g/liter CaCl2·2H2O, 0.61 g/liter N-(1,1-dimethyl-2-hydroxyethyl)-3-amino-2-hydroxypropanesulfonic acid, 0.001 g/liter resazurin, 3.0 g/liter yeast extract, 3.0 g/liter peptone, and 10.0 g/liter elemental sulfur. Trace elements and N-P mixture and Fe-EDTA solutions were added following Holden's method (1). To investigate the proteome during culture with CO, sodium formate, or soluble starch as substrates, we used modified M1 medium (7) that contained 35.0 g/liter NaCl, 0.7 g/liter KCl, 3.9 g/liter MgSO4, 0.4 g/liter CaCl2·2H2O, 0.3 g/liter NH4Cl, 0.15 g/liter Na2HPO4, 0.03 g/liter NaSiO3, 0.5 g/liter NaHCO3, 0.5 g/liter cysteine-HCl, 0.001 g/liter resazurin, and either CO (100% in the head space), 5.0 g/liter starch, or 5.0 g/liter formate. After autoclaving, medium was transferred into an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) and reduced by adding 0.005% (v/v) 5% (w/v) Na2S·9H2O, 1 ml/liter Holden's trace elements (1) and 1 ml/liter Balch's vitamin solution (19). The initial pH of the medium was adjusted to 8 at room temperature. T. onnurineus NA1 was cultured separately in four different batches for biological reliability, and the pooled samples were applied for proteome analysis.
Protein Extraction
Harvested cells were suspended in 20 mm Tris-HCl buffer (pH 8.0) and disrupted twice by sonication (Sonics Vibra Cell, Sonics & Materials INS, Newtown, CT) at 30 amplitude for 1 min. The supernatants (crude cell extracts) were collected by centrifugation at 15,000 rpm for 30 min and used for SDS-PAGE and LC-MS/MS.
SDS-PAGE and Trypsin Digestion
Protein content was quantified using a Bradford assay kit (Bio-Rad). Crude cell extracts were separated by 12% SDS-PAGE (Mini-Protean; Bio-Rad) according to a method described previously (20). The gels were stained with Coomassie Brilliant Blue R250, and 100 μg of proteins were loaded on the gel. The lanes were divided into eight fractions according to molecular weight, and slices were prepared for trypsin digestion (Promega, Madison, WI). The sliced gels were destained in a solution containing 50% ACN and 10 mm ammonium bicarbonate (ABC), and then rinsed with 100% ACN to stop the reaction. After drying in a speed vacuum concentrator (Mivac Quattro; Genevac Ltd.), the gels were incubated in a solution containing 10 mm DTT in 100 mm ABC at 56 °C for 1 h to reduce protein disulfide bonds and then in the same volume of 55 mm iodoacetamide in 100 mm ABC added to alkylate cysteines in the dark for 45 min. The gels were washed with three volumes of distilled water and dried using a speed vacuum concentrator. After swelling with 300 μl of 50 mm ABC, the proteins were digested with 15 μl of trypsin (0.1 μg/μl) at 37 °C for 18 h. The digested peptides were recovered by extracting twice with a solution containing 50 mm ammonium bicarbonate, 50% ACN, and 5% TFA. Peptide extracts were pooled at each gel line, lyophilized, and stored at −80 °C until required.
Analysis by Nano-UPLC-MSE Tandem Mass Spectrometry and Quantitative Analysis
Separations were performed on a nano-UPLC C18 RP column (75 μm × 250 mm; particle size, 1.7 μm) and an enrichment Symmetry C18 RP column (180 μm × 20 mm; particle size, 5 μm) using a nano-ACQUITY Ultra PerformanceTM chromatography system (Waters Corporation).
Tryptic-digested peptides (5 μl) were loaded onto the enrichment column with mobile phase A (3% acetonitrile in water with 0.1% formic acid). A step gradient was used at a flow rate of 300 nl/min. This included a 3–40% mobile phase B (97% acetonitrile in water with 0.1% formic acid) over 95 min and a 40–70% mobile phase B over 20 min, followed by a sharp increase to 80% B within 10 min. Sodium formate (1 μmol/min) was used to calibrate the TOF analyzer in the range of m/z 50–2000, and [Glu1]-fibrinopeptide (m/z 785.8426) was used for lock mass correction. During data acquisition, the collision energy of low energy MS mode and elevated energy mode (MSE) were set to 4 and 15–40 eV energy ramping mode, respectively. One cycle of MS and MSE was performed every 3.2 s. In each cycle, MS spectra were acquired for 1.5 s with a 0.1-s interscan delay (m/z 300–1990), and the ions exceeding 50 counts were selected for MSE fragmentation in the collision cell (m/z 50–2000).
LC-MSE data were processed and searched using ProteinLynx GlobalServer version 2.3.3 (Waters Corporation). Proteins were identified by searching the T. onnurineus NA1 database on the NCBI website (http://www.ncbi.nlm.nih.gov), including 4492 entries. For the ProteinLynx GlobalServer search, the parent ion tolerance was set at 100 ppm, and the fragment ion tolerance was set at 0.2 Da. Carbamidomethylation (+57 Da) of cysteine and methionine oxidation (+16 Da) were chosen as the fixed and variable modifications, respectively. Analysis of quantitative changes in protein abundance, based on measurements of peptide ion peak intensities observed in low collision energy mode (MS) in a triplicate set, was carried out using ExpressionTM software (version 2). To normalize comparative proteomic data, the “auto normalization” function of ProteinLynx GlobalServer was used. Protein identification was allowed only if the confidence was greater than 95% on the basis of the IDENTITYE algorithm (21). The Expression software generated results as an exact mass retention time table with quantitative protein and peptide values.
RESULTS AND DISCUSSION
H2-producing Growth on CO, Formate, and Starch
A previous study showed that T. onnurineus NA1 produced H2 when cultured with formate, CO, and starch as growth substrates (22). The highest level of growth and H2 production was observed in cultures grown on formate. The maximal yield of H2 was also achieved on formate or CO (22). These results indicate that formate and CO can serve as major energy sources for T. onnurineus NA1. We expected that T. onnurineus NA1 would exhibit specific responses to these single-carbon substrates, giving rise to changes in the proteome that would subsequently permit us to identify specific metabolic proteins involved in H2-producing growth.
Identification of Proteins Expressed during Growth on Different Substrates
To obtain a more complete picture of the metabolism involved in growth, a genome-wide proteome analysis of T. onnurineus NA1 grown on formate, CO, and starch was performed by one-dimensional SDS-PAGE sample fractionation and LC-MSE-based protein identification. Compared with previous approaches, the nano-UPLC-MSE method based on label-free quantitative analysis enables large scale comparison of all detectable proteins in complex samples separated by one-dimensional PAGE (21, 23).
Nano-UPLC-coupled Q-TOF tandem mass spectrometry analysis was used to identify, quantify, and compare individual proteins among CO-grown, formate-grown, and starch-grown cells. In total, 587 distinct proteins with more than two peptide fragmentations were identified at least twice across the experiment, which accounts for 29.7% of the 1976 genes predicted in the T. onnurineus NA1 genome. A list of identified proteins with quantitative data is provided in supplemental Table S1. Of the 587 proteins, we defined those whose expression was up-regulated ≥1.5 times or down-regulated ≤0.67 times with a p value < 0.05 as differentially regulated under the particular growth condition (Table I). A total of 447 proteins for starch versus CO-grown cells, 453 for starch versus formate-grown cells, and 422 for CO versus formate-grown cells were selected. Using our criterion (≥1.5-fold change), 91, 97, and 112 proteins were found to be differentially abundant in starch versus CO-grown, starch versus formate-grown, and CO versus formate-grown cells, respectively (Table I). These differentially abundant proteins were considered candidates for executing the predominant roles in the energy and carbon metabolisms induced by formate, CO, and starch.
Table I. Summary of quantitative proteomic analysis of Thermococcus onnurineus NA1.
| Substrate ratio (A/B) | CO/starch | Formate/starch | Formate/CO |
|---|---|---|---|
| p value < 0.05a | 282 | 278 | 252 |
| A/B ≥ 1.5 | 91 | 97 | 112 |
| A/B ≤ 0.67 | 148 | 140 | 99 |
| 1.5 ≥ A/B ≥ 0.67 | 43 | 41 | 41 |
| p value > 0.05 | 165 | 175 | 170 |
| Subtotal | 447 | 453 | 422 |
| Total | 587 |
a Proteins exclusively identified at a specific culture condition (CO or formate or starch) were included in the protein group of p values < 0.05.
The identified proteins were categorized by their predicted metabolic function (supplemental Table S1). The proteins were classified into 16 different categories based on the Kyoto Encyclopedia of Genes and Genomes database. Those that were significantly homologous with proteins of T. kodakaraensis KOD1 were classified into the same functional categories. However, the T. onnurineus NA1 proteome is not completely annotated; thus, 119 of the 587 identified proteins were assigned as “hypothetical.” Notably, large proportions (28, 21, and 27%) of the identified proteins in starch versus CO-grown, starch versus formate-grown, and CO versus formate-grown cells, respectively, were involved in energy metabolism. This result is consistent with the idea that T. onnurineus NA1 can grow under chemolithotrophic culture conditions by utilizing formate and/or CO as energy sources, because this organism possesses a large variety of metabolic pathways and enzymes involved in energy metabolism and H2 production.
Overview of General Metabolism in T. onnurineus NA1
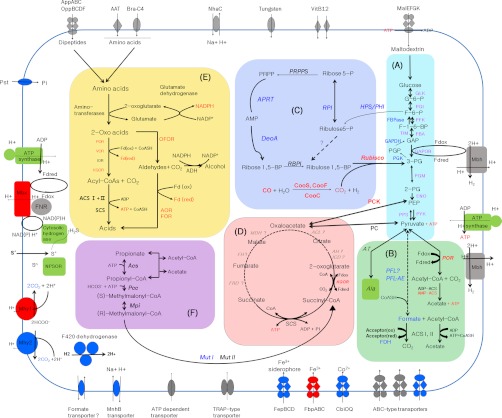
Comparative proteomics enabled us to unambiguously identify the major metabolic pathways utilized by T. onnurineus NA1 during growth on formate, CO, and starch as sources of energy and/or carbon. The proteome induced in starch was used as a control. An overview of the putative metabolic pathways is shown in Fig. 1. Our proteomic analysis identified most of the enzymes involved in the modified Embden-Meyerhof-Parnas pathway, the nonoxidative branch of the pentose phosphate pathway (PPP), amino acid degradation, aldehyde metabolism, and the pseudo-TCA cycle (Fig. 1 and Table II). These central metabolic pathways are similar to those proposed in genomic analyses of other Thermococcales (14, 15) but provide more information regarding which metabolic enzymes are specifically induced during growth on different substrates. In particular, the Embden-Meyerhof-Parnas pathway includes glyceraldehyde-3-phosphate dehydrogenase (TON_0639), 2-phosphoglycerate kinase (TON_0742), and FBPase (TON_1497), key enzymes in gluconeogenesis (24, 25); these enzymes were strongly up-regulated during growth on formate. Intriguingly, unlike the FBPase (Class V) of thermophilic Archaea including T. kodakaraensis KOD1 (26, 27), the recently characterized FBPase (TON_1497) from T. onnurineus NA1 has a different range of substrate specificities, although it has significant sequence similarity to T. kodakaraensis KOD1 (28). T. onnurineus NA1 also possesses key enzymes of the methylmalonyl pathway for propionate utilization, recently revealed in the thermophilic sulfur-reducing bacterium, Deferribacter desulfuricans SSM1 (29).
Fig. 1.
A putative scheme of the cellular metabolism in the hydrogen producing arachaeon T. onnurineus NA1. A, glycolysis and gluconeogenesis. B, pyruvate degradation. C, pentose phosphate synthesis and carbon dioxide fixation. D, pseudo-TCA cycle. E, amino acid degradation and aldehyde metabolism. F, propionate utilization. Dashed lines or faint italic letters represent pathways or enzymes not yet experimentally validated in Thermococcales species. Each protein with its predicted function in ion or solute transport, or H2 evolution or consumption, is illustrated on the membrane. Blue, red, and gray indicate proteins expressed prominently during growth on formate, CO, and starch, respectively. Major metabolic proteins deduced from the annotatable coding sequences (CDSs) were categorized by their induction on different substrates: formate (blue), CO (red), and starch (black). Other enzymes of the Embden-Meyerhof-Parnas pathway are designated by purple text, although they were mainly induced during growth on starch. Enzymes involved in sulfur reduction or ATP synthesis are shown in green, irrespective of their substrate specificity. Most proteins identified in T. onnurineus NA1 are shared by other Thermococcales: NPSOR (TON_0129), NADH:polysulfide oxidoreductase; FNR (TON_0056), ferredoxin-NADP(+) reductase; braC-4 (TON_0162), branched chain amino acid transporter; CbiOQ (TON_0243), ABC-type cobalt transport system; FepBCD (TON_0296), ABC-type Fe3+-siderophore transporter; FbpABC (TON_0983 and TON_0985), ABC-type Fe3+ transporter; Pst (TON_1464 and TON_1551), hypothetical phosphate transporter; NhaC (TON_1517), Na+/H+ antiporter; MnhB (TON_1577), multisubunit Na+/H+ antiporter; AppABC/OppBCDF (TON_1764 and TON_1768), ABC-type dipeptide/oligopeptide transporter; MalEFGK (TON_1791 and TON_1795), ABC-type maltodextrin transporter; AAT, additional amino acid ABC transporter; Fdred, reduced ferredoxin; Fdox, oxidized ferredoxin. The abbreviations of all of the identified proteins are presented separately in Table II and supplemental Table S1.
Table II. Relative abundance of the differentially expressed proteins in T. onnurineus NA1 cells grown on CO, formate, and starch.
S, starch; C, CO; F, formate; −, none detected in this experiment.
| Protein name | Gene identification | Ratioa |
p valueb |
||||
|---|---|---|---|---|---|---|---|
| CO/Starch | Formate/Starch | Formate/CO | CO/Starch | Formate/Starch | Formate/CO | ||
| Glycolysis and gluconeogenesis | |||||||
| ADP-dependent glucokinase (GLK) | TON_0246 | S | S | S | S | ||
| Glucose-6-phosphate isomerase (PGI) | TON_0247 | S | S | S | S | ||
| Phosphoenolpyruvate synthase (PPS) | TON_0311 | 0.54 | 0.57 | 1.05 | 0.00 | 0.00 | 0.88 |
| Cofactor-independent phosphoglycerate mutase (PGM) | TON_0442 | 0.64 | 0.84 | 1.32 | 0.00 | 0.11 | 0.94 |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | TON_0639 | C | F | 2.48 | C | F | 1.00 |
| 2-Phosphoglycerate kinase (PGK) | TON_0742 | F | F | - | F | F | |
| Pyruvate kinase (PYK) | TON_0785 | 0.56 | S | C | 0.15 | S | C |
| Fructose-bisphosphate aldolase (FBA) | TON_0903 | 0.06 | 0.12 | 1.92 | 0.00 | 0.00 | 1.00 |
| ADP-specific phosphofructokinase (PFK) | TON_1012 | S | S | S | S | ||
| Thermophile-specific fructose-1,6-bisphosphatase (FBPase) | TON_1497 | C | 1.73 | C | 1.00 | ||
| Tungsten-containing glyceraldehyde-3-phosphate:ferredoxin oxidoreductase (GAPOR) | TON_1498 | 0.12 | S | 0.00 | S | ||
| Phosphopyruvate hydratase (ENO) | TON_1613 | 0.59 | 0.65 | 1.11 | 0.00 | 0.00 | 0.96 |
| Triosephosphate isomerase (TIM) | TON_1640 | 0.32 | 0.44 | 1.39 | 0.00 | 0.00 | 0.98 |
| Pyruvate degradation | |||||||
| Glutamate dehydrogenase (GDH) | TON_0157 | 2.63 | 2.56 | 0.98 | 1.00 | 1.00 | 0.09 |
| Alanine aminotransferase (AT) | TON_0231 | 1.10 | 1.28 | 1.17 | 0.66 | 0.87 | 0.87 |
| Acetyl-CoA synthetase I (NDP forming), β subunit (ACSI_β) | TON_0327 | 0.98 | 1.27 | 1.30 | 0.40 | 1.00 | 1.00 |
| Pyruvate formate-lyase activating enzyme-related protein, radical SAM family (PFL-AE) | TON_0415 | S | S | S | S | ||
| Acetyl-CoA synthetase II (NDP forming), β subunit (ACSII_β) | TON_1002 | 1.27 | 1.39 | 1.11 | 1.00 | 1.00 | 0.91 |
| Acetyl-CoA synthetase I (NDP forming), α subunit (ACSI_α) | TON_1097 | 0.18 | 0.34 | 1.84 | 0.00 | 0.00 | 1.00 |
| Acetyl-CoA synthetase II (NDP forming), α subunit (ACSII_α) | TON_1313 | 1.43 | 1.82 | 1.27 | 1.00 | 1.00 | 1.00 |
| Pyruvate ferredoxin oxidoreductase subunit δ (POR_δ) | TON_1480 | C | C | C | C | ||
| Pyruvate ferredoxin oxidoreductase subunit α (POR_α) | TON_1481 | 1.04 | 0.70 | 0.68 | 0.88 | 0.00 | 0.00 |
| Pyruvate ferredoxin oxidoreductase subunit β (POR_β) | TON_1482 | 1.41 | 0.97 | 0.69 | 1.00 | 0.31 | 0.00 |
| Hypothetical formate dehydrogenase, α subunit (FDH_α) Fdh2 | TON_1563 | 0.74 | 12.50 | 17.46 | 0.00 | 1.00 | 1.00 |
| 4Fe-4S cluster-binding protein | TON_1564 | F | F | F | F | ||
| PFL-AE related proteins (radical SAM superfamily) | |||||||
| Hypothetical UPF0069 protein | TON_1269 | F | F | F | F | ||
| cmo tungsten-containing aldehyde ferredoxin oxidoreductase cofactor modifying protein | TON_1353 | F | F | F | F | ||
| Molybdenum cofactor biosynthesis protein A | TON_1410 | S | 1.59 | F | S | 0.97 | F |
| Hypothetical protein TON_1520 | TON_1520 | 0.41 | 0.58 | 1.39 | 0.00 | 0.00 | 0.99 |
| Pentose phosphate pathway | |||||||
| Adenine phosphoribosyltransferase (APRT) | TON_0120 | F | F | F | F | ||
| Ribose-5-phosphate isomerase A (RPI) | TON_0168 | S | 0.87 | F | S | 0.43 | F |
| Bifunctional d-arabino 3-hexulose-6-phosphate formaldehyde lyase/phosphohexuloisomerase (HPS/PHI) | TON_0336 | 0.21 | 0.48 | 2.27 | 0.00 | 0.00 | 0.99 |
| Thymidine phosphorylase (DeoA, AMP phosphorylase homolog) | TON_1062 | 2.22 | 2.27 | 1.02 | 1.00 | 1.00 | 0.64 |
| Ribulose bisophosphate carboxylase (RuBisCO) type III | TON_1234 | 1.19 | 1.04 | 0.87 | 1.00 | 0.73 | 0.00 |
| Translation initiation factor IF-2B subunit δ (RBPI Homolog) | TON_1296 | 0.70 | 0.56 | 0.79 | 0.00 | 0.00 | 0.02 |
| Ribose-phosphate pyrophosphokinase (PRPPS) | TON_1403 | S | S | S | S | ||
| TCA (CO2 fixation) | |||||||
| Phosphoenolpyruvate carboxykinase (PCK) | TON_0192 | 2.44 | 1.92 | 0.79 | 1.00 | 1.00 | 0.07 |
| 2-Oxoglutarate ferredoxin oxidoreductase subunit β (KGOR_β) | TON_0583 | 1.64 | S | C | 0.94 | S | C |
| 2-Oxoglutarate ferredoxin oxidoreductase subunit α (KGOR_α) | TON_0584 | C | C | C | C | ||
| 2-oxoglutarate ferredoxin oxidoreductase subunit β (KGOR_β) | TON_0586 | S | S | S | S | ||
| 2-oxoglutarate ferredoxin oxidoreductase subunit α (KGOR_α) | TON_0587 | C | C | C | C | ||
| Pyruvate carboxylase subunit B (PC) | TON_0904 | 0.54 | 0.53 | 0.98 | 0.02 | 0.18 | 0.47 |
| Ribulose bisophosphate carboxylase (RuBisCO) typeIII | TON_1234 | 1.19 | 1.04 | 0.87 | 1.00 | 0.73 | 0.00 |
| Pyruvate ferredoxin oxidoreductase subunit δ (POR_δ) | TON_1480 | C | C | C | C | ||
| Pyruvate ferredoxin oxidoreductase subunit α (POR_α) | TON_1481 | 1.04 | 0.70 | 0.68 | 0.88 | 0.00 | 0.00 |
| Pyruvate ferredoxin oxidoreductase subunit β (POR_β) | TON_1482 | 1.41 | 0.97 | 0.69 | 1.00 | 0.31 | 0.00 |
| Archaeal succinyl-CoA synthetase (NDP forming), large subunit (SCS) | TON_1665 | 0.74 | 0.88 | 1.21 | 0.00 | 0.15 | 0.92 |
| Propanoate metabolism | |||||||
| Methylmalonyl-CoA decarboxylase, α subunit (Pcc) | TON_0041 | 1.49 | 0.88 | 0.58 | 0.87 | 0.36 | 0.07 |
| Acetyl-CoA synthetase I (NDP forming), β subunit (ACSI_β) | TON_0327 | 0.98 | 1.27 | 1.30 | 0.40 | 1.00 | 1.00 |
| Methylmalonyl-CoA epimerase (Mpi) | TON_1074 | S | S | S | S | ||
| Hypothetical methylmalony-CoA mutase (MutI) | TON_1076 | F | F | F | F | ||
| Acetyl-CoA synthetase I (NDP forming), α subunit (ACSI_α) | TON_1097 | 0.18 | 0.34 | 1.84 | 0.00 | 0.00 | 1.00 |
| Methylmalonyl-CoA mutase, N terminus of large subunit (MutII) | TON_1110 | S | S | S | S | ||
| Amino acid degradation | |||||||
| Probable serine-glyoxylate aminotransferase, class V | TON_0061 | 1.43 | 1.85 | 1.28 | 0.98 | 1.00 | 0.95 |
| Glutamate dehydrogenase (GDH) | TON_0157 | 2.63 | 2.56 | 0.98 | 1.00 | 1.00 | 0.09 |
| Branched chain amino acid ABC transporter, periplasmic binding protein (braC-4) | TON_0162 | S | S | S | S | ||
| Alanine aminotransferase | TON_0231 | 1.10 | 1.28 | 1.17 | 0.66 | 0.87 | 0.87 |
| Acetyl-CoA synthetase I (NDP forming), β subunit (ACSI_β) | TON_0327 | 0.98 | 1.27 | 1.30 | 0.40 | 1.00 | 1.00 |
| Aspartate aminotransferase | TON_0565 | C | F | 1.14 | C | F | 0.73 |
| 2-Oxoglutarate ferredoxin oxidoreductase subunit β (KGOR_β) | TON_0583 | 1.64 | S | C | 0.94 | S | C |
| 2-Oxoglutarate ferredoxin oxidoreductase subunit α (KGOR_α) | TON_0584 | C | C | C | C | ||
| 2-Oxoglutarate ferredoxin oxidoreductase subunit β (KGOR_β) | TON_0586 | S | S | S | S | ||
| 2-Oxoglutarate ferredoxin oxidoreductase subunit α (KGOR_α) | TON_0587 | C | C | C | C | ||
| Acetyl-CoA synthetase II (NDP forming), β subunit (ACSII_β) | TON_1002 | 1.27 | 1.39 | 1.11 | 1.00 | 1.00 | 0.91 |
| Acetyl-CoA synthetase I (NDP forming), α subunit (ACSI_α) | TON_1097 | 0.18 | 0.34 | 1.84 | 0.00 | 0.00 | 1.00 |
| Hypothetical UPF0069 protein | TON_1269 | F | F | F | F | ||
| Multiple substrate aminotransferase | TON_1295 | 2.17 | 1.22 | 0.57 | 1.00 | 0.99 | 0.00 |
| Indolepyruvate oxidoreductase subunit β (IOR_β) | TON_1311 | 1.82 | 1.72 | 0.94 | 1.00 | 1.00 | 0.25 |
| Indolepyruvate:ferredoxin oxidoreductase, α subunit (IOR_α) | TON_1312 | 1.64 | 2.00 | 1.21 | 1.00 | 1.00 | 1.00 |
| Acetyl-CoA synthetase II (NDP forming), α subunit (ACSII_α) | TON_1313 | 1.43 | 1.82 | 1.27 | 1.00 | 1.00 | 1.00 |
| 2-Ketoisovalerate ferredoxin oxidoreductase subunit δ (VOR_δ) | TON_1477 | 1.92 | 1.02 | 0.53 | 1.00 | 0.52 | 0.00 |
| 2-Ketoisovalerate ferredoxin oxidoreductase subunit α (VOR_α) | TON_1478 | 1.32 | 0.94 | 0.72 | 1.00 | 0.07 | 0.00 |
| 2-Ketoisovalerate ferredoxin oxidoreductase subunit β (VOR_β) | TON_1479 | 1.37 | 0.95 | 0.69 | 1.00 | 0.22 | 0.00 |
| Pyruvate ferredoxin oxidoreductase subunit δ (POR_δ) | TON_1480 | C | C | C | C | ||
| Pyruvate ferredoxin oxidoreductase subunit α (POR_α) | TON_1481 | 1.04 | 0.70 | 0.68 | 0.88 | 0.00 | 0.00 |
| Pyruvate ferredoxin oxidoreductase subunit β (POR_β) | TON_1482 | 1.41 | 0.97 | 0.69 | 1.00 | 0.31 | 0.00 |
| 4-Aminobutyrate aminotransferase | TON_1605 | S | 3.23 | F | S | 1.00 | F |
| Archaeal succinyl-CoA synthetase (NDP forming), large subunit (SCS) | TON_1665 | 0.74 | 0.88 | 1.21 | 0.00 | 0.15 | 0.92 |
| ABC-type dipeptide/oligopeptide transport system (AppABC/OppBCDF) | TON_1764 | 0.39 | 0.64 | 1.63 | 0.00 | 0.00 | 1.00 |
| ABC-type dipeptide/oligopeptide transport system, ATPase component | TON_1768 | 0.92 | 1.41 | 1.54 | 0.48 | 0.78 | 0.75 |
| Diaminopimelate aminotransferase | TON_1785 | C | F | 1.67 | C | F | 0.85 |
| Aldehyde metabolism | |||||||
| Alcohol dehydrogenase (ADH) | TON_0544 | 0.58 | S | C | 0.00 | S | C |
| Formaldehyde:ferredoxin oxidoreductase (FOR) | TON_0749 | C | C | C | C | ||
| 2-Oxoacid:ferredoxin oxidoreductase, α subunit (OFOR_α) | TON_0860 | 1.89 | 1.43 | 0.76 | 1.00 | 0.97 | 0.04 |
| 2-Oxoacid ferredoxin oxidoreductase subunit β (OFOR_β) | TON_0861 | 2.50 | 1.20 | 0.48 | 1.00 | 0.82 | 0.00 |
| Aor-2 tungsten-containing aldehyde ferredoxin oxidoreductase (AOR) | TON_1354 | 4.76 | 0.54 | 0.11 | 1.00 | 0.00 | 0.00 |
| Formate-dependent hydrogenase and hydrogenase maturation system | |||||||
| ATPase involved in chromosome partitioning | TON_0262 | 0.60 | 0.88 | 1.48 | 0.00 | 0.12 | 0.98 |
| Hydrogenase maturation protease HycI | TON_0263 | F | F | F | F | ||
| Hydrogenase 4, component I or formate hydrogen lyase, subunit 7 (MhyI) | TON_0274 | S | S | S | S | ||
| Hydrogenase 4, component G or formate hydrogen lyase, subunit 5 (MhyI) | TON_0276 | 1.69 | C | 0.78 | C | ||
| Hydrogenase maturation protein HypF | TON_0286 | 0.70 | 0.44 | 0.64 | 0.00 | 0.00 | 0.00 |
| Hydrogenase expression/formation protein HypE | TON_0287 | 0.65 | 0.52 | 0.80 | 0.01 | 0.00 | 0.18 |
| Cysteine desulfurase | TON_0289 | 0.45 | 0.71 | 1.58 | 0.00 | 0.00 | 1.00 |
| Coenzyme F420 hydrogenase α subunit | TON_1559 | 0.44 | 2.00 | 4.57 | 0.00 | 1.00 | 1.00 |
| Coenzyme F420 hydrogenase/dehydrogenase β subunit | TON_1561 | 0.28 | 1.75 | 6.23 | 0.00 | 1.00 | 1.00 |
| TonB-dependent receptor protein:formate dehydrogenase, subunit FdhD | TON_1562 | F | F | F | F | ||
| Hypothetical formate dehydrogenase, α subunit (Fdh2) | TON_1563 | 0.74 | 12.50 | 17.46 | 0.00 | 1.00 | 1.00 |
| 4Fe-4S cluster-binding protein | TON_1564 | F | F | F | F | ||
| Hydrogenase 4, component G or formate hydrogen lyase, subunit 5 (MhyII) | TON_1569 | F | F | F | F | ||
| Formate hydrogen lyase subunit 6 (hydrogenase 3 component F) (MhyII) | TON_1570 | F | F | F | F | ||
| Hydrogenase 4, component I or formate hydrogen lyase, subunit 7 (MhyII) | TON_1571 | F | F | F | F | ||
| Hypothetical protein TON_1572 | TON_1572 | F | F | F | F | ||
| Hypothetical multisubunit Na+/H+ antiporter MnhB subunit (MnhB) | TON_1577 | F | F | F | F | ||
| CO-responsive hydrogenase and other membrane-bound hydrogenases | |||||||
| NADH dehydrogenase subunit D (Mbx) | TON_0487 | 0.70 | S | C | 0.22 | S | C |
| NADH dehydrogenase subunit C (Mbx) | TON_0488 | C | C | C | C | ||
| Cytosolic NiFe-hydrogenase, α subunit | TON_0534 | 1.75 | C | 1.00 | C | ||
| Cytosolic NiFe-hydrogenase, δ subunit | TON_0535 | C | C | C | C | ||
| Cytochrome-c3 hydrogenase subunit γ | TON_0536 | C | C | C | C | ||
| Sulfhydrogenase β subunit | TON_0537 | C | C | C | C | ||
| Hypothetical formate dehydrogenase, α subunit | TON_0539 | C | C | C | C | ||
| Oxidoreductase iron-sulfur protein | TON_0540 | C | C | C | C | ||
| Putative glutamate synthase subunit β | TON_0542 | 7.14 | C | 1.00 | C | ||
| 4Fe-4S cluster-binding protein | TON_0543 | C | C | C | C | ||
| Alcohol dehydrogenase (ADH) | TON_0544 | 0.58 | S | C | 0.00 | S | C |
| 4Fe-4S ferredoxin, iron-sulfur binding domain protein (CooF) | TON_1017 | C | 0.06 | C | 0.00 | ||
| Carbon-monoxide dehydrogenase, catalytic subunit (CooS) | TON_1018 | 100.00 | S | C | 1.00 | S | C |
| Hypothetical ATP-binding protein (CooC) | TON_1019 | C | C | C | C | ||
| Hydrogenase 4, subunit 5 (Mch) | TON_1023 | C | C | C | C | ||
| NADH dehydrogenase (ubiquinone), 20-kDa subunit (Mch) | TON_1024 | C | C | C | C | ||
| Membrane-bound hydrogenase, NiFe-hydrogenase large subunit 2 (Mbh) | TON_1593 | 0.44 | S | C | 0.00 | S | C |
| DNA metabolism | |||||||
| Hypothetical cell division control protein | TON_0043 | 2.50 | F | 0.88 | F | ||
| Archaeal histone A | TON_0185 | C | F | 0.71 | C | F | 0.00 |
| DNA primase | TON_0188 | 1.18 | 0.88 | 0.76 | 0.77 | 0.25 | 0.06 |
| Reverse gyrase | TON_0331 | F | 2.48 | F | 0.96 | ||
| Hypothetical AP endonuclease | TON_0382 | 0.82 | 1.08 | 1.31 | 0.23 | 0.59 | 0.81 |
| Single-stranded DNA-specific exonuclease | TON_0518 | F | F | F | F | ||
| MutS-like DNA mismatch repair ATPase | TON_0711 | F | F | F | F | ||
| DNA topoisomerase VI subunit B | TON_0764 | 0.65 | 0.68 | 1.05 | 0.15 | 0.22 | 0.58 |
| DNA polymerase sliding clamp (proliferating cell nuclear antigen homolog) | TON_0826 | 0.74 | 0.83 | 1.14 | 0.03 | 0.11 | 0.73 |
| Large helicase-related protein | TON_0937 | F | F | F | F | ||
| Predicted type II restriction endonuclease | TON_0989 | F | 1.31 | F | 0.67 | ||
| ERCC2/XPD/Rad3-related DNA repair helicase | TON_1040 | S | S | S | S | ||
| Hypothetical DNA methylase | TON_1221 | F | F | F | F | ||
| Type II restriction enzyme AccI | TON_1382 | F | F | F | F | ||
| ATPase involved in DNA replication | TON_1414 | 0.81 | 0.80 | 0.99 | 0.17 | 0.19 | 0.51 |
| Bipolar DNA helicase | TON_1420 | S | 1.27 | F | S | 0.80 | F |
| DNA repair exonuclease | TON_1421 | F | F | F | F | ||
| 5′-3′ nuclease | TON_1423 | F | F | F | F | ||
| Putative DNA-binding/iron metalloprotein/AP endonuclease | TON_1634 | F | F | F | F | ||
| DNA polymerase II large subunit | TON_1642 | S | 1.16 | S | 0.58 | ||
| DNA repair and recombination protein RadA | TON_1647 | 0.63 | 0.95 | 1.51 | 0.00 | 0.38 | 0.99 |
| DNA primase small subunit | TON_1777 | F | F | F | F | ||
| DNA helicase | TON_1882 | S | S | S | S | ||
| Hypothetical exonuclease SbcD | TON_1921 | F | F | F | F | ||
| Replication factor A complex, RPA32 subunit | TON_1952 | C | C | C | C | ||
| Replication factor A | TON_1954 | 0.68 | 1.12 | 1.67 | 0.03 | 0.78 | 1.00 |
| Endonuclease IV | TON_1961 | C | F | 3.90 | C | F | 0.98 |
a Ratio measured by MSE methods.
b p values calculated from individual peak intensities derived from all tryptic peptides detected per protein (see “Experimental Procedures”).
In the present study, we found that most hydrogenases, as well as enzymes involved in the stress response, were prominently expressed during growth on formate or CO (Table II and supplemental Table S1). Moreover, as recently described in other Thermococcales (14, 15), various ABC-type transporters, monovalent cation transporters that were differentially expressed during growth on formate, CO, and starch were identified. T. onnurineus NA1 harbors two TRAP-type transporter components (TON_0172 and TON_0174) in its genome, as does T. kodakaraensis KOD1 (14). In our proteomic analysis, only one TRAP-type transporter (TON_0174) was strongly up-regulated in cells grown on starch. The use of TRAP-type transporters might enable these bacteria to adapt to Na+-rich and nutritionally poor environments by lowering the energetic cost of catalyzing the uptake of specific substrates (30). In addition, a branched chain amino acid ABC transporter (braC-4, TON_0162) was expressed in T. onnurineus NA1 cells during growth on starch. As in T. gammatolerans (15), an ABC-type dipeptide/oligopeptide transporter system (TON_1764 and TON_1768) was also found in the T. onnurineus NA1 proteome.
In contrast, most of the proteins involved in DNA metabolism were up-regulated during growth on formate compared with starch (Table II and supplemental Table S1). Currently, we do not know why formate induces DNA repair proteins in T. onnurineus NA1. However, formic acid depurinates DNA in vitro, leading to the loss of a purine from the DNA backbone (31). In vivo, depurination causes the formation of apurinic or apyrimidinic sites in DNA, which can result in strand breaks or point mutations by adenine mispairing during DNA synthesis (32, 33). This depurination rate is greatly accelerated at low pH and elevated temperatures (34, 35). Thus, the addition of formate at a high temperature may lead to the inhibition of DNA synthesis by damaging the DNA molecule. However, the spontaneous mutation rates of thermophilic archaea are reported to be lower than those of other mesophilic bacteria, despite the higher chemical instability of their DNA (36, 37). Therefore, hyperthermophiles, including T. onnurineus NA1, seem to possess molecular strategies for DNA repair, to compensate for the intrinsic instability of DNA at their preferred growth temperatures and under acidic conditions (36, 38).
Gluconeogenesis and Pentose Phosphate Pathway
T. onnurineus NA1 can grow using formate as its sole source of energy, producing hydrogen gas as a by-product (9). However, it is not clear whether formate can be used as a carbon source, and little is known about the metabolic pathways of formate assimilation. In T. onnurineus NA1, a potential reaction for formate assimilation is direct or indirect conversion of formate and acetyl-CoA into pyruvate (Fig. 1), the reversible reaction catalyzed by pyruvate-formate lyase (PFL). PFL orthologs are ubiquitously distributed in bacteria, but PFL has only been found in archaea such as T. sibiricus (6), T. kodakaraensis KOD1 (14), D. desulfuricans SSM1 (29), Archaeoglobus fulgidus (39), and Methanobacterium thermoautotrophicum (40). Therefore, it is conceivable that assimilation of pyruvate may commence with formate and acetyl-CoA. This reaction is the first committed step of anaerobic glucose metabolism and is important in Escherichia coli and other facultative anaerobes (41, 42). Genomic analysis showed that no gene was annotated as PFL, suggesting that protein(s) with PFL activity in T. onnurineus NA1 may not be homologous to the PFLs of other archaea. Further study is necessary to identify PFL-like proteins in T. onnurineus NA1.
PFL is activated by pyruvate-formate lyase-activating enzyme (PFL-AE), which generates the stable glycyl radical at Gly-734 of PFL (43, 44). PFL-AE is a member of the novel enzyme family that utilizes S-adenosylmethionine to initiate radical catalysis (43, 44). TON_0415, which is a PFL-AE homolog belonging to the radical S-adenosylmethionine superfamily (EC 1.97.1.4), was strongly up-regulated during growth on starch, suggesting that TON_0415 contributes to glycolytic flux in the Embden-Meyerhof-Parnas pathway (Fig. 1 and Table II). This prompted us to inquire whether other isozymes of PFL-AE are present in T. onnurineus NA1. We pooled all of the identified proteins, investigated the domain assignments of the radical S-adenosylmethionine enzyme family in T. onnurineus NA1, and identified several possible PFL-AE candidates (TON_0789, TON_1269, TON_1353, TON_1410, and TON_1800). Of these, TON_1269, TON_1353, and TON_1410 were strongly up-regulated during growth on formate (Table II).
Pyruvate formed by the reverse reaction of PFL can then be converted into various biosynthetic intermediates by the action of enzymes involved in gluconeogenesis and the PPP. As shown in Fig. 1 and Table II, four enzymes involved in glycolysis were strongly up-regulated during growth on starch (GLK, TON_0246; PGI, TON_0247; PFK, TON_1012; GAPOR, TON_1498), whereas three enzymes involved in key steps of gluconeogenesis were robustly up-regulated in formate-grown cultures (glyceraldehyde-3-phosphate dehydrogenase, TON_0639; 2-phosphoglycerate kinase, TON_0742; FBPase, TON_1497). These data suggest that three gluconeogenic enzymes generate biosynthetic intermediates during growth on formate. More than seven enzymes of the PPP were identified (Fig. 1). Four related enzymes, ribose-5-phosphate isomerase (TON_0168), adenine phosphoribosyltransferase (TON_0120), AMP phosphorylase homolog DeoA (TON_1062), and HPS/PHI (TON_0336), were induced in response to formate (Fig. 1 and Table II), the exceptions being PRPPS (TON_1403) and an RBPI homolog (TON_1296). The relative levels of these enzymes among formate-, CO-, and starch-grown cells suggest that biosynthesis of PPP products is favored during growth on formate. These data collectively show that formate can be utilized as a source of organic carbon via gluconeogenesis and the PPP. However, HPS/PHI (TON_0336) was detected at a 2-fold lower abundance in formate-grown cells compared with starch-grown cells (Table II), indicating that HPS/PHI (TON_0336) expression during growth on formate can be suppressed when an energetically superior carbon source is present. Starch is a preferred substrate for growth so long as low levels of formate are provided. This process is reversible under certain conditions (e.g. glucose depletion) and may thus facilitate formate utilization for anabolic flux. Therefore, T. onnurineus NA1 can survive heterotrophically in environments with limited organic carbon, such as around hydrothermal vents. In addition, our observation that specific metabolic enzymes were elevated in response to formate provides the first evidence of the presence of a substrate-induced mechanism governing cellular metabolic flux in T. onnurineus NA1.
Pseudo Tricarboxylic Acid Cycle and Propionate Utilization
The T. onnurineus NA1 proteome indicated the presence of another metabolic pathway, the reductive tricarboxylic acid cycle. Two key enzymes are required to reverse the TCA cycle (reductive TCA cycle) in archaea: KGOR and ATP-dependent citrate lyase. Although ATP-dependent citrate lyase has not yet been detected in the T. onnurineus NA1 proteome, another key enzyme, KGOR (TON_0583–4 and TON_0586–7) was detected in this study. KGOR catalyzes the reductive carboxylation of succinyl-CoA to form 2-oxoglutarate using reduced ferredoxin. However, the reductive TCA cycle of T. onnurineus NA1 is incomplete because of the absence of malate dehydrogenase, fumarate hydratase, and fumarate reductase. Similar results were reported in genomic analyses of other Thermococcales (14, 15). Therefore, these cycles are called pseudo-TCA cycles. Despite the incompleteness of the reductive TCA cycle, our proteomic analysis showed significant up-regulation of proteins involved in CO-dependent anabolic flux for assimilation of CO2 via a reductive TCA cycle (29, 45–47), including pyruvate ferredoxin oxidoreductase (POR, TON_1480–82), phosphoenolpyruvae synthase (TON_0311), phosphoenolpyruvate carboxykinase (TON_0192), and KGOR (TON_0583–4 and TON_0586–7) as shown in Fig. 1. This CO-dependent anabolic flux appears to be important for T. onnurineus NA1, because it could assimilate CO2 into cellular carbon via the reductive TCA cycle. In contrast, SCS (TON_1665) and PC (TON_0904), enzymes of the pseudo-TCA cycle, were abundantly expressed in cells grown on starch (Fig. 1 and Table II).
As for the thermophilic sulfur-reducing bacterium D. desulfuricans (29), propionate can be fed directly into the TCA cycle via succinyl-CoA through the methylmalonyl pathway, including a propionyl-CoA carboxylase (TON_0041), a methylmalonyl-CoA epimerase (TON_1074), and a methylmalonyl-CoA mutase (methylmalonyl-CoA mutase I, TON_1076; methylmalonyl-CoA mutase II, TON_1110). Most protein components of the propionate pathway, except for methylmalonyl-CoA decarboxylase (TON_0041), were induced during growth on starch or formate (Fig. 1 and Table II).
In addition to key enzymes in the pseudo-TCA cycle, we found up-regulation of type III ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO, TON_1234) in CO-grown cells, as shown in Table II. RuBisCO activities were detected in T. kodakaraensis KOD1 (48, 49), A. fulgidus (50), and several other methanogenic archaea (51, 52). Intriguingly, type III RuBisCO from T. kodakaraensis KOD1 has been shown to complement a RuBisCO-deficient strain of the purple non-sulfur bacterium Rhodopseudomonas palustris and therefore may allow the Calvin-Benson-Bassham pathway mode (49). As recently described in other Thermococcales (14, 15), the CO2 produced from catabolism or CO oxidation may be a suitable substrate for the type III RuBisCO (TON_1234), which together with DeoA (TON_1062) and RBPI (TON_1296) replenish the 3-phosphoglycerate used in glycolysis and gluconeogenesis (Fig. 1). This CO-dependent functional plasticity of RuBisCO may be important for the survival and persistence of T. onnurineus NA1 living heterotrophically around hydrothermal vents where organic carbon is limited. Thus, this finding provides additional evidence that T. onnurineus NA1 can grow on CO as the sole carbon source.
Amino Acid Degradation and Aldehyde Metabolism
In T. onnurineus NA1, the metabolic fate of aldehydes produced during anaerobic fermentation is unclear. Interestingly, one unique alcohol dehydrogenase (ADH) and other key enzymes required to metabolize aldehydes into ethanol or carboxylic acids were detected in our proteomic analysis (Fig. 1 and Table II). In most anaerobic archaea (14, 15, 53), pyruvate is oxidatively decarboxylated into acetyl-CoA and CO2 via POR (TON_1480–82), which is then converted into acetate as an end product by the reverse reaction of acetyl-CoA synthetase (acetyl-CoA synthetase I, TON_1097 and TON_0327; and acetyl-CoA synthetase II, TON_1002 and TON_1313). Hence, a key question is how 2-oxoacids such as pyruvate are converted into ethanol in this bacterium. For hyperthermophilic archaea, an understanding of anaerobic fermentation will facilitate the development of a suitable enzyme system for producing ethanol at high temperatures with high efficiency. In addition to POR, three other types of 2-oxoacid ferredoxin oxidoreductase (KGOR, TON_0583–84 and TON_0586–87; IOR, TON_1311–12; VOR, TON_1477–79) metabolize the 2-oxoacids generated by transamination of amino acids to their corresponding acyl-CoA derivatives (Fig. 1). Alternatively, 2-oxoacids may also be metabolized by 2-oxoacid:ferredoxin oxidoreductase (TON_0860–61) into their corresponding aldehydes (53, 54). Thus, aldehydes would be subsequently oxidized into their corresponding carboxylic acids by two different tungsten-containing aldehyde ferredoxin oxidoreductases, aor-2-tungsten-containing aldehyde ferredoxin oxidoreductase (TON_1354), and formaldehyde:ferredoxin oxidoreductase (TON_0749). Therefore, we propose that 2-oxoacid:ferredoxin oxidoreductase (TON_0860–61) is the bridge linking the conversion of 2-oxoacid to its corresponding acyl-CoA or aldehyde derivatives in T. onnurineus NA1. The known steps of 2-oxoacid degradation are shown in Fig. 1 (solid arrows). Notably, these enzymes were robustly up-regulated in CO-grown cells (Table II), indicating that they could function in aldehyde metabolism during growth on CO. From these data, we infer that under CO-added conditions, the aldehyde produced from 2-oxoacids is oxidized into carboxylic acids by aldehyde ferredoxin oxidoreductase or formaldehyde:ferredoxin oxidoreductase, wherein the excess reductant (reduced ferredoxin) is utilized for H2S production, as proposed previously for other Thermococcales (54–56). ADH (TON_0544), which catalyzes aldehydes into ethanol, was also detected. However, this was more abundantly expressed in starch-grown cells than in CO-grown cells (Table II). In the case of T. kodakaraensis KOD1, when pyruvate or starch was added to the medium, the cells predominantly produced H2 rather than H2S (57). Under S0-limiting conditions, H2 and alcohol production increase dramatically, as does activation of ADH (54). Therefore, we propose that a fermentation process shift may occur under S0-limited and starch excess conditions, wherein both H2 and alcohol production increase but the amount of acids and H2S decrease, thereby serving to dispose of excess reductant.
Formate-dependent Hydrogenase and Hydrogenase Maturation System
Our proteomic analysis revealed a number of hydrogen clusters apparently associated with formate metabolism, consistent with previous results (9). These include two tripartite gene clusters containing formate dehydrogenases (Fdh): fdh1-mfh1-mnh1 (Hyg4-I) and fdh2-mfh2-mnh2 (Hyg4-III). In the comparative proteomic analysis, TON_1559–1577, belonging to the Hyg4-III cluster, was substantially abundant in formate-grown cells relative to other culture conditions (Table II and Fig. 2). In particular, Fdh2 (TON_1563) was highly induced on formate compared with both starch (>12.50-fold) and CO (>17.46-fold). Of these Hyg4 III components, the most strongly induced member was mfh2 (TON_1569–1572), which was also abundantly expressed in gene expression analysis (9). In a recent study, the hydrogenase encoded by mfh2 was proven to be responsible for growth and H2 production when formate was supplied as a substrate (9). Our proteomic results support the role of mfh2 in formate metabolism. Furthermore, the up-regulation of an Na+/H+ antiporter (TON_1577) in cells grown on formate is consistent with the previous report (9). Other Hyg4-III hydrogenases that were up-regulated under formate culture conditions were the F420 hydrogenases Frh TON_1559 and Frh TON_1561 and formate dehydrogenase FdhD (TON_1562). These protein complexes (FdhD and Frh), known as formate hydrogen lyase systems, convert formate into CO2 and H2. Therefore, during growth on formate, H2 can be produced by either the Fdh/Frh or fdh2-mfh2 Hyg4-III systems, probably in equilibrium with formate concentration.
Fig. 2.

Expression map of hydrogenase gene cluster proteins. The expression map was constructed using the proteome of T. onnurineus NA1 cultivated with formate, CO, and starch as carbon and/or energy sources. Blue, red, and black arrows indicate proteins expressed prominently during growth on formate, CO, and starch, respectively. The white arrow indicates an unidentified gene product. The numbers below the colored arrows indicate the gene identification numbers of hydrogenase induced during growth.
F420-reducing hydrogenase is found solely in methanogenic archaea (58–60), where it oxidizes H2 to provide reduced coenzyme F420 (F420H2), which acts as the electron donor for CO2 reduction via the methane metabolic pathway. Recently, this protein was also discovered in Thermococcus strains such as T. onnurineus NA1 and T. gammatolerans (8, 15). Although further examination of its role in T. onnurineus NA1 is necessary, generation of H2 by F420 hydrogenase may be required to maintain redox balance during formate-dependent growth. Coenzyme F420 may act as a low redox potential electron carrier, as proposed for methanogenic metabolism (58–61). In methane-producing cells, the intracellular concentration ratios of reduced and oxidized coenzyme F420 are in equilibrium with the H2 concentration in the medium (61). In view of this, formate metabolism may be modulated by the cellular concentration of H2, F420, and formate.
On the other hand, most Hyg4-I cluster proteins (TON_0266–0282) were differentially expressed during growth on CO and starch as well as formate, indicating that this cluster has a minor (or no) relationship with formate metabolism. This result is consistent with a previous gene expression analysis in which genes encoding the mfh1 cluster (TON_0273–78) were slightly down-regulated during growth on formate (9). Overall, the proteomic and gene expression data for the formate metabolic pathway studied correspond relatively well, indicating that this pathway is not subject to additional translational control. Along with the hydrogenase (Hyg4-I) cluster, we identified the known auxiliary proteins Hyc I (TON_0263), Hyp F (TON_0286), and Hyp E (TON_0287), which are involved in hydrogenase maturation (62, 63). Of these, Hyc I (TON_0263) was prominently up-regulated in cells grown on formate (Table II and Fig. 2). This finding is closely correlated with the fact that, in E. coli, expression of both the fdh gene and the hyc operon depends on formate and acidification of the growth medium (64). Also, the non-energy-conserving formate hydrogen lyase complex (Fdh-Mhy) is generally involved in formate dissimilation, thereby preventing acidification of the cytoplasm. Although formate dehydrogenase (FdhA, TON_0281) was not detected in cells grown on any substrates, the expression of Mhy1 (TON_0276) was up-regulated in cells grown on CO (Table II and Fig. 2). Presumably, the physiological functions of the FdhA-Mhy1 complex are linked to formate removal likely generated during amino acid catabolism, as proposed in Thermococcus litoralis (65).
CO-responsive Hydrogenase and Other Membrane-bound Hydrogenases
Previous studies revealed the presence of CO-dependent hydrogenase gene clusters in T. onnurineus NA1 (8, 18). In accordance with these studies, our proteomic analysis showed marked expression of hydrogenase gene clusters belonging to Hyg4_II during growth on CO (Table II and Fig. 2). In particular, both the CO dehydrogenase (CODH, TON_1017–19) and Mch hydrogenase (TON_1023–24) of the Hyg4_II cluster were substantially elevated in CO-grown cells, suggesting that both hydrogenases function during carboxydotrophic growth. This result is consistent with our previous microarray studies that showed that CODH and a large Mch subunit (TON_1023) were significantly up-regulated in CO-grown cells (data not shown). According to a previous report (66), most carboxydotrophic hydrogenogenic bacteria share a similar gene organization composed of CODH and an energy-converting hydrogenase. Although we detected no evidence of a functional energy-converting hydrogenase-like protein in T. onnurineus NA1, it is possible that CO-induced Mch hydrogenase clusters putatively encode a similar protein. Indeed, T. onnurineus NA1 encodes a putative membrane-bound hydrogenase (TON_1021) with NADH:quinone oxidoreductase activity, in addition to other Mch hydrogenase components (TON_1022–24). Even though it is membrane-bound, TON_1021 was not detected in our proteomic analysis; it is possible that this Mch hydrogenase may play a key role in carboxydotrophic hydrogenogenic metabolism (66, 67).
Also, in CO-grown cells, the marked expression of monofunctional CODH (TON_1018), homologous to CooS and CooF (TON_1017), indicates the possibility that T. onnurineus NA1 can evolve H2 during growth on CO, as displayed by Rhodospirillum rubrum (68) and Carboxydothermus hydrogenoformans (69, 70). In addition, it has been reported that energy-converting hydrogenase-type hydrogenases couple the formation of H2 to the generation of the proton motive force that drives ATP synthesis (66, 71). Likewise, coupling of CO oxidation to proton translocation in T. onnurineus NA1 may enable it to use CO as a sole energy source and generate H2.
Sulfhydrogenase-I was also up-regulated in cells grown on CO (Table II and Fig. 2). The CO-induced sulfhydrogenase-I cluster contains a number of genes that encode enzymes putatively involved in sulfur reduction and H2 recycling to provide NADPH for biosynthesis (72, 73). As shown in Table II and Fig. 2, cytoplasmic (Ni-Fe) hydrogenases (encoded by TON_0534 to TON_0536) homologous to the Hyh-I of T. kodakaraensis KOD1 were identified in the sulfhydrogenase-I cluster of T. onnurineus NA1. Like Hyh-I of T. kodakaraensis KOD1 (74), these enzymes may function as an H2 uptake hydrogenase during H2 production. In addition to sulfhydrogenase (TON_0537), a glutamate synthase β subunit (TON_0542) similar to that coded by KOD1-gltA was also present in T. onnurineus NA1. Glutamate synthase shows homology to the sulfide dehydrogenase that is involved in S0 reduction using NAD(P)H as the electron donor (75, 76). Up-regulation of formate dehydrogenase (TON_0539) was also detected in CO-grown cells but may be attributable to re-oxidation of formate rather than to formate production, as described in A. fulgidus (77). Although ADH (TON_0544) most likely functions in aldehyde reduction, it may also provide reductants for S0 reduction, as proposed in P. furiosus (78).
Furthermore, a membrane-bound oxidoreductase (Mbx, TON_0488) was also strongly up-regulated in CO-grown cells (Fig. 2). Mbx appears to be directly involved in oxidizing reduced ferredoxin to transfer electrons to elemental sulfur (79). However, expression of a membrane-bound hydrogenase (Mbh: TON_1593) was higher during growth on starch (Fig. 2), which is consistent with its attributed function in sugar metabolism as seen in other Thermococcales (5, 57, 63).
Conclusion
To the best of our knowledge, this is the most comprehensive quantitative analysis of T. onnurineus NA1, the first global survey of metabolic activities at the proteome level, and the first detailed proteomic investigation of anabolic reactions in hyperthermophilic archaea. We analyzed and compared the soluble proteomes of T. onnurineus NA1 cells grown on formate, CO, and starch using one-dimensional SDS-PAGE in combination with nano UPLC-MSE. Our data provide a solid basis for further studies of the metabolic characteristics of T. onnurineus NA1. One-carbon metabolism in the H2-producing hyperthermophilic archaeon T. onnurineus NA1 is an essential process that relies on at least two different one-carbon donor molecules: CO and formate. In particular, formate was metabolized via gluconeogenesis and the PPP into cellular carbon. On the other hand, CO2 formed from CO oxidation by CODH and CO-induced hydrogenases is a suitable substrate for carbon assimilation by type III RuBisCO. In addition, our data suggest that pyruvate formed from acetyl-CoA and CO2 by the reverse reaction of POR can enter the CO-dependent anabolic pathway, generating cellular carbon via the incomplete TCA cycle. Also, CO was utilized as an energy source for a CO oxidation-coupled electron transport mechanism that resulted in H2 production. Our data provide novel insights into the poorly understood metabolism of one-carbon compounds in hyperthermophilic archaea. Furthermore, identified proteins with known sequence or motif/domain homologies were classified into groups according to their known and putative biological functions. It is also intriguing to note that enzymes involved in energy metabolism were found as major proteins in our proteomic analysis. In addition to these key metabolic enzymes, we identified and quantified proteins involved in other cellular processes, including components of the translational machinery, amino acid metabolism, DNA metabolism, nucleotide metabolism, transcription, the stress response, and transporters. This study also provides a pool of potentially important proteins with unknown functions that are specific to hyperthermophilic archaea and adds to our understanding of their biological functions and metabolic adaptation in extreme environments. These proteins represent an attractive target for further directed genetic and biochemical analyses to shed light on the physiology of T. onnurineus NA1.
Footnotes
* This work was supported by the Development of Biohydrogen Production Technology using the Hyperthermophilic Archaea Program and the Marine and Extreme Genome Research Center Program of the Ministry of Land, Transport, and Maritime Affairs, and Grant K-MeP T31130 (to S. I. K.) and Creative Research Grant T31612 (to Y. H. C.) from the Korea Basic Science Institute.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- UPLC
- ultra performance liquid chromatography
- MSE
- high/low collision energy mass spectrometry
- TRAP
- tripartite ATP-independent periplasmic
- PFL
- pyruvate-formate lyase
- PFL-AE
- pyruvate-formate lyase activating enzyme
- FBPase
- fructose-1,6-bisphosphatase
- HPS/PHI
- bifunctional d-arabino 3-hexulose-6-phosphate formaldehyde lyase/phosphohexuloisomerase
- RuBisCO
- ribulose-1,5-bisphosphate carboxylase/oxygenase
- CODH
- carbon monoxide dehydrogenase
- Mch
- membrane-bound carbon monoxide-dependent hydrogenase
- ABC
- ammonium bicarbonate
- PPP
- pentose phosphate pathway
- ADH
- alcohol dehydrogenase
- POR
- pyruvate ferredoxin oxidoreductase
- KGOR
- 2-oxogutarate ferredoxin oxidoreductase
- Fdh
- formate dehydrogenase
- 2-DE
- Two-dimenstional gel electrophoresis
- cmo
- Cofactor modifying protein
- GLK
- Glucokinase
- PGI
- Glucose-6-phosphate isomerase
- PFK
- Pphosphofructokinase
- GAPOR
- Glyceraldehyde-3-phosphate:ferredoxin oxidoreductase
- PRPPS
- Ribose-phosphate pyrophosphokinase
- RBPI
- Ribose 1,5-bisphosphate isomerase
- SCS:
- Succinyl-CoA synthetase
- PC
- Pyruvate carboxylase
- IOR
- Indolepyruvate oxidoreductase
- VOR
- 2-ketoisovalerate ferredoxin oxidoreductase.
REFERENCES
- 1. Holden J. F., Takai K., Summit M., Bolton S., Zyskowski J., Baross J. A. (2001) Diversity among three novel groups of hyperthermophilic deep-sea Thermococcus species from three sites in the northeastern pacific ocean. FEMS Microbiol. Ecol. 36, 51–60 [DOI] [PubMed] [Google Scholar]
- 2. Xue Y., Xu Y., Liu Y., Ma Y., Zhou P. (2001) Thermoanaerobacter tengcongensis sp. nov., a novel anaerobic, saccharolytic, thermophilic bacterium isolated from a hot spring in Tengcong, China. Int. J. Syst. Evol. Microbiol. 51, 1335–1341 [DOI] [PubMed] [Google Scholar]
- 3. Miroshnichenko M. L., Hippe H., Stackebrandt E., Kostrikina N. A., Chernyh N. A., Jeanthon C., Nazina T. N., Belyaev S. S., Bonch-Osmolovskaya E. A. (2001) Isolation and characterization of Thermococcus sibiricus sp. nov. from a Western Siberia high-temperature oil reservoir. Extremophiles 5, 85–91 [DOI] [PubMed] [Google Scholar]
- 4. Bertoldo C., Antranikian G. (2006) The order Thermococcales, in The Prokaryotes (Dworkin M., Falkow S., Rosenberg E., Schleifer K. H., Stackebrandt E., eds) Vol. 3, 3rd Ed., pp. 69–81, Springer, New York [Google Scholar]
- 5. Sapra R., Bagramyan K., Adams M. W. (2003) A simple energy-conserving system: Proton reduction coupled to proton translocation. Proc. Natl. Acad. Sci. U.S.A. 100, 7545–7550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mardanov A. V., Ravin N. V., Svetlitchnyi V. A., Beletsky A. V., Miroshnichenko M. L., Bonch-Osmolovskaya E. A., Skryabin K. G. (2009) Metabolic versatility and indigenous origin of the archaeon Thermococcus sibiricus, isolated from a siberian oil reservoir, as revealed by genome analysis. Appl. Environ. Microbiol. 75, 4580–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sokolova T. G., Jeanthon C., Kostrikina N. A., Chernyh N. A., Lebedinsky A. V., Stackebrandt E., Bonch-Osmolovskaya E. A. (2004) The first evidence of anaerobic CO oxidation coupled with H2 production by a hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Extremophiles 8, 317–323 [DOI] [PubMed] [Google Scholar]
- 8. Lee H. S., Kang S. G., Bae S. S., Lim J. K., Cho Y., Kim Y. J., Jeon J. H., Cha S. S., Kwon K. K., Kim H. T., Park C. J., Lee H. W., Kim S. I., Chun J., Colwell R. R., Kim S. J., Lee J. H. (2008) The complete genome sequence of Thermococcus onnurineus NA1 reveals a mixed heterotrophic and carboxydotrophic metabolism. J. Bacteriol. 190, 7491–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim Y. J., Lee H. S., Kim E. S., Bae S. S., Lim J. K., Matsumi R., Lebedinsky A. V., Sokolova T. G., Kozhevnikova D. A., Cha S. S., Kim S. J., Kwon K. K., Imanaka T., Atomi H., Bonch-Osmolovskaya E. A., Lee J. H., Kang S. G. (2010) Formate-driven growth coupled with H2 production. Nature 467, 352–355 [DOI] [PubMed] [Google Scholar]
- 10. Bae S. S., Kim Y. J., Yang S. H., Lim J. K., Jeon J. H., Lee H. S., Kang S. G., Kim S. J., Lee J. H. (2006) Thermococcus onnurineus sp. nov., a hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent area at the PACMANUS field. J. Microbiol. Biotechnol. 16, 1826–1831 [Google Scholar]
- 11. Kawarabayasi Y., Sawada M., Horikawa H., Haikawa Y., Hino Y., Yamamoto S., Sekine M., Baba S., Kosugi H., Hosoyama A., Nagai Y., Sakai M., Ogura K., Otsuka R., Nakazawa H., Takamiya M., Ohfuku Y., Funahashi T., Tanaka T., Kudoh Y., Yamazaki J., Kushida N., Oguchi A., Aoki K., Kikuchi H. (1998) Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5, 55–76 [DOI] [PubMed] [Google Scholar]
- 12. Robb F. T., Maeder D. L., Brown J. R., DiRuggiero J., Stump M. D., Yeh R. K., Weiss R. B., Dunn D. M. (2001) Genomic sequence of hyperthermophile, Pyrococcus furiosus: Implications for physiology and enzymology. Methods Enzymol. 330, 134–157 [DOI] [PubMed] [Google Scholar]
- 13. Cohen G. N., Barbe V., Flament D., Galperin M., Heilig R., Lecompte O., Poch O., Prieur D., Quérellou J., Ripp R., Thierry J. C., Van der Oost J., Weissenbach J., Zivanovic Y., Forterre P. (2003) An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi. Mol. Microbiol. 47, 1495–1512 [DOI] [PubMed] [Google Scholar]
- 14. Fukui T., Atomi H., Kanai T., Matsumi R., Fujiwara S., Imanaka T. (2005) Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15, 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zivanovic Y., Armengaud J., Lagorce A., Leplat C., Guérin P., Dutertre M., Anthouard V., Forterre P., Wincker P., Confalonieri F. (2009) Genome analysis and genome-wide proteomics of Thermococcus gammatolerans, the most radioresistant organism known amongst the Archaea. Genome Biol. 10, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lim J. K., Kang S. G., Lebedinsky A. V., Lee J. H., Lee H. S. (2010) Identification of a novel class of membrane-bound [NiFe]-hydrogenases in Thermococcus onnurineus NA1 by in silico analysis. Appl. Environ. Microbiol. 76, 6286–6289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kwon S. O., Kang S. G., Park S. H., Kim Y. H., Choi J. S., Lee J. H., Kim S. I. (2009) Proteomic characterization of the sulfur-reducing hyperthermophilic archaeon Thermococcus onnurineus NA1 by 2-DE/MS-MS. Extremophiles 13, 379–387 [DOI] [PubMed] [Google Scholar]
- 18. Yun S. H., Kwon S. O., Park G. W., Kim J. Y., Kang S. G., Lee J. H., Chung Y. H., Kim S., Choi J. S., Kim S. I. (2011) Proteome analysis of Thermococcus onnurineus NA1 reveals the expression of hydrogen gene cluster under carboxydotrophic growth. J. Proteomics 74, 1926–1933 [DOI] [PubMed] [Google Scholar]
- 19. Balch W. E., Wolfe R. S. (1976) New approach to the cultivation of methanogenic bacteria: 2-Mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressureized atmosphere. Appl. Environ. Microbiol. 32, 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yun S. H., Choi C. W., Kwon S. O., Park G. W., Cho K., Kwon K. H., Kim J. Y., Yoo J. S., Lee J. C., Choi J. S., Kim S. I. (2011) Quantitative proteomic analysis of cell wall and plasma membrane fractions from multidrug-resistant Acinetobacter baumannii. J. Proteome Res. 10, 459–469 [DOI] [PubMed] [Google Scholar]
- 21. Yang H. Y., Kwon J., Cho E. J., Choi H. I., Park C., Park H. R., Park S. H., Chung K. J., Ryoo Z. Y., Cho K. O., Lee T. H. (2010) Proteomic analysis of protein expression affected by peroxiredoxin V knock-down in hypoxic kidney. J. Proteome Res. 9, 4003–4015 [DOI] [PubMed] [Google Scholar]
- 22. Bae S. S., Kim T. W., Lee H. S., Kwon K. K., Kim Y. J., Kim M. S., Lee J. H., Kang S. G. (2012) H2 production from CO, formate or starch using the hyperthermophilic archaeon, Thermococcus onnurineus. Biotechnol. Lett. 34, 75–79 [DOI] [PubMed] [Google Scholar]
- 23. Gao B. B., Stuart L., Feener E. P. (2008) Label-free quantitative analysis of one-dimensional PAGE LC/MS/MS proteome: Application on angiotensin II-stimulated smooth muscle cells secretome. Mol. Cell. Proteomics 7, 2399–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Verhees C. H., Kengen S. W., Tuininga J. E., Schut G. J., Adams M. W., De Vos W. M., Van Der Oost J. (2003) The unique features of glycolytic pathways in Archaea. Biochem. J. 375, 231–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsubara K., Yokooji Y., Atomi H., Imanaka T. (2011) Biochemical and genetic characterization of the three metabolic routes in Thermococcus kodakarensis linking glyceraldehyde 3-phosphate and 3-phosphoglycerate. Mol. Microbiol. 81, 1300–1312 [DOI] [PubMed] [Google Scholar]
- 26. Nishimasu H., Fushinobu S., Shoun H., Wakagi T. (2004) The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea. Structure 12, 949–959 [DOI] [PubMed] [Google Scholar]
- 27. Rashid N., Imanaka H., Kanai T., Fukui T., Atomi H., Imanaka T. (2002) A novel candidate for the true fructose-1,6-bisphosphatase in archaea. J. Biol. Chem. 277, 30649–30655 [DOI] [PubMed] [Google Scholar]
- 28. Lee Y. G., Kang S. G., Lee J. H., Kim S. I., Chung Y. H. (2010) Characterization of hyperthermostable fructose-1,6-bisphosphatase from Thermococcus onnurineus NA1. J. Microbiol. 48, 803–807 [DOI] [PubMed] [Google Scholar]
- 29. Takaki Y., Shimamura S., Nakagawa S., Fukuhara Y., Horikawa H., Ankai A., Harada T., Hosoyama A., Oguchi A., Fukui S., Fujita N., Takami H., Takai K. (2010) Bacterial lifestyle in a deep-sea hydrothermal vent chimney revealed by the genome sequence of the thermophilic bacterium Deferribacter desulfuricans SSM1. DNA Res. 17, 123–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mulligan C., Fischer M., Thomas G. H. (2011) Tripartite ATP-independent periplasmic (TRAP) transporters in bacteria and archaea. FEMS Microbiol. Rev. 35, 68–86 [DOI] [PubMed] [Google Scholar]
- 31. Tang E. P. (2006) Path to effective recovering of DNA from formalin-fixed biological samples in natural history collections: Workshop summary, National Research Council of the National Academies, National Academies Press, Washington, DC [Google Scholar]
- 32. Lindahl T., Andersson A. (1972) Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry 11, 3618–3623 [DOI] [PubMed] [Google Scholar]
- 33. Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature 362, 709–715 [DOI] [PubMed] [Google Scholar]
- 34. Lindahl T., Nyberg B. (1972) Rate of depurination of native deoxyribonucleic acid. Biochemistry 11, 3610–3618 [DOI] [PubMed] [Google Scholar]
- 35. Gates K. S., Nooner T., Dutta S. (2004) Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem. Res. Toxicol. 17, 839–856 [DOI] [PubMed] [Google Scholar]
- 36. Jacobs K. L., Grogan D. W. (1997) Rates of spontaneous mutation in an archaeon from geothermal environments. J. Bacteriol. 179, 3298–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mackwan R. R., Carver G. T., Kissling G. E., Drake J. W., Grogan D. W. (2008) The rate and character of spontaneous mutation in. Thermus thermophilus. Genetics 180, 17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grogan D. W. (1998) Hyperthermophiles and the problem of DNA instability. Mol. Microbiol. 28, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 39. Lehtiö L., Grossmann J. G., Kokona B., Fairman R., Goldman A. (2006) Crystal structure of a glycyl radical enzyme from Archaeoglobus fulgidus. J. Mol. Biol. 357, 221–235 [DOI] [PubMed] [Google Scholar]
- 40. Smith D. R., Doucette-Stamm L. A., Deloughery C., Lee H., Dubois J., Aldredge T., Bashirzadeh R., Blakely D., Cook R., Gilbert K., Harrison D., Hoang L., Keagle P., Lumm W., Pothier B., Qiu D., Spadafora R., Vicaire R., Wang Y., Wierzbowski J., Gibson R., Jiwani N., Caruso A., Bush D., Reeve J. N. (1997) Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: Functional analysis and comparative genomics. J. Bacteriol. 179, 7135–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kirkpatrick C., Maurer L. M., Oyelakin N. E., Yoncheva Y. N., Maurer R., Slonczewski J. L. (2001) Acetate and formate stress: Opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183, 6466–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kessler D., Knappe J. (1996) Anaerobic dissimilation of pyruvate in Escherichia coli and Salmonella: Cellular and molecular biology (Neidhardt F. C., Curtiss R., III, Ingraham J. L., Lin E. C., Low K. B., Magasanik B., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E., eds) 2nd Ed., pp. 199–205, ASM Press, Washington, DC [Google Scholar]
- 43. Buis J. M., Broderick J. B. (2005) Pyruvate formate-lyase activating enzyme: Elucidation of a novel mechanism for glycyl radical formation. Arch. Biochem. Biophys. 433, 288–296 [DOI] [PubMed] [Google Scholar]
- 44. Wang S. C., Frey P. A. (2007) S-Adenosylmethionine as an oxidant: The radical SAM superfamily. Trends Biochem. Sci. 32, 101–110 [DOI] [PubMed] [Google Scholar]
- 45. Fukuda W., Ismail Y. S., Fukui T., Atomi H., Imanaka T. (2005) Characterization of an archaeal malic enzyme from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. Archaea 1, 293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ikeda T., Ochiai T., Morita S., Nishiyama A., Yamada E., Arai H., Ishii M., Igarashi Y. (2006) Anabolic five subunit-type pyruvate:ferredoxin oxidoreductase from Hydrogenobacter thermophilus TK-6. Biochem. Biophys. Res. Commun. 340, 76–82 [DOI] [PubMed] [Google Scholar]
- 47. Furdui C., Ragsdale S. W. (2000) The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J. Biol. Chem. 275, 28494–28499 [DOI] [PubMed] [Google Scholar]
- 48. Sato T., Atomi H., Imanaka T. (2007) Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science 315, 1003–1006 [DOI] [PubMed] [Google Scholar]
- 49. Yoshida S., Inui M., Yukawa H., Kanao T., Tomizawa K., Atomi H., Imanaka T. (2006) Phototrophic growth of a RuBisCO-deficient mesophilic purple nonsulfur bacterium harboring a Type III RuBisCO from a hyperthermophilic archaeon. J. Biotechnol. 124, 532–544 [DOI] [PubMed] [Google Scholar]
- 50. Kreel N. E., Tabita F. R. (2007) Substitutions at methionine 295 of Archaeoglobus fulgidus ribulose-1,5-bisphosphate carboxylase/oxygenase affect oxygen binding and CO2/O2 specificity. J. Biol. Chem. 282, 1341–1351 [DOI] [PubMed] [Google Scholar]
- 51. Finn M. W., Tabita F. R. (2003) Synthesis of catalytically active form III ribulose 1,5-bisphosphate carboxylase/oxygenase in archaea. J. Bacteriol. 185, 3049–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Watson G. M., Yu J. P., Tabita F. R. (1999) Unusual ribulose 1,5-bisphosphate carboxylase/oxygenase of anoxic Archaea. J. Bacteriol. 181, 1569–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ma K., Hutchins A., Sung S. J., Adams M. W. (1997) Pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon, Pyrococcus furiosus, functions as a CoA-dependent pyruvate decarboxylase. Proc. Natl. Acad. Sci. U.S.A. 94, 9608–9613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ma K., Loessner H., Heider J., Johnson M. K., Adams M. W. (1995) Effects of elemental sulfur on the metabolism of the deep-sea hyperthermophilic archaeon Thermococcus strain ES-1: Characterization of a sulfur-regulated, non-heme iron alcohol dehydrogenase. J. Bacteriol. 177, 4748–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Adams M. W., Holden J. F., Menon A. L., Schut G. J., Grunden A. M., Hou C., Hutchins A. M., Jenney F. E., Jr., Kim C., Ma K., Pan G., Roy R., Sapra R., Story S. V., Verhagen M. F. (2001) Key role for sulfur in peptide metabolism and in regulation of three hydrogenases in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183, 716–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roy R., Mukund S., Schut G. J., Dunn D. M., Weiss R., Adams M. W. (1999) Purification and molecular characterization of the tungsten-containing formaldehyde ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus: The third of a putative five-member tungstoenzyme family. J. Bacteriol. 181, 1171–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kanai T., Imanaka H., Nakajima A., Uwamori K., Omori Y., Fukui T., Atomi H., Imanaka T. (2005) Continuous hydrogen production by the hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1. J. Biotechnol. 116, 271–282 [DOI] [PubMed] [Google Scholar]
- 58. Lupa B., Hendrickson E. L., Leigh J. A., Whitman W. B. (2008) Formate-dependent H2 production by the mesophilic methanogen Methanococcus maripaludis. Appl. Environ. Microbiol. 74, 6584–6590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Baron S. F., Ferry J. G. (1989) Purification and properties of the membrane-associated coenzyme F420-reducing hydrogenase from Methanobacterium formicicum. J. Bacteriol. 171, 3846–3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schauer N. L., Ferry J. G. (1980) Metabolism of formate in Methanobacterium formicicum. J. Bacteriol. 142, 800–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. de Poorter L. M., Geerts W. J., Keltjens J. T. (2005) Hydrogen concentrations in methane-forming cells probed by the ratios of reduced and oxidized coenzyme F420. Microbiology 151, 1697–1705 [DOI] [PubMed] [Google Scholar]
- 62. Forzi L., Sawers R. G. (2007) Maturation of [NiFe]-hydrogenases in Escherichia coli. Biometals 20, 565–578 [DOI] [PubMed] [Google Scholar]
- 63. Jenney F. E., Jr., Adams M. W. (2008) Hydrogenases of the model hyperthermophiles. Ann. N.Y. Acad. Sci. 1125, 252–266 [DOI] [PubMed] [Google Scholar]
- 64. Rossmann R., Sawers G., Böck A. (1991) Mechanism of regulation of the formate-hydrogen lyase pathway by oxygen, nitrate, and pH: Definition of the formate regulon. Mol. Microbiol. 5, 2807–2814 [DOI] [PubMed] [Google Scholar]
- 65. Takács M., Tóth A., Bogos B., Varga A., Rákhely G., Kovács K. L. (2008) Formate hydrogenlyase in the hyperthermophilic archaeon, Thermococcus litoralis. BMC Microbiol. 8, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hedderich R. (2004) Energy-converting [NiFe] hydrogenases from archaea and extremophiles: Ancestors of complex I. J. Bioenerg. Biomembr. 36, 65–75 [DOI] [PubMed] [Google Scholar]
- 67. Oelgeschläger E., Rother M. (2008) Carbon monoxide-dependent energy metabolism in anaerobic bacteria and archaea. Arch. Microbiol. 190, 257–269 [DOI] [PubMed] [Google Scholar]
- 68. Singer S. W., Hirst M. B., Ludden P. W. (2006) CO-dependent H2 evolution by Rhodospirillum rubrum: Role of CODH:CooF complex. Biochim. Biophys. Acta 1757, 1582–1591 [DOI] [PubMed] [Google Scholar]
- 69. González J. M., Robb F. T. (2000) Genetic analysis of Carboxydothermus hydrogenoformans carbon monoxide dehydrogenase genes cooF and cooS. FEMS Microbiol. Lett. 191, 243–247 [DOI] [PubMed] [Google Scholar]
- 70. Soboh B., Linder D., Hedderich R. (2002) Purification and catalytic properties of a CO-oxidizing:H2-evolving enzyme complex from Carboxydothermus hydrogenoformans. Eur. J. Biochem. 269, 5712–5721 [DOI] [PubMed] [Google Scholar]
- 71. Maness P. C., Huang J., Smolinski S., Tek V., Vanzin G. (2005) Energy generation from the CO oxidation-hydrogen production pathway in Rubrivivax gelatinosus. Appl. Environ. Microbiol. 71, 2870–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ma K., Weiss R., Adams M. W. (2000) Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J. Bacteriol. 182, 1864–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ma K., Adams M. W. (2001) Hydrogenases I and II from Pyrococcus furiosus. Methods Enzymol. 331, 208–216 [DOI] [PubMed] [Google Scholar]
- 74. Kanai T., Matsuoka R., Beppu H., Nakajima A., Okada Y., Atomi H., Imanaka T. (2011) Distinct physiological roles of the three [NiFe]-hydrogenase orthologs in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 193, 3109–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hagen W. R., Silva P. J., Amorim M. A., Hagedoorn P. L., Wassink H., Haaker H., Robb F. T. (2000) Novel structure and redox chemistry of the prosthetic groups of the iron-sulfur flavoprotein sulfide dehydrogenase from Pyrococcus furiosus: Evidence for a [2Fe-2S] cluster with Asp(Cys)3 ligands. J. Biol. Inorg. Chem. 5, 527–534 [DOI] [PubMed] [Google Scholar]
- 76. Dincturk H. B., Cunin R., Akce H. (2011) Expression and functional analysis of glutamate synthase small subunit-like proteins from archaeon Pyrococcus horikoshii. Microbiol. Res. 166, 294–303 [DOI] [PubMed] [Google Scholar]
- 77. Henstra A. M., Dijkema C., Stams A. J. (2007) Archaeoglobus fulgidus couples CO oxidation to sulfate reduction and acetogenesis with transient formate accumulation. Environ. Microbiol. 9, 1836–1841 [DOI] [PubMed] [Google Scholar]
- 78. Kelly R. M., Adams M. W. (1994) Metabolism in hyperthermophilic microorganisms. Antonie Van Leeuwenhoek. 66, 247–270 [DOI] [PubMed] [Google Scholar]
- 79. Schut G. J., Bridger S. L., Adams M. W. (2007) Insights into the metabolism of elemental sulfur by the hyperthermophilic archaeon Pyrococcus furiosus: Characterization of a coenzyme A-dependent NAD(P)H sulfur oxidoreductase. J. Bacteriol. 189, 4431–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]