Abstract
Most proteomic studies use liquid chromatography coupled to tandem mass spectrometry to identify and quantify the peptides generated by the proteolysis of a biological sample. However, with the current methods it remains challenging to rapidly, consistently, reproducibly, accurately, and sensitively detect and quantify large fractions of proteomes across multiple samples. Here we present a new strategy that systematically queries sample sets for the presence and quantity of essentially any protein of interest. It consists of using the information available in fragment ion spectral libraries to mine the complete fragment ion maps generated using a data-independent acquisition method. For this study, the data were acquired on a fast, high resolution quadrupole-quadrupole time-of-flight (TOF) instrument by repeatedly cycling through 32 consecutive 25-Da precursor isolation windows (swaths). This SWATH MS acquisition setup generates, in a single sample injection, time-resolved fragment ion spectra for all the analytes detectable within the 400–1200 m/z precursor range and the user-defined retention time window. We show that suitable combinations of fragment ions extracted from these data sets are sufficiently specific to confidently identify query peptides over a dynamic range of 4 orders of magnitude, even if the precursors of the queried peptides are not detectable in the survey scans. We also show that queried peptides are quantified with a consistency and accuracy comparable with that of selected reaction monitoring, the gold standard proteomic quantification method. Moreover, targeted data extraction enables ad libitum quantification refinement and dynamic extension of protein probing by iterative re-mining of the once-and-forever acquired data sets. This combination of unbiased, broad range precursor ion fragmentation and targeted data extraction alleviates most constraints of present proteomic methods and should be equally applicable to the comprehensive analysis of other classes of analytes, beyond proteomics.
Liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS)1 is considered the method of choice for the identification and quantification of proteins and proteomes (1–4) and for the analysis of metabolites, lipids, glycans, and many other types of (bio)molecules. For proteomics, two main LC-MS/MS strategies have been used thus far. They have in common that the sample proteins are converted by proteolysis into peptides, which are then separated by (capillary) liquid chromatography. They differ in the mass spectrometric method used. The first and most widely used strategy is known as shotgun or discovery proteomics. For this method, the MS instrument is operated in data-dependent acquisition (DDA) mode, where fragment ion (MS2) spectra for selected precursor ions detectable in a survey (MS1) scan are generated (5). The resulting fragment ion spectra are then assigned to their corresponding peptide sequences by sequence database searching (6, 7). The second main strategy is referred to as targeted proteomics. There, the MS instrument is operated in selected reaction monitoring (SRM) (also called multiple reaction monitoring) mode. With this method, a sample is queried for the presence and quantity of a limited set of peptides that have to be specified prior to data acquisition. SRM does not require the explicit detection of the targeted precursors but proceeds by the acquisition, sequentially across the LC retention time domain, of predefined pairs of precursor and product ion masses, called transitions, several of which constitute a definitive assay for the detection of a peptide in a complex sample (8). Data analysis in targeted proteomics essentially consists of computing the likelihood that a group of transition signal traces are derived from the targeted peptide (9). Both methods have different and largely complementary preferred uses and performance profiles that have been extensively discussed elsewhere (10). Specifically, shotgun proteomics is the method of choice for discovering the maximal number of proteins from one or a few samples. It does, however, have limited quantification capabilities on large sample sets because of stochastic and irreproducible precursor ion selection (11) and under-sampling (12). In contrast, targeted proteomics is well suited for the reproducible detection and accurate quantification of sets of specific proteins in many samples as is the case in biomarker or systems biology studies (13–15). At present, however, the method is limited to the measurements of a few thousands transitions per LC-MS/MS run (16). It therefore lacks the throughput to routinely quantify large fractions of a proteome.
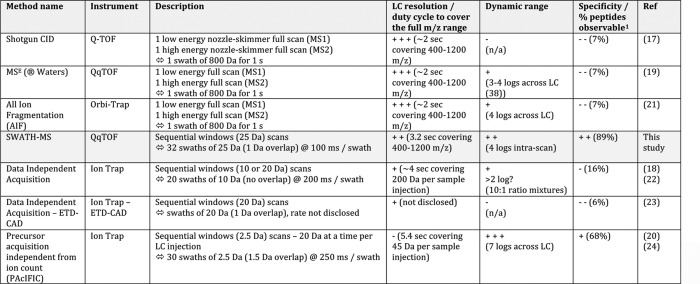
To alleviate the limitations of either method, strategies have been developed that rely on neither detection nor knowledge of the precursor ions to trigger acquisition of fragment ion spectra. Those methods operate via unbiased “data-independent acquisition” (DIA), in the cyclic recording, throughout the LC time range, of consecutive survey scans and fragment ion spectra for all the precursors contained in predetermined isolation windows. Various implementations of DIA methods have already been described using isolation windows of various widths, ranging from the complete m/z range to few Daltons (17–24) (Table I). Using such scans, the link between the fragment ions and the precursors from which they originate is lost, complicating the analysis of the acquired data sets. Also, large selection window widths increase the number of concurrently fragmented precursors and therefore the complexity of the acquired composite fragment ion spectra. To date, the composite spectra generated by DIA methods have been principally analyzed with the standard database searching tools developed for DDA, either by searching the composite MS2 spectra directly (18, 20) or by searching pseudo MS2 spectra reconstituted postacquisition based on the co-elution profiles of precursor ions (from the survey scans) and of their potentially corresponding fragment ions (22, 25–28).
Table I. LC time-resolved data-independent acquisition setups: description and current performance profiles.
1 The numbers reported here correspond to the percentages of peptides observable with at least five interference-free transitions in a yeast background, as reported in Fig. 2B.
Here, we report an alternative approach to proteome quantification that combines a high specificity DIA method with a novel targeted data extraction strategy to mine the resulting fragment ion data sets. For the data acquisition, we implement the sequential isolation window acquisition principle introduced by former DIA studies (18, 20) on a high resolution MS instrument. This time- and mass-segmented acquisition method generates, in a single injection, fragment ion spectra of all precursor ions within a user-defined precursor RT and m/z space and records the ensemble of these fragment ion spectra as complex fragment ion maps. Using computer simulations we show that the resulting maps achieve the highest fragment ion specificity of any DIA method described to date. We term this acquisition strategy “SWATH MS,” in reference to the swaths that are conceptually referred to designate the series of isolation windows acquired for a given precursor mass range across the LC.
To analyze the high specificity, multiplexed data sets generated by SWATH MS, we developed a novel data analysis strategy that fundamentally differs from the database search approaches used so far to identify peptides from DIA data sets. It consists of using a targeted data extraction strategy to query the acquired fragment ion maps for the presence and quantity of specific peptides of interest, using a priori information contained in spectral libraries. Practically, the fragment ion signals, their relative intensities, chromatographic concurrence, and other information accessible from a spectral library for each targeted peptide are used to mine the DIA fragment ion maps for constellations of signals that precisely correlate with the known coordinates of a targeted peptide, thus uniquely identifying the peptide in the map. The extraction of fragment ion traces from data-independently acquired samples sets has been reported for the quantification of formerly identified peptides (18); however, this strategy has never been purposely used to systematically search and identify peptides from the fragment ion maps of DIA data sets. Indeed, it is only with the increasing availability of proteome-wide spectral libraries that this targeted data extraction strategy becomes largely applicable to mine the acquired data sets for peptides never identified thus far with regular shotgun proteomics approaches.
We show that the combination of high specificity fragment ion maps and targeted data analysis using information from spectral libraries of complete organisms offers unprecedented possibilities for the qualitative and quantitative probing of proteomes. This approach should be applicable beyond proteomics to other “omics” measurements, including metabolomics and lipidomics, or to forensics or biomedical analytics fields, which require accurate quantitative analysis of as many analytes as possible from a LC-MS/MS single sample injection.
MATERIALS AND METHODS
LC-MS Sample Acquisition
A commercial 5600 TripleTOFTM (ABSciex, Concord, Canada) was used for all the experiments. The instrument was coupled with an Eksigent 1D+ Nano LC system (Eksigent, Dublin, CA) for the stable isotope dilution experiments or with an Eksigent NanoLC-2DPlus with nanoFlex cHiPLC system for the diauxic shift sample acquisition. The same solvents were used on both LC systems, with solvent A being composed of 0.1% (v/v) formic acid in water and solvent B comprising 95% (v/v) acetonitrile with 0.1% (v/v) formic acid. The serial dilution experiments were performed with a customer-packed emitter, which was created using a laser puller to an orifice of 4 μm and packed with 3-μm Zorbax C18 column using a pressure bomb. The samples were loaded directly onto this column from the nano LC system at a flow rate of 500 nl·min−1. The loaded material was eluted from this column in a linear gradient of 5% solvent B to 30% solvent B over 90 min. The column was regenerated by washing at 90% solvent B for 10 min and re-equilibrated at 5% solvent B for 10 min. The diauxic shift sample acquisitions were performed using a “trap and elute” configuration on the nanoFlex system. The trap column (200 μm × 0.5 mm) and the analytical column (75 μm × 15 cm) were packed with 3 μm ChromXP C18 medium. The samples were loaded at a flow rate of 2 μl·min−1 for 10 min and eluted from the analytical column at a flow rate of 300 nl·min−1 in a linear gradient of 5% solvent B to 35% solvent B in 155 min. The column was regenerated by washing at 80% solvent B for 10 min and re-equilibrated at 5% solvent B for 10 min.
For standard data-dependent analysis experiments, the mass spectrometer was operated in a manner where a 250-ms survey scan (TOF-MS) was collected, from which the top 20 ions were selected for automated MS/MS in subsequent experiments where each MS/MS event consisted of a 50-ms scan. The selection criteria for parent ions included intensity, where ions had to be greater than 150 counts/s with a charge state greater than 1+ and were not present on the dynamic exclusion list. Once an ion had been fragmented by MS/MS, its mass and isotopes were excluded for a period of 15 s. Ions were isolated using a quadrupole resolution of 0.7 Da and fragmented in the collision cell using collision energy ramped from 15 to 45 eV within the 50-ms accumulation time. In the instances where there were less than 20 parent ions that met the selection criteria, those ions that did were subjected to longer accumulation times to maintain a constant total cycle time of 1.25 s.
For SWATH MS-based experiments, the mass spectrometer was operated in a looped product ion mode. In this mode, the instrument was specifically tuned to allow a quadrupole resolution of 25 Da/mass selection. The stability of the mass selection was maintained by the operation of the Radio Frequency (RF) and Direct Current (DC) voltages on the isolation quadrupole in an independent manner. Using an isolation width of 26 Da (25 Da of optimal ion transmission efficiency + 1 Da for the window overlap), a set of 32 overlapping windows was constructed covering the mass range 400–1200 Da. Consecutive swaths need to be acquired with some precursor isolation window overlap to ensure the transfer of the complete isotopic pattern of any given precursor ion in at least one isolation window and thereby to maintain optimal correlation between parent and fragment isotopes peaks at any LC time point (supplemental Fig. S1, a–f). This overlap was reduced to a minimum of 1 Da, which experimentally matched the almost squared shape of the fragment ion transmission profile achieved through the specific quadrupole tuning developed for SWATH MS (supplemental Fig. S1, g and h). The windows setups used for these runs were as follows: Experiment 1: MS1 scan (see below); Experiment 2: 400–426; Experiment 3: 425–451… Experiment 33: 1175–1201. Those isolation windows of 26-Da width (25 Da + 1 Da) are the “nominal” windows used to compute the RF/DC voltages used to drive the isolation quadrupole during the acquisition. However, because the isolation windows are only “almost square shapes” (supplemental Fig S1, g and h), ∼0.3–0.5 Da of ion transmission can be estimated as being “lost” on either sides of the windows. The “100% efficient” transmission of precursor ions is therefore happening only for 25 Da effectively. In other words, the “effective” isolation windows can be considered as being 400.5–425.5, 425.5–450.5, etc. (plus the potential overlap left from the nominal window transmission). The collision energy for each window was determined based on the appropriate collision energy for a 2+ ion centered upon the window with a spread of 15 eV. This ensured optimal fragmentation for the broad range of precursors co-selected within the isolation windows. An accumulation time of 100 ms was used for each fragment ion scan and for the (optional) survey scans acquired at the beginning of each cycle. This results in a total duty cycle of 3.3 s (3.2 s total for stepping through the 32 isolation windows + 0.1 s for the optional survey scan). The mass resolution was between 15,000 and 30,000 for the MS/MS scans, depending on the mode used to record the SWATH MS data sets (high sensitivity or high resolution). For this study, the high sensitivity mode was used, which still allows accurate extraction of the fragment ion masses at 10–50 ppm accuracy (optimal extraction for the area under curve of the MS/MS profile signals at half peak width).
Fragment Ion Interference Simulations
To generate the background for our simulations, the Saccharomyces cerevisiae protein sequences were downloaded from ensembl.org (release 57_1j). The peptide set resulting from trypsin proteolysis (no missed cleavages) was generated in silico using carbamidomethyl cysteine as fixed modification. We then selected the peptides with theoretical precursor ion charge states 2+ and 3+ and with the monoisotopic and the first 13C isotopic masses (+0 and +1 Da) within the mass range of 400 to 1,200 m/z. For each of those precursor ions, the theoretical set of fragment ions was generated (all b and y ions of charge 1+ and 2+), giving rise to transition pairs. This data set contained 111,880 peptides (corresponding to 6,557 proteins) resulting in 194,314 doubly and triply charged precursors (388,781 overall, taking into account the monoisotope and first 13C isotope) and in 10,004,504 transitions altogether that constituted thus the background of our simulations. We also prepared a reduced data set that only contained the precursors of peptides that were reported in the PeptideAtlas (Yeast PeptideAtlas 200904 build, also containing the MS-identified modifications and nontryptic peptides). This reduced data set contained 48,087 peptides (corresponding to 3,898 proteins), resulting in 93,875 doubly and triply charged precursors (187,777 overall, taking into account the monoisotope and first 13C isotope) and in 5,476,964 transitions altogether that we used as a more realistic proteomic background. The retention times of the peptides were computed using the SSRCalc algorithm (50). To estimate the number of SRM interferences, we generated in silico query assays for all proteotypic peptides of the yeast genome as targets. We considered all singly charged b and y transitions of the monoisotopic 2+ precursor of those peptides as targets and ran them against the computed backgrounds (theoretical yeast digest or PeptideAtlas) and recorded an interference whenever a transition from the background (that did not belong to the query peptide) was within a specified distance of Q1, Q3, and RT from the target queried peptide. For each target peptide, the number of transitions that were interfered with was recorded and later used to compute the statistics. The detailed algorithm for the computation of the product ion interferences will be the subject of a separate study (51). This algorithm essentially expands on the principle of the “unique ion signatures” described by Sherman et al. (29) by taking into account peptide RT as an additional constraint for the calculation of fragment ion interferences. It should be acknowledged that the current algorithm does not simulate the peptide signal intensities. Even if, in theory, the different MS response factors of those peptides could be retrieved from, for example, the PeptideAtlas database, it is unlikely that those response factors can be extrapolated from one sample to another or from one study to the other, because of ion suppression effects during the ionization. However, it was not the aim of these theoretical simulations to perfectly depict the reality, but rather to give an impression about the overall ranking of the different Q1/Q3 scenarios. In this respect, the simulations are valid because upon increasing the background for the fragment ion simulations (from 93,875 to 194,314 precursors), the overall ranking of the scenarios is maintained. This means that those simulations may not capture the exact reality but can be perfectly used as a tool to compare the extent of fragment ion interference for different scenarios, exactly as it is applied here. Finally, a recent study has experimentally quantified the extent of fragment ion interference in a human cell lysate (12), and the results are in good agreement with our simulations overall.
Serial Dilution Samples for the Limit of Detection, Limit of Quantification, and Intrascan Dynamic Range Assessment
All of the isotopically labeled reference peptides were ordered from Thermo or Sigma-Aldrich with amino acid analysis-certified concentrations. The sequences of the 61 peptides used for the limit of detection experiment, as well as their precursor ion masses (defining the swath in which to extract the daughter ions) and fragment ion masses (used to extract the fragment ion chromatograms), are provided in supplemental Table 1. From those 61 peptides, 23 were kept at a constant concentration of 23.5 fmol/μl (47 fmol total amount loaded on column for a 2-μl injection) in all the samples, whereas the other 38 peptides were serially diluted from 23.5 fmol/μl to 45 amol/μl (from 47 fmol to 91 amol total amount loaded on column for a 2-μl injection) in 2-fold steps. The reference peptides were spiked in a 1 μg/μl concentrated 15N-labeled yeast tryptic digest as proteomic background (15N-labeled yeast trypsin digest was used as proteomic background to avoid the interference between the b fragment ions from endogenous 14N yeast peptides and those from the reference peptides). The peptides kept at the constant concentration through all the samples were used to estimate the coefficient of variance throughout the experiment, whereas the other 38 diluted peptides were used to estimate the limit of detection and limit of quantification of the SWATH MS acquisition method. The raw values (nondenoised, nonsmoothed) of the extracted peak areas for each of the fragment ions of the peptides in each of the samples are provided in the supplemental Table 1. For the limit of detection and quantification, a peak group composed of the three extracted fragment ion traces as well as a signal to noise ratio above 3 (respectively 10) for the considered peak were required. For the comparison with the MS1/label-free based analysis, the precursor ion traces were extracted from the survey scans of the SWATH MS acquisition. Because the accumulation time in SWATH MS mode was of 100 ms for each fragment ion scan and for the survey scans, the peak areas of the fragment ion traces (extracted from their swath) and of the precursor ion traces (extracted from the survey scan) were directly and fairly comparable. Increasing the acquisition time for the survey scan could have increased the signal of the precursors of interest but would have also equally increased the overall noise/interference signals, therefore not really affecting the detection/quantification of those (supplemental Fig. S7, a5–a8). Also, 100-ms acquisition time for MS1 scans is anyway in the range of what may be experimentally used during a typical shotgun experiment aiming at high numbers of precursor ion selection and identification while still allowing for MS1 label-free quantification (48).
For the comparison with shotgun analysis, the same serial dilution samples that had been acquired by SWATH MS were reacquired with a “top 20” DDA method and searched with Mascot for the peptides identification. The peptides used for the intrascan dynamic range experiment have already been used in a different context in our laboratory (49). They consist of the two following sequences: AADITSL*YK* (where L* indicates 13C6,15N, and K* indicates 13C6,15N2) for the doubly isotopically labeled peptide 1 and AADITSLYK* (where K* indicates 13C6,15N2) for the singly isotopically labeled peptide 2. For the intrascan dynamic range experiment, the singly isotopically labeled peptide 2 was kept as a constant concentration of 625 fmol/μl (1.25 fmol total amount loaded on column for a 2-μl injection) in all the samples, whereas the doubly isotopically labeled peptide 1 was serially diluted from 625 fmol/μl to 305 amol/μl (from 1.25 pmol to 610 amol total amount loaded on column for a 2-μl injection) in 2-fold steps. Those peptides were spiked in a 1 μg/μl concentrated yeast tryptic digest as proteomic background. The samples were acquired by SWATH MS. The fragment ion traces of the three most intense fragments were extracted from their swath (475–500 m/z in this case), and the precursor ion traces were extracted from the survey scans from the SWATH MS data sets. The raw values (nondenoised and nonsmoothed) of those extracted peak areas in each of the samples are provided in the supplemental Table 2. The fragment ion traces and MS/MS spectra around the y7 fragment at the RT of the peptides elution are provided as supplemental Fig. S6.
Data Analysis
As for SRM targeted acquisition, the three to five most intense fragments (of proteotypic peptides, as reported in the spectral libraries) were typically selected to perform the targeted data analysis of the SWATH MS data sets. Because those MS/MS spectra libraries were usually generated on low resolution instruments (i.e., triple quadrupoles or ion traps), the high mass accuracy value of those fragments was recalculated theoretically based on the amino acid sequence of the peptide. Those high mass accuracy fragment ion masses were then used as seeds for extracting ion chromatograms in the right LC-MS/MS swath map (indicated by the precursor ion mass). In the case of “borderline” peptides (i.e., peptides with precursor mass falling at an edge of an isolation window or in the zone of isolation window overlap), the fragment ion traces were extracted in the swath with borders furthest away from the precursor ion mass and/or containing most of the isotopic distribution of the precursor ion. All of the fragment ion chromatograms were extracted and automatically integrated with PeakView (v. 1.1.0.0). The raw peak areas as reported by PeakView were used for all the quantification calculations with no data processing (neither denoising nor smoothing, etc.) of any kind applied to the extracted ion chromatograms.
To assess the detection of a peptide, we used as a first pass the LC validation criteria for the extracted fragment ions traces suggested by Reiter et al. (9): co-elution, peak shape similarity, correlation of the relative intensities with reference spectra, correlation of the relative intensities with those of a spike-in reference peptide, co-elution with spiked-in reference, and peak shape similarity with spiked-in reference. The peptide retention time (predicted, e.g., with SSRCalc or experimental when available in spectral libraries) may be used to reduce the chromatographic space where to look for the targeted peak group (similarly to scheduled versus nonscheduled SRM). This is not absolutely necessary for intense signals, but the gain in identification validation can be important for lower intensity signals (or for noisy fragment ion signals over the LC space). To anticipate when a peptide is expected to elute, a simple retention time re-alignment for each gradient or column can be performed to recalculate the retention time relative to its RT available in the spectral library. This is typically done by using a set of reference peptides (relatively to which the retention time of the peptides of the spectra library were recorded), which are spiked into each sample prior to the SWATH MS measurement. Those peptides are used to recalibrate the retention times for each specific run and to help to restrict the extraction of peptides from the library to a reasonable elution time window in each SWATH MS run. In contrast to SRM measurements, the inclusion of such reference peptides does therefore not consume additional data acquisition time.
Finally, several SWATH MS-specific additional criteria may be used to confirm (or invalidate) the peptide identification: confirmation that the extracted fragment ions correspond to monoisotopic signals and verification of the charge state of those fragments in the full MS/MS spectra extracted from the swath around the apex of the candidate peak group, assessment of the mass accuracy (typically <5–10 ppm) of the extracted fragment ions, etc. A step-by-step tutorial describing the manual or automated targeted data analysis of SWATH MS data sets is provided in the supplemental materials.
Database Search of the Reference Peptides of the Dilution Series
The Mascot database search analysis for the reference peptide of the dilution series was performed on Mascot v. 2.4 with a self-compiled database comprising the 61 reference peptides grouped in three artificial proteins based on their abundances as reported in the Weissman list (32). The enzyme selected was trypsin with no missed cleavage. A search tolerance of 50 ppm was specified for the peptide mass tolerance and 0.05 Da for the MS/MS tolerance. The charges of the peptides to search for were set to 2+, 3+, and 4+. The search was set on monoisotopic mass. The instrument was set to ESI-QUAD-TOF. The following modifications were specified for the search: carbamidomethyl cysteines as fixed modification, C-terminal heavy lysine, C-terminal heavy arginine, and oxidized methionine as variable modification.
Diauxic Shift Samples for the Quantification Accuracy Assessment
The diauxic shift samples used in our experiments were the same than those analyzed by SRM in an earlier study (15). The sample set prepared for that study consisted in the tryptic digest of a mixture of (i) a lysate of yeast cells grown in regular 14N medium and sampled throughout the metabolic shift from fermentation to respiration spiked with (ii) a constant 15N-labeled yeast lysate background used as an internal standard for the fold change calculations. For our analysis, we reacquired, in SWATH MS mode, samples 1 and 8 prepared for that study, which constitute the start and end points of the diauxic shift (15). The biological triplicates of samples 1 and 8, prepared for the SRM study, were pooled for the SWATH MS reacquisition to reach enough volume for the sample injection. However, this pooling prevented us from comparing the standard deviations directly between the two studies (the SRM analysis contained the biological sample preparation variability from the triplicate analysis). Therefore, we decided not to present the error bars corresponding to the SRM and SWATH MS standard deviations in Fig. 4B, because they actually captured different information. The error bars were, however, reported individually for the SRM and the SWATH MS study in separated plots provided in supplemental Table 5 for the reader to appreciate the low standard deviations achieved by the SWATH MS quantification. The SWATH MS data analysis consisted of extracting the fragment ion traces of the precursors from the corresponding swath and in reporting the peak areas automatically integrated by PeakView for those extracted chromatograms. To not introduce any bias in the data analysis, the calculation of the abundance fold changes of the proteins was exactly copied from that of the SRM study (15). In short, for each peptide transition, the ratio of the light over heavy peak areas was calculated individually for each sample (samples 1 and 8); the abundance fold change for each transition was then calculated by dividing each transition ratio of sample 8 by the corresponding ratio of sample 1; the final abundance fold change of a protein was then calculated by averaging the individual abundance fold change of each of its transitions. The raw values (nondenoised and nonsmoothed) of those extracted peak areas in each of the samples are provided in supplemental Tables 4 and 6–8.
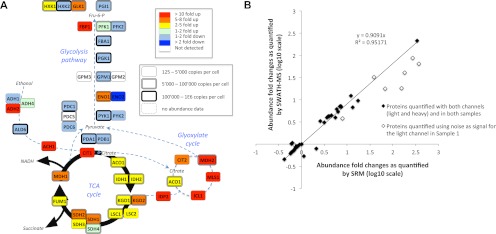
Fig. 4.
Quantification by SWATH MS of the abundance fold changes of 45 enzymes involved in the yeast central carbon metabolism during a diauxic shift experiment. A, schematic representation of the yeast central carbon metabolism network. The abundance fold changes of the enzymes quantified by SWATH MS (supplemental Tables 4–7) are coded with colors. The box shapes are indicative of the absolute abundances of the proteins determined for yeast in log phase growth (32). Those values constitute therefore an approximation of the absolute abundances of the enzymes at the beginning of the diauxic shift experiment, and based on which the fold changes are determined. B, correlation between the abundance fold changes of the same metabolic enzymes quantified by SRM (15) (abscissa) and SWATH MS (ordinate). The linear regression was calculated with the refined SWATH MS quantification values and without taking into account the fold changes for the proteins whose peptides presented light signal intensities below noise levels for the first time point of the diauxic shift (open diamonds), similar to the SRM study (15). Because the standard deviations for the SWATH MS and SRM quantifications are not directly comparable (see “Materials and Methods”), they are provided on separate plots in supplemental Table 5. The complete lists of peak areas for the quantified fragment ion traces and corresponding protein abundance fold changes are provided in supplemental Tables 4–7.
To query the SWATH MS data sets for the 60 yeast mitochondrial proteins involved in oxidative phosphorylation of the respiratory chain (as listed in the Kyoto Encyclopedia of Genes and Genomes website, http://www.genome.jp/kegg/), we used 287 proteotypic peptides assays that were available in our spectral libraries. The relatively low success rate for the identification of these mitochondrial proteins (36 proteins identified over 60) can be explained by the fact that the protocol used for the preparation of those samples was at that time originally devised for the quantification of the metabolic enzymes analyzed by SRM (15) and was therefore not optimized for the recovery of membrane mitochondrial proteins.
The raw (.wiff) files of the diauxic shift samples 1 and 8 acquired in SWATH MS mode may be downloaded from ProteomeCommons.org Tranche using the following hash code: He8q40Zqudc27nmV1fUpqMhPmhVzVVlYqDMNFKcI9dVSZzGkInuXjK9Mg7iBexSZ6eYmGCRkYYp5TgSD6FTqcC5qW2sAAAAAAAAGCg==.
RESULTS
We describe a new concept for the accurate, reproducible, high throughput identification and quantification of proteomes by mass spectrometry. It combines a high specificity data-independent LC-MS/MS acquisition method with a targeted data extraction and analysis strategy.
Data-independent Data Acquisition
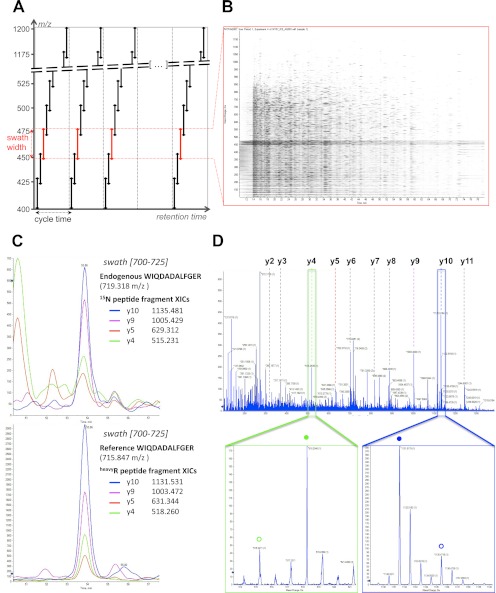
The acquisition method essentially extends the DIA approach initially described by Venable et al. (18). It consists of recording consecutive high resolution fragment ion spectral spectra of all precursors within a user-defined precursor ion window. This is achieved by stepping the precursor isolation window of a quadrupole-quadrupole TOF instrument in 25-Da increments (defining the swath width) recursively during the entire LC separation (Fig. 1A). At 100-ms accumulation time per swath, the quadrupole-accessible 400–1200 m/z range is covered in 32 steps for a total cycle time of 3.2 s, which is sufficient to reconstruct the ∼30-s chromatographic peak of each analyte for accurate quantification. The data structure can thus be conceptualized as 32 successive MS2 maps consisting of the composite fragment ion spectra from all the analytes fragmented in each swath (Fig. 1B). Similar to other windowed DIA methods (20, 23), consecutive swaths were acquired with some precursor isolation window overlap to ensure the transfer of the complete isotopic pattern of any given precursor ion in at least one isolation window and to thereby maintain optimal correlation between parent and fragment isotopes peaks at any LC time point (supplemental Fig. S1, a–f). This overlap was reduced here to a mere minimum of 1 Da. This value experimentally matched the almost square shape of the fragment ion transmission profile (supplemental Fig. S1, g and h), which was achieved through specific quadrupole tuning purposely developed for SWATH MS. Finally, to ensure optimal fragmentation for the broad range of precursors co-selected within each isolation window, a ±15 eV ramping of collision energy was used, centered around the optimal collision energy required to fragment a doubly charged precursor centered in the middle of the isolation window.
Fig. 1.
SWATH MS data-independent acquisition and targeted data analysis. A, the data-independent acquisition method consists of the consecutive acquisition of high resolution, accurate mass fragment ion spectra during the entire chromatographic elution (retention time) range by repeatedly stepping through 32 discrete precursor isolation windows of 25-Da width (black double arrows) across the 400–1200 m/z range. The series of isolation windows acquired for a given precursor mass range and across the LC is referred to as a “swath” (e.g., series of the red double arrows). The cycle time is defined as the time required to return to the acquisition of the same precursor isolation window. Note that the dotted line before the beginning of each cycle depicts the optional acquisition of a high resolution, accurate mass survey (MS1) scan. B, representation of the actual data acquired in one swath (450–475 m/z range) shown here as an MS2 map, with retention time as the abscissa, fragment ion m/z as the ordinate, and ion intensity represented by color intensity. The darker horizontal band visible between 450 and 475 m/z corresponds to residual precursor ions for this swath. The signals co-eluting in the vertical direction are likely fragment ions originating from the same precursor ion. C, the targeted data analysis consists of retrieving the most intense fragment ions of a peptide of interest from a spectral library (list of fragment masses for the 15N-labeled peptide WIQDADALFGER or the corresponding C-terminal isotopically labeled reference) and extracting those fragment ion traces in the appropriate 700–725 swath using a narrow m/z window (e.g., 10 ppm). These fragment ion traces can be plotted as overlaid extracted ion chromatograms, similarly to SRM transitions. The peak group displaying the best co-eluting characteristics and matching best to the peak group of extracted reference fragment ion traces identifies and quantifies the target peptide. D, the complete high resolution, accurate mass fragment ion spectra underlying the best candidate peak group can be extracted from the raw data. These spectra can be inspected to confirm that the extracted signals originate from mass accurate monoisotopic fragment ion with the right charge state (e.g., lower panel zooms on the y4 (green box) and y10 (blue box) fragment, with the endogenous and reference peptide fragments annotated with open or closed circles, respectively). They can also be extensively annotated to strengthen the identification of the peptide (top panel).
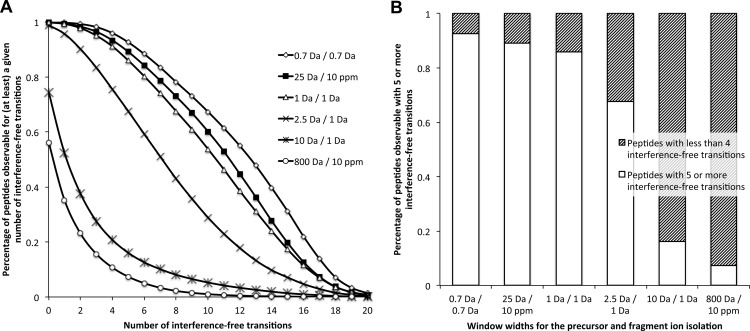
Like other DIA methods (17–24), SWATH MS performance is directly impacted by the width of the precursor isolation window. In principle, large isolation windows are preferable to cycle through a wider precursor mass range with faster cycling rates or with increased dwell times. However, large isolation widths increase the number of precursors concurrently fragmented in the respective window, increasing the likelihood of overlap of fragment ions from different precursors (fragment ion interference). The rate of fragment ion interference also depends on the mass accuracy and resolution of the fragment ion signals (supplemental Fig. S2). Using computer simulations, we assessed whether the signals in the complex fragment ion maps acquired by SWATH MS were sufficiently specific to support conclusive identification and quantification of peptides. As a benchmark, we used the specificity and accuracy levels of SRM, the gold standard MS quantification method. With the tool “SRM-Collider,”2 we computed the occurrence of fragment ion interferences for various combinations of precursor isolation window width and fragment ion mass accuracy. This tool extends the principle of the unique ion signatures described by Sherman et al. (29) by taking into account peptide RT as an additional constraint for the calculation of fragment ion interferences. As the basis for the simulations, we computed theoretical fragment ion spectra for 93,875 doubly and triply charged precursors corresponding to the tryptic peptides of 3,898 yeast proteins reported in the PeptideAtlas database (www.peptideatlas.org). Those represent essentially the complete yeast proteome observable by mass spectrometry (30) and constitute therefore a realistic proteomic background. Cumulative plots depicting the percentage of peptides observable with a given number of interference-free transitions as a measure for correct peptide identification and quantification are shown in Fig. 2A for SRM (0.7- and 1-Da isolation widths for precursor and fragment ions, respectively) and SWATH MS (25-Da swath width, 10-ppm fragment ion accuracy) scenarios. A histogram representing the percentage of peptides with five or more interference-free transitions is shown in Fig. 2B. Both figures show that SWATH MS provides a fragment ion specificity that is comparable with that achieved with standard SRM setups. Because the extensive shotgun data sets from yeast proteome mapping studies possibly underestimate the complexity of real samples, we compared the specificity of SWATH MS fragment ion maps with the specificity achievable by SRM in a more complex background. The simulations were repeated by including all of the doubly and triply charged precursors (194,314 precursors, corresponding to 6,557 proteins (data from ensembl.org) of a complete in silico yeast tryptic digest. As expected, the extent of fragment ion interferences with this more complex background was higher for the different scenarios. However, the relative specificity offered by SWATH MS versus SRM remained qualitatively the same (supplemental Fig. S3).
Fig. 2.
Simulated fragment ion interferences for various LC-MS/MS acquisition scenarios. A, fragment ion interference cumulative plots are computed as described under “Materials and Methods” by taking into account fragments ions from doubly charged yeast tryptic peptide precursors against the fragment ions from the doubly and triply charged yeast tryptic peptides reported in PeptideAtlas (www.peptideatlas.org). The distribution of peptides with specific numbers of interference-free transitions are shown for the following simulations (precursor and fragment ion isolation respectively): 0.7 Da/0.7 Da (open diamonds), 25 Da/10 ppm (black squares), 1 Da/1 Da (open triangles), 2.5 Da/1 Da (crosses), 10 Da/1 Da (asterisks), and 800 Da/10 ppm (open circles). Simulation plots for other background or acquisition scenarios are available in supplemental Fig. S3. B, the fraction of peptides observable with five or more interference-free transitions for the various acquisition scenarios is presented in the histogram with white bars. Accordingly, the shaded bars represent the fraction of peptides having less than four interference-free transitions.
As a comparison, we checked whether previous DIA methods would also provide sufficient fragment ion specificity to support the identification of peptides using a targeting data analysis strategy. We simulated the fragment ion interferences for various sequential windowed DIA methods on low resolution instruments (scenarios with 2.5-Da/1-Da, or 10-Da/1-Da swath width and fragment ion accuracy, respectively) or for DIA methods on high resolution instruments without isolation window (scenario with 800-Da swath width and 10-ppm fragment ion accuracy). Fig. 2 and supplemental Fig. S3 show that none of the former DIA methods are able to reach the level of fragment ion specificity of SRM or SWATH MS and are therefore not amenable to accurate targeted data mining without prior raw data filtering.
Targeted Data Analysis of SWATH MS Fragment Ion Maps
Using the same rationale used above for the simulation of fragment ion interferences in MS2 maps, we computed the overall precursor ion distribution in the LC-MS1 space. For this, we counted for each precursor the number of doubly and triply charged peptides concurrently coinciding within the 25-Da-wide swath and 20–30-s RT elution segment of that precursor. Using the 93,875 yeast tryptic precursors from the PeptideAtlas database, the simulations indicated that, for 75% of the peptides, more than 20 additional precursors (median = 40) were expected to be present in the specified window (supplemental Fig. S4). These numbers illustrate the extent of precursor co-selection, and by inference, the fragment ion spectral complexity that is generated when wide isolation windows are used. These simulations suggest that analyzing such data sets with traditional DIA database search strategies remains highly challenging.
To analyze the SWATH MS data sets, we therefore implemented a data mining strategy that is conceptually similar to targeted mass spectrometry by SRM. However, in contrast to SRM, the signals used for peptides identification and quantification are specified postacquisition and can therefore be flexibly adapted or optimized. The data analysis strategy is schematically illustrated in supplemental Fig. S5. The process starts by selecting, from reference spectral libraries such as SRMAtlas (31), a suitable set of fragment ions from peptides proteotypic for the proteins of interest. In SRM, those fragment ion masses are transition coordinates for the targeted acquisition. In SWATH MS, those fragment ion masses are used to extract ion chromatograms from the acquired data sets that are then combined into an identifying peak group. Fig. 1C provides an example of ion traces for the four most intense fragments of the endogenous peptide WIQDADALFGER that is proteotypic for yeast protein RIR2. The protein has an expected abundance of 500 copies per cell (32). The traces were extracted from a 15N-labeled yeast tryptic digest data set acquired by SWATH MS, specifically in the swath 700–725 that contained the 719.318 m/z doubly charged precursor. The data show that around the RT of 53.9 min, the extracted ion chromatograms form a peak group that identifies the queried peptide, based on the same criteria commonly used by automated SRM analysis tools (e.g., mProphet (9) or Skyline (33)) such as co-elution of the fragment ions traces, correlation of the relative fragment ion intensities with those of reference spectra, and more. The identification can be further strengthened by checking the co-elution with a reference peptide spiked into the sample (Fig. 1C) or by extensively annotating the full fragment ion spectra implicitly present in the SWATH MS data at that RT (Fig. 1D). As in SRM, the quantification is intrinsically linked to the peptide identification (supplemental Fig. S5) and proceeds by integration of the fragment ions traces across the chromatographic elution of the validated peak group, with the optional use of isotopically labeled references for relative or absolute quantification.
Performance of SWATH MS Coupled to Targeted Data Extraction
Limit of Detection, Limit of Quantification, and Intrascan Dynamic Range
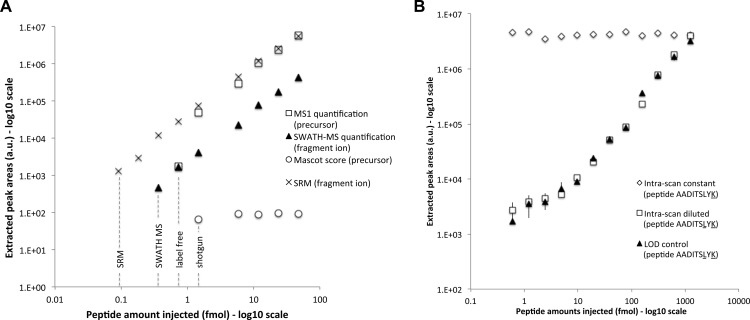
The LOD of the method was assessed by measuring dilution series of 61 reference peptides containing isotopically labeled lysine or arginine C termini, spiked into a background of a 15N-labeled yeast tryptic digest. Among those, 38 peptides were serially diluted, covering a range of 47 fmol to 91 amol, and 23 were kept constant at 47 fmol each. The samples were subjected to SWATH MS acquisition, and the ion traces for the three most intense fragment ions for each reference peptides were extracted and integrated. The resulting dilution plots show a limit of detection (signal to noise ratio above 3) and a limit of quantification (deviation from linearity above 30%) in the amol range for eight of the diluted peptides (Fig. 3A and supplemental Table 1). The coefficient of variance was estimated as 13.7% for the peptides spiked at constant concentrations (supplemental Table 1).
Fig. 3.
Limit of detection and intrascan dynamic range. A, the areas (y axis) of the precursor ion extracted from the survey scan (open squares) and of the most intense fragment ion extracted from the SWATH MS (closed triangles) and SRM (black crosses) quantifications are shown for the different serial dilution experiments (injected amounts of the peptide ELGQSGVDTYLQTK diluted in a yeast tryptic background in the x axis). The Mascot scores of the peptide identified in the same dilution series samples but acquired in DDA mode are shown as open circles. The limits of detection for the different methods are indicated with dotted lines. The complete series of LOD plots and corresponding lists of peak areas for the precursor and fragment ion traces quantified during these dilution series experiments are provided in supplemental Table 1 for the full set of 61 reference peptides. B, similar quantification plot for the doubly isotopically labeled peptide AADITSLYK serially diluted in a yeast tryptic background is shown here for the most intense fragment ion with closed triangles (“LOD control”). The intrascan dynamic range experiment consists of a dilution series of the same peptide AADITSLYK (open squares, “intrascan diluted”) in the presence of a constant amount of a singly isotopically labeled peptide AADITSLYK (open diamonds, “intrascan constant”), in the same yeast tryptic background. The complete lists of peak areas for the precursor and fragment ion traces quantified during the dilution series and intrascan dynamic range experiments are provided in supplemental Table 2. Screenshots of the quantified fragment ion traces and of the MS/MS spectra (zoomed around the y7 fragment) underlying the peptide peak apex are provided in supplemental Fig. S6 for the sample sets of the intrascan dynamic range experiment.
Next, we determined the intrascan dynamic range of the method, i.e., the fold change range separating the highest and lowest signal intensities concurrently observable within a same fragment ion spectrum. For this, an isotopically labeled peptide pair was chosen such that both (co-eluting) precursors were co-selected within the same swath. The samples consisted of a yeast tryptic digest spiked with one peptide at a constant amount of 1.25 pmol loaded on column, whereas the isotopic counterpart was diluted in a stepwise manner (supplemental Table 2 and supplemental Fig. S6). The data were acquired in SWATH MS mode and analyzed as described above. Fig. 3B shows that the diluted peptide species could be detected and quantified linearly through a dynamic range of almost 4 orders of magnitude. It is noteworthy that the signal did not demonstrate saturation even at the highest peptide concentration, suggesting that dynamic range could be further extended by using higher peptide concentrations. Thus, the sensitivity of the method seems so far limited by the chemical or electronic noise of the measurement itself rather than by intrascan dynamic range considerations.
We then compared the performance of SWATH MS with that of other standard proteomic strategies. For the comparison with DDA, the LOD dilution series samples described above were analyzed on the same MS instrument running in “top 20” shotgun mode. The data were searched with Mascot, and the identification score for the 61 reference peptides was reported on the same plots as those from the SWATH MS-extracted fragment intensities (supplemental Table 1). The results indicate that, for 26 of the 38 diluted peptides, the database searches failed to identify the reference peptides even when those were spiked at concentrations that were 2–10 fold higher than the respective LOD in the SWATH MS data sets. It is noteworthy that all the missing peptide identifications were actually due to nonselected signals for MS/MS sequencing. This experimentally demonstrates that precursor ion detection/picking in the MS1 scans is less reliable than fragment ion signal extraction from the MS/MS scans.
To compare the performance of SWATH MS with that of label-free workflows, we integrated the precursor ion traces extracted from the MS1 scans present in the exact same set of files acquired by SWATH MS for the dilution series samples. For the acquisition of this data set, a survey scan was carried out at the beginning of each swath cycle using the same periodicity (3.2 s) and accumulation time (100 ms) also applied per swath window (Fig. 1A), thus providing the closest quantification comparison possible. The MS1 areas were reported on the same plots as the SWATH MS-extracted fragment intensities (supplemental Table 1). The results show that, in half of the cases (for 19 of the 38 diluted reference peptides), SWATH MS quantification at the fragment ion spectra level offers a 2–8-fold gain in sensitivity compared with the LOD based on precursor ion signals detected in the MS1 maps. Supplemental Fig. S7 provides such an example of diluted peptide (ANLIPVIAK) whose precursor is only detectable until 1.5 fmol in the MS1 scans, whereas its fragment ions are still unambiguously identifiable and quantifiable down to 180 amol by targeted data extraction of the MS/MS scans. Finally, the LOD dilution series were analyzed on our most sensitive triple-quadrupole instrument operating in SRM mode (supplemental Table 3). This comparison showed that SRM was ∼10-fold more sensitive, placing SWATH MS coupled to targeted data extraction between SRM and MS1/label-free quantification workflows in terms of sensitivity.
Quantification Accuracy of SWATH MS-targeted Data Analysis
Next, we sought to benchmark the quantification accuracy of SWATH MS targeted analysis to that of SRM, the gold standard mass spectrometric quantification method. For this, we reacquired, via SWATH MS, samples 1 and 8 corresponding to the start and end points of a yeast diauxic shift experiment previously analyzed by SRM (15). Those samples consisted of tryptic digests of a mixture of (i) a lysate of yeast cells grown in regular 14N medium and sampled throughout the metabolic shift from fermentation to respiration and (ii) a constant 15N-labeled yeast lysate as internal standard for the fold change calculations. As a first pass analysis, the SWATH MS data set was mined with the exact same set of 476 transitions used to quantify the fold change of 80 peptides (45 metabolic enzymes) in the SRM study (15). From this initial data mining, mProphet automated analysis could validate 64 peptide identifications (1.5% false discovery rate; supplemental Fig. S8). Upon visual inspection of the extracted fragment ion traces, we could confirm the quantification for 40 proteins (72 peptides), whereas 5 proteins (8 peptides) were not convincingly detectable with this initial set of transitions (supplemental Tables 4 and 5).
Unlike SRM data, SWATH MS data sets contain transition signals different from those originally extracted and fragmentation information for other peptides than those originally targeted. Taking advantage of this, we re-extracted, from the exact same two files, additional or alternative peptide fragment ion traces for proteins whose identification and/or quantification was compromised because of fragment ion interferences or low signal to noise ratio during the primary data extraction. This straightforward data reanalysis rescued quantification information for three of the five undetected proteins, by quantifying nine novel peptides (supplemental Table 6) and significantly improved the quantification accuracy for the three proteins displaying the highest standard deviations in the primary analysis (supplemental Table 7). Fig. 4A summarizes the final quantification results and confirms that enzymes from the glycolysis pathway show a slight (maximum 2-fold) down-regulation, whereas those involved in the glyoxylate and citric acid cycles show between 10- and 300-fold up-regulations, consistent with the data of the SRM study. For a more direct comparison with the SRM results, we also plotted the proteins fold changes quantified with SWATH MS targeted analysis against those published in the SRM study. The correlation plot (Fig. 4B) shows an excellent linear correlation between the quantification results (slope > 0.9, r2 > 0.95) and benchmarks the quantification accuracy obtained by SWATH MS targeted analysis to the level of quality delivered by SRM data acquisition.
To demonstrate the effect of the fragment ion mass accuracy and resolution on the quantification performance, we artificially relaxed the resolution of the SWATH MS measurements, postacquisition, to mimic either a data-independent acquisition on a high resolution instrument but without isolation window or a windowed acquisition on a low resolution instrument (simulating the conditions of MSE/AIF (19, 21) or DIA (18) data sets, respectively, see Table 1). This was achieved in silico either by recombining the swaths prior to fragment ion chromatogram extraction at 10-ppm mass accuracy or by extracting the swaths data at 1-Da mass accuracy, respectively. The mProphet analysis results (supplemental Fig. S9) show that neither of those low specificity acquisition methods can match the number of identifications and quantification accuracy levels achieved by SWATH MS, especially for the proteins of low abundance.
Extending the Set of Quantified Proteins from SWATH MS Data Sets
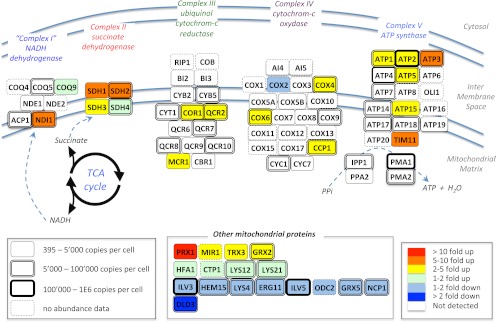
SWATH MS data sets implicitly contain a permanent fragment ion spectral record for all precursors within the mass and hydrophobicity range covered by specific LC-MS/MS acquisition conditions. This allows, in principle, for probing the data sets in silico for any new protein of interest suggested by a first pass biological review of the data, a situation that is common for systems biology studies. To illustrate this capability, the diauxic shift data sets were queried for 60 yeast mitochondrial proteins (287 peptides) involved in oxidative phosphorylation of the respiratory chain. These were not covered in the initial SRM study but were a posteriori considered relevant in the context of the switch from fermentation to respiration that occurs upon the diauxic shift. The data reanalysis consisted of extracting, from the same diauxic shift files, fragment ion traces of those targeted peptides for which we had assay records in our yeast spectral libraries. From the list of mitochondrial proteins, we could confidently quantify the abundance fold change for 36 proteins (103 peptides), 19 of which were membrane-associated proteins from the respiratory chain (Fig. 5 and supplemental Table 8). As for the previous analysis, the abundance fold change was measurable for proteins spanning a wide range of protein abundances (from 395 to 8.8E5 copies/cell (32)).
Fig. 5.
Extended quantification by SWATH MS of the abundance fold changes of mitochondrial enzymes during a diauxic shift experiment. Schematic representation of the respiratory chain and oxidative phosphorylation networks inspired by the Kyoto Encyclopedia of Genes and Genomes pathway representation (47). The abundance fold changes of the enzymes quantified by SWATH MS are coded with colors. The box shapes are indicative of the absolute abundances of the proteins with the same notices as those mentioned in Fig. 4. The complete list of peptides and of their fragment ions used to quantify those proteins, as well as the peak areas and corresponding protein abundance fold changes are provided in supplemental Table 7.
Identification of Post-translational Modifications
It is noteworthy that peptide modifications may also appear serendipitously as result of the targeted data extraction of SWATH MS data sets. When the fragment ion traces used to query a peptide are shared with modified forms of that peptide and when those are extracted in the same swath, multiple peak groups matching the original query can be observed. Fig. 6 illustrates such a case for the 14N-labeled (light) and 15N-labeled (heavy) forms of the endogenous peptide MIEIMLPVFDAPQNLVEQAK (proteotypic for protein PDC1), queried in the yeast diauxic shift sample 8 (late time point). Additional, nonshared fragment ions can then be re-extracted to distinguish which peak group corresponds to the nonmodified or modified peptides, respectively (supplemental Fig. S9). In cases where the modified peptide is fragmented in a different swath, the shared fragment ion masses may still be used to specifically query for the modified peptide form in that swath. These data illustrate the potential of SWATH MS targeted data extraction for unambiguous modification site assignment by extracting specific fragment ions characteristic of the modified peptide sequence. This opens completely novel opportunities to discover (and quantify) unanticipated modified peptide species from DIA data sets by a strategy that does not suffer from the combinatorial explosion of the search space usually experienced with traditional post-translational modification database search approaches.
Fig. 6.
Application of SWATH MS targeted analysis to identify peptide modifications. The six most intense fragment ion traces for the 14N-labeled (light) and 15N-labeled (heavy) forms of the peptide MIEIMLPVFDAPQNLVEQAK, extracted from the swath 750–775, are shown for the yeast diauxic shift sample y8 (late time point). None of the classical SRM criteria (fragments co-elution, light-heavy peptide co-elution, relative intensities of the fragment ions) can discriminate the three candidate peak groups found here. By extracting additional, nonshared fragment ion traces, the identification of the peptide can be confirmed, and the site of the oxidized methionine modification can be unambiguously assigned onto the peptide sequence (supplemental Fig. S10).
DISCUSSION
Among the various MS-based proteomic approaches, SRM is generally recognized as providing the most accurate and reproducible quantification results. The high degree of reproducibility is granted by the consistent recording, across the LC, of the intensities of predefined target fragment ions. This allows consistent tracking the abundance of specific peptides of interest across multiple samples. At present, however, SRM suffers from relatively slow analysis rates and lacks the capability to dynamically refine or expand the measured peptides/proteins for extensive proteome investigations. To alleviate most limitations of targeted data acquisition, we propose here a targeted data analysis strategy that brings the consistent and accurate quantification capabilities of SRM to a level of extensive proteome coverage by mining the complete fragment ion records generated during data-independent acquisition.
Not all DIA methods may be appropriate for targeted data extraction. To reach the quantification accuracy of SRM with targeted data extraction, the LC-MS/MS acquisition has to provide fragment ion data of a level of specificity that is comparable with that of SRM. Based on our fragment ion interference simulations (Fig. 2), we adopted a sequential window DIA method operating with 25-Da isolation width. On a fast, high resolution MS instrument, this setup allows documentation, in a single injection, of highly specific and time-resolved fragment ion data for all the precursors within the 400–1200 m/z mass and the monitored LC range (Fig. 1). The data thus generated constitute a series of extensive fragment ion maps ideally suited for proteome-wide investigation by targeted data analysis. DIA acquisition using consecutive swaths is not novel per se (18, 20). However, its rationally designed implementation on a fast, high resolution MS instrument provides, for the first time for a DIA method, the level of data quality necessary for targeted data extraction. This acquisition method is now commercially available on the ABSciex 5600 TripleTOF instrument under the SWATH MS denomination.
It should be noted that this SWATH MS setup (recording 32 swaths of 25 Da at 100-ms dwell time) is only one of many acquisition sets that can be applied. Like other mass spectrometric methods, SWATH MS operates within a space of interdependent parameters, including dwell time, duty cycle, and precursor isolation window width that affect the limit of detection, signal specificity, dynamic range, and quantification accuracy. Depending on the biological application or sample complexity, other parameters, including windows of variable widths throughout the LC gradient, might prove more efficient. Also, fragment ion specificities similar to those achieved by SWATH MS may very well be reached by other DIA methods, upon higher resolution of co-eluting analytes (e.g., using multidimensional protein identification technology (MudPIT) (18), ultrahigh pressure liquid chromatography (UPLC) (19), or ion mobility shift), although the gain in fragment ion specificity offered by extensive fractionations was recently questioned (12).
To mine the fragment ion maps recorded during SWATH MS acquisition, we devised a targeted data extraction strategy that conceptually transposes, to the data analysis, principles originating from SRM targeted acquisition. This targeted data analysis strategy differs fundamentally from the traditional search approaches described so far to analyze DIA data sets. Specifically, this type of analysis does not rely on precursor ion mass detection nor involve MS/MS spectra matching of any kind (neither using traditional database searching tools nor spectral matching algorithms). Instead, it consists of extracting, from the SWATH MS data sets, several fragment ion chromatograms for each peptide of interest. Collectively, these trace groups identify the targeted peptide, as in SRM analysis (Fig. 1C). Because both the peptide identification and quantification are performed at the MS/MS level, without the precursor ion signal having to be explicitly detected in the survey scans, this strategy allows extensive exploration of the multiplexed MS/MS DIA data sets to a level that was not possible with the traditional clustering/database approaches.
This targeted extraction strategy, like SRM, depends on spectral libraries as prior knowledge, to guide the selection of the optimal set of fragment ion signals. For several species, proteome-wide reference spectral libraries have been completed and will be made public in the near future. These libraries are S. cerevisiae,2 human,3 and Mycobacterium tuberculosis.4 Given that robust and high throughput methods for the generation of such libraries have been developed (e.g., by systematically recording MS/MS reference spectra of chemically synthesized proteotypic peptides (34)), we anticipate that proteomes of additional species will be equally mapped out in the near future. Alternatively, spectral libraries may be generated for any sample by extensive DDA analysis using the same instrument. To increase the reliability of such libraries, consensus spectra can be generated from repeated observations of the same peptide using freely available tools (35–37). The use of reference spectra as a priori information guiding the targeted extraction of DIA data sets may be less error-prone than approaches relying on clustering the fragment and precursor ions based on their LC elution profiles. Indeed, targeted data extraction can identify and quantify two exactly co-eluting peptides (e.g., light and heavy labeled peptide forms), even if they are present at vastly different abundance levels (Fig. 3B), a situation that challenges clustering approaches (26) and requires recursive search implementations to deconvolute the multiplexed spectra (38).
To evaluate the limit of detection of the method, a set of isotopically labeled serial dilution experiments was performed and showed that SWATH MS acquisition coupled to targeted data analysis could identify and quantify peptides down to the hundred amol range (Fig. 3). Even though the method in its current setup was slightly less sensitive than SRM, it remains to be determined whether the systematic optimization of the SWATH MS acquisition parameter sets, e.g., the use of dynamically adjusted window widths and increased dwell times, can further improve the LOD of the method. Generally, performance comparisons of methods are problematic if the comparisons include too many variables such as different samples or instrument types, instrument settings, etc. We therefore compared data acquired by SWATH MS with data generated by DDA and by MS1 quantification using aliquots of the same sample measured on the same ABSciex 5600 TripleTOF instrument. Overall, SWATH MS outperformed the two other methods for the consistent detection and quantification of low abundance precursors, especially if complex samples were analyzed (Fig. 3 and supplemental Figs. S6 and S7 and supplemental Tables 1 and 2). This result corroborates observations from previous DIA reports (18, 20, 22, 24) and can be explained by an increased signal to noise ratio in the fragment ion maps compared with the survey scans. This also emphasizes that unbiased acquisition methods such as SRM and DIA are particularly well suited for the detection of low level analytes in complex samples because the detection and quantification is based on fragment ion signals without the explicit need to detect the precursor ion in a survey scan above noise.
The intrascan dynamic range of the method was also experimentally assessed and was shown to cover almost 4 orders of magnitude (Fig. 3B). Such extent of identification (and quantification) of co-eluting peptides spanning 4 logs of concentration, reliably detected here with targeted data extraction (supplemental Fig. S6), may be more challenging to achieve with traditional DIA analysis approaches relying on fragment ion preclustering and/or regular database searches. To our knowledge, this is indeed the first attempt to objectively evaluate the intrascan dynamic range of peptide identification/quantification for a DIA approach, even though this parameter is of utmost importance for proteome analyses, in particular if wide precursor isolation windows are being used. It is noteworthy that the most abundant precursor actually limits the dynamic range only for its specific isolation window and therefore does not affect the detection sensitivity achievable simultaneously in other swaths. Thus, for SWATH MS, an even greater dynamic range may be anticipated throughout the 400–1200 m/z range at each time point and across the LC-MS range as a whole. A wide intrascan dynamic range achievable in flow-through instruments like the quadrupole-quadrupole TOF instrument used in this study might be difficult to achieve with trapping instruments. Their limited ion trapping capacity restricts the number of peptide species that can be concurrently analyzed without compromising performance through space charging. On quadrupole-quadrupole TOF instruments, the ions are transferred through a quadrupole to the collision cell and to the TOF analyzer, irrespective of the number or abundance of co-selected precursors, a feature that is critical for reaching a high intrascan dynamic range with DIA methods using large isolation windows that produce high ion fluxes. Also, an optimal “square shape” for the ion transmission efficiency (as achieved here by decoupling the DC and RF voltages of the isolation quadrupole; supplemental Figs. S1) might be difficult to maintain throughout the entire isolation window width on current trapping devices and may require larger overlaps between adjacent swaths to ensure consistent quantification of the analytes transmitted at the border of the isolation windows. Therefore, whereas the principles of data-independent acquisition with swaths can conceivably be implemented on different types of mass spectrometers, it appears that the characteristics of flow-through systems like quadrupole-quadrupole TOFs are currently the best match for the method.
More importantly, we evaluated the quantification reproducibility achievable by SWATH MS coupled to targeted data analysis and its potentials for proteome quantification for biology. Comparing SRM- and SWATH MS-derived quantitative values obtained from the same isotope labeled samples (two yeast diauxic shift samples previously analyzed by SRM (15)), both methods showed highly correlated values (Fig. 4B). Overall, SWATH MS coupled to targeted data extraction allowed consistent quantification of proteins spanning a wide range of concentrations, e.g., 125–106 copies/cell (Fig. 4A, box shapes). Unlike SRM data, SWATH MS data sets are permanent records of the fragment ion spectra of a sample that can be re-examined in silico without the need for further data acquisition. This characteristic, specific to DIA data sets, opens new possibilities to rescue missing quantification information and to improve the accuracy of initial quantification results simply through iterative targeted data reanalysis, as demonstrated here for several metabolic enzymes (supplemental Tables 5–7). It has been discussed that, for SRM measurements, interference of contaminating transitions, incomplete tryptic cleavage, or possible modifications of a peptide or other such artifacts may impede the accuracy of quantification (39, 40). The optimization of fragment ion sets for each targeted peptides by the iterative SWATH MS data analysis offers practical solutions to these important issues. Interfering transitions can be detected and eliminated using outlier detection algorithms, and the data set can be queried for other peptides from the targeted protein or for alternate peptides, e.g., derived by unspecific or partial cleavage or modified peptides covering the same segment of a protein. Once detected, such instances can be eliminated or taken into account to achieve higher quality data (supplemental Table 7).
The possibility of iteratively searching the SWATH MS data sets also supports ad libitum queries for protein sets. Although the diauxic shift samples used in this study were not originally intended for the recovery of mitochondrial membrane proteins (15), we could confidently quantify and assess the fold changes for 36 proteins involved in the oxidative phosphorylation and respiratory networks (Fig. 5). Those proteins were not covered in the initial analysis and would have required new targeted data acquisition of the samples by SRM. With the targeted data analysis strategy, the new protein set can simply be re-extracted in silico from the existing SWATH MS data files, without the need to reinject the sample. This dynamic extension of the search space applied to SWATH MS data sets is expected to be particularly attractive for systems biology studies where new query hypotheses are generated from mathematical models based on prior data analysis. Although it is in principle possible to probe SWATH MS data sets for the whole proteome of an organism at once, it is beyond the scope of this article to describe the exhaustive quantification of all the yeast proteins detectable in those diauxic shift samples by SWATH MS. Indeed, although the data are already analyzable with mProphet or Skyline, none of the currently available SRM analysis tools can so far fully exploit the information potential contained in SWATH MS data sets. For example, no SRM analysis pipeline takes into account the mass accuracy of the fragment ions, nor their isotopic distribution or charge state. Although those parameters are neither relevant nor accessible to quadrupole resolution used in SRM acquisition, they are instrumental to adequately mine SWATH MS data sets. Therefore, a more complete and specific targeted data analysis pipeline is required before attempting exhaustive qualitative and quantitative proteome characterization of SWATH MS data sets.
The concept of SWATH MS acquisition and targeted data analysis should be easily extendable to other classes of biomolecules such as metabolites, lipids, and more that are also frequently studied by LC-MS/MS and for which fragment ion spectral libraries have been developed (41–46). Also, the possibility to re-examine patterns in the SWATH MS data sets enables new opportunities for finding modified residues and search for the presence of previously unexpected analytes (Fig. 6).
In summary, we report a method for qualitative and quantitative proteome probing of a sample in a single LC-MS/MS injection. This is achieved by the combination of a sequential windowed DIA method, generating exhaustive high specificity fragment ion map records, coupled with a postacquisition targeted data analysis strategy. This method permits quantification of (at least) as many compounds as those typically identified by regular shotgun proteomics with the accuracy and reproducibility of SRM across many samples. The method also provides new possibilities for data analysis, allowing quantification refinement and dynamic protein probing by iteratively re-mining the once-and-forever acquired data sets.
Acknowledgments
We acknowledge Christine Carapito (CNRS, Strasbourg, France) for early contributions in evaluating the potentials of SWATH MS. We thank Paola Picotti (Institute of Biochemistry, ETH Zürich) for providing the diauxic shift samples originating from an earlier study (15). We thank Uwe Sauer and Ana Paula Oliveira (Institute of Molecular Systems Biology, ETH Zürich) for suggesting the set of respiratory chain proteins additionally quantified in the diauxic shift samples. We thank Lyle Burton (ABSciex) for active development of the PeakView software.
L. C. G., P. N., and R. A. designed the study. S. T. implemented and developed the acquisition method on the ABSciex 5600 TripleTOFTM instrument and performed the data acquisitions. H. R. computed the theoretical simulations of fragment ion interferences. N. S. performed the comparative measurement of the AQUA dilution series by SRM. L. R. helped implementing the SWATH MS analysis in mProphet. L. C. G. and P. N. performed the SWATH MS data analysis. R. A. and R. B. supervised the study.
Footnotes
* This work was supported by ABSciex; European Union FP7 Prospects Grant 201648; SystemsX.ch, the Swiss initiative for systems biology via the projects YeastX and PhosphonetX; ERC Proteomics v3.0 Grant 233226; and European Union FP7 “Unicellsys” Grant 201142. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
2 P. Picotti, et al., submitted for publication.
3 R. Aebersold, R. Moritz, et al., manuscript in preparation.
4 O. Schubert, J. Mouritsen, et al., manuscript in preparation.
1 The abbreviations used are:
- LC-MS/MS
- liquid chromatography coupled to tandem mass spectrometry
- DDA
- data-dependent acquisition
- DIA
- data-independent acquisition
- SRM
- single reaction monitoring
- RT
- retention time
- LOD
- limit of detection.
REFERENCES
- 1. Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 2. MacCoss M. J., Matthews D. L. (2005) Teaching a new dog old tricks. Anal. Chem. 77, 295A–302A [DOI] [PubMed] [Google Scholar]
- 3. Han X., Aslanian A., Yates J. R., 3rd (2008) Mass spectrometry for proteomics. Curr. Opin. Chem. Biol. 12, 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walther T. C., Mann M. (2010) Mass spectrometry-based proteomics in cell biology. J. Cell Biol. 190, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Domon B., Aebersold R. (2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 6. Kapp E., Schutz F. (2007) Overview of tandem mass spectrometry (MS/MS) database search algorithms, in Current Protocols in Protein Science, Chapter 25, pp. 25.2.1–25.2.19, John Wiley & Sons, Inc, Hoboken, New Jersey, USA: [DOI] [PubMed] [Google Scholar]
- 7. Nesvizhskii A. I. (2007) Protein identification by tandem mass spectrometry and sequence database searching. Methods Mol. Biol. 367, 87–119 [DOI] [PubMed] [Google Scholar]
- 8. Lange V., Picotti P., Domon B., Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst. Biol. 4:222, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reiter L., Rinner O., Picotti P., Hüttenhain R., Beck M., Brusniak M. Y., Hengartner M. O., Aebersold R. (2011) mProphet: Automated data processing and statistical validation for large-scale SRM experiments. Nat Methods 8, 430–435 [DOI] [PubMed] [Google Scholar]
- 10. Domon B., Aebersold R. (2010) Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 28, 710–721 [DOI] [PubMed] [Google Scholar]
- 11. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 12. Michalski A., Cox J., Mann M. (2011) More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J. Proteome Res. 10, 1785–1793 [DOI] [PubMed] [Google Scholar]
- 13. Addona T. A., Abbatiello S. E., Schilling B., Skates S. J., Mani D. R., Bunk D. M., Spiegelman C. H., Zimmerman L. J., Ham A. J., Keshishian H., Hall S. C., Allen S., Blackman R. K., Borchers C. H., Buck C., Cardasis H. L., Cusack M. P., Dodder N. G., Gibson B. W., Held J. M., Hiltke T., Jackson A., Johansen E. B., Kinsinger C. R., Li J., Mesri M., Neubert T. A., Niles R. K., Pulsipher T. C., Ransohoff D., Rodriguez H., Rudnick P. A., Smith D., Tabb D. L., Tegeler T. J., Variyath A. M., Vega-Montoto L. J., Wahlander A., Waldemarson S., Wang M., Whiteaker J. R., Zhao L., Anderson N. L., Fisher S. J., Liebler D. C., Paulovich A. G., Regnier F. E., Tempst P., Carr S. A. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 27, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cima I., Schiess R., Wild P., Kaelin M., Schüffler P., Lange V., Picotti P., Ossola R., Templeton A., Schubert O., Fuchs T., Leippold T., Wyler S., Zehetner J., Jochum W., Buhmann J., Cerny T., Moch H., Gillessen S., Aebersold R., Krek W. (2011) Cancer genetics-guided discovery of serum biomarker signatures for diagnosis and prognosis of prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 108, 3342–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kiyonami R., Schoen A., Prakash A., Peterman S., Zabrouskov V., Picotti P., Aebersold R., Huhmer A., Domon B. (2011) Increased selectivity, analytical precision, and throughput in targeted proteomics. Mol. Cell. Proteomics 10, M110.002931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Purvine S., Eppel J. T., Yi E. C., Goodlett D. R. (2003) Shotgun collision-induced dissociation of peptides using a time of flight mass analyzer. Proteomics 3, 847–850 [DOI] [PubMed] [Google Scholar]
- 18. Venable J. D., Dong M. Q., Wohlschlegel J., Dillin A., Yates J. R. (2004) Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 [DOI] [PubMed] [Google Scholar]
- 19. Plumb R. S., Johnson K. A., Rainville P., Smith B. W., Wilson I. D., Castro-Perez J. M., Nicholson J. K. (2006) UPLC/MS(E): A new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun. Mass Spectrom. 20, 1989–1994 [DOI] [PubMed] [Google Scholar]
- 20. Panchaud A., Scherl A., Shaffer S. A., von Haller P. D., Kulasekara H. D., Miller S. I., Goodlett D. R. (2009) Precursor acquisition independent from ion count: How to dive deeper into the proteomics ocean. Anal. Chem. 81, 6481–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Geiger T., Cox J., Mann M. (2010) Proteomics on an Orbitrap benchtop mass spectrometer using all ion fragmentation. Mol. Cell. Proteomics 9, 2252–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bern M., Finney G., Hoopmann M. R., Merrihew G., Toth M. J., MacCoss M. J. (2010) Deconvolution of mixture spectra from ion-trap data-independent-acquisition tandem mass spectrometry. Anal. Chem. 82, 833–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carvalho P. C., Han X., Xu T., Cociorva D., Carvalho Mda G., Barbosa V. C., Yates J. R., 3rd (2010) XDIA: Improving on the label-free data-independent analysis. Bioinformatics 26, 847–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Panchaud A., Jung S., Shaffer S. A., Aitchison J. D., Goodlett D. R. (2011) Faster, quantitative, and accurate precursor acquisition independent from ion count. Anal. Chem. 83, 2250–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wong J. W., Schwahn A. B., Downard K. M. (2009) ETISEQ: An algorithm for automated elution time ion sequencing of concurrently fragmented peptides for mass spectrometry-based proteomics. BMC Bioinformatics 10:244, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Geromanos S. J., Vissers J. P., Silva J. C., Dorschel C. A., Li G. Z., Gorenstein M. V., Bateman R. H., Langridge J. I. (2009) The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics 9, 1683–1695 [DOI] [PubMed] [Google Scholar]
- 27. Li G. Z., Vissers J. P., Silva J. C., Golick D., Gorenstein M. V., Geromanos S. J. (2009) Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9, 1696–1719 [DOI] [PubMed] [Google Scholar]
- 28. Blackburn K., Mbeunkui F., Mitra S. K., Mentzel T., Goshe M. B. (2010) Improving protein and proteome coverage through data-independent multiplexed peptide fragmentation. J. Proteome Res. 9, 3621–3637 [DOI] [PubMed] [Google Scholar]
- 29. Sherman J., McKay M. J., Ashman K., Molloy M. P. (2009) Unique ion signature mass spectrometry, a deterministic method to assign peptide identity. Mol. Cell. Proteomics 8, 2051–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 31. Picotti P., Lam H., Campbell D., Deutsch E. W., Mirzaei H., Ranish J., Domon B., Aebersold R. (2008) A database of mass spectrometric assays for the yeast proteome. Nat. Methods 5, 913–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 33. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Picotti P., Rinner O., Stallmach R., Dautel F., Farrah T., Domon B., Wenschuh H., Aebersold R. (2010) High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods 7, 43–46 [DOI] [PubMed] [Google Scholar]
- 35. Craig R., Cortens J. C., Fenyo D., Beavis R. C. (2006) Using annotated peptide mass spectrum libraries for protein identification. J. Proteome Res. 5, 1843–1849 [DOI] [PubMed] [Google Scholar]
- 36. Frewen B. E., Merrihew G. E., Wu C. C., Noble W. S., MacCoss M. J. (2006) Analysis of peptide MS/MS spectra from large-scale proteomics experiments using spectrum libraries. Anal. Chem. 78, 5678–5684 [DOI] [PubMed] [Google Scholar]
- 37. Lam H., Deutsch E. W., Eddes J. S., Eng J. K., King N., Stein S. E., Aebersold R. (2007) Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 7, 655–667 [DOI] [PubMed] [Google Scholar]
- 38. Huang X., Liu M., Nold M. J., Tian C., Fu K., Zheng J., Geromanos S. J., Ding S. J. (2011) Software for quantitative proteomic analysis using stable isotope labeling and data independent acquisition. Anal. Chem. 83, 6971–6979 [DOI] [PubMed] [Google Scholar]
- 39. Duncan M. W., Yergey A. L., Patterson S. D. (2009) Quantifying proteins by mass spectrometry: The selectivity of SRM is only part of the problem. Proteomics 9, 1124–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sherman J., McKay M. J., Ashman K., Molloy M. P. (2009) How specific is my SRM?: The issue of precursor and product ion redundancy. Proteomics 9, 1120–1123 [DOI] [PubMed] [Google Scholar]
- 41. Schmelzer K., Fahy E., Subramaniam S., Dennis E. A. (2007) The Lipid Maps Initiative in Lipidomics, po. 171–183, Elsevier Science Publishers B.V., Amsterdam: [DOI] [PubMed] [Google Scholar]
- 42. Blanksby S. J., Mitchell T. W. (2010) Advances in mass spectrometry for lipidomics. Annu. Rev. Anal. Chem. 3, 433–465 [DOI] [PubMed] [Google Scholar]
- 43. Smith C. A., O'Maille G., Want E. J., Qin C., Trauger S. A., Brandon T. R., Custodio D. E., Abagyan R., Siuzdak G. (2005) METLIN: A metabolite mass spectral database. Ther. Drug Monit. 27, 747–751 [DOI] [PubMed] [Google Scholar]
- 44. Horai H., Arita M., Kanaya S., Nihei Y., Ikeda T., Suwa K., Ojima Y., Tanaka K., Tanaka S., Aoshima K., Oda Y., Kakazu Y., Kusano M., Tohge T., Matsuda F., Sawada Y., Hirai M. Y., Nakanishi H., Ikeda K., Akimoto N., Maoka T., Takahashi H., Ara T., Sakurai N., Suzuki H., Shibata D., Neumann S., Iida T., Tanaka K., Funatsu K., Matsuura F., Soga T., Taguchi R., Saito K., Nishioka T. (2010) MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 45, 703–714 [DOI] [PubMed] [Google Scholar]
- 45. Dresen S., Gergov M., Politi L., Halter C., Weinmann W. (2009) ESI-MS/MS library of 1,253 compounds for application in forensic and clinical toxicology. Anal. Bioanal. Chem. 395, 2521–2526 [DOI] [PubMed] [Google Scholar]
- 46. Dresen S., Ferreirós N., Gnann H., Zimmermann R., Weinmann W. (2010) Detection and identification of 700 drugs by multi-target screening with a 3200 Q TRAP LC-MS/MS system and library searching. Anal. Bioanal. Chem. 396, 2425–2434 [DOI] [PubMed] [Google Scholar]
- 47. Kanehisa M., Goto S., Furumichi M., Tanabe M., Hirakawa M. (2010) KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38, D355–D360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Andrews G. L., Simons B. L., Young J. B., Hawkridge A. M., Muddiman D. C. (2011) Performance characteristics of a new hybrid quadrupole time-of-flight tandem mass spectrometer (TripleTOF 5600). Anal. Chem. 83, 5442–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wepf A., Glatter T., Schmidt A., Aebersold R., Gstaiger M. (2009) Quantitative interaction proteomics using mass spectrometry. Nat. Methods 6, 203–205 [DOI] [PubMed] [Google Scholar]
- 50. Krokhin O. V., Craig R., Spicer V., Ens W., Standing K. G., Beavis R. C., Wilkins J. A. (2004) An improved model for prediction of retention times of tryptic peptides in ion pair reversed-phase HPLC: Its application to protein peptide mapping by off-line HPLC-MALDI MS. Mol. Cell. Proteomics 3, 908–919 [DOI] [PubMed] [Google Scholar]
- 51. Rost H. L., Malmstrom L., Ruedi Aebersold R. (2012) A computational tool to detect and avoid redundancy in selected reaction monitoring. Mol. Cell. Proteomics, mcp.M111.013045. First Published on April 24, 2012, doi:10.1074/mcp.M111.013045 [DOI] [PMC free article] [PubMed] [Google Scholar]