Abstract
To broaden the range of tools available for proteomic research, we generated a library of 16,368 unique full-length human ORFs that are expressible as N-terminal GST-His6 fusion proteins. Following expression in yeast, these proteins were then individually purified and used to construct a human proteome microarray. To demonstrate the usefulness of this reagent, we developed a streamlined strategy for the production of monospecific monoclonal antibodies that used immunization with live human cells and microarray-based analysis of antibody specificity as its central components. We showed that microarray-based analysis of antibody specificity can be performed efficiently using a two-dimensional pooling strategy. We also demonstrated that our immunization and selection strategies result in a large fraction of monospecific monoclonal antibodies that are both immunoblot and immunoprecipitation grade. Our data indicate that the pipeline provides a robust platform for the generation of monoclonal antibodies of exceptional specificity.
Protein affinity reagents are fundamental tools of both basic and applied biomedical research. They are used for a wide range of applications, including measurement of protein expression levels, detection of protein-protein and protein-nucleic acid interactions, and detection of disease biomarkers (1). Currently, the most widely used protein affinity reagents are polyclonal and monoclonal antibodies. Monoclonal antibodies (mAbs)1 are indefinitely renewable and recognize a single protein epitope, making them the more desirable of the two reagents for most applications (2, 3). Indeed, with over 20 mAb-based drugs now in use and over 100 in clinical trials, they have also become a central pillar of the biopharmaceutical industry (4). However, despite their widespread use, well characterized protein affinity reagents are not available for the great majority of human proteins. This lack of characterization has led to a major bottleneck in analyzing protein expression and function, often making interpretation of data obtained using any class of protein affinity reagent problematic (5, 6). Several recent studies have suggested that many commercially available mAbs may not even recognize their purported targets and cross-react extensively with other cellular antigens (7). Antibody cross-reactivity is an even more pressing problem in diagnostic and therapeutic applications, as underlined by the recent withdrawal of several mAb-based pharmaceuticals from the market (8, 9).
Several large scale efforts are now underway to systematically identify high grade antibodies against much of the human proteome (4, 10–12). These approaches, which are primarily directed toward validation of polyclonal antibodies, rely heavily on immunocytochemistry and immunohistochemistry for validation, using both cell lines and tissue microarrays. Although these efforts provide a great deal of useful information, they neither cover all tissues nor confirm that the antibody in question is actually recognizing its target antigen in all of the tissues examined. To address this, it would be necessary to comprehensively measure cross-reactivity of any given antibody against the full proteome, something that is in principle possible using microarray-based analysis of antibody specificity (13–15).
A protein microarray approach has been previously used to analyze the specificity of antibodies generated against viral (16), microbial (17, 18), and mammalian (19–22) proteins and is already used on a small scale as part of the Human Protein Atlas project (11). However, existing human protein microarrays either are protein family-specific (22) or are comprised of only a minority of the human proteome (20, 23, 24). Although Goshima et al. (25) described fabrication of a more comprehensive human protein microarray that contained a total of 13,364 human proteins, these proteins were not purified away from the in vitro translation reaction mixtures used for protein synthesis, a fact that severely limits the potential usefulness of this reagent.
To remedy this situation, we have developed a microarray that includes nearly two-thirds of the annotated full-length human proteome. The proteins used to generate this microarray were purified under native conditions at low cost following galactose-induced expression from Saccharomyces cerevisiae (24, 26). Expressing recombinant eukaryotic proteins in yeast allows one to obtain higher success rate of purification and also improves the chances that proteins will retain biological activity relative to prokaryotic and in vitro-based expression systems (24, 26, 27). Furthermore, the use of an evolutionarily distant heterologous expression system like yeast minimizes the risk of contamination of recombinant human proteins with interacting cellular proteins, which is a potential complication that can result from the use of mammalian cells for protein expression.
A microarray with this level of coverage of the human proteome can potentially be used to identify antibodies that efficiently recognize proteins in their native conformation and that are thus useful for applications such as immunoprecipitation. We have used this tool as the backbone of an integrated platform that enables rapid and low cost identification of mAbs that both selectively bind a diverse assortment of human proteins and are useful in a wide range of experimental applications.
MATERIALS AND METHODS
Human Protein Microarray Construction
The cloning of human ORFs and protein purification was described previously (24). NanoPrint LM210 system (ArrayIT, Sunnyvale, CA) was used to print protein microarrays on Full Moon slides (Full Moon BioSystems, Sunnyvale, CA).
Generation of Hybridomas
Cultured human cells (SH-SY5Y, HepG2, HCT116, HeLa, HL-60, and MCF7) used for immunization were grown to mid-log phase in appropriate media, collected from culture by centrifugation, and washed three times with cold PBS. Equal volumes of pelleted cells and Titermax adjuvant were combined and emulsified by vortexing. 6–8-week-old BALB/c mice were immunized in rear footpad or hock area with volumes of this mixture containing the equivalent of 2.5 × 106 cells. Popliteal lymph nodes were harvested 14–16 days later and teased into single-cell suspensions. These immune cells were fused to SP-2/0 myeloma cells with 50% polyethylene glycol under standard conditions (3). Fusion reactions were plated into 60-mm Petri dishes containing DMEM, HAT, and 1% methyl cellulose and incubated at 37 °C under 5% CO2 atmosphere. Clonal colonies were visualized with an inverted stage dissecting microscope, harvested with microcapillary pipettes, and transferred into 96-well plates containing DMEM and HAT for expansion. Culture supernatants from individual wells were tested by ELISA for the presence of IgG, and antibody-positive wells were expanded into T-25 flasks to generate IgG-containing medium for subsequent antibody characterizations.
Immunocytochemistry (ICC) Prescreening of mAbs
Cultured cell lines used for immunization were grown to 80% confluence in appropriate media, harvested, and plated with fresh media onto untreated (for HeLa and HCT116 cells) or poly-l-lysine-coated (for SH-SY5Y, HepG2, MCF7, and HL-60 cells) 16-well glass slides. Slides receiving HL-60 cells were additionally centrifuged for 2 min at 200 r.c.f. to improve attachment. All of the slides were incubated overnight at 37 °C with 5% CO2 atmosphere. The cells were washed two times with PBS and fixed with 100 μl of 4% paraformaldehyde in PBS for 15 min at room temperature. The wells were blocked with 100 μl of a mixture of 3% goat serum, 3% BSA, 0.1% Triton X-100 in PBS for 1 h at room temperature. mAb-containing solutions were adjusted to10 μg/ml IgG in blocking buffer and incubated with cells for 2 h at room temperature or overnight at 4 °C. The wells were washed three times with 150 μl of PBS for 10 min. Secondary antibody (anti-mouse IgG:DyLight 488) was added to wells at room temperature for 2 h in the dark. Wells were washed three times with PBS, gaskets were removed, and the slides were mounted with antifade solution for viewing with a fluorescence microscope equipped with a computer-coupled camera.
Protein Microarray-based Analysis of mAb Specificity
Hybridoma supernatants (with antibodies at an average final concentration of 20 μg/ml in 1× PBS) were screened in batches of 144 as two-dimensional pools. Twelve horizontal and vertical pools of 300-μl volume were generated, consisting of equal mixtures of supernatants derived from 12 different hybridomas. These supernatants were combined such that each individual hybridoma was included in exactly one vertical and one horizontal pool, but such that no two hybridomas that are part of a given horizontal pool are found together in any vertical pool. The binding specificity of each horizontal and vertical pool was then profiled. Protein microarrays were then blocked for 2 h with 2% BSA in 1× PBS at room temperature, incubated with antibody pools for 1 h at room temperature along with rabbit anti-GST (Millipore) at 1:5000, washed three times for 15 min in 1× TBST, incubated with 1:800 Cy5-goat anti-mouse and 1:1000 Cy3 goat anti-rabbit (Invitrogen) in blocking buffer for 1 h at room temperature, washed three times for 15 min in 1× TBST, rinsed once in double-distilled H2O, spun dry, and scanned using a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA). Scanned images were analyzed using GenePix software, and proteins that bound to serum mouse IgG alone were excluded from analysis. Signal intensity was then calculated as the ratio of median foreground and median background signals in the Cy5 channel when analyzing bound mAb signal and the Cy3 channel when detecting GST.
To quantify the affinity of individual mAbs to specific proteins on the array, we first calculated the mean and standard deviation of the signal intensity across all spots on the chip. We obtain normalized signal intensity for any pair of spots, which we define as A, which is the mean z score for each duplicate pair of spotted proteins, where An = (In − m)/σ. Here, I is the ratio of median foreground and median background fluorescence for any given spot pair n, m is the median value for I for all spots on the array, and σ is the standard deviation for I. mAbs found in one unique row pool and one column pool that showed z scores greater than 2.8 for both duplicate spots of any given protein were then flagged for individual analysis.
mAbs identified as potentially specific using this pooling strategy were then tested individually against the entire array, and A was measured for each spotted protein. We next quantitatively evaluated the specificity of any individual mAb identified as potentially specific by means of this analysis. To do this, we calculated a value for specificity that we define as S, where S = A1 − A2. Here, A1 represents the spot pair on the array that shows the highest value of A, and A2 represents the spot pair with the second highest value of A.
Ascites Generation and Antibody Purification
6–8-week-old BALB/c mice were primed with 0.5 ml of Freund's incomplete adjuvant injected intraperitoneally and rested for 5–7 days. Hybridoma cells were grown to log phase and washed three times with PBS. Five million hybridoma cells were injected intraperitoneally with 0.5 ml of PBS. The mice were monitored daily for ascites production. Ascites was harvested a maximum of three times per animal, pooled, clarified by centrifugation at 6000 r.c.f. at 18 °C, and stored at −80 °C. Thawed ascites were centrifuged at 15,000 r.c.f. for 10 min at 4 °C to remove excess liquid. Protein G-Sepharose was added (125 μl of bead volume/ml of ascites) and rotated at 4 °C for 1 h. The beads were washed three times with cold 50 mm sodium phosphate, pH 7.0, and antibody was eluted with 300 μl of 100 mm glycine, pH 2.5. Eluted material was neutralized with 50 μl of 1 m Tris, pH 9.0, and dialyzed overnight at 4 °C against PBS.
Plasmid Preparation and Protein Expression in Human Cells
Target protein ORFs were transferred into pcDNA-3.1-V5 (Invitrogen) using LR reaction. The resulting expression constructs were digested with BsrGI to confirm clones. Sequencing further validated the identity of expression vectors.
Cell Culture and Transfection
HeLa cells were maintained in DMEM (10% heat-inactivated fetal bovine serum). SH-SY5Y neuroblastoma cell was maintained in a 1:1 mixture of DMEM/F12 medium supplemented with 10% heat-inactivated fetal bovine serum. The expression constructs were transfected into HeLa cell using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions.
shRNA Knockdown
Co-transfection was performed with 1:3 (expression construct: shRNA expression construct). Samples were collected 36 h after transfection for immunoblot. The following clones were used along with protein expression vectors: ANXA2 (TRCN0000056145), SRM (TRCN0000045729), SIRPB1 (TRCN0000029799), HNRPC (TRCN000006645), GMDS (TRCN000052471), and CEACAM6 (TRCN0000062302). TRC shRNA constructs were obtained from the Genome Resources core facility at the Johns Hopkins University School of Medicine.
Immunoblot and Immunoprecipitation
1 × 106 HeLa cells grown in 6-well plates were lysed in 500 μl of buffer consisting of 100 mm Tris HCl, pH 7.4, 150 mm NaCl, 15% glycerol, 1% Triton X-100, protease inhibitor tablet (Roche Applied Science), and MG-132 (final concentration, 100 μm). The cell lysate was then centrifuged to remove the insoluble fraction. Cleared lysates were used for immunoblotting with individual monoclonal antibodies. After being stripped, the same blots were probed using antibodies to actin and the V5 epitope tag to control for protein content and to confirm the expression of the transfected target protein. For IP assays, cleared lysate was aliquoted into equal volumes, and 2 μg of the appropriate antibody was added. Anti-V5 antibody was used in parallel as a positive control. As a negative control, primary antibody was excluded. The mix was incubated at 4 °C for 2 h. Twenty-five microliters of prewashed protein G Dynabeads (Invitrogen) was added to the mix and incubated for an additional 2 h at 4 °C. The IgG-protein complex was separated by a magnetic separator and washed two times with PBST (0.1% Tween 20). The samples were transferred to a fresh tube, and a final wash with PBST was followed. The samples were separated on SDS-PAGE and transferred to nitrocellulose membrane. After blocking with 5% skim milk in PBST, anti-V5 horseradish peroxidase conjugate (Invitrogen) was used for immunoblotting.
Analysis of Commercially Available mAbs
Only mouse mAbs reported to recognize unmodified human proteins were used for this analysis. Information on the vendor's website was used to determine the experimental uses of each mAb. To select 100 random mAbs from commercial suppliers, a random number generator was used to select a number between 1 and 26. This was then used to select the corresponding letter of the alphabet, which was then used to search the online catalogue for each supplier. mAbs were then selected for inclusion in descending alphabetical order. If more than one mAb recognizing a given protein was included in the catalogue, only the first listed mAb was included in the analysis.
Statistical Analysis
The values in this study are represented as the means ± S.D. Statistical comparisons were done using a two-tailed Student's t test, and p < 0.05 was considered to be statistically significant.
RESULTS
Construction of a Human Proteome Microarray
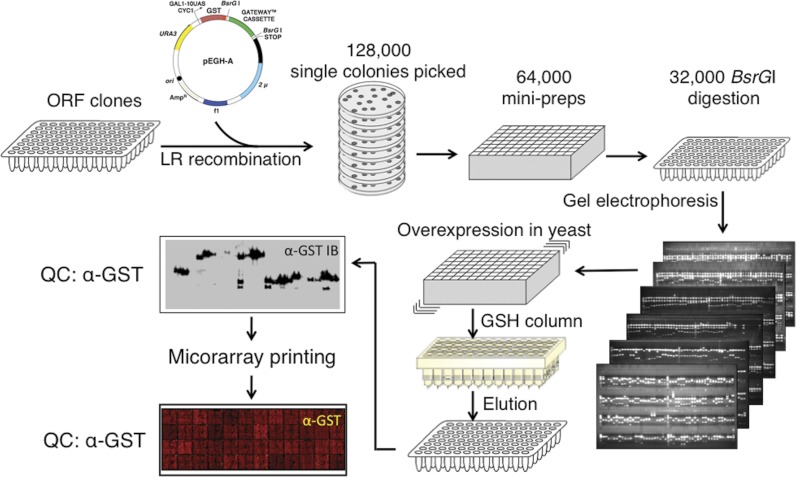
We constructed a proteome microarray consisting of 16,368 unique full-length human proteins, which represent a total of 12,586 different full-length genes, in all covering >60% of the annotated human proteome (28) (Fig. 1). To express and purify these proteins, we first subcloned the Invitrogen ultimate ORF collection (29), along with 445 additional full-length ORFs generated in our own labs, into a yeast expression vector, which allows galactose-dependent overexpression of N-terminal GST- and His6-tagged recombinant proteins as previously described (24) (supplemental Table 1). Protein quality was determined by GST immunoblotting of a subset of recombinant proteins (Fig. 1). Based on these results, we estimated that 98% of all proteins showed a major band of the expected size following purification (supplemental Fig. 1 and supplemental Table 2).
Fig. 1.
Construction of a human proteome microarray. A human ORF collection was mobilized into a yeast galactose-inducible GST fusion vector (pEGH-A) using Gateway-mediated site-specific recombination. Individual clones were verified to have correctly sized inserts by BsrGI digestion. Clones with confirmed identities were transformed into yeast, and large scale protein expression and purification were performed. Protein size and purity were tested by anti-GST immunoblotting. Protein samples were printed on a glass slide in duplicate, and printed spots were visualized by anti-GST antibody.
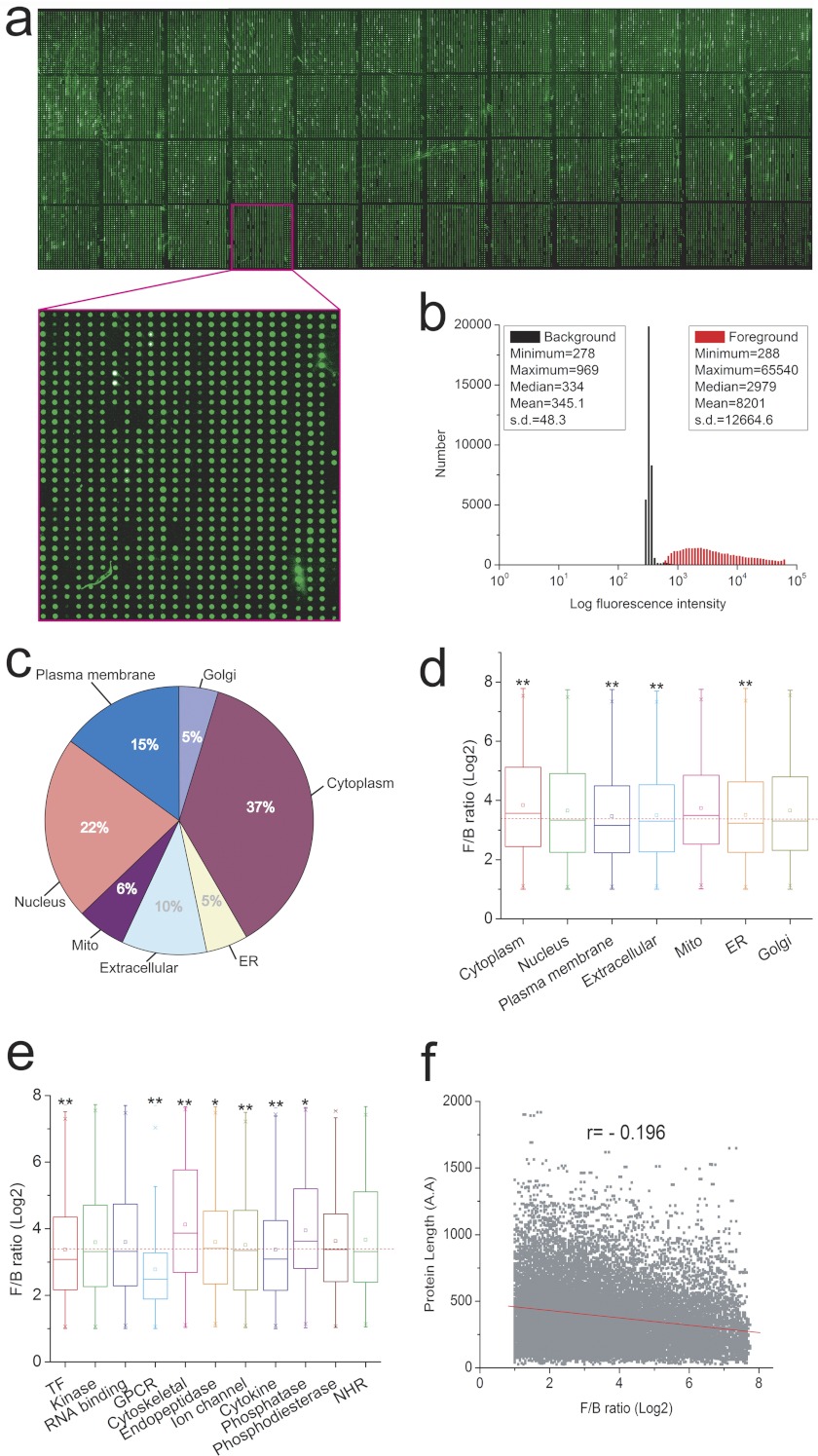
We next spotted the full set of purified recombinant proteins, in duplicate, along with a set of control proteins (e.g. GST, BSA, histones), on a single glass slide, and evaluated the quality of the resulting array by probing with an anti-GST antibody (Fig. 2a and inset). Histogram analysis showed that although the distribution of the foreground and background intensity exhibited typical bell-shaped curves, the distribution of the foreground intensity (Fig. 2b, red) is almost completely separated from that of the background (Fig. 2b, black), indicating that the great majority of printed spots contained substantial levels of recombinant protein. Indeed, >93% of spotted proteins showed a foreground/background signal (F/B) ratio of at least 1.5 (supplemental Table 3). As illustrated in Fig. 2c, the proteins spotted on the microarrays are expressed in a broad range of different subcellular compartments. To determine whether there is any bias against certain types of proteins, box plots of the F/B ratios versus subcellular compartment or protein family were generated (Fig. 2, d and e). The results indicated that although proteins localized to the cytoplasm (p = 1.27E-40) or mitochondria (p = 7.56E-8) showed significantly higher F/B ratios when compared with all proteins on the array, proteins localized to the plasma membrane are more challenging to produce. A similar trend was observed when F/B ratios were analyzed for individual protein families; both G-protein coupled receptors and transcription factors exhibited lower level of expression among protein families examined (Fig. 2e). Not unexpectedly, these data imply that the physical and functional properties of target proteins affect observed protein yield but also show that the majority of proteins in all major functional classes can be efficiently expressed and purified using this yeast-based expression system. Finally, to evaluate whether protein length affects the success of protein purification, we applied a scatter plot analysis but only observed an insignificant negative correlation (r = −0.196) between protein length and signal intensity (Fig. 2f). Taken together, these analyses indicate that the yeast expression system is suitable for high throughput protein purification in higher eukaryotes. The microarray produced from these recombinant proteins represents the most comprehensive protein microarray comprised of individually purified proteins reported to date.
Fig. 2.
Evaluation of human proteome microarray quality. a, a representative image of a protein microarray. A protein microarray was incubated with anti-GST antibody, and printed spots were identified by probing with an anti-rabbit Alexa Fluor 555 conjugate. b, a histogram of foreground and background signal intensities. The x axis represents log10 scale. c, endogenous distribution of target proteins. d, protein expression levels with different subcellular localization. e, protein expression level by different protein families. f, protein expression level by protein length. The small boxes in d and e represents mean values.
Generation of Hybridomas and Identification of Target Antigens
Ever since the early days of mAb production, highly complex mixtures of antigens such as whole cells or tissues have been used to generate ICC grade mAbs (30, 31). Generating mAbs in this so-called “shotgun” manner greatly increases the likelihood that they will recognize native protein epitopes, thus increasing the range of potential applications for the resultant mAbs. However, the major limitation of this approach has been the difficulty in identifying antigens preferentially recognized by a given mAb, which has typically only been possible through affinity purification coupled with mass spectrometry. By screening mAbs against human proteome microarrays, it becomes possible to directly identify the corresponding antigen recognized by a given mAb that has been generated by immunization with complex biological samples.
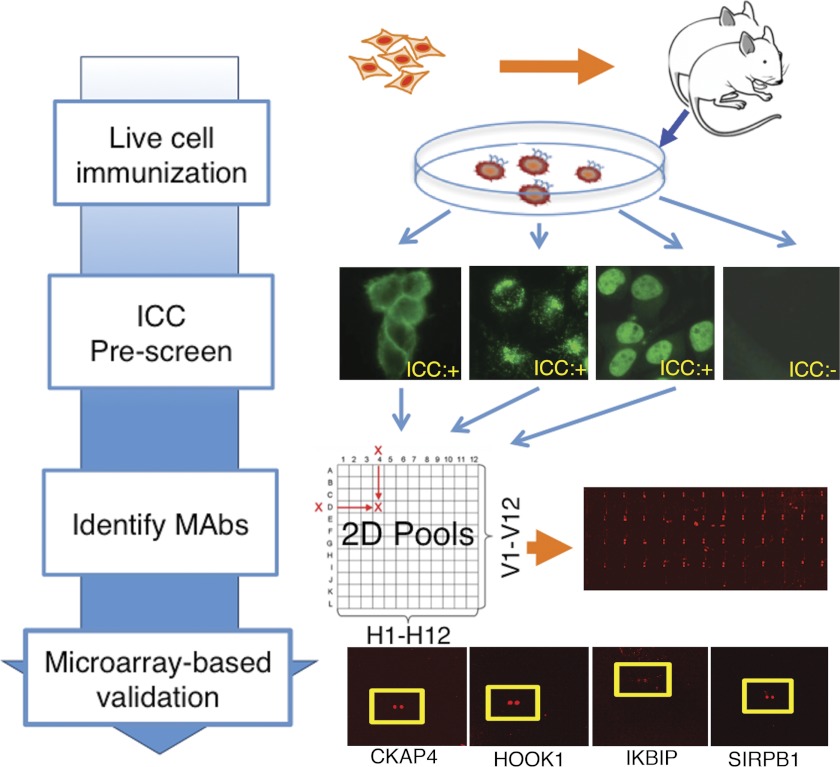
To generate mAbs that recognize a broad range of human proteins in their native conformations, we have immunized mice with a variety of different human cancer cell lines (Fig. 3). 2.5 × 106 cells were injected directly into footpad or hock areas, and 2 weeks later popliteal lymph nodes were collected, and lymphocytes were fused with a myeloma line. A total of 32,112 hybridomas were thus generated, of which 2326 both grew well and were IgG-positive. To enrich for useful mAbs, a high throughput ICC prescreening step against the cell line used for immunization was conducted, using supernatants from 2088 different IgG-positive hybridomas. A total of 643 ICC-positive hybridomas were then grown further, isotyped, and selected for microarray analysis. To determine whether this prescreening step indeed enriched for highly specific antibodies, a total of 332 randomly selected ICC-negative hybridomas. In addition, a total of 238 randomly selected hybridomas were used for microarray-based analysis of binding specificity without undergoing ICC prescreening. Individual supernatants were combined into sets of 12 × 12 two-dimensional pools, and these pools were then individually incubated on the human proteome microarrays. This was then followed by stringent washes and incubation with a Cy5-coupled anti-IgG secondary antibody to determine their binding profiles.
Fig. 3.
Strategy for identification of highly specific mAbs. Various different live human cell lines were used for immunization of BALB/c mice. The resulting hybridomas were tested for secretion of IgG and were used for ICC for the cell lines used. ICC-positive supernatants were combined in 12 × 12 two-dimensional pools, which were then used to probe the human proteome microarrays. Data from pooled samples were deconvoluted, and the candidate monospecific mAbs were then probed to the human proteome microarrays individually. Examples of antigens recognized by monospecific mAbs are shown.
Following scanning, we expressed the signal intensity for each spot as the ratio of median foreground to median background signals. To identify antigens bound by individual mAbs, a histogram of signal intensity of every protein spot on an array was plotted to determine the median signal intensity and S.D. value. Using a z score cutoff of ≥2.8, for both replicate spots, positive antigens were identified; mAbs found at the intersection between one unique row pool and one column pool that recognize a particular positive antigen were then flagged for individual analysis. mAbs identified as potentially specific using this pooling strategy were then tested again individually using a new array to reconfirm the reactive antigens. Positive antigens were identified as described above and ranked based on their signal intensity. When signal intensity of a top antigen is ≥6 S.D. above the median and ≥3 S.D. higher than the second best antigen, it is termed a monospecific mAb (mmAb) (supplemental Fig. 2). A total of 76 mmAbs were identified (supplemental Table 4). In many cases, mmAbs showed extremely high selectivity, with 15 displaying an S-score of >50, intensity between the top and second best antigens (supplemental Fig. 3). We also identified 27 mAbs that bound almost equally well to two different antigens on the array (S < 3), which we termed dispecific mAbs. Because the top two target proteins may be readily distinguishable if they show divergent cellular or subcellular expression, these dispecific mAbs were also selected for further analysis. Notably, three highly specific mAbs selectively recognized proteins for which the F/B ratio of the GST signal was less than 1.3, demonstrating their ability to readily detect relatively small quantities of recombinant protein.
To further characterize the utility and specificity of the mmAbs and dispecific mAbs identified in this initial screening step, we then purified antibodies from either hybridoma supernatants or, in some cases, ascites fluid from BALB/c mice injected with the hybridomas and used these to conduct a variety of different assays. We first repeated the ICC analysis on human cancer cell lines and collected images to assess the subcellular expression pattern of the endogenous target antigen. Of 79 tested mAbs, 70 produced specific staining in cultured cells (Fig. 4a and supplemental Fig. 3). Confirmation of the observed specificity of individual mAbs comes from the finding that 48 of 64 (75%) of all ICC patterns obtained in this study matched those previously reported for their target proteins, when these data were available (supplemental Table 5). The subcellular distribution of the proteins for which high quality mAbs were obtained showed no clear preference for any particular cellular compartment, confirming that shotgun immunization can generate mAbs to a wide range of cellular proteins (supplemental Fig. 4). A total of 19% (12 of 64) of the targets possessed a predicted/annotated transmembrane domain, as compared with 26% for all human proteins (32).
Fig. 4.
Analysis of highly specific mAbs in different research applications. To validate the specificities of individual mAbs, a series of experiments was performed. a, representative ICC data are shown for endogenous proteins in HeLa (XRCC5, RAB8A), HL-60 (DLAT), and HCT116 cells (ANXA2). b, shRNA knockdown of target antigens. Plasmids driving expression of target proteins tagged on the N terminus with the V5 epitope were then transfected either with corresponding shRNA or without shRNA expression constructs. The resulting cell lysates were analyzed by immunoblot with each mAb in question and anti-V5 antibody for validation of antigen specificity. In each sample, the first lane shows expression construct, the second lane shows co-transfection of expression and shRNA expression construct, and the third lane shows no transfection showing endogenous protein detection. c, IP assays. Immunoprecipitation was performed to test whether mAbs recognize native antigens. V5 fusion constructs were transfected in HeLa cells. Along with input cell lysate, IP was performed with or without mAbs (negative control). As a positive control, anti-V5 antibody was used to pull down target V5 fusion antigen proteins. First lane, input; second lane, mAb IP; third lane, no antibody (negative control); fourth lane, anti-V5 IP (positive control). d, assays for which highly specific mAbs were proven effective. The diagram summarizes the assays for which individual purified highly specific mAbs were confirmed to be specifics. IB, immunoblot.
Validation of Identified mAbs
Next, we analyzed the ability of the newly identified mAbs to detect their top target protein using immunoblot analysis. Individual target proteins were overexpressed in HeLa cells as N-terminal V5-tagged fusion proteins, and expression was confirmed via immunoblotting with anti-V5. Of 50 purified mAbs tested that recognized a total of 47 different proteins, 28 (56%) detected their target protein by immunoblotting, whereas 12 (24%) recognized endogenous untagged protein in these cells (supplemental Fig. 5). To further confirm the accuracy of these results, we co-transfected the V5-tagged ORF with individual shRNA constructs targeting the gene under investigation. In all six cases tested, we observed a substantial reduction in immunoblot signal, confirming the fidelity of these mAbs (Fig. 4b). We next tested whether these mAbs could work effectively for immunoprecipitation and thus recognize native proteins in cell homogenates. Remarkably, we found that of the 50 purified mAbs tested, 33 (66%) could efficiently precipitate transfected protein, which was then detected via immunoblotting with anti-V5 (Fig. 4c and supplemental Fig. 6). In all, 21 of 50 (42%) of all mAbs worked effectively for both immunoblotting and immunoprecipitation. We next investigated whether these mAbs were useful for a broader range of applications compared with those already available from other commercial suppliers. We first directly compared mAbs from 12 different suppliers generated to the same set of 47 antigens as those in this study (Table I). In all, based on the supplier's reports, 80% of commercially available mAbs work for immunoblotting, 55% are ICC or IHC grade, but only 18% are IP grade. In contrast, a much higher fraction of mAbs generated using the pipeline described here were ICC and IP grade, although a somewhat lower fraction were immunoblot grade. To eliminate any systematic bias introduced by selection of these antigens, we also investigated the performance of 100 randomly selected mAbs from a subset of major commercial suppliers and found that these numbers were very similar, with 86% of randomly chosen mAbs working for IB, 55% for IHC or ICC, 17% for IP, and 11% useful for all three applications (Table II). Furthermore, no commercially available antibodies were available for 10 of the proteins recognized by mmAbs identified in this study, whereas only a single antibody was available for six others (supplemental Table 5). We thus conclude that the protein microarray-based shotgun approach described here is able to generate highly specific mAbs to proteins that are both well and poorly targeted by the existing antibody repertoire and are useful for a broader range of applications than those currently available from commercial suppliers.
Table I. Summary of experimental uses for commercially available mAbs that recognize the same target antigens as the highly specific mAbs identified in this study.
The total number of mouse monoclonal antibodies that selectively bind proteins that are also recognized by the highly specific mAbs identified in this study are listed. All of the mouse mAbs from the indicated supplier that recognize these proteins are included. The total percentage of mAbs from each supplier reported to be usable for each of the indicated applications is shown. IB, immunoblot; ICC/IHC, immunocytochemistry or immunohistochemistry; IP, immunoprecipitation; Multiple, useful in at least two of these applications; All, useful in all three applications. The names of the individual suppliers have been concealed for this analysis.
| Supplier | Number of mAbs | IB (%) | ICC/IHC (%) | IP (%) | Multiple (%) | All (%) |
|---|---|---|---|---|---|---|
| Supplier A | 57 | 77 | 46 | 16 | 37 | 7 |
| Supplier B | 12 | 83 | 50 | 17 | 42 | 17 |
| Supplier C | 31 | 65 | 71 | 6 | 58 | 6 |
| Supplier D | 19 | 84 | 53 | 5 | 47 | 5 |
| Supplier E | 19 | 63 | 47 | 0 | 26 | 0 |
| Supplier F | 15 | 93 | 73 | 47 | 80 | 33 |
| Supplier G | 44 | 84 | 70 | 39 | 59 | 25 |
| Supplier H | 8 | 88 | 50 | 0 | 38 | 0 |
| Supplier I | 20 | 95 | 20 | 10 | 20 | 0 |
| Supplier J | 9 | 100 | 44 | 22 | 44 | 22 |
| Supplier K | 11 | 91 | 0 | 18 | 9 | 0 |
| Supplier L | 22 | 73 | 86 | 14 | 55 | 9 |
| All commercial mAbs | 267 | 80 | 55 | 18 | 58 | 11 |
| This study | 50 | 56 | 90 | 66 | 74 | 42 |
Table II. Summary of experimental uses for 100 randomly chosen mAbs from different commercial suppliers.
For seven major suppliers, 100 mAbs were randomly selected. The total percentage of mAbs from each supplier reported to be usable for each of the indicated applications is shown. IB, immunoblot; ICC/IHC, immunocytochemistry or immunohistochemistry; IP, immunoprecipitation; Multiple, useful in at least two of these applications; All, useful in all three applications. The names of the individual suppliers have been concealed for this analysis and match those in Table I.
| mAbs by supplier | IB (%) | IHC/ICC (%) | IP (%) | Multiple (%) | All (%) |
|---|---|---|---|---|---|
| Supplier A | 87 | 28 | 11 | 35 | 3 |
| Supplier B | 93 | 26 | 6 | 29 | 0 |
| Supplier C | 96 | 83 | 0 | 81 | 0 |
| Supplier D | 91 | 14 | 0 | 13 | 0 |
| Supplier E | 85 | 64 | 25 | 60 | 17 |
| Supplier F | 67 | 70 | 34 | 55 | 21 |
| Supplier G | 83 | 100 | 40 | 85 | 38 |
| Overall mean | 86 | 55 | 17 | 51 | 11 |
| This study | 56 | 90 | 66 | 74 | 42 |
Finally, to determine whether the ICC-prescreening step enriched for highly specific and broadly usable mAbs, we compared the quality of the antibodies analyzed from the ICC-positive and the ICC-negative groups. Overall, of the hybridomas that underwent ICC prescreening, we found that 10.3% (66 of 643) of ICC-positive hybridomas were highly specific, significantly higher than the 7.5% (25 of 332) positive rate observed in ICC-negative hybridomas (p < 0.03). Furthermore, we found that ICC-positive mAbs showed significantly higher signal intensity and specificity than did ICC-negative mAbs (p < 0.05). Although both pools generated similar proportions of IB and IP grade mAbs (supplemental Table 4), ICC prescreening results in a significant improvement in overall antibody quality and specificity.
DISCUSSION
We have constructed the most comprehensive human protein microarray to date that is comprised of individually purified proteins expressed in a eukaryotic cell system. A recent effort reported construction of a human proteome scale microarray using a wheat germ in vitro translation system for protein production (25). However, the use of crude translation reaction mixtures rather than individually purified proteins may limit the use of this type of protein microarray, because proteins in each spot are unavoidably contaminated with large amount of proteins present in the wheat germ system. Because assays conducted on protein microarrays are generally more sensitive than those performed in a traditional liquid-based format, it is harder to successfully carry out those assays that require high purity of the spotted proteins using such a reagent (33). The use of individually purified proteins to construct a functional protein microarray has been proven highly successful in characterizing a diverse range of protein functions over the past decade (11, 14, 15, 17, 22–24, 26, 27, 33). Furthermore, the fact that a majority of the full-length human proteome is printed on a single slide makes it possible to perform more comprehensive screens of protein function in a high throughput fashion.
Using the human proteome microarray as a central component, we built a pipeline for mmAb selection using random hybridomas generated by the shotgun approach. The pipeline for generation of mAbs described here has a number of unique advantages over existing approaches. First, the use of live cells as immunogens bypasses the cumbersome and costly step of antigen preparation and ensures that only proteins in their native conformation are detected by the host immune system. Second, the use of protein microarrays in the pipeline effectively allows ready determination of mAb specificity and also provides a direct measurement of specificity for every mAb analyzed. Third, the incorporation of an ICC prescreening step increases the likelihood of generating highly specific mAbs. Finally, mAbs identified using this approach are useful for a wide range of biochemical applications, most notably those requiring recognition of native epitopes such as immunoprecipitation, and outperform mAbs available from commercial suppliers. Because immunoprecipitation grade antibodies typically have Kd values no higher than 50 nm (34, 35), these mAbs typically show both high affinity and high specificity. The microarray-based analysis described here is both relatively low cost and high throughput and can also readily be used to evaluate the binding specificity of any protein affinity reagents, including recombinant antibodies and aptamers. This platform may prove particularly useful in evaluating the binding specificity of the many thousands of mAbs already used by the research and clinical communities and could be readily adapted to identify highly specific mAbs obtained following immunization with individual proteins. The development of large numbers of mAbs made using a single, consistent pipeline and exhibiting high specificity using the approaches described here may have major implications for both basic and clinical research.
Acknowledgments
We thank Alan Long and Sarah Zheng for expert assistance with laboratory automation and managing clone collections, Yu-Yi Lin for help with shRNA screening, Sheng-Ce Tao for help with protein purification, and S. Chen and T. Shimogori for comments on the manuscript.
S. B., H. Z., I. P., D. E., J. B., and J. D. B. are co-founders of CDI Laboratories and all hold stock in the corporation. I. P., J. B., E. A., D. B., L. R., Z. A. R., and C. T. are all current employees of CDI Laboratories.
Footnotes
* This work was supported by National Institutes of Health Grants R41MH088008 (to S. B. and I. P.) and 1R01EY017015 (to S. B.), by National Institutes of Health Common Fund Grant U54RR020839 (to H. Z. and J. D. B.), and by a grant from the Puerto Rico Science and Technology Trust. Additional support was obtained from the Institute for Cell Engineering and the Fund for Medical Discovery, Johns Hopkins University School of Medicine, and the Puerto Rico Science and Technology Trust. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- mAb
- monoclonal antibody
- DMEM
- Dulbecco's modified Eagle's medium
- ICC
- immunocytochemistry
- shRNA
- small hairpin RNA
- IP
- immunoprecipitation
- F/B
- foreground/background signal
- mmAb
- monospecific mAb
- QC
- quality control.
REFERENCES
- 1. Uhlén M., (2008) Affinity as a tool in life science. BioTechniques 44, 649–654 [DOI] [PubMed] [Google Scholar]
- 2. Strebhardt K., Ullrich A. (2008) Paul Ehrlich's magic bullet concept: 100 years of progress. Nat. Rev. Cancer 8, 473–480 [DOI] [PubMed] [Google Scholar]
- 3. Köhler G., Milstein C. (1975) Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497 [DOI] [PubMed] [Google Scholar]
- 4. Stoevesandt O., Taussig M. J. (2007) Affinity reagent resources for human proteome detection: Initiatives and perspectives. Proteomics 7, 2738–2750 [DOI] [PubMed] [Google Scholar]
- 5. Kalyuzhny A. E. (2009) The dark side of the immunohistochemical moon: Industry. J. Histochem. Cytochem. 57, 1099–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Couchman J. R., (2009) Commercial antibodies: The good, bad, and really ugly. J. Histochem. Cytochem. 57, 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jensen B. C., Swigart P. M., Simpson P. C. (2009) Ten commercial antibodies for α-1-adrenergic receptor subtypes are nonspecific. Naunyn-Schmiedebergs Arch. Pharmacol. 379, 409–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hughes B., (2010) Antibody-drug conjugates for cancer: Poised to deliver? Nat. Rev. Drug Discov. 9, 665–667 [DOI] [PubMed] [Google Scholar]
- 9. Berger J. R., Houff S. A., Major E. O. (2009) Monoclonal antibodies and progressive multifocal leukoencephalopathy. mAbs 1, 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berglund L., Björling E., Oksvold P., Fagerberg L., Asplund A., Szigyarto C. A., Persson A., Ottosson J., Wernérus H., Nilsson P., Lundberg E., Sivertsson A., Navani S., Wester K., Kampf C., Hober S., Pontén F., Uhlén M. (2008) A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol. Cell. Proteomics 7, 2019–2027 [DOI] [PubMed] [Google Scholar]
- 11. Uhlén M., Hober S. (2009) Generation and validation of affinity reagents on a proteome-wide level. J. Mol. Recognit. 22, 57–64 [DOI] [PubMed] [Google Scholar]
- 12. Colwill K., Gräslund S. (2011) A roadmap to generate renewable protein binders to the human proteome. Nat. Methods 8, 551–558 [DOI] [PubMed] [Google Scholar]
- 13. Mattoon D. R., Schweitzer B. (2009) Antibody specificity profiling on functional protein microarrays. Methods Mol. Biol. 524, 213–223 [DOI] [PubMed] [Google Scholar]
- 14. Predki P. F., Mattoon D., Bangham R., Schweitzer B., Michaud G. (2005) Protein microarrays: A new tool for profiling antibody cross-reactivity. Hum. Antibodies 14, 7–15 [PubMed] [Google Scholar]
- 15. Robinson W. H. (2006) Antigen arrays for antibody profiling. Curr. Opin. Chem. Biol. 10, 67–72 [DOI] [PubMed] [Google Scholar]
- 16. Davies D. H., Wyatt L. S., Newman F. K., Earl P. L., Chun S., Hernandez J. E., Molina D. M., Hirst S., Moss B., Frey S. E., Felgner P. L. (2008) Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus ankara is comparable to that of Dryvax. J. Virol. 82, 652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Michaud G. A., Salcius M., Zhou F., Bangham R., Bonin J., Guo H., Snyder M., Predki P. F., Schweitzer B. I. (2003) Analyzing antibody specificity with whole proteome microarrays. Nat. Biotechnol. 21, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 18. Keasey S. L., Schmid K. E., Lee M. S., Meegan J., Tomas P., Minto M., Tikhonov A. P., Schweitzer B., Ulrich R. G. (2009) Extensive antibody cross-reactivity among infectious Gram-negative bacteria revealed by proteome microarray analysis. Mol. Cell. Proteomics 8, 924–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kijanka G., Ipcho S., Baars S., Chen H., Hadley K., Beveridge A., Gould E., Murphy D. (2009) Rapid characterization of binding specificity and cross-reactivity of antibodies using recombinant human protein arrays. J. Immunol. Methods 340, 132–137 [DOI] [PubMed] [Google Scholar]
- 20. Lueking A., Possling A., Huber O., Beveridge A., Horn M., Eickhoff H., Schuchardt J., Lehrach H., Cahill D. J. (2003) A nonredundant human protein chip for antibody screening and serum profiling. Mol. Cell. Proteomics 2, 1342–1349 [DOI] [PubMed] [Google Scholar]
- 21. Büssow K., Nordhoff E., Lübbert C., Lehrach H., Walter G. (2000) A human cDNA library for high-throughput protein expression screening. Genomics 65, 1–8 [DOI] [PubMed] [Google Scholar]
- 22. Hu S., Li Y., Liu G., Song Q., Wang L., Han Y., Zhang Y., Song Y., Yao X., Tao Y., Zeng H., Yang H., Wang J., Zhu H., Chen Z. N., Wu L. (2007) A protein chip approach for high-throughput antigen identification and characterization. Proteomics 7, 2151–2161 [DOI] [PubMed] [Google Scholar]
- 23. Song Q., Liu G., Hu S., Zhang Y., Tao Y., Han Y., Zeng H., Huang W., Li F., Chen P., Zhu J., Hu C., Zhang S., Li Y., Zhu H., Wu L. (2010) Novel autoimmune hepatitis-specific autoantigens identified using protein microarray technology. J. Proteome Res. 9, 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu S., Xie Z., Onishi A., Yu X., Jiang L., Lin J., Rho H. S., Woodard C., Wang H., Jeong J. S., Long S., He X., Wade H., Blackshaw S., Qian J., Zhu H. (2009) Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139, 610–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goshima N., Kawamura Y., Fukumoto A., Miura A., Honma R., Satoh R., Wakamatsu A., Yamamoto J., Kimura K., Nishikawa T., Andoh T., Iida Y., Ishikawa K., Ito E., Kagawa N., Kaminaga C., Kanehori K., Kawakami B., Kenmochi K., Kimura R., Kobayashi M., Kuroita T., Kuwayama H., Maruyama Y., Matsuo K., Minami K., Mitsubori M., Mori M., Morishita R., Murase A., Nishikawa A., Nishikawa S., Okamoto T., Sakagami N., Sakamoto Y., Sasaki Y., Seki T., Sono S., Sugiyama A., Sumiya T., Takayama T., Takayama Y., Takeda H., Togashi T., Yahata K., Yamada H., Yanagisawa Y., Endo Y., Imamoto F., Kisu Y., Tanaka S., Isogai T., Imai J., Watanabe S., Nomura N. (2008) Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat. Methods 5, 1011–1017 [DOI] [PubMed] [Google Scholar]
- 26. Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Mitchell T., Miller P., Dean R. A., Gerstein M., Snyder M. (2001) Global analysis of protein activities using proteome chips. Science 293, 2101–2105 [DOI] [PubMed] [Google Scholar]
- 27. Hall D. A., Zhu H., Zhu X., Royce T., Gerstein M., Snyder M. (2004) Regulation of gene expression by a metabolic enzyme. Science 306, 482–484 [DOI] [PubMed] [Google Scholar]
- 28. Clamp M., Fry B., Kamal M., Xie X., Cuff J., Lin M. F., Kellis M., Lindblad-Toh K., Lander E. S. (2007) Distinguishing protein-coding and noncoding genes in the human genome. Proc. Natl. Acad. Sci. U.S.A. 104, 19428–19433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang F., Matrubutham U., Parvizi B., Yen J., Duan D., Mirchandani J., Hashima S., Nguyen U., Ubil E., Loewenheim J., Yu X., Sipes S., Williams W., Wang L., Bennett R., Carrino J. (2004) ORFDB: an information resource linking scientific content to a high-quality open reading frame (ORF) collection. Nucleic Acids Res. 32, D595–D599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McMichael A. J., Pilch J. R., Galfré G., Mason D. Y., Fabre J. W., Milstein C. (1979) A human thymocyte antigen defined by a hybrid myeloma monoclonal antibody. Eur. J. Immunol. 9, 205–210 [DOI] [PubMed] [Google Scholar]
- 31. Barnstable C. J., Bodmer W. F., Brown G., Galfre G., Milstein C., Williams A. F., Ziegler A. (1978) Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell 14, 9–20 [DOI] [PubMed] [Google Scholar]
- 32. Fagerberg L., Jonasson K., von Heijne G., Uhlén M., Berglund L. (2010) Prediction of the human membrane proteome. Proteomics 10, 1141–1149 [DOI] [PubMed] [Google Scholar]
- 33. Smith M. G., Jona G., Ptacek J., Devgan G., Zhu H., Zhu X., Snyder M. (2005) Global analysis of protein function using protein microarrays. Mech. Ageing Dev. 126, 171–175 [DOI] [PubMed] [Google Scholar]
- 34. Dyson M. R., Zheng Y., Zhang C., Colwill K., Pershad K., Kay B. K., Pawson T., McCafferty J. (2011) Mapping protein interactions by combining antibody affinity maturation and mass spectrometry. Anal. Biochem. 417, 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harlow E., Lane D. (1998) Using Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]