Abstract
N-terminal modifications play a major role in the fate of proteins in terms of activity, stability, or subcellular compartmentalization. Such modifications remain poorly described and badly characterized in proteomic studies, and only a few comparison studies among organisms have been made available so far. Recent advances in the field now allow the enrichment and selection of N-terminal peptides in the course of proteome-wide mass spectrometry analyses. These targeted approaches unravel as a result the extent and nature of the protein N-terminal modifications. Here, we aimed at studying such modifications in the model plant Arabidopsis thaliana to compare these results with those obtained from a human sample analyzed in parallel. We applied large scale analysis to compile robust conclusions on both data sets. Our data show strong convergence of the characterized modifications especially for protein N-terminal methionine excision, co-translational N-α-acetylation, or N-myristoylation between animal and plant kingdoms. Because of the convergence of both the substrates and the N-α-acetylation machinery, it was possible to identify the N-acetyltransferases involved in such modifications for a small number of model plants. Finally, a high proportion of nuclear-encoded chloroplast proteins feature post-translational N-α-acetylation of the mature protein after removal of the transit peptide. Unlike animals, plants feature in a dedicated pathway for post-translational acetylation of organelle-targeted proteins. The corresponding machinery is yet to be discovered.
As noted as far back as 25 years ago (1–3), protein N-α-acetylation (NAA)1 is one of the major protein modifications of eukaryotes. Up to 90% of the proteins that accumulate at the steady state display NAA (4–7). It was also identified, although to a lesser extent (18%), in several Archaea (8, 9). This situation in eukaryotes and Archaea is markedly different from prokaryotes, where only a few proteins undergo this modification (10). Thus, it has been suggested that there is a link between the degree of genome complexity of an organism and the NAA extent of the corresponding proteome (11). NAA, a co-translational modification occurring in close vicinity of the ribosome (12–14), consists of the transfer of an acetyl group from the acetyl coenzyme-A to the free N-terminal group of the nascent polypeptide. This reaction is catalyzed by several—at least six identified so far (15, 16)—N-α-acetyltransferase complexes (Nats). Although the corresponding genes were initially identified in yeast (17), their orthologs have been recently identified in humans (11, 18) and other eukaryotes (15), as well as in Archaea (19). The major Nats, i.e. NatA, NatB, and NatC, are specific to different substrates (10). NatA acts after the excision of N-terminal methionine (NME), whereas NatB and NatC target the initial Met (Met1) when it is retained and followed at position 2 by residues including Asp, Glu, and Asn for NatB and large hydrophobic residues such as Ile, Leu, or Phe for NatC. Several recent large scale analyses of N-α-acetylated (NAAed) proteins have been performed in Saccharomyces cerevisiae (11, 20), Homo sapiens (7, 11, 18), Drosophila melanogaster (6), Halobacterium salinarum, and Natronomonas pharaonis (8, 9). These studies have started to uncover this modification at the proteome scale leading to a better overview of the process.
NAA is an essential protein modification. Any gene inactivation of any components of the Nat complex is associated with oncogenesis and cell division defects in humans (21–24). Partial inactivation induces a number of metabolic disorders and apoptosis in mammalian cells (25–29). A recent study on recessive male infant lethality identifies a specific mutation of the NatA catalytic subunit (S37P variant) (24). This altered complex still displays 60–80% of the wild type NatA activity but appears to be lethal within few days after birth. Although NAA is associated to specific roles for several protein groups such as cytoskeleton protein binding (30–32) or protein targeting to a given compartment (33–35), its intrinsic function has remained elusive for the large majority of the proteins. Nevertheless, it was suggested that NAA is an important factor influencing protein half-life either globally or in some protein groups (4, 36–38). Most recently two studies have significantly revisited the field of acetylation function and regulation. First, it was suggested that NAA contributes to cell sorting by inhibiting protein to target the endoplasmic reticulum (39). Thus, proteins devoid of NAA signal would be preferentially guided to the secretory pathway in yeast. Next, it is now suggested that NAA is not only controlled by the level of the enzymes involved but mainly by the level of its substrate, namely acetyl-CoA (40). Such metabolic regulation would be crucial to promote, for instance, apoptosis in mammals.
NAA has been poorly studied in higher plants, and very little information concerning this modification is available so far. In this context, although large scale studies start to give a better picture of the N-α-acetylome for a limited number of species, 200–300 NAAed substrates have been described until now in higher plants (38, 41–43). Moreover, only one Nat catalytic subunit, i.e. NatC, has been identified to date in Arabidopsis thaliana. The associated NatC gene knock-out is specific to plants because it is associated to a chloroplast defect featured by decreasing effective quantum yield of photosystem II and, as a result, impairing plant growth (44).
Although this modification is reported to be one of the most frequent in higher eukaryotes (15), one wonders to what extent protein NAA is similar in plants because of the low number of substrates yet characterized. In this context, our analysis aimed to deepen our knowledge of NAA in the model plant A. thaliana and to compare it with an animal system, i.e. H. sapiens. Our main goal was to identify kingdom-specific differences, if any. For this purpose, we improved and used a strategy for the N terminus peptides enrichment based on strong cation exchange-liquid chromatography (SCX-LC) and tandem mass spectrometry (MS/MS), allowing simultaneous identification of both the protein and the associated modifications (6, 7, 9, 11, 18, 38, 45, 46). Our investigation provides the characterization of 1072 N terminus peptides related to 1007 proteins for the A. thaliana samples and 727 N terminus peptides over 715 proteins for the human sample.
Most of the NAAed peptides characterized in the A. thaliana sample are related to the expected co-translational modification, but interestingly, ∼30% of them are located further downstream of the expected protein N terminus. In contrast, less than 8% of the NAAed peptides from humans display such a behavior. For Arabidopsis, most of these cases appear to be related to chloroplast proteins encoded in the nuclear genome and translocated through a targeting signal. Because such NAA occurs at the neo-N terminus uncovered after the cleavage of the chloroplast transit peptide (cTP), this modification is clearly post-translational. Although a few similar cases have been described before (38, 42, 47–49), NAA of chloroplast proteins appears to be a widespread modification in this organelle that has never been described before to this extent. Substrate specificity appears to be close to the cytosolic NatA complex, suggesting a dedicated Nat occurring in this organelle. Our unprecedented large data sets from both Arabidopsis and human samples were further used to perform an investigation of prediction accuracy for a number of tools including (i) N terminus status prediction with TermiNator3 (41, 50), (ii) protein localization, (iii) cTP length prediction with TargetP (51–53), and (iv) determination of whether such prediction could be used to strengthen our experimental results.
EXPERIMENTAL PROCEDURES
Human and Arabidopsis Sample Preparation
Human A2780 cells were grown in T75 flasks to 70% confluence. After few washes in PBS, the cells were lysed adding in flask Nonidet P-40 lysis buffer (150 mm NaCl, 1% Nonidet P-40, 50 mm Tris, pH 8, and EDTA-free protease inhibitor). The cells were scraped off the walls, and the lysate was centrifuged to remove debris (54). The supernatant was stored at −20 °C.
A. thaliana cells (derived from Columbia ecotype) were used and produced as described previously (55). Cultured cells were harvested by centrifugation. Cells equivalent to 10 ml are mixed with 20 mg of SDS, 150 mg of DTT, 100 mg of sodium carbonate, and 5 ml of water and heated for 15 min at 95 °C. A. thaliana plant tissues were frozen in liquid nitrogen and ground in a 2-ml microcentrifuge tube containing 3- and 5-mm iron beads for 1 min each, using a MM 300 mixer mill at 30 Hz (Qiagen). The resulting fine powder was dissolved in 1 ml of lysis buffer (50 mm HEPES, pH 7.2, 150 mm NaCl, 15 mm MgCl2, 1 mm EDTA, 10% glycerol, 1% Triton X-100, 2 mm PMSF, and protease inhibitor mixture) as previously described in Ref. 56 (Buffer D). The homogenates were incubated at 4 °C for 20–45 min with shaking. The supernatants were separated from the insoluble fraction by centrifugation at 15,000 × g at 4 °C for 0.5–1 h. The resulting supernatants were used to measure protein concentration by the Bradford protocol (Bio-Rad) and were stored at −80 °C unless used immediately.
Sample Preparation for MS Analysis
The protein samples were precipitated with a 4× excess of acetone (v/v) at −20 °C for at least 1 h before centrifugation for 10 min at 13,000–15,000 × g. The pellets were resuspended and denaturated in 6 m guanidine-HCl, 50 mm Tris-HCl, pH 8, and 4 mm DTT, reduced for 15 min at 95 °C and finally alkylated by the addition of iodacetamide (55 mm) for 1 h at room temperature. Proteins were precipitated by the addition of four times the sample volume of cold acetone and centrifuge after 1 h at −20 °C to remove salt and reagents. The protein pellet was resuspended in 50 mm ammonium bicarbonate. Proteins were digested by the addition of 1/100th (w/w) of sequencing grade modified porcine trypsin (Promega, Madison, WI) for 1 h at 37 °C twice. The peptides were desalted by Sep-Pak (Waters, Milford, MA) solid phase extraction as recommended by the supplier. The retained material was eluted with 80% ACN and 0.1% TFA followed by evaporation to dryness using a vacuum centrifuge.
The peptide mixture was resuspended in SCX-LC buffer A (5 mm KH2PO4, 30% ACN, and 0.05% formic acid) and injected into a Summit LC system (Dionex, Sunnyvale, CA) equipped with polysulfoethyl A 200 × 2.1-mm 5-μm 200 Å column (PolyLC, Colombia, MD). The peptides were eluted with a gradient of increasing KCl (SCX-LC buffer B: 350 mm KCl in SCX-LC buffer A; 0–5 min, 0% B; 15–40 min, 5–26% B; 40–45 min, 26–35% B; and 45–50 min: 35–60% B). Fractions (detailed in supplemental Table S3) were collected along the 50-min elution gradient and then dried down before storage at −20 °C.
Sample Analysis by LC-MS
For each SCX-LC separation, the individual collected fractions eluting before 20–25 min or their combination (only used for the three technical replicates) were resuspended in 50 μl of 5% ACN and 0.1% formic acid and subjected to LC-MS/MS analysis (resumed in supplemental Table S3). The selected fractions (or their combination) were subjected to separation on an Ultimate 3000 nano-RP-LC (Dionex). Five μl of the preconcentrated sample were loaded onto a PepMap100 trap column at a flow of 20 μl/min of RP-LC buffer A (A: 5% ACN, 0.1% formic acid) and then separated using a PepMap C18 75-μm inner diameter × 15-cm analytical column over a 100-min gradient (nano-RP-LC buffer B: 80% ACN, 0.1% formic acid; 8–25% B in 70 min and then 25–50% B in 30 min) at a flow of 300 nl/min. The nano-RP-LC was coupled to a Q-Star XL (Applied Biosystems, Concorde, Canada), and Analyst software was used for data-dependant acquisition. The basic survey scan (1 s) from 400 to 1200 Da was followed by four Enhanced Precursor Ion scans (1 s). Acquired tandem mass spectra were exported from Analyst using the script Mascot.dll 1.6b23 (Matrix Science, London, UK). MS/MS of same precursor mass within 0.1 Da range were combined together for 60 s to avoid generating multiple fragmentation pattern of the same precursor within a single analysis.
Alternatively, some experiments were conducted with an OrbitrapTM Velos (Thermo Scientific, Courtaboeuf, France). This instrument was used for the technical replicates of the SCX-LC combined fraction (Preparation At5) and for the selected SCX-LC fractions of the plant tissues sample (Preparation At6) as detailed in supplemental Table 3. For both approaches, the survey scan was acquired by Fourier-Transform MS scanning 400–2000 Da at 60,000 resolution using an internal calibration (lock mass). For the first two technical replicates, the 20 most intense ions were subjected to CID MS (top-20) with a 30-s exclusion time. The third technical replicates targeted the 10 most abundant ions for high collision dissociation acquisition. Alternatively, each SCX-LC fraction (fractions 2–10) of preparation At6 was subjects of three repetitive acquisitions including a top-20 for parent ions with charge states +2 to +5 and a second with charge states +1 to +5 charges to favor the acquisition of the mono charge precursors that appears to be frequent for NAAed peptides. Both acquisitions were performed with an exclusion list related to the most frequent contaminants such as trypsin autolytic products, siloxanes, or polyethylene glycol (57). The third acquisition was performed applying high collision dissociation method on parent ions with charges +2 to +5. The raw data were exported with Proteome Discoverer (Thermo Scientific, Ver 1.1) in Mascot generic format files (*.mgf).
Data Processing and Result Validation
Exported files were submitted for protein identification and post-translational modification characterization using Mascot 2.0 software. The databases used were dependent of the sample: human subset of the UniprotKB/Swiss-Prot (Uniprot release 15.6, 20234 entries, www.uniprot.org) and The Arabidopsis Information Resource (TAIR) database for A. thaliana (TAIR version 10, 35,574 entries, www.arabidopsis.org) (58). The parent and fragment mass tolerance was defined to fit the observed error ranges and are available in supplemental Table S3. Because the proposed approach is commonly used for phosphopeptides enrichment but also NAAed peptides, a few different modifications should be investigated. After rapid prescreening of the data, we could conclude about the presence of NAAed peptides (protein N terminus but also at downstream position requiring a semi-tryptic search), phorphorylated peptides, pyroglutamylated peptides (especially for the peptides with a Gln at the N terminus), protein N-myristoylated peptides, methionine oxidation, and also cationized artifacts (especially sodium adducts combined with the protein C terminus peptides). Because of the limited number of peptides expected per protein, i.e. the NAAed peptide at least, any supplementary identification is highly valuable to strengthen the final protein identification list. Furthermore, we observed that the mass combination of the NAA modification (42.010565 Da) combined with the potassium cation (37.955882 Da) mimics the Mr of a phosphorylation modification (79.966447 instead of 79.966331 Da, ΔMr = 0.000116 Da) and is prone to false negative characterization of acetylated N termini. Thus, our data treatment was designed to harvest most of these peptides while preventing the tremendous increase of the calculation time using a two-pass strategy optimized for co- and post-translational modification characterization (supplemental Table S4). The second carbon isotope (13C) was considered only for the Q-Star-related data. MudPit scoring was applied, and only ions with a score higher than the identity threshold (determined for each data processing file individually) at less than 1% of false positive discovery rate (<1% false discovery rate using the decoy option in Mascot) were considered for further investigation. These two data processing passes were successfully used for the identification of various modification including NAA at the protein N terminus or downstream of the protein theoretical N terminus (dNAA) and N-Myr.
Mascot outputs were exported in XML format and submitted to an in-house script written in Python (version 2.5.2) using the library xml.dom.minidom. The parsing function searches for modification and identification of N or C terminus peptides. It collects all of the peptides with modifications like acetylation, pyro-glutamylation, or all free N and C terminus peptides at protein ends. For each protein, only the highest score was kept when the same peptide was identified several times with the same modification. For each peptide, the following information is collected: protein AC number, sequence, description, sequence of the peptide, and the associated modifications and positions.
Recent rules for experimental data validation in proteomics tend to require at least two peptides for relevant protein identification. Because the present approach is based on the enrichment of the NAAed peptides, in the worst case it could mean one protein identified by one peptide, making it extremely difficult to obey these rules. Although it has been shown that the protein N terminus sequence is highly discriminant (59, 60), we decided to increase the robustness of our study through strict false discovery rate lower than 1% and using analytical and biological replicates. Unique peptides were further manually checked and validated. The final protein lists highlight N-terminal peptides in the different replicates that increase further the confidence in the final protein list.
RESULTS
Tandem Mass Spectrometry Data Processing
SCX chromatography is able to discriminate peptides by their net charge. Most protein N-terminal peptides, which are most often blocked by NAA or to a lesser extent by N-Myr, are barely retained on such columns and can be segregated from internal peptides, which are unblocked and display an additional positive charge (see Ref. 45 for review). Using this fast and reliable strategy, we aimed to compare the modification profiles at the N termini of plant and human proteins. In the course of the analysis, new information relevant to other protein modifications, e.g. phosphorylation, pyroglutamylation, or protein C terminus peptides, were characterized. These data were not investigated further but are available in supplemental Table S5.
The A. thaliana samples were the subject of six experimental replicates, of which five preparations were obtained with suspension cells and one with whole seedling lysate. One of the suspension cell preparations was used to perform technical replicates for the MS analysis. The data of interest, i.e. N terminus peptides, related to the experimental and technical replicates are available in supplemental Table S1 (N termini worksheet). The human samples were subjected to two distinct preparations using the same cell lysate. Each of these preparations was fractionated by SCX-LC and then characterized by LC-MS/MS. All of the acquired data for a single preparation were pooled together and searched as a unique data file. The experimental replicates (supplemental Table S2, N termini worksheet) list the characterized N termini for each SCX-LC experiment.
Although the characterization of the N-terminal status of the protein and especially NAA is the main goal of this study, the approach used here has been previously used for phosphopeptide enrichment and as mentioned earlier for the characterization of C terminus peptides and N terminus pyroglutamylated peptides (61). In depth investigation of the remaining unmatched spectra uncovered other co- and post-translational modifications or numerous artifacts such as sodium and potassium peptide cationization (62). These modifications included N-Myr and NAA located downstream (dNAA) of the expected protein start position (i.e. the Met residue or the proximal residue at position 2 if the initial methionine undergoes NME) from genome annotation. A total of 5707 and 2431 peptides were characterized and extracted from the raw Mascot output considering all analyses of the Arabidopsis and human samples, respectively. Because of our approach related to the enrichment of the N terminus peptides, some of these proteins are related to a single nonredundant peptide that was frequently characterized in multiple SCX-LC fractions. These multiple characterizations related to distinct events, i.e. multiple acquisitions within a run or along different SCX-LC fractions or sample preparations, increase the reliability of such identifications. Thanks to these technical and experimental replicates, more than 80% of the proteins were identified by more than one unique peptide occurrence with a Mascot score higher than the identity threshold (false discovery rate < 1%).
N-terminal Modifications
A total of 1072 and 741 N termini for A. thaliana and H. sapiens, respectively (summarized in Table I from data listed in supplemental Table S1 and S2), were identified and assigned either to NME, NAA, unmodified N terminus, or N-Myr. The human NAAed characterized peptides were compared with the few recent studies targeting such modification (7, 11, 63) and the TopFind database, which lists N-terminome experimental data for a few organisms (64). A total of 635 of 703 acetylated peptides (NAAed and dNAAed) were previously characterized along these studies. Additionally, 73 NAAed peptides were newly characterized, of which 20 were located downstream of the expected protein start position. Altogether, a list of 1988 NAAed peptides were identified within these different studies, of which 232 are located downstream of the expected position (11.7%). This ratio appears to be slightly lower in this study, with only 55 dNAAed over 703 NAAed peptides (7.8%). This data comparison not only confirms the previous data in a one-shot analysis but also the robustness of our approach. In addition, this comparison points out the relevance of our workflow in the course of characterizing further new NAA in any organism. With Arabidopsis, a similar comparison performed against TopFind (only two instances so far) and other sources not yet indexed in this database (41, 42, 62, 65) revealed that the vast majority (i.e. 871 over 1007 proteins, 86.5%) were new (supplemental Table S1, subcellular localization worksheet).
Table I. Result summary for the different N termini characterized for both the Arabidopsis and the human sample.
The NME percentage is the ratio of the modified proteins over the number of identified proteins, whereas the percentage of the different N termini is the ratio over the sum of all of the N termini characterized. Because a few proteins were identified with more than a single downstream NAA peptide (especially for the Arabidopsis sample), percentage values exceed 100%.
| Modification | Modified peptides |
Identified proteins |
||||
|---|---|---|---|---|---|---|
| A. thaliana | H. sapiens | A. thaliana | H. sapiens | |||
| Hits | % mod | Hits | % mod | |||
| NME | 542 | 68.6% | 489 | 72.6% | 790 | 674 |
| Free NT | 13 | 1.6% | 36 | 4.9% | ||
| N-Myr | 5 | 0.6% | 3 | 0.4% | ||
| Protein NTA | 777 | 98.4% | 648 | 87.3% | ||
| dNTA | 277 | 25.8% | 55 | 7.4% | 224 | 55 |
| Total | 1072 | 742 | 1007 | 715 | ||
As expected, NME is a frequent modification in both organisms that covers ∼70% of the data set (Table I). Because of the SCX-LC enrichment step, NAA is another frequently encountered modification that covers most of the characterized N terminus peptides. In addition to these 777 Arabidopsis NAAed peptides, 277 peptides were found to be N-α-acetylated on a position of the proteins sequence downstream of annotated position 1 or 2 (depending of the NME status of the protein). Surprisingly numerous dNAA ranged from positions 2 to 409 for A. thaliana (277/1072 instances, i.e. 25.7%), and positions 2 to 344 for the human sample (53 of 727 instances, i.e. 7.3%) were characterized in this study. In Arabidopsis, if chloroplast imported protein could explain most of the dNAA (at least 184 of 224, 82.1%), alternative starts, protein splice variants, and other co- and post-translational modifications such as actin specific maturation (66) are likely to explain most of the remaining “unusual” modified peptides for the human sample as discussed later.
The present study also reveals a total of eight N-Myr proteins, of which glutathione peroxidase 5 (At3g63080, GPX5), a Vid27-related protein known as DEM2 (At3g19240), protein fructose-2,6-bisphosphatase (At1g07110 and ATF2KP), a phosphatidylserine decarboxylase (At4g25970 and PSD3), and a 26 S proteasome regulatory subunit 4 for both the Arabidopsis (At2g20140 and RPT2b) and the human samples (P62191 and RPT2). Of note, several of these proteins (At3g19240, At1g07110, and At2g20140) have been already proposed as N-Myr. This indirect conclusion derived from studies involving N-terminal octapeptides and the demonstration that they were substrates of NMT in vitro (67). In the human sample, we also identified a guanine nucleotide-binding protein G1 subunit α-2 (P04899 and GNAI2), a choline transporter-like protein 1 (Q8WWI5 and SLC44A1). It is interesting to note the identification of two homologous proteins sharing 77% of identity between Arabidopsis RPT2b and human RPT2. Although N-Myr at the N-terminal of the human RPT2 has been described before (68), our results show the conservation of this N terminus modification along different species such as humans, yeast (69, 70), and Arabidopsis. Thus, our results confirm even more strongly the conservation of such critical modification across distant species.
TermiNator3 N-terminal Co-translational Modification Predictions Accuracy
The Arabidopsis and human identified proteins were submitted to in silico prediction for the modifications that occur at their N termini using TermiNator3 (4) to assess the accuracy of the prediction. Only proteins for which the N-terminal was characterized at position 1 or 2 were retained for this investigation.
The overall TermiNator3 prediction for NME is highly accurate (Table II) for both the experimental Arabidopsis (100%) and human samples (97%). However, the human sample set showed few differences. Considering the 177 proteins that retain the initial Met (no NME or partial NME), a small set (13 hits with no-NME and 5 with partial NME), which was wrongly predicted, is related to proteins with Thr (10 of 18), Val (5 of 18), Ala (2 of 18), and Ser (1 of 18) at position 2. Indeed, Thr and Val residues are known to have a negative influence on NME efficiency and other amino acids beyond the penultimate residue strongly influence the cleavage (Thr, Pro, Glu, Asp…) making prediction sometimes difficult (50). In such cases, partial cleavage is expected as observed for proteins P12236, Q9GZS3, or Q9NPJ3. Interestingly, the Arabidopsis proteins with Val or Thr residues at the N terminus appear to be correctly processed in either the cell or the plantlet samples. Then the observed partial NME could be linked to the type of human cell lines that were used in this study. If it is not the case, this further supports the fact that MetAP2, the enzyme in charge of this cleavage, is more limiting in humans than in plants (71). This could explain why it is possible to inactivate this enzyme in plants and not in animals (72, 73). Of note, these polypeptides retaining the first Met, although they do have a Thr or a Val at position 2, were found to be NAAed on the Met residue that does not correspond to a clear Nat target (14).
Table II. Summary of the protein N terminus prediction using Terminator3 for NME, NAA, and N-Myr, applied to the experimentally characterized proteins from both the Arabidopsis and human samples.
| Modification | Experimental results |
Prediction |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| True positive | True negative | False positive | False negative | Hits | Specificity | Sensitivity | Accuracy | Matthews coefficient | |
| A. thaliana | |||||||||
| NME | 539 | 249 | 0 | 2 | 790 | 100% | 100% | 100% | 0.99 |
| NTA | 718 | 12 | 1 | 57 | 788 | 92% | 93% | 93% | 0.38 |
| N-Myr | 5 | 29 | 7 | 0 | 41 | 81% | 100% | 83% | 0.58 |
| H. sapiens | |||||||||
| NME | 479 | 177 | 13 | 4 | 673 | 93% | 99% | 97% | 0.94 |
| NTA | 588 | 29 | 2 | 37 | 656 | 94% | 94% | 94% | 0.62 |
| N-Myr | 3 | 4 | 2 | 0 | 9 | 67% | 100% | 78% | 0.63 |
Dealing with NAA predictions, the accuracy of TermiNator3 is higher than 93% for both species (Table II), which confirms the ability of this tool to predict correctly the status of both plant and human N termini at the proteome scale. For both the Arabidopsis and the human sample, the remaining proteins for which the prediction is not in agreement with the experimental result can be divided in two different subclasses: (i) N termini with NAAed Gly at position 2, which are predicted to be unmodified or N-Myr (supplemental Fig. S1) and (ii) potential NatC substrates, i.e. proteins retaining the first Met (supplemental Fig. S2). This divergence of the two predictions could be easily explained by the different modifications that could occur in parallel for the Gly residues, i.e. free N terminus, NAA, and N-Myr. This increases the difficulty to identify a clear pattern for the Gly residue, as well as a clear pattern for the NatC substrates penalized by their too low number to develop robust prediction rules.
As anticipated, a low number of proteins were characterized with N-Myr in both species, i.e. five for the Arabidopsis sample and three for the human sample; however, this is expected with the low frequency of this modification (0.6 and 0.4%, respectively). TermiNator was constructed to knowingly overpredict this modification by a factor of two (41, 67) (e.g. among the 12 potential candidates retrieved, only five could be experimentally verified as N-Myr). Although this modification is extremely difficult to predict so far, all experimentally characterized N-Myr proteins were correctly predicted with an accuracy of 83 and 78% for Arabidopsis and human sample, respectively. It is interesting to notice that all eight characterized N-Myr proteins have a Ser residue at position 6 (supplemental Fig. S3). This residue is known to strongly enhance the likelihood of modification as previously demonstrated by an in vitro investigation (67), which tends to confirm this in vivo result despite the low number of cases available.
Thus, the overall prediction for NME, NAA, and N-Myr using TermiNator is excellent and gives reliable information of the protein N terminus status in both Arabidopsis and human samples; however, a few scenarios need to be further improved, such as predicting NatC substrates as expected and previously noticed (41). Hence, because this type of substrate is rare, i.e. 10.8% (1205 of 11,117) and 12.1% (4252 of 35,095) for Arabidopsis and human, respectively (determined theoretically from the UniprotKB/Swiss-Prot human subset and TAIR10 protein sequences) and may be related to low abundant proteins (5), not enough experimental data were available to develop reliable rules that impact directly the accuracy for this prediction. Although the results of our study were initially expected to contribute to the improvement of prediction rules, the number of substrates identified is still too limited (24 of 790 for Arabidopsis and 29 of 674 for humans) and a negative training set, i.e. NatC substrate that are not NAAed, is still needed. To do so, naturally non-N-acetylated proteins should be recovered as well.
Subcellular Localization Prediction
Prediction for protein localization, e.g. secretion, chloroplast, or mitochondria, is still a challenging task. Because of the various maturation or import processes, the N terminus of the protein could be truncated. In our study, the fact that we were able to determine precisely the N terminus status of each identified protein and especially the position where the mature protein starts is highly informative. This is highly valuable information allowing us to distinguish whether a given protein is or is not subjected to targeting toward another compartment especially for the Arabidopsis proteins where chloroplast import mechanisms play a crucial role. Localization was predicted using TargetP (51) against both sample sets and ChloroP (53) against the Arabidopsis set to determine also the transit peptide length (Table III and supplemental Table S1 and S2).
Table III. Result summary of TargetP localization prediction and experimental localization extracted from UniProtKB for the human sample and various resources (PPDB, SUBA, TAIR9, and AT_CHLORO) for the Arabidopsis sample.
NA, not applicable.
| Experimental protein set |
Predictions |
|||||
|---|---|---|---|---|---|---|
| Hits | Percentage | Mitochondrion | Secreted | Chloroplast | Other | |
| A. thaliana | ||||||
| TargetP Predicted hits | 1007 | 52 | 37 | 231 | 687 | |
| % | 5.2% | 3.7% | 22.9% | 68.2% | ||
| Cytoplasm | 43 | 4.3% | 1 | 2 | 1 | 39 |
| Mitochondria confirmed | 26 | 2.6% | 5 | 2 | 1 | 18 |
| Secreted confirmed | 0 | 0.0% | 0 | 0 | 0 | 0 |
| Chloroplast confirmed | 193 | 19.2% | 16 | 1 | 155 | 21 |
| H. sapiens | ||||||
| TargetP Predicted hits | 727 | 41 | 16 | NA | 670 | |
| % | 5.6% | 2.2% | NA | 92.2% | ||
| Cytoplasm | 354 | 48.7% | 18 | 3 | NA | 333 |
| Mitochondria confirmed | 13 | 1.8% | 2 | 0 | NA | 11 |
| Secreted confirmed | 0 | 0.0% | 0 | 0 | NA | 0 |
For the human sample, 41 proteins (5.6%) were predicted to be mitochondrial, and 16 (2.2%) were secreted (Table III). If one relies on the available UniprotKB annotations, it appears that only a third is annotated as mitochondrial proteins (13 of 41), and none are known to be secreted. For the Arabidopsis samples, TargetP results highlight the predominance of chloroplast-targeted proteins (231 over 1007; 22.9%) versus mitochondrion (52; 5.2%) and secreted ones (37; 3.7%). Although the prediction for the secreted and mitochondrial proteins are not confirmed by the available annotation in PPDB (Plant Protein DataBase (74)), SUBA (SubCellular Proteomic Database (75)), AT_CHLORO (65), or TAIR, 150 of 231 have been shown experimentally to be localized in the chloroplast. If no conclusion could be drawn for TargetP predictions efficiency against mitochondrial and secreted proteins because of the low number of candidates, it appears to be reliable for our data set of chloroplast localized proteins with a prediction accuracy of 89% (specificity, 84%; sensitivity, 96%; Matthews correlation coefficient, 0.77; true positive = 155, true negative = 254, false positive = 47, false negative = 6). This favors the idea that most of the predicted chloroplastic proteins are indeed localized in the chloroplast and validates thereby that the occurrence of the cleavage of a transit peptide and the presence of a dNAA modification are highly relevant.
Thanks to the large number of predicted and confirmed chloroplast proteins combined with the experimental localization of the transit peptide cleavage site related to the chloroplast import mechanism (76), we determined the accuracy of ChloroP prediction for cTP length. Because we characterized some of these proteins with multiple cTP cleavage positions for a unique protein (up to five positions for At1g15500.1), we tried to determine whether the two subsets had significant cTP length differences. We defined two subsets in relation to this characteristic and compared the experimental cTP length to their predicted values. Because some of the proteins identified with a dNAA were not all related to chloroplast targeted protein, e.g. alternative starts or protein splice variants, and because of our wide distribution of cleavage site position, we decided to restrict the data set to proteins with cleavage in between positions 20 and 200 in accordance to previously published cTP average length (38, 42, 53) (supplemental Table S6). Thus, the average lengths of the transit peptide for both subsets are similar with 57 ± 21 (n = 164) and 62 ± 20 (n = 85) residues for single and multiple cleavage sites, respectively (Table IV).
Table IV. Result summary of the cTP length prediction using TargetP versus experimental characterized sites.
The experimentally characterized proteins are split in two categories related to the number of cTP cleavage sites found per protein. For each subgroup, we compare the average experimental length and the average TargetP prediction. These two experimental subgroups are not significantly different (Student's t test, p < 0.1).
| Number of proteins | Number of cleavage sites | Min cTP length | Max cTP length | Average experimental cTP length | TargetP predicted cTP length | Prediction discrepency (exp − Theo length) | |
|---|---|---|---|---|---|---|---|
| Single cleavage site | 164 | 164 | 20 | 159 | 57 ± 21 | 53 ± 16 | 4 ± 21 |
| Multiple cleavage sites | 37 | 85 | 24 | 123 | 62 ± 20 | 58 ± 14 | 4 ± 17 |
Note that the difference with the ChloroP prediction (supplemental Table S6) is four residues upstream of the experimentally characterized cleavage site for both the single and multiple cleavages protein subsets. Globally, only half of the predicted sites are within three residues around the experimentally characterized ones (supplemental Tables S1 and S6). Although TargetP tends to predict correctly chloroplast localization in A. thaliana, ChloroP determination of the cTP length is less accurate and subject to major discrepancy between the experimental results and the predicted values.
In Silico Identification of Arabidopsis N-Acetyltransferases
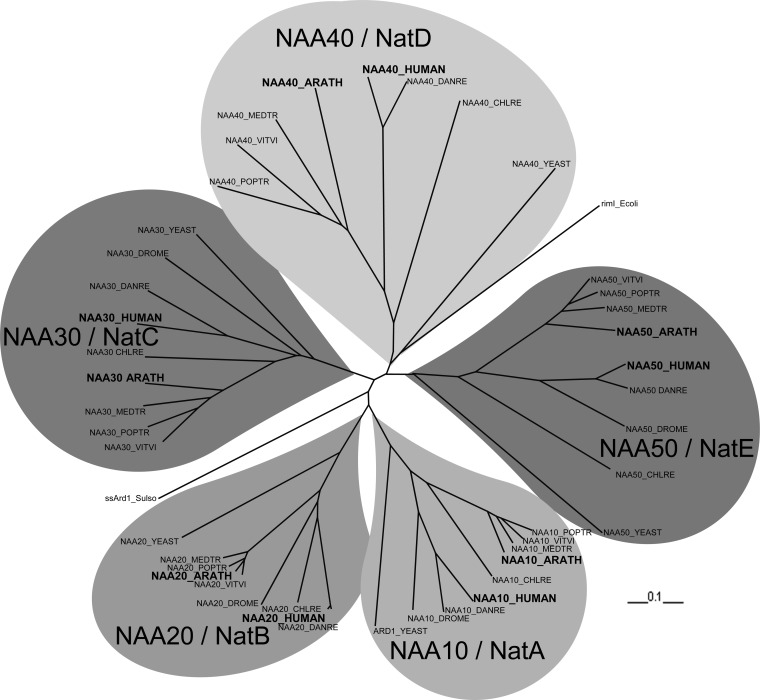
Because of the recent effort (15) to classify and define a relevant nomenclature of Nats, we decided to investigate and to identify the catalytic subunits of the main Nats in different plants including Populus trichocarpa, Chlamydomonas reinhardtii, Medicago truncatula, and Vitis vinifera. Based on the major human and yeast sequences, we extracted the orthologous genes using BLAST approaches for few relevant species closer to the human than the previously defined plants such as D. melanogaster or Danio rerio, highlighting sequence divergences along species evolution. We also included the sequence of the Nat characterized in Archaea (19) and those involved in the acetylation of Escherichia coli ribosomal proteins (77, 78). Finally, all of the in silico-predicted A. thaliana acetylases were also included in the final set of proteins (supplemental Table S7) and aligned with ClustalW. The associated phylogram (Fig. 1 and supplemental Fig. S4) highlights several subgroups clearly related to NatA, B, C, D, E, and F, including the selected gene products as probable plant acetylases candidates. It also appears clearly that the previously characterized NatC catalytic subunit (44) (or NAA30) clustered as expected with the NAA30 orthologous genes from various eukaryotes and especially the human and yeast copies.
Fig. 1.
Phylogram of the catalytic subunits of the main Nats, i.e. NAA10 (NatA), NAA20 (NatB), NAA30 (NatC), NAA40 (NatD), and NAA50 (NatE), for various species. The orthologous sequences of the human genes were extracted using BLAST searches against D. rerio (DANRO), D. melanogaster (DRONE), P. trichocarpa (POPTR), C. reinhardtii (CHLRE), V. vinifera (VITVI), M. truncatula (MEDTR), and A. thaliana (ARATH) and have been added, as well as the S. solfataricus ortholog of NAA10 and RIMI from E. coli.
DISCUSSION
Protein NAA is one of the major modifications found in mammals and yeast proteins, but so far, this modification has been characterized for only a limited number of plant proteins. Our simplified approach using SCX-LC and targeting blocked N-terminal peptides of both Arabidopsis and human samples highlights the presence of a large number of NAAed proteins in both species proving the conservation of this mechanism in plants. This approach allows for the first time the characterization of more than 1000 nonredundant plant proteins N termini, which corresponds to a strong boost of our knowledge of the process and makes comparisons now statistically relevant.
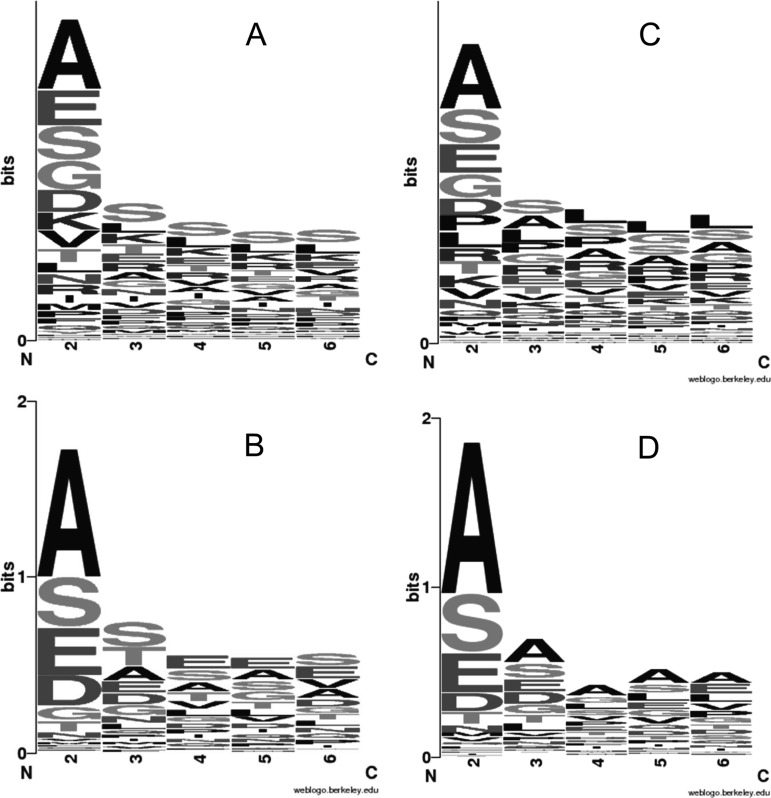
NAA has been investigated in few studies before (6–9, 11, 79), but this is the first report highlighting the characterization of so many proteins for A. thaliana carrying this modification. At the level of translated genes, WebLogo (80) visualization of the N-terminome for Arabidopsis (TAIR10; Fig. 2A and supplemental Fig. S5A) and humans (UniprotKB/Swiss-Prot: human subset; Fig. 2C) shows some similarities for the potential Nat substrates between both species. This is further confirmed experimentally with the characterized NAAed proteins for both the Arabidopsis (Fig. 2B) and the human sample (Fig. 2D). Clearly, the N-terminal sequence profile of TAIR10 (Fig. 2A and supplemental Fig. S5A) shows higher levels of hydroxylated residues and especially the Ser residues at the N-terminal side of the proteins compared with the corresponding human pattern (human subset of UniprotKB), where Ala is mainly favored (Fig. 2B). This visualization was produced with all predicted Arabidopsis proteins that includes the nuclear encoded chloroplastic proteins, of which the cTP is known to be rich in such residues (53, 81). Proteins experimentally characterized with NAA, usually located in the cytoplasm, do not show a more significant enrichment in Ser residues (supplemental Fig. S5B), unlike the proteins with dNAA, which are localized in the chloroplast (supplemental Fig. S5C). Although recent studies identified large differences of residue distribution at position 2 between species, e.g. D. melanogaster and humans (6, 7, 16), our results further confirm the tendency to observe acetylated Ala residues at the N terminus of human proteins regardless of the cell type (7, 11). Even though the chloroplast targeting signal displays a higher Ser residue frequency at the N-terminal side, the global NAA profile is more similar to that from humans than those profiles coming from other species such as Drosophila (6), yeast (11), or Archaea (9), which were strongly divergent. No major bias is visible on the residue distribution at position 2 between the proteome of both species (supplemental Fig. S6). This is unlike D. melanogaster where the Ser substrate is predominant over the Ala substrates; this effect is even more pronounced for the yeast or Halobacteria (Fig. 5 in Ref. 7). Thus, it appears that the NAAed substrates for Arabidopsis are similar to the human ones both theoretically with the predicted substrates (Fig. 2, A and C, and supplemental Fig. S6) and experimentally (Fig. 2, B and D).
Fig. 2.
WebLogo of the N-terminome (sequence at positions 2–6) of A. thaliana translated genes product (TAIR10) (A) and the experimentally characterized NAAed proteins (B) (n = 777). Similar representation have been produced for the human subset of (C) Uniprot/Swiss-Prot protein (human subset) versus (D) experimentally characterized NAAed proteins (n = 648).
These results tend to support the existence of similar Nats as the one characterized for human and yeast (15). Only a single one has been characterized through the identification of an A. thaliana phenotype affecting the efficiency of the photosystem II quantum yield (44) and associated to NatC activity. Our effort to identify the Nat catalytic subunits for Arabidopsis and other model plants was able to pinpoint NAA10, NAA20, NAA30, NAA40, and NAA50 orthologous genes (Fig. 1 and supplemental Fig. S4 and Table S7). Although NAA11 was not found in plant, which was expected because this protein has been described to be mammal-specific (21), NAA60 (16) on the other hand seems to be less specific because we found the orthologous gene in D. rerio, but we were unable to identify any relevant genes in plant, suggesting that this Nat is restricted to Euteleostomi (supplemental Fig. S4).
Although 777 over 1054 of the NAAed characterized peptides are related to the expected co-translational NAA, 277 peptides (associated to 224 nonredundant proteins) have a starting position located downstream the standard protein N terminus. Although chloroplast imported proteins have been previously described to be acetylated after the cleavage of the cTP (38, 42, 47), our current study gives for the first time an idea of the extent of this modification in this organelle with more than 220 characterized proteins. Although some previous studies (38, 42) suggest that the NAA is a post-translational modification on a limited number of chloroplast imported protein, the present results clearly highlight the high occurrence of this modification in the chloroplast compartment. Because these polypeptides are imported and processed to remove the cTP during their chloroplast translocation, it appears that the acetylation of these neo-N termini is a widespread protein post-translational modification in the chloroplast. The modified neo-N termini or dNAAed residues appear to be predominantly Ala, Ser, Val, and Thr (Fig. 3). These residues are also the preferential substrates of the conventional NatA with some restriction for the Val, which is a known substrate but is subject to numerous exceptions (5) during co-translation modification. This higher frequency for Val NAA is mostly linked to preferential localization of the cTP cleavage sites on such residues inducing a bias in frequency of the Val substrate but with still being compatible with the specificity of the NatA. Although several predicted acetylases appear to be imported into the chloroplast (supplemental Table S7), none of these gene products shared significant homology with the NatA group or similar catalytic member, e.g. E. coli RIM (ribosomal protein acetyltransferase) or Sulfolobus solfataricus Ard1 (supplemental Fig. S4). Therefore, it has not been possible to propose a potential candidate.
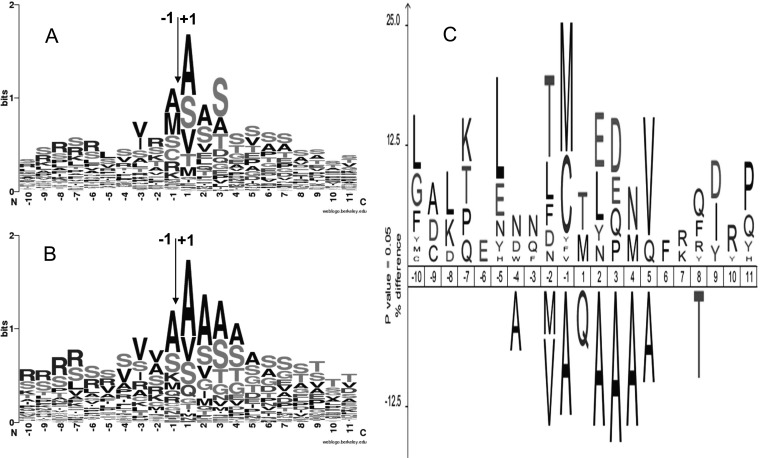
Fig. 3.
Sequence logos of the cTP cleavage sites related to the experimentally characterized dNAAed chloroplastic proteins. A, proteins with a single cTP cleavage site (n = 170). B, proteins with multiple cTP cleavage sites (n = 85). Each protein is represented with as many sequences as experimental cleavage sites characterized. C, IceLogo for the comparison of the sequences used in the two previous sequence logos, i.e. single (A) versus multiple (B) cleavage sites. cTP cleavage site occurs between residues −1 and +1.
dNAAed protein subset was a sample of choice to test the prediction accuracy of localization tools and transit peptide length such as TargetP and ChloroP. The overall TargetP prediction is correct with more than 90% accuracy for the chloroplast localization, but it appears that ChloroP performance to predict cTP length is not very accurate, with only half of the correctly predicted sites within a three-residue error margin (supplemental Tables S1 and S6). Interestingly, a third of the chloroplast targeted proteins characterized with dNAA were experimentally identified with more than a single cleavage site. If most of these proteins have two vicinal cleavage sites identified in close proximity, e.g. At4g18440 or At2g30390, few of them were identified with up to five different cleavage positions, i.e. At1g15500, distributed along a string of 10 residues, i.e. At3g01120. Sequence comparison of the single versus multiple cTP cleavage sites shows clear difference for the Ala distribution around the cleavage site (Fig. 3B) and especially between positions −1 to +4 (cleavage position defined between −1 and +1). Direct sequence comparison using IceLogo (82) (Fig. 3C) of these two protein sets confirms that Ala at positions −1, 2, 3, 4, and 5 are significantly more frequent in the “multiple cleavage” set (p > 0.05). Conversely, in the “single cleavage” set, residues such as Thr−2 (residue at position −2) or Thr1 are more frequently observed. It is difficult to determine whether the higher frequency of the Ala residues around the optimum sites tends to favor multiple cleavages or whether the absence of the hydroxylated or acidic residues, i.e. Ser/Thr−1, Thr1, Glu2, or Asp/Glu3 destabilizes the interaction with the endopeptidase involved in the cleavage. Although some proteins were characterized with multiple cleavage sites, this does not impact on the cTP length: 59 ± 26 for the single cleavage site and 63 ± 20 for the multiple sites. These cTP lengths are not significantly different and are comparable with previously published results (42). It is noteworthy that the cTP length determined recently for C. reinhardtii was ∼37 ± 14 (38), highlighting some important differences, suggesting potential divergences between chlorophytic species and their chloroplast import mechanism.
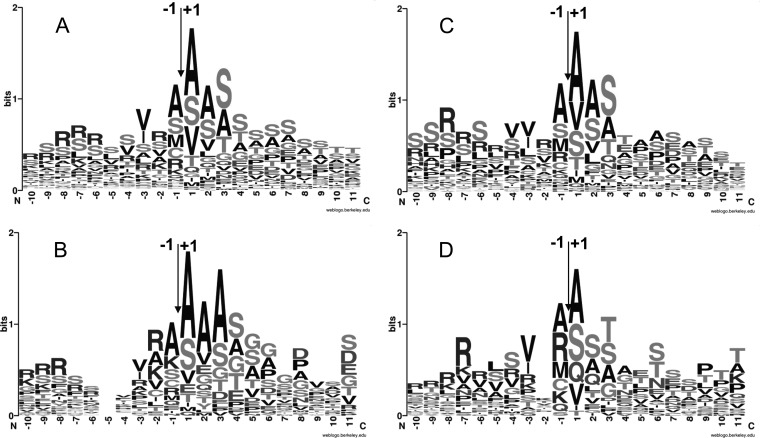
Because of the large number of proven chloroplast localized proteins (Fig. 4A), it was even possible to create subgroups targeting the inner integral envelope (Fig. 4B), the stroma (Fig. 4C), and the thylakoid (Fig. 4D). Despite the fact that the sequence upstream of the cTP cleavage site is directly linked to the import machinery (83), it appears that some residues are conserved and could be involved in alternative mechanism (Fig. 4A) such as Val−3 also found in C. reinhardtii (42). Furthermore, this specific residue is not present in the chloroplast inner integral envelope proteins (Fig. 4B), suggesting that this position could influence the protein translocation through the chloroplast envelope into the stroma. The Arabidopsis stromal protein pattern (Fig. 4C) exhibits some similarity with the logo obtained with C. reinhardtii (Fig. 2D in Ref. 42), stressing the influence of the Val−4-Val−3-Ala−1-Ala1, as well as Ser−9-Arg−8-Arg−7 for protein localization. Downstream of the cleavage position, one site appears to be preferentially populated with Ser3 (Fig. 4A) and was previously considered to be important for the mechanism and the specificity of the N-α-acetylation (42). Unfortunately, this residue is not conserved on integral envelope (Fig. 4B) and is weakly conserved in thylakoid subset (Fig. 4D), where Thr is also favored alternatively. Thus, this Ser3 may not be relevant for the acetylation mechanism but mainly for the protein localization, i.e. stroma (Fig. 4D), which corroborates the data of Zybailov et al. (42), in which only stromal protein were targeted.
Fig. 4.
Sequence logo of the cTP cleavage sites related to all experimentally characterized nuclear encoded chloroplastic proteins (n = 255) characterized with dNAA (cTP site position within 20–200) (A), chloroplast inner integral envelope proteins (n = 36) (B), plastid stroma proteins (n = 73) (C), and thylakoid proteins (n = 27) (D). cTP cleavage site occurs between residues −1 and +1.
Ala1, as well as Ser1 and Val1 in a lower extend, appears to be predominant in all subsets highlighting the crucial influence of these residues for the NAA of the neo-N termini. Substrates of this post-translational N-α-acetylation reaction appear to be similar to the conventional NatA substrates (supplemental Fig. S7) except for the Val, which is more frequent substrates for the dNAA. Because no known acetylases are apparently encoded in the chloroplast genome, the Nat involved in such modification must be an imported nuclear encoded chloroplastic protein.
Although a vast majority of the characterized dNAAed proteins are identified in Arabidopsis (chloroplast-located proteins), some of them remain unexplained, especially those identified in the human sample. Interestingly, for a large number of these human proteins, the dNAAed characterized peptides show some similarity to the formal N-terminal protein end with a Met or its vicinal residue uncovered after NME. For a significant number of human dNAAed proteins, these N termini suggest co-translational NAA on alternative protein starts. Protein splice variants are known sources for alternative protein N terminus and could explain the fifth of these dNAAed human proteins, whereas it deals only with few instances in Arabidopsis (supplemental Tables S1 and S2).
Another source of protein N terminus variability is dependent on alternative start mechanisms at the protein translation level (84). This phenomenon producing alternative N termini has been described with the so-called Kozak rules (85) for which consensus patterns are defined: i.e. a(a/c)(A/G)(A/C)aAUGGc for plants (86, 87) and n(A/G)nnAUGG for humans (84, 85). These rules could explain up to half of the characterized human dNAAed proteins, whereas it could justify only few candidates for the Arabidopsis sample, i.e. At4g16510.1, At3g44310.1, At5g13930.1, and At5g66100.1 (84–87). Although the human consensus pattern appears to fit well the coding sequence of our candidates (supplemental Table S2, mRNA Alt. start investigation worksheet), it does not seem to be that efficient with the Arabidopsis sequences where the consensus pattern still needs further improvement (supplemental Table S1, mRNA Alt. start investigation worksheet). Interestingly, almost half of the human protein characterized with dNAA were characterized among various experimental sources as detailed in supplemental Table S2 (mRNA and alternative start worksheet) (7) or UniprotKB, highlighting the value of such an approach to identify the mature form of the protein.
Recent development of the mass spectrometers allows large scale studies to characterize co- and post-translational modifications. Using SCX-LC, we enriched the NAAed peptides from whole cell lysate of various Arabidopsis sample and human cell lines with a total of 1072 and 727 nonredundant peptides characterized. First, the performances of the prediction tools for this type of modification were compared with our experimental results. Arabidopsis and humans show very similar theoretical and experimental substrate profiles for NAA and TermiNator3 prediction proved to be a very accurate prediction tool. Based on this knowledge, it was also possible to propose by homology all genes for the catalytic domain associated to this co-translational modification, which is also well conserved in plants.
Furthermore, one-third of the characterized NAAed peptides from the Arabidopsis sample were situated downstream of the conventional predicted start. Our investigation highlights that at least 82% of these proteins were targeted to the chloroplast (supplemental Table S1, subcellular localization worksheet). During the import mechanism, these proteins were processed and truncated, and the uncovered new N terminus was N-α-acetylated post-translationally. Although NAA was long considered to be a co-translational modification, this study highlights it as a post-translational modification for a large number of proteins, i.e. virtually all nuclear-encoded chloroplast protein could be involved by this modification. From these results, it appears that the Nat involved in such acetylation reaction has an activity close to the NatA because of its substrate type but was not identified. Other dNAA have been characterized and associated to protein alternative start or protein splice variants mainly for the human sample and wrong protein start prediction for the Arabidopsis sample.
The present proteomics study highlights the importance of protein N terminus characterization to better understand the important mechanisms involved for the organelles, e.g. chloroplast, and the interest of such experimental data for the improvement of our knowledge in protein maturation mechanism and particularly N-terminal co- and post-translational modifications.
Acknowledgments
We thank A. Bairoch, A. Estreicher, and M. Schneider (Swiss Institute of Bioinformatics, Geneva, Switzerland) for advice and help inserting the experimental data into the UniProtKB/Swiss-Prot database, W. Majeran and A. Bilsland for supplying cell lysates (Arabidopsis and human, respectively), J. Ho for performing some preliminary analyses of the Arabidopsis sample, and F. Frottin for useful discussions about NME. We also appreciate the use of the HPLC facilities at ICSN (Gif, France) and the facilities and expertise of IMAGIF (Centre de Recherche de Gif, www.imagif.cnrs.fr).
Footnotes
* The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- NAA
- N-α-acetylation
- NAAed
- N-α-acetylated
- cTP
- chloroplast transit peptide
- dNAA
- downstream N-α-acetylation
- dNAAed
- downstream N-α-acetylated
- SCX-LC
- strong cation exchange
- NME
- N-terminal methionine excision
- Nat
- N-α-acetyltransferase
- N-Myr
- N-myristoylation
- RP
- reverse phase.
REFERENCES
- 1. Persson B., Flinta C., von Heijne G., Jörnvall H. (1985) Structures of N-terminally acetylated proteins. Eur. J. Biochem. 152, 523–527 [DOI] [PubMed] [Google Scholar]
- 2. Driessen H. P., de Jong W. W., Tesser G. I., Bloemendal H. (1985) The mechanism of N-terminal acetylation of proteins. CRC Crit. Rev. Biochem. 18, 281–325 [DOI] [PubMed] [Google Scholar]
- 3. Tsunasawa S., Sakiyama F. (1984) Amino-terminal acetylation of proteins: An overview. Methods Enzymol. 106, 165–170 [DOI] [PubMed] [Google Scholar]
- 4. Meinnel T., Peynot P., Giglione C. (2005) Processed N-termini of mature proteins in higher eukaryotes and their major contribution to dynamic proteomics. Biochimie 87, 701–712 [DOI] [PubMed] [Google Scholar]
- 5. Meinnel T., Giglione C. (2008) Tools for analyzing and predicting N-terminal protein modifications. Proteomics 8, 626–649 [DOI] [PubMed] [Google Scholar]
- 6. Goetze S., Qeli E., Mosimann C., Staes A., Gerrits B., Roschitzki B., Mohanty S., Niederer E. M., Laczko E., Timmerman E., Lange V., Hafen E., Aebersold R., Vandekerckhove J., Basler K., Ahrens C. H., Gevaert K., Brunner E. (2009) Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster. PLoS Biol. 7, e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Helbig A. O., Gauci S., Raijmakers R., van Breukelen B., Slijper M., Mohammed S., Heck A. J. (2010) Profiling of N-acetylated protein termini provides in-depth insights into the N-terminal nature of the proteome. Mol. Cell. Proteomics 9, 928–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Falb M., Aivaliotis M., Garcia-Rizo C., Bisle B., Tebbe A., Klein C., Konstantinidis K., Siedler F., Pfeiffer F., Oesterhelt D. (2006) Archaeal N-terminal protein maturation commonly involves N-terminal acetylation: A large-scale proteomics survey. J. Mol. Biol. 362, 915–924 [DOI] [PubMed] [Google Scholar]
- 9. Aivaliotis M., Gevaert K., Falb M., Tebbe A., Konstantinidis K., Bisle B., Klein C., Martens L., Staes A., Timmerman E., Van Damme J., Siedler F., Pfeiffer F., Vandekerckhove J., Oesterhelt D. (2007) Large-scale identification of N-terminal peptides in the halophilic archaea Halobacterium salinarum and Natronomonas pharaonis. J. Proteome Res. 6, 2195–2204 [DOI] [PubMed] [Google Scholar]
- 10. Polevoda B., Sherman F. (2003) N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 325, 595–622 [DOI] [PubMed] [Google Scholar]
- 11. Arnesen T., Van Damme P., Polevoda B., Helsens K., Evjenth R., Colaert N., Varhaug J. E., Vandekerckhove J., Lillehaug J. R., Sherman F., Gevaert K. (2009) Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. U.S.A. 106, 8157–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gautschi M., Just S., Mun A., Ross S., Rücknagel P., Dubaquié Y., Ehrenhofer-Murray A., Rospert S. (2003) The yeast Nα-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol. Cell. Biol. 23, 7403–7414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raue U., Oellerer S., Rospert S. (2007) Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J. Biol. Chem. 282, 7809–7816 [DOI] [PubMed] [Google Scholar]
- 14. Meinnel T., Serero A., Giglione C. (2006) Impact of the N-terminal amino acid on targeted protein degradation. Biol. Chem. 387, 839–851 [DOI] [PubMed] [Google Scholar]
- 15. Polevoda B., Arnesen T., Sherman F. (2009) A synopsis of eukaryotic Nα-terminal acetyltransferases: Nomenclature, subunits and substrates. BMC Proc. 3, (Suppl. 6) S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Damme P., Hole K., Pimenta-Marques A., Helsens K., Vandekerckhove J., Martinho R. G., Gevaert K., Arnesen T. (2011) NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation. PLoS Genet. 7, e1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Polevoda B., Norbeck J., Takakura H., Blomberg A., Sherman F. (1999) Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J. 18, 6155–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Starheim K. K., Gromyko D., Velde R., Varhaug J. E., Arnesen T. (2009) Composition and biological significance of the human Nα-terminal acetyltransferases. BMC Proc. 3, (Suppl. 6) S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mackay D. T., Botting C. H., Taylor G. L., White M. F. (2007) An acetylase with relaxed specificity catalyses protein N-terminal acetylation in Sulfolobus solfataricus. Mol. Microbiol. 64, 1540–1548 [DOI] [PubMed] [Google Scholar]
- 20. Perrot M., Massoni A., Boucherie H. (2008) Sequence requirements for Nα-terminal acetylation of yeast proteins by NatA. Yeast 25, 513–527 [DOI] [PubMed] [Google Scholar]
- 21. Arnesen T., Betts M. J., Pendino F., Liberles D. A., Anderson D., Caro J., Kong X., Varhaug J. E., Lillehaug J. R. (2006) Characterization of hARD2, a processed hARD1 gene duplicate, encoding a human protein N-α-acetyltransferase. BMC Biochem. 7, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim S. H., Park J. A., Kim J. H., Lee J. W., Seo J. H., Jung B. K., Chun K. H., Jeong J. W., Bae M. K., Kim K. W. (2006) Characterization of ARD1 variants in mammalian cells. Biochem. Biophys. Res. Commun. 340, 422–427 [DOI] [PubMed] [Google Scholar]
- 23. Lim J. H., Park J. W., Chun Y. S. (2006) Human arrest defective 1 acetylates and activates β-catenin, promoting lung cancer cell proliferation. Cancer Res. 66, 10677–10682 [DOI] [PubMed] [Google Scholar]
- 24. Rope A. F., Wang K., Evjenth R., Xing J., Johnston J. J., Swensen J. J., Johnson W. E., Moore B., Huff C. D., Bird L. M., Carey J. C., Opitz J. M., Stevens C. A., Jiang T., Schank C., Fain H. D., Robison R., Dalley B., Chin S., South S. T., Pysher T. J., Jorde L. B., Hakonarson H., Lillehaug J. R., Biesecker L. G., Yandell M., Arnesen T., Lyon G. J. (2011) Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am. J. Hum. Genet. 89, 28–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arnesen T., Gromyko D., Pendino F., Ryningen A., Varhaug J. E., Lillehaug J. R. (2006) Induction of apoptosis in human cells by RNAi-mediated knockdown of hARD1 and NATH, components of the protein N-α-acetyltransferase complex. Oncogene 25, 4350–4360 [DOI] [PubMed] [Google Scholar]
- 26. Yu M., Ma M., Huang C., Yang H., Lai J., Yan S., Li L., Xiang M., Tan D. (2009) Correlation of expression of human arrest-defective-1 (hARD1) protein with breast cancer. Cancer Invest. 27, 978–983 [DOI] [PubMed] [Google Scholar]
- 27. Helbig A. O., Rosati S., Pijnappel P. W., van Breukelen B., Timmers M. H., Mohammed S., Slijper M., Heck A. J. (2010) Perturbation of the yeast N-acetyltransferase NatB induces elevation of protein phosphorylation levels. BMC Genomics 11, 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Starheim K. K., Arnesen T., Gromyko D., Ryningen A., Varhaug J. E., Lillehaug J. R. (2008) Identification of the human Nα-acetyltransferase complex B (hNatB): A complex important for cell-cycle progression. Biochem. J. 415, 325–331 [DOI] [PubMed] [Google Scholar]
- 29. Starheim K. K., Gromyko D., Evjenth R., Ryningen A., Varhaug J. E., Lillehaug J. R., Arnesen T. (2009) Knockdown of human N α-terminal acetyltransferase complex C leads to p53-dependent apoptosis and aberrant human Arl8b localization. Mol. Cell. Biol. 29, 3569–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Avdic V., Zhang P., Lanouette S., Voronova A., Skerjanc I., Couture J. F. (2011) Fine-tuning the stimulation of MLL1 methyltransferase activity by a histone H3-based peptide mimetic. FASEB J. 25, 960–967 [DOI] [PubMed] [Google Scholar]
- 31. Urbancikova M., Hitchcock-DeGregori S. E. (1994) Requirement of amino-terminal modification for striated muscle α-tropomyosin function. J. Biol. Chem. 269, 24310–24315 [PubMed] [Google Scholar]
- 32. Singer J. M., Shaw J. M. (2003) Mdm20 protein functions with Nat3 protein to acetylate Tpm1 protein and regulate tropomyosin-actin interactions in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 100, 7644–7649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Behnia R., Panic B., Whyte J. R., Munro S. (2004) Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat Cell Biol. 6, 405–413 [DOI] [PubMed] [Google Scholar]
- 34. Behnia R., Barr F. A., Flanagan J. J., Barlowe C., Munro S. (2007) The yeast orthologue of GRASP65 forms a complex with a coiled-coil protein that contributes to ER to Golgi traffic. J. Cell Biol. 176, 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hofmann I., Munro S. (2006) An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J. Cell Sci. 119, 1494–1503 [DOI] [PubMed] [Google Scholar]
- 36. Ciechanover A., Ben-Saadon R. (2004) N-terminal ubiquitination: More protein substrates join in. Trends Cell Biol. 14, 103–106 [DOI] [PubMed] [Google Scholar]
- 37. Hwang C. S., Shemorry A., Varshavsky A. (2010) N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bienvenut W. V., Espagne C., Martinez A., Majeran W., Valot B., Zivy M., Vallon O., Adam Z., Meinnel T., Giglione C. (2011) Dynamics of post-translational modifications and protein stability in the stroma of Chlamydomonas reinhardtii chloroplasts. Proteomics 11, 1734–1750 [DOI] [PubMed] [Google Scholar]
- 39. Forte G. M., Pool M. R., Stirling C. J. (2011) N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol. 9, e1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yi C. H., Pan H., Seebacher J., Jang I. H., Hyberts S. G., Heffron G. J., Vander Heiden M. G., Yang R., Li F., Locasale J. W., Sharfi H., Zhai B., Rodriguez-Mias R., Luithardt H., Cantley L. C., Daley G. Q., Asara J. M., Gygi S. P., Wagner G., Liu C. F., Yuan J. (2011) Metabolic regulation of protein N-α-acetylation by Bcl-xL promotes cell survival. Cell 146, 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martinez A., Traverso J. A., Valot B., Ferro M., Espagne C., Ephritikhine G., Zivy M., Giglione C., Meinnel T. (2008) Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics 8, 2809–2831 [DOI] [PubMed] [Google Scholar]
- 42. Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K. J. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3, e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baerenfaller K., Grossmann J., Grobei M. A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320, 938–941 [DOI] [PubMed] [Google Scholar]
- 44. Pesaresi P., Gardner N. A., Masiero S., Dietzmann A., Eichacker L., Wickner R., Salamini F., Leister D. (2003) Cytoplasmic N-terminal protein acetylation is required for efficient photosynthesis in Arabidopsis. Plant Cell 15, 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mohammed S., Heck A., Jr. (2011) Strong cation exchange (SCX) based analytical methods for the targeted analysis of protein post-translational modifications. Curr. Opin. Biotechnol. 22, 9–16 [DOI] [PubMed] [Google Scholar]
- 46. Gauci S., Helbig A. O., Slijper M., Krijgsveld J., Heck A. J., Mohammed S. (2009) Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal. Chem. 81, 4493–4501 [DOI] [PubMed] [Google Scholar]
- 47. Gómez S. M., Bil' K. Y., Aguilera R., Nishio J. N., Faull K. F., Whitelegge J. P. (2003) Transit peptide cleavage sites of integral thylakoid membrane proteins. Mol. Cell. Proteomics 2, 1068–1085 [DOI] [PubMed] [Google Scholar]
- 48. Wang B. C., Wang H. X., Feng J. X., Meng D. Z., Qu L. J., Zhu Y. X. (2006) Post-translational modifications, but not transcriptional regulation, of major chloroplast RNA-binding proteins are related to Arabidopsis seedling development. Proteomics 6, 2555–2563 [DOI] [PubMed] [Google Scholar]
- 49. Ferro M., Salvi D., Brugière S., Miras S., Kowalski S., Louwagie M., Garin J., Joyard J., Rolland N. (2003) Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics 2, 325–345 [DOI] [PubMed] [Google Scholar]
- 50. Frottin F., Martinez A., Peynot P., Mitra S., Holz R. C., Giglione C., Meinnel T. (2006) The proteomics of N-terminal methionine cleavage. Mol. Cell. Proteomics 5, 2336–2349 [DOI] [PubMed] [Google Scholar]
- 51. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 52. Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 53. Emanuelsson O., Nielsen H., von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bilsland A. E., Hoare S., Stevenson K., Plumb J., Gomez-Roman N., Cairney C., Burns S., Lafferty-Whyte K., Roffey J., Hammonds T., Keith W. N. (2009) Dynamic telomerase gene suppression via network effects of GSK3 inhibition. PLoS One 4, e6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Droillard M., Boudsocq M., Barbier-Brygoo H., Laurière C. (2002) Different protein kinase families are activated by osmotic stresses in Arabidopsis thaliana cell suspensions: Involvement of the MAP kinases AtMPK3 and AtMPK6. FEBS Lett. 527, 43–50 [DOI] [PubMed] [Google Scholar]
- 56. Espagne C., Martinez A., Valot B., Meinnel T., Giglione C. (2007) Alternative and effective proteomic analysis in Arabidopsis. Proteomics 7, 3788–3799 [DOI] [PubMed] [Google Scholar]
- 57. Keller B. O., Sui J., Young A. B., Whittal R. M. (2008) Interferences and contaminants encountered in modern mass spectrometry. Anal. Chim. Acta 627, 71–81 [DOI] [PubMed] [Google Scholar]
- 58. Huala E., Dickerman A. W., Garcia-Hernandez M., Weems D., Reiser L., LaFond F., Hanley D., Kiphart D., Zhuang M., Huang W., Mueller L. A., Bhattacharyya D., Bhaya D., Sobral B. W., Beavis W., Meinke D. W., Town C. D., Somerville C., Rhee S. Y. (2001) The Arabidopsis Information Resource (TAIR): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 29, 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wilkins M. R., Gasteiger E., Sanchez J. C., Appel R. D., Hochstrasser D. F. (1996) Protein identification with sequence tags. Curr. Biol. 6, 1543–1544 [DOI] [PubMed] [Google Scholar]
- 60. Wilkins M. R., Gasteiger E., Tonella L., Ou K., Tyler M., Sanchez J. C., Gooley A. A., Walsh B. J., Bairoch A., Appel R. D., Williams K. L., Hochstrasser D. F. (1998) Protein identification with N and C-terminal sequence tags in proteome projects. J. Mol. Biol. 278, 599–608 [DOI] [PubMed] [Google Scholar]
- 61. Thiede B., Lamer S., Mattow J., Siejak F., Dimmler C., Rudel T., Jungblut P. R. (2000) Analysis of missed cleavage sites, tryptophan oxydation and N-terminal pyroglutamination after in-gel tryptic digestion. Rapid Commun. Mass Spectrom. 14, 496–502 [DOI] [PubMed] [Google Scholar]
- 62. Newton K. A., McLuckey S. A. (2004) Generation and manipulation of sodium cationized peptides in the gas phase. J. Am. Soc. Mass Spectrom. 15, 607–615 [DOI] [PubMed] [Google Scholar]
- 63. Dormeyer W., Mohammed S., Breukelen B., Krijgsveld J., Heck A. J. (2007) Targeted analysis of protein termini. J. Proteome Res. 6, 4634–4645 [DOI] [PubMed] [Google Scholar]
- 64. Lange P. F., Overall C. M. (2011) TopFIND, a knowledgebase linking protein termini with function. Nat. Methods 8, 703–704 [DOI] [PubMed] [Google Scholar]
- 65. Ferro M., Brugière S., Salvi D., Seigneurin-Berny D., Court M., Moyet L., Ramus C., Miras S., Mellal M., Le Gall S., Kieffer-Jaquinod S., Bruley C., Garin J., Joyard J., Masselon C., Rolland N. (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics 9, 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cook R. K., Sheff D. R., Rubenstein P. A. (1991) Unusual metabolism of the yeast actin amino terminus. J. Biol. Chem. 266, 16825–16833 [PubMed] [Google Scholar]
- 67. Boisson B., Giglione C., Meinnel T. (2003) Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J. Biol. Chem. 278, 43418–43429 [DOI] [PubMed] [Google Scholar]
- 68. Wang X., Chen C. F., Baker P. R., Chen P. L., Kaiser P., Huang L. (2007) Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry 46, 3553–3565 [DOI] [PubMed] [Google Scholar]
- 69. Kimura Y., Saeki Y., Yokosawa H., Polevoda B., Sherman F., Hirano H. (2003) N-Terminal modifications of the 19S regulatory particle subunits of the yeast proteasome. Arch. Biochem. Biophys. 409, 341–348 [DOI] [PubMed] [Google Scholar]
- 70. Kimura Y., Takaoka M., Tanaka S., Sassa H., Tanaka K., Polevoda B., Sherman F., Hirano H. (2000) Nα-acetylation and proteolytic activity of the yeast 20 S proteasome. J. Biol. Chem. 275, 4635–4639 [DOI] [PubMed] [Google Scholar]
- 71. Xiao Q., Zhang F., Nacev B. A., Liu J. O., Pei D. (2010) Protein N-terminal processing: Substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry 49, 5588–5599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ross S., Giglione C., Pierre M., Espagne C., Meinnel T. (2005) Functional and developmental impact of cytosolic protein N-terminal methionine excision in Arabidopsis. Plant Physiol. 137, 623–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yeh J. R., Ju R., Brdlik C. M., Zhang W., Zhang Y., Matyskiela M. E., Shotwell J. D., Crews C. M. (2006) Targeted gene disruption of methionine aminopeptidase 2 results in an embryonic gastrulation defect and endothelial cell growth arrest. Proc. Natl. Acad. Sci. U.S.A. 103, 10379–10384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sun Q., Zybailov B., Majeran W., Friso G., Olinares P. D., van Wijk K. J. (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 37, D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Heazlewood J. L., Verboom R. E., Tonti-Filippini J., Small I., Millar A. H. (2007) SUBA: The Arabidopsis Subcellular Database. Nucleic Acids Res. 35, D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kessler F., Schnell D. (2009) Chloroplast biogenesis: Diversity and regulation of the protein import apparatus. Curr. Opin. Cell Biol. 21, 494–500 [DOI] [PubMed] [Google Scholar]
- 77. Tanaka S., Matsushita Y., Yoshikawa A., Isono K. (1989) Cloning and molecular characterization of the gene rimL which encodes an enzyme acetylating ribosomal protein L12 of Escherichia coli K12. Mol. Gen. Genet. 217, 289–293 [DOI] [PubMed] [Google Scholar]
- 78. Yoshikawa A., Isono S., Sheback A., Isono K. (1987) Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol. Gen. Genet. 209, 481–488 [DOI] [PubMed] [Google Scholar]
- 79. Van Damme P., Van Damme J., Demol H., Staes A., Vandekerckhove J., Gevaert K. (2009) A review of COFRADIC techniques targeting protein N-terminal acetylation. BMC Proc. 3, (Suppl. 6) S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Patron N. J., Waller R. F. (2007) Transit peptide diversity and divergence: A global analysis of plastid targeting signals. Bioessays 29, 1048–1058 [DOI] [PubMed] [Google Scholar]
- 82. Colaert N., Helsens K., Martens L., Vandekerckhove J., Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
- 83. Rimmer K. A., Foo J. H., Ng A., Petrie E. J., Shilling P. J., Perry A. J., Mertens H. D., Lithgow T., Mulhern T. D., Gooley P. R. (2011) Recognition of mitochondrial targeting sequences by the import receptors Tom20 and Tom22. J. Mol. Biol. 405, 804–818 [DOI] [PubMed] [Google Scholar]
- 84. Bazykin G. A., Kochetov A. V. (2011) Alternative translation start sites are conserved in eukaryotic genomes. Nucleic Acids Res. 39, 567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kozak M. (2005) Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361, 13–37 [DOI] [PubMed] [Google Scholar]
- 86. Rangan L., Vogel C., Srivastava A. (2008) Analysis of context sequence surrounding translation initiation site from complete genome of model plants. Mol. Biotechnol. 39, 207–213 [DOI] [PubMed] [Google Scholar]
- 87. Lukaszewicz M., Feuermann M., Jérouville B., Stas A., Boutry M. (2000) In vivo evaluation of the context sequence of the translation initiation codon in plants. Plant Sci. 154, 89–98 [DOI] [PubMed] [Google Scholar]