Abstract
This study examines the response of Symbiodinium sp. endosymbionts from the coral Stylophora pistillata to moderate levels of thermal “bleaching” stress, with and without trace metal limitation. Using quantitative high throughput proteomics, we identified 8098 MS/MS events relating to individual peptides from the endosymbiont-enriched fraction, including 109 peptides meeting stringent criteria for quantification, of which only 26 showed significant change in our experimental treatments; 12 of 26 increased expression in response to thermal stress with little difference affected by iron limitation. Surprisingly, there were no significant increases in antioxidant or heat stress proteins; those induced to higher expression were generally involved in protein biosynthesis. An outstanding exception was a massive 114-fold increase of a viral replication protein indicating that thermal stress may substantially increase viral load and thereby contribute to the etiology of coral bleaching and disease. In the absence of a sequenced genome for Symbiodinium or other photosymbiotic dinoflagellate, this proteome reveals a plethora of proteins potentially involved in microbial-host interactions. This includes photosystem proteins, DNA repair enzymes, antioxidant enzymes, metabolic redox enzymes, heat shock proteins, globin hemoproteins, proteins of nitrogen metabolism, and a wide range of viral proteins associated with these endosymbiont-enriched samples. Also present were 21 unusual peptide/protein toxins thought to originate from either microbial consorts or from contamination by coral nematocysts. Of particular interest are the proteins of apoptosis, vesicular transport, and endo/exocytosis, which are discussed in context of the cellular processes of coral bleaching. Notably, the protein complement provides evidence that, rather than being expelled by the host, stressed endosymbionts may mediate their own departure.
Key to the evolutionary success of reef-building corals that inhabit the often nutrient-poor waters of tropical oceans is the metabolic cooperation between the host coral and unicellular algae residing within its cells. These endosymbiotic algae typically are dinoflagellates of the genus Symbiodinium, which are often referred to as “zooxanthellae,” although this catch-all name established by Brandt in 1883 does include other resident taxa of microbial endosymbionts (1). In this photoautotrophic symbiosis, fixed organic carbon produced by Symbiodinium is released to the animal host for its nutrition, whereas the dinoflagellates benefit by acquiring inorganic waste from the host (2–4).
Breakdown of the coral symbiosis is manifested as bleaching, made apparent by the loss of photobionts (5) or destruction of their photosynthetic pigments (6). Because heterotrophic feeding by polyps does not provide sufficient nutrition for corals to survive (7), death of colonies may result from severe bleaching if the obligate association is not re-established, thereby threatening coral reef ecosystems (8). The symbiotic association typically fails when corals experience temperature anomalies as little as 2–3 °C above mean summer seawater temperatures (9–11), which in combination with high solar irradiance leads to the production of excessive levels of reactive oxygen species (ROS)1 from over-reduced photosystems of Symbiodinium that overwhelm the antioxidant defenses of the holobiont (12–14). In 1998, worldwide sea surface temperature anomalies associated with a strong El Niño Southern Oscillation resulted in a major global bleaching event, causing coral mortality on 16% of the world's coral reefs (15), which demonstrates the critical importance of symbiosis to coral reef health.
A consensus has emerged as to the biochemical mechanisms underlying ROS production in early stages of cnidarian bleaching (reviewed in Refs. 16 and 17). Briefly, under conditions of ambient light exposure, endosymbiotic algae liberate oxygen as a by-product of photosynthesis, causing host tissues to be hyperoxic on a diel cycle (18). In addition, bright sunlight and thermal stress can damage the photosynthetic reaction centers of Symbiodinium, leading to excessive levels of ROS production by generating the highly reactive hydroxyl radical (•OH) and hydrogen peroxide (H2O2) that by diffusion can overwhelm the antioxidant capacity of both the algal symbiont and host (19), thereby causing further cellular damage. Direct photoexcitation, particularly in the presence of solar ultraviolet radiation, may produce ROS in both the endosymbionts and the host cells (20). A major mechanism for detoxification of ROS is the production of the antioxidant enzymes superoxide dismutase (SOD) and catalase, which are found in both partners (21), and ascorbate peroxidase in the algal symbiont (22). Our understanding of the contribution that each partner makes to the physiological response of the holobiont to oxidative stress, however, remains limited (23), including the involvement of ROS sensing and oxidative signaling of apoptotic pathways in symbiotic cnidarians (14).
Ample evidence for stress-induced progression of programmed cell death and necrosis in partners of the coral symbiome has been reported (24–25), which corresponds with activation of apoptotic genes of the coral host (26–29) and in aposymbiotic coral larvae and embryos (30, 31). Apoptotic elements are similarly induced by hyperthermal stress in the host tissues of the symbiotic sea anemones Anemonia virdis (32), Aiptasia pallida (33, 34), and Anthopelura elegantissima (35). Elements of the NF-κB signaling pathway of apoptosis in cnidarians are highly conserved traits (36), including genes of the caspase cascade and the pro-apoptotic and anti-apoptotic B-cell lymphoma 2 (Bcl-2) family of proteins (33, 29) that likely predate the evolution of animal multicellularity via endosymbiotic incorporation of aerobic bacteria as the ancestral proto-mitochondria of unicellular eukaryotes (37).
Careful examination of the coral Agaricia agaricities at above normal temperatures under daylight exposure has revealed accumulation of H2O2 at a rate that exceeds its removal by cellular protection or loss by diffusion, thereby causing oxidative stress, damage to the calcium exclusion system, and exocytotic liberation of Symbiodinium into the coelenteron with subsequent fragmentation of the gastrodermis (38). Furthermore, the thermal tolerance of Symbiodinium spp. is demonstrably constrained by the production and accumulation of H2O2 (39) that would account for variation in the thermal sensitivity of Symbiodinium phylotypes (40). It has been suggested that excess H2O2 generated by endosymbiotic algae may provide the signal to initiate the expulsion of these photobionts from corals (41) and/or trigger apoptosis in cnidarian host tissues by activation of the transcription factor NF-κB that leads directly to host cell apoptosis and autophagy (42).
Alternatively, NF-κB can induce the expression of nitric-oxide synthase (NOS) to produce NO, a well known promoter of the pro-apoptotic transcription factor p53 that is the cell cycle gatekeeper of the caspase cascade. NOS activity in a sea anemone was first reported by Trapido-Rosenthal et al. (43), and NO production has since been linked to cnidarian bleaching (44, 45), presumably via up-regulation of NF-κB (46). Perez and Weis (47) found that the major source of stress-induced NO production was in the host cells of the symbiotic anemone A. pallida. Consistent with this finding, NOS activity was absent in symbiotic dinoflagellates isolated with rigorous cleansing from three phylogenetically distinct cnidarians and from their cultured algal symbionts, whereas significant activities were found in the host cytosol (48). Furthermore, cytosolic NO might combine with superoxide produced by heat-stressed Symbiodinium to afford the powerful oxidant peroxynitrite (47) known to damage host mitochondrial membranes and cause the release of cytochrome c to activate the intrinsic apoptotic pathway of caspase-induced cell death (49). Although expressed markers of apoptosis, including the Bcl-2 family of pro- and anti-apoptotic proteins, are observed in the response of freshwater Hydra to stress (50), a direct link between molecular regulation of the Bcl-2 family of apoptotic mediators to activate cell death in cnidarian bleaching has yet to be demonstrated (29).
Other cellular mechanisms leading to bleaching that have been described include in situ symbiont degradation and autophagy (42), host cell detachment (51), host cell necrosis (24), and exocytosis (5, 52, 53). It is generally agreed that cnidarian bleaching involves multiple complex processes that depend on the type, synergy, and duration of the bleaching stress. For instance, in a recent publication, we have shown that thermal stress contributing to bleaching of the coral Stylophora pistillata is exacerbated by a limitation in the bioavailability of trace metals (specifically, iron) essential as cofactors to major metabolic processes, perhaps including antioxidant enzymes, in both the host and dinoflagellate partners (54). Conversely, iron hyperenrichment can also be detrimental to coral reef health arising from increased algal abundance and enhanced microbial activity as found in shipwreck-affected coral atoll mesocosms (55).
Although genomic techniques are yielding valuable insights into how the partners of coral symbiosis respond to environmental stress at transcription, the use of mRNA abundance as a correlation proxy to protein abundance and its functional activity has limitations in assessing phenotypic adaptation to abiotic stress (56). Accordingly, inroads to stress assessment at the proteome level of cnidarian symbioses have been progressing (57–62), as has understanding of the proteomic response of other marine organisms to environmental stress (63). In an attempt to understand the physiological response of symbiotic corals to the stresses of high irradiance, temperature, and trace metal limitation, we construct for the first time, using a quantitative high throughput platform, a comprehensive profile of the major proteins and peptides in a Symbiodinium-enriched fraction obtained from a reef-building coral under experimental conditions of environmental stress. Herein, we report the results of trace metal restriction at different temperatures in S. pistillata leading to a bleaching response in this coral.
EXPERIMENTAL PROCEDURES
Collection and Maintenance of Corals
Colonies of the reef-building coral S. pistillata (Esper, 1797) were collected at Pith Reef (18°13.434′S 147°01.052′E), Great Barrier Reef, Australia in January 2010 (Australian Institute of Marine Science Trip 4912). Collection depth ranged from 7 to 9 m at an average water temperature of 28 °C. Following collection, the corals were kept in shaded plastic tanks (64 liters) with flow-through seawater until transferred to the Australian Institute of Marine Science. On arrival at the Australian Institute of Marine Science, the corals were moved to a shaded 1000-liter tank with aeration and flow-through seawater (∼2 liters min−1, 28 °C ± 0.5 °C, salinity 30–32). Incoming seawater was filtered to 5 μm, and the corals received intensities of photosynthetically available radiation (PAR) ranging between 60 and 100 μmol photons m−2 s−1. Coral colonies were cut into 3–5-cm pieces and prepared for experimental procedures as described by Shick et al. (54). Only corals without obvious endolithic organisms were chosen for experimentation. Coral fragments were then transferred to indoor holding tanks for recovery and acclimation (28 °C ± 0.5 °C, PAR 200 μmol photons m−2 s−1, salinity 30–32) for 4 weeks prior to their exposure to experimental treatments.
Manipulative Trace Metal Experiment
Experimental treatments were conducted in an indoor aquarium room where water parameters, air temperature, and irradiance were controlled. Four 32-liter tanks were set in line, serving as temperature-controlled water baths. Each tank was fitted with a submersible pump to generate constant water circulation (1000 liters h−1) and its own seawater inlet from a temperature-controlled reservoir set to a flow rate of 400 ± 25 ml min−1. Incident PAR was subsaturating, 200 μmol photons m−2 s−1 (400 W Metal Halide Fixture Complete; Lamp Technologies International Pty. Ltd., Nunawading, Australia) at the water's surface, and illumination was provided for 12 h/day for the first 6 days. On day 6, the irradiance was increased to 425 ± 75 μmol photons m−2 s−1 (sufficient to saturate photosynthesis in this coral) for the remaining 7 days of the experiment. The irradiance inside the treatment chambers (see below) was ∼87% of incident irradiance. There was no detectable UVB radiation beneath the chamber lids, and UVA was only 25 μW cm−2 (54). Water temperature was regulated using 2-kW heating bars controlled by a CR1000 measurement and control datalogger (Campbell Scientific, Hyde Park, Australia) regulated to within ± 0.1 °C by a computer controlled system. The room's air temperature was set to 25 °C and circulated across the exposed tops of the experimental treatment chambers to minimize radiative heating.
The experiments were conducted in low density polyethylene containers (3 liters; Sistema, Dandenong, Australia) including 2 liters of treatment media and sealed by lids with silicone gaskets. Prior to use, treatment containers and fittings were acid-cleaned thoroughly following the protocol described by Shick et al. (54). Coral fragments were suspended in the experimental media using a nylon monofilament line secured to polyethylene bolts and wing nuts attached to the container lid. Seawater inside the experimental chambers was mixed and aerated by a flow of air through a 0.45-μm filter delivered from an aquarium pump; air exited the sealed chamber via a 0.40-μm filter pressure-fitted into the lid. The chambers were externally ballasted and immersed in the temperature-regulated water baths to a level just below the sealing gasket. The experimental medium (treated seawater) in each chamber was changed daily in a room clean of trace metals as described by Shick et al. (54).
Trace metal-free seawater was prepared by oxidizing seawater sent through a 0.5-μm filter by exposure to UV radiation in a purpose-built, flow-through UV reactor using a medium pressure mercury vapor lamp similar in design to that described by Achterberg and van den Berg (64) to release trace metals bound in dissolved organic complexes. The UV-treated seawater was then passed through a metal chelating resin (Chelex® 100; Bio-Rad) at a flow rate of 8 ml min−1 to remove total trace metals (65). A nutritional suite of essential trace metals and vitamins was added back to the UV-Chelex-treated seawater at concentrations used to obtain optimal growth for most phytoplankton (66) (supplemental information Data Set 1). Trace metal and vitamin amendments were added to 45 ml of UV-Chelex-treated seawater passed through a 0.5-μm filter prior to daily medium changes. Amendments sat for a maximum of 3 h prior to addition to experimental chambers, where they were brought up to 2 liters with UV-Chelex-treated seawater passed through a 0.5-μm filter.
The trace metal-replete medium was prepared by adding all metals and vitamins listed in supplemental information Data Set 1, whereas the effects of iron- and manganese-limiting conditions were tested by individually omitting their additions. The effects of trace metal limitation and temperature on S. pistillata were examined by exposing coral fragments to five treatments: 1) low temperature (28 °C) plus all metals (LT +M), 2) low temperature minus iron (LT −Fe), 3) low temperature minus manganese (LT −Mn), 4) high temperature (31 °C) plus all metals (HT +M), and 5) high temperature minus iron (HT −Fe). An additional treatment using high temperature minus manganese was also included in the experimental design, but coral fragments in this treatment died suddenly during the experiment, before endosymbiotic algae could be harvested. Three fragments from each of two coral colonies were placed in each treatment chamber. Water temperature in high temperature treatments was increased gradually (0.75 °C/h) from 28 to 31 °C on the 10th day of the experiment.
Isolation of Endosymbiotic Dinoflagellates
Endosymbiotic dinoflagellates were isolated on day 0 and following termination of the experiment at day 13. The day 0 sample for one of the coral colonies was designated replicate A, whereas the day 0 samples from the second colony was divided to become replicates B and C. Replicates A, B, and C at t = 0 represented the base-line proteome prior to stress against which the effects of the treatments were compared. Isolation was carried out under low light conditions and on ice. Coral tissue was removed from the skeleton of S. pistillata using an airgun and filtered (0.2 μm, Absolute D, Cuno Pacific Pty. Ltd., NSW, Australia) seawater. The resulting slurry was homogenized in a hand-held homogenizer (PRO 250; PRO Scientific, Oxford, CT) for 45 s and passed through a 25-μm Nitex mesh. The filtered slurry was then centrifuged for 20 min at 500 × g and 4 °C to sediment the algae. The supernatant containing the host tissue was discarded, and the pellet containing the freshly isolated endosymbiotic dinoflagellates was resuspended in Moore's calcium-free artificial seawater (www.mbl.edu/BiologicalBulletin/COMPENDIUM/CompTab2.html) containing 0.02% (w/v) SDS to remove any adhering host phagosomal membrane and to minimize clumping and again centrifuged for 20 min at 500 × g and 4 °C. To remove SDS, the algal pellet was washed three times with artificial seawater without SDS and centrifuged for 20 min at 500 × g and 4 °C after each wash. Following the final wash, the pellet was resuspended in 1 ml of artificial seawater and transferred to vials of a known weight. A small subsample was microscopically examined to assess and confirm the purity of the algal isolate. The suspension of cleaned endosymbiotic dinoflagellates was again centrifuged for 10 min at 16,000 × g and 4 °C, the supernatant was aspirated, and the wet weight of the pellet was recorded. A minimum of 200 mg of wet algae were freeze-dried. Samples having a dry weight between 3 and 110 mg were subsequently prepared for protein extraction.
Protein Extraction and Tandem Mass Tag Labeling
A volume of 0.5 ml of 50 mm triethylammonium bicarbonate (TEAB) buffer (pH 7.5) was added to each freeze-dried sample in a microcentrifuge tube. Then ∼300 μl of a suspension of acid-washed and rinsed glass beads (400–625-μm diameter; Sigma) was added. Each sample was vortexed for 1 min and then placed on ice for 1 min. This procedure was repeated 10 times. Lysis was visually verified by green (chlorophyll) discoloration of the lysis buffer. The beads were allowed to settle, and the supernatant was transferred to a fresh microcentrifuge tube and centrifuged at 12,000 × g for 10 min at 4 °C. The supernatant was then assayed with Bradford reagent (Sigma) to ensure that sufficient protein was available for labeling with amine-reactive tandem mass tag (TMT) reagents to enable the identification and quantification of extracted proteins by mass spectrometry (67). Proteins from our five treatments plus a t = 0 sample representing the unstressed proteome were labeled using a TMT sixplex tagging kit (Thermo Scientific, East Riding, UK) comprising an isobaric set of six mass tags with five isotopic substitutions. Each tag produces a unique reporter ion during MS/MS fragmentation analysis enabling concurrent identification and quantification of proteins in different samples by labeling of digested proteins at the peptide level (bottom-up strategy) (68) or by labeling at the protein level followed by to tryptic digestion (top-down strategy) (69). Simultaneous processing of six samples minimizes analytical variation introduced by differences in gel loading or extraction and eliminates the need for six individual analyses that would be affected by LC-MS/MS signal fluctuations between operations. Extracted proteins were labeled with TMT126–TMT130 reagents for each respective treatment, and the process was repeated for the three replicates (A–C) from each experimental treatment. For the unstressed t = 0 samples representing the basal proteome, TMT131 reagent was used for labeling. Thus, in total, 18 protein samples were labeled.
Procedures for protein (top-down) TMT labeling were as follows. Briefly, each TMT tag was reconstituted in 24 μl of acetonitrile and equilibrated at room temperature before adding it to each sample. Protein quantities were adjusted to reduce variability between replicates. Appropriate volumes of the protein extract from each treatment sample or pretreatment t = 0 sample equivalent to 25 μg were lyophilized and reconstituted in 50 μl of 50 mm TEAB containing 0.04% SDS for resolubilization. The samples were then reduced by adding 5 μl of 9 mm tris(2-carboxyethyl)phosphine hydrochloride in 50 mm TEAB at 55 °C for 1 h and then alkylated with 5 μl of 16 mm iodoacetamide in 50 mm TEAB at room temperature for 30 min in darkness. A volume of 8 μl of 5% hydroxylamine (w/w) was added according to TMT procedures to terminate the reaction. Combined TMT-tagged samples for each treatment replicate A1–5, B1–5, and C1–5 were mixed individually with their corresponding TMT-tagged, pretreatment samples A(t = 0), B(t = 0), and C(t = 0), respectively. The samples were then lyophilized and reconstituted in 20 μl of 50 mm TEAB and 20 μl of Laemmli buffer (70). In these samples, excess TMT reagents were removed during protein SDS-PAGE separation.
One-dimensional Gel SDS-PAGE and In-gel Digestion
The sixplex TMT-labeled samples (A–C) were boiled for 5 min before loading across a 4–12% NuPAGE Novex Bis-Tris gel (Invitrogen) with MES (2-(N-morpholino)ethanesulfonic acid) buffer alongside Novex® SeeBlue® Plus2 prestained standards (Invitrogen). Electrophoresis was carried out for 100 min at 150 V. The gel was fixed, Coomassie Blue-stained, destained, and visualized with an ImageQuant (GE Healthcare) biomolecular imager. The bands were then selected for excision and in-gel digestion (supplemental information Data Set 2). The three gel lanes were sectioned into 15 portions of ∼2 mm2 in area. In-gel reduction, alkylation, and proteolytic digestion with trypsin were performed prior to subsequent analysis by mass spectrometry (71) as follows. Briefly, cysteine residues were reduced with 10 mm dithiothreitol and alkylated with 55 mm iodoacetamide in 100 mm ammonium bicarbonate to form stable carbamidomethyl derivatives. Trypsin digestion of each section was carried out overnight at 37 °C in 50 mm ammonium carbonate buffer, and the supernatant was retained. The peptides were extracted from the excised gel portions by two washes with 50 mm ammonium bicarbonate and acetonitrile. Each wash involved shaking gel sections for 15 min before collecting the peptide-containing extract. The extract was pooled with the initial digestion supernatant and then lyophilized. The samples were resuspended in 46 μl of 50 mm ammonium bicarbonate per band prior to LC-MS/MS analysis on injection of half the sample volume.
LC-MS/MS
Chromatographic separations of each band were performed using an Ultimate LC system (Dionex Ltd., East Riding of Yorkshire, UK). The peptides were resolved by reversed phase chromatography on an Acclaim® PrepMap100 C18, 3-μm particle, 100 Å (75 μm × 150 mm) column (Thermo Scientific/Dionex). Depending on the intensity of the gel band, gradients of 30–180 min of acetonitrile in 0.05% formic acid were delivered to elute the peptides at a flow rate of 200 nl/min. Peptides were ionized by electrospray ionization using a Z-spray source fitted to a QToF-micro quadrupole orthogonal acceleration time-of-flight tandem mass spectrometer (Waters Corp., Milford, MA). The instrument was set to operate in automated switching mode, selecting the two most intense precursor ions for sequencing by collision-induced fragmentation. Precursor ions were surveyed across a range of 400–1800 m/z. The MS/MS analyses were conducted across a range of 100–2000 m/z using collision energy profiles that were chosen based on the mass-to-charge ratio and the charge state of the peptide. Each precursor mass was excluded from further selection from the mass list for 90–120 s following its fragmentation depending on the length of the gradient. Quality control tests with 200 fmol of BSA were analyzed between each set of 15 gel sections to ensure that coverage of the protein was ≥30%.
Data Analysis
Tandem mass spectra were extracted into peak lists by Masslynx version 4.0 and Protein Lynx Global Server 2.2.5 (Waters Corporation, Milford, MA). Charge state deconvolution and deisotoping were not performed. All of the MS/MS spectra were analyzed using Mascot software (version 2.2.03; Matrix Science, London, UK). Mascot was developed to search entries of the uniprot_sprot_100713 database (selected for eukaryotes, 518,415 entries), assuming use of the digestion enzyme trypsin and up to three missed cleavages. The data were searched with a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 1.2 Da. The following were stated as variable modifications: N-terminal acetylation, deamidation of asparagine and glutamine, N-terminal glutamine to pyroglutamine, oxidation of methionine, iodoacetamide derivative of cysteine, and phosphorylation of serine, threonine, and tyrosine. Mascot Daemon was used to perform merged searches for the 15 peak list files per replicate set. The raw data were recalibrated against internal tryptic peptides where necessary to produce a mass accuracy of less than 50 ppm for most peptides.
A complete sequence for the genome of Symbiodinium is unavailable, and only incomplete and fragmentary information for this genus exists in the Uniprot protein database. Therefore, an all-protein search was chosen to match orthologs from other species. To maximize the sweep of coverage of the Symbiodinium-enriched protein extract, all of the labeled peptides marked as bold (most likely assignment) by Mascot were used regardless of the probability-based expected value (supplemental information Data Set 4), and data assigned to peptides but not to Mascot protein hits were individually annotated by a manual search of the Uniprot database (supplemental information Data Set 5). All of the Mascot hits of any peptide score and probability-based expected value were accepted meeting the criteria: all peptides rank 1, lysine-containing peptides flagged as bold with one or more tag(s) plus TMT131 detected. Grouping of peptides matched to multiple database entries was not reported. Instead, the assignment declared first in the Mascot peptide summary report was used. Also, using rank 1, peptides marked as bold red ensured that peptides reported here were based on the highest scoring assignments. The Mascot peptide summary report resulting from the merged data was exported as a comma-separated values (.csv) file. An in-house PerlScript program was then used to merge the .csv file with a tab-separated values (.tsv) file resulting from the search that contains the TMT tag intensities. The resulting .csv file was then used for sorting and processing data in Microsoft Excel. False discovery rates were estimated by again doing the merged searches against a Mascot decoy database. The decoy database contains randomized sequences, and therefore any matching peptides would be highlighted as a false positive.
Peptide data for each replicate set were annotated with the sample representing the t = 0 base-line proteome prior to stress category (A–C) and combined into one file. The data were then sorted according to protein accession number and replicate category. Proteins present in two or three replicates were then carried forward for further analysis. Data assigned to peptides but not to Mascot protein hits were then individually annotated by Mascot's peptide view (therefore including all eukaryotic, prokaryotic, Achaean, and viral proteins). Quantification was determined for peptides that were present in at least two of the three replicates. TMT tag intensities giving x-fold changes of 0 or negative values between treatment and the unstressed t = 0 proteome taken prior to treatment were discarded. Peptides without detectable tags do not allow relative quantification and were also eliminated. TMT tag intensities for each assigned peptide present in at least two of the replicates were used to calculate the analysis of variance between treatments relative to the pretreatment t = 0 proteome. Experimental condition had a statistically significant effect on the average TMT tag intensity when p < 0.05. In these cases, a Student's t test was also calculated between individual pairs of samples (i.e. between each individual treatment and its t = 0 base line and between individual sample/treatments) to confirm that the experimental condition produced a significant effect (72, 73).
RESULTS
Experiments were performed wherein the coral S. pistillata in seawater either replete or lacking in iron or manganese was exposed to photosynthetically saturating levels (>400 μmol of photons m−2 s−1) of PAR at nonstressful seasonal and at elevated water temperatures. Under these conditions, the dark-adapted photosynthetic efficiency of the symbiotic algae in all treatments declined significantly (paired t tests, p < 0.05) from day 1 to the end of the experiment, as evinced by reduced maximum quantum fluorescence yields (ratio of variable to maximal chlorophyll fluorescence in photosystem II, Fv:Fm) of the photosynthetic pigments of photosystem II (supplemental information Data Set 3), which is consistent with the symbiotic photophysical dysfunction that occurs during coral bleaching (74).
The MS/MS spectra for single-peptide-based protein identifications are shown in supplemental information Data Set 6. The average x-fold change of the TMT tag intensities for each treatment relative to the unstressed t = 0 proteome was calculated from all of the peptides assigned to the same protein across two, or better three, replicates (supplemental information Data Set 7). Values <1 or >1 indicate either a reduction or an increase in tag intensity as compared with the pretreatment t = 0 proteome, whereas a value equal to 1 indicates no change in protein abundance. Of the 109 peptides meeting our stringent criteria for quantification, 83 (76%) did not show any significant change in TMT tag intensity compared with the unstressed t = 0 proteome. Fig. 1 shows the remaining 26 assigned peptides where differences in TMT tag intensities between treatments relative to the pretreatment t = 0 proteome were found to be statistically significant. Of these, 20 of 26 peptides have a significant reduction in TMT tag intensity compared with the unstressed t = 0 proteome in at least one of the low temperature treatments. Even in the relatively benign treatment of low temperature (i.e. unchanged from the day 0 temperature) in seawater replete with trace metals, 17 of 26 peptides had a significant reduction in tag intensity, perhaps a response to the more than doubling of irradiance on day 6, which was manifested in an immediate decrease in photosynthetic efficiency, such that photosynthetic efficiency in all treatments was lower at the end of the experiment on day 13 than immediately before the start of the experiment on day 1 (supplemental information Data Set 3). Only 12 of 26 assigned peptides showed a statistically significant increase in TMT tag intensity compared with the unstressed t = 0 proteome; these 12 peptides were all associated with high temperature treatments and were generally involved in protein biosynthesis (perhaps reflecting a general accelerating effect of the increased temperature on metabolism) with the exception of proteins Q9J4Z9 and P27551, which are of viral origin. The only statistically significant increase in TMT tag intensity observed at low temperature was caused by manganese depletion (LT −Mn) to enhance the abundance of a protein from the TCA cycle (B3EN95, succinyl-CoA ligase) and that of the abundance of a protein of viral origin (P27551, viral replication protein E1). Only 5 of 26 proteins showed varying TMT tag intensities under both temperature conditions that were statistically significant compared with the pretreatment t = 0 proteome. No statistically significant decrease in TMT tag intensity was associated with treatments at the higher temperature treatment. Overall, it appeared not to be metal limitation per se that influenced TMT tag intensity but rather the maintenance temperature of the coral, regardless of the presence or absence of iron. That is, at the higher temperature, significant increases in TMT tag intensities occurred in five proteins in both HT −Fe and HT +M conditions, an additional four proteins showed significant increases only in HT +M, and a further three proteins only in HT −Fe. Corals in the HT −Mn treatment perished before their symbiotic algae could be harvested.
Fig. 1.
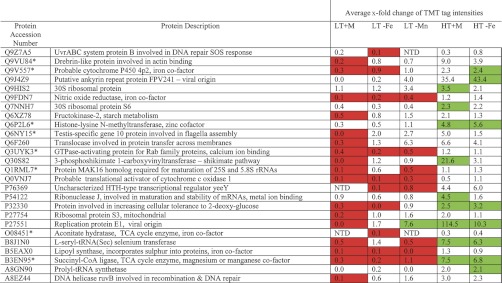
Statistically significant average fold change of the TMT tag intensities for each treatment relative to control t = 0. The treatments are low temperature (28 °C) plus all metals (LT +M), low temperature minus iron (LT −Fe), low temperature minus manganese (LT −Mn), high temperature (31 °C) plus all metals (HT +M), and high temperature minus iron (HT −Fe). Corals in the treatment high temperature minus manganese (HT −Mn) became necrotic before the endosymbiotic algae could be harvested. Values of <1 or >1 would indicate either a reduction or increase in tag intensity compared with the control, whereas a value of 1 indicates no change. Statistically significant changes are indicated by shading red for fold decrease and green for fold increase. NTD, no tag detected; *, proteins also detected in coral genome sequences.
Protein analysis (supplemental information Data Set 7) gave the presence of antioxidant, metabolic redox, and cytoprotective enzymes (Table I), which were expected to change in response to stress treatments. Surprisingly, only changes in nitric-oxide reductase (Q9FDN7) and a probable thiol peroxidase (Q8NRG3) (supplemental information Data Set 7) could be quantified (Fig. 1), with the TMT tag intensity of the nitric-oxide reductase being significantly repressed in the low temperature treatments. The intensity of the thiol peroxidase TMT tag did not change significantly compared with the pretreatment t = 0 proteome in any experimental condition (supplemental information Data Set 7). Interestingly, this protein requires an iron cofactor, as does nitric-oxide reductase, but the TMT tag intensity in the high temperature treatment with iron limitation did not change significantly compared with the unstressed t = 0 proteome. Thermal stress enhances the effects of high irradiance on photosystems I and II of the algal symbiont, leading to increased production of ROS (40) having a variety of cytotoxic effects, including DNA damage. Although a variety of DNA repair enzymes were detected in the data set (Table II and supplemental information Data Set 7), surprisingly, only the two proteins Q9Z7A5 and A8EZ44 that belong to the Uvr and Ruv DNA repair systems, respectively, were detected in the quantified data set (Fig. 1) that is potentially linked to damage caused by oxidative stress. The TMT tag intensity of neither protein increased under the experimental stress conditions compared with the unstressed t = 0 proteome. Indeed, the tag intensities were significantly reduced under low temperature conditions compared with the unstressed t = 0 proteome.
Table I. Antioxidants, metabolic redox enzymes, and cytoprotective heat shock proteins detected in the Symbiodinium-enriched fraction of Stylophora pistillata.
| Protein accession number | Protein description |
|---|---|
| B2SAW2 | Alkyl hydroperoxide reductase |
| P21688 | β-Carotene hydroxylase |
| P29422a, P12365a, B1W5Q9, B1W5Q9, B1W5Q9, A1SWM1, B3EBB7, B4SAT7, C0ZSW0, P0C8F0, Q0U324, Q1M498, Q3KE62, Q87WL6, A0K4Q2, O08404, Q8EIV5 | Catalase/Catalase-peroxidase |
| P52649a | Cytochrome b245 heavy chain |
| Q5N5B0a | Cytochrome b6-f complex iron-sulfur subunit |
| P50686a, P07255 | Cytochrome c oxidase |
| Q6C0Z6, Q6URB0 | Cytochrome c peroxidase |
| O09164, P81527, P09213 | Superoxide dismutase |
| Q3TQB2a | FAD-dependent oxidoreductase |
| Q2GCZ2, A4YER6, Q55318a, B2GEF7, B2U9D2 | Ferredoxin-NADP reductase |
| P00390a, P0AC65 | Glutathione reductase |
| Q7N809 | Hydroxyacylglutathione hydrolase |
| Q03751 | Cysteine-string protein (HSP-binding) |
| Q54J68 | Heat shock protein, 10 kDa |
| P22774a, Q01889, Q05746a, P12078a, P08418a | Heat shock protein, 70 kDa |
| P33125a | Heat shock protein, 82 kDa |
| P40292a | Heat shock protein, 90 kDa |
| P41887a | Heat shock protein, 90 homolog |
| Q06068a | Heat shock protein, 97 kDa |
| O94641a, P31539 | Heat shock protein, 104 kDa |
| P04792a | Heat shock protein, β1 |
| Q10284a | Heat shock protein, Sks2 |
| Q96VB8 | Heat shock protein, Sse1 |
| P38530a | Heat shock factor, protein 2 |
| Q42592 | l-Ascorbate peroxidase S |
| A2BQ24, A3PBR8, Q85A72 | Light-independent protochlorophyllide reductase |
| C0IW58 | Low redox potential peroxidase |
| Q84TF4 | Monothiol glutaredoxin-S13 |
| Q0MQI7a, Q08822a, Q2KIG0a | NADH dehydrogenase (ubiquinone) flavoprotein |
| A1ALP0a, A1BF33a, A8ERL5a, B2V988a, B3QP57a, Q0BSK3a, Q89AU5a, Q1IPE7a, Q1IS59a, P56912a, A1SE34a, A0QJV5a, A4SLN5, Q68WJ7a | NADH-quinone oxidoreductase |
| A6MMH7a, Q09FR3a, Q32126a, Q6EW03a, Q7YJS8, Q8DLN5 | NAD(P)H-quinone oxidoreductase |
| Q43644a, Q07500a, Q2I3H4a, Q70Y29a, Q9B8C8a, P05510a, P12776a | NADH-ubiquinone oxidoreductase |
| Q9FDN7 | Nitric oxide reductase |
| O48677 | Peroxidase |
| Q5HNJ2, Q8NRG3 | Probable thiol peroxidase |
| B2SK49 | Putative NADH dehydrogenase/NAD(P)H nitroreductase |
| Q58065 | Putative NADH oxidase |
| Q7Q161 | Putative oxidoreductase |
| Q11W68, Q488Q1 | Putative reductase |
| Q9FJD6, Q9LZU9 | Putative respiratory burst oxidase |
| Q82ZW4 | Redox-sensing transcriptional repressor rex 2 |
| O81209, Q5ZAJ0, O81210, O81211 | Respiratory burst oxidase |
| Q87L91 | Sulfite reductase (NADPH) hemoprotein |
| Q86GL4a | Superoxide-generating NADPH oxidase |
| Q6P902, O48737, Q655X0, P0AA25a, Q8CDN6a | Thioredoxin |
| Q69PS6, P52215, Q9ZL18 | Thioredoxin reductase |
| Q10216a, P0AG84a | Uncharacterized oxidoreductase |
| Q58698 | Uncharacterized polyferredoxin-like protein |
| P81701 | Vanadium-dependent bromoperoxidase |
a Proteins also detected in coral genome sequences.
Table II. DNA repair enzymes detected in the Symbiodinium-enriched fraction of Stylophora pistillata.
None of the proteins were detected in coral genome sequences.
The most extreme increase in protein expression was observed for the viral replication protein E1 (P27551), with a massive 114-fold increase in TMT tag intensity under high temperature, metal-replete conditions (Fig. 1 and supplemental information Data Set 7). The only protein associated with apoptosis that could be quantifiably measured (supplemental information Data Set 7) was the Tax1-binding protein (Q86VP1), which actually inhibits TNF-induced apoptosis (75) and is a known transactivator of viral replication and transformation (76). However, the TMT tag intensity of this protein did not change significantly compared with the pretreatment t = 0 proteome under any condition of experimental treatment. Likewise, the only protein associated with exocytosis was a Ras GTPase-activating protein (4Q6PFQ7), the TMT tag intensity of which also did not change significantly compared with the unstressed t = 0 proteome (supplemental information Data Set 7).
Despite not finding a greater range of proteins differentially expressed in our experimental treatments in response to experimental stress, our analysis gave an exceptional depth of proteome coverage to reveal a comprehensive profile of proteins relevant to the metabolic interactions of coral symbiosis. Quantification of proteins can be achieved by several nonradioactive isotope labeling approaches, notably by stable isotope labeling and by stable isotope tagging. The former method affords protein labeling by the incorporation of isotopically enriched constituent amino acids in culture (77) or by in vivo metabolic labeling in whole organisms (78). Both metabolic labeling and isobaric chemical tagging are generally capable of accurate, precise, and reproducible quantification capable of deep proteome coverage (79). Tagging includes the use of TMTs and isobaric tags for relative and absolute quantitation (iTRAQ). Both approaches involve differential labeling of protein mixtures with isobaric (equal mass) reagents that bond to primary amine groups that fragment during tandem MS into unique reporter ions corresponding to the original labeled samples. The use of labeling with stable isotope-substituted metabolites, such as the approach of stable isotope labeling with amino acids in cell culture (SILAC), is limited in application because not all model biological systems are amenable to these methods. Furthermore, isobaric tagging enables a higher throughput than a stable isotope labeling approach with multiplexing of up to six and eight protein mixtures by TMT and iTRAQ reagents, respectively. Our rationale in choosing TMT labeling at the protein level was to reduce the complexity of the samples to maximize the number of digested peptides that could be resolved in sufficient detail to be identified and quantified on a high throughput analytical platform. However, there was trade-off between gaining the resolution required to separate peptides in sufficient detail to be identified and to achieve quantitative precision. This limitation arises because only lysine and protein N-terminal residues are labeled by TMT reagents. Also, fewer peptides are produced by tryptic digestion because most lysine residues are modified by TMT reagents, resulting in missed cleavages at TMT-labeled lysine sites, thus reducing the efficiency of detection.
Nevertheless, high resolution TMT-labeled protein analysis of the symbiont-enriched fraction of S. pistillata has yielded valuable information on the protein structure of coral symbiosis. In addition to the quantified data set, there were a total of 8098 MS/MS events relating to individual peptides that could be separated to allow sufficient amino acid sequence to be used to search Uniprot. Only ∼1% of peptides were labeled in this experiment, although this is not uncommon for an experiment of this nature but adequate for peptide detection by the model instrument of LC-MS/MS used. Of the 8098 MS/MS spectra acquired, 7790 peptides could be further assigned a biochemical function based on KEGG gene ontology descriptors (supplemental information Data Set 8, with the peptide sequences for the remaining 308 unassigned proteins listed in supplemental information Data Set 9). The qualitative data set (supplemental information Data Set 8) was searched for antioxidant enzymes using the KEGG biological process descriptors “oxidation-reduction process” and “cell redox homeostasis,” as well as the KEGG molecular function descriptors “oxidoreductase activity” and “peroxidase activity” (Table I). To this table we have also added the cytoprotective heat shock proteins extracted from supplemental information Data Set 8. Proteins associated with repair to DNA following ROS damage were searched in the qualitative supplemental information Data Set 8 using the descriptor “DNA” (Table II). Both the qualitative and quantitative data sets were searched also for effectors of bleaching using the KEGG biological process descriptors “vesicle-mediated transport,” “exocytosis,” “apoptosis,” “autophagy,” and the molecular function descriptor “NF-κB” (Table III). While going through the qualitative data set, we noticed a striking number of proteins that are not usually associated with algal metabolism. These included oxygen-binding globins (Table IV), nitrogen fixation and metabolizing enzymes (Table V), and an astonishing complement of biotoxins (Table VI); the last we thought might arise from bacterial consorts or contamination by coral nematocysts (stinging capsules of cnidarians). We were intrigued by the massive increase of protein expression in the viral replication protein E1 (P7551) and viral ankyrin repeat protein (Q9J4Z9) in high temperature treatments (Fig. 1), and a greater complement of viral proteins were uncovered from supplemental information Data Set 8 that are involved in viral replication and protein biosynthesis (Table VII). Photosystem proteins were present as expected for Symbiodinium, and the additional presence of phycoerythrin and phycocyanin (Table VIII) gives evidence that our corals harbored cyanobacteria as either consorts (80) or pathogens (81). Interestingly, proteins analogous to photo-system proteins could be found in the coral genome sequences which warrants further investigation. The presence of surface-associated unicellular cyanobacteria also is consistent with a strong indication of nitrogen-fixing enzyme systems (82).
Table III. Major proteins detected in the Symbiodinium-enriched fraction of Stylophora pistillata related to coral bleaching: apoptosis, vesicle transport, and endo/exocytosis.
| Protein accession numbers | Protein function | Protein description |
|---|---|---|
| O04093 | Apoptosis | Putative inactive disease susceptibility protein LOV1 |
| O42354a | Negative regulation of apoptosis | E3 ubiquitin-protein ligase Mdm2 |
| O60125 | Apoptosis | BAG family molecular chaperone regulator 1A |
| O64789 | Apoptosis | Probable disease resistance protein |
| O92529 | Apoptosis | Genome polyprotein |
| P58801a | Positive regulation of apoptosis | Receptor-interacting serine/threonine-protein kinase 2 |
| Q12933a | Regulation of apoptosis | TNF receptor-associated factor 2 |
| Q32NG6 | Induction of apoptosis | Tumor necrosis factor receptor type 1-associated DEATH domain protein |
| Q6UXS9a | Apoptosis | Inactive caspase-12 |
| Q6ZNE5 | Positive regulation of autophagy | Beclin 1-associated autophagy-related key regulator |
| Q8RXS5 | Apoptosis | Probable disease resistance protein At5g63020 |
| Q8W3K3 | Apoptosis | Putative disease resistance protein At1g58400 |
| Q8YTC2a | Apoptosis | Uncharacterized WD repeat-containing protein Alr2800 |
| Q95KV0a | IκB kinase activity | Inhibitor of nuclear factor κB kinase subunit β |
| Q99KF0 | Regulation of apoptosis | Caspase recruitment domain-containing protein 14 |
| Q9BZR8 | Regulation of apoptosis | Apoptosis facilitator Bcl-2-like protein 14 |
| Q9FL92 | Apoptosis | Probable WRKY transcription factor 16 |
| Q9LVT1 | Apoptosis | Putative disease resistance protein At5g47280 |
| Q9QAX1 | Apoptosis | Genome polyprotein |
| Q9SZA7 | Apoptosis | Probable disease resistance protein At4g33300 |
| Q86VP1a | Inhibits TNF-induced apoptosis | Tax1-binding protein 1 |
| Q3UHC7a | Inhibits CHUK-NF-κB signaling | Disabled homolog 2-interacting protein |
| Q54DK4a | ARF GTPase activator activity | α-Protein kinase 1 |
| Q8H100a | ARF GTPase activator activity | Probable ADP-ribosylation factor GTPase-activating protein AGD8 |
| O14234a | Calcium ion transport | Calcium-channel protein Cch1 |
| Q0VD05 | Calcium ion transport | Voltage-dependent calcium channel γ-3 subunit |
| Q8NC96a | Endocytosis | Adaptin ear-binding coat-associated protein 1 |
| Q9UU81a | Endocytosis | AP-1 complex subunit γ-1 |
| Q2KJ81a | Endocytosis | AP-1 complex subunit μ-1 |
| Q9Y6Q5a | Endocytosis | AP-1 complex subunit μ-2 |
| O00203a | Endocytosis | AP-3 complex subunit β-1 |
| A2RV61a | Endocytosis | GTPase-activating protein and VPS9 domain-containing protein 1 |
| P90761a | Endocytosis | Putative stoned B-like protein |
| Q681Q7 | Endocytosis | Uncharacterized protein At1g03900 |
| Q9Z1C7a | Guanine nucleotide exchange factor | Rap guanine nucleotide exchange factor 4 |
| Q8C0Q9a | Guanine nucleotide exchange factor | Rap guanine nucleotide exchange factor 5 |
| Q06836 | Nucleotide exchange from the GDP- to the GTP-bound form | Arf guanine nucleotide exchange factor SYT1 |
| P47102 | Nucleotide exchange from the GDP- to the GTP-bound form | ARF guanine-nucleotide exchange factor 1 |
| Q8NEV8 | Rab effector protein in vesicle trafficking | Exophilin-5 |
| Q8R3D1a | Rab GTPase activator activity | TBC1 domain family member 13 |
| Q8BYJ6a | Rab GTPase activator activity | TBC1 domain family member 4 |
| O95759a | Rab GTPase activator activity | TBC1 domain family member 8 |
| Q0IIM8a | Rab GTPase activator activity | TBC1 domain family member 8B |
| Q3UYK3a | Rab GTPase activator activity | TBC1 domain family member 9 |
| P47709a | Rab GTPase binding | Rabphilin-3A |
| Q9EQZ7a | Rab GTPase binding | Regulating synaptic membrane exocytosis protein 2 |
| Q9QXG2a | Rab-protein activation | Rab proteins geranylgeranyltransferase component A1 |
| Q8MLZ5 | Regulation of small GTPase mediated signal transduction | Ras GTPase-activating protein Gap-2 |
| Q9UJF2a | Regulation of small GTPase mediated signal transduction | Ras GTPase-activating protein nGAP |
| Q3T000a | v-SNARE | Synaptobrevin homolog YKT6 |
| Q9R0N5a | v-SNARE | Synaptotagmin-5 |
| Q8TDW5a | v-SNARE | Synaptotagmin-like protein 5 |
| Q8N4C7 | t-SNARE | Syntaxin-19 |
| Q39233 | t-SNARE | Syntaxin-21 |
| Q94KK7 | t-SNARE | Syntaxin-52 |
| Q9Y2D4a | Vesicle docking involved in exocytosis | Exocyst complex component 6B |
| Q9LXX6 | Vesicle docking involved in exocytosis | Probable exocyst complex component 6 |
a Proteins also detected in coral genome sequences.
Table IV. Globin hemoproteins in the Symbiodinium-enriched fraction of Stylophora pistillata.
| Protein accession number | Protein description |
|---|---|
| P0ABD3a, O68926 | Bacterioferritin |
| P13578 | Extracellular globin-2B |
| P12549 | Globin CTT-VIIB-6 |
| P19645 | Hemoglobin subunit α |
| Q9PVU6 | Hemoglobin embryonic subunit α |
| Q1AGS4 | Hemoglobin subunit α-2 |
| P04444 | Hemoglobin subunit β-H1 |
| Q94FT8a | Nonsymbiotic hemoglobin 3 |
| Q47952a | Hemoglobin-haptoglobin binding protein |
| Q9DGJ0, P02193 | Myoglobin |
| P42578a | Yolk ferritin (snail) |
a Proteins also detected in coral genome sequences.
Table V. Major proteins of nitrogen and purine metabolism in the Symbiodinium-enriched fraction of Stylophora pistillata.
| Protein accession number | Protein description |
|---|---|
| A0RHQ8a, A9WSA8a, B1W463a, C5C687a, Q2FHN5a, Q5FJC0a, Q87WP4a, Q8FT42a, Q8Y665a, Q9K8V7a | Carbamoyl-phosphate synthase |
| O61608a, Q92037a, Q9TUX8a | Nitric-oxide synthase |
| Q8H157a, Q9M173a | Nitrate transporter |
| P36859a Q7VJT5a, P43101a, P49050a, P22945a, A8LLY9a | Nitrate reductase (NADH) |
| P32712a | Formate-dependent nitrite reductase NrfG |
| P30667a | Nif-specific regulatory protein NifA |
| B2J5Z9 | Nitrogenase-stabilizing/protective protein NifW |
| P10996a | Nitrogen cofactor biosynthetic protein NifE |
| P26248a, B7KG76a P25768a | Nitrogenase iron protein NifH |
| P00468, P10996, P25313, B7KG76, P26248, P16267a | Nitrogenase molybdenum-iron protein NifD |
| Q9X192a | NifU-like protein |
| P10577, P17429, O13415, P30667, P28349a | Nitrogen assimilation regulatory protein |
| A3LYV8a, Q0CQ46a | Nitrogen permease regulator |
| P17429a, O13415a | Nitrogen regulatory protein |
| P26050a | Nod factor export ATP-binding protein I |
| P32288a, Q42689a, Q9HU65 | Glutamine synthetase |
| Q0DG35a | Glutamate synthase |
| P46011 | Bifunctional nitrilase/nitrile hydratase NIT4 |
| B0TT70a, Q3IRZ6a, Q2FEK3a, Q79VJ3a, Q8XXT1a | Urease |
| A1U501, Q144D8, Q4KJ05, A6X1Q4, Q1MCW3, Q826R6 | Urease accessory proteins |
| B2G592a; Q2FEZ3a; Q5DYV8a | Purine nucleoside phosphorylase |
| A7GCI1a | Xanthine phosphoribosyltransferase |
| P77165a | Xanthine dehydrogenase |
| Q8MKJa | Uricase (urate oxidase) |
a Proteins also detected in coral genome sequences.
Table VI. Proteins and peptide toxins detected in the Symbiodinium-enriched fraction of Stylophora pistillata.
None of the proteins were detected in coral genome sequences.
| Protein accession number | Protein description |
|---|---|
| Q45894 | Botulinum neurotoxin type A (bacterium) |
| P46081 | Botulinum neurotoxin type C1 (bacterium) |
| P19321 | Botulinum neurotoxin type D (bacterium) |
| Q00496 | Botulinum neurotoxin type E (bacterium) |
| P81242 | Nonhemolytic enterotoxin (bacterium) |
| P55981 | Vacuolating cytotoxin autotransporter (bacterium) |
| Q01886 | HC-toxin synthetise (fungal plant pathogen) |
| Q9TWG1 | Potassium channel toxin kaliseptin (sea anemone) |
| Q9BPG0 | Conotoxin Pn-B0151 (marine cone snail) |
| P0CH23 | Conotoxin Pu3.5 (marine cone snail) |
| Q9XZK2 | ο-Conotoxin SO-3 (marine cone snail) |
| P58917 | ο-Conotoxin CVIA (marine cone snail) |
| P0C8U3 | μ-Conotoxin-like SxIIB (marine cone snail) |
| D5GSJ8 | Toxin CpTx1 (spider) |
| D2Y284 | Hainantoxin-XVI-14 (tarantula) |
| D2Y2G1 | Hainantoxin-XI-10 (tarantula) |
| P0C8D5 | Scolopendra toxin (centipede) |
| Q86QU3 | Potassium channel toxin γ-KTx 4.10 (scorpion) |
| P60209 | Potassium channel toxin α-KTx 9.4 (scorpion) |
| A0ZSK4 | Neoverrucotoxin subunit β (reef stonefish) |
| P0CAR3 | Short neurotoxin E1 (coral snake) |
| A2CKF6 | Neurotoxin 3FTx-8a (banded krait) |
| Q9WVC2 | Ly-6/neurotoxin-like protein (mouse) |
Table VII. Viral proteins detected in the Symbiodinium-enriched fraction of Stylophora pistillata.
None of the proteins were detected in coral genome sequences.
| Protein accession number | Protein description |
|---|---|
| Q8V2M9 | A-type inclusion protein, viral reproduction |
| O93182, P03344, Q00071 | Gag polyprotein, viral reproduction |
| O89940, P0C211, P18042 | Gag-Pol polyprotein, viral reproduction |
| Q6QLN1, Q8JJX1 | Nonstructural polyprotein pORF1, viral genome reproduction |
| P06163, Q66T64, P32530 | Phosphoprotein, viral genome replication |
| P36309, O73556, Q6XW15, Q99D35, P33515, Q85197, Q85197, Q9QEJ5, Q9QAX1, P29152, O92529, O92529, Q66474, P03302, A0AUJ5, Q1X881, P89509, P19724, Q01901, Q83883, P07564, Q04544, Q5UCB8, Q6J3P1, P03303 | Polyprotein, viral genome reproduction |
| Q5NDM9 | Protein P2-P3, viral reproductive process |
| P03168 | Protein X, viral genome replication |
| A7IXI8 | Regulation of viral transcription |
| P36796 | Regulatory protein E2, viral reproduction |
| P0C6U3, Q68772, P19811, P0C6Y5, Q88920, P0C6W4, P0C6V9, P0C6Y4, P0C6Y0, P0C6W0, P0C6V8, O36966 | Replicase polyprotein, viral genome replication |
| P17779, Q8UZB6, P17965 | RNA replication protein, viral |
| Q1HVH8 | Protein BOLF1, virion assembly |
| Q64746 | Protein involved in viral genome replication |
| Q9H4T2, A6NJL1, Q5RJ54, A6QNZ0, Q9Z2K3 | Zinc finger proteins involved in viral reproduction |
| Q9J566 | Late transcription factor VLTF-3, viral protein |
| P29044 | Putative RNA-directed RNA polymerase, viral |
| Q85431, P03594, A8C8X1, Q05318, Q6WB93, Q9IWW8, P31332, Q8JPX5, Q88434, P35341, Q8B0H5, P41357, O91940, P12577, Q6UY63, Q91DR9, A0PJ24, Q6UY61, P13615, Q6XQH7, P27316, P22956 | RNA-dependent RNA polymerase, viral |
| P68966 | Late 100-kDa protein, intracellular transport of viral proteins |
| Q9DUD9 | Fusion glycoprotein F0, viral penetration |
| O10685 | Major surface glycoprotein G, viral entry |
| P17287 | Protein Vpr, viral infection |
| P36278, P07234, Q9QEE7, P33427, P15100 | Capsid protein, viral envelope |
| Q9WC60, P12430, P03388, Q9QBZ8, P05878 | Envelope glycoprotein gp160 |
| P36711 | Fiber protein, viral envelope |
| P28882 | Hemagglutinin glycoprotein, viral envelope |
| P03465 | Hemagglutinin-esterase-fusion glycoprotein, viral envelope |
| Q05138, P09510, P06794, | Major capsid protein L1 |
| P26536, P16715 | Major core protein P4a, viral polyprotein |
| Q5VKP0 | Matrix protein, viral envelope |
| P28959 | Membrane protein UL43 homolog, viral tegument |
| P41483, Q83953 | Occlusion-derived virus envelope protein |
| Q9E779, P12436 | Outer capsid glycoprotein, viral envelope |
| P33422 | Protein VP6, viral capsid |
| Q02385, Q0Q466 | Viral envelope spike glycoprotein |
| P06490 | Viral capsid assembly protein |
| Q91HK5, P13561 | Viral capsid RNA2 polyprotein |
| Q02385, Q0Q466 | Viral envelope spike glycoprotein |
| P21945, Q82680, Q7T6S8, P27318, Q27YE3, Q9DK04, Q06927 | Viral nucleocapsid |
Table VIII. Major photosystem proteins detected in the Symbiodinium-enriched fraction of Stylophora pistillata.
None of the proteins were detected in coral genome sequences.
| Protein accession number | Protein description |
|---|---|
| Q9MSC2, A2C057 | Photosystem Q(B) protein |
| A6MMC3 | Photosystem I assembly protein ycf3 |
| Q3AZ40 | Photosystem I assembly protein ycf4 |
| O04006 | Photosystem I reaction center subunit VI |
| P17229 | Photosystem I reaction center subunit IX |
| Q6EW23 | Photosystem II reaction center subunit H |
| P51874 | Peridinin-chlorophyll a-binding protein |
| P51873 | Peridinin-chlorophyll a-binding protein 2 |
| Q01922 | R-phycoerythrin β chain |
| P28557, P0032 | C-phycocyanin α chain |
DISCUSSION
Study of the molecular basis for the physiological response of coral-algal symbiosis to environmental stress has centered on transcriptomics using various methods of differential gene expression to assess the genetic response of corals to stress (Ref. 83 and references therein). Those studies used high throughput sequencing to obtain large libraries of expressed sequence tags representing host, symbiont, or holobiont transcripts; smaller scale differential gene expression libraries originating from isolated algal endosymbionts; or cDNA microarray analysis of endosymbiont gene expression in hospite. Although these studies have enhanced considerably our understanding of host, symbiont, and holobiont molecular responses during environmental stress, transcriptome analyses are limited in that they do not represent the true phenotype of the symbiosis at the protein level of microbial-host interaction. This is because the relative abundance of transcripts is low and attempts to amplify the sequences by PCR can introduce bias. Furthermore, as in transcription, the translation of mRNA can be controlled by a number of processes, mostly at the level of initiation, either by induction or repression. In marine biology, transcriptomics is further hampered by poor sequence annotation because many organisms of marine origin are under-represented in databases, and it appears from comparative analysis that DNA sequences from marine and terrestrial organisms are highly divergent (84).
A more accurate reflection of biological function may be gained from the proteome because proteins generally are of higher abundance and have a longer half-life within cells than do transcripts. Also, many proteins are post-translationally modified, which cannot be detected in the transcriptome. It has been estimated that 80% of a cell's phenotype can be described by the proteome compared with only 30% by the transcriptome (56). A very limited number of studies have examined the proteome of both symbiotic and aposymbiotic cnidarians using two-dimensional SDS-PAGE analysis (57–62). The success of such attempts was restricted by the applicability of low throughput protein analyses. Herein we report the use of high resolution, quantitative high throughput protein analysis to obtain the first comprehensive proteome expression profile of symbiont-enriched fractions taken from an intact coral holobiont to examine the response of the endosymbiont to varied conditions of environmental stress. Although differential expression of proteins proved low in our experiments, relative yet quantifiable measurements of expressed proteins were attained by use of TMT reagents for protein level amino group labeling to provide an isotopic and isobaric tag enabling coelution of peptides from multiplexed samples during liquid chromatography with sample differentiation provided by MS/MS fragmentation within the mass reporter region. Protein quantities used for TMT labeling were standardized, and all three symbiont replicates obtained from individual coral treatments were analyzed in parallel. The loading of proteins from all treatments and the unstressed t = 0 proteome in a single sample eliminated problems arising from variation in the consistency of sample loading, the in-gel digestion process, and LC-MS/MS performance between sample runs, to afford protein expression profiles with a high degree of accuracy.
Many clades of Symbiodinium can be grown in pure culture, although often with considerable difficulty. However, exposing these pure cultures to conditions of environmental stress would not necessarily provide a true picture of the response of the endosymbiont in hospite. To achieve this, we extracted and rapidly cleaned the Symbiodinium fraction obtained from coral that had been exposed to conditions designed to elicit a bleaching response. However, absolute removal of host tissue contamination by repeated washing and centrifugation is nearly impossible given the abundance and robust nature of host nematocysts and phagosomal membranes that typically persist as contaminants. Differentially expressed proteins of this fraction from experimental treatments are presented under “Results”; thus, significant elements of the qualitative expression profile remain to be discussed.
Protein and Peptide Toxins
Despite direct microscopic examination of our cleaned algal isolates to ensure that contamination was removed as far as possible, we acknowledge that this host-derived contamination was evident particularly by the presence of non-dinoflagellate toxins (Table VI) that appear in the qualitative protein data set (supplemental information Data Set 8), which are thought to be derived either from microbial consorts or from the venome of coral nematocysts. To identify contaminating host toxins in our analysis data sets, protein sequences were compared against the very recently available genome sequences for the corals Acropora millipora (http://coralbase.org/) and Acropora digitifera (85), mindful that these corals are of a different genus and harbor different clades of Symbiodinium. To our surprise, we were unable to find homolog toxin sequences encoded in either of the published coral genomes, although a sequences homologous to botulinum toxin substrate was found in the A. digitifera genome (supplemental information Data Set 10). It is intriguing that so many in our data set are related to toxins from such dissimilar organisms (e.g. bacteria, fungi, invertebrates, and vertebrates). The venoms of many species of cnidarians contain a highly complex mixture of peptides, proteins, phospholipids, phospholipases, glycoproteins, bioactive amines, and carbohydrates that, with the exception of the sea anemones and several dangerous species of jellyfish, are largely uncharacterized (86).
The sequence of one toxin (G9TWG1) from our Symbiodinium-enriched coral fraction was similar to the K+ channel peptide toxin kaliseptine (36 amino acids) obtained from the sea anemone Anemonia sulcata (87). Isolated also from this anemone are the kalicludines (58–60-amino acid peptides) that are structurally related to the neurotoxins produced by mamba snakes (88). Further studies on the provenance, biological properties, and evolutionary significance of early metazoan toxins are clearly warranted, especially the botulinum-like zinc-metalloprotease toxins (Table VI) that may serve as virulence agents of coral disease (89). Additionally, marine venoms are a rich source of neuroactive agents; sea anemone toxins are being applied to the treatment of multiple sclerosis (90), whereas analog toxins of a marine cone snail are in clinical use for the suppression of neuropathic pain (91).
Antioxidant and Cellular Redox Enzymes and Globin Hemoproteins
A major mechanism for detoxification of ROS is the production of the antioxidant enzymes SOD and catalase, which have previously been reported in both partners (21, 22), and ascorbate peroxidase, reported only in the algal symbiont (22). All three detoxification enzymes were detected in our data set (Table I), as were other peroxidases and thiol reductases. Not surprisingly, mining of coral sequences for the first three enzymes revealed only SOD and catalase encoded in these genomes, confirming previous experimental findings that ascorbate peroxidase is of algal origin, although a plant-like ascorbate peroxidase gene has been reported in host tissue of the symbiotic cnidarian Hydra viridis (92). The only quantifiable proteins that had antioxidant functions were a thiol peroxidase and surprisingly nitric-oxide reductase (Fig. 1), the latter serving as a redox enzyme for NO detoxification. NOS and its encoding gene have previously been detected in only host cnidarians (44, 48) but not their endosymbionts.
NOS transcription is activated by NF-κB, which itself is up-regulated by H2O2 arising from stress-induced overproduction by the algal symbiont. By this process, it is thought that excess H2O2 of the endosymbiont is released by diffusion to the host cnidarian where NOS induction affects a NO-mediated response of the host to cause the initiation of cnidarian bleaching (44, 47). Although NOS was detected in our data set (Table V), likely as a contaminant, the gene encoding this enzyme was found in the published coral genome sequences, whereas nitric-oxide reductase was not. That TMT tagging is proportional to protein concentration and that nitric-oxide reductase was tagged at high yield suggest this protein is produced by the algal partner to protect against the damaging effects of the nitric oxide generated by the host. Nitric oxide can be produced independently in the symbiont by nitrite reductase (P32712) but intriguingly also via the redox-regulated nitrite reductase activity of the highly conserved heme superfamily of proteins (93, 94) that are abundant in coral endosymbionts (Table IV). These hemoproteins may function as post-translationally activated redox-regulated nitrite reductases that could mediate the NO response to cause cnidarian bleaching. Separate from the hemoproteins of metabolism and photosynthesis, these oxygen-binding globins may serve additionally as O2 sensors to activate cellular defense pathways by regulating the transcription of protective oxygen-responsive genes (95), particularly those factors activated in response to hypoxia (96), an usual condition of coral tissues during dark respiration (18). Expressed also are the ubiquitous ferritin intracellular iron storage proteins providing iron homeostasis in heme metabolism (Table IV). These proteins are reported to be transcriptionally up-regulated by corals during thermal bleaching, possibly in response to iron release caused by ROS disruption to heme-ferritin binding (97).
DNA Repair Enzymes
Although exposure to sunlight is an absolute requirement for maintaining phototrophic symbiosis in reef-building corals, excessive exposure can lead not only to direct damage of UV-sensitive cellular components, but in combination with thermal stress can lead to damaging or catastrophic effects characteristic of bleaching from excessive production of ROS. The damaging effects of UV are especially pronounced at the structural level of DNA. All of the main elements ascribed for DNA repair from damage caused by UV exposure, including that indirectly from ROS production, were detected in this study, including photolyase, base excision repair, and recombination mechanisms (Table II). Although we note that UV radiation was not a factor in our experiments, these findings suggest either that elements of the cellular UV damage control metabolism (stochastic) remain active weeks (>4) after exposure or that their presence is mainly attributable to the ROS generated from excess electron flow in photosynthesis under the elevated level of PAR used during the experiment. It is noteworthy that we found significant repression of the UvrABC system excinuclease B protein (Q9Z75A) under low temperature, metal-free conditions (Fig. 1), such being a highly conserved DNA repair enzyme that requires manganese ions.
Enzymes of Nitrogen Fixation
Evident in our data (Table V) are proteins associated with nitrogen fixation, which is not known in algal endosymbionts such as Symbiodinium, and with the exception of NifU, these proteins could not be found encoded in coral genomes, suggesting either that Symbiodinium can fix dissolved nitrogen or more likely that these nitrogenases are from coral-associated cyanobacteria (80, 81) or from diazotrophic bacteria (predominantly γ-proteobacteria) that in coral are strongly correlated with dinoflagellate abundance (98), suggesting a tight physiological relationship between these heterotrophic and phototrophic partners. Present also (Table V) is a nodulation factor export ATP-binding protein (P26050), which may provide molecular recognition for this bacteria-dinoflagellate affinity, as is required for sympathogenesis of N2-fixing rhizobia in plant legumes (99). This bacterial association may be endosymbiotic (as reported in Ref. 80) or instead may include surface-associated cyanobacteria that fix nitrogen at night when photosynthetic production of nitrogenase-destructive O2 has ceased (100).
Enzymes of Nitrogen Metabolism
Nitrogen assimilation is essential to metabolic interactions between resident algal endosymbionts and their cnidarian hosts (101). Ammonium, nitrate, and free amino acids are taken up by cnidarians, and nitrogen-availability and uptake have been linked to increases in endosymbiont population densities (102–104). Essential amino acid translocation from the symbiotic algae to the animal host is a core element of cnidarian nitrogen-recycling (105). The coral host accumulates waste urea in its tissues as an important store of nitrogen for translocation to its symbiotic partners (106), providing NH3 as a nitrogen source for photosynthesis-based anabolism on release by urease enzymes, which are plentiful in the Symbiodinium-enriched fraction of S. pistillata (Table V). Glutamate synthase and glutamine synthetase, both implicated in ammonia assimilation in Symbiodinium (see Ref. 107, pages 178–179 for a brief overview), are present in our database (Table V). Clode et al. (108) have shown that accretions of uric acid, thought previously to be calcium oxalate (109), are deposited throughout the algal cytosol and within vacuoles of Symbiodinium spp. to provide a store of nitrogen when normal routes of cnidarian nitrogen-availability are limited. Additionally, uric acid is a strong reducing agent and potent antioxidant, reacting freely with oxyradicals but not H2O2. Uric acid is an end product of purine catabolism by xanthine oxidase from xanthine and hypoxanthine while producing both superoxide anion and hydrogen peroxide. We did find in our data set xanthine dehydrogenase (Table V), which is a xanthine oxidoreductase enzyme that is converted to xanthine oxidase by reversible sulfhydryl oxidation or by irreversible proteolytic modification (110). Present also (Table V) is uricase (urate oxidase) that might be utilized to extract nitrogen stored as uric acid for algal metabolism.
Proteins of Apoptosis, Vesicular Transport, and Endo/exocytosis
There are five separate cellular mechanisms postulated for the loss of algal endosymbionts from cnidarian host tissues in the thermal stress response of coral bleaching: in situ symbiont degradation and digestion, symbiont exocytosis, host cell detachment, host cell necrosis, and host cell and symbiont apoptosis (reviewed in Ref. 16). Although no single mechanism of bleaching may be applicable to all conditions of stress causing cnidarians to bleach, it is widely held that cellular changes associated with bleaching are a result of host cell apoptosis. Although ample evidence obtained under controlled experimental conditions supports this view, evidence from the environment suggests that the rates of apoptosis might only reflect that expected from normal cell cycling, as indicated by Starcevic et al. (53) and likewise noted by Pernice et al. (29) on up-regulation of Bcl-2 as an anti-apoptotic response followed by a decrease in caspase-dependent apoptosis soon after the initial onset of coral bleaching. In contrast, algal endosymbionts may exit host cells under moderate stress early in the bleaching process, after which induced apoptosis comes only as a terminal response to prolonged and severe stress for initiation of cell death (13, 14). We found no strong evidence for proteins that mediate apoptosis in this study, and indeed only inactive forms of apoptotic mediators or positive regulators were detected (Table III). Interestingly, none of these proteins were detected as being significantly enhanced by thermal treatment in the quantitative data set (Fig. 1), including Tax1 (Q86VP1.2), which is an inhibitor of NF-κB-induced apoptosis, even though the host corals exhibited signs of early tissue necrosis immediately prior to harvesting. However, the TMT tag intensity of this protein was not significantly different in any of the treatments compared with the t = 0 base-line proteome prior to stress (supplemental information Data Set 7). The significant decreases in dark-adapted Fv:Fm that occurred in all treatments in the present study (supplemental information Data Set 3), similar to those seen in our earlier work (54), indicated moderate bleaching under our experimental conditions, but these proteomic data suggest that apoptosis was not a contributing cellular mechanism.
Histological examination establishes that coral host cells are not significantly degraded during bleaching (25), and those intact dinoflagellates in hospite are released by exocytosis from the endoderm during coral bleaching (5, 52). Consistent with exocytosis of endosymbionts, we have previously found transcriptomic evidence for differential expression of genes encoding not only Rab-like proteins that are known to regulate vesicle transport in eukaryotes, but also SNARE and calcium transporters that direct exocytosis (53). In this study we provide proteomic evidence for the existence of proteins required for exocytosis, which are expressed during the early stages of coral bleaching. Indeed, the TMT tag intensity of the GTPase activator for calcium binding Rab proteins (Q3UYK3) at the higher temperature is not statistically different from the t = 0 base-line proteome prior to stress but is less in the low temperature treatment than at the higher temperature (Fig. 1).
It is becoming increasingly clear from numerous studies on microbial pathogenicity that intracellular pathogens can mediate their own entry into and exit from host cells or lysosomes by encoding their own SNARE proteins (111, 112). These SNAREs can both enhance and inhibit exocytosis (113). Furthermore, an emerging theme (114) provides that SNARE protein function is regulated by complexation with cysteine string protein α (Q03751 in Table I; note that this protein is not encoded in the coral genomes available to search) and Hsc70, a cognate member of the 70-kDa family of stress-induced heat shock proteins, which is a required chaperone of SNARE assembly for exocytosis to occur (115). A range of these proteins (Table III) can be detected when supplemental information Data Set 4 is searched, and opposing vesicular and target SNARE proteins that are necessary for vesicle transport appear to be encoded either by the host (vesicular SNARE, for example, synaptotagmin proteins Q9R0N5 and Q8TDW5) or symbiont (target SNARE, for example, syntaxin proteins Q8N4C7, Q39233, and Q94KK7). We posit that under conditions of stress, heat shock protein chaperones cysteine string protein-Hsc70-SNARE assembly utilizing target SNARE proteins of the endosymbiont, enabling stress-sensitive endosymbionts to mediate their own exit from the host, which departs from customary thinking that the host solely expels the symbiont. This model provides a molecular mechanism for symbiont shuffling (116, 117) whereby thermally sensitive Symbiodinium clades are self-displaced during coral bleaching, allowing residual symbionts of a heat-resistant clade to repopulate the symbiosis.
Viral Proteins
A remarkable finding from this quantitative study of the proteome expression profile of the endosymbiont-enriched fraction of coral symbiosis was the massive 114-fold increase in viral replication protein E1 levels that occurred under metal-replete conditions at high temperature (Fig. 1). A review of the qualitative data (supplemental information Data Set 8) for viral proteins reveals a comprehensive complement of proteins required for viral replication, production of new viral particles, and viral-host interactions (Table VII), especially RNA-dependent polymerase indicating the presence of negative strand RNA viruses, which are present in seawater and in marine virus-host assemblages (118). It would seem from the proteomics results that virulence is high during coral stress, which has been reported previously (119–121) when trace metals are replete. Viral replication requires new viral particles to be transmitted to new host cells; these processes include viral budding or host cell lysis in either the infected coral tissue or its endosymbionts. Large scale perturbation of the host cell membrane or host cell lysis has been associated with severe coral bleaching in the past. We suggest that coral recovery post-bleaching not only depends on the retention (or acquisition) of heat tolerant endosymbionts but must also reduce the viral load of the coral to prebleaching levels. Although viruses have been implicated as a cause of coral disease and bleaching (122), the complexity of coral-viral interactions is poorly understood (123), particularly in connection with environmental change (124), which clearly warrants future investigation. Our proteomics data thus provide unique insight to how coral endosymbionts and their microbial consorts respond to conditions of elevated temperature, with and without trace metal limitation, affecting a significant loss of photochemical efficiency in the bleaching response of S. pistillata.
In summary, quantitative high resolution TMT-labeled protein analysis of a symbiont-enriched fraction of S. pistillata has yielded valuable information on the protein structure of coral symbiosis. Although the sample proteome has revealed a vast complement of proteins involving the metabolic interactions of coral symbiosis, it is the unexpected finding of a massive increase in a key viral replication protein that highlights the potential significance of the coral virome in coral bleaching and disease progression caused by environmental stress. Quantitative high resolution protein analyses in future studies of the intact coral symbiome shall reveal more of the complex and interactive processes that regulate the function of cnidarian symbioses.
Acknowledgments
W. Dunlap gratefully thanks the Institute of Pharmaceutical Science at King's College London for appointment as Honorary Professor. We gratefully thank the master and crew of the RV Cape Ferguson for shipboard field support. Christopher Rigaud, Diving Safety Officer of the University of Maine, provided refresher training for M. Wells and M. Shick. In using the genome data from A. millipora (http://coralbase.org/), we acknowledge that these sequence data were produced by the Australian Genome Research Facility and the Australian Research Council Centre of Excellence for Coral Reef Studies in collaboration with the user community.
Footnotes
* This work was supported by National Science Foundation Grant 0648478 (Biological Oceanography), by the National Institutes of Health Research Biomedical Research Centre for Mental Health (King's College London, UK), by Ministry of Science, Education and Sports, Republic of Croatia Grant 058-0000000-3475, and by the Australian Institute of Marine Science. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
1 The abbreviations used are:
- ROS
- reactive oxygen species
- Bcl-2
- B-cell lymphoma 2
- NF-κB
- nuclear factor-κB
- NOS
- nitric-oxide synthase
- MES
- 2-(N-morpliolino) ethanesufonic acid
- PAR
- photosynthetically active radiation (400–700 nm)
- SOD
- superoxide dismutase
- SNARE
- soluble NSF (N-ethylmaleimide-sensitive factor) attachment receptor proteins
- TCEP
- tris(2-carboxyethyl)phosphine hydrochloride
- TEAB
- triethylammonium bicarbonate
- TMT
- tandem mass tag
- KEGG
- Kyoto Encyclopedia of Genes and Genomes.
REFERENCES
- 1. Stambler N. (2011) Zooxanthellae: The yellow symbionts inside animals. In Coral Reefs: An Ecosystem in Transition (Dubinski Z., Falkowski P., eds) pp. 107–118, Springer-Verlag, Berlin [Google Scholar]
- 2. Muscatine L. (1990) The role of symbiotic algae in carbon and energy flux in reef corals. In Coral Reefs: Ecosystems of the World (Dubinsky Z., ed) No. 25, pp. 75–84, Elsevier, Amsterdam [Google Scholar]
- 3. Stanley G. D., Jr. (2006) Photosymbiosis and the evolution of modern coral reefs. Science 312, 857–858 [DOI] [PubMed] [Google Scholar]
- 4. Dubinski Z., Falkowski P. (2011) Light as a source of information and energy in zooxanthellate corals. In Coral Reefs: An Ecosystem in Transition (Dubinski Z., Falkowski P., eds) pp. 107–118, Springer-Verlag, Berlin [Google Scholar]
- 5. Brown B. E., Le Tissier M. D., Bythell J. C. (1995) Mechanisms of bleaching deduced from histological studies of reef corals samples during a natural bleaching event. Mar. Biol. 122, 655–663 [Google Scholar]
- 6. Takahashi S., Whitney S., Itoh S., Maruyama T., Badger M. (2008) Heat stress causes inhibition of de novo synthesis of antenna proteins and photobleaching in cultured Symbiodinium. Proc. Natl. Acad. Sci. U.S.A. 105, 4203–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Falkowski P. G., Dubinsky Z., Muscatine L., Porter J. (1984) Light and the bioenergetics of a symbiotic coral. Bioscience 34, 705–709 [Google Scholar]
- 8. Glynn P. W. (1996) Coral reef bleaching: Facts, hypotheses and implications. Global Change Biol. 2, 495–509 [Google Scholar]
- 9. Hoegh-Guldberg O. (1999) Climate change, coral bleaching and the future of the world's coral reefs. Mar. Freshwat. Res. 50, 839–866 [Google Scholar]
- 10. Lough J. M. (2000) 1997–1998: Unprecedented thermal stress to coral reefs? Geophys. Res. Lett. 27, 3901–3904 [Google Scholar]
- 11. Fitt W. F., Brown B. E., Warner M. E., Dunne R. P. (2001) Coral bleaching: Interpretation of thermal tolerance limits and thermal thresholds in tropical corals. Coral Reefs 20, 51–65 [Google Scholar]
- 12. Jokiel P. L. (2004) Temperature stress and coral bleaching. In Coral Health and Disease (Rosenberg E., Loya Y., eds) pp. 401–425, Springer-Verlag, Berlin [Google Scholar]
- 13. Lesser M. P. (2006) Oxidative stress in marine environments: Biochemistry and physiological ecology. Annu. Rev. Physiol. 68, 253–278 [DOI] [PubMed] [Google Scholar]
- 14. Lesser M. P. (2011) Coral bleaching: Causes and mechanisms. In Coral Reefs: An Ecosystem in Transition (Dubinsky Z., Stambler N., eds) pp. 405–420, Springer-Verlag, Berlin [Google Scholar]
- 15. Wilkinson C., Linden O., Cesar H., Hodgson G., Rubens J., Strong A. E. (1999) Ecological and socioeconomic impacts of 1998 coral mortality in the Indian Ocean: An ENSO impact and warning of future change? Ambio 28, 188–196 [Google Scholar]
- 16. Weis V. M. (2008) Cellular mechanisms of cnidarian bleaching: Stress causes the collapse of symbiosis. J. Exp. Biol. 211, 3059–3066 [DOI] [PubMed] [Google Scholar]
- 17. Weis V. M. (2010) The susceptibility and resilience of corals to thermal stress: Adaptation, acclimatization or both? Mol. Ecol. 19, 1515–1517 [DOI] [PubMed] [Google Scholar]
- 18. Kühl M., Cohen Y., Dalsgaard T., Jørgensen B., Revsbech N. P. (1995) Microenvironment and photosynthesis of zooxanthellae in scleractinian corals studied with microsensors for O2, pH and light. Mar. Ecol. Prog. Ser. 117, 159–172 [Google Scholar]
- 19. Baird A. H., Bhagooli R., Ralph P. J., Takahashi S. (2009) Coral bleaching: The role of the host. Trends Ecol. Evol. 24, 16–20 [DOI] [PubMed] [Google Scholar]
- 20. Dykens J. A., Shick J. M., Benoit C., Buettner G. R., Winston G. W. (1992) Oxygen radical production in the sea anemone Anthopleura elegantissima and its endosymbiotic algae. J. Exp. Biol. 168, 219–241 [Google Scholar]
- 21. Shick J. M., Dykens J. A. (1985) Oxygen detoxification in algal-invertebrate symbioses from the Great Barrier Reef. Oecologia 66, 33–41 [DOI] [PubMed] [Google Scholar]
- 22. Shick. J. M., Lesser M. P., Dunlap W. C., Stochaj W. R., Chalker B. E., Wu Won J. (1995) Depth-dependent responses to solar ultraviolet radiation and oxidative stress in the zooxanthellae of the coral Acropora microphthalma. Mar. Biol. 122, 41–51 [Google Scholar]
- 23. Weis V. M., Allemand D. (2009) What determines coral health? Science 324, 1153–1155 [DOI] [PubMed] [Google Scholar]
- 24. Dunn S. R., Thomason J. R., Le Tissier M. D., Bythell J. C. (2004) Heat stress induces different forms of cell death in sea anemones and their endosymbiotic algae depending on temperature and duration. Cell Death Differ. 11, 1–10 [DOI] [PubMed] [Google Scholar]
- 25. Strychar K. B., Sammarco P. W. (2009) Exaptation in corals to high seawater temperatures: Low concentrations of apoptotic and necrotic cells in host coral tissues under bleaching conditions. J. Exp. Mar. Biol. Ecol. 369, 31–42 [Google Scholar]
- 26. Edge S. E., Morgan M. B., Gleason D. F., Snell T. W. (2005) Development of a coral cDNA array to examine gene expression profiles in Montastraea faveolata exposed to environmental stress. Mar. Pollut. Bull. 51, 507–523 [DOI] [PubMed] [Google Scholar]
- 27. Ainsworth T. D., Hoegh-Guldberg O., Heron S. F., Skirving W. J., Leggat W. (2008) Early cellular changes are indicators of pre-bleaching thermal stress in the coral host. J. Exp. Mar. Biol. Ecol. 364, 63–71 [Google Scholar]
- 28. DeSalvo M. K., Voolstra C. R., Sunagawa S., Schwarz J. A., Stillman J. H., Coffroth M. A., Szmant A. M., Medina M. (2008) Diffferential gene expression during therma stress and bleaching in the Caribbean coral Montastraea faveolata. Mol. Ecol. 17, 3952–3971 [DOI] [PubMed] [Google Scholar]
- 29. Pernice M., Dunn S. R., Miard T., Dufour S., Dove S., Hoegh-Guldberg O. (2011) Regulation of apoptotic mediators reveals dynamic responses to thermal stress in the reef building coral Acropora millepora. PLoS One 6, e16095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Voolstra C. R., Schnetzer J., Peshkin L., Randall C. J., Szmant A. M., Medina M. (2009) Effects of temperature on gene expression in embryos of the coral Monstastraea faveolata. BMC Genomics 10, 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Polato N. R., Voolstra C. R., Schnetzer J., DeSalvo M. K., Randall C. J., Szmant A. M., Medina M., Baums I. B. (2010) Location-specific responses to thermal stress in larvae of the reef-building coral Montastraea faveolata. PLoS One 5, e11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Richier S., Sabourault C., Courtiade J., Zucchini N., Allemand D., Furla P. (2006) Oxidative stress and apoptotic events during thermal stress in the symbiotic sea anemone, Anemonia virdis. FEBS J. 273, 4186–4198 [DOI] [PubMed] [Google Scholar]
- 33. Dunn S. R., Phillips W. S., Spatafora J. W., Green D. R., Weis V. M. (2006) Highly concerved caspase and Bcl-2 homologues from the sea anemone Aiptasia pallida: Lower metazoans for the study of apoptosis evolution. J. Mol. Evol. 63, 95–107 [DOI] [PubMed] [Google Scholar]
- 34. Sunagawa S., Choi J., Forman H. J., Medina M. (2008) Hyperthermic stress-induced increase in the expression of glutamate-cysteine ligase and glutathione levels in the symbiotic sea anemone Aiptasia pallida. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 151, 133–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rodriguez-Lanetty M., Phillips W. S., Weis V. M. (2006) Transcriptome analysis of a cnidarians-dinoflagellate mutualism reveals complex modulation of host gene expression. BMC Genomics 7, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wolenski F. S., Garbati M. R., Lubinski T. J., Traylor-Knowles N., Dresselhaus E., Stefanik D. J., Goucher H., Finnerty J. R., Gilmore T. D. (2011) Characterization of core elements of the NF-κB signalling pathway of the sea anemone Nematostella vectensis. Mol. Cell. Biol. 31, 1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kroemer G. (1997) Mitochondrial implication in apoptosis: Towards an endosymbiont hyporthesis of apoptosis evolution. Cell Death Differ. 4, 443–456 [DOI] [PubMed] [Google Scholar]
- 38. Sandeman I. M. (2006) Fragmentation of the gastrodermis and detachment of zooxanthellae in symbiotic cnidarians: A role for hydrogen peroxide and Ca2+ in coral bleaching and algal density control. Rev. Biol. Trop. 54, 79–96 [Google Scholar]
- 39. Suggett. D. J., Warner M. E., Smith D. J., Davey P., Hennige S., Baker N. R. (2008) Photosynthesis and production of hydrogen peroxide by Symbiodinium (Pyrrhophyta) phylotypes with different thermal tolerances. J. Phycol. 44, 948–956 [DOI] [PubMed] [Google Scholar]
- 40. Ragni M., Airs R. L., Hennige S. J., Suggett D. J., Warner M. E., Geider R. J. (2010) PSII photoinhibition and photorepair in Symbiodinium (Pyrrhophyta) differs between thermally tolerant and sensitive phylotypes. Mar. Ecol. Prog. Ser. 406, 57–70 [Google Scholar]
- 41. Smith D. J., Suggett D. J., Baker N. R. (2005) Is photoinhibition of zooxanthellae photosynthesis the primary cause of thermal bleaching in corals? Global Change Biol. 11, 1–11 [Google Scholar]
- 42. Dunn S. R., Schnitzler C. E, Weis V. M. (2007) Apoptosis and autophagy as mechanisms of dinoflagellate symbiont release during cnidarian bleaching: Every which way you lose. Proc. R. Soc. Lond. B Biol. Sci. 274, 3079–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Trapido-Rosenthal H. G., Sharp K. H., Galloway T. S., Morrall C. E. (2001) Nitric oxide and cnidarians-dinoflagellate symbioses: Pieces of a puzzle. Am. Zool. 41, 247–257 [Google Scholar]
- 44. Trapido-Rosenthal H., Zielke S., Owen R., Buxton L., Boeing B., Bhagooli R., Archer J. (2005) Increased zooxanthellae nitric oxide synthase activity is associated with coral bleaching. Biol. Bull. 208, 3–6 [DOI] [PubMed] [Google Scholar]
- 45. Bouchard J. N., Yamasaki H. (2008) Heat stress stimulates nitric oxide production in Symbiodinium microadriaticum: A possible linkage between nitric oxide and coral bleaching phenomenon. Plant Cell Physiol. 49, 641–652 [DOI] [PubMed] [Google Scholar]
- 46. Souter P., Bay L. K., Andreakis N., Császár N., Seneca F. O., van Oppen M. J. (2011) A multilocus, temperature stress-related gene expression profile assay in Acropora millepora, a dominant reef-building coral. Mol. Ecol. Resour. 11, 328–334 [DOI] [PubMed] [Google Scholar]
- 47. Perez S., Weis V. M. (2006) Nitric oxide and cnidarians bleaching: An eviction notice mediates breakdown of a symbiosis. J. Exp. Biol. 209, 2804–2810 [DOI] [PubMed] [Google Scholar]
- 48. Safavi-Hemami H., Young N. D., Doyle J., Llewellyn L., Klueter A. (2010) Characterisation of nitric oxide synthase in three cnidarian-dinoflagellate symbioses. PLoS One 5, e10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jiang X, Wang X. (2004) Cytochrome c-mediated apoptosis. Annu. Rev. Biochem. 73, 87–106 [DOI] [PubMed] [Google Scholar]
- 50. Lasi M., Pauly B., Schmidt N., Cikala M., Stiening B., Käsbauer T., Zenner G., Popp T., Wagner A., Knapp R. T., Huber A. H., Grunert M., Söding J., David C. N., Böttger A. (2010) The molecular cell death machinery in the simple cnidarian Hydra includes the expanded caspase family and pro- and anti-apoptotic Bcl-2 proteins. Cell Res. 20, 812–825 [DOI] [PubMed] [Google Scholar]
- 51. Gates R. D., Baghdasarian G., Muscatine L. (1992) Temperature stress causes host cell detachment in symbiotic cnidarians: Implications for coral bleaching. Biol. Bull. 182, 324–332 [DOI] [PubMed] [Google Scholar]
- 52. Bhagooli R., Hidaka M. (2004) Release of zooxanthellae with intact photosynthetic activity by the coral Galaxea fascicularis in response to high temperature stress. Mar. Biol. 145, 329–337 [Google Scholar]
- 53. Starcevic A., Dunlap W. C., Cullum J., Shick J. M., Hranueli D., Long P. F. (2010) Gene expression in the scleractinian Acropora microphthalma exposed to high solar irradiance reveals elements of photoprotection and coral bleaching. PLoS One 5, e13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shick J. M., Iglic K., Wells M. L., Trick C. G., Doyle J., Dunlap W. C. (2011) Responses to iron limitation in two colonies of Stylophora pistillata exposed to high temperature: Implications for coral bleaching. Limnol. Oceanogr. 56, 813–828 [Google Scholar]
- 55. Kelly L. W., Barott K. L., Dinsdale E., Friedlander A. M., Nosrat B., Obura D., Sala E., Sandin S. A., Smith J. E., Vermeij M. J., Williams G. J., Willner D., Rohwer F. (2012) Black reefs: Iron-induced phase shifts on coral reefs. ISME J. 6, 638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Feder M. E., Walser J. C. (2005) The biological limitations of transcriptomics in elucidating stress and stress response. J. Evol. Biol. 18, 901–910 [DOI] [PubMed] [Google Scholar]
- 57. Weis V. V., Levine R. (1996) Differential protein profiles reflect the different lifestyles of symbiotic and aposymbiotic Anthopleura elegantissima, a sea anemone from temperate waters. J. Exp. Biol. 199, 883–892 [DOI] [PubMed] [Google Scholar]
- 58. Weis V. M., Reynolds W. S. (1999) Carbonic anhydrase expression and synthesis in the sea anemone Anthopleura elegantissima are enhanced by the presence of dinoflagellate symbionts. Physiol. Biochem. Zool. 72, 307–316 [DOI] [PubMed] [Google Scholar]
- 59. Barneah O., Benayahu Y., Weis V. M. (2006) Comparative proteomics of symbiotic and aposymbiotic juvenile soft corals. Mar. Biotechnol. 8, 11–16 [DOI] [PubMed] [Google Scholar]
- 60. deBoer M. L., Krupp D. A., Weis V. M. (2007) Proteomics and transcriptional analyses of coral larvae newly engaged in symbiosis with dinoflagellates. Comp. Biochem. Physiol. D Genom. Proteom. 2, 63–73 [DOI] [PubMed] [Google Scholar]
- 61. Pollack K., Balazs K., Ogunseitan O. (2009) Proteomic assessment of caffeine effects on coral symbionts. Environ. Sci. Technol. 43, 2085–2091 [DOI] [PubMed] [Google Scholar]
- 62. Peng S. E., Wang Y. B., Wang L. H., Chen W. N., Lu C. Y., Fang L. S., Chen C. S. (2010) Proteomic analysis of the symbiosome membranes in Cnidaria-dinoflagellate endosymbiosis. Proteomics 10, 1002–1016 [DOI] [PubMed] [Google Scholar]
- 63. Tomanek L. (2011) Environmental proteomics: Changes in the proteome of marine organisms in response to environmental stress, pollutants, infection, symbiosis, and development. Annu. Rev. Mar. Sci. 3, 373–399 [DOI] [PubMed] [Google Scholar]
- 64. Achterberg E, P., van den Berg C. M. (1994) In-line ultraviolet-digestion of natural seawater samples for trace metal determination using and automated voltammetric system. Anal. Chim. Acta 291, 213–232 [Google Scholar]
- 65. Davey E. W., Gentile J. H., Erickson S. J., Betzer P. (1970) Removal of trace metals from marine culture media. Limnol. Oceanogr. 15, 486–488 [Google Scholar]
- 66. Price N. M., Harrison G. I., Hering J. G., Hudson R. J., Nivel M. V., Palenic D., Morel F. M. (1988) /89) Preparation and chemistry of the artificial algal culturing medium Aquil. Biol. Oceanogr. 6, 443–461 [Google Scholar]
- 67. Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., Hamon C. (2003) Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 68. Dayon L., Hainard A., Licker V., Turck N., Kuhn K., Hochstrasser D. F., Burkhard P. R., Sanchez J. C. (2008) Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal. Chem. 80, 2921–2931 [DOI] [PubMed] [Google Scholar]
- 69. Hung C. W., Tholey A. (2012) Tandem mass tag protein labeling for top-down identification and quantification. Anal. Chem. 84, 161–170 [DOI] [PubMed] [Google Scholar]
- 70. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 71. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 72. Bhattacharyya G. K., Johnson R. A. (1977) Statistical Concepts and Methods, Wiley, New York [Google Scholar]
- 73. Daly F., Hand D. J., Jones M. C., Lunn A. D., McConway K. J. (1995) Elements of Statistics, Addison Wesley, Wokingham, UK [Google Scholar]
- 74. Warner M. E., Fitt W. K., Schmidt W. (1996) The effects of elevated temperature on the photosynthetic efficiency of zooxanthellae in hospite from four different species of reef coral: A novel approach. Plant Cell Environ. 19, 291–299 [Google Scholar]
- 75. Verstrepen L., Verhelst K., Carpentier I., Beyaert R. (2011) TAX1BP1, a ubiquitin-binding adaptor protein in innate immunity and beyond. Trends Biochem. Sci. 36, 347–354 [DOI] [PubMed] [Google Scholar]
- 76. Mireskandari A., Reid R. L., Kashanchi F., Dittmer J., Li W. B., Brady J. N. (1996) Isolation of a cDNA clone, TRX encoding a human T-cell lymphotrophic virus type-I Tax1 binding protein. Biochim Biophys. Acta 1306, 9–13 [DOI] [PubMed] [Google Scholar]
- 77. Sharma K., Weber C., Bairlein M., Greff Z., Kéri G., Cox J., Olsen J. V., Daub H. (2009) Proteomics strategy for quantitative protein interaction profiling in cell extracts. Nat. Meth. 6, 741–744 [DOI] [PubMed] [Google Scholar]
- 78. Gouw J. W., Krijgsveld J., Heck A. J. (2010) Quantitative proteomics by metabolic labeling of model organisms. Mol. Cell. Proteomics 9, 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Li Z., Adams R. M., Chourey K., Hurst G. B., Hettich R. L., Pan C. (2012) Systematic comparison of label-free, metabolic labeling, and isobaric chemical labeling for quantitative proteomics on LTQ Orbitrap velos. J. Proteome Res. 11, 1582–1590 [DOI] [PubMed] [Google Scholar]
- 80. Lesser M. P., Mazel C. H., Gorbunov M. Y., Falkowski P. G. (2004) Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305, 997–1000 [DOI] [PubMed] [Google Scholar]
- 81. Frias-Lopez J., Bonheyo G. T., Jin Q., Fouke B. W. (2003) Cyanobacteria associated with coral black band disease in Caribbean and Indo-Pacific reefs. Appl. Environ. Microbiol. 69, 2409–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zehr J. P., Waterbury J. B., Turner P. J., Montoya J. P., Omoregie E., Steward G. F., Hansen A., Karl D. M. (2001) Unicellular cyanobacteria fix N2 in the subtropical North Pacific Ocean, Nature 412, 635–638 [DOI] [PubMed] [Google Scholar]
- 83. DeSalvo M. K., Sunagawa S., Voolstra C. R., Medina M. (2010) Transcriptomic responses to heat stress and bleaching in the elkhorn coral Acropora palmata. Mar. Ecol. Prog. Ser. 402, 97–113 [Google Scholar]
- 84. Dawson M. N., Hamner W. M. (2008) A biophysical perspective on dispersal and the geography of evolution in marine and terrestrial systems. J. R. Soc. Interface 5, 135–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shinzato C., Shoguchi E., Kawashima T., Hamada M., Hisata K., Tanaka M., Fujie M., Fujiwara M., Koyanagi R., Ikuta T., Fujiyama A., Miller D. J., Satoh N. (2011) Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323 [DOI] [PubMed] [Google Scholar]
- 86. Messerli S. M., Greenberg R. M. (2006) Cnidarian toxins acting on voltage-gated ion channels. Mar. Drugs 4, 70–81 [Google Scholar]
- 87. Schweitz H., Bruhn T., Guillemare E., Moinier D., Lancelin J. M., Béress L., Lazdunski M. (1995) Kalicludines and kaliseptine: Two different classes of sea anemone toxins for voltage sensitive K+ channels. J. Biol. Chem. 270, 25121–25126 [DOI] [PubMed] [Google Scholar]
- 88. Lewis R. J., Garcia M. L. (2003) Therapeutic potential of venom peptides. Nat. Rev. Drug Discov. 2, 790–802 [DOI] [PubMed] [Google Scholar]
- 89. Santos Ede O., Alves N., Jr., Dias G. M., Mazotto A. M., Vermelho A, Vora G. J., Wilson B., Beltran V. H., Bourne D. G., Le Roux F., Thompson F. L. (2011) Genomic and proteomic analyses of the coral pathogen Vibrio coraliilyticus reveal a diverse virulence repertoire. ISME J. 5, 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Norton R. S., Pennington M. W., Wulff H. (2004) Potassium channel blockade by the sea anemone toxin ShK for the treatment of multiple sclerosis and other autoimmune diseases. Curr. Med. Chem. 11, 3041–3052 [DOI] [PubMed] [Google Scholar]
- 91. McGivern J. G. (2007) Ziconotide: A review of its pharmacology and use in the treatment of pain. Neuropychiatr. Dis. Treat. 3, 69–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Habetha M., Bosch T. C. (2005) Symbiotic Hydra express a plant-like peroxidase gene during oogenesis. J. Exp. Biol. 208, 2157–2165 [DOI] [PubMed] [Google Scholar]
- 93. Hardison RC. (1996) A brief history of hemoglobins: Plant, animal, protist, and bacteria. Proc. Natl. Acad. Sci. U.S.A. 93, 5675–5679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tiso M., Tejero J., Basu S., Azarov I., Wang X., Simplaceanu V., Frizzell S., Jayaraman T., Geary L., Shapiro C., Ho C., Shiva S, Kim-Shapiro D. B., Gladwin M. T. (2011) Human neuroglobin functions as a redox-regulated nitrite reductase. J. Biol. Chem. 286, 18277–18289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Bailey-Serres J., Chang R. (2005) Sensing and signalling in response to oxygen deprivation in plants and other organisms. Ann. Bot. 96, 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Levy O., Kaniewska P., Alon S., Eisenberg E., Karako-Lampert S., Bay L. K., Reef R., Rodriguez-Lanetty M., Miller D. J., Hoegh-Guldberg O. (2011) Complex diel cycles of gene expression in coral-algal symbiosis. Science 331, 175. [DOI] [PubMed] [Google Scholar]
- 97. Császár N. B., Seneca F. O., van Oppen M. J. (2009) Variation in antioxidant gene expression in the scleractinian coral Acropora millepora under laboratory thermal stress. Mar. Ecol. Prog. Ser. 392, 93–102 [Google Scholar]
- 98. Olsen N. D., Ainsworth T. D., Gates R. D., Takabayashi M. (2009) Diazotrophic bacteria associated with Hawaiian Montipora corals: Diversity and abundance in correlation with symbiotic dinoflagellates. J. Exp. Mar. Biol. Ecol. 371, 140–146 [Google Scholar]
- 99. Spaink H. P. (1995) The molecular basis of infection and nodulation by rhizobia: The ins and outs of sympathogenesis. Annu. Rev. Phytopathol. 33, 345–368 [DOI] [PubMed] [Google Scholar]
- 100. Berman-Frank L., Quigg A., Finkel Z. V., Irwin A. J., Haramaty L. (2007) Nitrogen-fixation strategies and Fe requirements in cyanobacteria. Limnol. Oceanogr. 52, 2260–2269 [Google Scholar]
- 101. Yellowlees D., Rees T. A., Leggat W. (2008) Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ. 31, 679–694 [DOI] [PubMed] [Google Scholar]
- 102. Muller-Parker G., McCloskey M. L., Hoegh-Guldberg O., McAuley P. (1994) Effect of ammonium enrichment on animal and algal biomass of the coral Pocillopora damicornis. Pac. Sci. 48, 273–283 [Google Scholar]
- 103. Marubini F., Davies P. S. (1996) Nitrate increases zooxantheallae population density and reduces skeletogenesis in corals. Mar. Biol. 127, 319–328 [Google Scholar]
- 104. Grover R., Maguer J. F., Allemand D., Ferrier-Pagès C. (2003) Nitrate uptake in the scleractinian coral Stylophora pistillata. Limnol. Oceanogr. 48, 2266–2274 [Google Scholar]
- 105. Wang J. T., Douglas A. E. (1999) Essential amino acid synthesis and nitrogen recycling in an alga-invertbrate symbiosis. Mar. Biol. 135, 219–222 [Google Scholar]
- 106. Grover R., Maguer J. F., Allemand D., Ferrier-Pagès C. (2006) Urea uptake by the scleractinian coral Stylophora pistillata. J. Exp. Mar. Biol. 332, 216–225 [Google Scholar]
- 107. Shick J. M. (1991) A Functional Biology of Sea Anemones, pp. 198–199, Chapman & Hall, London [Google Scholar]
- 108. Clode P. L., Saunders M., Maker G., Ludwig M., Atkins C. A. (2009) Uric acid deposits in symbiotic marine algae. Plant Cell Environ. 32, 170–177 [DOI] [PubMed] [Google Scholar]
- 109. Taylor D. L. (1968) In situ studies on the chemistry and ultrastructure of a symbiotic marine dinoflagellate. J. Mar. Biol. Assoc. U. K. 48, 349–366 [Google Scholar]
- 110. Nishino T., Okamoto K., Eger B. T., Pai E. F., Nishino T. (2008) Mammalian xanthine oxidoreductase: Mechanism of transition from xanthine dehydrogenase to xanthine oxidase. FEBS J. 275, 3278–3289 [DOI] [PubMed] [Google Scholar]
- 111. Delevoye C., Nilges M., Dehoux P., Paumet F., Perrinet S., Dautry-Varsat A., Subtil A. (2008) SNARE protein mimicry by an intracellular bacterium. PLoS Pathog. 4, e1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Paumet F., Wesolowski J., Garcia-Diaz A., Delevoye C., Aulner N., Shuman H. A., Subtil A., Rothman J. E. (2009) Intracellular bacteria encode inhibitory SNARE-like proteins. PLoS One 4, e7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wesolowski J., Paumet F. (2010) SNARE motif: A common motif used by pathogens to manipulate membrane fusion. Virulence 1, 319–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Sharma M., Burré J., Südhof T. C. (2011) CSPα promotes SNARE-complex assembly by chaperoning SNAP-25 during synaptic activity. Nat. Cell Biol. 13, 30–39 [DOI] [PubMed] [Google Scholar]
- 115. Zinsmaier K. E., Bronk P. (2001) Molecular chaperones and the regulation of neurotransmitter exocytosis. Biochem. Physiol. 62, 1–11 [DOI] [PubMed] [Google Scholar]
- 116. Berkelmans R., van Oppen M. J. (2006) The role of zooxanthellae in the thermal tolerance of corals: “A nugget of hope” for coral reefs in an era of climate change. Proc. R. Soc. Lond. B Biol. Sci. 273, 2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Jones A. M., Berkelmans R., van Oppen M. J., Mieog J. C., Sinclair W. (2008) A community change in the algal endosymbionts of a scleractinian coral following a natural bleaching event: Field evidence of acclimatization. Proc. R. Soc. Lond. B Biol. Sci. 275, 1359–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lang A. S., Rise M. L., Culley A. I., Steward G. F. (2009) RNA viruses in the sea. FEMS Microbiol. Rev. 33, 295–323 [DOI] [PubMed] [Google Scholar]
- 119. Wilson W. H., Francis I., Ryan K., Davy K. (2001) Temperature induction of viruses in symbiotic dinoflagellates. Aquat. Microb. Ecol. 25, 99–102 [Google Scholar]
- 120. Lohr J., Munn C. B., Wilson W. H. (2007) Characterization of a latent virus-like infection of symbiotic zooxanthellae. Appl. Environ. Microbiol. 73, 2976–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Vega-Thurber R. L., Barott K. L., Liu H., Rodriguez-Mueller B., Desnues C., Edwards R. A., Haynes M., Angly F. E., Wegley L., Rohwer L. (2008). Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc. Natl. Acad. Sci. U.S.A. 105, 18413–18418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wilson W. H., Dale A. L., Davey J. E., Davey S. K. (2005) An enemy within? Observations of virus-like particles in reef corals. Coral Reefs 24, 145–148 [Google Scholar]
- 123. Marhaver K. L., Edwards R. A., Rohwer F. (2008) Viral communities associated with healthy and bleaching corals. Environ. Microbiol. 10, 2277–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. van Oppen M. J., Leong J. A., Gates R. D. (2009) Coral-virus interactions: A double-edged sword? Symbiosis 47, 1–8 [Google Scholar]


