Abstract
In the past 5–10 years, the power of the green fluorescent protein (GFP) and its numerous derivatives has been harnessed toward the development of genetically encoded fluorescent biosensors. These sensors are incorporated into cells or organisms as plasmid DNA, which leads the transcriptional and translational machinery of the cell to express a functional sensor. To date, over 100 different genetically encoded biosensors have been developed for targets as diverse as ions, molecules and enzymes. Such sensors are instrumental in providing a window into the real-time biochemistry of living cells and whole organisms, and are providing unprecedented insight into the inner workings of a cell.
Introduction
Several fluorescent sensors and probes are available that enable researchers to monitor ions, molecules, enzyme activities and channel conformational changes with exquisite spatial and temporal resolution in live cells. These probes facilitate not only measurement of steady-state concentrations or activity levels, but also observation and quantification of in vivo flux and kinetics. Genetically encoded fluorescent biosensors in particular hold great promise for enabling researchers to examine biochemical processes within the complex cellular context. Such sensors can be targeted to specific locations within a cell, expressed within specific cell types in a transgenic organism, and incorporated into organisms for long-term imaging. The sheer diversity of sensor platforms and targets is expanding rapidly, making a comprehensive review challenging. Therefore, this review focuses only on biosensors that are entirely genetically encoded (Box 1).
Box 1. Constructing a genetically encoded sensor.
Genetically encoded sensors are generated by translation of a nucleic acid sequence and incorporation into cells, tissues or organisms by transfection, electroporation or viral transduction of plasmid DNA (Figure I). Chemical transfection is generally the least labor-intensive method for introducing plasmid DNA into cells. For cell lines that are difficult to transfect (e.g. primary cells, post-mitotic cells), electroporation and viral transduction are more effective means of incorporating DNA. On incorporation of DNA, the cell then transcribes and translates the template into a fully functional protein-based sensor (Figure I). These sensors rely on the use of one or more fluorescent or luminescent proteins to generate a signal that is detectable above cellular autofluorescence. As a general design principle, these sensors convert a molecular event, such as the binding of a molecule to a sensing domain or a signal-induced change in protein conformation, into a change in fluorescence or luminescence. These changes include localization of fluorescence (Figure 1a), intensity of the fluorescence (Figure 1b,c), fluorescence resonance energy transfer (FRET) between two different FPs (Figure 1d–f), or bioluminescent resonance energy transfer (Figure 2). Sensors have been created for ions (e.g. Ca2+, Zn2+, H+, I−), molecules (e.g. NO, cAMP, ATP, phosphoinositides, glutamate, metabolites), enzyme activity (e.g. protease, kinase, GTPase, phosphatase) and other cellular events.
To provide insight into the overall design of genetically encoded fluorescent sensors and to emphasize properties of successful sensors, we highlight critical lessons learned over the past decade in optimizing genetically encoded FRET-based Ca2+ sensors. We also summarize various sensor platforms, noting classical examples of each, and provide important considerations for selecting the appropriate sensor for a given application. Interested readers are also pointed to recent reviews of genetically encoded sensors that focus on different aspects of sensor design and use [1–3].
Overview of sensor design platforms
The goal of fluorescence-based sensors used for live-cell imaging is to convert a molecular event into an optical signal that can be detected by a microscope. Over 100 different genetically encoded sensors have been developed for cellular targets as diverse as ions; molecules; enzymatic activity; oxidation-reduction events; changes in membrane potential and channel conformation; and phases of the cell cycle. The technical adaptability of genetically encoded sensors is exemplified by the fact that this vast array of sensors can be encompassed by a handful of straightforward design platforms (Figure 1), including translocation-based probes, intensity-based single FP probes, ratio-metric-based single FP probes, and FRET-based probes, where FRET can be altered by conformational change, enzyme activity, or enzymatic cleavage.
Figure 1.
Schematics of design platforms for fluorescent sensors. (a) A translocation-based probe for detecting the plasma membrane PtdIns(4,5)P2 concentration [48]. The PH domain of PLC-δ1, which can selectively recognize PtdIns(4,5)P2, is fused with GFP. When PtdIns(4,5)P2 in the plasma membrane is decreased, the sensor translocates from the plasma membrane to the cytosol, which increases cytosolic fluorescence. Examples include sensors for phosphoinositides. (b) An intensity-based single FP probe GCaMP2 [49,50]. GCaMP2 consists of the M13 fragment from myosin light chain kinase (shown in purple), a circularly permutated EGFP (shown in green) and calmodulin (CaM, shown in red). Ca2+ binding promotes the binding of M13 to CaM, which alters the protonation state of the chromophore, leading to an increase in fluorescence intensity. Classic examples include sensors for Ca2+, Cl− and H+. (c) A ratiometric single-FP-based redox sensor, roGFP [51]. The relative fluorescence intensity of the two excitation maxima of roGFP1 shifts depending on the redox state: reduction causes a decrease in the excitation at 400 nm and an increase in the excitation at 480 nm (arrows). Classic examples include the ratiometric H+ sensor pHlorin, the ratiometric Ca2+ sensor pericam, and sensors for cGMP and membrane potential. (d) A FRET sensor, ZapCY, activated by conformational change [52]. Conformational change of the Zn2+-binding domain (zinc fingers 1 and 2 of transcription factor Zap1) in the presence of Zn2+ leads to an increase in FRET between CFP and YFP. Examples of this sensing platform include cameleon Ca2+ sensors, as well as sensors for sugars, glutamate, Zn2+, cAMP, cGMP, NO and membrane potential. (e) The FRET-based sensor Phocus for kinase activity [53]. On phosphorylation of the substrate domain (shown in pink) by protein kinase, the adjacent phosphorylation recognition domain (shown in purple) binds to the phosphorylated substrate domain, which causes a change in FRET between CFP and YFP. Classic examples include sensors for kinases and GTPase activity (e.g. Raichu probes). (f) Schematic of a protease-activated FRET biosensor for caspase [54]. During apoptosis, activated caspase cleaves the DEVD amino acid sequence, which reduces the FRET between GFP and BFP. Examples of this sensor platform include those for caspases and matrix metalloproteases.
In addition to fluorescence, bioluminescence can be harnessed to generate sensors (Figure 2). Such sensors rely on bioluminescent proteins, such as Renilla luciferase, in which light is generated by a chemical reaction, the oxidation of coelenterazine, thus obviating the need for external excitation light [4]. The first bioluminescent sensor was the Ca2+ sensor aequorin, derived from the bioluminescent hydrozoan jellyfish Aequorea victoria. In fact, aequorin was the first sensor to be genetically targeted, in this case to mitochondria, and facilitated direct measurement of mitochondrial Ca2+ [5]. Luminescence can be used in combination with fluorescence in the case of bioluminescence resonance energy transfer (BRET) between a donor luciferase and an acceptor FP, typically a YFP variant such as citrine. This platform has been used in the cAMP biosensor depicted in Figure 2b [6]. In addition, luminescence can be used in lieu of fluorescence in sensors based on split luciferase, such as in a reporter of AKT kinase activity [7]. Although luminescence signals are weaker than corresponding fluorescence signals, necessitating sensitive signal detection, luminescence avoids the high background of cellular autofluorescence, and hence has a better overall signal-to-noise ratio. Luminescence-based sensors might be a more appropriate choice for high-throughput screening and in vivo imaging [4].
Figure 2.
Examples of sensors that use bioluminescence. (a) Aequorin senses Ca2+ [55]. First, coelenterazine is added to apo-aequorin to form the intermediate activated aequorin. Subsequently, on binding to Ca2+, the coelenterazine is oxidized, leading to emission of blue light at 480 nm. (b) Schematic of a BRET sensor CAMYEL used to measure cAMP concentrations [6]. The inactive cytosolic mutant form of the cAMP-binding protein human Epac-1 (shown in green) was flanked by citrine (in yellow) and Renilla luciferase (in grey). On oxidation of its cell-permeable substrate coelenterazine h, the luciferase emits light at 480 nm. Changes in cellular cAMP levels alter BRET between luciferase and citrine (excitation at 513 nm, emission at 530 nm) such that increases in cAMP cause BRET to increase.
Case study of Ca2+ sensors: lessons for sensor design
One of the oldest and most optimized families of genetically encoded sensors is that of the FRET-based Ca2+ sensors known as cameleons. These sensors were named after calmodulin (on which the original sensors were based) and also because they resemble chameleons by changing color on Ca2+ binding and their mechanism of action involves movement of a long ‘tongue’ (the peptide M13) in and out of the mouth of calmodulin, much like a chameleon capturing a fly. A historical account of the evolution of these sensors highlights the trials and tribulations of sensor optimization and provides numerous considerations for sensor design.
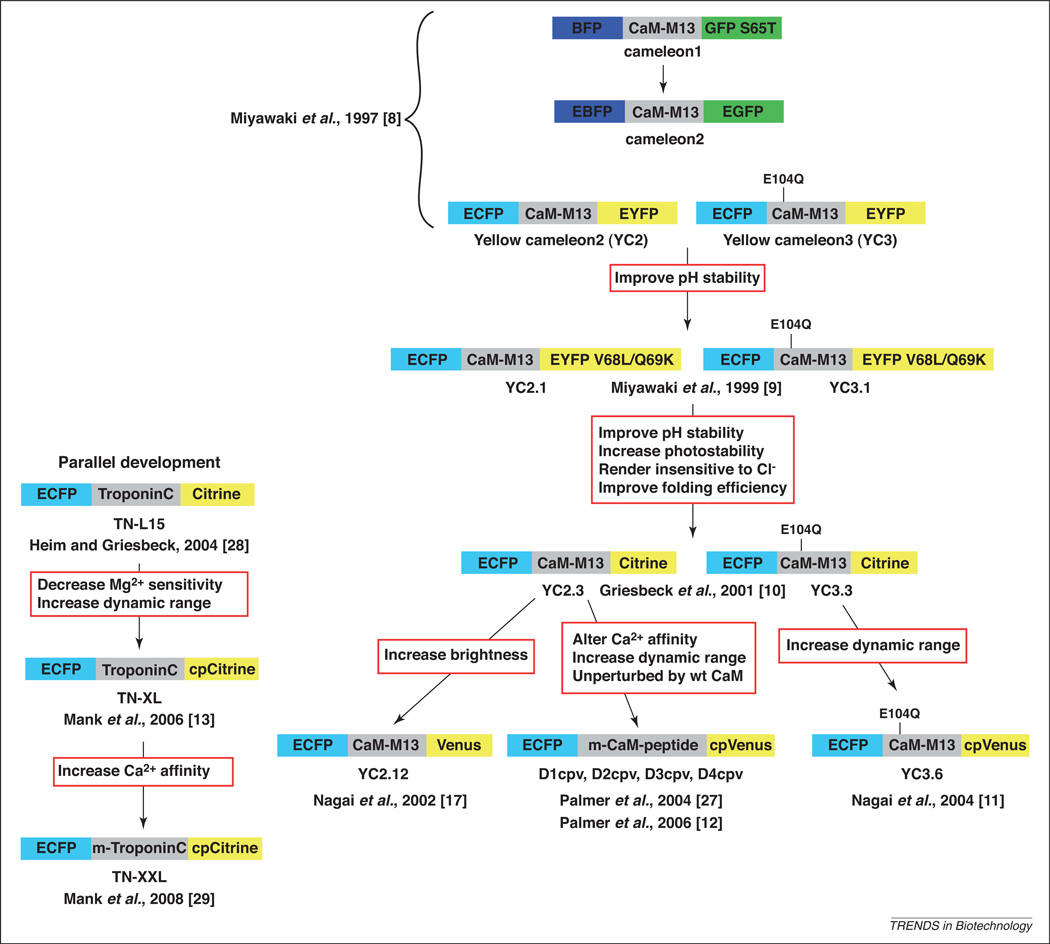
Figure 3 shows the cameleon family tree and outlines modifications that have been made to these sensors over the last 10 years or more. The first intramolecular FRET-based calcium sensor, dubbed cameleon 1, comprised calmodulin and a calmodulin-binding peptide sandwiched between the fluorescent proteins BFP and S65T GFP [8]. The red-shifted FRET pair ECFP and EYFP had more favorable properties for cellular imaging and led to the development of yellow cameleon 2 [8], commonly referred to as the YC family of cameleons. Since 1997, these cameleons have gone through iterative improvements involving replacement of EYFP with YFP variants that have a lower pKa, lower Cl− sensitivity, and lower susceptibility to photobleaching [9,10]. In more recent years, YFP has been replaced with circularly permuted (cp) YFP proteins (either cpCitrine or cpVenus) [11–13]. Circular permutation involves covalently linking the N and C termini through a peptide linker and cleaving the protein backbone at an alternative location, generating new N and C termini [14]. This transformation leads to the first critical lesson for design of genetically encoded sensors: the identity of the fluorescent proteins can have a profound impact on the sensor properties, and not necessarily in predictable ways. Certainly, it is logical that replacement of EYFP with more stable YFP variants (EYFP containing amino acid substitutions V68L/Q69K and, eventually, citrine) leads to sensors that are less perturbed by H+ and Cl−, and have enhanced photostability [9,10]. Surprisingly, however, replacement of ECFP with the substantially brighter Cerulean [15] actually led to diminished Ca2+-induced FRET changes in the troponin C family of cameleons [13]. This unexpected result is attributed to the fact that the Ca2+ response is reported as the increase in FRET divided by the decrease in donor emission and the decrease in Cerulean emission on Ca2+ binding was smaller than for ECFP emission, which leads to a smaller FRET change overall (130% vs 400%) [13]. Similarly, incorporation of the CyPet–YPet FP pair that is engineered for high FRET [16] does not yield improvements in the FRET response of D-family cameleons, perhaps owing to problems with fluorescent protein folding at 37 °C in mammalian cells (A. Palmer, unpublished data).
Figure 3.
Historical evolution of the cameleon family of Ca2+ sensors. These sensors use Ca2+ binding to a specific domain to induce a conformational change and to alter the energy transfer between two FPs. The original Ca2+-binding domain is derived from Xenopus calmodulin and the 26-residue M13 calmodulin-binding peptide from skeletal muscle myosin light chain kinase. The E104Q mutation in the yellow cameleon (i.e. YC family) decreases Ca2+ affinity.
In addition to affecting the photophysical properties of the sensor, the nature of the FP impacts biochemical properties, such as protein folding and hence stability, within a cell. Replacement of EYFP V68L/Q69K with either Citrine [10] or Venus [17] has led to enhanced folding at 37 °C and an increase in the effective brightness of the sensor. In addition, the order of FPs in a fusion construct (i.e. which FP is at the N terminus and which is at the C terminus) can affect the quality of FP folding and maturation (i.e. chromophore formation) and consequently fluorescence brightness and FRET response [18]. Different subcellular locations have unique chemical environments, such as high pH in mitochondria, low pH in vesicles and an oxidizing environment in the secretory pathway, and thus cellular location can affect FP folding and the sensor response. The resulting lesson is that the ideal FRET pair is capricious and likely to depend on fusion with the sensing domain and on the intended cellular use. Various combinations of donors (ECFP or Cerulean or circularly permuted variants thereof) with various acceptors (Citrine, Venus, or circularly permuted variants thereof) will probably need to be tested empirically.
This lesson might be particularly relevant as researchers move beyond cyan–yellow FRET pairs to green–red or orange–red pairs. These red-shifted fluorescent proteins are less influenced by cellular autofluorescence, which leads to increased contrast, and are also less prone to scatter, which makes them more appropriate for tissue and organism imaging [19]. Moreover, sensors with more diverse color palettes would facilitate efforts to use two or more sensors within the same cell. However, red-shifted FPs are used less in FRET-based biosensors. One complication is that RFPs are more susceptible to aggregation, are prone to incomplete chromophore formation, and perform more poorly in fusions than FP variants derived from A. victoria GFP [19]. Perhaps another challenge is the somewhat daunting list of RFPs and a lack of consensus over which FP is an optimal FRET partner. Interestingly, several successful sensors have been generated with alternative FRET pairs [20–25], with each research team selecting different FPs for the resulting sensor, which suggests that there is unlikely to be one optimal green–red, green–orange or orange–red FRET pair.
To create cameleon 1, Miyawaki and Tsien explored 28 different mutants with insertions, deletions and amino acid substitutions in the boundary regions between the FPs and the Ca2+-sensing domains [18] and found that subtle changes in the number and identity of amino acids has a profound impact on the FRET change. They postulated that FP orientation (as opposed to distance) might have the greatest impact on the overall dynamic range, defined as the maximum FRET ratio (Rmax) divided by the minimum FRET ratio (Rmin). This prediction was validated by the cameleons, because replacement of the acceptor FP with a circularly permuted variant, which alters the orientation of the acceptor FP with respect to the donor FP, led to a >300% increase in the dynamic range of a number of cameleon sensors [11–13]. This approach has also served to enhance the dynamic range of other FRET-based sensors [2], which suggests that it might be a universal mechanism for improving sensors. In an alternative approach, explicit promotion of reversible dimerization between two FP variants can increase the dynamic range and impact the apparent affinity for a targeted ligand [26]. Dimerization between FPs in the ligand-bound state might reduce the off-rate and hence increase the fraction of protein molecules in the high FRET state, thus enhancing the signal change. Although there are no published reports that apply this approach to other FRET-based sensors, it is reasonable to postulate that promotion of intramolecular FP dimerization could be a general mechanism for enhancing the dynamic range. Given that the vast majority of FRET-based sensors exhibit modest (15–30%) FRET ratio changes (and these are often not sensitive enough for use in transgenic organisms, as discussed below), these explicit methods for optimizing the dynamic range could improve overall sensor functionality.
Over the years it has become apparent that in designing sensors it might be important to reduce the likelihood that a sensor can be perturbed by and/or perturbing to the cellular environment. The FRET-based cameleons based on calmodulin are a classic example. Calmodulin is an attractive platform for sensor design because it converts a Ca2+ binding event into a conformational change that increases its affinity for target peptides. However, calmodulin is a highly abundant, ubiquitous protein and has numerous binding targets within a cell. Indeed, it has been suggested that YC cameleons targeted to some subcellular locations, in certain cell types and in long-term expression in transgenic organisms, lead to aberrant responses [12]. It was postulated that the calmodulin-binding sensor proteins were partially to blame. Engineering of calmodulin–peptide pairs that do not bind cellular proteins such as endogenous calmodulin has apparently fixed this problem, and has led to the development of improved cameleon sensors [12,27]. In an alternative approach, calmodulin was replaced with a less widely expressed Ca2+-binding domain, troponin C, and a series of optimizations generated a robust cameleon (termed TN-XXL) that is fully functional in neurons and is not perturbed by the cellular environment [13,28,29].
Incorporating sensors into transgenic organisms
One of the more exciting applications of genetically encoded sensors is the potential to create transgenic organisms for long-term investigation of signaling processes during development, disease progression and aging. To this end, a variety of genetically encoded sensors have been optimized for in vivo applications (Table 1). Ideally, genetically encoded sensors facilitate observation of the physiology and pathology of organisms without perturbing them. However, optimization of sensors for in vivo applications has not been straightforward, as discussed in recent reviews [30,31]. One common difficulty is that the signal-to-noise ratio and dynamic range of genetically encoded biosensors decrease significantly in vivo. This is partly because the majority of sensors use CFP–YFP; blue–yellow light is therefore sub-optimal for in vivo optical imaging owing to poor tissue penetration, interference from cellular autofluorescence, and increased scatter compared to lower-energy red light [19]. This problem might be partly overcome by using genetically encoded sensors with red-shifted FPs, particularly those that absorb and emit in the far-red visible spectrum [32].
Table 1.
Selected examples of organisms expressing genetically encoded sensors
| Target molecule or cellular signal | Sensor name or family | Species | Refs |
|---|---|---|---|
| Ca2+ | Cameleon family | Mus musculus | [56–59] |
| Danio rerio | [38,60,61] | ||
| Drosophila melanogaster | [62–64] | ||
| Caenorhabditis elegans | [39] | ||
| Arabidopsis thaliana | [35,36,65,66] | ||
| Nicotiana tabacum | [66] | ||
| Medicago truncatula | [67,68] | ||
| Ca2+ | GCaMP family | M. musculus | [33,69,70] |
| D. rerio | [64,71,72] | ||
| D. melanogaster | [33,40] | ||
| C. elegans | [33,73] | ||
| Ca2+ | Troponin C-based sensor | M. musculus | [29,74] |
| D. melanogaster | [29,64] | ||
| Ca2+ | Pericam family | D. rerio | [75] |
| D. melanogaster | [64] | ||
| cAMP | HCN2-camps | M. musculus | [76] |
| cAMP | GFP-PKA FRET sensor | D. melanogaster | [77] |
| Cell cycle | zFucci (Invitrogen) | D. rerio | [34] |
| Cl− | Clomeleon | M. musculus | [37,78–81] |
| A. thaliana | [82] | ||
| M. truncatula | [83] | ||
| H+ (i.e. pH) | synaptopHluorin | M. musculus | [84] |
| D. melanogaster | [64] | ||
| Hydrogen peroxide (H2O2) | HyPer (Evrogen) | D. rerio | [85] |
| A. thaliana | [35] | ||
| Myosin light chain kinase (MLCK) activation | MLCK FRET sensor | M. musculus | [86–88] |
| Phosphatidylinositol 4-phosphate | YFP-PH | A. thaliana | [83] |
| M. truncatula | [83] | ||
| Rac1 and RhoA activation | Rac1 and RhoA FRET sensors | D. rerio | [89] |
| Redox state | roGFP2 | A. thaliana | [65] |
| Voltage | hVOS | D. melanogaster | [90] |
An alternative approach would be to use sensors with luminescent rather than fluorescent output, because these probes circumvent the need for excitation light that is scattered by tissue, which limits the penetration depth. However, bioluminescent sensors require incorporation of the coelenterazine substrate, which might complicate in vivo studies. Another challenge associated with incorporation of genetically encoded sensors into transgenic organisms is that over-expression of some sensors has led to abnormal behavior or development. For example, GCaMP1 and GCaMP2, but not GCaMP3, led to a behavioral defect in Caenorhabditis elegans involving decreased local search turning [33]. Occasionally, species-specific optimizations are necessary: the Fucci cell cycle sensor, based on human proteins, has to be modified to retain functionality in developing zebrafish embryos (dubbed zFucci for zebrafish) (Table 1) [34]. Conversely, several sensors have been expressed in plants, such as Arabidopsis thaliana, with little species-specific modification (Table 1) [35,36]. In addition, targeted expression in specific cell types, particularly in the mammalian brain, can be difficult, as evidenced by thy1-driven expression of various sensors [31,37].
Although the creation of transgenic organisms expressing genetically encoded sensors has involved some challenges, continual optimization of sensors for in vivo imaging has led to impressive successes in a variety of organisms (Table 1). Yellow cameleon YC2.1 was unsuccessful in measuring Ca2+ transients in the olfactory bulb of transgenic zebrafish [38], however an improved YC6.1 has been successful in moving transgenic C. elegans to observe physiologic pulses of calcium [39]. More recently, GCaMP3 has facilitated observation of calcium transients in the cortex of live walking Drosophila [40] and functional imaging of hippocampal neurons in mice during virtual navigation [41]. In such studies, the power of genetically encoded sensors to visualize the neuronal circuitry and how it is modified during behavioral tasks is beginning to be realized.
Identifying the right sensor for a given application
Given the plethora of both genetically encoded and small-molecule sensors for cellular imaging, researchers must consider several criteria to determine the appropriate sensor for a given application. Despite the many advantages of each type of sensor, no single sensor or sensing platform is likely to address all possible applications. Instead, the optimal sensor often depends on the l experimental considerations highlighted below, including ease of use, sensitivity, kinetics, signal location and quantification.
Ease of use
One important consideration in choosing a sensor is the availability of equipment. For example, ratiometric- and FRET-based sensors require collection of data at two wavelengths, which necessitates instruments with multiple filters and the hardware to rapidly (or simultaneously) collect data from two fluorescence channels. Conversely, intensity-based sensors require data collection at only one wavelength, so simpler instrumentation is necessary for data collection. The availability of supplies must also be considered; small-molecule-based probes must be continually purchased, whereas genetically encoded sensors (Box 1) are infinitely renewable because DNA can easily be re-amplified. In addition, some small-molecule staining protocols can be onerous, particularly if the sensor is not cell-permeable and requires microinjection. Finally, many bioluminescent probes require the addition of a substrate, such as coelenterazine [4]. This is a small hydrophobic molecule that readily crosses the cell membrane, but the extra step is a consideration is in deciding which sensor possesses the most desirable properties.
Sensitivity
It is essential to match the sensitivity of a probe to a given application. For example, to measure steady-state concentrations of a molecule or ion, it is advisable to select a sensor with an apparent dissociation constant (Kd′) at or near the expected concentration. An even more rigorous approach is to measure the resting concentration with several sensors, each with a slightly different (Kd′) value, as was done in measuring Zn2+ levels in pancreatic β cells [42]. If the goal is to measure a change in concentration or activity when the signal is expected to be small, the probe with the greatest sensitivity will be that with the highest dynamic range and a Kd′ value such that the concentration change is within ±5 × Kd′ (i.e. the steep part of the binding curve). For genetically encoded Ca2+ sensors, optimization of the dynamic range and consideration of the Kd′) value have been essential steps toward facilitating measurement of single action potentials in neurons [33,43].
Kinetics
Because cellular signals occur over a range of time scales, the kinetics of the process being studied will also dictate the optimal sensor to use. Small molecules often offer faster on and off rates (kon and koff) and thus give better resolution of events that occur on rapid time scales. Even within families of genetically encoded sensors, individual sensors can differ significantly in their kinetics [13,27,33]. The general rule of thumb is that the sensor kinetics should be able to faithfully reproduce the biological signal. However, there might be some advantage in a slow koff in facilitating integration and hence detection of very weak signals [33].
Signal location
Genetically encoded sensors can easily be targeted to specific organelles or cellular domains by incorporation of localization signals, and such sensors have provided critical insight into the spatial heterogeneity of cellular signaling processes. Targeting of a fluorescent reporter of protein kinase C (PKC) to the cytosol, nucleus, plasma membrane, mitochondrial membrane and Golgi membrane has revealed differences in the magnitude and duration of PKC activity in different locations within a cell [44]. It is important to recognize that the environment (e.g. pH, redox state) of a given organelle might limit the available sensors. For example, endocytic vesicles maintain an acidic pH (<6), which quenches the fluorescence of YFP and thus precludes the use of CFY-YFP sensors in such acidic compartments.
Quantification
For events that require quantification, ratiometric- or FRET-based probes are generally preferred over intensity-based probes (Figure 1). Although intensity-based sensors typically have greater dynamic ranges, the intensity depends on the target molecule, sensor expression levels within a cell or organelle, and cell thickness in the region of interest. Therefore, these sensors are at a disadvantage for quantifying steady-state levels or concentration changes.
To compare sensor responses between one cell and another or one stimulus and another, the probe should be calibrated in situ to measure the minimum and maximum signals (either fluorescence intensity or FRET ratio, depending on the probe) in each individual cell. Although individual signals can vary from cell to cell, the overall dynamic range (max. signal divided by min. signal) should be consistent. For quantitative experiments, care should be taken to minimize photobleaching by using the lowest excitation intensity and/or exposure time necessary to achieve an acceptable signal-to-noise ratio. For a 16-bit camera, a good rule of thumb is to ensure the lowest signal is at least 1000 counts above the background. For all sensors, it is important to ensure that the sensor response is independent of the amount of sensor expressed within the cell. For ratio-metric probes, such as those relying on intra-molecular FRET, the sensor concentration can be directly estimated from the intensity in the acceptor FP fluorescence channel. The ratio or response can then be plotted as a function of sensor concentration. Researchers should endeavor to select cells with sensor concentrations in a range for which there is no strong correlation between the ratio or response and the sensor concentration, because strong correlation could lead to misinterpretation of cellular data.
Outlook
Genetically encoded sensors are rapidly transforming our understanding of cellular signaling processes and opening up new avenues of research in whole organisms, such as imaging of signal transduction during behavioral tasks in awake animals [40,41]. Sensors for virtually any target can be created from a few general platforms. Although first-generation sensors might have modest responses, further optimization can enhance output. These sensors have been developed primarily to explore fundamental cell biology; however, exciting future applications include the use of sensors in high-throughput screening, including genome-wide screens to identify genes involved in regulatory and signaling processes, as well as drug discovery screens to identify drug-like compounds with bioactivity. Both of these applications are beginning to be realized.
Genetically encoded Ca2+ sensors (e.g. pericam, mitochondrial-targeted aequorin) have recently been used in screens to identify highly elusive proteins in mitochondrial Ca2+ regulation [45,46]. Similarly, genetically encoded Cl− sensors have been used to identify activators, inhibitors, potentiators and correctors of the defective Cl− channel cystic fibrosis transmembrane conductance regulator (CTFR) [47] and identified chlorine channels as potential drug targets. The advantage of genetically encoded sensors in such screens is that they can be stably incorporated into cells to facilitate the generation of a screening cell line. The greatest challenge for such screens is to obtain a favorable Z′ factor (Z′ >0.5), which compares the dynamic range of the assay to variability in the data. With the expansion and improvement of red-shifted FPs, opportunities are ripe for developing red-shifted sensors to examine multiple signaling pathways in parallel and to improve applications in transgenic organisms, for example. A recent study used red-shifted sensors for cAMP and protein kinase A (PKA) activity in conjunction with traditional Ca2+ indicators to examine Ca2+–cAMP–PKA oscillations; complex feedback circuits in insulin-secreting MIN6 cells were identified as a result [25]. All of these future applications would greatly benefit from a better understanding of systematic principles for sensor optimization and high-throughput ways to screen and select optimal sensors.
Figure I.
Steps involved in engineering a genetically encoded sensor.
Acknowledgements
We would like to acknowledge the following sources of financial support: Signaling and Cell Cycle Regulation Training Grant (NIH T32 GM08759 to J.E.M.), NIH GM084027 to A.E.P., and the University of Colorado.
References
- 1.Ibraheem A, Campbell RE. Designs and applications of fluorescent protein-based biosensors. Curr. Opin. Chem. Biol. 2010;14:30–36. doi: 10.1016/j.cbpa.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 2.Herbst KJ, et al. Dynamic visualization of signal transduction in living cells: from second messengers to kinases. IUBMB Life. 2009;61:902–908. doi: 10.1002/iub.232. [DOI] [PubMed] [Google Scholar]
- 3.Okumoto S. Imaging approach for monitoring cellular metabolites and ions using genetically encoded biosensors. Curr. Opin. Biotechnol. 2010;21:45–54. doi: 10.1016/j.copbio.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfleger KD, Eidne KA. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET) Nat. Methods. 2006;3:165–174. doi: 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
- 5.Rizzuto R, et al. Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin. Nature. 1992;358:325–327. doi: 10.1038/358325a0. [DOI] [PubMed] [Google Scholar]
- 6.Jiang LI, et al. Use of a cAMP BRET sensor to characterize a novel regulation of cAMP by the sphingosine 1-phosphate/G13 pathway. J. Biol. Chem. 2007;282:10576–10584. doi: 10.1074/jbc.M609695200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, et al. Molecular imaging of Akt kinase activity. Nat. Med. 2007;13:1114–1119. doi: 10.1038/nm1608. [DOI] [PubMed] [Google Scholar]
- 8.Miyawaki A, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 9.Miyawaki A, et al. Dynamic and quantitative Ca2+ measurements using improved cameleons. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2135–2140. doi: 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griesbeck O, et al. Reducing the environmental sensitivity of yellow fluorescent protein. J. Biol. Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 11.Nagai T, et al. Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer AE, et al. Ca2+ indicators based on computationally-redesigned calmodulin-peptide pairs. Chem. Biol. 2006;13:521–530. doi: 10.1016/j.chembiol.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Mank M, et al. A FRET-based calcium biosensor with fast signal kinetics and high fluorescence change. Biophys. J. 2006;90:1790–1796. doi: 10.1529/biophysj.105.073536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luger K, et al. Correct folding of circularly permuted variants of a beta alpha barrel enzyme in vivo. Science. 1989;243:206–210. doi: 10.1126/science.2643160. [DOI] [PubMed] [Google Scholar]
- 15.Rizzo MA, et al. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 17.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 18.Miyawaki A, Tsien RY. Monitoring protein conformations and interactions by fluorescence resonance energy transfer between mutants of green fluorescent protein. Methods Enzymol. 2000;327:472–500. doi: 10.1016/s0076-6879(00)27297-2. [DOI] [PubMed] [Google Scholar]
- 19.Davidson MW, Campbell RE. Engineered fluorescent proteins: innovations and applications. Nat. Methods. 2009;6:713–717. doi: 10.1038/nmeth1009-713. [DOI] [PubMed] [Google Scholar]
- 20.Ai HW, et al. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat. Methods. 2008;5:401–403. doi: 10.1038/nmeth.1207. [DOI] [PubMed] [Google Scholar]
- 21.Niino Y, et al. Simultaneous live cell imaging using dual FRET sensors with a single excitation light. PLoS ONE. 2009;4:e6036. doi: 10.1371/journal.pone.0006036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piljic A, Schultz C. Simultaneous recording of multiple cellular events by FRET. ACS Chem. Biol. 2008;3:156–160. doi: 10.1021/cb700247q. [DOI] [PubMed] [Google Scholar]
- 23.Grant DM, et al. Multiplexed FRET to image multiple signaling events in live cells. Biophys. J. 2008;95:L69–L71. doi: 10.1529/biophysj.108.139204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouyang M, et al. Simultaneous visualization of protumorigenic Src and MT1-MMP activities with fluorescence resonance energy transfer. Cancer Res. 2010;70:2204–2212. doi: 10.1158/0008-5472.CAN-09-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ni Q. Signaling diversity of PKA achieved via a Ca2+–cAMP–PKA oscillatory circuit. Nat. Chem. Biol. 2011;7:34–40. doi: 10.1038/nchembio.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotera I, et al. Reversible dimerization of Aequorea victoria fluorescent proteins increases the dynamic range of FRET-based indicators. ACS Chem. Biol. 2010;5:215–222. doi: 10.1021/cb900263z. [DOI] [PubMed] [Google Scholar]
- 27.Palmer AE, et al. Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17404–17409. doi: 10.1073/pnas.0408030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heim N, Griesbeck O. Genetically encoded indicators of cellular calcium dynamics based on troponin C and green fluorescent protein. J. Biol. Chem. 2004;279:14280–14286. doi: 10.1074/jbc.M312751200. [DOI] [PubMed] [Google Scholar]
- 29.Mank M, et al. A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat. Methods. 2008;5:805–811. doi: 10.1038/nmeth.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barth AL. Visualizing circuits and systems using transgenic reporters of neural activity. Curr. Opin. Neurobiol. 2007;17:567–571. doi: 10.1016/j.conb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Misgeld T, Kerschensteiner M. In vivo imaging of the diseased nervous system. Nat. Rev. Neurosci. 2006;7:449–463. doi: 10.1038/nrn1905. [DOI] [PubMed] [Google Scholar]
- 32.Shcherbo D, et al. Near-infrared fluorescent proteins. Nat. Methods. 2010;7:827–829. doi: 10.1038/nmeth.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugiyama M, et al. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc. Natl. Acad. Sci. U.S.A. 2009;106:20812–20817. doi: 10.1073/pnas.0906464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costa A, et al. H2O2 in plant peroxisomes: an in vivo analysis uncovers a Ca2+-dependent scavenging system. Plant J. 2010;62:760–772. doi: 10.1111/j.1365-313X.2010.04190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rincón-Zachary M, et al. Fluorescence resonance energy transfer-sensitized emission of yellow cameleon 3.60 reveals root zone-specific calcium signatures in Arabidopsis in response to aluminum and other trivalent cations. Plant Physiol. 2010;152:1442–1458. doi: 10.1104/pp.109.147256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berglund K, et al. Imaging synaptic inhibition throughout the brain via genetically targeted Clomeleon. Brain Cell Biol. 2008;36:101–118. doi: 10.1007/s11068-008-9031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yaksi E, et al. Topological reorganization of odor representations in the olfactory bulb. PLoS Biol. 2007;5:e178. doi: 10.1371/journal.pbio.0050178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nehrke K, et al. Intestinal Ca2+ wave dynamics in freely moving C. elegans coordinate execution of a rhythmic motor program. Am. J. Physiol. Cell Physiol. 2008;294:C333–C344. doi: 10.1152/ajpcell.00303.2007. [DOI] [PubMed] [Google Scholar]
- 40.Seelig JD, et al. Two-photon calcium imaging from head-fixed Drosophila during optomotor walking behavior. Nat. Methods. 2010;7:535–540. doi: 10.1038/nmeth.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dombeck DA, et al. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat. Neurosci. 2010;13:1433–1440. doi: 10.1038/nn.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vinkenborg JL, et al. Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat. Methods. 2009;6:737–740. doi: 10.1038/nmeth.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasan MT, et al. Functional fluorescent Ca2+ indicator proteins in transgenic mice under TET control. PLoS. 2004;2:763–775. doi: 10.1371/journal.pbio.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallegos LL, et al. Targeting protein kinase C activity reporter to discrete intracellular regions reveals spatiotemporal differences in agonist-dependent signaling. J. Biol. Chem. 2006;281:30947–30956. doi: 10.1074/jbc.M603741200. [DOI] [PubMed] [Google Scholar]
- 45.Jiang D, et al. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science. 2009;326:144–147. doi: 10.1126/science.1175145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perocchi F, et al. MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake. Nature. 2010;467:291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verkman AS, Galietta LJ. Chloride channels as drug targets. Nat. Rev. Drug Discov. 2009;8:153–171. doi: 10.1038/nrd2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stauffer TP, et al. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- 49.Wang Q, et al. Structural basis for calcium sensing by GCaMP2. Structure. 2008;16:1817–1827. doi: 10.1016/j.str.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akerboom J, et al. Crystal structures of the GCaMP calcium sensor reveal the mechanism of fluorescence signal change and aid rational design. J. Biol. Chem. 2009;284:6455–6464. doi: 10.1074/jbc.M807657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dooley CT, et al. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J. Biol. Chem. 2004;279:22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 52.Qiao W, et al. Zinc binding to a regulatory zinc-sensing domain monitored in vivo by using FRET. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8674–8679. doi: 10.1073/pnas.0600928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato M, et al. Fluorescent indicators for imaging protein phosphorylation in single living cells. Nat. Biotechnol. 2002;20:287–294. doi: 10.1038/nbt0302-287. [DOI] [PubMed] [Google Scholar]
- 54.Xu Q, Reed JC. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol. Cell. 1998;1:337–346. doi: 10.1016/s1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- 55.Deng L, et al. Crystal structure of a Ca2+-discharged photoprotein: implications for mechanisms of the calcium trigger and bioluminescence. J. Biol. Chem. 2004;279:33647–33652. doi: 10.1074/jbc.M402427200. [DOI] [PubMed] [Google Scholar]
- 56.Hara M, et al. Imaging endoplasmic reticulum calcium with a fluorescent biosensor in transgenic mice. Am. J. Physiol. Cell Physiol. 2004;287:C932–C938. doi: 10.1152/ajpcell.00151.2004. [DOI] [PubMed] [Google Scholar]
- 57.Nyqvist D, et al. Pancreatic islet function in a transgenic mouse expressing fluorescent protein. J. Endocrinol. 2005;186:333–341. doi: 10.1677/joe.1.06204. [DOI] [PubMed] [Google Scholar]
- 58.Tsujino N, et al. Cholecystokinin activates orexin/hypocretin neurons through the cholecystokinin A receptor. J. Neurosci. 2005;25:7459–7469. doi: 10.1523/JNEUROSCI.1193-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kotlikoff MI. Genetically encoded Ca2+ indicators: using genetics and molecular design to understand complex physiology. J. Physiol. 2007;578:55–67. doi: 10.1113/jphysiol.2006.120212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Higashijima S, et al. Imaging neuronal activity during zebrafish behavior with a genetically encoded calcium indicator. J. Neurophysiol. 2003;90:3986–3997. doi: 10.1152/jn.00576.2003. [DOI] [PubMed] [Google Scholar]
- 61.Tsuruwaka Y, et al. Real-time monitoring of dynamic intracellular Ca2+ movement during early embryogenesis through expression of yellow cameleon. Zebrafish. 2008;4:253–260. doi: 10.1089/zeb.2007.0519. [DOI] [PubMed] [Google Scholar]
- 62.Fiala A, et al. Genetically expressed cameleon in Drosophila melanogaster is used to visualize olfactory information in projection neurons. Curr. Biol. 2002;12:1877–1884. doi: 10.1016/s0960-9822(02)01239-3. [DOI] [PubMed] [Google Scholar]
- 63.Liu L, et al. Identification and function of thermosensory neurons in Drosophila larvae. Nat. Neurosci. 2003;6:267–273. doi: 10.1038/nn1009. [DOI] [PubMed] [Google Scholar]
- 64.Reiff DF, et al. In vivo performance of genetically encoded indicators of neural activity in flies. J. Neurosci. 2005;25:4766–4778. doi: 10.1523/JNEUROSCI.4900-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meyer AJ, et al. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 2007;52:973–986. doi: 10.1111/j.1365-313X.2007.03280.x. [DOI] [PubMed] [Google Scholar]
- 66.Iwano M, et al. Fine-tuning of the cytoplasmic Ca2+ concentration is essential for pollen tube growth. Plant Physiol. 2009;150:1322–1334. doi: 10.1104/pp.109.139329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miwa H, et al. Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J. 2006;48:883–894. doi: 10.1111/j.1365-313X.2006.02926.x. [DOI] [PubMed] [Google Scholar]
- 68.Sieberer BJ, et al. A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol. 2009;151:1197–1206. doi: 10.1104/pp.109.142851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fletcher ML, et al. Optical imaging of postsynaptic odor representation in the glomerular layer of the mouse olfactory bulb. J. Neurophysiol. 2009;102:817–830. doi: 10.1152/jn.00020.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tallini YN, et al. Imaging cellular signals in the heart in vivo: cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4753–4758. doi: 10.1073/pnas.0509378103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chi NC, et al. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008;6:e109. doi: 10.1371/journal.pbio.0060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dreosti E, et al. A genetically encoded reporter of synaptic activity in vivo. Nat. Methods. 2009;6:883–889. doi: 10.1038/nmeth.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsunozaki M, et al. A behavioral switch: cGMP and PKC signaling in olfactory neurons reverses odor preference in C. elegans. Neuron. 2008;59:959–971. doi: 10.1016/j.neuron.2008.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heim N, et al. Improved calcium imaging in transgenic mice expressing a troponin C-based biosensor. Nat. Methods. 2007;4:127–129. doi: 10.1038/nmeth1009. [DOI] [PubMed] [Google Scholar]
- 75.Li J, et al. Early development of functional spatial maps in the zebrafish olfactory bulb. J. Neurosci. 2005;25:5784–5795. doi: 10.1523/JNEUROSCI.0922-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nikolaev VO, et al. Cyclic AMP imaging in adult cardiac myocytes reveals far-reaching beta1-adrenergic but locally confined beta2-adrenergic receptor-mediated signaling. Circ. Res. 2006;99:1084–1091. doi: 10.1161/01.RES.0000250046.69918.d5. [DOI] [PubMed] [Google Scholar]
- 77.Lissandron V, et al. Transgenic fruit-flies expressing a FRET-based sensor for in vivo imaging of cAMP dynamics. Cell Signal. 2007;19:2296–2303. doi: 10.1016/j.cellsig.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 78.Berglund K, et al. Imaging synaptic inhibition in transgenic mice expressing the chloride indicator. Clomeleon. Brain Cell Biol. 2006;35:207–228. doi: 10.1007/s11068-008-9019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haverkamp S, et al. The primordial, blue-cone color system of the mouse retina. J. Neurosci. 2005;25:5438–5445. doi: 10.1523/JNEUROSCI.1117-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pond BB, et al. The chloride transporter Na+-K+-Cl− cotransporter isoform-1 contributes to intracellular chloride increases after in vitro ischemia. J. Neurosci. 2006;26:1396–1406. doi: 10.1523/JNEUROSCI.1421-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Duebel J, et al. Two-photon imaging reveals somatodendritic chloride gradient in retinal ON-type bipolar cells expressing the biosensor Clomeleon. Neuron. 2006;49:81–94. doi: 10.1016/j.neuron.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 82.Lorenzen I, et al. Salt stress-induced chloride flux: a study using transgenic Arabidopsis expressing a fluorescent anion probe. Plant J. 2004;38:539–544. doi: 10.1111/j.0960-7412.2004.02053.x. [DOI] [PubMed] [Google Scholar]
- 83.Vermeer JEM, et al. Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. Plant J. 2009;57:356–372. doi: 10.1111/j.1365-313X.2008.03679.x. [DOI] [PubMed] [Google Scholar]
- 84.Wyatt RM, Balice-Gordon RJ. Heterogeneity in synaptic vesicle release at neuromuscular synapses of mice expressing synaptopHluorin. J. Neurosci. 2008;28:325–335. doi: 10.1523/JNEUROSCI.3544-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Niethammer P, et al. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Isotani E, et al. Real-time evaluation of myosin light chain kinase activation in smooth muscle tissues from a transgenic calmodulin-biosensor mouse. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6279–6284. doi: 10.1073/pnas.0308742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Raina H, et al. Activation by Ca2+/calmodulin of an exogenous myosin light chain kinase in mouse arteries. J. Physiol. 2009;587:2599–2612. doi: 10.1113/jphysiol.2008.165258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wier WG, et al. A technique for simultaneous measurement of Ca2+, FRET fluorescence and force in intact mouse small arteries. J. Physiol. 2008;586:2437–2443. doi: 10.1113/jphysiol.2008.151522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kardash E, et al. A role for Rho GTPases and cell–cell adhesion in single-cell motility in vivo. Nat. Cell Biol. 2010;12:47–53. doi: 10.1038/ncb2003. [DOI] [PubMed] [Google Scholar]
- 90.Sjulson L, Miesenböck G. Rational optimization and imaging in vivo of a genetically encoded optical voltage reporter. J. Neurosci. 2008;28:5582–5593. doi: 10.1523/JNEUROSCI.0055-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]