Abstract
Preclinical biodistribution studies with INGN 007, an oncolytic adenovirus (Ad) vector, supporting an early stage clinical trial were conducted in Syrian hamsters, which are permissive for Ad replication, and mice, which are a standard model for assessing toxicity and biodistribution of replication-defective (RD) Ad vectors. Vector dissemination and pharmacokinetics following intravenous administration were examined by real-time PCR in nine tissues and blood at five time points spanning 1 year. Select organs were also examined for the presence of infectious vector/virus. INGN 007 (VRX-007), wild-type Ad5 and AdCMVpA (an RD vector) were compared in the hamster model, whereas only INGN 007 was examined in mice. DNA of all vectors was widely disseminated early after injection, but decayed rapidly in most organs. In the hamster model, DNA of INGN 007 and Ad5 was more abundant than that of the RD vector AdCMVpA at early times after injection, but similar levels were seen later. An increased level of INGN 007 and Ad5 DNA but not AdCMVpA DNA in certain organs early after injection, and the presence of infectious INGN 007 and Ad5 in lung and liver samples at early times after injection, strongly suggests that replication of INGN 007 and Ad5 occurred in several Syrian hamster organs. There was no evidence of INGN 007 replication in mice. In addition to providing important information about INGN 007, the results underscore the utility of the Syrian hamster as a permissive immunocompetent model for Ad5 pathogenesis and oncolytic Ad vectors.
Keywords: adenovirus, biodistribution, oncolytic, Syrian hamster, mice, preclinical
Introduction
An emerging modality for the treatment of cancer is the use of oncolytic (replication competent, RC) viral vectors whose therapeutic principle is multiple rounds of lytic vector replication resulting in widespread tumor cell destruction.1 Vectors from many different virus families are being explored as oncolytic agents including those based on wild-type (wt) human adenovirus (Ad) serotype 5 (Ad5). Most oncolytic Ad vectors, including the best-characterized ONYX-015,2 are genetically engineered to replicate preferentially in neoplastic cells as opposed to normal cells. However, these genetic alterations frequently attenuate vector replication, which is in opposition to the therapeutic principle. Although Ad can be safely administered to humans,3 promising preclinical studies have not been translated into similar efficacy in cancer patients. Efficacy has been modest even when combined with radiation or chemotherapy. These clinical data underscore the need for oncolytic vectors with improved efficacy to achieve successful clinical translation of this new treatment.
The oncolytic Ad vector INGN 007 (VRX-007) was designed to maximize vector replication. INGN 007, which is based on Ad5, does not contain a genetic alteration to restrict replication to malignant cells. However, INGN 007 was engineered to overexpress the Ad-encoded protein named adenovirus death protein (ADP; formerly named E3-11.6K).4 This viral protein, which is required for efficient release of Ad at the culmination of the infection cycle,5–8 enhances the cell to cell spread of vectors in which it is overexpressed and improves efficacy in tumor xenograft models.9–15 Other groups have also incorporated ADP into their oncolytic Ad vectors with beneficial results.16–23
Enhanced efficacy, however, cannot be achieved at the expense of vector safety. The safety characteristics of a vector are determined in part by the distribution of the vector within the host. Biodistribution studies with RC Ad vectors have largely been conducted in mice.24–29 However, the utility of this model with respect to RC Ad vectors is questionable because Ad replication in normal mouse tissues is inefficient at best.30–34 Biodistribution and safety studies with oncolytic vectors require a permissive, immunocompetent animal model in which the effect of vector replication in normal host tissues and the immune system response to infection can be examined. Although animal models other than the mouse have been explored, these models are only semipermissive, expensive or difficult to work with.35–43 Our laboratory has recently developed the golden Syrian hamster (Mesocricetus auratus) as a permissive, immunocompetent animal model for efficacy studies of oncolytic Ad vectors44,45 and as a model for Ad pathogenicity and testing of anti-Ad drugs.46 We now report the first comprehensive biodistribution study of an oncolytic Ad vector in the Syrian hamster model. In addition, INGN 007 biodistribution was examined in C57BL/6 mice, a model in which the biodistribution of previous Adbased vectors has been examined. The results from both models indicate that Ad-based vectors are widely distributed shortly after intravenous administration, that vector DNA decays rapidly but persists for as long as a year in some organs and that INGN 007 and wt Ad5 replicate in certain Syrian hamster organs but not mouse organs.
Materials and methods
Cells and viruses
Human HEK-293, human A549 lung carcinoma, Syrian hamster HaK kidney and mouse mammary adenocarcinoma JC cells were grown in Dulbecco’s modified essential medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (100 units per ml) and streptomycin (100 μg per ml). INGN 007, an oncolytic Ad vector, has been described previously (Figures 1a and b).11 In addition, the reference strain of wt Ad5 was used in this study (GenBank accession number AY339865). The replication-defective (RD) vector AdCMVpA is an E1-deleted vector based on dl309 in which an empty expression cassette was placed into the deleted portion of E3 (see Figures 1a and b). Vectors and virus propagated on HEK-293 cells were purified by column chromatography by Introgen Therapeutics Inc. (Houston, TX). All three stocks had a concentration of greater than 1.0 × 1012 virus particles (vp) per ml.
Figure 1.
Genomic structure of INGN 007, Ad5 and AdCMVpA and the QPCR assays to detect viral DNA. (a) Schematic of vectors used in this study. The genomes of wild-type Ad5, INGN 007 and AdCMVpA are depicted. The early transcription units 1 (E1), 2 (E2), 3 (E3) and 4 (E4) are shown as arrows. Exons 1, 2 and 3 of the tripartite leader as well as the late transcription units L1, L2, L3, L4, ADP and L5 are shown above the Ad5 genome. The E3 region of INGN 007 is deleted and replaced by the ADP gene. AdCMVpA, which was derived from dl309, lacks all of E1 and a portion of the E3 transcription unit. AdCMVpA does not contain a transgene. (b) Schematic of the differences between the E3 region of each of the genomes. The amplicon of the INGN 007 QPCR assay is shown above the INGN 007 genome. The amplicon of the Ad5/AdCMVpA QPCR assay is shown below the Ad5 genome. Performance characteristics of the INGN 007 (c) and Ad5/AdCMVpA (d) assays were derived from three independent experiments.
Animals
A total of 200 golden Syrian hamsters (115 males and 115 females, 5–6 weeks of age and weighing 80–100 g) were purchased from Harlan Sprague Dawley (Indianapolis, IN) for the hamster biodistribution study. A total of 100 C57BL/6 mice (60 males and 60 females, 5–6 weeks of age and weighing 18–25 g) were purchased from Harlan Sprague Dawley for the mouse biodistribution study. All animals were housed in the Department of Comparative Medicine at the Saint Louis University School of Medicine and were cared for in compliance with the animal protocol approved by the Animal Care Committee of Saint Louis University and in accordance with the Guideline for the Care and Use of Laboratory Animals (NIH publication number 85-23, 1996).
Syrian hamster study
One day before vector/virus injection, animals were randomized by weight into groups of 10 animals (5 each males and females) (Table 1). The mean body weight for males was 96.95 g and that for females was 88.44 g. Treatment groups consisted of INGN 007, Ad5, AdCMVpA and vehicle (10 mM Tris-HCl (pH 8.2), 10% glycerol). On day 1, animals were anesthetized and then administered a single intrajugular injection of vector/virus (1.9 × 1012 vp per kg) or vehicle. Animals (five per sex per group) were killed for necropsy on days 2, 7, 29, 92 and 372 (371 for females).
Table 1.
Experimental design of the Syrian hamster biodistribution study
| Group | Vector/virus | Dose (vp per kg) | Sexes | N |
|---|---|---|---|---|
| 1 | Vehicle | – | 2 | 50 |
| 2 | AdCMVpA | 1.9×1012 | 2 | 50 |
| 3 | Ad5 | 1.9×1012 | 2 | 50 |
| 4 | INGN 007 | 1.9×1012 | 2 | 50 |
Mouse study
Mice were randomized by weight into groups of 10 animals (5 males and 5 females) the day before vector/ virus injection (Table 2). The mean body weight for males was 23.27 g and that for females was 19.85 g. Treatment groups in the mouse study were INGN 007 and vehicle (10 mM Tris-HCl (pH 8.2), 10% glycerol). On day 1, animals received a single intravenous (tail vein) injection of INGN 007 (1.5 × 1011 vp per kg) or vehicle. Animals (five per sex per group) were killed for necropsy on days 2, 7, 29, 92 and 365.
Table 2.
Experimental design of the mouse biodistribution study
| Group | Vector/virus | Dose (vp per kg) | Sexes | N |
|---|---|---|---|---|
| 1 | Vehicle | – | 2 | 50 |
| 2 | INGN 007 | 1.5×1011 | 2 | 50 |
Necropsy, organ collection and homogenization
Groups of animals were anesthetized with CO2 and then killed by exsanguination through cardiac puncture. Animals were processed in the following order: vehicle control group, AdCMVpA group (hamster study only), Ad5 group (hamster study only) and INGN 007 group. Samples/organs were collected in the following order: blood, gonads, brain, lymph nodes (mesenteric), spleen, kidneys, adrenal glands (separated from the kidneys after dissection), bone marrow (flushed from the femur in phosphate-buffered saline, PBS), heart, lungs/bronchi and liver. Blood was collected in EDTA tubes and flash frozen in liquid nitrogen. Organs were trimmed of connective tissue and then flash frozen. In the hamster study, only a portion of some organs was frozen as follows: the proximal half of the right testis, the right half of the brain and the right lateral lobe of the liver. Only a portion of the liver (the right lateral lobe) was collected in the mouse study. All samples were stored at 80 °C until processed further. Samples (excluding blood and bone marrow) were thawed and then homogenized in PBS using 3 mm tungsten carbide beads (Qiagen, Valencia, CA) and a bead-beater type homogenizer (TissueLyser; Qiagen). To reduce the possibility of cross contamination among organs from a single animal and between different animals, each organ was removed and trimmed with a different set of instruments. In addition, gloves and bench paper covering the work surface were changed between animals, and disposable lab coats were changed between groups.
Purification and quantification of DNA
Genomic DNA was purified from a portion of each homogenate with a Magtration 12GC automated DNA isolation instrument and the Magtration-MagaZorb DNA Kit-200 High Yield (both from Precision System Science USA Inc., Livermore, CA). DNA was eluted in 200 μl of water and then quantified in triplicate with the Quant-iT PicoGreen dsDNA kit (Invitrogen, Carlsbad, CA). DNA quantification assays were set up using a Biomek 2000 Laboratory Automation Workstation (Beckman-Coulter, Fullerton, CA) located in a biological safety cabinet. Aerosol barrier tips were used for all liquid-handling steps. The assays were read with a BioTek Synergy HT (BioTek, Winooski, VT) fluorescence plate reader outfitted with fluorescein isothiocyanate excitation and emission filters. Software provided with the Synergy HT plate reader was used to generate a standard curve and calculate the concentration of each sample.
Real-time PCR assays
Two quantitative, TaqMan-based real-time PCR (QPCR) assays were developed; one that preferentially detects INGN 007 and one that detects both wt Ad5 and AdCMVpA, but is incapable of detecting INGN 007. Table 3 shows the primers and probes used for the assays. Primers and probes were synthesized by Integrated DNA Technologies (Coralville, IA). All hamster and mouse samples were assayed in triplicate, one of which was spiked with 100 copies of the appropriate viral DNA (INGN 007 or Ad5 genomic DNA for the INGN 007 or Ad5/AdCMVpA assays). All real-time PCR assays were set up in 96-well PCR plates (Applied Biosystems Inc., Foster City, CA) using a Biomek 2000 located in a biological safety cabinet. Assays contained 1 universal PCR master mix (Applied Biosystems Inc.), 250 nM of forward and reverse primers, 250 nM of probe and up to 1 μg of DNA in a total reaction volume of 50 μl. All assays were performed using an ABI model 7500 genetic analyzer with the following cycling parameters: 1 cycle at 50 °C for 2 min, 1 cycle at 95 °C for 10 min and 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Data were analyzed using Sequence Detection System software (Applied Biosystems Inc.) with the threshold set to 0.128 for all INGN 007 assays or 0.200 for all Ad5/AdCMVpA assays. Controls included with every assay consisted of a ‘no template control’ (no DNA added), an ‘animal genomic DNA control’ (only hamster or mouse liver genomic DNA), a ‘nontarget DNA control’ (Ad5 for INGN 007 assays and INGN 007 for Ad5/AdCMVpA assays) and standards from 1 × 102 to 1 × 106 copies of the appropriate viral genomic DNA (purified INGN 007 or Ad5 viral genomic DNA for INGN 007 or Ad5/AdCMVpA assays) diluted in Syrian hamster (or mouse) liver genomic DNA. The large majority of reactions contained 1 μg of organ genomic DNA, but the data for those reactions that contained less genomic DNA were normalized to 1 μg. All samples in which the mean copy number of the duplicate reactions was <100 were considered to be negative for viral genomic DNA and were treated as having zero copies for the purpose of calculating the average number of copies for that particular sample type. When all organs from a group at a particular time point were negative for viral genomic DNA, then that organ from that group was not evaluated at subsequent time points.
Table 3.
Real-time PCR amplicons for detection of INGN 007, Ad5 and AdCMVpA
| Amplicon | Size (bp) | Function | Sequence |
|---|---|---|---|
| INGN 007 | 139 | Forward primer | 5′-AACGCGCCCGACCAC-3′ |
| Probea | 5′-TGCTACACCCAAACAATGATGGAATCCA-3′ | ||
| Reverse primer | 5′-AATTCCGTCCATTTCTAGATCTCAT-3′ | ||
| Ad5/AdCMVpA | 74 | Forward primer | 5′-CCGGTCATTTCCTGCTCAATA-3′ |
| Probea | 5′-CATTCCCCTGAACAATTGACTCTATGTGGG-3′ | ||
| Reverse primer | 5′-AGGTTGTAGCGCTGGAGCATA-3′ |
Probe was modified with the fluorophore 6-FAM at the 5′ end and the quencher TAMRA at the 3′ end.
Infectious titer assay
The level of infectious vector/virus in select Syrian hamster organs was measured using a tissue culture 50% infectious dose (TCID50) assay. Organ homogenates of lymph nodes (mesenteric) and QPCR-positive livers, lungs and gonads were subjected to three freeze-thaw cycles, sonicated for 15 min and then clarified by centrifugation. The clarified homogenates were serially diluted and then select dilutions were inoculated onto HEK-293 cells in a 96-well culture dish. Seven replicates were tested for each dilution. HEK-293 cells were used because they complement the E1 deletion in the AdCMVpA vector, thus allowing it to replicate. Each individual organ homogenate was assayed in a separate plate. Each plate also contained multiple negative (that is, no homogenate) and positive control wells. Positive control wells consisted of a known amount of vector/ virus spiked into the homogenate dilution; these wells served to evaluate inhibition of vector/virus replication by the homogenate. After 2 weeks of incubation at 37 °C, each well was examined for cytopathic effect (CPE). Viral titers per organ were calculated by the Reed–Muench method taking into account the lowest dilution of homogenate in which at least one of the spiked wells exhibited CPE. This dilution was used to calculate the threshold of calculability. The threshold of calculability determined for each organ was 2.0 × 104 TCID50/liver, 2.8 × 103 TCID50/lung, 4.0 × 102 × TCID50/ovaries, 102 × 2.4 × 103 TCID /testes and 2.7 × 50 TCID50 per ml of lymph node homogenate. Homogenates that yielded positive wells (that is, wells that showed CPE) below the threshold of calculability were scored as ‘positive but not quantifiable’. Homogenates for which no positive wells were evident were scored as ‘undetected’.
Immunohistochemistry
Tissue sections from the toxicology study47 were mounted on glass slides. Deparaffinized sections were subjected to antigen retrieval using DIVA Decloaker solution (Biocare Medical, Concord, CA) and then stained using an antifiber monoclonal antibody 4D2 (Lab Vision, Freemont, CA) and horseradish-peroxidase-conjugated secondary antibody (Dako, Carpinteria, CA).
Infectivity ratio determination
Human A549, Syrian hamster HaK and mouse JC cells were infected at a multiplicity of infection (MOI) of 100 plaque-forming units (PFU) per cell. After an adsorption period of 1 h, cells were washed three times with PBS and then supplemented with 2 ml of complete growth medium. Cells plus medium were harvested at 0 (immediately after the rinses following the adsorption period), 1, 2, 3 and 4 days after infection by freezing plates at −80 °C. Lysates were subjected to three freeze-thaw cycles. DNA purified (Magtration 12GC system) from a portion of each lysate was quantified (Quant-iT PicoGreen dsDNA kit) and then subjected to the Ad5/AdCMVpA QPCR assay. A portion of each lysate was assayed by the TCID50 assay. Both values were normalized to account for the number of cells originally infected.
Statistical analyses
Statistical analyses were performed using nonparametric tests due to the variance within groups. First, an analysis of variance between groups was conducted using the Kruskal–Wallis test. Pair-wise comparisons between the AdCMVpA-injected group and the INGN 007- or Ad5-injected groups were carried out using the Mann–Whitney U-test.
Results
The INGN 007 and Ad5/AdCMVpA real-time PCR assays are specific and sensitive
Two TaqMan-based QPCR assays were developed to detect INGN 007, Ad5 and AdCMVpA. Because the INGN 007 genome differs from the wt Ad5 genome only by the deletion of two regions (Figures 1a and b), sequences in INGN 007 are juxtaposed that are not normally adjacent in Ad5. This juxtaposition forms the basis of specificity of the INGN 007 QPCR assay because the reverse primer spans one of the deletion junctions (Figure 1b). As a result, the INGN 007 QPCR assay preferentially detects INGN 007; the assay is >1000 times less sensitive for Ad5 than for INGN 007 (data not shown). A second QPCR assay was developed to detect both Ad5 and AdCMVpA genomic DNA. The specificity of this assay resides in the fact that the amplicon is present in the Ad5 and AdCMVpA genomes but is deleted in the INGN 007 genome (Figure 1b).
The performance characteristics of both QPCR assays were thoroughly investigated. Both assays yielded a single product of the appropriate size (data not shown) and were linear in the range of 1 × 101 to 1 × 107 copies of purified viral genomic DNA (Figures 1c and d). The Ad5/ AdCMVpA assay performed equally well with Ad5 and AdCMVpA viral genomic DNA as the template (data not shown). The INGN 007 assay had similar performance characteristics in the presence of 1 μg of Syrian hamster or mouse genomic DNA (data not shown). Furthermore, 100 copies of viral genomic DNA were reproducibly detectable in the presence of 1 μg background genomic DNA purified from various Syrian hamster (INGN 007 and Ad5/AdCMVpA assays) or mouse (INGN 007 assay only) organs (data not shown). The single exception was genomic DNA isolated from Syrian hamster lymph nodes, which interfered with the QPCR assay (data not shown). These data indicate that both QPCR assays are specific, sensitive and reproducible.
Syrian hamster organs contain more DNA from replication-competent viruses than from a replication-defective vector
The experimental design used for the Syrian hamster biodistribution study is shown in Table 1. After injection (defined as day 1), animals were monitored daily for mortality. All hamsters in the vehicle and AdCMVpA groups remained healthy throughout the yearlong course of study. No female hamsters died during the study but four INGN 007-treated and five Ad5-treated male hamsters were found dead. With one exception, the cause of death was undetermined and the animals died on or before day 10 of the study; one of the Ad5-treated animals died on day 270 because of kidney failure; this death was deemed not related to treatment.
Groups of 10 hamsters (5 males and 5 females) from each treatment group were killed on days 2, 7, 29, 92 and 372 (371 for females). Because the day of injection was day 1, this corresponds to 1, 6, 28, 91 and 371 (370) days after infection. Select organs were harvested from each hamster at the time of necropsy. Purified genomic DNA was subjected to the INGN 007 (vehicle and INGN 007 groups) or the Ad5/AdCMVpA (vehicle, Ad5 and AdCMVpA groups) QPCR assay. Each sample was assayed in triplicate, one of which served as a spike control. This control reaction assured that samples evaluated as having fewer than 100 copies did not contain a substance that inhibited the PCR. On the basis of the spike controls, none of the samples exhibited inhibition in the QPCR assay. Importantly, only a single organ from the vehicle control group yielded a signal in the INGN 007 or Ad5/AdCMVpA assays. In addition, a positive PCR signal was not detected in any of the ‘no template’ or ‘animal genomic DNA’ control reactions, and only a single nontarget DNA control reaction yielded a signal. These results indicate that there was little or no cross contamination during vector/virus injection, necropsy, the in-life observation period, sample processing or QPCR assay setup.
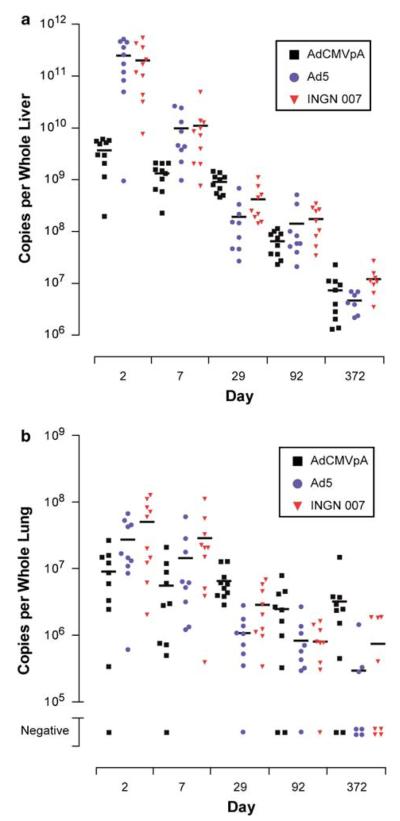
INGN 007, Ad5 and AdCMVpA DNA were detected in all 11 organs examined indicating that all three genomes are widely distributed after intravenous injection (Tables 4, 5 and 6). The primary target organ for all three vectors/viruses was the liver because this organ contained the most viral DNA at 1 day after injection and at all subsequent time points (Tables 4, 5 and 6). Other organs generally contained a much lower level of viral DNA.
Table 4.
Mean copy number of INGN 007 DNA in various Syrian hamster organs
| Day | Blood | Bone marrow | Brain | Heart | Adrenal glands | Kidney | Liver | Lungs/Bronchi | Spleen | Testes | Ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 121 515a | 13 701 | 365 | 5399b | 10 496b | 2024c | 18 339 870c | 20 176b | 93 970 | 101 | 20 255 |
| 7 | 7342c | 5918 | 769b | 11 003b | 66 159a | 14 796b | 1 005 784a | 11 488b | 15 439c | 91 | 6794 |
| 29 | 352b | 1411 | 0 | 1646 | 2093 | 2517 | 38 146a | 1138b | 3364 | 28 | 513 |
| 92 | 0 | 156b | NT | 736 | 530 | 390 | 15 873b | 319 | 1871b | 0 | 26 |
| 372d | NT | 0 | NT | 306 | 160 | 161 | 1093 | 295a | 981 | NT | 0 |
Abbreviation: NT, not tested because all samples from the previous time point had <100 copies.
Copies per 1 μg of DNA.
P≤0.01 INGN 007 vs AdCMVpA (Mann–Whitney U-test).
P<0.05 INGN 007 vs AdCMVpA (Mann–Whitney U-test).
P≤0.001 INGN 007 vs AdCMVpA (Mann–Whitney U-test).
Males were killed on day 372, females were killed on day 371.
Table 5.
Mean copy number of Ad5 DNA in various Syrian hamster organs
| Day | Blood | Bone marrow | Brain | Heart | Adrenal glands | Kidney | Liver | Lungs/Bronchi | Spleen | Testes | Ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 41 202a | 15 222 | 234 | 1837 | 15 000b | 1795 | 22 768 720c | 10 882b | 85 320 | 0 | 1675 |
| 7 | 2342c | 5874 | 173b | 4638a | 12 863c | 5101a | 900 544a | 5760 | 8406 | NT | 2513 |
| 29 | 0 | 1296 | 0 | 421a | 860 | 1448 | 16 329c | 428c | 2200 | NT | 854a |
| 92 | NT | 168 | NT | 1054 | 732 | 576 | 12 850 | 329 | 2851a | NT | 0 |
| 372d | NT | 0 | NT | 68a | 146 | 42 | 420 | 117b | 1101 | NT | NT |
Abbreviation: NT, not tested because all samples from the previous time point had <100 copies.
Copies per 1 μg of DNA.
P≤0.01 Ad5 vs AdCMVpA (Mann–Whitney U-test).
P<0.05 Ad5 vs AdCMVpA (Mann–Whitney U-test).
P≤0.001 Ad5 vs AdCMVpA (Mann–Whitney U-test).
Males were killed on day 372, females were killed on day 371.
Table 6.
Mean copy number of AdCMVpA DNA in various Syrian hamster organs
| Day | Blood | Bone marrow | Brain | Heart | Adrenal glands | Kidney | Liver | Lungs/Bronchi | Spleen | Testes | Ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 1300 | 13 966 | 40 | 1596 | 2667 | 660 | 337 890 | 3598 | 83 730 | 50 | 809 |
| 7 | 61 | 4962 | 0 | 1065 | 1033 | 442 | 120 540 | 2216 | 6182 | 0 | 680 |
| 29 | 0 | 1541 | NT | 1322 | 1451 | 596 | 72 590 | 2599 | 2326 | NT | 138 |
| 92 | NT | 318 | NT | 562 | 841 | 289 | 5891 | 982 | 1008 | NT | 0 |
| 372a | NT | 20 | NT | 314 | 287 | 109 | 664 | 1079 | 1437 | NT | NT |
Abbreviation: NT, not tested because all samples from the previous time point had <100 copies.
Copies per 1 μg of DNA.
Males were killed on day 372, females were killed on day 371.
Because Syrian hamsters are permissive for Ad replication, it was noteworthy that several Syrian hamster organs contained statistically significantly more INGN 007 and Ad5 DNA than AdCMVpA DNA early after vector/virus injection (Figures 2a and b; Tables 4, 5 and 6). In the liver, there was at least 50-fold more INGN 007 or Ad5 DNA than AdCMVpA DNA on day 2. This difference declined to less than 10-fold on day 7, and by day 29 there was actually more AdCMVpA DNA than either INGN 007 or Ad5 DNA (Figure 2a). Although the difference between RC and RD Ad DNA levels was particularly evident in the liver, it was also apparent in adrenal glands, blood, heart (only day 7 for Ad5), kidneys (only day 7 for Ad5) and lungs (only day 2 for Ad5). By day 29, most other organs had comparable levels of viral DNA, although there were still some statistically significant differences (in some cases there was more AdCMVpA DNA than INGN 007 or Ad5 DNA).
Figure 2.
Genome copies in Syrian hamster liver and lung samples. The number of genome copies of AdCMVpA, Ad5 and INGN 007 in whole Syrian hamster liver (a) and lungs (b) was calculated. The horizontal bar represents the mean copy number. Tissue samples that were negative for viral genomic DNA were considered as zero for the purposes of calculating the mean copy number.
With the exceptions noted below, the peak quantity of viral genomic DNA for most organs was observed on day 2, after which the signal generally decreased at each successive time point such that by 1 year viral DNA was no longer detectable in blood, bone marrow (except AdCMVpA), brain, testes and ovaries. With the RC vector/virus, the level of viral DNA increased from day 2 to day 7 such that the peak DNA level in heart, kidneys, adrenal glands (INGN 007 only) and ovaries (Ad5 only) occurred on day 7. The RD vector AdCMVpA did not exhibit this pattern; the peak viral DNA level for all organs was day 2.
Recovery of infectious vector/virus from Syrian hamster organs
Select organs from the Syrian hamster biodistribution study were assessed for the presence of infectious vector/ virus using a TCID50 assay. All liver, lung and gonad samples that were positive in the PCR assay were tested for infectious vector/virus. Infectious Ad5 and/or INGN 007 were detected in liver, lung and testes but all ovary samples tested were negative. In contrast, none of the liver, lung or gonad samples from AdCMVpA-injected animals contained infectious vector, even on day 2 when the mean copy number in liver samples was >300 000 per μg of genomic DNA. As anticipated from the QPCR results, liver harbored the greatest level of infectious INGN 007 and Ad5; however, infectious INGN 007 or Ad5 was not detected in liver beyond day 7 (Figure 3). Lung samples from days 2 and 7 did contain infectious vector/virus but at a much lower level and frequency compared to liver samples (Table 7). The testes of only two animals were found positive, both of which were from the INGN 007 group on day 2.
Figure 3.
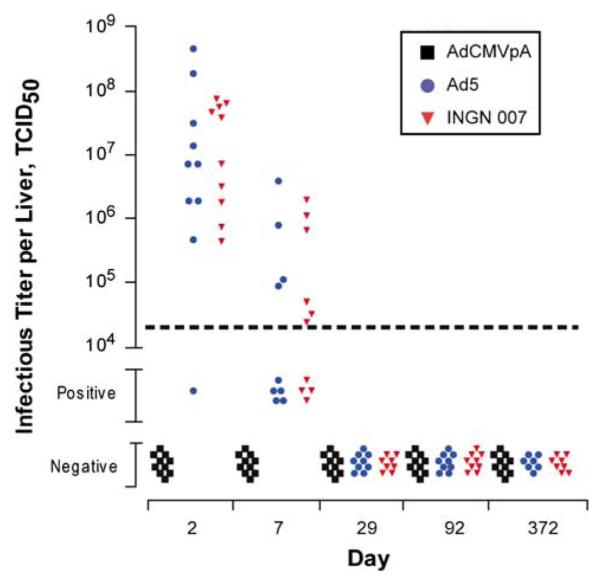
Infectious titer of INGN 007, Ad5 and AdCMVpA in Syrian hamster liver samples. A portion of each liver sample was homogenized and then subjected to a TCID50 assay on HEK-293 cells to determine the amount of infectious vector/virus present. The lower limit of sensitivity of the assay with liver homogenates is shown by the horizontal dashed line. Samples that contained detectable infectious vector/virus but whose titer could not be calculated are shown as being positive. Samples where vector/virus was not detectable are shown as being negative.
Table 7.
TCID50 assay results from Syrian hamster lung samples
| Group | Day | Number PCR Pos. |
Number quantifiable |
Titer (TCID50/ organ) |
Number positive |
Number negative |
|---|---|---|---|---|---|---|
| AdCMVpA | 2 | 9 | 0 | NA | 0 | 10 |
| 7 | 9 | 0 | NA | 0 | 8 | |
| 29 | 10 | 0 | NA | 0 | 10 | |
| 92 | 8 | 0 | NA | 0 | 7 | |
| 372 | 8 | 0 | NA | 0 | 8 | |
| Ad5 | 2 | 10 | 1 | 509 | 1 | 7 |
| 7 | 9 | 0 | NA | 0 | 4 | |
| 29 | 8 | 0 | NA | 0 | 8 | |
| 92 | 8 | 0 | NA | 0 | 7 | |
| 372 | 3 | 0 | NA | 0 | 3 | |
| INGN 007 | 2 | 10 | 2 | 1800 7310 |
1 | 4 |
| 7 | 10 | 0 | NA | 0 | 10 | |
| 29 | 8 | 0 | NA | 0 | 7 | |
| 92 | 8 | 0 | NA | 0 | 8 | |
| 372 | 4 | 0 | NA | 0 | 4 |
Abbreviation: NA, not applicable.
Immunohistochemical analysis of liver samples for Ad fiber protein supported the conclusions from the infectivity assays that infectious vector/virus was present in the liver of animals injected with the RC vector/virus but not the RD vector. Because there was no remaining liver samples from the biodistribution study that could be used for immunohistochemistry, livers from the companion toxicity47 study were used instead. The hamsters from the toxicity study were injected with the same dose of vector/ virus and necropsied on the same day (day 2 or 1 day after infection). Ad fiber protein was detected in hepatocytes from INGN 007- (Figures 4c and d) and Ad5-injected (data not shown) animals. The signal seen in these animals likely represents newly formed vector/virus and not simply input vector/virus inasmuch as fiber protein was not detected in the liver sections of AdCMVpA-injected animals (Figure 4b). Also, the staining is predominantly localized to the nuclei, the site of Ad assembly. No signal was observed in samples from vehicle-injected hamsters (Figure 4a) or when liver sections from all treatment groups were stained with an isotype-matched control antibody (data not shown), demonstrating the specificity of the assay.
Figure 4.

Immunohistochemistry of liver samples for adenovirus (Ad) fiber protein. Liver samples from the companion toxicology study47 were prepared for immunohistochemical staining and then stained using an antifiber antibody. Representative fields from the mock-injected (a), AdCMVpA-injected (b) and INGN 007-injected (c, d) Syrian hamsters are shown. (d) A magnified portion of the field shown in c. The black bar represents 100 μm (a, b and c) or 10 μm (d).
Lymph nodes were also assessed in the TCID50 assay because this organ could not be examined in the QPCR assay (see Materials and methods). Although some lymph nodes were positive for vector/virus, none of the samples contained sufficient vector/virus to be quantified (Table 8). Whereas infectious vector/virus was present in lymph nodes from all three groups on day 2, only samples from the INGN 007 and Ad5 groups were positive on day 7. By day 29, no infectious vector/virus was present in any of the samples examined, so additional time points were not tested.
Table 8.
Number of TCID50-positive Syrian hamster lymph node samples
| Virus per vector | Day |
||
|---|---|---|---|
| 2 | 7 | 29 | |
| AdCMVpA | 2 (10)a | 0 (10) | 0 (10) |
| Ad5 | 5 (10) | 1(10) | 0 (10) |
| INGN 007 | 3 (10) | 1(10) | 0 (10) |
() total number of hamsters tested.
INGN 007 DNA is much less abundant in mouse tissues
The experimental design used for the mouse biodistribution study is shown in Table 2. After injection with vehicle or INGN 007, animals were monitored daily for mortality. No treatment-related mortality was observed in this study, although two male mice from the INGN 007 group died (one on day 28 and one on day 135).
For the mouse biodistribution study, groups of 10 mice (5 each males and 5 females) were killed on days 2, 7, 29, 92 and 365, and then purified tissue genomic DNA was subjected to the INGN 007 QPCR assay. All of the negative control (no template, mouse genomic DNA only and nontarget viral DNA) reactions and all of the organs from the vehicle control group were negative in the QPCR assay indicating lack of cross contamination throughout the study. Furthermore, based on the spike controls, none of the samples in the mouse biodistribution study exhibited inhibition in the QPCR assay.
INGN 007 DNA was widely distributed in mice on day 2. However, unlike in hamsters, INGN 007 DNA was detected in only 10 out of 12 organs (Table 9). In addition, several organs contained only a low level of viral DNA. With few exceptions, the level of INGN 007 DNA decreased in each organ at each successive time point such that by 1 year nine out of twelve organs were negative for viral DNA. Similar to the Syrian hamster study, and in agreement with numerous other studies in mice, the liver was the primary target of distribution in mice (Table 9).24,30,48–51 The viral DNA copy number diminished rapidly in the liver such that at 1 year six of the nine liver samples were negative for INGN 007 DNA (data not shown). Although there was a relatively high level of INGN 007 DNA in the blood on day 2, that level decreased dramatically by day 7 and there was no detectable viral DNA in blood by day 29. Despite a high mean level of INGN 007 viral DNA present in liver on day 2, infectious vector was not detected in these samples, nor was infectious vector detected in any day-7 liver samples.
Table 9.
Mean copy number of INGN 007 in various mouse organs
| Day | Blood | Bone marrow | Brain | Heart | Adrenal glands | Kidney | Liver | Lungs/Bronchi | Lymph nodes | Spleen | Testes | Ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 135 990 | 771 | 21 | 1485 | 0 | 445 | 1 899 103 | 1682 | 162 | 6952 | 0 | 29 |
| 7 | 718 | 263 | 0 | 450 | NT | 66 | 91 456 | 331 | 372 | 2888 | NT | 31 |
| 29 | 0 | 245 | NT | 0 | NT | 34 | 4604 | 185 | 0 | 2779 | NT | 0 |
| 92 | NT | 190 | NT | NT | NT | 43 | 2052 | 164 | NT | 1898 | NT | NT |
| 365 | NT | 0 | NT | NT | NT | 0 | 60 | 15 | NT | 266 | NT | NT |
Abbreviation: NT, not tested.
Copies per 1 μg of DNA.
Infectivity ratios in vivo and in vitro are high
There was a great excess of vector/viral genomic DNA present in Syrian hamster liver and lungs compared to the amount of infectious vector/virus present in those organs. The mean infectivity ratio (the ratio of vector/viral DNA to infectious vector/virus) for INGN 007 in liver was 2.4 × 104 and 5.0 × 104 on days 2 and 7, respectively. For Ad5, the infectivity ratio was 1.3 × 105 and 4.5 × 104 on days 2 and 7, respectively. For the few lung samples that contained a titratable amount of vector/virus on day 2, the ratio was 2.7 × 104 for INGN 007 (mean of two samples) and 2.9 × 104 for Ad5 (only one sample).
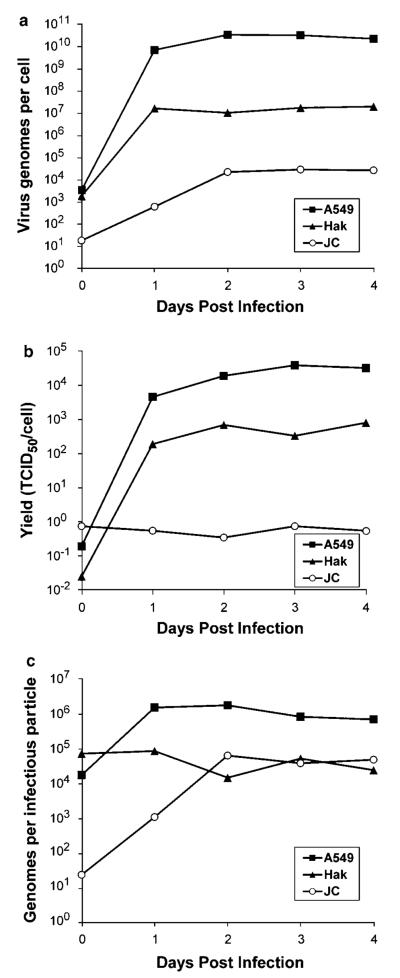
To determine if the infectivity ratio was also high in vitro, Ad5-infected cells were harvested at various times after infection and then the samples were assayed by QPCR to quantify the amount of genomic DNA and by TCID50 to measure the amount of infectious virus. Cell lines from three different species were examined: human A549 cells that are highly permissive for Ad5 replication, Syrian hamster HaK cells that were previously shown to support Ad5 replication44 and mouse JC cells that were reported to be among the more permissive mouse cell lines.52 In A549 and HaK cells, Ad5 genomic DNA and infectious virus were initially present at low levels, increased dramatically over the course of 1–2 days and then leveled off by about 2 days after infection (Figures 5a and b). The infectivity ratio in A549 cells leveled off at about 1 × 106, whereas that in HaK cells leveled off at nearly 1 × 105 (Figure 5c). It is quite interesting that the infectivity ratio for Ad5 and INGN 007 in hamsters, which ranged from 2.4 × 104 to 1.3 × 105 on was that days 2 and 7, similar to of Ad5 in hamster HaK cells (1 × 105). With JC cells, genomic DNA increased for 2 days before leveling off at about 1 × 104 genomes per cell (Figure 5a). However, little or no infectious Ad5 was produced in JC cells (Figure 5b) yielding a maximum infectivity ratio of 6.5 × 104 (Figure 5c). We could not ascertain whether the QPCR assay detected complete genomes, merely DNA fragments or both types of DNAs.
Figure 5.
Ratio of Ad5 genomic DNA copies to infectious virus in different cell lines. Human A549, Syrian hamster HaK and mouse JC cells infected at a multiplicity of infection (MOI) of 100 PFU per cell were harvested on the indicated days. Each lysate was subjected to the TCID50 assay (a) and the Ad5/AdCMVpA QPCR assay (b). The Ad5 copy number was divided by the TCID50 titer and plotted vs days post infection (c).
Discussion
Two nonclinical biodistribution studies were conducted to examine the dissemination and pharmacokinetics of the oncolytic Ad vector INGN 007 following intravenous administration, one in Syrian hamsters and the other in C57BL/6 mice. Both studies demonstrated that viral DNA is widely distributed shortly after administration. However, high levels of viral DNA were not sustained as evidenced by a rapid decline in DNA levels in most organs between days 2 and 7. Hamster blood, brain, testes, ovaries and bone marrow (INGN 007 and Ad5 only) were negative by 1 year after injection. The decrease in INGN 007 DNA in mouse organs was even more dramatic; seven organs were negative by day 29 and nine out of twelve organs were negative after 1 year.
Ad viral DNA was principally localized to the liver in Syrian hamsters and mice, which is in agreement with previous reports with the mouse and other animal models.24,30,39,44,46,48,51 Using various model systems, distribution to and uptake into liver cells has been ascribed to direct binding to liver cells and a variety of interactions between Ad virions and blood cells/proteins (for review see Baker et al.53). In the Syrian hamster model, all three vector/viral DNAs were rapidly cleared from the blood and were most abundant in the liver. Additional studies are needed to determine the mechanism(s) underlying rapid clearance from the blood and distribution to the liver in this animal model.
The Syrian hamster has been described as a permissive animal model for RC Ads and data from the hamster study provided further evidence for this view. INGN 007 and Ad5 DNA were more abundant than that of AdCMVpA in several hamster organs early after injection. This was particularly evident in the liver where INGN 007 and Ad5 DNA were >50- and ~7-fold more abundant than AdCMVpA DNA on day 2 and day 7, respectively (Tables 4, 5 and 6; Figure 2a). In addition, Ad fiber protein, as detected by immunohistochemistry, was only detected in INGN 007 (and Ad5)-injected hamsters (Figure 4). Furthermore, infectious INGN 007 and Ad5, but not AdCMVpA, were recovered from liver samples on days 2 and 7. Interestingly, data from the hamster biodistribution study are concordant with the extent of liver toxicity observed in the parallel toxicology study: INGN 007 and Ad5 resulted in more liver damage than AdCMVpA on day 7.47 Together, these data strongly support the liver as a site of Ad replication in Syrian hamsters and that replication is associated with enhanced liver damage.
Syrian hamster organs other than liver also likely supported Ad replication. Compared to the amount of AdCMVpA DNA, 3- and 5-fold more Ad5 and INGN 007 genomic DNA, respectively, was present in lungs on day 2 (Tables 4, 5 and 6; Figure 2b). In addition, infectious INGN 007 and Ad5 were detected in lungs on day 2, whereas no infectious AdCMVpA was detected in any lung samples (Table 7). These data suggest that the lung is a site of replication following intravenous injection and agree with previously published results where Ad was administered either intratracheally or intranasally.32,44 The heart, kidneys and adrenal glands may also be a site of replication. These organs contained more INGN 007 and Ad5 DNA than AdCMVpA DNA on day 2 (statistically significant for all three organs for INGN 007 vs AdCMVpA and for adrenal glands for Ad5 vs AdCMVpA) (Tables 4, 5 and 6). This difference was also apparent on day 7 (statistically significant for all three organs and both RC viruses vs AdCMVpA) (Tables 4, 5 and 6). Furthermore, the amount of INGN 007 and Ad5 DNA increased from day 2 to day 7 in heart, kidneys and adrenal glands (INGN 007 only). Two pieces of data suggest that this increase likely represents replication in these organs. First, AdCMVpA DNA did not exhibit a similar increase. Second, levels of INGN 007 and Ad5 DNA in other organs did not increase from day 2 to day 7.
Although INGN 007 and Ad5 were able to replicate in certain Syrian hamster organs, the burst of replication seen early after vector/virus injection appeared to be only transient. No infectious vector/virus was detected beyond day 7 despite positive QPCR results, and viral DNA in all organs was greatly diminished by day 29. Control of vector/virus replication by the immune system probably accounts for these results. Serum samples were not available from the biodistribution study to confirm this hypothesis, but data from the companion acute toxicology study demonstrated that hamsters did mount an anti-Ad humoral immune response by day 29.47 Further support for this proposition comes from the observation that Ad5 and INGN 007 exhibit enhanced replication in immunosuppressed hamsters.45,46 Also, in studies in which INGN 007 was injected into subcutaneous HaK tumors growing in immunocompetent Syrian hamsters, anti-Ad antibodies could be detected in as little as 7 days after injection (D Dhar and W Wold, unpublished results).
Results obtained in the Syrian hamster biodistribution study with respect to bone marrow may help explain the results obtained for this organ in the toxicity study. Histopathological lesions and defects in bone marrow function described for Ad5- and INGN 007-injected hamsters could be attributed to replication because these aberrations were not evident in AdCMVpA-injected animals.47 However, the biodistribution study provided no evidence for Ad5 or INGN 007 replication in bone marrow, suggesting another cause for bone marrow toxicity, such as gene expression.
In contrast to the results seen in Syrian hamsters, the biodistribution study in mice provided no evidence that INGN 007 was able to replicate in this species. Despite the high burden of vector DNA in mouse liver on day 2, infectious INGN 007 was not recovered from this organ on day 2 or 7 (data not shown). In addition, the peak copy number for INGN 007 for all mouse organs was on day 2 and generally decreased at each successive time point in each organ (Table 9). It may well be that viral DNA replicates to some extent in mouse organs, but it appears that very little if any infectious virus is produced. Viral DNA replication without reproduction was observed in the mouse JC cell line (Figure 5). These data support previously published reports indicating that normal mouse tissues are poorly permissive for Ad replication at best.30–34
Less infectious vector/virus was detected in Syrian hamster liver and lung than might be expected based on the large quantity of genomic DNA detected in these organs. The infectivity ratio in these organs was 1 × 104 to 1 × 105 genome copies per infectious particle. Similar infectivity ratios were found in cell lines from three different species suggesting that both in vivo and in vitro far more viral DNA is produced than is packaged into infectious particles.
This study expands our understanding and highlights the utility of this new, valuable animal model for RC Ads. Specifically, this report confirmed that Ad is capable of replicating in Syrian hamster liver and lungs and that other organs such as heart, kidneys and adrenal glands may also represent a target for Ad replication. Further studies are required to confirm these results. On the basis of these data and the lack of evidence for replication in normal mouse tissues, this study provides further evidence that the Syrian hamster is the small animal model of choice for examining Ad-mediated pathogenicity, immune responses to RC Ads and efficacy of oncolytic Ad vectors.
Acknowledgements
This research was supported by grants CA118022, CA108335 and CA81829 from the National Institutes of Health to WSMW. Funding for this work was also supported by a research and development agreement to VirRx Inc. from Introgen Therapeutics Inc.
Footnotes
Disclosure/Conflict of interest WSMW, KT, AET, KD, MK and Introgen Therapeutics Inc. own shares of VirRx Inc.
References
- 1.Everts B, van der Poel H. Replication-selective oncolytic viruses in the treatment of cancer. Cancer Gene Ther. 2005;12:141–161. doi: 10.1038/sj.cgt.7700771. [DOI] [PubMed] [Google Scholar]
- 2.Bischoff JR, Kirn DH, Williams A, Heise C, Horn S, Muna M, et al. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 3.Lichtenstein DL, Wold WSM. Experimental infections of humans with wild-type adenoviruses and with replication-competent adenovirus vectors: replication, safety, and transmission. Cancer Gene Ther. 2004;11:819–829. doi: 10.1038/sj.cgt.7700765. [DOI] [PubMed] [Google Scholar]
- 4.Wold WSM, Cladaras C, Magie SC, Yacoub N. Mapping a new gene that encodes an 11 600-molecular-weight protein in the E3 transcription unit of adenovirus 2. J Virol. 1984;52:307–313. doi: 10.1128/jvi.52.2.307-313.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tollefson AE, Ryerse JS, Scaria A, Hermiston TW, Wold WSM. The E3-11.6 kDa adenovirus death protein (ADP) is required for efficient cell death: characterization of cells infected with adp mutants. Virology. 1996;220:152–162. doi: 10.1006/viro.1996.0295. [DOI] [PubMed] [Google Scholar]
- 6.Tollefson AE, Scaria A, Hermiston TW, Ryerse JS, Wold LJ, Wold WSM. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J Virol. 1996;70:2296–2306. doi: 10.1128/jvi.70.4.2296-2306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tollefson AE, Scaria A, Ying B, Wold WSM. Mutations within the ADP (E3-11.6K) protein alter processing and localization of ADP and the kinetics of cell lysis of adenovirus infected cells. J Virol. 2003;77:7764–7778. doi: 10.1128/JVI.77.14.7764-7778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ying B, Wold WSM. Adenovirus ADP protein (E3-11.6K), which is required for efficient cell lysis and virus release, interacts with human MAD2B. Virology. 2003;313:224–234. doi: 10.1016/s0042-6822(03)00287-3. [DOI] [PubMed] [Google Scholar]
- 9.Doronin K, Toth K, Kuppuswamy M, Ward P, Tollefson AE, Wold WSM. Tumor-specific, replication-competent adenovirus vectors overexpressing the adenovirus death protein. J Virol. 2000;74:6147–6155. doi: 10.1128/jvi.74.13.6147-6155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doronin K, Kuppuswamy M, Toth K, Tollefson AE, Krajcsi P, Krougliak V, et al. Tissue-specific, tumor-selective, replication-competent adenovirus vector for cancer gene therapy. J Virol. 2001;75:3314–3324. doi: 10.1128/JVI.75.7.3314-3324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doronin K, Toth K, Kuppuswamy M, Krajcsi P, Tollefson AE, Wold WSM. Overexpression of the ADP (E3-11.6K) protein increases cell lysis and spread of adenovirus. Virology. 2003;305:378–387. doi: 10.1006/viro.2002.1772. [DOI] [PubMed] [Google Scholar]
- 12.Habib NA, Mitry R, Seth P, Kuppuswamy M, Doronin K, Toth K, et al. Adenovirus replication-competent vectors (KD1, KD3) complement the cytotoxicity and transgene expression from replication-defective vectors (Ad-GFP, Ad-Luc) Cancer Gene Ther. 2002;9:651–654. doi: 10.1038/sj.cgt.7700481. [DOI] [PubMed] [Google Scholar]
- 13.Kuppuswamy M, Spencer JF, Doronin K, Tollefson AE, Wold WS, Toth K. Oncolytic adenovirus that overproduces ADP and replicates selectively in tumors due to hTERT promoter-regulated E4 gene expression. Gene Therapy. 2005;12:1608–1617. doi: 10.1038/sj.gt.3302581. [DOI] [PubMed] [Google Scholar]
- 14.Toth K, Tarakanova V, Doronin K, Ward P, Kuppuswamy M, Locke JL, et al. Radiation increases the activity of oncolytic adenovirus cancer gene therapy vectors that overexpress the ADP (E3-11.6K) protein. Cancer Gene Ther. 2003;10:193–200. doi: 10.1038/sj.cgt.7700555. [DOI] [PubMed] [Google Scholar]
- 15.Toth K, Djeha H, Ying BL, Tollefson AE, Kuppuswamy M, Doronin K, et al. An oncolytic adenovirus vector combining enhanced cell-to-cell spreading, mediated by the ADP cytolytic protein, with selective replication in cancer cells with deregulated Wnt signaling. Cancer Res. 2004;64:3638–3644. doi: 10.1158/0008-5472.CAN-03-3882. [DOI] [PubMed] [Google Scholar]
- 16.Barton KN, Paielli D, Zhang Y, Koul S, Brown SL, Lu M, et al. Second-generation replication-competent oncolytic adenovirus armed with improved suicide genes and ADP gene demonstrates greater efficacy without increased toxicity. Mol Ther. 2006;13:347–356. doi: 10.1016/j.ymthe.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Bauzon M, Castro D, Karr M, Hawkins LK, Hermiston TW. Regulated, multi-gene expression from a replicating adenovirus using native viral promoters. Mol Ther. 2003;7:526–534. doi: 10.1016/s1525-0016(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 18.Kohno S, Nakagawa K, Hamada K, Harada H, Yamasaki K, Hashimoto K, et al. Midkine promoter-based conditionally replicative adenovirus for malignant glioma therapy. Oncol Rep. 2004;12:73–78. [PubMed] [Google Scholar]
- 19.Oosterhoff D, Pinedo HM, Witlox MA, Carette JE, Gerritsen WR, Van Beusechem VW. Gene-directed enzyme prodrug therapy with carboxylesterase enhances theanticancer efficacy of the conditionally replicating adenovirus AdDelta24. Gene Therapy. 2005;12:1011–1018. doi: 10.1038/sj.gt.3302492. [DOI] [PubMed] [Google Scholar]
- 20.Ramachandra M, Rahman A, Zou A, Vaillancourt M, Howe JA, Antelman D, et al. Re-engineering adenovirus regulatory pathways to enhance oncolytic specificity and efficacy. Nat Biotechnol. 2001;19:1035–1041. doi: 10.1038/nbt1101-1035. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki K, Alemany R, Yamamoto M, Curiel DT. The presence of the adenovirus E3 region improves the oncolytic potency of conditionally replicative adenoviruses. Clin Cancer Res. 2002;8:3348–3359. [PubMed] [Google Scholar]
- 22.Yu DC, Chen Y, Seng M, Dilley J, Henderson DR. The addition of adenovirus type 5 region E3 enables calydon virus 787 to eliminate distant prostate tumor xenografts. Cancer Res. 1999;59:4200–4203. [PubMed] [Google Scholar]
- 23.Zhu M, Bristol JA, Yuefong X, Mina M, Ji H, Forry-Schaudies S, et al. Linked tumor-selective virus replication and transgene expression from E3-containing oncolytic adenoviruses. J Virol. 2005;79:5455–5465. doi: 10.1128/JVI.79.9.5455-5465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernt KM, Ni S, Gaggar A, Li ZY, Shayakhmetov DM, Lieber A. The effect of sequestration by nontarget tissues on anti-tumor efficacy of systemically applied, conditionally replicating adenovirus vectors. Mol Ther. 2003;8:746–755. doi: 10.1016/j.ymthe.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Huang D, Pereboev AV, Korokhov N, He R, Larocque L, Gravel C, et al. Significant alterations of biodistribution and immune responses in Balb/c mice administered with adenovirus targeted to CD40(+) cells. Gene Therapy. 2008;15:298–308. doi: 10.1038/sj.gt.3303085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, Zhuang L, Cao Y, Gao Q, Han Z, Tang D, et al. Biodistribution and kinetics of the novel selective oncolytic adenovirus M1 after systemic administration. Mol Cancer Ther. 2008;7:1624–1632. doi: 10.1158/1535-7163.MCT-07-2134. [DOI] [PubMed] [Google Scholar]
- 27.Kanerva A, Wang M, Bauerschmitz GJ, Lam JT, Desmond RA, Bhoola SM, et al. Gene transfer to ovarian cancer versus normal tissues with fiber-modified adenoviruses. Mol Ther. 2002;5:695–704. doi: 10.1006/mthe.2002.0599. [DOI] [PubMed] [Google Scholar]
- 28.Paielli DL, Wing MS, Rogulski KR, Gilbert JD, Kolozsvary A, Kim JH, et al. Evaluation of the biodistribution, persistence, toxicity, and potential of germ-line transmission of a replication-competent human adenovirus following intraprostatic administration in the mouse. Mol Ther. 2000;1:263–274. doi: 10.1006/mthe.2000.0037. [DOI] [PubMed] [Google Scholar]
- 29.Stone D, Liu Y, Li ZY, Tuve S, Strauss R, Lieber A. Comparison of adenoviruses from species B, C, E, and F after intravenous delivery. Mol Ther. 2007;15:2146–2153. doi: 10.1038/sj.mt.6300319. [DOI] [PubMed] [Google Scholar]
- 30.Duncan SJ, Gordon FC, Gregory DW, McPhie JL, Postlethwaite R, White R, et al. Infection of mouse liver by human adenovirus type 5. J Gen Virol. 1978;40:45–61. doi: 10.1099/0022-1317-40-1-45. [DOI] [PubMed] [Google Scholar]
- 31.Ginsberg HS, Moldawer LL, Sehgal PB, Redington M, Kilian PL, Chanock RM, et al. A mouse model for investigating the molecular pathogenesis of adenovirus pneumonia. Proc Natl Acad Sci USA. 1991;88:1651–1655. doi: 10.1073/pnas.88.5.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hjorth RN, Bonde GM, Pierzchala WA, Vernon SK, Wiener FP, Levner MH, et al. A new hamster model for adenoviral vaccination. Arch Virol. 1988;100:279–283. doi: 10.1007/BF01487691. [DOI] [PubMed] [Google Scholar]
- 33.Liu TC, Hallden G, Wang Y, Brooks G, Francis J, Lemoine N, et al. An E1B-19 kDa gene deletion mutant adenovirus demonstrates tumor necrosis factor-enhanced cancer selectivity and enhanced oncolytic potency. Mol Ther. 2004;9:786–803. doi: 10.1016/j.ymthe.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 34.Oualikene W, Gonin P, Eloit M. Short and long term dissemination of deletion mutants of adenovirus in permissive (cotton rat) and non-permissive (mouse) species. J Gen Virol. 1994;75:2765–2768. doi: 10.1099/0022-1317-75-10-2765. [DOI] [PubMed] [Google Scholar]
- 35.Lubeck MD, Davis AR, Chengalvala M, Natuk RJ, Morin JE, Molnar-Kimber K, et al. Immunogenicity and efficacy testing in chimpanzees of an oral hepatitis B vaccine based on live recombinant adenovirus. Proc Natl Acad Sci USA. 1989;86:6763–6767. doi: 10.1073/pnas.86.17.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lubeck MD, Natuk R, Myagkikh M, Kalyan N, Aldrich K, Sinangil F, et al. Long-term protection of chimpanzees against high-dose HIV-1 challenge induced by immunization. Nat Med. 1997;3:651–658. doi: 10.1038/nm0697-651. [DOI] [PubMed] [Google Scholar]
- 37.Ni S, Bernt K, Gaggar A, Li ZY, Kiem HP, Lieber A. Evaluation of biodistribution and safety of adenovirus vectors containing group B fibers after intravenous injection into baboons. Hum Gene Ther. 2005;16:664–677. doi: 10.1089/hum.2005.16.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pacini DL, Dubovi EJ, Clyde WA., Jr A new animal model for human respiratory tract disease due to adenovirus. J Infect Dis. 1984;150:92–97. doi: 10.1093/infdis/150.1.92. [DOI] [PubMed] [Google Scholar]
- 39.Page JG, Tian B, Schweikart K, Tomaszewski J, Harris R, Broadt T, et al. Identifying the safety profile of a novel infectivity-enhanced conditionally replicative adenovirus, Ad5-delta24-RGD, in anticipation of a phase I trial for recurrent ovarian cancer. Am J Obstet Gynecol. 2007;196:389.e1–9. doi: 10.1016/j.ajog.2006.12.016. discussion 389.e9-10. [DOI] [PubMed] [Google Scholar]
- 40.Prince GA, Porter DD, Jenson AB, Horswood RL, Chanock RM, Ginsberg HS. Pathogenesis of adenovirus type 5 pneumonia in cotton rats (Sigmodon hispidus) J Virol. 1993;67:101–111. doi: 10.1128/jvi.67.1.101-111.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torres JM, Alonso C, Ortega A, Mittal S, Graham F, Enjuanes L. Tropism of human adenovirus type 5-based vectors in swine and their ability to protect against transmissible gastroenteritis coronavirus. J Virol. 1996;70:3770–3780. doi: 10.1128/jvi.70.6.3770-3780.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toth K, Spencer JF, Tollefson AE, Kuppuswamy M, Doronin K, Lichtenstein DL, et al. Cotton rat tumor model for the evaluation of oncolytic adenoviruses. Hum Gene Ther. 2005;16:139–146. doi: 10.1089/hum.2005.16.139. [DOI] [PubMed] [Google Scholar]
- 43.Wildner O, Morris JC. Subcutaneous administration of a replication-competent adenovirus expressing HSV-tk to cotton rats: dissemination, persistence, shedding, and pathogenicity. Hum Gene Ther. 2002;13:101–112. doi: 10.1089/10430340152712656. [DOI] [PubMed] [Google Scholar]
- 44.Thomas MA, Spencer JF, La Regina MC, Dhar D, Tollefson AE, Toth K, et al. Syrian hamster as a permissive immunocompetent animal model for the study of oncolytic adenovirus vectors. Cancer Res. 2006;66:1270–1276. doi: 10.1158/0008-5472.CAN-05-3497. [DOI] [PubMed] [Google Scholar]
- 45.Thomas MA, Spencer JF, Toth K, Sagartz JE, Phillips NJ, Wold WSM. Immunosuppression leads to increased oncolytic adenovirus efficacy and regression of large tumors in the Syrian hamster model. Mol Ther. 2008;16:1665–1673. doi: 10.1038/mt.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toth K, Spencer JF, Dhar D, Sagartz JE, Buller RM, Painter GR, et al. Hexadecyloxypropyl-cidofovir, CMX001, prevents adenovirus-induced mortality in a permissive, immunosuppressed animal model. Proc Natl Acad Sci USA. 2008;105:7293–7297. doi: 10.1073/pnas.0800200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lichtenstein DL, Spencer JF, Patra D, Meyer J, Shashkova EV, Kuppuswamy M, et al. An acute toxicology study with INGN 007, an oncolytic adenovirus vector, in permissive Syrian hamsters and nonpermissive mice; comparisons with wild-type Ad5 and a replication-defective vector. Cancer Gene Ther. 2008 doi: 10.1038/cgt.2009.5. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shayakhmetov DM, Li ZY, Ni S, Lieber A. Analysis of adenovirus sequestration in the liver, transduction of hepatic cells, and innate toxicity after injection of fiber-modified vectors. J Virol. 2004;78:5368–5381. doi: 10.1128/JVI.78.10.5368-5381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shayakhmetov DM, Gaggar A, Ni S, Li ZY, Lieber A. Adenovirus binding to blood factors results in liver cell infection and hepatotoxicity. J Virol. 2005;79:7478–7491. doi: 10.1128/JVI.79.12.7478-7491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood M, Perrotte P, Onishi E, Harper ME, Dinney C, Pagliaro L, et al. Biodistribution of an adenoviral vector carrying the luciferase reporter gene following intravesical or intravenous administration to a mouse. Cancer Gene Ther. 1999;6:367–372. doi: 10.1038/sj.cgt.7700090. [DOI] [PubMed] [Google Scholar]
- 51.Worgall S, Wolff G, Falck-Pedersen E, Crystal RG. Innate immune mechanisms dominate elimination of adenoviral vectors following in vivo administration. Hum Gene Ther. 1997;8:37–44. doi: 10.1089/hum.1997.8.1-37. [DOI] [PubMed] [Google Scholar]
- 52.Hallden G, Hill R, Wang Y, Anand A, Liu TC, Lemoine NR, et al. Novel immunocompetent murine tumor models for the assessment of replication-competent oncolytic adenovirus efficacy. Mol Ther. 2003;8:412–424. doi: 10.1016/s1525-0016(03)00199-0. [DOI] [PubMed] [Google Scholar]
- 53.Baker AH, Mcvey JH, Waddington SN, Di Paolo NC, Shayakhmetov DM. The influence of blood on in vivo adenovirus bio-distribution and transduction. Mol Ther. 2007;15:1410–1416. doi: 10.1038/sj.mt.6300206. [DOI] [PubMed] [Google Scholar]