Abstract
Anapleurosis is the filling of the TCA cycle with four-carbon units. The common substrate for both anapleurosis and glucose phosphorylation in bacteria is the terminal glycolytic metabolite, phosphoenolpyruvate (PEP). Here we show that E. coli quickly and almost completely turns off PEP consumption upon glucose removal. The resulting build-up of PEP is used to quickly import glucose if it becomes re-available. The switch-like termination of anapleurosis results from depletion of fructose-1,6-bisphosphate (FBP), an ultrasensitive allosteric activator of PEP carboxylase. E. coli expressing an FBP-insensitive point mutant of PEP carboxylase grow normally on steady glucose. However, they fail to build-up PEP upon glucose removal, grow poorly on oscillating glucose, and suffer from futile cycling at the PEP node on gluconeogenic substrates. Thus, bacterial central carbon metabolism is intrinsically programmed with ultrasensitive allosteric regulation to enable rapid adaptation to changing environmental conditions.
Introduction
The fate of PEP is among the most important governors of overall cellular metabolic activity, dictating the relative flux of glycolysis versus gluconeogenesis1. In eukaryotes, pyruvate kinase is the canonical PEP-catabolizing enzyme. Its importance is indicated by the existence in humans of four isozymes, one of which plays a central role in oncogenesis2. In prokaryotes, PEP can be catabolized by two additional routes: It is used as the phosphate donor for import of glucose and related sugars via the phosphotransferase system (PTS, net reaction: glucoseextracellular + PEP → glucose-6-phosphateintracellular + pyruvate). It is also the direct substrate for anapleurosis in some bacteria including E. coli, providing four carbon units to the TCA cycle via PEP carboxylase (pyruvate is used for anapleurosis in other bacteria, yeast, mammals3). In E. coli growing freely on glucose as the sole carbon source, ~ 82% of PEP is consumed for glucose import and ~ 12% for oxaloacetate synthesis4, with pyruvate kinase inessential for cell growth5.
In natural environments, microbes face of myriad of potential carbon sources whose availability varies. This places a premium on the ability to adapt to changing access to carbon. The mechanisms that enable rapid adaptation, on a time-scale faster than transcription, remain largely unproven. Metabolomics—the systems level analysis of metabolites—has opened a new window on such rapid regulation6,7. A striking finding, conserved across both E. coli and yeast, is that, while upper glycolytic compounds (from glucose to FBP) drop as expected upon glucose removal, PEP rises the most of any canonical metabolite8. In E. coli and other prokaryotes, this rise can in part be accounted for by PEP consumption by the PTS terminating immediately upon glucose removal. The regulation of other reactions, however, is clearly required to produce and sustain the dramatic rise in PEP. Here we show that PEP carboxylase shuts off in a switch-like manner upon glucose removal via ultrasensitive allosteric regulation by FBP, and that this regulation is essential for E. coli to utilize effectively gluconeogenic substrates or intermittently available glucose.
Results
Glucose removal results in PEP build-up
We measured the short term (within 90 minutes) metabolome response of E. coli to sudden switches from glucose to no carbon, acetate, succinate, or glycerol. E. coli cells were grown dispersed on filters on top of an agarose-media mixture, fed by diffuse of media components from below and oxygen from above. This culture method allowed rapid, non-disruptive glucose removal, isotope-switches, and metabolome harvesting by transfer to cold organic solvent (40:40:20 acetonitrile:methanol:water at −20 °C9). Metabolites were extracted in the cold organic solvent mixture and extracts analyzed by liquid chromatography – mass spectrometry (LC-MS), with compound identities verified by mass and retention time match to authenticated standards.
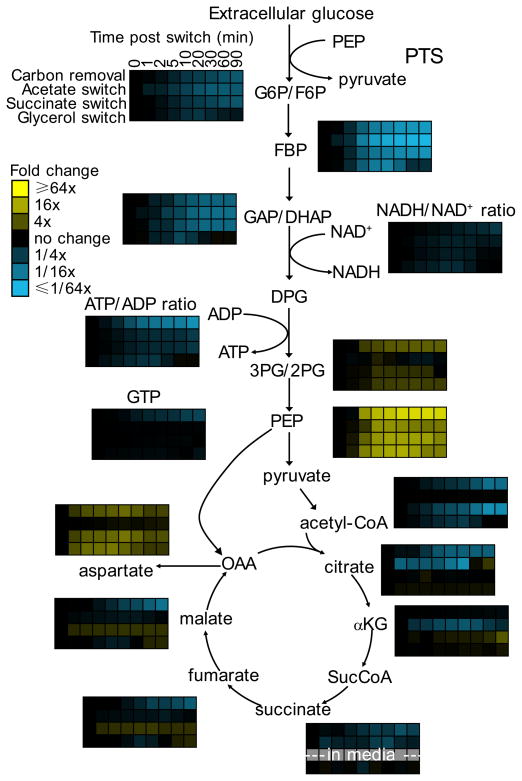
Within the first 10 minutes after glucose removal, under each of the four examined conditions, hexose phosphate concentrations decreased by 5- to 10-fold; FBP and GAP/DHAP decreased by 15- to 30-fold; and PEP increased by at least 10-fold (Fig. 1; Supplementary Results, Supplementary Fig. 1 – 3; Supplementary Dataset 1). The increase in PEP suggests a rapid shut-off of PEP consumption or rapid onset of gluconeogenesis. To evaluate the possibility that the PEP pool is being fed from TCA components, we switched cells from unlabeled glucose to U-13C-acetate and observed substantial malate labeling (> 80%) but no detectable PEP labeling (< 1%) over the first 45 minutes following glucose removal (Supplementary Fig. 4). This rules out substantial PEP formation by gluconeogenesis or the PEP-glyoxylate cycle10 on this time scale, and indicates that PEP consumption is blocked.
Fig. 1. Glucose removal results in PEP build-up.
E. coli cells growing freely in glucose minimal media were switched to no carbon, acetate, succinate, or glycerol as indicated. After the indicated duration of glucose removal, the metabolome was quantitated by LC-MS. Data are shown in heat map format, with each line reflecting the dynamics of a particular compound in a particular culture condition. Metabolite levels of biological duplicates were averaged, normalized to cells growing steadily in glucose (time zero), and the resulting fold changes log transformed. See Supplementary Fig. 1 for absolute concentration changes for key metabolites and Supplementary Fig. 2 for a heat map of all measured metabolites. Note that the decreased ATP/ADP and NADH/NAD+ ratios upon glucose removal thermodynamically drive carbon towards the bottom of glycolysis (i.e. towards PEP) (Supplementary Fig. 3). G6P, glucose-6-phosphate; F6P. fructose-6-phosphate; FBP, fructose-1,6-bisphosphate; GAP, glyceraldehyde-3-phosphate; DHAP, dihydroxyacetone phosphate; DPG, 1,3-diphosphateglycerate; 3PG, 3-phosphoglycerate; 2PG, 2-phosphoglycerate; OAA, oxaloacetate; αKG, α-ketoglutarate; SucCoA, succinyl-coenzyme A. G6P and F6P, GAP and DHAP, and 3PG and 2PG were not differentiated by employed LC-MS method.
Routes of PEP consumption
In E. coli, the phosphotransferase system (PTS) and PEP carboxylase (anapleurosis) are two major consumers of PEP. Assuming optimal aerobic carbon utilization for biomass production, these two reactions account for > 94% of PEP consumption (Supplementary Fig. 5). The other significant consumer of PEP during optimal growth on glucose is aromatic amino acid biosynthesis (~ 2%), with fluxes to cell wall and outer membrane biosynthesis having a combined contribution of only ~ 0.5%11,12.
While pyruvate kinase activity is not required for optimal growth on glucose, it has been recently been reported to account for ~ 15% of PEP consumption in batch culture13,14. However, pyruvate kinase activity decreases with oxygenation, and filter cultures have direct exposure to the aerobic environment15. To assess pyruvate kinase flux, we performed metabolic flux analysis using 1-13C-glucose as the tracer. In filter cultures, we find that pyruvate kinase flux accounts for less than 5% of PEP consumption (Supplementary Fig. 6, Supplementary Methods). Consistent with pyruvate kinase making only a minor contribution, side-by-side comparison of wild type and ΔpykF/ΔpykA double knockout cells revealed no differences in in growth rate, PEP concentration, or 1-13C-glucose labeling (Supplementary Fig. 6 and 7), and only small changes in metabolome response to glucose removal (Supplementary Fig. 8).
Having ruled out a primary contribution of pyruvate kinase, we considered how the other routes of PEP consumption might be regulated upon glucose removal. As glucose is a substrate of the PTS, glucose removal immediately stops this largest route of PEP consumption. Absence of glucose also stops protein synthesis; this led to build-up of tyrosine, tryptophan, and phenylalanine (Supplementary Fig. 5), which feedback-inhibit aromatic amino acid biosynthesis16. Moreover, absence of glucose decreased the concentration of substrates involved in the PEP-consuming reactions of cell wall and outer membrane synthesis; such depletion provides a potential route for turning off these minor fluxes (Supplementary Fig. 5). Thus, in order to explain the increase in PEP concentration upon glucose removal in our culture system, the major question regards the mechanism of PEP carboxylase inhibition.
FBP is a known positive regulator of PEP carboxylase17,18, and its depletion could potentially decrease PEP carboxylase activity and thus cause PEP accumulation. Existing biochemical literature data suggested, however, that the observed change in FBP concentration would lead to only a five-fold decrease in PEP carboxylase activity19,20. This seemed insufficient for PEP to build-up, given that the PEP carboxylase flux in cells growing steadily in glucose minimal media is 15 mM/min (see Methods), which would consume the observed PEP in 15 s. Other known regulators of PEP carboxylase include acetyl-CoA and GTP (positive regulators) and aspartate and malate (negative regulators)18. These species showed inconsistent and comparatively mild changes in the various carbon switches (Fig. 1); thus, it seemed unlikely that they are the main drivers of the universal spike in PEP.
Quantitation of residual PEP consumption flux
To measure directly the extent to which PEP consumption is cut off in vivo, we conducted a metabolomics experiment involving transient feeding of U-13C-glucose followed by glucose removal. By labeling glycolytic intermediates but not macromolecule stores (protein, glycogen)21, such transient labeling allowed us to determine the extent to which PEP production via macromolecule breakdown contributes to the high PEP pool size following glucose removal. For example, degradation of glycogen, glycerolipids, or nucleic acids could in theory feed unlabeled carbon into the PEP pool. As PEP remained largely labeled after the removal of U-13C-glucose (Fig. 2a and b; Supplementary Fig. 9), the PEP build-up is predominantly from glycolytic intermediates existing at the time of glucose removal. Such build-up requires an almost complete termination of PEP consumption, whose extent can be quantitated based on the labeling dynamics.
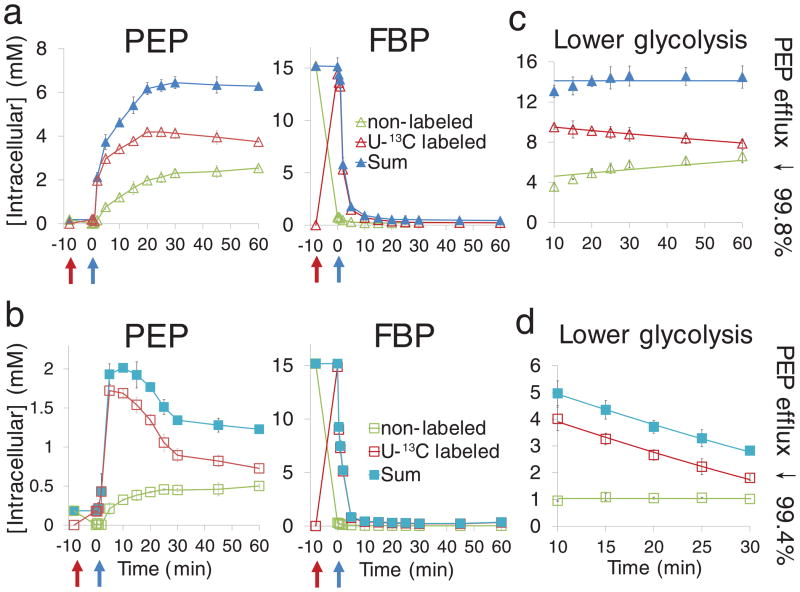
Fig. 2. PEP carboxylase is inhibited by 99% upon glucose removal.
(a, b). Concentrations of unlabeled and U-13C-labeled PEP and FBP, showing rapid initial conversion from unlabeled to labeled forms upon the switch to U-13C-glucose, and then, upon glucose removal, accumulation of unlabeled PEP and depletion of labeled FBP. Cells were grown on glucose minimal media as in Fig. 1. Glycolytic intermediates were labeled by switching cells to U-13C-glucose for 8 min. Thereafter, glucose was removed and replaced with either no carbon (a, b) or acetate (c, d). Green = nonlabeled, red = U-13C labeled, blue = sum of nonlabeled and U-13C labeled. (c, d) Concentrations of unlabeled and U-13C-labeled lower glycolytic intermediates (sum of all measured compounds from FBP to PEP) starting 10 min after glucose removal. Points reflect data (mean ± range of N = 2 biological replicates); lines represent a fit of the data to equations described in Methods. The fitting enabled estimation of the decrease in PEP carboxylase flux relative to glucose-fed cells.
In the case of the switch to no carbon source, the total concentration of lower glycolytic intermediates (from FBP to PEP) remained roughly stable from 10 – 60 minutes after glucose removal. The changing labeling patterns can accordingly be interpreted assuming equal total triose production and consumption fluxes during this period (Fig. 2c and Supplementary Fig. 10a). The resulting fluxes after glucose removal, based on the rate of decrease of the labeled form, were 0.051 mM/min (99.6% less than PEP carboxylase flux in the glucose-fed cells; see Methods). In the case of the switch to acetate, the total lower glycolytic pool dropped while the unlabeled pool remained steady. This is consistent with steady consumption of lower glycolytic intermediates during the first 10 – 30 minutes after the switch from glucose to acetate (Fig. 2d and Supplementary Fig. 10b) at a rate of 0.15 mM/min, requiring a 99.0% inhibition of PEP carboxylase. These decreases far exceed what would be expected based on prior biochemical literature19,20,22.
The above analysis indicates that PEP consumption in carbon starvation is yet lower than in the acetate switch condition. This is logical, as carbon starvation triggers depletion of acetyl-CoA, a PEP carboxylase activator, and accumulation of aspartate, a PEP carboxylase inhibitor. While aspartate is produced in a single step from oxaloacetate, the isotope labeling pattern of the accumulated aspartate indicates that it did not come from anapleurosis. A likely source is instead from glutamate, which declined in parallel with the rise in aspartate (Supplementary Fig. 11). Given that acetyl-CoA and aspartate do not change substantially during the acetate switch (Fig. 1), it is particularly striking that PEP carboxylase flux nevertheless terminates 99%.
FBP activation of PEP carboxylase is ultrasensitive
Given the almost complete elimination of PEP carboxylase flux, we hypothesized that the enzyme might be degraded or covalently modified. To examine these possibilities, we quantitated PEP carboxylase protein concentration and activity before and after glucose removal. Targeted LC-MS revealed no change in PEP carboxylase protein concentration following glucose removal (Supplementary Fig. 12a, Supplementary Methods). Similarly, PEP carboxylase activity in cell lysates remained constant, which is inconsistent with PEP carboxylase inactivation by a stable covalent modification event23 (Supplementary Fig. 12b, Supplementary Methods). To further rule-out potential regulation at the level of protein concentration, we inducibly over-expressed PEP carboxylase in wild type E. coli. While PEP carboxylase over-expression modestly reduced the basal PEP concentration in glucose-fed cells, it did not block the PEP spike after glucose removal (Supplementary Fig. 12c).
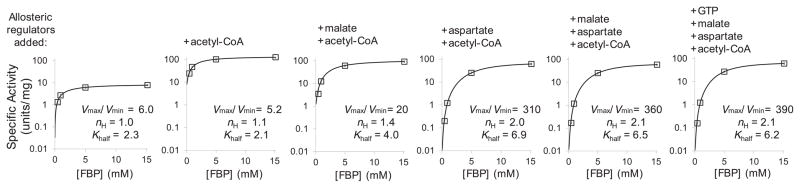
Given the above evidence that PEP carboxylase is present but not actively consuming PEP following glucose removal, we re-examined PEP carboxylase’s allosteric regulation. A deficiency of prior literature is that PEP carboxylase activity was measured one effector at a time, not always using the effectors’ physiological concentrations19,20,22,24,25. Given our evidence from metabolomics that FBP appears to be the primary regulator, we examined FBP’s effect over its physiological range, alone and in the presence of additional regulators (Fig. 3 and Supplementary Fig. 13). FBP alone showed only modest control of PEP carboxylase activity (6-fold), and its effect was not ultrasensitive (Hill coefficient 1.0). However, in the presence of both acetyl-CoA and aspartate, the effects of FBP were magnified (310-fold) and ultrasensitive (Hill coefficient 2.1). This ultrasensitive regulation is in principle sufficient to produce rapidly the observed > 99% decrease in PEP carboxylase activity upon glucose removal (Supplementary Fig. 14).
Fig. 3. Activation of PEP carboxylase by FBP is enhanced by physiological levels of acetyl-CoA and aspartate.
Each small plot shows activity of purified His-tagged PEP carboxylase as a function of FBP concentration. The six small plots differ in terms of the additional PEP carboxylase effectors present, building from FBP alone (left-most) to all known effectors (right-most) as indicated. The concentration of PEP (2 mM), as well as effectors acetyl-CoA (0.63 mM), aspartate (5.2 mM), malate (1.1 mM), and GTP (4.6 mM) were selected to match the physiological concentration in cells switched from glucose to acetate for 10 minutes (Supplementary Table 1; see Supplementary Fig. 13 for other conditions). Reactions were carried out at 30°C and pH 8.5 in the presence of 10 mM bicarbonate and magnesium. In the plots, the x axis represents different FBP concentrations, and the logarithmic y axis represents specific PEP carboxylase activity in enzyme activity Units (μmol of product produced per minute) per mg of enzyme. Experimental data (mean ± range of N = 2) were fit to Hill-equations (lines). Vmax/Vmin represents observed range of PEP carboxylase activity as a function of FBP concentration, where Vmax was measured at 15 mM FBP, matching the physiological concentration in cells growing on glucose, and Vmin was measured at 0.45 mM FBP, matching the physiological FBP concentration in cells switched from glucose to acetate for 10 minutes. nH and Khalf represent the Hill-coefficient for FBP and the FBP concentration producing half-maximal activation.
We were also interested in whether allosteric regulation of PEP carboxylase by FBP is in principle sufficient for PEP to rise when FBP falls. To this end, we developed a simple mathematical model of glycolytic regulation (Supplementary Methods). The model includes FBP production, its reversible interconversion with PEP, and PEP consumption by the PTS and PEP carboxylase, with PEP carboxylase flux proportional to [FBP]h. Using the model, we examined the steady-state concentrations of FBP and PEP as a function of glycolytic flux (F). We find that FBP always decreases when F decreases. This matches experimental observations. For h ≤ 1, the same is true for PEP. For h > 1, however, PEP can increase when F decreases (Supplementary Fig. 15). Thus, ultrasensitive regulation of PEP carboxylase is a prerequisite for PEP to accumulate when glycolytic flux decreases, as is observed experimentally.
PEP accumulation results from the depletion of FBP
To determine the physiological significance of PEP carboxylase regulation by FBP, we sought a PEP carboxylase point mutant desensitized to FBP activation (Fig. 4). While the FBP binding site on E. coli’s PEP carboxylase has not been determined, an allosteric regulatory site for glucose-6-phosphate on maize PEP carboxylase has been identified crystallographically26. This site involves four arginine residues. Although E. coli PEP carboxylase is not activated by G6P or F6P (Supplementary Fig. 16), all four of these arginine residues are conserved in the E. coli protein. We thus hypothesized that a phosphate group of FBP might interact with one or more of these residues. Site-directed mutagenesis led to a mutant form of E. coli PEP carboxylase, R313Q, which was more active than native PEP carboxylase in the absence of FBP and desensitized to FBP activation (Fig. 4a). For kinetic parameters of the altered enzyme, see Supplementary Table 2 and Supplementary Fig. 17
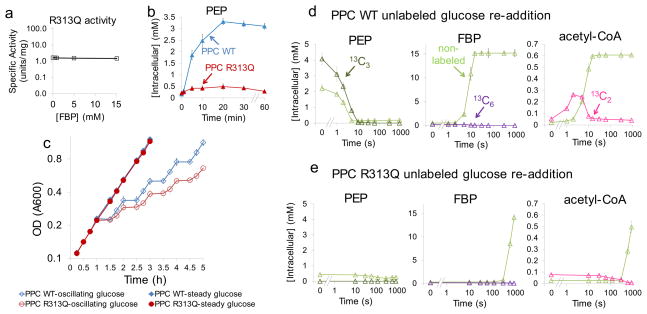
Fig. 4. PEP accumulation upon glucose removal enables rapid import of glucose when it reappears.
(a). PEP carboxylase mutant R313Q has higher activity than wild type in the absence of FBP and is desensitized to FBP activation. The concentration of PEP and effectors and the y-axis units match the right-most plot in Fig. 3. (b). Expression of PEP carboxylase R313Q ablates the PEP spike upon carbon starvation (see Supplementary Fig. 18a for switch from glucose to acetate). E. coli strains in which endogenous PEP carboxylase has been replaced with inducible expression of wild type enzyme (Δppc/pCA24N-ppc, shown as PPC WT) or the FBP-insensitive R313Q mutant (Δppc/pCA24N-ppcR313Q, shown as PPC R313Q) were switched to no carbon. (c). Growth curves of Δppc/pCA24N-ppc and Δppc/pCA24N-ppcR313Q. Cells were grown in either steady glucose (closed symbols) or alternating between glucose minimal media and no carbon minimal media every 30 min (open symbols). The x axis represents time in hours, and the logarithmic y axis represents optical density (A600). (d, e). Cells were switched to U-13C-glucose for 8 minutes, followed by no carbon for 30 minutes, and finally re-addition of unlabeled glucose. Upon glucose re-addition, samples were collected and intracellular metabolites (labeled and unlabeled) quantitated by LC-MS. The x axis represents seconds after glucose re-addition (in logarithmic scale), and the y axis represents absolute intracellular concentration (mean ± range of N = 2 biological replicates); (d) Δppc/pCA24N-ppc, (e) Δppc/pCA24N-ppcR313Q. Throughout (b) to (e), experiments were performed in the presence of 100 μM IPTG.
We constructed an E. coli strain that lacked native PEP carboxylase (Δppc) and inducibly expressed the R313Q mutant. In the presence of inducer, these cells (Δppc/pCA24N-ppcR313Q) grew normally on glucose, with an intracellular PEP concentration slightly higher than wild type cells (0.21 mM versus 0.16 mM). Upon glucose removal, despite a normal decline in FBP, the mutant cells failed to show a PEP spike (Fig. 4b and Supplementary Fig. 18 and 19). Expression of wild type enzyme in the same manner (Δppc/pCA24N-ppc) resulted in a normal PEP spike. Thus, the FBP-insensitive R313Q mutant resulted in inappropriate anapleurotic flux that drained PEP.
Accumulated PEP rapidly imports glucose when it reappears
The FBP-insensitive mutant allowed us to test the physiological function of PEP accumulation. We hypothesized that a reserve of PEP is useful for rapidly importing glucose if it becomes re-available. To test this possibility, we labeled glycolytic intermediates with uniformly 13C-glucose, removed carbon for 30 minutes, and then added unlabeled glucose. For Δppc/pCA24N-ppc, glucose addition resulted in consumption of the pre-existing 13C3-PEP within 10 s (Fig. 4d). Consistent with this PEP being used for glucose import, the labeled carbon appeared in 13C3-pyruvate within 5 seconds. This pyruvate was then quickly oxidized by pyruvate dehydrogenase to yield 13C2-acetyl-CoA. The resulting import of glucose resulted in a rapid rise in FBP, which reached its normal steady state level within 15 seconds. This increase in FBP activated anapleurosis: malate increased to its normal level within 1 minute. The labeling pattern of malate confirmed that it was produced via anapleurosis (Supplementary Fig. 20a). In contrast, for Δppc/pCA24N-ppcR313Q, due to the lack of PEP to jump-start glucose assimilation, FBP did not reach normal levels until 15 minutes after glucose re-addition, 60-fold more slowly than with wild type PEP carboxylase (Fig. 4e). The slow increase of FBP also caused a slow turn-on of anapleurosis (Supplementary Fig. 20b).
The above metabolomic data suggested that the PEP accumulation resulting from allosteric regulation of PEP carboxylase might be required for normal resumption of growth after glucose re-addition. To test this possibility, we grew Δppc/pCA24N-ppcR313Q and Δppc/pCA24N-ppc on steady versus oscillating glucose. While the two strains grew identically on steady glucose, the R313Q PEP carboxylase mutant strain was deficient in growing on oscillating glucose (Fig. 4c). Thus, the ultrasensitive regulation of PEP carboxylase enables cellular adaptation to varying carbon availability.
FBP regulation inhibits futile cycle in gluconeogenesis
In the presence of non-hexose carbon sources, E. coli will eventually transition to gluconeogenesis. For carbon sources feeding into the TCA cycle (e.g., succinate or acetate), this requires production of PEP via either PEP carboxykinase or malic enzyme-PEP synthase27, both of which consume ATP. If PEP carboxylase is simultaneously active, ATP will be wasted by futile cycling. Because gluconeogenic growth is associated with reduced FBP levels28, we hypothesized that another physiological function of PEP carboxylase regulation is to prevent futile cycling during gluconeogenesis. During steady growth on acetate, PEP carboxylase protein is present intracellularly at > 25% of the protein level of glucose grown cells15 (Supplementary Fig. 21). In wild type cells, however, futile cycling is so low as to be undetectable29.
To examine the role of PEP carboxylase allosteric regulation in limiting futile cycling, we measured metabolite levels and growth yields in succinate- or acetate-fed E. coli Δppc/pCA24N-ppcR313Q and Δppc/pCA24N-ppc at various levels of inducer. Addition of 10 μM IPTG resulted in expression of PEP carboxylase to a similar level as that found in wild type cells growing on acetate (Supplementary Fig. 21b). Increasing expression of wild type PEP carboxylase had only a slight effect on cellular metabolite levels (Fig. 5a). Growth rate, succinate and acetate uptake rates, and growth yields decreased only modestly with higher PEP carboxylase expression (Fig. 5b). In contrast, expression of the R313Q mutant resulted in substantial depletion of glycolytic intermediates and build-up of TCA cycle four-carbon units, consistent with marked PEP carboxylase activity and associated futile cycling. Moreover, growth rate fell monotonically with increasing PEP carboxylase R313Q expression, approaching zero at high expression levels. This decrease in growth rate reflected a combination of lower acetate and succinate uptake (presumably due to TCA cycle overloading impairing net carbon import) and worse yield per carbon consumed (presumably due to futile cycling).
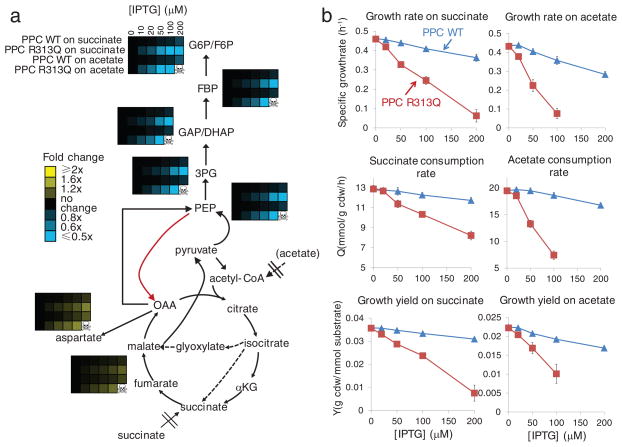
Fig. 5. The FBP-insensitive variant of PEP carboxylase causes futile cycling on gluconeogenic media.
PEP carboxylase does not productively contribute to growth on acetate or succinate, and thus optimal regulation would eliminate its activity. Persistent activity under these conditions would be expected to deplete PEP and upstream metabolites, to elevate TCA intermediates, and to reduce growth yields. (a) Intracellular metabolite concentrations in Δppc/pCA24N-ppc (PPC WT) and Δppc/pCA24N-ppcR313Q (PPC R313Q) as a function of added IPTG. Data are shown in heat map format, with each line a particular strain and carbon source. Metabolite levels were averaged, normalized to cells without recombinant enzyme expression (IPTG = 0), and the resulting fold changes log transformed. Data for Δppc/pCA24N-ppcR313Q at 200 μM IPTG is missing because the strain would not grow. (b) Growth parameters of Δppc/pCA24N-ppc and Δppc/pCA24N-ppcR313Q growing on succinate (left) and acetate (right). The x axis represents different IPTG concentration in μM, and the y axis represents parameters shown in the plots. Points reflect data (mean ± range of N = 2 biological replicates).
Discussion
Here we show that ultrasensitive allosteric activation of PEP carboxylase by FBP plays a major role in controlling central metabolic activity in E. coli. At the protein level, such regulation involves sensitization of PEP carboxylase to FBP by physiological concentrations of acetyl-CoA and aspartate. At the systems level, it enables accumulation of PEP upon glucose removal, which in turn provides an advantage in reinitiating glycolysis when glucose reappears.
A key question is whether the ultrasensitive nature of the regulation is truly required to achieve PEP accumulation. A simple mathematical model of glycolytic regulation demonstrates that the observed ultrasensitivity is a prerequisite for PEP showing inverse concentration changes to FBP (Supplementary Fig. 15), and thus PEP accumulating when glycolytic flux decreases, as has been observed experimentally in E. coli and other organisms30,31. Like PEP carboxylase, the pyruvate kinase isozyme which is expressed in the presence of glucose (PykF) is activated by FBP32. We anticipate that, in the presence of physiological metabolite concentrations, PykF will also respond ultrasensitively to FBP, rendering PEP accumulation a robust physiological response to glucose removal.
The effects of PEP carboxylase and pyruvate kinase regulation may be augmented by allosteric inactivation of phosphofructokinase by PEP33: initial depletion of FBP and build-up of PEP will promote further inactivation of both enzymes, thereby locking glycolysis in an off state until a suitable glycolytic substrate reappears.
A putative advantage of allosteric regulation is speed34. This seems paramount for shut off of PEP carboxylase and pyruvate kinase upon glucose removal, where termination of glycolytic egress must out race depletion of upstream glycolytic compounds. In contrast, in cases where enzymes in E. coli are inactivated via covalent modification35, there is not a corresponding need for fast regulation. For example, the central nitrogen metabolic enzyme glutamine synthetase is turned off by covalent modification upon ammonia upshift36; in this case, the delay introduced by the covalent modification step may be advantageous, favoring uptake of a useful initial burst of nitrogen.
An unexpected feature of PEP carboxylase’s allosteric activation is its ability to produce switch-like flux control that overrides the effects of substrate levels. PEP carboxylase is a tetrameric enzyme, compromised of a four identical catalytic units37. The same is true of PykF32. A likely reason for the evolution of multimeric enzymes is to enable ultrasensitivity, to substrate38–42, to feedback inhibitors in biosynthesis43,44, or, as shown here, to feed forward activation in central carbon metabolism. The existence of such intrinsically ultrasensitive enzymes confers metabolism with substantial capabilities for self-regulation.
Methods
Microbial culture conditions and extraction
Filter cultures and extraction of metabolites in E. coli were carried out as described previously28,45. Briefly, E. coli K-12 strain NCM3722 was grown at 37°C in minimal salts media46 with 10 mM ammonium chloride as the nitrogen source and 0.4% (w/v) glucose as the carbon source resulting in an exponential growth rate of 0.74 h−1. For glucose removal, filters were then switched from glucose plates to plates containing no carbon source, 0.4% (w/v) acetate, 0.4% (w/v) succinate, or 0.4% (w/v) glycerol, and extracted at indicated min after the switch. For transient labeling experiments, cells were first grown on unlabeled glucose (0.4%) plates and then switched to the plates containing 0.4% U-13C-glucose as the carbon source for 2 or 8 minutes21. After that, filters were switched to the plates containing either no carbon or 0.4% acetate as the carbon source. For growth curves on oscillating glucose (Fig. 4c), cells were switched between glucose and no carbon plates (a new plate each time), spending half an hour on each plate. For steady glucose growth, cells were switched every 30 min to a new glucose plate. For details, see Supplementary Methods.
Metabolite and flux measurement
Cell extracts were analyzed by reversed phase ion-pairing liquid chromatography (LC) coupled by electrospray ionization (ESI) (negative mode) to either a high-resolution, high-accuracy mass spectrometer (Exactive; Thermo Fisher) operated in full scan mode, or a Thermo TSQ Quantum triple quadrupole mass spectrometer operating in multiple reaction monitoring mode47, with compound identities verified by mass and retention time match to authenticated standard48 (Supplementary Table 3 and 4; Supplementary Fig. 22). The mean and range of measured metabolite concentrations were presented. For details, see Supplementary Methods.
Steady-state fluxes in glucose-fed, exponentially growing cells were inferred based on measured nutrient uptake and waste excretion, growth rate and associated requirements for biomass production, and metabolic flux analysis with 1-13C-glucose tracer. For details, see Supplementary Methods. Substrate uptake and waste excretion rates were measured by a combination of LC-MS and NMR.
Preparation of recombinant PEP carboxylase protein
E. coli PEP carboxylase was purified from bacteria that carried the plasmid pCA24N (from the ASKA library), which encodes His-tagged PEP carboxylase49, using Ni-NTA spin columns (QIAGEN). The purification protocol was optimized to obtain nearly homogenous expression of tetrameric enzyme. For details, see Supplementary Methods.
Measurement of PEP carboxylase activity
PEP carboxylase activity was measured by coupling the PEP carboxylase reaction to the malate dehydrogenase (MDH) reaction as described previously19. However, instead of using a spectrophotometer to measure NADH absorbance, we used LC-MS to directly measure the reaction product malate. This enabled more direct and accurate measurement of PEP carboxylase activity separate from other potential NADH consuming reactions. For details, see Supplementary Methods
Mutant construction and site-directed mutagenesis
P1vir transduction was used to move the necessary alleles from the Keio collection50 strains in which ppc had been replaced with a kanamycin (kan) resistance cassette into the NCM3722 background. The kan cassette was removed using FLP recombinase to create unmarked, in-frame ppc deletion strain (Δppc) in the NCM3722 background.
The plasmid pCA24N from the ASKA library containing PEP carboxylase was used as the template for site-directed mutagenesis reactions following the protocol of Quickchange Site-Directed Mutagenesis (Stratagene). For details, see Supplementary Methods.
Quantitation of PEP consumption flux based on data obtained from transient 13C-glucose labeling followed by glucose removal
In the case of the 8 min switch to no carbon, the total PEP pool is roughly stable from 20 – 60 min, and steadily becomes less labeled during this time. This is consistent with steady production of unlabeled PEP from macromolecule degradation and matching steady consumption PEP (with labeled and unlabeled PEP consumed in proportion to their fraction of the total PEP pool). To quantitate the flux from 10 – 60 min, we used the total pool of lower glycolytic intermediates (FBP + GAP/DHAP + DPG/3PG/2PG + PEP, which are interconverted via reversible reactions). This pool is roughly steady from 10 – 60 minutes, and has the same labeling dynamics as PEP (Fig. 2c). As the source of labeled carbon is depleted after 10 minutes, production of unlabeled PEP from macromolecules (glycogen, glycerolipids, or nucleic acids) and total consumption of PEP (labeled + unlabeled) were balanced. This steady-state flux is given by the rate of conversion from labeled to unlabeled carbon by kinetic flux profiling45. Briefly, the kinetics of 13C lower glycolytic pool XL is determined by
| (1) |
where f is the steady-state flux and XT is the total (unlabeled + labeled) lower glycolytic pool size. The solution of this ordinary differential equation (ODE) starting at t0 = 10 min after carbon removal is
| (2) |
where XL0 is the 13C pool size at t0. The plot (XT ln(XL0/XL)) vs. (t − t0) gave a straight line (Supplementary Fig. 10a), consistent with the above analysis. Least square fitting results in f = 0.051 mM/min, which is 99.6% less than PEP carboxylase flux in the glucose-fed cells.
In the acetate switch case, the total lower glycolytic pool size drops after 10 min. However, total pool size of unlabeled species is steady (Fig. 2d). Accordingly, the kinetics of the 13C-labeled lower glycolytic pool XL is determined by
| (3) |
where fex is the steady PEP consumption flux (efflux) and XU is the constant unlabeled lower glycolytic pool size. The solution of this ODE starting at t0 = 10 min is
| (4) |
where XL0 is the 13C pool size at t0. The plot (XU ln XL + XL) vs. (t − t0) gave a straight line (Supplementary Fig. 10b) at 10 – 30 min, consistent with the above analysis. Least square results in fex = 0.15 mM/min, which corresponds to a 99.0% inhibition of PEP carboxylase.
Supplementary Material
Acknowledgments
We thank Chris Doucette for the ppc deletion strain, Alison Hottes for the pCA24N-ppc plasmid, Junyoung Park for the flux calculations, Allison Michaelis and Zemer Gitai for the microscopy, David Perlman, Wenyun Lu, Kyin Saw and Henry Shwe for protein mass spectrometry, and the above colleagues plus Ned Wingreen for helpful discussions. This research was funded by NSF CAREER award MCB-0643859, Joint DOE-AFOSR Award DOE DE-SC0002077 - AFOSR FA9550-09-1-0580, the NIH Center for Quantitative Biology Award P50 GM071508, and NSF grant CBET-0941143. M.L.R. was supported by NSF Graduate Research Fellowship DGE-0646086.
Footnotes
Author Contributions
Y.F.X. and J.D.R. designed experiments, analyzed data, and wrote the paper. D.A.N. and M.L.R. performed preliminary experiments and contributed to discussion. X.J. F. contributed to modeling.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Kotte O, Zaugg JB, Heinemann M. Bacterial adaptation through distributed sensing of metabolic fluxes. Molecular Systems Biology. 2010;6:355. doi: 10.1038/msb.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 3.Sauer U, Eikmanns BJ. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. Fems Microbiology Reviews. 2005;29:765–794. doi: 10.1016/j.femsre.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Palsson BO, et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Molecular Systems Biology. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siddiquee KA, Arauzo-Bravo MJ, Shimizu K. Effect of a pyruvate kinase (pykF-gene) knockout mutation on the control of gene expression and metabolic fluxes in Escherichia coli. Fems Microbiology Letters. 2004;235:25–33. doi: 10.1016/j.femsle.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Ishii N, et al. Multiple high-throughput analyses monitor the response of E-coli to perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- 7.Fiehn O. Metabolomics - the link between genotypes and phenotypes. Plant Molecular Biology. 2002;48:155–171. [PubMed] [Google Scholar]
- 8.Brauer MJ, et al. Conservation of the metabolomic response to starvation across two divergent microbes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19302–19307. doi: 10.1073/pnas.0609508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabinowitz JD, Kimball E. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Analytical Chemistry. 2007;79:6167–6173. doi: 10.1021/ac070470c. [DOI] [PubMed] [Google Scholar]
- 10.Fischer E, Sauer U. A novel metabolic cycle catalyzes glucose oxidation and anaplerosis in hungry Escherichia coli. Journal of Biological Chemistry. 2003;278:46446–46451. doi: 10.1074/jbc.M307968200. [DOI] [PubMed] [Google Scholar]
- 11.Salleh HM, Patel MA, Woodard RW. Essential cysteines in 3-deoxy-D-manno-octulosonic acid 8-phosphate synthase from Escherichia coli: Analysis by chemical modification and site-directed mutagenesis. Biochemistry. 1996;35:8942–8947. doi: 10.1021/bi952373w. [DOI] [PubMed] [Google Scholar]
- 12.Kim DH, et al. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry. 1996;35:4923–4928. doi: 10.1021/bi952937w. [DOI] [PubMed] [Google Scholar]
- 13.Siddiquee KA, Arauzo-Bravo MJ, Shimizu K. Metabolic flux analysis of pykF gene knockout Escherichia coli based on C-13-labeling experiments together with measurements of enzyme activities and intracellular metabolite concentrations. Applied Microbiology and Biotechnology. 2004;63:407–417. doi: 10.1007/s00253-003-1357-9. [DOI] [PubMed] [Google Scholar]
- 14.van Rijsewijk BRBH, Nanchen A, Nallet S, Kleijn RJ, Sauer U. Large-scale (13)C-flux analysis reveals distinct transcriptional control of respiratory and fermentative metabolism in Escherichia coli. Molecular Systems Biology. 2011;7:477. doi: 10.1038/msb.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng L, Shimizu K. Global metabolic regulation analysis for Escherichia coli K12 based on protein expression by 2-dimensional electrophoresis and enzyme activity measurement. Applied Microbiology and Biotechnology. 2003;61:163–178. doi: 10.1007/s00253-002-1202-6. [DOI] [PubMed] [Google Scholar]
- 16.Herrmann KM. The Shikimate Pathway as an Entry to Aromatic Secondary Metabolism. Plant Physiology. 1995;107:7–12. doi: 10.1104/pp.107.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanwal BD, Maeba P. Regulation of Activity of Phosphoenolypyruvate Carboxylase by Fructose Diphosphate. Biochemical and Biophysical Research Communications. 1966;22:194–199. doi: 10.1016/0006-291x(66)90431-1. [DOI] [PubMed] [Google Scholar]
- 18.Izui K, Matsumura H, Furumoto T, Kai Y. Phosphoenolpyruvate carboxylase: A new era of structural biology. Annual Review of Plant Biology. 2004;55:69–84. doi: 10.1146/annurev.arplant.55.031903.141619. [DOI] [PubMed] [Google Scholar]
- 19.Morikawa M, Izui K, Taguchi M, Katsuki H. Regulation of Escherichia-Coli Phosphoenolpyruvate Carboxylase by Multiple Effectors Invivo.1. Estimation of the Activities in the Cells Grown on Various Compounds. Journal of Biochemistry. 1980;87:441–449. doi: 10.1093/oxfordjournals.jbchem.a132764. [DOI] [PubMed] [Google Scholar]
- 20.Izui K, Taguchi M, Morikawa M, Katsuki H. Regulation of Escherichia-Coli Phosphoenolpyruvate Carboxylase by Multiple Effectors Invivo.2. Kinetic-Studies with a Reaction System Containing Physiological Concentrations of Ligands. Journal of Biochemistry. 1981;90:1321–1331. doi: 10.1093/oxfordjournals.jbchem.a133597. [DOI] [PubMed] [Google Scholar]
- 21.Yuan J, Rabinowitz JD. Differentiating metabolites formed from de novo synthesis versus macromolecule decomposition. Journal of the American Chemical Society. 2007;129:9294–5. doi: 10.1021/ja072997c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kodaki T, Fujita N, Kameshita I, Izui K, Katsuki H. Phosphoenolpyruvate Carboxylase of Escherichia-Coli - Specificity of Some Compounds as Activators at the Site for Fructose-1,6-Bisphosphate, One of the Allosteric Effectors. Journal of Biochemistry. 1984;95:637–642. doi: 10.1093/oxfordjournals.jbchem.a134652. [DOI] [PubMed] [Google Scholar]
- 23.Garnak M, Reeves HC. Phosphorylation of Isocitrate Dehydrogenase of Escherichia-Coli. Science. 1979;203:1111–1112. doi: 10.1126/science.34215. [DOI] [PubMed] [Google Scholar]
- 24.Wohl RC, Markus G. Phosphoenolpyruvate Carboxylase of Escherichia-Coli -Purification and Some Properties. Journal of Biological Chemistry. 1972;247:5785–92. [PubMed] [Google Scholar]
- 25.Silverst R, Willis MS. Concerted Regulation in-Vitro of Phosphoenolpyruvate Carboxylase from Escherichia-Coli. Journal of Biological Chemistry. 1973;248:8402–8407. [PubMed] [Google Scholar]
- 26.Takahashi-Terada A, et al. Maize phosphoenolpyruvate carboxylase - Mutations at the putative binding site for glucose 6-phosphate caused desensitization and abolished responsiveness to regulatory phosphorylation. Journal of Biological Chemistry. 2005;280:11798–11806. doi: 10.1074/jbc.M408768200. [DOI] [PubMed] [Google Scholar]
- 27.Oh MK, Rohlin L, Kao KC, Liao JC. Global expression profiling of acetate-grown Escherichia coli. Journal of Biological Chemistry. 2002;277:13175–13183. doi: 10.1074/jbc.M110809200. [DOI] [PubMed] [Google Scholar]
- 28.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nature Chemical Biology. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J, Baba T, Mori H, Shimizu K. Effect of zwf gene knockout on the metabolism of Escherichia coli grown on glucose or acetate. Metabolic Engineering. 2004;6:164–174. doi: 10.1016/j.ymben.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Schaub J, Reuss M. In Vivo Dynamics of Glycolysis in Escherichia coli Shows Need for Growth-Rate Dependent Metabolome Analysis. Biotechnology Progress. 2008;24:1402–1407. doi: 10.1002/btpr.59. [DOI] [PubMed] [Google Scholar]
- 31.Theobald U, Mailinger W, Baltes M, Rizzi M, Reuss M. In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae.1. Experimental observations. Biotechnology and Bioengineering. 1997;55:305–316. doi: 10.1002/(SICI)1097-0290(19970720)55:2<305::AID-BIT8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 32.Zhu T, Bailey MF, Angley LM, Cooper TF, Dobson RCJ. The quaternary structure of pyruvate kinase type 1 from Escherichia coli at low nanomolar concentrations. Biochimie. 2010;92:116–120. doi: 10.1016/j.biochi.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa T, Mori H, Tomita M, Yoshino M. Inhibitory effect of phosphoenolpyruvate on glycolytic enzymes in Escherichia coli. Research in Microbiology. 2007;158:159–163. doi: 10.1016/j.resmic.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Kern D, Zuiderweg ER. The role of dynamics in allosteric regulation. Curr Opin Struct Biol. 2003;13:748–57. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Walsh K, Koshland DE. Branch Point Control by the Phosphorylation State of Isocitrate Dehydrogenase - a Quantitative Examination of Fluxes during a Regulatory Transition. Journal of Biological Chemistry. 1985;260:8430–8437. [PubMed] [Google Scholar]
- 36.Stadtman ER. The story of glutamine synthetase regulation. Journal of Biological Chemistry. 2001;276:44357–44364. doi: 10.1074/jbc.R100055200. [DOI] [PubMed] [Google Scholar]
- 37.Kai Y, Matsumura H, Izui K. Phosphoenolpyruvate carboxylase: three-dimensional structure and molecular mechanisms. Archives of Biochemistry and Biophysics. 2003;414:170–179. doi: 10.1016/s0003-9861(03)00170-x. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez CF, et al. Molecular Basis of Formaldehyde Detoxification CHARACTERIZATION OF TWO S-FORMYLGLUTATHIONE HYDROLASES FROM ESCHERICHIA COLI, FrmB AND YeiG. Journal of Biological Chemistry. 2006;281:14514–14522. doi: 10.1074/jbc.M600996200. [DOI] [PubMed] [Google Scholar]
- 39.Brown G, et al. Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli. Journal of Biological Chemistry. 2009;284:3784–3792. doi: 10.1074/jbc.M808186200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee M, Chan CW, Guss JM, Christopherson RI, Maher MJ. Dihydroorotase from Escherichia coli: Loop movement and cooperativity between subunits. Journal of Molecular Biology. 2005;348:523–533. doi: 10.1016/j.jmb.2005.01.067. [DOI] [PubMed] [Google Scholar]
- 41.Song WJ, Jackowski S. Kinetics and Regulation of Pantothenate Kinase from Escherichia-Coli. Journal of Biological Chemistry. 1994;269:27051–27058. [PubMed] [Google Scholar]
- 42.Bhasin M, Billinsky JL, Palmer DRJ. Steady-state kinetics and molecular evolution of Escherichia coli MenD [(1R,6R)-2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase], an anomalous thiamin diphosphate-dependent decarboxylase-carboligase. Biochemistry. 2003;42:13496–13504. doi: 10.1021/bi035224j. [DOI] [PubMed] [Google Scholar]
- 43.Broglie KE, Takahashi M. Fluorescence studies of threonine-promoted conformational transitions in aspartokinase I using the substrate analogue 2′(3′)-O-(2,4,6-trinitrophenyl)adenosine 5′-triphosphate. J Biol Chem. 1983;258:12940–6. [PubMed] [Google Scholar]
- 44.Eisenstein E, Yu HD, Schwarz FP. Cooperative binding of the feedback modifiers isoleucine and valine to biosynthetic threonine deaminase from Escherichia coli. J Biol Chem. 1994;269:29423–9. [PubMed] [Google Scholar]
- 45.Yuan J, Bennett BD, Rabinowitz JD. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nat Protoc. 2008;3:1328–40. doi: 10.1038/nprot.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gutnick D, Calvo JM, Klopotow T, Ames BN. Compounds Which Serve as Sole Source of Carbon or Nitrogen for Salmonella Typhimurium Lt-2. Journal of Bacteriology. 1969;100:215–219. doi: 10.1128/jb.100.1.215-219.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu W, Bennett BD, Rabinowitz JD. Analytical strategies for LC-MS-based targeted metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871:236–42. doi: 10.1016/j.jchromb.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rabinowitz JD, et al. Metabolomic Analysis via Reversed-Phase Ion-Pairing Liquid Chromatography Coupled to a Stand Alone Orbitrap Mass Spectrometer. Analytical Chemistry. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (A complete Set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Research. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 50.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular Systems Biology. 2006:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.