Abstract
The Culex pipiens complex includes two widespread mosquito vector species, Cx. pipiens and Cx. quinquefasciatus. The distribution of these species varies in latitude, with the former being present in temperate regions and the latter in tropical and subtropical regions. However, their distribution range overlaps in certain areas and interspecific hybridization has been documented. Genetic introgression between these species may have epidemiological repercussions for West Nile virus (WNV) transmission. Bayesian clustering analysis based on multilocus genotypes of 12 microsatellites was used to determine levels of hybridization between these two species in Macaronesian islands, the only contact zone described in West Africa. The distribution of the two species reflects both the islands' biogeography and historical aspects of human colonization. Madeira Island displayed a homogenous population of Cx. pipiens, whereas Cape Verde showed a more intriguing scenario with extensive hybridization. In the islands of Brava and Santiago, only Cx. quinquefasciatus was found, while in Fogo and Maio high hybrid rates (∼40%) between the two species were detected. Within the admixed populations, second-generation hybrids (∼50%) were identified suggesting a lack of isolation mechanisms. The observed levels of hybridization may locally potentiate the transmission to humans of zoonotic arboviruses such as WNV.
Keywords: Culex pipiens, Culex quinquefasciatus, hybridization, Macaronesian islands, West Nile virus
Introduction
The biological diversity of islands with recent volcanic origin and high isolation from mainland is a result of the colonizers ability to break the isolation and survive the island's environmental conditions. The highly stochastic nature of colonizing events means that only a very limited number of taxa may be present in each archipelago (Gillespie and Roderick 2002). For example, in Hawaii, only 15% of the known insect families were observed (Howarth and Ramsay 1991), and a similar scenario occurs in the Macaronesian region (Juan et al. 2000; Gillespie and Roderick 2002). This region is formed by four archipelagos of volcanic islands located in the northern hemisphere of the Atlantic Ocean: Azores, Canary Islands, Cape Verde, and Madeira. Isolation and low colonization rates in these islands promote divergence by adaptive radiation, leading to a higher proportion of neoendemic species than in regions with lower levels of genetic isolation (Gillespie and Roderick 2002). In Macaronesia, there are several examples of adaptive radiations in vertebrate species such as lizards (Gallotiinae, Gekkonidae, and Scincidae; Carranza et al. 2001, 2002; Carranza and Arnold 2006; Cox et al. 2010) and invertebrates such as beetles (Calathus, Meladema, Pimelia, Tarphius), butterflies (Gonepteryx), and spiders (Pholcus; Brunton and Hurst 1998; Emerson et al. 2000a,b; Contreras-Diaz et al. 2003; Ribera et al. 2003; Dimitrov et al. 2008). However, rates of island endemism appear to be lower for mosquitoes (Diptera: Culicidae). Of the 11 mosquito species/subspecies found in the Canary Islands, only two are endemic for Macaronesia and these are shared with Madeira (Capela 1982; Báez and Oromí 2010). This contrasts with the nearly 50% endemism rate among terrestrial invertebrate species in Canary Islands (Juan et al. 2000). A similarly low proportion of endemic mosquitoes is observed in other volcanic islands such as Cape Verde and Hawaii (Shroyer 1981; Alves et al. 2010). The reason for the relative paucity of adaptive radiation in island mosquito populations is that they are very recent colonizers often as a result of multiple human-mediated introductions (Fonseca et al. 2000; Lounibos 2002; Bataille et al. 2009).
Invasions of certain mosquito species can have a negative impact in vertebrates and humans due to their ability to serve as transmission vectors of diseases (Lounibos 2002; Delatte et al. 2011). A remarkable example was the decline of native bird' populations in Hawaii associated with avian malaria and avian pox virus transmitted by the introduced mosquito vector Culex quinquefasciatus Say, 1823 (Fonseca et al. 2000; Lapointe et al. 2012).
The Culex pipiens complex (Fig. 1) comprises mosquito vectors responsible for the transmission of lymphatic filariasis and neurotropic arboviruses from the Japanese encephalitis serogroup including the West Nile virus (WNV) to humans (Smith and Fonseca 2004; Solomon 2004). The nominal species of the complex, Culex pipiens Linnaeus, 1758 sensu stricto (hereafter termed Cx. pipiens) and Cx. quinquefasciatus are the most common and widespread species. The former is found primarily in temperate zones, whereas the latter occurs in tropical and subtropical zones. Cx. pipiens has a greater ecological range with populations found from the low subarctic of Siberia and Scandinavian countries to the semidesert regions of the Maghreb (Vinogradova 2000). Cx. quinquefasciatus is confined to warmer tropical and subtropical regions with a higher degree of humidity (Subra 1981; Fonseca et al. 2006). However, it is possible to find regions where both species coexist sympatrically and where hybrids of the two species have been observed (Urbanelli et al. 1995; Humeres et al. 1998; Kothera et al. 2009; Alves et al. 2010).
Figure 1.

Mosquito (female) of the Culex pipiens complex. Photograph with a digital camera SC30 (OLYMPUS, Tokyo, Japan) under a stereomicroscope OLYMPUS SZ61 (12× magnification).
In North America, morphological identification of males based on the length of the dorsal and ventral arms of the phallosome, namely the DV/D ratio, revealed the presence of only Cx. pipiens at latitudes above 39ºN while Cx. quinquefasciatus was the only species found at latitudes below 36ºN (Barr 1957). In the areas between 36ºN and 39ºN, a hybrid zone between the two species has been described (Barr 1957; Savage et al. 2008). Females are morphologically indistinguishable, and several molecular methods have been described to identify these sibling species (Farajollahi et al. 2011). Of these, the PCR assay based on species-specific polymorphisms in the intron-2 of the acetylcholinesterase-2 gene (ACE-2) has been one of the most widely used (Smith and Fonseca 2004). Allozyme studies confirmed the latitudinal cline between the two species (Cornel et al. 2003). A recent microsatellite-based study extended the geographic limits of “Barr's hybrid zone” suggesting a wider area between 30ºN and 40ºN (Kothera et al. 2009).
In contrast with the American continent, isolation between Cx. pipiens from Mediterranean Europe and Cx. quinquefasciatus from the northern hemisphere of Africa appears to be absolute. The most plausible explanation for this isolation is the presence of the Sahara desert. This inhospitable region lying between 15ºN and 33ºN acts as a barrier to gene flow not only for insects but also for other organisms (Douady et al. 2003; Kodandaramaiah and Wahlberg 2007). An exception is likely to be found in the Macaronesian region. Madeira, the Canary Islands, and Cape Verde locate within the latitudinal interval of the Saharan desert. In spite of the influence of the Saharan winds, the islands that compose these archipelagos have quite varying climates, ranging from temperate with dry summers (Madeira: Csb, Köppen Classification System) to arid with hot temperatures (Cape Verde: BWh; Peel et al. 2007). Importantly, many of these islands display environmental conditions for sustaining mosquito populations.
Populations of Cx. pipiens have been identified in the four archipelagos, and Cx. quinquefasciatus has been recorded in Cape Verde (Capela 1982; Alves et al. 2010; Báez and Oromí 2010; Vieira et al. 2010). The observation of intermediate DV/D values for the male genitalia of some specimens from Cape Verde suggested the presence of hybrids (Ribeiro et al. 1980). Similarly, in a recent update on the mosquito fauna of Cape Verde, molecular identification based on the ACE-2 marker suggested hybrid frequencies of 39–67% in two islands of the archipelago (Alves et al. 2010). However, the extent of hybridization and genetic introgression between Cx. pipiens and Cx. quinquefasciatus in these islands is still largely unknown.
There are several examples of species expansion mediated by human activity that have broken the geographic isolation between sibling species of insects and other organisms (Pinto et al. 2005; Steeves et al. 2010). The lack of other isolation mechanisms between these species may allow introgression leading to species assimilation or erosion of species boundaries. There is evidence supporting that two isolation mechanisms between Cx. pipiens and Cx. quinquefasciatus are likely to occur: (1) prezygotic isolation may result from differences in species distribution and in mating behavior; and (2) intrinsic postzygotic may result from cytoplasmic incompatibility that creates unviable hybrids (Vinogradova 2000; Cornel et al. 2003). A role of extrinsic postzygotic mechanisms linked to hybrid fitness (McBride and Singer 2010) in the isolation of the Cx. pipiens complex members remains unclear.
Both species also display important bio-ecological differences. Culex quinquefasciatus is generally considered a more synanthropic urban mosquito compared with a more rural Cx. pipiens (Ribeiro et al. 1980; Subra 1981). In Brazil and in Africa, Cx. quinquefasciatus displays a strong preference for mammals (including humans; Subra 1981; Muturi et al. 2008; Lorosa et al. 2010). In North America, there are differences in host preference among populations of Cx. quinquefasciatus, with some preferring mammals (Zinser et al. 2004; Molaei et al. 2007) and others birds (Savage et al. 2007; Molaei et al. 2010). Culex pipiens preferentially feeds upon birds (Kilpatrick et al. 2006, 2007; Molaei et al. 2006). Hybridization between members of the Cx. pipiens complex with different host preferences may promote a more opportunistic feeding behavior increasing the importance of the host availability (Fonseca et al. 2004; Balenghien et al. 2011). Consequently, this population with more catholic feeding behavior would have an increased potential as a bridge vector between bird and humans for the transmission of WNV (Molaei et al. 2007; Savage et al. 2007; Kilpatrick 2011).
In this study, Bayesian model-based methods were applied to multilocus microsatellite genotypes to infer the genetic structure of the Cx. pipiens complex in Madeira and in four islands of Cape Verde. The aims were (1) to determine the degree of genetic differentiation among island populations; (2) to measure rates of hybridization and genetic introgression between the sibling species; and (3) to infer about the colonization process and their impacts in Macaronesian region.
Materials and Methods
Mosquito collections
Indoor resting collections of adult mosquitoes using mechanical aspirators were carried out in four localities of Madeira Island in September 2006 and in June 2007 (Fig. 2). Given the very low adult mosquito densities found in Cape Verde (Pinto et al. 1999), collections of immature culicids were undertaken using dippers and pipettes, between November and December 2007, in four islands of Cape Verde: Brava, Fogo, Santiago, and Maio (Fig. 2). Immature collections were made on a wide range of breeding sites such as ponds, pools, swamps, pits, water tanks, and septic tanks. Information on the localities in which Cx. pipiens s.l. larvae were sampled in Cape Verde is shown in Table S1 (Supporting information; see also Alves et al. 2010). Collected larvae were transported to a laboratory in Praia (Santiago Island) and reared to adulthood. Adult mosquitoes were killed by freezing and identified to species/complex using morphological keys (Ribeiro and Ramos 1995, 1999). Samples were stored over silica gel until DNA extraction.
Figure 2.
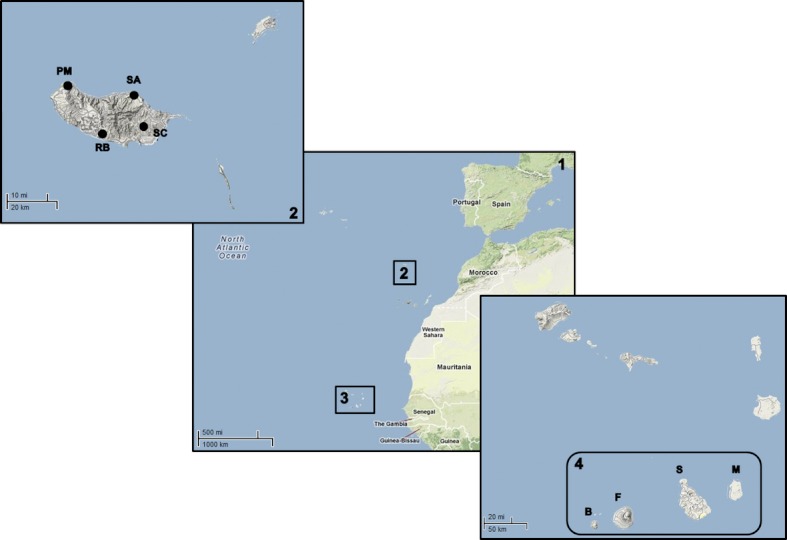
Maps of the North Atlantic region showing the localities/islands sampled. (1) North Atlantic region including West Africa and Mediterranean region; (2) Madeira archipelago, PM, Porto Moniz; RB, Ribeira Brava; SC, Santa Cruz; SA, Santana; (3) Cape Verde archipelago; (4) Islands sampled in Cape Verde; B, Brava; F, Fogo; S, Santiago; M, Maio. Images collected in Google Maps – ©2012 Google (http://maps.google.com/).
Molecular analyses
DNA extraction from individual female mosquitoes was performed using the method of Collins et al. (1987). Each specimen was identified to species by a multiplex PCR assay targeting species-specific polymorphisms in intron-2 of the ACE-2 gene using primers specific for Cx. pipiens, Cx. quinquefasciatus, and Cx. torrentium (Smith and Fonseca 2004).
Twelve microsatellites (Fonseca et al. 1998; Keyghobadi et al. 2004; Smith et al. 2005) were genotyped (see Table S2). For each specimen, each locus was amplified separately in a 20 μL PCR reaction that contained 1X GoTaq® Flexi Buffer (Promega, Madison, Wisconsin), 2.5 mm MgCl2, 0.25 mm dNTPs, 0.20 mg/mL Bovine Serum Albumin, 0.20 μm of each primer, and 0.5 U GoTaq® Flexi DNA polymerase (Promega). For each locus, one of the primers was fluorescently labeled (NED, HEX our 6-FAM; Applied Biosystems, Carlsbad, California). Thermocycling conditions included an initial denaturation step of 5 min at 96ºC, followed by 30 cycles each of 96ºC for 30 sec, annealing at 52ºC-56ºC (locus-dependent) for 30 sec and 72ºC for 30 sec. After a final extension step of 5 min at 72ºC, reactions were stopped at 4ºC.
Amplified products were separated by capillary electrophoresis in a genetic analyzer ABI3730 (Applied Biosystems) at Yale DNA Analysis Facility (USA). Fragment sizes and genotypes were scored using the software GeneMarker 1.4. (Softgenetics, State College, Pennsylvania).
Data analysis
Genetic diversity at each microsatellite locus was characterized by estimates of expected heterozygosity (Nei 1987) and inbreeding coefficient (FIS). Significance of FIS values was assessed by randomization tests. These analyses were performed using FSTAT v. 2.9.3.2. (Goudet 1995). Estimates of allele richness (AR), a measure of allele diversity adjusted for the lowest sample size, were obtained by the statistical rarefaction approach implemented in HP-RARE (Kalinowski 2005). Departures from Hardy–Weinberg proportions were tested by exact tests available in ARLEQUIN v.3.11 (Excoffier et al. 2005). The same software was used to perform exact tests of linkage equilibrium between pairs of loci based on the expectation-maximization approach described by Slatkin and Excoffier (1996). The software Micro-Checker 2.2.3. (Van Oosterhout et al. 2004) was used to test for the presence of null alleles (99% confidence interval) at each locus/sample.
Bayesian clustering analysis as implemented by STRUCTURE 2.3.3 (Pritchard et al. 2000) was used to infer population substructure/ancestry from the data set without prior information of sampling groups under the conditions of admixture (α allowed to vary between 0 and 10), and allele frequencies correlated among populations (λ was set at 1, default value). Ten independent runs with 105 iterations and replications were performed for each value of K (K = 1–10 clusters). The inference of the number of genetic clusters (K) in the Bayesian method implemented by STRUCTURE is not straightforward, and it is normally performed by ad hoc approaches: an estimation of ln[Pr(X|K)], described in the original publication (Pritchard et al. 2000) and the ΔK statistic (Evanno et al. 2005). We used a combination of these approaches with a sequential procedure in which data were analyzed at three levels: (1) all samples, (2) each archipelago, and (3) each island. Following the suggestions of Vähä and Primmer (2006), individual genetic assignment to clusters was based on a minimum posterior probability threshold (Tq) of 0.90. Individuals displaying 0.1 ≤ qi ≤ 0.90 were considered of admixed ancestry. The information from the outputs of each K (10 runs) was aligned by the Greedy method implemented in CLUMPP (Jakobsson and Rosenberg 2007).
The Bayesian method implemented by NEWHYBRIDS 1.1. (Anderson and Thompson 2002) was used to assign individuals into six classes: two pure (parental Cx. pipiens and Cx. quinquefasciatus) and four hybrid (F1, F2, and backcrosses with the parental populations). The approach of uniform priors was used because it reduces the influence of low-frequency alleles thus which may result from sampling and genotyping errors in closely related populations. Results were based on the average of five independent runs of 105 iterations. Following the suggestions of Anderson and Thompson (2002), individual genetic assignment to classes was based on a minimum posterior probability threshold (Tq) of 0.50.
A neighbor-joining (NJ) tree based on Cavalli-Sforza and Edwards (1967) chord distance (Dc) was used to represent the relationships among genetic clusters and geographic samples. Individuals with an admixed genetic background (i.e. with a probability of assignment not attributable to any of the purebred or hybrid clusters) were excluded from this analysis. A consensus tree was obtained by bootstrapping (1000 replicates) distance values over loci. Calculations were performed with the program Populations 1.2.30 (Langella 1999). The software Treeview (Page 1996) was used to visualize the tree.
Whenever multiple testing was performed, the nominal significance level of rejection of the null hypothesis (α = 0.05) was corrected by the sequential Bonferroni procedure (Holm 1979).
Results
ACE-2 molecular identification
A total of 374 females (Madeira: 190 and Cape Verde: 184, distributed as follows, Brava: 31, Fogo, 36, Santiago: 54, Maio: 63) were analyzed by the molecular assay ACE-2 (Smith and Fonseca 2004; Table 1). Of these, 203 were identified as Cx. pipiens and were collected in Madeira (N = 190) and in Maio (N = 13). Culex quinquefasciatus was found in the four islands of Cape Verde (N = 115), and it was the only member of the complex present in the collections from Brava (N = 31) and Santiago (N = 54). Fifty-six mosquitoes displayed a heterozygous pattern for ACE-2 and were collected in Fogo (N = 14) and Maio (N = 42). The island of Maio was the only island where the two species and putative hybrids were found in sympatry.
Table 1.
Molecular identification of Culex pipiens complex species based on the molecular assay in the ACE-2
| Localities | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Madeira | Cape Verde | ||||||||
| N | PM | RB | SC | SA | B | F | S | M | |
| Culex pipiens | 203 | 66 (100.0) | 39 (100.0) | 34 (100.0) | 51 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 13 (20.6) |
| Hybrids | 56 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 14 (38.9) | 0 (0.0) | 42 (66.7) |
| Culex quinquefasciatus | 115 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 31 (100.0) | 22 (61.1) | 54 (100.0) | 8 (12.7) |
N, number of individuals; PM, Porto Moniz; RB, Ribeira Brava; SC, Santa Cruz; SA, Santana; B, Brava island; F, Fogo island; S, Santiago island; M, Maio island.
Values in parenthesis refer to the relative genotypic frequencies (in percentage) within each locality.
Clustering analysis
Genetic diversity estimates for the 12 microsatellites in whole sample (N = 374) and subsamples determined by ACE-2 identification and geographic location are shown in Table S3. Loci CQ26 and CQ41 exhibited heterozygote deficits in all subsamples from Madeira, possibly reflecting locus-specific effects, such as null alleles or selection. The analysis performed by the Micro-Checker software confirmed the possibility of null alleles at loci CQ26 and CQ41 in samples from Madeira island (see Table S3). These loci were therefore excluded from Bayesian assignment and genetic differentiation analyses.
The Bayesian analysis implemented in STRUCTURE and the two ad hoc approaches to define the number of clusters revealed a homogeneous population in Madeira (K = 1) and the intriguing scenario of Cape Verde with three possible subdivisions (K = 2, K = 3, or K = 4; Fig. 3, see Figs. S1, S2). The sequential procedure under the three levels of organization (whole sample, archipelago, and island) highlighted a further subdivision within the islands of Maio and Fogo providing support for K = 3 in the archipelago of Cape Verde and consequently a K = 4 for the whole sample (Fig. 3, see Fig. S3).
Figure 3.
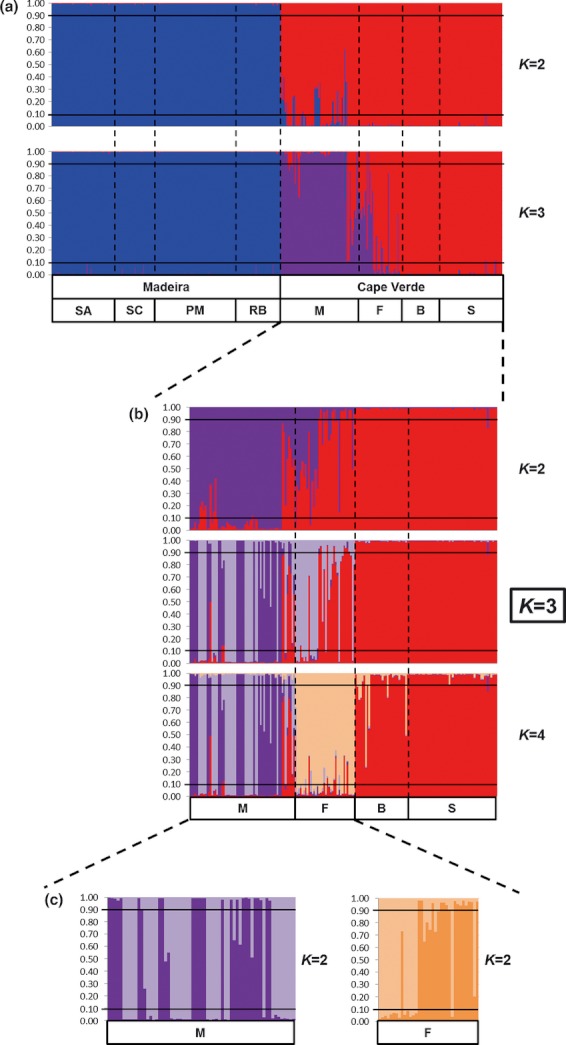
Bayesian cluster analysis conducted by STRUCTURE at three different levels. (a) all samples, (b) Cape Verde samples, (c) Maio and Fogo islands. K, number of clusters. Columns correspond to the multilocus genotype of each individual, partitioned in different colors representing the probability of ancestry (qi) to each cluster. Individuals were ordered according to their geographic information. Lines indicate the qi threshold used to determine admixed individuals (see Methods).
The combination of the Bayesian clustering results with the ACE-2 identification clarified the separation of sampled mosquitoes into four different clusters (Table 2): Cluster 1 (C1) grouped all the 190 Cx. pipiens from Madeira, while the other three clusters were restricted to Cape Verde. Cluster 4 (C4) was the most abundant in the archipelago with 91 specimens from three islands (Brava, Fogo and Santiago), all identified as Cx. quinquefasciatus by ACE-2 PCR. Cluster 2 (C2) was the smallest cluster with 25 specimens from Maio Island being classified as Cx. pipiens or hybrid by ACE-2 PCR. Cluster 3 (C3) includes individuals from Fogo and Maio Islands, and the majority (87.2%) of the specimens were identified as hybrids by ACE-2 PCR. Twenty-nine specimens, the majority of which from Maio and Fogo, were not assigned to any of the four clusters and were thus considered admixed.
Table 2.
ACE-2 PCR species composition and relative distribution per locality/island of each genetic cluster revealed by STRUCTURE
| ACE-2 | Localities | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Madeira | Cape Verde | |||||||||||
| N | P | H | Q | PM | RB | SC | SA | B | F | S | M | |
| Cluster 1 | 190 | 190 (100.0) | 0 (0.0) | 0 (0.0) | 66 (34.7) | 39 (20.5) | 34 (17.8) | 51 (26.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Cluster 2 | 25 | 9 (36.0) | 16 (64.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 25 (100.0) |
| Cluster 3 | 39 | 1 (2.6) | 34 (87.2) | 4 (10.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 14 (35.9) | 0 (0.0) | 25 (64.1) |
| Cluster 4 | 91 | 0 (0.0) | 0 (0.0) | 91 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 31 (34.1) | 7 (7.7) | 53 (58.2) | 0 (0.0) |
| Admixed | 29 | 3 (10.3) | 6 (20.7) | 20 (69.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 15 (51.6) | 1 (3.4) | 13 (44.8) |
N, number of individuals; P, Culex pipiens by ACE-2 identification; Q, Culex quinquefasciatus by ACE-2 identification; H, hybrids between Culex pipiens and Culex quinquefasciatus by ACE-2 identification; PM, Porto Moniz; RB, Ribeira Brava; SC, Santa Cruz; SA, Santana; B, Brava island; F, Fogo island; S, Santiago island; M, Maio island.
Values in parenthesis refer to the frequencies (in percentage) within each cluster.
The analysis with NEWHYBRIDS confirmed the homogeneity of the Madeira population (C1). In Cape Verde, all the samples from C4 were classified as pure Cx. quinquefasciatus, while the majority of the individuals of C2 (96.0%) were classified as pure Cx. pipiens and one individual was classified as a backcross with Cx. pipiens (BxP). The majority of individuals of C3 (87.1%) were classified as hybrids. Of these, 10 (nine in Fogo, one in Maio) were classified as F2 hybrids and nine individuals from Maio were backcrosses with Cx. pipiens (BxP; Table 3).
Table 3.
Frequencies purebred and hybrid individuals detected by NEWHYBRIDS in each of the ancestry clusters revealed by STRUCTURE
| NEWHYBRIDS | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| H | |||||||||
| N | P | Q | H | F1 | F2 | BxP | BxQ | H′ | |
| Cluster 1 | 190 | 189 (99.4) | 0 (0.0) | 1 (0.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.6) |
| Cluster 2 | 25 | 24 (96.0) | 0 (0.0) | 1 (4.0) | 0 (0.0) | 0 (0.0) | 1 (4.0) | 0 (0.0) | 0 (0.0) |
| Cluster 3 | 39 | 4 (10.3) | 1 (2.6) | 34 (87.1) | 0 (0.0) | 10 (25.6) | 9 (23.1) | 0 (0.0) | 15 (38.4) |
| Cluster 4 | 91 | 0 (0.0) | 91 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Admixed | 29 | 3 (10.4) | 13 (44.8) | 13 (44.8) | 0 (0.0) | 8 (27.6) | 0 (0.0) | 0 (0.0) | 5 (17.2) |
N, number of individuals; P, pure Culex pipiens; Q, pure Culex quinquefasciatus; H, hybrids between the pure groups (Culex pipiens and Culex quinquefasciatus); F1, hybrid first generation; F2, hybrids second generation; BxP, backcross Culex pipiens; BxQ, backcross Culex quinquefasciatus, H′, hybrids defined by the sum of assignment probabilities for all hybrid classes.
Values in parenthesis refer to the frequencies (in percentage) within each cluster.
The Dc-based NJ tree was consistent with the presence of the four clusters identified in the analysis performed by STRUCTURE (Fig. 4). Culex pipiens samples from Madeira (C1) and Maio (C2) displayed a high genetic distance but were still grouped in a common cluster separated from the remaining samples. Samples from cluster C3, composed mainly by hybrid individuals, displayed an intermediate position in the topology of the tree. Culex quinquefasciatus samples from cluster C4 shared the same cluster, but it was possible to observe significant divergence between the populations of the three islands (Brava, Fogo, and Santiago).
Figure 4.
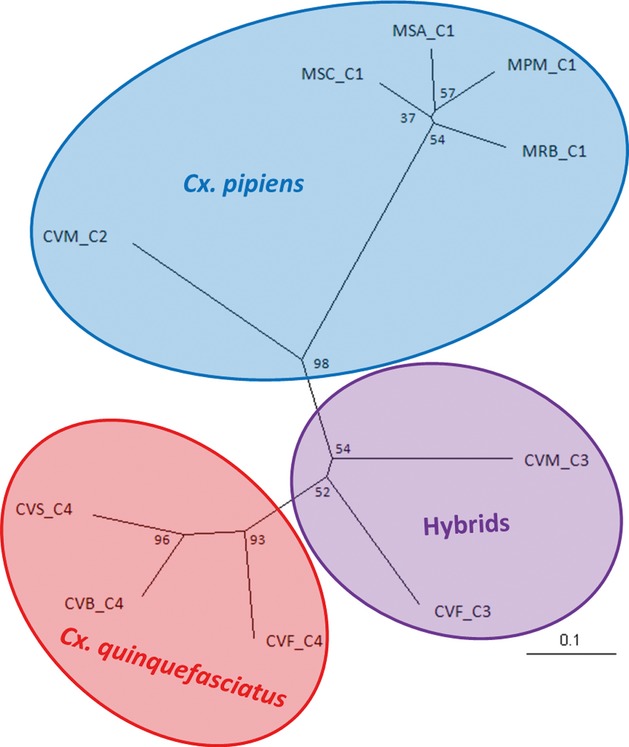
Phylogenetic tree of the Culex pipiens complex in Madeira and Cape Verde. CVM, Maio; CVF, Fogo; CVB, Brava; CVS, Santiago; MSC, Santa Cruz (Madeira); MSA, Santana (Madeira); MPM, Porto Moniz (Madeira); MRB, Ribeira Brava (Madeira); C1, cluster 1; C2, cluster 2; C3, cluster 3 and C4, cluster 4.
Discussion
In this study, the distribution and levels of hybridization between Cx. pipiens and Cx. quinquefasciatus were found to differ among islands of the Macaronesian region. Madeira showed a genetically homogenous Cx. pipiens population. In Cape Verde, it was possible to identify monospecific populations of Cx. quinquefasciatus in Brava and Santiago, while admixed populations between both species were observed in Maio and Fogo. The species diagnostic ACE-2 PCR was effective in the identification of each species in the allopatric populations of Madeira, Brava, and Santiago. However, in sympatric populations with interspecific admixture such as those of Maio and Fogo, repeated introgression and recombination lead to a disruption of the linkage between the diagnostic alleles and the respective genetic backgrounds of each species. Under these conditions, a more cautious interpretation of the results obtained by a single diagnostic marker such as the ACE-2 is needed for the correct identification of each species and hybrids (McAbee et al. 2008; Fonseca et al. 2009).
The presence of a monospecific population of Cx. pipiens in Madeira agrees with previous reports (Capela 1981; Fonseca et al. 2004). This volcanic island locates in the temperate zone of the North Atlantic, 700 km off the coast of Morocco and 850 km from continental Portugal. In both countries, only Cx. pipiens has been identified (Trari et al. 2002; Almeida et al. 2008). Since its discovery in the 15th century, Madeira has been an important port-of-call in the Atlantic so that the introduction of Cx. pipiens could have resulted from human-mediated passive dispersal (Lounibos 2002). The Mediterranean temperate climate of this island should also be compatible with the establishment of Cx. pipiens. The absence of Cx. quinquefasciatus may reflect a lower tolerance of this species to more temperate climates with lower temperatures during winter. The possibility of this vector having never been introduced into this island is probably less likely in spite of the ca. 2000-km distance between Madeira island and the sub-Saharan African coast. Migration by human-mediated dispersal in Cx. pipiens complex includes long-distance introductions that spread organophosphate insecticides resistance genes between continents and established invasive populations in isolated archipelagos such as Hawaii (Chevillon et al. 1999; Fonseca et al. 2006).
The distribution of the two members of the Cx. pipiens complex in Cape Verde is more intricate and reflects both bio-geographic features and historical aspects of the human peopling of the islands. The apparent predominance of Cx. quinquefasciatus on the archipelago agrees with the bio-geographic context of the islands, which lie in the tropical zone of the North Atlantic. The most likely origin of this species would be the West African continent. However, Fonseca et al. (2006) in a worldwide genetic survey of Cx. quinquefasciatus provided evidence for a recent introduction of Cx. quinquefasciatus in West Africa from the New World. Given the geographic intermediate location and the strategic importance of Cape Verde in maritime routes, one cannot exclude the possibility of introduction of mosquitoes of New World origin. The occurrence of Cx. pipiens most likely reflects the historical relationship of the archipelago with the Mediterranean region of the European continent. The islands were discovered by Portuguese sailors in the 15th century, and the subsequent peopling was made by migrants of both European and African origin. The Portuguese traders used the archipelago as a port-of-call for ship provisioning during travels between Europe and the African continent and also as a Senegambian slave outpost for the Atlantic (Brehm et al. 2002). The considerable movement of ships between the islands and both continents could have provided the opportunity for the introduction of both mosquito species. It remains to be determined whether the present mosquito populations in Cape Verde islands result from multiple introduction events of one or both species. The analysis of mainland samples and of other molecular markers (e.g. mtDNA) would be required for this purpose (Hardouin et al. 2010).
In Maio and Fogo, a considerable number of individuals were assigned as hybrids and yet one of the parental species was absent from the samples (Cx. quinquefasciatus in Maio; Cx. pipiens in Fogo). A possible explanation for this apparent contradiction could be insufficient sampling of the least abundant species. Factors that may have contributed for an insufficient sampling were the collection method used (immature captures) coupled with the low number of breeding sites positive for Cx. pipiens s.l. larvae. In Cape Verde, very low adult mosquito densities preclude the use of collection methods targeting adult mosquitoes for sampling sufficient numbers of individuals (Ribeiro et al. 1980; Pinto et al. 1999). However, other explanations may be proposed for these observations. Maio is the driest of the islands sampled and a lower density or virtual absence of a stable Cx. quinquefasciatus population agrees with its lower tolerance to aridity. Fogo has a very steep topography marked by the presence of a volcanic cone. A similar scenario to that of Madagascar, where Cx. quinquefasciatus predominated in the lowland urban areas and Cx. pipiens were found at altitudes above 1300 m (Urbanelli et al. 1995), may occur in this island. Insufficient sampling could also explain the apparent absence of Cx. pipiens in Brava and Santiago, although in these cases there was no evidence of admixture. While the absence of this species agrees with previous surveys in Brava, the same does not hold for Santiago. In this island, Cx. pipiens prevailed over Cx. quinquefasciatus in larval collections performed in the late 1970s (Ribeiro et al. 1980). This apparent inversion in the relative abundance of both species may be associated with an increase in urbanization of this island over the past recent years. Such an increase in urbanization could confer a greater adaptive advantage for Cx. quinquefasciatus over Cx. pipiens.
High hybridization rates between Cx. pipiens and Cx. quinquefasciatus were detected in two islands of Cape Verde. These rates (Fogo: 39%; Maio: 40%) are comparable with those recorded in the hybrid zone of North America (Savage et al. 2008; Kothera et al. 2009) and contrast with the pattern of sympatry without hybridization observed in southeast Africa (Cornel et al. 2003). The lack of hybridization in southeast Africa was justified by the presence of Wolbachia pipientis only in Cx. quinquefasciatus, whereas in North America, both species are infected with the same strain (Cornel et al. 2003; Rasgon and Scott 2003). Wolbachia pipientis infection can induce sterility by cytoplasmic incompatibility (an intrinsic postzygotic isolation mechanism) between infected males and uninfected females or females infected by incompatible strains (Atyame et al. 2011). In West Africa and in the Mediterranean region, both Cx. pipiens and Cx. quinquefasciatus populations share closely related strains of W. pipientis (Atyame et al. 2011). Assuming a putative origin of both species from those regions, the introduction of mosquito populations possessing similar strains of W. pipientis (or no infection) may explain the high levels of hybridization in Cape Verde. Molecular analysis of W. pipientis in these mosquito populations would help clarifying this hypothesis.
Isolation between close species can be promoted by several mechanisms that may act in simultaneous. The lack or incomplete action of prezygotic (e.g. mating behavior) and intrinsic postzygotic (e.g. cytoplasmic incompatibility) mechanisms allows hybridization creating first generation (F1) hybrids. However, an extrinsic postzygotic mechanism such as hybrid sterility or hybrid low fitness can restrict gene flow to one generation avoiding introgression (Bono and Markow 2009; McBride and Singer 2010; Muñoz et al. 2010). The analysis performed by NEWHYBRIDS in Cape Verde samples showed the presence of ∼50% of second generation hybrids (25.6% F2 and 23.1% BxP; Table 3) within the hybrid cluster. The repeated hybridization and backcrossing with Cx. pipiens indicate mating success of F1 individuals (males and females) suggesting a low effect of extrinsic postzygotic isolation mechanisms between the Cx. pipiens and Cx. quinquefasciatus in Cape Verde.
Macaronesia is a passage route and breeding region for migratory birds (Garcia-del-Rey 2011). These birds may potentially introduce parasites and viruses that are known to be transmitted by the Cx. pipiens complex such as Plasmodium relictum (avian malaria), avian pox virus, Usutu virus and WNV. The introduction of these pathogens may place the endemic bird populations in danger (Kilpatrick 2011; Savini et al. 2011; Lapointe et al. 2012). Furthermore, the high levels of hybridization between Cx. pipiens and Cx. quinquefasciatus may promote a more opportunistic feeding behavior increasing the chance for the accidental transmission of WNV to humans. A serologic survey in 1980s showed 40% positive cases of WNV in Cape Verdean children (Vieira 1985). However, it has been recognized that WNV serologic surveys of the last century had considerable false positives due to cross-reactivity with other flavivirus (Tardei et al. 2000). Even so, the possibility of disease outbreaks should not be neglected given the outcome of the introduction of WNV to the western hemisphere (Kilpatrick 2011), or the more recent dengue epidemic in Cape Verde in 2009 (Franco et al. 2010), highlighting the receptivity of a territory once a suitable vector is present.
Acknowledgments
We are grateful to the inhabitants of Madeira and Cape Verde for their collaboration in this study. We acknowledge the logistic support in Cape Verde given by the General Department of Health (J. Pereira), the delegates and technicians of the Health Care Units of Praia, St. Cruz, Tarrafal, S. Miguel, Órgãos, Rª Grande de Santiago and St. Catarina, in Santiago island,, and of Maio, Fogo and Brava islands. We thank the support of Regina Rodrigues (National Malaria Program) and João Silva (CMDT/IHMT) in the field collections. This study was funded by Fundação para a Ciência e a Tecnologia, Portugal (POCI/BIA-BDE/57650/2004 and PPCDT/BIA-BDE/57650/2004; PPCDT/SAU-ESP/55110/2004). Joana Alves and Bruno Gomes were funded by a PhD fellowship of Fundação para a Ciência e Tecnologia/MCTES (SFRH/BD/153451/2005, SFRH/BD/36410/2007).
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Localities positive for Culex pipiens complex in Cape Verde.
Table S2. Microsatellite loci used in the analysis.
Table S3. Genetic diversity at microsatellite loci of Culex pipiens complex from Macaronesian islands.
Figure S1. Graphics of ad hoc approaches to infer the number of clusters (K) in STRUCTURE analysis with all samples, Cape Verde and Madeira.
Figure S2. Bayesian cluster analysis conducted by STRUCTURE in Madeira.
Figure S3. Graphics of ad hoc approaches to inference the number of clusters (K) in STRUCTURE analysis in each island of Cape Verde.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Almeida APG, Galão RP, Sousa CA, Novo MT, Parreira R, Pinto J, et al. Potential mosquito vectors of arboviruses in Portugal: species, distribution, abundance and West Nile infection. Trans. R Soc. Trop. Med. Hyg. 2008;102:823–832. doi: 10.1016/j.trstmh.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Alves J, Gomes B, Rodrigues R, Silva J, Arez AP, Pinto J, et al. Mosquito fauna on the Cape Verde Islands (West Africa): an update on species distribution and a new finding. J. Vector Ecol. 2010;35:307–312. doi: 10.1111/j.1948-7134.2010.00087.x. [DOI] [PubMed] [Google Scholar]
- Anderson EC, Thompson EA. A model-based method for identifying species hybrids using multilocus genetic data. Genetics. 2002;160:1217–1229. doi: 10.1093/genetics/160.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atyame CM, Delsuc F, Pasteur N, Weill M, Duron O. Diversification of Wolbachia Endosymbiont in the Culex pipiens Mosquito. Mol. Biol. Evol. 2011;28:2761–2772. doi: 10.1093/molbev/msr083. [DOI] [PubMed] [Google Scholar]
- Báez M, Oromí P. Diptera. In: Arechavaleta M, Rodríguez N, Zurita N, García A, editors. Lista de especies silvestres de Canarias. Hongos, plantas y animales terrestres. Santa Cruz de Tenerife, Tenerife: Gobierno de Canarias; 2010. pp. 318–342. [Google Scholar]
- Balenghien T, Fouque F, Sabatier P, Bicout DJ. Theoretical formulation for mosquito host-feeding patterns: application to a West Nile virus focus of Southern France. J. Med. Entomol. 2011;48:1076–1090. doi: 10.1603/me10097. [DOI] [PubMed] [Google Scholar]
- Barr AR. The distribution of Culex p. pipiens and C. p. quinquefasciatus in North America. Am. J. Trop. Med. Hyg. 1957;6:153–165. doi: 10.4269/ajtmh.1957.6.153. [DOI] [PubMed] [Google Scholar]
- Bataille A, Cunningham AA, Cedeño V, Cruz M, Eastwood G, Fonseca DM, et al. Evidence for regular ongoing introductions of mosquito disease vectors into the Galápagos Islands. Proc. Biol. Sci. 2009;276:3769–3775. doi: 10.1098/rspb.2009.0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono JM, Markow TA. Post-zygotic isolation in cactophilic Drosophila: larval viability and adult life-history traits of D. mojavensis/D. arizonae hybrids. J. Evol. Biol. 2009;22:1387–1395. doi: 10.1111/j.1420-9101.2009.01753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A, Pereira L, Bandelt H-J, Prata MJ, Amorim A. Mitochondrial portrait of the Cabo Verde archipelago: the Senegambian outpost of Atlantic slave trade. Ann. Hum. Genet. 2002;66:49–60. doi: 10.1017/S0003480001001002. [DOI] [PubMed] [Google Scholar]
- Brunton C, Hurst G. Mitochondrial DNA phylogeny of Brimstone butterflies (genus Gonepteryx) from the Canary Islands and Madeira. Biol. J. Linn. Soc. Lond. 1998;63:69–79. [PubMed] [Google Scholar]
- Capela RA. Contribution to the study of mosquitoes (Diptera: Culicidae) from the Archipelago of Madeira and the Salvages I – Madeira. Arq. Mus. Bocage. 1981;1:45–66. Série A. [Google Scholar]
- Capela RA. Contribuição para o conhecimento dos mosquitos (Diptera, Culicidae) dos arquipélagos da Madeira e das Selvagens. Bol. Mus. Munich. Funchal. 1982;34:105–123. [Google Scholar]
- Carranza S, Arnold EN. Systematics, biogeography, and evolution of Hemidactylus geckos (Reptilia: Gekkonidae) elucidated using mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2006;38:531–545. doi: 10.1016/j.ympev.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Carranza S, Arnold EN, Mateo JA, López-Jurado LF. Parallel gigantism and complex colonization patterns in the Cape Verde scincid lizards Mabuya and Macroscincus (Reptilia: Scincidae) revealed by mitochondrial DNA sequences. Proc. Biol. Sci. 2001;268:1595–1603. doi: 10.1098/rspb.2001.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carranza S, Arnold EN, Mateo JA, Geniez P. Relationships and evolution of the North African geckos, Geckonia and Tarentola (Reptilia: Gekkonidae), based on mitochondrial and nuclear DNA sequences. Mol. Phylogenet. Evol. 2002;23:244–256. doi: 10.1016/S1055-7903(02)00024-6. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Edwards AW. Phylogenetic analysis. Models and estimation procedures. Am. J. Hum. Genet. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
- Chevillon C, Raymond M, Guillemaud T, Lenormand T, Pasteur N. Population genetics of insecticide resistance in the mosquito Culex pipiens. Biol. J. Linn. Soc. Lond. 1999;68:147–157. [Google Scholar]
- Collins FH, Mendez MA, Rasmussen MO, Mehaffey PC, Besansky NJ, Finnerty V. A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. Am. J. Trop. Med. Hyg. 1987;37:37–41. doi: 10.4269/ajtmh.1987.37.37. [DOI] [PubMed] [Google Scholar]
- Contreras-Diaz HG, Moya O, Oromi P, Juan C. Phylogeography of the endangered darkling beetle species of Pimelia endemic to Gran Canaria (Canary Islands) Mol. Ecol. 2003;12:2131–2143. doi: 10.1046/j.1365-294x.2003.01884.x. [DOI] [PubMed] [Google Scholar]
- Cornel AJ, Mcabee RD, Rasgon J, Stanich MA, Scott TW, Coetzee M. Differences in extent of genetic introgression between sympatric Culex pipiens and Culex quinquefasciatus (Diptera: Culicidae) in California and South Africa. J. Med. Entomol. 2003;40:36–51. doi: 10.1603/0022-2585-40.1.36. [DOI] [PubMed] [Google Scholar]
- Cox SC, Carranza S, Brown RP. Divergence times and colonization of the Canary Islands by Gallotia lizards. Mol. Phylogenet. Evol. 2010;56:747–757. doi: 10.1016/j.ympev.2010.03.020. [DOI] [PubMed] [Google Scholar]
- Delatte H, Bagny L, Brengue C, Bouetard A, Paupy C, Fontenille D. The invaders: phylogeography of dengue and chikungunya viruses Aedes vectors, on the South West islands of the Indian Ocean. Infect. Genet. Evol. 2011;11:1769–1781. doi: 10.1016/j.meegid.2011.07.016. [DOI] [PubMed] [Google Scholar]
- Dimitrov D, Arnedo MA, Ribera C. Colonization and diversification of the spider genus Pholcus Walckenaer, 1805 (Araneae, Pholcidae) in the Macaronesian archipelagos: evidence for long-term occupancy yet rapid recent speciation. Mol. Phylogenet. Evol. 2008;48:596–614. doi: 10.1016/j.ympev.2008.04.027. [DOI] [PubMed] [Google Scholar]
- Douady CJ, Catzeflis F, Raman J, Springer MS, Stanhope MJ. The Sahara as a vicariant agent, and the role of Miocene climatic events, in the diversification of the mammalian order Macroscelidea (elephant shrews) Proc. Natl. Acad. Sci. USA. 2003;100:8325–8330. doi: 10.1073/pnas.0832467100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson BC, Oromí P, Hewitt GM. Interpreting colonization of the Calathus (Coleoptera: Carabidae) on the Canary Islands and Madeira through the application of the parametric bootstrap. Evolution. 2000a;54:2081–2090. doi: 10.1111/j.0014-3820.2000.tb01251.x. [DOI] [PubMed] [Google Scholar]
- Emerson BC, Oromí P, Hewitt GM. Tracking colonization and diversification of insect lineages on islands: mitochondrial DNA phylogeography of Tarphius canariensis (Coleoptera: Colydiidae) on the Canary Islands. Proc. Biol. Sci. 2000b;267:2199–2205. doi: 10.1098/rspb.2000.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Goudet J, Regnaut S. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Farajollahi A, Fonseca DM, Kramer LD, Kilpatrick AM. “Bird biting” mosquitoes and human disease: a review of the role of Culex pipiens complex mosquitoes in epidemiology. Infect. Genet. Evol. 2011;11:1577–1585. doi: 10.1016/j.meegid.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca DM, Atkinson CT, Fleischer RC. Microsatellite primers for Culex pipiens quinquefasciatus, the vector of avian malaria in Hawaii. Mol. Ecol. 1998;7:1617. [PubMed] [Google Scholar]
- Fonseca DM, Lapointe DA, Fleischer RC. Bottlenecks and multiple introductions: population genetics of the vector of avian malaria in Hawaii. Mol. Ecol. 2000;9:1803–1814. doi: 10.1046/j.1365-294x.2000.01070.x. [DOI] [PubMed] [Google Scholar]
- Fonseca DM, Keyghobadi N, Malcolm CA, Mehmet C, Schaffner F, Mogi M, et al. Emerging vectors in the Culex pipiens complex. Science. 2004;303:1535–1538. doi: 10.1126/science.1094247. [DOI] [PubMed] [Google Scholar]
- Fonseca DM, Smith JL, Wilkerson RC, Fleischer RC. Pathways of expansion and multiple introductions illustrated by large genetic differentiation among Worldwide populations of the Southern house mosquito. Am. J. Trop. Med. Hyg. 2006;74:284–289. [PubMed] [Google Scholar]
- Fonseca DM, Smith JL, Kim H-C, Mogi M. Population genetics of the mosquito Culex pipiens pallens reveals sex-linked asymmetric introgression by Culex quinquefasciatus. Infect. Genet. Evol. 2009;9:1197–1203. doi: 10.1016/j.meegid.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco L, Di Caro A, Carletti F, Vapalahti O, Renaudat C, Zeller H, et al. Recent expansion of dengue virus serotype 3 in West Africa. Euro. Surveill. 2010;15:19490. [PubMed] [Google Scholar]
- Garcia-del-Rey E. Field guide to the birds of Macaronesia. Azores, Madeira, Canary Islands, Cape Verde. Bellaterra, Barcelona: Lynx Edicions; 2011. [Google Scholar]
- Gillespie RG, Roderick GK. Arthropods on islands: colonization, speciation, and conservation. Annu. Rev. Entomol. 2002;47:595–632. doi: 10.1146/annurev.ento.47.091201.145244. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT (version 1.2): a computer program to calculate F-statistics. J. Hered. 1995;86:485–486. [Google Scholar]
- Hardouin EA, Chapuis J-L, Stevens MI, Quillfeldt JB, van Vuuren P, Scavetta RJ, et al. House mouse colonization patterns on the sub-Antarctic Kerguelen Archipelago suggest singular primary invasions and resilience against re-invasion. BMC Evol. Biol. 2010;10:325. doi: 10.1186/1471-2148-10-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Statist. 1979;6:65–70. [Google Scholar]
- Howarth FG, Ramsay GW. The conservation of island insects and their habitats. In: Collins NM, Thomas JA, editors. The conservation of insects and their habitat. Lond: Academic Press; 1991. pp. 71–107. [Google Scholar]
- Humeres SG, Almirón WR, Sabattini MS, Gardenal CN. Estimation of genetic divergence and gene flow between Culex pipiens and Culex quinquefasciatus (Diptera: Culicidae) in Argentina. Mem. Inst. Oswaldo Cruz. 1998;93:57–62. doi: 10.1590/s0074-02761998000100011. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Juan C, Emerson BC, Oromí P, Hewitt GM. Colonization and diversification: towards a phylogeographic synthesis for the Canary Islands. Trends Ecol. Evol. 2000;15:104–109. doi: 10.1016/s0169-5347(99)01776-0. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST. HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes. 2005;5:187–189. [Google Scholar]
- Keyghobadi N, Matrone MA, Ebel GD, Kramer LD, Fonseca DM. Microsatellite loci from the northern house mosquito (Culex pipiens), a principal vector of West Nile virus in North America. Mol. Ecol. Notes. 2004;4:20–22. [Google Scholar]
- Kilpatrick AM. Globalization, land use, and the invasion of West Nile virus. Science. 2011;334:323–327. doi: 10.1126/science.1201010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick AM, Kramer LD, Jones MJ, Marra PP, Daszak P. West Nile virus epidemics in North America are driven by shifts in mosquito feeding behavior. PLoS Biol. 2006;4:e82. doi: 10.1371/journal.pbio.0040082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick AM, Kramer LD, Jones MJ, Marra PP, Daszak P, Fonseca DM. Genetic influences on mosquito feeding behavior and the emergence of zoonotic pathogens. Am. J. Trop. Med. Hyg. 2007;77:667–671. [PubMed] [Google Scholar]
- Kodandaramaiah U, Wahlberg N. Out-of-Africa origin and dispersal-mediated diversification of the butterfly genus Junonia (Nymphalidae: Nymphalinae) J. Evol. Biol. 2007;20:2181–2191. doi: 10.1111/j.1420-9101.2007.01425.x. [DOI] [PubMed] [Google Scholar]
- Kothera L, Zimmerman EM, Richards CM, Savage HM. Microsatellite characterization of subspecies and their hybrids in Culex pipiens complex (Diptera: Culicidae) mosquitoes along a north–south transect in the Central United States. J. Med. Entomol. 2009;46:236–248. doi: 10.1603/033.046.0208. [DOI] [PubMed] [Google Scholar]
- Langella O. Populations 1.2.30. 1999. Available at http://bioinformatics.org/∼tryphon/populations/ (accessed September 25, 2009)
- Lapointe DA, Atkinson CT, Samuel MD. Ecology and conservation biology of avian malaria. Ann. N. Y. Acad. Sci. 2012;1249:211–226. doi: 10.1111/j.1749-6632.2011.06431.x. [DOI] [PubMed] [Google Scholar]
- Lorosa ES, Faria MS, Alencar LCM, De Oliveira J, Marcondes CB. Blood meal identification of selected mosquitoes in Rio De Janeiro, Brazil. J. Am. Mosq. Control Assoc. 2010;26:18–23. doi: 10.2987/09-5914.1. [DOI] [PubMed] [Google Scholar]
- Lounibos LP. Invasions by insect vectors of human disease. Annu. Rev. Entomol. 2002;47:233–266. doi: 10.1146/annurev.ento.47.091201.145206. [DOI] [PubMed] [Google Scholar]
- McAbee RD, Green EN, Holeman J, Christiansen J, Frye N, Dealey K, et al. Identification of Culex pipiens complex mosquitoes in a hybrid zone of West Nile virus transmission in Fresno County, California. Am. J. Trop. Med. Hyg. 2008;78:303–310. [PubMed] [Google Scholar]
- McBride CS, Singer MC. Field studies reveal strong postmating isolation between ecologically divergent butterfly populations. PLoS Biol. 2010;8:e1000529. doi: 10.1371/journal.pbio.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaei G, Andreadis TG, Armstrong PM, Anderson JF, Vossbrinck CR. Host feeding patterns of Culex mosquitoes and West Nile virus transmission, Northeastern United States. Emerging Infect. Dis. 2006;12:468–474. doi: 10.3201/eid1203.051004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaei G, Andreadis TG, Armstrong PM, Bueno R, Jr, Dennett JA, Real SV, et al. Host feeding pattern of Culex quinquefasciatus (Diptera: Culicidae) and its role in transmission of West Nile virus in Harris County, Texas. Am. J. Trop. Med. Hyg. 2007;77:73–81. [PubMed] [Google Scholar]
- Molaei G, Cummings RF, Su T, Armstrong PM, Williams GA, Cheng M-L, et al. Vector-host interactions governing epidemiology of West Nile virus in Southern California. Am. J. Trop. Med. Hyg. 2010;83:1269–1282. doi: 10.4269/ajtmh.2010.10-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz AG, Salazar C, Castaño J, Jiggins CD, Linares M. Multiple sources of reproductive isolation in a bimodal butterfly hybrid zone. J. Evol. Biol. 2010;23:1312–1320. doi: 10.1111/j.1420-9101.2010.02001.x. [DOI] [PubMed] [Google Scholar]
- Muturi EJ, Muriu S, Shililu J, Mwangangi JM, Jacob BG, Mbogo C, et al. Blood-feeding patterns of Culex quinquefasciatus and other culicines and implications for disease transmission in Mwea rice scheme, Kenya. Parasitol. Res. 2008;102:1329–1335. doi: 10.1007/s00436-008-0914-7. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Colombia University Press; 1987. [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Peel MC, Finlayson BL, McMahon TA. Updated world map of the Köppen-Geiger climate classification. Hydrol. Earth Syst. Sci. 2007;4:1633–1644. [Google Scholar]
- Pinto J, Sousa CA, Arez AP, Alves J, Modiano D, Petrarca V, et al. Assessment of malaria transmission in an area with very low mosquito density. Res. Rev. Parasitol. 1999;59:23–26. [Google Scholar]
- Pinto MA, Rubink WL, Patton JC, Coulson RN, Johnston JS. Africanization in the United States: replacement of feral European honeybees (Apis mellifera L.) by an African hybrid swarm. Genetics. 2005;170:1653–1665. doi: 10.1534/genetics.104.035030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasgon JL, Scott TW. Wolbachia and cytoplasmic incompatibility in the California Culex pipiens mosquito species complex: parameter estimates and infection dynamics in natural populations. Genetics. 2003;165:2029–2038. doi: 10.1093/genetics/165.4.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro H, Ramos HC. Guia ilustrado para a identificação dos mosquitos de Angola. 1995. Suplemento n° 4 ao Boletim da Sociedade Portuguesa de Entomologia, Lisboa.
- Ribeiro H, Ramos HC. Identification keys of the mosquitoes (Diptera: Culicidae) of Continental Portugal, Acores and Madeira. Eur. Mosq. Bull. 1999;3:1–13. [Google Scholar]
- Ribeiro H, Ramos HC, Capela RA, Pires CA. Os mosquitos de Cabo Verde (Diptera: Culicidae) – sistemática, distribuição, bioecologia e importância médica. Lisboa: Junta de Investigações Científica do Ultramar; 1980. [Google Scholar]
- Ribera I, Bilton DT, Vogler AP. Mitochondrial DNA phylogeography and population history of Meladema diving beetles on the Atlantic Islands and in the Mediterranean basin (Coleoptera, Dytiscidae) Mol. Ecol. 2003;12:153–167. doi: 10.1046/j.1365-294x.2003.01711.x. [DOI] [PubMed] [Google Scholar]
- Savage HM, Aggarwal D, Apperson CS, Katholi CR, Gordon E, Hassan HK, et al. Host choice and West Nile virus infection rates in blood-fed mosquitoes, including members of the Culex pipiens complex, from Memphis and Shelby County, Tennessee, 2002–2003. Vector Borne Zoonotic Dis. 2007;7:365–386. doi: 10.1089/vbz.2006.0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage HM, Anderson M, Gordon E, McMillen L, Colton L, Delorey M, et al. Host-seeking heights, host-seeking activity patterns, and West Nile virus infection rates for members of the Culex pipiens Complex at different habitat types within the hybrid zone, Shelby County, TN, 2002 (Diptera: Culicidae) J. Med. Entomol. 2008;45:276–288. doi: 10.1603/0022-2585(2008)45[276:hhhapa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Savini G, Monaco F, Terregino C, Bano A, Di Gennaro L, Pinoni C, et al. Usutu virus in ITALY: an emergence or a silent infection? Vet. Microbiol. 2011;151:264–274. doi: 10.1016/j.vetmic.2011.03.036. [DOI] [PubMed] [Google Scholar]
- Shroyer DA. Establishment of Wyeomyia mitchellii on the island of Oahu, Hawaii. Mosq. News. 1981;41:805–806. [Google Scholar]
- Slatkin M, Excoffier L. Testing for linkage disequilibrium in genotypic data using the Expectation-Maximization algorithm. Heredity. 1996;76:377–383. doi: 10.1038/hdy.1996.55. [DOI] [PubMed] [Google Scholar]
- Smith JL, Fonseca DM. Rapid assays for identification of members of the Culex (Culex) pipiens complex, their hybrids, and other sibling species (Diptera: Culicidae) Am. J. Trop. Med. Hyg. 2004;70:339–345. [PubMed] [Google Scholar]
- Smith JL, Keyghobadi N, Matrone MA, Escher RL, Fonseca DM. Cross-species comparison of microsatellite loci in the Culex pipiens complex and beyond. Mol. Ecol. Notes. 2005;5:697–700. [Google Scholar]
- Solomon T. Flavivirus encephalitis. N. Engl. J. Med. 2004;351:370–378. doi: 10.1056/NEJMra030476. [DOI] [PubMed] [Google Scholar]
- Steeves TE, Maloney RF, Hale ML, Tylianakis JM, Gemmell NJ. Genetic analyses reveal hybridization but no hybrid swarm in one of the world's rarest birds. Mol. Ecol. 2010;19:5090–5100. doi: 10.1111/j.1365-294X.2010.04895.x. [DOI] [PubMed] [Google Scholar]
- Subra R. Biology and control of Culex pipiens quinquefasciatus Say, 1823 (Diptera, Culicidae) with special reference to Africa. Int. J. Trop. Insect Sci. 1981;1:319–338. [Google Scholar]
- Tardei G, Ruta S, Chitu V, Rossi C, Tsai TF, Cernescu C. Evaluation of immunoglobulin M (IgM) and IgG enzyme immunoassays in serologic diagnosis of West Nile virus infection. J. Clin. Microbiol. 2000;38:2232–2239. doi: 10.1128/jcm.38.6.2232-2239.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trari B, Dakki M, Himmi O, El Agbani MA. Les moustiques (Diptera Culicidae) du Maroc. Revue bibliographique (1916–2001) et inventaire des espèces. Bull. Soc. Pathol. Exot. 2002;95:329–334. [PubMed] [Google Scholar]
- Urbanelli S, Silvestrini F, Sabatinelli G, Raveloarifera F, Petrarca V, Bullini L. Characterization of the Culex pipiens complex (Diptera: Culicidae) in Madagascar. J. Med. Entomol. 1995;32:9. doi: 10.1093/jmedent/32.6.778. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson W, Wills D, Shipley P. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004;4:535–538. [Google Scholar]
- Vähä JP, Primmer CR. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol. Ecol. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. [DOI] [PubMed] [Google Scholar]
- Vieira HdeS. 1985. Relatório do grupo de trabalho do Instituto de Higiene e Medicina Tropical (luta contra os culicídeos) em colaboração com a Brigada de Luta contra o Paludismo da República de Cabo Verde: período de 10 de Abril a 09 de Maio de 1985, Lisboa.
- Vieira V, Diaz S, Báez M. Diptera (other families) In: Borges P, Costa A, Cunha R, Gabriel R, Gonçalves V, Martins AF, Melo I, Parente M, Raposeiro P, Rodrigues P, Santos RS, Silva L, Vieira P, Vieira V, editors. A list of the terrestrial and marine biota from the Azores. Cascais: Princípia; 2010. pp. 219–221. [Google Scholar]
- Vinogradova EB. Sofia: Pensoft; 2000. Culex pipiens pipiens mosquitoes: taxonomy, distribution, ecology, physiology, genetics, applied importance and control. [Google Scholar]
- Zinser M, Ramberg F, Willott E. Culex quinquefasciatus (Diptera: Culicidae) as a potential West Nile virus vector in Tucson, Arizona: blood meal analysis indicates feeding on both humans and birds. J. Insect Sci. 2004;4:20. doi: 10.1093/jis/4.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


