Abstract
Background
The ATP-binding cassette transporter ABCC6 gene is located on chromosome 16 between its two pseudogenes (ABCC6P1 and ABCC6P2). Previously, we have shown that ABCC6P1 is transcribed and affects ABCC6 at the transcriptional level. In this study we aimed to determine copy number variations of ABCC6, ABCC6P1 and ABCC6P2 in different populations. Moreover, we sought to study the transcription pattern of ABCC6 and ABCC6 pseudogenes in 39 different human tissues.
Findings
Genomic DNA from healthy individuals from five populations, Chinese (n = 24), Middle East (n = 20), Mexicans (n = 24), Caucasians (n = 50) and Africans (n = 24), were examined for copy number variations of ABCC6 and its pseudogenes by pyrosequencing and quantitative PCR. Copy number variation of ABCC6 was very rare (2/142; 1.4%). However, one or three copies of ABCC6P1 were relatively common (3% and 8%, respectively). Only one person had a single copy of ABCC6P2 while none had three copies. In Chinese, deletions or duplications of ABCC6P1 were more frequent than in any other population (9/24; 37.5%). The transcription pattern of ABCC6P2 was highly similar to ABCC6 and ABCC6P1, with highest transcription in liver and kidney. Interestingly, the total transcription level of pseudogenes, ABCC6P1 + ABCC6P2, was higher than ABCC6 in most tissues, including liver and kidney.
Conclusions
Copy number variations of the ABCC6 pseudogenes are quite common, especially in populations of Chinese ancestry. The expression pattern of ABCC6P2 in 39 human tissues was highly similar to that of ABCC6 and ABCC6P1 suggesting similar regulatory mechanisms for ABCC6 and its pseudogenes.
Keywords: Copy number variation, ABCC6, Pseudogenes, Pyrosequencing, Transcription
Findings
Background
The ATP-binding cassette transporter ABCC6 belongs to a large family of membrane proteins (ABC transporters) that are a highly conserved and present in all organisms from bacteria to man [1,2]. The ABCC6 gene (Entrez Gene ID 368) is located on the short arm of chromosome 16 along with two shorter, almost identical (> 99% sequence identity), pseudogenes; ABCC6P1 (Entrez Gene ID 653190) and ABCC6P2 (Entrez Gene ID 730013) (Figure 1) [3]. Pseudogenes are generally defined as non-functional genes, meaning that they usually do not produce a transcript or a functional protein [4,5]. Transcription of ABCC6 pseudogenes have been described [3,6,7], and recently we found strong evidence for a regulatory interdependency between ABCC6 and its pseudogene ABCC6P1[7].
Figure 1.
Genomic organization of ABCC6, ABCC6P1 and ABCC6P2 . ABCC6 is located on chromosome 16p13 between its two pseudogenes, ABCC6P1 and ABCC6P2, at a distance of 2.3 Mb and 1.3 Mb, respectively. The boxes indicate the size of the genes and the arrows indicate the direction for transcription. Nucleotide difference in exon 2 between ABCC6 (G), ABCC6P1 (G) and ABCC6P2 (A) is indicated in red colour while nucleotide difference in intron 7 between ABCC6 (C) and ABCC6P1 (T) is indicated in orange colour.
Mutations and deletions in ABCC6 are known to cause the rare (prevalence between 1:25,000 and 1:100,000), autosomal recessive disease pseudoxanthoma elasticum (PXE, OMIM 264800), a metabolic disorder characterized by ectopic mineralization of soft connective tissues [8,9]. ABCC6 is located on chromosome 16, a known hotspot of chromosomal instability, showing several genomic duplications and deletions (generally called copy number variations) [10,11]. We therefore hypothesized that ABCC6 pseudogenes would be liable to chromosomal rearrangements and thereby subject to copy number variations. Having less or more copies of ABCC6 pseudogenes is likely to influence the expression level of these pseudogenes, and thus, may have an impact on the parent gene ABCC6, including the genetic message, the protein level and the function of the protein.
Estimation of copy number variations and transcription of pseudogenes is generally difficult because of the high sequence similarity between pseudogenes and their parent genes. However, pyrosequencing has recently shown to be a helpful tool to differentiate between highly similar genes by only one nucleotide difference [12]. Therefore, principally by the use of pyrosequencing, the aim of this study was to determine copy number variations of ABCC6ABCC6P1 and ABCC6P2. Moreover, we sought to study the mRNA transcription pattern of ABCC6ABCC6P1 and ABCC6P2 in 39 different human tissues.
Results
Copy number variations in ABCC6 and its pseudogenes in human samples
Copy number variation of ABCC6 was very rare (2/142; 1.4%) (Table 1) in healthy individuals. No individuals had deletions of ABCC6. Deviation in copy number was more frequent for ABCC6 pseudogenes. In Chinese, deletions or duplications of ABCC6P1 were more frequent than in any other population (9/24; 37.5%). Furthermore, in the total population (n = 142), one or three copies of ABCC6P1 was relatively common (3% and 8%, respectively). Only one person had one copy of ABCC6P2 while none had three copies. In Africans, however, no copy number variation was found for ABCC6 pseudogenes. All the members of two Centre d’Etude du Polymorphisme Humain (CEPH) pedigrees had two copies of ABCC6, ABCC6P1 and ABCC6P2 (data not shown). As copy number variation was analyzed in short specific regions of ABCC6 and ABCC6 pseudogenes, small deletions/insertions in other regions of these genes cannot be excluded.
Table 1.
Copy number variation in different populations
|
Genes |
Copies |
Caucasians (N = 50) |
Mexicans (N = 24) |
Middle-East (N = 20) |
Africans (N = 24) |
Chinese (N = 24) |
Total (N = 142) |
|---|---|---|---|---|---|---|---|
| N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | ||
|
ABCC6 |
1 |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
| |
2 |
50 (100) |
23 (96) |
20 (100) |
24 (100) |
23 (96) |
140 (99) |
| |
3 |
0 (0) |
1 (4) |
0 (0) |
0 (0) |
1 (4) |
2 (1) |
|
ABCC6P1 |
1 |
0 (0) |
0 (0) |
1 (5) |
0 (0) |
4 (17) |
5 (3) |
| |
2 |
47 (94) |
23 (96) |
17 (85) |
24 (100) |
15 (62) |
126 (89) |
| |
3 |
3 (6) |
1 (4) |
2 (10) |
0 (0) |
5 (21) |
11 (8) |
|
ABCC6P2 |
1 |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
1 (4) |
1 (1) |
| |
2 |
50 (100) |
24 (100) |
20 (100) |
24 (100) |
23 (96) |
141 (99) |
| 3 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
Gene expression of ABCC6 and its pseudogenes in human samples
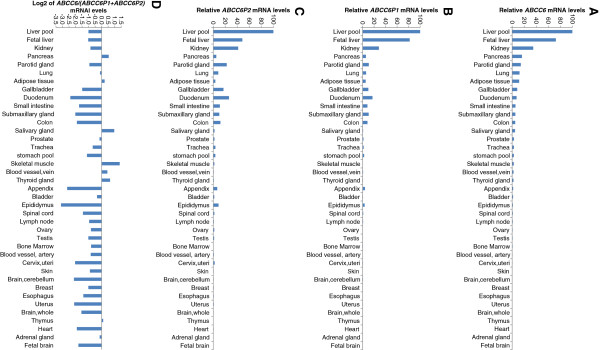
ABCC6 and ABCC6 pseudogene expression profiling was performed in various human tissues (not corresponding to individuals analyzed for copy number variations). The transcription pattern of ABCC6P2 was highly similar to ABCC6 and ABCC6P1, with highest transcription in liver and kidney (Figure 2 A-C). Interestingly, the total mRNA level of ABCC6 pseudogenes, ABCC6P1 + ABCC6P2, was higher than ABCC6 mRNA levels in most tissues, including liver and kidney (Figure 2 D). However, it should be noted that in tissues with low ABCC6 mRNA levels, differences in expression between ABCC6 and its pseudogenes may result from stochastic effects during the PCR.
Figure 2.
mRNA expression of ABCC6 and its pseudogenes ABCC6P1 and ABCC6P2 in a variety of human tissues. Relative normalized transcription of ABCC6 (A), ABCC6P1 (B) and ABCC6P2 (C) in 39 human tissues. The tissue with the highest expression (liver pool) was used as reference tissue (100%). (D) Expression of ABCC6 relative to ABCC6P1 and ABCC6P2. Ratios are shown as a logarithmic scale (base 2).
Discussion
In this study, we found copy number variation of ABCC6 pseudogenes to be frequent, especially in populations of Chinese ancestry. However, as expected in healthy populations, no individuals had deletions of ABCC6. ABCC6 and its pseudogenes, with their high sequence identity (>99%), represent a low copy repeat (LCR). LCRs are paralogue segments of usually >10 kb with >97% sequence identity and can act as substrates for nonallelic homologous recombination which may lead to deletion, duplication or inversion of the intervening sequence [13,14]. Phylogenetic trees on human and chimpanzee sequences show that ABCC6 pseudogenes have occurred independently several times in these species, which further demonstrate the high mobility of these genes [15]. Our results confirm the hypothesis that ABCC6 pseudogenes would be liable to genomic instability and thereby copy number variations.
Six percent of Caucasians are expected to have three copies of ABCC6P1 (Table 1). However, none of the 35 members of two 3-generation-pedigrees (CEPH pedigrees) had deviation from the normal copy of two of ABCC6 or ABCC6 pseudogenes, indicating a Mendelian transmission of these copy numbers and that de novo deletion/duplication of pseudogenes do not arise frequently.
ABCC6 and its pseudogenes share highly similar proximal promoter sequences (> 98.5% sequence identity) [7]. Furthermore, the hepatocyte nuclear factor 4α (HNF4α) binding site located at −166/-154 of the ABCC6 promoter, which is crucial for tissue-specific expression pattern of ABCC6[16,17], is also present in the ABCC6P1 and ABCC6P2 promoters. Thus, the finding of similar expression pattern for ABCC6 and both pseudogenes in human tissues strongly imply similar regulatory mechanisms for ABCC6 and its pseudogenes. On the other hand, these results also suggest that both ABCC6- and ABCC6 pseudogene transcripts have similar half-lives, which is surprising since they do not share the same mRNA 3’-ends. Pseudogenes were for a long time assumed to be “junk DNA”. However, recent studies have shown that many pseudogenes are functionally active and that they may influence their parent gene [7,18-23], and potential mechanisms of pseudogene function have been suggested. Copy number variation in pseudogenes has previously also been identified for the Neutrophil cytosolic factor 1 (NCF1) pseudogenes [24-26]. NCF1 is a component of NADPH oxidase and having fewer or more copies of NCF1 pseudogenes seem to influence the production of reactive oxygen intermediates [24,26]. It would therefore be interesting to investigate whether different copy numbers of ABCC6 pseudogenes also influences the expression of the ABCC6 gene. Unfortunately, the expression of ABCC6ABCC6P1 and ABCC6P2 in various human lymphoblastoid cell lines with one, two or three copies of ABCC6P1, which could be used for these studies, was too low to be detected by reverse transcription - quantitative real-time PCR (RT-qPCR) or pyrosequencing (data not shown).
Methods
Human samples
Genomic DNA from the National Institute of General Medical Science (NIGMS) Human Variation Panels was purchased from the Coriell Cell Repositories (Camden, USA): The Caucasian Panel (n = 50); The Han People of Los Angeles Panel (n = 24); The Middle Eastern Panel, version 1 and 2, (n = 20); The Mexican-American Community of Los Angeles Panel (n = 24), The African-American Panel (n = 24) and two pedigrees (CEPH/Utah Pedigree 1331 and CEPH/Amish Pedigree 884). High quality RNA samples of the Human Total Master panel II and the Human Adult Normal Tissue Total RNA (39 tissues in total) were purchased from Clontech (Mountain View, USA) and BioCat Gmbh (Heidelberg, Germany) respectively.
Copy number variation analysis
For absolute copy number determination of ABCC6, the TaqMan® Copy Number Assay was used targeting ABCC6 specifically in intron 11 (Hs03952142_cn; Applied Biosystems, Foster City, USA). Rnase P, which is known to be present in two copies in the human genome, was used as endogenous reference gene (TaqMan® Copy Number Reference Assay Rnase P, Applied Biosystems). The ABCC6 assay (labeled with FAM), the RnaseP assay (labeled with VIC), sample DNA and 2xTaqMan Universal PCR Master Mix was combined in 20 μl reactions and ran in quadruplicate on a 7900HT Fast Real-Time PCR System using standard conditions (Applied Biosystem). The absolute copy number of ABCC6 was thereby calculated using CopyCaller™Software v1.0 (Applied Biosystems).
For pyrosequencing, two sets of PCR primers were designed to amplify ABCC6, ABCC6P1 and ABCC6P2. The first set of primers targeted intron 7 of ABCC6 and ABCC6P1 only, while the other set of primers targeted exon 2 of all three genes (Table 2 and Figure 1). The genes were amplified from ~100 ng of genomic DNA in 25 μL reactions using 1 x PyroMark PCR Master Mix, 1 x CoralLoad Concentrate (Qiagen, Venlo, The Netherlands) and 0.2 μM primers. Cycling conditions were an initial enzyme activation step at 95°C for 15 min and 45 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, and a final extension cycle of 72°C for 10 min. Twenty micro liter of PCR products were added to 40 μL Binding Buffer (Qiagen), 2 μL streptavidin sepharose high-performance beads (GE Healthcare, Little Chalfont, United Kingdom) and 18 μL water and stirred for 5–10 min at 1400 rpm on a mixer. Single stranded biotinylated templates were isolated using PyroMark Vacuum Prep WorkStation (Qiagen) and dispensed onto PyroMarkQ24 plate containing 25 μL of 0.3 μM sequencing primer and Annealing Buffer (Qiagen). The plates were incubated for 2 min at 80°C and subsequently cooled at room temperature for at least 5 min. Sequencing was performed using a PyroMark Q24 instrument with PyroGold reagents (Qiagen). In the sequencing reaction, one and one nucleotide is added at the time, and peak heights are propotional to the amount of nucleotide molecules incorporated. Therefore, relative copy numbers were calculated using ratios of pyrogram peak heights: ABCC6/(ABCC6 + ABCC6P1) = C5/C8 (C5 and C8 being the peak heights at dispensation 5 and 8, respectively for assay #1) and ABCC6P2/(ABCC6 + ABCC6P1 + ABCC6P2) = T8/T6 (T6 and T8 being the peak heights at dispensation 6 and 8, respectively for assay # 2, note; reverse sequencing) (Table 2).
Table 2.
Primer sequences and nucleotide dispensation order used in pyrosequencing assays
|
Assay # |
Genes |
Template |
Primer |
Sequence |
Dispensation order |
Amplicon size (bp) |
|---|---|---|---|---|---|---|
| 123456789 | ||||||
| 1 |
ABCC6-ABCC6P1 |
gDNA |
Forward |
5’-TGAGGGAGCCAGGCTAGA-3’ |
|
129 |
| |
|
|
Reverse |
5’-Biotin-GAGGGGAAGGGAGAGATTAGC-3’ |
|
|
| |
|
|
Sequencing |
5’-GCCTGGCCCTGCCGC-3’ |
GTAGCTGCT |
|
| 2 |
ABCC6-ABCC6P1-ABCC6P2 |
gDNA |
Forward |
5’-Biotin-TCCCATCTACCTCCTCTTCATC-3’ |
|
76 |
| |
|
|
Reverse |
5’-ATCTTGGCTTTGAAGAGTGG-3’ |
|
|
| |
|
|
Sequencing |
5’-TGGCTTTGAAGAGTGG-3’ |
CGACATCT |
|
| 3 |
ABCC6-ABCC6P1-ABCC6P2 |
cDNA |
Forward |
5’-Biotin-CGGGGCAGGGGGTCTGGAAC-3’ |
|
195 |
| |
|
|
Reverse |
5’-ATCTTGGCTTTGAAGAGTGG-3’ |
|
|
| Sequencing | 5’-TGGCTTTGAAGAGTGG-3’ | CGACATCT |
The absolute copy number for each allele (ABCC6, ABCC6P1 and ABCC6P2) was finally deduced from the TaqMan® Copy Number Assay and the two pyrosequencing assays.
Gene expression analysis
cDNA was synthesized from total RNA from 39 tissues (1 μg) by reverse-transcription using Omniscript RT kit (Qiagen) in the presence of oligo-dT and random hexamer primers (Applied Biosystems) in 20 μL reactions.
Previously, the mRNA expression pattern of ABCC6 and ABCC6P1 in 20 different tissues was described by our group [7]. In this study, we determined the mRNA expression pattern of ABCC6 and ABCC6P1 in a total of 39 tissues by RT-qPCR on a 7900HT Fast Real-Time PCR System (Applied Biosystems). Five of twelve reference genes were estimated by geNorm [27] to give the most reliable normalization factors (see [28] for assay IDs): GAPDH = PGK1 > SDHA > CTBP1 > GOLGA1 . The expression of ABCC6 and ABCC6P1 was normalized to these factors for each tissue.
Due to methodological issues, the mRNA expression pattern of ABCC6P2 was not investigated in our previous study [7]. In this study, the relative mRNA expression pattern of ABCC6P2 was analyzed by pyrosequencing. Primers for mRNA expression were designed to amplify ABCC6ABCC6P1 and ABCC6P2 expressed genes (Table 2). The relative transcription of ABCC6P2 to ABCC6 and ABCC6P1 in the same 39 tissues was quantified using the same equation as above: ABCC6P2/(ABCC6 + ABCC6P1 + ABCC6P2) = T8/T6; Assay #3, Table 2), and by using the expressional data (RT-qPCR) of ABCC6 and ABCC6P1. All PCR samples were run in parallels. No parallels varied more than 0.5 quantification cycles in qPCR experiments or 0.1 in pyrosequencing peak height ratios.
Availability of supporting data
The data set supporting the results of this article are included within the article in Additional file 1 Table S1.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MKK, AP and JPB participated in the study design. MKK, CS and RMG carried out the experimental work. MKK and CS did the interpretation of data. MKK drafted the manuscript with assistance from CS and AP. All authors read and approved the final manuscript.
Supplementary Material
Copy numbers of ABCC6, ABCC6P1 and ABCC6P2.
Contributor Information
Marianne K Kringen, Email: m.k.kringen@medisin.uio.no.
Camilla Stormo, Email: camilla.stormo@medisin.uio.no.
Runa M Grimholt, Email: r.m.grimholt@medisin.uio.no.
Jens P Berg, Email: j.p.berg@medisin.uio.no.
Armin P Piehler, Email: armin.piehler@medisin.uio.no.
Acknowledgements
This project has been financed with aid from the South-Eastern Norway Regional Health Authority and grant from the Clinic of Diagnostics and Intervention, Oslo university hospital, Oslo, Norway.
References
- Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Holland IB, Cole SPC, Kuchler K, Higgins CF. ABC Proteins: From Bacteria to Man. 1. Academic Press, London; 2002. [Google Scholar]
- Pulkkinen L, Nakano A, Ringpfeil F, Uitto J. Identification of ABCC6 pseudogenes on human chromosome 16p: implications for mutation detection in pseudoxanthoma elasticum. Hum Genet. 2001;109:356–365. doi: 10.1007/s004390100582. [DOI] [PubMed] [Google Scholar]
- Mighell AJ, Smith NR, Robinson PA, Markham AF. Vertebrate pseudogenes. FEBS Lett. 2000;468:109–114. doi: 10.1016/S0014-5793(00)01199-6. [DOI] [PubMed] [Google Scholar]
- Balakirev ES, Ayala FJ. Pseudogenes: are they “junk” or functional DNA? Annu Rev Genet. 2003;37:123–151. doi: 10.1146/annurev.genet.37.040103.103949. [DOI] [PubMed] [Google Scholar]
- Aranyi T, Ratajewski M, Bardoczy V, Pulaski L, Bors A, Tordai A. et al. Identification of a DNA methylation-dependent activator sequence in the pseudoxanthoma elasticum gene, ABCC6. J Biol Chem. 2005;280:18643–18650. doi: 10.1074/jbc.M501139200. [DOI] [PubMed] [Google Scholar]
- Piehler AP, Hellum M, Wenzel JJ, Kaminski E, Haug KB, Kierulf P. et al. The human ABC transporter pseudogene family: Evidence for transcription and gene-pseudogene interference. BMC Genomics. 2008;9:165. doi: 10.1186/1471-2164-9-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costrop LM, Vanakker OO, Van LL, Le SO, Martin L, Chassaing N. et al. Novel deletions causing pseudoxanthoma elasticum underscore the genomic instability of the ABCC6 region. J Hum Genet. 2010;55:112–117. doi: 10.1038/jhg.2009.132. [DOI] [PubMed] [Google Scholar]
- Uitto J, Li Q, Jiang Q. Pseudoxanthoma elasticum: molecular genetics and putative pathomechanisms. J Invest Dermatol. 2010;130:661–670. doi: 10.1038/jid.2009.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA. et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD. et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian A, Gharizadeh B, Gustafsson AC, Sterky F, Nyren P, Uhlen M. et al. Single-nucleotide polymorphism analysis by pyrosequencing. Anal Biochem. 2000;280:103–110. doi: 10.1006/abio.2000.4493. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S. et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/S0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Symmons O, Varadi A, Aranyi T. How segmental duplications shape our genome: recent evolution of ABCC6 and PKD1 Mendelian disease genes. Mol Biol Evol. 2008;25:2601–2613. doi: 10.1093/molbev/msn202. [DOI] [PubMed] [Google Scholar]
- Ratajewski M, de Boussac H, Pulaski L. Liver-specific enhancer in ABCC6 promoter-Functional evidence from natural polymorphisms. Biochem Biophys Res Commun. 2009;383:73–77. doi: 10.1016/j.bbrc.2009.03.131. [DOI] [PubMed] [Google Scholar]
- de Boussac H, Ratajewski M, Sachrajda I, Koblos G, Tordai A, Pulaski L. et al. The ERK1/2-hepatocyte nuclear factor 4alpha axis regulates human ABCC6 gene expression in hepatocytes. J Biol Chem. 2010;285:22800–22808. doi: 10.1074/jbc.M110.105593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pink RC, Wicks K, Caley DP, Punch EK, Jacobs L, Carter DR. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA. 2011;17:792–798. doi: 10.1261/rna.2658311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y. et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S. et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehler AP, Haug KB, Wenzel JJ, Kierulf PB, Kaminski WE. ABCA-transporters: regulators of cellular lipid transport. Tidsskr Nor Laegeforen. 2007;127:2930–2933. [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- Muro EM, Mah N, Andrade-Navarro MA. Functional evidence of post-transcriptional regulation by pseudogenes. Biochimie. 2011;93:1916–1921. doi: 10.1016/j.biochi.2011.07.024. [DOI] [PubMed] [Google Scholar]
- Greve B, Hoffmann P, Vonthein R, Kun J, Lell B, Mycko MP. et al. NCF1 gene and pseudogene pattern: association with parasitic infection and autoimmunity. Malar J. 2008;7:251. doi: 10.1186/1475-2875-7-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunson T, Wang Q, Chambers I, Song Q. A copy number variation in human NCF1 and its pseudogenes. BMC Genet. 2010;11:13. doi: 10.1186/1471-2156-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson LM, Nerstedt A, Lindqvist AK, Johansson SC, Medstrand P, Olofsson P. et al. Copy number variation of the gene NCF1 is associated with rheumatoid arthritis. Antioxid Redox Signal. 2012;16:71–78. doi: 10.1089/ars.2011.4013. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, de Preter K, Pattyn F, Poppe B, van Roy N, de Paepe A. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3: . doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehler AP, Grimholt RM, Ovstebo R, Berg JP. Gene expression results in lipopolysaccharide-stimulated monocytes depend significantly on the choice of reference genes. BMC Immunol. 2010;11:21. doi: 10.1186/1471-2172-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Copy numbers of ABCC6, ABCC6P1 and ABCC6P2.