Abstract
N-methyl-d-aspartate (NMDA) receptor activation involves a dynamic series of structural rearrangements initiated by glutamate binding to glycine-loaded receptors and culminates with the clearing of the permeation pathway, which allows ionic flux. Along this sequence, three rate-limiting transitions can be quantified with kinetic analyses of single-channel currents, even though the structural determinants of these critical steps are unknown. In inactive receptors, the major permeation barrier resides at the intersection of four M3 transmembrane helices, two from each GluN1 and GluN2 subunits, at the level of the invariant SYTANLAAF sequence, known as the lurcher motif. Because the A7 but not A8 residues in this region display agonist-dependent accessibility to extracellular solutes, they were hypothesized to form the glutamate-sensitive gate. We tested this premise by examining the reaction mechanisms of receptors with substitutions in the lurcher motifs of GluN1 or GluN2A subunits. We found that, consistent with their locations relative to the proposed activation gate, A8Y decreased open-state stability, whereas A7Y dramatically stabilized open states, primarily by preventing gate closure; the equilibrium distribution of A7Y receptors was strongly shifted toward active states and resulted in slower microscopic association and dissociation rate constants for glutamate. In addition, for both A8- and A7-substituted receptors, we noticed patterns of kinetic changes that were specific to GluN1 or GluN2 locations. This may be a first indication that the sequence of discernible kinetic transitions during NMDA receptor activation may reflect subunit-dependent movements of M3 helices. Testing this hypothesis may afford insight into the activation mechanism of NMDA receptors.
INTRODUCTION
NMDA receptors represent one of four classes of ionotropic glutamate receptors (iGluRs) responsible for fast excitation in central neurons. Despite overall common tetrameric structures, each class generates currents with characteristic waveforms and fulfills nonoverlapping physiological roles. The structural details that support class-specific functions are largely unknown (Traynelis et al., 2010).
Like all iGluR subunits, NMDA receptor subunits (GluN) have modular architecture. Extracellular residues form two genetically separable N-terminal (NTD) and ligand-binding (LBD) domains (Wo and Oswald, 1995). In tetrameric receptors, eight such modules adopt globular shapes and stack as discernible NTD and LBD layers (Sobolevsky et al., 2009). Each LBD connects through three short linkers to the M1, M3, and M4 transmembrane helices but not to the intervening M2 segment, which loops only halfway through the membrane from the cytoplasmic side. The permeation pathway is lined on its extracellular half by M3 residues and on its cytoplasmic half by M2 residues, which also form the narrowest portion of the open pore (Kuner et al., 1996; Beck et al., 1999; Sobolevsky et al., 2002).
Arguments based on structural homology and patterns of accessibility to extracellular reagents support the hypothesis that in iGluRs, the agonist-controlled gate resides at the intersection of M3 helices. In functional iGluRs, the M1–M3 segments superimpose well with the pore structure of the KcsA channel, whose ligand-dependent constriction is made by the crossing of four M3-like helices (Doyle et al., 1998; Jiang et al., 2002; Armstrong, 2003; Kuner et al., 2003; Sobolevsky et al., 2009). In the corresponding region, iGluRs have a conserved nine-residue stretch, SYTANLAAF, and the A8 of this motif corresponds to the bundle crossing of KcsA (Chang and Kuo, 2008). Consistent with a location at or internal to the agonist-controlled gate, glutamate increases the accessibility of extracellular reagents to A7, whereas the more superficial A8 is accessible regardless of agonist presence (Beck et al., 1999). Functional evidence from A7- and A8-substituted receptors revealed stark differences in function for these two positions in NMDA versus non-NMDA receptors.
When present in δ-type iGluRs, A8T substitutions cause gross gating modifications that result in cerebellar neurodegeneration and ataxic behaviors in the eponymous Lurcher mice (Zuo et al., 1997). Similarly, when introduced in AMPA- or kainate-type iGluRs, the lurcher substitution (A8T) produces substantial constitutive activity and slows desensitization and deactivation kinetics of agonist-activated whole-cell currents (Schwarz et al., 2001). In contrast, in NMDA receptors, lurcher mutations cause only mild kinetic changes whether present on GluN1 (N1) or GluN2 (N2) subunits (Zuo et al., 1997; Kohda et al., 2000; Hu and Zheng, 2005a,b). Instead, A7Y mutations (lurcher-like) and other bulky side chains introduced at A7 sites render receptors constitutively open (Jones et al., 2002; Yuan et al., 2005; Blanke and VanDongen, 2008).
If indeed the adjacent A7 and A8 residues have different relationships to the activation gate of the NMDA receptors, substitutions at these positions should affect the gating reaction with distinct kinetic mechanisms. To test this corollary, we determined the gating mechanism of NMDA receptors carrying lurcher (A8T) or lurcher-like (A7Y) mutations in N1 or N2A subunits. We found that at A8, T had no effect and Y caused receptors to close faster, whereas at A7, Y acted as a kinetic break on channel closure and shifted the entire equilibrium distribution such that the association and dissociation rate constants for glutamate were slower, and a substantial fraction of channels was open in the absence of agonist. Our results are consistent with the hypothesis that A7 residues of N1 and N2 subunits form the agonist-controlled gate of NMDA receptors.
MATERIALS AND METHODS
Cell culture and receptor expression
Rat GluN1-1a (U08261) (N1) or GluN2A (M91561) (N2) was subcloned into pcDNA3.1(+), and the following substitutions were introduced in N1: A653T (N18T), A653Y (N18Y), or A652Y (N17Y); and in N2: A651T (N28T), A651Y (N28Y), or A650Y (N27Y). These constructs were transiently expressed in HEK293 cells (CRL-1573; American Type Culture Collection) along with GFP in a 1:1:1 ratio to produce the following NMDA receptors: N1/N2 (wild type [WT]), N18T/N2, N1/N28T, N18Y/N2, N1/N28Y, N17Y/N2, or N1/N27Y.
Electrophysiological recordings and analyses
Stationary single-channel currents were recorded from cell-attached patches with glass pipettes filled with extracellular solution containing (in mM): 150 NaCl, 2.5 KCl, 1 EDTA, 10 N-(2-hydroxyethyl) piperazine-N′-(4-butanesulfonic acid) (HEPBS), and agonists (Glu, Gly) as indicated, adjusted to pH 8.0 (NaOH). Currents were amplified and low-pass filtered (10 kHz) with an Axopatch 200B (Molecular Devices), digitized (20 kHz) with an A/D board (PCI-6229; National Instruments), and acquired and stored as digital files with QuB software. Files that reflected activity from a single active receptor throughout were selected for processing. Idealization and modeling were done with QuB functions as described in detail by Kussius et al. (2009).
Overall kinetic parameters and gating rate constants were estimated by fitting state models to the entire sequence of events in each file; results are presented as the average value for each dataset (mean ± SEM). Statistical differences were evaluated with Student’s t test or with one-way ANOVA and were considered significant for P < 0.05. Agonist binding and dissociation rate constants were estimated by globally fitting kinetic models directly to data obtained at several agonist concentrations, as described in detail by Popescu et al. (2004). Values were rounded to the 90th percentile.
Macroscopic responses were recorded as whole-cell currents with glass pipettes containing intracellular solution (in mM): 135 CsF, 33 CsOH, 2 MgCl2, 1 CaCl2, 10 HEPES, and 11 EGTA, adjusted to pH 7.4 (CsOH). Cells were perfused with extracellular solution containing (in mM): 150 NaCl, 2.5 KCl, 0.5 CaCl2, 0.01 EDTA, 10 HEPBS, and 0.1 Gly, adjusted to pH 8.0 (NaOH). Responses were elicited by switching into solutions with added Glu (1 mM). Currents were amplified and analogue filtered (2 kHz; Axopatch 200B), sampled (5 kHz; Digidata 1440A), acquired with pClamp software (Molecular Devices), and analyzed offline.
Simulated macroscopic currents were calculated from the specified kinetic models, expanded as necessary with glutamate-binding steps. Responses were initiated by positioning all channels (n = 100; 10 pA/each) in the glutamate-free state C0 and stepping Glu concentrations instantaneously from 0 to 1 mM. Responses were represented as time-dependent occupancies of the aggregated open state and were analyzed as the experimentally recorded currents (Popescu et al., 2004).
Energy diagrams were calculated using the rate constants specified in each model and the relationship ΔG0 = −RT(lnKeq), where R is the molar gas constant, T is the absolute temperature, and Keq is the equilibrium constant of the transition considered, calculated as the ratio of the forward to reverse rate constants. Barrier heights were calculated with the relationship E‡ = ΔG0 + (10 − lnk+) and were arbitrarily illustrated at equal distances along the reaction coordinate (Kussius and Popescu, 2009).
Online supplemental material
Table S1 shows closed kinetic components for lurcher and lurcher-like N1/N2A receptors. Table S2 lists open kinetic components for lurcher and lurcher-like N1/N2A receptors. Fig. S1 illustrates rate constants for NMDA receptors with lurcher and lurcher-like mutations estimated with 5C4O kinetic models. The online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201210786/DC1.
RESULTS
To test the hypothesis that A7 but not A8 of the SYTANLAAF motif forms the agonist-controlled gate of NMDA receptors, we set out to delineate the reaction mechanism of GluN1/GluN2A receptors carrying A8T or A7Y substitutions in GluN1 (N1) or GluN2A (N2) subunits (Fig. 1, A–C). We recorded on-cell currents from patches containing exactly one active receptor in the presence of maximally effective agonist concentrations (1 mM Glu and 0.1 mM Gly), low proton concentrations (pH 8; 10 mM HEPBS), and the absence of divalent cations (150 mM NaCl, 2.5 mM KCl, and 1 mM EDTA) (Figs. 2 A and 3 A). In these conditions, the recorded traces mark the stochastic flux of sodium across the membrane and faithfully chronicle receptor excursions into and out of ion-permeable conformations (Popescu and Auerbach, 2003). Such traces inform directly about each receptor’s unitary current amplitude and after kinetic analyses and modeling of dwell times can help define the receptor’s activation mechanism (Kussius et al., 2009).
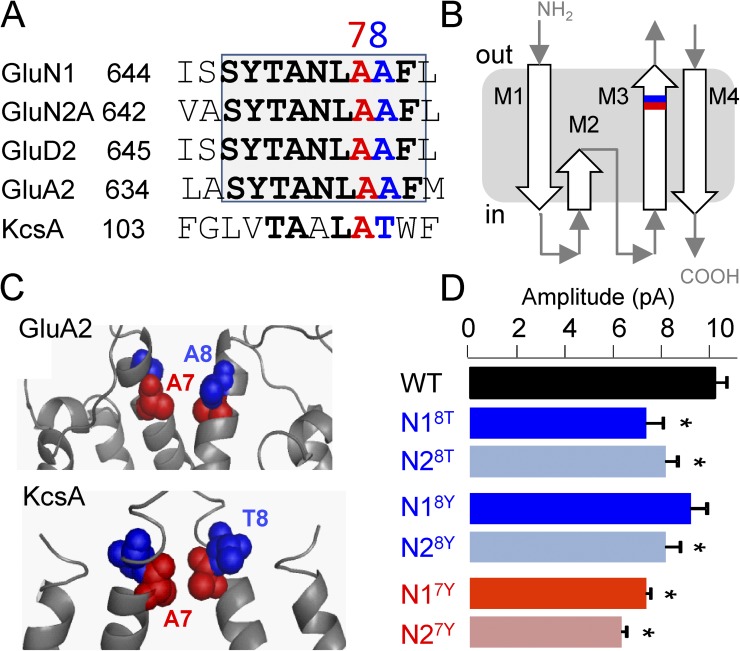
Figure 1.
Lurcher-motif residues control NMDA receptor unitary current amplitude. (A) Sequence alignment of the external segment of M3 highlights the conserved lurcher motif; the residues tested here are in red (A7) and blue (A8). This color scheme is used in all subsequent panels. (B) Cartoon of iGluR subunit membrane topology. (C) Partial model of two diagonally situated subunits based on an AMPA receptor (Protein Data Bank accession no. 3KG2) and on KcsA (Protein Data Bank accession no. 1BL8) structures. (D) Unitary current amplitudes of NMDA receptors carrying lurcher (A8T) or lurcher-like (A7Y) substitutions in the N1 or N2A subunits. *, statistically significant differences relative to WT (P < 0.05; one-way ANOVA).
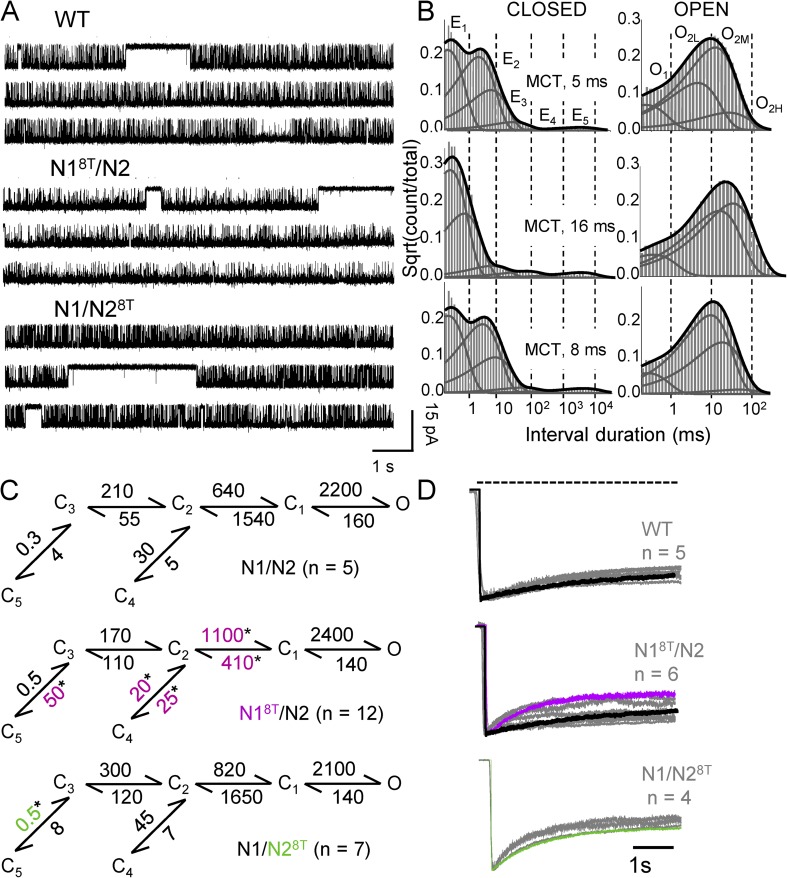
Figure 2.
Gating effects of the lurcher mutation A8T on NMDA receptors. (A) Continuous 30-s segments were selected from minutes-long recordings obtained from one WT or one lurcher (N18T/N2 or N1/N28T) receptor (on-cell; open is down). (B) Dwell-time distributions calculated from the records illustrated in A are overlaid with probability distribution functions calculated from 5C4O models (thick) and individual closed or open kinetic components (thin). (C) Kinetic models optimized by fits to the entire sequence of closed and open intervals in each record; for each postulated transition, rate constants are given as rounded means for each dataset (in s−1). *, significant difference relative to WT (P < 0.05; Student’s t test). All states (C, O) represent fully liganded (2 Glu, 2 Gly) receptors. (D) Whole-cell currents recorded during 5-s application of 1 mM Glu (gray) are overlaid with the trace simulated with the corresponding kinetic model in C (black, purple, or green). Traces were normalized to peak.
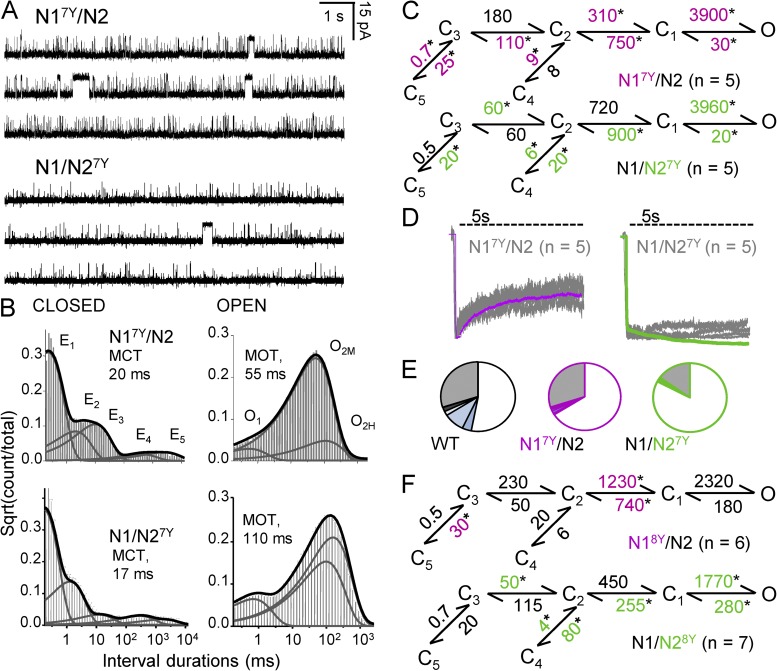
Figure 3.
Gating mechanism of A7Y and A8Y NMDA receptors. (A) Continuous 30-s traces produced by one N17Y/N2 (top) or one N1/N27Y receptor (bottom); open is down. (B) Dwell-time histograms for the records shown in A; overlaid are probability density functions calculated with a 5C3O model (thick line) and kinetic components (thin lines). (C) Reaction mechanisms derived from fits to the entire event sequence in each file; rate constants (s−1) are given as the rounded mean for each dataset. *, significant differences relative to WT (P < 0.05; Student’s t test). All states (C, O) represent fully liganded (2 Glu, 2 Gly) receptors. (D) Whole-cell responses to 5-s applications of 1 mM Glu were recorded from multiple cells (gray) and are superimposed with traces simulated with models in C (purple and green). (E) Occupancy plots calculated from the corresponding models in C. (F) Reaction mechanisms derived from records produced by NMDA receptors with A8Y substitutions.
A8 and A7 substitutions decreased NMDA receptor unitary current amplitudes
In these records, we noted that regardless of the subunit in which they were introduced, A8T and A7Y substitutions resulted in significantly smaller unitary currents (Fig. 1 D and Table 1). Previously, Kohda et al. (2000) reported that A8T produced macroscopic currents with lower noise than WT and concluded that NMDA receptors carrying the lurcher mutation may have “at least one low-conductance open state” (Kohda et al., 2000). Similarly, when channels with cysteine substitutions at A7 of N2A subunits were modified with chemical reagents, the mean single-channel current amplitudes decreased relative to the parent A7C mutant (Yuan et al., 2005). Here, we show that substitutions with natural residues at A7 or A8 of the lurcher motif also significantly decreased NMDA receptor unitary current amplitude. In addition, we noted that when all four subunits contained A7Y substitutions, the amplitude of unitary currents was not further reduced relative to the single-subunit substitutions (not depicted). These results highlight a role of SYTANLAAF residues in controlling the large conductance characteristic for NMDA receptors and advocate for a systematic investigation into the mechanism by which this control occurs.
Table 1.
Kinetic attributes of NMDA receptors with A7 and A8 substitutions
| Mutation | Amplitude | Po | Mean open time | Mean closed time | n | Duration | Events ×106 | |
| pA | ms | ms | min | |||||
| WT | N1/N2 | 10.0 ± 0.6 | 0.54 ± 0.03 | 9 ± 1 | 8 ± 1 | 5 | 244 | 1.5 |
| A8T | N18T/N2 | 7.3 ± 0.3a | 0.48 ± 0.06 | 17 ± 4 | 21 ± 5 | 12 | 552 | 2.8 |
| N1/N28T | 7.5 ± 0.5a | 0.56 ± 0.05 | 11 ± 1 | 10 ± 2 | 7 | 266 | 1.7 | |
| A8Y | N18Y/N2 | 8.3 ± 0.8 | 0.56 ± 0.1 | 9 ± 2 | 8 ± 3 | 6 | 441 | 3.1 |
| N1/N28Y | 7.1 ± 0.3a | 0.11 ± 0.03a | 5 ± 0.4a | 56 ± 14a | 7 | 198 | 0.46 | |
| A7Y | N17Y/N2 | 7.2 ± 0.2a | 0.7 ± 0.03a | 65 ± 13a | 28 ± 6 | 5 | 184 | 0.23 |
| (−/Gly) | 5.9 ± 0.4ab | 0.24 ± 0.04ab | 30 ± 2ab | 112 ± 25ab | 5 | 184 | 0.19 | |
| (−/−) | 5.8 ± 0.2ab | 0.06 ± 0.01ab | 77 ± 8a | 1,400 ± 400ab | 5 | 370 | 0.03 | |
| N1/N27Y | 6.2 ± 0.2a | 0.81 ± 0.06a | 110 ± 10a | 29 ± 13 | 5 | 190 | 0.18 |
Significant difference relative to WT; P < 0.05 (Student’s t test).
Significant difference relative to N17Y/N2 with Glu/Gly; P < 0.05 (Student’s t test).
Aside from precise information about unitary current amplitudes, single-channel traces also illustrate how long receptors dwell in nonconductive (closed [C]) and conductive (open [O]) conformations, and reveal the precise sequence in which the receptor moves between closed and open structures. We used statistical methods to extract this kinetic information, and we present results for lurcher and lurcher-like mutations below.
A8T substitutions had minimal effect on NMDA receptor gating
We measured open probability (Po), mean open times, and mean closed times from currents produced by WT (n = 5) and receptors carrying the lurcher mutation A8T. These parameters were not statistically different across the three datasets, regardless of whether the mutation was introduced in the N1 (N18T/N2, n = 12 and P > 0.05) or the N2A subunit (N1/N28T, n = 7 and P > 0.05) (Table 1). For N18T/N2, we noticed a trend toward longer closures (21 vs. 10 ms) and longer openings (17 vs. 11 ms), but these differences did not achieve statistical significance even for the larger size of this dataset.
Next, we used statistical methods to determine the number of kinetically distinct conformations visited by receptors during gating and to estimate gating rate constants (Fig. 2, B and C). Single NMDA receptors generate current traces whose open and closed intervals have complex distributions, each containing several components (Howe et al., 1988; Gibb and Colquhoun, 1991, 1992). Previous studies concluded that in the conditions used here (high agonist concentration and absence of divalent cations), interval durations have five closed components and up to four open components (Popescu and Auerbach, 2003; Kussius et al., 2009). Closures reflect three kinetically distinct preopen states (C3–1) and two desensitized states (C4 and C5) (Banke and Traynelis, 2003; Popescu and Auerbach, 2003; Auerbach and Zhou, 2005; Kussius et al., 2009). Openings reflect the two obligatory components present in each mode (O1 and O2), with additional multiplicity resulting from the distinct durations of O2 in each mode (O2L, O2M, and O2H) (Jahr and Stevens, 1987; Popescu and Auerbach, 2003, 2004; Magleby, 2004; Amico-Ruvio and Popescu, 2010; Popescu, 2012).
The records we obtained from N18T/N2 and N1/N28T had interval distributions that were very similar to those described previously for WT receptors, an indication that, like WT, these receptors followed a basic kinetic mechanism with five closed and two open states, and also experienced modal gating (Fig. 2 B and Tables S1 and S2). As described previously, the recorded activity can be comprehensively described with a simplified 5C1O-state model, where each C represents a kinetically distinct closed state and O represents collectively all observed open states. In this simplified model, the rate constants for entry into and exit from this aggregate state report the kinetics of several microscopic transitions, including mode switching (Fig. 2 B) (Kussius et al., 2009). Consistent with the mean parameters we estimated for A8T mutants, kinetic modeling results indicated that relative to WT receptors, gating changes were small for N18T/N2 and negligible for N1/N28T receptors (Figs. 2 C and S1). We noted that for N18T/N2, rate changes clustered around the C2 state (with faster exit rates C2→C1 and C2→C4 and slower entry rates C2←C1 and C2←C4); in addition, entry into the principal desensitized state C5 was also faster (C3→C5). In contrast, for N1/N28T the only rate change we observed was the resensitization rate C3←C5, which was faster.
The deduced kinetic models predicted macroscopic responses that were comparable with experimentally measured whole-cell currents elicited with long (5-s) glutamate pulses (in the presence of glycine) (Fig. 2, C and D). For WT and N1/N28T, we observed a nearly perfect match between model predictions, which were deduced from cell-attached recordings, and experimentally measured traces, which were obtained from whole-cell recordings. For N18T/N2, the simulated response overlapped with only a set of the whole-cell traces measured. However, we noted that the single-channel records of N18T/N2 receptors also displayed considerable patch-to-patch variability, and this was consistent with the range of whole-cell traces recorded. Based on these results and observations, we conclude that overall, the A8T substitution had no observable effect on NMDA receptor gating kinetics when introduced in N2 subunits, and had only a modest effect when introduced in N1 subunits. Kohda et al. (2000) reported changes in macroscopic desensitization and deactivation for lurcher mutants. However, when trace amounts of contaminant Zn2+ were removed, as is the case with our recordings, these changes were substantially attenuated (Hu and Zheng, 2005a). Collectively with these previous reports, our results support the conclusion that lurcher mutations have minimal effect on NMDA receptor gating.
Lurcher-like mutations redistribute NMDA receptors into open states
In contrast with the small gating effects we observed for A8T mutations, A7Y produced robust and reproducible kinetic changes regardless of the subunit in which it was introduced. We noted that the patterns of change were subunit dependent, and overall, the effects were larger when the mutation was in the N2 subunit (Fig. 3, A–C and Table 1). Relative to WT, A7Y channels had substantially higher Po. The increased activity was entirely caused by substantially longer openings (6-fold or 10-fold). In both preparations, closures were also longer, but these differences were smaller and not statistically significant (approximately threefold; P > 0.05) (Table 1). Unlike the nonadditive effect observed for changes in unitary current amplitudes, gating changes appeared to add up such that open probabilities for the double mutant N17Y/N27Y were in excess of 0.99 (not depicted). We conclude that the observed changes in unitary current amplitude and channel gating are not correlated, an indication that these effects may arise by separate mechanisms.
Analyses of interval durations revealed that in all A7Y records, closed interval distributions had five components, whereas open distributions had only three components, lacking the open component indicative of low mode (τL) (Fig. 3 B and Tables S1 and S2). Relative to WT, the principal open components (τM and τH) were five- to sevenfold longer for both N17Y/N2 and N1/N27Y. We conclude that for both A7Y mutants, the dramatic increase in mean open durations occurred by two mechanisms: a substantial increase in the duration of openings (τf, τM, and τH) and an absence of low-mode openings.
To organize this information into a coherent even if simplified reaction mechanism, we used the same 5C1O scheme described above for A8T mutants. The rate constants estimated with this approach indicated that although both A7Y mutations affected almost all rate constants, the increased Po originated primarily from substantially slower closing rates: approximately fivefold and approximately eightfold for N1 and N2 mutants, respectively (Fig. 3 C). Macroscopic responses simulated with the deduced models predicted well the experimentally recorded whole-cell current relaxations during prolonged glutamate application (Fig. 3 D). This close match indicated that the kinetic models deduced from single-channel traces captured correctly the salient changes in activation mechanism produced by the A7Y mutations. Specifically, the mechanisms postulate that A7Y mutants had dramatically more stable open states, resulting in considerable increased open occupancies (125 or 155% relative to WT) at the expense of preopen (C3–C1) and desensitized (C4, C5) states (Fig. 3 E). Thus, consistent with their hypothesized distinct physical locations relative to the activation gate, A7Y and A8T affected gating with distinct mechanisms: A7 but not A8 substitutions prevented gate closure.
We considered whether the observed effects reflected the difference in the residues’ substitutions, threonine at A8 versus tyrosine at A7, rather than the residues’ position along the M3 helix. To examine whether tyrosine can prevent channel closure when inserted at A8, we introduced A8Y substitution in N1 and N2 subunits and determined the gating reaction mechanism for N18Y/N2 and N1/N28Y receptors. Results show that when introduced at A8, tyrosine residues had no effect (N18Y/N2, P > 0.05) or accelerated channel closure twofold (N1/N28Y, P < 0.05) (Fig. 3 F). We conclude that bulky substitutions at A7 but not at A8 stabilize receptors in open conformations. These results support the proposed location of A7 residues at the agonist-controlled gate of NMDA receptors and suggest a foot-in-the-door mechanism for how voluminous residues prevent channel closure.
When we compared the C3→C2→C1→O activation sequences of the six NMDA receptor mutants for which we developed detailed reactions mechanisms, we were surprised to note patterns of change that were common for the N1 or the N2 mutants but different between the two groups (Figs. 2 C and 3, C and F). For example, the N1 but not the N2 mutations affected the C2–C1 equilibrium for all mutations examined, whereas N2 but not N1 mutations affected preferentially the C3→C2 transition. The simplest interpretation of these observations is that exit from the kinetic states C3 and C2 is controlled by rate-limiting events that reflect rearrangements at the level of the A7 residues within N2 and N1 subunits, respectively. However, because the changes in closed components were relatively small and the inherent variability of single-molecule measurements reduced the resolution with which differences could be reliably ascertained, this hypothesis is highly speculative at this time.
Constitutive activation mechanism of N17Y/N2 receptors
The experiments described above were deliberately performed in the presence of supra-maximal concentrations of glycine (0.1 mM) and glutamate (1 mM) to focus on transitions experienced by liganded receptors. The results demonstrated that A7Y substitutions stabilized receptors in open conformations. However, they did not inform about any changes that may have occurred in glutamate-binding kinetics. Similar to Blanke and VanDongen (2008), we also observed substantial leak currents when measuring whole-cell activity from N17Y/N2 or N1/N27Y receptors, an indication of constitutive activity. To investigate the mechanism by which A7Y substitutions produced spontaneous openings, we set out to delineate the activation reaction of agonist-free N17Y/N2 receptors.
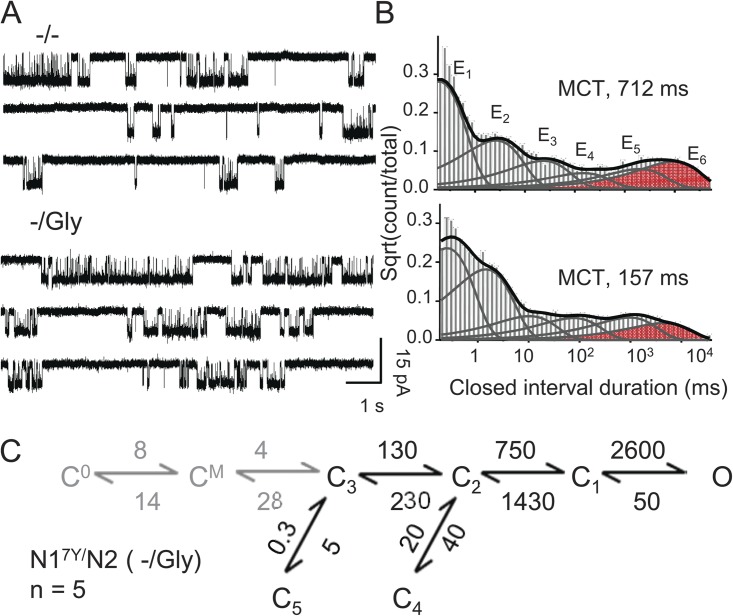
We recorded one-channel activity from N17Y/N2 channels in the absence of added agonists (−/−) and with 0.1 mM glycine in the absence of added glutamate (−/Gly) (Fig. 4 A). These records revealed measurable Po in both conditions: 0.06 ± 0.01 (n = 5) and 0.24 ± 0.04 (n = 5), respectively, with no statistical change in mean open durations relative to activity recorded from this mutant in high concentrations of both glycine and glutamate (P > 0.05) (Table 1). Thus, relative to recordings obtained in high agonist concentrations, the lower activities observed in these two conditions were entirely the result of longer closed periods, caused primarily by the presence of an additional closed component (E6), which was more prominent in the (−/−) condition (Fig. 4 B). As described previously, this additional closed component most likely reflects dwells in nonconducting receptor species that have at least one vacant ligand-binding site (Popescu et al., 2004).
Figure 4.
Constitutive activity of N17Y/N2 receptors. (A) Continuous activity (30 s) selected from recordings obtained from one receptor with no agonists added (−/−) or no glutamate added and 0.1 mM Gly (−/Gly). (B) Histograms of closed events observed in the corresponding records shown in A; probability distribution function (thick line) and kinetic components (E1–E6, thin lines) calculated with a 6C1O model; the concentration-dependent component areas are in red. (C) Kinetic model used to fit the entire sequence of events in each (−/Gly) record; rate constants (s−1) are given as the rounded means for the entire dataset. All states (C, O) represent glycine-bound glutamate-free receptors.
The ubiquitous incidence of glycine contamination makes it problematic to ascertain the presence of glycine-free NMDA receptors and prevents us from measuring gating rate constants for agonist-free receptors. Instead, we determined the gating reaction of glycine-bound glutamate-free N17Y/N2 channels. For this, we fit the records obtained in high glycine concentrations and no added glutamate (−/Gly) with models that included receptor states corresponding to the apo- and mono-liganded conformations of the WT receptors (C0 and CM). Note however that although in this model receptors transition between conformations designated C0, CM, C, and O, at all times the N2 agonist-binding sites are vacant. Results show that the unitary current amplitude and mean open duration were similar to those measured for fully liganded N17Y/N2 receptors, and the much lower Po in the absence of glutamate was entirely caused by slower progress into C3, the first liganded-like conformation. Once the receptor has surmounted this energetic barrier, it gates with rates similar to those experienced by fully liganded receptors (Fig. 4 C and Table 1).
N17Y/N2 receptors have higher affinity for glutamate
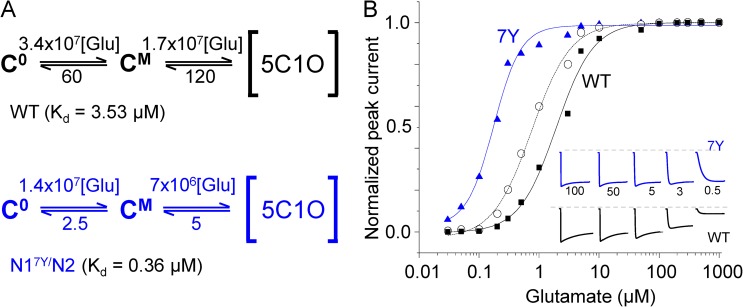
Encouraged by this result, we aimed to measure the agonist-binding kinetics of N17Y/N2 receptors. At present, we cannot extract kinetic information about the glycine-binding reaction because a model for glycine biding to NMDA receptors is not available. Instead, we set out to examine the glutamate-binding reaction to glycine-bound receptors. For this we collected six additional records from on-cell one-channel patches with 0.1 mM glycine present and three concentrations of glutamate (0.5, 1, and 3 µM; n = 2 for each). These records displayed the additional closed component E6, whose time constant was concentration dependent, becoming shorter as glutamate concentration was increased. To estimate glutamate on- and off-rates, to these six files we fit globally a state model that, in addition to the 5C1O sequence inferred from measurements in high glutamate concentration, also included glutamate-binding steps (Fig. 5 A). The results revealed a 2.5-fold slower glutamate on-rate (kon, 7 × 106 M−1s−1) and a 25-fold slower glutamate off-rate (koff, 2.5 s−1) for glycine-bound N17Y/N2 receptors relative to the rate constants reported previously for WT receptors (Fig. 5 A) (Popescu and Auerbach, 2004; Kussius and Popescu, 2010). The rate constants measured here indicate a 10-fold increase in the calculated microscopic affinity for glutamate (koff/kon, 0.36 µM vs. 3.53 µM for WT). Simulations with the measured rates predicted a 10-fold shift in glutamate potency (EC50, 0.17 µM vs. 1.9 µM for WT) (Fig. 5 B), as reported previously by Blanke and VanDongen (2008) for receptors expressed in Xenopus laevis oocytes (EC50, 0.45 µM vs. 4.05 µM for WT).
Figure 5.
N17Y/N2 NMDA receptors have increased affinity for glutamate. (A) Reaction schemes include explicit glutamate-binding steps appended to the [5C1O]-liganded sequences represented in Figs. 2 C (WT, black) and 3 C (A7Y, blue). At all times receptors are fully occupied with glycine. Rate constants for WT (top) are from Popescu et al. (2004); rate constants for N17Y/N2 (bottom) were estimated from global fits to records obtained in high glycine concentrations (0.1 mM) and three separate low glutamate concentrations (0.5, 1, and 3 µM; n = 2 for each). Except for the association rate constants, which are in M−1s−1, all others are in s−1. (B) Dose–response curves were calculated for WT (▪) and N17Y/N2 (▴) receptors from responses generated with the kinetic models illustrated in A, and again for N17Y/N2 receptors (○) with the 5C1O model in Fig. 3 C and assuming glutamate-binding rates reported for WT (A, top). Inset illustrates simulated macroscopic traces used to construct the dose–response curves.
Importantly, the detailed reaction mechanism we developed affords us the insight needed to separate the observed change in glutamate EC50 produced by the A7Y mutation into its constituent components, i.e., changes in macroscopic affinity and changes in macroscopic efficacy (Colquhoun, 1998). To this end, we constructed a glutamate dose–response curve using a hybrid model that combined the rates we determined here for gating of fully liganded A7Y receptors and the binding rates reported previously for WT receptors (Fig. 5 B). By simulating dose–response curves with this hybrid model, we determined that if the mutation had changed only gating, the EC50 should have been 0.74 µM, which represents only a 2.5-fold change relative to WT receptors (Fig. 5 B, middle trace). This result indicates that the fourfold change in macroscopic affinity made the major contribution to the 10-fold shift in glutamate EC50 produced by the A7Y mutation.
N17Y mutation energizes the resting states of NMDA receptors
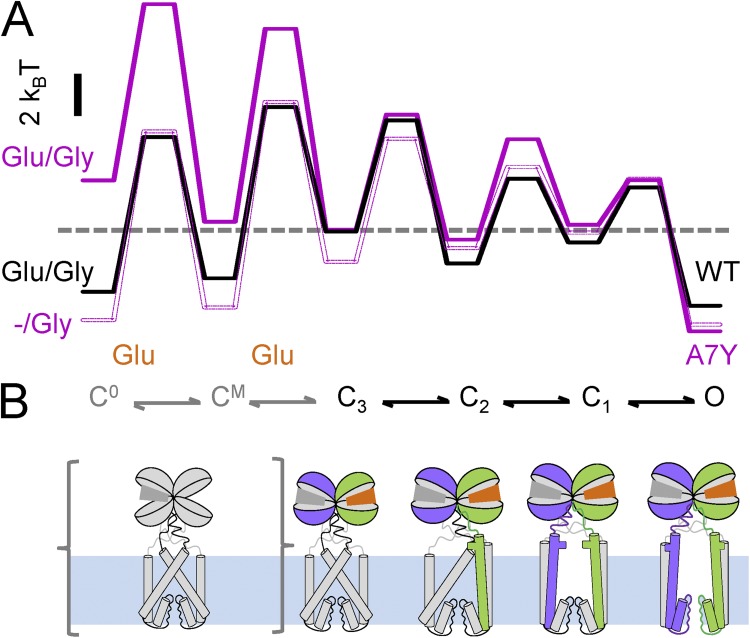
The kinetic models we developed enabled us also to calculate relative fluctuations in the free energies of WT and A7Y receptors during activation (Fig. 6 A). To compare these energetic profiles, we made the following assumptions. First, based on the observation that the channel open durations were similar for A7Y when measured in (Glu/Gly) and in (−/Gly), we aligned the calculated energy profiles for these two conditions at the open states. Second, based on the observations that the C3–C2 transition had similar kinetics for WT and liganded A7Y receptors, we aligned these two profiles at the energy level corresponding to C3. This alignment also reflected correctly the more stable open states we observed for A7Y relative to WT. Keeping these assumptions in mind, the calculated energy diagram postulates that, in addition to stabilizing the aggregated open state, the A7Y mutation also energized the resting states (C0 and CM) of glutamate-free receptors. The free energy contributed by the mutation was comparable in value to the free energy provided by glutamate binding to WT receptors, such that glutamate-bound WT and glutamate-free A7Y receptors differed by only ∼2 kBT. In addition, regardless of whether glutamate was bound or not, the mutation stabilized the open state by a similar ∼2-kBT amount. This analysis implies that the structures responsible for agonist binding and channel gating remain highly coupled in A7Y mutants; estimates the energetic contributions of the A7Y mutation to the resting and open states of the receptor; and illustrates that regardless of glutamate presence, the equilibrium point for A7Y receptors is shifted to the right, or further along the activation pathway, as compared with WT receptors.
Figure 6.
Proposed NMDA receptor activation sequence. (A) Free energy profiles for WT (black) and A7Y (purple) were calculated from the kinetic models given for WT in Fig. 5 A, constitutive N17Y/N2 gating (dotted line) in Fig. 4 C, and glutamate-activated gating (filled line) in Fig. 5 A; the resulting traces were aligned based on the measured differences in open-state stabilities for the three conditions. (B) A possible sequence of structural changes that accompany NMDA receptor activation and its correspondence to the rate-limiting steps identified in single-molecule traces; for clarity, desensitization steps are omitted. Only one glycine-bound heterodimer is represented, and NTD, M1, and CTD modules are omitted. The reaction is initiated from the glycine-bound resting state (C0, gray) and proceeds toward active conformations represented in purple for N1 and green for N2.
DISCUSSION
We examined single-channel activity of NMDA receptors with substitutions at positions A8 and A7 of N1 or N2 subunits and determined their activation mechanisms. These data and results allowed us to identify gating changes caused by substitutions at A8 or A7, which may reflect discrete locations for these residues relative to the activation gate (external vs. internal); to note gating changes caused by substitutions in N1 or N2 subunits, which may reflect subunit-specific contributions to specific steps within the gating reaction (earlier vs. later); and to measure separately the effects of A7Y substitution on microscopic glutamate affinity (10-fold lower Kd) from those on receptor gating (1.3-fold higher Po). In addition, we observed that mutations at A7 and A8 produced changes in unitary current amplitudes that were independent of the effects on gating.
We noted that A8 and A7 substitutions had opposite effects on open durations. Relative to WT, Y at A8 produced a decrease (for N1/N28Y) or no change (for N18Y/N2) in Po, and this change was caused in part by shorter openings. In contrast, Y at A7 produced an increase in Po for both N1 and N2 locations, and the change was primarily caused by dramatically longer openings (Table 1 and Figs. 2 and 3). These results are consistent with the hypothesized locations for A8 as external and for A7 as internal or coincident with the glutamate-controlled channel gate. This functional topology was previously proposed by Chang and Kuo (2008) based on accessibility experiments. It is also supported by homology models based on structures of inactive KcsA and GluA2 channels (Doyle et al., 1998; Armstrong, 2003; Sobolevsky et al., 2009). Still, it remains unclear why, despite similar structures, non-NMDA receptor A8T (lurcher), which is external to the gate, impacts function as dramatically as A7Y does in NMDA receptors (Kohda et al., 2000; Klein and Howe, 2004).
Second, we noted asymmetric effects of identical substitutions when introduced in N2 or N1subunits. Across all conditions tested, the following pattern emerged: substitutions in N2 slowed progression for the first gating transition, C3→C2, whereas the same substitutions in N1 affected the second step, C2→C1. Although the changes were modest, the distinct contributions observed may indicate that the M3 helices of N2 and N1 subunits move sequentially. Over the past decade, several studies have addressed the issue of subunit-dependent gating. So far, results show that perturbations in the ligand-binding layer and in the linkers connecting LBDs with transmembrane helices have modest or no subunit-dependent effects on gating (Banke and Traynelis, 2003; Kussius and Popescu, 2009, 2010; Talukder and Wollmuth, 2011). Overall, these results are consistent with the view that gating is initiated by the agonist-induced closure of the four LBD clefts followed by, at least, the repositioning of individual LBDs within dimers, movements in the LBD–transmembrane domain linkers, and the separation of M3 transmembrane helices (Armstrong and Gouaux, 2000; Kussius and Popescu, 2010; Borschel et al., 2011; Talukder and Wollmuth, 2011). Our observation that mutations in M3 of N2 affected the C3→C2 transition, and mutations in M3 of N1 affected preferentially the C2→C1 transition, may reflect that all structural transitions preceding the separation of M3 helices also precede the C3→C2 transition and thus most likely they are amalgamated within the long-lived state C3 (Fig. 6). Clearly, to substantiate these suggestions and to establish the trajectory of conformational change during NMDA receptor gating with the detail accomplished for peripheral excitatory transmitters, additional, more focused investigations are necessary (Grosman et al., 2000; Auerbach, 2010).
Third, we report for the first time equilibrium open probabilities for agonist-free A7Y receptor (Po, 0.06) and for glycine-bound glutamate-free N17Y/N2 receptor activity (Po, 0.24), and we show that in both cases, the gating reaction was largely similar to that of fully liganded receptors (Figs. 3 and 4). These results demonstrate that grafting a tyrosine side chain at A7 of N1 subunits provided sufficient energy to gate NMDA receptors with agonist-free N2 subunits and allowed us to estimate that this energy was only ∼2 kBT shy of the energy provided by glutamate binding to WT receptors (Fig. 6). Also, we measured microscopic glutamate association and dissociation rate constants for glycine-bound N17Y/N2 receptors and determined that both were substantially decreased (2.5- and 25-fold, respectively). Based on these results, we estimate that N17Y-containing receptors have an ∼10-fold greater microscopic affinity for glutamate and suggest that, like in WT receptors, conformational changes at the LBD of N2 subunits (responsible for changes in glutamate affinity) remain highly coupled with conformational changes of the M3 transmembrane helix of N1 subunits (responsible for moving the gate). This coupling may explain these receptors’ previously reported decreased sensitivity to competitive antagonists and to allosteric inhibitors such as protons, zinc, and ethanol (Low et al., 2003; Hu and Zheng, 2005a; Gielen et al., 2009; Xu et al., 2012). Our results explain the constitutive activation of N17Y/N2 receptors as a shift of the entire activation equilibrium toward active states, caused by increased energetic barriers to channel closing and tight allosteric coupling between the membrane-embedded gate and the extracellular LBDs, which causes changes in affinity.
Last, our results demonstrate that, when introduced at A7 of N1 or N2 subunits, tyrosine substitutions decreased unitary current amplitudes by ∼30% (Table 1). This decrease was not correlated with gating changes, and also unlike gating changes, it was not accentuated when substitutions were introduced simultaneously in N1 and N2. A role for lurcher-motif residues in controlling NMDA receptor channel conductance has been suggested previously, although it has not been quantified, and the mechanism by which it occurs is unknown (Kohda et al., 2000; Yuan et al., 2005).
In summary, probing the activation sequence of NMDA receptors with perturbations in the lurcher region enabled us to gather additional support for the idea that A7 forms part of the activation gate and to show that introducing a tyrosine residue at this position in the N1 subunit has two major consequences on receptor function. First, it energizes resting receptors to a level close to that of glutamate-bound WT receptors, resulting in substantial constitutive activity; and second, it stabilizes open receptors by a mechanism that prevents gate closure, resulting in very high open probabilities within bursts. Overall, the mutation shifts the reaction equilibrium toward active states, decreases both the glutamate-binding and the glutamate dissociation rates, and increases glutamate affinity by an order of magnitude. In addition, we observed patterns of kinetic changes that were subunit specific, an indication that residues in the M3 of N1 and N2 may affect separate rate-limiting steps along the activation reaction. Collectively, these results offer a mechanistic and quantitative platform on which to build toward the broader goal of resolving the temporal trajectory of structural changes that make the NMDA receptor activation reaction.
Supplementary Material
Acknowledgments
We thank Morgan Preziosi for help with electrophysiological recordings.
This work was supported by National Institute of Neurological Disorders and Stroke (grant 052669 to G.K. Popescu).
Author contributions: S.E. Murthy collected the majority of the data and performed all of the analyses; T. Shogan and J.C. Page contributed to single-channel data collection; E.M. Kasperek carried out all of the molecular biology; and S.E. Murthy and G.K. Popescu designed the experiments, interpreted the results, and prepared the manuscript.
Angus C. Nairn served as editor.
Footnotes
Abbreviations used in this paper:
- iGluR
- ionotropic glutamate receptor
- LBD
- ligand-binding domain
- N1
- GluN1
- N2
- GluN2
- NTD
- N-terminal domain
- Po
- open probability
- WT
- wild type
References
- Amico-Ruvio S.A., Popescu G.K. 2010. Stationary gating of GluN1/GluN2B receptors in intact membrane patches. Biophys. J. 98:1160–1169 10.1016/j.bpj.2009.12.4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong C.M. 2003. Voltage-gated K channels. Sci. STKE. 2003:re10 10.1126/stke.2003.188.re10 [DOI] [PubMed] [Google Scholar]
- Armstrong N., Gouaux E. 2000. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 28:165–181 10.1016/S0896-6273(00)00094-5 [DOI] [PubMed] [Google Scholar]
- Auerbach A. 2010. The gating isomerization of neuromuscular acetylcholine receptors. J. Physiol. 588:573–586 10.1113/jphysiol.2009.182774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach A., Zhou Y. 2005. Gating reaction mechanisms for NMDA receptor channels. J. Neurosci. 25:7914–7923 10.1523/JNEUROSCI.1471-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banke T.G., Traynelis S.F. 2003. Activation of NR1/NR2B NMDA receptors. Nat. Neurosci. 6:144–152 10.1038/nn1000 [DOI] [PubMed] [Google Scholar]
- Beck C., Wollmuth L.P., Seeburg P.H., Sakmann B., Kuner T. 1999. NMDAR channel segments forming the extracellular vestibule inferred from the accessibility of substituted cysteines. Neuron. 22:559–570 10.1016/S0896-6273(00)80710-2 [DOI] [PubMed] [Google Scholar]
- Blanke M.L., VanDongen A.M. 2008. The NR1 M3 domain mediates allosteric coupling in the N-methyl-D-aspartate receptor. Mol. Pharmacol. 74:454–465 10.1124/mol.107.044115 [DOI] [PubMed] [Google Scholar]
- Borschel W.F., Murthy S.E., Kasperek E.M., Popescu G.K. 2011. NMDA receptor activation requires remodelling of intersubunit contacts within ligand-binding heterodimers. Nat. Commun. 2:498 10.1038/ncomms1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.R., Kuo C.C. 2008. The activation gate and gating mechanism of the NMDA receptor. J. Neurosci. 28:1546–1556 10.1523/JNEUROSCI.3485-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun D. 1998. Binding, gating, affinity and efficacy: the interpretation of structure-activity relationships for agonists and of the effects of mutating receptors. Br. J. Pharmacol. 125:923–947 10.1038/sj.bjp.0702164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., MacKinnon R. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77 10.1126/science.280.5360.69 [DOI] [PubMed] [Google Scholar]
- Gibb A.J., Colquhoun D. 1991. Glutamate activation of a single NMDA receptor-channel produces a cluster of channel openings. Proc. Biol. Sci. 243:39–45 10.1098/rspb.1991.0007 [DOI] [PubMed] [Google Scholar]
- Gibb A.J., Colquhoun D. 1992. Activation of N-methyl-D-aspartate receptors by L-glutamate in cells dissociated from adult rat hippocampus. J. Physiol. 456:143–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M., Siegler Retchless B., Mony L., Johnson J.W., Paoletti P. 2009. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 459:703–707 10.1038/nature07993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosman C., Zhou M., Auerbach A. 2000. Mapping the conformational wave of acetylcholine receptor channel gating. Nature. 403:773–776 10.1038/35001586 [DOI] [PubMed] [Google Scholar]
- Howe J.R., Colquhoun D., Cull-Candy S.G. 1988. On the kinetics of large-conductance glutamate-receptor ion channels in rat cerebellar granule neurons. Proc. R. Soc. Lond. B Biol. Sci. 233:407–422 10.1098/rspb.1988.0030 [DOI] [PubMed] [Google Scholar]
- Hu B., Zheng F. 2005a. Differential effects on current kinetics by point mutations in the lurcher motif of NR1/NR2A receptors. J. Pharmacol. Exp. Ther. 312:899–904 10.1124/jpet.104.077388 [DOI] [PubMed] [Google Scholar]
- Hu B., Zheng F. 2005b. Molecular determinants of glycine-independent desensitization of NR1/NR2A receptors. J. Pharmacol. Exp. Ther. 313:563–569 10.1124/jpet.104.080168 [DOI] [PubMed] [Google Scholar]
- Jahr C.E., Stevens C.F. 1987. Glutamate activates multiple single channel conductances in hippocampal neurons. Nature. 325:522–525 10.1038/325522a0 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Cadene M., Chait B.T., MacKinnon R. 2002. The open pore conformation of potassium channels. Nature. 417:523–526 10.1038/417523a [DOI] [PubMed] [Google Scholar]
- Jones K.S., VanDongen H.M., VanDongen A.M. 2002. The NMDA receptor M3 segment is a conserved transduction element coupling ligand binding to channel opening. J. Neurosci. 22:2044–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R.M., Howe J.R. 2004. Effects of the lurcher mutation on GluR1 desensitization and activation kinetics. J. Neurosci. 24:4941–4951 10.1523/JNEUROSCI.0660-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohda K., Wang Y., Yuzaki M. 2000. Mutation of a glutamate receptor motif reveals its role in gating and delta2 receptor channel properties. Nat. Neurosci. 3:315–322 10.1038/73877 [DOI] [PubMed] [Google Scholar]
- Kuner T., Wollmuth L.P., Karlin A., Seeburg P.H., Sakmann B. 1996. Structure of the NMDA receptor channel M2 segment inferred from the accessibility of substituted cysteines. Neuron. 17:343–352 10.1016/S0896-6273(00)80165-8 [DOI] [PubMed] [Google Scholar]
- Kuner T., Seeburg P.H., Guy H.R. 2003. A common architecture for K+ channels and ionotropic glutamate receptors? Trends Neurosci. 26:27–32 10.1016/S0166-2236(02)00010-3 [DOI] [PubMed] [Google Scholar]
- Kussius C.L., Popescu G.K. 2009. Kinetic basis of partial agonism at NMDA receptors. Nat. Neurosci. 12:1114–1120 10.1038/nn.2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussius C.L., Popescu G.K. 2010. NMDA receptors with locked glutamate-binding clefts open with high efficacy. J. Neurosci. 30:12474–12479 10.1523/JNEUROSCI.3337-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussius C.L., Kaur N., Popescu G.K. 2009. Pregnanolone sulfate promotes desensitization of activated NMDA receptors. J. Neurosci. 29:6819–6827 10.1523/JNEUROSCI.0281-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low C.M., Lyuboslavsky P., French A., Le P., Wyatte K., Thiel W.H., Marchan E.M., Igarashi K., Kashiwagi K., Gernert K., et al. 2003. Molecular determinants of proton-sensitive N-methyl-D-aspartate receptor gating. Mol. Pharmacol. 63:1212–1222 10.1124/mol.63.6.1212 [DOI] [PubMed] [Google Scholar]
- Magleby K.L. 2004. Modal gating of NMDA receptors. Trends Neurosci. 27:231–233 10.1016/j.tins.2004.03.001 [DOI] [PubMed] [Google Scholar]
- Popescu G.K. 2012. Modes of glutamate receptor gating. J. Physiol. 590:73–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu G., Auerbach A. 2003. Modal gating of NMDA receptors and the shape of their synaptic response. Nat. Neurosci. 6:476–483 [DOI] [PubMed] [Google Scholar]
- Popescu G., Auerbach A. 2004. The NMDA receptor gating machine: lessons from single channels. Neuroscientist. 10:192–198 10.1177/1073858404263483 [DOI] [PubMed] [Google Scholar]
- Popescu G., Robert A., Howe J.R., Auerbach A. 2004. Reaction mechanism determines NMDA receptor response to repetitive stimulation. Nature. 430:790–793 10.1038/nature02775 [DOI] [PubMed] [Google Scholar]
- Schwarz M.K., Pawlak V., Osten P., Mack V., Seeburg P.H., Köhr G. 2001. Dominance of the lurcher mutation in heteromeric kainate and AMPA receptor channels. Eur. J. Neurosci. 14:861–868 10.1046/j.0953-816x.2001.01705.x [DOI] [PubMed] [Google Scholar]
- Sobolevsky A.I., Rooney L., Wollmuth L.P. 2002. Staggering of subunits in NMDAR channels. Biophys. J. 83:3304–3314 10.1016/S0006-3495(02)75331-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky A.I., Rosconi M.P., Gouaux E. 2009. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 462:745–756 10.1038/nature08624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder I., Wollmuth L.P. 2011. Local constraints in either the GluN1 or GluN2 subunit equally impair NMDA receptor pore opening. J. Gen. Physiol. 138:179–194 10.1085/jgp.201110623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis S.F., Wollmuth L.P., McBain C.J., Menniti F.S., Vance K.M., Ogden K.K., Hansen K.B., Yuan H., Myers S.J., Dingledine R. 2010. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 62:405–496 10.1124/pr.109.002451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wo Z.G., Oswald R.E. 1995. Unraveling the modular design of glutamate-gated ion channels. Trends Neurosci. 18:161–168 10.1016/0166-2236(95)93895-5 [DOI] [PubMed] [Google Scholar]
- Xu M., Smothers C.T., Trudell J., Woodward J.J. 2012. Ethanol inhibition of constitutively open N-methyl-D-aspartate receptors. J. Pharmacol. Exp. Ther. 340:218–226 10.1124/jpet.111.187179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Erreger K., Dravid S.M., Traynelis S.F. 2005. Conserved structural and functional control of N-methyl-D-aspartate receptor gating by transmembrane domain M3. J. Biol. Chem. 280:29708–29716 10.1074/jbc.M414215200 [DOI] [PubMed] [Google Scholar]
- Zuo J., De Jager P.L., Takahashi K.A., Jiang W., Linden D.J., Heintz N. 1997. Neurodegeneration in Lurcher mice caused by mutation in delta2 glutamate receptor gene. Nature. 388:769–773 10.1038/42009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.