Abstract
Replication Protein A (RPA) is known to interact with G-rich sequences that adopt G-quadruplex (GQ) structures. Most studies in the literature have been performed on GQ formed by homogenous sequences, such as the human telomeric repeat, and RPA’s ability to unfold GQ structures of differing stability is not known. We compared the thermal stability of three potential GQ forming DNA sequences (PQS) to their stability against RPA mediated unfolding using single molecule FRET and bulk biophysical and biochemical experiments. One of these sequences is the human telomeric repeat and the other two located in the promoter region of tyrosine hydroxylase gene are highly heterogeneous sequences, which better represent PQS in the genome. The three GQ constructs have thermal stabilities that are significantly different from each other. Our measurements showed that the most thermally stable structure (Tm= 86 °C) was also the most stable against RPA mediated unfolding, although the least thermally stable structure (Tm= 69 °C) had at least an order of magnitude higher stability against RPA mediated unfolding compared to the structure with intermediate thermal stability (Tm= 78 °C). The significance of this observation becomes more evident when considered within the context of cellular environment where protein-DNA interactions can be an important determinant of GQ viability. Considering these, we conclude that thermal stability is not necessarily an adequate criterion for predicting physiological viability of GQ structures. Finally, we measured the time it takes for an RPA molecule to unfold a GQ from a fully folded to a fully unfolded conformation using a single molecule stopped-flow type method. All three GQ structures were unfolded within Δt≈0.30±0.10 sec, a surprising result as the unfolding time does not correlate with thermal stability or stability against RPA mediated unfolding. These results suggest that the limiting step in G-quadruplex unfolding by RPA is simply the accessibility of the structure to the RPA protein.
INTRODUCTION
Potential G-Quadruplex forming sequences (PQS) have been identified in both telomeric and non-telomeric genomic DNA1–4. Bioinformatics studies have identified 375 000 distinct and non-overlapping PQS in the human genome5 and mapped their distribution 6–10. Non-telomeric PQS are over represented in or near gene promoters9, suggesting their potential role might be involved in regulation of gene expression at the transcription level 11,12. Similarly RNA GQs located in the 5’-UTR region13 have been demonstrated to regulate gene expression at the translational level14–16. Recently, the GQs located in the promoter regions of several oncogenes have been targeted by specific small molecules that stabilize the GQ structure in order to modulate gene expression17,18.
Despite the abundance of PQS in eukaryotic genomes and the numerous in vitro demonstrations of such structures, it has been challenging to directly demonstrate their existence and function in vivo19. However, recent genome-wide studies have presented indirect but increasingly convincing evidence for their in vivo significance. For instance, absence of Pif1, a GQ unfolding helicase, causes severe retardation in DNA replication and a significant increase in the number of DNA breaks in the GQ forming segments of the genome20. These data have been interpreted as evidence for formation of GQs in vivo, and the need for their unfolding by proteins. Since GQs are typically thermodynamically more stable than Watson-Crick base paired duplex DNA21, they would need to be unwound for effective progression of cellular processes that require unwinding of DNA, such as replication and transcription19,22. Various helicases including Pif1, WRN, BLM, and FANC-J, have been shown to perform this task22.
Single strand DNA (ssDNA) binding proteins are also known to unfold GQ structures. Replication Protein A (RPA) is the most abundant ssDNA binding protein in eukaryotes (≈1 μM concentration in vivo23). RPA has three subunits (RPA70, RPA32, and RPA14), and is involved in DNA replication, repair and recombination24,25. RPA has about three orders of magnitude higher affinity for ssDNA (kd≈10 nM under physiologically relevant conditions) and GQ structures compared to double stranded DNA (dsDNA)26–28. RPA binding unwinds GQs formed by human telomeric repeat sequence (TTAGGG)n26,29–31, and it is known that RPA, along with another ssDNA binding protein Pot1, is involved in telomere maintenance32. Interestingly, it has been shown that Cdc13, Stn1, and Ten1, which are proteins specialized for maintaining telomeric DNA, have extensive structural and functional homology to subunits of RPA33,34. It has been proposed that Cdc13, Stn1, and Ten1 form an RPA-like complex that is involved in the maintenance of telomeres. These observations combined with the ubiquitous nature of both RPA and PQS demonstrate the physiological relevance and significance of RPA-GQ interactions.
In the present study, we compared three GQ constructs in terms of their thermal stability and stability against RPA mediated unfolding. The thermal stabilities of GQs formed by human telomeric and non-telomeric sequences and RPA mediated unfolding of GQ formed by human telomeric sequence have been measured 35,36. However, a comparative study that relates thermal stability and unfolding by RPA of multiple GQ constructs has not been performed. The non-telomeric sequences studied in this work are highly heterogeneous, and the interaction of RPA with such sequences has not been studied before. In particular, whether higher thermal stability also implies higher stability against protein mediated unfolding is a question that has not been addressed before. Additionally, interaction of RPA with any GQ construct, including that formed by human telomeric repeat, has not been studied at the single molecule level.
METHODS
We used circular dichroism (CD), DMS foot-printing, and UV-thermal melting to characterize the GQs and single molecule FRET to study RPA-GQ interactions. The experimental methods and protocols followed to perform these measurements are provided as Supporting Information.
RESULTS AND DISCUSSION
RPA has very high affinity to bind to ssDNA and the non-tetrad portions, e.g. loops, of the GQ can act as natural substrates for RPA. Therefore, we hypothesized that a long loop in the GQ structure could dramatically reduce the stability of the GQ against RPA mediated unfolding, while the effect of such a long loop on thermal stability may not be as significant. The critical step in this process is finding a GQ structure that has a relatively high melting temperature as well as a long loop within the structure. Structures with both of these attributes can be difficult to locate, as longer loops have been shown to decrease thermal stability35. Identification of a GQ structure with a long loop and high thermal stability motivated us to address this question using a small set of GQ structures.
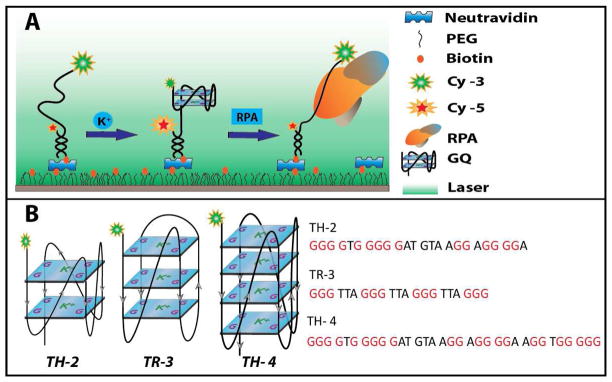
Such a GQ structure is formed by a non-telomeric and heterogeneous sequence (will be referred to as TH-2) that forms a two layer GQ and could potentially have a 6–9 nucleotide long loop while maintaining a relatively high melting temperature of Tm=78 °C. To compare, we studied two other GQ structures as well: a homogenous telomeric repeat that forms a three layer GQ (TR-3) and a heterogeneous non-telomeric construct that potentially forms a four layer GQ (TH-4). TH-2 and TH-4 sequences are located within the promoter region of the tyrosine hydroxylase gene37. The TH-2 sequence is embedded within TH-4 sequence, which has an additional segment that enables a four layer GQ structure (see Figure 1-B for sequences and a possible folding pattern for each GQ).
Figure 1.
(A) Schematic of single molecule FRET experiments. GQ forming DNA constructs are immobilized on a poly ethylene glycol coated quartz slide via a neutravidin biotin linker. Adding potassium stabilizes the GQ structure whereas RPA unfolds the structure. (B) Sequences and a possible folding conformation for each of the three GQ constructs studied in this work. TH-2 and TH-4 are located within the promoter region of the tyrosine hydroxylase gene and they can form a two and four layer GQ, respectively. TR-3 is the human telomeric repeat that forms a three layer GQ.
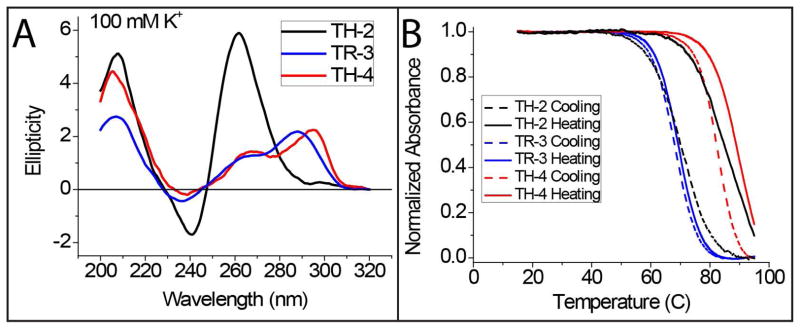
CD measurements were performed to confirm GQ formation in each of the three PQS at 100 mM K+ concentration as summarized in Figure 2-A (control measurements at 100 mM Li+ are shown in Supporting Figure S1). The CD spectrum of TH-2 showed a peak at 260 nm and a trough at 240 nm which is consistent with parallel GQ conformation. On the other hand, TH-4 has a shoulder around 260 nm in addition to another peak at 295 nm, suggesting a mix of parallel and anti-parallel GQs or a hybrid conformation. TR-3 had a spectrum similar to that of TH-4, with the exception of the second peak being at 288 nm rather than 295 nm, consistent with a hybrid conformation38,39. It should be mentioned that CD data are used as an evidence for GQ formation rather than identifying the exact conformation of GQ, as there are exceptions to the presented interpretations on conformations40. In this study, we focused on comparing the average thermal stability of GQs with the RPA mediated unfolding activity of such structures. The thermal stabilities of the GQs were determined via UV-melting measurements, and the following melting temperatures were obtained: Tm(TH-2)=78 °C; Tm(TR-3)=69 °C; and Tm(TH-4)=86 °C (Figure 2-B).
Figure 2.
(A) Circular Dichroism (B) UV thermal melting measurements on the three GQs studied in this work. Both measurements were performed under similar conditions to smFRET measurements for consistency. CD measurements demonstrate the GQ formation for all three constructs. The UV thermal melting measurements are used to characterize the thermal stability of the GQ structures.
Dimethyl sulfate (DMS) footprinting experiments were performed to distinguish the guanines that are part of the G-tetrad structure, which would protect them from DMS modification, from those that are within the loop regions, and therefore are not protected 41. The results of these measurements are reported in Supporting Figure S2. Based upon the DMS protection pattern, one of the many possible GQs is shown for each sequences in Figure 1-B. The DMS footprinting data suggest that there is not a unique set of guanines that participate in the GQ structure for TH-2 and TH-4. Redundancy in the sequences allows for different subsets of contiguous guanines to participate in GQ formation, resulting in a variety of possible structures.
smFRET measurements were performed in prism-type total internal reflection configuration, as schematically shown in Figure 1-A 42–44. Proper folding into GQ structure was monitored by measuring the FRET efficiency between the donor and acceptor fluorophores. Increasing K+ concentration results in stabilization of the GQ, thereby decreasing the distance between the donor-acceptor fluorophores and resulting in higher FRET efficiency (see Supporting Figure S3 for K+ titration data on TH-2). At 100 mM K+ all three constructs showed a single high FRET peak suggesting stable folding (Figure 3). It should be noted that a single FRET population does not necessarily mean a single GQ folding conformation as the distance between the donor-acceptor fluorophores could be similar for different conformations. In order to minimize the interaction between the fluorophores and the GQ structure, thymidine spacers were placed between the fluorophores and GQ (see Supporting Information for sequences). We confirmed that these spacers do not have a significant effect on the thermal stability of the GQs and that constructs with and without the spacers had similar melting temperatures (data not shown). Finally, smFRET measurements are performed at picomolar DNA concentrations thereby excluding possibility of intermolecular GQ formation.
Figure 3.
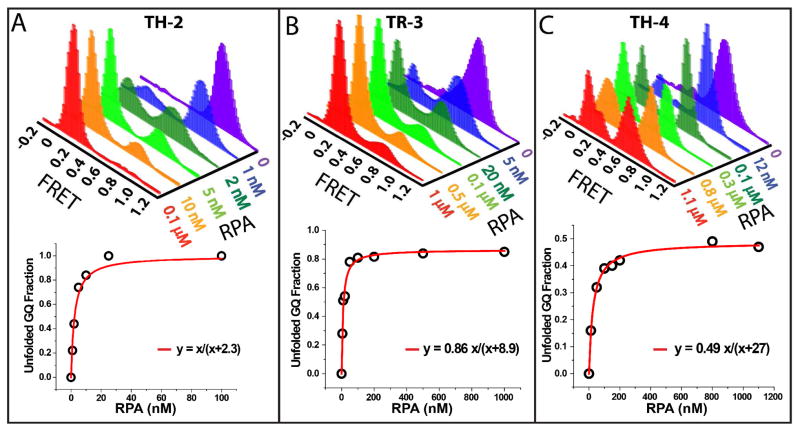
Single molecule FRET experiments probing GQ unfolding at different RPA concentrations for (A) TH-2; (B) TR-3; and (C) TH-4. When GQ is folded, the donor and acceptor dyes are close to each other, resulting in high-FRET efficiency. RPA binding to DNA induces GQ unfolding, separating the fluorophores away from each other and resulting in low FRET efficiency. As the RPA concentration is increased, the population of the low FRET peak gradually increases as more GQ constructs are unfolded. For TR-3 and TH-4 the fraction of unfolded GQ structures saturates at certain value and complete unfolding of all molecules cannot be attained. The panels at the bottom show a Langmuir isotherm fit to the unfolding data.
The smFRET data in Figure 3 summarizes the unfolding of the GQs at 100 mM K+ by different RPA concentrations. Among the three GQs, the stability against RPA mediated unfolding varies by orders of magnitude. TH-2 is the least stable against RPA and completely unfolds in the presence of about 25 nM RPA. TR-3 and TH-4 demonstrate an unexpected unfolding pattern: they are both significantly unfolded at relatively low RPA concentrations (<100 nM) but maintain a steady state unfolded/folded proportion up to physiological level RPA concentrations (≈1 μM). For TR-3, the steady state unfolded population constitutes about 86% of the total population whereas for TH-4 it is about 49%. The bottom panels in Figure 3A and 3B show that RPA mediated unfolding activity can be fitted well by a Langmuir isotherm of the form y=x/(x+kD), where y describes the fraction of unfolded GQ structures, x is the concentration of RPA in the solution, and kD is the dissociation constant. This expression is justified as the surface concentration of GQ (sub nanomolar) is much lower than the protein concentration in solution. In order to accommodate for incomplete unfolding of GQ for TR-3 and TH-4, the equation is multiplied by a normalization constant y=α[RPA]/([RPA]+kD), where α essentially describes the saturating unfolded fraction of GQ. It should be mentioned that this equation assumes that binding of RPA to GQ is equivalent to unfolding of GQ. A more complete description would require separating the two processes however; given the number of data points we have and the quality of the fits based on the simplified model, we believe we would not be able to distinguish the two models. Two parameters are extracted from these fits: α and kD which are the fraction of unfolded GQs at saturation (up to physiological RPA concentration which is about 1 μM) and the dissociation constant of binding/unfolding process, respectively. The results of the fits are: y=x/(x+2.3) for TH-2, y=0.86x/(x+8.9) for TR-3, and y=0.49x/(x+27) for TH-4. These results mean that RPA can bind and unfold all of TH-2 with a kD=2.3 nM. However, in the case of TR-3 86% and TH-4 49% of the GQ molecules can be unfolded at saturating RPA concentrations. The corresponding dissociation constants are kD=8.9 nM for RPA-TR-3 interaction and kD=27 nM for RPA-TH-4 interaction. Despite its lower thermal melting temperature, TR-3 was significantly more stable against RPA mediated unfolding compared to TH-2 as it had about 4 times higher kD. In addition, about 14% of TR-3 molecules were able to maintain a folded structure even in the presence of 1 μM RPA concentration, while all of TH-2 molecules were unfolded at less than 100 nM RPA concentration.
The DNA constructs we used for this study have a double stranded stem sequence (see Figure 2), and the donor/acceptor fluorophores are placed at the ends of the GQ structure but are on separate ssDNA strands. RPA is known to have helix destabilization activity however, at near physiological salt conditions (100 mM KCl and 5 mM MgCl2), the affinity of RPA to dsDNA was measured to be about three orders of magnitude smaller than its affinity to ssDNA27. It is also known that RPA has indistinguishable affinity for GQ and ssDNA26. Furthermore, Fan et al. showed that the affinity of RPA to the duplex formed by a GQ forming sequence and its complementary strand is negligible compared to its affinity to the GQ forming sequence alone26. Therefore, the possibility of unfolding of the dsDNA stem by RPA can be discarded. All three DNA constructs we studied have the same dsDNA stem, and therefore it would be the GQ constructs that are the cause of the significant variation in the stability against RPA mediated unfolding.
Interestingly, the unfolded population of TH-4 shows a double peak, with one present in the intermediate FRET region, shown in Figure 3-C. This suggests that the unfolded state of the TH-4 could still maintain some folded secondary structure. RPA has four active DNA binding domains (DBD), and can bind an 8–30 nucleotide long ssDNA45. Given that TH-4 is 37 nucleotide long (including thymidine spacer), it could accommodate an RPA molecule and some folded secondary structure or two RPA molecules at once. If the latter scenario is assumed, the low FRET peak would represent two RPA molecules binding to the same DNA molecule simultaneously. If that were the case, the population of lower FRET peak would be expected to systematically increase with increasing RPA concentration between 100 nM-1 μM RPA. However, such a systematic increase is not observed making this scenario unlikely. The more likely alternative is that the lower FRET peak represents a completely unfolded DNA molecule that is bound by a single RPA molecule. In this case, the intermediate FRET peak would represent an RPA bound DNA molecule that continues to maintain some folded secondary structure, but not necessarily a GQ.
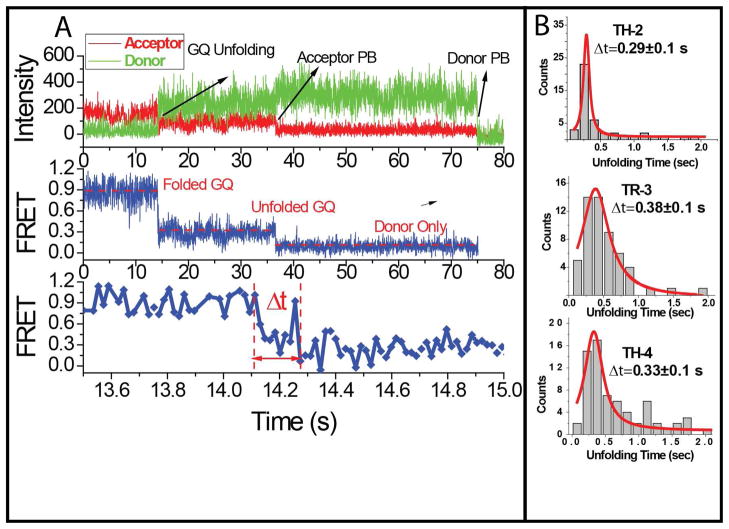
Finally, we monitored the time it takes for RPA to unfold GQ using a single molecule assay that resembles the ensemble level stopped-flow assay. In this assay, we incubate the DNA constructs in a buffer containing 100 mM K+ for 20 minutes to attain stably folded GQ structures. Then we inject a buffer that contains 100 mM K+ and 100 nM RPA, while simultaneously recording the data. As a result we obtain single molecule FRET time trajectories as shown in Figure 4-A, which was acquired at 0.018 sec integration time. The initial high FRET state corresponds to the folded GQ conformation and the following low FRET state corresponds to the RPA-bound unfolded state. By measuring the transition time between these two states we obtain the time it takes for RPA to unfold the GQ. Several points should be mentioned about this assay to distinguish it from ensemble level stopped-flow assay. First of all, uncertainty in mixing time is not an issue for this single molecule assay as we monitor the interaction of a single DNA and single protein molecules in real time. We can determine the exact time, within our time resolution, that a GQ starts unfolding and completes the unfolding process. Hence, the start and end of the unfolding process are determined independently for every GQ molecule included in our analysis. Secondly, the protein concentration used for the measurements does not affect the transition time. The GQ is unfolded by a single RPA molecule and the start and end of the GQ unfolding can be determined for each molecule. Having a higher protein concentration would provide larger number of unfolding events and better statistics. To be consistent we maintained 100 nM RPA concentration for all our measurements. Finally, we can distinguish between photobleaching and GQ unfolding and were able to exclude the photobleaching events during data analysis. Fluorophore photobleaching could arise because of buffer exchange at the time of RPA flow in the chamber. As shown in Figure 4-A, the photobleaching and the unfolding transition can be clearly distinguished. Apart from temporal separation between the two events, the FRET level upon GQ unfolding (~0.25) and the FRET level upon acceptor photobleaching (~0.10 for our setup and dye pair) are significantly different and can be easily distinguished. These criteria have been applied to all the data included in the histograms shown in Figure 4-B.
Figure 4.
(A) Stopped-flow type single molecule measurements characterizing the unfolding of GQ by RPA. 0.018 sec acquisition time was used for these data and the transition from the folded GQ to unfolded structure upon RPA addition is monitored in real time. GQ unfolding, donor photobleaching, and acceptor photobleaching are indicated on the top graph. Δt represents the unfolding time of GQ by RPA. (B) Histograms of unfolding times obtained using this method. The histograms are fitted with Lorentzian functions to determine the characteristic GQ unfolding time.
The histograms of the unfolding time and Lorentzian fits to these histograms are shown in Figure 4-B. While constructing these histograms, the unfolding events that were within few seconds of RPA injection were used for TH-2 and TH-4, whereas for TR-3 we had to consider later events as well in order to obtain reasonable statistics. Characteristic unfolding times were determined to be (based on Lorentzian fits): Δt=0.29±0.10 sec for TH-2; Δt=0.38±0.10 sec for TR-3; and Δt=0.33±0.10 sec for TH-4. The image acquisition time for the measurements was 0.035 sec, about an order of magnitude smaller than the duration of unfolding transition. We also performed control measurements on the TR-3 construct using 0.018 sec acquisition time which was achieved by using half of the CCD sensor for imaging. An average unfolding time of Δt=0.27±0.05 sec was obtained from these measurements (Figure S4), which is consistent with the data presented in Figure 4 within the uncertainty of the method. The most important contribution to the uncertainty in determining the characteristic times is due to uncertainty in selecting the beginning and end of the transition time. We estimate this uncertainty to be about 3 data points (images), increasing the uncertainty in the measurement to about 0.10 sec for 0.035 sec image acquisition time, and to about 0.050 sec for 0.018 sec image acquisition time. Given this uncertainty, all three GQ structures essentially have the same unfolding time despite significant variations in their melting temperatures and steady state stabilities against RPA mediated unfolding. This novel result suggests that the limiting step in unfolding of GQ structures by RPA is the binding of RPA to GQ. Once RPA binds and the unfolding of GQ starts, it takes similar times for RPA to unfold GQ structures with otherwise very different stabilities. How this binding is influenced by GQ structure requires systematic studies in which RPA mediated unfolding is studied with GQ structures with differing loop lengths, sequences, and number of G-tetrad layers. Such a systematic study is beyond the scope of the current work although our results suggest that having a long loop in the GQ structure may be the determining factor in the stability (against protein mediated unfolding) of certain structures such as the one adopted by TH-2. TH-2 can have a 6–9 nucleotide loop, depending on which guanines participate in the G-tetrad formation. RPA can efficiently bind this loop and destabilize the GQ. In the case of TR-3, the loops are all 3 nucleotide long which may be the reason why TR-3 is more stable than TH-2. However, loop length does not describe the entire complexity of the process as TH-4 has a similar long loop in its structure, 9–10 nucleotide long, but it is very stable against RPA mediated unfolding. The important difference between TH-2 and TH-4 is of course the number of their G-tetrad layers: TH-2 has only two tetrad layers whereas TH-4 has four tetrad layers. Interestingly, the stability against RPA mediated unfolding correlates with the number of layers, i.e. the four layered TH-4 is the most stable and the two layered TH-2 is the least stable. However, understanding how these different structural elements influence the stability of a GQ structure would require systematic studies varying these parameters. However, this study clearly shows that thermal stability alone is not an adequate criterion to determine the stability of a GQ against protein unfolding.
CONCLUSION
We have demonstrated that the relative stability of GQs against RPA mediated unfolding can be significantly different from their relative thermal stability. Hence, thermal stability alone should not be used as a gauge for putative physiological viability of GQs, given the presence of various unfolding proteins in vivo. It should be emphasized that in vitro RPA-GQ interactions are not an adequate gauge for in vivo viability either as numerous other molecular factors could affect the stability of a GQ. However, our measurements demonstrate the potential problems that would be encountered if in vivo viability is directly correlated with thermal stability. Potential variables such as the sequence, loop length and number of G-tetrads could have a dramatic influence on protein-GQ interactions compared to their influence on thermal stability of the GQ. In addition, we found that the unfolding time of the three GQs by RPA to be very similar, Δt≈0.30±0.10 sec, despite their significantly different melting temperatures. To our knowledge, this quantity has not been measured before and its similarity between GQ structures with different stabilities suggests that binding of RPA to GQ may be the limiting step in this process. Once RPA binds to GQ, the unfolding event takes a place within a very similar time frame for GQs that otherwise have very different stabilities.
Supplementary Material
Acknowledgments
We thank Prof. Aziz Sancar and his group members for their help in RPA preparation, and Dr. Erdal Toprak for the control UV-thermal melting studies. This work is supported by the start-up funds from KSU to HB and SB and by NIH Grant 1R15GM096285 to SB and HB. MHQ thanks ICAM for their travel support that enabled RPA purification.
Footnotes
SUPPORTING INFORMATION. Details of experimental procedures, DMS data, control CD experiments in Li+, data on RPA mediated unfolding of GQ imaged at 18 msec image integration time, and DNA sequences are available as Supporting Information. This information is available free of charge via the Internet at http://pubs.acs.org/.
G-Quadruplex Unfolding Efficiency of RPA
References
- 1.Blackburn EH. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Gellert M, Lipsett MN, Davies DR. Proc Natl Acad Sci USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert DE, Feigon J. Curr Opin Struct Biol. 1999;9:305–314. doi: 10.1016/S0959-440X(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 4.Williamson JR. Annu Rev Biophys Biomol Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 5.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eddy J, Maizels N. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eddy J, Maizels N. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huppert JL, Balasubramanian S. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huppert JL, Balasubramanian S. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todd A, Johnston M, Neidle S. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du Z, Zhao Y, Li N. Genome Res. 2008;18:233–241. doi: 10.1101/gr.6905408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin Y, Hurley LH. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. Nat Chem Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris MJ, Basu S. Biochemistry. 2009;48:5313–5319. doi: 10.1021/bi900498z. [DOI] [PubMed] [Google Scholar]
- 16.Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. J Am Chem Soc. 2010;132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- 17.Balasubramanian S, Hurley LH, Neidle S. Nat Rev Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balasubramanian S, Neidle S. Curr Opin Chem Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipps HJ, Rhodes D. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Paeschke K, Capra JA, Zakian VA. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar N, Sahoo B, Varun KA, Maiti S. Nucleic Acids Res. 2008;36:4433–4442. doi: 10.1093/nar/gkn402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paeschke K, McDonald KR, Zakian VA. FEBS Lett. 2010;584:3760–3772. doi: 10.1016/j.febslet.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim C, Wold MS. Biochemistry. 1995;34:2058–2064. doi: 10.1021/bi00006a028. [DOI] [PubMed] [Google Scholar]
- 24.Oakley GG, Patrick SM. Front Biosci. 2010;15:883–900. doi: 10.2741/3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wold MS. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 26.Fan JH, Bochkareva E, Bochkarev A, Gray DM. Biochemistry. 2009;48:1099–1111. doi: 10.1021/bi801538h. [DOI] [PubMed] [Google Scholar]
- 27.Lao Y, Lee CG, Wold MS. Biochemistry. 1999;38:3974–3984. doi: 10.1021/bi982371m. [DOI] [PubMed] [Google Scholar]
- 28.Walther AP, Gomes XV, Lao Y, Lee CG, Wold MS. Biochemistry. 1999;38:3963–3973. doi: 10.1021/bi982370u. [DOI] [PubMed] [Google Scholar]
- 29.Masuda-Sasa T, Polaczek P, Peng XP, Chen L, Campbell JL. J Biol Chem. 2008;283:24359–24373. doi: 10.1074/jbc.M802244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salas TR, Petruseva I, Lavrik O, Bourdoncle A, Mergny JL, Favre A, Saintome C. Nucleic Acids Res. 2006;34:4857–4865. doi: 10.1093/nar/gkl564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prakash A, Kieken F, Marky LA, Borgstahl GE. J Nucleic Acids. 2011:529828. doi: 10.4061/2011/529828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flynn RL, Centore RC, O'Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L. Nature. 2011;471:532–536. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 34.Gelinas AD, Paschini M, Reyes FE, Heroux A, Batey RT, Lundblad V, Wuttke DS. Proc Natl Acad Sci U S A. 2009;106:19298–19303. doi: 10.1073/pnas.0909203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guedin A, Gros J, Alberti P, Mergny JL. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran PL, Mergny JL, Alberti P. Nucleic Acids Res. 2010;39:3282–3294. doi: 10.1093/nar/gkq1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romano G, Suon S, Jin H, Donaldson AE, Iacovitti L. J Cell Physiol. 2005;204:666–677. doi: 10.1002/jcp.20319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phan AT. FEBS J. 2009;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 40.Masiero S, Trotta R, Pieraccini S, De Tito S, Perone R, Randazzo A, Spada GP. Org Biomol Chem. 2010;8:2683–2692. doi: 10.1039/c003428b. [DOI] [PubMed] [Google Scholar]
- 41.Sun D, Hurley LH. Methods Mol Biol. 2010;608:65–79. doi: 10.1007/978-1-59745-363-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jena PV, Shirude PS, Okumus B, Laxmi-Reddy K, Godde F, Huc I, Balasubramanian S, Ha T. J Am Chem Soc. 2009;131:12522–12523. doi: 10.1021/ja903408r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JY, Okumus B, Kim DS, Ha T. Proc Natl Acad Sci U S A. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okumus B, Ha T. Methods Mol Biol. 2010;608:81–96. doi: 10.1007/978-1-59745-363-9_6. [DOI] [PubMed] [Google Scholar]
- 45.Cai L, Roginskaya M, Qu Y, Yang Z, Xu Y, Zou Y. Biochemistry. 2007;46:8226–8233. doi: 10.1021/bi7004976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.