Abstract
MicroRNAs (miRNAs) are a class of small (~22 nucleotides) non-coding RNAs involved in the regulation of gene expression at the post-translational level. It is estimated that 30–90% of human genes are regulated by microRNAs, which makes these molecules of great importance for cell growth, activation and differentiation. Microglia are CNS-resident cells of a myeloid lineage that play an important role in immune surveillance and are actively involved in many neurologic pathologies. Although the exact origin of microglia remains enigmatic, it is established that primitive macrophages from a yolk sac populate the brain and spinal cord in normal conditions throughout development. During various pathological events such as neuroinflammation, bone marrow derived myeloid cells also migrate into the CNS. Within the CNS, both primitive macrophages from the yolk sac and bone marrow derived myeloid cells acquire a specific phenotype upon interaction with other cell types within the CNS microenvironment. The factors that drive differentiation of progenitors into microglia and control the state of activation of microglia and bone marrow-derived myeloid cells within the CNS are not well understood. In this review we will summarize the role of microRNAs during activation and differentiation of myeloid cells. The role of miR-124 in the adaptation of microglia and macrophages to the CNS microenvironment will be further discussed. We will also summarize the role of microRNAs as modulators of activation of microglia and microphages. Finally, we will describe the role of miR-155 and miR-124 in the polarization of macrophages towards classically and alternatively activated phenotypes.
Keywords: Microglia, macrophages, myeloid cells, microRNAs, activation, differentiation, neuroinflammation
INTRODUCTION
Since their discovery by del Rio-Hortega in 1932, microglia were recognized as one of the four types of the cells in the CNS along with oligodendrocytes, astrocytes and neurons (Lynch, 2009). In contrast to the latter cell types which have exclusive CNS-specific phenotypes and functions, microglia share many markers of myeloid cells from other organs: expression of macrophage markers CD11b and F4/80, Fc receptors, CD45 (pan leukocyte marker) as well as exhibiting phagocytic properties (Prinz et al., 2011). Therefore, microglia are often referred to as CNS-resident macrophages, since these cells are similar to other macrophages located in other tissues such as liver, peritoneum, and lungs. The first phagocytic cells of a myeloid lineage appear in the CNS of rodents starting at day 8.5 of embryogenesis and these cells predominantly originate from the yolk sac. A large expansion of microglia occurs within two weeks after birth (Cuadros and Navascues, 2001; Ginhoux et al., 2010; Sorokin et al., 1992). It remains uncertain whether a small proportion of microglia might derive from circulating progenitors which enter the CNS during late-embryonic or early-postnatal life. The molecular mechanisms of microglial cell differentiation during pre- and postnatal development are not well understood, although certain transcriptional factors and microRNA (miRNA) that regulate these factors contribute to this process which is discussed later in this review.
In a normal adult, CNS microglia exhibit a specific morphology with long processes with branches (ramified microglia) and a non-activated or “resting” phenotype, which is determined by low levels of expressions of CD45, MHC class II, co-stimulatory molecules (CD80, CD86) and several integrins such as CD11c (Lynch, 2009; Ponomarev et al., 2005b; Prinz et al., 2011). The low levels of expression of these markers in resting resident microglia in normal CNS differentiate the microglia from peripheral macrophages (e.g. inflammatory or splenic macrophages), which typically express high levels of CD45, MHC class II, co-stimulatory molecules and integrins (Ponomarev et al., 2005b). During neuroinflammation, resident microglia become activated as defined by upregulation of a number of activation markers such as CD45, MHC class II, CD86 etc.; activated cells change morphology from ramified to “amoeboid” (when microglia lose processes and increase in size) and become phenotypically and morphologically undistinguishable from peripheral macrophages (Ponomarev et al., 2005b).
Although activated resident microglia are similar to peripheral macrophages in their expression of surface markers and attempts to find specific molecules at mRNA or protein levels that would definitively distinguish activated microglia from other types of macrophages has failed so far, there are several differences in the behavior of resident microglial cells. It was found that activated microglia produce a lower level of superoxide dismutase (SOD) and a very low level of NO during EAE when compared to peripheral macrophages (Enose et al., 2005; Ponomarev et al., 2007a). When we compared the level of expression of CD45, MHC class II, CD40, CD86 and CD11c in activated microglia vs. peripheral macrophages in the CNS of mice with experimental autoimmune encephalitis (EAE), we found that microglia had lower levels of expression of all of these markers in comparison (Ponomarev et al., 2005a; Ponomarev et al., 2006). These results suggested that expression of activation markers in microglia vs. peripheral macrophages differed quantitatively but not qualitatively. In other words, certain genes (e.g. CD45, MHC class II) along with other genes that are discussed later, were downregulated in microglia, which provided the basis for our study to perform expression profiling of microRNAs in microglia vs. peripheral macrophages. We hypothesized that miRNAs serve as epigenetic factors contributing to the microglial phenotype, since it is known that a single miRNA could quantitatively modulate expression of many genes during inflammation (reviewed by (Dai and Ahmed, 2011)). The role of miRNA in induction and maintenance of the specific phenotype and function of microglia and macrophages in the CNS are further discussed in this review.
miRNA AS A NEW CLASS OF REGULATORS OF GENE EXPRESSION
MiRNAs were first discovered in the early 1990s as molecules that control the development of nematode C. Elegans (Lee et al., 1993). However, substantial interest in miRNAs emerged a decade later in the early 2000s after the discovery of the phenomenon of RNA interference (RNAi). Discovery of RNAi led to the re-evaluation of the role of miRNA as natural analogs for small interfering RNA (siRNA) molecules that complement and bind to specific mRNAs (mRNAs of “target” genes) and promote degradation of miRNA-mRNA duplexes and/or prevent translation of these genes. As a result of the action of siRNA or miRNA overexpression in the cell, the expression of the target gene(s) is downregulated in the protein and to some extent in mRNA levels (Kutter and Svoboda, 2008). Nowadays, more than a thousand miRNAs have been identified in mammals including mice and humans as well as in more than 100 other species (Griffiths-Jones et al., 2006). According to bioinformatic estimates, a single miRNA has the potential to regulate hundreds of genes and 30% to 92% of genes in humans are potentially regulated by various miRNAs (Lewis et al., 2005; Miranda et al., 2006).
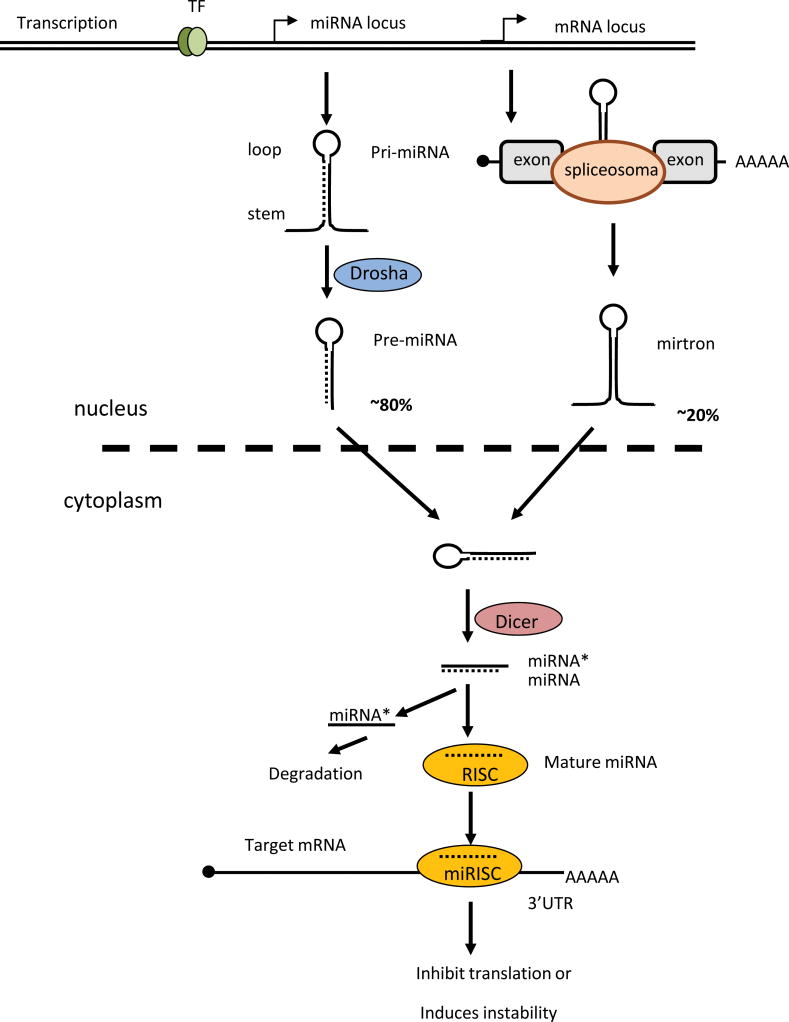
Most miRNAs (~80%) are encoded in intron regions of the genome and are transcribed by RNA polymerase II into rather long (~100–1000 nucleotides) primary transcripts called “pre-miRNA”. In a typical or canonical miRNA maturation process, pre-miRNA is cleaved by a nuclear enzymatic complex (“microprocessor complex”) that consists of nuclear RNAase III and the adapter protein Dorsha. These cleaved primary transcripts are called ‘pre-miRNA”. In an alternative or non-canonical pathway of miRNA biosynthesis (~20% of all miRNAs), other types of primary miRNA transcripts called “mirtrons” are produced independently of the microprocessor nuclear complex (Kim et al., 2009). Pre-miRNAs or mirtrons are transported from the nucleus to the cytoplasm and become further processed by a specific Dicer enzyme that belongs to the RNase III family. The Dicer processes pre-miRNAs or mirtrons into imperfectly matched, short (~22 nucleotides) miRNA/miRNA* duplexes, which later form a RNA-induced silencing complex (RISC) with the help of adapter proteins such as Argonaute. In the RISC, one strain of a miRNA/miRNA* duplex remains to perform binding to mRNA of target genes (this strain is called mature miRNA), while the other complementary non-functional miRNA* strain is degraded (Dai and Ahmed, 2011; Kim et al., 2009). Therefore, mature miRNA is expressed mainly in the cytoplasm where it binds to mRNA and silences the target genes (Fig. 1).
Figure 1. Biosynthesis of microRNA.
MicroRNAs are transcribed as longer precursors (pri-miRNAs or mirtrons), which then become processed to a shorter form called pre-miRNA and then further processed to mature miRNA. Regulation of expression of miRNA occurs at the level of transcription of pri-miRNA or at the level of regulation of the processing of miRNA precursors into mature miRNA. Specific enzymes Dorsha and Dicer are involved in the processing of miRNA. Dorsha processes pri-miRNA into pre-miRNA in the nucleus and then Dicer process pre-miRNA into miRNA/miRNA* duplex in the cytoplasm. In the cytoplasm, one functional strain of miRNA/miRNA* duplex (mature miRNA) becomes part of RISC (RNA-induced silencing complex), while miRNA* degrades. RISC (with integrated miRNA) binds to mRNA of target genes and prevents translation of these mRNAs or promote degradation of this mRNAs resulting in decreased expression of the target gene on the protein level.
To perform silencing of target genes, miRNA in the RISC complex needs to bind to the mRNA of the target gene. There are specific sites on mRNA on the target genes (also called “seeds”) which are crucial for miRNA binding. Weak miRNA-regulated genes have one miRNA-binding site, while strongly regulated target genes have multiple miRNA-binding sites (3–5 sites or even more) on their 3′-untranslated region (3′-UTR) of mRNA. Revolutionary advances in the field of bioinformatics allowed scientists to use specialized software (e.g. free online program “TargetScan”) to predict target genes based on the sequence and structure of both miRNA and mRNA of gene of interests (Thomson et al., 2011). Although the in silico discovery of new target genes that are regulated by miRNAs is an innovative technology, these predicted targets must be validated in vitro using reporter systems and site-directed mutagenesis of miRNA-binding sites, and later on validated in vivo.
Advances in the field of miRNA clearly demonstrate that these molecules play an important role in cancer, neurodegneration, inflammation, as well as both branches of adaptive and innate immunity (Alam and O’Neill, 2011; Dai and Ahmed, 2011; El Gazzar and McCall, 2011; Junn and Mouradian, 2012; Lovat et al., 2011). In this review we will primarily discuss the role of miRNA in functions of myeloid cells and to some extent in neuronal cells, since microglia belong to cells of myeloid origin and reside in the CNS microenvironment under constant influence of the cells of neuronal tissue.
FUNCTIONS OF MICROGLIA IN NORMAL CNS: RESTING OR ALTERNATIVELY ACTIVATED?
Despite the significant efforts of scientists during the last decade who have investigated the role of microglia during inflammation and neurodegeneration, little is known about the phenotype and function of microglia in normal CNS. We still have insufficient knowledge about the exact origin of microglial cells, their role during development of the CNS, the heterogeneity of microglial subsets in various regions of the brain and spinal cord, as well as the nature of paracrine interactions of microglia with oligodendrocytes, astrocytes and neurons.
Historically it has been assumed that microglia play a passive role in adult CNS having a “resting” non-active state and perform their functions only when they become activated by pathological events such as neuronal damage or infection. This point of view results in many scientists focusing on microglial activation during pathology rather than investigating the physiological role of these cells in normal CNS. Only experiments using modern technologies such as intravital microscopy clearly demonstrated that microglia are very active cells in normal CNS and produce many dynamic contacts with neurons (Nimmerjahn et al., 2005; Tremblay et al., 2011). The reasons for these microglia-neuronal contacts are largely unknown for normal CNS, but recently it was shown that these contacts are required for normal synaptic activity of neurons in the visual cortex and the pruning of neuronal axons during development (Tremblay et al., 2011). When we investigated microglia in normal CNS, we found that these cells do not have a completely non-activated phenotype but already exhibited properties of activated macrophages. However, microglia in normal CNS did not exhibit the phenotype of classically activated macrophages, but had the properties of alternatively activated macrophages (Ponomarev et al., 2007a).
There are two main pathways for macrophage activation: the classical pathway (also referred to as M1) and the alternative pathway (also referred to as M2). The M1 pathway is mediated by Th1 cytokines such IFN-γ and Toll-like receptor (TLR) agonists such as LPS. Classically-activated macrophages express high levels of MHC class II and CD86, efficiently process and present antigen to T-cells and produce large amounts of reactive oxygen species and nitric oxide (NO) to eliminate intracellular pathogens. M1 macrophages are potent mediators of inflammatory responses associated with substantial tissue damage of neuronal tissue by secretion of large amounts of TNF-α IL–6 and NO. Alternatively activated M2 macrophages (also called M2a) are induced by Th2-producing cytokines IL-4 and IL-13 (Gordon, 2003; Martinez et al., 2008). Recently a new class of innate helper type 2 (iH2) effector cells were described. Innate helper type 2 cells produce IL-13 and can also contribute to the activation of macrophages towards M2 (Price et al., 2010). Alternatively activated M2 macrophages have low levels of MHC class II and CD86, poorly stimulate or even inhibit T-cell proliferation, but express a number of proteins important for pinocytosis of carbohydrate-rich parasitic products and promote tissue repair. Typical proteins that are abundantly expressed by M2 but not M1 macrophages include CD206 (mannose receptor), Ym-1 (heparin-binding lectin) and Arg 1 (arginine metabolism enzyme), and FIZZ1. M2 macrophages also release anti-inflammatory factors such as IL-10 and TGFβ and Th1-antagonizing cytokines IL-4 and IL-13. Macrophages can be also deactivated by immune complexes (also called M2b), or by glucocorticoids and by cytokines mainly produced by regulatory T-cells such as TGF-β and IL-10 (also called M2c). Deactivated macrophages (also called M2b and M2c) have a phenotype that overlaps with M2a macrophages, but are not identical to alternative activated macrophages induced by Il-4 or IL-13 (Table I) (Fairweather and Cihakova, 2009; Gordon, 2003; Mantovani et al., 2009; Martinez et al., 2008).
Table I.
Comparison of Types of Activation of Macrophages and Microglia
| Type of Activation | Inducers | Type of effectors | Upregulalated Markers | Downregulated Markers | Immunological Function |
|---|---|---|---|---|---|
| Classically Activated Macrophages (M1) | TLR agonists, IFN-γ, GM-CSF, TNF-α | Th1 | MHC class II, CD86, iNOS (NO), TNF-α, IL-1, IL-6 CXCL9 |
IL-10, TGF-β | Inflammation Tissue Damage |
| Alternatively Activated Macrophages (M2) | IL-4, IL-13 (M2a) | Th2 iH2 |
CD204, Ym-1, FIZZ1, Arg 1, TGF-β, IL-10 CXCL22, CXCL17 |
CD86, iNOS (NO), TNF-α | Humoral and antiparasitic responses Tissue Repair |
| Deactivated Macrophages | Immune complexes (M2b) | Tumor cells Tregs |
Scavenger receptors, Ym-1, FIZZ1, Arg 1 | MHC class II, CD86, iNOS (NO), TNF-α, IL-1, IL-6, CD11b, CD11c | Resolution of Inflammation Tissue repair |
| Gluco-corticoids, IL-10, TGF-β (M2c) | |||||
| Resting Microglia | Unknown, IL-4, IL-13 (?) | Astrocytes, Neurons (?) | Ym-1, IL-4, IL-10, TGF-β1 | CD45, MHC class II, CD86, iNOS (NO), TNF-α, IL-1, IL-6, CD11b, CD11c, F4/80 | Immunoregulation Synaptic transmission (?) Development Tissue repair |
| Activated Microglia | TLR agonists, IFN-γ, GM-CSF, TNF-α | Th1, Th17 | Ym-1, FIZZ1, CD45, MHC class II, CD40, CD86, CD11c IL-4, IL-1, TNF-α, IL-6 |
IL-10, TGF-β | Inflammation Phagocytosis Tissue Damage (?) |
We found that in the normal CNS microglia produced IL-4 (Ponomarev et al., 2007a) and TGF-β1 (unpublished observation). In addition, microglia expressed Ym-1 on mRNA and protein levels and do not produce actual NO in vivo. We also found that during EAE the expression of Ym-1 in activated microglia was further increased, and in contrast to peripheral macrophages, activated resident microglia failed to produce NO. Thus, our data indicated that microglia in the normal CNS exhibit a phenotype of M2-like cells (Ponomarev et al., 2007a). Surprisingly M2 features of microglia were even enhanced during EAE when microglia up-regulated expression of IL-4 and Ym-1 (Ponomarev et al., 2007a), a disease which was induced in our model by the adoptive transfer of Th1 cells that are known to secrete IFN-γ, TNF-α and GM-CSF that typically result in activation of macrophages towards M1 (Ponomarev et al., 2007b). We found that expression of Ym-1 in microglia was IL-4 dependent, since this molecule was significantly downregulated in microglia from IL-4 and IL-4/13 receptor deficient mice (Table I). Using chimeric animals expressing IL-4 only by CNS-resident cells we concluded that the expression of Ym-1 in microglia a CNS-derived source of IL-4 is required (Ponomarev et al., 2007a). These facts led us to postulate that the M2-like phenotype of microglia is induced by the CNS microenvironment (e.g. IL-4 produced by microglia in autocrine manner or produced by astrocytes), which was further explored when we investigated the role of miRNA in contributing to the microglia phenotype.
miRNA PROFILING IN MICROGLIA VS. PERIPHERAL MACROPHAGES
Since we found that microglia exhibited an M2-like phenotype in normal CNS and during EAE, we proposed that there are specific microRNAs that contribute to this phenotype. As we mentioned above, during EAE induced in chimeric mice (to distinguish microglia from peripheral macrophages), activated microglia expressing the M2 marker Ym-1 did not produce NO (Ponomarev et al., 2007a). However, microglia upregulated certain M1 markers such as MHC class II, CD45 and CD86, but the level of expression of these M1 markers was still lower than that of macrophages (Ponomarev et al., 2005b). Thus, even activated microglia exhibited a quantitatively different phenotype from peripheral macrophages that are located at the same sites of inflammatory lesions in CNS during EAE (Ponomarev et al., 2005b).
Recent advances in deep sequencing technologies of miRNA expression in various tissues of mammalian species enabled us to perform selected profiling of miRNA, which is known to be expressed in myeloid cells and in the CNS (Landgraf et al., 2007), since microglia belong to myeloid lineage and reside in the CNS. We found that during EAE activated microglia expressed CNS-specific miR-124, which is absent or expressed at low levels in the peripheral macrophages. We also found that miR-124 was expressed only in normal CNS-resident microglia, but not in thioglicolate-induced inflammatory macrophages or CD11b+F4/80+ macrophages from spleen, bone marrow, peritoneal cavity or liver tissues (Ponomarev et al., 2011a).
MiR-124 has been found to be the most abundant microRNA expressed in neurons (Mishima et al., 2007). It was shown that miR-124 promotes neuronal differentiation by targeting the mRNA of the anti-neural function protein SCP1, which represses neuronal gene expression in non-neuronal cell types (Makeyev et al., 2007). Several studies suggested that miR-124 could be used as an anti-cancer therapeutic agent (Sonntag et al., 2011). Specifically, miR-124 induced differentiation of mouse neuroblastoma cell lines. The mechanism of suppression of proliferation by miR-124 involves targeting cell cycle dependent kinases CDK4 and CDK6 resulting in a decrease in the proliferation of tumor cells (Ponomarev et al., 2011b). In contrast to neuronal cells where miR-124 targeted SCP1 results in neuronal maturation and differentiation, in microglia and macrophages miR-124 targeted CEBPα, a master transcription factor important for the development of myeloid cells. The differentiation of monocytic lineage cells is controlled by two main transcription factors CEBPα and PU.1, which will be further discussed in the section describing the role of miRNA in development of myeloid cells. Briefly, CEBPα induces PU.1 and both of these factors are sufficient for the differentiation of non-myeloid cells such as fibroblasts into macrophages (Cai et al., 2008; Feng et al., 2008). The mRNA of the CEBPα gene consists of a short 5′UTR contained region and a large 3′-UTR with three sites for binding miR-124 (Ponomarev et al., 2011a). Resting microglial cells are known to express low levels of CEBPα, but CEBPα is upregulated upon microglial activation (Walton et al., 1998). Thus, our finding suggested that miR-124 contributed to resting phenotype of microglia by targeting the CEBPα/PU.1 pathway.
After discovering expression of miR-124 in microglia, we further investigated two main questions: 1) what induced miR-124 expression in microglia and 2) what is the role of miR-124 in the phenotype and function of microglia and macrophages.
IT IS ALL ABOUT ENVIRONMENT!
When we investigated the level of expression of miR-124 in microglia and peripheral macrophages using chimeric mice we found that there was a reciprocal correlation between the level of expression of M1 activation markers and expression of miR-124. The highest level of miR-124 expression was found in CD45lowMHC class IIlow non-activated resident microglia. When microglia become activated during EAE or are treated with pro-inflammatory cytokines IFN-γ and GM-CSF, microglia unregulated CD45, MHC class II and downregulated miR-124. An opposite situation was found for peripheral macrophages. During the peak of EAE, infiltrating peripheral macrophages in the CNS were CD45hiMHC class IIhi and were negative for miR-124, while during the recovery phase of EAE, macrophages dowregulated CD45, MHC class II and upregulated miR-124 (Ponomarev et al., 2011a). Thus, we found that during active inflammation in the CNS both microglia and macrophages expressed M1 markers and had low levels of miR-124, while in normal CNS or during disease recovery macrophages downregulated M1 markers such as MHC class II and upregulated miR-124. Interestingly, during EAE peripheral macrophages in the CNS exhibited a dually activated or M1/M2 mixed phenotype expressing NO (M1 marker) and high levels of Ym-1 (M2 marker) (Ponomarev et al., 2007a). Given the fact that during EAE activated microglia upregulated M1 markers (CD45 and MHC class II) (Ponomarev et al., 2005b), but still expressed M2 markers (Ym-1), we conclude that activated microglia also exhibited a mixed M1/M2 phenotype. Similar results were reported for peripheral macrophages infiltrating the CNS in a spinal cord injury model that exhibited a M2-like phenotype and promoted CNS repair during resolution of inflammation (Schwartz, 2010). Thus, in normal CNS and during resolution of inflammation microglia and macrophages often exhibited an M2 phenotype, while during inflammation microglia and macrophages also upregulated M1 markers.
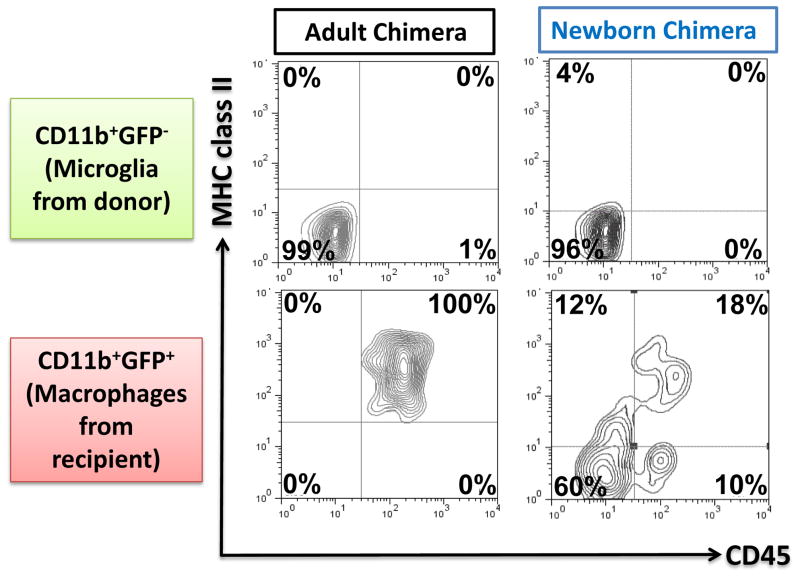
In addition to inflammatory conditions, a similar situation with microglial phenotype and miR-124 expression was observed in microglia isolated from CNS during development. Within the first two weeks after birth, microglia had CD45hiMHC class IIhimiR-124low phenotype, while adult microglia had CD45lowMHC class IIlow miR-124hi phenotype (Ponomarev et al., 2011a). We assume that under certain circumstances bone marrow derived myeloid cells can also migrate into the brain from periphery during post-natal period and acquire a MHC class IIlowCD45low microglia-like phenotype similar to appearance of CD45lowMHC class IIlow bone-marrow derived myeloid cells during recovery phase of EAE in chimeras. To investigate this possibility we created new born mixed chimeras by transplanting bone marrow from ActB-GFP transgenic mice that ubiquitously expressed GFP under the actin promoter into sublethally irradiated newborn mice (P1-P3). When these chimeric mice became adults (8 weeks old), engrafted GFP positive CD11b+ cells were found in the CNS and 60% of them had MHC class IIlowCD45low phenotype similarly to GFP negative microglia of recipient origin (Fig. 2, Newborn Chimera). When we compared the phenotype of CD11b+GFP+ cells from the donor in the CNS after 8 weeks of reconstitution in adult chimera, we found that all of them were activated with high levels of CD45 and MHC class II (Fig. 2, Adult Chimera). Thus, at least in chimeras, BM-derived cells in the CNS acquired MHC class IIlowCD45low phenotype during development or resolution of inflammation. Based on these data we suggested that in a normal CNS a MHC class IIlowCD45low miR-124hi phenotype could be induced in myeloid cells under the influence of CNS microenvironment.
Figure 2. Comparison of phenotype of bone marrow-derived myeloid cells in adult and newborn chimeras.
An adult (at 8 weeks) (a) or a newborn (2–3 days old) (b) mice were irradiated and transplanted with bone marrow from transgenic mice ubiquitously expressing GFP under an actin promoter. After 8 weeks of reconstitution, mononuclear cells were isolated from the CNS and CD11b+GFP− (resident microglia) and CD11b+GFP+ (bone marrow-derived myeloid cells) were analyzed for the expression of CD45 and MHC class II by four color flow cytometry. In adult chimera bone marrow derived myeloid cells exhibited activated CD45hiMHC class IIhi (a), while 60% of bone marrow derived myeloid cells in newborn chimeras exhibit CD45low MHC class IIlow phenotype similar to resident microglia (b).
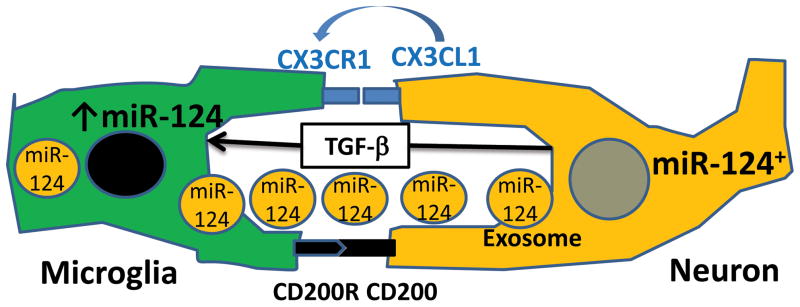
We directly tested our hypothesis by co-culturing bone-marrow derived macrophages with astroglial and neuronal cell lines and found that a co-culture of bone marrow-derived macrophages grown in the presence of M-CSF with a neuronal cell line and to less extent with an astroglial line resulted in downregulation of MHC class II and CD45 and upregulation of miR-124. The mechanisms of induction of such a phenotype remain to be determined, but there are three possibilities: 1) cell-cell contact; 2) the presence of soluble factors, and 3) direct transfer of miR-124 from miR-124+ neuronal cells into macrophages through exosomal shuttle vesicles (Fig. 3). There are several examples of deactivation of microglia in the CNS upon interaction with neurons mediated by cell-cell contact. It was shown in CD200 deficient mice that microglia exhibited an activated phenotype and the EAE disease course was exacerbated (Hoek et al., 2000). Since CD200 is expressed in neurons and CD200R is expressed by microglia, it is believed that cell-cell contact with neurons is required for the maintenance of their quiescent phenotype. Similar results were observed in mice deficient in the Fractalkine receptor CX3CR1 (Cardona et al., 2006). Both membrane-bound and soluble forms of Fractalkine (CX3CL1) are expressed by neurons, while receptors for Fractalkine (CX3CR1) are expressed in microglia. Microglia in CX3CR1 deficient mice have elevated levels of activation, suggesting that Fractalkine could be a membrane-bound and soluble factor that contribute to the MHC class IIlowCD45lowmiR-124hi phenotype in microglia. Other pairs of ligand-receptor proteins were suggested to play a role in contact deactivation of microglia by neurons including CD45-CD22 and CD172-CD47 (Ransohoff and Cardona, 2010). An example of a soluble factor that is highly expressed in the CNS by astrocytes and neurons and that deactivates macrophages is TGF-β(Bottner et al., 2000), which could also regulate miR-124 expression in microglia. In addition to factors that regulate expression of miRNAs such as miR-124, other examples of microRNAs transferred from IL-4-treated macrophages to tumor cells in secreted vesicles have been recently reported (Yang et al., 2011). There is also the possibility that multiple miR-124 containing vesicles secreted by neuronal cells could be transferred to microglia; however whether these processes take place and how effective they are, remains to be defined.
Figure 3. The proposed mechanisms of induction of miR-124 expression in microglia and macrophages interacting with neuronal cells.
Increased expression of miR-124 in macrophages in the CNS is influenced by many factors of paracrine interactions between neuronal cells and microglia/macrophages: cell-cell contacts such as CD200-CD200R or CX3CR1-CX3CL, the presence of soluble factors secreted by neurons such as soluble form of CX3CL1 and TGF-β, and direct transfer of miR-124 from miR-124+ neuronal cells into macrophages through exosomal shuttle vesicles.
miRNA IN DEVELOPMENT OF MYELOID CELLS
Even though the exact origin of microglia is still debatable, there is agreement that myeloid cells in the CNS could originate from two distinct periods of hemopoiesis: the first, during primitive hematopoiesis that occurred primarily in extraembryonic sites such as the yolk sac, and during the second wave of hematopoiesis that persists throughout adulthood and occurs primarily in bone marrow. It was recently established that microglia originate from primitive macrophages in the yolk sac (Ginhoux et al., 2010), although there are examples in which bone marrow-derived myeloid cells can be engrafted into the CNS at least under pathological conditions (Eglitis and Mezey, 1997; Soulet and Rivest, 2008).
Despite a difference in origin of primitive macrophages and bone-marrow derived monocytic cells, two transcription factors PU.1 and CEBPα are critical for the development of yolk sac derived primitive macrophages and resident microglia as well as bone marrow derived myeloid cells. Indeed PU.1 deficient mice do not have microglia, but transplantation in newborn PU.1-deficient mice with wild type bone marrow resulted in a population of CNS with resident macrophages with all the phenotypic features of microglia that are obviously derived from bone marrow (Beers et al., 2006). We also demonstrated that the CD45lowMHC class IIlow bone marrow derived myeloid cells can be engrafted into the CNS of mixed newborn chimeras (Fig. 2) suggesting a contribution of BM-derived cells to the total pool of myeloid cells with a microglial phenotype in an adult CNS of chimeric animals. In addition to PU.1, it was also clearly demonstrated that CEBPα initiated a primitive myelopoiesis in pluropotent embryonic stem cells (Chen et al., 2009). In both cases of primitive and adult hemopoiesis of monocytic cells, the role of M-CSF (also known as CSF-1R) and its analog IL-34 (both acting through the same M-CSF receptor) is critical for the development of monocytic cells and microglia, since deficiency in the M-CSFR inhibited population of CNS by primitive macrophages suggests a similarity of the processes of microglial origins during embryogenesis or through the development of monocytic cells in bone marrow during adulthood (Ginhoux et al., 2010; Herbomel and Levraud, 2005). The known difference between primitive and adult myelopoiesises is that in case of primitive hemopoiesis the process starts with pluripotent embryonic stem cells (ESC) and in case of adult hemopoiesis the process starts from hematopoietic stem cells (HSC). However, mechanisms by which transcription factors PU.1, CEBPα and M-CSFR drive the processes of development of primitive macrophages and BM-derived myeloid cells appear to be quite similar (Saijo and Glass, 2011). Thus, we hypothesize that the microRNAs that modulate expression of the PU.1, CEBPα and M-CSFR pathways affect microglia originating from primitive macrophages as well as bone marrow derived myeloid cells in the same way.
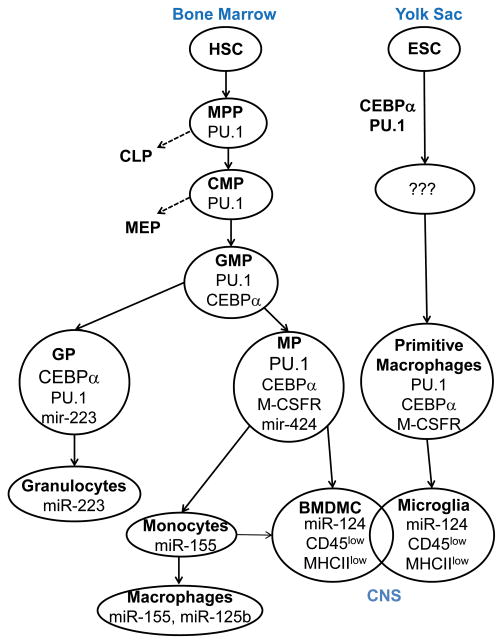
As we mentioned, adult hematopoietic linage progenitors arise from HSC. HSC have the ability for self renewal or can differentiate into multipotent progenitors (MPP) that have limited proliferating capacity. MPP give rise to common lymphoid lineage progenitors (CLP) and common myeloid progenitors (CMP). CMP then further differentiate into megakaryocyte-erythroid progenitors (MEP) and granulocyte-macrophages progenitors (GMP), which later differentiate into granulocytic progenitor (GP) and monocytic progenitor (MP) cells (El Gazzar and McCall, 2011). Thus, development of bone marrow-derived cells (BMDMC) with microglial phenotypes can be viewed as a sequence of events of differentiation by: HSC→MPP→CMP→GMP→MP→BMDMC (Fig. 4). The myelopoiesis of primitive macrophages that give rise to microglia is not well characterized when compared to adult myelopoiesis, but we believe that similar or equivalent stages of differentiation take place in this process involving the same three CEBPα, PU.1 and M-CSFR pathways (Fig. 4). We know that microglia originating from the yolk sac and bone marrow derived myeloid cells in the CNS share many morphological phenotypic features, although we do not know whether these two subsets become completely identical in the CNS (Fig. 4).
Figure 4. Role of miRNA in development of myeloid cells and microglia.
This figure demonstrates the developmental relationship between microglia and bone-marrow derived myeloid cells and the role of transcriptional factors CEBPα and PU. 1, the M-CSFR pathway, miR-424, miR-155 and miR-124. Bone marrow derived myeloid cells (BMDMC) arise from hematopoietic stem cells (HSC) and have the ability to migrate into CNS after birth and adult life under pathological conditions. Microglia arise from the yolk sac most likely from embryonic stem cells (EST) and migrate into the CNS before birth, during development starting from day 8.5. CEBPα, PU. 1, M-CSFR pathway and miR-124 are critical for the development of microglia and CNS BMDMC. Both microglia and CNS bone-marrow derived myeloid cells have similar morphology and CD45low MHC class IIlow phenotype indicating that these two subsets have overlapping properties and functions in the CNS.
CEBPα is required for transition from MPP to CMP. PU.1 expression is induced in MPP and continues to be expressed in CMP and GMP. PU.1 promotes transition from CMP to GMP. One of the factors that induces PU.1 is CEBPα, therefore CEBPα is often called a master transcription factor for the development of monocytic cells. However, high levels of PU.1 when compared to CEBPα levels need to be maintained in GMP to promote differentiation of GMP towards the monocytic lineage. If the level of PU.1 is relatively low when compared to CEBPα, GMP differentiate towards granulocytes (Friedman, 2007). The M-CSFR (CSF-1R) pathway plays a critical role in skewing GMP towards the monocytic lineage, since PU.1 directly up-regulates M-CSFR, which increases responsiveness of GMP to M-CSF and induces proliferation of MP (Friedman, 2007; Yeamans et al., 2007).
Various microRNAs are actively involved in the regulation of transcription factors PU.1 and CEBPα and, vice versa certain miRNAs are regulated by CEBPα and PU.1 (El Gazzar and McCall, 2011). In this review, we will concentrate on the role of miRNAs in late events of differentiation of myeloid cells from GMP to monocytic cells, since at these stages microglial progenitors most likely differentiate into mature microglia in the CNS. One of the good examples is miR-223, which is critical for the differentiation of granulocytes. MiR-223 is induced by CEBPα and targets the transcription factor NFI-A (NFI-A is upregulated by both CEBPα and PU.1), which is required for the differentiation of GMP into mature granulocytes. MiR-223 decreases proliferation of GP, promoting their differentiation into mature granulocytes (Fazi et al., 2005). As with miR-223, miR-424 is upregulated by PU.1 and then targets NFI-A, which results in the upregulation of M-CSFR and the differentiation of GMP into MP (Forrest et al., 2010; Rosa et al., 2007). Thus, miR-223 and miR-424 promote differentiation of the GMP into granulocytes and monocytes in bone marrow, respectively. Our data suggests that miR-124 most likely affects the maturation of immature progenitors into mature microglia or bone marrow derived myeloid cells in the CNS. Similar to miR-424 in the bone marrow, miR-124 restricts the proliferation of monocytic cells by targeting CEBPα and most likely CDK4/CDK6. The decrease in expression of CEBPα results in the downregulation of PU.1 and its downstream target M-CSFR, further restricting proliferation and most likely promoting differentiation of MP into CNS bone marrow derived myeloid cells and primitive macrophages into mature adult microglia (Ponomarev et al., 2011a) (Fig. 4). Another important microRNA which is expressed in T, B and monocytic cells is miR-155 (Forrest et al., 2010). MiR-155 overexpression in bone marrow increases the number of immature granulocytes, similar to its effect in miR-223 deficient mice (O’Connell et al., 2008). MiR-155 seems to oppose miR-223 and leads to the skewing of differentiation of GMP towards monocytes in the bone marrow. The main role of miR-155 during classical M1 activation of macrophages (Fig. 4) will be described later.
miRNA IN ACTIVATION OF MONOCYTIC CELLS
As we described above, miR-414, miR-155 and miR-124 significantly contribute to the development of monocytic cells in bone marrow and most likely in the CNS. However, certain miRNAs are also involved in the processes of activating macrophages and microglia during inflammation. The action of miRNA on macrophages can be subdivided into three categories: pro-inflammatory, participating in the resolution of inflammation and anti-inflammatory (Table II).
Table II.
MiRNAs That Modulate Activation of Macrophages and Microglia
| Group of microRNAs | miRNA | Inducers of miRNA expression | Targets genes | Contribution to Phenotype |
|---|---|---|---|---|
| Pro-inflammatory | miR-155 | LPS, Poly I:C, IFN-γ, TNF-α | FADD, SOCS-1, IKK, IL13Rα1, SMAD-2, CEBPβ | Decreased apoptosis, up-reugulation of IL-1, IL-6, TNF-α, iNOS; downregulation of IL-10, Arg 1, iNOS, IL13R, TGF-βR pathway proteins |
| miR-125 | TLR agonists | IRF4 | Upregulation of MHC class II, CD40, CD80, CD86 | |
| miR-101 | TLR agonists | MAPK phosphatase-1 | Upregulation of IL-1, IL-6, TNF-α | |
| Resolution of inflammation | miR-146 | TLR agonists | TLR and TNF-α pathways | Douwnregulation of IL-1, IL-6, CCL5, CXCL8 |
| miR-21 | TLR agonists | PDCD4, FasL, PTEN | Decreased apoptosis, upregulation of IL-10 | |
| Anti-inflammatory | miR-124 | IL-4/IL-13(?) IL-10 (?) |
CEBPα, CDK4/6 | Decreased proliferation, upregulation of Arg 1, FIZZ1, TGF-β1; downregulation of iNOS, TNF-α, IL-6, MHC class II, CD86, CD11b, M-CSFR |
It was found that overexpression of miR-155 in bone marrow resulted in a myeloproliferative phenotype similar to systemic administration of LPS (O’Connell et al., 2008). Further investigations demonstrated that miR-155 was significantly upregulated in macrophages in response to LPS, suggesting their important role in macrophage activation (O’Connell et al., 2007). Recently, it was demonstrated that miR-155 was upregulated in monocytes, macrophages and in a microglial line in response to various M1-activating stimuli such as TLR agonists, IFN-γ, and TNF-α. MiR-155 has been shown to target several pro-apoptotic and anti-inflammatory proteins such as Fas-associated death domain proteins (FADD), suppressors of cytokine signaling (SOCS-1), and IκB kinases (IKK) (Bala et al., 2011; Louafi et al., 2010; Martinez-Nunez et al., 2011; Wang et al., 2010). Thus it has been demonstrated that miR-155 plays a pro-inflammatory role in promoting classical M1 path of activation of macrophages.
Another pro-inflammatory miRNA, miR-101 is also induced by M1 stimuli such as TLR agonists and targeted MAPK phosphatase-1, which deactivates MAPK. Overexpression of miR-101 in macrophages results in MAPK activation and in the secretion of M1-associated pro-inflammatory cytokines IL-1, IL-6, and TNF-α (Zhu et al., 2010).
As a last example of pro-inflammatory miRNAs, miR-125 significantly increases M1 activation of macrophages by targeting IRF4, increasing responsiveness to IFN-γ, and most importantly for antigen presentation, by upregulating MHC class II, CD40, CD80, CD86 (Chaudhuri et al., 2011).
Similar to other pro-inflammatory miRNAs, miR-146 is induced by M1 stimuli such as LPS. However, miR-146 provides a negative signaling feedback in the resolution of inflammation by targeting signaling molecules downstream of TLRs and IL-1R including IL-1R receptor-associated kinase 1 and TNF receptor associated factor 6. This inhibition decreases the production of M1 pro-inflammatory cytokines IL-1, IL-6 and several chemokines including CCL5 and CXCL8. It was demonstrated that HIV-infected human microglia expressed miR-146 that targeted chemokine CCL8, suggesting an important role for this miRNA in the activation of microglia in humans (Alam and O’Neill, 2011; Rom et al., 2010).
MiR-21 is also induced by LPS in macrophages; miR-146 and miR-21 promote the resolution of inflammation. MiR-21 inhibits PDCD4, an inhibitor of IL-10 production. Therefore, miR-21 promotes IL-10 expression in macrophages. miR-21 also targets several pro-apoptotic genes including FasL and PTEN which could potentially attenuate apoptosis of LPS-treated myeloid cells (Alam and O’Neill, 2011).
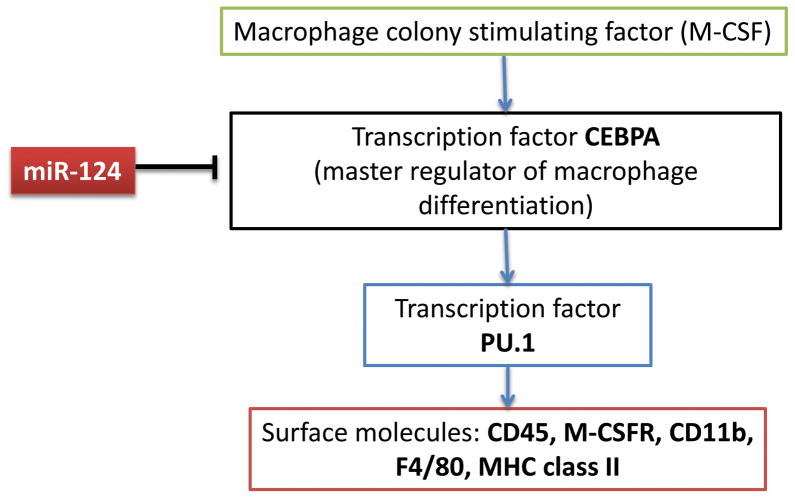
Finally, we found that miR-124 decreased activation of bone-marrow derived and splenic macrophages by downregulating MHC class II and CD86 and decreasing production of TNF-α and IL-6 (Ponomarev et al., 2011a). As we mentioned, miR-124 targeted CEBPα, results in the downregulation of PU.1 and the further downregulation of its downstream targets such as M-CSFR, CD45, MHC class II, CD11b, and F4/80 (Fig. 5).
Figure 5. Deactivation of macrophages by miR-124 via targeting CEBPα/PU.1 pathway.
Macrophage growth and differentiation is driven by M-CSF that acts through M-CSFR on macrophages and involves induction of expression of CEBPα. CEBPα directly upregulates PU.1, which, in turn, directly upregulates several macrophage activation markers including M-CSFR, CD45, CD11b, F4/80 and MHC class II. MiR-124 directly inhibits CEBPα which results in downregulation of PU.1 and its downstream genes M-CSFR, CD45, CD11b, F4/80 and MHC class II. Downregulation of M-CSFR also resulted in the decrease of proliferation driven by M-CSF.
miRNA AS REGULATORS OF M1 AND M2 POLARIZATION
Initially it was assumed that miR-155 is a pro-inflammatory miRNA that contributes to macrophage activation by targeting anti-inflammatory genes. However, more recent studies demonstrate that miR-155 promotes skewing toward the M1 phenotype by targeting M2-associated genes. It was demonstrated that miR-155 downregulates the IL-13 receptor (IL13Rα1). Moreover miR-155 also targets SMAD2 involved in signal transduction in the TGF-β pathway, an anti-inflammatory cytokine associated with the M2 phenotype (Louafi et al., 2010; Martinez-Nunez et al., 2011). Lastly, miR-155 inhibits CEBPβ(not to be confused with CEBPα), which was recently shown to be important for the expression of a number of M2-asociated markers such as Arg 1, IL-10, IL13Rα1 and CD206 (Ruffell et al., 2009). Thus, miR-155 clearly targets multiple genes associated with the M2 phenotype and is induced by M1 stimuli such as IFN-γ and TLR agonists (Table II).
As with miR-155, we initially assumed that miR-124 deactivated macrophages by decreasing the expression of CD45, CD11b, F4/80, MHC class II and CD86. However, more detailed gene expression profiling in miR-124 transfected macrophages revealed that miR-124 downregulated M1-associated markers IL-6, TNFα, iNOS (an inducible form of NO synthetase) and upregulated expression of M2-associated markers TGF-β1, Arg 1 and FIZZ1 (Ponomarev et al., 2011a). Thus, miR-124 contributed to the polarization of macrophages toward the M2 phenotype (Table II).
Since M1 and M2 stimuli potentially upregulate multiple miRNAs, the question remains whether miR-155 and miR-124 are primarily miRNAs related to M1/M2 polarization. The analysis of multiple miRNAs in the mouse macrophage line RAW 264.7 shows that treatment of these cells with LPS results in marked up regulation of miR-155 within six hours after treatment (Garmire et al., 2010). We found that treatment of RAW 264.7 cells with IL-4 resulted in upregulation of miR-124 as early as four hours after treatment with IL-4 (an unpublished observation), suggesting that miR-155 and miR-124 are likely to be miRNAs directly induced by M1 and M2 stimuli (Table II). Further analysis should establish the role of miR-155 and miR-124 in macrophage polarization in vitro and in vivo and whether other microRNAs contribute to this process.
PROSPECTS FOR USAGE OF miRNAs AS MODULATORS OF MACROPHAGE FUNCTIONS AND IMMUNOMODULATORY THERAPEUTICS
Macrophages and monocytes play an active role in the pathogenesis of many inflammatory diseases including rheumatoid arthritis, type I diabetes, and autoimmune colitis. Depletion of monocytes/macrophages in the EAE completely prevents disease: which is not surprising considering the fact that ~70% of infiltrating cells in the CNS inflammatory lesions are macrophages (Tran et al., 1998). Activation of microglia is also a feature of many neuroinflammatory and neurodegenerative diseases (Prinz et al., 2011). Therefore, the application of anti-inflammatory microRNAs such as miR-124 and inhibitors for pro-inflammatory microRNAs such as miR-155 that modulate macrophage activation could be useful for the treatment of CNS inflammatory diseases such as MS. In the case of Alzheimer’s disease when insufficient activation of macrophages results in increased amyloid deposition and neurotoxicity (Frenkel et al., 2005), use of pro-inflammatory microRNAs such as miR-155, miR-125b and miR-21 might be beneficial to promote clearing of amyloid plaques. One of the main difficulties in using microRNAs to modulate microglia and macrophages is the delivery of miRNA or miRNA inhibitors. To effectively deliver 20 b.p. oligonucleotides such as microRNA, siRNA or miRNA inhibitors in the cytoplasm of the target cells in vitro, cationic liposomes that fuse with cell plasma membrane are widely used (Liu et al., 1997). We found that i.v. injection of liposomes with miRNA was effectively engulfed by macrophages in the spleen (Ponomarev et al., 2011a). It has been demonstrated that macrophages selectively take up liposomes and other particles primarily in the spleen, liver, lungs and the site of inflammation (Deissler et al., 2008; Liu et al., 1997). Delivery of liposomes with miRNAs to the microglia in the CNS is especially difficult due to the blood-brain barrier. However, several studies have suggested that liposomes could be delivered to the sites of inflammation where the blood-brain barrier is compromised (Cavaletti et al., 2009; Kizelsztein et al., 2009). Recently, it was demonstrated that siRNA was effectively delivered to splenic macrophages using glucan particles that were administrated orally (Aouadi et al., 2009). This would allow macrophages to take up these particles through the mannose receptor (CD206), which is highly expressed in M2 macrophages such as microglia or CNS perivascular macrophages. Nevertheless, a problem remains when liposomes with miRNA are degraded in lysosomal compartments of macrophages activated towards M1. The usage of the targeted delivery to M2 macrophages via CD206 or scavenger receptors and the stabilization of miRNAs using synthetically modified olionucleotides that are not sensitive to lysosomal degradation could potentially increase the efficacy of delivery of miRNA to macrophages in various organs including the CNS.
CONCLUDING REMARKS
The immune system is a dynamic and multifunctional system that not only fights infection, but also plays an active role in development, metabolic processes and tissue remodeling and repair. The heterogeneity of the adaptive branch of the immune system such as T-cells, appears to be more dynamic and complex than we initially imagined. Macrophages appear to be much more dynamic and “plastic” than T-cells. Monocytic cells are not as terminally differentiated as T-cells and appear to switch between activated and deactivated states to M1 and M2 phenotypes and back, often exhibiting dually activated M1/M2 phenotype. Thus, macrophages could dynamically adapt to the local microenvironment especially in tissues such as the CNS, which could be critical for the regulation of the inflammatory response at the site of inflammation. Therefore, more attention should be paid in the future to investigate the interactions of macrophages with local stromal cells in tissues; particularly in the case of the CNS, and in the interaction of microglia and infiltrating macrophages with astrocytes, neurons and oligodendrocytes. In the context of interactions of resident macrophages with stromal tissue cells, microRNAs could significantly contribute to the adaptation of macrophages to the local environment and the regulation of inflammation in situ.
Acknowledgments
Grant sponsors: The work was supported by NIH R01 NS071039-01A1 research grant.
The work that was referred to and reported here was supported by NIH R01 NS071039-01A1 research grant.
Abbreviations
- BMDMC
bone marrow derived myeloid cells
- CDK
cyclin-dependent kinase
- CEBP
CCAAT/enhancer binding protein
- CLP
common lymphoid progenitors
- CMP
common myeloid progenitors
- EAE
experimental autoimmune encephalitis
- ESC
embryonic stem cells
- FADD
Fas-associated death domain protein
- GP
granulocytic progenitors
- HSC
hematopoietic stem cells
- LPS
lipopolysaccharide
- MEP
megakaryocyte-erythroid progenitors
- MHC
major histocompatibility complex
- miRNA
microRNA
- MP
monocytic progenitors
- MPP
multipotent progenitors
- RISC
RNA-induced silencing complex
- siRNA
small interfering RNA
- SOCS
suppressors of cytokine signaling
- SOD
superoxide dismutase
- TLR
toll-like receptor
- UTR
untranslated terminal region
References
- Alam MM, O’Neill LA. MicroRNAs and the resolution phase of inflammation in macrophages. Eur J Immunol. 2011;41:2482–2485. doi: 10.1002/eji.201141740. [DOI] [PubMed] [Google Scholar]
- Aouadi M, Tesz GJ, Nicoloro SM, Wang M, Chouinard M, Soto E, Ostroff GR, Czech MP. Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation. Nature. 2009;458:1180–1184. doi: 10.1038/nature07774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bala S, Marcos M, Kodys K, Csak T, Catalano D, Mandrekar P, Szabo G. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2011;286:1436–1444. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers DR, Henkel JS, Xiao Q, Zhao W, Wang J, Yen AA, Siklos L, McKercher SR, Appel SH. Wild-type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2006;103:16021–16026. doi: 10.1073/pnas.0607423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottner M, Krieglstein K, Unsicker K. The transforming growth factor-betas: structure, signaling, and roles in nervous system development and functions. J Neurochem. 2000;75:2227–2240. doi: 10.1046/j.1471-4159.2000.0752227.x. [DOI] [PubMed] [Google Scholar]
- Cai DH, Wang D, Keefer J, Yeamans C, Hensley K, Friedman AD. C/EBP alpha:AP-1 leucine zipper heterodimers bind novel DNA elements, activate the PU.1 promoter and direct monocyte lineage commitment more potently than C/EBP alpha homodimers or AP-1. Oncogene. 2008;27:2772–2779. doi: 10.1038/sj.onc.1210940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona AE, Pioro EP, Sasse ME, Kostenko V, Cardona SM, Dijkstra IM, Huang D, Kidd G, Dombrowski S, Dutta R, Lee JC, Cook DN, Jung S, Lira SA, Littman DR, Ransohoff RM. Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci. 2006;9:917–924. doi: 10.1038/nn1715. [DOI] [PubMed] [Google Scholar]
- Cavaletti G, Cassetti A, Canta A, Galbiati S, Gilardini A, Oggioni N, Rodriguez-Menendez V, Fasano A, Liuzzi GM, Fattler U, Ries S, Nieland J, Riccio P, Haas H. Cationic liposomes target sites of acute neuroinflammation in experimental autoimmune encephalomyelitis. Mol Pharm. 2009;6:1363–1370. doi: 10.1021/mp8001478. [DOI] [PubMed] [Google Scholar]
- Chaudhuri AA, So AY, Sinha N, Gibson WS, Taganov KD, O’Connell RM, Baltimore D. MicroRNA-125b potentiates macrophage activation. J Immunol. 2011;187:5062–5068. doi: 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Costa RM, Love NR, Soto X, Roth M, Paredes R, Amaya E. C/EBPalpha initiates primitive myelopoiesis in pluripotent embryonic cells. Blood. 2009;114:40–48. doi: 10.1182/blood-2008-11-189159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadros MA, Navascues J. Early origin and colonization of the developing central nervous system by microglial precursors. Prog Brain Res. 2001;132:51–59. doi: 10.1016/S0079-6123(01)32065-4. [DOI] [PubMed] [Google Scholar]
- Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011;157:163–179. doi: 10.1016/j.trsl.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deissler V, Ruger R, Frank W, Fahr A, Kaiser WA, Hilger I. Fluorescent liposomes as contrast agents for in vivo optical imaging of edemas in mice. Small. 2008;4:1240–1246. doi: 10.1002/smll.200701069. [DOI] [PubMed] [Google Scholar]
- Eglitis MA, Mezey E. Hematopoietic cells differentiate into both microglia and macroglia in the brains of adult mice. Proc Natl Acad Sci U S A. 1997;94:4080–4085. doi: 10.1073/pnas.94.8.4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Gazzar M, McCall CE. MicroRNAs regulatory networks in myeloid lineage development and differentiation: regulators of the regulators. Immunol Cell Biol. 2011 doi: 10.1038/icb.2011.74. [DOI] [PubMed] [Google Scholar]
- Enose Y, Destache CJ, Mack AL, Anderson JR, Ullrich F, Ciborowski PS, Gendelman HE. Proteomic fingerprints distinguish microglia, bone marrow, and spleen macrophage populations. Glia. 2005;51:161–172. doi: 10.1002/glia.20193. [DOI] [PubMed] [Google Scholar]
- Fairweather D, Cihakova D. Alternatively activated macrophages in infection and autoimmunity. J Autoimmun. 2009;33:222–230. doi: 10.1016/j.jaut.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest AR, Kanamori-Katayama M, Tomaru Y, Lassmann T, Ninomiya N, Takahashi Y, de Hoon MJ, Kubosaki A, Kaiho A, Suzuki M, Yasuda J, Kawai J, Hayashizaki Y, Hume DA, Suzuki H. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia. 2010;24:460–466. doi: 10.1038/leu.2009.246. [DOI] [PubMed] [Google Scholar]
- Frenkel D, Maron R, Burt DS, Weiner HL. Nasal vaccination with a proteosome-based adjuvant and glatiramer acetate clears beta-amyloid in a mouse model of Alzheimer disease. J Clin Invest. 2005;115:2423–2433. doi: 10.1172/JCI23241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman AD. C/EBPalpha induces PU.1 and interacts with AP-1 and NF-kappaB to regulate myeloid development. Blood Cells Mol Dis. 2007;39:340–343. doi: 10.1016/j.bcmd.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmire LX, Shen Z, Briggs S, Yeo G, Subramaniam S, Glass C. Regulatory network of microRNAs in RAW 264.7 macrophage cells. Conf Proc IEEE Eng Med Biol Soc. 2010;2010:6198–6201. doi: 10.1109/IEMBS.2010.5627742. [DOI] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P, Levraud JP. Imaging early macrophage differentiation, migration, and behaviors in live zebrafish embryos. Methods Mol Med. 2005;105:199–214. doi: 10.1385/1-59259-826-9:199. [DOI] [PubMed] [Google Scholar]
- Hoek RM, Ruuls SR, Murphy CA, Wright GJ, Goddard R, Zurawski SM, Blom B, Homola ME, Streit WJ, Brown MH, Barclay AN, Sedgwick JD. Down-regulation of the macrophage lineage through interaction with OX2 (CD200) Science. 2000;290:1768–1771. doi: 10.1126/science.290.5497.1768. [DOI] [PubMed] [Google Scholar]
- Junn E, Mouradian MM. MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol Ther. 2012;133:142–150. doi: 10.1016/j.pharmthera.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kizelsztein P, Ovadia H, Garbuzenko O, Sigal A, Barenholz Y. Pegylated nanoliposomes remote-loaded with the antioxidant tempamine ameliorate experimental autoimmune encephalomyelitis. J Neuroimmunol. 2009;213:20–25. doi: 10.1016/j.jneuroim.2009.05.019. [DOI] [PubMed] [Google Scholar]
- Kutter C, Svoboda P. miRNA, siRNA, piRNA: Knowns of the unknown. RNA Biol. 2008;5:181–188. doi: 10.4161/rna.7227. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Liu Y, Mounkes LC, Liggitt HD, Brown CS, Solodin I, Heath TD, Debs RJ. Factors influencing the efficiency of cationic liposome-mediated intravenous gene delivery. Nat Biotechnol. 1997;15:167–173. doi: 10.1038/nbt0297-167. [DOI] [PubMed] [Google Scholar]
- Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-{beta} J Biol Chem. 2010;285:41328–41336. doi: 10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovat F, Valeri N, Croce CM. MicroRNAs in the pathogenesis of cancer. Semin Oncol. 2011;38:724–733. doi: 10.1053/j.seminoncol.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Lynch MA. The multifaceted profile of activated microglia. Mol Neurobiol. 2009;40:139–156. doi: 10.1007/s12035-009-8077-9. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Garlanda C, Locati M. Macrophage diversity and polarization in atherosclerosis: a question of balance. Arterioscler Thromb Vasc Biol. 2009;29:1419–1423. doi: 10.1161/ATVBAHA.108.180497. [DOI] [PubMed] [Google Scholar]
- Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor alpha1 (IL13Ralpha1) J Biol Chem. 2011;286:1786–1794. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Mishima T, Mizuguchi Y, Kawahigashi Y, Takizawa T. RT-PCR-based analysis of microRNA (miR-1 and -124) expression in mouse CNS. Brain Res. 2007;1131:37–43. doi: 10.1016/j.brainres.2006.11.035. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev ED, Maresz K, Tan Y, Dittel BN. CNS-derived interleukin-4 is essential for the regulation of autoimmune inflammation and induces a state of alternative activation in microglial cells. J Neurosci. 2007a;27:10714–10721. doi: 10.1523/JNEUROSCI.1922-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev ED, Novikova M, Maresz K, Shriver LP, Dittel BN. Development of a culture system that supports adult microglial cell proliferation and maintenance in the resting state. J Immunol Methods. 2005a;300:32–46. doi: 10.1016/j.jim.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Ponomarev ED, Shriver LP, Dittel BN. CD40 expression by microglial cells is required for their completion of a two-step activation process during central nervous system autoimmune inflammation. J Immunol. 2006;176:1402–1410. doi: 10.4049/jimmunol.176.3.1402. [DOI] [PubMed] [Google Scholar]
- Ponomarev ED, Shriver LP, Maresz K, Dittel BN. Microglial cell activation and proliferation precedes the onset of CNS autoimmunity. J Neurosci Res. 2005b;81:374–389. doi: 10.1002/jnr.20488. [DOI] [PubMed] [Google Scholar]
- Ponomarev ED, Shriver LP, Maresz K, Pedras-Vasconcelos J, Verthelyi D, Dittel BN. GM-CSF production by autoreactive T cells is required for the activation of microglial cells and the onset of experimental autoimmune encephalomyelitis. J Immunol. 2007b;178:39–48. doi: 10.4049/jimmunol.178.1.39. [DOI] [PubMed] [Google Scholar]
- Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med. 2011a;17:64–70. doi: 10.1038/nm.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev ED, Veremeyko T, Barteneva NS. Visualization and quantitation of the expression of microRNAs and their target genes in neuroblastoma single cells using imaging cytometry. BMC Res Notes. 2011b;4:517. doi: 10.1186/1756-0500-4-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AE, Liang HE, Sullivan BM, Reinhardt RL, Eisley CJ, Erle DJ, Locksley RM. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc Natl Acad Sci U S A. 2010;107:11489–11494. doi: 10.1073/pnas.1003988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz M, Priller J, Sisodia SS, Ransohoff RM. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat Neurosci. 2011;14:1227–1235. doi: 10.1038/nn.2923. [DOI] [PubMed] [Google Scholar]
- Ransohoff RM, Cardona AE. The myeloid cells of the central nervous system parenchyma. Nature. 2010;468:253–262. doi: 10.1038/nature09615. [DOI] [PubMed] [Google Scholar]
- Rom S, Rom I, Passiatore G, Pacifici M, Radhakrishnan S, Del Valle L, Pina-Oviedo S, Khalili K, Eletto D, Peruzzi F. CCL8/MCP-2 is a target for mir-146a in HIV-1-infected human microglial cells. FASEB J. 2010;24:2292–2300. doi: 10.1096/fj.09-143503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Ballarino M, Sorrentino A, Sthandier O, De Angelis FG, Marchioni M, Masella B, Guarini A, Fatica A, Peschle C, Bozzoni I. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci U S A. 2007;104:19849–19854. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, Nerlov C. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci U S A. 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo K, Glass CK. Microglial cell origin and phenotypes in health and disease. Nat Rev Immunol. 2011;11:775–787. doi: 10.1038/nri3086. [DOI] [PubMed] [Google Scholar]
- Schwartz M. “Tissue-repairing” blood-derived macrophages are essential for healing of the injured spinal cord: from skin-activated macrophages to infiltrating blood-derived cells? Brain Behav Immun. 2010;24:1054–1057. doi: 10.1016/j.bbi.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Sonntag KC, Woo TU, Krichevsky AM. Converging miRNA functions in diverse brain disorders: A case for miR-124 and miR-126. Exp Neurol. 2011 doi: 10.1016/j.expneurol.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin SP, Hoyt RF, Jr, Blunt DG, McNelly NA. Macrophage development: II. Early ontogeny of macrophage populations in brain, liver, and lungs of rat embryos as revealed by a lectin marker. Anat Rec. 1992;232:527–550. doi: 10.1002/ar.1092320410. [DOI] [PubMed] [Google Scholar]
- Soulet D, Rivest S. Bone-marrow-derived microglia: myth or reality? Curr Opin Pharmacol. 2008;8:508–518. doi: 10.1016/j.coph.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EH, Hoekstra K, van Rooijen N, Dijkstra CD, Owens T. Immune invasion of the central nervous system parenchyma and experimental allergic encephalomyelitis, but not leukocyte extravasation from blood, are prevented in macrophage-depleted mice. J Immunol. 1998;161:3767–3775. [PubMed] [Google Scholar]
- Tremblay ME, Stevens B, Sierra A, Wake H, Bessis A, Nimmerjahn A. The Role of Microglia in the Healthy Brain. J Neurosci. 2011;31:16064–16069. doi: 10.1523/JNEUROSCI.4158-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton M, Saura J, Young D, MacGibbon G, Hansen W, Lawlor P, Sirimanne E, Gluckman P, Dragunow M. CCAAT-enhancer binding protein alpha is expressed in activated microglial cells after brain injury. Brain Res Mol Brain Res. 1998;61:11–22. doi: 10.1016/s0169-328x(98)00169-7. [DOI] [PubMed] [Google Scholar]
- Wang P, Hou J, Lin L, Wang C, Liu X, Li D, Ma F, Wang Z, Cao X. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol. 2010;185:6226–6233. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- Yang M, Chen J, Su F, Yu B, Lin L, Liu Y, Huang JD, Song E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeamans C, Wang D, Paz-Priel I, Torbett BE, Tenen DG, Friedman AD. C/EBPalpha binds and activates the PU.1 distal enhancer to induce monocyte lineage commitment. Blood. 2007;110:3136–3142. doi: 10.1182/blood-2007-03-080291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QY, Liu Q, Chen JX, Lan K, Ge BX. MicroRNA-101 targets MAPK phosphatase-1 to regulate the activation of MAPKs in macrophages. J Immunol. 2010;185:7435–7442. doi: 10.4049/jimmunol.1000798. [DOI] [PubMed] [Google Scholar]