Abstract
Purpose
To develop and evaluate image registration methodology for automated re-identification of tumor-suspicious foci from pre-procedural MR exams during MR-guided transperineal prostate core biopsy.
Materials and Methods
A hierarchical approach for automated registration between planning and intra-procedural T2-weighted prostate MRI was developed and evaluated on the images acquired during 10 consecutive MR-guided biopsies. Registration accuracy was quantified at image-based landmarks and by evaluating spatial overlap for the manually segmented prostate and sub-structures. Registration reliability was evaluated by simulating initial mis-registration and analyzing the convergence behavior. Registration precision was characterized at the planned biopsy targets.
Results
The total computation time was compatible with a clinical setting, being at most 2 minutes. Deformable registration led to a significant improvement in spatial overlap of the prostate and peripheral zone contours compared to both rigid and affine registration. Average in-slice landmark registration error was 1.3±0.5 mm. Experiments simulating initial mis-registration resulted in an estimated average capture range of 6 mm and an average in-slice registration precision of ±0.3 mm.
Conclusion
Our registration approach requires minimum user interaction and is compatible with the time constraints of our interventional clinical workflow. The initial evaluation shows acceptable accuracy, reliability and consistency of the method.
Keywords: Prostate cancer, image-guided interventions, prostate biopsy, image registration, non-rigid registration, mutual information, performance characterization
INTRODUCTION
Magnetic Resonance Imaging (MRI) plays an important role in the detection and staging of prostate cancer (PCa). There is increasing evidence of significantly improved detection (1,2) and characterization of PCa (3) through imaging at 3 Tesla (3T) and the use of a combination of various multi-parametric MRI (mpMRI) techniques in addition to the T2-weighted (T2W) MRI. Such mpMRI techniques now include diffusion-weighted imaging (DWI), dynamic contrast-enhanced MRI (DCE) and magnetic resonance spectroscopy imaging (MRSI). The serum prostate-specific antigen (PSA) is a biomarker commonly used for prostate cancer screening, but unfortunately it has high rate of false-positives, and can lead to over-diagnosis and over-treatment (4). Recent publications have raised concerns over its use in screening (5). As always, histological analysis of core biopsy sample is required for the accurate diagnosis of PCa and subsequent Gleason grading. Traditional diagnostic biopsy approaches use sextant/octant systematic sampling using an 18-gauge core biopsy needle, under the guidance of transrectal ultrasound (TRUS). This approach is limited and can miss cancer in up to 30% of cases (6). Targeted biopsy using ultrasound imaging alone is not reliable, since up to 50% of PCa lesions are isoechoic to non-cancerous tissue (7). Hence, there is a population of men with rising PSA levels in spite of repeated negative sextant-based TRUS-guided biopsies. It is difficult to correctly diagnose such patients, which can lead to under-treatment and reduced survival, or over-treatment and unnecessary negative effects (stress and anxiety) on the patient quality of life.
This unsatisfactory situation has led to the development of improved image guided biopsy approaches that use mpMRI for detecting suspicious lesions or tumor suspected regions (TSR) and targeting to improve the accuracy of prostate cancer detection. Unfortunately, both the acquisition and post-processing of mpMRI are time-consuming and complex, requiring specialized expertise and equipment. As a result, targeted prostate biopsies typically use mpMRI processed in advance of the procedure to identify TSRs (8,9). At the time of biopsy, a fast, but lower resolution imaging modality can be used to visualize the prostate anatomy and biopsy needle. Both MRI (8-11) and TRUS (12,13) have been explored as imaging methods for such intra-procedural visualization. Accurate targeting of TSRs requires spatial correlation of the pre-procedural MRI with the intra-procedural anatomical imaging, which can be particularly complicated when an endorectal coil (ERC) is used during the pre-procedural imaging, but not during the biopsy procedure (14,15). This is due to the inevitable deformations induced by the inflated coil, which can be more extreme than standard TRUS deformation. To the best of our knowledge, even the most recent studies use manual alignment or visual assessment to recover the locations of the suspected areas during MR-guided biopsy (8,9). Such operator-centric approaches can be time-consuming and require specialized training in interpretation of prostate MRI, leading to increased complexity of the procedure, inter-rater variability and difficulties in accounting for prostate deformation. The goal of this study was to develop and evaluate a registration pipeline for automated re-identification of tumor-suspicious foci during MR-guided transperineal prostate core biopsy.
MATERIALS AND METHODS
Patient Population and MRI Acquisition
This study was performed using MR images collected from 10 consecutive patients who underwent MR-guided transperineal prostate biopsy as part of an on-going prospective clinical trial. All patients gave informed consent. The study was HIPPA compliant and approved by the local institutional review board. The choice of the transperineal over the more common transrectal approach was based on technical advantages, ease of access and the clinical considerations that include lower risk of infection, improved core coverage of the peripheral zone and better tolerance by the patients. The enrollment criteria included men with high clinical suspicion of prostate cancer (palpable abnormality and/or rising PSA), with at least 2 negative TRUS biopsies or inability to access the rectum by TRUS usually due to prior rectal surgery such as an abdomino-perineal resection.
The pre-procedural mpMRI imaging protocol included a T1-weighted (T1W), T2W, DWI and DCE series acquired with the patient in the supine position on a 3T GE SignaHdx scanner (GE Medical Systems, Milwakee, WI). In 8 out of 10 patients a combination of ERC (MEDRAD, Warrendale, PA) with body and surface coils was used. In 2 patients the ERC was not used due to the patient’s rectal condition. The T2W series being registered was acquired using a 2D fast recovery fast spin echo (FRFSE) sequence with the following parameters: repetition time (TR) 3083 ms, echo time (TE) 106 ms, field of view (FOV) 160 mm, matrix 384×224, slice thickness 3 mm, pixel spacing 0.3 mm. The individual MR series were reviewed prior to the procedure by a radiologist to identify the TSR foci.
The MR-guided biopsy procedure was performed in a 70 cm bore 3T Siemens Magnetom Verio scanner (Siemens Medical Systems, Erlangen, Germany). The patient was placed in the lithotomy position using custom MR-compatible stirrups. T2W axial images of the prostate gland were obtained using a combination of Body Matrix (anteriorly placed) and Spine surface (posteriorly located) coil elements (Siemens Medical Systems, Erlangen, Germany), using turbo spin echo (TSE) sequence (TR/TE 5250/100 ms, FOV 140 mm, matrix 320×224, slice thickness 3 mm, pixel spacing 0.4 mm).
Registration Methodology
All image preprocessing and registration steps were performed within the 3D Slicer open source environment1. Approximate contours of the prostate gland were manually prepared in each slice of both the diagnostic and intra-procedural T2W images. Signal intensity inhomogeneity was corrected using the N4ITK method (16). The initial transformation was computed as the offset in the coordinates of the segmented gland centroids refined by a sparse search of the minimum value of the negated mutual information (MI) metric in the small parameter range of the initial transformation. The resulting transformation was used to initialize the automatic hierarchical registration. At each level of this hierarchy, we used an increasingly flexible transformation model that was initialized with the final transformation computed at the previous step, starting from 6 degrees of freedom (DOF) rigid transformation up to free form B-splines transformation (17) parameterized by 3×3×3 control points uniformly spaced over the bounding box that covers the union of the binary masks of the prostate glands after the initial alignment.
The MI metric was calculated over the region defined by the binary masks of the prostate glands. Optimization was achieved using the gradient descent optimizer for all the transformation hierarchy levels except the B-spline transformation. The B-spline registration used Limited memory Broyden Fletcher Goldfarb Shannon optimization with simple bounds (L-BFGS-B) (18). The registration approach was implemented as a module within 3D Slicer software by introducing application-specific modifications described above to the 3D Slicer BRAINSFit module (19). Identical parameters were used in all of the experiments.
Performance Evaluation Methodology
Accuracy
Our accuracy evaluation included two components: 1) Landmark Registration Error (LRE) estimated at the image features that can be identified in both images and 2) overlap of the prostate gland and its sub-regions between the intra-procedural and registered pre-procedural images. Image landmarks were defined by the naturally occurring image-specific features of the prostate (e.g., calcifications and BPH nodules). LRE after rigid, affine and B-spline steps of the registration was calculated as the distance between the corresponding points after applying the respective transformations recovered by the registration.
Spatial overlap between the contoured structures was evaluated using Dice similarity coefficient (DSC) (20). The total gland (TG), central gland (CG) and peripheral zone (PZ) of the prostate were contoured in the pre- and intra-procedural T2W MRI by two abdominal radiologists (F.F. and K.T.) each with over 10 years of experience in prostate MRI interpretation. The masks defined for the pre-procedural scans were resampled to the voxel grid of the intra-procedural images using the transformations produced by the rigid, affine and B-spline registration steps. DSC between the registered pre-procedural and intra-procedural masks was calculated, and the average DSC value over two raters was reported for TG, CG and PZ. Two-sided paired t-test on the logit-transformed DSC values (20) was used to establish statistical significance.
Robustness and precision
Experimental estimation of the registration capture range was performed by perturbing the initialization transformation. A series of random perturbation vectors was generated by uniformly sampling the vector direction over unit sphere, and uniform sampling of the magnitude between 0 and 10 mm. A total of 500 perturbed initial transformations were generated for each of the 10 registration cases. The registration procedure was initialized with each of these transformations. Successful registration experiments were defined as those that converged and resulted in high overlap between the registered glands as measured by DSC on a per-case basis. The B-spline component of registration was deemed successful when the maximum LRE was less than the largest voxel dimension (3 mm for our data). Robustness of the registration to the perturbations in the initial orientation was evaluated in a separate experiment by introducing a random perturbations of up to ±10° in yaw, roll and pitch (200 experiments per rotation angle for each of the analyzed datasets).
The metrics used to summarize registration reliability included success rate (proportion of the successful registration experiments), capture range (maximum magnitude of the initial misalignment that did not result in registration failure), and non-convergence rate (proportion of the experiments where optimization did not converge). Registration precision was estimated for the locations of the warped biopsy targets. Registration consistency was summarized with the maximum Inter-quartile Range (IQR) for the X and Y coordinates of these mapped biopsy targets. We were mostly concerned with the axial in-slice consistency of registration (X and Y coordinates of the mapped biopsy location), since the typical biopsy core is approximately 10 mm in length.
RESULTS
Feasibility and accuracy
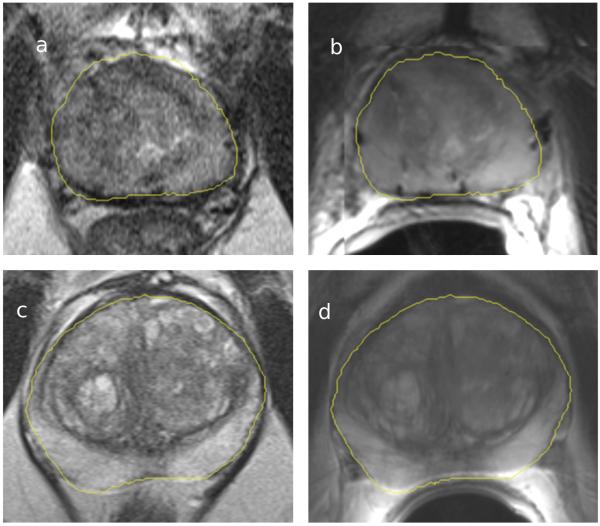
We registered data sets collected during 10 consecutive MR-guided prostate biopsy procedures. Manual contouring of the prostate gland used for registration initialization was prepared within 2-3 minutes using the Editor module of 3D Slicer. Registration convergence was achieved in all 10 cases. The results were visually inspected by the interventional radiologist performing the MRI-guided prostate biopsy (K.T., over 10 years experience in interventional MRI) and found to be satisfactory based on the alignment of the image features after the registration, see Fig.1. The average computation time was 70 sec (range 42 to 123 sec). The B-spline registration step accounted on average for 42 (maximum 88) sec. During the first 5 cases, registration was done in a “shadow” mode (registration results were not used for biopsy guidance) for the initial evaluation of the approach. During the last 5 cases, registration was completed and used during the course of the procedure.
Figure 1.
Registration results for patients 2 (top row) and 5 (bottom row). Images on the left (a and c) show an intra-procedural axial T2W image, images on the right (b and d) show the same slice of the registered pre-procedure T2W MRI. Yellow contour corresponds to the outline of the prostate gland segmented in the diagnostic image and warped to the intra-procedural image space by means of registration. Images for Patient 2 show brachytherapy seeds artifacts, as the patient previously underwent radiation therapy.
Quantitative evaluation of the registration accuracy used a total of 34 landmarks identified for the registered image pairs (between 3 and 4 landmarks per patient). Mean±SD (maximum) LRE over all the landmarks in the 10 patients was 1.6±0.8 (4.6) mm, 1.4±0.7 (2.9) mm and 1.3±0.5 (2.3) mm after the rigid, affine and B-spline registration stages, respectively. Statistically significant reduction in the LRE was observed between the rigid and B-spline registration results (p<0.05). Evaluation of the improvement in the overlap for the manually contoured prostate regions (TG, CG and PZ) augmented our LRE-based analysis. Statistically significant improvement in DSC for TG and PZ was observed (rigid vs affine and affine vs B-spline) based on two-sided paired t-test (p<0.05), see Table 1.
Table 1.
Summary statistics (mean and standard deviation, SD) of the label overlap (Dice similarity coefficient, DSC) between the segmentations of the total gland (TG), peripheral zone (PZ) and central gland (CG) in the intra-procedural image, and the segmentations in the diagnostic image resampled to the fixed image space following registration. Inter-rater agreement is defined as the average DSC between the masks produced by the two raters for the same structure in the pre- and intra-procedural images.
| Mean (SD) | ||||
|---|---|---|---|---|
| Structure of interest |
Mean (SD) inter-rater agreement |
Rigid | Affine (* p<0.05 relative to Rigid) |
B-spline (* p<0.05 relative to Affine) |
| TG | 0.91 (0.03) | 0.85 (0.04) | 0.86 (0.04) * | 0.88 (0.03) * |
| CG | 0.86 (0.04) | 0.83 (0.05) | 0.84 (0.05) | 0.85 (0.04) |
| PZ | 0.80 (0.03) | 0.67 (0.06) | 0.71 (0.06) * | 0.74 (0.05) * |
Robustness and precision
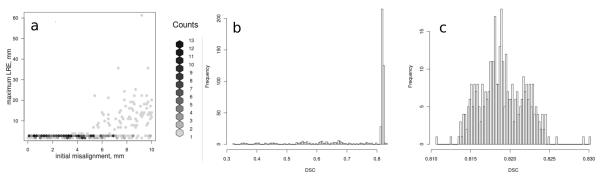
A total of 500 randomly perturbed initial transformation configurations were generated and applied to each of the registration cases resulting in the total of 5000 registration experiments over 10 patients. Rigid and affine steps of the registration hierarchy converged successfully in all of the registration experiments. Non-convergence rate of the B-spline registration component across the registration experiments for all the patients was at most 4%. Representative plots that summarize the registration results in terms of LRE and DSC as a function of the initial misalignment magnitude for one of the patients are shown in Fig. 2. In all of the cases we observed that most of the registration experiments resulted in the DSC values for the total gland grouped around some case-specific maximum value, which was selected to separate successful instances of affine registration. Capture range varied from 3.4 to 8.8 mm. All of the experiments evaluating the robustness of the method to rotational perturbations resulted in successful registration. Between 2 and 5 suspected cancer locations were selected for each of the patients (total of 36 targets) during biopsy planning. Maximum in-slice IQRs calculated at these biopsy locations for each patient were at most 2.6 mm (see Table 3).
Figure 2.
Summary of the LRE and DSC for the affine registration results with various values of the initial misalignment for the registration case with the smallest capture range (patient 2, n=500 registration experiments). (a) Binned histogram of the maximum LRE as a function of the initial misalignment magnitude. As the initial misalignment magnitude exceeds 4 mm, it becomes more likely for the registration to converge to a local minimum. (b) Histogram of the DSC values over the 500 experiments. Registration success was concluded when DSC exceeded 0.8 so that the results corresponding to the local minima (n=129) are discarded. (c) Histogram of the DSC values for the registration experiments results deemed successful.
Table 3.
Summary of the registration precision for the registered targets. The inter-quartile range (IQR) of the transverse slice coordinates was computed for each of the registered biopsy targets.
| Patient | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Number of biopsy targets | 2 | 3 | 3 | 4 | 5 | 4 | 4 | 3 | 5 | 3 |
| Maximum IQR, mm | 1.5 | 2.6 | 1.3 | 2.6 | 2.2 | 0.9 | 0.4 | 0.4 | 0.9 | 0.4 |
DISCUSSION
This study of 3D image data sets demonstrates that a fast non-rigid intra-procedural registration leads to statistically significant improvement in the overlap for both the total gland and peripheral zone of the prostate, as compared to rigid/affine registration. This observation is consistent with the previously published results (20) performed from 1.5T to 0.5T exams. We also observed statistically significant reduction of the LRE between the rigid and B-spline registration, with deformable registration reducing the LRE by more than 2 mm in some of the cases. Our experiments in estimating capture range of the registration led us to conclude that on average the registration method can tolerate translational variability in the initialization of 6 mm in magnitude and perturbations in rotation of 10°. Based on our simulations, over- or under-segmenting the prostate gland in our patient population by up to 3 slices could lead to a shift of the center of mass at most by 1.6 mm, which is within the capture range for all of the patients. The registration results are consistent, as the perturbations of the initialization do not lead to large variation in the locations of the mapped biopsy targets.
A commonly used approach to prostate MRI registration relies on finite element method (FEM) (20,21) instead of the image intensities. Since the FEM-based registration approaches are driven by the displacements of organ surfaces, intensity-based methods may enable more accurate registration by aligning features inside the gland, since each internal structure contributes to the calculation of the image similarity metric. FEM-based methods require accurate parameterization of the biomechanical properties and boundary conditions. Existing studies rely on population-averaged or experimental estimation of the prostate material properties (20,21). Benign and malignant neoplasms common in the prostates of such patients, as well as prior radiation treatment, can cause high variability in tissue properties, leading to potential errors (22). Finally, segmentation and matching of the segmentation surfaces introduces another source of error (23).
Our registration approach relies on maximization of mutual information similarity metric to recover the alignment between the two MR studies. In the past, mutual information has been successfully applied to a variety of image registration problems, including registration of prostate MRI (24,25). Unlike the earlier studies at 1.5T and 0.5T of image-based registration for prostate MRI, this current study was based on imaging obtained using 3T magnets from 2 different vendors, was applied during the biopsy to assist in needle guidance during the procedure, and paid attention to several aspects that are important for interventional applications, such as robustness, consistency and precision.
The presented registration workflow has several limitations. Currently, manual definition of the approximate prostate gland contour is required, which can potentially be alleviated by using a robust automated segmentation technique. The computation time of ~2 minutes is certainly more than acceptable in our clinical workflows but may not be suitable for all interventional applications. The processing time may be further reduced by using a parallelized implementation, as all the results we reported used single CPU for the computation.
The results of this early study of the proposed registration pipeline are very promising. Compared to the approaches suggested previously for integrating mpMRI into the process of target selection during MRI-guided biopsy (8,9), our registration-based methodology is highly automated, can be applied by a non-clinical operator and accounts for prostate deformation. We believe the software technology we described can be easily adopted and evaluated by the interested research groups involved in MRI-guided interventions, since it does not require any specialized hardware or commercial components, and is based on a well-documented portable platform of 3D Slicer and Insight Toolkit (26,27). Following the principles of open science (28), we are making the software and datasets used in this study publicly available2,3.
In conclusion, we presented an automated approach to non-rigid registration of 3T prostate MRI that facilitates the use of diagnostic images for re-identification of TSRs and targeting needle insertions during 3T MRI-guided prostate biopsy. Compared to manual approaches, our technology helps reduce the time and expertise required for mapping biopsy targets, while increasing reproducibility and accuracy. Our evaluation of reliability and consistency of the registration pipeline showed that it is tolerant to initialization errors, and provides mapping of the suspicious foci with the in-plane precision of one pixel on average.
Table 2.
Summary of the registration convergence and reliability for the affine and B-spline registration stages. Success rate corresponds to the portion of the experiments that led to successful registration based on Dice similarity coefficient (DSC) and Landmark registration error (LRE) measures. 95% capture range corresponds to the maximum initial mis-alignment that led to successful completion of 95% of the registration experiments.
| Patient | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Affine registration | ||||||||||
| Success rate, % | 98.2 | 74.2 | 68.0 | 93.0 | 85.4 | 87.6 | 92.8 | 93.4 | 84.8 | 79.8 |
| Capture range, mm | 8.8 | 3.4 | 4.1 | 7.5 | 5.6 | 6.3 | 6.9 | 6.8 | 4.7 | 5.7 |
| 95% capture range, mm | 10.0 | 5.5 | 5.5 | 9.5 | 7.5 | 8.0 | 9.5 | 9.0 | 8.0 | 7.5 |
| B-spline registration | ||||||||||
| Non-convergence rate, % | 1.0 | 3.8 | 4.0 | 2.0 | 4.0 | 1.0 | 2.0 | 3.8 | 2.2 | 0.4 |
| Overall success rate, % | 97.2 | 70.4 | 64.0 | 91.0 | 81.4 | 86.6 | 90.8 | 89.6 | 82.2 | 79.4 |
Acknowledgments
Grant support: R01 CA111288, U54 EB005149, P41 RR019703, P41 RR013218, P01 CA067165 and U01 CA151261. This research was supported in part by the National Science Foundation through TeraGrid resources provided by NCSA under grant number TG-ASC090061.
Footnotes
MR-guided Prostate Biopsy Pre- and Intra-procedural MRI Registration Evaluation Dataset: http://www.na-mic.org/publications/item/view/2147
Image Registration for MRg Prostate Biopsy, software usage instructions: http://wiki.namic.org/Wiki/index.php/Image_Registration_for_MRg_Prostate_Biopsy
REFERENCES
- 1.Langer DL, van der Kwast TH, Evans AJ, Trachtenberg J, Wilson BC, Haider MA. Prostate cancer detection with multi-parametric MRI: logistic regression analysis of quantitative T2, diffusion-weighted imaging, and dynamic contrast-enhanced MRI. Journal of magnetic resonance imaging. 2009;30:327–334. doi: 10.1002/jmri.21824. [DOI] [PubMed] [Google Scholar]
- 2.Turkbey B, Pinto PA, Mani H, et al. Prostate cancer: value of multiparametric MR imaging at 3 T for detection--histopathologic correlation. Radiology. 2010;255:89–99. doi: 10.1148/radiol.09090475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hambrock T, Somford DM, Huisman HJ, et al. Relationship between Apparent Diffusion Coefficients at 3.0-T MR Imaging and Gleason Grade in Peripheral Zone Prostate Cancer. Radiology. 2011;259:453–461. doi: 10.1148/radiol.11091409. [DOI] [PubMed] [Google Scholar]
- 4.Schröder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. The New England journal of medicine. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 5.Chou R, Croswell J, Dana T, et al. [Retrieved December 30, 2011];Screening for prostate cancer: a review of the evidence for the U.S. preventive services task force. 2011 from http://www.uspreventiveservicestaskforce.org/uspstf12/prostate/prostateart.htm.
- 6.Patel U. TRUS and prostate biopsy: current status. Prostate cancer and prostatic diseases. 2004;7:208–210. doi: 10.1038/sj.pcan.4500728. [DOI] [PubMed] [Google Scholar]
- 7.Heijmink SWTPJ, Fütterer JJ, Strum SS, et al. State-of-the-art uroradiologic imaging in the diagnosis of prostate cancer. Acta oncologica. 2011;50(Suppl 1):25–38. doi: 10.3109/0284186X.2010.578369. [DOI] [PubMed] [Google Scholar]
- 8.Franiel T, Stephan C, Erbersdobler A, et al. Areas Suspicious for Prostate Cancer: MR-guided Biopsy in Patients with at Least One Transrectal US-guided Biopsy with a Negative Finding--Multiparametric MR Imaging for Detection and Biopsy Planning. Radiology. 2011;259:162–172. doi: 10.1148/radiol.10101251. [DOI] [PubMed] [Google Scholar]
- 9.Hambrock T, Fütterer JJ, Huisman HJ, et al. Thirty-two-channel coil 3T magnetic resonance-guided biopsies of prostate tumor suspicious regions identified on multimodality 3T magnetic resonance imaging: technique and feasibility. Investigative radiology. 2008;43:686–694. doi: 10.1097/RLI.0b013e31817d0506. [DOI] [PubMed] [Google Scholar]
- 10.Hata N, Jinzaki M, Kacher D, et al. MR imaging-guided prostate biopsy with surgical navigation software: device validation and feasibility. Radiology. 2001;220:263–268. doi: 10.1148/radiology.220.1.r01jl44263. [DOI] [PubMed] [Google Scholar]
- 11.Pondman KM, Fütterer JJ, ten Haken B, et al. MR-guided biopsy of the prostate: an overview of techniques and a systematic review. European urology. 2008;54:517–527. doi: 10.1016/j.eururo.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Xu S, Kruecker J, Turkbey B, et al. Real-time MRI-TRUS fusion for guidance of targeted prostate biopsies. Computer aided surgery : official journal of the International Society for Computer Aided Surgery. 2008;13:255–264. doi: 10.1080/10929080802364645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Natarajan S, Marks LS, Margolis DJA, et al. Clinical application of a 3D ultrasound-guided prostate biopsy system. Urologic oncology. 2011;29:334–342. doi: 10.1016/j.urolonc.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heijmink SWTPJ, Fütterer JJ, Hambrock T, et al. Prostate cancer: body-array versus endorectal coil MR imaging at 3 T--comparison of image quality, localization, and staging performance. Radiology. 2007;244:184–195. doi: 10.1148/radiol.2441060425. [DOI] [PubMed] [Google Scholar]
- 15.Heijmink SWTPJ, Scheenen TWJ, van Lin ENJT, et al. Changes in prostate shape and volume and their implications for radiotherapy after introduction of endorectal balloon as determined by MRI at 3T. International journal of radiation oncology, biology, physics. 2009;73:1446–1453. doi: 10.1016/j.ijrobp.2008.06.1491. [DOI] [PubMed] [Google Scholar]
- 16.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: Improved N3 Bias Correction. IEEE transactions on medical imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rueckert D, Sonoda L, Hayes C, Hill D, Leach M, Hawkes D. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Transactions on medical imaging. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 18.Byrd RH, Lu P, Nocedal J, Zhu C. A Limited Memory Algorithm for Bound Constrained Optimization. SIAM Journal on Scientific Computing. 1995;16:1190. [Google Scholar]
- 19.Johnson HJ, Harris G, Williams K. BRAINSFit: Mutual Information Registrations of Whole-Brain 3D Images, Using the Insight Toolkit. Insight Journal. 2007 Jul-Dec; [Google Scholar]
- 20.Bharatha A, Hirose M, Hata N, et al. Evaluation of three-dimensional finite element-based deformable registration of pre- and intraoperative prostate imaging. Medical physics. 2001;28:2551–2560. doi: 10.1118/1.1414009. [DOI] [PubMed] [Google Scholar]
- 21.Hensel JM, Ménard C, Chung PWM, et al. Development of multiorgan finite element-based prostate deformation model enabling registration of endorectal coil magnetic resonance imaging for radiotherapy planning. International journal of radiation oncology, biology, physics. 2007;68:1522–1528. doi: 10.1016/j.ijrobp.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Chi Y, Liang J, Yan D. A material sensitivity study on the accuracy of deformable organ registration using linear biomechanical models. Medical physics. 2006;33:421–433. doi: 10.1118/1.2163838. [DOI] [PubMed] [Google Scholar]
- 23.Liang J, Yan D. Reducing uncertainties in volumetric image based deformable organ registration. Medical Physics. 2003;30:2116. doi: 10.1118/1.1587631. [DOI] [PubMed] [Google Scholar]
- 24.Wu X, Dibiase SJ, Gullapalli R, Yu CX. Deformable image registration for the use of magnetic resonance spectroscopy in prostate treatment planning. International journal of radiation oncology, biology, physics. 2004;58:1577–1583. doi: 10.1016/j.ijrobp.2003.09.072. [DOI] [PubMed] [Google Scholar]
- 25.Oguro S, Tokuda J, Elhawary H, et al. MRI signal intensity based B-spline nonrigid registration for pre- and intraoperative imaging during prostate brachytherapy. Journal of magnetic resonance imaging. 2009;30:1052–1058. doi: 10.1002/jmri.21955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pieper S, Lorensen B, Schroeder W, Kikinis R. The NA-MIC Kit: ITK, VTK, Pipelines, Grids and 3D Slicer as An Open Platform for the Medical Image Computing Community. Proc. of 3rd IEEE International Symposium on Medical Imaging.2006. pp. 698–701. [Google Scholar]
- 27.Ibanez L, Schroeder W. The ITK Software Guide 2.4. Kitware, Inc.; 2005. p. 804. [Google Scholar]
- 28.Ince DC, Hatton L, Graham-Cumming J. The case for open computer programs. Nature. 2012;482:485–488. doi: 10.1038/nature10836. [DOI] [PubMed] [Google Scholar]