Abstract
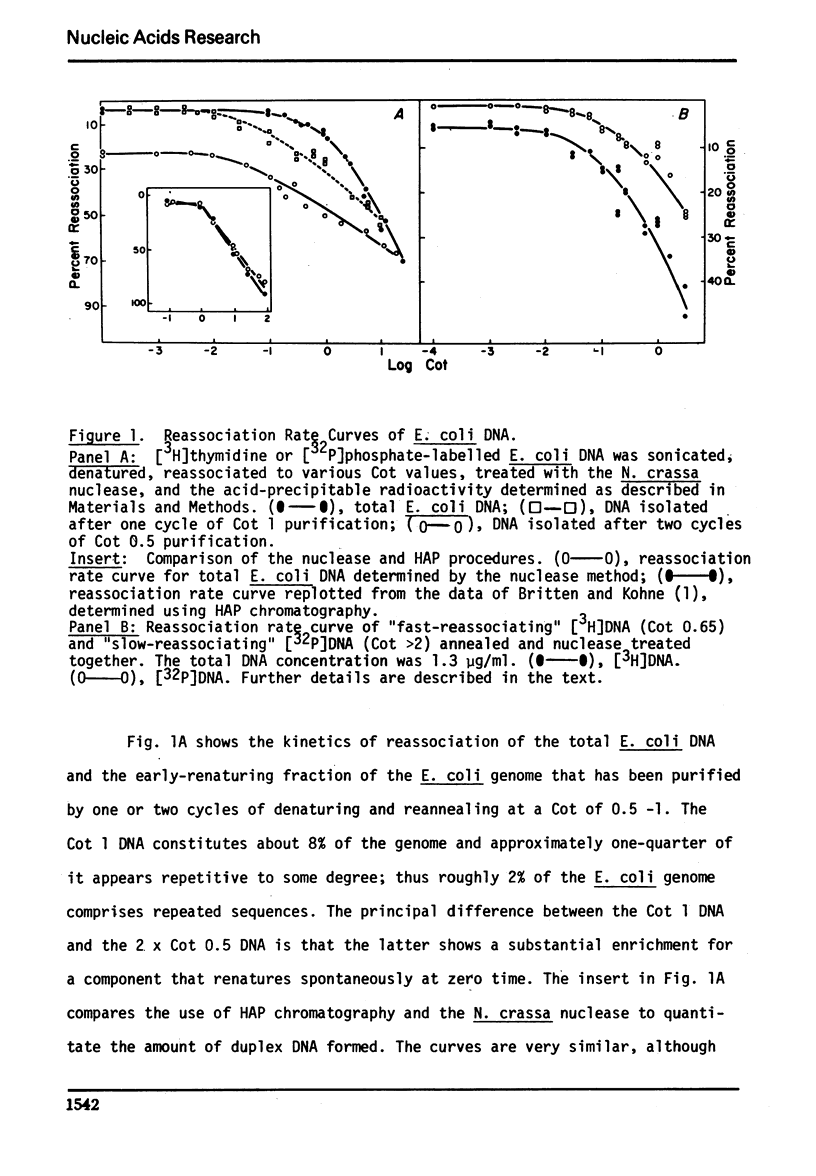
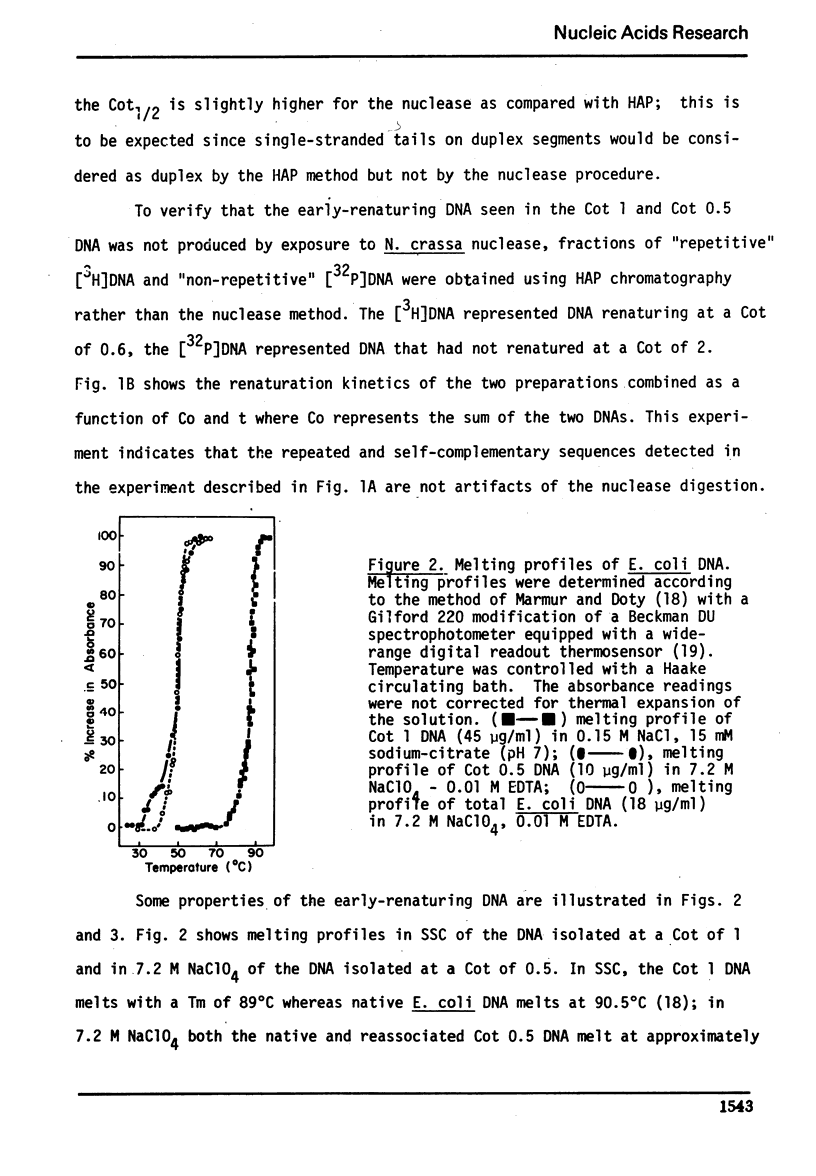
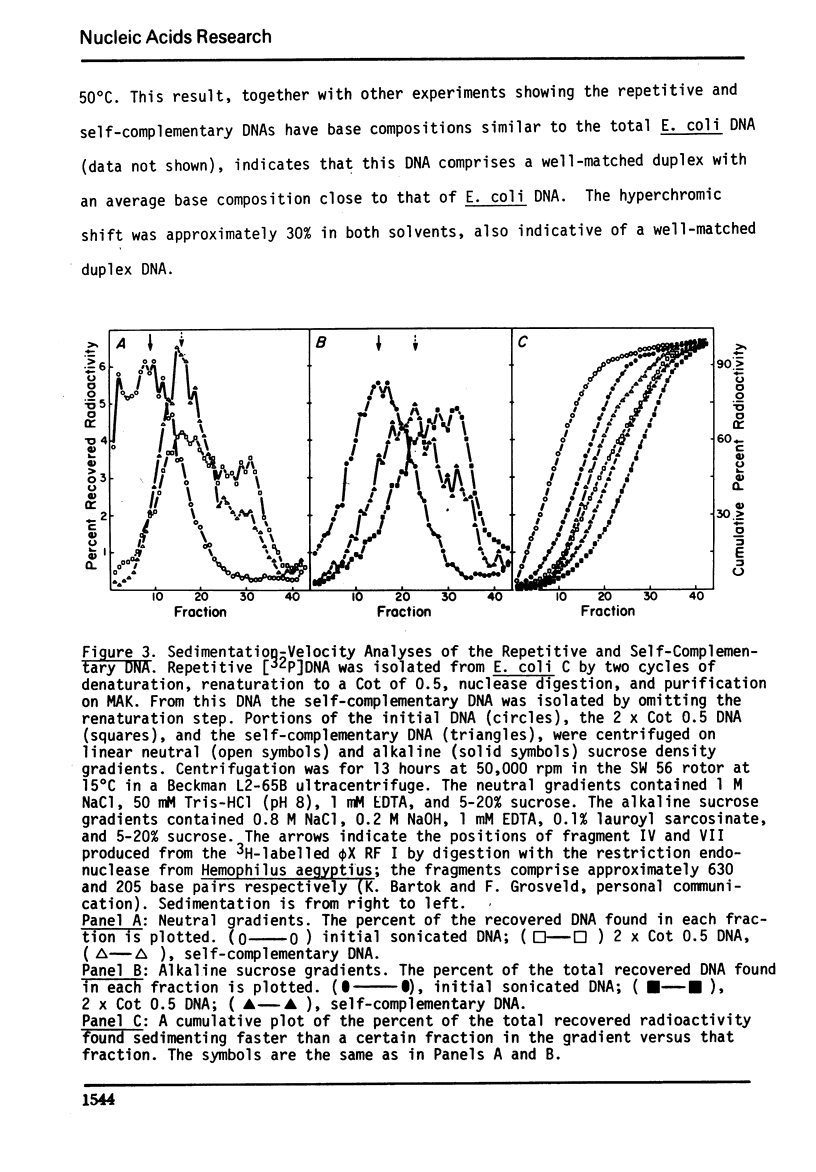
We have used the single-strand specific nuclease from Neurospora crassa and chromatography on methylated albumin-kieselguhr to purify and characterize repeated and self-complementary sequences from Escherichia coli DNA. Approximately 0.5% of the genome renatures spontaneously at zero time and another 2% renatures somewhat more rapidly than the total DNA. The early renaturing DNA has a base composition and a Tm similar to the total DNA and contains on the average 100 base pairs; the self-complementary DNA also has a base composition like E. coli but contains a mean of 170 base pairs. No evidence was obtained for the presence of a highly redundant sequence.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M., Doty P. Characterization of a naturally occurring, cross-linked fraction of DNA. 1. Nature of the cross-linkage. J Mol Biol. 1968 Mar 14;32(2):379–403. doi: 10.1016/0022-2836(68)90017-x. [DOI] [PubMed] [Google Scholar]
- Brahic M., Fraser M. J. Isolation and properties of a rapidly renaturing fraction of mammalian DNA. Biochim Biophys Acta. 1971 Jun 17;240(1):23–36. doi: 10.1016/0005-2787(71)90508-9. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Rosenfeld A., Hearst J. E. Characterization of the most rapidly renaturing sequences in mouse main-band DNA. J Mol Biol. 1973 Dec 15;81(3):299–325. doi: 10.1016/0022-2836(73)90143-5. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A theory of DNA replication. J Theor Biol. 1972 Mar;34(3):487–508. doi: 10.1016/0022-5193(72)90137-3. [DOI] [PubMed] [Google Scholar]
- Kato A. C., Bartok K., Fraser M. J., Denhardt D. T. Sensitivity of superhelical DNA to a single-strand specific endonuclease. Biochim Biophys Acta. 1973 Apr 21;308(7):68–78. doi: 10.1016/0005-2787(73)90123-8. [DOI] [PubMed] [Google Scholar]
- Kohne D. E. Evolution of higher-organism DNA. Q Rev Biophys. 1970 Aug;3(3):327–375. doi: 10.1017/s0033583500004765. [DOI] [PubMed] [Google Scholar]
- Kohne D. E. Isolation and characterization of bacterial ribosomal RNA cistrons. Biophys J. 1968 Oct;8(10):1104–1118. doi: 10.1016/S0006-3495(68)86542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Mace M. L., Jr, Clark R. W., Arrighi F. E. Isolation of repetitive DNA from two mammalian species using S1 nuclease. Biochem Biophys Res Commun. 1974 Jan;56(1):9–13. doi: 10.1016/s0006-291x(74)80308-6. [DOI] [PubMed] [Google Scholar]
- Rabin E. Z., Preiss B., Fraser M. J. A nuclease from Neurospora crassa conidia specific for single-stranded nucleic acids. Prep Biochem. 1971;1(4):283–307. doi: 10.1080/00327487108081946. [DOI] [PubMed] [Google Scholar]
- Rabin E. Z., Tenenhouse H., Fraser M. J. An exonuclease of Neurospora crassa specific for single-stranded nucleic acids. Biochim Biophys Acta. 1972 Jan 18;259(1):50–68. doi: 10.1016/0005-2787(72)90473-x. [DOI] [PubMed] [Google Scholar]
- Saedler H., Heiss B. Multiple copies of the insertion-DNA sequences IS1 and IS2 in the chromosome of E. coli K-12. Mol Gen Genet. 1973 May 9;122(3):267–277. doi: 10.1007/BF00278602. [DOI] [PubMed] [Google Scholar]
- Schekman R. W., Iwaya M., Bromstrup K., Denhardt D. T. The mechanism of replication of phi X174 single-stranded DNA. 3. An enzymic study of the structure of the replicative form II DNA. J Mol Biol. 1971 Apr 28;57(2):177–199. doi: 10.1016/0022-2836(71)90340-8. [DOI] [PubMed] [Google Scholar]
- Sobell H. M. The stereochemistry of actinomycin binding to DNA and its implications in molecular biology. Prog Nucleic Acid Res Mol Biol. 1973;13:153–190. doi: 10.1016/s0079-6603(08)60103-8. [DOI] [PubMed] [Google Scholar]
- Spadari S., Ritossa F. Clustered genes for ribosomal ribonucleic acids in Escherichia coli. J Mol Biol. 1970 Nov 14;53(3):357–367. doi: 10.1016/0022-2836(70)90071-9. [DOI] [PubMed] [Google Scholar]
- Sutton W. D., Walker P. M. Self-renaturing fractions in the separated strands of mouse satellite deoxyribonucleic acid. Biochem J. 1972 Jun;128(2):193–198. doi: 10.1042/bj1280193a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szybalski W., Kubinski H., Sheldrick P. Pyrimidine clusters on the transcribing strand of DNA and their possible role in the initiation of RNA synthesis. Cold Spring Harb Symp Quant Biol. 1966;31:123–127. doi: 10.1101/sqb.1966.031.01.019. [DOI] [PubMed] [Google Scholar]
- Wilson D. A., Thomas C. A., Jr Palindromes in chromosomes. J Mol Biol. 1974 Mar 25;84(1):115–138. doi: 10.1016/0022-2836(74)90216-2. [DOI] [PubMed] [Google Scholar]
- Yu M. T., Vermeulen C. W., Atwood K. C. Location of the genes for 16S and 23S ribosomal RNA in the genetic map of Escherichia coli. Proc Natl Acad Sci U S A. 1970 Sep;67(1):26–31. doi: 10.1073/pnas.67.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]


