Abstract
Interferon-stimulated gene 56 (ISG56) family members play important roles in blocking viral replication and regulating cellular functions, however, their underlying molecular mechanisms are largely unclear. Here, we present the crystal structure of ISG54, an ISG56 family protein with a novel RNA-binding structure. The structure shows that ISG54 monomers have 9 tetratricopeptide repeat-like motifs and associate to form domain-swapped dimers. The C-terminal part folds into a super-helical structure and has an extensively positively-charged nucleotide-binding channel on its inner surface. EMSA results show that ISG54 binds specifically to some RNAs, such as adenylate uridylate (AU)-rich RNAs, with or without 5′ triphosphorylation. Mutagenesis and functional studies show that this RNA-binding ability is important to its antiviral activity. Our results suggest a new mechanism underlying the antiviral activity of this interferon-inducible gene 56 family member.
Keywords: structure biology, ISG54
Introduction
Type I interferon (IFN) can block virus replication by stimulating the expression of various antiviral genes termed interferon-stimulated genes (ISG), including IFN-stimulated gene 56 family members (also known as IFN-induced tetratricopeptide repeats, IFITs)1,2,3,4,5,6. The human ISG56 family includes 4 members, ISG54 (IFIT2), ISG56 (IFIT1), ISG58 (IFIT5) and ISG60 (IFIT3), which have overlapping functions but distinct induction patterns7,8,9.
ISG56 family members have been predicted to be tetratricopeptide repeat (TPR)-containing proteins10. TPR-containing proteins are well known for their ability to mediate protein-protein interactions. The TPR motif is defined by the presence of 34 amino acids containing two anti-parallel helices, helix-A and helix-B11. Eight loosely conserved residues, W4, L7, G8, Y11, A20, F24, A27, and P32, at stringently conserved positions, mediate the interaction between helix-A and helix-B11,12. Tandem anti-parallel helix-turn-helix arrays generate a right-handed helical channel; helix-A is situated inside the channel while helix-B is on the outside of the channel, providing a complementary binding surface for other proteins13. The TPR motifs form a super helical structure and the amino acids in the channel often change considerably, determining the specificity of the protein for its protein substrates11. Many TPR-containing proteins have been shown to specifically recognize their partners and form complexes such as molecular chaperones, anaphase promoting complexes, and protein transport complexes14,15,16.
Studies investigating the mechanism of this family of proteins have yielded inconsistent results. Early results indicated that ISG54 and ISG56 can interact with eIF3e and eIF3c to inhibit translation10,17. In contrast, it was later found that ISG56 family proteins inhibit viruses by recognizing viral mRNA lacking 2′-O methylation18. Recently, it was reported that ISG56 inhibits viral replication through its 5′-triphosphate RNA-binding ability19. In addition, it has been suggested that ISG56 family proteins are involved in cellular processes such as IFN signaling20, inhibition of apoptosis and cell migration, and regulation of the production of cytokines in the inflammation process20,22,23, but the mechanisms are largely unknown. Detailed structural information is required to elucidate the molecular mechanisms underlying all these functions.
Here, we report the crystal structure of human ISG54. ISG54 monomers have 9 TPR-like motifs and form domain-swapped dimers. It has an exclusively positively-charged C-terminus for RNA binding. Mutation of the residues that are important for the RNA-binding ability of ISG54 disrupts its antiviral activity. We show that ISG54 can specifically bind to RNAs such as adenylate uridylate (AU)-rich RNAs in vitro. In addition, we found that ISG54 can bind some cellular RNA motifs such as AU-rich elements (AREs)24, suggesting the cellular function of this protein. Our results shed light on how ISG54 functions as an antiviral protein and carries out its cellular functions.
Results
Overall fold of ISG54
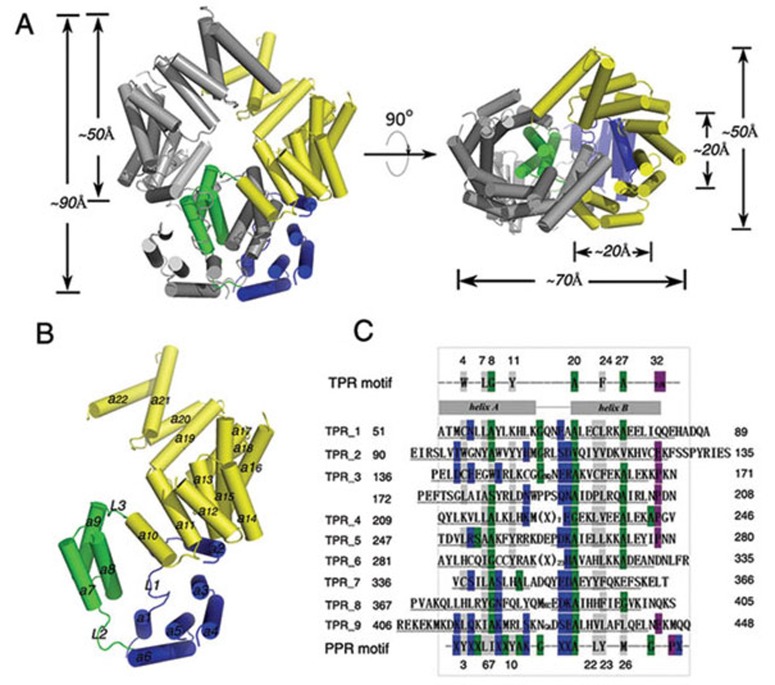
We determined the 2.8 Å crystal structure of human ISG54 by the single-wavelength anomalous dispersion (SAD) method using a selenomethionine (SeMet)-derivative protein crystal (Figure 1 and Table 1). Most residues of the protein, except for the first 5 residues at the N-terminus, the last 22 residues at the C-terminus and amino acids on several loops, fit well to the electron density map. ISG54 forms a dimer in our structure (Figure 1A). Each ISG54 monomer consists of 22 tandem anti-parallel α-helices, most of which are involved in the formation of TPR-like motifs. Helices 3 to 6, 7 and 8, and 11 to 22 can be recognized as a TPR-like motif (Figure 1B and 1C). However, ISG54, unlike some known TPR proteins, does not form a continuous extended super-helix as predicted by previous reports8,19, but rather is grouped into three regions; helices 1-6 form the N-terminal region, helices 7-9 the middle region and helices 10-22 the C-terminal region (Figure 1B). These three regions are linked by long loops, L2 (between helices 6 and 7) and L3 (between helices 9 and 10), and have few interactions between them. They form a triangle, leaving an empty space in the center. In the N-terminal region, helix 1 is short, covering only six residues (residues 10-15) and interacts with both helices 5 and 6 at the bottom of the protein structure. Many conserved hydrophobic residues in helix 1 and loop L1 (between helices 1 and 2), such as L9, L13, L16, H19, F20, W22 and L24, interact with residues Y59, Y105, V117 and V120 in helices 3-6 to stabilize the N-terminal conformation (Figure 2). Helix 2 is located on top of this region and functions as a bridge to interact with both the N-terminal and the C-terminal regions. The middle region is linked with the N- and C-terminal regions by loops L2 and L3 but is located at some distance from them. In this region, helices 7-9 interact with each other to form an anti-parallel helix bundle-like structure. The distance between helices 7 and 9 is greater than that between helices 7 and 8 or between helices 8 and 9. The C-terminal region covers helices 10-22, which are involved in forming a partial right-handed super-helix.
Figure 1.
Overall structure of ISG54. (A) Cartoon diagram of the ISG54 dimer, showing the front and top view. The dimensions of the protein, the C-terminus, and the RNA-binding channel are given. The N-terminal region (blue), domain-swapped region (green) and C-terminal region (yellow) are labeled in monomer A and monomer B is in gray. (B) Cartoon diagram of the ISG54 monomer A. The secondary structural elements are labeled. (C) Sequence alignment of ISG54 TPR-like motifs with typical TPR and PPR motifs. The 172-208 sequence forms a swapped TPR-like motif as described in the main text. The conserved residues in the standard TPR/PPR motifs are labeled in gray, blue, green and purple representing the hydrophobic, charged, small and terminal residues, respectively.
Table 1. Data collection and refinement statistics (SAD).
| Native | Se Derivative | |
|---|---|---|
| Data collection | ||
| Beam Line | SSRF_BL17U | KEK_BL5A |
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 80.02,95.21,155.90 | 79.65, 94.69, 153.34 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50-2.8(2.85-2.80) * | 50-3.0 (3.05-3.00) |
| Rmerge | 0.08 (0.49) | 0.10 (0.79) |
| I/σI | 27.7 (1.95) | 23.7 (4.17) |
| Completeness (%) | 98.9 (87.1) | 100 (100) |
| Redundancy | 7.0 (4.7) | 13.7 (13.9) |
| Refinement | ||
| Resolution (Å) | 40-2.8 | |
| No. reflections | 296917 | |
| Rwork / Rfree | 23.1% / 28.9% | |
| No. atoms | 6810 | |
| Protein | 6783 | |
| Ligand/ion | 0 | |
| Water | 27 | |
| B-factors | ||
| Protein | 68.67 | |
| Ligand/ion | 0 | |
| Water | 51.47 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.009 | |
| Bond angles (°) | 1.20 | |
*Values in parentheses are for the highest-resolution shell.
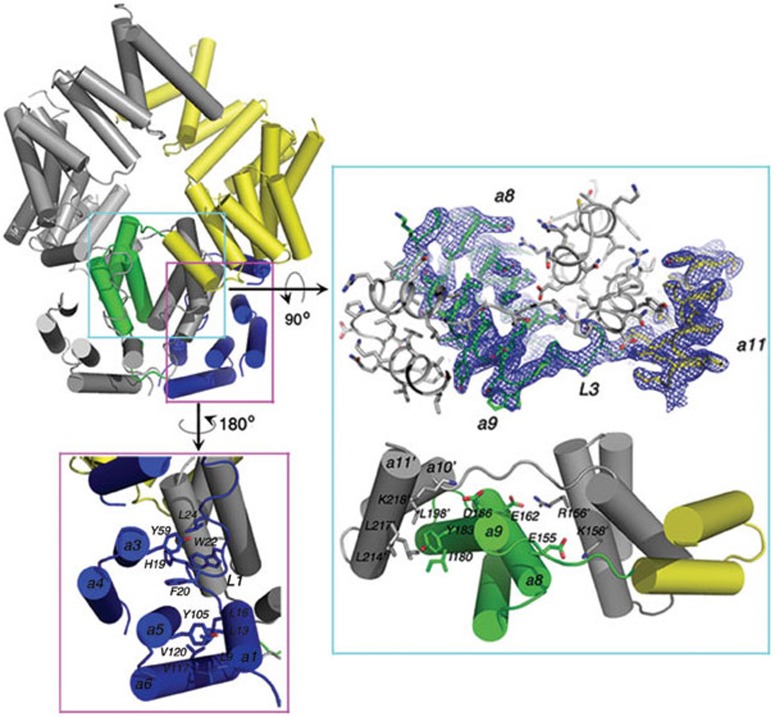
Figure 2.
Close-up views of the interactions in the ISG54 structure. The conserved residues in the N-terminal region are shown as sticks (panel in purple box). Residues L9, L13, L16, H19, F20, W22 and L24 on Loop 1 are stabilized by residues Y59, Y105, V117 and V120 from helices 3-6 in the N-terminal domain. The domain-swapped region in ISG54 are shown in cyan box. The blue net represents the 2Fo-Fc electron density map contoured at 1.0 σ (upper panel). Residues are shown as sticks and helices are shown as cylinders. Conserved residues in helices 8, 9, 10 and 11 are shown as sticks and labeled (bottom panel). The helices and residues in monomer B are indicated with a prime. For clarity, only half of the duplicate interactions between monomers A and B are shown.
ISG54 forms a swapped dimer
In contrast to previously described TPRs that exist as monomers in solution13,15, ISG54 monomers in our structure formed a tightly associated homodimer (Figure 1A). The N-terminal region and the middle region are involved in the interaction between monomers. In the ISG54 dimer, the middle region from one monomer inserts into the empty space surrounded by the three regions in the other monomer, forming a domain-swapped conformation. In total, the swap interface is about 4 240 Å2 as computed by PISA25, suggesting a very strong interaction between the monomers. The dimer structure of ISG54 looks like a binocular telescope, an eyepiece consisting of the two N-terminal regions and the two middle regions (helices 1-9) from two monomers, and an objective lens consisting of the C-terminal spiral region (helices 10-22). In the swapped dimer of ISG54, the two middle regions interact with each other through their helix 8 (Figure 2). Helix 9 of the middle region from one monomer interacts in an anti-parallel manner with helix 10 of the C-terminal region from the other monomer, linking the two regions as a continuous hybrid super-helix running from helix 7 to helix 22. Both electrostatic and hydrophobic interactions play important roles in the domain-swapped dimer conformation (Figure 2). Two pairs of electrostatic interactions, E155/K158′ and E162/R156′ between helix 8 of each monomer, where E155 and R162 are from monomer A and K158′ and E156′ are from monomer B, are observed. A strong interaction between D186 in helix 9 from monomer A and K218′ in helix 11 from monomer B is present. Hydrophobic interactions, for example, between residues I180 and Y183 in helix 9 of monomer A and L198′, L214′ and L217′ in helices 10 and 11 of monomer B, also seem to be important.
ISG54 is a novel RNA-binding protein
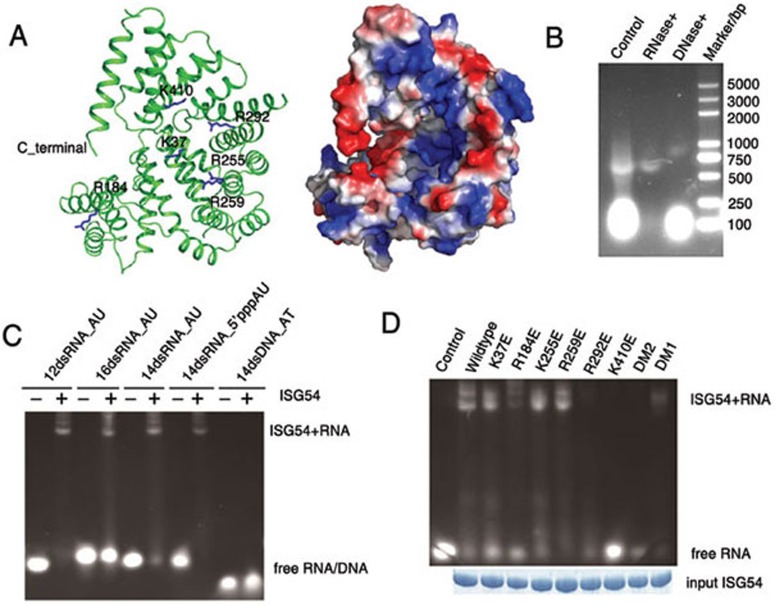
The tertiary structure of the continuous hybrid super-helix that constitutes almost three-quarters of a turn includes helices 7-9 of one monomer and helices 10-22 of the other monomer. The super-helix surrounds an open cylindrical space, which is approximately 50 × 20 × 20 Å3 in size. In a domain-swapped dimer, the open sides of the C-terminus stand face to face and have no interactions with each other. Interestingly, the inner side of the super-helix is exclusively positively charged and involves many basic residues (Figure 3A). Some conserved positively-charged residues, such as K37, R184, K255, R259, R292 and K410, are located along the super-helix. They form a continuous long positively-charged region. This feature strongly implies that ISG54 probably serves as a nucleotide-binding protein. In our initial experiments, we noted that large amounts of host cell polynucleotides were extracted together with the recombinant GST-fused wild-type protein during the purification process when a low-salt buffer (20 mM HEPES, pH 7.4, 150 mM NaCl) was used. When the protein sample was treated with nucleases, nucleic acids were largely digested by RNase but not by DNase (Figure 3B). We tested the RNA-binding ability of ISG54 and found that it could pull-down poly (IC), a synthetic viral RNA analogue (Supplementary information, Figure S3). Electrophoretic mobility shift assay (EMSA) was also performed using 12-, 14- and 16-nt model poly (AU) RNAs and a 12-nt poly (AT) DNA. A 5′ triphosphorylated 14-nt poly (AU) was also tested. Our results showed that ISG54 binds to poly (AU) but not model DNA, showing no selectivity for the 5′ phosphate state (Figure 3C). On the basis of these results, we suggest that ISG54 is an RNA-binding protein.
Figure 3.
RNA-binding ability of ISG54. (A) The ISG54 structure shows a nucleotide-binding region. Positively-charged residues along the channel of monomer A (cartoon diagram, green), which are thought to be involved in RNA binding, are labeled and shown as sticks (blue). The electrostatic potential surface of monomer A is shown in the right panel (red, negative charge; blue, positive charge; white, neutral). (B) A large amount of host cell RNA and a small amount of DNA were extracted with recombinant ISG54 during the purification process. After treatment with RNase or DNase and incubation at room temperature for 30 min, the samples were separated by a 1% agarose gel and the nucleotides were stained with ethidium bromide. (C) ISG54 binds to poly (AU) independent of its 5′ triphophosphate state (sequences shown in Supplementary information, Table S1). Only the RNA used in lane 8 labeled as 14dsRNA_5′pppAU was triphosphorylated. (D) The effect of mutations on the RNA-binding ability of ISG54. Purified ISG54 wild type (lane 2) and mutants (lanes 3-10) were pre-incubated with annealed synthetic 14-nt poly (AU). RNA alone was loaded in lane 1. Equal amounts of protein were loaded in each lane, as shown at the bottom of the panel.
To find the RNA-binding sites of ISG54, we mutated the positively-charged residues K37, R184, K255, R259, R292, and K410 on the inner surface of the super-helix to negatively-charged glutamic acids, and constructed two double mutants, K255E/R259E (DM1) and R292E/K410E (DM2). Using 14-nt poly (AU) as a substrate RNA, we found that RNA-binding ability was abolished in the ISG54 mutants R184E, R292E, K410E and double mutant DM2 (Figure 3D). Single mutations at K255 and R259 had little effect on RNA-binding ability; however, the double mutation K255E/R259E (DM1) impaired RNA-binding activity significantly. Based on our structure, the fragment covering the last three TPR-like motifs (helices 17-22) has a higher overall B factor than other parts of the protein (Supplementary information, Figure S4A), and tends to be less restrained by the other parts of the protein. We therefore made a truncation at residue 334, which separates ISG54 into two mutants, N-334 and 335-C. The RNA-binding ability of 335-C was significantly reduced (Figure 5B). Although we did not test the RNA-binding ability of mutant N-334 as it cannot be expressed and well purified from E. coli, we suppose that it should also lose its RNA-binding ability since the residue K410 in ISG54 is important for RNA binding. Taken together, these results suggest that ISG54 can bind RNA and that the residues on the inner face of the ISG54 super-helix are important for this function.
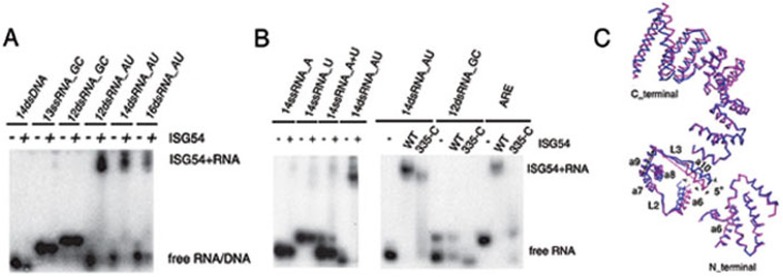
Figure 5.
The RNA-binding specificity of ISG54. (A) EMSA results of ISG54 binding with synthetic model RNAs (sequences shown in Supplementary information, Table S1). ISG54 binds to Poly (AU) but not to GC-rich RNAs and poly (AT). (B) ISG54 cannot bind to 14ssRNA_A, 14ss RNA_U, or the annealed 14ssRNA_A+U (left panel). ISG54 binds to the ARE sequence, using a 14dsRNA_AU as a positive control and 12dsRNA_GC as a negative control (right panel). RNAs alone are shown in lanes 1, 4 and 7. Wild-type ISG54 (lanes 2, 5 and 8) and mutant 335-C (lanes 3, 6 and 9) were pre-incubated with poly (AU), 12dsRNA_GC and ARE, respectively. The RNA-binding ability of truncation mutant 335-C with both 14dsRNA_AU and ARE (lanes 3 and 9) was destroyed. (C) Flexibility of ISG54 structures. Superimposition of the N_terminus (residues 31-91), domain-swapped regions (residues 136-187) and C_terminus (residues 194-334) of monomers A (purple) and B (blue) in the ISG54 dimer. The superimposition was performed using the helix 7-9 bundle as a basis and shows an angle between the two monomers at both helices 6 and 10, which alters the positions of the N- and C-terminus. The superimposition was performed with COOT46 and the angles were measured with PyMOL (http://www.pymol.org/).
RNA-binding ability is involved in the antiviral activity of ISG54
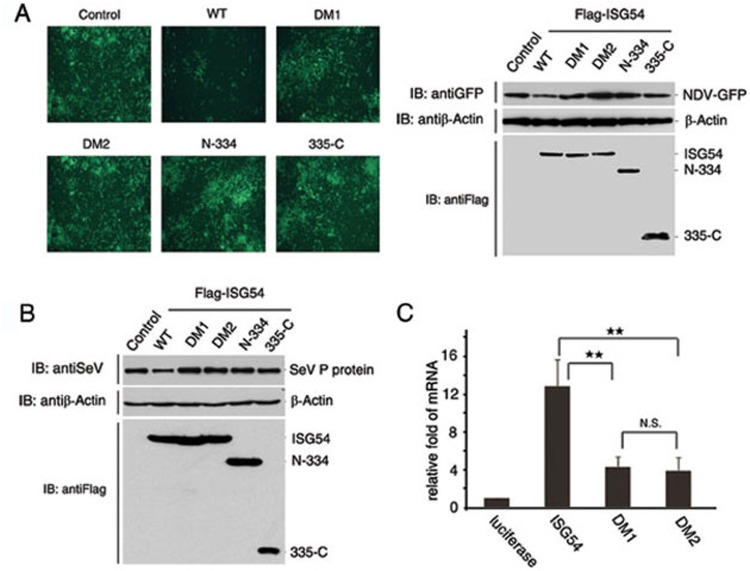
To test whether the RNA-binding ability of ISG54 is related to its antiviral function, we transfected a GFP-labeled Newcastle virus into HEK293T cells overexpressing ISG54 or its mutants (DM1, DM2, N-334 and 335-C). The GFP fluorescence of the cells was measured after 48 h of viral infection to determine viral production. Viral replication was partially inhibited in wild-type ISG54-transfected cells. The fluorescence of GFP in HEK293T cells transfected with mutant ISG54 proteins whose RNA-binding ability was destroyed, however, increased significantly, suggesting increased replication of the virus (Figure 4A). To explore whether ISG54's antiviral mechanism is the same for different viruses, we tested the effect of ISG54 on inhibiting the replication of the Sendai virus. Translation levels of Sendai viral phosphate protein (P protein) were monitored in ISG54-expressing HEK293T cells. Western blotting showed that overexpression of the ISG54 mutants cannot effectively inhibit the production of P protein (Figure 4B). Our result suggests that the RNA-binding ability of ISG54 is involved in its antiviral activity.
Figure 4.
ISG54's antiviral activity is related to its RNA-binding ability. (A) Replication of the GFP-labeled Newcastle virus in HEK293T cells overexpressing ISG54 and its mutants. The GFP fluorescence of the cells was monitored by microscopy 48 h after viral infection (left panel). Protein expression levels were quantified by western blotting (right panel). (B) Production of Sendai viral P protein in cells overexpressing ISG54 and its mutants as detected by immunoblotting. (C) ISG54 binds to the mRNA of the P protein of NDV. RNA-binding ability was destroyed by DM1 and DM2.
Since ISG54 is an RNA-binding protein, we speculate that its antiviral activity relies on its ability to bind viral RNA directly. To test whether ISG54 can bind to virus RNA directly and independently, we developed an in vitro pull-down assay. The mRNA of P protein from Newcastle disease virus was prepared using an in vitro transcription system. The purified GST-tagged ISG54 and its mutants DM1 and DM2 were loaded onto the GST binding beads using a GST-tagged luciferase fusion protein as a control. After incubating with viral mRNA, the GST beads were extensively washed to remove overloaded mRNA. The RNA pulled down by ISG54 proteins was separated by 1% agarose gel electrophoresis, stained by ethidium bromide, and quantified using ImageQuant™ software. The results showed that ISG54 can bind to the viral mRNA (Figure 4C). RNA-binding ability was destroyed in DM1 and DM2, consistent with the loss of antiviral functions in DM1 and DM2. Thus, we suggest that ISG54 probably inhibits viral replication through its ability to bind to viral RNAs, such as viral mRNAs.
RNA-binding specificity of ISG54
Recently, it was reported that ISG54 cannot directly bind to 5′ triphosphorylated RNA, and may associate with RNA indirectly by forming a complex with ISG5619. Our results, however, suggest that ISG54 can directly bind to RNA. In contrast to ISG56, the binding ability of ISG54 does not depend on the triphosphorylation of the 5′ end of the RNA (Figure 3C). To further clarify the molecular mechanism of ISG54, we attempted to examine whether it recognizes viral RNA with specific sequences.
To study the specificity of ISG54 for substrate RNA, we performed EMSA using model RNAs that had previously been applied in RIG-I RNA-binding analysis26,27 (Supplementary information, Table S1). In contrast to RIG-I, which exhibits no sequence selectivity for dsRNA26,27, our results show that ISG54 does not bind to the tested GC-rich RNA or dsDNA, but instead tends to bind to poly (AU) RNA (Figure 5A). While it is impossible to test all the RNA-binding possibilities of ISG54 using an in vitro EMSA assay, our results indicate that ISG54 has a previously unknown RNA selective mechanism, which is quite different from RIG-I and ISG5619,27. We propose that this protein can specifically recognize some viral RNAs in vivo.
ISG54 can bind ARE sequences in vitro
It has been reported that ISG54 can regulate the production of cell cytokines, such as TNFα and IL6, promote apoptosis and inhibit the migration of oral cancer cells21,22,23, but the mechanisms are unknown. We hypothesize that these cellular functions of ISG54 are also correlated with its RNA-binding ability. Unique RNA fragment AREs, which are reported to mediate the rapid degradation of host cell mRNA24, attracted our attention since ISG54 tends to bind to AU-rich RNA. AREs are found at the 3′ ends of the mRNAs of many genes in eukaryotic cells and are characterized by a tandem repeat AUUUA sequence. More importantly, ARE-containing mRNAs encode proteins that regulate both cell growth and acute responses, such as inflammatory factors TNFα and interleukins, apoptotic factor Bcl2 and cell growth factors24,28,29. These reports are consistent with the reported cellular functions of ISG54.
The ARE sequence, poly(A) and poly(U) were also tested as AU rich model RNAs in the ISG54 RNA-binding experiments. Surprisingly, EMSA results showed that ISG54 can bind to the ARE sequence but not to poly(A), poly(U) or annealed dsRNA poly(A) and poly(U) (poly(A+U)) (Figure 5B). Our results indicate that ISG54 may bind motifs in RNA such as the ARE sequence and thereby regulate cytokine production. Although this needs to be elucidated further, our results imply a possible mechanism underlying the cellular functions of ISG54.
Discussion
The ISG56 family proteins are well-known interferon-stimulated antiviral host factors with additional cellular functions; however, their underlying functional mechanisms are largely unknown. From its structure, we see that ISG54, quite different from other known TPR-containing proteins, has a positively-charged spiral that we show here to be an RNA-binding region. Our biochemical experiments suggest that ISG54 has high RNA-binding specificity. It selectively binds to AU-rich RNA. We proposed a new mechanism for ISG54 in which its high RNA-binding ability and selectivity is related to its antiviral and cellular functions.
The domain-swapped dimer conformation may exist in other ISG56 family members
Different from other reported TPR-containing proteins15,30, ISG54 is a domain-swapped dimer in solution. Results from both gel filtration and ultra-centrifugation experiments support this conclusion (Supplementary information, Figure S1). Since the residues in the domain-swapped interface are conserved in ISG56 family members (Supplementary information, Figure S2), we suggest that the swapped architecture observed in ISG54 may also exist in some other ISG56 family proteins. Recent reports have suggested that ISG54 can form multi-protein complexes with ISG5619,21. We also observed a putative dimer-dimer interaction surface in the structure (Supplementary information, Figure S1C). Based on this structural evidence, we suggest that the dimer-dimer interaction surface may also mediate interactions between ISG56 family members.
Different mechanisms for the antiviral functions of ISG54
Members of the ISG56 family proteins have different functions and their underlying mechanisms are both unclear and controversial. Early results indicated that ISG54 and ISG56 can interact with eIF3 to inhibit translation17,31. However, a recent report showed that ISG54 does not inhibit translation even at a concentration of 35 μM19. Consistent with this result (Supplementary information, Figures S5 and S6), we suggest that rather than recognizing eIF3 and inhibiting translation, ISG54 binds to viral RNA directly and has an as-yet-undiscovered mechanism underlying its antiviral activity.
In addition, one recent report has suggested that ISG54 may associate with RNA indirectly by forming a complex with ISG56, since it cannot directly bind to 5′-triphoshpate RNA19. It has also been reported that ISG54 recognizes viral 5′ mRNAs from all viruses except those producing 2′-O methylated mRNAs18. However, we did not detect direct interactions of ISG54 with m7GTP analogs in vitro using Isothermal Titration Calorimetric (ITC) and Surface Plasmon Resonance (SPR) (data not shown). Since we found that ISG54 can bind to specific AU-rich RNA directly and that the antiviral function of ISG54 is lost if RNA-binding sites are mutated, we suggest that ISG54 may bind directly in cells to viral RNA with special sequences.
ISG54 may function as a PPR-like protein
It has been predicted that members of the ISG56 family are TPR motif-containing proteins. TPR proteins have been reported to mediate protein-protein interactions. However, quite different from other TPR proteins, ISG54 has a positively-charged spiral that has been shown here to be an RNA-binding region. Pentatricopeptide repeat (PPR) motif-containing proteins, which have TPR-like motifs, have been identified mainly in plants and are reported to be sequence-specific RNA-binding proteins, which probably play important roles in post-transcriptional regulation32,33. No detailed structural information is available yet for PPR proteins, although computational methods have predicted that PPR and TPR motifs have very similar structural elements34,35 (Figure 1C). Just like PPR proteins, ISG54 has the same RNA-binding characteristics and tends to assemble into homodimers in the functional state. We thus hypothesize that ISG54 might function as a PPR protein in post-transcriptional regulation.
The RNA-binding model of ISG54
Based on the structure, we have shown that ISG54 is a novel RNA-binding protein. The inner channel of the C-terminus is involved in RNA binding. From the alignment of the two domain-swapped monomers by superimposing helices 7-9 from the two monomers (Figure 5C) we can see that the N-terminal region and C-terminal spiral have different opening angles. We suggest that the C-terminus of ISG54 may have the ability to open or close using the L3 loops as hinges when it binds to RNA. Helix 22 has low density and a high model B factor (Supplementary information, Figure S4). The interface between the last two helices, helix 21 and helix 22, is smaller and the cross angle between them is larger than that of other TPR motifs in the protein. This implies that helix 22 tends to move away from the position shown in the crystal structure and thus open or close the entrance of the cylinders when ISG54 binds to target RNAs. How RNA binds to the ISG54 dimer is still a mystery. Since residues that are involved in RNA binding locate along the positively-charged track of the ISG54 super-helix, we propose two possible models for the RNA binding of ISG54 protein. In the first model, RNA is inserted into the opening super helical cylinders along the positively-charged track; while in the second model, RNA is clamped by the two C-terminal helical domains. Further study is needed to determine which model is correct.
In summary, we have presented the first crystal structure of an ISG56 family protein, ISG54. We have shown that this protein is a novel RNA-binding protein. Its selective RNA-binding ability serves as an important molecular basis for both its antiviral activity and cellular functions. This work provides not only a novel structure for RNA-binding proteins, but also a platform for further study of the functional mechanisms of the ISG56 family.
Materials and Methods
Constructs, protein expression and purification
The full-length ISG54 gene was cloned into a pGEX6p-1 vector (Invitrogen) with an N-terminal GST tag. Mutations were introduced using PCR with primers containing mutated residues and were confirmed by plasmid sequencing. The plasmid was transformed into E. coli strain BL21 (DE3) cells. Cells were cultured in LB medium at 37 °C with 100 mg/L ampicillin, and when the OD600 reached 0.6-0.8, they were induced at 16 °C with 0.5 mM isopropyl-thio-D-glactosidase (IPTG). Cells were harvested after 20 h of incubation by centrifugation at 5 000 rpm for 10 min. The selenomethionyl-derivative protein was derived using the same construct and E. coli strain, grown in cultures containing 50 μg/ml SeMet36. Cells were resuspended in a high-salt buffer (20 mM HEPES, pH 7.4, 1 M NaCl) and lysed by sonication. The lysate was separated by centrifugation at 16 000 rpm for 30 min and the supernatant was applied to a GST affinity column (GE Healthcare) followed by intensive washing with binding buffer. The GST tag was removed by precision digestion on the column at 4 °C overnight in low-salt buffer (20 mM HEPES, pH 7.4 and 150 mM NaCl). The ISG54 eluted from the GST column was further purified by anion exchange and then by Superdex-200 gel filtration chromatography (GE Healthcare).
Crystallization and structure determination
Protein samples used for crystallization were concentrated to about 10 mg/ml. A crystallization trial was set up using the hanging-drop vapor diffusion method. Crystals were obtained in buffer containing 0.2 M NaSCN and 20% PEG3350, and were flash-frozen with 60% glycerol as a cryoprotectant. 2.8 Å native diffraction data was collected at the Shanghai Synchrotron Radiation Facility beamline BL17U (SSRF, China, λ = 0.979 Å), and 3.0 Å anomalous diffraction data was collected at the High Energy Accelerator Research Organization beamline BL5A (KEK-PF, Japan, λ= 0.978 Å), using a selenomethionyl-derivative crystal. Both sets of data were processed using HKL200037. Phasing was performed using SAD method with Se-derivative data. A total of 16 heavy atoms were located by SHELXD38. The program Phenix39 was used to calculate the initial phase, and the improved phase. Approximately 60% of the residues were traced automatically by Phenix using 2.8 Å native data. The resulting protein model was completed manually by COOT40 and refined using Phenix. In the Ramachandran plot generated using PROCHECK41, 92.3% of the residues in the final model were in the favored regions and 7.4% residues were in the allowed region. The atomic coordinates and diffraction data have been deposited in the Protein Data Bank (4G1T). All the structural figures were prepared by COOT and PyMOL (http://www.pymol.org/).
EMSA assay
Model RNAs were purchased from Takara Biotechnology Inc. and DNA was chemically synthesized by Sangon Biotech Co., Ltd. (Shanghai). Double-stranded DNA and RNAs were annealed by heating and slow cooling. The wild-type and mutant proteins were expressed and purified as described above. For results in Figure 3, annealed poly (AU) was mixed with ISG54 at a molar ratio of protein to RNA of 1:2, i.e., about 200 pmol : 400 pmol. The mixture was kept at room temperature for 30 min and then loaded on a pre-run native 8% acrylamide:bisacrylamide (29:1) gel in boric acid buffer without EDTA, which was run at 12 V cm-1, keeping the temperature under 60 °C. The gel was stained with ethidium bromide and visualized using a gel imaging system (Tanon 1600, Tanon Inc.). For results in Figure 5, we used γ-32P-labeled RNA in the EMSA since single-stranded RNAs are not as clear as double-stranded RNAs when stained with ethidium bromide. Annealed nucleotides were radio-labeled at their 5′ ends with γ-32P ATP using T4 polynucleotide kinase (NEB) and mixed with purified protein at a molar ratio of 5 pmol : 20 pmol. The mixture was kept at room temperature for 30 min and then loaded on a pre-run native 6% acrylamide:bisacrylamide (29:1) gel in boric acid buffer without EDTA. Radioactive signals were detected after electrophoresis at 12 V cm-1 for 2 h.
NDV-GFP assay
HEK293T cells (∼1 × 105) were seeded in 6-well plates overnight and then transfected with 1.0 μg of ISG54 plasmids. The culture medium was replaced 18 h after transfection by serum-free DMEM (Invitrogen), and NDV-GFP42 was added to the medium. After 1 h, the medium was removed and cells were cultured in DMEM containing 10% FBS (ExCell). NDV-GFP replication in HEK293T cells was visualized by fluorescence microscopy (Olympus). Immunoblotting was wlso performed to test the production of viral protein as previously described43. The GFP polyclonal antibody and the mouse monoclonal antibody for actin were purchased from Sigma.
Immunoblotting of Sendai virus
HEK293T cells (∼2 × 105) were seeded in 6-well plates overnight and then transfected with 1 μg of ISG54 plasmids by a standard calcium phosphate precipitation method44. The Sendai virus was added 20 h after transfection. After 24 h, immunoblotting was performed to test the production of viral P protein as previously described43. The rabbit polyclonal antibody for the Sendai virus were purchased from Sigma.
In vitro pull-down assay
The cDNA of ZJ1-NDV was a kind gift from Dr XF Liu45. The cDNA sequence of P protein was cloned into pET-28a (Novagen) for amplification. The purified plasmid was digested with the restriction enzyme HindIII to give linear DNA and was used for in vitro transcription. In vitro transcription experiments were performed using a Large Scale T7 RNA Production System kit (Promega RiboMAX™). GST-tagged ISG54 proteins were pre-purified using high-salt buffer (20 mM HEPES, pH 7.4, 1 M NaCl) to remove non-specifically bound RNA from E. coli and bound to GST beads (GE Healthcare). An excess amount of transcribed RNA was added to the proteins and incubated for 30 min at 4 °C in a low-salt buffer (20 mM HEPES, pH 7.4, 150 mM NaCl) before the beads were centrifuged and extensively washed with low-salt buffer. The RNA pulled down by the ISG54 proteins was separated by 1% agarose gel electrophoresis, stained by ethidium bromide, and quantified using ImageQuant™ software (GE).
Acknowledgments
We thank our colleagues Drs Hongfei Wang and Lijie Wu for crystal data collection. We thank Yuanyuan Chen and Xiaoxia Yu in core facility center (Institute of Biophysics, CAS) for technical assistance with the ITC, SPR and ultra-centrifugation experiments. We thank colleagues at the Photon factory, KEK (Japan) and SSRF (China) for assistance in the use of the synchrotron resource. We also thank Drs Xiufan Liu and Shunlin Hu (Yangzhou University) for providing the cDNA of ZJ1-NDV. This work was supported by grants to YL from the Ministry of Science and Technology (863 Project 2006AA02A314; 973 Programs 2007CB914303, 2011CB910304 and 2012CB910204) and the National Natural Science Foundation of China (30925011, 31030024 and 31021062).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
The aggregation state of ISG54.
Sequence alignment of ISG56 family proteins.
GST-fused ISG54 was incubated with excess poly (IC).
A) The B factor distribution of ISG54.
In vitro translation assay.
Co-immunoprecipitation shows that ISG54 cannot bind to eIF3c.
Sequences of model RNAs used for ISG54 binding
References
- Chebath J, Merlin G, Metz R, Benech P, Revel M. Interferon-induced 56,000 Mr protein and its mRNA in human cells: molecular cloning and partial sequence of the cDNA. Nucleic Acids Res. 1983;11:1213–1226. doi: 10.1093/nar/11.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathelet M, Moutschen S, Defilippi P, et al. Molecular cloning, full-length sequence and preliminary characterization of a 56-kDa protein induced by human interferons. Eur J Biochem. 1986;155:11–17. doi: 10.1111/j.1432-1033.1986.tb09452.x. [DOI] [PubMed] [Google Scholar]
- Kessler DS, Levy DE, Darnell JE., Jr Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc Natl Acad Sci USA. 1988;85:8521–8525. doi: 10.1073/pnas.85.22.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Tong JH, Mao M, et al. Cloning of a gene (RIG-G) associated with retinoic acid-induced differentiation of acute promyelocytic leukemia cells and representing a new member of a family of interferon-stimulated genes. Proc Natl Acad Sci USA. 1997;94:7406–7411. doi: 10.1073/pnas.94.14.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Veer MJ, Sim H, Whisstock JC, Devenish RJ, Ralph SJ. IFI60/ISG60/IFIT4, a new member of the human IFI54/IFIT2 family of interferon-stimulated genes. Genomics. 1998;54:267–277. doi: 10.1006/geno.1998.5555. [DOI] [PubMed] [Google Scholar]
- Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GC, Sarkar SN. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr Top Microbiol Immunol. 2007;316:233–250. doi: 10.1007/978-3-540-71329-6_12. [DOI] [PubMed] [Google Scholar]
- Fensterl V, Sen GC. The ISG56/IFIT1 gene family. J Interferon Cytokine Res. 2011;31:71–78. doi: 10.1089/jir.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensterl V, Wetzel JL, Ramachandran S, et al. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 2012;8:e1002712. doi: 10.1371/journal.ppat.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Sen GC. Characterization of the interaction between the interferon-induced protein P56 and the Int6 protein encoded by a locus of insertion of the mouse mammary tumor virus. J Virol. 2000;74:1892–1899. doi: 10.1128/jvi.74.4.1892-1899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR. Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Cortajarena AL, Regan L. Ligand binding by TPR domains. Protein Sci. 2006;15:1193–1198. doi: 10.1110/ps.062092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Dye BT, Rajashankar KR, Kurinov I, Schulman BA. Insights into anaphase promoting complex TPR subdomain assembly from a CDC26-APC6 structure. Nat Struct Mol Biol. 2009;16:987–989. doi: 10.1038/nsmb.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiski CL, Harwich M, Jain S, et al. AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure. 2010;18:265–273. doi: 10.1016/j.str.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzi F, Hui DJ, Merrick WC, Sen GC. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J Biol Chem. 2006;281:34064–34071. doi: 10.1074/jbc.M605771200. [DOI] [PubMed] [Google Scholar]
- Daffis S, Szretter KJ, Schriewer J, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Lassnig C, Eberle CA, et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- Li Y, Li C, Xue P, et al. ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc Natl Acad Sci USA. 2009;106:7945–7950. doi: 10.1073/pnas.0900818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawowczyk M, Van Scoy S, Kumar KP, Reich NC. The interferon stimulated gene 54 promotes apoptosis. J Biol Chem. 2011;286:7257–7266. doi: 10.1074/jbc.M110.207068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai KC, Chang KW, Liu CJ, Kao SY, Lee TC. IFN-induced protein with tetratricopeptide repeats 2 inhibits migration activity and increases survival of oral squamous cell carcinoma. Mol Cancer Res. 2008;6:1431–1439. doi: 10.1158/1541-7786.MCR-08-0141. [DOI] [PubMed] [Google Scholar]
- Berchtold S, Manncke B, Klenk J, Geisel J, Autenrieth IB, Bohn E. Forced IFIT-2 expression represses LPS induced TNF-alpha expression at posttranscriptional levels. BMC Immunol. 2008;9:75. doi: 10.1186/1471-2172-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Chen CY, Shyu AB. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ludwig J, Schuberth C, et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Xu H, Ranjith-Kumar CT, et al. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabar KS. Post-transcriptional control during chronic inflammation and cancer: a focus on AU-rich elements. Cell Mol Life Sci. 2010;67:2937–2955. doi: 10.1007/s00018-010-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey AJ, Russell LC, Chinkers M. C-terminal sequences of hsp70 and hsp90 as non-specific anchors for tetratricopeptide repeat (TPR) proteins. Biochem J. 2009;423:411–419. doi: 10.1042/BJ20090543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Hui DJ, Merrick WC, Sen GC. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 2000;19:6891–6899. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R. Pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Mol Cell. 2011;42:106–117. doi: 10.1016/j.molcel.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ID, Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- Raczynska KD, Augustyniak H. [Family of pentatricopeptide repeat proteins] Postepy Biochem. 2005;51:440–446. [PubMed] [Google Scholar]
- Guerrero SA, Hecht HJ, Hofmann B, Biebl H, Singh M. Production of selenomethionine-labelled proteins using simplified culture conditions and generally applicable host/vector systems. Appl Microbiol Biotechnol. 2001;56:718–723. doi: 10.1007/s002530100690. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W.Processing of X-ray diffraction data collected in oscillation mode Methods in Enzymology 1997276Carter Jr CW, Sweet RM, Eds. Macromolecular Crystallography (part A), 1997; 307-326. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski AR, Macarthur WM, Moss SD, Thornton MJ. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Park MS, Shaw ML, Munoz-Jordan J, et al. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J Virol. 2003;77:1501–1511. doi: 10.1128/JVI.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Jordan M, Schallhorn A, Wurm FM. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wan HQ, Liu HQ, Wu YT, Liu XF. Genomic sequence of an isolate of Newcastle disease virus isolated from an outbreak in geese: a novel six nucleotide insertion in the non-coding region of the nucleoprotein gene. Brief Report. Arch Virol. 2004;149:1445–1457. doi: 10.1007/s00705-004-0297-8. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The aggregation state of ISG54.
Sequence alignment of ISG56 family proteins.
GST-fused ISG54 was incubated with excess poly (IC).
A) The B factor distribution of ISG54.
In vitro translation assay.
Co-immunoprecipitation shows that ISG54 cannot bind to eIF3c.
Sequences of model RNAs used for ISG54 binding