MicroRNAs (miRNAs) are about 22-nucleotide noncoding RNAs that, in general, downregulate target mRNA expression. Their function ensures precise protein abundance and protein distribution under diverse cellular and developmental conditions 1. miRNA are transcribed by RNA polymerase II, processed in nucleus and matured in the cytoplasm. Mature miRNA are then packaged in a functional ribonucleoprotein (RNP) complex. This RNP is referred to as the RNA-induced silencing complex or RISC 2. In essence, the miRNA itself provides RISC with transcript specificity and thereby tethers protein factors that elicit the transcript silencing. The mRNA 3′UTR is plastic in nature and is therefore often the preferred site of RISC recruitment.
Mechanistically, miRNAs have been shown to inhibit multiple steps of protein synthesis, but are also suggested to impact message stability 3, 4. Indeed, in a report from David Bartel's lab, the overall effect of miRNA on gene expression was argued to be at the level of message decay 5. Integrating results from transcriptome ribosome profiling, mRNA-seq analysis, and proteomic analysis, Guo et al. 5 observed that when the exogenous miR-1 or miR-155 is expressed in HeLa cells, the decrease of target mRNAs' level correlates well with the reduction of the corresponding proteins' level. Similar results were also found for the endogenous miR-223 in mouse neutrophils. It appears, therefore, that mammalian miRNAs silence gene expression by predominantly promoting transcript destruction. Importantly, however, these experiments address the overall effect of miRNA-mediated gene silencing that, in general, is transcript destruction. The miRNA's primary effect on its targets, however, could still be upstream of transcript destruction.
Critically, mRNA degradation and translation are intimately connected events thus modulating one can result in a corresponding and dramatic change in the other 6. The question remains, therefore, what is the sequence of events that occurs during miRNA-mediated gene silencing? Does rapid mRNA decay cause perceived translational repression or does translational quiescence ultimately result in an ensuing transcript destabilization? Now back-to-back publications in Science attempt to unravel these mysteries using kinetic analysis in flies and zebrafish 7, 8. In both studies, the overall conclusion is that miRNA elicits translational control, which thereby triggers mRNA destruction.
In Drosophila S2 cells, Rachel Green's lab monitored miRNA effects as a function of time 7. Importantly, they used a luciferase reporter under the control of the inducible metallothionein promoter. This allows them to modulate the reporter's expression by changing the media's copper concentration. The reporter's 3′UTR contains either the Bantam, miR-9b, or miR-279 miRNA-binding sites. Together, this system allows them to monitor both miRNA-mediated translation repression, as determined by the changes of luciferase activity, and miRNA-induced mRNA degradation, as measured by quantitative RT-PCR. Reporter synthesis was allowed to reach steady state and then transcription was shut off. Following transcriptional inhibition, time points were taken and at each interval both protein and mRNA levels were monitored. Importantly, the effect of each distinct miRNA was measured in this manner. In all three cases, the Green lab observed that translational repression always occurred before mRNA degradation. In addition, they observed that the removal of poly(A) tail (deadenylation), the initiating and rate-limiting step for the degradation of most mRNAs, is not required for the translational repression of the luciferase reporter. Consistently, a reporter lacking a poly(A) tail can be translationally repressed by a miRNA. Finally, Djuranovic and colleagues defined where miRNA is working in the translation cycle. By inserting a lysine tract in the reporter's ORF, the mRNA was cleaved as it experienced a codon-induced slowing in translational elongation. Thus if repression occurs upstream of translational elongation, the reporter will not be cleaved efficiently. This is precisely what was observed for all miRNAs tested. Together, these data argue that translation repression, presumably at the level of initiation, is the initial event of miRNA-mediated gene silencing and the ultimate degradation of the target is a secondary consequence.
In a parallel study from Antonio Giraldez's lab, translational repression is also the trigger event for miR-430-mediated gene silencing in zebrafish embryos 8. In the fish, miR-430 is dramatically induced at the onset of zygotic transcription (about 4 hours post fertilization) and therefore facilitates the clearance of maternal mRNAs. Taking the advantage of this temporal expression pattern, Bazzini et al. analyzed the translational status and the stability of all the miR-430 target mRNAs at several time points before and after the induction of miR-430 in the embryos. Specifically, they used ribosome profiling analysis to monitor the translation status and mRNA-seq to determine the stability of mRNAs. Bioinformatic analysis of these high throughput sequencing data obtained at different time points revealed that during the induction of miR-430, the target mRNAs first experienced translational repression, then mRNA decay ensued. Thus, inhibition of translation is the trigger event of miR-430-mediated gene silencing in zebrofish embryo. mRNA decay analysis during this developmental process also supports this notion. In detail, they observed that the miR-430 target mRNAs still had intact poly(A) tails when their translation repression had occurred. In addition, slowing deadenylation of miR-430 target mRNA did not influence the translational repression. Together, these data argue that translation repression is the cause, while the mRNA degradation is the effect of the gene silencing mediated by miR-430 during early fish development. Like the Green lab, this work also suggests that repression may occur predominantly at translational initiation.
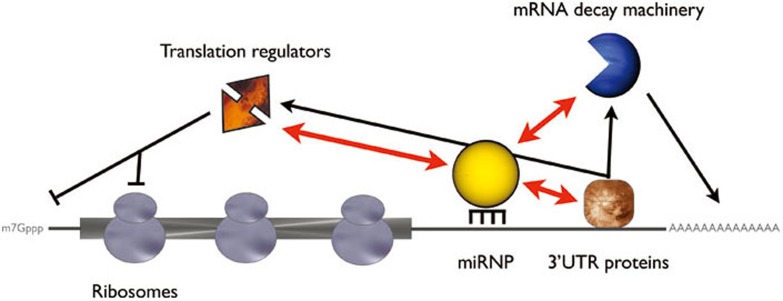
So how do we reconcile the differences seen in these two recent studies vs. the mammalian work? Perhaps it is simply the way that the studies were conducted. For instance, the earliest time point analyzed in the mammalian work was 12 hours post transfection of the exogenous miRNAs 5. Conversely, the earliest time point measured is 2 hours in the fish and fly studies 7, 8. If translational repression stimulates robust transcript decay in mammalian systems, then this effect might be simply missed by taking late time points. Alternatively, and perhaps more provocatively is the possibility that miRNAs function the same but the readout depends on context. For instance, if miRNA stimulates deadenylation (albeit directly or indirectly) the effect on a target depends on its environment. In systems where robust decay occurs after deadenylation, the transcript is destroyed rapidly and a stable translational quiescent transcript is not observed. In systems where mRNA decay is not as prevalent following deadenylation, miRNA targets may persist in a stored state. Also possible is that miRNA have distinct effect on target mRNA that is dependent on context (Figure 1). For example, the miRNA ribonucleoprotein complex (miRNP) interacts with mRNA decay factors, such as the deadenylase and decapping complex, but it also binds translational regulators 9. Moreover, some decay factors can function as translation regulators 10. Thus the event triggered by a miRNA might be dictated by the target mRNA's specific 3′UTR environment. Since 3′UTRs can be littered with protein factors having positive or negative effects on stability, translation, transport, localization, and polyadenylation/deadenylation 11, the overall effect of the miRNP is integrated into these other events. Thus the primary function of a miRNA may be one critical and still poorly understood event, but the final readout of a miRNA is most likely complex and malleable to the biological system.
Figure 1.
The trigger event of miRNA-mediated gene silencing may be determined by the interactions among miRNP, mRNA decay machinery, translation regulators, and the proteins associated with the 3′UTR of the target mRNA.
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight. Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]