Abstract
Hepatitis C virus (HCV) infection is a major cause of chronic liver disease worldwide. Although novel drugs against HCV are under development, the current standard therapy consists principally of interferon (IFN). To improve the response to IFN treatment by enhancing interferon-stimulated response element (ISRE)-mediated gene transcription, we screened 75 microRNAs highly expressed in hepatocytes for their ability to modulate ISRE activity. Overexpression of microRNA-122 (miR122) significantly suppressed ISRE activity. Conversely, silencing of miR122 function enhanced IFN-induced ISRE activity, by decreasing expression of suppressor of cytokine signaling 3 (SOCS3). This decrease in SOCS3 level was not mediated by microRNA target gene suppression, but rather by enhanced methylation at SOCS3 gene promoter. Taken together, our data, along with the fact that antisense oligonucleotides of miR122 also directly inhibit HCV replication, suggest that a combination therapy comprising IFN and silencing of miR122 function may be a promising therapeutic option in the near future.
More than 170 million individuals worldwide are chronically infected with hepatitis C virus (HCV), which results in hepatic inflammation, hepatic fibrosis, and liver cirrhosis1. End-stage liver diseases as well as hepatocellular carcinoma attributable to chronic hepatitis C are increasingly serious problems2. For almost a decade, the standard of care in patients with chronic hepatitis C has consisted of pegylated interferon-α2a (pegIFN-α2a) or pegIFN-α2b in combination with the guanosin analog ribavirin. However, this eradicates HCV in only about half of those infected with HCV genotype 1, the most common genotype globally. Moreover, severe adverse events are associated with IFN therapy, such as myelo-suppression and flu-like syndrome. Because these effects are dose-limiting, many patients are unable to receive a higher dose of IFN that might more effectively inhibit HCV replication3. While recent licensing of HCV protease inhibitors for the treatment of patients with chronic hepatitis C as part of a triple therapy with pegIFN-α and ribavirin is expected to increase the sustained viral response (SVR) rate, IFN currently remains the principal drug for the eradication of HCV.
Type I interferons (IFNs), such as IFN-α and IFN-β, bind to the type I IFN receptor4. One major pathway in type I IFN signaling involves the Jak-STAT signaling cascade5. Activated tyrosine kinases phosphorylate STAT-1 and STAT-2 proteins, which bind to p48, a member of the IFN regulatory family (IRF), to form interferon-stimulated gene factor-3 (ISGF3). ISGF3 translocates to the nucleus and binds to the interferon-stimulated response element (ISRE) in the promoter region of IFN target genes, which code for antiviral proteins such as double-stranded RNA-activated protein kinase (PKR) and 2′5′-oligoadenylate synthethase (OAS1). On the other hand, regulatory molecules are also involved in the IFN pathway. The suppressor of cytokine signaling (SOCS) protein is a negative regulator of the Jak-STAT cascade6. The SOCS family includes eight members (SOCS-1 to SOCS-7 and CIS), all sharing a central SH2 domain and a C-terminal SOCS box. SOCS-1 and SOCS-3 are the most effective members of this family, and act as negative regulators of several intracellular pathways, particularly the Jak–STAT pathway. In hepatic cells, inhibition of IFN-α-induced STAT-1 activation by HCV core protein overexpression is associated with induction of SOCS-3 mRNA expression7. Therefore, increased SOCS3 protein expression during HCV infection may be a mechanism of IFN resistance8,9,10. Such regulatory functions may also be important determinants of the efficacy of anti-HCV IFN therapy.
MicroRNAs are short, single-stranded, non-coding RNAs. They are expressed in most organisms, ranging from plants to vertebrates11, and are involved in the regulation of target gene expression. Different microRNAs are responsible for the control of various biological processes12,13,14. In this context, a number of microRNAs have recently been shown to regulate the function of intracellular signaling intermediates, such as p53 and NF-κB pathways, by regulating expression of their target genes15,16,17,18.
Primary microRNAs, which possess stem-loop structures, are processed into mature microRNAs by Drosha and Dicer RNA polymerase III. These mature microRNAs then associate with the RNA-induced silencing complex (RISC), and the resulting complex binds directly to the 3′-untranslated regions (3′-UTRs) of target mRNAs to suppress translation and gene expression post-transcriptionally. While this is undoubtedly the main action of microRNAs, recent studies have demonstrated that microRNAs can enter the nucleus19, and are involved in establishing DNA methylation20,21,22. In addition, microRNAs may also regulate chromatin structure by regulating key histone modifiers23. Taken together, these results suggest that microRNAs are important players in epigenetic and post-transcriptional control of gene expression20.
The aim of this study was to determine the possible role of microRNAs in IFN signaling. We focused on microRNAs expressed in the liver because we were interested in regulators of IFN signaling during HCV treatment. We screened a subset of microRNAs for their ability to modulate ISRE activity to develop a more effective IFN-based therapy against chronic hepatitis C infection.
Results
Screening for microRNAs regulating ISRE activities
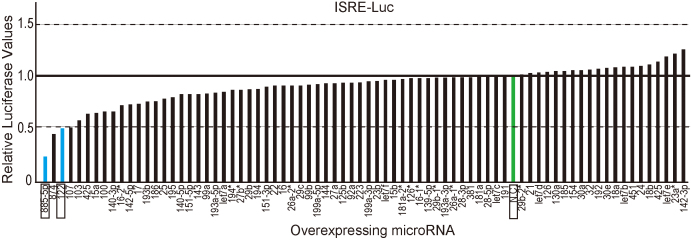
We initially screened for microRNAs that affected ISRE-mediated gene transcription using stable ISRE activity reporter cell lines and by transiently overexpressing 75 mature synthetic microRNAs, as we did previously to screen for microRNAs that affect NF-κB activity15. Because we were interested in IFN-mediated intracellular signaling in the liver, the microRNAs examined were selected on the basis of their hepatic expression level24. In addition, we used non-liver 293T cells for the initial screening to determine the effects of the microRNA overexpression. The data suggested differential effects of microRNAs on ISRE activity in response to IFN-α stimulation (Fig. 1). Of the microRNAs examined, we chose miR122 and miR885-5p for further investigation because they suppressed ISRE activity significantly and reproducibly in two independent screens.
Figure 1. MiR122 and miR885-5p suppress ISRE activity.
Functional screening for liver microRNAs that modulate ISRE activity. Seventy-five mature microRNA oligonucleotides known to be highly expressed in the liver were transiently reverse-transfected into stable 293T cell-derived ISRE reporter cell lines, followed by IFN-α stimulation. Reporter activities were normalized to the values of negative control RNA oligonucleotides (N.C. in green) and ranked in ascending order. Determinations were performed in duplicate, and a representative result is shown. MiR122 and miR885-5p, which were chosen for further analyses, are in blue and represented as rectangles.
Silencing of miR122 enhances ISRE activities
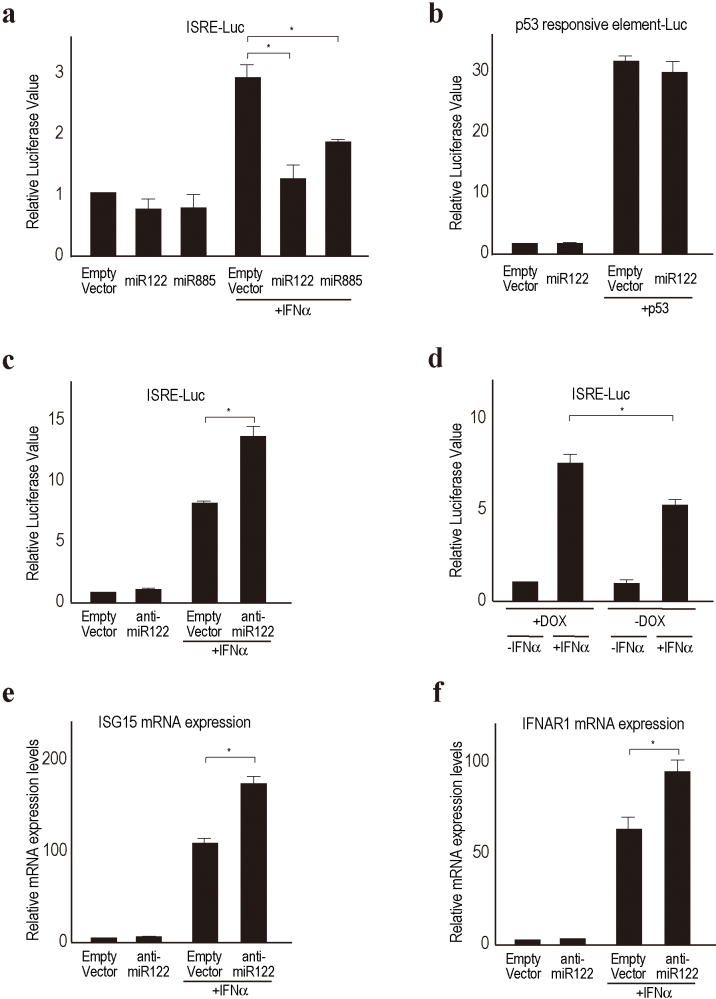
To confirm the suppressive effects of miR122 and miR885 overexpression on ISRE activities, we first performed a reporter assay to monitor ISRE activities with plasmid-based miR-overexpressing constructs. While both miR122 and miR885 suppressed ISRE activities induced by IFN-α stimulation in 293T cells, the effect of miR122 was more significant (Fig. 2a). For this reason, and because miR122 is the most abundant microRNA in the liver24, we further focused on miR122. The suppressive effect of miR122 was ISRE-specific, because it had no effect on p53-mediated transcriptional activities (Fig. 2b). Next, to examine the effects of silencing miR122 function on ISRE activity in hepatoma cell lines, we transiently transfected plasmid-based anti-miR122 constructs into Huh7 cells, in which miR122 is highly expressed25. The silencing of miR122 function resulted in about two-fold augmentation of IFN-α-induced ISRE activity (Fig. 2c), suggesting that miR122 is also involved in ISRE activity in hepatoma cell lines during IFN-α treatment. To further confirm these effects, we examined Hela-Tet-Off-miR122 cells, in which the expression of miR122 precursors can be shut off by doxycyclin treatment. In these cells, ISRE activity was more highly induced by IFN-α treatment when the expression of miR122 precursors was suppressed by doxycyclin treatment (Fig. 2d). Interferon stimulated genes, such as ISG15 and IFNAR1, were induced to a greater extent by IFN-α treatment in miR122-silenced Huh7 cells than in control cells (Fig. 2e and 2f). These data suggest that silencing miR122 can enhance IFN-α-ISRE activities.
Figure 2. MiR122 modulates ISRE activities.
(a) Overexpression of the selected microRNA precursors suppresses ISRE activity following IFN-α stimulation. ISRE reporter plasmids were transiently transfected, with or without selected microRNA precursor-expressing plasmids, into Huh7 cells. Luciferase values were normalized to those of cells transfected with an empty vector and without IFN, which were set to 1. *, p < 0.05. Data represent the means ± standard deviations (SD) of three independent determinations. Similar results were obtained using HepG2 cells. (b) p53 activities were unaffected by miR122 expression. Reporter assays were performed with p53 reporter and p53 expressing plasmids with or without miR122 precursor expression in Huh7 cells. Data represent the means ± SD of three independent determinations. (c) Silencing of miR122 function enhances ISRE activity. Anti-miR122 expressing plasmids were used to perform an ISRE reporter assay in Huh7 cells. *, p < 0.05. Data represent the means ± SD of three independent determinations. (d) Hela-Tet-Off-miR122 precursor cell lines were used to measure ISRE activity. Cells were transfected with ISRE reporter constructs with (DOX+) or without (DOX-) doxycyclin. DOX+ shut off the miR122 expression in these cells. The reporter activities after 6 h of IFN-α stimulation were compared. *, p < 0.05. Data represent the means ± SD of three independent determinations. (e, f) IFN-inducible genes (e, ISG15; f, IFNAR1) were more induced in miR122-silenced Huh7 cells, determined by quantitative RT-PCR using RNA after 6 h of IFN-α stimulation. *, p < 0.05. Data represent the means ± SD of three independent determinations.
Silencing miR122 suppresses SOCS3 expression by methylation of its promoter
To gain insight into the mechanisms underlying the suppression of ISRE activity by miR122, we searched the Gene Expression Omnibus (GEO) database regarding the effect of silencing miR122 on changes in IFN pathway-related gene expression (DataSet Record GDS1729)26. Consistent with our results (Fig. 2), the expression of several known ISRE-mediated IFN-stimulated genes, such as OAS1, interferon α and β receptor 1 (Ifnar1), interferon α-inducible protein 27-like 2A (Ifi27l2a), and interferon regulatory factor 6 (IRF6), were indeed up-regulated by silencing miR122 function in the mouse liver. However, the expression of regulatory genes involved in the IFN signaling pathway from the receptor to the nucleus, such as STAT1, STAT2, JAK1, and JAK2, were unchanged. Although we searched for potential miR122 target genes related to IFN signaling in several microRNA target databases, including TargetScan (http://www.targetscan.org), no major IFN-related genes were found.
Because epigenetic changes induced by microRNAs have been reported20,21,22, we compared the comprehensive methylation levels of 27,578 promoter-associated CpG sites using an Illumina Infinium methylation assay (Human Methylation27 BeadChip) between control and stably miR122-silenced Huh7 cell lines. Although the methylation levels of most CpG sites were unchanged, those of a number of CpG sites were altered by silencing miR122 (Table 1). While the methylation of most of these decreased, the CpG sites of a few genes were more methylated by miR122 silencing. The most significantly increased methylation levels were observed in the promoter of the SOCS3 gene, which is a negative regulator of IFN signaling. The enhanced methylation levels in silencing miR122 were confirmed by bisulphite sequencing at the CpG island in the SOCS3 promoter, from −556 to −335 relative to the transcriptional start site (Fig. 3a and b). Thus, we hypothesized that the greater methylation of SOCS3 induced by miR122 silencing results in decreased expression of SOCS3 protein, which may, in turn, enhance IFN-α signaling.
Table 1. Top 30 genes with promoters differentially methylated in control and miR122-silenced Huh7 cells. Values indicate methylation levels. Values in ‘Differences' indicate quantitative differences in methylation levels of target gene CpG sites. Higher values indicate greater methylation levels. Genes in bold and highlighted in gray means greater methylation; others were less methylated in miR122-silenced Huh7 cells.
| SYMBOL | Control | miR122-silenced | Difference | CpG ISLAND LOCATIONS |
|---|---|---|---|---|
| GPC3 | 0.4242135 | 0.11596 | 0.3082535 | X:132946499-132947763 |
| FLJ30058 | 0.4155016 | 0.1428133 | 0.2726883 | X:130019792-130020537 |
| EIF3S6IP | 0.672594 | 0.4012034 | 0.2713906 | 22:36574299-36576077 |
| BHLHB9 | 0.3481071 | 0.07687688 | 0.27123022 | X:101862184-101862707 |
| PORCN | 0.6389685 | 0.3697215 | 0.269247 | |
| MSI2 | 0.4593301 | 0.1955636 | 0.2637665 | 17:52688030-52689554 |
| DNTT | 0.6114385 | 0.3489618 | 0.2624767 | 10:98054238-98054478 |
| SEMA3B | 0.3901392 | 0.1301293 | 0.2600099 | 3:50285369-50286362 |
| RYR2 | 0.4331115 | 0.1756537 | 0.2574578 | 1:235271651-235273428 |
| TCEAL7 | 0.6519198 | 0.4006093 | 0.2513105 | |
| NR0B2 | 0.3711949 | 0.1234481 | 0.2477468 | |
| SOCS3 | 0.1831344 | 0.4273662 | 0.2442318 | 17:73866027-73868731 |
| TAS2R16 | 0.5821649 | 0.3380581 | 0.2441068 | |
| BHLHB9 | 0.3021148 | 0.06171004 | 0.24040476 | X:101862184-101862707 |
| OR12D3 | 0.5266486 | 0.2884241 | 0.2382245 | |
| ADAMTSL1 | 0.4537211 | 0.2196532 | 0.2340679 | |
| TBX6 | 0.5031447 | 0.2695281 | 0.2336166 | 16:30010427-30011656 |
| ICAM4 | 0.4990334 | 0.2668367 | 0.2321967 | 19:10258558-10259935 |
| C9orf125 | 0.2387415 | 0.4707038 | 0.2319623 | 9:103287963-103289481 |
| ELN | 0.1332373 | 0.3629701 | 0.2297328 | 7:73080116-73080605 |
| ANKRD30A | 0.898423 | 0.6711555 | 0.2272675 | 10:37453955-37454965 |
| OR10J1 | 0.5324367 | 0.3094136 | 0.2230231 | |
| TP73 | 0.3596564 | 0.1386476 | 0.2210088 | 1:3596889-3597535 |
| CNNM1 | 0.6313883 | 0.4106784 | 0.2207099 | 10:101078809-101080801 |
| DHH | 0.2955738 | 0.07619943 | 0.21937437 | 12:47774151-47774653 |
| CTAG2 | 0.5303309 | 0.3110451 | 0.2192858 | X:153536438-153536702 |
| GNB4 | 0.3091451 | 0.09039365 | 0.21875145 | 3:180651316-180652496 |
| PNPLA2 | 0.4458646 | 0.2272328 | 0.2186318 | 11:808884-809164 |
| FGF7 | 0.6298472 | 0.4128944 | 0.2169528 | |
| NTE | 0.4557684 | 0.2400911 | 0.2156773 | 19:7504300-7507429 |
Figure 3. CpG methylation status in the SOCS3 promoter.
(a) The CpG island in the SOCS3 promoter is indicated by double underlines. The numbers are the positions relative to the transcription start site. Primers used for the bisulphate sequences in this study are indicated by arrows. (b) Methylation status in the CpG island of the SOCS3 promoter in control and miR122-silenced Huh7 cells, determined by the bisulphate sequences. Seven clones each were sequenced. Circles represent CpG sites. Black circles, methylated CpG sites.
MiR122 silencing enhances STAT3 activation by decreasing SOCS3 expression
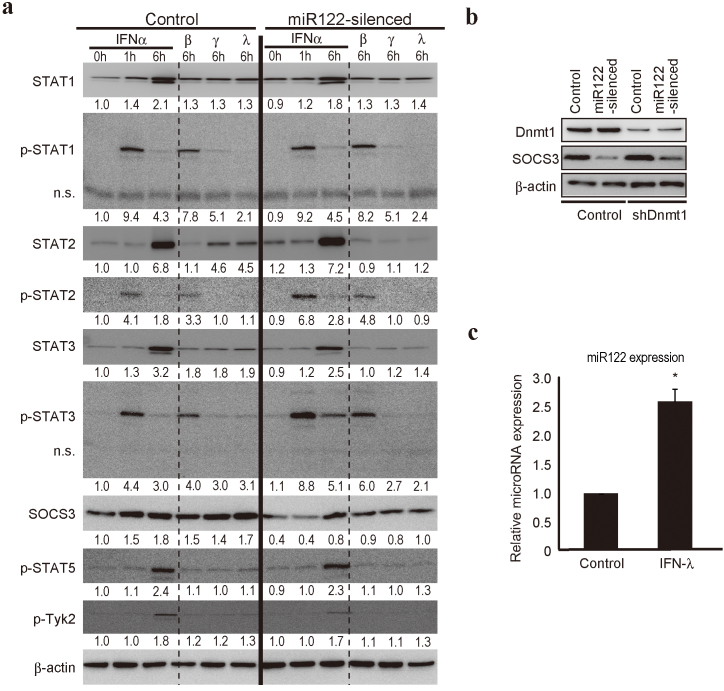
To confirm the above results, we examined SOCS3 expression and IFN signaling-related molecules in miR122-silenced Huh7 cells (Fig 4a). While the GEO database contained no direct data regarding SOCS3 cDNA expression, we found decreased SOCS3 protein levels in miR122-silenced cells, consistent with promoter hyper-methylation (Fig. 4a). Although the mechanisms underlying the altered methylation induced by miR122 silencing remain unknown, it did not depend on DNA methyltransferase 1 (Dnmt1), which is a key mediator of DNA methylation that catalyzes the methylation of CpG dinucleotides in genomic DNA27, because the decreased SOCS3 expression by miR122 silencing was also present in Dnmt1 knockdown cells (Fig. 4b).
Figure 4. SOCS3 expression is decreased by miR122 silencing.
(a) Protein levels of the indicated IFN signaling-related genes were determined using control and miR122-silenced Huh7 cells after IFN-α, β, γ, or λ stimulation at the indicated time points. A representative of three independent determinations is shown. n.s. indicates non-specific bands. The band intensities were quantitated and adjusted by the expression levels of β-actin. The calculated ratios are indicated below each panel after setting the value of control cells at 0 h as 1.0. (b) Dnmt1 was not involved in the SOCS3 promoter methylation induced by miR122 silencing. SOCS3 protein levels were determined using control, miR122-silenced, Dnmt1 knocked-down, and Dnmt1 knocked-down with miR122-silenced Huh7 cell lysates. A representative of three independent determinations is shown. (c) MiR122 expression levels after IFN-λ stimulation for 6 h in Huh7 cells were determined by quantitative RT-PCR. Results were calculated by normalizing to U6 amounts and the relative ratio was determined by setting the value of unstimulated cells as 1. Data represent the mean ± SD of three independent determinations. *, p < 0.05.
Because SOCS3 is a potent inhibitor of STAT3 activation6, and because type I IFNs induce STAT3 as well as STAT1 and STAT2 activation5,28, we examined the phosphorylation status of STAT proteins after IFN treatment in Huh7 control and miR122-silenced cells (Fig. 4a). While the STAT1, 2, and 3 protein levels and the phosphorylation levels of STAT1 before and after IFN-α stimulation in these two cell lines did not differ significantly, STAT3 phosphorylation levels were higher in miR122-silenced cells 1 and 6 h after IFN-α stimulation, consistent with the decreased expression of SOCS3 (Fig. 4a). Furthermore, STAT2 phosphorylation was slightly higher in miR122-silenced cells (Fig. 4a). Similar tendencies were also observed after treatment with IFN-β, another type I IFN. These data suggest that IFN-α treatment induced greater STAT3 activation in miR122-silenced cells, probably due to decreased SOCS3 expression.
In contrast, whereas IFN-γ (type II IFN) and IFN-λ (type III IFN) induced slight phosphorylation of STAT1 and STAT3, the levels in control and miR122-silenced cells were comparable. STAT2 protein levels were significantly lower in miR122-silenced cells than in controls after treatment with these cytokines (Fig. 4a). No induction of SOCS3 after treatment with IFN-α/β, IFN-γ, or IFN-λ was detected in miR122-silenced cells, probably due to promoter methylation (Fig. 4a), whereas SOCS3 protein was induced by all IFNs in control cells (Fig. 4a). Because increased expression of miR122 was detected after IFN-λ stimulation (Fig. 4c), this might be responsible for the increased SOCS3 expression induced by IFN-λ stimulation.
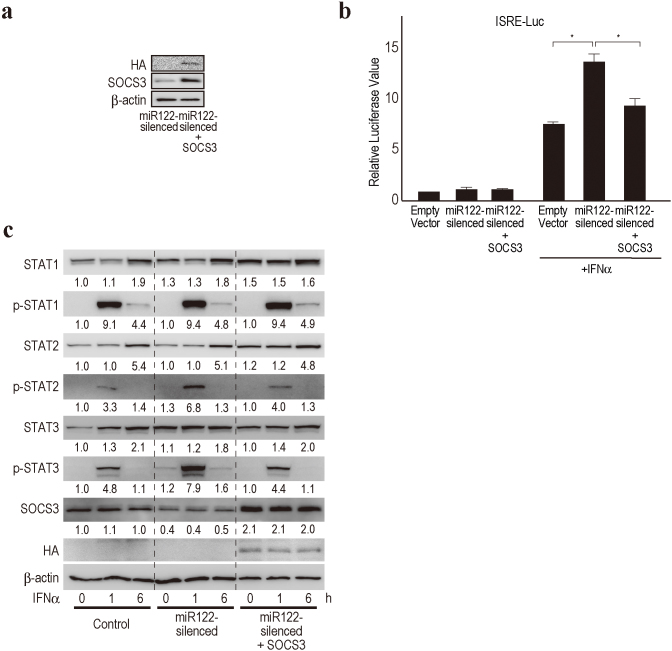
To confirm whether the induction of the enhanced ISRE activity in miR122-silencing was dependent on the decreased expression of SOCS3, we investigated whether the restoration of SOCS3 expression in miR122-silencing could reduce the ISRE activity in a reporter assay. The overexpression of SOCS3 in miR122-silenced Huh7 cells reduced the induction of ISRE activity caused by miR122-silencing (Fig. 5a and b), although we could not fully exclude the possibility that other mechanisms were also involved because the reversal of ISRE activity did not completely reach the level of the control. To support this, we confirmed the restoration of STAT2 and STAT3 phosphorylation levels induced by IFN-α treatment in miR122-silenced cells that stably overexpressed SOCS3 (Fig. 5c). These results suggest that the enhanced ISRE activity in miR122-silenced cells is mostly, if not completely, dependent on the reduced expression of SOCS3.
Figure 5. SOCS3 overexpression in miR122-silenced Huh7 cells reverses enhanced ISRE activities.
(a) HA-tagged SOCS3 was overexpressed in miR122-silenced Huh7 cells. Representative blotting images are shown. (b) SOCS3 overexpression suppresses enhanced ISRE activities in miR122-silenced Huh7 cells by a reporter assay. *, p < 0.05. Data represent the means ± SD of three independent determinations. (c) SOCS3 overexpression restored STAT2 and STAT3 phosphorylation levels induced by IFN-α in miR122-sileced Huh7 cells. A representative of three independent determinations is shown. The band intensities were quantitated and adjusted by the expression levels of β-actin. The calculated ratios are indicated below each panel after setting the value of control cells at 0 h as 1.0.
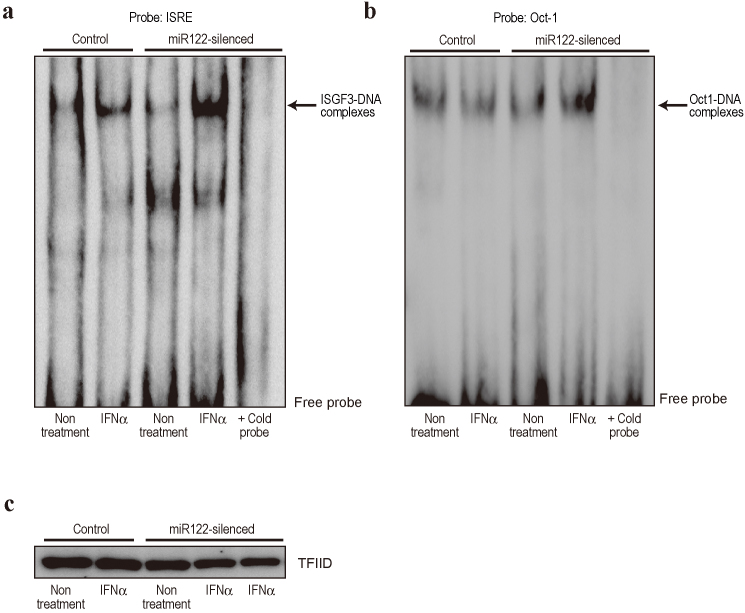
MiR122 silencing enhances ISGF3-DNA binding
Type I IFNs (IFN-α and -β) activate STATs by phosphorylation, followed by formation of the ISGF3 complex, which is composed of STAT1, STAT2, and IRF9. The importance of ISGF3 in antiviral responses is well established29. In contrast, the precise role of STAT3 in type I IFN signaling is not completely understood30. However, numerous clinicopathological results suggest that increased SOCS3 expression in the liver is closely related to a poor response to IFN therapy for HCV eradication8,9. This in turn suggests that high SOCS3 expression and low STAT3 activation may be related to impaired ISGF3 complex activation. In addition, STAT3 activation supports the ISGF3-dependent induction of antiviral genes in vitro31. Based on these reports, and because SOCS3 expression was lower in miR122-silenced cells (Fig. 4), we hypothesized that the level of ISGF3 complex after IFN-α treatment is higher in miR122-silenced cells, leading to greater ISRE activation (Fig. 2). While the levels of Oct-1-DNA binding as a loading control were not changed, activation of ISGF3 binding to an ISRE-containing oligonucleotide after IFN-α treatment was significantly greater in miR122-silenced cells (Fig. 6a, b, c), consistent with the fact that ISRE activities were enhanced in miR122-silenced cells in a reporter assay (Fig. 2). These data suggest that miR122-silencing in hepatocytes results in low SOCS3 expression via promoter methylation, which may subsequently enhance the induction of IFN-stimulated gene expression by increasing ISGF3-ISRE binding activities triggered by type I IFN treatment.
Figure 6. ISGF3-ISRE DNA binding activity is increased in miR122-silenced cells.
(a) DNA-binding ability of ISGF3 was determined by gel shift assay using ISRE consensus oligonucleotides. Nuclear extracts from control and miR122-silenced Huh7 cells with and without IFN-α stimulation for 1 h were used. Arrow indicates ISGF3-DNA complexes. The specificity of the DNA–protein complex was tested by adding unlabeled ISRE probe (cold probe) to IFN-α-stimulated miR122-silenced Huh7 nuclear extracts. A representative of three independent determinations is shown. (b) DNA-binding ability of Oct-1 was determined as a control for equal loading of nuclear extract. (c) TFIID amounts were examined by Western blotting to confirm equal nuclear extract loading. A representative of two independent determinations is shown.
Discussion
Although the treatment options for HCV infection are changing due to the introduction of HCV protease inhibitors and DAAs, the principal drug for HCV therapy remains IFN. In this study, we demonstrated that the reduced expression of miR122 contributes to decreased SOCS3 expression via promoter methylation and, subsequently, enhanced ISRE activity results after IFN-α stimulation. These data provide a molecular rationale for, and a method for increasing the efficacy of, IFN therapy for HCV infection.
MicroRNAs are involved in various biologically important intracellular signaling pathways15,16,17,18. Regarding the convergence of microRNAs and IFN signaling, some microRNAs are reported to be involved in endogenous IFN production in the innate immune response induced by pathogen infection32. In addition, several microRNAs that may regulate genes that have anti-pathogen effects are induced by IFN stimulation33. In this study, however, the level of miR122 expression seemed to determine the efficacy of the signaling triggered by the exogenous IFN used as an anti-HCV therapy.
MiR122 is the most abundant microRNA in the liver24, where it has many important biological roles, such as in fatty acid metabolism26,34 and circadian rhythms35 under normal conditions. Further, it is also a determinant of the biological aggressiveness of hepatocellular carcinoma36 in the pathological state. In general, microRNAs act as repressors of target gene expression37. However, regarding miR122 and HCV, miR122 somehow enhanced HCV RNA replication in an in vitro replicon system38. Although the precise molecular mechanisms underlying this phenomenon remain unknown, antisense miR122 has been developed as a therapeutic drug for HCV based on in vitro data39. Indeed, treatment of chronically HCV-infected chimpanzees with a locked nucleic acid (LNA)-modified oligonucleotide (SPC3649) complementary to miR122 leads to long-lasting suppression of HCV viremia39. Development of this drug for human use is now in Phase IIa trials40. Because our results showed that lower expression of miR122 leads to augmentation of the intracellular signaling induced by IFN, a combination therapy consisting of an antisense of miR122 and type I IFNs represents a promising and realistic therapeutic option. In addition, because IFN-α/β was reported to suppress miR122 expression, which is considered one of the mechanisms of action of IFN against HCV41, the effects of exogenous IFN may be self-augmented by the decreased expression of SOCS3 due to decreased miR122 expression. High expression levels of SOCS3 in the liver are negative predictors of IFN treatment of HCV infection8,9. This may reflect suppression of IFN signaling by the high miR122 levels in the liver, as suggested by our data.
Our results indicate that reduced miR122 function leads to promoter methylation and decreased SOCS3 expression, which is possibly not the direct target of miR122, because no predictable sites for miR122 interaction in its 3′UTR were found in a computational search. A hypothesis involving an epigenetics-microRNA regulatory circuit has emerged recently20. While a number of microRNAs are regulated epigenetically, others simultaneously regulate epigenetic pathway-related molecules. Taken together, these results suggest that post-transcriptional regulation by microRNAs and transcriptional control machinery by epigenetics cooperate to determine the global gene expression profile and to maintain physiological functions in cells20. In our genome-wide study, methylation levels of a subset of gene CpG islands were aberrantly induced by miR122 silencing. While our data suggest that SOCS3 methylation was not mediated by Dnmt1, the precise mechanisms of the aberrant methylation induced by miR122, including whether it operates through regulation of the expression of other genes or directly in the nucleus, remains to be elucidated. Nonetheless, our results indicate that aberrant functions of some miRNAs may lead to changes in the methylation levels of a subset of gene CpG islands.
Recent genome-wide association studies have discovered a significant association between the response to pegIFN and ribavirin therapy and common single nucleotide polymorphisms in the vicinity of IL-28 (IFN-λ) genes42,43. In addition, carriers of the alleles associated with resolution of HCV infection have increased serum IFN-λ levels43. Our results demonstrate that IFN-λ stimulation increases SOCS3 and miR122 expression, which may block innate type I IFN signaling. This seems inconsistent with the fact that patients with high IFN-λ levels have better responses to pegIFN and ribavirin therapy. However, we speculate that blockade of innate IFN-α signaling by high IFN-λ through SOCS3 expression may prevent the chronically low levels of innate IFN-α, which may increase the sensitivity to exogenous IFN-α when applied in therapeutic quantities. Although further work is required, it is consistent with the fact that low expression of hepatic IFN-stimulated genes (ISGs) is strongly associated with a better response to IFN treatment and genetic variation in IL-28B44,45.
Higher expression of α-fetoprotein (AFP) is also known as a poor prognostic factor for IFN treatment in HCV therapy46,47. As we described previously, decreased expression levels of miR122 are linked to increased expression of AFP36. Therefore, in cases with higher AFP levels, miR122 levels in hepatocytes may be low and thus, innate IFN signaling may be high through SOCS3 promoter methylation. These may provide a molecular explanation of the poor response to IFN therapy in cases with high AFP levels.
In summary, our study provides information on the involvement of miR122 in the regulation of ISRE activity through the modulation of SOCS3 expression via gene promoter methylation. Our results provide a molecular rationale that will facilitate more effective use of IFN as an anti-HCV combination therapy, specifically by including modulators of miR122 function.
Methods
Cell culture
The human hepatocellular carcinoma cell lines Huh7 and HepG2 were obtained from the Japanese Collection of Research Bioresources (JCRB, Osaka, Japan). The human embryonic kidney cell line 293T was obtained from the American Type Culture Collection (ATCC, Rockville, MD). MiR122 functionally knocked-down Huh7 (miR122-silenced Huh7) cell lines were as described previously36. Hela-Tet-Off cell lines were purchased from Clontech (Mountain View, CA). Huh7-Tet-Off cell lines were established by transfecting with the pTet-Off vector (Clontech) and selecting with 400 μg/ml G418 (Wako, Osaka, Japan). All cells were maintained in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum.
Reagents
Human recombinant IFN-α, IFN-β, IFN-γ, and IL-28B (IFN-λ3) were purchased from R&D Systems (Minneapolis, MN) and used at final concentrations of 100 U/ml, 100 U/ml, 10 ng/mL, and 100 ng/mL, respectively. Doxycyclin and Tet-system fetal bovine serum were purchased from Clontech. Hygromycin was purchased from EMD Chemicals (Darmstadt, Germany).
Stable reporter cell lines and microRNA screening
To establish ISRE-luc reporter cell lines, 293T cells were transfected with the reporter plasmid pISRE-luc (Clontech) with a linear hygromycin resistance marker (Clontech). Single clones were selected in the presence of 250 μg/mL hygromycin. To screen for microRNAs that modulate ISRE activity, 75 microRNAs that are highly expressed in the liver34, were reverse-transfected as described previously15. Briefly, synthetic mature microRNAs and, as a negative control, double-stranded RNA with artificial sequences (B-Bridge, Sapporo, Japan) were applied, with transfection reagents, to 96-well plates. The reporter cells were seeded to the plates, reverse-transfected, and then incubated for 48 h. ISRE-derived luciferase activity was measured using a GloMax 96 Microplate Luminometer (Promega, Madison, WI) after exposure to IFN-α for 12 h. Determinations were performed in duplicate, and the data were compared to those of the negative control.
Plasmids and tetracycline-inducible stable cell lines
The miR122 precursor expression plasmid was described previously36. The miR885 precursor expression plasmid was constructed by inserting a polymerase chain reaction (PCR)-generated 500 bp sequence encoding the miR885 precursor into the pCDH vector (System Biosciences, Mountain View, CA) using the XbaI and NotI sites. The resulting plasmid drives expression of miR885 under control of the CMV promoter. Plasmids expressing Halo-tagged SOCS3 were purchased from Promega. HA-tagged SOCS3 cDNA was amplified by PCR using the Halo-tagged SOCS3 plasmid as a template and the product was cloned into the pCDH-neo vector (System Biosciences) at the NotI site using In-fusion cloning method (Clontech). Plasmids expressing p53 were described previously48. To construct a tetracycline-regulated miR122 precursor expression plasmid, a PCR-amplified 500 bp region encoding themiR122 precursor was inserted into the pTRE2 vector (Clontech) using the BamHI and SalI sites. To establish tetracycline-regulated miR122 expressing stable cell lines, pTRE2miR122 plasmids were transfected into Hela-Tet-Off or Huh7-Tet-Off cell lines with a linear hygromycin marker (Clontech), followed by selection with hygromycin. Cells were cultured with or without 2.5 µg/ml doxycyclin. To establish SOCS3 overexpressing stable cells, HA-tagged SOCS3 expressing lentiviral particles were produced as described previously36 and transduced into miR122-silenced Huh7 cells, followed by selection with G418 (Sigma, St. Louis, MO).
Lentiviral transduction
Huh7 cells were transduced with Dnmt1-shRNA and control-shRNA lentiviral particles (Santa Cruz Biotechnology, Santa Cruz, CA) and then selected on puromycin.
Western blot analysis and antibodies
Western blotting was performed as described previously49. All antibodies were purchased from Cell Signaling Technology (Danvers, MA), except anti-HA (Roche Applied Science, Indianapolis, IN) and anti-β-actin (Sigma).
Transfection and luciferase assay
Plasmid transfection was performed using FuGENE6 (Boehringer Mannheim, Mannheim, Germany) according to the manufacturer's protocol36. Luciferase activities were measured using a Dual Luciferase Reporter Assay System (Promega) as described previously50.
Genome-wide DNA methylation analysis
Comprehensive DNA methylation analyses were performed by the outsourcing company MBL (Nagoya, Japan) using the Illumina Infinium methylation assay (Human Methylation27 BeadChip), which provides quantitative methylation levels at 27,578 promoter-associated CpG sites. Genomic DNA extracted from control and miR122-silenced Huh7 cells using QIAamp DNA mini kit (QIAGEN, Hilden, Germany) was used for this assay. Promoters with differential methylation levels were determined based on the standard criteria that differential values above 0.15 indicate significant differences in methylation status51.
Bisulphite sequence analyses
Bisulphite sequence analyses were performed to check the methylation status of the SOCS3 promoter. Genomic DNA was extracted by QIAamp DNA mini kit (Qiagen) and bisulphate modified using MethylEasy Xceed Bisulphite Modification Kit (Human Genetic Signatures, North Ryde, Australia). The sequences of the SOCS3 promoter region were extracted as the upstream sequence of the transcriptional start site. The region of CpG islands and the PCR primers were determined using the web-based software MethPrimer52. PCR was performed using EpiTaq HS enzyme (TaKaRa, Shiga, Japan), according to the manufacturer's instructions. The PCR products were gel-purified using the QIAquick Gel Extraction Kit (Qiagen) and cloned into the TA vector using TOPO-TA cloning (Invitrogen). Seven clones from each sample were selected and the sequences were analyzed using a 3700 DNA analyzer (Applied Biosystems, Foster City, CA). The results were summarized using the web-based software, QUMA (http://quma.cdb.riken.jp/top/quma_main_j.html).
RNA extraction and quantitative RT-PCR analysis of microRNA expression levels
Total RNA was isolated from cells using Trizol Reagent (Invitrogen, Carlsbad, CA). To determine the levels of miR122 expression, cDNA was synthesized from RNA, and quantitative PCR was performed using an Mir-X miRNA First-Strand Synthesis and SYBR qRT-PCR Kit (Clontech). Relative expression values of microRNAs were calculated by the CT-based calibrated standard curve method. These values were then normalized to that of the U6 snRNA.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared as described previously53. Five micrograms of nuclear extract were incubated with a double-stranded biotin-labeled DNA probe containing ISRE sites (5′-GAT CCA TGC CTC GGG AAA GGG AAA CCG AAA CTG AAG CC-3′)54 or Oct-1 sites (Affymetrix, Santa Clara, CA) plus 1-μg poly (dI-dC) in a binding buffer (50 mM Tris [pH 7.5], 250 mM NaCl, 2.5 mM DTT, 2.5 mM EDTA, 5 mM MgCl2, and 20% glycerol) at 15°C for 30 min. DNA–protein complexes were separated on 6% non-denaturing polyacrylamide gels in 0.5× TBE (25 mM Tris base, 24.25 mM boric acid, and 1 mM disodium EDTA) by electrophoresis at 120 V, 4°C for 30 min, and then transferred to Presoak Pall Biodyne B nylon membranes (Hybond-N+; GE Healthcare Life Sciences) by electrophoresis in the same buffer for 30 min at 300 mA. Oligonucleotides were fixed using a UV cross-linker, and visualized using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific, Rockford, IL), according to the manufacturers' instructions.
Statistical analysis
Statistically significant differences between groups were determined using Student's t-test when variances were equal. When variances were unequal, Welch's t-test was used. P-values less than 0.05 were considered statistically significant.
Author Contributions
T.Y., A.T., and M.O. planned the research and wrote the paper. T.Y., A.T., M.O., and T.K. performed the experiments. H.Y. analyzed the data. K.K. supervised the entire project.
Acknowledgments
This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan (#22390058, #24590956 and #20390204) (to M.O., H.Y. and K.Koike), by Health Sciences Research Grants of The Ministry of Health, Labour and Welfare of Japan (Research on Hepatitis) (to K.Koike) and grants from the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Yakult Bio-Science Foundation and the Cell Science Research Foundation (to M.O.).
References
- Hofmann W. P. & Zeuzem S. A new standard of care for the treatment of chronic HCV infection. Nat Rev Gastroenterol Hepatol 8, 257–264 (2011). [DOI] [PubMed] [Google Scholar]
- Di Bisceglie A. M. et al. Prolonged therapy of advanced chronic hepatitis C with low-dose peginterferon. N Engl J Med 359, 2429–2441 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoofnagle J. H. A step forward in therapy for hepatitis C. N Engl J Med 360, 1899–1901 (2009). [DOI] [PubMed] [Google Scholar]
- Stetson D. B. & Medzhitov R. Type I interferons in host defense. Immunity 25, 373–381 (2006). [DOI] [PubMed] [Google Scholar]
- Schindler C. Teaching resources. Cytokine receptors and Jak-STAT signaling. Sci STKE 2006, (2006). [DOI] [PubMed] [Google Scholar]
- Yoshimura A., Naka T. & Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol 7, 454–465 (2007). [DOI] [PubMed] [Google Scholar]
- Funaoka Y. et al. Analysis of interferon signaling by infectious hepatitis C virus clones with substitutions of core amino acids 70 and 91. J Virol 85, 5986–5994 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. A. et al. Hepatic SOCS3 expression is strongly associated with non-response to therapy and race in HCV and HCV/HIV infection. J Hepatol 50, 705–711 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaaki H. et al. Predictive value of suppressor of cytokine signal 3 (SOCS3) in the outcome of interferon therapy in chronic hepatitis C. Hepatol Res 39, 850–855 (2009). [DOI] [PubMed] [Google Scholar]
- Shao R. X. et al. Suppressor of cytokine signaling 3 suppresses hepatitis C virus replication in an mTOR-dependent manner. J Virol 84, 6060–6069 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington J. & Ambros V. Role of microRNAs in plant and animal development. Science 301, 336–338 (2003). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature 431, 350–355 (2004). [DOI] [PubMed] [Google Scholar]
- Lu J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838 (2005). [DOI] [PubMed] [Google Scholar]
- Takata A. et al. MicroRNA-22 and microRNA-140 suppress NF-κB activity by regulating the expression of NF-κB coactivators. Biochem Biophys Res Commun 411, 826–831 (2011). [DOI] [PubMed] [Google Scholar]
- Park S. Y., Lee J. H., Ha M., Nam J. W. & Kim V. N. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol 16, 23–29 (2009). [DOI] [PubMed] [Google Scholar]
- Kasinski A. L. & Slack F. J. Potential microRNA therapies targeting Ras, NFkappaB and p53 signaling. Curr Opin Mol Ther 12, 147–157 (2010). [PubMed] [Google Scholar]
- Ma X., Becker Buscaglia L. E., Barker J. R. & Li Y. MicroRNAs in NF-{kappa}B signaling. J Mol Cell Biol 3, 159–166 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H. W., Wentzel E. A. & Mendell J. T. A hexanucleotide element directs microRNA nuclear import. Science 315, 97–100 (2007). [DOI] [PubMed] [Google Scholar]
- Wu L. et al. DNA methylation mediated by a microRNA pathway. Mol Cell 38, 465–47 (2010). [DOI] [PubMed] [Google Scholar]
- Sato F., Tsuchiya S., Meltzer S. J. & Shimizu K. MicroRNAs and epigenetics. FEBS J 278, 1598–1609 (2011). [DOI] [PubMed] [Google Scholar]
- Fabbri M. & Calin G. A. Epigenetics and miRNAs in human cancer. Adv Genet 70, 87–99 (2010). [DOI] [PubMed] [Google Scholar]
- Saetrom P., Snøve O. & Rossi J. J. Epigenetics and microRNAs. Pediatr Res 61, 17R–23R (2007). [DOI] [PubMed] [Google Scholar]
- Landgraf P. et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S., Habermacher R., Martine U., Closs E. & Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–1124 (2006). [DOI] [PubMed] [Google Scholar]
- Esau C. et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3, 87–98 (2006). [DOI] [PubMed] [Google Scholar]
- Li E., Beard C. & Jaenisch R. Role for DNA methylation in genomic imprinting. Nature 366, 362–365 (1993). [DOI] [PubMed] [Google Scholar]
- Brierley M. M. & Fish E. N. Stats: multifaceted regulators of transcription. J Interferon Cytokine Res 25, 733–744 (2005). [DOI] [PubMed] [Google Scholar]
- Platanias L. C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 5, 375–386 (2005). [DOI] [PubMed] [Google Scholar]
- Icardi L. et al. Opposed regulation of type I IFN-induced STAT3 and ISGF3 transcriptional activities by histone deacetylases (HDACS) 1 and 2. FASEB J 26, 240–9 (2011). [DOI] [PubMed] [Google Scholar]
- Ho H. H. & Ivashkiv L. B. Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J Biol Chem 281, 14111–14118 (2006). [DOI] [PubMed] [Google Scholar]
- Gantier M. P. New perspectives in MicroRNA regulation of innate immunity. J Interferon Cytokine Res 30, 283–289 (2010). [DOI] [PubMed] [Google Scholar]
- David M. Interferons and microRNAs. J Interferon Cytokine Res 30, 825–828 (2010). [DOI] [PubMed] [Google Scholar]
- Krützfeldt J. et al. Silencing of microRNAs in vivo with ‘antagomirs'. Nature 438, 685–689 (2005). [DOI] [PubMed] [Google Scholar]
- Gatfield D. et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev 23, 1313–1326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K. et al. MicroRNA122 is a key regulator of α-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. Nat Commun 2, 338 (2011). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C., Yi M., Lancaster A., Lemon S. & Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309, (2005). [DOI] [PubMed] [Google Scholar]
- Lanford R. E. et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327, 198–201 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C. New Merck and Vertex drugs raise standard of care in hepatitis C. Nat Biotechnol 29, 553–554 (2011). [DOI] [PubMed] [Google Scholar]
- Pedersen I. M. et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449, 919–922 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppiah V. et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet 41, 1100–1104 (2009). [DOI] [PubMed] [Google Scholar]
- Tanaka Y. et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 41, 1105–1109 (2009). [DOI] [PubMed] [Google Scholar]
- Pillai R. S., Bhattacharyya S. N. & Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 17, 118–126 (2007). [DOI] [PubMed] [Google Scholar]
- Honda M. et al. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology 139, 499–509 (2010). [DOI] [PubMed] [Google Scholar]
- Akuta N. et al. Predictors of viral kinetics to peginterferon plus ribavirin combination therapy in Japanese patients infected with hepatitis C virus genotype 1b. J Med Virol 79, 1686–1695 (2007). [DOI] [PubMed] [Google Scholar]
- Akuta N. et al. Substitution of amino acid 70 in the hepatitis C virus core region of genotype 1b is an important predictor of elevated alpha-fetoprotein in patients without hepatocellular carcinoma. J Med Virol 80, 1354–1362 (2008). [DOI] [PubMed] [Google Scholar]
- Dharel N. et al. Potential contribution of tumor suppressor p53 in the host defense against hepatitis C virus. Hepatology 47, 1136–1149 (2008). [DOI] [PubMed] [Google Scholar]
- Otsuka M. et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27, 123–134 (2007). [DOI] [PubMed] [Google Scholar]
- Otsuka M. et al. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J Clin Invest 118, 1944–1954 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M. et al. Genome-wide DNA methylation profiling using Infinium assay. Epigenomics 1, 177–200 (2009). [DOI] [PubMed] [Google Scholar]
- Li L. C. & Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18, 1427–1431 (2002). [DOI] [PubMed] [Google Scholar]
- Schreiber E., Matthias P., Müller M. & Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts', prepared from a small number of cells. Nucleic Acids Res 17, 6419 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L. H., Hatzinisiriou I., Devenish R. J. & Ralph S. J. IFN-gamma priming up-regulates IFN-stimulated gene factor 3 (ISGF3) components, augmenting responsiveness of IFN-resistant melanoma cells to type I IFNs. J Immunol 160, 5475–5484 (1998). [PubMed] [Google Scholar]