Abstract
We present a new model of evolutionary dynamics in one-dimensional space. Individuals are arranged on a cycle. When a new offspring is born, another individual dies and the rest shift around the cycle to make room. This rule, which is inspired by spatial evolution in somatic tissue and microbial colonies, has the remarkable property that, in the limit of large population size, evolution acts to maximize the payoff of the whole population. Therefore, social dilemmas, in which some individuals benefit at the expense of others, are resolved. We demonstrate this principle for both discrete and continuous games. We also discuss extensions of our model to other one-dimensional spatial configurations. We conclude that shift dynamics in one dimension is an unusually strong promoter of cooperative behavior.
1. Introduction
The impact of population structure on ecological and evolutionary dynamics is a topic of great interest (Wright, 1943; Slatkin, 1981; Nowak & May, 1992; Barton, 1993; Durrett & Levin, 1994; Hassell et al., 1994; Dieckmann et al., 2000; Hauert & Doebeli, 2004; Rousset, 2004; Nowak et al., 2010a). Spatial and social structure can promote the evolution of cooperation (Nowak & May, 1992; Nakamaru et al., 1997; Killingback & Doebeli, 1996; van Baalen & Rand, 1998; Mitteldorf & Wilson, 2000; Le Galliard et al., 2003; Nowak, 2006b; Traulsen & Nowak, 2006; Szabó & Fáth, 2007; Helbing & Yu, 2009; Tarnita et al., 2009a), though this effect depends on the underlying details of the evolutionary process (Wilson et al., 1992; Taylor, 1992; Nowak et al., 1994; Hauert & Doebeli, 2004; Ohtsuki et al., 2006; Lion & van Baalen, 2008; Roca et al., 2009a; Korolev & Nelson, 2011; van Veelen et al., 2012).
Evolutionary graph theory provides an elegant framework for studying the effect of fixed population structure on evolutionary dynamics (Lieberman et al., 2005; Santos & Pacheco, 2005; Ohtsuki et al., 2006; Taylor et al., 2007; Szabó & Fáth, 2007; Santos et al., 2008; Roca et al., 2009b; Fowler & Christakis, 2010; Perc & Szolnoki, 2010). In this framework, individuals are located at the nodes of a graph. Interaction and reproduction occur along edges. The one-dimensional cycle is a particularly simple graph topology for which many results can be obtained analytically (Ohtsuki & Nowak, 2006; van Veelen & Nowak, 2012). More generally, there may be two different graphs, one for interaction and one for replacement (Ohtsuki et al., 2007). There are also various approaches for studying the effect of dynamical population structure on social evolution, where individuals can adjust their social ties either at random (Wu et al., 2010b) or depending on game actions and payoffs (Skyrms & Pemantle, 2000; Pacheco et al., 2006a,b; Fu et al., 2008; Antal et al., 2009; Helbing & Yu, 2009; Tarnita et al., 2009a; Perc & Szolnoki, 2010; Poncela et al., 2009; Fehl et al., 2011; Rand et al., 2011).
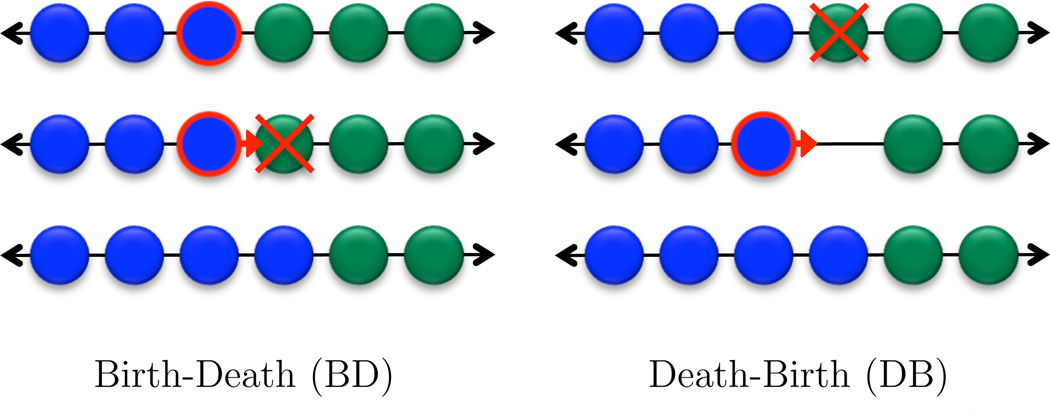
In any evolutionary model, the “update rule” determines how reproduction occurs. One particular rule is called “Birth-Death” (BD) updating. Here an individual is chosen for reproduction (proportional to payoff) and the offspring replaces a randomly chosen neighbor (in the replacement graph). Another rule is called “Death-Birth” (DB) updating. Here a random individual dies and the neighbors compete for the empty site proportional to their payoff. Different update rules can lead to different evolutionary outcomes (Ohtsuki & Nowak, 2006; Ohtsuki et al., 2006).
Here we introduce a new rule, which we call “shift” updating. One individual is chosen for reproduction. The offspring is placed next to the parent. Elsewhere one individual dies. The individuals shift around to redistribute themselves evenly in space. Shift updating is particularly simple in one dimension, which is the topic of this paper. Generalizations to higher dimensions or more complicated graphs are possible and will be studied in subsequent papers.
We find that shift updating is an unusually strong promoter of cooperative behavior, compared to other update rules that have been studied previously. For shift updating, the abundance of cooperators increases primarily because cooperators reproduce deep inside clusters. The clusters then expand according to the shift rule. In contrast, for other update rules such as BD and DB, a cooperator cluster can only expand through reproduction of cooperators that are located near the boundary between cooperators and defectors (Ohtsuki & Nowak, 2006, 2008). But there cooperators can be at a disadvantage relative to the nearby defectors who exploit them.
Shift updating is inspired by cellular reproduction. The idea is that a cell divides and subsequently the population readjusts to accommodate the new cell. A particular example is the organization of epithelial layers in human tissue. The human colon, for instance, contains crypts where cell division leads to a movement of cells along the crypt. It has been argued that this architecture reduces the rate of cancer initiation (Nowak et al., 2003; Michor et al., 2004b). Another example is microbial colonies growing on a at surface. In such colonies, since existing individuals are generally fixed in place, competition dynamics occur primarily at the expanding one-dimensional circumference of the colony (Hallatschek et al., 2007; Hallatschek & Nelson, 2010; Korolev et al., 2010; Nadell et al., 2010). As new offspring arise, the colony boundary expands in a manner similar to the shift dynamic we consider here. We note, however, that each of these biological systems differs in certain ways from our model. In the human colon model of Nowak et al. (2003), cells are arranged in a directed line segment, with stem cells at one end, rather than a cycle. In growing microbial colonies, the boundary is always increasing in size, whereas our model holds population size fixed.
Our paper is organized as follows. Section 2 formally presents our model of evolutionary shift dynamics. Section 3 provides general formulas for fixation probability. Section 4 examines the case of constant selection, showing that evolution in this model neither amplifies nor suppresses selection. We then turn to evolutionary games in Section 5. We obtain simple closed-form results in two cases: the large population limit (Section 5.1) and the weak selection limit (Section 5.2). In each case we obtain the result that, for sufficiently large population sizes, evolution favors the type that maximizes whole-population payoff. Section 6 introduces the possibility of mutation, and shows how this weakens the spatial benefit to cooperation. In Section 7 we turn to continuous games. Using the adaptive dynamics approach (Nowak & Sigmund, 1990; Hofbauer & Sigmund, 1990; Dieckmann & Law, 1996; Metz et al., 1996), we study how the shift dynamics model affects the long-term evolutionary trajectories of continuous traits. Finally, in Section 8 we compare the results obtained from the shift rule with other update rules on the cycle.
2. Model
We consider a cycle with N positions, each occupied by a single individual. In each time-step, each individual is assigned a fecundity (reproductive rate). Fecundities may be constant, depending only on the individual’s type—a case we explore in Section 4. For most of this work, however, we consider fecundities to be determined through game interactions with neighbors, as we discuss in Section 5.
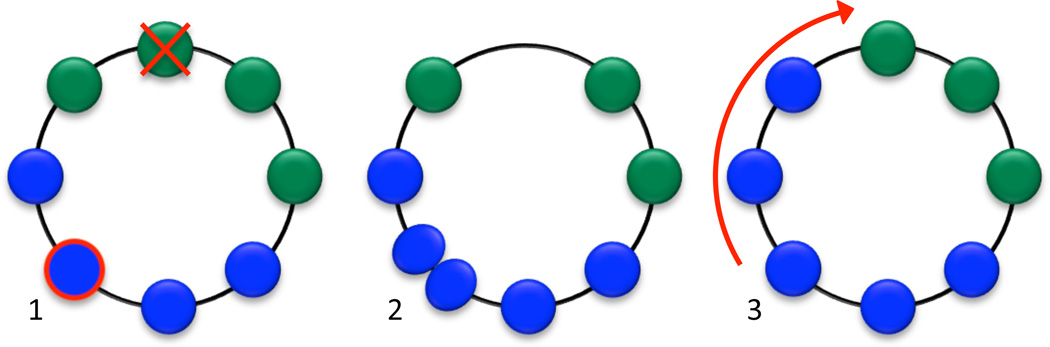
After fecundities are determined, an individual is chosen for reproduction, with probability proportional to fecundity. Another individual is chosen for death at random, with uniform probability. The new individual appears in a position neighboring the parent, and the remaining individuals shift around the cycle until the position of the deceased individual is filled. Since the cycle has rotational symmetry, it does not matter in which direction the birth and shifts occur. It may happen that the parent is itself chosen for death, in which case the new offspring simply replaces the parent. This update rule is illustrated in Figure 1.
Figure 1.
Shift dynamics on a cycle. (1) First, an individual is chosen to reproduce with probability proportional to fecundity, and an individual is chosen for death with uniform probability. (2) The new offspring appears adjacent to the parent. (3) The remaining individuals shift along the cycle until the position of the deceased individual is filled. It does not matter which direction the reproduction and shifts occur—the resulting state is the same (up to a rotation of the cycle).
Though this model could be applied to any number of competing types, we focus here on competition between two types A and B. For the majority of this work we assume that offspring always inherit the type of their parent; the exception is Section 6, where the possibility of mutation is added.
3. Fixation probability
We quantify evolutionary success using the fixation probabilities ρA and ρB (Nowak et al., 2004; Nowak, 2006b). Here, ρA denotes the probability that, starting from a population with one A and (N − 1) B, type A will eventually occupy all N positions. ρB denotes the analogous probability with the roles of A and B reversed. We say that selection favors A if ρA > ρB.
We observe that, absent mutation, any population founded by one individual of one type and (N − 1) individuals of the opposite type will always consist of two “clusters”, each comprised of only one type. In other words, the two types remain spatially segregated. The state of the evolutionary system can therefore be completely characterized by the number k of A individuals.
Because of this, the fixation probability of A can be expressed as (Taylor et al., 2004; Nowak, 2006a)
| (1) |
Here (resp., ) denotes the probability that the number of As will increase (resp., decrease) in the next evolutionary step, when there are currently k individuals of type A. We can express these probabilities as
Using these expressions, we can simplify the ratio to
Substituting into (1) yields the formula
| (2) |
Here fk (resp., gk) denotes the average fecundity of A’s (resp., B’s), when the number of A individuals is k.
Combining the above arguments with the analysis of Taylor et al. (2004), we can express the ratio ρA/ρB as
| (3) |
4. Constant selection
We first consider constant selection. We suppose type B has constant fecundity 1, and A has constant fecundity 1 + r. Thus fk = 1 + r and gk = 1 for each k.
In this case, the birth and death probabilities of each individual are exactly the same as they would be in the well-mixed Moran (1958) process. In other words, this particular spatial structure has no effect on evolutionary dynamics when selection is constant. Using (2), we can express the fixation probability of A as
Since this is the same fixation probability as would be found in the Moran process, we conclude that the shift model is neither an amplifier nor a suppressor of selection, in the language of Lieberman et al. (2005).
5. Discrete games
We now turn to evolutionary games. We begin with the case of two competing types, A and B, who interact according to the game matrix
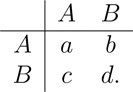 |
(4) |
Individuals play this game with each neighbor. For each individual, we retain the average of the two payoffs obtained from the neighbors.
This average payoff must then be transformed into fecundity (reproductive rate). For this, we consider a positive, differentiable, increasing function F(x), and a real number δ > 0 quantifying the intensity of selection. The mapping from payoff to fecundity is given by π ↦ F (δπ). Specific examples of payoff-to-fecundity mappings are the linear map π ↦ 1 + δπ (with the constraint δπ > −1; see Nowak et al., 2004; Ohtsuki et al., 2006) and the exponential map π ↦ eδπ (Traulsen et al., 2008). Wu et al. (2010a) provide further discussion of payoff-to-fecundity mappings. Unless otherwise specified, the results presented here are independent of the particular choice of F(x).
In general, the expressions (2) and (3) for ρA and ρA/ρB do not reduce to closed-form expressions when considering evolutionary games. However, for two important cases, we can obtain simple conditions for which type is favored. These cases are the large population regime, N ≫ 1, and the weak selection regime, Nδ ≪ 1.
5.1. Large populations
For large population size, the condition for A to be favored is simply a > d. In other words, A must obtain a higher payoff from other A’s than B’s do from other B’s. This result can be interpreted as saying that, in the large population limit, only same-type interactions matter.
This result is intuitive: when population size is large, most individuals reside in the interiors of the two segregated clusters. Thus most A’s have payoff a, and most B’s have payoff d. Since, according to (2), the evolutionary dynamics depend on the average fecundities of the two types, the success of the two types depends predominantly on the same-type payoffs a and d.
We state this result in the following theorem, the proof of which appears in Appendix A.
Theorem 1. For any positive, differentiable, increasing payoff-to-fecundity mapping F(x), and any selection intensity δ > 0, we have: if a > d, then
Thus A is favored (ρA > ρB) in the large population limit if and only if a > d. In short, evolution favors the type that maximizes the whole-population payoff.
As an example, for any Prisoner’s Dilemma (c > a > d > b), Snowdrift (c > a > b > d), or Stag Hunt (a > c ≥ d > b) game, the cooperative strategy is favored over the non-cooperative strategy as long as population size is sufficiently large.
This result readily extends to multiplayer games (Gokhale & Traulsen, 2010) played with nearest neighbors on the cycle (van Veelen & Nowak, 2012). For such games, strategy A is selected over strategy B in large populations if and only the payoff to A interacting with other A’s exceeds the payoff to B interacting with other Bs. Thus, as in two-player games, selection favors the type that obtains the higher payoff from its own kind.
5.2. Weak selection
For weak selection, Tarnita et al. (2009b) proved that to every population structure is associated a “structure coefficient” σ, which is independent of the game played in the population (see also Nowak et al., 2010a; Tarnita et al., 2011). For any particular pairwise game of the form (4), A is favored over B (ρA > ρB) if and only if
| (5) |
The value of σ quantifies the relative importance of same-type versus opposite-type interactions for evolutionary success in a given structure.
In Appendix B we compute the structure coefficient for shift dynamics on a cycle of size N, obtaining
| (6) |
Here hN is the Nth harmonic number:
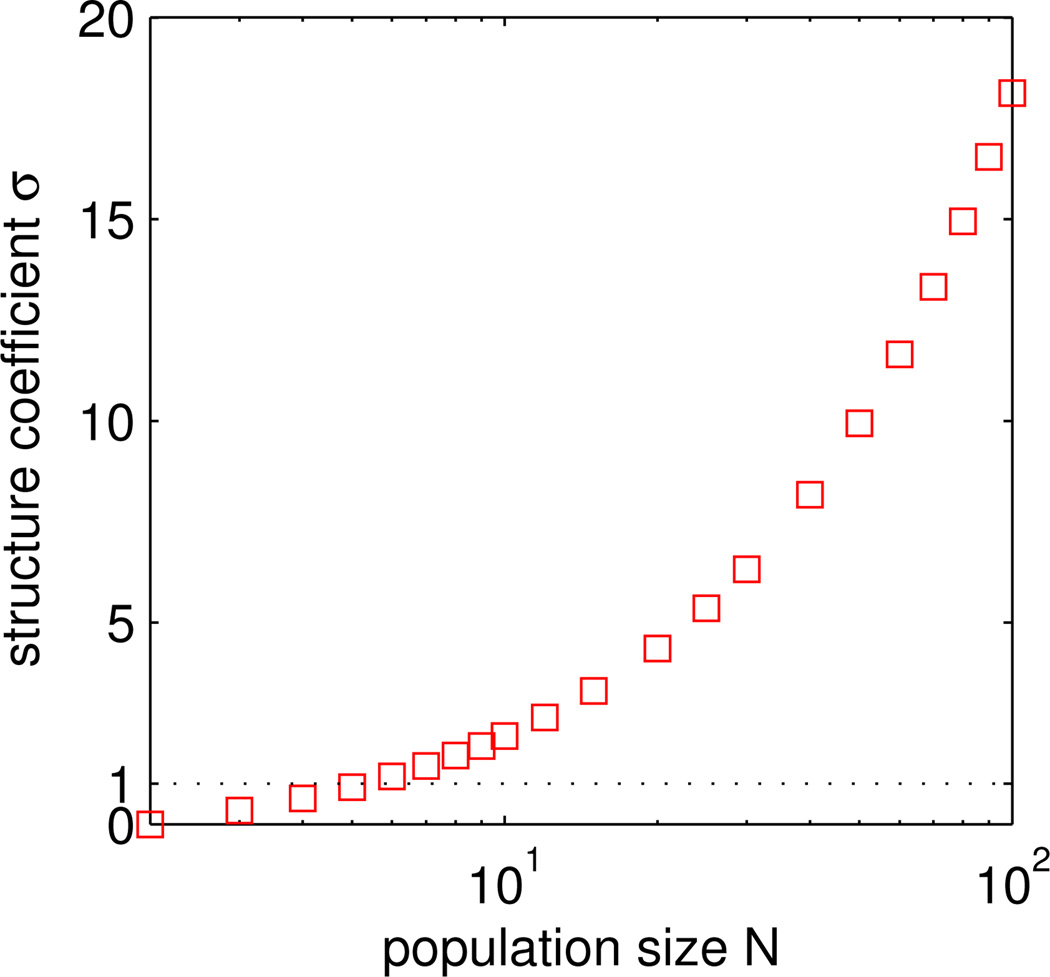
The harmonic number hN can be approximated as γ + ln N, where γ ≈ 0:577 is the Euler-Mascheroni constant. The value of σ therefore grows asymptotically as N/ln N, as shown in Figure 2. This growth reflects the fact that same-type interactions grow increasingly important as N increases.
Figure 2.
The structure coefficient σ for the shift rule, as a function of the population size N, according to (6). A logarithmic scale is used for the horizontal axis. For N = 2 we have σ = 0. The value of σ exceeds one for N ≥ 6. Asymptotically, as N increases, σ increases as N/ln N.
We observe from (6) that σ = 0 for a population of size two. This is because the only relevant state in the N = 2 case consists of one individual of each type. Consequently, selection favors whichever type obtains a higher payoff from the other—b versus c in the payoff matrix (4). For populations of size six or larger, the value of σ exceeds 1, indicating that cooperative strategies are aided by the population structure.
6. Mutation
We can also analyze how mutation affects evolutionary success under shift dynamics. We incorporate mutation into our model by supposing that, whenever a new individual is born, that individual has probability u of having a mutation. An offspring with a mutation has an equal chance of being born as either of the two competing types.
With nonzero mutation, the evolutionary process is ergodic. This means there is a stationary distribution of states to which the time-averaged behavior of the evolutionary process converges. We consider type A to be favored if it has greater than 50% abundance in this stationary distribution, following Antal et al. (2009).
Before continuing, we adopt the following notation: We index the individuals in the population by the integers modolu N (thus i = −1 and i = N − 1 refer to the same individual). We designate individual 0 as a focal individual (by symmetry, this choice does not affect the results). The type of individual i will be denoted si ∈ {A, B}. The fecundity of individual i will be denoted Fi. For simplicity, we assume that the payoff-to-fecundity function F(x) satisfies F(0) = F′ (0) = 1, though our results hold for any positive, differentiable, increasing function F(x).
In the limit of weak selection, Nowak et al. (2010b, Appendix A) showed that, if death probabilities are constant over all states (as they are in our shift model), type A is favored over type B if and only if
| (7) |
Above, b0 is the reproduction probability (per time-step) of the focal individual. The notation indicates an expectation over the stationary distribution of states of the neutral (δ = 0) process, conditioned on the focal individual being of type A. The partial derivative in (7) is also assumed to be taken at δ = 0.
For the shift model in particular, the left-hand side of condition (7) can be expanded as
| (8) |
using the fact that Fi = 1 for all i when δ = 0.
We now restrict our attention to the additive Prisoner’s Dilemma game, with payoff matrix
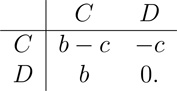 |
(9) |
We will use this game to compute the structure coefficient σ for the shift model with arbitrary mutation rate u. This value of σ can then be used to obtain success conditions for any 2 × 2 game, using relation (5).
For this game, using F′(0) = 1, we have
| (10) |
We obtain the quantities using the method of stationary identity-by-descent probabilities (Rousset & Billiard, 2000; Taylor et al., 2007; Antal et al., 2009; Tarnita et al., 2009a; Allen et al., 2012b). Specifically, we compute the probabilities qk that two individuals located k positions apart on the cycle are identical-by-descent (IBD). Two individuals are IBD if no mutations have occurred in either of their lineages since they diverged from their common ancestor. These probabilities can be obtained using the coalescent method, in which the evolutionary process is traced backwards through time. In Appendix C we use this method to derive recurrence relations for the qk, k = 0;…,N − 1. Though these recurrence relations do not appear to have a general closed-form solution, the qk can be obtained numerically, for any combination of N and u, using Gaussian elimination.
Since mutation is equally likely to result in either type, the IBD probabilities qk are related to the quantities by
Thus (10) can be rewritten as
Combining with (7) and (8), we obtain the result that C is favored if and only if
Here q1 is the probability that two nearest neighbors are IBD. The quantity
is the average IBD probability of two randomly chosen individuals. We conclude that cooperation is favored in the Prisoner’s Dilemma under weak selection if and only if
| (11) |
This condition can be interpreted as a form of Hamilton’s rule. Many studies of inclusive fitness theory (e.g. Rousset & Billiard, 2000; Rousset, 2004; Taylor et al., 2007) use
as a formula for relatedness between neighbors; with this definition, condition (11) becomes equivalent to bR > c.
We note, however, that the condition for successful cooperation takes this simple form only because the shift rule is a “global update rule”, a term that we define and discuss in Section 8.2. For other update rules that do not possess this property, including BD and DB (defined above and in Figure 3), the condition for successful cooperation cannot be written as bR > c with R representing relatedness; see Nowak et al. (2010b).
Figure 3.
Two update rules introduced by Ohtsuki & Nowak (2006). For Birth-Death updating, at each time-step, an individual is chosen to reproduce, with probability proportional to fecundity. The new offspring displaces a neighbor of the parent, chosen randomly with uniform probability. For Death-Birth updating, in each time-step, an individual is chosen to die, with uniform probability. This death leaves a vacancy. The neighbors of this vacant site then compete for the opportunity to reproduce. One of them is chosen, with probability proportional to fecundity, and the offspring of the chosen neighbor fills the vacant site.
Combining (11), and (5), we can express the structure coefficient for the shift rule as
| (12) |
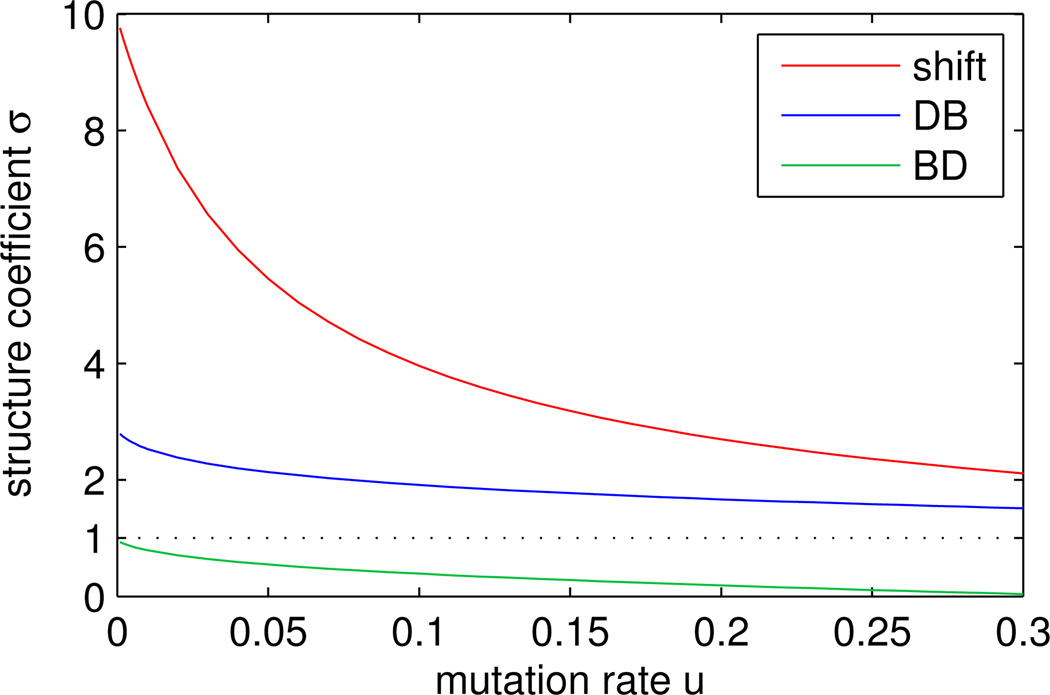
The dependence of σ on u is illustrated in Figure 6.
Figure 6.
The structure coefficient σ as a function of mutation rate u for shift, DB, and BD updating on a cycle of size N = 50. In each case, mutation hinders cooperative strategies by diluting assortment between cooperators. We note, however, that the σ-value for the shift rule, even for the relatively high mutation rate of u = 0.15, exceeds the maximum possible σ-value for DB.
7. Continuous games
We can also consider continuous games. In this case, an individual’s strategy or trait value is represented by a real number (or, for multivariate traits, a vector). The payoff to an individual of trait value x interacting with a partner of trait value y is given by the value of a payoff function G(x; y).
For continuous games, the course of evolution can be studied by considering the long-term trajectory of the trait value x, as it evolves through a sequence of invasion and fixation events. We assume that mutation in x is both rare and incremental. These assumptions allow us to apply the adaptive dynamics approach (Nowak & Sigmund, 1990; Hofbauer & Sigmund, 1990; Dieckmann & Law, 1996; Metz et al., 1996). We also assume that the payoff function G is positive and differentiable in both arguments, and that each individual’s fecundity is equal to the average payoff from interacting with both neighbors.
Though adaptive dynamics is traditionally formulated for well-mixed populations, a recent study (Allen et al., 2012a) has extended the adaptive dynamics approach to a broad class of structured population models. This class includes the shift dynamics model considered here. Under the above assumptions, this work derived the following equation for expected evolutionary trajectories under any model in this class:
Here,
N is population size,
Ne is an effective population size (discussed below),
u is the mutation probability per birth,
v is the variance in the distribution of mutational steps,
σ is the structure coefficient.
The effective population size Ne for a particular model is obtained by comparing the fixation probability of a trait with constant selective advantage to the analogous fixation probability in the baseline Wright-Fisher model. Since for constant selection, fixation probabilities in the shift model are the same as those in the Moran process (see Section 4), the effective population size for the shift model is equal to that of the Moran process, which is N/2.
The expected evolutionary trajectory of the trait value x under the shift model is therefore given by
| (13) |
We observe that, as N → ∞, the equation for expected evolutionary trajectories (13) approaches
This implies that, as N → ∞, evolutionary trajectories perform gradient ascent on the average population payoff G(x; x). In other words, evolution acts to optimize the payoff of the whole population. This stands in stark contrast to evolution in well-mixed or otherwise structured populations, in which social dilemmas can cause whole-population payoff to evolve downward, possibly even leading to extinction of the population (Gyllenberg & Parvinen, 2001).
Equation (13) for adaptive dynamics with the shift model can readily be extended to multivariate (vector-valued) traits and more general mutation schemes (Allen et al., 2012a). An important open question is to understand how shift dynamics affect the possibility of evolutionary branching (Geritz et al., 1997; Dieckmann & Doebeli, 1999; Doebeli & Dieckmann, 2003; Doebeli et al., 2004), in which multiple trait values coexist and evolve in different directions.
8. Comparison to other update rules
In their study of evolutionary games on cycles Ohtsuki & Nowak (2006) considered a number of update rules, all of which had the feature that individuals remain fixed in place until they are replaced or updated. Of the update rules they considered, two in particular have been extensively studied in subsequent research: the death-birth (DB) rule and the birth-death (BD) rule. We define and illustrate these update rules in Figure 3.
8.1. Conditions for success in evolutionary games
Different update rules can lead to strikingly different outcomes for evolutionary games. For example, for the additive Prisoner’s Dilemma game (9), cooperation is never favored under BD updating. For DB updating, cooperation is favored on a large cycle if b/c > 2. In contrast, for the shift rule, cooperation is favored on a large cycle whenever b > c.
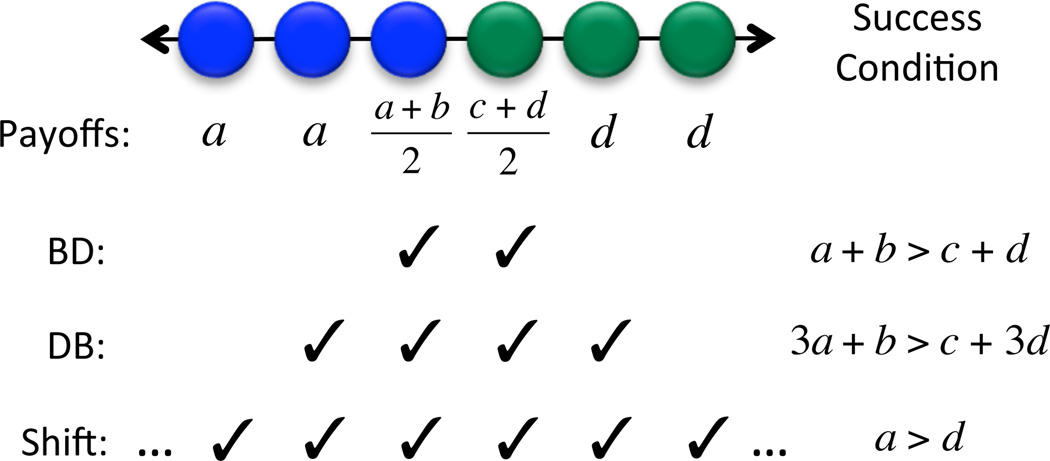
We can understand the different outcomes of these rules by examining the dynamics of clusters—connected chains of individuals who share a common type. For all three update rules (BD, DB, and shift), if there is no further mutation after an initial mutant is introduced, then the population will be comprised of two clusters until one type reaches fixation. The update rules differ in the the mechanisms by which these clusters expand or contract (Figure 4):
For BD updating, clusters change size only if an individual at an boundary (where the two types meet) is selected to reproduce. Thus evolutionary success is determined by the payoffs of individuals at cluster boundaries. To see whether type A is favored in the general evolutionary game (4), we compare the payoff of an A individual at a boundary, (a + b)/2, to that of a B individual at a boundary, (c + d)/2, yielding the condition a + b > c + d.
For DB updating, for a cluster to change size, an individual an a boundary must be selected to die. Competition then occurs between the two neighbors of this now-vacant site. We observe that one of these neighbors is at a boundary, while the other is in the interior of its respective cluster. Thus evolutionary success is determined by comparing the payoffs of an individual at a boundary versus another of the opposing type in a cluster interior. There are therefore two relevant comparisons, a versus (c + d)/2, and (a + b)/2 versus d. Combining these linearly (which is meaningful for weak selection or exponential payoff-to-fecundity map) yields the condition 3a + b > c + 3d.
For shift updating, changes in cluster size can result from any reproduction event, not just those at or near boundaries. Thus evolutionary success depends on the payoffs of all individuals. For large populations, in a generic state of the evolutionary process, most individuals reside in a cluster interior. This implies the success condition a > d.
Figure 4.
Cluster expansion under different update rules. Payoffs are given in terms of the entries of the game matrix (4). The check marks indicate the individuals whose payoffs matter for cluster expansion. For Birth-Death (BD) updating, success is determined by comparing the payoffs of individuals at cluster boundaries. For Death-Birth (DB) updating, an individual at an interface must be compared to an individual of an opposing type in a cluster interior. For shift updating, success depends on the payoffs of all individuals. If the population size is large then most individuals have payoff a or d. These comparisons lead to the success conditions shown at right, which are valid in the limit of large population size, for the exponential payoff-to-fecundity mapping π ↦ eδπ with any selection strength δ > 0.
Though the above calculations are heuristic, the conditions they yield are correct in the limit of large population size, for the exponential payoff-to-fecundity mapping π ↦ eδπ with any selection strength δ > 0. If instead a linear payoff-to-fecundity mapping is used, the conditions for BD and shift remain the same, while the condition for DB becomes nonlinear and varies with the selection strength; see van Veelen & Nowak (2012).
8.2. Local vs. global updating
The shift rule is a global update rule, in the terminology of Nathanson et al. (2009). This means that the birth and death probabilities of an position i depend only on its fecundity, Fi, and the total fecundity of the population, (using the notation introduced in Section 6). In contrast, BD and DB are local update rules, meaning that the birth and/or death probabilities of position i also depend on the fecundities of i’s neighbors. Local update rules may induce additional forms of local interaction, beyond those represented by the game. For example, under BD updating, for example, it is disadvantageous to be located adjacent to a high-fecundity individual, for then one is more likely to be displaced during the next transition.
For global update rules, the structure coefficient σ quantifies the degree of spatial assortment between like types arising from the evolutionary dynamics; see (12) and related formulas obtained by Nathanson et al. (2009). For local update rules, the value of σ is also affected by any additional local interactions induced by the update rule.
8.3. Dependence on population size
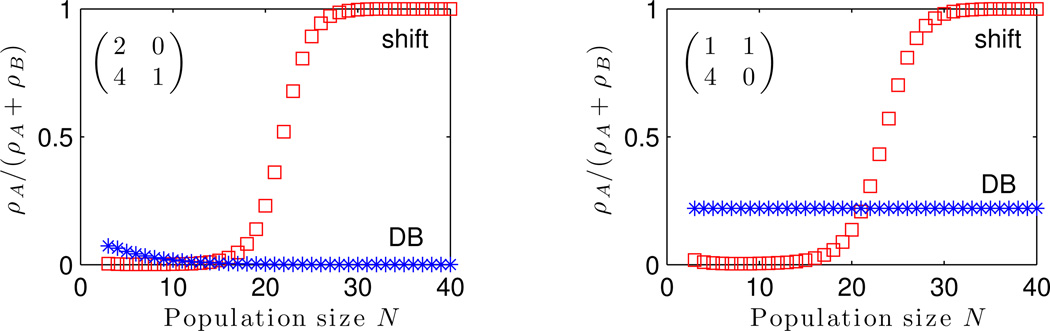
Combining (3) with previous analyses of the DB process on the cycle (Ohtsuki & Nowak, 2006; van Veelen & Nowak, 2012), we can see how the success of cooperation varies with the game matrix, update rule, and population size. Figure 5 illustrates this for a Prisoner’s Dilemma and a snowdrift game. In each case, for sufficiently large N, shift dynamics favor the type that maximizes the whole-population payoff, as guaranteed by Theorem 1. The same is not necessarily true for DB updating. However, for small populations, DB can sometimes favor cooperative behaviors more than shift does.
Figure 5.
The success of cooperative strategies on the cycle for shift and death-birth updating, as a function of population size N. On the left is a Prisoner’s Dilemma, on the right is a Snowdrift game. We use the exponential payoff-to-fecundity mapping, π ↦ eδπ, with selection strength δ = 1. In each case, for sufficiently large N, shift dynamics favor the strategy that maximizes whole-population payoff (while the same is not necessarily true for DB). However, for small population sizes, DB provides a larger benefit to cooperation compared to the shift rule in the examples shown here.
8.4. Dependence on mutation rate
In previous work (Allen et al., 2012b), we investigated how nonzero mutation rates affect success in spatial evolutionary games under the DB and BD update rules. We found that mutation tends to weaken the spatial benefit to cooperation, by diluting the clustering between cooperators.
For shift dynamics, we find the same qualitative effect: the structure coeffiecient σ—and hence the success of cooperative strategies—decreases with u (Figure 6).
9. Variations on the one-dimensional model
Our main result—that that selection favors A over B, in the large population limit, if and only if a > d—depends on two qualitative features of our model. First, due to one-dimensional geometry, the population is typically composed of clusters of like cells. Thus, in a typical configuration of a large population, most A’s receive payoff a and most B’s receive payoff d. Second, since shift is a global update rule (see Section 8.2), selection favors the type with the larger average payoff, which is type A if and only if a > d.
These two basic features—one-dimensional geometry and global updating—are also shared by a number of variants of our model, including the following:
Changing cycle size: One can consider variations in which birth and death events are decoupled, so that the total population size varies stochastically. The cycle expands with each division and contracts with each death. One can then study the evolutionary dynamics conditioned on non-extinction of the population. To prevent the population size from growing arbitrarily large, a density limitation can be introduced so that division rates decrease as the population grows.
Empty sites: One can also consider a cycle of fixed size in which some sites may be empty. A death results in an empty site, and empty sites are filled by reproduction (with shifts). Again, the dynamics can be studied conditioned on non-extinction of the population.
Chains with endpoints: Instead of a cycle, one can consider a linear chain of cells. Shits might occur in both directions, or they might occur in only one direction, as in the colon cancer model of Nowak et al. (2003). There are also different conventions for cell death: one might suppose death affects random cells within the chain, or only cells that are shifted past the end of the chain (which again corresponds to the colon cancer model of Nowak et al., 2003).
Cell stacking: One might suppose that sometimes, instead of appearing to the side of the parent and shifting other cells aside, a newly created cell may stack on top of its parent. It is possible to incorporate such stacking into our model while maintaining one-dimensional spatial geometry. For example, one could suppose that each cell plays the game with all other cells in the same or in neighboring stacks. A newly created cell can join its parent’s stack or initiate a new stack to either side of the parent (resulting in shifts). Stacks shift as a single unit, and disappear when their last cell dies.
Since the above variations maintain one-dimensional geometry and global updating, the argument at the beginning of this section applies in principle to them as well. Thus we expect the result that selection favors A, in the large population limit, if and only if a > d to hold for these variations. We caution, however, that the correctness of this argument may depend on certain details of how these variations on the model are implemented (for example, how game payoffs are calculated in the presence of empty sites, whether stacks are allowed to grow indefinitely, etc.).
10. Outlook for higher dimensions
So far we have considered only one-dimensional spatial geometry. One may also ask to what extent shift dynamics can promote cooperation in two- or three-dimensional space. This question is made complicated by the fact that there are a number of different update rules that can be considered higher-dimensional analogues of the shift rule.
We have studied a number of higher-dimensional analogues of shift dynamics in preliminary simulations. These include
Lattice-structured populations in which shifts occur in a randomly chosen direction from the reproducer,
Populations structured as a regular graph, in which shifts occur along a randomly determined path from the reproducer,
Populations located in continuous space, in which physical forces push cells away from their neighbors.
In our preliminary simulations of models based on (i) and (ii), we have observed only a weak spatial benefit to cooperative strategies. We traced this weakness to the fact that spatial clusters of cooperators are often disrupted by shifts arising from reproductions elsewhere. In contrast, for continuous-space models (iii), our simulations show only a minimal disruption effect, and consequently, a strong spatial benefit to cooperation. We hope to present these results in future work.
11. Discussion
We have studied a new update rule of spatial evolution. Offspring do not replace their parents’ neighbors, but shift them aside. Shift updating is a strong promoter of cooperation in one dimensional models. In the limit of large population size, social dilemmas are avoided entirely: selection favors the type that maximizes the payoff of the whole population. Here natural selection favors what is good for the population.
Our work may have implications for social evolution in microbes (Crespi, 2001; Velicer, 2003; Fiegna et al., 2006; Santorelli et al., 2008; Gore et al., 2009; Damore & Gore, 2011). While microbial colony evolution differs in important ways from our shift model, the two systems share the feature that reproductions do not necessarily co-occur with death of a neighbor (Hallatschek et al., 2007). This lack of direct spatial competition between neighbors is what enables the unusually high degree of success for cooperators in our model. Thus the spatial benefit to cooperation in microbes may be stronger than traditional evolutionary game theory models suggest (see, for example, Gore, 2010). Since cooperative dilemmas in microbes have been shown to favor the evolution of multicellular clumping (Koschwanez et al., 2011), models with shift dynamics may be useful in understanding evolutionary transitions from unicellularity to multicellularity (Maynard Smith & Szathmáry, 1997; Grosberg & Strathmann, 2007; Knoll, 2011).
Shift dynamics may also be relevant in somatic evolution, including cancer development (Moolgavkar & Knudson, 1981; Wysocki et al., 1986; Nowak et al., 2002; Michor et al., 2004a; Boland & Goel, 2005; Michor et al., 2005; Wodarz & Komarova, 2005; Iwasa et al., 2006; Merlo et al., 2006; Beerenwinkel et al., 2007; Gatenby & Gillies, 2008; Jones et al., 2008; Meza et al., 2008; Attolini & Michor, 2009; Bozic et al., 2010; Durrett & Moseley, 2010; Yachida et al., 2010). Spatial tumor structure, including with shift-type dynamics (Nowak et al., 2003; Michor et al., 2004b), has been shown to significantly affect the waiting time to cancer (Komarova, 2006; Martens et al., 2011). While somatic evolution of cancer is typically not viewed through the lens of game theory, there are opportunities for cancer cells to cooperate by sharing resources or inuencing the tumor microenvironment (Axelrod et al., 2006; Bidard et al., 2008; Nadell et al., 2010). Spatial evolution with shift dynamics may promote cooperation among neighboring cells in healthy or cancerous tissue.
We emphasize, however, that toy models of shift dynamics, such as the model considered here, may overlook important features of spatial evolution in microbial colonies and somatic tissues. One particularly interesting consideration is that the spatial geometry may change through time. For example, an initially two-dimensional colony or tissue may become three-dimensional as cells layer on top of one another. Incorporating these spatial dynamics into future models can provide a closer interplay between theory and experiment, and may reveal further nuances in how spatial structure affects cooperative behavior in cells.
We investigate a new model of one-dimensional spatial evolution among cells
When a cell divides, it does not replace a neighbor, but shifts neighbors aside
We find a very strong benefit to cooperative strategies
For large populations, selection maximizes whole population fitness
Acknowledgements
We thank Jeff Gore for conversations that inspired this work. We also thank Kyle A. Ward, José Reyes, and Anna S. Roth for their work on higher-dimensional analogues of the shift model. B. A. is supported by the Foundational Questions in Evolutionary Biology initiative of the John Templeton Foundation.
Appendix A. Evolutionary games in the large population limit
This appendix provides the proof of Theorem 1, which states that, for any game matrix
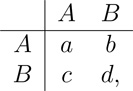 |
fecundity function F, and selection intensity δ > 0, if a > d then
Though we consider only two-player games, the extension to multiplayer games (Gokhale & Traulsen, 2010; van Veelen & Nowak, 2012) is straight-forward.
Proof. We begin by calculating payoffs in each state. We adopt the shorthand notation for the fecundity of a type X individual interacting with types Y and Z, with X, Y, Z ∈ {A, B}. So, for example, and
Suppose there are k cooperators a population of size N ≥ 3. We first calculate the average payoff to A’s:
- For k ≥ 2, there are two A’s that have one neighbor of each type, and the rest of the A’s have two A neighbors. The average payoff to A’s is
(A.1) For k = 1, the lone A has payoff .
Now we calculate the average payoff to B’s:
- For k ≤ N − 2, there are two B’s that have one neighbor of each type, and the rest of the B’s have two B neighbors. The average payoff to B’s is
(A.2) For k = N − 1, the lone B has payoff
For the remainder of the proof we restrict to the case a > d; the case a < d will follow by interchanging the roles of A and B. Note that since F is increasing, a > d implies .
Turning first to the fixation probability ρA, we observe that that, according to equation (2) of the main text, limN→∞ ρA > 0 if and only if
| (A.3) |
We will show that this limit converges by establishing the following two bounds on gk/fk:
Bound 1 : There exists an M ≥ 1 such that gk/fk ≤ M for all N ≥ 1 and 1 ≤ k ≤ N − 1.
Bound 2 : There exists a r, 0 < r < 1, and integers K1 ≥ 1, K2 ≥ 2 (independent of N and k) such that for all N ≥ 1, K1 < k ≤ N − K2 implies gk/fk < r.
Once these two bounds have been established, we show that the limit (A.3) converges—and thus limN→∞ ρA > 0—by the following argument:
The third line above uses the facts that M ≥ 1 (hence Mx is nondecreasing in x) and r < 1 (hence rx is decreasing in x).
For Bound 1, we simply let
To establish Bound 2, we define
which is less than 1 since . We define K1 and K2 by
where ⌊ ⌋ and ⌈ ⌉ respectively denote the oor function (the greatest integer less than or equal to the argument) and the ceiling function (the least integer greater than or equal to the argument). These values of K1 and K2 are chosen so that
| (A.4) |
and
| (A.5) |
Therefore, if K1 < k ≤ N − K2, then applying (A.1), (A.2), (A.4), and (A.5), we have
as required in the statement of Bound 2. This completes the proof that limN→∞ ρA > 0.
To show that limN→∞ ρB = 0, we look at the ratio of fixation probabilities ρB/ρA. Combining formula (3) from the main text with Bounds 1 and 2 yields
The right-hand side above converges to zero as N → ∞, since r < 1. Therefore, limN→∞ ρB/ρA = 0: Since ρA is bounded between 0 and 1, ρB must converge to zero.
Appendix B. Evolutionary games with weak selection
In this appendix we calculate the value of the structure coefficient σ for the shift model. Since the value of σ is independent of the evolutionary game, its value can be computed by considering any particular game (Tarnita et al., 2009b). We consider the additive Prisoner’s Dilemma game:
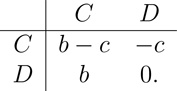 |
The value of σ is also independent of the choice of the fecundity function F (Tarnita et al., 2009b). This is because the values F(0) and F′(0) suffice to determine fixation probabilities under weak selection (Wu et al., 2010a). We therefore use the linear payoff-to-fitness mapping π ↦ 1 + δπ (that is, we choose F(x) = 1 + x).
The formulas for the average payoffs fk and gk, obtained in Appendix A, simplify greatly in this case, due to the additivity of the game and linearity of F. For each k = 1;…, N − 1, the average payoffs to cooperators and defectors, respectively, simplify to
Applying (main text, 3), we compute the ratio ρC/ρD as
The approximation in the second line above is valid in the weak selection regime Nδ ≪ 1.
Using this result, cooperation is favored in the sense ρC/ρD > 1 if and only if
| (B.1) |
The value of the structure coefficient σ can be obtained from the critical benefit-to-cost ratio (b/c)* by
Substituting the right-hand side of (B.1) for (b/c)* we calculate:
Appendix C. Identity-by-descent probabilities
To obtain recurrence relations between the IBD probabilities qk, we consider the evolutionary process backwards in time. The reverse of a single time-step in the model can be described as follows:
Two pairs of neighboring individuals are chosen at random (the same pair can be chosen both times).
A new individual appears between the first pair of individuals (this is the reverse of a death).
The second pair of individuals fuse together (this is the reverse of a birth).
If the same pair is chosen twice, then a new individual appears and instantly fuses with one of the two neighbors (this is the reverse of an individual reproducing and dying in the same time-step). In each fusion event, one individual is randomly designated the parent, and the other is designated the offspring.
In the neutral (δ = 0) process, the two neighboring pairs are chosen independently with uniform probability, and the designations of parent and offspring occur with equal probability for the two fusing individuals.
We introduce mutation into this reverse-evolutionary process by supposing that, with each fusion event, the offspring has probability u of being a mutant (thus destroying any identity by descent with any other individual).
Consider two individuals, labeled α and β, alive in the current state of the system. α and β are IBD if and only if
Neither α nor β is a new offspring, and α and β were IBD in the previous state,
α is a new offspring, α’s parent was IBD to β in the previous state, and α was born without a mutation, or
β is a new offspring, β’s parent was IBD to α in the previous state, and β was born without a mutation.
We must also incorporate the possibility that the distance between α and β may change over the course of a time step, due to shifts. To this end, consider two positions i and j in the current state of the system, where i and j are integers modulo N. (We emphasize that i and j represent positions, in contrast to α and β above, which represent individuals.) Using our description of the reverse process, we can say that the current occupants of i and j are IBD if and only if
-
An appearance occurs in between one of the pairs (i, i + 1),…, (j − 1, j), and
A fusion occurs in one of the pairs (i + 1; i + 2),…,(j − 2, j − 1), and positions i and j are IBD in the previous time-step.
A fusion occurs in one of the pairs (j + 1, j + 2),…, (i − 2, i − 1), and positions i and j + 1 are IBD in the previous time-step.
A fusion occurs in one of the pairs (i, i + 1) or (j − 1, j), positions i and j are IBD in the previous time-step, and neither of the current occupants of i or j is a new mutant offspring.
A fusion occurs in one of the pairs (i − 1, i) or (j, j + 1), positions i and j + 1 are IBD in the previous time-step, and neither of the current occupants of i or j is a new mutant offspring.
- An appearance occurs in between one of the pairs (j, j + 1),…,( i − 1, i), and
- A fusion occurs in one of the pairs (i + 1, i + 2),…,(j − 2, j − 1), and positions i and j − 1 are IBD in the previous time-step.
- A fusion occurs in one of the pairs (j + 1, j + 2), …, (i − 2, i − 1), and positions i and j are IBD in the previous time-step.
- A fusion occurs in one of the pairs (i, i + 1) or (j − 1, j), positions i and j − 1 are IBD in the previous time-step, and neither of the current occupants of i or j is a new mutant offspring.
- A fusion occurs in one of the pairs (i − 1; i) or (j, j + 1), positions i and j are IBD in the previous time-step, and neither of the current occupants of i or j is a new mutant offspring.
Special consideration must also be given to the case in which i and j are neighboring positions, and thus their occupants may fuse during a reverse time-step.
Noting that the stationary IBD probabilities depend only on relative, rather than absolute positions, we use the above case-by-case analysis to obtain the following recurrence relations between the stationary IBD probabilities qk:
Though these relations do not appear to admit a general closed-form solution, numerical values of qk can be obtained for arbitrary u, N and k via Gaussian elimination.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting galley proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen B, Nowak MA, Dieckmann U. Adaptive dynamics with interaction structure. 2012a doi: 10.1086/670192. Preprint,. [DOI] [PubMed] [Google Scholar]
- Allen B, Traulsen A, Tarnita CE, Nowak MA. How mutation affects evolutionary games on graphs. Journal of Theoretical Biology. 2012b;299:97–105. doi: 10.1016/j.jtbi.2011.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antal T, Ohtsuki H, Wakeley J, Taylor PD, Nowak MA. Evolution of cooperation by phenotypic similarity. Proceedings of the National Academy of Sciences of the USA. 2009;106(21):8597–8600. doi: 10.1073/pnas.0902528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attolini CS-O, Michor F. Evolutionary theory of cancer. Annals of the New York Academy of Sciences. 2009;1168(1):23–51. doi: 10.1111/j.1749-6632.2009.04880.x. [DOI] [PubMed] [Google Scholar]
- Axelrod R, Axelrod DE, Pienta KJ. Evolution of cooperation among tumor cells. Proceedings of the National Academy of Sciences. 2006;103(36):13474–13479. doi: 10.1073/pnas.0606053103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH. The probability of fixation of a favoured allele in a subdivided population. Genetical Research. 1993;62:149–157. [Google Scholar]
- Beerenwinkel N, Antal T, Dingli D, Traulsen A, Kinzler KW, Velculescu VE, Vogelstein B, Nowak MA. Genetic progression and the waiting time to cancer. PLoS Comput Biol. 2007;3(11):e225. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidard F-C, Pierga J-Y, Vincent-Salomon A, Poupon M-F. A “class action” against the microenvironment: do cancer cells cooperate in metastasis? Cancer and Metastasis Reviews. 2008;27:5–10. doi: 10.1007/s10555-007-9103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland CR, Goel A. Somatic evolution of cancer cells. Seminars in Cancer Biology. 2005;15(6):436–450. doi: 10.1016/j.semcancer.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, Karchin R, Kinzler KW, Vogelstein B, Nowak MA. Accumulation of driver and passenger mutations during tumor progression. Proceedings of the National Academy of Sciences. 2010;107(43):18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi BJ. The evolution of social behavior in microorganisms. Trends in Ecology & Evolution. 2001;16(4):178–183. doi: 10.1016/s0169-5347(01)02115-2. [DOI] [PubMed] [Google Scholar]
- Damore JA, Gore J. Understanding microbial cooperation. Journal of Theoretical Biology. 2011 doi: 10.1016/j.jtbi.2011.03.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann U, Doebeli M. On the origin of species by sympatric speciation. Nature. 1999;400(6742):354–357. doi: 10.1038/22521. [DOI] [PubMed] [Google Scholar]
- Dieckmann U, Law R. The dynamical theory of coevolution: a derivation from stochastic ecological processes. Journal of Mathematical Biology. 1996;34(5):579–612. doi: 10.1007/BF02409751. [DOI] [PubMed] [Google Scholar]
- Dieckmann U, Law R, Metz JAJ. The Geometry of Ecological Interactions: Simplifying Spatial Complexity. Cambridge: Cambridge University Press; 2000. [Google Scholar]
- Doebeli M, Dieckmann U. Speciation along environmental gradients. Nature. 2003;421(6920):259–264. doi: 10.1038/nature01274. [DOI] [PubMed] [Google Scholar]
- Doebeli M, Hauert C, Killingback T. The evolutionary origin of cooperators and defectors. Science. 2004;306(5697):859–862. doi: 10.1126/science.1101456. [DOI] [PubMed] [Google Scholar]
- Durrett R, Levin S. The importance of being discrete (and spatial) Theoretical Population Biology. 1994;46(3):363–394. [Google Scholar]
- Durrett R, Moseley S. Evolution of resistance and progression to disease during clonal expansion of cancer. Theoretical Population Biology. 2010;77(1):42–48. doi: 10.1016/j.tpb.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Fehl K, van der Post DJ, Semmann D. Co-evolution of behaviour and social network structure promotes human cooperation. Ecology Letters. 2011 doi: 10.1111/j.1461-0248.2011.01615.x. in press. [DOI] [PubMed] [Google Scholar]
- Fiegna F, Yu YTN, Kadam SV, Velicer GJ. Evolution of an obligate social cheater to a superior cooperator. Nature. 2006;441(7091):310–314. doi: 10.1038/nature04677. [DOI] [PubMed] [Google Scholar]
- Fowler JH, Christakis NA. Cooperative behavior cascades in human social networks. Proceedings of the National Academy of Sciences. 2010;107(12):5334–5338. doi: 10.1073/pnas.0913149107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu F, Hauert C, Nowak MA, Wang L. Reputation-based partner choice promotes cooperation in social networks. Physical Review E. 2008;78(2):026117. doi: 10.1103/PhysRevE.78.026117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nature Reviews Cancer. 2008;8(1):56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- Geritz SAH, Kisdi E, Meszéna G, Metz JAJ. Evolutionarily singular strategies and the adaptive growth and branching of the evolutionary tree. Evolutionary Ecology. 1997;12(1):35–57. [Google Scholar]
- Gokhale CS, Traulsen A. Evolutionary games in the multiverse. Proceedings of the National Academy of Sciences of the USA. 2010;107(12):5500. doi: 10.1073/pnas.0912214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore J. American Physical Society March Meeting. vol. 55. American Physical Society; 2010. Games microbes play: the game theory behind cooperative sucrose metabolism in yeast. [Google Scholar]
- Gore J, Youk H, Van Oudenaarden A. Snowdrift game dynamics and facultative cheating in yeast. Nature. 2009;459(7244):253–256. doi: 10.1038/nature07921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosberg RK, Strathmann RR. The evolution of multicellularity: a minor major transition? Annual Review of Ecology and Systematics. 2007;38:621–654. [Google Scholar]
- Gyllenberg M, Parvinen K. Necessary and sufficient conditions for evolutionary suicide. Bulletin of Mathematical Biology. 2001;63:981–993. doi: 10.1006/bulm.2001.0253. [DOI] [PubMed] [Google Scholar]
- Hallatschek O, Hersen P, Ramanathan S, Nelson DR. Genetic drift at expanding frontiers promotes gene segregation. Proceedings of the National Academy of Sciences. 2007;104(50):19926–19930. doi: 10.1073/pnas.0710150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallatschek O, Nelson DR. Life at the front of an expanding population. Evolution. 2010;64(1):193–206. doi: 10.1111/j.1558-5646.2009.00809.x. [DOI] [PubMed] [Google Scholar]
- Hassell MP, Comins HN, May RM. Species coexistence and self-organizing spatial dynamics. Nature. 1994;370:290–292. [Google Scholar]
- Hauert C, Doebeli M. Spatial structure often inhibits the evolution of cooperation in the snowdrift game. Nature. 2004;428(6983):643–646. doi: 10.1038/nature02360. [DOI] [PubMed] [Google Scholar]
- Helbing D, Yu W. The outbreak of cooperation among success-driven individuals under noisy conditions. Proceedings of the National Academy of Sciences of the USA. 2009;106(10):3680–3685. doi: 10.1073/pnas.0811503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer J, Sigmund K. Adaptive dynamics and evolutionary stability. Applied Mathematics Letters. 1990;3(4):75–79. [Google Scholar]
- Iwasa Y, Nowak MA, Michor F. Evolution of resistance during clonal expansion. Genetics. 2006;172(4):2557–2566. doi: 10.1534/genetics.105.049791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Chen W, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, Traulsen A, Nowak MA, Siegel C, Velculescu VE, Kinzler KW, Vogelstein B, Willis J, Markowitz SD. Comparative lesion sequencing provides insights into tumor evolution. Proceedings of the National Academy of Sciences. 2008;105(11):4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killingback T, Doebeli M. Spatial evolutionary game theory: hawks and doves revisited. Proceedings of the Royal Society B: Biological Sciences. 1996;263(1374):1135–1144. [Google Scholar]
- Knoll AH. The multiple origins of complex multicellularity. Annual Review of Earth and Planetary Sciences. 2011;39:217–239. [Google Scholar]
- Komarova N. Spatial stochastic models for cancer initiation and progression. Bulletin of Mathematical Biology. 2006;68:1573–1599. doi: 10.1007/s11538-005-9046-8. [DOI] [PubMed] [Google Scholar]
- Korolev KS, Avlund M, Hallatschek O, Nelson DR. Genetic demixing and evolution in linear stepping stone models. Reviews of Modern Physics. 2010;82:1691–1718. doi: 10.1103/RevModPhys.82.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev KS, Nelson DR. Competition and cooperation in one-dimensional stepping-stone models. Phys. Rev. Lett. 2011;107:088103. doi: 10.1103/PhysRevLett.107.088103. [DOI] [PubMed] [Google Scholar]
- Koschwanez JH, R. Foster K, W. Murray A. Sucrose utilization in budding yeast as a model for the origin of undifferentiated multicellularity. PLoS Biol. 2011;9(8):e1001122. doi: 10.1371/journal.pbio.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Galliard J-F, Ferrièere R, Dieckmann U, Tonsor S. The adaptive dynamics of altruism in spatially heterogeneous populations. Evolution. 2003;57(1):1–17. doi: 10.1111/j.0014-3820.2003.tb00211.x. [DOI] [PubMed] [Google Scholar]
- Lieberman E, Hauert C, Nowak M. Evolutionary dynamics on graphs. Nature. 2005;433(7023):312–316. doi: 10.1038/nature03204. [DOI] [PubMed] [Google Scholar]
- Lion S, van Baalen M. Self-structuring in spatial evolutionary ecology. Ecology Letters. 2008;11(3):277–295. doi: 10.1111/j.1461-0248.2007.01132.x. [DOI] [PubMed] [Google Scholar]
- Martens EA, Kostadinov R, Maley CC, Hallatschek O. Spatial structure increases the waiting time for cancer. New Journal of Physics. 2011;13(11):115014. doi: 10.1088/1367-2630/13/11/115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Szathmáry E. The Major Transitions in Evolution. USA: Oxford University Press; 1997. [Google Scholar]
- Merlo LMF, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nature Reviews Cancer. 2006;6(12):924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- Metz JAJ, Geritz SAH, Meszéna G, Jacobs FA, van Heerwaarden JS. Adaptive dynamics, a geometrical study of the consequences of nearly faithful reproduction. In: van Strien SJ, Lunel SMV, editors. Stochastic and Spatial Structures of Dynamical Systems. Amsterdam: KNAW Verhandelingen, Afd; 1996. pp. 183–231. [Google Scholar]
- Meza R, Jeon J, Moolgavkar SH, Luebeck EG. Age-specific incidence of cancer: phases, transitions, and biological implications. Proceedings of the National Academy of Sciences. 2008;105(42):16284–9. doi: 10.1073/pnas.0801151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michor F, Iwasa Y, Nowak MA. Dynamics of cancer progression. Nature Reviews Cancer. 2004a;4(3):197–205. doi: 10.1038/nrc1295. [DOI] [PubMed] [Google Scholar]
- Michor F, Iwasa Y, Rajagopalan H, Lengauer C, Nowak MA. Linear model of colon cancer initiation. Cell Cycle. 2004b;3(3):358–362. [PubMed] [Google Scholar]
- Michor F, Iwasa Y, Vogelstein B, Lengauer C, Nowak MA. Can chromosomal instability initiate tumorigenesis? Seminars in Cancer Biology. 2005;15(1):43–49. doi: 10.1016/j.semcancer.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Mitteldorf J, Wilson DS. Population viscosity and the evolution of altruism. Journal of Theoretical Biology. 2000;204(4):481–496. doi: 10.1006/jtbi.2000.2007. [DOI] [PubMed] [Google Scholar]
- Moolgavkar S, Knudson A. Mutation and cancer: a model for human carcinogenesis. Journal of the National Cancer Institute. 1981;66(6):1037–1052. doi: 10.1093/jnci/66.6.1037. [DOI] [PubMed] [Google Scholar]
- Moran PAP. Random processes in genetics. Mathematical Proceedings of the Cambridge Philosophical Society. 1958;54(01):60–71. [Google Scholar]
- Nadell CD, Foster KR, Xavier JB. Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput Biol. 2010;6(3):e1000716. doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamaru M, Matsuda H, Iwasa Y. The evolution of cooperation in a lattice-structured population. Journal of Theoretical Biology. 1997;184(1):65–81. doi: 10.1006/jtbi.1996.0243. [DOI] [PubMed] [Google Scholar]
- Nathanson C, Tarnita C, Nowak M. Calculating evolutionary dynamics in structured populations. PLoS computational biology. 2009;5(12):e1000615. doi: 10.1371/journal.pcbi.1000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M, Sigmund K. The evolution of stochastic strategies in the prisoner’s dilemma. Acta Applicandae Mathematicae. 1990;20(3):247–265. [Google Scholar]
- Nowak MA. Evolutionary Dynamics. Cambridge, MA, USA: Harvard University Press; 2006a. [Google Scholar]
- Nowak MA. Five rules for the evolution of cooperation. Science. 2006b;314(5805):1560–1563. doi: 10.1126/science.1133755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, Bonhoeffer S, May RM. Spatial games and the maintenance of cooperation. Proceedings of the National Academy of Sciences. 1994;91(11):4877–4881. doi: 10.1073/pnas.91.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, Komarova NL, Sengupta A, Jallepalli PV, Shih I-M, Vogelstein B, Lengauer C. The role of chromosomal instability in tumor initiation. Proceedings of the National Academy of Sciences. 2002;99(25):16226–16231. doi: 10.1073/pnas.202617399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, May RM. Evolutionary games and spatial chaos. Nature. 1992;359(6398):826–829. [Google Scholar]
- Nowak MA, Michor F, Iwasa Y. The linear process of somatic evolution. Proceedings of the National Academy of Sciences. 2003;100(25):14966–14969. doi: 10.1073/pnas.2535419100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, Sasaki A, Taylor C, Fudenberg D. Emergence of cooperation and evolutionary stability in finite populations. Nature. 2004;428(6983):646–650. doi: 10.1038/nature02414. [DOI] [PubMed] [Google Scholar]
- Nowak MA, Tarnita CE, Antal T. Evolutionary dynamics in structured populations. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010a;365(1537):19. doi: 10.1098/rstb.2009.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, Tarnita CE, Wilson EO. The evolution of eusociality. Nature. 2010b;466(7310):1057–1062. doi: 10.1038/nature09205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki H, Hauert C, Lieberman E, Nowak MA. A simple rule for the evolution of cooperation on graphs and social networks. Nature. 2006;441:502–505. doi: 10.1038/nature04605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki H, Nowak MA. Evolutionary games on cycles. Proceedings of the Royal Society B: Biological Sciences. 2006;273(1598):2249–2256. doi: 10.1098/rspb.2006.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki H, Nowak MA. Evolutionary stability on graphs. Journal of Theoretical Biology. 2008;251(4):698–707. doi: 10.1016/j.jtbi.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki H, Nowak MA, Pacheco JM. Breaking the symmetry between interaction and replacement in evolutionary dynamics on graphs. Physical Review Letters. 2007;98(10):108106. doi: 10.1103/PhysRevLett.98.108106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco JM, Traulsen A, Nowak MA. Coevolution of strategy and structure in complex networks with dynamical linking. Physical Review Letters. 2006a;97(25):258103. doi: 10.1103/PhysRevLett.97.258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco JM, Traulsen A, Nowak MA. Active linking in evolutionary games. Journal of Theoretical Biology. 2006b;243(3):437–443. doi: 10.1016/j.jtbi.2006.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perc M, Szolnoki A. Coevolutionary games–A mini review. BioSystems. 2010;99(2):109–125. doi: 10.1016/j.biosystems.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Poncela J, nes JG-G, Traulsen A, Moreno Y. Evolutionary game dynamics in a growing structured population. New Journal of Physics. 2009;11(8):083031. [Google Scholar]
- Rand DG, Arbesman S, Christakis NA. Dynamic social networks promote cooperation in experiments with humans. Proceedings of the National Academy of Sciences. 2011;108(48):19193–19198. doi: 10.1073/pnas.1108243108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca CP, Cuesta JA, Sánchez A. Effect of spatial structure on the evolution of cooperation. Physical Review E. 2009a;80(4):046106. doi: 10.1103/PhysRevE.80.046106. [DOI] [PubMed] [Google Scholar]
- Roca CP, Cuesta JA, Sánchez A. Evolutionary game theory: Temporal and spatial effects beyond replicator dynamics. Physics of Life Reviews. 2009b;6(4):208–249. doi: 10.1016/j.plrev.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Rousset F. Genetic structure and selection in subdivided populations. Princeton: Princeton University Press; 2004. [Google Scholar]
- Rousset F, Billiard S. A theoretical basis for measures of kin selection in subdivided populations: finite populations and localized dispersal. Journal of Evolutionary Biology. 2000;13(5):814–825. [Google Scholar]
- Santorelli LA, Thompson CRL, Villegas E, Svetz J, Dinh C, Parikh A, Sucgang R, Kuspa A, Strassmann JE, Queller DC, Shaulsky G. Facultative cheater mutants reveal the genetic complexity of cooperation in social amoebae. Nature. 2008;451(7182):1107–1110. doi: 10.1038/nature06558. [DOI] [PubMed] [Google Scholar]
- Santos FC, Pacheco JM. Scale-free networks provide a unifying framework for the emergence of cooperation. Physical Review Letters. 2005;95(9):98104. doi: 10.1103/PhysRevLett.95.098104. [DOI] [PubMed] [Google Scholar]
- Santos FC, Santos MD, Pacheco JM. Social diversity promotes the emergence of cooperation in public goods games. Nature. 2008;454(7201):213–216. doi: 10.1038/nature06940. [DOI] [PubMed] [Google Scholar]
- Skyrms B, Pemantle R. A dynamic model of social network formation. Proceedings of the National Academy of Sciences. 2000;97(16):9340–9346. doi: 10.1073/pnas.97.16.9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. Fixation probabilities and fixation times in a subdivided population. Evolution. 1981;35(3):477–488. doi: 10.1111/j.1558-5646.1981.tb04911.x. [DOI] [PubMed] [Google Scholar]
- Szabó G, Fáth G. Evolutionary games on graphs. Physics Reports. 2007;446(4–6):97–216. [Google Scholar]
- Tarnita CE, Antal T, Ohtsuki H, Nowak MA. Evolutionary dynamics in set structured populations. Proceedings of the National Academy of Sciences of the USA. 2009a;106(21):8601. doi: 10.1073/pnas.0903019106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnita CE, Ohtsuki H, Antal T, Fu F, Nowak MA. Strategy selection in structured populations. Journal of Theoretical Biology. 2009b;259(3):570–581. doi: 10.1016/j.jtbi.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnita CE, Wage N, Nowak MA. Multiple strategies in structured populations. Proceedings of the National Academy of Sciences of the USA. 2011;108(6):2334–2337. doi: 10.1073/pnas.1016008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor C, Fudenberg D, Sasaki A, Nowak M. Evolutionary game dynamics in finite populations. Bulletin of Mathematical Biology. 2004;66:1621–1644. doi: 10.1016/j.bulm.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Taylor P. Altruism in viscous populations — an inclusive fitness model. Evolutionary Ecology. 1992;6:352–356. [Google Scholar]
- Taylor PD, Day T, Wild G. Evolution of cooperation in a finite homogeneous graph. Nature. 2007;447(7143):469–472. doi: 10.1038/nature05784. [DOI] [PubMed] [Google Scholar]
- Traulsen A, Nowak MA. Evolution of cooperation by multilevel selection. Proceedings of the National Academy of Sciences of the USA. 2006;103(29):10952–10955. doi: 10.1073/pnas.0602530103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traulsen A, Shoresh N, Nowak MA. Analytical results for individual and group selection of any intensity. Bulletin of Mathematical Biology. 2008;70(5):1410–1424. doi: 10.1007/s11538-008-9305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baalen M, Rand DA. The unit of selection in viscous populations and the evolution of altruism. Journal of Theoretical Biology. 1998;193(4):631–648. doi: 10.1006/jtbi.1998.0730. [DOI] [PubMed] [Google Scholar]
- van Veelen M, García J, Rand DG, Nowak MA. Direct reciprocity in structured populations. Proceedings of the National Academy of Sciences. 2012 doi: 10.1073/pnas.1206694109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veelen M, Nowak MA. Multi-player games on the cycle. Journal of Theoretical Biology. 2012;292(0):116–128. doi: 10.1016/j.jtbi.2011.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velicer GJ. Social strife in the microbial world. Trends in Microbiology. 2003;11(7):330–337. doi: 10.1016/s0966-842x(03)00152-5. [DOI] [PubMed] [Google Scholar]
- Wilson D, Pollock G, Dugatkin L. Can altruism evolve in purely viscous populations? Evolutionary Ecology. 1992;6:331–341. [Google Scholar]
- Wodarz D, Komarova NL. Computational biology of cancer: lecture notes and mathematical modeling. World Scientific Pub Co Inc; 2005. [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28(2):114. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Altrock PM, Wang L, Traulsen A. Universality of weak selection. Phys. Rev. E. 2010a;82(4):046106. doi: 10.1103/PhysRevE.82.046106. [DOI] [PubMed] [Google Scholar]
- Wu B, Zhou D, Fu F, Luo Q, Wang L, Traulsen A. Evolution of cooperation on stochastic dynamical networks. PLoS ONE. 2010b;5(6):e11187. doi: 10.1371/journal.pone.0011187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki L, Manser T, Gefter ML. Somatic evolution of variable region structures during an immune response. Proceedings of the National Academy of Sciences. 1986;83(6):1847–1851. doi: 10.1073/pnas.83.6.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, Velculescu VE, Kinzler KW, Vogelstein B, Iacobuzio-Donahue CA. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467(7319):1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]