Abstract
Despite their homology, IκB kinase α (IKKα) and IKKβ have divergent roles in NF-κB signaling. IKKβ strongly activates NF-κB while IKKα can downregulate NF-κB under certain circumstances. Given this, identifying independent substrates for these kinases could help delineate their divergent roles. Peptide substrate array technology followed by bioinformatic screening identified TRAF4 as a substrate for IKKα. Like IKKα, TRAF4 is atypical within its family because it is the only TRAF family member to negatively regulate innate immune signaling. IKKα's phosphorylation of serine-426 on TRAF4 was required for this negative regulation. Binding to the Crohn's disease susceptibility protein, NOD2, is required for TRAF4 phosphorylation and subsequent inhibition of NOD2 signaling. Structurally, serine-426 resides within an exaggerated β-bulge in TRAF4 that is not present in the other TRAF proteins, and phosphorylation of this site provides a structural basis for the atypical function of TRAF4 and its atypical role in NOD2 signaling.
INTRODUCTION
Genetic mutations in pathways affecting NF-κB activity cause a myriad of diseases, including both immunodeficiencies and autoinflammatory disorders (11, 44). In no pathway is this more evident than in the inflammatory disease-associated NOD2 signaling pathway. The CARD15 gene, which encodes NOD2, was the first identified Crohn's disease susceptibility gene, and its normal function is to recognize cytoplasmic exposure to bacteria and dictate an appropriate cytokine response (15, 20, 21, 27, 36). Loss-of-function polymorphisms in NOD2 can cause Crohn's disease, a granulomatous inflammatory disorder of the gastrointestinal tract, while gain-of-function mutations cause early-onset sarcoidosis (EOS) and Blau syndrome (3, 8, 23). Like Crohn's disease, EOS and Blau syndromes are also characterized by granulomatous inflammation, only at different anatomic locations in the body. Since both gain-of-function and loss-of-function mutations and polymorphisms in NOD2 cause granulomatous inflammation, there must be a delicate balance between the activation and termination of NOD2 signaling. When this balance is disrupted, inflammatory disease results.
Upon activation, the NOD2-RIP2 signaling complex initiates a series of ubiquitination and phosphorylation events that ultimately allow a coordinated cytokine response (6, 26, 45). This response is largely mediated through NF-κB and, by extension, the I kappa kinase (IKK) signalosome (10, 11, 32). The IKK signalosome consists of the kinases IKKα and IKKβ held together by the scaffolding protein IKKγ (NEMO, for NF-κB essential modifier). IKKα and IKKβ are highly homologous kinases (9, 11). Of the two, IKKβ has been much better studied. Upon activation, IKKβ phosphorylates the NF-κB inhibitory protein, IκBα, to target it for degradation (9, 11, 32). Beyond IκBα, it is clear that IKKβ has a number of additional substrates in the cell. Peptide substrate array technology outlined the preferred amino acid phosphorylation sequence for IKKβ (19), and many of the substrates identified match this array motif. These substrates include Bcl10 (50), CYLD (41), A20 (19), FOXO3a (14), and p65 (42) and, in general, help to direct much of the positive NF-κB signaling in the cell.
In contrast, much less is known about IKKα. Fewer substrates have been identified, and of these substrates, most are also phosphorylated by IKKβ, suggesting a possible redundancy between these two kinases. However, this redundancy has been challenged recently by genetic mouse models suggesting that IKKα has a more nuanced role in the cell. In particular, under certain circumstances, IKKα may actually help limit NF-κB activation (28, 29). For instance, knock-in mice harboring a kinase-inactive IKKα are viable but have exacerbated inflammation at a variety of mucosal surfaces. In these mice, IKKα inactivation results in greatly increased expression of proinflammatory and antiapoptotic genes (28). This negative regulatory role of IKKα has also recently been shown to be crucial for signaling in the A20 ubiquitin-editing complex. IKKα, but not IKKβ, phosphorylates the ubiquitin binding protein TAX1BP1. This phosphorylation is essential for the formation of the A20 ubiquitin-editing complex and is required for proper termination of tumor necrosis factor (TNF)- and interleukin-1 (IL-1)-dependent NF-κB signaling (43). Given the enigmatic nature of IKKα and the surprising role that it plays in negatively regulating NF-κB signaling, the identification of IKKα-specific substrates may provide great insight into its unique cellular functions.
In this work, we perform peptide substrate array analysis on IKKα and identify the atypical TRAF family member protein, TRAF4, as a novel substrate for IKKα. Members of the TNF receptor-associated factor (TRAF) superfamily have been well characterized as downstream adaptor molecules in a variety of proinflammatory signaling pathways, yet the role of phosphorylation of the TRAF proteins has not been studied (5). Of the TRAF proteins, TRAF4 is an atypical member of the TRAF superfamily both structurally and functionally (25). Unlike other TRAF family members that work to positively regulate inflammatory signaling downstream of agonists like TNF, lipopolysaccharide (LPS), or IL-1, TRAF4 negatively regulates innate immune signaling. TRAF4 binds directly to the Nod-like receptor (NLR) family member, NOD2, to inhibit NF-κB activation (34). Additionally, TRAF4 also binds to TRAF6 and TRIF to dampen Toll-like receptor (TLR)-mediated NF-κB and interferon (IFN) activation (47). The reason TRAF4 functions so much differently than the other TRAFs is unknown. In this work, we identify an IKKα-inducible phosphorylation site on TRAF4 that is required for its inhibitory role. Mapping of this phosphorylation shows that it resides within a unique extended β-bulge in the MATH domain of TRAF4 that is not present in the other TRAF family members. Phosphorylation of this β-bulge increases TRAF4 stability and activity to allow TRAF4 to negatively regulate downstream NOD2-induced NF-κB activity. Thus, in this work, we generate proteomic tools to help identify IKKα substrates. We identify a phosphorylation site on TRAF4 that helps explain TRAF4's unique function among the TRAF family proteins as well as identifying a novel mechanism of IKKα-mediated negative regulation of inflammatory signaling. In total, this work helps outline a novel negative regulatory circuit for an innate immune signaling pathway.
MATERIALS AND METHODS
Cell culture, transfection, IP, and Western blotting.
HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 5% fetal bovine serum (FBS; HyClone). RAW264.7 macrophages and HCT116 cells were maintained in DMEM containing 10% FBS (HyClone). All media contained antibiotic/antimycotic solution (Invitrogen). Stable cell lines were generated by retroviral infection of the indicated cells followed by neomycin selection (300 μg/ml; InvivoGen) in their respective media. Clones (>1,000) were pooled. Calcium phosphate precipitation transfections in HEK293T cells were carried out as previously described (1). Immunoprecipitations (IPs) were conducted in Cell Signaling lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% Triton X-100, 1 mM EGTA, 1 mM EGTA, 2.5 mM sodium orthophosphate, 1 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM NaVO4, 10 nM calyculin A in the presence of protease inhibitor cocktail [Sigma]). Protein G Sepharose beads (Invitrogen) were added to lysates, and IPs were washed five times in lysis buffer prior to Western blotting. Western blotting was completed on nitrocellulose membranes (Bio-Rad) as previously described (2).
Antibodies, plasmids, and reagents.
RIP2, Omni, and TRAF4 antibodies were purchased from Santa Cruz Technology. Glutathione S-transferase (GST), phospho-IκBα, and total IκBα antibodies were purchased from Cell Signaling Technology. Antihemagglutinin (anti-HA; HA-11) was purchased from Covance. Phospho-IKK motif antibody was generated as previously described (19). NTAP-TRAF4, Omni-TRAF4, HA-NOD2, GST-IKKβ, and GST-IKKε were used as previously described (18, 19, 34, 37). GST-IKKα was generated by PCR cloning into the BamHI and NotI sites of pEBG. The GST-IKKα K44A, Omni-S426A TRAF4, NTAP-S426A TRAF4, and Omni-S426D TRAF4 mutants were generated by QuikChange site-directed mutagenesis (Stratagene). Point mutations were verified by sequence analysis. Recombinant full-length human IKKα containing an N-terminal GST tag was purchased from Millipore. A 5′ Flag-fused retroviral TRAF4 was generated as previously described for generation of stable macrophages and HT29 cells (34). Flag beads (M2) and streptavidin agarose were purchased from Sigma. Muramyl dipeptide (MDP) and LPS were obtained from Invivogen. IL-1β was purchased from R&D Systems.
Positional scanning peptide library assay.
Recombinant GST-IKKα and GST-IKKα K44A were generated as described above, and a positional scanning peptide library assay was carried out as described previously (17, 48). Briefly, the kinase buffer contained 50 mM Tris (pH 7.5), 12 mM MgCl2, 1 mM glycerophosphate, 0.1% Tween, 100 μM ATP, and 5 μCi of [γ-32P]ATP per reaction mix. Kinase reaction mixes were incubated at 30°C for 2 h before being spotted on streptavidin-coated nitrocellulose. After extensive washing, including washes in 1% SDS and 2 M NaCl, phosphorimager analysis was performed. Position-specific scoring matrix (PSSM) plots were generated by the quantification of relative 32P incorporation into each peptide and were normalized within a particular amino acid position as a ratio of individual amino acid signal intensity relative to the average signal intensity for all of the other amino acids at that position. For example, proline at the −5 position was quantitated and ranked against the average signal of the 21 amino acids at that position.
In vitro kinase assays.
NTAP-TRAF4 and NTAP-S426A TRAF4 were isolated from HEK293T cell lysates using streptavidin-agarose as described above. Recombinant full-length human GST-IKKα (150 ng; Millipore) and 10 μCi of [γ-32P]ATP per reaction mix were used. The kinase buffer contained 25 mM Tris (pH 7.5), 1 mM β-glycerophosphate, 1 mM dithiothreitol (DTT), 1 mM Na3VO4, 10 mM MgCl2, and 100 μM ATP. Reaction mixes were incubated at 30°C for 30 min. The reaction was stopped by the addition of 2× SDS-PAGE sample buffer, followed by boiling. The reaction product was then subjected to SDS-PAGE and visualized by autoradiography.
Luciferase reporter gene assay.
HEK293T cells were plated in triplicate at 4 × 104 cells/well. Reporter plasmids and expression constructs were transfected with Polyfect (Qiagen) as described previously (13). Cells were lysed in 1× reporter lysis buffer (Promega) and assayed for luciferase and β-galactosidase activities. Luciferase values were normalized to β-galactosidase activity.
Quantitative reverse transcription-PCR (RT-PCR).
Total RNA was extracted using an RNeasy kit (Qiagen) according to the manufacturer's instructions from the cell lines indicated in the text and figure legends. DNA synthesis was performed using Qiagen's Quantitect reverse transcription kit according to the manufacturer's instructions. Real-time PCR was carried out using primers against mouse TNF-α (forward, 5′-GGTGCCTATGTCTCAGCCTCTT-3′; reverse, 5′-GCCATAGAACTGATGAGAGGGAG-3′), MIP2α (forward, 5′-CCACCAACCACCAGGCTACAGGGGC-3′; reverse, 5′-AGGCTCCTCCTTTCCAGGTCAGTTAGC-3′), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (forward, 5′-AGGCCGGTGCTGAGTATGTC-3′; reverse, 5′-TGCCTGCTTCACCACCTTCT-3′), along with the iQ SYBR green Supermix (Bio-Rad) and detection using a Bio-Rad iCycler. Data shown are normalized to GAPDH.
Salmonella infection and gentamicin protection assay.
Salmonella cells were grown overnight in LB broth at 30°C at 180 rpm. The following day, Salmonella cells were diluted 1:8 and allowed to grow for another 45 min until they reached exponential growth phase. Salmonella was diluted in DMEM lacking antibiotics, and cells were infected at a multiplicity of infection (MOI) of 10:1. After 30 min, Salmonella-containing medium was replaced with fresh medium containing gentamicin sulfate (50 μg/ml). After 1 h, lysates were placed on ice for 5 min in 1% Triton X-100 in phosphate-buffered saline (PBS). Lysates were then diluted and plated on LB agar plates and incubated overnight at 30°C. CFU were counted the following day.
Statistics.
Data are presented as means, with error bars representing standard error of the mean (SEM) of at least three different experiments for each condition. Statistical analysis was completed using Prism, version 5, software. Significance was determined by P values of <0.05 after one-way analysis of variance (ANOVA).
Structural analyses.
Clustal W2 was used for multiple sequence alignment of protein sequences obtained from BLASTP. TRAF4 TRAF domain protein structure homology modeling was done using SWISS-MODEL workspace based on TRAF2 template 1ca9B which was solved at 2.3Å with an E-value of 2.9E−43 (39) and Q mean Z-score of −0.143. DeepView-Swiss-PdbViewer was used for visualization of the model structure.
RESULTS
Determination of IKKα phosphorylation motif and novel substrates.
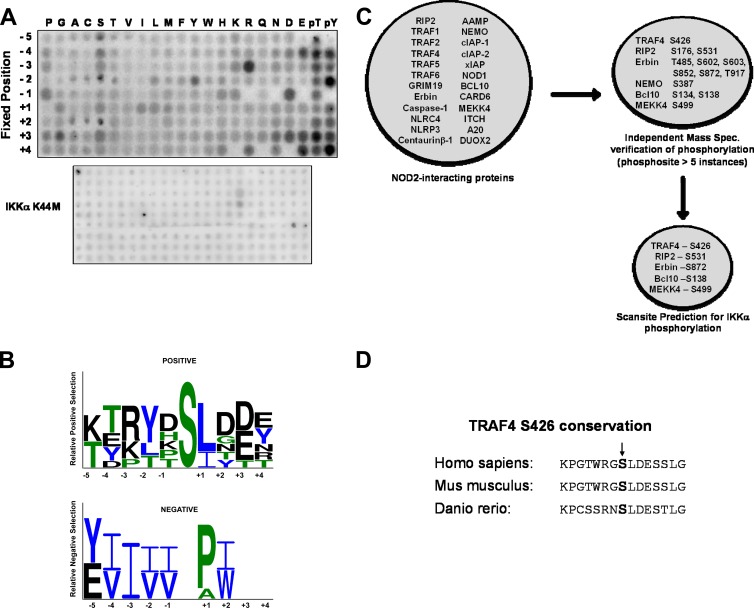
Many innate immune signaling pathways converge at the level of the IKK signalosome, the core of which consists of the catalytic subunits IKKα and IKKβ and the regulatory scaffolding protein IKKγ (NEMO) (9, 11). IKKβ, but not IKKα, is required for IκBα phosphorylation and subsequent NF-κB activation (32). IKKα is instead emerging as a negative regulator of canonical NF-κB activation (28, 29, 43). To elucidate the kinase-dependent mechanisms by which IKKα inhibits NF-κB, it is important to identify novel substrates of IKKα that are not also substrates of IKKβ. To this end, we performed a proteomic and bioinformatic screen to identify novel substrates for IKKα. Positional scanning peptide array technology was used to identify the preferred phosphorylation motif for IKKα. Biotinylated peptide libraries were subjected to kinase assays using recombinant GST-IKKα and [γ-32P]ATP. The libraries consisted of peptides that have a centrally fixed Ser or Thr residue flanked by a degenerate mixture of amino acids on either side, except for one position which is fixed to a single amino acid (17). Phosphothreonine or phosphotyrosine residues were included in the fixed positions to screen for priming phosphorylation events within the preferred substrate motif for IKKα. Peptides were captured on a streptavidin-coated membrane, and relative incorporation of [γ-32P]ATP was measured for each amino acid at each position relative to the phosphorylation site. As a control for kinase specificity, a catalytically inactive IKKα (K44M IKKα) was utilized. This analysis revealed that IKKα, but not kinase-dead K44M IKKα, displayed both positive and negative sequence specificity at several positions relative to the central phosphoacceptor (Fig. 1A). Selection preferences for IKKα are more evident in the fixed amino acids in the C-terminal side of the peptide. Acidic residues and phosphorylated residues are strongly preferred at the +2 and +3 positions, and branched hydrophobics are preferred at the +1 position. IKKα has a preference for phosphorylated substrates in both the +3 and +4 and the −3 and −4 positions. In fact, the elevated phosphothreonine at the −4 position is consistent with whole-cell mass spectrometry experiments that suggest threonine at −4 is phosphorylated in TRAF4 (http://www.phosida.de) (16, 38). PSSM plots show the relative positive and negative selections and were used to generate a matrix for bioinformatic searching of potential substrates (Fig. 1B). There is a strongly preferred phosphorylation sequence between IKKβ (19) and IKKα, which is not unexpected given the high degree of homology within their kinase domains.
Fig 1.
Determination of IKKα preferred phosphorylation motif and target substrates. (A) Kinase assays were performed on 198 peptide libraries with recombinant IKKα or, as a control, kinase-dead IKKα. The general sequence for these libraries was Y-A-X-X-X-Z-X-S/T-X-X-X-X-A-G-K-K-biotin (Z, fixed amino acid; X, equimolar mixture of amino acids) with a fixed amino acid at the indicated position. Relative incorporation of 32P was measured after the individual degenerate phosphorylated peptides were captured on a streptavidin-coated membrane. While IKKα (upper panel) gave a strong, consistent motif, no motif could be identified using a kinase-inactive (K44A) IKKα. (B) PSSM plot of positive and negative amino acid selections for IKKα. There is a general, strong preference for charged and phosphorylated amino acids surrounding the phosphoacceptor, with a general negative selection from branched, hydrophobic amino acids. (C) Bioinformatics strategies were used to decipher potential substrates of IKKα downstream in the NOD2 signaling pathway. Phosphosite and Phosida mass spectrometry-verified phosphorylation databases were searched with proteins known to either bind NOD2 or proximally affect NOD2 signaling. Phosphorylation cutoffs were phosphorylation sites identified more than five times in independent experiments. Of the 24 proteins searched, 6 had phosphorylation sites identified more than five times in mass spectrometry databases (13 total phosphorylation sites). These sites were then subjected to Scansite analysis, using the IKKα preferential phosphorylation motif identified in a matrix. Of the 13 potential sites identified, 5 of these passed Scansite screening as potential IKKα phosphorylation sites. (D) The potential IKKα phosphorylation sites identified were analyzed for evolutionary conservation. TRAF4 showed the highest degree of evolutionary conservation.
After determining this preferred phosphorylation motif for IKKα, we conducted a tiered bioinformatic screen to narrow our results and to decrease the number of potential false-positive substrates. Given our lab's long-term interest in NOD2 signaling, we first narrowed our search for potential IKKα substrates to proteins known to bind to NOD2 or known to proximally affect NOD2 signaling (45). Twenty-four of these proteins were subjected to Phosphosite (www.phosphosite.org) and Phosida (www.phosida.de) searches to identify proteins that contain phosphorylation sites verified by independent mass spectrometry analysis. A cutoff of five independent instances of mass spectrometry identification in Phosphosite or Phosida was set as the threshold for positivity. A Scansite (http://scansite.mit.edu/) matrix search using IKKα's phosphorylation motif further narrowed the data set to five proteins that passed the first two filters and also contained the preferred IKKα phosphorylation motif (Fig. 1C). Of these proteins, we focused on TRAF4. Similar to the atypical nature of IKKα, TRAF4 is an atypical member of the TRAF superfamily of proteins (25). Unlike the other six TRAF family members, TRAF4 plays a negative regulatory role in NF-κB activation (34, 47). Since S426 within the predicted IKKα phosphorylation motif of TRAF4 is also conserved across species (Fig. 1D), we sought to validate TRAF4 as an IKKα substrate.
IKKα phosphorylates TRAF4 at S426.
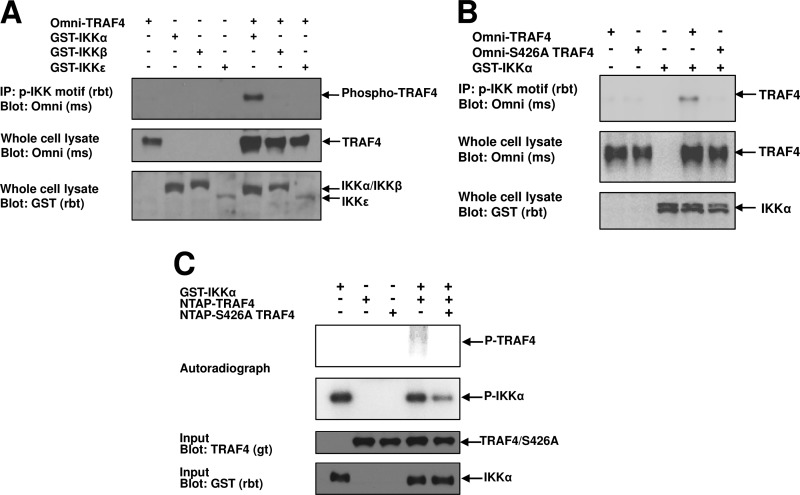
To determine if IKKα in fact phosphorylates TRAF4 and to determine the specificity of this phosphorylation motif compared to other IKK family members, we cotransfected HEK293T cells with Omni-TRAF4 and the IKK family members IKKα, IKKβ, and IKKε. Using an antibody that immunoprecipitates proteins containing a phosphorylated IKK motif (19), phospho-TRAF4 was immunoprecipitated in the presence of IKKα but not in the presence of IKKβ or IKKε (Fig. 2A). To verify that IKKα phosphorylation occurred at S426, the point mutation S426A was introduced into TRAF4. IKKα phosphorylation of TRAF4 was not detected in HEK293T cells cotransfected with GST-IKKα and Omni-S426A TRAF4 (Fig. 2B). In order to verify that TRAF4 could be phosphorylated directly by IKKα at S426, Omni-tagged TRAF4 or S426A TRAF4 was immunoprecipitated from HEK293T cells and incubated with recombinant full-length human GST-IKKα and [γ-32P]ATP. We found that IKKα could phosphorylate TRAF4 but not S426A TRAF4 in vitro (Fig. 2C). Taken together, these data strongly suggest that IKKα phosphorylates TRAF4 at S426.
Fig 2.
IKKα phosphorylates TRAF4 at S426. (A) Omni-TRAF4 was cotransfected into HEK293T cells with GST-IKKα, IKKβ, or IKKε. Proteins containing a phosphorylated IKK (p-IKK) motif were immunoprecipitated and subjected to Western blotting for TRAF4. Only IKKα induced TRAF4 phosphorylation. (B) Omni-TRAF4 or S426A TRAF4 was cotransfected into HEK293 cells with GST-IKKα. TRAF4 immunoprecipitated with the phospho-IKK motif antibody while the S426A TRAF4 did not. (C) In vitro kinase assays were performed using recombinant full-length GST-IKKα and purified NTAP-TRAF4 or NTAP-S426A TRAF4 from transfected HEK293T cells. GST-IKKα induced phosphorylation of TRAF4 but not S426A TRAF4 as shown by incorporation of [γ-32P]ATP. rbt, rabbit; ms, mouse; gt, goat.
Innate immune activation induces phosphorylation of endogenous TRAF4.
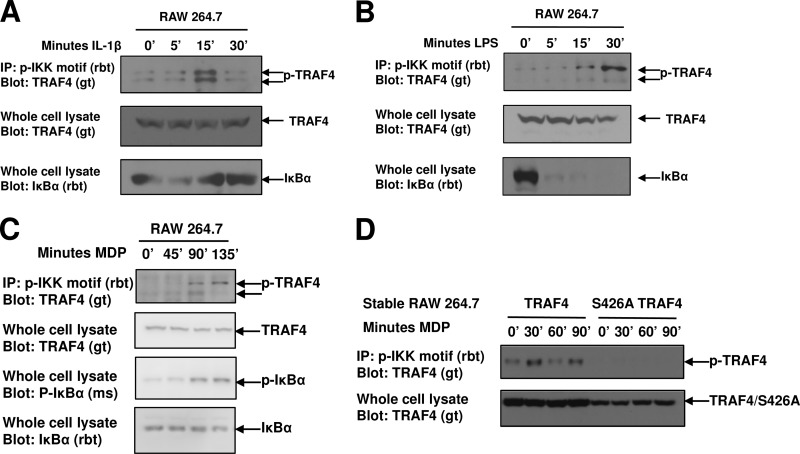
Endogenous TRAF4 phosphorylation was detected in a time- and stimulus-dependent manner. RAW264.7 macrophages were treated with IL-1β (Fig. 3A) or lipopolysaccharide (LPS) (Fig. 3B), and endogenous TRAF4 was phosphorylated between 15 and 30 min after agonist addition (Fig. 3A and B). Double banding of phosphorylated TRAF4 is likely due to the fact that upon treatment with IL-1 or LPS, a variety of kinases in addition to IKKα are activated. Mass spectrometry phosphorylation databases such as Phosphosite or Phosida show at least eight independent phosphorylation sites on TRAF4. Given this, it is likely that upon treatment with agonists, which activate a variety of kinases, endogenous TRAF4 is phosphorylated by other kinases, and the double banding represents multiphosphorylated TRAF4 (for instance, a potential example would include phospho-S426 alone or phospho-T422/phospho-S426 double phosphorylation; both of these bands could be visualized by this motif antibody). In all, phosphorylation of S426 has been identified in over five independent mass spectrometry studies (7, 16, 38); phosphorylation of S426 was identified as a potential substrate of IKKα by peptide substrate array technology, and in both in vitro overexpression systems (Fig. 2) and in response to IKKα agonists in endogenous systems (Fig. 3), TRAF4 is phosphorylated.
Fig 3.
Endogenous TRAF4 is phosphorylated in both a time- and stimulus-dependent manner. (A to C) RAW264.7 Macrophages were stimulated with IL-1β (A), LPS (B), or MDP (C) for the indicated times. Lysates were subjected to immunoprecipitation with a phospho-IKK motif antibody. Endogenous phosphorylation of TRAF4 (p-TRAF4) was detected following each these stimuli. (D) RAW264.7 macrophages were retrovirally transduced with viruses expressing either WT TRAF4 or S426A TRAF6. After neomycin selection, MDP-inducible TRAF4 phosphorylation was detected in macrophages expressing WT TRAF4 but not in macrophages expressing the S426A TRAF4.
Because TRAF4 is a NOD2 binding protein and negatively affects NOD2 signaling (34), we tested whether activation of NOD2, which is upstream of the IKK signalosome (1, 20), also induced TRAF4 phosphorylation. RAW264.7 macrophages were stimulated with the NOD2 agonist, muramyl dipeptide (MDP), for 45, 90, or 135 min. A longer time course is required for MDP-induced NF-κB activation since it must be internalized before the intracellular cytosolic sensor, NOD2, is activated (45). Similar to the results of IL-1β and LPS stimulation, endogenous TRAF4 phosphorylation was detected following MDP-induced NF-κB (Fig. 3C). To ensure that TRAF4 phosphorylation following MDP stimulation occurs at S426, RAW264.7 macrophages were stably transduced with a Flag-TRAF4 retrovirus or a Flag-S426A TRAF4 retrovirus. Stable expression of TRAF4 increased both basal and MDP-induced TRAF4 phosphorylation, while neither basal nor stimulus-dependent TRAF4 phosphorylation was detected in macrophages stably expressing S426A TRAF4 (Fig. 3D).
IKKα phosphorylation of TRAF4 following NOD2 activation requires NOD2 binding.
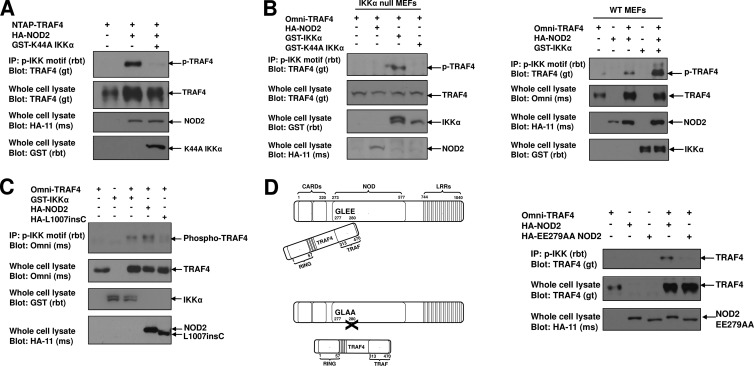
Since MDP activation of NOD2 induced TRAF4 phosphorylation, we suspected that NOD2 overexpression would also induce TRAF4 phosphorylation in cells expressing functional IKKα. Overexpression of NOD2 in HEK293T cells provided a model system that we could manipulate to determine if NOD2-induced TRAF4 phosphorylation was dependent on IKKα. HA-NOD2 cotransfection with NTAP-TRAF4 was sufficient for TRAF4 phosphorylation. Importantly, the presence of the kinase-dead GST-K44A IKKα was able to inhibit NOD2-induced TRAF4 phosphorylation (Fig. 4A) while NOD2 overexpression in IKKα−/− murine embryonic fibroblasts (MEFs) failed to induce TRAF4 phosphorylation (Fig. 4B, left panel). As a control, NOD2-induced phosphorylation of TRAF4 in wild-type (WT) MEFs was included (Fig. 4B, right panel). We next sought to determine if NOD2-induced phosphorylation of TRAF4 was dependent on NOD2 activity and/or TRAF4 binding to NOD2. We first compared the ability of wild-type NOD2 or the most common Crohn's disease-associated variant, L1007insC NOD2, to phosphorylate TRAF4. The L1007insC NOD2 contains a missense mutation that introduces a premature stop codon leading to the production of a truncated protein lacking the last 33 amino acids in the leucine rich repeat region (36). The L1007insC is a loss-of-function mutation in that it has decreased capacity for IKK signalosome activation and subsequent NF-κB activation (27, 36, 49). We have previously shown that TRAF4 maintains binding to L1007insC NOD2 (34). However, cotransfection of HEK293T cells with Omni-TRAF4 and HA-L1007insC failed to induce TRAF4 phosphorylation (Fig. 4C) suggesting that NOD2 activity is necessary for TRAF4 phosphorylation. Collectively, these findings suggest that in addition to activating IKKβ for IκBα phosphorylation, NOD2 activation induces downstream IKKα activation to promote TRAF4 phosphorylation at S426.
Fig 4.
NOD2 activation and binding to TRAF4 induce TRAF4 phosphorylation, and this is dependent on IKKα. (A) HA-NOD2 was cotransfected into HEK293T cells with NTAP-TRAF4 in the absence or presence of GST-K44A IKKα, which lacks kinase activity. NOD2 induced TRAF4 phosphorylation; however, this phosphorylation was lost when the kinase-dead K44A IKKα was present. (B) IKKα−/− MEFs were cotransfected with Omni-TRAF4 or HA-NOD2 (left panel). NOD2 overexpression was insufficient to induce TRAF4 phosphorylation in the absence of IKKα. Reconstitution of the MEFs with IKKα induced TRAF4 phosphorylation, while the K44A IKKα failed to do so. NOD2 induced TRAF4 phosphorylation in wild-type MEFs (right panel). (C) HEK293T cells were cotransfected with Omni-TRAF4 and HA-NOD2 or HA-L1007insC NOD2. Both IKKα and NOD2 induced TRAF4 phosphorylation, while the L1007insC NOD2 failed to do so. (D) To determine if TRAF4 binding is required for NOD2-induced phosphorylation, HEK293T cells were cotransfected with Omni-TRAF4 and HA-NOD2 or HA-EE279AA NOD2, which has previously been shown to be unable to bind TRAF4 (34). Wild-type NOD2 induced TRAF4 phosphorylation while the EE279AA NOD2 did not, suggesting that binding of NOD2 to TRAF4 is important for S426 phosphorylation.
The TRAF4 binding motif of NOD2 has been mapped to a GLEE motif at the N terminus of the NOD domain of NOD2 (amino acids 277 to 280). Mutation of the two glutamates to alanines, EE279AA, prevents TRAF4 binding to NOD2 and prevents TRAF4 from inhibiting NOD2 signaling (34). To determine if this binding is required for NOD2-induced TRAF4 phosphorylation, we utilized the EE279AA NOD2 mutant that cannot bind TRAF4. This NOD2 mutant was unable to induce TRAF4 phosphorylation upon cotransfection experiments (Fig. 4D). Since the EE279AA NOD2 mutant has increased basal NF-κB-inducing activity, this result suggests that IKK signalosome and NF-κB activation is not sufficient for NOD2-induced phosphorylation of TRAF4 (34). These findings suggest a mechanism whereby NOD2 both activates IKKα and also localizes TRAF4 such that IKKα has access to it as a substrate.
TRAF4 phosphorylation increases TRAF4 stability and activity.
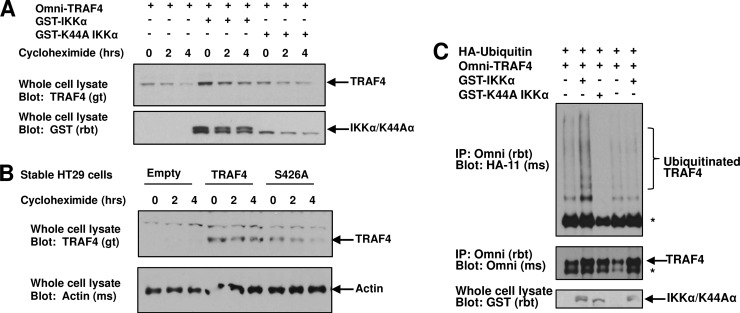
Given that the TRAF family members have been shown to be regulated by proteasomal-mediated degradation (22, 30, 31), the effect of phosphorylation on TRAF4 stability and activity was tested. The half-life of TRAF4 was measured in transfected HEK293T cells treated with the protein biosynthesis inhibitor cycloheximide in the absence or presence of IKKα or K44A IKKα. Alone, TRAF4 protein is degraded between 2 and 4 h after the start of cycloheximide treatment. Cotransfection of IKKα increases TRAF4 expression while the kinase-inactive K44A IKKα did not (Fig. 5A). Similarly, HT29 cells that stably express WT TRAF4 or S426A TRAF4 were treated with cycloheximide. In the stably transduced cell lines, WT TRAF4 was not degraded as rapidly; however, the half-life of S426A TRAF4 was markedly decreased (Fig. 5B). These results indicate that phosphorylation at S426 by IKKα increases the stability of TRAF4. The substrate for TRAF4 remains unknown; however, since TRAF4 is an E3 ubiquitin ligase, its autoubiquitination can be used as a surrogate measure of activity. Cotransfection of HEK293T cells with ubiquitin, TRAF4, and IKKα increased TRAF4 autoubiquitination. IKKα cotransfection with S426A TRAF4 failed to induce autoubiquitination (Fig. 5C). Furthermore, cotransfection of K44A IKKα inhibited basal levels of TRAF4 autoubiquitination (Fig. 5C). Taken together, these results support a model whereby IKKα phosphorylation at S426 increases the stability and activity of TRAF4.
Fig 5.
IKKα phosphorylation of TRAF4 at S426 increases TRAF4 stability and activity. (A and B) Cycloheximide time courses were performed to assess TRAF4 stability in HEK293T cells that were transfected with TRAF4 alone or in the presence of IKKα or K44A IKKα (A) and in HT29 cells stably expressing TRAF4 or S426A TRAF4 (B). Cells were treated with cycloheximide for indicated times. Lysates were standardized for total protein concentration and expression levels of TRAF4 were detected by Western blotting. IKKα, but not K44A IKKα, increased TRAF4 expression and stability (A). S426A TRAF4 had decreased stability in stable HT29 cells (B). (C) Autoubiquitination and activity of TRAF4 were detected in HEK293T cells transfected with Omni-TRAF4, HA-ubiquitin, and either IKKα or K44A IKKα. IKKα but not K44A IKKα increased autoubiquitination of TRAF4. IKKα did not increase autoubiquitination of S426A TRAF4.
S426 lies in a structural domain unique to TRAF4.
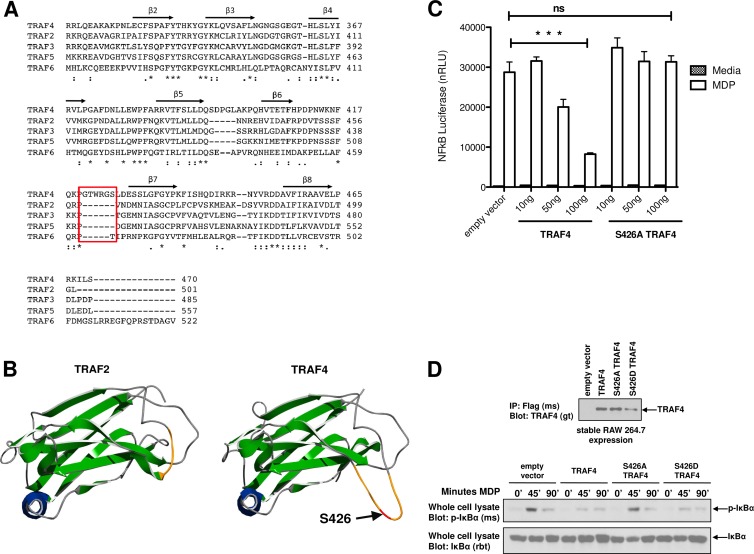
TRAF4 is an atypical member of the TRAF family of proteins. Unlike other TRAF proteins, TRAF4 has not been found to bind to classical TNF receptors (25); instead, TRAF4 is the only TRAF family member to inhibit TLR and NLR signal transduction pathways (34, 47). To determine if phosphorylation of S426 could contribute to the novel function of TRAF4, we examined whether this residue contributed to the unique structure of TRAF4 relative to the other TRAF family members. Structural differences of TRAF4 have largely been characterized in regard to the N-terminal RING domain and to the zinc fingers (5, 25). The structural relationship of the TRAF domain, which contains S426 and which confers substrate binding, has not been examined. Given this, we first examined sequence homology alignment of each of the TRAF domains (Fig. 6A). The crystal structures of TRAF2 and TRAF6 have been determined (4, 39, 52, 53). Residues of the β-sheets that constitute the antiparallel β-sandwich of the TRAF2 TRAF domain are indicated in arrows above the corresponding TRAF4 residues (Fig. 6A). Of note, S426 resides within an amino acid stretch that is not present in any of the other TRAFs (Fig. 6A, red box) and structurally resides in an extended β-bulge found between β-sheets 6 and 7 that is unique to TRAF4 (Fig. 6B). Importantly, S426 is placed prominently within this exaggerated loop with clear surface accessibility (Fig. 6B), raising the possibility that phosphorylation of S426 on TRAF4 may underlie its unique properties relative to the other TRAF family members.
Fig 6.
S426 resides with a loop that is unique to TRAF4 to allow the atypical inhibition of NOD2-induced NF-κB activation. (A) Sequence homology alignment of TRAF4 with other TRAF family members reveals that S426 is within an amino acid stretch that is unique to TRAF4 between strands β6 and β7. This stretch adds 6 amino acid residues (boxed in red) to the linker between β6 and β7. Asterisk, single, fully conserved residue; colon, conservation between groups of strongly similar properties; period, conservation between groups of weakly similar properties. (B) Sequence homology models for TRAF4 based on the published TRAF2 crystal structure (39, 53). The left panel shows ribbon diagram of the TRAF2 TRAF domain crystal structure while the right panel shows the predicted structure of the TRAF4 TRAF domain. S426 is indicated by an arrow and resides within an exaggerated β-bulge that connects the antiparallel β-sandwich sheets. This 6-amino-acid insertion in TRAF4 relative to the other TRAF proteins creates an extended, surface-accessible linker that contains the S426 phosphorylation site. (C) To determine if phosphorylation of this extended loop had functional consequences, NF-κB luciferase assays were performed. These showed that TRAF4 inhibits NOD2-induced NF-κB in a dose-dependent manner while the point mutation S426A does not. Data are represented as mean ± SEM. (D) To show this in a stimulus-dependent manner, RAW264.7 macrophages retrovirally transduced to stably express TRAF4, S426A TRAF4, or S426D TRAF4 were subjected to MDP stimulation. NF-κB activation was assessed by Western blotting lysates for IκBα phosphorylation. Macrophages expressing TRAF4 or the phospho-mimetic S426D TRAF4 had reduced NF-κB activation while the S426A-expressing macrophages maintained intact signaling.
To test whether TRAF4 phosphorylation of this unique β-bulge augments TRAF4 function, as would be predicted from the stability and activity results, NF-κB luciferase reporter assays were conducted in HEK293T cells cotransfected with NOD2 and either Omni-TRAF4 or Omni-S426A TRAF4. TRAF4 was able to inhibit MDP-induced NF-κB reporter activity in a dose-dependent manner while the S426A TRAF4 did not (Fig. 6C). MDP time course experiments in RAW264.7 macrophages recapitulated this result. RAW264.7 macrophages stably expressing TRAF4 had dampened NF-κB responses, indicated by decreased IκBα phosphorylation, while macrophages expressing S426A TRAF4 maintained a response similar to empty vector-transduced macrophages treated with MDP (Fig. 6D). These results suggest that S426 phosphorylation by IKKα is required for TRAF4 to function as a negative regulator. If this is true, we might expect a potential phosphomimetic S426D TRAF4 to maintain or enhance the inhibitory role of TRAF4. In fact, RAW264.7 macrophages stably expressing S426D TRAF4 also inhibited MDP-induced NF-κB activation (Fig. 6D).
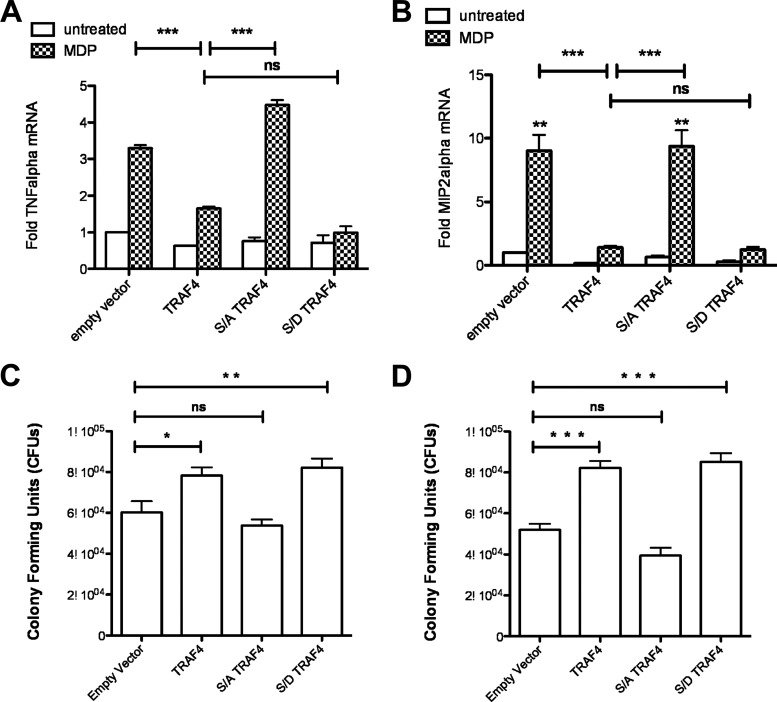
To then determine if TRAF4 phosphorylation and inhibition of NF-κB signaling had a physiological impact, we examined mRNA levels of two genes regulated by NF-κB, TNF-α, and MIP2α. These genes were chosen because expression of these cytokines is increased in macrophages derived from kinase-inactive IKKα knock-in mice (28). RAW264.7 macrophages stably expressing TRAF4 exhibited decreased TNF-α (Fig. 7A) and MIP2α (Fig. 7B) both basally and following stimulation with MDP compared to cells expressing empty vector. In contrast, expression of S426A TRAF4 failed to decrease expression of TNF-α and MIP2α induction, while expression of S426D TRAF4 inhibited expression of these genes as well as or better than wild-type TRAF4 expression (Fig. 7A and B).
Fig 7.
TRAF4 phosphorylation is required for inhibition of NOD2-induced target genes and Salmonella killing. (A and B) RAW264.7 macrophages stably expressing TRAF4, S426A TRAF4, or S426D TRAF4 were stimulated with MDP for 4 h. RNA was isolated and subjected to quantitative RT-PCR for TNF-α (A) or MIP2 (B) mRNA. Macrophages expressing TRAF4 or the phosphomimetic TRAF4 inhibited MDP-induced TNF-α and MIP2 mRNA levels while S426A-expressing macrophages did not. (C and D) Gentamicin protection assays were performed to determine the effect of TRAF4 phosphorylation on NOD2-induced Salmonella killing. HCT116 cells, a cell line that endogenously expresses NOD2, were transfected with an empty vector, TRAF4, S426A TRAF4, or S426D TRAF4 (C). TRAF4 and the phosphomimetic S426D TRAF4 inhibited MDP-induced Salmonella killing while the S426A TRAF4 maintained MDP-induced Salmonella killing. TRAF4 and S426D TRAF4 also inhibited Salmonella killing in gentamicin protection assays performed in retrovirally transduced RAW264.7 macrophages (D), while S426A TRAF4-expressing macrophages maintained MDP-induced Salmonella killing. Data are represented as mean ± SEM.
NOD2 activation plays an important role in bacterial clearance, and this has been studied using the intracellular Gram-negative bacterium Salmonella as a model. MDP stimulation during Salmonella infection enhances bacterial clearance in a NOD2-dependent manner (12, 13, 35, 51), and TRAF4 has been shown to inhibit NOD2-enhanced Salmonella killing by a mechanism dependent on a physical interaction of TRAF4 with NOD2 (34). We therefore tested whether TRAF4 phosphorylation was critical for inhibition of NOD2 antibacterial activity. Gentamicin protection assays were performed in HCT116 cells transfected with either TRAF4, S426A TRAF4, or S426D TRAF4. Expression of TRAF4 or the S426D TRAF4 mutant inhibited MDP-induced Salmonella clearance, while the S426A TRAF4 mutant did not affect NOD2 antibacterial activity (Fig. 7C). Similar results were observed in gentamicin protection assays performed in RAW264.7 macrophages stably expressing TRAF4, S426A TRAF4, or S426D TRAF4. MDP-enhanced Salmonella clearance was inhibited by TRAF4 and S426D TRAF4 but not affected by S426A TRAF4 (Fig. 7D). Collectively, these results indicate that TRAF4 contains a functional IKKα phosphorylation site, located in the extended β-bulge between β-sheets 6 and 7 unique to this TRAF family member, which is required for TRAF4 inhibition of the NOD2 signaling pathway.
DISCUSSION
Both genetic and biochemical work suggests that the kinases of the IKK signalosome, IKKα and IKKβ, have opposing in vivo roles. Due to the high level of homology between these kinases, however, it has been difficult to differentiate their respective substrates despite their known in vivo phenotypic divergence. IKKβ appears to be the major kinase mediating activation of NF-κB while IKKα has emerged as a negative regulator of NF-κB signaling (28, 29, 32, 33, 40, 43). Given this dichotomy, we sought to identify additional substrates for IKKα using a proteomic/bioinformatic approach. To this end, we performed peptide substrate array analysis followed by bioinformatic screening to identify the E3 ubiquitin ligase, TRAF4, as a potential IKKα substrate (Fig. 1). We found that IKKα, but not IKKβ, phosphorylates TRAF4 at S426 following IKKα and subsequent NF-κB activation (Fig. 2). Functionally, we showed that IKKα phosphorylation of TRAF4 is required for TRAF4 to act as a negative regulator. IKKα phosphorylates S426 of TRAF4, and the point mutation, S426A, renders TRAF4 unable to inhibit NOD2-induced NF-κB activation (Fig. 6) and Salmonella killing (Fig. 7). NOD2-induced phosphorylation is dependent on IKKα activity as neither kinase-inactive K44A IKKα nor cells genetically deficient for IKKα allow NOD2-induced phosphorylation of TRAF4 (Fig. 3). NOD2-induced IKKα phosphorylation of TRAF4 was also dependent on NOD2 activation as the loss-of-function Crohn's disease-associated variant, L1007insC, had a markedly decreased ability to induce TRAF4 phosphorylation (Fig. 3). These findings identify the atypical TRAF family member, TRAF4, as a novel substrate of IKKα and suggest that phosphorylation by IKKα plays a key role in the negative regulation of the NOD2 signaling pathway.
The finding that IKKα phosphorylates TRAF4 to inhibit innate signaling and bacterial clearance is consistent with our previous findings that there is increased bacterial killing and, correspondingly, decreased amounts of intracellular Salmonella bacteria when endogenous TRAF4 expression is inhibited by small interfering RNA (siRNA) (34). This is also consistent with prior genetic studies showing increased bacterial clearance in the absence of IKKα or in the presence of a catalytically inactive IKKα (28, 29). IKKα knockout mice die shortly after birth (46), but macrophages derived from knockout embryos show enhanced phagocytic clearance of bacteria and elevated levels of NF-κB target genes (28). Similarly, knock-in mice harboring a kinase-inactive IKKα also have elevated NF-κB target genes and bacterial clearance (29). Increased bacterial clearance could be beneficial to the host; however, the importance of having negative regulators to counterbalance immune activation and phagocytosis is clearly highlighted in the kinase-inactive IKKα knock-in mice. While the kinase-inactive IKKα mice are viable and able to clear a bacterial challenge, they have unchecked and exacerbated inflammation, which ultimately increases their mortality rates (28). In humans, loss of a negative regulator can also contribute to disease pathogenesis and/or severity, and this may help explain the inflammation seen in inflammatory bowel disease (3). It has been difficult to conceptualize how Crohn's disease-associated NOD2 alleles can lose the ability to activate NF-κB but still give rise to a disease characterized by inflammation. Our results suggest the possibility that in addition to loss of acute NF-κB activity, the loss-of-function NOD2 allele, L1007insC, also loses the ability to activate negative regulators of NF-κB, such as IKKα and TRAF4. Loss of TRAF4 phosphorylation could uncouple the coordination of signaling events that occur following NOD2 activation and could exacerbate inflammatory disease.
TRAF4 itself is highly conserved, with 97% identity between mouse and human orthologues. Evolutionarily, the only TRAF proteins described in Danio rerio are TRAF4a and TRAF4b (77% and 68% identity with human TRAF4). Of the two, TRAF4a is more homologous to TRAF4, while TRAF4b more closely resembles TRAF6. This high degree of evolutionary conservation, coupled with the fact that TRAF4 and TRAF6 precursor genes have arisen earliest during evolution, implies that TRAF4 serves a very important biological role (24, 25). In fact, there are unique structural elements in TRAF4 that are not present in any of the other TRAFs. IKKα's TRAF4 phosphorylation site lies in a 6-amino-acid insertion that is not present in any of the other TRAFs. This insertion is present in Danio rerio TRAF4a but is absent in the only other conserved TRAF (TRAF6/TRAF4b) of this species. This suggests a functionally relevant evolutionary divergence of TRAF4. The presence of these extra amino acids confers a unique structure to TRAF4 by creating an exaggerated β-bulge that connects β-sheets 6 and 7 within the TRAF domain (Fig. 6). This unique β-bulge, with its corresponding phosphorylation, allows TRAF4 to then function as a negative regulator of NOD2 signaling. In light of our previous findings showing that the binding of TRAF4 to NOD2 is required for inhibition of NOD2 signaling, we propose a model here whereby both the initial binding of TRAF4 to NOD2 and the subsequent phosphorylation of TRAF4 by IKKα are required for it to function as a negative regulator of NOD2 signaling. This is an induced proximity model in which the binding of TRAF4 to NOD2 serves to nucleate TRAF4 with the IKK signaling complex for subsequent phosphorylation of TRAF4 by IKKα and thus allows the stabilization of TRAF4 (Fig. 5) such that it can have an increased inhibitory function. In this model, TRAF4 first binds to NOD2. This is supported by time course experiments from our previous study showing that MDP-induced TRAF4 binding to NOD2 occurs between 30 and 60 min after stimulation (34), while time courses from the present study (Fig. 3C) show MDP-induced phosphorylation occurring slightly later, between 45 and 90 min. The requirement for TRAF4 to bind NOD2 preceding its phosphorylation is supported by data in which the NOD2 mutant that is unable to bind TRAF4 fails to induce TRAF4 phosphorylation (Fig. 4D). Thus, the unique β-bulge structural feature, with its phosphorylation by a negative regulator of NF-κB, could confer TRAF4's ability to function differently than the other TRAF proteins.
In summary, we have revealed a structural basis for the unique function of TRAF4. We highlight the importance of TRAF4's structural divergence from the other TRAFs as TRAF4 contains a phosphorylation site for the atypical IKK family member, IKKα. Phosphorylation of this unique site confers TRAF4's ability to act as a negative regulator in innate signaling and confers the ability to negatively regulate a key signaling pathway in inflammatory disease pathogenesis.
ACKNOWLEDGMENTS
This work was supported by NIH research grants R01GM86550-01 (D.W.A.), P01DK091222 (D.W.A.), R01DK082437 (C.M.), and R01GM056203 (L.C.C.) and a Burroughs Wellcome Career Award for Biomedical Scientists (10061206.01 to D.W.A.).
We thank George Dubyak (Case Western Reserve University, Cleveland, OH) and George Stark and XiaoXia Li (Cleveland Clinic Foundation, Cleveland, OH) for helpful comments on the project.
Footnotes
Published ahead of print 30 April 2012
REFERENCES
- 1. Abbott DW, Wilkins A, Asara JM, Cantley LC. 2004. The Crohn's disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr. Biol. 14: 2217–2227 [DOI] [PubMed] [Google Scholar]
- 2. Abbott DW, et al. 2007. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol. Cell. Biol. 27: 6012–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Casanova JL, Abel L. 2009. Revisiting Crohn's disease as a primary immunodeficiency of macrophages. J. Exp. Med. 206: 1839–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chung JY, Lu M, Yin Q, Lin SC, Wu H. 2007. Molecular basis for the unique specificity of TRAF6. Adv. Exp. Med. Biol. 597: 122–130 [DOI] [PubMed] [Google Scholar]
- 5. Chung JY, Park YC, Ye H, Wu H. 2002. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 115: 679–688 [DOI] [PubMed] [Google Scholar]
- 6. Coll RC, O'Neill LA. 2010. New insights into the regulation of signalling by toll-like receptors and nod-like receptors. J. Innate Immun. 2: 406–421 [DOI] [PubMed] [Google Scholar]
- 7. Dephoure N, et al. 2008. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 105: 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. 2006. Nod-like proteins in immunity, inflammation and disease. Nat. Immunol. 7: 1250–1257 [DOI] [PubMed] [Google Scholar]
- 9. Ghosh S, Karin M. 2002. Missing pieces in the NF-κB puzzle. Cell 109(Suppl):S81–S96 [DOI] [PubMed] [Google Scholar]
- 10. Hacker H, Karin M. 2006. Regulation and function of IKK and IKK-related kinases. Sci. STKE 2006: re13 doi:10.1126/stke.3572006re13 [DOI] [PubMed] [Google Scholar]
- 11. Hayden MS, Ghosh S. 2008. Shared principles in NF-κB signaling. Cell 132: 344–362 [DOI] [PubMed] [Google Scholar]
- 12. Hisamatsu T, et al. 2003. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology 124: 993–1000 [DOI] [PubMed] [Google Scholar]
- 13. Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. 2010. ATG16L1 and NOD2 interact in an autophagy-dependent, anti-bacterial pathway implicated in Crohn's disease pathogenesis. Gastroenterology 139: 1630–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu MC, et al. 2004. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell 117: 225–237 [DOI] [PubMed] [Google Scholar]
- 15. Hugot JP, et al. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411: 599–603 [DOI] [PubMed] [Google Scholar]
- 16. Hutchins AP, Robson P. 2009. Unraveling the human embryonic stem cell phosphoproteome. Cell Stem Cell 5: 126–128 [DOI] [PubMed] [Google Scholar]
- 17. Hutti JE, et al. 2004. A rapid method for determining protein kinase phosphorylation specificity. Nat. Methods 1: 27–29 [DOI] [PubMed] [Google Scholar]
- 18. Hutti JE, et al. 2009. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKε promotes cell transformation. Mol. Cell 34: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hutti JE, et al. 2007. IkappaB kinase beta phosphorylates the K63 deubiquitinase A20 to cause feedback inhibition of the NF-κB pathway. Mol. Cell. Biol. 27: 7451–7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Inohara N, et al. 2000. An induced proximity model for NF-κB activation in the Nod1/RICK and RIP signaling pathways. J. Biol. Chem. 275: 27823–27831 [DOI] [PubMed] [Google Scholar]
- 21. Inohara N, et al. 2003. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J. Biol. Chem. 278: 5509–5512 [DOI] [PubMed] [Google Scholar]
- 22. Kalkan T, Iwasaki Y, Park CY, Thomsen GH. 2009. Tumor necrosis factor-receptor-associated factor-4 is a positive regulator of transforming growth factor-beta signaling that affects neural crest formation. Mol. Biol. Cell 20: 3436–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanazawa N, et al. 2005. Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-kappaB activation: common genetic etiology with Blau syndrome. Blood 105: 1195–1197 [DOI] [PubMed] [Google Scholar]
- 24. Kedinger V, et al. 2005. Spatial and temporal distribution of the traf4 genes during zebrafish development. Gene Expr. Patterns 5: 545–552 [DOI] [PubMed] [Google Scholar]
- 25. Kedinger V, Rio MC. 2007. TRAF4, the unique family member. Adv. Exp. Med. Biol. 597: 60–71 [DOI] [PubMed] [Google Scholar]
- 26. Kim YG, et al. 2008. The cytosolic sensors Nod1 and Nod2 are critical for bacterial recognition and host defense after exposure to Toll-like receptor ligands. Immunity 28: 246–257 [DOI] [PubMed] [Google Scholar]
- 27. Kobayashi K, et al. 2002. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416: 194–199 [DOI] [PubMed] [Google Scholar]
- 28. Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. 2005. IKKα limits macrophage NF-κB activation and contributes to the resolution of inflammation. Nature 434: 1138–1143 [DOI] [PubMed] [Google Scholar]
- 29. Li Q, et al. 2005. Enhanced NF-κB activation and cellular function in macrophages lacking IκB kinase 1 (IKK1). Proc. Natl. Acad. Sci. U. S. A. 102: 12425–12430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li S, et al. 2010. Ubiquitin ligase Smurf1 targets TRAF family proteins for ubiquitination and degradation. Mol. Cell. Biochem. 338: 11–17 [DOI] [PubMed] [Google Scholar]
- 31. Li X, Yang Y, Ashwell JD. 2002. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature 416: 345–347 [DOI] [PubMed] [Google Scholar]
- 32. Li ZW, et al. 1999. The IKKβsubunit of IκB kinase (IKK) is essential for nuclear factor kappaB activation and prevention of apoptosis. J. Exp. Med. 189: 1839–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luo JL, et al. 2007. Nuclear cytokine-activated IKKalpha controls prostate cancer metastasis by repressing Maspin. Nature 446: 690–694 [DOI] [PubMed] [Google Scholar]
- 34. Marinis JM, Homer CR, McDonald C, Abbott DW. 2011. A novel motif in the Crohn's disease susceptibility protein, NOD2, allows TRAF4 to down-regulate innate immune responses. J. Biol. Chem. 286: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oehlers SH, et al. 2011. The inflammatory bowel disease (IBD) susceptibility genes NOD1 and NOD2 have conserved anti-bacterial roles in zebrafish. Dis. Model. Mech. 4: 832–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ogura Y, et al. 2001. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411: 603–606 [DOI] [PubMed] [Google Scholar]
- 37. Ogura Y, et al. 2001. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J. Biol. Chem. 276: 4812–4818 [DOI] [PubMed] [Google Scholar]
- 38. Olsen JV, et al. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3: ra3 doi:10.1126/scisignal.2000475 [DOI] [PubMed] [Google Scholar]
- 39. Park YC, Burkitt V, Villa AR, Tong L, Wu H. 1999. Structural basis for self-association and receptor recognition of human TRAF2. Nature 398:533–538 [DOI] [PubMed] [Google Scholar]
- 40. Razani B, et al. 2010. Negative feedback in noncanonical NF-κB signaling modulates NIK stability through IKKα-mediated phosphorylation. Sci. Signal. 3: ra41 doi:10.1126/scisignal.2000778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reiley W, Zhang M, Wu X, Granger E, Sun SC. 2005. Regulation of the deubiquitinating enzyme CYLD by IκB kinase gamma-dependent phosphorylation. Mol. Cell. Biol. 25:3886–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. 1999. IκB kinases phosphorylate NF-κB p65 subunit on serine 536 in the transactivation domain. J. Biol. Chem. 274: 30353–30356 [DOI] [PubMed] [Google Scholar]
- 43. Shembade N, Pujari R, Harhaj NS, Abbott DW, Harhaj EW. 2011. The kinase IKKα inhibits activation of the transcription factor NF-κB by phosphorylating the regulatory molecule TAX1BP1. Nat. Immunol. 12: 834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smahi A, et al. 2002. The NF-κB signalling pathway in human diseases: from incontinentia pigmenti to ectodermal dysplasias and immune-deficiency syndromes. Hum. Mol. Genet. 11: 2371–2375 [DOI] [PubMed] [Google Scholar]
- 45. Strober W, Watanabe T. 2011. NOD2, an intracellular innate immune sensor involved in host defense and Crohn's disease. Mucosal Immunol. 4: 484–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takeda K, et al. 1999. Limb and skin abnormalities in mice lacking IKKα. Science 284: 313–316 [DOI] [PubMed] [Google Scholar]
- 47. Takeshita F, et al. 2005. TRAF4 acts as a silencer in TLR-mediated signaling through the association with TRAF6 and TRIF. Eur. J. Immunol. 35: 2477–2485 [DOI] [PubMed] [Google Scholar]
- 48. Turk BE, Hutti JE, Cantley LC. 2006. Determining protein kinase substrate specificity by parallel solution-phase assay of large numbers of peptide substrates. Nat. Protoc. 1: 375–379 [DOI] [PubMed] [Google Scholar]
- 49. Watanabe T, et al. 2006. Nucleotide binding oligomerization domain 2 deficiency leads to dysregulated TLR2 signaling and induction of antigen-specific colitis. Immunity 25: 473–485 [DOI] [PubMed] [Google Scholar]
- 50. Wegener E, et al. 2006. Essential role for IκB kinase beta in remodeling Carma1-Bcl10-Malt1 complexes upon T cell activation. Mol. Cell 23: 13–23 [DOI] [PubMed] [Google Scholar]
- 51. Yamamoto-Furusho JK, Barnich N, Hisamatsu T, Podolsky DK. 2010. MDP-NOD2 stimulation induces HNP-1 secretion, which contributes to NOD2 antibacterial function. Inflamm. Bowel Dis. 16: 736–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ye H, et al. 2002. Distinct molecular mechanism for initiating TRAF6 signalling. Nature 418: 443–447 [DOI] [PubMed] [Google Scholar]
- 53. Ye H, Park YC, Kreishman M, Kieff E, Wu H. 1999. The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol. Cell 4: 321–330 [DOI] [PubMed] [Google Scholar]