Abstract
ISWI is an evolutionarily conserved ATPase that catalyzes nucleosome remodeling in different macromolecular complexes. Two mammalian ISWI orthologs, SNF2H and SNF2L, are thought to have specialized functions despite their high sequence similarity. To date, the function of SNF2L in human cells has not been a focus of research. Newly established specific monoclonal antibodies and selective RNA interference protocols have now enabled a comprehensive characterization of loss-of-function phenotypes in human cells. In contrast to earlier results, we found SNF2L to be broadly expressed in primary human tissues. Depletion of SNF2L in HeLa cells led to enhanced proliferation and increased migration. These phenomena were explained by transcriptome profiling, which identified SNF2L as a modulator of the Wnt signaling network. The cumulative effects of SNF2L depletion on gene expression portray the cell in a state of activated Wnt signaling characterized by increased proliferation and chemotactic locomotion. Accordingly, high levels of SNF2L expression in normal melanocytes contrast with undetectable expression in malignant melanoma. In summary, our data document an inverse relationship between SNF2L expression and features characteristic of malignant cells.
INTRODUCTION
The nucleosome, a 147-bp segment of DNA wrapped around a histone octamer, is the universal basis of chromatin organization in all eukaryotic genomes. The nucleosome itself, the folding of nucleosomal fibers, and the association of nonhistone proteins package, organize, and protect the precious genetic information. This packaging necessitates being transiently reverted if regulatory factors need to “read” the DNA, to find regulatory sites for transcription, or to denature the DNA double strand for templating reactions. The reversible opening of chromatin is not left to chance but is selectively achieved by a dedicated class of enzymes, the so-called nucleosome-remodeling factors. These enzymes bind to nucleosomes and detach segments of DNA from the histone octamer surface by a series of conformation changes that are driven by ATP hydrolyzation cycles. The basic nucleosome-remodeling reaction can be tuned to affect the sliding histone octamers on DNA, to disassemble nucleosomes by transferring histones to chaperones, or to exchange histones for variants. All of these reactions are reversible (10, 15).
Nucleosome-remodeling factors usually are multisubunit machineries. The ATPases directly responsible for the remodeling can be grouped into different families due to their domain organization. ATPases of the INO80/SWR1, Mi-2/CHD, SWI/SNF, and ISWI classes have been conserved during evolution from yeast to humans (19).
ISWI, one of the best-studied nucleosome-remodeling ATPases, was originally identified in Drosophila (13). In vitro, ISWI can cause nucleosomes to slide along DNA, a basic reaction that may create access to DNA or close gaps in the nucleosomal array by equalizing nucleosome spacing (52). ISWI is part of several remodeling factors with vital functions in gene expression, DNA replication, and repair (13). Mutations in Drosophila ISWI are associated with chromosome condensation abnormalities, possibly due to diminished loading of histone H1 on chromatin (12, 14). In addition, transcriptional defects have been described, and a recent study supports this notion by showing enriched ISWI binding near gene promoters (38).
The two mammalian orthologs of ISWI, SNF2H and SNF2L (also known as SMARCA5 and SMARCA1), share a high degree of amino acid sequence homology but appear to have different functions, as judged, for example, by their expression profiles (24). SNF2H resides in several structurally and functionally different remodeling complexes, such as CHRAC, ACF, WICH, NoRC, and RSF (7, 26, 27, 34, 45). In contrast, SNF2L has so far been found in the context of only two complexes, human NURF (hNURF) and CERF (1, 3). The SNF2L and SNF2H genes have diverged remarkably, as several alternative splice forms have been reported for SNF2L (2, 25), but not for SNF2H.
Early reports related an unbalanced expression of SNF2H to pathological cell proliferation. Stopka et al. explored the relationship between SNF2H expression and hematopoietic progenitor cell differentiation and found higher levels of SNF2H in CD34+ progenitors of acute myeloid leukemia (AML) patients, which decreased after complete hematologic remission of the tumor (44). Later studies showed that SNF2H is essential for proliferation of adult hematopoietic progenitors, in line with the first observations (43). More recently, SNF2H was identified as a fusion partner of EWSR1 in Ewing sarcoma/primitive neuroectodermal tumors, and the tumorigenic potential of the chimeric protein was documented (46). Furthermore, higher levels of SNF2H have been reported in gastric cancer than in normal mucosa, suggesting a role in malignancy (17).
Very little is known about the physiological functions of SNF2L. In mice, SNF2L expression has been reported to be limited to neuronal and gonadal tissues (1, 3). Using novel antibodies, our current study reveals SNF2L as a widely expressed modulator of the canonical Wnt/β-catenin signaling pathway. In this highly conserved relay network, β-catenin serves as the major transducer of Wnt signals to effect cellular responses. Wnt signaling is important during development, but also for cellular homoeostasis, since dysfunction results in developmental defects, as well as diseases like cancer (for a review, see references 11, 23, and 30). The signaling cascade is initiated by binding of Wnt ligands to receptors of the Frizzled and low-density lipoprotein-related receptor (LRP) family at the cell surface (6, 33). This activation of the receptors leads to the disassembly of the so-called destruction complex in the cytoplasm, consisting of β-catenin, Axin2, and the adenomatous polyposis coli gene product (APC) (31). Free β-catenin translocates to the nucleus, where it binds to TCF/LEF-type transcription factors and regulates genes controlling differentiation, apoptosis, cell proliferation, and migration (5). Transcription activation crucially involves chromatin remodeling (51). In the absence of signal, β-catenin is sequestered in the destruction complex, where it is sequentially phosphorylated by casein kinase 1 and glycogen synthase kinase 3 (28). The phosphorylated form is then recognized by the β-transducin-like enhancer of split product (β-Trcp), ubiquitinated, and subsequently degraded by the proteasome (20). Apart from the destruction complex, β-catenin can be part of complexes with cadherins and thereby also participates in adherens junctions (22).
In this study, we generated and characterized monoclonal antibodies (MAbs) that reliably distinguish SNF2H and SNF2L and used them to monitor the expression of both orthologs in human tissues. Refuting expectations raised in the literature, SNF2L was found to be broadly expressed but functionally very distinct from the SNF2H ortholog. Ablation of SNF2L in HeLa cells enhances proliferation and modulates the expression of 349 genes. Many responding genes are involved in locomotion and chemotaxis, and indeed, SNF2L-depleted cells showed enhanced migration behavior. Many genes involved in locomotion are known targets of Wnt signaling, and their transcription is stimulated upon SNF2L depletion, which leads to β-catenin stabilization. In addition, genes involved in the regulation of angiogenesis are affected by the depletion of SNF2L. As the responding gene groups and the described phenotype represent typical features of cancer cells, we monitored SNF2L expression in normal and malignant cells. Using melanoma as a model system, we indeed found strong SNF2L expression in normal primary melanocytes, in contrast to markedly reduced SNF2L expression in malignant melanoma.
MATERIALS AND METHODS
Cell culture.
HeLa and melanoma cell lines were cultured under a humidified atmosphere of 5% CO2 at 37°C in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal calf serum (FCS) (Invitrogen) and penicillin-streptomycin. Human primary melanocytes derived from neonatal foreskin were cultivated in melanocyte medium MGM-3 (Gibco) under a humidified atmosphere of 5% CO2 at 37°C (42). The cells were used between passages 3 and 6.
For cell counting during maintenance and for growth curves, the CASY counter (model TT; Innovatis) was used.
Antibodies.
For generation of MAbs against SNF2H, a peptide comprising amino acids 58MEEIFDDASPGKQKEIQEPPD76-C of SNF2H was synthesized and coupled to bovine serum albumin (BSA) and ovalbumin (OVA). For generation of MAbs against SNF2L, a glutathione S-transferase (GST) fusion protein (amino acids [aa] 571 to 772) was used. Rats were immunized subcutaneously and intraperitoneally with 50 μg peptide-OVA or 50 μg GST fusion protein, 5 nmol cytosine-guanosine (CPG) oligonucleotide (Tib Molbiol), 500 μl phosphate-buffered saline (PBS), and 500 μl incomplete Freund's adjuvant. A boost without adjuvant was given 6 weeks after the primary injection. Fusion was performed using standard procedures. Supernatants were tested by a differential enzyme-linked immunosorbent assay (ELISA) on SNF2H-OVA or SNF2L-GST and an irrelevant control with the same carrier. MAbs that reacted specifically with the SNF2H or SNF2L peptide were further analyzed in Western blot experiments. SNF 2C4 (rat IgG2b) against SNF2L and SNF-P 6F12 (rat IgG2b) against SNF2H were used in the study.
In addition, the following antibodies were used: GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Santa Cruz; sc-25778), tubulin (Sigma; T9026), lamin B (C-20; Santa Cruz), beta-actin (AC-74; Sigma), and β-catenin (610153; BD Biosciences).
Immunohistochemistry of a human tissue microarray.
The tissue microarray was constructed as described previously (40) and contained normal human tissue from 24 different organs. Human biopsy specimens were collected at the Institute of Pathology, University Hospital Regensburg, Regensburg, Germany. As prescribed by the medical ethical committee of the University of Regensburg, data privacy was protected, and safeguards were put in place to protect the identities of the subjects used in the study.
The paraffin-embedded biopsy specimens were dewaxed and rehydrated. The biopsy specimens were then boiled for 30 min in citrate buffer (pH 6.0). Endogenous peroxidase activity was blocked in 3% (vol/vol) H2O2 in PBS for 10 min. Sections were placed in a humidified chamber and covered with blocking solution (Zytomed) for 5 min. The sections were incubated with SNF2H or SNF2L antibody in 1-in-5 dilution in Tris-buffered saline (TBS)–Tween (0.1%) overnight at 4°C. After washing with TBS-Tween, the sections were incubated with biotinylated secondary antibody (Zytomed) for 20 min before incubation with streptavidin-horseradish peroxidase (HRP) conjugate for 20 min and 3-amino-9-ethylcarbazole (AEC) for 30 min (Zytomed) according to the manufacturers' instructions. All slides were then counterstained with routine hematoxylin.
siRNA and DNA transfections.
Small interfering RNAs (siRNAs) (MWG) were transfected using Oligofectamine and OptiMEM (Invitrogen) according to the suppliers' protocols and as described previously (39). The sense sequences of the oligonucleotides used were as follows: luciferase (siCTRL), CUUACGCUGAGUACUUCGA; SNF2H (siSNF2H), GGAAUGGUAUACUCGGAUA; and SNF2L (siSNF2L), CUGAAACACUACCGAAAUA, UAACAUAGCUCGAGAGGUA, and GGACAUUGAACAAAGUUUA.
For reporter gene assays, HeLa cells were seeded into 6-well dishes at 80% confluence. Five hundred nanograms of TOP-Flash or FOP-Flash reporter plasmids, together with 100 ng Renilla luciferase reporter plasmid, was transfected into each well using Fugene HD (Roche), and luciferase activity was measured utilizing the dual-luciferase reporter 1000 assay system (Promega).
RNA extraction and reverse transcription (RT)-PCR.
Total RNA was isolated from HeLa cells employing the RNeasy Kit (Qiagen) with additional RNase-free DNase (Qiagen) on column digestion. One microgram of RNA was subjected to subsequent cDNA synthesis using the Retroscript Kit (Ambion) following the manufacturer's instructions. Quantitative real-time PCR analysis was implemented with SYBR green 2× Master Mix (Applied Biosystems) and the ABI7000 (Applied Biosystems) detection system. Primer sequences are available upon request. GAPDH and actin served as internal controls for normalization. Amplifications were done in triplicate. Mean values were calculated according to the ΔΔCT quantification method, and error bars indicate standard deviations.
Total RNA for microarray experiments was also quality checked, utilizing the Bioanalyzer platform (Agilent).
Microarray analysis.
Total RNA labeling and hybridization to Human Gene 1.0 ST Arrays (Affymetrix) were performed according to the Human Gene 1.0 ST Array kit protocol (Affymetrix). The raw microarray data were processed in R/Bioconductor (http://www.bioconductor.org) as follows: gene-based expression values were calculated using the robust multichip average (RMA) method provided by the oligonucleotide package. Genes that had a log2 expression value of at least 4 under at least one of the treatment conditions were kept for downstream analyses. Differential expression estimation was based on a moderated t statistic (limma package) with subsequent calculation of the local false-discovery rate (lfdr) (locfdr package). Genes were classified as responders with an lfdr cutoff of 0.2. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using a hypergeometric distribution test supplied by the GOstats package with a P value cutoff of 0.001.
Migration assay.
Migration assays were performed using Boyden chambers containing polycarbonate membranes with 8-μm pore size (Neuro Probe). The filters were coated with gelatin (Sigma-Aldrich). The lower compartment was filled with fibroblast-conditioned medium, used as a chemoattractant. HeLa cells were harvested by trypsinization for 2 min, resuspended in DMEM without FCS at a density of 3 × 104 cells/800 μl/Boyden chamber, and placed in the upper compartment of the chamber. After incubation at 37°C for 4 h, the filters were collected, and the cells adhering to the lower surface were fixed, stained, and counted. Each assay was repeated at least three times.
Cell extract preparation and Western blot analysis.
For fractionated extraction of proteins, cells were washed with PBS, lysed in cytoplasmic buffer (20 mM Tris-Cl, pH 8, 85 mM KCl, 0.5% NP-40), and incubated on ice for 15 min. The subsequent centrifugation (800 × g; 4°C; 5 min) separated the cytoplasmic from the nuclear fraction, and the resulting pellet was resuspended in nuclear lysis buffer (50 mM Tris-Cl, pH 8, 10 mM EDTA, 1% SDS). Whole-cell extracts were prepared by lysing cells in NET buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, protease inhibitor cocktail [Roche]), with subsequent homogenization using a syringe.
To check for specific depletion of a protein after siRNA transfections, cells were counted and directly lysed in urea sample buffer (9 M urea, 1% SDS, 25 mM Tris-Cl, pH 6.8, 1 mM EDTA, 0.02% bromophenol blue, 200 mM dithiothreitol [DTT]) at concentrations of 103 cells/μl.
For Western blotting, proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes (Whatman) with successive blocking and antibody incubations. Secondary antibodies were compatible with infrared fluorescence detection on a Li-Cor Odyssey imaging system.
Microarray data expression number.
Raw microarray data were deposited at the National Center for Biotechnology Information (NCBI) gene expression omnibus (GEO) (data series GSE30471).
RESULTS
Generation and characterization of SNF2H- and SNF2L-specific antibodies.
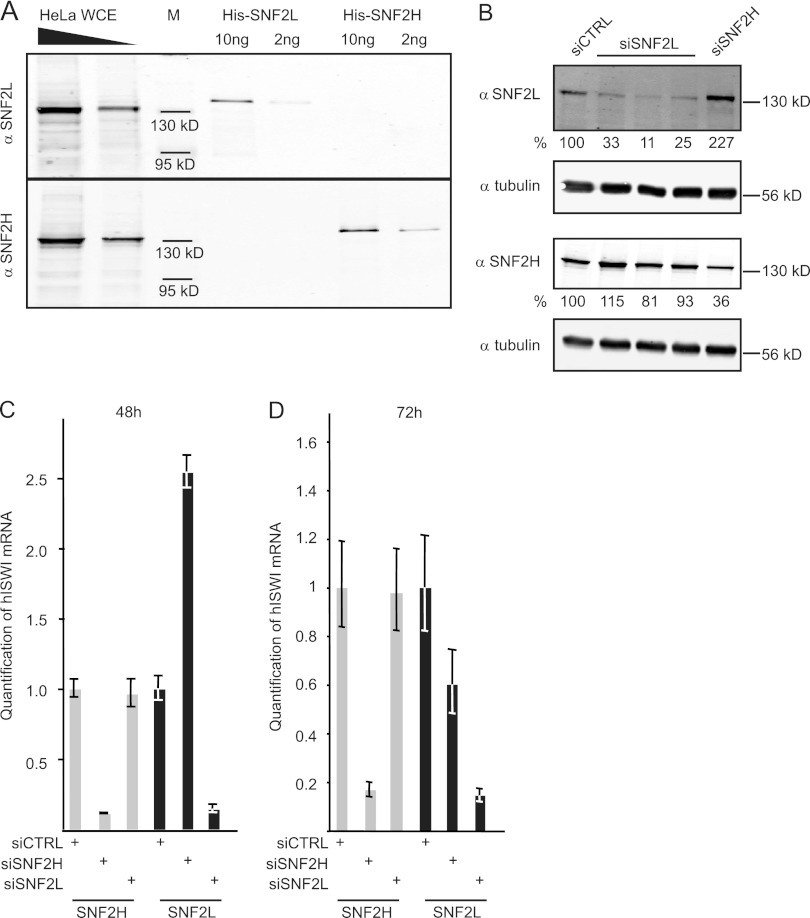
A consistent analysis of human ISWI function in vivo is still lacking, mainly because no antibodies were commercially available that could distinguish the two highly related human ISWI orthologs, SNF2H and SNF2L. We generated rat monoclonal antibodies against the two remodelers in order to fill this knowledge gap. The SNF2H-specific antibody recognized recombinant purified SNF2H, but not SNF2L, whereas the SNF2L-specific antibody detected only SNF2L, but not SNF2H, on a Western blot (Fig. 1A). Both antibodies marked single proteins with molecular masses of about 130 kDa, the predicted size of both remodelers, in a whole-HeLa-cell extract (Fig. 1A). Minor, faster-migrating bands detected by the SNF2L antibody likely represent SNF2L splice variants.
Fig 1.
SNF2L is transiently upregulated upon SNF2H depletion. (A) New antibodies against SNF2H and SNF2L were characterized by probing recombinant His-tagged SNF2H and SNF2L and increasing amounts of HeLa whole-cell extract (WCE) in duplicate Western blot experiments. M, molecular mass standards. (B) HeLa cells were transfected with the indicated siRNAs (top), and proteins were extracted 48 h after transfection. Western blots were probed with the antibodies shown on the left. The numbers below the blots represent quantification normalized to tubulin. (C) RNA from HeLa cells (treated as in panel B) was analyzed for expression of SNF2H and SNF2L. The data are given as means ± standard deviations (SD) (3 replicates). (D) As in panel C, but 72 h after transfection.
The antibody specificities were confirmed by selective ablation of SNF2H or SNF2L using siRNAs. For specific depletion of SNF2L, we tested six different siRNAs targeting different regions of the mRNA. Three siRNAs that showed strong and comparable knockdown efficiencies were chosen for further analysis (Fig. 1B). They were transfected into HeLa cells, and proteins were extracted 48 h later and analyzed by Western blotting. A clear reduction in signal was observed with the antibody raised against SNF2L only when cells had been transfected with SNF2L-specific siRNAs. Likewise, the SNF2H antibody revealed a lower signal only in extracts from cells treated with SNF2H-specific siRNAs (Fig. 1B). Interestingly, when SNF2H was depleted from HeLa cells, a reproducible ∼2-fold increase in the SNF2L signal was observed after 48 h. The faithful depletion of the respective mRNAs and the unexpected upregulation of SNF2L expression after RNA interference (RNAi) against SNF2H were confirmed at the level of mRNA by quantitative RT-PCR (Fig. 1C). In contrast, depleting SNF2L did not influence the amount of SNF2H RNA or protein. The increased SNF2L expression in response to SNF2H depletion was transient, as lower SNF2L RNA levels (about 50% those of control siRNA-treated cells) were observed 72 h after SNF2H knockdown (Fig. 1D). Further experiments confirmed this finding and revealed that the decreased expression is also reflected at the protein level. Off-target effects of the RNAi directed against SNF2H can be excluded because of the transient upregulation of SNF2L upon reduction of SNF2H as a primary response.
In conclusion, we generated new antibodies with specificity for their respective antigens and have established gene-specific RNA interference protocols to selectively ablate one of the two SNF2 isoforms.
Expression of SNF2H and SNF2L in human tissues.
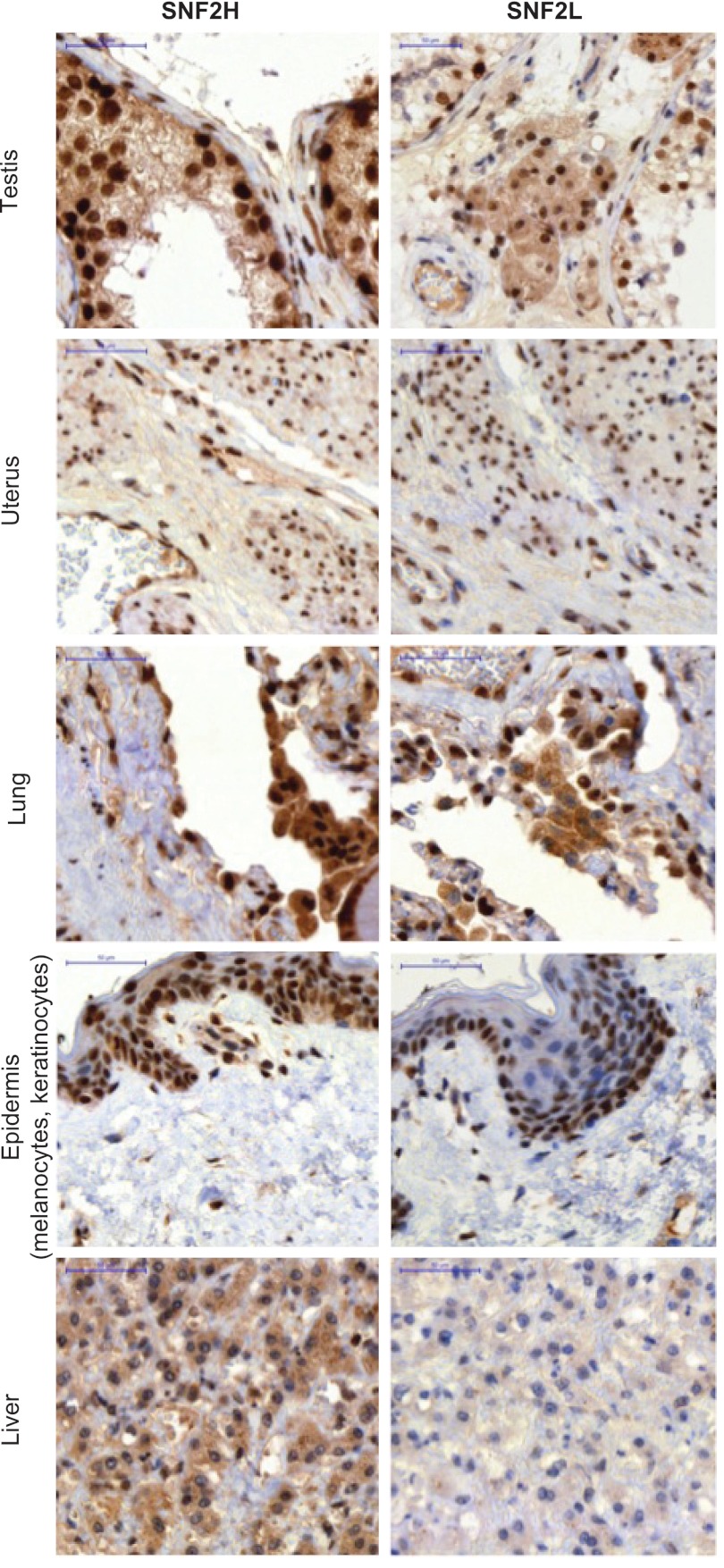
The common view is that the expression of SNF2L is restricted to terminally differentiated cells, mainly of neuronal or gonadal origin, whereas SNF2H is ubiquitously expressed (24). This view is mainly grounded in studies in the murine system. Ye et al. previously pointed out differences between the expression patterns in mouse versus human tissues, based on RT-PCRs and expressed sequence tags (ESTs) posted on the NCBI website (53). Taken together, SNF2L expression in humans was solely monitored on the RNA level and not on the protein level. We used the new pair of specific monoclonal antibodies to probe a human tissue array for the expression of SNF2H and SNF2L by immunohistochemistry and hematoxylin counterstaining. This approach also allowed us to distinguish different cell types in one tissue sample.
All tissues showed strong immunoreactivity for SNF2H. SNF2L, however, was expressed in testis, uterus, lung, epidermis, ovary, kidney, heart, parotid gland, and salivary gland, but not in liver, stomach, tonsil, breast, colon, or pancreas (Fig. 2; see Fig. S1 in the supplemental material). Specific cell types were stained in the following tissues: spleen, duodenum, appendix, adrenal gland, placenta, endometrium, prostate, and cortex (brain) (for a description, see Fig. S1).
Fig 2.
SNF2L and SNF2H expression in human tissues. The same antibodies as in Fig. 1 were used to stain a human tissue microarray monitoring the expression pattern of SNF2H and SNF2L. Hematoxylin was used for counterstaining. Representative examples of the array are shown. Scale bars, 50 μm. For further tissues, see Fig. S1 in the supplemental material.
In summary, SNF2H and SNF2L showed distinct expression in a variety of human tissues. Notably, SNF2L appears to be more widely expressed than has been reflected in the literature.
Depletion of SNF2H and that of SNF2L affect cell proliferation differently.
To explore the functions of SNF2H and SNF2L in human cells, we transiently depleted HeLa cells of each protein by RNA interference and compared their proliferation to the growth of cells treated with control siRNAs. The depletion of SNF2H resulted in an apoptotic phenotype leading to reduced cell numbers after only 48 h (Fig. 3A), as shown in our previous publication (39). We used siRNAs with lower knockdown efficiencies in our study in order to be able to grow HeLa cells further with lower SNF2H levels. Nevertheless, cell numbers were always reduced in comparison to parallel cultures of control cells. Strikingly, depletion of SNF2L with two different siRNAs for 72 h led to increased cell numbers compared to control cells (Fig. 3A). Figure 3B illustrates the statistical significance of the observed proliferation differences. The increased proliferation of SNF2L-depleted HeLa cells was not accompanied by obvious changes in the cell cycle profiles measured by flow cytometry (data not shown).
Fig 3.
Depletion of SNF2L enhances cell proliferation. (A) Representative growth curves of siRNA-treated HeLa cells. Knockdown efficiencies were similar to the ones shown in Fig. 4A and 5B for SNF2H and SNF2L, respectively. siCTRL, control siRNA; siSNF2H, SNF2H siRNA; siSNF2L, SNF2L siRNA. (B) Numbers of SNF2H- and SNF2L-depleted cells were determined 72 h after siRNA transfection. The bars represent cell numbers relative to cells treated with control siRNAs, which were set to 100%, ± standard errors of the mean (SEM). n, number of biological replicates. The P values were calculated using two-sided, unpaired Student t tests.
Taken together, the depletion of SNF2H abets apoptosis, whereas depletion of SNF2L promotes cell proliferation.
Involvement of SNF2L in regulating cell adhesion and locomotion.
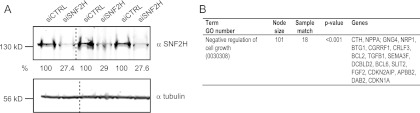
We conducted gene expression profiling using Affymetrix Human Gene 1.0 ST arrays covering 28,869 genes in order to see whether the different HeLa cell phenotypes resulting from depletion of SNF2H or SNF2L were reflected at the level of gene transcription. Three biological replicates of RNA interference showing comparable reductions of SNF2H levels 72 h after siRNA transfection were combined for subsequent analysis (Fig. 4A). Reducing the SNF2H levels resulted in the increased expression of 535 genes and lower expression of 567 genes. We next tested for overrepresentation of these responders (including up- and downregulated genes) in distinct biological processes based on GO classifications. A given GO term was considered to be overrepresented when the observed number was significantly higher than the number expected by chance. Although the term “apoptosis” was not found to be enriched, a slight overrepresentation of genes involved in “negative regulation of cell growth” (GO 0030308) was noted when all responding genes were considered (Fig. 4B). As expected, SNF2L was among the responders, confirming our initial results (Fig. 1D). Because the effects of SNF2H depletion on gene expression may be partly due to the indirect effect of reduced SNF2L levels, we did not engage in further analysis of the SNF2H depletion data.
Fig 4.
Ablation of SNF2H affects the expression of specific gene groups. (A) Western blot of extracts from the three independent replicate siRNA depletion experiments used for gene expression analysis. SNF2H-specific antibody confirmed knockdown after siRNA treatment, and tubulin was used as a loading control. The dashed lines indicate that intervening lanes were spliced out. (B) Simplified GO analysis of genes responding to SNF2H knockdown.
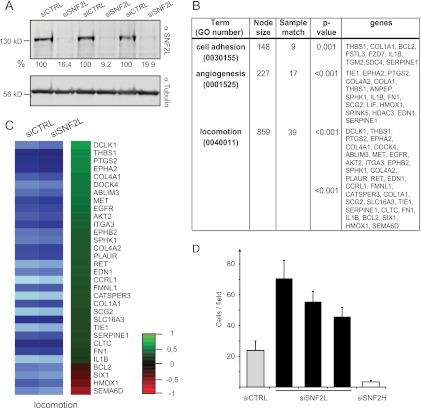
We also determined the gene expression profile after SNF2L depletion. As for SNF2H, we combined three biological replicates with strong knockdown efficiencies for global gene expression profiling (Fig. 5A). Overall, the analyses revealed only small changes in expression, with the SNF2L gene itself being the strongest responder. Nevertheless, 102 genes displayed significantly decreased expression and 247 genes showed significantly increased expression after SNF2L ablation. Interestingly, a search for affected gene groups according to the classification of the KEGG pathways yielded the maps of focal adhesion (identifier [ID] 04510) and extracellular matrix (ECM)-receptor interaction (ID 04512). Furthermore, our data map to pathways in cancer (ID 05200). We also analyzed the complete list of SNF2L-responding genes according to the GO classification. In agreement with the KEGG analysis, genes involved in the “regulation of cell adhesion” (GO 0030155), “cell-substrate adhesion” (GO 0031589), and “focal adhesion assembly” (GO 0048041) were enriched (Fig. 5B). In addition, overrepresentation of genes involved in “angiogenesis” (GO 0001525) and different terms subsumed under the highest hierarchy level, “locomotion” (GO 0040011), were among the genes whose expression was increased as SNF2L was depleted (Fig. 5B and C). “Locomotion” is defined as self-propelled movement of a cell leading to a change in location and includes terms like cell migration and chemotaxis. This phenomenon is clearly accompanied by changes in cell-substrate adhesion and its regulation. SNF2L-depleted cells may thus show increased migration behavior. To directly test whether this was indeed the case, we performed migration assays in Boyden chambers. HeLa cells were transfected with control siRNAs or three unrelated oligonucleotides directed against SNF2L. In addition, we ablated SNF2H. Three days later, identical numbers of cells were seeded into the Boyden chamber system. Two- to 3-fold more cells with reduced SNF2L levels than control cells migrated. Remarkably, SNF2H depletion led to the opposite behavior, as cells lacking the remodeler migrated about 5-fold less (Fig. 5D).
Fig 5.
Ablation of SNF2L affects the expression of specific gene groups. (A) Western blot of extracts from the three independent replicate siRNA depletion experiments used for gene expression analysis. The SNF2L-specific antibody was used to confirm the knockdown after siRNA treatment. Tubulin served as a loading control. The numbers below the blots indicate the knockdown efficiencies normalized to that of tubulin. (B) Simplified GO analysis of genes responding to SNF2L knockdown. (C) Heat map illustrating the change in expression after SNF2L depletion of 28 upregulated (green) and four downregulated (red) genes belonging to the category “locomotion” according to GO. (D) Reduction of SNF2L increases chemotaxis. Boyden chamber experiments were performed 72 h after siRNA transfection using fibroblast-conditioned medium as a chemoattractant. The bars represent the numbers of cells that migrated after 4 h (means plus SEM).
In summary, SNF2L depletion in HeLa cells led to unexpected expression changes of gene groups important for cell adhesion and locomotion. The significance of these changes was validated by the experimental demonstration of increased chemotaxis of SNF2L-depleted cells.
Activation of Wnt/β-catenin signaling upon SNF2L depletion.
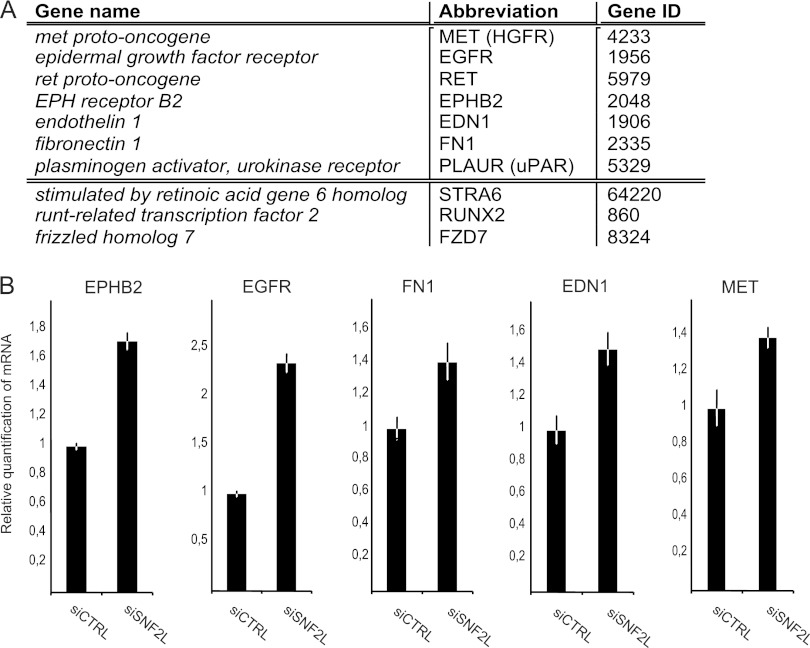
Efficient depletion of SNF2L results in changes in proliferation and migration behavior, two cellular aspects that are regulated by the canonical Wnt-signaling pathway. This pathway is activated when Wnt ligands bind to their cell surface receptor. The signal transduction is mainly achieved by the cytoplasmic stabilization of the transcription factor β-catenin, which translocates into the nucleus, where it regulates the expression of target genes. Many of these genes are involved in processes like proliferation, adhesion, and migration. To explore the relationship between the SNF2L ablation phenotype and Wnt signaling, we searched the list of genes that were affected by SNF2L depletion for Wnt target genes. A comparison of SNF2L responders in the transcriptome analysis to the Wnt target genes listed on the Wnt homepage (http://www.stanford.edu/group/nusselab/cgi-bin/wnt/target_genes) yielded a list of 10 coincident genes (Fig. 6A). The expression of 9 out of these 10 genes was increased upon reduction of SNF2L levels. Strikingly, 7 of these had an influence on cell locomotion according to the GO classification (Fig. 6A, upper part of the table). We confirmed the transcriptional activation of five representative genes upon SNF2L depletion by quantitative reverse transcription-PCR (Fig. 6B).
Fig 6.
SNF2L depletion influences Wnt/β-catenin target gene expression. (A) Wnt target genes responding to SNF2L depletion. The upper part of the table comprises genes involved in locomotion according to GO classification. (B) qPCR validation of selected Wnt target genes after SNF2L depletion in HeLa cells. For normalization, primer sets amplifying actin and GAPDH were used (means ± SD; 3 replicates).
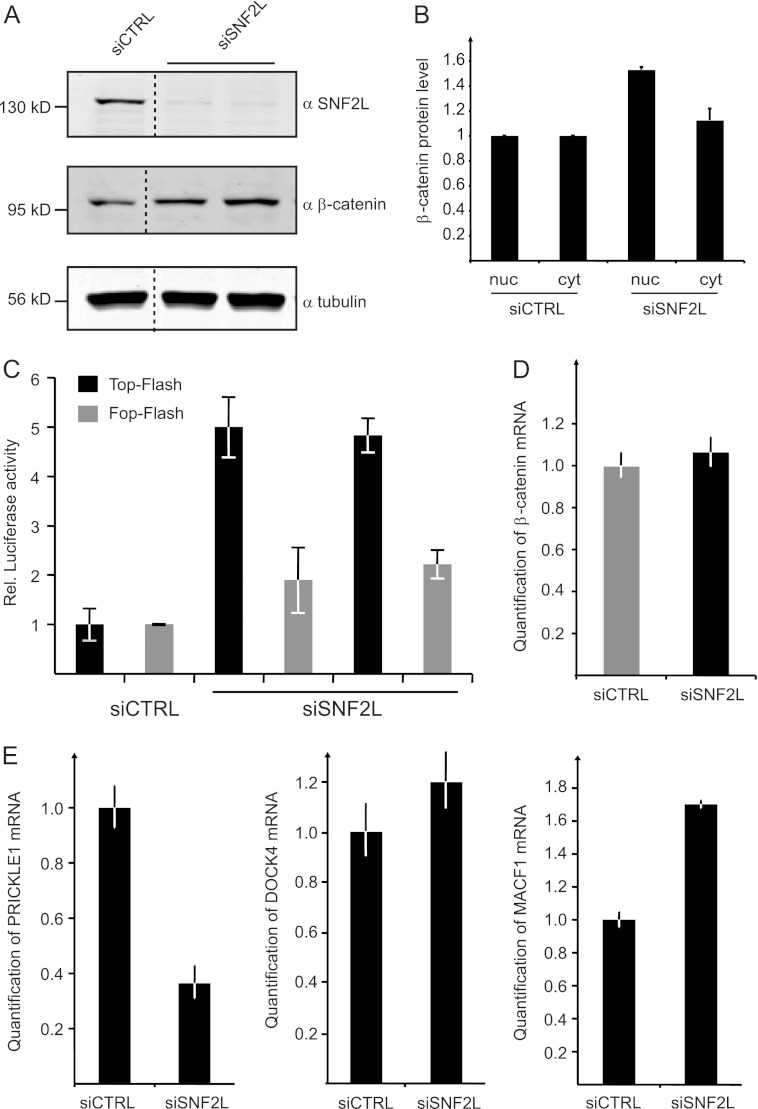
To test if the activation of these genes correlated with activation of the Wnt pathway, we analyzed β-catenin as the main mediator of canonical Wnt signaling. Depletion of SNF2L for 72 or 96 h indeed led to a clear increase in β-catenin levels in whole-cell extracts (Fig. 7A). The enhanced β-catenin levels in cells with lower SNF2L levels should be nuclear in order to be transcriptionally relevant. Extracts were prepared from siRNA-treated cells, separating the cytoplasm from the nuclear fraction. Probing the fractions for β-catenin by quantitative Western blotting not only confirmed the increased levels of β-catenin in SNF2L-depleted cells, but also revealed that the relative distribution of the signal transducer had shifted to the nucleus compared to control cells (Fig. 7B).
Fig 7.
Wnt/β-catenin signaling is activated in SNF2L-depleted cells. (A) Western blot experiments probing β-catenin and SNF2L after knockdown of SNF2L in whole-cell HeLa extracts. Tubulin staining served as a loading control. The dashed lines indicate that intervening lanes were spliced out. (B) Proteins from HeLa cells (treated with siRNA as indicated) were fractionated into cytoplasmic (cyt) and nuclear (nuc) parts. Subsequent Western blots using β-catenin, tubulin, and lamin antibodies were quantified in the Li-Cor imaging system. The values for β-catenin staining were normalized to nuclear lamin and to cytoplasmic tubulin, respectively. The bars represent the fold increase in β-catenin amounts compared to the control cells. (C) HeLa cells were treated with two different siRNAs against SNF2L and control siRNA before TOP-Flash and FOP-Flash reporter plasmids were transfected. A Renilla luciferase reporter plasmid was introduced in parallel for normalization. Luciferase activity was measured 72 h after siRNA treatment and calculated as relative activity compared to control values, which were set to 1 (mean ± SD). (D) HeLa cells were depleted of SNF2L for 72 h. RNA was isolated, and selected RNAs were quantitated by RT-PCR. β-Catenin mRNA was measured in quantitative PCR (qPCR) experiments using actin mRNA for normalization (means ± SD; 3 replicates). (E) mRNA levels of PRICKLE1, DOCK4, and MACF1 were determined as for panel A.
Further support for the idea that SNF2L depletion leads to activation of Wnt signaling via increasing the β-catenin levels was obtained from a functional reporter gene assay. The TOP- and FOP-Flash plasmids harbor six multimerized canonical or mutated TCF/β-catenin binding sites, respectively, that control the transcription of a firefly luciferase gene (36, 50). TOP- and FOP-Flash plasmids were transiently transfected into HeLa cells that had been depleted of SNF2L for 48 h. The transfection efficiencies were normalized to the activity of a cotransfected Renilla luciferase-encoding plasmid. Luciferase activities 24 h after transfection of the reporter system (72 h after siRNA transfection) provided a measure for activated Wnt/β-catenin signaling. In cells with reduced SNF2L levels, the transcription of the luciferase gene from the TOP-Flash plasmid was strongly activated relative to control cells (Fig. 7C). Mutating the TCF/β-catenin binding sites (FOP-Flash) largely diminished this activation, demonstrating the direct relationship between reduced SNF2L levels, stabilization of β-catenin, and the activation of target genes.
In summary, the ablation of SNF2L results in increased canonical Wnt signaling and, consequently, in the expression of Wnt/β-catenin-responsive genes.
Indirect regulation of β-catenin by SNF2L.
We interrogated our expression array data in order to see if the increase in β-catenin protein after SNF2L knockdown was due to enhanced transcription. The β-catenin gene was not among the responders in our array, and we confirmed this result by RT-PCR (Fig. 7D). Clearly, the β-catenin gene is not a direct transcriptional target of SNF2L. We conclude that the regulation of β-catenin occurs on the posttranscriptional level, presumably by modulation of the protein's stability.
Many proteins had been shown to directly influence Wnt signaling and β-catenin stability. We therefore searched our gene expression profiles again for known regulators of the Wnt signaling pathway or proteins that are themselves part of the signaling cascade and whose expression changes in response to SNF2L depletion. We found, for example, the gene encoding the porcupine homologue (PORCN) that is involved in the processing of Wnt proteins in the endoplasmic reticulum (47). The upregulation of this protein could directly change the outcome of an activated Wnt signaling cascade by increasing the number of (biologically active) Wnt molecules.
Furthermore, we identified several genes in our list of responders whose changes in expression upon SNF2L depletion may contribute to the observed activation of Wnt signaling. Transcription of the prickle homologue (PRICKLE1) gene is reduced upon SNF2L knockdown, a result that was confirmed by quantitative RT-PCR (Fig. 7E). The PRICKLE1 protein negatively modulates Wnt signaling by inducing the degradation of the Dishevelled (Dvl) proteins (8, 16). Upon canonical Wnt signaling, Dvl is normally assembled with Frizzled receptors, in addition to Axin recruitment to LRPs, and is responsible for the release of β-catenin from the destruction complex.
Moreover, the dedicator of cytokinesis 4 (DOCK4) gene shows stronger expression after SNF2L depletion. DOCK4 enhances the release of β-catenin from the destruction complex consisting of APC and Axin (49). Similarly, the microtubule-actin cross-linking factor 1 (MACF1) gene is upregulated in SNF2L-depleted cells (Fig. 7E). MACF1 affects the β-catenin level, presumably by the translocation of the destruction complex to the plasma membrane (9), which leads to the release of β-catenin and thereby to its stabilization. The decrease in PRICKLE1 and the increase in DOCK4 and MACF1 are expected to cooperate in increasing the stability of β-catenin.
In addition, we spotted increased expression of the transcription coactivator p300 and decreased expression of the corepressor transducin-like enhancer of split 4 (TLE4) after SNF2L depletion (data not shown). Even though these cofactors do not act solely in the Wnt signaling pathway, they are described as being involved in the expression of Wnt-responsive genes (21, 37). Thus, the modulation of their expression may contribute to tipping the balance toward the transcription of Wnt targets.
In summary, many of the genes that respond to the depletion of SNF2L with modest expression changes participate in the Wnt signaling cascade itself or in its regulation. These data place the nucleosome remodeler SNF2L within the Wnt signaling network. Depletion of SNF2L leads to many small changes in the expression of a variety of genes. The cumulative effects of these modulatory rather than decisive changes puts the cell in a state of activated Wnt signaling characterized by increased proliferation and chemotactic locomotion.
SNF2L is strongly expressed in normal melanocytes but is almost absent in melanoma.
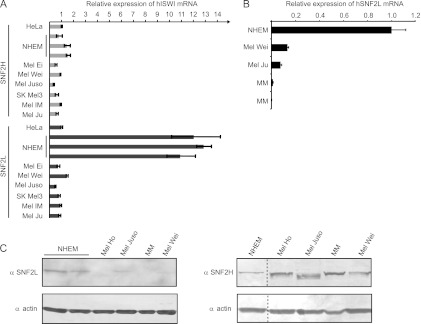
Increased proliferation and chemotactic locomotion upon SNF2L depletion, along with changes in a group of genes regulating angiogenesis, are typical characteristics of tumors and metastasis. SNF2L expression, therefore, should inversely correlate with tumors and metastasis. Since SNF2L is strongly expressed in normal skin with melanocytes, we used this cell system to compare the SNF2L transcript levels in normal human epidermal melanocytes (NHEM) to cell lines derived from primary melanoma (Mel Ei, Mel Wei, or Mel Juso) or metastasis (SK Mel3, Mel Im, or Mel Ju). For comparison, we measured the levels of SNF2H mRNA. SNF2H showed similar expression in the tested samples and levels comparable to those of HeLa cells, which we used as a reference (Fig. 8A). Strikingly, SNF2L expression in normal melanocytes was more than an order of magnitude higher than the expression in HeLa cells or any other cell line derived from primary tumors or metastasis (Fig. 8A).
Fig 8.
SNF2L, but not SNF2H, is reduced in malignant melanoma. (A) Primer sets amplifying SNF2H and SNF2L sequences were used to measure mRNA levels in normal primary melanocytes (NHEM), cell lines derived from primary melanoma (Mel Ei, Mel Wei, and Mel Juso), or melanoma metastasis (SK Mel3, Mel IM, and Mel Ju). The mRNA level in HeLa cells was used as a reference and set at 1 (means plus SD; 3 replicates). (B) qPCR experiments as in panel A, including RNA extracted from microdissected tissue samples from MM. NHEM values were used as references and set to 1 (mean plus SD; 3 replicates). (C) Western blot duplicates detecting SNF2L and SNF2H in extracts from different samples (A and B). Actin antibody served as a loading control. The dashed lines indicate that intervening lanes have been spliced out.
As a further test for the detected lower SNF2L expression in tumor-derived cell lines compared to primary normal melanocytes, we also monitored SNF2L transcript levels in freshly isolated primary melanoma cells from two different donors (MM1 and MM2). Remarkably, we observed even lower expression in the malignant melanoma samples than in the melanoma-derived cell lines Mel Wei and Mel Ju (Fig. 8B). This result was validated at the protein level by Western blotting using the SNF2L-specific antibody and an actin antibody as a loading control. We probed extracts from NHEM; cell lines like Mel Juso, Mel Wei, and Mel Ho; and malignant melanoma-derived samples (MM). As expected, we found SNF2L highly expressed in NHEM, less so in Mel Juso, and undetectable in Mel Ho, Mel Wei, and the malignant melanoma material (Fig. 8C). The extracts were also probed with the SNF2H-specific antibody, and the result confirms our initial finding that SNF2H is ubiquitously expressed and correlates with the RNA levels (Fig. 8C).
SNF2L affects the migratory potential of melanoma cells.
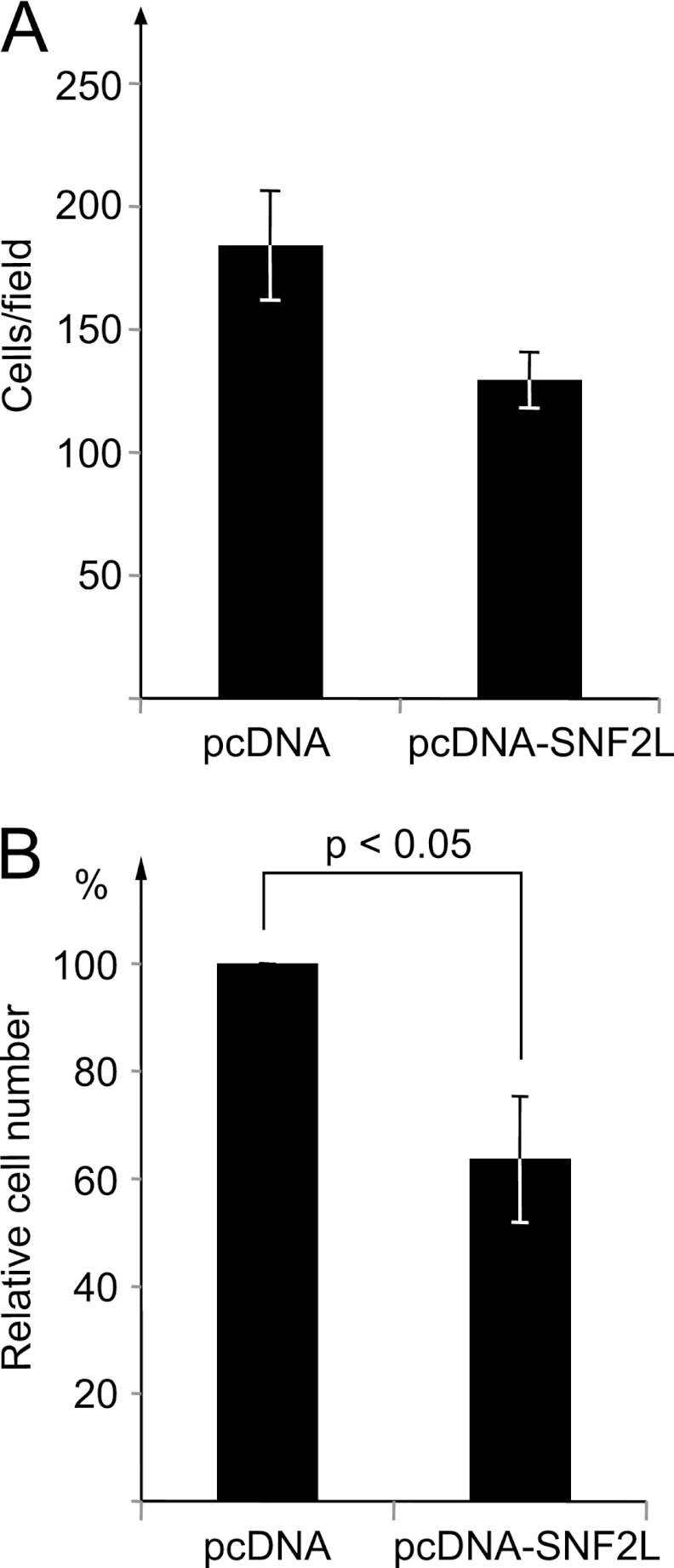
The results of SNF2L depletion so far established an inverse correlation between SNF2L levels and HeLa cell migration potential, as well as with melanoma malignancy. Forced expression of SNF2L in melanoma cells, therefore, may decrease their migration potential. To test this hypothesis, we transfected Mel Im cells with an SNF2L expression vector or, as a control, with the empty vector (pcDNA). When these cell populations were tested in parallel for their migration potential in Boyden chambers, a significant decrease in migration behavior upon SNF2L overexpression was observed (Fig. 9A and B).
Fig 9.
Overexpression of SNF2L in melanoma cells reduces the migration potential. (A) Mel IM cells were transfected with an SNF2L expression vector or the empty vector (pcDNA) as a control, and the two populations were assayed for their migration potential in Boyden chambers using fibroblast-conditioned medium as a chemoattractant. The bars represent the numbers of cells that had migrated after 4 h (means ± SEM). (B) The bars represent the relative numbers of cells that migrated in three independent experiments of the kind shown in panel A. The number of control cells was set to 100% ± SEM. The indicated P values were calculated using two-sided, unpaired Student t tests.
In summary, our data reveal an inverse correlation between SNF2L expression and features that characterize malignant cells. SNF2L is widely expressed in many normal human tissues. In a series of melanocyte/melanoma isolates, SNF2L levels inversely correlate with malignancy. At least part of this phenomenon can be explained by its modulatory role in the Wnt signaling pathway.
DISCUSSION
Widespread expression of SNF2L in human tissues.
The current view of the two mammalian ISWI orthologs, SNF2H and SNF2L, proposes two remodeling enzymes that are differently expressed and reside in different complexes (19). Many studies focused on SNF2H, which is considered the major ISWI-type remodeler in the cell, as it is ubiquitously expressed and is part of abundant chromatin-remodeling complexes that are functionally diverse, ranging from chromatin assembly to rDNA transcriptional repression (7, 55). In contrast, SNF2L is known only for its important role in neurogenesis (1, 24). Overexpression of SNF2L leads to neurite outgrowth in neuronal precursor cell lines (3). Interestingly, the GO classification deduced from our set of downregulated genes upon SNF2L depletion yielded an overrepresentation of genes involved in “neuron apoptosis” (GO 0051402), in agreement with a report showing increased cell numbers in the brains of SNF2L knockout mice (54).
Considering this emphasis on neuronal phenotypes in the literature, we were surprised to observe fairly broad expression of SNF2L in human tissues. It is possible, that the SNF2L expression patterns in human and mouse tissues differ, as was previously suggested based on mRNA analyses (53). In this study, the authors found nearly ubiquitous expression of SNF2L in the tested human samples. Nevertheless, the use of whole organs (comprising different cell types) for the isolation of RNA could be one reason for this result and the discrepancies with our study. Characterization at the protein level was possible due to the generation of monoclonal antibodies that clearly discriminate between the two ISWI orthologs. Our analysis suggests that hSNF2L must be considered an important player in cellular programs in a broad variety of human tissues.
Functional diversification of the two hSNF2 isoforms.
Our study documents very obvious differences in the responses of HeLa cells to depletion of the two SNF2 isoforms. HeLa cells displayed opposite phenotypes with respect to proliferation and migration behavior when challenged with depletion of SNF2H or SNF2L. However, several observations indicate that the contrasting results are not easily explained by clear-cut antagonistic roles of the two remodelers. For example, the global transcriptome analysis revealed that the expression of only two genes (SLC2A3 and PLCL1) was modulated in opposite directions after depletion of SNF2H and SNF2L. The opposing outcomes of the two isoform depletions may reflect their different roles in a complex gene expression network that integrates many modest primary, but presumably also numerous indirect, effects. Notably, SNF2L levels were transiently increased about 2 days after the depletion of SNF2H was initiated. This phenomenon may reflect an attempt of the cell population to compensate for a lethal depletion of SNF2H, a futile attempt, considering that we found the SNF2L gene to be downregulated upon prolonged depletion of SNF2H.
Our study presents the first global gene expression profiling of human cells depleted of either of the two SNF2 isoforms. In both cases, similar numbers of genes were up- as well as downregulated. Whether the remodelers are directly involved in gene activation and repression events (13) can be assessed only in the context of in vivo interaction.
SNF2L modulates the Wnt signaling network.
Ablation of SNF2L led to changes in proliferation and chemotactic migration that are consistent with the observed gene expression changes. Most of these changes were rather modest in magnitude, despite the respectable knockdown efficiencies we achieved. Conceivably, residual low-level expression of the remodelers may ameliorate the effects of the depletion. In any case, the many small gene expression changes all contribute to a consistent phenotype that is characterized by stimulated Wnt signaling. We found the key regulator of the canonical Wnt signaling pathway, β-catenin, to be elevated in the nucleus if SNF2L was reduced. The elevated nuclear β-catenin levels are likely the explanation for the enhanced migration potential of SNF2L-depleted cells. Strikingly, 7 out of the 10 Wnt target genes that responded to SNF2L depletion are involved in locomotion, according to gene ontology classification, consistent with the increased migration behavior. The specification of Wnt target genes is highly context dependent, but the underlying mechanisms are unclear (35).
In this context, a review of the relevant studies performed in the Drosophila model may be informative, as relationships between Wnt signaling and ISWI-containing remodeling complexes have been noted (29, 41). ISWI in complex with ACF1 (in the form of CHRAC or ACF complexes) was found to be involved in basal repression of Wnt target genes (23). On the other hand, ISWI in the context of the NURF complex has been shown to activate Wnt target genes upon Wnt signaling (29, 41). This is in agreement with a reported dependency of the expression of human engrailed genes, known Wnt targets, by hNURF (3). The prevailing view in the literature is that in mammals a division of labor coincided with the evolution of the two ISWI orthologs. SNF2L is mainly found in hNURF complexes, whereas SNF2H resides in several other assemblies, including hACF/hCHRAC (19). Whereas the aforementioned studies would have predicted a role for SNF2L as an activator of Wnt target genes, we found 9 out of 10 genes that responded to SNF2L depletion were upregulated. Their moderate activation of gene expression may be explained by indirect effects rather than by specific, direct coactivator functions of SNF2L.
Chromatin remodeling appears to be crucial for the regulation of Wnt target genes, as nucleosome-remodeling complexes are known to be involved. For example, the remodeling ATPases Brg1 and Chd8 were shown to modulate Wnt target gene expression as cofactors (4, 48). Interestingly, Brg1 apparently also influences the level of β-catenin by a transcription-independent mechanism (18). This indirect regulation is reminiscent of the phenomenon we observed for SNF2L and documents the complex relationship between Wnt signaling and chromatin remodeling.
Inverse correlation of SNF2L and melanoma malignancy.
The phenotype achieved by ablation of SNF2L in HeLa cells is characterized by increased proliferation and migration behavior. However, the loss-of-function situation imposed on the cells by RNA interference is far from physiological. To what extent are SNF2L levels an indication of a normal, healthy tissue phenotype? Probing a range of tissues for SNF2L expression using the novel diagnostic monoclonal antibody, we found SNF2L well expressed in a range of human tissues. Since aberrant activation of Wnt signaling is known to play a major role in tumorigenesis and has been discussed as one reason for the onset of melanoma (32), we analyzed melanocytes and melanoma cells in more detail and found a strong inverse correlation of SNF2L expression and malignancy. Apparently, the phenotype associated with malignant cells also includes low SNF2L levels. The RNA interference experiments performed in HeLa cells are consistent with a causal relationship. It will be interesting to see whether our basic observation in models will also apply to other tissues and tumor types.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Lamm for technical assistance. We thank A. Schepers, who initiated the generation of the monoclonal SNF2H/L antibodies; F. Müller-Planitz for recombinant SNF2H/L proteins; D. Martin and K. Maier of the Gene Center Affymetrix Microarray Platform for help with microarray experiments; and D. J. Picketts for providing the SNF2L overexpression construct.
This work was supported by grants from the Deutsche Forschungsgemeinschaft to P.B.B. through SFB 684 and the Leibniz Programme.
Footnotes
Published ahead of print 16 April 2012
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1. Banting GS, et al. 2005. CECR2, a protein involved in neurulation, forms a novel chromatin remodeling complex with SNF2L. Hum. Mol. Genet. 14: 513–524 [DOI] [PubMed] [Google Scholar]
- 2. Barak O, Lazzaro MA, Cooch NS, Picketts DJ, Shiekhattar R. 2004. A tissue-specific, naturally occurring human SNF2L variant inactivates chromatin remodeling. J. Biol. Chem. 279: 45130–45138 [DOI] [PubMed] [Google Scholar]
- 3. Barak O, et al. 2003. Isolation of human NURF: a regulator of Engrailed gene expression. EMBO J. 22: 6089–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barker N, et al. 2001. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 20: 4935–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Behrens J, et al. 1996. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382: 638–642 [DOI] [PubMed] [Google Scholar]
- 6. Bhanot P, et al. 1996. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature 382: 225–230 [DOI] [PubMed] [Google Scholar]
- 7. Bozhenok L, Wade PA, Varga-Weisz P. 2002. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 21: 2231–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan DW, Chan CY, Yam JW, Ching YP, Ng IO. 2006. Prickle-1 negatively regulates Wnt/beta-catenin pathway by promoting Dishevelled ubiquitination/degradation in liver cancer. Gastroenterology 131: 1218–1227 [DOI] [PubMed] [Google Scholar]
- 9. Chen HJ, et al. 2006. The role of microtubule actin cross-linking factor 1 (MACF1) in the Wnt signaling pathway. Genes Dev. 20: 1933–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78: 273–304 [DOI] [PubMed] [Google Scholar]
- 11. Clevers H. 2006. Wnt/beta-catenin signaling in development and disease. Cell 127: 469–480 [DOI] [PubMed] [Google Scholar]
- 12. Corona DF, et al. 2007. ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol. 5: e232 doi: 10.1371/journal.pbio.0050232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Corona DF, Tamkun JW. 2004. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim. Biophys. Acta 1677: 113–119 [DOI] [PubMed] [Google Scholar]
- 14. Deuring R, et al. 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5: 355–365 [DOI] [PubMed] [Google Scholar]
- 15. Flaus A, Owen-Hughes T. 2011. Mechanisms for ATP-dependent chromatin remodelling: the means to the end. FEBS J. 278: 3579–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fujimura L, Watanabe-Takano H, Sato Y, Tokuhisa T, Hatano M. 2009. Prickle promotes neurite outgrowth via the Dishevelled dependent pathway in C1300 cells. Neurosci. Lett. 467: 6–10 [DOI] [PubMed] [Google Scholar]
- 17. Gigek CO, et al. 2011. SMARCA5 methylation and expression in gastric cancer. Cancer Invest. 29: 162–166 [DOI] [PubMed] [Google Scholar]
- 18. Griffin CT, Curtis CD, Davis RB, Muthukumar V, Magnuson T. 2011. The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proc. Natl. Acad. Sci. U. S. A. 108: 2282–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hargreaves DC, Crabtree GR. 2011. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 21: 396–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hart M, et al. 1999. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr. Biol. 9: 207–210 [DOI] [PubMed] [Google Scholar]
- 21. Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 19: 1839–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huber O, Krohn M, Kemler R. 1997. A specific domain in alpha-catenin mediates binding to beta-catenin or plakoglobin. J. Cell Sci. 110: 1759–1765 [DOI] [PubMed] [Google Scholar]
- 23. Klaus A, Birchmeier W. 2008. Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer 8: 387–398 [DOI] [PubMed] [Google Scholar]
- 24. Lazzaro MA, Picketts DJ. 2001. Cloning and characterization of the murine Imitation Switch (ISWI) genes: differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J. Neurochem. 77: 1145–1156 [DOI] [PubMed] [Google Scholar]
- 25. Lazzaro MA, et al. 2008. Characterization of novel isoforms and evaluation of SNF2L/SMARCA1 as a candidate gene for X-linked mental retardation in 12 families linked to Xq25-26. BMC Med. Genet. 9: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. LeRoy G, Loyola A, Lane WS, Reinberg D. 2000. Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem. 275: 14787–14790 [DOI] [PubMed] [Google Scholar]
- 27. LeRoy G, Orphanides G, Lane WS, Reinberg D. 1998. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science 282: 1900–1904 [DOI] [PubMed] [Google Scholar]
- 28. Liu C, et al. 2002. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108: 837–847 [DOI] [PubMed] [Google Scholar]
- 29. Liu YI, et al. 2008. The chromatin remodelers ISWI and ACF1 directly repress Wingless transcriptional targets. Dev. Biol. 323: 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Logan CY, Nusse R. 2004. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20: 781–810 [DOI] [PubMed] [Google Scholar]
- 31. Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. 1995. Regulation of intracellular beta-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc. Natl. Acad. Sci. U. S. A. 92: 3046–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'Connell MP, Weeraratna AT. 2009. Hear the Wnt Ror: how melanoma cells adjust to changes in Wnt. Pigment Cell Melanoma Res. 22: 724–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. 2000. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature 407: 535–538 [DOI] [PubMed] [Google Scholar]
- 34. Poot RA, et al. 2000. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 19: 3377–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Railo A, et al. 2009. Genomic response to Wnt signalling is highly context-dependent—evidence from DNA microarray and chromatin immunoprecipitation screens of Wnt/TCF targets. Exp. Cell Res. 315: 2690–2704 [DOI] [PubMed] [Google Scholar]
- 36. Roose J, et al. 1999. Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science 285: 1923–1926 [DOI] [PubMed] [Google Scholar]
- 37. Roose J, et al. 1998. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395: 608–612 [DOI] [PubMed] [Google Scholar]
- 38. Sala A, et al. 2011. Genome-wide characterization of chromatin binding and nucleosome spacing activity of the nucleosome remodelling ATPase ISWI. EMBO J. 30: 1766–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sanchez-Molina S, et al. 2011. Role for hACF1 in the G2/M damage checkpoint. Nucleic Acids Res. 39: 8445–8456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simon R, Mirlacher M, Sauter G. 2003. Tissue microarrays in cancer diagnosis. Expert Rev. Mol. Diagn. 3: 421–430 [DOI] [PubMed] [Google Scholar]
- 41. Song H, Spichiger-Haeusermann C, Basler K. 2009. The ISWI-containing NURF complex regulates the output of the canonical Wingless pathway. EMBO Rep. 10: 1140–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spangler B, Vardimon L, Bosserhoff AK, Kuphal S. 2011. Post-transcriptional regulation controlled by E-cadherin is important for c-Jun activity in melanoma. Pigment Cell Melanoma Res. 24: 148–164 [DOI] [PubMed] [Google Scholar]
- 43. Stopka T, Skoultchi AI. 2003. The ISWI ATPase Snf2h is required for early mouse development. Proc. Natl. Acad. Sci. U. S. A. 100: 14097–14102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stopka T, et al. 2000. Chromatin remodeling gene SMARCA5 is dysregulated in primitive hematopoietic cells of acute leukemia. Leukemia 14: 1247–1252 [DOI] [PubMed] [Google Scholar]
- 45. Strohner R, et al. 2001. NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 20: 4892–4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sumegi J, et al. 2011. A novel t(4;22)(q31;q12) produces an EWSR1-SMARCA5 fusion in extraskeletal Ewing sarcoma/primitive neuroectodermal tumor. Mod. Pathol. 24: 333–342 [DOI] [PubMed] [Google Scholar]
- 47. Tanaka K, Okabayashi K, Asashima M, Perrimon N, Kadowaki T. 2000. The evolutionarily conserved porcupine gene family is involved in the processing of the Wnt family. Eur. J. Biochem. 267: 4300–4311 [DOI] [PubMed] [Google Scholar]
- 48. Thompson BA, Tremblay V, Lin G, Bochar DA. 2008. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol. Cell. Biol. 28: 3894–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Upadhyay G, et al. 2008. Molecular association between beta-catenin degradation complex and Rac guanine exchange factor DOCK4 is essential for Wnt/beta-catenin signaling. Oncogene 27: 5845–5855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van de Wetering M, et al. 1997. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88: 789–799 [DOI] [PubMed] [Google Scholar]
- 51. Willert K, Jones KA. 2006. Wnt signaling: is the party in the nucleus? Genes Dev. 20: 1394–1404 [DOI] [PubMed] [Google Scholar]
- 52. Yadon AN, Tsukiyama T. 2011. SnapShot: chromatin remodeling: ISWI. Cell 144: 453–453.e451 doi: 10.1016/j.cell.2011.01.019 [DOI] [PubMed] [Google Scholar]
- 53. Ye Y, et al. 2009. Inhibition of expression of the chromatin remodeling gene, SNF2L, selectively leads to DNA damage, growth inhibition, and cancer cell death. Mol. Cancer Res. 7: 1984–1999 [DOI] [PubMed] [Google Scholar]
- 54. Yip DJ, Coulombe MJJ, Rudnicki M, Picketts DJ. 2006. SNF2L-mediated control of cell number in the developing brain. 16th Biennial meeting of the International Society for Developmental Neuroscience. Int. J. Dev. Neurosci. 24: 586 [Google Scholar]
- 55. Zhou Y, Santoro R, Grummt I. 2002. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J 21: 4632–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.