Abstract
The eyegone (eyg) gene encodes Eyg, a transcription factor of the Pax family with multiple roles during Drosophila development. Although Eyg has been shown to act as a repressor, nothing is known about the mechanism by which it represses its target genes. Here, we show that Eyg forms a protein complex with heterochromatin protein 1a (HP1a). Both proteins bind to the same chromatin regions on polytene chromosomes and act cooperatively to suppress variegation and mediate gene silencing. In addition, Eyg binds to a wingless (wg) enhancer region, recruiting HP1a to assemble a closed, heterochromatin-like conformation that represses transcription of the wg gene. We describe here the evidence that suggests that Eyg, encoded by eyegone (eyg), represses wingless (wg) during eye development by association with HP1a. We show that Eyg forms a protein complex with HP1a and both proteins colocalize on salivary gland polytene chromosomes. Using position effect variegation (PEV) experiments, we demonstrated that eyg has a dose-dependent effect on heterochromatin gene silencing and identified a genetic interaction with HP1a in this process. We further demonstrated that HP1a binds to the same wg enhancer element as Eyg. DNase I sensitivity assays indicated that this enhancer region has a closed heterochromatin-like conformation, which becomes open in eyg mutants. In these mutants, much less HP1a binds to the wg enhancer region, as shown by ChIP experiments. Furthermore, as previously described for Eyg, a reduction in the amount of HP1a in the eye imaginal disc derepresses wg. Together, our results suggest a model in which Eyg specifically binds to the wg enhancer region, recruiting HP1a to that site. The recruitment of HP1a prevents transcription by favoring a closed, heterochromatin-like structure. Thus, for the first time, we show that HP1a plays a direct role in the repression of a developmentally regulated gene, wg, during Drosophila eye development.
INTRODUCTION
One mechanism used by organisms from yeast to mammals to repress gene expression is gene silencing. Silencing inactivates chromosome domains that contain key regulatory genes by packaging them into a specialized chromatin structure that is inaccessible to DNA-binding proteins (14, 32). Several Drosophila genes are required for silencing, including the Su(var)205 gene, which encodes heterochromatin protein 1a (HP1a) (19). HP1a has been shown to bind to methylated histone H3 lysine 9 (H3K9) and to recruit H3K9 methyltransferase to chromatin (1, 24, 41). This sequential process is thought to mediate the spreading of H3K9 methylation and the formation of heterochromatin. HP1-like proteins, in addition to HP1a, have been described in Drosophila, but whereas HP1b and HP1c are ubiquitously expressed, HP1d (Rhino) and HP1e are expressed only in the germ line (45).
eyegone (eyg) encodes a homeodomain Pax protein (22). Pax proteins, which are present throughout the animal kingdom, are transcription factors that bind DNA through their paired domains (44). In vertebrates, nine Pax genes that fulfill different roles in development have been described to date. Mutations in these genes lead to several diseases, including a variety of cancers (for a review, see references 4 and 31). In Drosophila, eyg is part of the genetic network activated during eye development (16, 17); it also is involved in the development of the salivary gland ducts (21) and plays a role in the genetic subdivision of the thorax (3). Although the functions of Pax proteins during Drosophila development have been studied extensively, very little is known about their molecular modes of action. Nevertheless, in all cases studied so far, they have been shown to act as transcriptional repressors (47).
The most extensively studied aspect of eyg is its role in eye development. Proliferative growth in the eye-antennal disc is continuous from the late first-instar to the late second-/early third-instar stages. A key proliferative signal is provided by the Notch pathway, which is active only along the dorsoventral compartment boundary of the eye disc. It has been shown that the action of Notch in stimulating eye growth is mediated by Eyg (10). Thus, eyg is expressed from the second instar in a wedge within the eye primordium straddling the dorsal-ventral boundary; this expression is under the control of Notch and is a downstream requirement for eye growth (10). wingless (wg) is also expressed at these stages at the anterior-dorsal and anterior-ventral disc margins. Thus, Eyg represses wg expression, thereby promoting eye growth and preventing head capsule formation (16, 20). As a result, viable eyg alleles develop without eyes and with expanded head tissue (18).
Here, we describe the repression mechanism of Eyg. We show that Eyg acts as a suppressor of variegation, suggesting a role for Eyg in heterochromatin-mediated gene silencing. We show that Eyg colocalizes with HP1a to heterochromatin regions on polytene chromosomes. Eyg and HP1a also colocalize to some euchromatin regions, including the wg genomic region. We demonstrate that Eyg and HP1a bind to the wg enhancer, where they silence wg activity by generating a closed, transcriptionally inactive chromatin structure. Our data thus support a role for Eyg in repressing its targets through a heterochromatin gene-silencing mechanism.
MATERIALS AND METHODS
Eye pigmentation measurement.
The heads of 25 female flies (raised at 25°C) of each genotype were homogenized in methanol (1 ml, acidified with 0.1% HCl). Eye pigmentation was expressed as the absorbance of the supernatant at 480 nm.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were carried out with DNA obtained from 200 yw eye-antennal discs or 0.6 g of yw Drosophila melanogaster embryos and 200 eyg20MD1/+ heterozygous mutant eye-antennal discs, as previously described (29), with some minor modifications. Homogenized discs were sonicated five times (10-s continuous pulses; power amplitude, 7 μm) in an MSE Soniprep 150 Sonifier with a microtip probe at 4°C, with 30 s cooling on ice between pulses, yielding predominantly 200- to 700-bp DNA fragments. For immunoprecipitation, the following antibodies were used: a 1:20 dilution of polyclonal guinea pig anti-Eyg antibody (3) and its respective preimmune serum, a 1:25 dilution of monoclonal mouse anti-HP1a (Developmental Hybridoma Bank), and anti-2meH3K9 (Cell Signaling) at a 1:25 dilution. The immunoprecipitated DNA was used for PCR amplification of 320 bp of the 1360 satellite sequence using the primers 1360fw (5′-TGT ATC GTT TTT AAA AAA TTG TC-3′) and 1360rv (5′-GTG GAC CTG TAA TAT ATG CTC T-3′). A 410-bp wg enhancer region was amplified using the primers wg1fw (5′-GCG TGT AGT TCG AGG CCT AAG C-3′) and wg1rv (5′-GCT TGA CGG CCA AAC GGG GCT TG-3′). A 210-bp wg promoter region was amplified using the following primers: wgpromfw (5′-GCG GAA TTA ATC GCA CAA AT-3′) and wgpromrv (5′-TTT ATC TGT TCG ACG GCA CA-3).
The control labial region was amplified using the primers labfw (5′-GGC GGG AAG TGC CCC ATC CCA AC-3′) and labrv (5′-CGC GTC AAG TAG CGA TTG AAG TGG-3′).
Quantitative PCR (qPCR) was performed using Roche Light Cycler equipment and accessories, as described previously (5). To ensure that only a single product was amplified, we evaluated PCR products by agarose gel electrophoresis and performed dissociation curve analyses. For each ChIP assay, the reference sample, termed Mock, corresponds to a ChIP assay performed at the same time without the addition of any specific antibody; negative controls used preimmune serum. Data are presented as the amount of DNA enrichment normalized to the input (100% value) diluted 1:100. All ChIP experiments were performed as three independent events, and the results are presented as the average of each experiment ± the standard deviation.
DNase I sensitivity assay.
DNase I sensitivity assays were performed as previously described (50) with minor modifications. Samples (2 ml each) were analyzed by qPCR using Roche Light Cycler equipment. The cycle threshold (CT) value for each locus was obtained using Roche Molecular Biochemical-Light Cycler Software (version 3.5). The euchromatin actin (act5c) locus was amplified using the primers Ac5Cfw (5′-CAC GGT ATC GTG ACC AAC TG-3′) and Ac5Crv (5′-GCC ATC TCC TGC TCA AAG TC-3′), and the heterochromatin locus was amplified using the primers H23fw (5′-CCA AGT TGG CCA GTT TTG AT-3′) and H23rv (5′-AGT TCA AGC CCG GGT ATT CT-3′). The wg enhancer region was amplified using the primers described above.
Fly strains and genetic crosses.
All crosses were carried out at 25°C on standard cornmeal-agar medium, unless otherwise specified. The yw Drosophila strain was used as the wild-type (WT) strain in this study. For position-effect variegation (PEV) analyses, P[white+] (from M. Calleja), DX1 (from L. Wallrath), Su(var)205 and His2AV810 (from Bloomington Stock Center), and T(2;3)Sb[V] (from J. A. Birchler) strains were used. The eyg mutant lines used were eygSA2 and eyg20MD1 (3, 10). The following transgenes and Gal4 lines were used: HP1aRNAi (from Vienna Drosophila RNAi Center), UAS-shmiR-dppmutant (15), 248GAL4 (37), ywhsFLP;act>y+>Gal4UAS-GFP (from Bloomington), and P{PZ}wg[r0727] (wglacZ). UAS transgenes were expressed in random clones by heat shocking the larvae for 10 min at 34°C and dissecting them at the third-instar larval stage. The P{PZ}wg[r0727] line was used for fluorescence in situ hybridization (FISH) experiments on polytene chromosomes.
Antibodies and immunostaining.
Polytene chromosomes were prepared according to standard procedures (40). The antibodies used were mouse monoclonal anti-HP1a (C1A9; 1:75; Developmental Hybridoma Bank) and guinea pig polyclonal anti-Eyg (1:200). Topro3 was used to counterstain nuclei. Combined FISH and immunostaining of polytene chromosomes was performed as described previously (25) using the PZ vector as a biotin-labeled probe. For staining of eye-antennal imaginal discs, third-instar larvae were fixed in 4% paraformaldehyde for 25 min, washed, and blocked in BBT (phosphate-buffered saline [25 mM NaCl] containing 0.1% Triton X-100 and 1% bovine serum albumen). The primary antibodies used were mouse monoclonal anti-Wg (CD4; 1:50; Developmental Hybridoma Bank) and chicken anti-β-galactosidase (1:200); incubations with primary antibodies were performed overnight in BBT at 4°C. After 1-h washes in BBT, the appropriate fluorescent secondary antibody was added and incubated for 1 h at room temperature. After extensive washes in BBT, the imaginal discs were mounted in Vectashield (Vector Laboratories). Images were obtained with a SPE Leica Confocal microscope and subsequently processed using Adobe Photoshop.
Western blotting and immunoprecipitation experiments.
For coimmunoprecipitation experiments, salivary gland extracts (1 mg protein) from yw larvae were homogenized in lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 1 mM EDTA) containing protease inhibitors (Roche). The cell pellet was discarded, and the protein extract was incubated overnight with 6 mg protein A-Sepharose (Sigma). Precleared lysates were incubated with 6 mg protein A-Sepharose for 2 h, and antibody (1:500 guinea pig anti-Eyg, 1:500 guinea pig preimmune serum, 1:75 mouse anti-HP1a, 1:1,000 mouse anti-α-tubulin, or 1:2,000 mouse antihemagglutinin (anti-HA) (HA.11 from Covance) was added and incubated overnight at 4°C. After two washes in lysis buffer, proteins in immunoprecipitated extracts were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a nitrocellulose membrane, and incubated with guinea pig anti-Eyg antibody (1:750) and mouse anti-HP1a antibody (1:500). Immunoreactive proteins were detected with the appropriate horseradish peroxidase-conjugated secondary antibodies, and the signal was developed using the ECL Western Blotting Analysis System (Amersham Pharmacia).
Generation of eygPSSLN and stable S2 cells.
eygPSSLN was generated by PCR using the primers PSNfw (5′-GGT CCG CCA CCT TCG TCG CTG AAT CAC CCA CTG CAT GGT-3′) and PSNrv (5′-ACC ATG CAG TGG GTG ATT CAG CGA CGA AGG TGG CGG ACC-3′). The template vector tubulin>Flag-HA-dAgo1 was kindly provided by S. Cohen (11a). The dAgo1 coding sequence was excised using NotI/XhoI and replaced with eygPSVLL and eygPSSLN. Transgenic Schneider S2 cells were established as described by Easow et al. (11a) with slight modifications. Briefly, an empty pRmHA vector containing a puromycin resistance gene was cotransfected with Flag-HA-eygPSVLL or -eygPSSLN, and puromycin was applied 24 h after transfection. Puromycin-resistant cells were grown to confluence and used for coimmunoprecipitation experiments as described above.
RESULTS
Eyg is a suppressor of variegation.
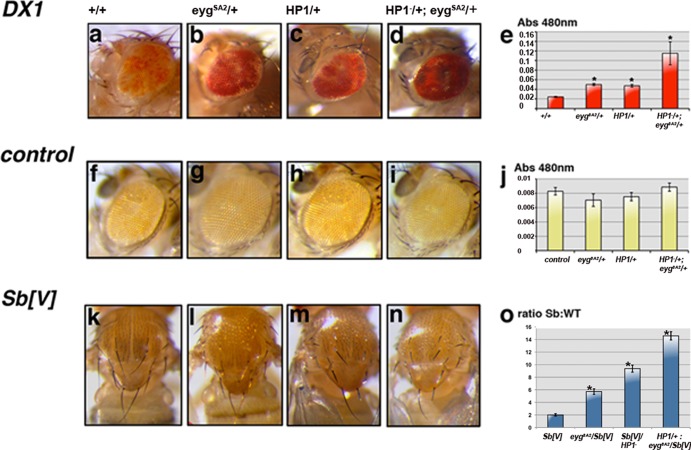
Eyg, a Pax protein encoded by the eyg gene, has been reported to act as a transcriptional repressor (46–48). We therefore assessed whether Eyg might repress its targets through heterochromatic gene silencing. The effects of reducing the eyg dosage on heterochromatic gene silencing were examined by PEV using two independent variegated lines: DX1 and T(2;3)Sb[V]. DX1 contains seven tandem copies of a P[white+] reporter transgene inserted in a euchromatic region that induces heterochromatin formation at the insertion locus (Fig. 1a) (11). Reducing the eyg dosage using a null allele (eygSA2) (see Materials and Methods) strongly suppressed variegation (Fig. 1b), causing a marked derepression of the white gene and a substantial increase in eye pigmentation.
Fig 1.
Eyg is required for heterochromatic gene silencing. Effects of mutations in eyg and Su(var)205 on the white+ variegation induced by DX1 transgene repeats (a to e) or a control transgene (f to j), are shown as changes in red eye pigmentation. Altering the dosage of eyg (b) or HP1a (c) suppresses variegation in DX1 flies. Note the increased red eye pigmentation in DX1 flies (d) heterozygous for both eyg and Su(var)205 genes. (e and j) Changes in pigmentation were determined by measuring absorbance at a wavelength of 480 nm. The data are expressed as means ± standard errors of the mean (SEM) (*, P < 0.001 compared with +/+ control; Student's t test; n = 60 per group). control is a single P[white+] element inserted in a euchromatic region that does not induce heterochromatin formation. (k to o) Variegation of Sb in a T(2;3)Sb[V] rearranged chromosome (7) is suppressed by partial depletion of Eyg and/or HP1a. (o) Suppression of Sb variegation is shown as changes in the Sb-to-WT macrochete ratio. The data are expressed as means ± SEM (*, P < 0.005 compared with an Sb[V] control; Student's t test; n = 40 per group).
One of the best-studied proteins involved in heterochromatic gene silencing is HP1, encoded by Su(var)205 in Drosophila (8, 23, 26). HP1 also acts as a dosage-dependent modifier of PEV; therefore, we analyzed its relationship with eyg. The suppression of variegation observed in heterozygous HP1−/+ flies (Fig. 1c) was enhanced when combined with eygSA2/+ heterozygous flies in the DX1 variegated line (Fig. 1d). Derepression of the white gene was quantified by measuring red pigment absorbance levels at 480 nm (Fig. 1e) (35a). We found a statistically significant difference under each mutant condition compared to the control situation. Heterozygosity of eyg did not affect the expression of a control P(white+) transgene (compare Fig. 1f with g). The same result was obtained in HP1−/+ (Fig. 1h) and HP1−/+eygSA2/+ flies (Fig. 1i), and quantification of eye pigment levels showed no statistically significant difference from the control situation (Fig. 1j).
We also examined the influence of halving the eyg dosage on the T(2;3)Sb[V] variegated line. T(2;3)Sb[V] is caused by a chromosomal translocation that juxtaposes the Sb gene to a heterochromatin region (7). Silencing of Sb caused a variegated increase in bristle length toward the wild-type phenotype (Fig. 1k). Flies heterozygous for eygSA2/+ developed more Sb bristles in the notum than did control flies (compare Fig. 1k with l). HP1−/+ heterozygous flies also suppressed variegation of Sb (Fig. 1m), and again, double-mutant HP1−/+ eygSA2/+ flies displayed enhanced suppression of variegation (Fig. 1n). The ratio between Sb and wild-type bristles was calculated for all phenotypes and was shown to be statistically significant (Fig. 1o) for each heterozygous mutant condition. Similar results were obtained with the eyg20MD1/+ allele (not shown).
We did not observe any genetic interaction of eyg with His2Av, a gene known to be essential for heterochromatin assembly in Drosophila (reference 43 and data not shown), suggesting that Eyg acts specifically with HP1a and not in general with heterochromatin proteins. Taken together, these results indicate that Eyg is a suppressor of variegation that plays a role in heterochromatin-mediated gene silencing and suggest that Eyg represses gene expression through heterochromatic gene silencing. We also found that eyg and HP1 variegating phenotypes were additive, suggesting possible cooperation in the gene-silencing process.
Eyg colocalizes and coimmunoprecipitates with HP1a.
We next examined the localization of Eyg and HP1a in wild-type salivary gland polytene chromosomes. Eyg was located at the chromocenter (Fig. 2c, arrow, and c′), where the HP1a protein was also detected (Fig. 2b, arrow, and b′), and was also found in other heterochromatin regions, such as that on chromosome 4 (not shown). We also detected colocalization of Eyg with HP1a in some bands along the chromosomal arms (Fig. 2e and f, arrows) that could be targets of Eyg. In this context, we found that HP1a localizes to the wg genomic region, a known target of Eyg during Drosophila eye development (16, 20) (Fig. 2g to i).
Fig 2.
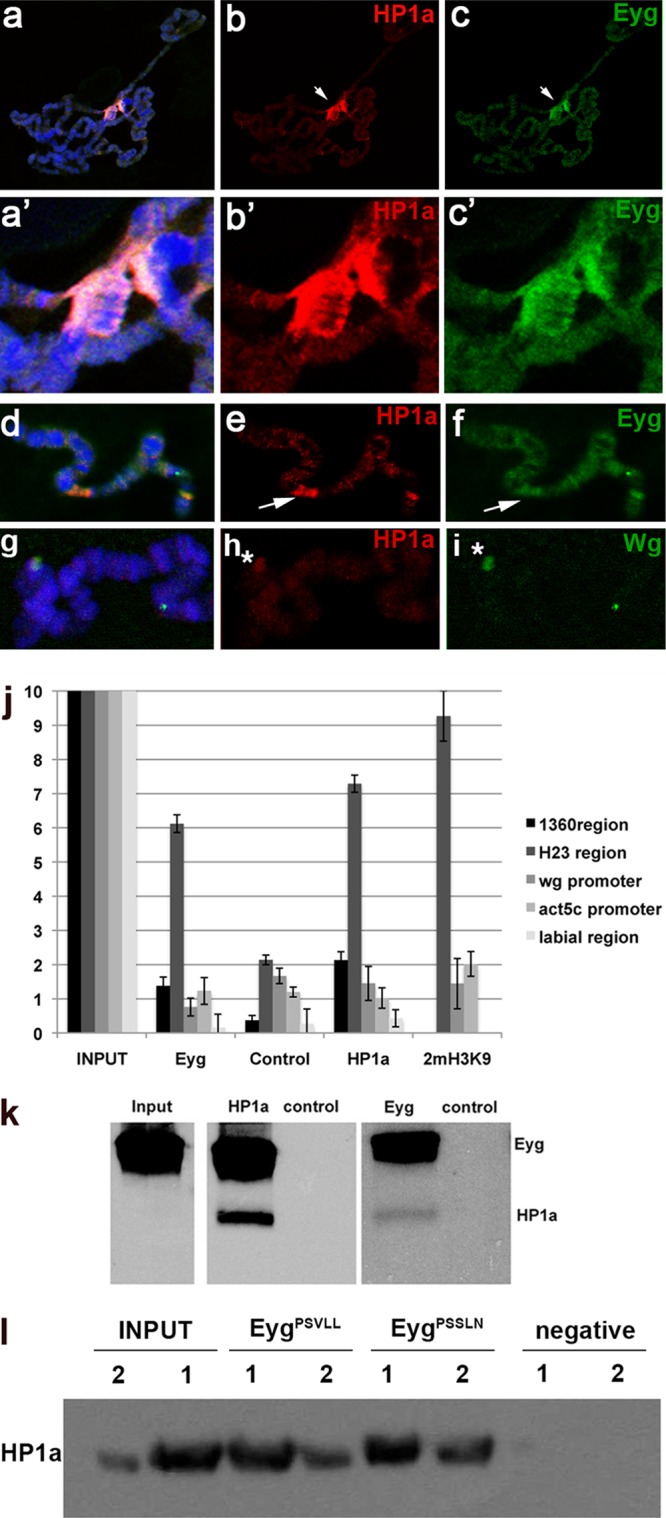
Eyg and HP1a colocalize to heterochromatic regions. (a to c) Eyg and HP1a colocalize to the heterochromatic regions of salivary gland polytene chromosomes (arrows in panels b and c). (a′ to c′) Enlargement of the chromocenter of the chromosome. (d to f) Eyg and HP1a also colocalize in some interband regions of salivary gland polytene chromosomes (arrows in panels e and f). (g to i) HP1a localizes to the wg genomic region, marked by a fluorescent DNA probe (asterisks in panels h and i). (j) Enrichment of constitutive heterochromatic regions 1360 and H23 when immunoprecipitated with anti-Eyg, expressed as the percentage of input (1:100 dilution) in ChIP experiments. HP1a binding to the same heterochromatic regions was also observed. Control refers to chromatin immunoprecipitated with guinea pig preimmune serum. An isotype control for mouse antibody gave similar results (not shown). The same immunoprecipitated DNA was amplified with primers for the labial genomic region, the wg promoter, and the act5c promoter region, which were used as negative controls. (k) Eyg and HP1a belong to the same protein complex. Western blot analyses detected HP1a protein in salivary gland extracts immunoprecipitated with anti-Eyg antibody and Eyg protein in salivary gland extracts immunoprecipitated with anti-HP1a antibody. In the control lanes, salivary gland extracts were immunoprecipitated with guinea pig preimmune serum or with a monoclonal anti-α-tubulin antibody (control). (l) An EygPSSLN mutant for the PSVLL motif is able to bind HP1a in S2 cells, as does wild-type Eyg (EygPSVLL). negative, no antibody. 1 and 2 refer to two independent coimmunoprecipitation experiments.
To further confirm that Eyg is associated with heterochromatin, we performed ChIP assays using the transposable 1360 element as a heterochromatin marker (42). We used this transposable element because the terminal inverted-repeat 1360 element initiates heterochromatic domains (9, 42). Enrichment of HP1 binding to 1360 sequences was described previously (9), and we found similar enrichment of Eyg binding to these 1360 elements (expressed as a percentage of the input) (Fig. 2j), suggesting that both Eyg and HP1a bind to the 1360 sequence.
A similar degree of enrichment was found in the heterochromatic H23 locus (region 22000 to 24000 of chromosome 2 [chr2] heterochromatin) (Fig. 2j) (41). We used the promoter region of labial as a negative control, because neither Eyg nor HP1a binds to it (Fig. 2j).
We also found that Eyg and HP1a coimmunoprecipitated from salivary gland extracts (Fig. 2k), indicating that Eyg and HP1a are parts of the same protein complex. The consensus pentapeptide sufficient for interaction with HP1 has been identified as (PL)(WRY)V(MIV)(MLV) (39). Eyg does not contain an exact consensus motif but does contain a PSVLL domain in the C-terminal part of the protein (residues 472 to 476). A previous characterization of Eyg functional domains showed that the C-terminal region is not necessary for the repressive function of Eyg (47). In order to test if the PSVLL domain of Eyg was necessary for binding to HP1a, we generated an Eyg mutant in which PSVLL was replaced by PSSLN (as described in reference 28) and assayed its binding to HP1a in stably transfected S2 cells. High endogenous levels of HP1a and undetectable expression of Eyg in S2 were confirmed using the Drosophila Schneider S2 cell microarray data set generated and described previously (35). We found that both EygPSSLN and EygPSVLL coimmunoprecipitated with HP1a, indicating that the PSVLL of Eyg is dispensable for the binding of HP1a (Fig. 2l).
HP1a mediates wg repression in the eye imaginal disc through heterochromatic gene silencing.
Because Eyg is a known transcriptional repressor, we speculated that its repressor activity might be executed by recruiting HP1a to chromatin, triggering gene silencing. The wg gene is a known target of Eyg during eye development (16, 20), and the minimal wg enhancer region that drives wg expression in the eye imaginal disc has been defined (33) (Fig. 3a, wg 2.10). To determine whether wg repression by Eyg was mediated by gene silencing and generated changes in its chromatin structure, we performed DNase I sensitivity assays to analyze the chromatin structure of this wg enhancer region in eye imaginal discs. Quantitative PCR was performed on native (undigested; 0 U) chromatin samples and on samples digested with DNase I (50 U). The CT value was calculated as a function of the amount of DNA at the outset (unless otherwise stated, ΔCT values refer to CT [50 U] − CT [0 U]).
Fig 3.
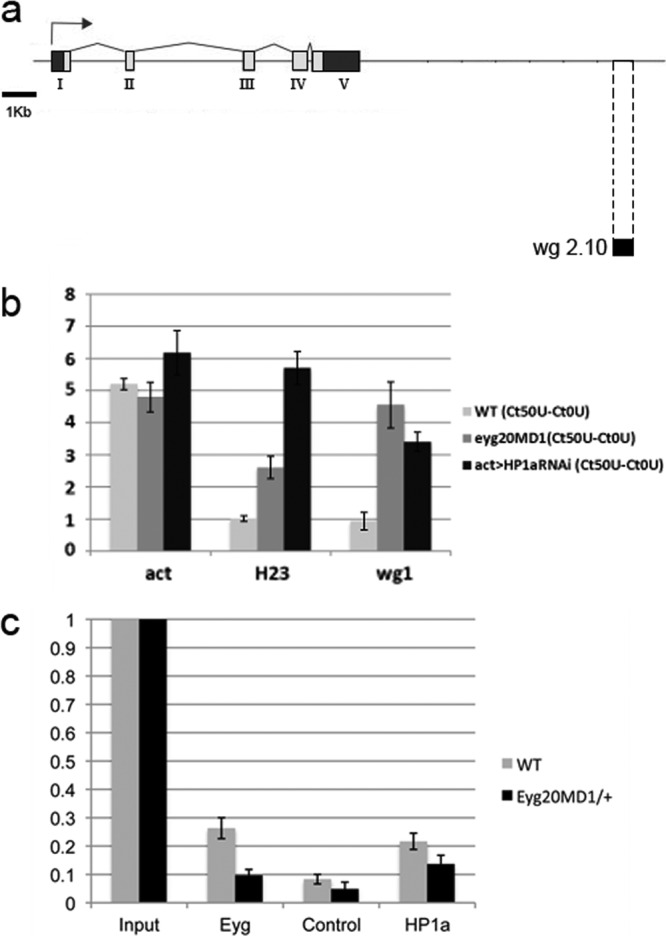
Eyg recruits HP1a and mediates the assembly of a heterochromatic-like structure in the wg enhancer region. (a) Schematic view of the wg genomic region. The wg enhancer region that drives Wg expression in the eye imaginal disc is located at the 3′ end of the genomic region (wg 2.10, black bar). (b) Formation of a DNase I-resistant structure in the wg enhancer region. The act5c and H23 loci were used as controls for open and closed genomic regions, respectively. The bars represent changes in CT values (ΔCT) in the wg enhancer region of wild-type, eyg20MD1/+, and HP1a mutant act>HP1aRNAi eye-antennal disc extracts after treatment with 50 U DNase I. In eyg20MD1/+ heterozygous discs, as well as in act>HP1aRNAi, the wg enhancer region became almost as sensitive to DNase I as the euchromatic actin locus. The H23 locus remained mostly closed for wild-type control and eyg20MD1/+ and open and DNase I sensitive for HP1 mutant extracts. (c) Analysis of HP1a and Eyg binding to the wg enhancer region using ChIP. A similar enrichment in binding to the wg enhancer region was found in wild-type control chromatin immunoprecipitated with an anti-Eyg or an anti-HP1a antibody. Binding of Eyg and, to some extent, HP1a was reduced in the chromatin of eyg20MD1/+ eye-antennal disc extracts.
For the transcribed region of actin (act5c), a constitutively active gene used as a positive control for the “open” chromatin state, the ΔCT value was around 5 in wild-type, eyg20MD1/+ heterozygous and HP1a mutant (act>HP1aRNAi) eye-antennal disc extracts (Fig. 3b). We used the Gal4-UAS binary system to eliminate HP1 via RNA interference (RNAi), because HP1 mutations are recessive lethal (see below). In contrast, the heterochromatic H23 region (22,000 to 24,000 of chromosome 2 heterochromatin) (50), used as a negative control for the “closed” chromatin state, was refractory to DNase I treatment, exhibiting a lower ΔCT value in both wild-type and eyg20MD1/+ heterozygous eye-antennal disc extracts (Fig. 3b). In act>HP1aRNAi extracts, the H23 region became sensitive to DNase I, supporting a role for HP1a in constitutive heterochromatin formation/maintenance.
We found that in wild-type control eye-antennal imaginal discs, the wg enhancer region behaved like heterochromatin, with a ΔCT between 0 and 2 (Fig. 3b). However, the same genomic region was almost as sensitive to DNase I as the act5c locus region in eyg20MD1/+ and act>HP1aRNAi mutant extracts, with a ΔCT value between 3.5 and 5 (Fig. 3b). Halving the dosage of eyg was sufficient to change the sensitivity to digestion by DNase I of the wg enhancer region. Consistent with this, in some heterozygous eyg20MD1/+ eye discs, the change in chromatin structure led to the derepression of wg described previously (16, 47) and gave rise to a small-eye phenotype (not shown). This result indicates the high sensitivity achieved with the technique and also suggests that there might be other mechanisms regulating wg expression.
Using ChIP experiments, we confirmed that Eyg and HP1a bind similarly to this wg region (Fig. 3c), but not to the wg promoter region or to the act5c promoter used as controls (Fig. 2j). The amount of HP1a protein bound to the wg enhancer region in eyg20MD1/+ was reduced (Fig. 3c), but to a lesser extent than the Eyg protein. This result suggests that not all HP1a binding to this enhancer region depends on Eyg. We believe that Eyg may recruit HP1a to the wg enhancer region to form a heterochromatin structure that most likely represses wg expression in the eye. To further analyze this hypothesis, we tried to determine the localization of HP1a in the polytene chromosomes of eyg mutant larvae. However, eyg mutant salivary glands are very small and contain fragile chromosomes, which we were unable to squash.
Loss of HP1a leads to wg misexpression in the eye and wing disc.
To confirm that HP1a binding to the wg enhancer region in the eye is indeed functionally important, we expressed a specific interfering RNA against HP1a in random clones of cells in eye imaginal disc using the actin-Gal4 Drosophila line. The Gal4-UAS system combined with the flippase recombination target (FRT) system was used to generate the RNAi-expressing clones. In summary, flies of the genotype hsflp122;actFRTstopFRTGal4UASGFP/Cyo were crossed to flies containing a UASHP1RNAi construct. Second-instar larvae were then heat shocked for 10 min and dissected in the third-instar period to visualize the discs. Because of the FRT cassette, only clones of cells in which recombination has occurred (and that no longer have the cassette) will activate Gal4 and, consequently, express the interfering RNA that reduces or eliminates the HP1a product.
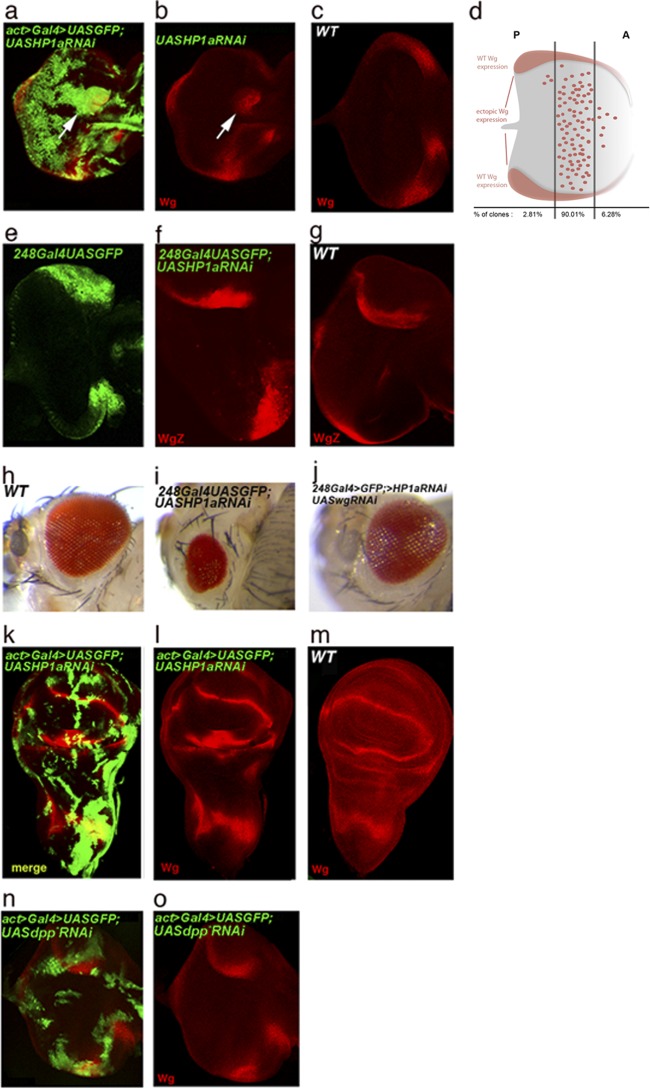
Reduction of HP1a levels resulted in ectopic activation of wg (Fig. 4a, b, and f), as previously described for eyg mutant eye discs (16).
Fig 4.
HP1a controls Wg expression in the eye imaginal disc and in the hinge region of the wing disc. (a and b) wg expression (red) in third-instar eye imaginal disc clones of cells with reduced HP1a levels induced using the act>Gal4 line (green). The arrow points to ectopic wg activation. (c) Expression of wg in a wild-type third-instar eye disc. (d) Schematic depiction of a third-instar eye imaginal disc showing the disc domains in which ectopic activation of wg was more frequent after reducing HP1a levels. Of the UAS-HP1aRNAi clones that showed ectopic wg activation, 90% of ectopic activation sites lay in the middle portion of the disc. (e) Expression domain of the 248Gal4 line in the eye disc (green). (f) wglacZ activity in an eye disc of the genotype 248Gal4<UAS-HP1aRNAi. (g) Expression of wglacZ in a wild-type eye imaginal disc. (h) Wild-type eye. (i) Eye phenotype after driving UAS-HP1aRNAi;UAS-GFP under the control of the 248Gal4 line. (j) Adult eye of a fly in which both HP1a and Wg levels were reduced in the same domain. Note that the eye size is rescued to almost wild-type levels (compare with panel i). (k to m) Generation of HP1aRNAi clones (green) in the wing disc. (l) Reducing HP1a levels in the cells of the wing hinge activated Wg expression (red). However, the distribution of Wg protein was not altered in the notum. (m) Distribution of Wg protein in a wild-type wing disc. (n and o) No ectopic wg expression (red) was observed in clones of cells (green fluorescent protein [GFP]-positive cells) expressing a mutated form of dpp, UAS-dpp*RNAi (15), commonly used as an RNAi control line.
Ectopic activation of wg in eye discs of HP1aRNAi clones did not occur in all parts of the disc domain and was more specific for the middle region. We found that 32% of HP1aRNAi clones (n = 278) were positive for ectopic wg activation, and more than 90% of these positive clones showed ectopic expression in the middle portion of the third-instar eye imaginal disc (Fig. 4d). These results suggest a positional restriction for ectopic wg activation in the eye disc, as has also been described for eyg mutants (16).
Interestingly, the HP1RNAi clones generated in the wing imaginal disc also exhibited derepression of wg in the corresponding hinge region, where eyg has been shown to regulate wg (20) (Fig. 4k and l), but not in the notum, where wg expression does not depend on eyg.
HP1a has been shown to control gene expression at the posttranscriptional level (34). To determine if the observed regulation of wg expression occurs at the transcriptional level, we expressed HP1a RNAi under the control of the 248Gal4 driver (which expresses Gal4 in a discrete domain of the eye disc) by crossing the UAS-HP1aRNAi line with the 248Gal4 line in a wgLacZ background. Reducing HP1a levels resulted in ectopic wgLacZ expression (Fig. 4f), suggesting that wg activation actually occurs at the transcriptional level.
To rule out the possibility that derepression of wg by HP1a RNAi is a more general effect of the RNAi machinery, we performed a series of experiments in which shmiR-dpp2HBmutant (15) was expressed in clones of cells in the eye imaginal disc. In contrast to the effects of expressing HP1aRNAi, wg was not derepressed by expressing shmiR-dpp2HBmutant (Fig. 4n and o), indicating that derepression of wg in the eye disc is not a general effect of the RNAi machinery on heterochromatin assembly, but rather a specific effect of reducing HP1a levels.
On the basis of these data, we conclude that HP1a is involved in wg repression in the eye imaginal disc and in the hinge region of the wing disc, where wg is a target of Eyg. We did not detect any alteration of wg expression in the notum region of the wing disc (Fig. 4l), where Eyg is also expressed and was shown to be functional (3) but does not control wg expression (20).
We also sought to analyze the phenotypic outcome in adult flies with reduced HP1a levels in the eye disc. To this end, we expressed UAS-HP1aRNAi together with UAS-GFP (see below) under the control of the above-mentioned 248Gal4 driver, which allows flies to develop to adulthood (Fig. 4e) (see Materials and Methods). Adult flies carrying this genetic combination showed a reduction in eye size (Fig. 4i). To further demonstrate that this was due, at least in part, to ectopic activation of wg in the eye field, we performed an experiment in which we coexpressed UAS-HP1aRNAi and UAS-wgRNAi under the control of the 248Gal4 driver. The flies were allowed to develop at 29°C until adulthood. The transgenes were balanced on different chromosomes, so that we were able to distinguish flies that carried either or both transgenes in the same cross. Flies of the genotype 248Gal4;UAS-wgRNAi;UAS-HP1aRNAi showed some head defects, but eye size was rescued to almost normal (compare Fig. 4i to j). This result strongly suggests that the reduction in eye size that we observed following reduction of HP1a levels depends on ectopic wg activation. Introducing a second UAS transgene (UAS-GFP) when expressing UAS-HP1aRNAi allowed us to eliminate the possibility of a Gal4 protein titration effect in the experiment.
DISCUSSION
The Pax protein Eyg, a product of the eyg gene, is a transcriptional repressor known to function during all developmental stages in Drosophila. One known target of Eyg is wg. It has been previously reported that Eyg represses wg in the eye and that this repression is a critical function of Eyg (16). In this work, we investigated the molecular mechanism underlying the repressive function of Eyg, examining repression of wg using the 3′ cis-regulatory region known to control wg expression in the Drosophila eye as an assay to study Eyg's mode of action. On the basis of our findings, we conclude that Eyg acts through heterochromatic gene silencing.
PEV experiments demonstrated that Eyg is a suppressor of variegation that is required for heterochromatic gene silencing and suggested that Eyg represses its target genes through this mechanism. We also demonstrated a genetic interaction between eyg and HP1a in the modification of PEV, suggesting that the products of these two genes act together in the gene-silencing process. Consistent with this interpretation, we found that Eyg colocalized with HP1a protein on polytene chromosomes in the chromocenter, on chromosome 4, in telomeres, and in some discrete bands along the chromosomes. The localization of Eyg on polytene chromosomes suggests that the two other ubiquitously expressed HP1-like proteins, HP1b and HP1c, are not Eyg partners because we did not detect Eyg protein on the many euchromatic bands where HP1b and HP1c localize on polytene chromosomes (13, 49a) (Fig. 2c).
Eyg contains a PSVLL motif at residues 472 to 476 located in the C-terminal domain of the protein. This is similar to the conserved pentapeptide motif present in the HP1 chromodomain and in other known HP1-interacting proteins (6, 38). However, when this domain was mutated in the Eyg protein, the interaction with HP1a was not affected, suggesting that the PSVLL motif in Eyg is dispensable for interaction with HP1a. This consensus sequence is absent in other well-known HP1 interactors, like INCENP (2), Su(var)3-9 (1), and the lamin B receptor (49; for a review, see reference 39). Eyg could therefore be another example of an HP1 interactor that engages HP1 differently from proteins that contain the consensus; alternatively, it could interact with HP1a indirectly. This result is consistent with a previous report by Yao and Sun, who showed that the C-terminal domain of Eyg is not essential for the repression of wg in the eye disc (47). The Eyg protein domain responsible for HP1a binding and the identity of the scaffold protein(s) that complexes Eyg and HP1a are still to be determined.
The results of ChIP experiments designed to determine if Eyg and HP1a bind to the same DNA sequences in vivo showed that both proteins bound to the same terminal inverted-repeat 1360 element, which initiates heterochromatic domains (9, 42); to the H23 locus, a region of heterochromatin of chromosome 2 (Fig. 2j); and to the wg enhancer region known to be responsible for trans-activation of the wg gene in the eye (and where binding sites for Pax-type paired-domain transcription factors have been previously reported [33]). However, both chromatin domains behaved differently in the absence of Eyg. Using DNase I sensitivity assays, we showed that, in eyg mutants, the 1360 element remained considerably more refractory to DNase I treatment and thus more closed (although not as closed as in the wild type) (data not shown). It should also be noted that, for all the “closed” genomic regions tested in our DNase I sensitivity assays, none remained as closed in eyg mutants after DNase I treatment as they did in the wild type; even the H23 locus was more open in the mutant background than in the wild type (Fig. 3b). This result points to a more general role for Eyg in heterochromatin formation and/or maintenance that warrants further analysis. In this context, Eyg function does not seem to depend on HP1a, because eliminating HP1a results in a ΔCT value of 5 to 6 in the DNase I sensitivity assay on the H23 locus (Fig. 3b).
The wg enhancer region behaves differently. In eyg mutants, it adopted an open chromatin state (with ΔCT values between 4 and 5), as it does in the absence of HP1a (Fig. 3b). In this region, Eyg and HP1a cooperate to silence gene expression. Binding of Eyg and HP1a is not a general feature of the wg locus, because we did not observe any binding to the wg promoter.
HP1a binding to the wg enhancer region was greatly reduced in eyg heterozygous mutant extracts, but to a lesser extent than it was the Eyg protein, suggesting that not all of the HP1a binding to that region depends on Eyg. However, the derepression of wg was evident in the eye imaginal disc of heterozygous eyg mutants, resulting in a small-eye phenotype in the adult fly (not shown). One possible explanation for this observation is that the HP1a that remains bound to the wg locus is insufficient to repress wg expression or that it has another, as yet undetermined role.
In yeast, it has been demonstrated that HP1 proteins form distinct complexes with different heterochromatin-related functions (27). In Drosophila, HP1-mediated silencing has been reported to operate in both a Su(Var)3-9-dependent and -independent manner (8). HP1 silencing of up to 1.9 kb has been shown to be Su(Var)3-9 independent, whereas long-range silencing depends upon the presence of Su(var)3-9. We propose that HP1-dependent wg silencing is Eyg dependent but Su(Var)3-9 independent, whereas for the heterochromatinization of other regions (e.g., H23 or the 1360 element), the presence of Eyg is not crucial, but H3K9 methylation is. Indeed, HP1a and 3mH3K9 colocalize to constitutive heterochromatin regions in salivary gland nuclei of wild-type larvae (12) and still do so in eyg mutant larvae (not shown). However, 3mH3K9 is never detected on the chromosomal bands where HP1a and Eyg colocalize (reference 12 and our observations).
HP1a has recently been shown to have a role in the posttranscriptional upregulation of many euchromatic genes (34) owing to the association of its chromodomain with different transcripts. This unexpected role of HP1a indicates that HP1a is a multifunctional protein whose versatility lies in its capacity to interact with different partners in distinct contexts. It has been proposed that the main function of HP1a is nucleic acid compaction, by interacting either with modified histones or with specific hnRNP proteins (34). Our results point to the same conclusion in that the interaction of HP1a with Eyg leads to compaction of the DNA at the wg enhancer region and subsequent silencing of the gene. In this way, the HP1a protein could fulfill at least two different functions: assembly of the constitutive heterochromatin present in telomeres and pericentromeres and on chromosome 4 by binding to methylated histone H3K9 and (once targeted to regulatory regions) specific euchromatic gene repression by interacting with transcription factors to form a repressive chromatin structure. In the latter case, the resulting chromatin structure has a heterochromatin appearance and exerts heterochromatin-like repression.
In conclusion, we describe a physical and functional interaction between Eyg and HP1a that regulates wg expression in the eye. Although HP1 has been shown to interact with other transcription factors, like those encoded by retinoblastoma (Rb) or Krab (30, 36), mediating the repression of their targets, a direct role of HP1a in this process has not previously been described. Our experiments show that HP1a is necessary for the repression of wg in the eye imaginal disc, endowing HP1a with a restricted function in development. In the same way that Eyg represses wg in the eye disc, other repressors could also interact with HP1a or other heterochromatin proteins to mediate spatial and/or temporal repression of their targets through heterochromatic gene silencing. This possibility opens new avenues that link specific transcription factors with general modifiers of chromatin.
ACKNOWLEDGMENTS
We thank Ginés Morata, Ernesto Sánchez-Herrero, Marco Milán, and Héctor Herranz for helpful discussions in the course of this work. We also thank all the Morata and Sánchez-Herrero laboratory members for their continuous support, as well as Elise Corsetti and Sara Mederos for help with the experiments.
This work was supported by grants from the Ministerio Español de Ciencia y Tecnología (BFU2009-12152), the Consolider program (CSD2007-00008), and the Fundación Ramón Areces.
Footnotes
Published ahead of print 30 April 2012
REFERENCES
- 1. Aagaard L, et al. 1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 18:1923–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ainsztein AM, Kandels-Lewis SE, Mackay AM, Earnshaw WC. 1998. INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J. Cell Biol. 143:1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aldaz S, Morata G, Azpiazu N. 2003. The Pax-homeobox gene eyegone is involved in the subdivision of the thorax of Drosophila. Development 130:4473–4482 [DOI] [PubMed] [Google Scholar]
- 4. Chi N, Epstein JA. 2002. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 18:41–47 [DOI] [PubMed] [Google Scholar]
- 5. Comet I, et al. 2006. PRE-mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev. Cell 11:117–124 [DOI] [PubMed] [Google Scholar]
- 6. Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ. 2000. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol. 10:517–525 [DOI] [PubMed] [Google Scholar]
- 7. Csink AK, Linsk R, Birchler JA. 1994. The Lighten up (Lip) gene of Drosophila melanogaster, a modifier of retroelement expression, position effect variegation and white locus insertion alleles. Genetics 138:153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Danzer JR, Wallrath LL. 2004. Mechanisms of HP1-mediated gene silencing in Drosophila. Development 131:3571–3580 [DOI] [PubMed] [Google Scholar]
- 9. De Lucia F, Ni JQ, Vaillant C, Sun FL. 2005. HP1 modulates the transcription of cell-cycle regulators in Drosophila melanogaster. Nucleic Acids Res. 33:2852–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dominguez M, Ferres-Marco D, Gutierrez-Avino FJ, Speicher SA, Beneyto M. 2004. Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat. Genet. 36:31–39 [DOI] [PubMed] [Google Scholar]
- 11. Dorer DR, Henikoff S. 1994. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 77:993–1002 [DOI] [PubMed] [Google Scholar]
- 11a. Easow G, Teleman AA, Cohen SM. 2007. Isolation of microRNA targets by miRNP immunopurification. RNA 13:1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ebert A, Lein S, Schotta G, Reuter G. 2006. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 14:377–392 [DOI] [PubMed] [Google Scholar]
- 13. Font-Burgada J, Rossell D, Auer H, Azorin F. 2008. Drosophila HP1c isoform interacts with the zinc-finger proteins WOC and Relative-of-WOC to regulate gene expression. Genes Dev. 22:3007–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grewal SI, Moazed D. 2003. Heterochromatin and epigenetic control of gene expression. Science 301:798–802 [DOI] [PubMed] [Google Scholar]
- 15. Haley B, Hendrix D, Trang V, Levine M. 2008. A simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev. Biol. 321:482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hazelett DJ, Bourouis M, Walldorf U, Treisman JE. 1998. decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development 125:3741–3751 [DOI] [PubMed] [Google Scholar]
- 17. Hunt DM. 1970. Lethal interactions of the eye-gone and eyeless mutants in Drosophila melanogaster. Genet. Res. 15:29–34 [DOI] [PubMed] [Google Scholar]
- 18. Inoue Y. 1980. Report of new mutants-Drosophila melanogaster. Drosoph. Inf. Serv. 55:206 [Google Scholar]
- 19. James TC, et al. 1989. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 50:170–180 [PubMed] [Google Scholar]
- 20. Jang CC, et al. 2003. Two Pax genes, eye gone and eyeless, act cooperatively in promoting Drosophila eye development. Development 130:2939–2951 [DOI] [PubMed] [Google Scholar]
- 21. Jones NA, Kuo YM, Sun YH, Beckendorf SK. 1998. The Drosophila Pax gene eye gone is required for embryonic salivary duct development. Development 125:4163–4174 [DOI] [PubMed] [Google Scholar]
- 22. Jun S, Wallen RV, Goriely A, Kalionis B, Desplan C. 1998. Lune/eye gone, a Pax-like protein, uses a partial paired domain and a homeodomain for DNA recognition. Proc. Natl. Acad. Sci. U. S. A. 95:13720–13725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kwon SH, Workman JL. 2008. The Heterochromatin Protein 1 (HP1) family: put away a bias toward HP1. Mol. Cells 26:217–227 [PubMed] [Google Scholar]
- 24. Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120 [DOI] [PubMed] [Google Scholar]
- 25. Lavrov S, Dejardin J, Cavalli G. 2004. Combined immunostaining and FISH analysis of polytene chromosomes. Methods Mol. Biol. 247:289–303 [DOI] [PubMed] [Google Scholar]
- 26. Lu BY, Ma J, Eissenberg JC. 1998. Developmental regulation of heterochromatin-mediated gene silencing in Drosophila. Development 125:2223–2234 [DOI] [PubMed] [Google Scholar]
- 27. Motamedi MR, et al. 2008. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol. Cell 32:778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murzina N, Verreault A, Laue E, Stillman B. 1999. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell 4:529–540 [DOI] [PubMed] [Google Scholar]
- 29. Negre N, Lavrov S, Hennetin J, Bellis M, Cavalli G. 2006. Mapping the distribution of chromatin proteins by ChIP on chip. Methods Enzymol. 410:316–341 [DOI] [PubMed] [Google Scholar]
- 30. Nielsen SJ, et al. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561–565 [DOI] [PubMed] [Google Scholar]
- 31. Noll M. 1993. Evolution and role of Pax genes. Curr. Opin. Genet. Dev. 3:595–605 [DOI] [PubMed] [Google Scholar]
- 32. Paro R. 1993. Mechanisms of heritable gene repression during development of Drosophila. Curr. Opin. Cell Biol. 5:999–1005 [DOI] [PubMed] [Google Scholar]
- 33. Pereira PS, Pinho S, Johnson K, Couso JP, Casares F. 2006. A 3′ cis-regulatory region controls wingless expression in the Drosophila eye and leg primordia. Dev. Dyn. 235:225–234 [DOI] [PubMed] [Google Scholar]
- 34. Piacentini L, et al. 2009. Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet. 5:e1000670 doi:10.1371/journal.pgen.1000670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rehwinkel J, et al. 2006. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol. Cell. Biol. 26:2965–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a. Reuter G, Wolff Y. 1981. Isolation of dominant suppressor mutations for position-effect variegation in Dorosophila melanogaster. Mol. Gen. Genet. 182:516–519 [DOI] [PubMed] [Google Scholar]
- 36. Ryan RF, et al. 1999. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 19:4366–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sanchez L, Casares F, Gorfinkiel N, Guerrero I. 1997. The genital disc of Drosophila melanogaster. Dev. Genes Evol. 207:219–241 [DOI] [PubMed] [Google Scholar]
- 38. Smothers JF, Henikoff S. 2001. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol. Cell. Biol. 21:2555–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smothers JF, Henikoff S. 2000. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol. 10:27–30 [DOI] [PubMed] [Google Scholar]
- 40. Stephens GE, Craig CA, Li Y, Wallrath LL, Elgin SC. 2004. Immunofluorescent staining of polytene chromosomes: exploiting genetic tools. Methods Enzymol. 376:372–393 [DOI] [PubMed] [Google Scholar]
- 41. Stewart MD, Li J, Wong J. 2005. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol. Cell. Biol. 25:2525–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun FL, et al. 2004. cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol. Cell. Biol. 24:8210–8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Swaminathan J, Baxter EM, Corces VG. 2005. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 19:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Treisman J, Harris E, Desplan C. 1991. The paired box encodes a second DNA-binding domain in the paired homeo domain protein. Genes Dev. 5:594–604 [DOI] [PubMed] [Google Scholar]
- 45. Vermaak D, Henikoff S, Malik HS. 2005. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 1:96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang LH, Chiu SJ, Sun YH. 2008. Temporal switching of regulation and function of eye gone (eyg) in Drosophila eye development. Dev. Biol. 321:515–527 [DOI] [PubMed] [Google Scholar]
- 47. Yao JG, Sun YH. 2005. Eyg and Ey Pax proteins act by distinct transcriptional mechanisms in Drosophila development. EMBO J. 24:2602–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yao JG, et al. 2008. Differential requirements for the Pax6(5a) genes eyegone and twin of eyegone during eye development in Drosophila. Dev. Biol. 315:535–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ye Q, Worman HJ. 1996. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J. Biol. Chem. 271:14653–14656 [DOI] [PubMed] [Google Scholar]
- 49a. Zhang D, Wan D, Sun F. 2011. Drosophila melanogaster heterochromatin protein HP1b plays important roles in transcriptional activation and development. Chromosoma 120:97–108 [DOI] [PubMed] [Google Scholar]
- 50. Zhang Y, et al. 2008. Epigenetic blocking of an enhancer region controls irradiation-induced proapoptotic gene expression in Drosophila embryos. Dev. Cell 14:481–493 [DOI] [PMC free article] [PubMed] [Google Scholar]