Abstract
The microRNA miR-519 robustly inhibits cell proliferation, in turn triggering senescence and decreasing tumor growth. However, the molecular mediators of miR-519-elicited growth inhibition are unknown. Here, we systematically investigated the influence of miR-519 on gene expression profiles leading to growth cessation in HeLa human cervical carcinoma cells. By analyzing miR-519-triggered changes in protein and mRNA expression patterns and by identifying mRNAs associated with biotinylated miR-519, we uncovered two prominent subsets of miR-519-regulated mRNAs. One subset of miR-519 target mRNAs encoded DNA maintenance proteins (including DUT1, EXO1, RPA2, and POLE4); miR-519 repressed their expression and increased DNA damage, in turn raising the levels of the cyclin-dependent kinase (cdk) inhibitor p21. The other subset of miR-519 target mRNAs encoded proteins that control intracellular calcium levels (notably, ATP2C1 and ORAI1); their downregulation by miR-519 aberrantly elevated levels of cytosolic [Ca2+] storage in HeLa cells, similarly increasing p21 levels in a manner dependent on the Ca2+-activated kinases CaMKII and GSK3β. The rises in levels of DNA damage, the Ca2+ concentration, and p21 levels stimulated an autophagic phenotype in HeLa and other human carcinoma cell lines. As a consequence, ATP levels increased, and the level of activity of the AMP-activated protein kinase (AMPK) declined, further contributing to the elevation in the abundance of p21. Our results indicate that miR-519 promotes DNA damage, alters Ca2+ homeostasis, and enhances energy production; together, these processes elevate the expression level of p21, promoting growth inhibition and cell survival.
INTRODUCTION
Cell homeostasis is maintained through tightly regulated gene expression patterns. Among the mechanisms of gene regulation, posttranscriptional processes critically influence the catalog of proteins expressed in the cell. MicroRNAs (miRNAs) (22-nucleotide [nt]-long noncoding RNAs) have emerged as key posttranscriptional regulators of gene expression (29). They are integral components of the RNA-induced silencing complexes (RISCs), which also include Argonaute (Ago) proteins. As part of the Ago/RISC machinery, microRNAs interact with mRNAs (typically at their 3′-untranslated regions [3′UTRs]) with partial complementarity and generally reduce mRNA stability and/or translation (25). By controlling the expression of subsets of target mRNAs, microRNAs potently influence the collection of expressed proteins. In this manner, microRNAs can influence many cellular processes, including cell proliferation, differentiation, senescence, and the response to damaging agents, as well as disease processes like carcinogenesis, neurodegeneration, and cardiovascular pathologies (7, 13, 17, 20, 32, 49, 50).
Previous studies using both transformed and untransformed cells showed that the microRNA miR-519 accelerated cellular senescence and repressed cell division and tumor development (2, 4, 34). These effects were due at least in part to the miR-519-mediated decrease in the level of expression of the RNA-binding protein HuR (human antigen R), a key regulator of gene expression (4). miR-519 selectively reduced HuR translation and diminished the abundance of HuR, in turn repressing HuR-elicited processes like cell cycle progression and tumorigenesis and enabling HuR-suppressed events like cellular senescence (34).
However, it is increasingly being appreciated that microRNAs do not elicit their specific phenotypes by acting on a single mRNA but by associating with and affecting the fates of multiple mRNAs (for example, see reference 31). In this investigation, we sought to identify the effectors of miR-519 actions systematically by taking a three-pronged approach. First, we studied miR-519-dependent changes in protein composition using the proteomics method SILAC (stable isotope labeling with amino acids in cell culture). Second, we detected global changes in mRNA levels as a function of miR-519 abundance using microarray analysis. Third, we identified miR-519-interacting mRNAs by transfecting biotinylated miR-519 (biot-miR-519) and pulling down endogenous mRNAs associated with biot-miR-519. From this combined analysis, two main pathways emerged. Through one pathway, miR-519 repressed the production of DNA repair and maintenance proteins and thus triggered DNA damage. Through the second pathway, miR-519 reduced the production of proteins that control calcium homeostasis, augmented cytosolic [Ca2+] concentrations, and activated calcium-dependent kinases. Both pathways converged on the upregulation of the cyclin-dependent kinase (cdk) inhibitor p21, which was found to play an essential role in stimulating autophagy. We propose that microRNAs such as miR-519, by acting simultaneously on multiple mRNA subsets, are particularly well suited to implement critical cellular processes like growth arrest in a robust and tightly controlled manner.
MATERIALS AND METHODS
Cell culture, treatments, transfection, small RNAs, and plasmids.
Human cervical carcinoma HeLa cells, colon carcinoma RKO cells, and lung carcinoma A549 and H1299 cells were cultured in Dulbecco's modified essential medium (DMEM; Invitrogen), and HCT116 human colon carcinoma cells (including parental wild-type HCT116 [HCT116 wt] cells and HCT116 cells bearing somatic deletions of both p21 alleles [HCT116 p21−/− cells]) were cultured in McCoy's 5A medium (Invitrogen). The media were supplemented with 10% fetal bovine serum and antibiotics. miR-519 and the antagomir antisense miR-519 [(AS)miR-519] were obtained from Ambion, the control (Ctrl) small interfering RNA (siRNA) was obtained from Qiagen, and the siRNAs directed toward p21, p53, ATM (ataxia-telangiectasia mutated), ATR (ATM related), NBS, ATP2B1, ATP2B4, ATP2C1, and ORAI1 were obtained from Santa Cruz; all small RNAs were used at 50 nM. The reporter plasmids used were an enhanced green fluorescent protein (EGFP)-LC3 reporter plasmid (Addgene), psiCHECK2-TOP2A (51), and newly engineered plasmids psiCHECK2-DUT1, -EXO1, -RPA2, -POLE4, -PMR1, and -ORAI1, which were constructed by cloning the corresponding PCR-amplified 3′UTR segments after the Renilla luciferase (RL) coding region. Small RNAs and plasmids were transfected with Lipofectamine 2000 (Invitrogen). The chemicals 3-methyladenine (3-MA), 2-[N-(2-hydroxyethyl)]-N-(4-methoxybenzenesulfonyl)]amino-N-(4-chlorocinnamyl)-N-methylbenzylamine (KN-93),, and indirubin-3′-monoxime (I3MO) were obtained from Enzo Life Sciences; 2,4-dinitrophenol (DNP) was obtained from Sigma; and EGTA was obtained from Quality Biological. Luciferase activity assays were performed by using the Dual-Luciferase reporter assay system (Promega). Viable and nonviable cells were counted by using a Bio-Rad TC10 automated cell counter.
Ca2+ measurements.
HeLa cells were loaded with 20 μM calcium indicator Fluo-4 AM (Invitrogen), a Ca2+ indicator, for 30 min at 25°C and then incubated in a solution containing 137 mM NaCl, 4.9 mM KCl, 1.2 mM NaH2PO4, 15 mM glucose, 20 mM HEPES, 1 mM CaCl2, and 1.2 mM MgSO4 (pH 7.4). Confocal images were obtained every 20 s by using a Zeiss LSM 510 microscope (Carl Zeiss) in the frame scan mode. Cytosolic Ca2+ transients were normalized to the basal fluorescence intensity (F0) and expressed as F/F0. Image processing was performed with Matlab (Mathworks). All measurements were performed at 25°C.
Microscopy.
For electron microscopy, cells were fixed with 2% glutaraldehyde in 0.1 M cacodylate buffer with 3% sucrose and 3 mM CaCl2 (1 h at 27°C). After rinsing in a solution containing 0.1 M cacodylate, 3% sucrose, and 3 mM CaCl2, cells were incubated with OsO4 for 1 h (at 4°C), washed with water, and stained with 2% uranyl acetate (1 h). Following dehydration with progressively more concentrated ethanol (50 to 100%) and incubation with fresh Epon resin for an additional 24 h, samples were placed at 37°C for 48 h and at 60°C for 16 h.
For fluorescence microscopy to detect γ-H2AX foci, cells were fixed with 2% formaldehyde, permeabilized with 0.2% Triton X-100, and blocked with 5% bovine serum albumin (BSA), as previously described (1). After incubation with a primary antibody recognizing γ-H2AX (Santa Cruz), an Alexa 488-conjugated secondary antibody (Invitrogen) was used to detect primary antibody-antigen complexes. Images were acquired by using an Axio Observer microscope (Zeiss) with AxioVision 4.7 Zeiss image processing software or with LSM 510 Meta (Zeiss). The Z-sectioning mode was employed with 15 slices and 0.4-μm spacing and merged using maximum intensity.
Protein analysis.
For Western blot analysis, whole-cell lysates were prepared by using radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.1% SDS, and 1 mM dithiothreitol). Proteins were separated by electrophoresis in SDS-containing polyacrylamide gels and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore). Primary antibodies were used to detect LC3-II, calmodulin kinase II (CaMKII), phosphorylated CaMKII (p-CaMKII), phosphorylated AMP-activated protein kinase (p-AMPK), or AMPK (polyclonal; Cell Signaling); Ung (polyclonal; Abcam); Brca1, p53, or Top2A (monoclonal; Santa Cruz); p21 (monoclonal; Upstate); or β-actin (monoclonal; Abcam). After incubations with the appropriate secondary antibodies conjugated with horseradish peroxidase (HRP) (GE Healthcare), signals were detected by using enhanced chemiluminescence.
Stable amino acid labeling in culture.
For proteomic analyses by stable amino acid labeling in culture (SILAC) using a ThermoElectron LTQ-Orbitrap XL instrument, HeLa cells were cultured with both isotopic arginine (R) and lysine (K) (Cambridge Isotope Laboratories, Andover, MA) for 3 days to achieve 93 to 95% stable amino acid incorporation prior to extraction and analysis. Label swapping of the Ctrl siRNA and miR-519 siRNA groups with the two label states, medium (K4 and R6) and heavy (K8 and R10), was used to reduce artifactual label bias. SILAC was carried out, as previously described (42, 43), by using HeLa cells (300 μg from the Ctrl siRNA group and 300 μg from the miR-519 group).
KEGG signaling pathway analysis.
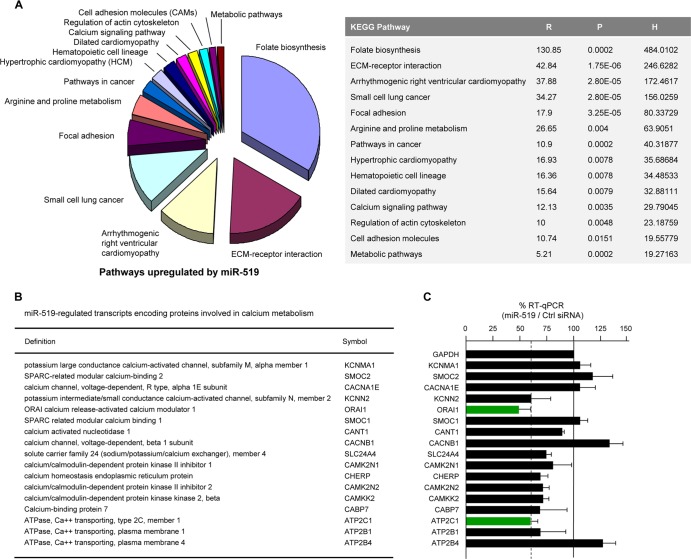
The proteins identified by SILAC as being significantly up- or downregulated in response to the ectopic expression of miR-519 were analyzed by using annotational clustering (http://www.genome.jp/kegg/) and WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/), a Web-based gene set analysis tool kit (9). This application allows the expression frequency of a specific protein in the experimental set to be compared to its expression frequency in a background set that is maintained by WebGestalt. The functional clustering of proteins into KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways yielded two indices. The numerical degree of set enrichment (R) is represented as a ratio of the expected number (E) (scaled to the size of the input data set size compared to the background whole-proteome data set) of proteins that would cluster into this pathway using a randomly selected protein set compared to the number of proteins observed (O) to actually populate this pathway from the input experimental data set (R = O/E). The probability (P) that this enrichment of multiple proteins in the specific KEGG pathway would occur by chance, based on a scalar comparison of the input experimental data set to a whole-proteome data set, was also derived. A cutoff of at least 2 proteins populating a KEGG pathway and a probability value of ≤0.05 were used. To generate a combined appreciation of R and P, we created a “hybrid” score (H) by multiplying the negative log10(P) by R.
Total RNA analysis.
Whole-cell RNA was prepared from whole cells 48 h after transfection with either Ctrl siRNA or miR-519 (using TRIzol; Invitrogen). Changes in total mRNA levels were analyzed by using microarrays (Illumina). The levels of individual mRNAs were quantified by reverse transcription (RT) using random hexamers and SSII reverse transcriptase (Invitrogen), followed by quantitative PCR (qPCR) analysis using SYBR green PCR master mix (Kapa Biosystems) and gene-specific primer sets (see below). RT-qPCR analysis was performed on Applied Biosystems model 7300 and 7900 instruments.
Analysis of mRNAs associated with biotinylated miR-519.
The pulldown of biotinylated miR-519 (biot-miR-519) and biotinylated control microRNA from Caenorhabditis elegans (cel-miR-67 [here named biot-Ctrl-miR]) was performed as previously reported (31). Briefly, HeLa cells (2.5 × 105 cells) were transfected with 20 nM biot-miR-519 or biot-Ctrl-miR using Lipofectamine 2000. Twenty-four hours after transfection, cells were rinsed and lysed in a buffer containing 20 mM Tris-HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.3% NP-40, 50 U of RNase Out (Invitrogen), and a protease cocktail inhibitor (Roche Applied Science). The cytoplasmic lysate was cleared by centrifugation at 10,000 × g for 10 min. Streptavidin Dynabeads (Invitrogen) were preincubated in lysis buffer containing yeast tRNA (1 mg/ml) and BSA (1 mg/ml) for 2 h at 4°C, rinsed, and incubated with cytoplasmic extract for 4 h at 4°C. After washes with lysis buffer, the RNA bound to the beads (“pulldown RNA”) and the RNA in 50 μl of cytoplasmic extract (“input”) were isolated by using TRIzol LS reagent (Invitrogen). The mRNAs associated with the biotinylated miRNAs were subsequently identified either en masse using microarray analysis (as described in reference 3) or individually by RT-qPCR using the primer pairs listed below.
PCR primer pairs used.
Following RT, several mRNAs were measured by qPCR using the following primer pairs (forward and reverse, respectively, in each case): GCTGGTTCTAGCCCTGAGTG and AACCCACAAGTGTCCAGAGG for ATP2B4, TGTTGGATTTGCAATGGGTA and CGTCCCCACATAACTGCTTT for ATP2B1, TGAGTCCAGCTTGACAGGTG and CTTTGCTTTGCCACATCTGA for ATP2C1, CGAATTGCACAGCTCATTTG and GTGGAACCAAAACCTCCTGA for DUT, TCAAGCTCGGCTAGGAATGT and TGCCTTTGCTAATCCAATCC for EXO1, TTAAGATCATGCCCCTGGAG and ATAGGTGCTCTCCCTGCTGA for RPA2, TGTGAATCATGTGCTGCTGA and TCCGATTGAGGTTTCTGGTC for ERCC6L, GAGTGAAGGCCTTGGTGAAG and AGCGCAACAGTAGGCATCTT for POLE4, GCCCACTGGTGGTAGTGTCT and GGTGCTAGTGGCATTTGGAT for RAD51AP1, CTGTAGCTGCCACGTTTTGA and CACAGGCCTACCACCAATCT for FANCG, CTCAAGGAACCAGGGATGAA and CGCTTCTCAGTGGTGTTCAA for BRCA1, GTGGAGATGACCCCTGAGAA and GTCAGCATGAGCTTGGCATA for FANCE, CCCACACCAAGTCTTCACCT and GAGCCCGTGAGCTTGATTAG for UNG, CGCTGCGTTGAGTTTCATAA and TGGGACAATAGGCATCACAA for FANCB, TTTTTGCACTTGACGCTTTG and ACTGGCAGAGGAAGTGTGCT for FANCA, TGGGCCTTCTATCTTGGATG and GCAAGGCTGACACCACTGTA for TP53INP1, GGAGGATGCCTCGAATATCA and AGCTCGGGATGTTTAGCAGA for KCNMA1, GTCGGCAGAACAAAACCAAT and CACCACATTGTCGTTCTTGG for SMOC2, AGTGGGGCCTCATCATACAG and TGTCAGCCCCAGCTCTTACT for CACNA1E, GCGTCGCTGTATTCCTTAGC and TCCAGTCATCTGCTCCATTG for KCNN2, ACGTGCACAATCTCAACTCG and AGCACCACCTCAGCTAGGAA for ORAI1, CACCACTGACATGGTTCAGG and GTTGCTGCTATTGCTGTCCA for SMOC1, TGCTGTGGAAAGACTTCACG and TGGCCATTCTTACTCCCTTG for CANT1, AAAGCCATAGCATGGACACC and GCTCACTCCTCAGGGTTCTG for CACNB1, GCCTTCTTTGCTGTCACTCC and TTCAGGAGGGGAAAAGTCCT for SLC24A4, CTGTTGTGTGAGCAGCCTGT and ATGCCTTTTGGCATTTCAAG for CAMK2N1, CAGTGGAGGCCTTCTACAGC and CTCTTCTCCTGGCCTTTCCT for CHERP, CTGGCAGATAGGATGGGTGT and GCCCTTGGAAGGTGATGATA for CAMK2N2, AAGGAGGGATTTGCACAGG and CCCAGATAGGCTGCTGTCTC for CABP7, CGAGTGTGCTGGTCACTAA and ACAATTGGCCGCTAAACTTG for TOP2A, TTTGCTTGAGGCTGATCCTT and GATTGACTCTGCAGCCAACA for ATM, CTCTGGTCCAAGGGTGATGT and GCATAGCTCGACCATGGATT for ATR, AATGGCTTTTCCCGAACTTT and CAAGAAGAGCATGCAACCAA for NBS, GTAACGCTGCCTCCAGCTAC and TGATCTTGTCTTGGTGCTCGTA for RL, and GGACATCACCTATGCCGAGTACT and AAGCCCTGGTAGTCGGTCTTAG for firefly luciferase (FL).
RESULTS
Analysis of the global influence of miR-519.
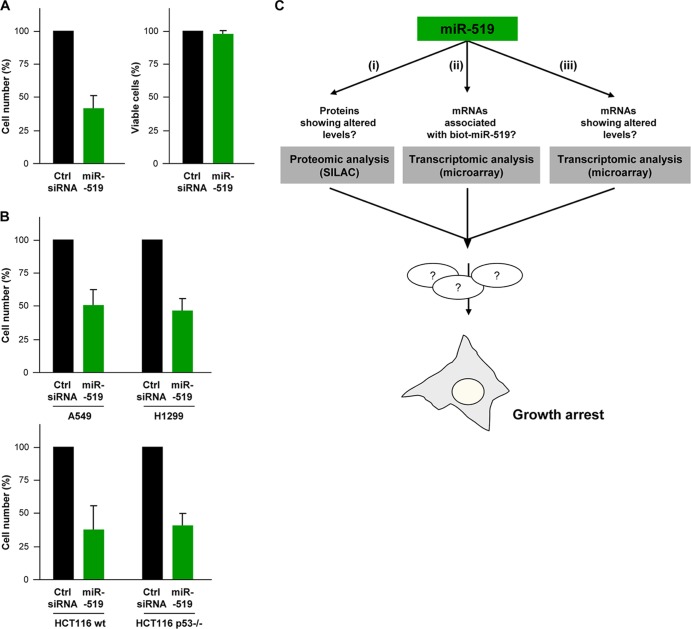
Forty-eight hours after the transfection of human cervical carcinoma HeLa cells with miR-519b-3p (here called miR-519), cell proliferation ceased, as determined by the ∼60% fewer cells in the miR-519-expressing population than control (Ctrl siRNA) cells (Fig. 1A); miR-519 expression did not affect cell viability, as determined by trypan blue exclusion. Although HeLa cells express a wild-type p53 protein, they are rendered deficient in p53 function due to the expression of the human papillomavirus (HPV) protein E6, which accelerates p53 degradation. Thus, we sought to study whether the effect of miR-519 was dependent on the p53 status by investigating the responses of two pairs of p53-deficient and -proficient cell lines. As shown in Fig. 1B, comparisons of human lung carcinoma A549 (p53-proficient) and H1299 (p53-deficient) cells as well as parental HCT116 (wt p53) human colon carcinoma cells and HCT116 human colon carcinoma cells bearing somatic deletions of both p53 alleles (HCT116 p53−/− cells) indicated that miR-519 was growth inhibitory regardless of the p53 status.
Fig 1.
Analysis of miR-519-triggered growth arrest. (A) Forty-eight hours after the transfection of HeLa cells with either Ctrl siRNA or miR-519, cell numbers and cell viability were determined by direct cell counts and by analyzing trypan blue-positive cells. The graph represents the means and standard deviations (SD) of data from 3 independent experiments. (B) Forty-eight hours after the transfection of A549 (p53-proficient) and H1299 (p53-deficient) human lung carcinoma cells (top) and HCT116 wt and HCT116 p53−/− human colon carcinoma cells (bottom) with either Ctrl siRNA or miR-519, cell numbers were determined by direct cell counting. The graph represents the means and SD of data from 3 independent experiments. (C) Schematic of efforts undertaken to investigate miR-519-regulated gene expression, including an analysis of proteins (i) and mRNAs (iii) showing altered abundances as a function of miR-519 levels and the identification of putative mRNA targets by using biotinylated miR-519 (ii).
We set out a systematic, three-arm strategy to analyze the changes in gene expression implemented by miR-519 (Fig. 1C): (i) global changes in protein profiles elicited by miR-519 were studied by a proteomic approach (SILAC); (ii) the collection of miR-519-interacting mRNAs was studied by transfecting biotinylated miR-519 (biot-miR-519), pulling down the associated endogenous mRNAs with streptavidin beads, and identifying them by microarray analysis; and (iii) the effect of miR-519 on transcriptome-wide changes in mRNA levels was also investigated by a microarray analysis of steady-state mRNA levels (Fig. 1C). From these analyses, we sought to identify mRNAs that were miR-519 targets whose abundances and/or whose encoded proteins' abundances were modulated by miR-519.
miR-519 inhibits DNA repair pathways.
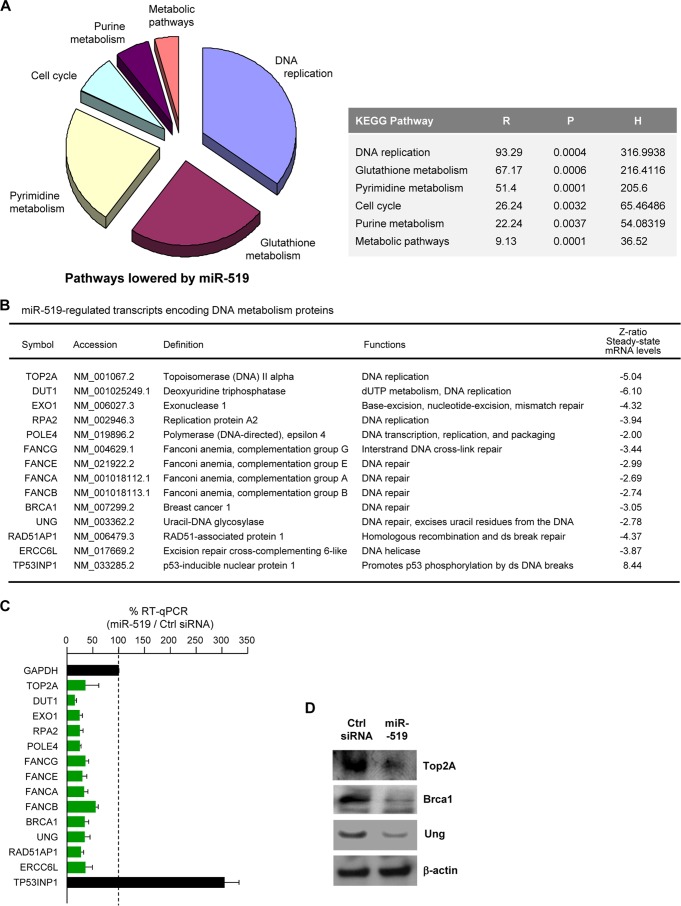
To investigate the molecular mediators of miR-519-induced growth inhibition, we first performed SILAC analysis with HeLa cells 48 h after transfection with miR-519 or Ctrl siRNA. SILAC followed by KEGG analysis (see Materials and Methods) identified proteins in 6 distinct functionally related pathways that were selectively reduced in miR-519-transfected cells. The pathways were identified by their KEGG signaling pathway hybrid scores (H), which measure the strength and significance of the association of the input protein set with the specific signaling pathway (Fig. 2A, and see Table S1 in the supplemental material). Several functional pathways that were inhibited in the miR-519 group were involved in DNA repair and metabolism (Fig. 2A).
Fig 2.
miR-519 inhibits DNA repair pathways. (A) Pathways of proteins that were downregulated after miR-519 transfection. Forty-eight hours after the transfection of HeLa cells with either Ctrl siRNA or miR-519, SILAC followed by KEGG proteomic analysis (see Materials and Methods) was performed to identify the families of proteins collectively influenced by miR-519. R, numerical degree of set enrichment of subsets of proteins; P, probability that the enrichment in the specific KEGG pathway would not occur by chance; H, hybrid score obtained by multiplying the negative log10(P) by R. (B) Forty-eight hours after the transfection of HeLa cells with Ctrl siRNA or miR-519, total RNA was extracted and used for microarray hybridization. Several mRNAs encoding DNA metabolism/repair whose steady-state abundances were reduced in miR-519-expressing cells are listed (“Z-ratio Steady-state mRNA levels” column). Array data were compiled from data from 3 independent experiments. (C and D) Validation of microarray-identified changes in steady-state mRNA levels after the expression of miR-519. Forty-eight hours after Ctrl siRNA or miR-519 transfection, total RNA was isolated and analyzed by RT-qPCR using mRNA-specific primer pairs (C), and the levels of several encoded proteins were assessed by Western blot analysis (D). Data in panel C represent the means and SD of data from 3 independent experiments. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To identify possible direct effects of miR-519 on protein levels, we searched for parallel, miR-519-triggered changes in mRNAs by microarray analysis using total RNA prepared from HeLa cells 48 h after the transfection of either miR-519 or Ctrl siRNA. It is important to note that this strategy may miss instances in which miR-519 primarily (or exclusively) represses the translation of target mRNAs without affecting their stability. As shown in Fig. 2B, many mRNAs encoding DNA metabolism, structure, and repair proteins (including several Fanconi anemia [FANC] proteins and other DNA replication and repair proteins) showed reduced abundances in miR-519-expressing cells; as anticipated, TP53INP1 mRNA (encoding a p53-inducible protein) was upregulated (Fig. 2B) (complete microarray results are shown in Table S2 in the supplemental material). The influence of miR-519 on the steady-state levels of these mRNAs was assessed by reverse transcription (RT) followed by real-time, quantitative PCR (qPCR) amplification using gene-specific primer pairs (see Materials and Methods). As shown in Fig. 2C, all of the individual mRNA changes were verified by RT-qPCR analysis. These differences were also verified by Western blot analysis of some of the proteins implicated in DNA metabolism and repair, including Top2A (topoisomerase 2α), Ung (uracil-DNA glycosylase), Brca1 (breast cancer 1), and the repair protein Dut1 (deoxyuridine 5′-triphosphate nucleotidohydrolase) (Fig. 2D).
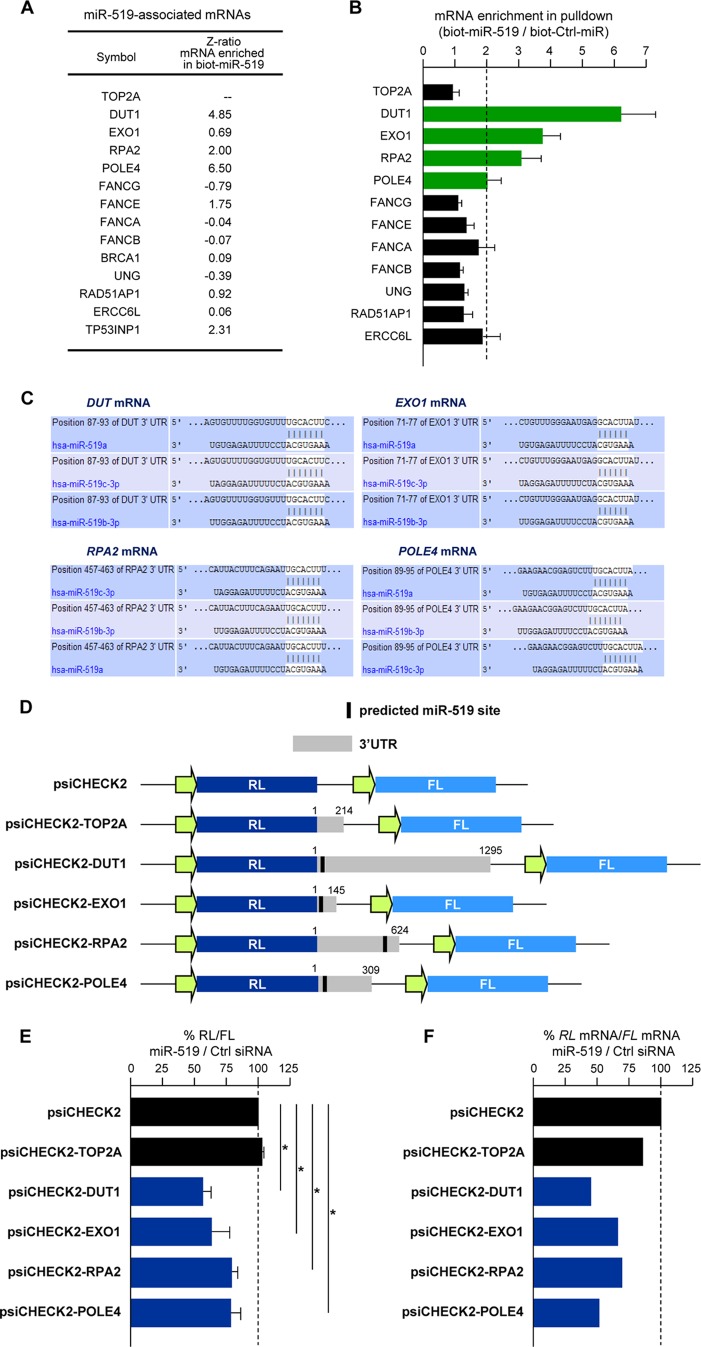
Next, we assessed the possible association of miR-519 with putative targets using a high-throughput method described recently (31). Twenty-four hours after the transfection of HeLa cells with biot-miR-519 or a control microRNA (biot-Ctrl-miR), biotin-RNA-containing complexes were pulled down by using streptavidin beads. The RNA bound to the beads was isolated and used for microarray analysis (Fig. 3A) (complete results are shown in Table S3 in the supplemental material). Candidate miR-519 target mRNAs were validated by RT-qPCR analysis of the pulled-down RNA (Fig. 3B). This analysis showed that many mRNAs encoding DNA repair proteins (including DUT1 mRNA, RPA2 mRNA [encoding replication protein A2], and POLE4 mRNA [encoding RNA polymerase E4]) were putative direct miR-519 interaction targets. EXO1 mRNA, encoding exonuclease 1, was not identified in biot-miR-519 pulldown microarrays but was identified as a biot-miR-519-associated mRNA when tested individually by the more sensitive RT-qPCR analysis (Fig. 3B). Other transcripts (notably the FANC mRNAs) were enriched less than 2-fold in the biot-miR-519 pulldown.
Fig 3.
miR-519 induces DNA damage. (A) Forty-eight hours after the transfection of HeLa cells with either biotinylated Ctrl-miR (biot-Ctrl-miR) or biot-miR-519, mRNAs bound to each biotinylated transcript were isolated by using streptavidin beads (see Materials and Methods) and identified by microarray hybridization (“Z-ratio mRNA enriched in biot-miR-519” column). Array data were compiled from data from 3 independent experiments. (B) The validation of microarray-identified mRNAs associated with biot-miR-519 was performed by RT-qPCR using mRNA-specific primer pairs. (C) TargetScan prediction of the complementarity between miR-519 and the four target mRNAs shown in Fig. 2 (DUT mRNA, EXO1 mRNA, RPA2 mRNA, and POLE4 mRNA). (D and E) Schematic of reporter constructs that were prepared in order to test if miR-519 influenced the expressions of TOP2A, DUT1, EXO1, RPA2, and POLE4 mRNAs through their respective 3′UTRs. (D) Each 3′UTR (gray rectangles, with predicted miR-519 sites indicated by a black bar) was amplified by PCR and cloned into plasmid psiCHECK2, as shown. Twenty-four hours after the transfection of HeLa cells with either Ctrl siRNA or miR-519, each reporter plasmid was transfected, and the ratio of Renilla luciferase (RL) activity to the activity of the internal control firefly luciferase (FL) was calculated 24 h after that. (E) The relative RL/FL ratio of miR-519-transfected cells versus the RL/FL of Ctrl siRNA-transfected cells is indicated. (F) RL mRNA and FL mRNA levels for each reporter plasmid were calculated by RT-qPCR analysis. The relative RL mRNA/FL mRNA ratio for miR-519-transfected cells versus the RL mRNA/FL mRNA ratio for Ctrl siRNA-transfected cells is indicated. Data in panels B and E represent the means and SD of data from 3 independent experiments. *, P < 0.05. Data in panel F are the averages of data from two experiments showing comparable results.
The presence of predicted miR-519 sites at the 3′UTRs of the enriched targets (DUT, EXO1, RPA2, and POLE4 mRNAs) was examined by using TargetScan (Fig. 3C). To investigate further the effects of miR-519 on these targets, a heterologous luciferase reporter vector (psiCHECK2) was used to prepare several reporter constructs (Fig. 3D, left) bearing the respective 3′UTRs. The ratio of Renilla luciferase (RL) (encoded by the reporter transcript bearing the test sequences) to firefly luciferase (FL) (encoded by an internal control reporter transcript) was set as 1 for the parent vector (psiCHECK2). RL/FL ratios for miR-519-transfected cells relative to those for Ctrl siRNA-transfected cells are shown in Fig. 3E (right). In this analysis, the reporter psiCHECK2-TOP2A, which lacked any predicted miR-519 sites, showed no significant difference with the parent vector; in contrast, miR-519 significantly repressed luciferase expression from reporters that contained predicted miR-519 sites (psiCHECK2-DUT1, psiCHECK2-EXO1, psiCHECK2-RPA2, and psiCHECK2-POL4) (Fig. 3E, right), supporting the hypothesis that miR-519 reduced the expression levels of these genes by interacting with their 3′UTRs. Since microRNAs can reduce the stability of a target mRNA, its translation, or both of these processes, we examined the influence of miR-519 on the expressions of reporter mRNAs. As shown in Fig. 3F, in the cases of DUT1, EXO1, RPA2, and POLE4 reporters, there was a parallel relative reduction in RL mRNA levels compared with FL mRNA levels in miR-519-expressing cells compared with Ctrl siRNA-expressing cells. These data suggest that mRNA turnover likely contributes to the repression of the target DUT1, EXO1, RPA2, and POLE4 mRNAs.
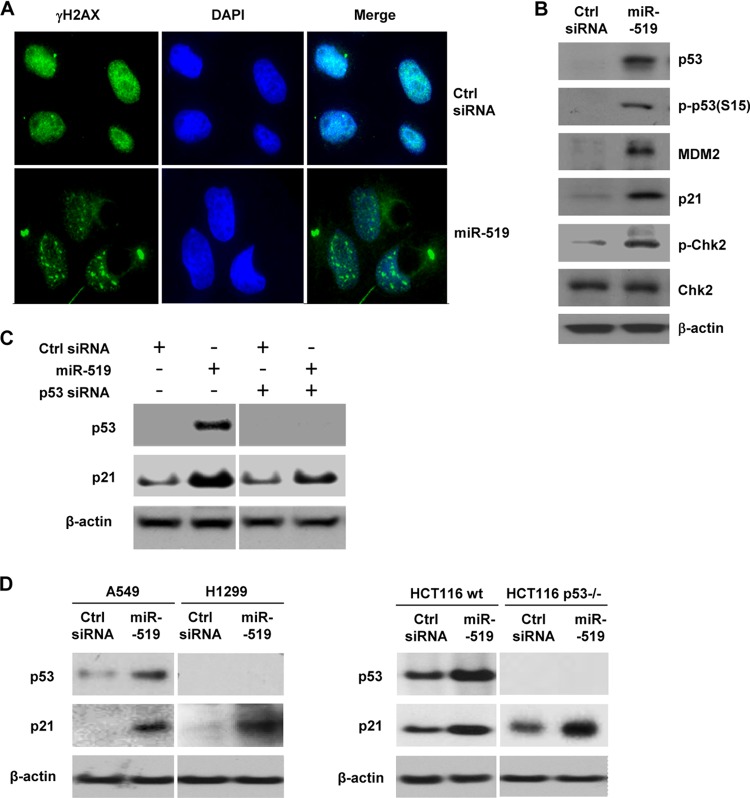
In keeping with the view that miR-519 reduces the expression levels of several DNA repair genes either directly (via miR-519–mRNA interaction) or indirectly, miR-519-overexpressing cells exhibited features of DNA damage. Fluorescence microscopy revealed that levels of nuclear γ-H2AX foci, an indicator of DNA damage, were markedly elevated in miR-519-transfected cells (Fig. 4A), while Western blot analysis (Fig. 4B) showed that miR-519 upregulated several DNA damage marker proteins, including the tumor suppressor and transcription factor p53 (the level of which is very low in HeLa cells but is detectable by using a sensitive antibody), p53 phosphorylated at Ser-15 (a modification that blocks p53 degradation via MDM2), the cdk inhibitor and potent growth suppressor p21 (a transcriptional target of p53), and phosphorylated checkpoint kinase 2 (Chk2), a DNA damage-inducible kinase (Fig. 4B). To investigate whether the increase in p21 expression levels by miR-519 was due to the upregulation of p53 levels, 48 h after the transfection of HeLa cells with p53 siRNA to reduce endogenous p53 levels, p53 protein levels were drastically reduced, but p21 was still substantially induced in miR-519-expressing cells (Fig. 4C). Moreover, in two pairs of p53-deficient and -proficient cells (A549 and H1299 cells as well as HCT116 wt and p53−/− cells), miR-519 upregulated p21 (Fig. 4D), indicating that the increase in p21 levels by miR-519 did not depend on the miR-519-elicited DNA damage and increase in p53 levels. In sum, miR-519 inhibits the expressions of numerous DNA repair and DNA metabolism proteins, triggering a robust DNA damage response and inducing an abundance of p21 in a manner that was not strictly dependent on p53 upregulation.
Fig 4.
(A) Immunofluorescence micrographs depicting γ-H2AX foci, indicative of DNA damage, 48 h after the transfection of HeLa cells with Ctrl siRNA or miR-519. DAPI, 4′,6-diamidino-2-phenylindole. (B) Western blot analysis of proteins or phosphoproteins implicated in the response to DNA damage. (C) Forty-eight hours after the transfection of HeLa cells with the small RNAs indicated, the levels of p53 and p21 were assessed by Western blot analysis. (D) Forty-eight hours after the transfection of A549 and H1299 (left) or HCT116 wt and HCT116 p53−/− (right) cells with the small RNAs indicated, the levels of p53 and p21 were assessed by Western blot analysis. Data are representative of data from three independent experiments.
miR-519 modulates proteins implicated in Ca2+ metabolism.
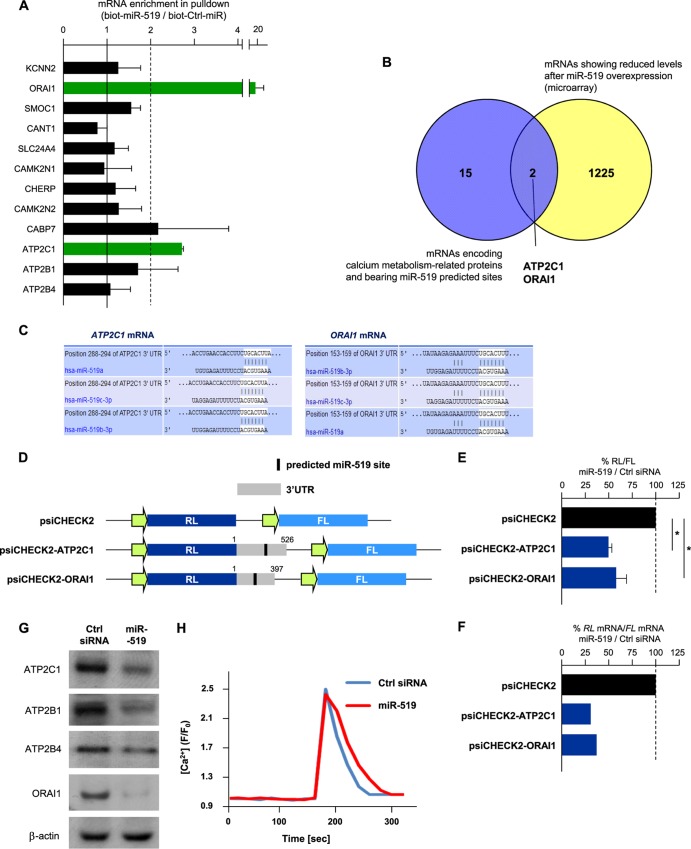
Among the proteins identified by SILAC as being upregulated by miR-519, several were included in pathways associated with aberrant calcium levels, such as cardiomyopathies, and pathways affecting cell adhesion and interaction with the extracellular matrix (Fig. 5A). A number of mRNAs enriched in biot-miR-519 pulldowns also encoded Ca2+-regulatory proteins (see Table S3 in the supplemental material). To narrow down the possible direct involvement of some of these targets, we carried out several complementary experiments. First, an analysis of the abundances of mRNAs enriched in biot-miR-519 pulldowns revealed two mRNAs whose levels were particularly lower after miR-519 transfection: the mRNAs encoding ORAI1 (calcium release-activated calcium modulator 1) and ATP2C1 (ATPase, Ca2+ transporting, type 2C, member 1) (also named PMR1) (Fig. 5B). These changes were verified by individual RT-qPCR analyses of the mRNAs (Fig. 5C).
Fig 5.
miR-519 upregulates pathways affecting Ca2+ homeostasis. (A) Pathways of proteins that were upregulated after miR-519 transfection. Forty-eight hours after the transfection of HeLa cells with either Ctrl siRNA or miR-519, SILAC followed by KEGG proteomic analysis (see Materials and Methods) was performed to identify the families of proteins collectively influenced by miR-519, as explained in the legend of Fig. 2A. ECM, extracellular matrix. (B and C) Among the mRNAs showing differential abundances after the expression of miR-519 (as identified in Fig. 2B), several encoded proteins were implicated in the maintenance of intracellular calcium (B); the steady-state levels of these mRNAs were individually validated by using RT-qPCR and specific primer pairs (C), as explained in the legend of Fig. 2C. Green bars indicate miR-519 target transcripts (ATP2C1 and ORAI1 mRNAs) whose steady-state abundances were particularly low after the expression of miR-519.
We then tested the enrichment of these target transcripts by individual mRNA amplification in biot-miR-519 relative to that in biot-Ctrl-miR; here too, ORAI1 and ATP2C1 mRNAs were specifically enriched (Fig. 6A). Moreover, of the mRNAs showing reduced levels after the overexpression of miR-519 (see Table S2 in the supplemental material) and the computationally identified mRNAs encoding Ca2+ metabolism-related proteins bearing 3′UTR miR-519 sites, the only transcripts present in both groups were ATP2C1 and ORAI1 mRNAs (Fig. 6B and C). To test if ectopically expressed miR-519 directly regulated their expressions, we engineered heterologous reporters derived from plasmid psiCHECK2, bearing the 3′UTRs of either ATP2C1 or ORAI1 (each with a putative miR-519 interaction site) (Fig. 6D). As shown, miR-519 significantly reduced the activity (RL/FL) of each reporter relative to the activity of the parent vector, psiCHECK2 (Fig. 6E). As shown in Fig. 6F, for the ATP2C1 and ORAI1 reporters, there was a parallel relative reduction in RL mRNA levels compared with FL mRNA levels in miR-519-expressing cells compared with Ctrl siRNA-expressing cells, suggesting that the turnover of ATP2C1 and ORAI1 mRNAs likely contributed to their lower expression levels after the transfection of miR-519. ATP2B1 and ATP2B4 3′UTRs were not included in the reporter analysis because they did not appear to be interaction targets of miR-519 (Fig. 6A, and see Table S3 in the supplemental material); however, they might be indirectly regulated by miR-519, since the overexpression of miR-519 lowered the levels of the endogenous proteins ATP2C1, ATP2B1, ATP2B4, and ORAI1 (Fig. 6G).
Fig 6.
miR-519 represses Ca2+ transport proteins and delays cytoplasmic Ca2+ clearance. (A) Forty-eight hours after Ctrl siRNA or miR-519 transfection, the enrichment of mRNAs identified in Fig. 5A in biotin-miR-519 pulldown materials was individually validated by using RT-qPCR and specific primer pairs, as explained in the legend of Fig. 3A. The miR-519 target transcript, ATP2C1 and ORAI1 mRNAs (green) showed low steady-state abundances after the expression of miR-519 and were highly enriched in the biot-miR-519 pulldown. (B) Venn diagram illustrating the overlap between the mRNAs encoding calcium metabolism-related proteins bearing 3′UTR miR-519 sites and mRNAs showing reduced abundances after the overexpression of miR-519 (see Table S2 in the supplemental material). The two transcripts present in both groups were ATP2C1 and ORAI1 mRNAs. (C) TargetScan prediction of the complementarity of miR-519 with ORAI1 mRNA and ATP2C1 mRNA. (D) Schematic of reporter constructs that were engineered in order to test whether miR-519 influenced the expression of ATP2C1 or ORAI1 through their respective 3′UTRs. (E) Twenty-fours after the transfection of HeLa cells with either Ctrl siRNA or miR-519, each reporter was transfected, and the influence of miR-519 on their expressions was calculated 24 h after that, as explained in the legend of Fig. 3E. (F) RL mRNA and FL mRNA levels for each reporter plasmid were calculated by RT-qPCR analysis. The relative RL mRNA/FL mRNA ratio for miR-519-transfected cells versus the RL mRNA/FL mRNA ratio for Ctrl siRNA-transfected cells is indicated. (G) The expression of the Ca2+ transport proteins shown was assessed by Western blot analysis 48 h after transfection with Ctrl siRNA or miR-519. (H) Forty-eight hours after the transfection of HeLa cells with Ctrl siRNA or miR-519, cells were loaded with 20 μM Fluo-4 AM. Following stimulation with histamine (100 μM), confocal images were obtained every 20 s, quantified, and plotted (see Materials and Methods). Data in panels B and E represent the means and SD of data from 3 independent experiments. *, P < 0.05. Data in panel F are the averages of data from two experiments showing comparable results.
The repressive influence of miR-519 on the expression of proteins that control intracellular Ca2+ levels led us to hypothesize that miR-519 might affect cytosolic Ca2+ concentrations. To test this possibility, HeLa cells (transfected with either Ctrl siRNA or miR-519) were loaded with Fluo-4 AM and then stimulated with histamine (100 μM), an agonist of Gαq-coupled receptors, which led to the production of 1,4,5-trisphosphate (IP3) and rapidly increased cytosolic [Ca2+] concentrations. The robust and rapid rise in cytosolic Ca2+ concentrations returned to baseline levels after ∼2 min, but the time required for the decline of cytosolic [Ca2+] concentrations was significantly longer for the mRNA-519 group than for the Ctrl siRNA group (0.2 ± 0.03 compared to 0.27 ± 0.04 liter/min) (Fig. 6H). In sum, these results highlight several miR-519-repressed proteins that control intracellular Ca2+ levels, ATP2C1, ATP2B1, ATP2B4, and ORAI1, at least two of them by a putative direct interaction of miR-519 with their cognate mRNAs (ATP2C1 and ORAI1). In turn, miR-519 affects cytosolic calcium levels.
miR-519-influenced Ca2+ signaling impacts p21 expression.
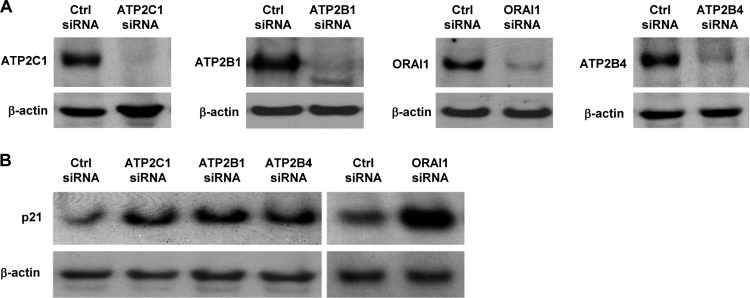
Since the increase in p21 expression levels by miR-519 was only partly dependent on p53 levels (Fig. 4C and D), we hypothesized that the miR-519-impaired Ca2+ levels could contribute to the increase in the abundance of p21. To investigate this possibility, ATP2C1, ATP2B1, ATP2B4, and ORAI1 were individually silenced by using siRNAs (Fig. 7A); under these conditions, p21 levels were constitutively upregulated (Fig. 7B), supporting the notion that Ca2+-regulated pathways did contribute to the increase in p21 expression levels by miR-519.
Fig 7.
miR-519 represses levels of Ca2+ transport proteins. Forty-eight hours after the individual silencing of ATP2C1, ATP2B1, ATP2B4, or ORAI1 (A), the levels of p21 were assessed by Western blot analysis (B). The data are representative of data from at least 3 independent experiments.
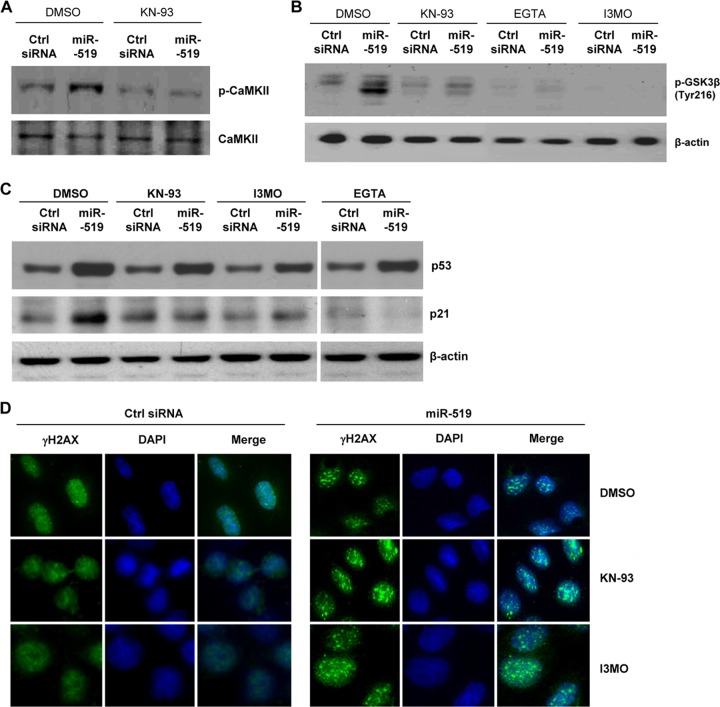
In response to heightened Ca2+ levels, a number of signaling pathways are activated in the cell. The role of Ca2+-activated protein kinase C (PKC) in the upregulation of both the transcription and stability of p21 mRNA is well established (for example, see references 8 and 44). We identified Ca2+-activated calmodulin kinase II (CaMKII) as well as glycogen synthase kinase 3β (GSK3β) as kinases responsible for the upregulation of p21. Treatments with KN-93 to block CaMKII phosphorylation (Fig. 8A), with indirubin-3′-monoxime (I3MO) to block GSK3β phosphorylation (Fig. 8B), and with EGTA to chelate Ca2+ each inhibited the miR-519-triggered increase in p21 levels (Fig. 8C). In support of the view that miR-519 elevated p21 levels via Ca2+-regulated events and not only by triggering DNA damage, the inhibition of CaMKII and GSK3β prevented p21 upregulation by miR-519 (Fig. 8C) but did not prevent DNA damage (Fig. 8D).
Fig 8.
Blocking of Ca2+ signaling inhibits miR-519-upregulated p21 but not miR-519-triggered DNA damage. (A) Western blot analysis of phosphorylated CaMKII (p-CaMKII) and total CaMKII 48 h after transfection of HeLa cells with Ctrl siRNA or miR-519 in the presence of KN-93 (6 μM). (B) The levels of phosphorylated GSK3β (p-GSK3β) at residue Tyr-216 (an inhibitory modification) were assessed by Western blot analysis 48 h after the transfection of HeLa cells with the small RNAs indicated, in the presence of dimethyl sulfoxide (DMSO), KN-93, EGTA, or I3MO (10 μM). (C and D) Forty-eight hours after the transfection of HeLa cells with Ctrl siRNA or miR-519 in the presence of either DMSO, KN-93, I3MO, 3-MA, or EGTA, the levels of p53, p21, and β-actin were assessed by Western blot analysis (C), and DNA damage was assessed by the visualization of γ-H2AX distribution patterns (D), as described in the legend of Fig. 3F. Data are representative of data from three independent experiments.
miR-519 triggers autophagy and elevates p21 levels in a Ca2+-dependent manner.
While studying the function of miR-519, we discovered that it caused the appearance of large cytoplasmic vacuoles (Fig. 9A). Since both DNA damage and Ca2+ can promote autophagy (10, 47), we asked if these vacuoles might be associated with enhanced cellular autophagy, a process of self-digestion of large cytoplasmic components (11, 35, 38, 45, 59).
Fig 9.
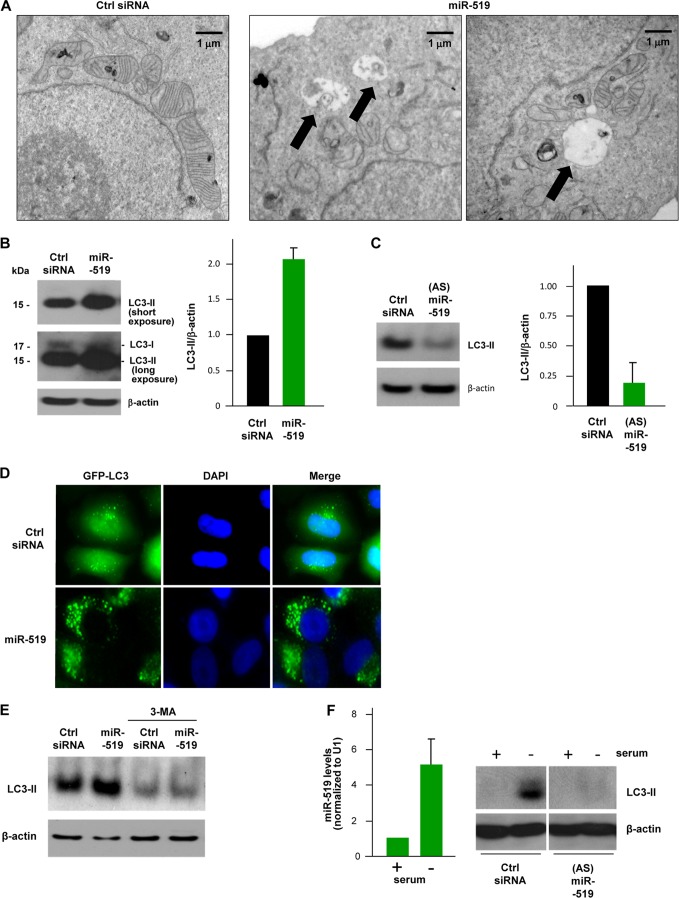
miR-519 triggers the formation of autophagic vesicles. (A) Forty-eight hours after the transfection of HeLa cells with either Ctrl siRNA or miR-519, electron microscopy was used to visualize the cytoplasm of HeLa cells. Arrows indicate autophagic vacuoles. (B) Western blot analysis of the autophagy marker LC3 in HeLa cells. Although two bands are seen after a long exposure of the blot, a strong ∼16-kDa LC3-II band and a faint ∼18-kDa LC3-I band, the predominant LC3-II band was visualized as a marker of autophagy throughout the study. The graph shows the quantification of the LC3-II intensity. (C) HeLa cells were transfected with a plasmid that expresses GFP-LC3, a fluorescent fusion protein that is recruited to autophagosomes, and with either Ctrl siRNA or miR-519. Forty-eight hours later, GFP-LC3 signals were visualized by fluorescence microscopy. DNA was stained with DAPI in order to visualize nuclei. (D) Western blot analysis (left) and quantification (right) of the levels of LC3-II in HeLa cells, as assessed by Western blot analysis 48 h after transfection with antisense miR-519 [(AS)miR-519] RNA. (E) HeLa cells were transfected with either Ctrl siRNA or miR-519 in the presence or absence of the autophagy inhibitor 3-MA (1 mM). Forty-eight hours later, Western blot analysis was used to assess LC3-II levels. (F) RT-qPCR detection of miR-519 levels and LC3-II detection by Western blot analysis of HeLa cells transfected with Ctrl siRNA or (AS)siRNA and then subjected to 72 h of serum starvation or normal culture. Data are representative of data from at least 3 independent experiments; in panels B, C, and F, graphs represent the means and SD from at least 3 independent experiments. β-Actin was included as a loading control.
Several lines of evidence were investigated in order to study this possibility. First, electron microscopy was used to visualize the cytoplasm of miR-519-expressing cells, where we observed numerous membrane-enclosed structures resembling autophagic vesicles (autophagosomes), which were absent in the Ctrl siRNA group (Fig. 9A). Second, we examined microtubule-associated protein 1 light chain 3 (LC3) (33); the conversion of LC3-I (18 kDa) to LC3-II (16 kDa) is broadly used to monitor autophagy, as the amount of LC3-II correlates with the number of autophagosomes. As assessed by Western blot analysis using an antibody that preferentially recognizes the processed form of LC3 (LC3-II), miR-519-transfected HeLa cells exhibited increased levels of the autophagy marker LC3-II compared with levels in Ctrl siRNA-transfected cells (Fig. 9B). Moreover, supporting the notion that miR-519 enhances autophagy, a reduction in the level of activity of endogenous miR-519 (which is readily expressed in HeLa cells) by transfection with an antisense transcript directed toward miR-519 [“antagomir” (AS)miR-519] lowered LC3-II signals (Fig. 9C). Third, since LC3-II remains on mature autophagosomes until after fusion with lysosomes, the pattern of intracellular LC3-II dots (or “puncta”) is commonly used to monitor autophagosomes (28). As shown in Fig. 9D, the transfection of HeLa cells with a plasmid that expresses a green fluorescent protein (GFP)-tagged LC3 (26) revealed a diffuse pattern of the GFP-LC3 signal in control cells; in contrast, miR-519 expression caused the accumulation of distinct GFP-LC3 puncta. Fourth, treatment with 3-methyladenine (3-MA), which blocks autophagosome formation by inhibiting type III phosphatidylinositol 3-kinases (PI3Ks), potently diminished LC3-II signals (Fig. 9E). In addition, while serum starvation of HeLa cells for 72 h increased autophagy, as determined by the monitoring of LC3-II levels, the lowering of miR-519 function by (AS)miR-519 blocked this increase (Fig. 9F).
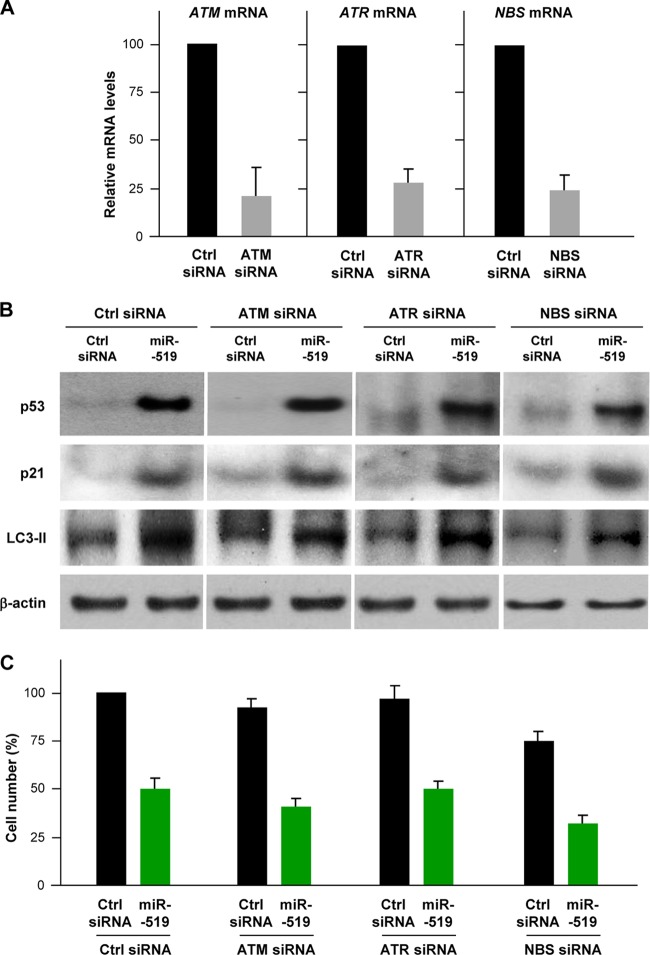
To investigate whether the effects of miR-519 on autophagy and p21 induction involved the DNA damage-activated kinases ATM (ataxia-telangiectasia mutated) or ATR (ATM related) or the DNA damage response complex MRN (Mre11/Rad50/Nbs), we silenced ATM, ATR, or NBS1 in HeLa cells (Fig. 10A). A Western blot analysis 48 h after siRNA transfection revealed that ATM, ATR, and NBS were dispensable for the miR-519-mediated upregulation of p53, p21, and LC3-II levels (Fig. 10B). Similarly, these genes did not appear to be necessary for the growth-inhibitory influence of miR-519 (Fig. 10C). Taken together, these data indicate that miR-519 enhances autophagy independently of DNA damage.
Fig 10.
ATM, ATR, and NBS are not required for the effects of miR-519. (A) Effect of transfection of HeLa cells with ATM, ATR, or NBS siRNAs on the expression of target mRNAs, as assessed by RT-qPCR analysis 48 h after transfection of the siRNAs. (B) In cells that were transfected, as explained above for panel A, together with either Ctrl siRNA or miR-519, the abundances of p53, p21, and LC3-II were assessed by Western blot analysis. Representative data from three independent experiments are shown. (C) The effect of miR-519 on the proliferation of cells with normal or reduced ATM, ATR, or NBS levels was assessed by monitoring changes in cell numbers 48 h after the transfection of small RNAs.
Central role of p21 in the phenotype triggered by miR-519.
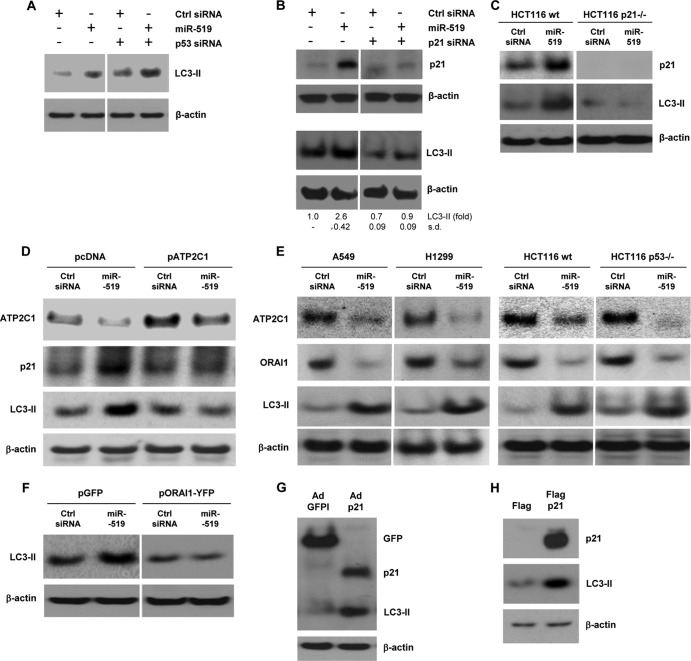
While p53 silencing did not prevent miR-519-triggered autophagy, p21 silencing did (Fig. 11A and B). Similarly, in HCT116 colon cancer cells bearing somatic deletions of both p21 alleles (HCT116 p21−/− cells), LC3-II levels were constitutively low and did not increase after the overexpression of miR-519 (Fig. 11C). The Ca2+-triggered elevation of p21 levels was linked to miR-519-enhanced autophagy, since the overexpression of ATP2C1 prevented the miR-519-mediated accumulation of p21 and LC3-II (Fig. 11D); in contrast, p53 did not appear to be necessary for the accumulation of LC3-II, since miR-519 elicited comparable increases in LC3-II levels in both p53-proficient and p53-deficient cells (A549 versus H1299 cells as well as HCT116 wt versus HCT116 p53−/− cells). Similar to the elevation of ATP2C1 levels (Fig. 11D), the overexpression of ORAI1 also blunted the miR-519-mediated increase in LC3-II levels (Fig. 11F). Conversely, the overexpression of p21 in colon carcinoma cells (RKO) using an adenovirus or in HeLa cells using a plasmid vector increased the levels of LC3-II (Fig. 11G and H). These results support the view that the miR-519-triggered increase in p21 levels promotes autophagy.
Fig 11.
p53 is dispensable and Ca2+-induced p21 is essential for miR-519-triggered autophagy. (A and B) HeLa cells were transfected with the small RNAs indicated, and 48 h later, the levels of LC3-II (A and B), p21 (B), and β-actin were assessed by Western blot analysis. p53 levels were reduced, as shown in Fig. 4C. (C) Parental (wt) and p21-null (p21−/−) HCT116 cells were transfected with Ctrl siRNA or miR-519. Forty-eight hours later, the p21 and LC3-II signals were assessed by Western blot analysis. (D) HeLa cells were transfected with Ctrl siRNA and miR-519 together with the parent vector (pcDNA) or pATP2C1. Forty-eight hours later, p21 and LC3-II levels were assessed by Western blot analysis. (E) Forty-eight hours after the transfection of A549, H1299, HCT116 wt, and HCT116 p53−/− cells with the small RNAs indicated, the levels of ATP2C1, ORAI1, LC3-II, and the loading control β-actin were assessed by Western blot analysis. (F) HeLa cells were transfected with Ctrl siRNA and miR-519 together with either the parent vector (pGFP) or pORAI1-GFP. Forty-eight hours later, LC3-II levels were assessed by Western blot analysis. (G) RKO (colon cancer) cells were infected with 100 PFU of either control AdGFP or Adp21, and 48 h later, the levels of GFP, p21, LC3-II, and β-actin were assessed by Western blot analysis. (H) HeLa cells were transfected with plasmid pFlag-p21 to overexpress tagged p21 or with the parent vector pFlag. Forty-eight hours later, the levels of p21, LC3-II, and β-actin were assessed by Western blot analysis. The data represent the means and SD of data from 3 independent experiments.
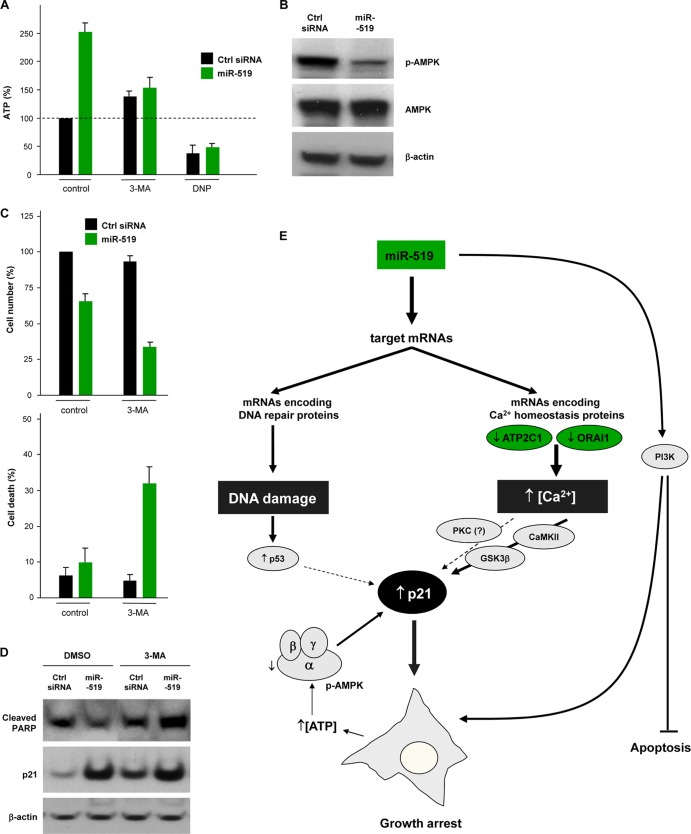
In agreement with the fact that autophagy can elevate the intracellular ATP concentration, ATP levels were higher in miR-519-transfected HeLa cells but not in cells pretreated with 3-MA; the ATP-depleting drug DNP was included as a control (Fig. 12A). miR-519 markedly lowered the activity of AMPK (AMP-activated protein kinase, an enzyme exquisitely sensitive to relative ATP/AMP levels), as revealed by the reduced phosphorylation of threonine 172 of the AMPK α subunit (Fig. 12B). The inhibition of AMPK likely contributed to the upregulation of p21, since the blocking of AMPK activity robustly induced p21 (54). According to the protective influence afforded by autophagy and p21, the ectopic increase of the miR-519 level was cytostatic and failed to increase cell death, as measured by trypan blue exclusion (Fig. 12C) or by measuring the cleavage of poly(ADP-ribose) polymerase (PARP), a marker of apoptosis. Importantly, however, the protective actions of miR-519 were abrogated by the use of the autophagy inhibitor 3-MA (Fig. 12D). In sum, by reducing the levels of production of specific subsets of proteins, miR-519 increases genotoxic damage and alters Ca2+ homeostasis; the ensuing elevation of p21 levels exerts a potent antiapoptotic influence, while the blocking of miR-519-triggered autophagy enhances cell death.
Fig 12.
miR-519-engendered growth arrest and survival are linked to p21 upregulation. (A) ATP levels in cells were quantified 48 h after transfection with Ctrl siRNA or miR-519 in the presence or absence of 3-MA. DNP was included as a negative control. (B) Western blot analysis of the levels of total AMPK and active AMPK (p-AMPK, AMPK phosphorylated at Thr-172 of subunit α). (C and D) The influence of miR-519 on cell numbers and survival was assessed 48 h after the transfection of Ctrl siRNA or miR-519 by measuring remaining cells (C, top) and trypan blue-positive (dead) cells (bottom) and by assessing PARP cleavage by Western blot analysis (D). (E) General model depicting the events whereby miR-519 triggers growth arrest by inducing p21. The molecules showing a direct implication in the miR-519-elicited increase in p21 expression levels are shown in green. The data in panels A and C are the means and SD of data from at least three independent experiments.
DISCUSSION
The finding that miR-519 potently triggered growth arrest prompted us to analyze the mediators of this phenotype. Several en masse approaches were undertaken, including the collective (microarray-based) identification of mRNAs associated with biotinylated miR-519 and mRNAs showing altered steady-state abundances after miR-519 overexpression, the bioinformatic analysis of miR-519 target mRNAs, and the high-throughput analysis of proteins (using SILAC) displaying changes in abundance after the overexpression miR-519. The search strategy described here was stringent, given the limitations of the experimental approach (e.g., some miR-519–mRNA interactions were not reflected in the array after the pulldown), the limited overlap among existing algorithms (e.g., TargetScan and miR databases do not identify an identical set of putative miR-519 target mRNAs), and the fact that algorithms do not predict all occurrences of miRNA-mRNA interactions (e.g., some miRNA-mRNA hybrids occur without interactions via miRNA “seed” sequences). Nonetheless, at the intersection of these searches were two main cellular pathways controlled by miR-519 target mRNAs: a pathway of DNA damage and a pathway of maintenance of intracellular calcium levels (Fig. 12E).
miR-519 causes genotoxic stress.
miR-519-triggered DNA damage was facilitated by the downregulation of miR-519 targets encoding DNA repair/maintenance proteins, such as DUT1, EXO1, RPA2, and POLE4, as well as numerous other mRNAs that were indirectly downregulated (encoding the proteins FANCA, FANCB, FANCG, FANCE, and BRCA1). Accordingly, miR-519-expressing cells displayed extensive DNA damage and increased expression levels of the DNA damage response protein p53 and its downstream effector p21, also a DNA damage-inducible protein (19). Indeed, p53 was not essential for miR-519 to upregulate p21, as miR-519 triggered comparable increases in p21 levels in p53-deficient cells (H1299 and HCT116 p53−/− cells) and p53-proficient cells (A549 and HCT116 wt cells) (Fig. 4D). Other DNA damage response proteins (ATM, ATR, and NBS) also appeared not to be essential for the effects of miR-519 on growth arrest or gene expression (Fig. 10C). While p53 can induce p21 levels transcriptionally, many other mechanisms can also upregulate p21 expression transcriptionally and posttranscriptionally (discussed below). A search for such alternative inducers of p21 in miR-519-transfected cells led us to discover a second miR-519-modulated pathway.
miR-519 perturbs intracellular calcium levels.
The miR-519-elicited change in intracellular calcium levels was mediated by the influence of miR-519 on proteins that affect calcium homeostasis, including ORAI1 and ATP2C1, ATP2B1, and ATP2B4. The modulation of intracellular calcium levels, particularly via changes in the abundances of ORAI1 and ATP2C1, led directly to the upregulation of p21 and promoted autophagy (Fig. 7, 8, and 11). The link between calcium and autophagy was first supported by evidence that autophagy could be provoked by endoplasmic reticulum (ER) stress (6, 40, 60). This association was later strengthened by evidence that calcium mobilization induced autophagy linked to the activation of calcium/calmodulin kinase kinase β (CamKKβ), an upstream kinase of AMPK, which in turn inhibits the target of rapamycin (mTOR), a negative regulator of autophagy (22, 23). Since then, there has been increasing recognition that calcium plays a role in stimulating autophagy and that these processes may be important for cell survival in different stress situations (21). Our studies have identified two miR-519 target proteins, ORAI1 and ATP2C1, which are essential for the maintenance of cytosolic calcium homeostasis. ATP2C1 is an ATPase localized on the Golgi membrane that pumps Ca2+ and Mn2+ from the cytosol into the lumen of secretory pathway vesicles; these vesicles carry proteins destined for secretion from the rough ER through the Golgi apparatus and finally fuse with the plasma membrane to release the proteins outside the cell. A deficiency in ATP2C1 activity reduces Ca2+ accumulation in the Golgi apparatus and ER and sensitizes cells to ER stress (46). In humans, the loss of one ATP2C1 allele results in Hailey-Hailey disease, also known as familial benign pemphigus (24, 52), characterized by the development of blistering skin erosions and attributed to the defective adhesion of keratinocytes.
ORAI1, a member of a family that also comprises ORAI2 and ORAI3, plays an essential role in store-operated Ca2+ entry (SOCE) and distributes uniformly on the plasma membrane, forming dimers in the inactive state (53). ORAI1 is a Ca2+ release-activated Ca2+ (CRAC) channel subunit, which mediates Ca2+ influx following the depletion of intracellular Ca2+ stores and channel activation by the Ca2+ sensor STIM (stromal interaction molecule). STIM activation induces the tetramerization of ORAI proteins and subsequent STIM-ORAI colocalization, which forms the active store-operated calcium channel. STIM-ORAI channels are the underlying molecular mechanism of SOCE, which allows rapid Ca2+ efflux from the ER after intracellular Ca2+ stores are emptied. Thus, ORAI1 links Ca2+ entry through plasma membrane store-operated Ca2+ channels and the ER Ca2+ pump (5). STIM-ORAI1 channels are important for T lymphocyte activation, and the loss of ORAI1 causes severe combined immunodeficiency (SCID) in humans (14).
Many pathways upregulate p21.
The expression of p21 is controlled by many different mechanisms (27). Besides well-studied transcription factors such as p53, AP1, Myc, Sp1/Sp3, and E-box-binding proteins (16, 19), numerous proteins can control the half-life and translation of p21 mRNA, including HuR, AUF1, calreticulin, hnRNP K, Musashi-1, CUGBP1, and NF-90 (reviewed in reference 61). Similarly, microRNAs such as miR-208 and the miR-17-92 cluster (58, 62) reduce p21 levels posttranscriptionally. The p21 protein is also subject to posttranslational control in a number of ways, including ubiquitin-proteasome-modulated p21 stability (55). In particular, Ca2+-regulated kinases like PKC can upregulate both the transcription and stability of p21 mRNA (8, 44), indicating additional ways whereby miR-519-triggered calcium upregulation can elevate p21 levels. Moreover, the miR-519-mediated reduction in AMPK levels could also directly influence p21 levels, since active AMPK promotes the importin-1α-mediated nuclear import of HuR, thus depleting from the cytoplasm a major posttranscriptional inducer of p21 mRNA stabilization and p21 accumulation in the cell (54, 56, 57).
p21 and miR-519 in cytoprotection, senescence, and tumor suppression.
The convergence of these processes on the upregulation of p21 is significant, given its role in the cellular response to various genotoxic and metabolic stresses. The overexpression of p21 is cytoprotective, rendering cells resistant to a variety of stress agents; this prosurvival influence was attributed in part to the inhibition of cell growth by p21, which might allow cells time to repair damage (15, 18). Increased p21 levels can also bring about cellular senescence, as does the elevation of miR-519 levels; likewise, the overexpression of either p21 or miR-519 can repress tumor growth (2, 12, 37, 39). Since cellular senescence represents an antitumor mechanism (30, 41), these two events could be functionally linked in the reduction of tumorigenesis. However, autophagy has also proven to be a prosurvival process (36, 48), suggesting that miR-519 and p21 could elicit their actions at least in part by triggering autophagy. Future studies are needed in order to delineate the events downstream of p21 that promote autophagy and how the absence of p21 impairs it (Fig. 11).
In conclusion, a single microRNA, miR-519, acting upon subsets of target mRNAs, can collectively influence the levels of functionally related proteins, DNA repair proteins and calcium homeostasis proteins, and hence affects the processes that these proteins govern (genotoxic stress and intracellular calcium concentrations). The spectrum of functions performed by these proteins can functionally converge on the regulation of major cellular processes like growth arrest, autophagy, and cytoprotection. Our study underscores the importance of systematically analyzing microRNA functions on a global scale and from different angles: identifying its endogenous targets, elucidating the mRNAs and proteins affected by the microRNA, and examining the cellular pathways affected by the target proteins. The collective information from these approaches can illuminate the complex, robust, and versatile manners in which microRNAs function to maintain cell homeostasis.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. J. Morin, B. Valle, W. H. Wood III, and S. S. Subaran (NIA) and A. M. Cuervo (Albert Einstein College of Medicine) for reagents and expertise.
This work was supported in part by the NIA-IRP, NIH.
Footnotes
Published ahead of print 30 April 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Abdelmohsen K, et al. 2010. miR-375 inhibits differentiation of neurites by lowering HuD levels. Mol. Cell. Biol. 30: 4197–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abdelmohsen K, et al. 2010. miR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle 9: 1354–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abdelmohsen K, et al. 2009. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 28: 1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M. 2008. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc. Natl. Acad. Sci. U. S. A. 105: 20297–20302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alonso MT, Manjarrés IM, García-Sancho JJ. 2012. Privileged coupling between Ca(2+) entry through plasma membrane store-operated Ca(2+) channels and the endoplasmic reticulum Ca(2+) pump. Mol. Cell. Endocrinol. 353: 37–44 [DOI] [PubMed] [Google Scholar]
- 6. Bernales S, McDonald KL, Walter P. 2006. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4: e423 doi:10.1371/journal.pbio.0040423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bueno MJ, Malumbres M. 2011. MicroRNAs and the cell cycle. Biochim. Biophys. Acta 1812: 592–601 [DOI] [PubMed] [Google Scholar]
- 8. Cerda SR, et al. 2006. Protein kinase C delta inhibits Caco-2 cell proliferation by selective changes in cell cycle and cell death regulators. Oncogene 25: 3123–3138 [DOI] [PubMed] [Google Scholar]
- 9. Chadwick W, Brenneman R, Martin B, Maudsley S. 2010. Complex and multidimensional lipid raft alterations in a murine model of Alzheimer's disease. Int. J. Alzheimers Dis. 2010: 604792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Decuypere JP, Bultynck G, Parys JB. 2011. A dual role for Ca(2+) in autophagy regulation. Cell Calcium 50: 242–250 [DOI] [PubMed] [Google Scholar]
- 11. Deretic V. 2011. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol. Rev. 240: 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fang L, et al. 1999. p21Waf1/Cip1/Sdi1 induces permanent growth arrest with markers of replicative senescence in human tumor cells lacking functional p53. Oncogene 18: 2789–2797 [DOI] [PubMed] [Google Scholar]
- 13. Farazi TA, Spitzer JI, Morozov P, Tuschl T. 2011. miRNAs in human cancer. J. Pathol. 223: 102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feske S, et al. 2006. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 441: 179–185 [DOI] [PubMed] [Google Scholar]
- 15. Garner E, Raj K. 2008. Protective mechanisms of p53-p21-pRb proteins against DNA damage-induced cell death. Cell Cycle 7: 277–282 [DOI] [PubMed] [Google Scholar]
- 16. Gartel AL, Radhakrishnan SK. 2005. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 65: 3980–3985 [DOI] [PubMed] [Google Scholar]
- 17. Gorospe M, Abdelmohsen K. 2011. MicroRegulators come of age in senescence. Trends Genet. 27: 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gorospe M, Wang X, Guyton KZ, Holbrook NJ. 1996. Protective role of p21(Waf1/Cip1) against prostaglandin A2-mediated apoptosis of human colorectal carcinoma cells. Mol. Cell. Biol. 16: 6654–6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gorospe M, Wang X, Holbrook NJ. 1999. Functional role of p21 during the cellular response to stress. Gene Expr. 7: 377–385 [PMC free article] [PubMed] [Google Scholar]
- 20. Han M, Toli J, Abdellatif M. 2011. MicroRNAs in the cardiovascular system. Curr. Opin. Cardiol. 26: 181–189 [DOI] [PubMed] [Google Scholar]
- 21. Harr MW, Distelhorst CW. 2010. Apoptosis and autophagy: decoding calcium signals that mediate life or death. Cold Spring Harb. Perspect. Biol. 2: a005579 doi:10.1101/cshperspect.a005579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Høyer-Hansen M, et al. 2007. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase beta, and Bcl-2. Mol. Cell 25: 193–205 [DOI] [PubMed] [Google Scholar]
- 23. Høyer-Hansen M, Jäättelä M. 2007. AMP-activated protein kinase: a universal regulator of autophagy? Autophagy 3: 381–383 [DOI] [PubMed] [Google Scholar]
- 24. Hu Z, et al. 2000. Mutations in ATP2C1, encoding a calcium pump, cause Hailey-Hailey disease. Nat. Genet. 24: 61–65 [DOI] [PubMed] [Google Scholar]
- 25. Huntzinger E, Izaurralde E. 2011. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12: 99–110 [DOI] [PubMed] [Google Scholar]
- 26. Jackson WT, et al. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3: e156 doi:10.1371/journal.pbio.0030156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jung YS, Qian Y, Chen X. 2010. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell. Signal. 22: 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kabeya Y, et al. 2004. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117: 2805–2812 [DOI] [PubMed] [Google Scholar]
- 29. Krol J, Loedige I, Filipowicz W. 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11: 597–610 [DOI] [PubMed] [Google Scholar]
- 30. Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. 2010. The essence of senescence. Genes Dev. 24: 2463–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lal A, et al. 2011. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 7: e1002363 doi:10.1371/journal.pgen.1002363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leung AK, Sharp PA. 2010. MicroRNA functions in stress responses. Mol. Cell 40: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mann SS, Hammarback JA. 1994. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J. Biol. Chem. 269: 11492–11497 [PubMed] [Google Scholar]
- 34. Marasa BS, et al. 2010. MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging 2: 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mathew R, White E. 2011. Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr. Opin. Genet. Dev. 21: 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mathew R, Karantza-Wadsworth V, White E. 2007. Role of autophagy in cancer. Nat. Rev. Cancer 7: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McConnell BB, Starborg M, Brookes S, Peters G. 1998. Inhibitors of cyclin-dependent kinases induce features of replicative senescence in early passage human diploid fibroblasts. Curr. Biol. 8: 351–354 [DOI] [PubMed] [Google Scholar]
- 38. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. 2008. Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Noda A, Ning Y, Venable SF, Pereira-Smith OM, Smith JR. 1994. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp. Cell Res. 211: 90–98 [DOI] [PubMed] [Google Scholar]
- 40. Ogata M, et al. 2006. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 26: 9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ohtani N, Mann DJ, Hara E. 2009. Cellular senescence: its role in tumor suppression and aging. Cancer Sci. 100: 792–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ong SE, Mann M. 2006. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1: 2650–2660 [DOI] [PubMed] [Google Scholar]
- 43. Park SS, Maudsley S. 2011. Discontinuous pH gradient-mediated separation of TiO2-enriched phosphopeptides. Anal. Biochem. 409: 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park JW, Jang MA, Lee YH, Passaniti A, Kwon TK. 2001. p53-independent elevation of p21 expression by PMA results from PKC-mediated mRNA stabilization. Biochem. Biophys. Res. Commun. 280: 244–248 [DOI] [PubMed] [Google Scholar]
- 45. Rabinowitz JD, White E. 2010. Autophagy and metabolism. Science 330: 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ramos-Castañeda J, et al. 2005. Deficiency of ATP2C1, a Golgi ion pump, induces secretory pathway defects in endoplasmic reticulum (ER)-associated degradation and sensitivity to ER stress. J. Biol. Chem. 280: 9467–9473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rodriguez-Rocha H, Garcia-Garcia A, Panayiotidis MI, Franco R. 2011. DNA damage and autophagy. Mutat. Res. 711: 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rosenfeldt MT, Ryan KM. 2011. The multiple roles of autophagy in cancer. Carcinogenesis 32: 955–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sayed D, Abdellatif M. 2011. MicroRNAs in development and disease. Physiol. Rev. 91: 827–887 [DOI] [PubMed] [Google Scholar]
- 50. Sonntag KC. 2010. MicroRNAs and deregulated gene expression networks in neurodegeneration. Brain Res. 1338: 48–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Srikantan S, et al. 2011. Translational control of TOP2A influences doxorubicin efficacy. Mol. Cell. Biol. 31: 3790–3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vanoevelen J, Dode L, Raeymaekers L, Wuytack F, Missiaen L. 2007. Diseases involving the Golgi calcium pump. Subcell. Biochem. 45: 385–404 [DOI] [PubMed] [Google Scholar]
- 53. Vig M, et al. 2006. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 312: 1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang W, et al. 2002. AMP-activated kinase regulates cytoplasmic HuR. Mol. Cell. Biol. 22: 3425–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang W, Nacusi L, Sheaff RJ, Liu X. 2005. Ubiquitination of p21Cip1/WAF1 by SCFSkp2: substrate requirement and ubiquitination site selection. Biochemistry 44: 14553–14564 [DOI] [PubMed] [Google Scholar]
- 56. Wang W, et al. 2004. AMP-activated protein kinase-regulated phosphorylation and acetylation of importin alpha1: involvement in the nuclear import of RNA-binding protein HuR. J. Biol. Chem. 279: 48376–48388 [DOI] [PubMed] [Google Scholar]
- 57. Wang W, Yang X, López de Silanes I, Carling D, Gorospe M. 2003. Increased AMP:ATP ratio and AMP-activated protein kinase activity during cellular senescence linked to reduced HuR function. J. Biol. Chem. 278: 27016–27023 [DOI] [PubMed] [Google Scholar]
- 58. Wong P, et al. 2010. The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. 70: 3833–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang Z, Klionsky DJ. 2010. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yorimitsu T, Nair U, Yang X, Klionsky DJ. 2006. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 281: 30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang J, Chen X. 2008. Posttranscriptional regulation of p53 and its targets by RNA-binding proteins. Curr. Mol. Med. 8: 845–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang Y, et al. 2011. Insulin promotes vascular smooth muscle cell proliferation via microRNA-208-mediated downregulation of p21. J. Hypertens. 29: 1560–1568 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.