Abstract
For a range of eukaryote transcripts, the initiation of degradation is coincident with the addition of a short pyrimidine tag at the 3′ end. Previously, cytoplasmic mRNA tagging has been observed for human and fungal transcripts. We now report that Arabidopsis thaliana mRNA is subject to 3′ tagging with U and C nucleotides, as in Aspergillus nidulans. Mutations that disrupt tagging, including A. nidulans cutA and a newly characterized gene, cutB, retard transcript degradation. Importantly, nonsense-mediated decay (NMD), a major checkpoint for transcript fidelity, elicits 3′ tagging of transcripts containing a premature termination codon (PTC). Although PTC-induced transcript degradation does not require 3′ tagging, subsequent dissociation of mRNA from ribosomes is retarded in tagging mutants. Additionally, tagging of wild-type and NMD-inducing transcripts is greatly reduced in strains lacking Upf1, a conserved NMD factor also required for human histone mRNA tagging. We argue that PTC-induced translational termination differs fundamentally from normal termination in polyadenylated transcripts, as it leads to transcript degradation and prevents rather than facilitates further translation. Furthermore, transcript deadenylation and the consequent dissociation of poly(A) binding protein will result in PTC-like termination events which recruit Upf1, resulting in mRNA 3′ tagging, ribosome clearance, and transcript degradation.
INTRODUCTION
Modulation of mRNA function within the cytoplasm is critical to the control of gene expression, function being determined primarily by the balance between transcript degradation and translation. Two cotranscriptional modifications, the 5′ cap and the 3′ poly(A) tail, are essential to both mRNA transcript stability and translation (15, 81, 83), and integral to this is their association mediated by an array of proteins which modulate function (6, 71).
The poly(A) tail, a homopolymeric sequence at the 3′ end of transcripts, is added by the canonical poly(A) polymerase in the nucleus. Within the cytoplasm, controlled deadenylation of this tail to ∼A15 in fungi and ∼A15 to A25 in mammals generally precedes decapping and subsequent 5′-3′ and/or exosome-dependent 3′-5′ decay (17, 20, 60, 64, 66). In specific instances, e.g., during embryonic development, deadenylated transcripts remain translationally dormant and can be reactivated by cytoplasmic adenylation (45). Deadenylation is a key control point through which rapid physiological change can occur. However, the mechanisms that underlie this switch between translation and mRNA turnover remain poorly understood.
It has recently been discovered that poly(A) tails in Aspergillus nidulans and Schizosaccharomyces pombe are modified in the cytoplasm with the addition of nontemplated U or C/U-rich tags at the point of mRNA decapping, and this increases the rate of transcript degradation (60, 76). Human H2a and H3.3 mRNAs, which are not polyadenylated, also undergo 3′ uridylation prior to decapping and degradation (67). This 3′ tagging of mRNA is conducted by noncanonical ribonucleotidyltransferases (5, 60, 76, 77). Generally, these enzymes do not include recognizable RNA-binding motifs and may therefore operate within complexes, although the human PAPD5, a noncanonical poly(A) polymerase, contains an unusual RNA-binding motif (75). This class of enzymes seems to be conserved throughout eukaryotes, with Saccharomyces cerevisiae being an apparent exception (76). In animals and plants, short noncoding RNAs (microRNA [miRNA], small interfering RNA [siRNA], and Piwi-interacting RNA [piRNA]) and the products of miRNA-directed degradation are also modified with U and A, resulting in either stabilization (36, 44, 52, 89) or destabilization (1, 30, 31, 37, 43, 50, 80, 93) of the RNA molecules. Such tagging can also influence the function of noncoding RNA (41, 42, 56, 78), RNA 3′ tagging being an important means to control both maturation and function (68, 97). However, it is not yet known whether polyadenylated transcripts of higher eukaryotes are subject to 3′ tagging.
The role of cytoplasmic mRNA 3′ tagging appears to run against the accepted paradigm, in which addition of a poly(A) tail protects mRNA from degradation and promotes translation. However, within the nucleus the addition of short (oligonucleotide) A tracts is associated with RNA degradation (95), as it is in prokaryotes (51, 59, 73). The addition of U nucleotides increases the affinity of the Lsm-Pat1 complex for the modified RNA (12, 84). Once loaded onto the RNA, this complex both represses translation and promotes transcript decapping and degradation, in an intricate process involving multiple factors, including decapping, surveillance, and translation factors. Consistent with this model, 3′ pyrimidine tagging occurs concurrently with the initiation of mRNA degradation, and disruption of tagging leads to retarded transcript decay (60, 67, 76), although the effect on degradation rates varies widely between transcripts. In A. nidulans, mRNA tagging occurs predominantly when the poly(A) tail is degraded to ∼15 nucleotides, which is precisely the point at which decapping is triggered (60) and supports the hypothesis that pyrimidine tagging earmarks the transcript for degradation. Interestingly, deletion of a single nucleotidyltransferase does not completely abolish tagging (60, 76, 77). This implies that more than one nucleotidyltransferase may be involved in the modification of specific transcripts.
Previously, we demonstrated that disruption of the ribonucleotidyltransferase CutA led to a dramatic reduction in mRNA 3′ tagging and retarded transcript degradation in A. nidulans (60, 61). To further characterize the molecular mechanism, we deleted a second putative ribonucleotidyltransferase coding gene, cutB. The mutant phenotype was similar to that of ΔcutA, and in the ΔcutA ΔcutB double mutant, no 3′ tagging was observed. To assess the possible role of mRNA 3′ tagging in other mRNA degradation processes, we investigated nonsense-mediated decay (NMD). Although NMD was shown to induce a high frequency of 3′ tagging, this was not required for NMD-induced transcript degradation. However, we observed that pyrimidine tagging is associated with NMD-induced poly(A)-independent decapping, and importantly, it contributes to the dissociation of transcripts with premature termination codons (PTC) from ribosomes; mutations disrupting either CutA or CutB result in a significantly increased proportion of PTC-containing transcripts associated with ribosomes. Finally, we demonstrated that mRNA 3′ pyrimidine tagging occurs in plants. These data are consistent with mRNA 3′ pyrimidine tagging being a conserved eukaryotic modification associated with mRNA degradation and dissociation from ribosomes. We propose that the deadenylation of transcripts, and the consequent dissociation of poly(A) binding protein (PAB1), will result in an NMD-like translational termination event. Unlike normal termination, where translational reinitiation is facilitated, this would provide a quality control mechanism which through 3′ tagging promotes transcript degradation and ribosome clearance, thus preventing further translation.
MATERIALS AND METHODS
A. nidulans strains and genetic techniques.
A. nidulans markers and the genetic techniques used have been described previously (13, 14). Growth media were as described by Cove (18). The ΔcutB and Δupf1 strains were constructed in A1149 (pyrG89 pyroA4 nkuA::argB) by direct transformation of recombinant PCR constructs utilizing Aspergillus fumigatus pyrG as the selectable marker, as described in reference 86. The fidelity of the knockout strains was confirmed by PCR and Southern hybridization. The cutA point mutations (N310D, D95A, and D97A) were produced by fusion PCR using overlapping oligonucleotides incorporating the appropriate sequence changes. The oligonucleotides used, with the mutant nucleotides indicated (underlined), were N95AFor, CTAGCGACTCGGCTGGTGTGTATTTCTTTGGGC; N95ARev, GAAATACACACCAGCCGAGTCGCTAGAACACAG; N97AFor, GAGACAGTCGCCATTTGTATAACTACAACCTG; N97ARev, GTTATACAAATGGCGACTGTCTCGGGTCAGTAAG; N310DFor, AAGAGCCTTTCGACACAATACGAAATCTGGGT; and N310DRev, TCGTATTGTGTCGAAAGGCTCTTCCACGCAAAG.
The mutant allele was introduced into the original ΔcutA strain by replacing the pyrG cassette integrated at the cutA locus (60). Transformants were selected as being 5-fluoroorotic acid resistant (62). The strains assayed were constructed by standard genetic crosses and had the following genotypes: nmdA1 pantoB100, nmdA1 uaZ14 pantoB100, uaZ14 pantoB100, ΔcutA riboB2 pabaA1, ΔcutB pyroA4, ΔcutA ΔcutB pyroA4, Δupf1, Δupf1 uaZ14 pyroA4, hxA5 pantoB100, nmdA1 hxA5 pantoB100, ΔcutA hxA5 pabaA1, ΔcutB hxA5, ΔcutA ΔcutB hxA5 pabaA1, cutA-N310D pyroA4, and cutA-N310D uaZ14.
Transcript analysis.
Growth of mycelia, RNA preparation, and quantitative Northern analysis were as described previously (11, 62). Briefly, overnight cultures were incubated with ammonium as the nitrogen source, washed, and transferred to nitrogen-free medium for 1 h prior to sampling. For Northern analysis, proflavin was added to the cultures to inhibit transcription, 10 min prior to the first sample being taken (60). Circularized reverse transcriptase PCR (cRT-PCR) of gdhA, uaZ, and CCR2 mRNA was conducted according to the method in reference 60. For the analysis of naturally decapped transcripts, there was no pretreatment of RNA. For total (capped and uncapped) transcripts, the RNA was pretreated with tobacco acid pyrophosphatase (TAP). For uaZ, first-strand cDNA synthesis utilized oligonucleotide uaZ_5′RT (CGGTCATTTCAGTGACGG), and subsequent PCR utilized a second oligonucleotide, uaZ_3′For (CAGACAAACCCTAACGGA). For CCR2, the equivalent oligonucleotides were CCR2_rev1_RT (TGAAGGTGACGAATCCGA) and For CCR2 (TTACGGAGGAAGCGGTGGT), and a second round of PCR was then conducted with nested primers (TCCCCTTGATCTTCCAGTC and AGGATGGTAATTCCTTTA). The PCR products were cloned into pGEM-T Easy (Promega), and random clones were selected and sequenced. The nonnormal distributions of poly(A) tail lengths were analyzed using the nonparametric Mann-Whitney test and the Gini coefficient (StatsDirect) for significance.
Polysome analysis.
Ribosomal fractionation in both the presence and the absence of 10 mM EDTA was based on the method of Arava et al. (4). The cultures were grown overnight at 30°C, as described for transcript analysis. Ten minutes prior to harvesting, cycloheximide was added at a final concentration of 0.1 mg ml−1 to trap elongating ribosomes. Harvested mycelia were washed with 20 mM Tris-HCl, pH 8.0, 140 mM KCl, 1.5 mM MgCl2, 0.1 mg ml−1 cycloheximide, and 0.1 mM dithiothreitol (DTT); frozen in liquid nitrogen; and stored for up to 24 h at −80°C. Approximately 0.25 g mycelia was ground in liquid nitrogen with a mortar and pestle, and the powder was resuspended in 1 ml ice-cold polysome extraction buffer (20 mM Tris-HCl, pH 8.0, 140 mM KCl, 1.5 mM MgCl2, 0.1 mg ml−1 cycloheximide, 1% Triton X-100, 1 mg ml−1 heparin, and 0.5 mM DTT). Excess cell debris was removed by sedimentation at 4,000 × g for 5 min at 4°C. The supernatant was clarified by further centrifugation (8,000 × g, 5 min, 4°C). Eight hundred micrograms of each RNA sample (optical density [OD] of 20 at A254) was loaded onto a sucrose gradient (10 ml, 15 to 50% [wt/vol]; Sorvall OTD-Combi ultracentrifuge, Beckman tubes, catalog no. 331372) and sedimented at 37,000 rpm at 4°C in an RPS40T rotor for 170 min. Sucrose solutions were prepared in polysome extraction buffer lacking Triton. The gradient was fractionated with the BioLogic LP system at a flow speed of 0.8 ml min−1 (Bio-Rad). Fractions of 1.1 ml were collected from the bottom of the gradient directly into 1 ml 6 M guanidine hydrochloride. RNA was precipitated by adding 2 volumes of 100% ethanol, redissolved in H2O, and treated with phenol-chloroform. The RNA was again precipitated by addition of 10% 3 M sodium acetate (pH 5.2) and 2.5 volumes of 100% ethanol and redissolved in 10 mM HEPES-KOH buffer, pH 7.0, containing SDS (2%) and EDTA (5 mM). The resulting samples were subjected to Northern analysis.
Western analysis.
Whole lysates were prepared as described in reference 96. Total protein extracts were loaded into each lane, fractionated by SDS-PAGE, and then electroblotted to Hybond ECL for 4 h under conditions recommended by the manufacturer (GE Healthcare). Specific primary polyclonal antibody against the N-terminal sequence of UaZ (positions 16 to 30, VYKVHKDPKTGVQTV) was obtained (Eurogentec). Binding and washing conditions for antibodies were as described in reference 96. The washing solution after binding with horseradish peroxidase (HRP)-conjugated secondary antibody contained 0.1% Tween 20 to increase the specificity of the analysis using the enhanced chemiluminescence kit from GE Healthcare. Rabbit β-actin antibody (A2066; Sigma) was used as a loading control.
Growth and analysis of Arabidopsis.
Growth and analysis of Arabidopsis were conducted using the Columbia (Col-0) strain. Seeds were surface sterilized as described previously (26). Once sterilized, the seed was kept at 4°C in the dark for 2 to 4 days and then grown at 22°C in 12-h-light–12-h-dark (12L:12D) cycles of white light (80 μmol m−2 s−1) on Murashige and Skoog (MS) medium containing 3% sucrose and 2% agar. Plants were entrained for 7 days before transfer to constant blue light at 12°C. From 72 h to 96 h of constant conditions, seedlings were harvested every 4 h and frozen immediately in liquid nitrogen. RNeasy plant minikits (Qiagen, Crawley, United Kingdom) were used to extract total RNA from each sample according to the manufacturer's instructions. This was done for two biological repeats. Using the QuantiTect reverse transcription kit (Qiagen, Crawley, United Kingdom), cDNA was produced from 1 μg total RNA. A liquid-handling robot (Freedom Evo; Tecan, Reading, United Kingdom) was used to set up the quantitative reverse transcription-PCR (qRT-PCR) mixes. The 10-μl reaction mixture contained 1 μl cDNA (1/5 dilution), 5 μl LightCycler 480 SYBR green master mix (Roche), and 0.3 μM forward and reverse primers (CCR2 F, GATGACAGAGCTCTTGAGAC; CCR2 R, CCTTGATCTTCCAGTCTCAC; UBQ10 F, CACACTCCACTTGGTCTTGCGT; UBQ10 R, TGGTCTTTCCGGTGAGAGAGTCTT). The LightCycler 480 system (Roche, Burgess Hill, United Kingdom) was used to measure CCR2 transcript levels normalized to UBQ10. Each cDNA was assayed in triplicate. Data were analyzed using the Roche LightCycler 480SW 1.5 software with the second derivative maximum method.
RESULTS
Identification of a second nucleotidyltransferase involved in mRNA tagging, CutB.
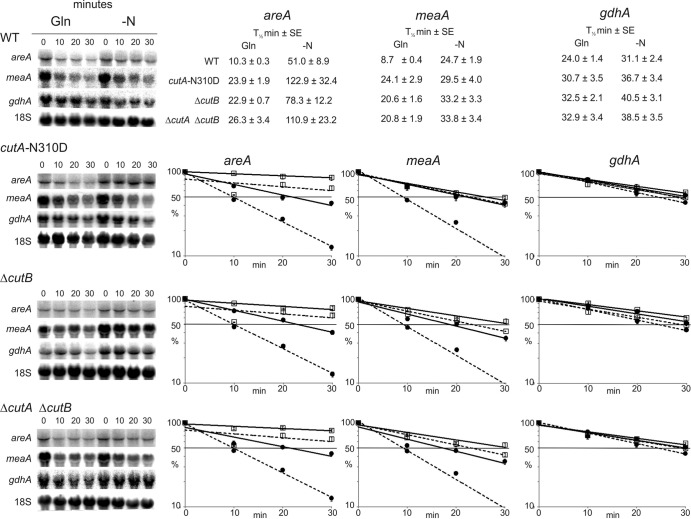
We have previously demonstrated that CutA was important for mRNA 3′ tagging with C and U nucleotides in A. nidulans. However, a residual level of tagging still occurred in a ΔcutA strain (60). Interestingly, the residual activity had a uridine bias. In order to identify the enzyme responsible, we deleted AN5694, which we had identified as encoding a putative poly(A) RNA polymerase, based on its phylogenetic relationship to S. cerevisiae Pap2 and S. pombe Cid14 (60). The gene, designated cutB, was deleted, and the mutant strain was characterized on the basis of the RNA degradation profile for a range of transcripts using quantitative Northern analysis (Fig. 1). Additionally, we undertook a circularization RT-PCR (cRT-PCR) analysis of the gdhA (Fig. 2) and uaZ (Fig. 3) transcripts. This technique allows the mRNA 3′ and 5′ ends to be sequenced simultaneously, thus defining poly(A) tail length, the extent of any degradation, and the sequence of nontemplated tags. From these data, it is clear that deletion of cutB leads to significant retardation of mRNA degradation for a number of transcripts (Fig. 1) and that this phenotype correlates with reduced 3′ tagging (Fig. 2 and 3). This phenotype is very similar to that of the ΔcutA mutant (60).
Fig 1.
Deletion of cutB retards the degradation of several transcripts. Northern blot analysis of areA (encoding a global transcription factor mediating nitrogen regulation), meaA (encoding a high-affinity ammonium transporter), and gdhA (encoding NADP-linked glutamate dehydrogenase) is shown. Quantitative data from multiple (≥3) experiments are presented graphically, comparing the wild type (WT) (dashed line) with mutant strains (solid line) deleted for cutB or both cutA and cutB, or bearing the point mutation cutA-N310D, as indicated. Transcript levels were monitored under conditions of nitrogen sufficiency (Gln) (□) or nitrogen starvation (−N) (●) over a 30-min time course after transcription was inhibited, to monitor degradation rates. These two nitrogen regimes were chosen as both areA and meaA transcript stability is differentially regulated under these conditions (11). 18S rRNA was used as a loading control. The calculated half-lives (min) derived by regression analysis are also presented (± standard error [SE]).
Fig 2.
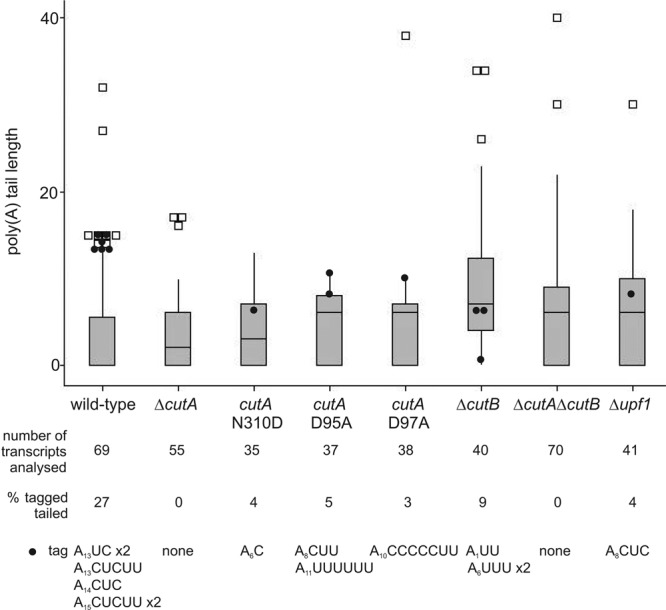
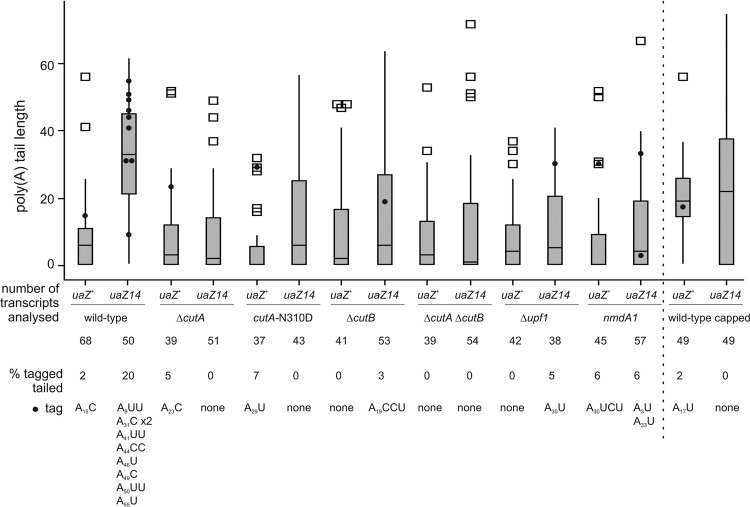
cRT-PCR analysis of naturally decapped gdhA mRNA. Poly(A) tail length was determined by cRT-PCR and sequencing of RNA samples derived from the wild-type, ΔcutA (60), cutA-N310D, cutA-D95A, cutA-D97A, ΔcutB, ΔcutA ΔcutB, and Δupf1 strains. The distribution of the poly(A) tail lengths is displayed using a box plot, where the top and bottom of the box represent limits of the upper and lower quartiles, with the median being indicated by the horizontal line which lies within the box. For the wild type, the majority of decapped transcripts were fully deadenylated, and the median poly(A) tail length is therefore zero. The whiskers show the highest and lowest readings within 1.5 times the interquartile range. All the outliers (□) represent 2 to 12% of transcripts with the longest poly(A) tails analyzed in each strain. Projected onto this is the distribution of transcripts that include 3′ pyrimidine tags on the poly(A) tail (●). The percentage (%) of adenylated transcripts which have a 3′ tag is given. The sequences of these tag modifications and their respective poly(A) tail lengths are shown. In the cutA-D97A strain, one deadenylated transcript was modified with the addition of a single C nucleotide. These data are derived from three separate experiments, and sequences were obtained by cRT-PCR of 35 to 70 transcripts from each strain.
Fig 3.
cRT-PCR analysis of uaZ+ and uaZ14 mRNAs. Poly(A) tail length of naturally decapped transcripts of uaZ+ (encoding urate oxidase) and uaZ14 (containing an ochre mutation terminating the UaZ protein prematurely after residue 131 in exon 2) was determined by cRT-PCR and sequencing. Capped transcripts were also identified for uaZ+ and uaZ14 in the wild-type background. For this, RNA was treated with alkaline phosphatase prior to decapping with tobacco acid pyrophosphatase. RNA samples were derived from the wild-type, ΔcutA, cutA-N310D, ΔcutB, ΔcutA ΔcutB, Δupf1, and nmdA1 strains. The distribution of the poly(A) tail lengths is shown as in Fig. 2. Sequences were obtained by cRT-PCR of 37 to 68 transcripts from each strain. ΔcutA uaZ14 and Δupf1 uaZ+ did not show 3′ tagging of adenylated transcripts but revealed modification to a degraded (deadenylated) transcript for ΔcutA uaZ14 with one C modification and to two transcripts for Δupf1 uaZ+ with a single C and a single U, respectively. Calculation of Gini coefficients of inequality for the distributions of tail lengths in all strains indicated a significant difference between wild-type uaZ14 (0.30; standard error, 0.040) and those of all other strains (e.g., wild-type uaZ [0.57; standard error, 0.043]). Gini coefficients for all other strains lie between 0.59 ± 0.05 and 0.79 ± 0.05.
To test whether CutA and CutB are together responsible for all 3′ tagging, the ΔcutA ΔcutB double mutant was constructed. This showed a phenotype similar to those of the single mutants with respect to mRNA degradation (Fig. 1) (60). Consistent with these two enzymes being responsible for all the mRNA 3′ tagging in A. nidulans, no modifications of gdhA or uaZ mRNA were observed by cRT-PCR analysis (Fig. 2 and 3). Furthermore, in both ΔcutA and ΔcutB mutants the frequency of uaZ14 tagging is reduced by >85% compared to the wild type, which is consistent with the enzymes acting together in vivo.
To investigate the role of the putative catalytic domain of CutA, mutations were designed based on the most closely related crystal structures. Mutations D95A and D97A were chosen to eliminate key interactions with catalytically essential bound metal: the corresponding changes abolish activity in a Trypanosoma brucei enzyme (85). A third cutA mutation, N310D, was designed to change nucleotide specificity according to present hypotheses by which the polarity of the water-mediated hydrogen bonding network at the catalytic site contributes to substrate selection (85). All three mutations significantly reduced the frequency of gdhA mRNA 3′ tagging (Fig. 2). The cutA-N310D allele, which was selected for further analysis, was also shown to result in reduced 3′ tagging of the uaZ transcript and generally retarded transcript degradation (Fig. 1 and 3).
3′ tagging is induced during nonsense-mediated decay.
Previously, mRNA 3′ tagging has been linked to mRNA degradation, being coincident with decapping and the initiation of transcript degradation (60, 76). NMD is a well-conserved degradation process, which rapidly eliminates transcripts containing a PTC, thus preventing accumulation of truncated proteins (69). Previously, we have shown that uaZ14, a mutant allele of the urate oxidase-encoding gene, is subject to NMD and that this is dependent on the fidelity of NmdA/Upf2 (63). To determine whether 3′ tagging is associated with NMD, we undertook cRT-PCR analysis of both uaZ+ and uaZ14 transcripts (Fig. 3). The proportion of the adenylated uaZ+ transcripts which were tagged in the wild-type background is very low (2%) compared to gdhA+ (27%), but it is not possible to determine if this is due to the frequency of tagging or the subsequent removal of tags. From Fig. 3, it can be seen that uaZ14 mRNA is subject to a significantly higher frequency of tagging than is the wild-type transcript and that the majority of tagging is associated with relatively long poly(A) tails (>30 nucleotides). The poly(A) tail length of natively decapped uaZ14 transcripts is also significantly greater than that in the wild type (Gini coefficient; Fig. 3 legend). These data indicate that the presence of a PTC induces both tagging and decapping independently of poly(A) tail shortening in the wild-type background. In the cutA-N310D, cutA-D95A, cutA-D97A, ΔcutA, ΔcutB, or ΔcutA ΔcutB double mutant strain, little or no modification occurred for uaZ+ or uaZ14 mRNA, consistent with both CutA and CutB being required and, with respect to the cutA-N310D strain, specifically implicating the catalytic activity of CutA. Furthermore, the decapping of the uaZ14 transcript prior to poly(A) shortening is lost in these mutants; the median poly(A) tail length of decapped uaZ14 mRNA in cutA and cutB mutants, like that of Δupf1 and nmdA1 strains, being statistically indistinguishable from that of uaZ+ in the wild-type background (Mann-Whitney test, >0.05; Fig. 3). With respect to capped transcripts, the median poly(A) tail lengths of uaZ+ and uaZ14, in a wild-type background, are not significantly different (Mann-Whitney test, >0.05) and the frequency of tagging observed is very low for both. These data are consistent with mRNA 3′ tagging directly promoting mRNA decapping.
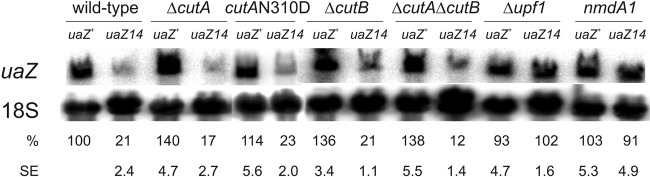
To determine whether mRNA 3′ tagging is essential for NMD, we undertook Northern hybridization analysis to compare uaZ+ and uaZ14 transcript levels in cutA and/or cutB mutant strains. From these data (Fig. 4), it is clear that neither CutA nor CutB is required for NMD, since the uaZ14 transcript is not stabilized by disruption of either gene. These observations were confirmed through failure to suppress NMD in a second transcript, hxA5 (see Fig. S1 in the supplemental material) (63).
Fig 4.
Disruption of mRNA tagging does not suppress NMD. Northern blot analysis was conducted to monitor the level of uaZ+ and uaZ14 transcripts in different genetic backgrounds: wild type, ΔcutA, cutA-N310D, ΔcutB, ΔcutA ΔcutB, Δupf1, and nmdA1. The strains were grown overnight for 16 h in the presence of ammonium as the nitrogen source. The overnight cultures were washed and transferred to media containing uric acid (10 mM) for 2 h to induce expression of uaZ. 18S rRNA was used as a loading control. Multiple Northern blots (≥3) were quantified, and the average level of expression was indicated for each strain (%) with the standard error (SE).
To establish whether the high frequency of tagging observed for uaZ14 is induced by NMD, we investigated two highly conserved components of the NMD machinery, Upf1 and NmdA/Upf2 (54). We have already demonstrated that the nmdA1 mutation, which disrupts nmdA, leads to suppression of NMD (63) (Fig. 4), and we exploited the A. nidulans genome sequence (23) to locate and then delete the UPF1 ortholog (AN0646). Northern analysis confirmed that, as expected, deletion of upf1 suppressed NMD with respect to uaZ14 (Fig. 4). Both nmdA1 and Δupf1 strains were subsequently utilized in cRT-PCR analysis of uaZ+ and uaZ14 transcripts, to determine if the enhanced rate of mRNA 3′ tagging induced by a PTC is dependent on NmdA and Upf1 (Fig. 3). From these data, it is clear that that the high frequency of uaZ14 mRNA 3′ tagging is suppressed by both nmdA1 and Δupf1, consistent with PTC-activated tagging being induced as part of the NMD response. Interestingly, the frequency of tagging of the decapped gdhA+ transcripts is also significantly lower in the Δupf1 strain (Fig. 2), suggesting that Upf1 may play a role in this process that extends beyond NMD and the degradation of nonadenylated transcripts such as human histone mRNA (67).
mRNA 3′ tagging is required for efficient dissociation of mRNA from ribosomes after premature termination.
Previously, we postulated that mRNA 3′ tagging may be associated with the transfer of mRNA away from ribosomes prior to the formation of processing (P) bodies and degradation, P bodies being absent from the ΔcutA strain (61). To determine whether CutA and CutB are implicated in ribosome dissociation, we undertook Northern analysis of polysome fractionated samples (Fig. 5) (see Fig. S2 in the supplemental material). The polysome profiles and the distribution of a control mRNA, gdhA+ (see Fig. S2), appeared to be unaltered in any of the mutant strains. In the wild-type, Δupf1, and nmdA1 backgrounds, the relative proportion of mRNA associated with the ribosome compared to the observed levels of total mRNA, as determined by conventional Northern analysis (Fig. 4), did not differ greatly between uaZ+ and uaZ14 (Fig. 5). Critically, the proportion of PTC-containing uaZ14 transcript associated with ribosomes dramatically increased in strains mutated for cutA and/or cutB, ranging from 7-fold enrichment for the cutA-N310D strain to >12-fold enrichment for both the ΔcutA strain and the ΔcutA ΔcutB double mutant. We obtained equivalent results for hxA+ and hxA5 in that the proportion of PTC-containing mRNA (hxA5) associated with the polysome fractions was significantly enriched compared to that of the wild type (hxA+) in the ΔcutA and ΔcutB strains (see Fig. S3 in the supplemental material). Interestingly in ΔcutA, cutA-N310D, ΔcutB, and ΔcutAΔcutB strains, the uaZ14 transcript predominates in the lighter ribosomal fractions compared with the heavier fractions derived from Δupf1 and nmdA1 strains. This effect was not as marked for hxA5 (see Fig. S3) and may therefore relate to the location of the PTC-induced termination event, uaZ14 being at codon 131 and hxA5 being at codon 369.
Fig 5.
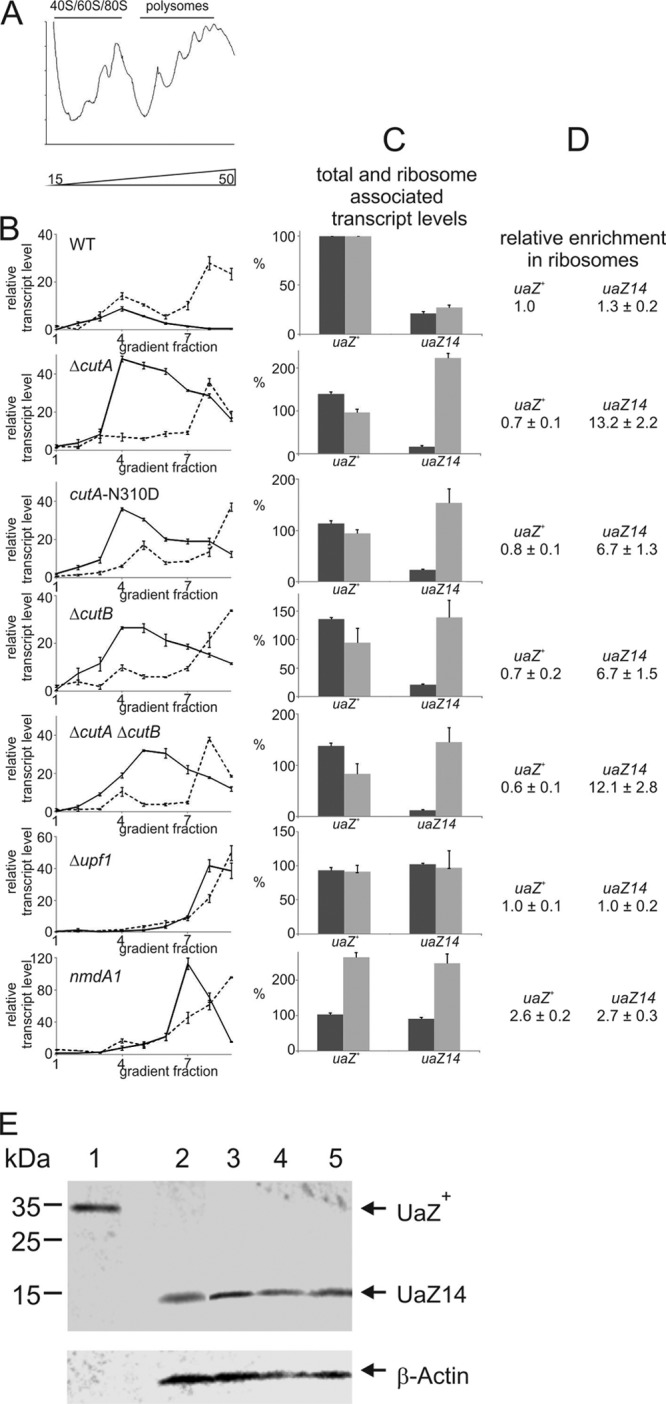
CutA and CutB are implicated in ribosome dissociation from uaZ14 mRNA. (A) Representative UV absorbance profile of cellular extracts after velocity sedimentation through a 15 to 50% sucrose gradient. The positions of 40S and 60S ribosomal subunits as well as monosomes (80S) and polyribosomes are shown above the UV profiles. Each experiment was repeated at least three times, and representative data are shown. (B) Distribution of polyribosome-associated uaZ+ (dashed line) and uaZ14 (solid line) mRNAs from different strains (wild-type [WT], ΔcutA, cutA-N310D, ΔcutB, ΔcutA ΔcutB, Δupf1, and nmdA1 strains). A 10-ml gradient was separated into nine equal fractions, and RNA was extracted from each fraction and subjected to Northern analysis for uaZ transcripts. The data points represent the intensity of each fraction relative to the total combined intensity of all fractions for each gradient. These are scaled to the total combined intensity observed for uaZ+ in the wild-type background. Error bars show standard errors. (C) The total and the ribosome-associated uaZ transcript levels are shown for each strain. The Northern data for total mRNA (black bars) derived from Fig. 4 were compared to the ribosome-associated signal (gray bars) of pooled aliquots from each gradient fraction shown in panel B. To facilitate comparison, the data were scaled relative to uaZ+ in the wild-type background. The normalized signals are from independent biological replicates (≥3). Error bars show standard errors. (D) The relative proportions of uaZ+ and uaZ14 mRNAs in ribosome-associated fractions versus total mRNA derived from panel C are given. Mean values with standard errors are shown from three replicate experiments. (E) Western analysis of 10 μg protein from the wild type (lane 1) and 200 μg protein uaZ14 (lane 2), ΔcutA uaZ14 (lane 3), ΔcutB uaZ14 (lane 4), and cutA-N310D uaZ14 (lane 5). Protein concentration was determined by Bradford assays and confirmed by Coomassie blue staining of SDS-PAGE, and β-actin was used as a loading control. No actin band was observed in the wild type due to the small amount of protein loaded. Bands of ∼34 and ∼14 kDa were detected with the UaZ-specific antibody, consistent with the expected sizes of UaZ+ and UaZ14 proteins, respectively. Based on three separate experiments, no consistent variation in UaZ14 expression was observed between the different genetic backgrounds. Representative images from one experiment are given. As mutations in either cutA or cutB did not enhance uaZ14 expression, we conclude that the enrichment of uaZ14 transcript in the ribosomal fraction is not associated with enhanced translation.
To confirm that the mRNA detected within the density gradient was directly associated with polysomes, extracts were treated with 10 mM EDTA. After EDTA treatment, polysomes were lost and both uaZ and gdhA mRNAs were found in the lighter, 40S/60S/80S fractions of the gradient (see Fig. S4 in the supplemental material). These observations are consistent with the mRNA detected being bound to polysomes rather than forming non-rRNA-protein complexes.
From these data, it is apparent that, although 3′ mRNA tagging is not required for NMD-mediated degradation of PTC-containing transcripts, it is implicated in a key aspect, the efficient dissociation of mRNA from ribosomes after premature termination. To determine if polysome enrichment of uaZ14 mRNA in cutA and cutB mutant strains is associated with increased translation, Western analysis of UaZ14 protein expression was conducted (Fig. 5E). From these data, there was no evidence that mutations in either cutA or cutB enhanced translation of uaZ14, consistent with the high levels of polysome-associated uaZ14 mRNA representing accumulation after termination at the PTC.
mRNA 3′ tagging occurs in plants.
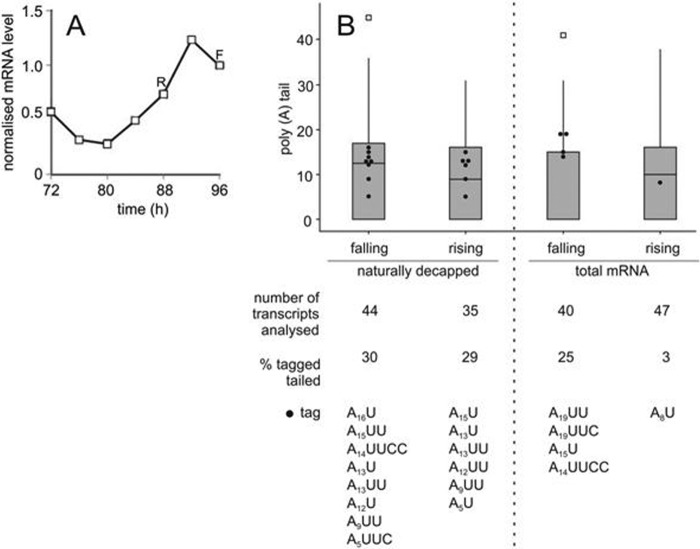
The 3′ tagging of adenylated transcripts has so far been reported only in fungi (60, 76). However, in human cells the histone H2a- and H3.3-encoding transcripts, which are nonadenylated, are also modified immediately prior to transcript decapping and degradation (67). Additionally, 3′ pyrimidine tagging of small RNA molecules, primarily involving uridylation, has been observed in both animal and plant systems (72) and appears to correlate with control of degradation. To determine whether polyadenylated plant transcripts are also subject to 3′ tagging, we investigated the CCR2 transcript from Arabidopsis thaliana. Gene expression of this transcript is under circadian control with a peak in abundance toward the end of the day (29). Utilizing cRT-PCR analysis of both natively decapped and total (capped and uncapped) mRNA, we monitored 3′-end tagging at two phases within the circadian cycle (Fig. 6), one where the abundance of the CCR2 transcript was at a rising phase of gene expression (88 h after transfer to constant conditions [R]) and the second where transcript abundance was at a phase of falling gene expression (96 h after transfer to constant conditions [F]). We identified 3′ tagging, with 29% of natively decapped transcripts being modified at both time points, consistent with 3′-end modified mRNA being targeted for degradation. The median poly(A) tail length was ∼14 nucleotides, strikingly similar to the situation in fungi, where deadenylation to ∼A15 is known to trigger both mRNA tagging and decapping (20, 60, 64). Among the total pool of transcripts (capped and uncapped), the distribution of 3′ tags differed dramatically between the two samples, with 25% of adenylated transcripts being tagged when transcript levels were falling but only 3% when the transcript levels were rising. This demonstrates a strong correlation between mRNA tagging and downregulation of a specific plant transcript. Although the modifications are primarily U nucleotides, 19% were C nucleotides, which is intermediate between the uridylation observed in S. pombe and C/U modification in A. nidulans (60, 76).
Fig 6.
CCR2 mRNA expression and modification. (A) Arabidopsis thaliana seedlings were grown under entrainment conditions (12-h-light/12-h-dark cycle at 22°C) before transfer to constant blue light at 12°C. Seedlings were harvested every 4 h from 72 to 96 h. RNA was extracted, and cDNA was produced. The cDNA was used in qRT-PCR to measure normalized transcript levels of CCR2 relative to UBQ10. (B) cRT-PCR was used to monitor CCR2 transcript poly(A) tail length and 3′ tagging for both naturally decapped and total (capped and uncapped) mRNA. These data are represented as in Fig. 2.
DISCUSSION
The first reports of cytoplasmic RNA 3′ tagging were for small noncoding RNAs and their degradation products, where it was shown to be associated with degradation, maturation, and the modulation of function (27, 31, 37, 41, 48, 49, 52, 80). Subsequent in vitro analysis determined that the addition of a short uridine tag to RNA molecules increased their affinity for the LSM complex (12, 84), which, through interactions with other proteins, including Dhh1 and Pat1, promotes RNA degradation (71, 74). The human H2a and H3.3 histone coding transcripts were the first mRNAs reported to be uridylated (67). 3′ tagging of these nonadenylated transcripts occurs as a precursor to decapping and degradation. Interestingly, Upf1, which is a key player in NMD, is directly involved in histone mRNA degradation as well as the turnover of a significant proportion of other transcripts which do not include a PTC (70).
The 3′ tagging of polyadenylated transcripts has previously been reported only in fungi (60, 76). In both S. pombe and A. nidulans, the pyrimidine tagging of mRNA is coincident with decapping and associated with efficient transcript degradation. It was therefore important to determine if this also occurs in plants. We chose the circadian-regulated transcript CCR2 (29) in Arabidopsis for investigation. From our data, it is clear that the transcript is subject to 3′ tagging, with 29% of decapped adenylated transcripts being modified at a median poly(A) tail length of 14 nucleotides. As in fungi, these observations are consistent with mRNA 3′ pyrimidine tagging being associated with transcript degradation. From these data, it appears that 3′ modification of transcripts is a common eukaryotic phenomenon, which is consistent with the broad distribution of noncanonical terminal transferases throughout eukaryotes (72).
In A. nidulans, we previously identified CutA as being primarily responsible for 3′ tagging of mRNA. However, a low level of 3′ modification persisted in ΔcutA strains. We have now identified a second enzyme, CutB, as the source of this residual activity. C and U nucleotide tags persist in both ΔcutA and ΔcutB single mutants, demonstrating that neither activity is nucleotide specific. This was particularly surprising in respect of cutB as it shows strong sequence similarity to known nuclear adenylases which form part of the TRAMP complex. However, PAPD5, which has a role in human histone mRNA uridylation (67), is closely related to S. cerevisiae Pap2p/Trf4 and Trf5p, which are both involved in RNA adenylation within the nucleus (79). Furthermore, a nucleotidyltransferase, Star-PAP, has previously been shown to both adenylate and uridylate RNA (57, 90).
A key finding of this work is that 3′ tagging is associated with NMD. The presence of a PTC within the uaZ14 mRNA led to a dramatic increase in 3′ tagging, which, in addition to being dependent on the terminal transferases CutA and CutB, also requires the NMD components Upf1 and NmdA. Mutations disrupting components of the NMD mechanism are highly pleiotropic, with a significant proportion of the transcriptome and proteome being differentially expressed (e.g., references 28, 55, and 58). It is formally a possibility that the effects of Δupf1 and nmdA1 alleles on mRNA 3′ tagging and poly(A) profiles could be indirect, as is the case for NMD-associated loss of termination fidelity, which appears to result indirectly due to increased intracellular Mg2+ levels in S. cerevisiae (40). Additionally, the high frequency of 3′ tagging associated with NMD may be a general consequence of accelerated mRNA degradation. As previous reports have linked transcript 3′ modification with transcript degradation, it was surprising that mutations which disrupt tagging did not have a significant effect on the stability of PTC-containing transcripts. However, disruption of either cutA or cutB did inhibit decapping of uaZ14 mRNA with long poly(A) tails. These data support the hypothesis that deadenylation-independent decapping is promoted by 3′ tagging (60). Furthermore, as NMD persists in strains where deadenylation-independent decapping is suppressed, we conclude that at least one additional RNA degradation mechanism must be involved in effecting this surveillance mechanism in A. nidulans, as is the case for metazoans (38).
Premature termination of translation has previously been shown in vitro to differ significantly from general termination, a key difference being inefficient release of the terminating ribosome (2). Our in vivo analysis of uaZ and hxA distribution within polysomes supports this, as both cutA and cutB mutants specifically increase the proportion of PTC-containing transcripts retained within mono- and polysomes (Fig. 5). However, there was no corresponding increase in the translation of uaZ14. This is consistent with mRNA 3′ tagging facilitating the clearance of NMD-inducing transcripts from ribosomes. Previously, it has been demonstrated that pyrimidine tags facilitate recruitment of the LSM-Pat1 complex (12, 84), which is known to promote decapping and repress translational initiation by limiting the formation of a 48S preinitiation complex. Our data are consistent with the possibility that the LSM-Pat1 complex, once recruited to the tagged mRNA, may also promote the dissociation of transcripts from the PTC-induced termination complex. Critically, there is a fundamental difference between premature and normal termination, which is consistent with distinct mechanisms being involved (2). Transcripts undergoing normal termination are generally retained in order to allow repeated rounds of translation (9, 91, 94). However, premature termination is concomitant with transcript degradation, preventing rather than facilitating further translation. The association of mRNA 3′ tagging with NMD is consistent with the role of this surveillance mechanism in effecting transcript specific repression and promoting ribosome clearance and mRNA degradation.
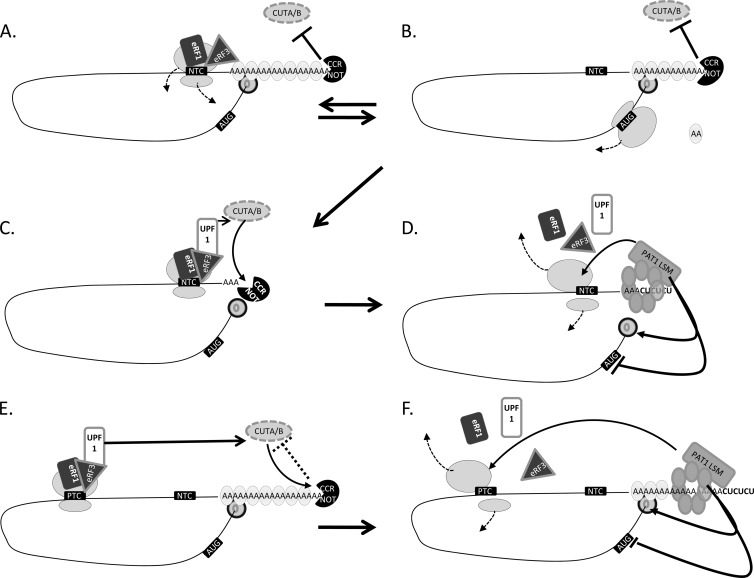
It was surprising to observe that disruption of upf1 led to low levels of mRNA 3′ tagging of wild-type as well as PTC-containing transcripts. Interestingly, Upf1 plays a direct role in the 3′ tagging and degradation of human histone mRNAs (67). Furthermore, Upf1 is known to interact directly with the translation release factor eRF3 (19, 39), which is also the target by which poly(A) binding protein (PAB1) promotes efficient termination and ribosome dissociation (16, 25, 32, 47, 53, 91). One intriguing possibility is that Upf1 and perhaps other components of the NMD machinery are directly involved in the termination of deadenylated transcripts, effecting mRNA surveillance at the level of poly(A) length. There is a significant body of evidence to suggest that NMD is triggered when the ribosomes encounter a stop codon which is not in close proximity to PAB1 (2, 7, 65, 82). One possibility is that wild-type transcripts which have undergone poly(A) tail degradation to ∼A15, and consequently dissociation of PAB1, could similarly undergo an “NMD-like” termination event due to the absence of the PAB1-eRF3 interaction, which will potentially include the recruitment of Upf1. This model (Fig. 7) would predict that Upf1 and/or the absence of PAB1 promotes mRNA 3′ tagging, which facilitates degradation of deadenylated transcripts. According to this model, deadenylated mRNA or aberrant transcripts with a PTC elicit the same cellular response, with two consequences. First, reinitiation of translation is prevented to avoid production of either unwanted or truncated proteins. Second, ribosome clearance would occur, allowing effective and efficient mRNA decay to take place. We propose that the ribosomal termination complex triggers these events in deadenylated transcripts, as is the case for NMD.
Fig 7.
Translational termination of deadenylated transcripts promotes an NMD-like response. Translational termination of poly(A) transcripts is known to involve recruitment of eRF1 and the GTPase eRF3 (46) (A). Critical to efficient ribosome dissociation is the interaction between eRF3 and poly(A) binding protein (PAB1) (oval with “AA”). The association of the 5′-end cap structure (thick gray zero in a circle) with PAB1 facilitates reinitiation of translation (21) (B). During translation, the poly(A) tail is gradually shortened by deadenylase complexes (16, 22, 32–34, 60, 61); in the case of A. nidulans, this has been shown to involve the CCR4-Not complex (60, 61). Under these circumstances, with a long poly(A) tail bound by PAB1 and an active deadenylase, CUTA/B-mediated 3′ tagging is inhibited (60). After deadenylation has progressed to a critical point of ∼15A nucleotides, poly(A) binding protein dissociates from the tail and the processivity of the deadenylase complex is altered (C). In the absence of PAB1, we propose that the terminating ribosome will recruit Upf1 through its interaction with eRF3, as is known to occur during NMD (E and F), where a premature termination codon (PTC) is encountered sufficiently distant from the poly(A) tail and associated PAB1 (2). Under these circumstances, ribosome dissociation is inhibited (53, 70) and the NMD pathway is triggered. Based on our data, efficient clearance of this termination complex requires 3′ tagging. Consistent with this, pyrimidine tagging generally occurs in transcripts with short poly(A) tails (D) (60) or in the case of NMD is poly(A) independent (F). The other well-characterized example is for the nonadenylated human histone mRNAs upon S-phase arrest (67). In A. nidulans, 3′ tagging of short poly(A) tails is not completely lost in Δupf1 strains, suggesting that other factors are involved in facilitating the process, possibly the loss of PAB1 and/or altered interactions with the deadenylase complex. Generally, 3′ tagging is a precursor to mRNA decapping; one possibility is that a pyrimidine tract enhances affinity for the LSM-PAT1 complex (12, 84), recruitment of which (D and E) is known to initiate a cascade of events, promoting transcript decapping and degradation as well as the inhibition of translational initiation (71). We propose that recruitment of the LSM-PAT1 complex will also facilitate dissociation of the mRNA from the relatively stable termination complex which is formed in the absence of PAB1, facilitating efficient clearance from the polysomes.
These data provide a new insight into the interplay between translation and transcript degradation. Currently, there are significant discrepancies in the literature concerning the localization of degrading transcripts. Recent work in S. cerevisiae suggests that a proportion of degradation is cotranslational (35), contradicting substantial data suggesting that mRNA dissociates from ribosomes at the point of degradation and is sequestered to P bodies prior to decapping and degradation (10, 88). This discrepancy could represent a consequence of assessing the integrity of transcripts after polysome fractionation, which may lead to limited mRNA degradation during sample preparation. Alternatively, cotranslational degradation may be specific to S. cerevisiae as it does not appear to undertake cytoplasmic uridylation of transcripts (76) and interestingly has a unique cellular component, Ski7, a GTPase with some sequence similarity to eRF3 (8), which plays an essential role in NMD (3, 24, 87, 92).
Cytoplasmic 3′ tagging has been linked to decapping and RNA degradation, and we have now shown an additional link to polysome exclusion. We therefore propose that 3′ pyrimidine tagging represents an important trigger for the coordinated repression of transcripts: promoting the dissociation of transcripts from the polysomes and simultaneously initiating decapping and degradation, thus preventing the reinitiation of translation. Through these mechanisms, mRNA 3′ tagging coordinates the rapid and controlled termination of a transcript's expression. We propose that, unlike translational termination in transcripts with a long poly(A) tail, termination in deadenylated transcripts specifically elicits 3′ tagging as part of an NMD-like response, preventing the further translation of unwanted transcripts.
Supplementary Material
ACKNOWLEDGMENTS
I.Y.M. and P.D.G. were supported by BBSRC grants to M.X.C. (BB/E017657/1) and A.J.W.H. (BB/F005318/1).
We thank J. Wood and J. Forman for technical assistance.
Footnotes
Published ahead of print 30 April 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Ameres SL, et al. 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328:1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amrani N, et al. 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432:112–118 [DOI] [PubMed] [Google Scholar]
- 3. Araki Y, et al. 2001. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 20:4684–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arava Y, et al. 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 100:3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aravind L, Koonin EV. 1999. DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 27:1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balagopal V, Parker R. 2009. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol. 21:403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. 2007. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 26:1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benard L, Carroll K, Valle RC, Masison DC, Wickner RB. 1999. The ski7 antiviral protein is an EF1-alpha homolog that blocks expression of non-poly(A) mRNA in Saccharomyces cerevisiae. J. Virol. 73:2893–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Borman A, Michel YM, Kean KM. 2000. Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5′-end. Nucleic Acids Res. 28:4068–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brengues M, Teixeira D, Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310:486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caddick MX, et al. 2006. Opposing signals differentially regulate transcript stability in Aspergillus nidulans. Mol. Microbiol. 62:509–519 [DOI] [PubMed] [Google Scholar]
- 12. Chowdhury A, Mukhopadhyay J, Tharun S. 2007. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 13:998–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clutterbuck AJ. 1993. Aspergillus nidulans, 6th ed, vol 3 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 14. Clutterbuck AJ. 1974. Aspergillus nidulans, vol 1 Plenum Press, New York, NY [Google Scholar]
- 15. Coldwell MJ, Gray NK, Brook M. 2010. Cytoplasmic mRNA: move it, use it or lose it! Biochem. Soc. Trans. 38:1495–1499 [DOI] [PubMed] [Google Scholar]
- 16. Cosson B, et al. 2002. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol. Cell. Biol. 22:3301–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T. 1997. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 94:5628–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cove DJ. 1966. Induction and repression of nitrate reductase in fungus Aspergillus nidulans. Biochim. Biophys. Acta 113:51–56 [DOI] [PubMed] [Google Scholar]
- 19. Czaplinski K, et al. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 12:1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Decker CJ, Parker R. 1993. A turnover pathway for both stable and unstable messenger-RNAs in yeast—evidence for a requirement for deadenylation. Genes Dev. 7:1632–1643 [DOI] [PubMed] [Google Scholar]
- 21. Derry MC, Yanagiya A, Martineau Y, Sonenberg N. 2006. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harbor Symp. Quant. Biol. 71:537–543 [DOI] [PubMed] [Google Scholar]
- 22. Funakoshi Y, et al. 2007. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 21:3135–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Galagan JE, et al. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A-fumigatus and A-oryzae. Nature 438:1105–1115 [DOI] [PubMed] [Google Scholar]
- 24. Garneau NL, Wilusz J, Wilusz CJ. 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8:113–126 [DOI] [PubMed] [Google Scholar]
- 25. Ghosh S, Ganesan R, Amrani N, Jacobson A. 2010. Translational competence of ribosomes released from a premature termination codon is modulated by NMD factors. RNA 16:1832–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gould PD, et al. 2009. Delayed fluorescence as a universal tool for the measurement of circadian rhythms in higher plants. Plant J. 58:893–901 [DOI] [PubMed] [Google Scholar]
- 27. Hagan JP, Piskounova E, Gregory RI. 2009. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 16:1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He F, et al. 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12:1439–1452 [DOI] [PubMed] [Google Scholar]
- 29. Heintzen C, Nater M, Apel K, Staiger D. 1997. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 94:8515–8520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heo I, et al. 2008. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell 32:276–284 [DOI] [PubMed] [Google Scholar]
- 31. Heo I, et al. 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138:696–708 [DOI] [PubMed] [Google Scholar]
- 32. Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 274:16677–16680 [DOI] [PubMed] [Google Scholar]
- 33. Hoshino S, et al. 1998. Molecular cloning of a novel member of the eukaryotic polypeptide chain-releasing factors (eRF). Its identification as eRF3 interacting with eRF1. J. Biol. Chem. 273:22254–22259 [DOI] [PubMed] [Google Scholar]
- 34. Houseley J, Tollervey D. 2009. The many pathways of RNA degradation. Cell 136:763–776 [DOI] [PubMed] [Google Scholar]
- 35. Hu W, Petzold C, Coller J, Baker KE. 2010. Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 17:244–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ibrahim F, Rohr J, Jeong WJ, Hesson J, Cerutti H. 2006. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science 314:1893. [DOI] [PubMed] [Google Scholar]
- 37. Ibrahim F, et al. 2010. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc. Natl. Acad. Sci. U. S. A. 107:3906–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Isken O, Maquat LE. 2008. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 9:699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 27:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johansson MJ, Jacobson A. 2010. Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake. Genes Dev. 24:1491–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jones MR, et al. 2009. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat. Cell Biol. 11:1157–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kable ML, Seiwert SD, Heidmann S, Stuart K. 1996. RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science 273:1189–1195 [DOI] [PubMed] [Google Scholar]
- 43. Kamminga LM, et al. 2010. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J. 29:3688–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Katoh T, et al. 2009. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 23:433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim JH, Richter JD. 2006. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell 24:173–183 [DOI] [PubMed] [Google Scholar]
- 46. Kisselev LL, Frolova L. 1995. Termination of translation in eukaryotes. Biochem. Cell Biol. 73:1079–1086 [DOI] [PubMed] [Google Scholar]
- 47. Kozlov G, et al. 2001. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl. Acad. Sci. U. S. A. 98:4409–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Landgraf P, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lehrbach NJ, et al. 2009. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 16:1016–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li J, Yang Z, Yu B, Liu J, Chen X. 2005. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15:1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li Z, Pandit S, Deutscher MP. 1998. Polyadenylation of stable RNA precursors in vivo. Proc. Natl. Acad. Sci. U. S. A. 95:12158–12162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lu S, Sun YH, Chiang VL. 2009. Adenylation of plant miRNAs. Nucleic Acids Res. 37:1878–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mangus DA, Evans MC, Jacobson A. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4:223 doi:10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maquat LE. 2004. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 5:89–99 [DOI] [PubMed] [Google Scholar]
- 55. McGlincy NJ, et al. 2010. Expression proteomics of UPF1 knockdown in HeLa cells reveals autoregulation of hnRNP A2/B1 mediated by alternative splicing resulting in nonsense-mediated mRNA decay. BMC Genomics 11:565 doi:10.1186/1471-2164-11-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. McManus MT, Adler BK, Pollard VW, Hajduk SL. 2000. Trypanosoma brucei guide RNA poly(U) tail formation is stabilized by cognate mRNA. Mol. Cell. Biol. 20:883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mellman DL, et al. 2008. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature 451:1013–1017 [DOI] [PubMed] [Google Scholar]
- 58. Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36:1073–1078 [DOI] [PubMed] [Google Scholar]
- 59. Mohanty BK, Kushner SR. 1999. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 34:1094–1108 [DOI] [PubMed] [Google Scholar]
- 60. Morozov IY, Jones MG, Razak AA, Rigden DJ, Caddick MX. 2010. CUCU modification of mRNA promotes decapping and transcript degradation in Aspergillus nidulans. Mol. Cell. Biol. 30:460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Morozov IY, et al. 2010. Distinct roles for Caf1, Ccr4, Edc3 and CutA in the co-ordination of transcript deadenylation, decapping and P-body formation in Aspergillus nidulans. Mol. Microbiol. 76:503–516 [DOI] [PubMed] [Google Scholar]
- 62. Morozov IY, Martinez MG, Jones MG, Caddick MX. 2000. A defined sequence within the 3′ UTR of the areA transcript is sufficient to mediate nitrogen metabolite signalling via accelerated deadenylation. Mol. Microbiol. 37:1248–1257 [DOI] [PubMed] [Google Scholar]
- 63. Morozov IY, et al. 2006. Nonsense-mediated mRNA decay mutation in Aspergillus nidulans. Eukaryot. Cell 5:1838–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Muhlrad D, Decker CJ, Parker R. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 8:855–866 [DOI] [PubMed] [Google Scholar]
- 65. Muhlrad D, Parker R. 1999. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5:1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Muhlrad D, Parker R. 1992. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 6:2100–2111 [DOI] [PubMed] [Google Scholar]
- 67. Mullen TE, Marzluff WF. 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 22:50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Newman MA, Mani V, Hammond SM. 2011. Deep sequencing of microRNA precursors reveals extensive 3′ end modification. RNA 17:1795–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nicholson P, Muhlemann O. 2010. Cutting the nonsense: the degradation of PTC-containing mRNAs. Biochem. Soc. Trans. 38:1615–1620 [DOI] [PubMed] [Google Scholar]
- 70. Nicholson P, et al. 2010. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell. Mol. Life Sci. 67:677–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nissan T, Rajyaguru P, She M, Song H, Parker R. 2010. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell 39:773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Norbury CJ. 2010. 3′ uridylation and the regulation of RNA function in the cytoplasm. Biochem. Soc. Trans. 38:1150–1153 [DOI] [PubMed] [Google Scholar]
- 73. O'Hara EB, et al. 1995. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 92:1807–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Parker R, Song HW. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11:121–127 [DOI] [PubMed] [Google Scholar]
- 75. Rammelt C, Bilen B, Zavolan M, Keller W. 2011. PAPD5, a noncanonical poly(A) polymerase with an unusual RNA-binding motif. RNA 17:1737–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rissland OS, Norbury CJ. 2009. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol. 16:616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schmidt MJ, West S, Norbury CJ. 2011. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA 17:39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Seiwert SD, Heidmann S, Stuart K. 1996. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 84:831–841 [DOI] [PubMed] [Google Scholar]
- 79. Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG. 2010. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 11:106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shen B, Goodman HM. 2004. Uridine addition after microRNA-directed cleavage. Science 306:997 doi:10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 81. Shyu AB, Wilkinson MF, van Hoof A. 2008. Messenger RNA regulation: to translate or to degrade. EMBO J. 27:471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Singh G, Rebbapragada I, Lykke-Andersen J. 2008. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 6:e111 doi:10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Song MG, Kiledjian M. 2007. 3′ terminal oligo U-tract-mediated stimulation of decapping. RNA 13:2356–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. 2007. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. J. Mol. Biol. 366:882–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Szewczyk E, et al. 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1:3111–3120 [DOI] [PubMed] [Google Scholar]
- 87. Takahashi S, Araki Y, Sakuno T, Katada T. 2003. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J. 22:3951–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Trippe R, et al. 2006. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 12:1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Trippe R, Sandrock B, Benecke BJ. 1998. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res. 26:3119–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Uchida N, Hoshino S, Imataka H, Sonenberg N, Katada T. 2002. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J. Biol. Chem. 277:50286–50292 [DOI] [PubMed] [Google Scholar]
- 92. van Hoof A, Frischmeyer PA, Dietz HC, Parker R. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295:2262–2264 [DOI] [PubMed] [Google Scholar]
- 93. van Wolfswinkel JC, et al. 2009. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139:135–148 [DOI] [PubMed] [Google Scholar]
- 94. Wells SE, Hillner PE, Vale RD, Sachs AB. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2:135–140 [DOI] [PubMed] [Google Scholar]
- 95. West S, Gromak N, Norbury CJ, Proudfoot NJ. 2006. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol. Cell 21:437–443 [DOI] [PubMed] [Google Scholar]
- 96. Wilson JB, et al. 2001. The Chinese hamster FANCG/XRCC9 mutant NM3 fails to express the monoubiquitinated form of the FANCD2 protein, is hypersensitive to a range of DNA damaging agents and exhibits a normal level of spontaneous sister chromatid exchange. Carcinogenesis 22:1939–1946 [DOI] [PubMed] [Google Scholar]
- 97. Wyman SK, et al. 2011. Post-transcriptional generation of miRNA variants by multiple nucleotidyltransferases contributes to miRNA transcriptome complexity. Genome Res. 21:1450–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.