Abstract
Proneural NEUROG2 (neurogenin 2 [Ngn2]) is essential for neuronal commitment, cell cycle withdrawal, and neuronal differentiation. Although NEUROG2's influence on neuronal commitment and differentiation is beginning to be clarified, its role in cell cycle withdrawal remains unknown. We therefore set out to investigate the molecular mechanisms by which NEUROG2 induces cell cycle arrest during spinal neurogenesis. We developed a large-scale chicken embryo strategy, designed to find gene networks modified at the onset of NEUROG2 expression, and thereby we identified those involved in controlling the cell cycle. NEUROG2 activation leads to a rapid decrease of a subset of cell cycle regulators acting at G1 and S phases, including CCND1, CCNE1/2, and CCNA2 but not CCND2. The use of NEUROG2VP16 and NEUROG2EnR, acting as the constitutive activator and repressor, respectively, indicates that NEUROG2 indirectly represses CCND1 and CCNE2 but opens the possibility that CCNE2 is also repressed by a direct mechanism. We demonstrated by phenotypic analysis that this rapid repression of cyclins prevents S phase entry of neuronal precursors, thus favoring cell cycle exit. We also showed that cell cycle exit can be uncoupled from neuronal differentiation and that during normal development NEUROG2 is in charge of tightly coordinating these two processes.
INTRODUCTION
One important challenge in neurobiology is to understand how different types of postmitotic neurons, with distinct cellular and physiological properties, are generated in the developing central nervous system (CNS) from a pool of dividing neural progenitors. The embryonic spinal cord is a good model to tackle these issues, because the role of extracellular signals and transcription factors in neuron specification and differentiation is relatively well defined. This structure is derived from the neural tube, a single pseudoepithelium that will sequentially give rise to a large variety of neurons and glial cells dedicated to serve specific functions in the adult. Neurogenesis is achieved via a succession of steps that follow a stereotypic temporal order. A neural progenitor is committed to a neuronal fate at the expense of a glial fate and becomes a neuronal precursor. Concomitantly, this neural progenitor is destined to differentiate into a specific neuronal subtype. Soon after, neuronal precursors stop cycling and initiate their differentiation to give rise to postmitotic differentiated neurons.
The main positive regulators of vertebrate neurogenesis are proneural transcription factors of the neural basic helix-loop-helix (bHLH) family, including neurogenins (NeuroG1/2/3) (5, 35). They control different steps of neurogenesis, such as neuronal commitment, cell cycle exit, subtype specification, and neuronal differentiation (5, 35, 42). In the spinal cord, loss-of-function studies have shown that NEUROG2 is involved in the acquisition of motoneuron and interneuron fates (46). Together with NEUROG1, NEUROG2 also controls neuronal differentiation as shown by the loss of neurons in NeuroG1/2 double knockout mice and by the presence of ectopic neurons, when NEUROG2 is misexpressed in the proliferative zone of the neural tube (35, 38, 42). Proneural proteins also trigger cell cycle exit of neural progenitors. Hence, overexpression of NEUROG2 in the chick neural tube leads to premature cell cycle arrest as revealed by the lack of BrdU incorporation in NEUROG2 misexpressing cells (38, 40). This proliferation arrest is always linked to neuronal differentiation, making it difficult to know whether cell cycle exit is necessary or sufficient to trigger neuronal differentiation or whether it is an independent event directly controlled by NEUROG2.
Control of these different cellular processes by NEUROG2 implies that it regulates a large panel of genes performing different functions. Neurogenins are transcriptional activators that dimerise with the ubiquitous bHLH proteins E12 or E47 to bind to the E-box consensus DNA motifs in the regulatory regions of their target genes (19). They can also exert their regulatory activity independently of DNA binding, via a protein-protein interaction with CBP/p300 as described in cortical cell migration or gliogenesis (17, 49). NEUROG1/2's earliest action is to trigger the NOTCH signaling pathway and the lateral inhibition process, in order to control the balance between progenitor and differentiating states (25). Hence, it upregulates NOTCH ligands such as DLL1, JAG1, and JAG2, which trigger neuronal commitment in the cells in which they are expressed, and activates the NOTCH signaling cascade in neighboring cells. NEUROG2 also represses genes expressed in neural progenitors, including the pan-neural genes SOX1 to SOX3 and genes involved in subtype specification such as PAX6 and OLIG2. Indeed, these genes participate in the neuronal fate acquisition via NEUROG2-positive regulation. However, they antagonize subsequent neuronal differentiation as long as they are expressed in committed cells, thus maintaining a pool of immature neuronal precursors (3, 29, 40, 48). Their repression by negative feedback from increasing NEUROG2 levels is thus the gate to neuronal differentiation. In addition, NEUROG1/2 activates the expression of a cascade of differentiation genes such as NEUROD and NEUROD4 (7, 13, 35) while suppressing gliogenesis by sequestering CBP/p300 (49). NEUROG2 also participates in the correct expression of neuronal subtype-specific homeodomains, such as the interneuron markers Lim1/2 or the MN markers Hb9 (29, 46). NEUROG2 thus acts at different molecular levels to affect neuronal commitment, specification, and differentiation, and as data start accumulating, we are identifying the molecular links between proneural genes and gene networks involved in specification and differentiation.
On the other hand, the molecular mechanisms by which proneural genes trigger cell cycle arrest remain elusive. Progression through the cell cycle is driven by cyclin-dependent kinases (CDK) and their activating cyclin (CCN) partners. Specific combinations of CDK/cyclin heterodimers allow progression through specific phases of the cell cycle. CDK/cyclin activity is suppressed by interactions with two main groups of inhibitor proteins belonging to the INK4 and CIP/Kip families. The rate of cell cycle progression is determined by the relative abundance of these positive and negative regulators. A recent study conducted in the cortex shows that Ascl1 sequentially activates positive and negative cell cycle regulators such as Cdk1, Cdk2, or Cdc25B and Gadd45 or Ccng2, respectively. This reveals an unexpected role for Ascl1 in cell cycle progression, and such a role has never been described before for other proneural genes (10). In the neural tube, cells overexpressing NEUROG2 accumulate high levels of cyclin-dependent kinase inhibitors (CKI) p27Kip1 and p57KIP2 (20, 40), but this observation has been made in differentiated neurons, making it difficult to conclude whether CKI upregulation is the molecular event that initiates the proliferation arrest of neuronal precursors.
The goal of our study was to examine the molecular mechanisms by which NEUROG2 drives cell cycle arrest of spinal progenitors. Our strategy was to identify NeuroG2 early response genes by using a large-scale approach and to identify those involved in cell cycle control. We combined gain of function in the developing spinal cord with a global detection of transcript levels using microarrays and identified 942 genes regulated by NEUROG2 in a short time frame. We analyzed core cell cycle regulators and found that NEUROG2 triggers cell cycle exit by a rapid downregulation of a subset of cyclins acting at the G1 and S phases of the cell cycle; the expression of associated cyclin-dependent kinases (CDK) or cyclin-dependent kinase inhibitors (CKI) remains unaffected. We demonstrate that this repression prevents S phase entry of neural cells, thus favoring cell cycle withdrawal. Finally, we show that in the developing spinal cord, cell cycle exit and neuronal differentiation can be experimentally uncoupled and that NEUROG2, by controlling both events, coordinates them.
MATERIALS AND METHODS
Embryos.
Fertile hen's eggs (from Gallus gallus) obtained from a local supplier were incubated at 38°C in a humidified incubator for appropriate periods to yield embryos of Hamburger and Hamilton stage 9 (HH9) to HH11 (day E1.5 of development) (21).
Electroporation of DNA.
DNA constructs were electroporated on the left side of the neural tube of stage HH10 chick embryos as described in reference 24. Chick NEUROG2 constructs were derived from a NEUROG2-Flag in pADRSV, a gift from P. Charnay. The NEUROG2AQ mutation is a two-amino-acid (aa) substitution at the C terminus of the NEUROG2 basic domain (aa 89 and 90 changed from NR to AQ) designed according to reference 17; the mutated DNA used to perform the substitution was synthesized by ATG:biosynthetics and replaced into pADRSVNEUROG2flag. Both NEUROG2 forms were cut from pADRSV and cloned into the pCIG vector (a gift from A. McMahon). Mouse NEUROG2 is a gift from Y. E. Sun (17). pCIG-NEUROG2EnR and pCIG-NEUROG2VP16 are a gift from C. Schuurmans (Hospital Dr NW, Calgary). pCIG-CCND1-internal ribosome entry site (IRES)-green fluorescent protein (GFP) (31) and pCIG-CCNE-IRES-GFP were obtained by cloning the CCND1 or CCNE cDNA (a gift from V. J. Kidd) into the pCIG vector. Control experiments have been performed by electroporation of the pCIG vector alone. For the loss-of-function experiments, we injected two NEUROG2 small interfering RNA (siRNA) at 50 μM (commercial source, Eurogentec). Targeted sequences were as follows: 5′-GGTTAGAAGTCATTGTATA-3′ and 5′-CCAACAACCGCGAGCGCAA-3′. The siRNA CCND1 was a gift from D. Anderson (34). The constructs were coelectroporated with pCIG to detect electroporated cells by fluorescence.
mRNA expression profiling.
Stage HH10 to HH11 embryos (11 to 15 somites) were electroporated with a control vector (pGIG-GFP) or a NEUROG2- or NEUROG2AQ-expressing vector (pCIGNEUROG2-GFP and pCIGNEUROG2AQ-GFP). We performed 4 biological replicates for each experimental condition. For each biological replicate, neural tubes from 20 embryos were pooled for GFP-positive cell collection using a fluorescence-activated cell sorter (FACS) (Epics Altra HSS cell sorter, Toulouse Rio platform). Total RNA was prepared using trireagent (Molecular Research Center) and stored at −80°C. The RNA quality and concentration were controlled by Nanodrop 1000 (Thermo Scientifics) spectrometry and a 2100 Bioanalyzer (Agilent). The preparation of labeled cRNAs and hybridization were performed at the IGBMC Microarray and Sequencing Platform, Strasbourg, France (http://www-microarrays.u-strasbg.fr/index.php). cRNAs were prepared according to the Affymetrix recommendations (without amplification) and hybridized on the GeneChip chicken genome array (Affymetrix), which contains 32,773 transcripts corresponding to over 28,000 chicken genes. All samples were normalized and summarized by the robust multichip analysis (RMA) normalization method using the Affymetrix package in Bioconductor version release (2.4.1). Gene annotations were completed by using the Manteia database (41, 51). Significant differences in gene expression were identified using the analysis of variance (ANOVA) and a false discovery rate (FDR) of 0.01 (4, 26). Under these conditions, a significant ANOVA result suggests that the means across the three groups are different at least between two of the three groups. The hierarchical clustering was performed by using the Hierarchical Clustering Explorer software (HCE; version 3.5; http://www.cs.umd.edu/hcil/hce/).
In situ hybridization and immunohistochemistry.
In situ hybridization was performed as described previously (2). Riboprobes have been synthesized from isolated specific PCR fragments to avoid any corresponding plasmid regions in the probes (sequences of primers used are available on request). Probes used were PAX6 (44), NEUROD4 (45), DLL1 and HES6 (22), ID1 and SMAD3 (15), NHLH1 (50), FGFR3 (43), and GLI3 (36); FOXM1 (clone 8e118r4) and MYCN (clone 24e15r1) (purchased from the Helmholtz zentrum münchen [GmbH]); CCND1, CCND2, CCND3, and CCNB2 (gifts from V. J. Kidd); and CCNE2 (obtained by PCR on E3.5 cDNA). Immunohistochemistry was performed as described previously (31). Staining was performed on 45-μm transversal sections performed with a vibratome (Leica VT1000S). Detailed references of used antibodies are available on request. Acquisition of images and analyses were done with a Leica TCS SP-2 spectral confocal microscope.
Proliferation rate analysis.
For proliferation analysis, 10 μl of 48 mM bromodeoxyuridine (BrdU) solution or of 40 mM 5-ethynyl-2′ deoxyuridine (EdU) solution was deposed on the heart and neural tube of electroporated embryos 30 min before fixation. BrdU was detected by immunohistochemistry, and EdU detection was performed according to the manufacturer's protocol (Invitrogen). The percentage of transfected neural precursor cells in S phase was determined by counting the number of total GFP nuclei per section and the number of nuclei expressing both GFP and BrdU or EdU on optic confocal sections (0.7-μm axial resolution; images acquired at 3-μm steps). The data correspond to the analysis of 3 to 5 sections (45-μm thickness) of at least 6 embryos processed in at least 3 independent experiments for each conditions (more than 2,000 cells were counted for each condition; analyses of variance were calculated using the appropriate t test method.
FACS.
Stage HH10 embryos (11 to 15 somites) were electroporated with the different constructs. For each experimental point, neural tubes from 16 embryos were dissected 6 h following electroporation, incubated for 10 min in trypsin-EDTA to obtain a single-cell suspension, and fixed for 30 minutes in 4% paraformaldehyde. Cell suspensions were incubated in 1 mg/ml RNase A and 40 μg/ml propidium iodure (IP) in phosphate-buffered saline (PBS)-0.5% Triton X-100 before analysis. IP and GFP fluorescence were quantified using a FACScan cytometer, and DNA content analysis was performed with the Cyflogic software.
Microarray data accession number.
Microarray raw data sets have been submitted to NCBI GEO with accession number GSE37782.
RESULTS
Setting up an experimental strategy to identify NeuroG2 early response genes.
To identify overall NEUROG2 early response genes, we combined a gain of function (GOF) strategy using in ovo electroporation in the chick neural tube, allowing for the targeting of a thousand spinal neural cells per embryo at a precise time, with a global measure of transcript levels using chicken microarrays. We first determined which time window would allow identification of early response genes in our experimental system. Based on our previous experiments, showing that DLL1 is upregulated and PAX6 is downregulated 6 h following NEUROG2 electroporation in the neural tube (3), we performed a time course of PAX6 extinction between 4 h and 6 h following NEUROG2 electroporation. PAX6 transcripts start to be notably diminished 5 h after electroporation and are strongly repressed 6 h after electroporation (see Fig. S1 in the supplemental material). We then checked the differentiation status of NEUROG2 electroporated cells 6 h after electroporation: NEUROG2 cells have activated markers of young neurons, such as NEUROD4 (or NEUROM), but we do not detect the neuronal class III β-tubulin protein (TUBB3 or Tuj1) whose expression starts in differentiating neurons (see Fig. S1). This suggests that NEUROG2 cells are engaged in the neuronal program but are not yet differentiated. Therefore, a 6-h time window appears to be quite appropriate to identify NEUROG2 early response genes.
To be able to distinguish between targets for NEUROG2 transcriptional activity (i.e., E-box binding) versus those that are independent of DNA binding, we generated a chick version of the NEUROG2 mutant form NEUROG2AQ. This mutant form cannot bind DNA and is unable to activate the neurogenic cascade in the cortex (17, 49). However, it retains the ability to interact with protein partners such as CBP/p300. We verified that as in the cortex, NEUROG2AQ is unable to promote neurogenesis in the neural tube (see Fig. S1 in the supplemental material). The use of this NEUROG2 mutant form thus allows selecting specifically NEUROG2 targets involved with transcriptional activity.
Identification of NEUROG2 early response genes in the developing spinal cord.
To compare the transcriptome of neural precursors overexpressing (i) only GFP, (ii) NEUROG2, or (iii) NEUROG2AQ, we transfected the neural tube of E1.5 chick embryos by electroporation with expression vectors coding for GFP, NEUROG2-GFP, or NEUROG2AQ-GFP. GFP-positive cells were collected 6 h later using FACS and processed for RNA probe preparation and hybridization on Affymetrix microarrays (Fig. 1A; see also Materials and Methods). Expression profiling of neural cells electroporated with control, NEUROG2, or NEUROG2AQ identified 1,038 probe sets corresponding to 942 unique genes, revealing significantly modified expression with a 0.01 false-discovery rate (FDR) (false-positive rate accepted, ≤1%) (Fig. 1B; see also Table S1 in the supplemental material). As expected for a transcription factor acting mainly as a transcriptional activator, 670 probe sets were upregulated by NEUROG2. However, we also detected 368 probe sets downregulated by NEUROG2. This suggests that an important fraction of NEUROG2 activity also goes to the repression of transcriptional targets. Of these 1,038 probe sets, only 75 have their expression similarly modified by NEUROG2 and NEUROG2AQ, indicating that in the neural tube, NEUROG2 function requires mainly DNA binding.
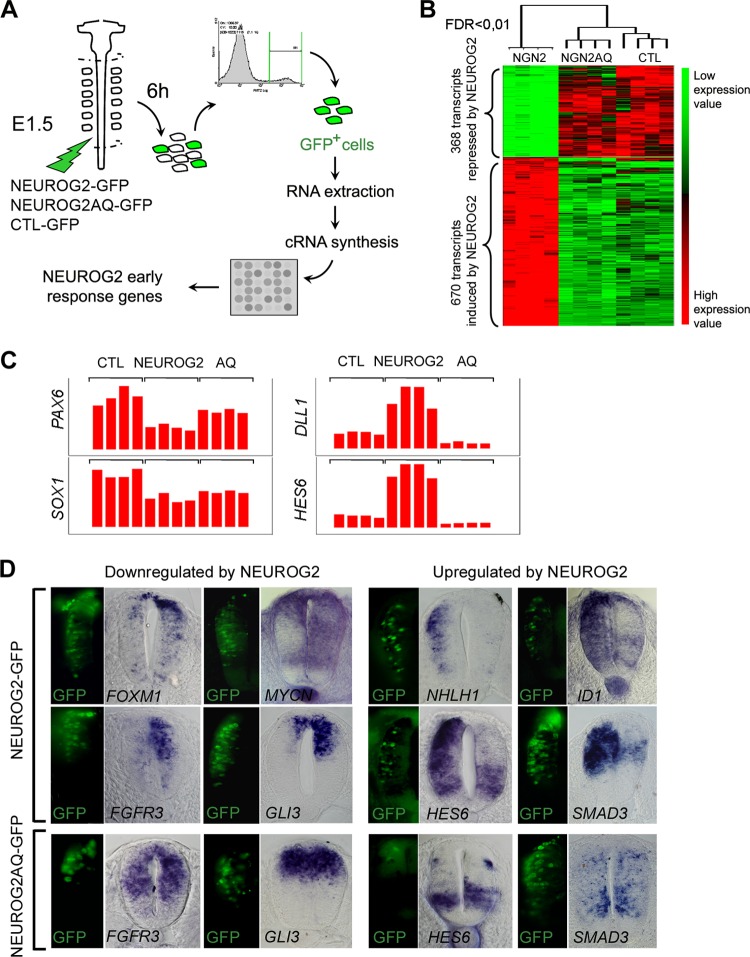
Fig 1.
Identification of NEUROG2 early target genes in the neural tube. (A) Schematic representation of the step-by-step protocol. Neural tubes were electroporated at stage E1.5 with NEUROG2-GFP, NEUROG2AQ-GFP, or CTL-GFP and dissected 6 h later. GFP-positive dissociated neural cells were processed for RNA extraction and cRNA synthesis before hybridization on chick Affymetrix arrays. Four biological replicates were performed for each experimental condition. (B) Hierarchical clustering of the expression values in each replicate for each probe set modified with an FDR of <0.01 (1,038 probe sets). Probe sets are displayed in lines and array slides in columns. The two main clusters correspond to genes downregulated (368 probe sets) or upregulated (670 probe sets) by NEUROG2. On top of the dendrogram are shown the schematic distances of the 4 replicates for the 3 experimental conditions, right to left: NEUROG2, NEUROG2AQ, and CTL. Expression signal intensities are shown in red and green, indicating high and low expression, respectively. (C) Histogram representing the expression values of PAX6, SOX1, DLL1, and HES6 in the 4 replicates of each biological condition. (D) Expression analysis of predicted NEUROG2 targets on neural tube transversal sections, 6 h following NEUROG2 or NEUROG2AQ electroporation. In the left panels, GFP expression indicating NEUROG2 or NEUROG2AQ expression; in the right panels, detection of transcripts by in situ hybridization. Note that the use of digoxigenin NBT/BCIP complexes to detect the riboprobes masks the GFP signal when gene expression is high (see, for example, NEUROD4). Hence, the GFP signal is just indicative of the electroporated domain.
As expected, many of the already-known NEUROG2 transcriptional targets have their expression consistently modified in our screen. For example, PAX6, SOX1, DLL1, LHX1 (see introduction), and HES6 (14), known to be regulated by NEUROG2 in the neural tube, are found significantly upregulated (DLL1, LHX1, and HES6) or downregulated (PAX6, SOX1) in our experimental context (Fig. 1C and Table 1). Moreover, we compared our data with those of screens already performed to identify NEUROG2 targets in mouse embryonic cortex (18, 37) or in the Xenopus ectoderm (47). We retrieved by our approach 25 out of the 183 genes that were identified in the cortex and 9 out of the 59 that were identified in the Xenopus neurectoderm. Among them are the NHLH1, ELAV4, SNAIL, ZNF238, HES5, or CXCR4 genes that are known to be expressed in the neural tube. Furthermore, we analyzed randomly selected putative targets, and all of them are expressed in the neural tube and have their expression modified 6 h after NEUROG2 electroporation (Fig. 1D and not shown). As our experimental set up was efficient in retrieving classical NEUROG2 targets, it is likely that a large proportion of the novel genes identified are bona fide spinal progenitor NEUROG2 targets.
Table 1.
Examples of some NEUROG2 targets according to gene ontology analysis
| Gene | Gene title | Description | FDR | NGN2/CTL ratio | NGN2/AQ ratio | CTL/AQ ratio |
|---|---|---|---|---|---|---|
| Nervous system development (BP) | ||||||
| Gga.556.1.S1_at | PAX6 | Paired box gene 6 | 5.89E−04 | 0.418 | 0.578 | 1.381 |
| Gga.621.1.S1_at | HOXA2 | Homeobox A2 | 7.32E−03 | 0.532 | 0.639 | 1.202 |
| Gga.789.1.S1_at | LHX1 | LIM homeobox 1 | 4.52E−03 | 1.802 | 1.813 | 1.006 |
| Gga.165.1.S1_at | SOX1 | SRY (sex determining region Y) box 1 | 7.60E−04 | 0.528 | 0.807 | 1.529 |
| Gga.2703.1.S1_at | HES6 | Hairy and enhancer of split 6 | 3.36E−06 | 4.937 | 13.270 | 2.688 |
| Gga.6180.1.S2_at | SMAD3 | SMAD family member 3 | 9.10E−06 | 2.917 | 3.831 | 1.313 |
| Gga.141.1.S1_at | NHLH1 | Nescient helix-loop-helix 1 | 9.61E−06 | 5.953 | 8.086 | 1.358 |
| Notch signaling (KP) | ||||||
| Gga.2894.1.S1_at | JAG1 | Jagged 1 | 2.64E−04 | 2.125 | 4.611 | 2.170 |
| Gga.4932.1.S1_at | JAG2 | Jagged 2 | 3.73E−04 | 2.182 | 2.662 | 1.220 |
| Gga.2283.1.S1_at | DLL1 | Delta-like 1 | 2.97E−05 | 3.348 | 9.069 | 2.709 |
| Gga.3180.1.S2_a_at | LFNG | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase | 2.99E−03 | 1.077 | 1.627 | 1.511 |
| Gga.3754.1.S1_a_at | HES1 | Hairy and enhancer of split 1 | 2.16E−03 | 0.536 | 0.565 | 1.054 |
| Gga.11762.1.S1_at | HES5 | Similar to hairy and enhancer of split 5 | 2.87E−04 | 2.167 | 2.578 | 1.190 |
| Axon guidance (KP) | ||||||
| Gga.2807.1.S1_at | SEMA6B | Semaphorin 6B | 1.69E−05 | 15.456 | 23.337 | 1.510 |
| GgaAffx.21262.1.S1_s_at | SEMA6D | Semaphorin 6D | 2.61E−03 | 2.504 | 3.087 | 1.233 |
| Gga.694.1.S1_at | EPHB1 | EPH receptor B1 | 1.66E−03 | 1.634 | 1.723 | 1.054 |
| Chemotaxis (KP) | ||||||
| Gga.11252.1.S1_at | CCL19 | Chemokine (C-C motif) ligand 19 | 2.16E−03 | 1.948 | 1.943 | 0.997 |
| Gga.2305.1.S1_at | CXCR4 | Chemokine (C-X-C motif) receptor 4 | 5.67E−04 | 0.384 | 0.555 | 1.444 |
| Gga.9513.1.S2_at | CXCL12 | Chemokine (C-X-C motif) ligand 12 | 2.02E−04 | 3.423 | 5.898 | 1.723 |
| Glutamate receptor activity (KP) | ||||||
| GgaAffx.9910.1.S1_s_at | GRIN3A | Glutamate receptor, ionotropic, N-methyl-d-aspartate 3A | 2.09E−04 | 3.738 | 3.300 | 0.883 |
| GgaAffx.1387.1.S1_s_at | GRIK3 | Glutamate receptor, ionotropic, kainate 3 | 6.09E−04 | 2.494 | 2.906 | 1.165 |
| GgaAffx.4124.1.S1_at | GRIK4 | Glutamate receptor, ionotropic, kainate 4 | 7.82E−03 | 1.839 | 1.799 | 0.979 |
| Synaptic transmission (KP) | ||||||
| Gga.19588.1.S1_at | KCNMA1 | Potassium large conductance calcium-activated channel | 7.96E−03 | 1.863 | 1.987 | 1.067 |
| Gga.12259.1.S1_at | KCNMB4 | Potassium large conductance calcium-activated channel | 2.15E−03 | 2.267 | 2.012 | 0.888 |
| Cell cycle (BP) | ||||||
| Gga.1030.1.S1_at | MYC | v-myc viral oncogene homolog | 1.13E−03 | 0.321 | 0.413 | 1.289 |
| Gga.5109.1.S1_s_at | N-MYC | v-myc, neuroblastoma derived | 1.86E−05 | 0.404 | 0.549 | 1.357 |
| Gga.4129.1.S1_at | CCNA2 | Cyclin A2 | 4.50E−03 | 0.587 | 0.576 | 0.982 |
| GgaAffx.11332.1.S1_s_at | CCNE1 | Cyclin E1 | 7.21E−03 | 0.653 | 0.855 | 1.310 |
| GgaAffx.11513.1.S1_s_at | CCNE2 | Cyclin E2 | 2.55E−03 | 0.575 | 0.484 | 0.841 |
| Gga.3039.1.S1_at | CCND1 | Cyclin D1 | 2.09E−03 | 0.575 | 0.562 | 0.977 |
| Gga.3974.1.S2_at | CCND2 | Cyclin D2 | 6.86E−03 | 1.101 | 1.305 | 1.185 |
| Gga.7180.1.S1_s_at | BUB1 | Budding uninhibited by benzimidazole 1 homolog | 4.92E−03 | 0.667 | 0.624 | 0.936 |
| GgaAffx.12026.1.S1_s_at | FOXM1 | Forkhead box M1 | 2.33E−03 | 0.440 | 0.471 | 1.069 |
| Gga.9936.1.S1_at | AURKA | Aurora kinase A | 4.50E−04 | 0.486 | 0.438 | 0.902 |
| GgaAffx.21171.1.S1_s_at | BTG2 | BTG family, member 2 | 7.82E−06 | 3.405 | 4.598 | 1.350 |
| GgaAffx.12080.1.S1_s_at | DCK | Deoxycytidine kinase | 1.90E−03 | 0.491 | 0.501 | 1.018 |
| Gga.1927.1.S1_at | GADD45A | Growth arrest and DNA-damage-inducible, alpha | 1.38E−04 | 2.644 | 3.364 | 1.273 |
| Gga.13515.1.S1_at | LBR | Lamin B receptor | 1.17E−03 | 0.666 | 0.608 | 0.912 |
| Gga.1984.2.A1_a_at | MAD2L1 | MAD2 mitotic arrest deficient-like 1 | 4.92E−03 | 0.715 | 0.716 | 1.002 |
| Gga.4036.1.S1_at | PCNA | Proliferating cell nuclear antigen | 1.78E−05 | 0.662 | 0.663 | 1.002 |
| Gga.2303.1.S1_at | RAD51 | RAD51 homolog (reca homolog, Escherichia coli) | 1.45E−03 | 0.658 | 0.566 | 0.860 |
| Gga.4068.1.S1_at | RANP1 | RAN, member RAS oncogene family pseudogene 1 | 1.04E−03 | 0.849 | 0.854 | 1.005 |
| GgaAffx.12545.1.S1_s_at | RPA1 | Replication protein A1, 70 kDa | 8.73E−05 | 0.634 | 0.610 | 0.962 |
| Gga.1820.1.S1_s_at | RPA2 | Replication protein A2, 32 kDa | 2.01E−03 | 0.773 | 0.734 | 0.950 |
| Gga.13504.1.S1_at | RRM2 | Ribonucleotide reductase M2 polypeptide | 7.91E−05 | 0.587 | 0.496 | 0.846 |
| Gga.421.2.S1_a_at | TK1 | Thymidine kinase 1. soluble | 5.11E−04 | 0.430 | 0.396 | 0.922 |
| Gga.12234.1.S1_at | TYMS | Thymidylate synthetase | 2.72E−03 | 0.700 | 0.699 | 0.998 |
| Gga.9320.1.S1_at | WEE1 | WEE1 homolog | 1.01E−04 | 0.444 | 0.446 | 1.004 |
NEUROG2 rapidly modulates the expression of major players of neural cell proliferation.
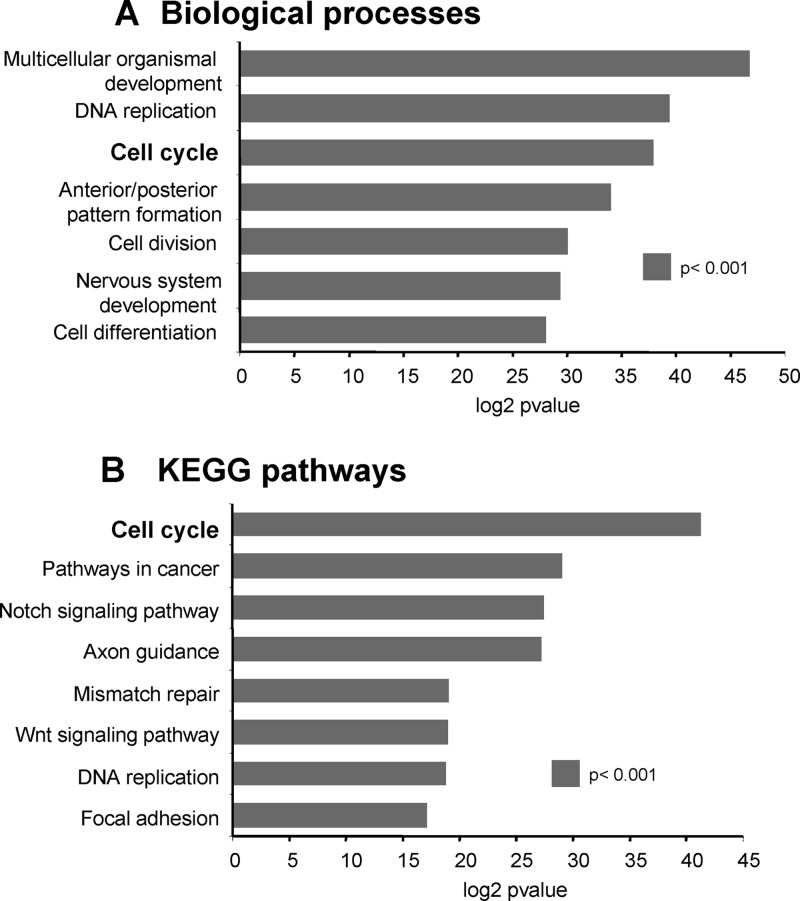
To extract from our data set all the genes related to cell cycle control, we performed a global analysis of the main biological processes modified after NEUROG2 expression using the Genecodis software (9, 39) (http://genecodis.dacya.ucm.es/) (Table 1 and Fig. 2). Among the 942 NEUROG2 targets, 669 annotated genes were processed further. A total of 624 were assigned one or more GO terms: biological process (BP; Fig. 2A) or KEGGS pathways (Kyoto Encyclopedia of Genes and Genomes; KP; Fig. 2B). A large set of targets is involved in biological processes such as cell differentiation (P = 3.87e−08, 34 genes) or nervous system development (P = 3.08e−07, 30 genes, e.g., PAX6, HES6, NHLH1, or LHX1). As expected, numerous members of the NOTCH signaling pathway are significantly regulated (P = 4.30e−06, 10 genes, e.g., JAG1, JAG2, DLL1, LNFNG, or HES1). Several genes related to neuronal function are also enriched in the NEUROG2 transcriptome, such as genes associated with axon guidance (SEMA6B, SEMA6D, EPHB1), synaptic transmission (KCNMB4, KCNMA1), chemotaxis (CCL19, CXCR4, CXCL12), or glutamate receptor activity (GRIN3A, GRIK4, GRIK3). Thus, this global analysis allows identifying gene groups in coherence with known NEUROG2 functions.
Fig 2.
Ontological classification of genes modified by NEUROG2. Graphs showing the number of genes per singular ontological annotation determined by GeneCodis 2.0. (A) Biological processes; (B) KEGG pathways (B) (Kyoto Encyclopedia of Genes and Genomes). The statistical significance is determined by calculating the P value.
Importantly, NEUROG2 also regulates a high proportion of genes involved in the cell cycle, allowing us to examine the molecular mechanisms by which NEUROG2 induces cell cycle arrest (Table 1, Cell cycle). Indeed, cell cycle is one of the most enriched biological processes in KEGGS pathways (BP, P = 1.63e−09, 38 genes; KP, P = 4.06e−09, 19 genes). This subset of NEUROG2 targets includes transcription factors influencing cell proliferation, such as MYCN or FOXM1, as well as genes involved in the core cell cycle machinery, such as WEE1, AURKA, and BUB1, or direct cell cycle regulators, such as CCND1 or CCNE1/2. All these putative targets involved in promoting proliferation are downregulated by NEUROG2. Thus, this global analysis indicates that one of the first functions of NEUROG2 is to repress the expression of positive players of cell proliferation.
Core regulators of the G1 and S phases of the cell cycle are rapidly downregulated by NEUROG2.
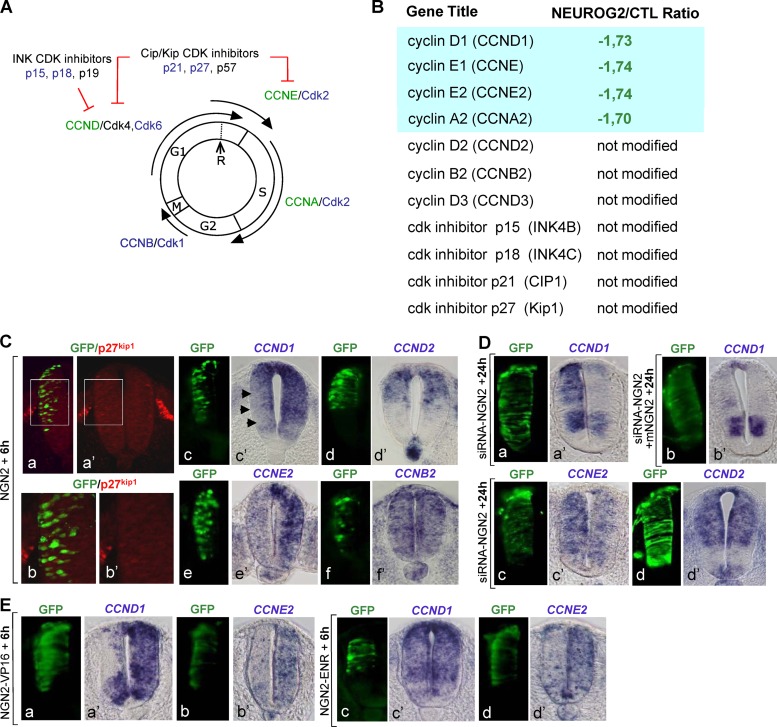
Many cell cycle-related genes are rapidly downregulated after NEUROG2 overexpression, and we analyzed in detail the expression of core cell cycle regulators present in the array (Fig. 3A). First, we found that p27kip1 expression is not modified by NEUROG2 overexpression (Fig. 3B). We confirmed this result by immunohistochemistry showing that p27kip1 protein is not activated 6 h postelectroporation (Fig. 3C), whereas, as previously reported, it clearly accumulated in NEUROG2-transfected cells 48 h hours postelectroporation (see Fig. S2 in the supplemental material) (20, 40). We made similar observations for other inhibitors belonging to the CIP/Kip or the INK families (Fig. 3B). Thus, upregulation of CKIs is not an early response to NEUROG2.
Fig 3.
G1 and S cyclins are rapidly downregulated by NEUROG2. (A) Schematic drawing of the relationship between core regulators of the different phases of the cell cycle. Specific combinations of CDK/cyclin heterodimers allow progression in the different phases of the cell cycle. Two families of cyclin-dependent kinase inhibitors (CKIs) promote cell cycle withdrawal by blocking the activity of cyclin/cyclin-dependent kinase complexes: the Cip/Kip family, including p21cip1, p27kip1, p57kip2, and the INK4 family, including p15 Ink-4b, p18 Ink-4c, and p19 Ink-4d (p16 Ink-4a is not found in the chicken genome). In blue, genes being annotated in the Affymetrix array but not modified; in black, genes not found on the array; in green, genes downregulated by NEUROG2. (B) Table listing the NEUROG2/CTL expression ratio of the core cell cycle regulators present on the array. (C) Analysis of the expression of some cell cycle regulators 6 h following NEUROG2 (NGN2) electroporation of HH10 embryos. The electroporated side is on the left on transversal sections (GFP in green). p27Kip1 protein was detected by immunohistochemistry (panels b and b′ are close-ups of panels a and a′, respectively). See that no electroporated cells (in green) express p27Kip1 protein (in red). Transcripts of CCND1 (c′), CCND2 (d′), CCNE2 (e′), and CCNB2 (f′) were detected by in situ hybridization. As predicted by the microarray data, CCND1 and CCNE2 are downregulated by NEUROG2. (D) Analysis of the expression of CCND1 (a and a′), CCNE2 (c and c′), and CCND2 (d and d′) 24 h following NEUROG2 downregulation by siRNA-NEUROG2 electroporation (siRNA-NGN2). CCND1 and CCNE2 transcripts are upregulated (a′ and c′), confirming that NEUROG2 is necessary for their repression. (b and b′) Coelectroporation of a cDNA expressing a mouse NEUROG2 (mNGN2) with the siRNA-NEUROG2 restores a normal expression of CCND1. (E) Analysis of the expression of CCND1 and CCNE2 6 h following electroporation of NEUROG2-VP16 (NGN2-VP16) or NEUROG2-EnR (NGN2-ENR). CCND1 and CCNE2 are downregulated by NEUROG2-VP16 (a′ and b′). CCND1 transcripts are not reduced by NEUROG2-ENR (c and c′), whereas CCNE2 transcripts are significantly reduced (d and d′).
Instead, we found that CCND1, CCNE1, CCNE2, and CCNA2, acting at the G1 and S phases of the cell cycle, are downregulated within 6 h (Fig. 3B). The expression of their associated CDKs is not modified under these conditions (Fig. 3B). Moreover, the expression of CCNB2 involved in the G2/M transition is not changed (Fig. 3B). From the three D-type cyclins (CCND) involved in G1 progression, only CCND1 is downregulated, with CCND2 and CCND3 being unaffected (Fig. 3B). Analysis of our data set therefore suggests that among direct cell cycle regulators, only a subtype of cyclins has its expression modified by NEUROG2, but not by NEUROG2AQ, indicating that this regulation is DNA binding dependent. We validated these data obtained with our transcriptomic approach by analyzing in more detail the regulation of two of these cyclins: CCND1 and CCNE2. Using in situ hybridization, we confirmed that CCND1 and CCNE2 are rapidly downregulated following NEUROG2 gain of function (Fig. 3C). To ascertain the specificity of NEUROG2 action on these cell cycle regulators, we analyzed their expression in loss-of-function conditions, using two different siRNAs to target endogenous NEUROG2 (see Materials and Methods). We started verifying that our siRNAs extinguish specifically NEUROG2 transcript and lead to a reduction of neuronal differentiation (see Fig. S3 in the supplemental material). We then tested the expression of CCND1 and CCNE2 24 h after siRNA electroporation. As seen on Fig. 3D, the acute reduction of endogenous NEUROG2 provokes an accumulation of CCND1 and CCNE2 transcripts without affecting CCND2 expression as expected from our transcriptomic data. This upregulation doesn't occur when the siRNA is coelectroporated with a vector expressing the mouse NeuroG2, confirming the specificity of the siRNAs (Fig. 3D). NEUROG2 is thus necessary for CCND1 and CCNE2 downregulation in spinal progenitors. Hence, among the core regulators controlling the cell cycle, only a subset of cyclins governing the G1 and S phases are rapidly and specifically downregulated in response to NEUROG2.
Although NEUROG2 has not been shown to act as a transcriptional repressor, we nevertheless tested whether this repression could be direct. We used two NEUROG2 modified forms: a NEUROG2VP16 form acting as a constitutive activator and a NEUROG2EnR form acting as a constitutive repressor in the cerebral cortex (C. Schuurmans, personal communication). We assumed that if NEUROG2 acts as a transcriptional activator, NEUROG2VP16 would mimic the effect of NEUROG2, NEUROG2EnR having no or the opposite effect. We validated these constructs by looking at PAX6 expression that we know to be indirectly repressed by NEUROG2 in the neural tube (M. Lacomme, unpublished observation). As expected, PAX6 expression is strongly repressed by NEUROG2VP16 but not significantly affected by NEUROG2EnR in the neural tube (see Fig. S4 in the supplemental material). We tested the expression of CCND1 and CCNE2 in the same experimental conditions. We found that CCND1 is strongly repressed by NEUROG2VP16, NEUROG2EnR having little or no effect (Fig. 3E), indicating that CCND1 is repressed indirectly by NEUROG2. However, the regulation of CCNE2 appears to be more complex, as we observed a reduction of CCNE2 transcripts with both NEUROG2VP16 and NEUROG2EnR (Fig. 3E). Hence, these experiments indicate that NEUROG2 represses CCND1 and CCNE2 in an indirect manner but open the possibility that CCNE2 is also repressed by a direct mechanism (see Discussion).
NEUROG2 rapidly impedes cell cycle reentry of spinal progenitors.
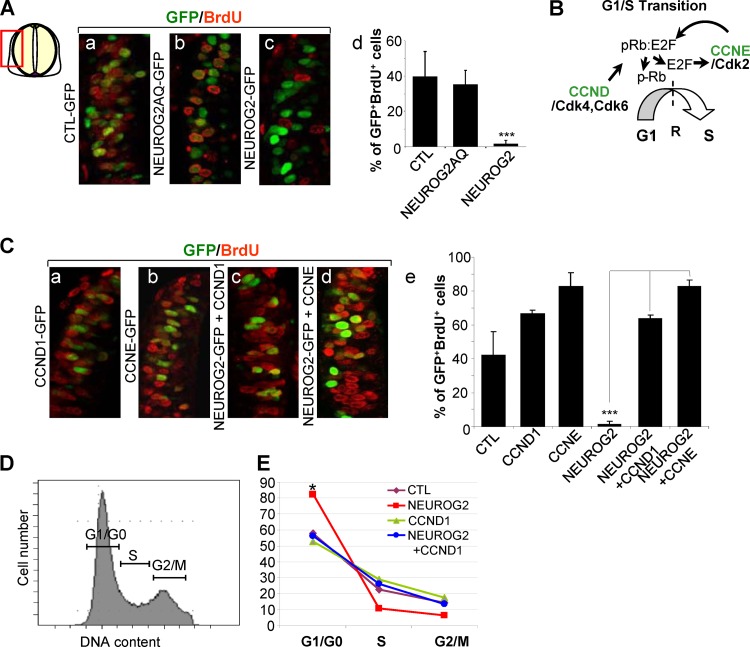
The rapid downregulation of G1/S cyclins suggests that within 6 h, neural cells overexpressing NEUROG2 no longer enter S phase. To test this hypothesis, we measured the percentage of cells in S phase in transfected populations overexpressing NEUROG2, NEUROG2AQ, or CTL 6 h after electroporation, by applying a 30-min BrdU pulse just before collecting the embryos (Fig. 4A). As expected from the array data, NEUROG2AQ does not modify the BrdU incorporation rate (38.2% ± 8% of the NEUROG2AQ cells incorporated BrdU compared with 41.8% ± 14% in the CTL condition). In contrast, we observed a dramatic fall of this percentage upon NEUROG2 overexpression (2.1% ± 2%) (Fig. 4A), indicating that cells overexpressing NEUROG2 are retained in some cell cycle phases. If NEUROG2 controls the G1/S transition, neural cells overexpressing NEUROG2 should be kept in G1. We thus performed FACS analysis to determine the distribution of transfected cells in the different phases of the cell cycle 6 h after NEUROG2 overexpression. We observed that NEUROG2-positive cells clearly start to accumulate at the G1/G0 phase of the cell cycle (Fig. 4E) (74.6% ± 6.4% of the NEUROG2-positive cells versus 60.6% ± 2.9% in CTL cells; P < 0.05). As these cells do not yet express p27kip1 or the β tubulin protein (see Fig. S1 in the supplemental material), our results indicate that NEUROG2 upregulation causes a rapid arrest in G1 preceding the upregulation of CKIs.
Fig 4.
NEUROG2 blocks S phase reentry via the repression of CCND1 and CCNE. (Aa to Ac and Ca to Cd) Close-ups on transversal sections of embryos electroporated with the mentioned constructs and incubated 30 min with BrdU just before harvesting, 6 h following electroporation. GFP in green; BrdU in red. (Ad and Ce) Histograms presenting the quantification of the BrdU incorporation rate in the mentioned experimental conditions (***, P < 0.001; Student t test). (B) Schematic representation of G1/S transition, which is controlled by two families of cyclin: CCND and CCNE. An important restriction point (R) is located at the end of G1 phase. The passage of this point leads to nonreversible passage through S phase. Phosphorylation of pRb by CDK4/6 cyclin D leads to E2F release. E2F promotes cyclin E transcription that in turn further phosphorylates pRb releasing more E2F. High levels of E2F promote the passage through the restriction point in late G1 and activate CCNA transcription that drives cells through S phase and replication. (D) Flow cytometry analysis of cell cycle phase distribution: representative example of the profile of cells expressing control DNA. (E) Curves showing the repartition of the cells in G1/G0, S, and G2/M phases measured on FACS profiles of GFP-positive cells 6 h following electroporation with CTL, NEUROG2, CCND1, or NEUROG2 and CCND1 (*, P < 0.05; Student t test).
Shutting down CCND1 and CCNE is needed to hold cells in G1 downstream of NEUROG2.
If the cell accumulation in G1 phase that we observed following NEUROG2 electroporation is directly linked to the repression of G1/S cyclin expression, maintaining one of these cyclins in otherwise NEUROG2-expressing cells should restore cell cycle progression. To test this, we electroporated NEUROG2 alone or in conjunction with CCND1 or CCNE and 6 h after performed a 30-min BrdU pulse just before collecting the embryos (Fig. 4C). Cotransfection of either CCND1 or CCNE with NEUROG2 fully rescues the lack of BrdU incorporation observed with NEUROG2 alone, to a level similar to that observed with CCND1 or CCNE alone (Fig. 4C) (63.8% ± 2% of GFP+ BrdU+ cells with NEUROG2 and CCND1 and 87% ± 8% with NEUROG2 and CCNE versus 2.1% ± 2% with NEUROG2 alone). In addition, FACS analysis shows that when coelectroporated with NEUROG2, CCND1 prevents cell accumulation in the G1/G0 phases observed with NEUROG2 alone (Fig. 4E) (74.6% ± 6.4% of the NEUROG2+ cells versus 57% ± 6.5% in NEUROG2+ CCND1+ cells; P < 0.05). This finding links the early NEUROG2-driven G1 accumulation to the repression of G1 and S cyclins.
NEUROG2 coordinates cell cycle exit with neuronal differentiation.
The results presented so far show that NEUROG2 represses specifically a subset of cell cycle regulators that drive cell cycle arrest in G1. Given that NEUROG2 also represses patterning genes and initiates differentiation genes, we wondered if these different events were dependent on each other or could be controlled independently by NEUROG2. It has been proposed that in the developing cortex, CCND1 downregulation is sufficient to trigger neuronal differentiation, suggesting that neuronal differentiation could be a consequence of cell cycle exit. We checked to see whether in our experimental conditions CCND1 downregulation was sufficient to induce precocious neuronal differentiation. The downregulation of CCND1, using a short hairpin RNA (shRNA) construct (RNAiD1) (34), leads to a significant reduction of BrdU incorporation as seen 24 h postelectroporation (see Fig. S5 in the supplemental material) (27% ± 4.6% of RNAiD1+ BrdU+ cells and 37.5% ± 7.4% of GFP+ BrdU+ cells in a control). However, it is not accompanied by an increase of TUJ1+ cells, indicating that reducing the CCND1 level is not sufficient to promote neuronal differentiation (see Fig. S5) (5.9% ± 1.5% of RNAiD1+ TUJ1+ cells and 6.1% ± 1.3% of GFP+ TUJ1+ cells in a control).
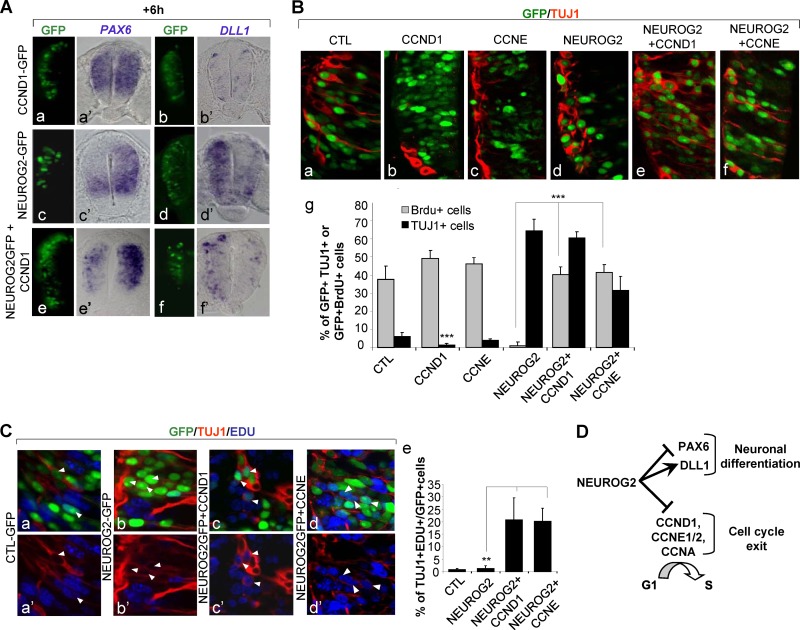
We then examined if, on the contrary, the maintenance of one G1/S cyclin would impede the capacity of NEUROG2 to push cells out of the progenitor state and trigger neuronal differentiation. We started analyzing the expression pattern of PAX6 6 h following NEUROG2 and CCND1 electroporation (Fig. 5A). PAX6 transcripts expression is repressed by NEUROG2 even in the presence of CCND1. We also observed that DLL1 is upregulated in NEUROG2 and CCND1 transfected cells (Fig. 5A). These data show that NEUROG2 cells, despite maintaining CCND1 expression, are still able to extinguish progenitor genes and to commit to the neuronal fate. We asked if these NEUROG2 and CCND1 transfected cells were able to initiate neuronal differentiation by analyzing the differentiation status of cells overexpressing NEUROG2, CCND1, CCNE, or combinations of them 24 h postelectroporation. As already described, electroporation of NEUROG2 alone leads to precocious neuronal differentiation (Fig. 5B) (64.3% ± 6.7% of NEUROG2+ cells express TUJ1 instead of 6.2% ± 1.9% in a control). Interestingly, we also detect many GFP+ TUJ1+ cells in the ventricular zone of the neural tube electroporated either with NEUROG2 and CCND1 or NEUROG2 and CCNE (Fig. 5B) (60.4% ± 3.7% or 31.7% ± 7.5%, respectively), whereas neither of these cyclins alone is able to induce an early expression of TUJ1 (1.2% ± 0.9% of GFP+ TUJ1+ cells for CCND1 and 1% ± 0.5% for CCNE). This indicates that NEUROG2 can promote expression of differentiation markers independently of G1/S cyclin repression.
Fig 5.
NEUROG2 can control differentiation independently of cell cycle exit. (A) Detection of PAX6 (a′, c′, and e′) and DLL1 (b′, d′, and f′) transcripts on transversal sections by in situ hybridization 6 h following electroporation with CCND1 (a and b′), NEUROG2 (c and d′), or NEUROG2 and CCND1 (e and f′). PAX6 and DLL1 transcripts are modified by NEUROG2 even in the presence of high levels of cyclin D1. (B) Detection of GFP+ TUJ1+ cells 24 h postelectroporation with the mentioned constructs. (a and f) Close-up on transversal sections of embryos electroporated with the mentioned constructs 24 h following electroporation and analysis of TUBB3 (Tuj1). GFP in green; Tuj1 in red. (g) Histograms presenting the T quantification of GFP+ Tuj1+ cells (in black) and GFP+ BrdU+ cells (in gray) in the mentioned experimental conditions, 24 h following electroporation (**, P < 0.01; ***, P < 0.001; Student t test). (C) Detection of GFP+ TUJ1+ EDU+ cells 24 h postelectroporation with the mentioned constructs. (a to d′) Close-up on transversal sections; GFP in green, EdU in blue, TUJ1 in red. (e) Histograms presenting the quantification of TUJ1+ EDU+ cells among the GFP+ cells in the mentioned experimental conditions (**, P < 0.01; ***, P < 0.001; Student t test). (D) NEUROG2 can control neuronal differentiation independently of cell cycle exit.
This could be due to the fact that cells coelectroporated with NEUROG2 and CCND1 or CCNE manage to escape the cell cycle to differentiate. To check if these cells are still cycling, we performed a 30-min BrdU pulse 24 h postelectroporation. Cells misexpressing NEUROG2 alone do not incorporate BrdU at 24 h as expected from data obtained at 6 h (Fig. 5B) (0.93% ± 2.3% of NEUROG2+ BrdU+ cells). However, cells transfected with NEUROG2 and CCND1 or NEUROG2 and CCNE are still proliferating 24 h postelectroporation (Fig. 5B) (37.5% ± 7.3% of GFP+ BrdU+ control cells, 40% ± 4.6% of NEUROG2+ CCND1+ BrdU+ cells, and 41.5% ± 4.4% of NEUROG2+ CCNE+ BrdU+ cells). This suggests that a large proportion of cells being kept in the cell cycle are able to initiate their differentiation.
We further analyzed in these different conditions the number of cells both cycling and expressing TUJ1. To circumvent incompatibility between antibodies, we used EdU instead of BrdU to mark proliferating cells. We applied a 30-min pulse of EdU before harvesting the embryos and measured differentiating neurons that incorporated EdU (Fig. 5C). When electroporating a control vector, 0.77% ± 0.48% transfected cells are EdU+ TUJ1+, indicating that few cells start to express neuronal markers before they accomplish their last division. Similarly when electroporating NEUROG2 alone, only 1.1% ± 1.1% of transfected cells are EdU+ TUJ1+, consistent with the fact that NEUROG2 rapidly triggers cell cycle exit. In contrast, the proportion of EdU+ TUJ1+ cells is increased by 20-fold in the GFP+ population when NEUROG2 is coelectroporated either with CCND1 or CCNE (20.7% ± 8.8% and 20.2% ± 5.2% of EDU+ TUJ1+ cells, respectively). This indicates that a large proportion of NEUROG2 and CCND1 or NEUROG2 and CCNE cells start to differentiate despite being still in the cell cycle.
All together, these observations show that in the developing neural tube NEUROG2 can trigger expression of markers of neuronal differentiation independently of cell cycle arrest. This indicates that cell cycle exit is not an indirect consequence of neuronal differentiation but rather an event which is controlled in parallel by NEUROG2 (Fig. 5D). Hence, in the developing spinal cord, cell cycle exit can be uncoupled from neuronal differentiation, and a main function of NEUROG2, in the course of normal development, is to coordinate these two separable events.
DISCUSSION
In this study, we set up a global approach that allowed us to define how NEUROG2 induces cell cycle arrest in spinal progenitors. We found that NEUROG2 represses specifically some cyclins, including CCND1 and CCNE1/2, acting at the G1 and S phases of the cell cycle. We showed that the downregulation of these cyclins prevents S phase entry and favors withdrawal from the cell cycle. Conversely, maintaining CCND1 or CCNE expression was sufficient to prevent the cell cycle arrest mediated by NEUROG2, without affecting the capacity of NEUROG2 to promote neuronal differentiation. All together, our study unmasks the mechanisms by which NEUROG2 triggers cell cycle exit and indicates that cell cycle withdrawal and neuronal differentiation are two independent processes regulated in parallel by NEUROG2.
NEUROG2 specifically represses G1 and S phase cyclins to induce a rapid G1 phase arrest.
We showed that NEUROG2 specifically represses some G1 and S phase cyclins and that this repression occurs prior to p27KIP1 activation. This is in accordance with the work of Gui et al., who proposed that the decision to withdraw from the cell cycle occurs before CKI activation in the spinal cord, at the end of mitosis or early G1 (20). Here, we identified a molecular mechanism that accounts for this early cell cycle arrest (Fig. 6). It has been shown that later on, nascent neurons express CKI of the CIP/KIP family (20, 40). All together, these data argue for a multistep process of cell cycle withdrawal, involving early repression of G1/S phase cyclins by NEUROG2 for the initial decision to stop cycling and a later action of CKIs such as p27KIP1or p57KIP2 that could, by blocking residual CDK/cyclin activity, reinforce blockade of S phase entry, thereby rendering cell cycle withdrawal irreversible (Fig. 6). A recent study performed in a Xenopus embryo shows that CDK activity can phosphorylate NEUROG2, thereby destabilizing the protein and preventing neurogenesis (1). The rapid downregulation of CCND1, CCNE, and CCNA, by reducing the level of CDK activity, may contribute to stabilizing NEUROG2 protein, thus turning the balance toward neuronal differentiation.
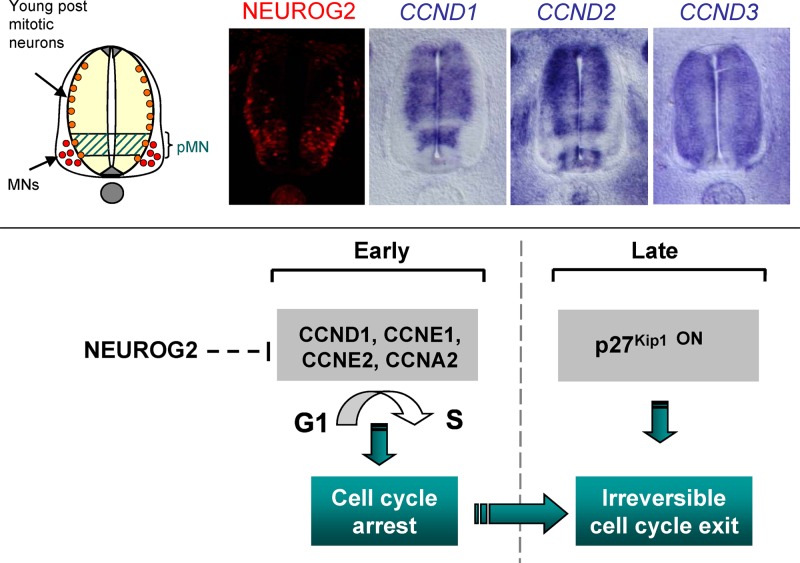
Fig 6.
Cell cycle arrest downstream of NEUROG2 is a multistep process initiated by G1 and S cyclin repression. (Top) Expression of the NEUROG2 protein compared with CCND1, CCND2, and CCND3 transcripts on neural tube section of E3 chick embryos (HH18 and HH19). (Bottom) Scheme illustrating that NEUROG2 controls cell cycle arrest by a multistep process: NEUROG2 initially triggers cell cycle arrest via the repression of G1 and S cyclins. In a second phase, activation of a CKI such as p27KIP1 leads to irreversible cell cycle withdrawal in differentiated neurons.
NEUROG2-positive cells are rapidly arrested in G1. The G1 phase is a critical phase allowing response to extracellular cues that induce either a further round of division through reentry into the S phase or withdrawal from the cell cycle (23). It is long known that, in the cortex, neurogenesis correlates with a lengthening of G1 phase (8, 28, 33). It has been recently proposed that this G1 phase lengthening may be required to provide enough time for cell fate determinants to be effective (8). Hence, time may be a limiting factor for cell fate change to occur, and a relatively long G1 phase may allow a switch to neurogenesis. Thus, in addition to favoring cell cycle exit, NeuroG2-induced G1 arrest could allow spinal neuronal precursors to accumulate proteins necessary for their differentiation. This could be a good way to couple cell cycle exit with neuronal differentiation.
It has been shown recently that the proneural Ascl1 gene has a positive action both on G1/S and G2/M core regulators, as it induces Cdk1, Cdk2, E2f1, and Cdc25B expression in cortical cells (10). We didn't detect any induction of positive players of the cell cycle with NEUROG2, but we cannot exclude that low levels of NEUROG2, prior to triggering cell cycle exit of neuronal precursors, could also promote proliferation of neural progenitors.
Mode of regulation of G1 and S cyclins by NEUROG2.
One remaining question is how NEUROG2 represses CCND1 and CCNE expression. D type cyclins are the targets of extracellular signals and major effectors of G1 phase progression (27, 31). Their repression causes a reduction of the hyperphosphorylated form of pRb, leading to a decrease of the E2F-dependent transcription of CCNE1/2 and CCNA1/2 and thereby triggering cell cycle arrest. Cyclin D1 repression could be thus sufficient to account for the CCNE/A rapid downregulation we observed. Alternatively, NEUROG2 could repress CCND1 and CCNE1/2 all at once to ensure a more efficient block of the S phase entry. NEUROG2 acts mainly as an activator, but it is also able to repress some genes by sequestering CBP/p300. The observation that NEUROG2AQ doesn't mimic the NEUROG2 effect on these cyclins argues against a repression through CBP/p300 sequestration. By using NEUROG2VP16 and NEUROG2EnR, we show that the repression of CCND1 is indirect and requires NEUROG2 activator's activity, suggesting the involvement of an intermediary gene. For CCNE2, we also observe a reduction of CCNE2 transcript level with NEUROG2VP16, indicating an indirect repression. This could be the result of CCND1 downregulation and of the resulting decrease of the E2F-dependent transcription of CCNE2 as described above. However, we observe as well a reduction of CCNE2 transcript with NEUROG2EnR, suggesting that NEUROG2 could repress CCNE2 directly, thereby allowing a sharp cell cycle arrest. Consistent with this result, we found a putative NEUROG2-specific EBOX in the vicinity of the CCNE2 promoter (data not shown). Resolving the mechanism of CCND1 and CCNE regulation by NEUROG2 is important but will require in-depth investigation.
Cyclin D1 is the only D-type cyclin to be repressed by NEUROG2.
An intriguing finding is that among the three D-type cyclins, only CCND1 is inhibited. All three G1 cyclins interact with CDK4/6 to trigger G1 progression. One possibility is that CCND2 and CCND3, in addition to controlling the cell cycle, also play a role in postmitotic neurons. Indeed, CCND3 is barely expressed in progenitors and seems to accumulate preferentially in differentiating neurons, and CCND2 is reinforced at the basal side of the neural tube (Fig. 6), suggesting that it could be expressed in young postmitotic neurons. These cyclins could be required in these cells to sustain proliferation arrest, as it has been described for CCND3 in differentiated muscle cells (11). Another rationale for repressing specifically CCND1 could come from the fact that CCND1 can also control proliferation via a transcriptional function, directly binding to the NOTCH upstream regulatory sequences where it recruits CBP/p300 to trigger proliferation (6). NOTCH signaling is involved in maintaining or even stimulating the proliferative state (32) and is also rapidly modulated by NEUROG2. Repressing CCND1 would thus be an efficient way for NEUROG2 to rapidly block cell cycle progression, directly via CCND1 and indirectly via the NOTCH signaling pathway.
NEUROG2 coordinates cell cycle exit with neuronal differentiation.
Whether neuronal differentiation mediated by NEUROG2 can be controlled independently of cell cycle exit is a long-standing debate. We show that maintaining CCND1, despite keeping NEUROG2+ cells cycling, doesn't prevent premature expression of markers of neuronal differentiation triggered by NEUROG2. These observations indicate that in the spinal cord, neuronal differentiation can be controlled independently of cell cycle exit by NEUROG2. This is in accordance with studies showing that cell cycle exit is not a prerequisite for neuronal differentiation (30) and is not sufficient to trigger neuronal differentiation (12, 16). Moreover, it indicates that cell cycle withdrawal is not a consequence of neuronal differentiation. However, in the course of normal development, these two events have to be properly coordinated in order to avoid the production of aberrant cycling neurons. Here, we identify an early effect of NEUROG2 on cell cycle arrest, via the specific repression of cyclin of the G1 and S phases of the cell cycle. This finding provides molecular evidence that NEUROG2, while regulating specification and differentiation genes, also actively controls cell cycle regulators, thereby coupling neuronal differentiation to cell cycle withdrawal.
Supplementary Material
ACKNOWLEDGMENTS
We thank François Medevielle for providing efficient technical assistance, the Plateforme Biostatistique, Genopôle Toulouse Midi-Pyrénées, in particular Sébastien Déjean from the Institut de Mathematiques de Toulouse and Magali SanCristobal from the Laboratoire de Genetique Cellulaire, and the Imagerie Platform TRI Toulouse and in particular Fatima L'faqihi and Valérie Duplan for cell sorting and FACS analysis. We also thank O. Tassy and O. Pourquié for giving us access to the Manteia database and C. Schuurmans for sharing unpublished constructs. Finally, we thank members of the lab for helpful discussions and E. Cau, A. Davy, V. Lobjois, C. Monod-Wissler, S. Plaza, and A. Vincent for critical reading of the manuscript.
M.L. was supported by a grant from CNRS and region Midi-Pyrenees.
Footnotes
Published ahead of print 30 April 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Ali F, et al. 2011. Cell cycle-regulated multi-site phosphorylation of neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development 138:4267–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bel-Vialar S, Itasaki N, Krumlauf R. 2002. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development 129:5103–5115 [DOI] [PubMed] [Google Scholar]
- 3. Bel-Vialar S, Medevielle F, Pituello F. 2007. The on/off of Pax6 controls the tempo of neuronal differentiation in the developing spinal cord. Dev. Biol. 305:659–673 [DOI] [PubMed] [Google Scholar]
- 4. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B (Methodol.) 57:289–300 [Google Scholar]
- 5. Bertrand N, Castro DS, Guillemot F. 2002. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3:517–530 [DOI] [PubMed] [Google Scholar]
- 6. Bienvenu F, et al. 2010. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature 463:374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blader P, Fischer N, Gradwohl G, Guillemot F, Strahle U. 1997. The activity of neurogenin1 is controlled by local cues in the zebrafish embryo. Development 124:4557–4569 [DOI] [PubMed] [Google Scholar]
- 8. Calegari F, Haubensak W, Haffner C, Huttner WB. 2005. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J. Neurosci. 25:6533–6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. 2007. GENECODIS: a Web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 8:R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Castro DS, et al. 2011. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 25:930–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cenciarelli C, et al. 1999. Critical role played by cyclin D3 in the MyoD-mediated arrest of cell cycle during myoblast differentiation. Mol. Cell. Biol. 19:5203–5217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. El Wakil A, Francius C, Wolff A, Pleau-Varet J, Nardelli J. 2006. The GATA2 transcription factor negatively regulates the proliferation of neuronal progenitors. Development 133:2155–2165 [DOI] [PubMed] [Google Scholar]
- 13. Farah MH, et al. 2000. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development 127:693–702 [DOI] [PubMed] [Google Scholar]
- 14. Fior R, Henrique D. 2005. A novel hes5/hes6 circuitry of negative regulation controls Notch activity during neurogenesis. Dev. Biol. 281:318–333 [DOI] [PubMed] [Google Scholar]
- 15. Garcia-Campmany L, Marti E. 2007. The TGFbeta intracellular effector Smad3 regulates neuronal differentiation and cell fate specification in the developing spinal cord. Development 134:65–75 [DOI] [PubMed] [Google Scholar]
- 16. Garcia-Dominguez M, Poquet C, Garel S, Charnay P. 2003. Ebf gene function is required for coupling neuronal differentiation and cell cycle exit. Development 130:6013–6025 [DOI] [PubMed] [Google Scholar]
- 17. Ge W, et al. 2006. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc. Natl. Acad. Sci. U. S. A. 103:1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gohlke JM, et al. 2008. Characterization of the proneural gene regulatory network during mouse telencephalon development. BMC Biol. 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gradwohl G, Fode C, Guillemot F. 1996. Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev. Biol. 180:227–241 [DOI] [PubMed] [Google Scholar]
- 20. Gui H, Li S, Matise MP. 2007. A cell-autonomous requirement for Cip/Kip cyclin-kinase inhibitors in regulating neuronal cell cycle exit but not differentiation in the developing spinal cord. Dev. Biol. 301:14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hamburger V. 1992. The stage series of the chick embryo. Dev. Dyn. 195:273–275 [DOI] [PubMed] [Google Scholar]
- 22. Henrique D, et al. 1995. Expression of a delta homologue in prospective neurons in the chick. Nature 375:787–790 [DOI] [PubMed] [Google Scholar]
- 23. Ho A, Dowdy SF. 2002. Regulation of G(1) cell-cycle progression by oncogenes and tumor suppressor genes. Curr. Opin. Genet. Dev. 12:47–52 [DOI] [PubMed] [Google Scholar]
- 24. Itasaki N, Bel-Vialar S, Krumlauf R. 1999. “Shocking” developments in chick embryology: electroporation and in ovo gene expression. Nat. Cell Biol. 1:E203–E207 doi:10.1038/70231 [DOI] [PubMed] [Google Scholar]
- 25. Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. 2008. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat. Neurosci. 11:1247–1251 [DOI] [PubMed] [Google Scholar]
- 26. Kang G, Ye K, Liu N, Allison DB, Gao G. 2009. Weighted multiple hypothesis testing procedures. Stat. Appl. Genet. Mol. Biol. 8:Article23 doi:10.2202/1544-6115.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kenney AM, Rowitch DH. 2000. Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol. Cell. Biol. 20:9055–9067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lange C, Huttner WB, Calegari F. 2009. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell 5:320–331 [DOI] [PubMed] [Google Scholar]
- 29. Lee SK, Lee B, Ruiz EC, Pfaff SL. 2005. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 19:282–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lobjois V, Bel-Vialar S, Trousse F, Pituello F. 2008. Forcing neural progenitor cells to cycle is insufficient to alter cell-fate decision and timing of neuronal differentiation in the spinal cord. Neural Dev. 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lobjois V, Benazeraf B, Bertrand N, Medevielle F, Pituello F. 2004. Specific regulation of cyclins D1 and D2 by FGF and Shh signaling coordinates cell cycle progression, patterning, and differentiation during early steps of spinal cord development. Dev. Biol. 273:195–209 [DOI] [PubMed] [Google Scholar]
- 32. Louvi A, Artavanis-Tsakonas S. 2006. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 7:93–102 [DOI] [PubMed] [Google Scholar]
- 33. Lukaszewicz A, et al. 2005. G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron 47:353–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lukaszewicz AI, Anderson DJ. 2011. Cyclin D1 promotes neurogenesis in the developing spinal cord in a cell cycle-independent manner. Proc. Natl. Acad. Sci. U. S. A. 108:11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma Q, Kintner C, Anderson DJ. 1996. Identification of neurogenin, a vertebrate neuronal determination gene. Cell 87:43–52 [DOI] [PubMed] [Google Scholar]
- 36. Marigo V, Johnson RL, Vortkamp A, Tabin CJ. 1996. Sonic hedgehog differentially regulates expression of GLI and GLI3 during limb development. Dev. Biol. 180:273–283 [DOI] [PubMed] [Google Scholar]
- 37. Mattar P, et al. 2004. A screen for downstream effectors of Neurogenin2 in the embryonic neocortex. Dev. Biol. 273:373–389 [DOI] [PubMed] [Google Scholar]
- 38. Mizuguchi R, et al. 2001. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron 31:757–771 [DOI] [PubMed] [Google Scholar]
- 39. Nogales-Cadenas R, et al. 2009. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 37:W317–W322 doi:10.1093/nar/gkp416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Novitch BG, Chen AI, Jessell TM. 2001. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 31:773–789 [DOI] [PubMed] [Google Scholar]
- 41. Ozbudak EM, Tassy O, Pourquie O. 2010. Spatiotemporal compartmentalization of key physiological processes during muscle precursor differentiation. Proc. Natl. Acad. Sci. U. S. A. 107:4224–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perez SE, Rebelo S, Anderson DJ. 1999. Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Development 126:1715–1728 [DOI] [PubMed] [Google Scholar]
- 43. Philippe JM, Garces A, deLapeyiere O. 1998. Fgf-R3 is expressed in a subset of chicken spinal motorneurons. Mech. Dev. 78:119–123 [DOI] [PubMed] [Google Scholar]
- 44. Pituello F, Yamada G, Gruss P. 1995. Activin A inhibits Pax-6 expression and perturbs cell differentiation in the developing spinal cord in vitro. Proc. Natl. Acad. Sci. U. S. A. 92:6952–6956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roztocil T, Matter-Sadzinski L, Alliod C, Ballivet M, Matter JM. 1997. NeuroM, a neural helix-loop-helix transcription factor, defines a new transition stage in neurogenesis. Development 124:3263–3272 [DOI] [PubMed] [Google Scholar]
- 46. Scardigli R, Schuurmans C, Gradwohl G, Guillemot F. 2001. Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron 31:203–217 [DOI] [PubMed] [Google Scholar]
- 47. Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL. 2007. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 26:5093–5108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sugimori M, et al. 2007. Combinatorial actions of patterning and HLH transcription factors in the spatiotemporal control of neurogenesis and gliogenesis in the developing spinal cord. Development 134:1617–1629 [DOI] [PubMed] [Google Scholar]
- 49. Sun Y, et al. 2001. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell 104:365–376 [DOI] [PubMed] [Google Scholar]
- 50. Theodorakis K, Kyriakopoulou K, Wassef M, Karagogeos D. 2002. Novel sites of expression of the bHLH gene NSCL1 in the developing nervous system. Mech. Dev. 119(Suppl 1:S103–S106 [DOI] [PubMed] [Google Scholar]
- 51. Zhang SO, Mathur S, Hattem G, Tassy O, Pourquie O. 2010. Sex-dimorphic gene expression and ineffective dosage compensation of Z-linked genes in gastrulating chicken embryos. BMC Genomics 11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.