Abstract
Meiotic recombination hot spot locus (mrhl) RNA is a nuclear enriched long noncoding RNA encoded in the mouse genome and expressed in testis, liver, spleen, and kidney. mrhl RNA silencing in Gc1-Spg cells, derived from mouse spermatogonial cells, resulted in perturbation of expression of genes belonging to cell adhesion, cell signaling and development, and differentiation, among which many were of the Wnt signaling pathway. A weighted gene coexpression network generated nine coexpression modules, which included TCF4, a key transcription factor involved in Wnt signaling. Activation of Wnt signaling upon mrhl RNA downregulation was demonstrated by beta-catenin nuclear localization, beta-catenin–TCF4 interaction, occupancy of beta-catenin at the promoters of Wnt target genes, and TOP/FOP-luciferase assay. Northwestern blot and RNA pulldown experiments identified Ddx5/p68 as one of the interacting proteins of mrhl RNA. Downregulation of mrhl RNA resulted in the cytoplasmic translocation of tyrosine-phosphorylated p68. Concomitant downregulation of both mrhl RNA and p68 prevented the nuclear translocation of beta-catenin. mrhl RNA was downregulated on Wnt3a treatment in Gc1-Spg cells. This study shows that mrhl RNA plays a negative role in Wnt signaling in mouse spermatogonial cells through its interaction with p68.
INTRODUCTION
In recent years, there has been an explosion in the discovery of several classes of noncoding RNAs which constitute a huge repertoire of gene regulatory molecules in higher eukaryotes (10). The discovery of these molecules has made a tremendous impact in our understanding of higher-order genome regulation and the complexity of an organism itself. The small noncoding RNAs such as microRNA (miRNA) and small interfering RNA (siRNA) play critical roles in transcriptional and posttranscriptional gene regulation and also in fine-tuning the level of expression of cognate mRNAs (1, 12). Another class of regulatory noncoding RNAs is long noncoding RNAs, which are of various sizes between 200 bp and several kilobases (41). Many studies have uncovered a plethora of processes in which these RNAs participate within the living cell, such as dosage compensation by XIST (14) and roX (30), imprinting by AIR (37) and Kcnq1ot1 (24, 29), an architectural role by NEAT1 (6), RNA localization by CTN1 (31), etc. Many such functions are mediated through their protein binding partners wherein the noncoding RNA is an indispensable component of such complexes. For example, small RNAs (siRNA and miRNA) are often found in association with the RISC (RNA-induced silencing complex), which consist of Argonaute and other proteins (11). In the case of P-element-induced wimpy testis (Piwi)-associated RNAs (piRNAs), which are involved in transposon regulation, the RNA component is associated with Piwi proteins such as Miwi or Mili in mouse and Hiwi in humans (2). Long noncoding RNAs also have functional protein partners. For example, XIST RNA is associated with macroH2A during the process of X chromosome inactivation (32). The Kcnq1ot1 RNA interacts with PRC2 component members Ezh2 and Suz12 (40).
Wnt signaling is a highly conserved developmental signaling pathway involving the major effector protein beta-catenin (27). The canonical Wnt signaling is activated upon binding of Wnt ligands to its receptors Frizzled/LRP, which results in stabilization of beta-catenin in the cytoplasm. The stabilized beta-catenin translocates to the nucleus, whereby it binds to the TCF/LEF family of transcription factors and activates the Wnt target genes. In the Wnt-uninduced condition, beta-catenin is phosphorylated by the glycogen synthase kinase 3β (GSK-3β)/axin/adenomatous polyposis coli complex, which targets beta-catenin to ubiquitin-mediated proteolysis (27). The Wnt signaling is a conserved and essential pathway in development and cell proliferation, and ES cell differentiation and dysregulation of Wnt signaling have been associated with many cancers (15).
p68, also referred to as Ddx5, is a founding member of a large family of DEAD box helicases. In addition to the DEAD box motif, they also possess other conserved sequences, including an ATPase domain and an RNA helicase domain (7). These proteins have been shown to play important roles in diverse biological processes such as development, regulation of transcription, RNA processing, and ribosome biogenesis and also in the miRNA pathway (7, 36). DEAD box proteins act by promoting the formation of optimal secondary structure in RNA and mediate RNA-protein association or dissociation. Apart from its transcriptional coactivator function, p68 has been shown to play a role in signaling events. During the epithelial-to-mesenchymal transition that is induced by platelet-derived growth factor (PDGF) stimulation, p68, which gets phosphorylated at tyrosine 593, was shown to translocate to the cytoplasm, where it stabilizes beta-catenin; subsequently p68–beta-catenin translocates back to the nucleus, bringing about downstream transcriptional regulation (43).
mrhl is a 2.4-kb noncoding RNA, identified in our laboratory, encoded in the mouse genome embedded within a meiotic recombination hot spot locus on mouse chromosome 8 (26). mrhl RNA is an intronic long noncoding RNA (lncRNA) that is transcribed by RNA polymerase II as an independent transcription unit from the intron of the phkb gene. It is a polyadenylated and unspliced transcript. mrhl RNA is expressed in liver, kidney, spleen, and testes but not in brain, heart, lung, and muscle tissues (26). The 2.4-kb primary transcript is nuclear restricted, localizes to the nucleolus, and gets processed to an 80-nucleotide (nt) intermediate RNA by the Drosha machinery (13). Although Dicer can further process the 80-nt intermediate RNA to 22 nt in an in vitro reaction, the mature 22-nt miRNA derived from mrhl RNA was not detected in vivo, suggesting a nuclear regulatory role for this noncoding RNA. In an effort to understand the function of mrhl RNA in vivo, we have now carried out siRNA-mediated silencing of mrhl RNA in mouse spermatogonial cells (Gc1-Spg) and carried out an expression array analysis to study the genes that are perturbed upon mrhl RNA silencing. We have also constructed a weighted gene coexpression network from the perturbed gene list, and we find representation of many of the important transcription factors and genes involved in the Wnt signaling pathway, including TCF4, which, in association with beta-catenin, regulates the expression of wnt downstream targets. A series of experiments described here demonstrate that beta-catenin does translocate into the nucleus under mrhl RNA downregulated conditions and interacts with TCF4 at the promoter of TCF4 target genes. Subsequently we have observed that p68/Ddx5 interacts with mrhl RNA and p68 translocates to the cytoplasm under mrhl RNA downregulated conditions. Further, we have shown that p68 is indeed essential in translocating beta-catenin to the nucleus upon mrhl RNA downregulation. Interestingly, there was a significant decrease in steady-state mrhl RNA levels in Gc1-Spg cells upon stimulation with Wnt3a ligand. Thus, here we report a functional role of a long noncoding mrhl RNA and its protein partner p68 in Wnt signaling in mouse male germ cells.
MATERIALS AND METHODS
Antibodies, plasmids, and siRNAs.
The sources of antibodies were as follows: rabbit anti-p68 antibody, Novus Biologicals; rabbit anti-beta-catenin antibody and phospho-Y142 beta-catenin antibody, Abcam; rabbit anti-phospho-Ser33/37/Thr41 beta-catenin antibody, Cell Signaling; mouse anti-beta-catenin antibody for chromatin immunoprecipitation (ChIP), BD Transduction; mouse anti-TCF4 antibody for ChIP, Upstate; protein A-horseradish peroxidase (HRP) conjugated secondary antibody, Bangalore Genei; rabbit anti-GAPDH, rabbit antilamin, and rabbit anti-phospho-Y593 p68 antibodies, Abcam. TOPFLASH and FOPFLASH reporter plasmids were purchased from Addgene (plasmid 12456 and 12457, respectively; courtesy of Randall Moon). Control siRNA pool and mrhl siRNAs were purchased from Dharmacon, and p68 siRNAs were purchased from Sigma.
The sequences of the siRNAs are as follows: mrhl siRNA 1, 5′GCACAUACAUACAUACACAUAUAUU3′ and 5′ GUGAAAUGACUGUGCUUUAUU3′; mrhl siRNA 2, 5′CAAGUUGACUGCUGAUUUAUU3′ and 5′GGAGAAACCCUCAAAAGUAUU3′.
The siRNA IDs of p68 siRNAs from Sigma are as follows: p68 siRNA 1, SASI_Mm01_00096788 and SASI_Mm01_00096789; p68 siRNA 2, SASI_Mm01_00096792 and SASI_Mm01_00096793.
Cell lines and cell fractionation.
All fine chemicals were purchased from Sigma-Aldrich, USA, or Invitrogen, USA. Gc1-Spg, HeLa, and MCF7 cells were obtained from the American Type Culture Collection (ATCC) and were cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (Invitrogen) and 100 U/ml penicillin-streptomycin solution (Sigma) at 37°C in a humidified 5% CO2 atmosphere. Nuclear and cytoplasmic proteins were prepared by using the NE-PER reagent (Pierce) per the manufacturer's protocol.
RNA isolation, qRT-PCR, and in vitro transcription.
Total RNA from mouse testis and cultured cells were isolated by using TRIzol reagent (Invitrogen) per the manufacturer's protocol. Approximately 2 to 3 μg of RNA was taken and reverse transcribed by using oligo(dT) reverse primers or random primers. About 1/20 of the reverse transcription (RT) reaction product was used for PCR using gene-specific primers. For real-time quantitative PCR (qPCR), the cDNA was diluted to 1:10, added to Sybr green mix (Bio-Rad), and analyzed in a Rotor-Gene 6000 machine (Corbett Life Sciences). Primer pair sequences are given in Table S4 at http://www.jncasr.ac.in/faculty_templates/default/index.php?menu_id=17&user_id=21&page_id=1562. The 2.4-kb mrhl cloned in the pGem vector under the T7 promoter was linearized using XhoI, purified, and used as a template for in vitro transcription. The template was incubated with 1 mM (each) ATP, CTP, GTP, and UTP or [α-32P]UTP and T7 RNA polymerase in 1× transcription buffer. The in vitro transcribed RNA was purified using the RNeasy purification kit (Qiagen, Germany). For labeling the transcripts with biotin, the UTP concentration was reduced to 0.7 mM and biotin UTP was added to a final concentration of 0.3 mM.
siRNA-mediated silencing of mrhl RNA and global gene expression.
The Gc1-Spg cells were grown a day prior to transfection to reach up to 60 to 70% confluence. Cells were then transfected with 100 nM mrhl siRNA pool (Dharmacon) using Lipofectamine 2000 (Invitrogen). The cells were harvested 48 h posttransfection. Downregulation of mrhl RNA expression was checked using the RT-qPCR method. For control experiments, scrambled siRNA pool (Dharmacon) was used. Three independent experiments were subjected to microarray expression profiling using Affymetrix mouse 43.0 expression arrays. The arrays after hybridization and washes were scanned, and the preliminary analyses were done by using Array assist software using the probe logarithmic intensity error (PLIER) algorithm. The genes were grouped based on P value and fold change. Most of the analysis was performed using the genes that showed a P value of ≤0.05 and fold change of ≥1.5. The gene ontology analysis was done using DAVID EASE (8). Hierarchical clustering was done using Cluster 3.0 (9) and visualized in JAVA Tree View software (34).
siRNA-mediated silencing of p68 expression.
Gc1-Spg cells were seeded a day prior to transfection to reach 60 to 70% confluence. Cells were then transfected with a p68 siRNAs (Sigma) or control siRNA pool (Dharmacon) (final siRNA concentration, 100 nM). Cells were harvested 48 h posttransfection, and silencing was assessed by qRT-PCR and Western blotting.
Coexpression network construction and module identification.
We used weighted coexpression network analysis methodology developed by Horvath and colleagues (20, 46). The weighted coexpression network is constructed by calculating a pairwise correlation matrix between all probe sets across microarray samples. The resulting Pearson correlation matrix was transformed into an adjacency matrix. Microarray data can be noisy, and the number of samples is often small, so we weighed the Pearson correlations by taking their absolute value and raising them to the power β. Thus, the connection strength (adjacency) a (i,j) between gene expressions xi and xj is defined as a(i,j) = [cor(xi, xj)]β. The nodes of coexpression network correspond to gene expressions, and edges between genes are determined by the correlations between gene expressions. Using scale-free topology criteria, the soft threshold function was set at a slightly higher power, β = 7, due to the smaller size of the microarray data set, which resulted in a weighted network. Genes were organized into modules by using topological overlap measure (1 topological overlap) as a robust measurement of interconnectedness in hierarchical cluster analysis, with the merge height cut as 0.10 and the minimum module size as 30 using the automatic detection algorithm. VisANT integrative visual analysis tool software was used to identify which pairs of genes are connected within the network structure (17).
Immunoprecipitation and immunofluorescence.
Cells lysed in IP buffer (20 mM HEPES-KOH, pH 7.4, 100 mM NaCl, 10% glycerol, 2 mM dithiothreitol [DTT], 5 mM MgCl2, 0.1 mM phenylmethylsulfonyl fluoride [PMSF], 0.5% Triton X-100) were precleared and incubated with primary antibody (rabbit polyclonal p68) or rabbit preimmune sera for 4 h, followed by incubation for 2 h with protein A-agarose. Immunoprecipitated proteins were separated on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane. The membrane, after blocking in 5% skim milk in phosphate-buffered saline (PBS), was incubated with primary antibody overnight at 4°C. The membrane was then washed thrice in PBS-Tween (PBST) followed by incubation in horseradish peroxidase (HRP)-conjugated secondary antibody. After washes, the blot was subjected to chemiluminescence using luminol as the substrate. Tris-buffered saline (TBS) was used instead of PBS for detection of phosphoproteins. For immunofluorescence experiments, cells grown on coverslips were fixed for 20 min in PBS containing 4% paraformaldehyde, washed three times with PBS, permeabilized with 0.1% Triton X-100 for 15 min, and blocked with 1% bovine serum albumin (BSA) for 1 h. Subsequently, cells were incubated at room temperature with primary antibody (β-catenin or phospho-Y142 β-catenin antibody or p68 antibody, 1:100 dilution in 0.1% BSA) for 45 min followed by three washes with PBS containing 0.1% Tween 20 (0.1% PBST). This was followed by incubation with an appropriate secondary antibody (1:400 dilution in 0.1% BSA) conjugated with Alexa Fluor for 45 min at room temperature. The cells were washed thrice with 0.1% PBST and subsequently stained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) and then washed twice with 0.1% PBST. The coverslips were mounted on 60% glycerol. The images were acquired in an LSM 10 Meta confocal microscope (Carl Zeiss) and analyzed by image analysis software provided by Carl Zeiss.
Luciferase reporter assay.
Cells were seeded (20,000 cells per well) in a 24-well plate 24 h prior to transfection and transfected with 100 nM mrhl siRNA pool or 100 nM control or scrambled siRNA. At 24 h after siRNA transfection, cells were transfected with 1 μg of reporter (FOPFLASH or TOPFLASH) and 0.2 μg of internal control plasmid (CMV-βGal). Transfection was carried out using Lipofectamine 2000. At 24 h after plasmid transfection (48 h after siRNA transfection), total cell extracts were assayed for luciferase activity according to the manufacturer's instructions (Promega).
Northwestern blotting.
The proteins from total lysates from testis or Gc1-Spg cells that were separated on a 10% SDS-polyacrylamide gel were transferred to a nitrocellulose membrane. They were allowed to renature on the membrane by overnight incubation in renaturation buffer containing 10 mM HEPES, pH 7.9, 40 mM KCl, 5% glycerol, 0.2% NP-40, 3 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 5 mg/ml BSA. The membrane was incubated in an RNA binding buffer (buffer A) (10 mM HEPES, pH 7.9, 150 mM KCl, 5 mM MgCl2, 5% glycerol, 0.2 mM DTT, and 0.05% NP-40) containing 50 μg/ml tRNA, 500 μg/ml heparin, and 50,000 cpm of probe prepared by in vitro transcription. The membrane was incubated for 3 h at 20°C, washed twice in buffer A, and exposed to a phosphorimager.
Mass spectrometry.
The SDS-PAGE gel after separation of proteins was stained with EZBlue (Sigma) or silver stained. The bands that were present were excised along with the control where no proteins were present. In-gel tryptic digestion was performed according to the protocol provided by Bruker. The tryptic digest was analyzed in a Bruker mass spectrometer. Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry was done initially to identify the peptide mass fingerprint. Specific peaks were lifted further to perform tandem mass spectrometry (MS-MS) analysis to identify the peptide sequence. Proteins were identified using MASCOT software. Carbamidation was set as the fixed modification, and methionine oxidation was set as the variable modification. The threshold cutoff was set as 0.6 kDa. Proteins for which at least two peptides matched were considered to be significant.
RNA affinity pulldown.
The in vitro transcribed 2.4-kb RNA that was biotin labeled was denatured at 65°C and slowly cooled to room temperature. Freshly harvested Gc1-Spg cells were washed once in ice-cold PBS and lysed in lysis buffer containing 10 mM Tris, pH 8.0, 150 mM NaCl, 5 mm MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.5% Triton X-100, and 5% glycerol containing 1 mM PMSF and 1× protease inhibitor cocktail (1,000× stock; Sigma). The cell lysate was precleared for about 1 h at 4°C in streptavidin agarose beads (Invitrogen) equilibrated in lysis buffer. The precleared cell lysate was incubated with in vitro transcribed and biotinylated RNA along with 0.5 U/μl RNase inhibitor (Promega)–80 μg/ml tRNA overnight followed by incubation with equilibrated streptavidin agarose beads for 2 h. The beads containing the RNA protein complex were washed thrice in the lysis buffer and then were directly boiled in SDS-gel loading dye and loaded onto a 10% SDS-PAGE gel. For in vivo RNA pulldown experiments, we used the S1 aptamer approach as described by Srisawat and Engelke (38). The S1 aptamer sequence was ligated to the 3′end of 2.4-kb mrhl and was cloned in the pCDNA 3.1 vector downstream of the cytomegalovirus (CMV) promoter. The construct was transfected into Gc1-Spg cells, and 36 h posttransfection, the cells were harvested and lysed in lysis buffer. The cell lysate, along with RNAsin and tRNA, was incubated with equilibrated streptavidin beads overnight, and the washes were performed as described above. The RNA-bound proteins were subjected to 10% SDS-PAGE analysis.
Chromatin immunoprecipitation (ChIP) and PCR.
Gc1-Spg cells were cross-linked with 1% formaldehyde for 20 min at room temperature. The reaction was quenched with glycine at a final concentration of 0.125 M. The cells were washed with PBS, resuspended in ChIP incubation buffer (0.1% SDS, 0.5% Triton X-100, 20 mM Tris-HCl, pH 8, and 150 mM NaCl), and sheared using a Bioruptor sonicator (Diagenode). Sonication efficiency was checked, and sonicated DNA was found to be enriched in a range from 600 bp to 2 kb. The sonicated chromatin was centrifuged for 15 min and precleared using protein A-agarose beads (Invitrogen). For β-catenin and TCF4 ChIP the precleared chromatin was incubated with 3 μg of the respective antibody or 3 μg of mouse IgG as a negative control for 12 h at 4°C. The immune complex was captured with protein A beads for 2 h at 4°C. The beads were successively washed 2 times with buffer 1 (0.1% SDS, 0.1% deoxycholate, 1% Triton X-100, 0.15 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES [pH 7.6]), once with buffer 2 (0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, 0.5 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES [pH 7.6]), and once with buffer 3 (0.25 M LiCl, 0.5% sodium deoxycholate, 0.5% NP-40, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES [pH 7.6]) for 5 min each at 4°C. The precipitated chromatin was eluted by incubation of the beads with elution buffer (1% SDS, 0.1 M NaHCO3) at room temperature for 20 min, de-cross-linked by incubation at 65°C for 5 h in the presence of 200 mM NaCl, extracted with phenol-chloroform, and precipitated. PCR was done using gene-specific primers and standard conditions for 30 cycles. Primers used for ChIP-PCR are listed in Table S3 at http://www.jncasr.ac.in/faculty_templates/default/index.php?menu_id=17&user_id=21&page_id=1562.
Wnt3a treatment and mrhl RNA expression analysis.
Gc1-Spg cells were transfected with the 2 μg of TOPFLASH and FOPFLASH reporter plasmids and 0.4 μg of the internal control plasmid (CMV-βGal) in a six-well plate. At 24 h posttransfection, cells were treated with Wnt3a ligand (procured from R & D Systems) for 12 h. After Wnt3a treatment (150 ng/ml), cells were processed for luciferase assay and RNA isolation. cDNA synthesis was carried out using 1.5 μg total RNA, and mrhl RNA expression was analyzed by qRT-PCR.
Microarray accession number.
The microarray gene expression data have been submitted to the NCBI Gene Expression Omnibus database under accession number GSE19355.
RESULTS
Downregulation of mrhl RNA and global gene expression.
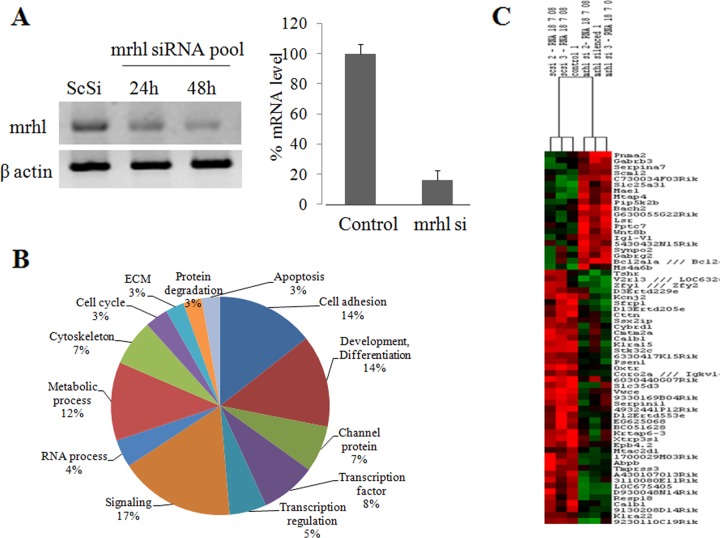
In an attempt toward understanding the biological function(s) of mrhl noncoding RNA, we decided to silence this RNA and analyze its effect on global gene expression. Since mrhl RNA is abundantly expressed in mouse testis, we used the Gc1-Spg cell line, which is derived from mouse spermatogonia expressing 2.4-kb mrhl noncoding RNA. It is generally believed that siRNA-mediated downregulation of its target mRNA is a cytoplasmic event. Although the noncoding RNA and its processed 80-nt intermediate are nuclear restricted, we were encouraged by recent reports that many nuclear RNAs could be downregulated through RNA interference, though the mechanism of this downregulation is unclear at present (33). We designed four siRNAs targeting four different regions of the mrhl 2.4-kb primary transcript, and the siRNAs were searched against a mouse genome database using BLAST to verify the specificity of the designed sequences. Initial siRNA experiments were carried out using individual siRNA and then as a pool with different concentrations. The effective concentration was found to be 100 nM using siRNA pools, and at 48 h posttransfection we observed nearly 70 to 80% efficiency of downregulation (Fig. 1A).
Fig 1.
mrhl RNA downregulation and global gene expression profiling in mouse Gc1-Spg cells. (A) Semiquantitative RT-PCR showing downregulation of mrhl RNA after 24 h and 48 h in cells treated with a pool of four siRNAs compared to the control scrambled siRNA (ScSi). Bottom panel is the quantitative real-time PCR analysis showing the percentage of mRNA decrease after 48 h in the siRNA experiment cells compared to the control siRNA-treated cells. The experiment was done in triplicate, with three technical replicates. Error bars represent standard deviations. (B) Classification of genes based on GO terms showing an overrepresented class of genes in the microarray experiment. (C) Representative heat map of microarray data. Normalized log 2 transformed expression ratios of 652 genes from 6 samples (three ScSi control and three silenced samples of mrhl RNA). Up- and downregulated genes are colored red and green, respectively.
Total RNA samples obtained from three biological replicates were processed further for expression profiling using the Affymetrix 43K mouse genome expression array. The values of all the negative and positive controls of three sets of the experiments were within the limits of Affymetrix standards and quality. We then carried out a thorough analysis of the differentially regulated genes using the PLIER algorithm of Array assist software. Around 652 genes showed a differential pattern of regulation across all three experiments and were perturbed more than 1.5-fold, with a P value of < 0.05 (GEO accession number GSE19355; see also Table S1 at http://www.jncasr.ac.in/faculty_templates/default/index.php?menu_id=17&user_id=21&page_id=1562). The overrepresented class of perturbed genes were classified based on its GO annotation and biological function, and the pie chart generated, of the genes grouped under various functional categories, is given in Fig. 1B. Nearly 45% of the genes that are perturbed belong to cell adhesion (14%), signaling (17%), and development and differentiation (14%). Interestingly 13% of the perturbed genes belong to the transcription process, comprising transcription factors (8%) and transcription regulation (5%). We did not find any enrichment of genes in any particular chromosome (data not shown). A representative heat map of coregulated and clustered genes is shown in Fig. 1C. Genes from siRNA-treated samples grouped together separately, whereas the control scrambled-siRNA-treated samples fell into a separate group without any overlap between them. The expression of the phkb gene, which harbors mrhl in one of its introns, was also not affected, thus showing that mrhl RNA downregulation does not perturb the expression of the gene locus in which it is encoded.
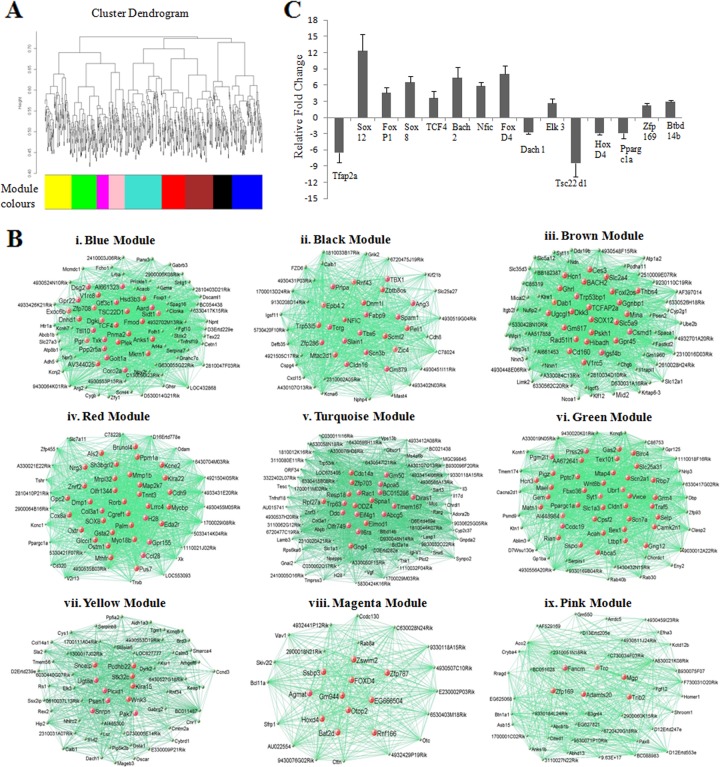
Recently, new approaches have been designed to analyze the gene expression data to identify gene-gene interactions termed coexpression modules. One such approach, namely, weighted gene coexpression network analysis (WGCNA), has been used to construct expression modules in many disease models and also in human brain (20, 28, 35, 46). We subjected our gene expression data (652 genes) to WGCNA, which is based on topological overlap (TO) between genes and which calculates possible correlation values between all the genes and converts them into measures of connection strengths by taking absolute values and raising them to a power β. Genes are clustered based on TO, and those with similar expression patterns are grouped together. This resulted in nine well-separated and clustered coexpression modules (Fig. 2A). These modules represented genes belonging to different cellular pathways (Fig. 2B). These include Wnt signaling and transcription (blue, 79 genes), cell differentiation and development process (black, 48 genes), cell adhesion and apoptosis (brown, 73 genes), G-coupled receptor signaling and ion channel activity (red, 61 genes), cell signaling and development (turquoise, 91 genes), receptor/ion channels (green, 67 genes), transcription regulation (yellow, 60 genes), transcription (magenta, 29 genes), and cell adhesion (pink, 45 genes). The gene list in each of the coexpression module and the number of connections between each gene nodes is given in Table S2 at http://www.jncasr.ac.in/faculty_templates/default/index.php?menu_id=17&user_id=21&page_id=1562. We identified 12 transcription factors and three coactivators and validated their perturbation of expression by real-time PCR analysis (Fig. 2C). The transcription factors Sox8, Sox12, FoxD4, FoxP1, Bach2, and Nfic were upregulated severalfold upon silencing of mrhl RNA. Sox8 and Sox12 are involved in the developmental processes (19), while FoxD4 and Fox P1 are members of the Fox head group of transcription factors (21). On the other hand, transcription factors Ap2a and Odz4 were downregulated 6- and 9-fold, respectively, upon silencing of mrhl RNA. Among the coactivators, Zfp169 and Btbd14b were upregulated by about 2-fold and Ppargc1a was downregulated 3-fold.
Fig 2.
Coexpression modules and their functional classification. (A) The dendrogram represents network modular organization. The expression data of mrhl RNA-silenced samples were organized into nine modules. Each gene represented by the vertical line in x axis and the dissimilarity in expression between neighboring genes on y axis modules was assigned a different color. (B) Visualization of coexpression modules from weighted gene coexpression network analysis. Functional classification of the modules. Blue, Wnt signaling and signal transduction; black, cell differentiation and developmental process; brown, cell adhesion and apoptosis; red, G-coupled receptor signaling and cell cycle; turquoise, cell signaling and development; green, receptor/ion channels; yellow, transcription regulation; magenta, transcription; pink, cell adhesion. (The gene list in each of these modules can be seen in Table S2 at http://www.jncasr.ac.in/faculty_templates/default/index.php?menu_id=17&user_id=21&page_id=1562.) (C) Real-time PCR validation of the transcription factors and coactivators identified from the microarray analysis. The experiment was done in triplicate, with three technical replicates. Error bars represent standard deviations.
Activation of Wnt signaling in mrhl RNA-silenced cells.
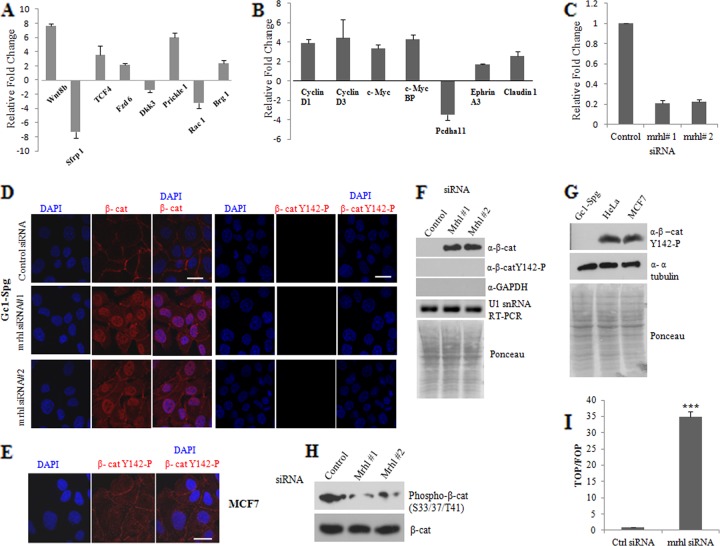
TCF4, which was found in the Wnt signaling and transcription module (Fig. 2B), is an important transcription factor in the Wnt signaling pathway which, upon association with beta-catenin as a coactivator, regulates the expression of downstream target genes (23). Since our microarray data showed perturbation of gene expression of several of the Wnt signaling pathway genes, we carried out real-time qPCR analysis of a subset of these genes, and the results are shown in Fig. 3A. Wnt8b was significantly upregulated both in microarray as well as real-time qPCR, while Wnt negative regulators Sfrp1 and Dkk3 were significantly downregulated. Two other genes that are part of noncanonical Wnt signaling, namely, Prickle1 and Rac1, were also found to be differentially regulated in mrhl RNA-silenced cells. Smarca4 or Brg1, which promotes TCF target gene activation by facilitating chromatin remodeling (3), was also found to be upregulated in mrhl RNA downregulated conditions.
Fig 3.
Nuclear translocation of beta-catenin and increased beta-catenin/TCF4 transcription activity upon mrhl RNA downregulation. (A and B) Real-time PCR validation of Wnt pathway genes and Wnt target genes, respectively. (C) qRT-PCR analysis showing downregulation of mrhl RNA expression upon treatment with mrhl siRNA1 and mrhl siRNA2. (D) Nuclear staining of beta-catenin in mrhl siRNA-treated cells compared to membranous staining in control siRNA-treated Gc1-Spg cells. Lack of staining using beta-catenin Y142-P antibody in control and mrhl siRNA-treated Gc1-Spg cells. (E) Immunofluorescence with beta-catenin Y142-P antibody in MCF7 cells showing the efficiency of the antibody. (F) Western blot analysis showing the presence of beta-catenin and absence of phospho-Y142 beta-catenin in nuclear extracts upon mrhl RNA silencing. The Ponceau-stained blot shows equal loading. (G) Western blot analysis using total cell lysates of Gc1-Spg, HeLa, and MCF7 cell lines with Y142-P beta-catenin antibody showing the efficiency of the antibody. (H) Western blot analysis using total cell lysates of Gc1-Spg cells with beta-catenin antibody (lower panel) and phospho-S33/S37/T41 beta-catenin antibody (upper panel). (I) Transcriptional activation of TCF4/beta-catenin-dependent luciferase reporter expression (TOP/FOP ratio) upon mrhl RNA downregulation using a pool of four siRNAs. Images shown in panels A to I are representative of three independent biological replicates, each done in duplicate. Error bars in panels A, B, C, and I represent standard deviations. Scale bar (D and E), 25 μm. ***, P < 0.0001 by t test.
Interestingly, we also find in our microarray data many Wnt target genes that were differentially regulated in the mrhl RNA-silenced cells. The real-time PCR validation of some of these genes is given in Fig. 3B. These included the known targets of the Wnt/beta-catenin pathway, namely, cyclin D1, cyclin D3, c-Myc, c-Myc binding protein (cMycBP), claudin 1, and ephrin A3, which were upregulated, and proto-cadherin alpha 11 (Pcdha11), which was downregulated upon mrhl RNA silencing. From the results presented so far we could infer that the Wnt signaling, one of the major developmental signaling pathways, was activated upon mrhl RNA silencing. Beta-catenin is an important Wnt effector molecule, and its regulation is mediated predominantly at the level of protein rather than at the transcription level. Hence we examined the stability of beta-catenin in mrhl RNA-downregulated cells as monitored by immunofluorescence using anti-beta-catenin antibody. For these experiments, we divided the four-siRNA pool into two subgroups, mrhl siRNA1 and mrhl siRNA2, each containing two siRNAs, to make sure that the results are reproducible when the siRNAs are targeted to different regions of mrhl RNA. The qRT-PCR analysis shows about 80% downregulation of mrhl RNA on treatment with these two independent siRNA pools (Fig. 3C). The confocal images in Fig. 3D show cytoplasm-stabilized as well as nucleus-localized beta-catenin in mrhl siRNA-treated cells compared to the control siRNA-treated cells, which show uniform staining of beta-catenin only in the cell membrane, as expected. Beta-catenin is also a part of the adherens junctions, as it links E-cadherin to alpha-catenin, and the choice of beta-catenin between cell adhesion and Wnt signaling can be affected by other signals (4). Phosphorylation of tyrosine residue 142 of beta-catenin induces loss of beta-catenin binding to alpha-catenin and promotes the interaction of beta-catenin with BCL9-2. The beta-catenin–BCL9-2 complex translocates to the nucleus and, by associating with the LEF/TCF DNA binding proteins, regulates the transcription of crucial target genes. In order to determine the source of nuclear translocated beta-catenin under mrhl RNA downregulation, i.e., whether it is from membrane (Y142 phosphorylated) or is cytoplasmic stabilized (unphosphorylated), we carried out immunofluorescence with phospho-Y142 beta-catenin antibody. In this experiment, there was no staining in control or mrhl siRNA-treated cells, suggesting the absence of the phosphorylation modification in Gc1-Spg cells (Fig. 3D). The efficiency of phospho-Y142 beta-catenin antibody was checked using MCF7 cells, in which staining was observed (Fig. 3E). The immunofluorescence result was also substantiated by Western blot analysis using nuclear extract of Gc1-Spg cells treated with mrhl siRNA or control siRNA which showed the presence of beta-catenin but not phospho-Y142 beta-catenin only in the nuclear extract of mrhl siRNA-treated cells (Fig. 3F). The efficiency of phospho-Y142 beta-catenin antibody for Western blotting was checked using total cell lysates of HeLa and MCF7 cells (Fig. 3G). We then checked for phosphorylation of beta-catenin mediated by GSK-3β at the N-terminal residues Ser 33, Ser37, and Thr 41, which mark beta-catenin for degradation (22). Western blot analysis in Fig. 3H shows that phosphorylation at these residues was drastically reduced in mrhl RNA-silenced cells compared to control cells, substantiating the fact that beta-catenin is stabilized upon mrhl RNA downregulation. Finally, we carried out the standard TOP FLASH/FOP FLASH reporter assay to examine whether TCF/beta-catenin-dependent transcription was enhanced upon mrhl RNA silencing. The TOP/FOP ratio increased about 33-fold with mrhl RNA-silenced cells compared to the control cells (Fig. 3I). All these results thus demonstrate an activation of Wnt signaling that involves cytoplasmic stabilization and nuclear translocation of beta-catenin and increased TCF/beta-catenin-mediated transcription activity in mrhl RNA-silenced cells.
Recruitment of beta-catenin at a subset of TCF4 occupied gene promoters.
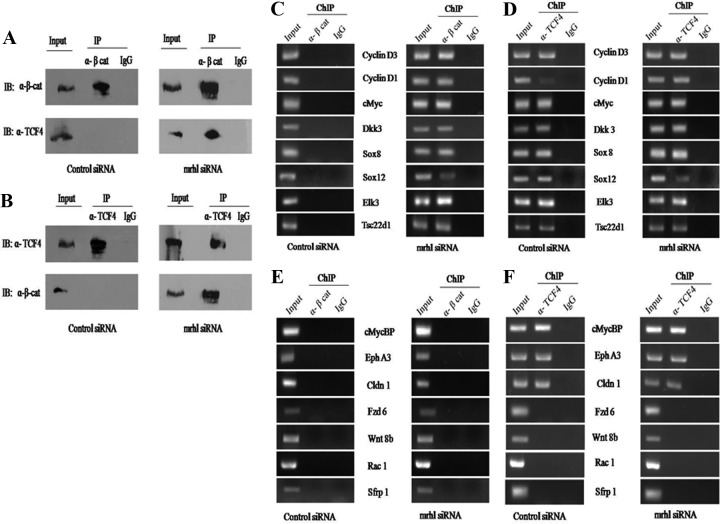
During active Wnt signaling, the nuclear translocated beta-catenin interacts with TCF4 on displacing the Groucho/TLE1 repressor. On interaction with TCF4, beta-catenin is recruited to the promoters of TCF4 target genes (23). Beta-catenin and TCF4 bind to the Wnt responsive element (WRE) (A/T-A/T-C-A-A-A-G consensus motif) and activate or repress the transcription of target genes by association with specific coactivators or corepressors. TCF4 is bound to the WRE of the target genes even in the absence of Wnt signaling. To determine whether beta-catenin and TCF4 interact upon mrhl RNA silencing, we carried out coimmunoprecipitation experiments with beta-catenin and TCF4 antibodies. As seen in Fig. 4A and B, beta-catenin and TCF4 coimmunoprecipitated in mrhl siRNA-treated cells but not in control cells. We then analyzed whether the beta-catenin-TCF4 complex is present at the promoters of Wnt pathway-related genes. We selected 15 genes which included the known targets of TCF4 (cyclin D1, cyclin D3, cMyc, cMycBP, ephrin A3 and claudin1), transcription factors known to regulate Wnt or be regulated by Wnt (Sox 8, Sox 12, Tsc22d1, Elk3), Wnt pathway genes, and Wnt modulators (Wnt 8b, Fzd6, Rac1, Dkk3, and Sfrp1). For the selected genes, a region of about 5 kb upstream of the transcription start site (TSS) was retrieved from the ENSEMBL database and primers were designed flanking the WRE or the TCF4 consensus motif (see Table S3 at http://www.jncasr.ac.in/faculty_templates/default/index.php?menu_id=17&user_id=21&page_id=1562). Beta-catenin ChIP and TCF4 ChIP were carried out in Gc1-Spg cells treated with control siRNA or a pool of four mrhl siRNAs (48 h after siRNA transfection). PCR was done with the gene-specific primers. Out of the 15 genes, the promoters of 8 genes, namely, cyclin D1, cyclin D3, cMyc, Dkk3, Sox8, Sox12, Tsc22d1, and Elk3, showed beta-catenin occupancy only upon mrhl RNA downregulation (Fig. 4C). Promoters of these 8 genes also showed TCF4 occupancy in both control siRNA as well as mrhl siRNA-treated cells (Fig. 4D), consistent with the literature showing the occupancy of TCF4 even in the absence of Wnt signaling. ChIP (beta-catenin) PCR for the remaining 7 genes did not show beta-catenin occupancy on their promoters (Fig. 4E), while TCF4 was present only on the promoters of genes which are known Wnt targets (cMycBP, ephrin A3, and claudin1) in control as well as mrhl siRNA-treated cells (Fig. 4F). These results clearly show that beta-catenin is recruited to promoters of genes related to the Wnt signaling pathway selectively upon mrhl RNA downregulation.
Fig 4.
Interaction of beta-catenin and TCF4 and recruitment of beta-catenin to a subset of Wnt target gene promoters on mrhl RNA downregulation. (A and B) Coimmunoprecipitation of beta-catenin and TCF4 from total cell lysates of Gc1-Spg cells seen in mrhl siRNA-treated cells but not in control siRNA-treated cells (IB, immunoblot; IP, immunoprecipitation). 10% input was loaded. (C) ChIP (beta-catenin) PCR of indicated genes showing the occupancy of beta-catenin on their promoters in mrhl-silenced conditions but not in control siRNA-treated cells. (D) ChIP (TCF4) PCR of indicated genes showing the occupancy of TCF4 on their promoter in control siRNA-treated cells as well as mrhl siRNA-treated cells. (E) ChIP (beta-catenin) PCR of indicated genes showing lack of occupancy of beta-catenin on their promoters. (F) ChIP (TCF4) PCR of indicated genes showing the occupancy of TCF4 on promoters of known Wnt target genes. Images shown in panels A to F are representative of three independent biological replicates, each done in duplicate.
mrhl RNA interacts with Ddx5/p68, a DEAD box helicase both in vitro and in vivo.
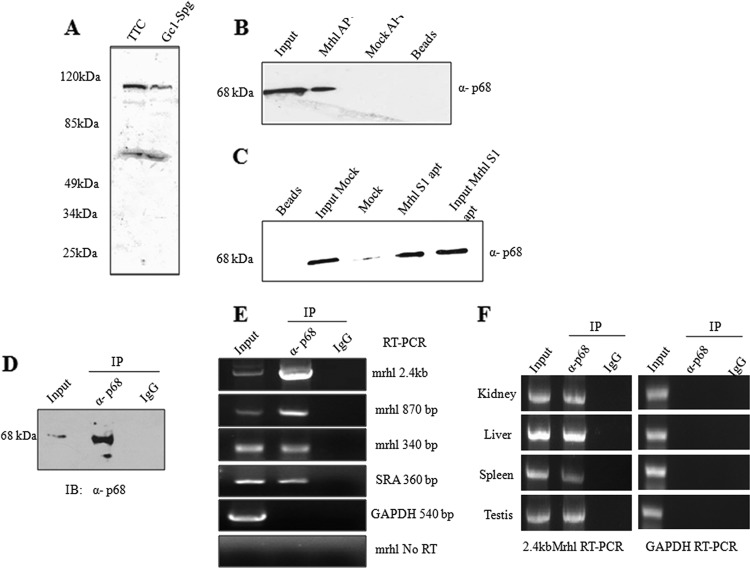
Since many of the RNA functions are mediated through its interaction with proteins, we were curious to identify the mrhl RNA interacting partner(s). For this purpose, we carried out a Northwestern analysis with the full-length 2.4-kb mrhl RNA. As shown in Fig. 5A, we identified at least three interacting proteins, around ∼115 kDa, ∼100 kDa, and ∼65 kDa, among which the 115-kDa and 65-kDa proteins were the prominent ones. This experiment was done in both total testicular lysate and the Gc1-Spg cell lysate. In order to identify these proteins, we resorted to RNA affinity pulldown. The 2.4-kb mrhl transcript was in vitro transcribed in the sense orientation from the transcript cloned in the pcDNA3.1 vector under the T7 promoter in the presence of biotinylated UTP. The pulldown was performed using total cell lysate of Gc1-Spg cells. The mock pulldown was done only with the streptavidin beads. The proteins that were pulled down were resolved by SDS-PAGE, and proteins were visualized after silver staining (see Fig. S1 at http://www.jncasr.ac.in/faculty_templates/default/index.php?menu_id=17&user_id=21&page_id=1562). The protein bands marked with arrows were subjected to MALDI-TOF mass spectrometric analysis. At least four proteins (Lim domain protein, Ddx5, beta tubulin, and Dusp11) could be identified by MS-MS analysis. The proteins, with their respective molecular masses, and the peptides that were identified are given in Table 1. Though the peptides match p68, one cannot rule out the possibility of p72 interaction as well, since these two proteins comigrate and share more than 80% sequence homology and act in a concerted way. We further confirmed the interaction of p68 with mrhl RNA by performing a Western analysis using anti-p68 antibody on the mrhl RNA affinity pulldown proteins. We could see that p68 was present in the input cell lysate and the mrhl RNA pulldown sample whereas it was not observed in a mock pulldown where antisense mrhl RNA or only the beads were used (Fig. 5B). This observation was extended further to check for an in vivo interaction between mrhl RNA and p68 by doing an S1 aptamer pulldown experiment. S1 aptamers are high-affinity streptavidin aptamers that can be tagged to RNA/DNA sequences, and pulldown can then be performed using streptavidin beads (38). The mrhl gene was tagged to the S1 aptamer and was cloned in the pCDNA 3.1 mammalian expression vector. The mrhl-S1 fusion construct was transfected in Gc1-Spg cells, and the RNasin and protease inhibitors in the cell lysate after 36 h of transfection were directly bound to streptavidin agarose beads. Controls were performed using cell line transfected with empty vector and the beads only. The proteins were eluted and subjected to Western analysis with anti-p68 antibody. As can be seen from Fig. 5C, p68 is present in the input cell lysate of mrhl S1- and mock-transfected cells and in the affinity pulldown proteins of mrhl S1-transfected cells only whereas it is absent in the mock-transfected pulldown proteins. Beads which were bound to the lysate served as a negative control and did not yield any signal. We also carried out the reverse experiment to validate the interaction of p68 with mrhl RNA. The nuclear lysate from Gc1-Spg was immunoprecipitated with anti-p68 antibody, and total RNA was extracted from the immunoprecipitate. The presence of mrhl RNA was scored by RT-PCR. Figure 5D shows the specificity of the immunoprecipitation reaction, and the RT-PCR results are shown in Fig. 5E. We could demonstrate the presence of both full-length 2.4-kb mrhl RNA amplicons as well as two internal amplicons. As a positive control, we also observed the presence of SRA RNA, a known interactor of p68 (5), in these immunoprecipitates. GAPDH and preimmune IgG served as negative controls. The lack of amplification with mrhl RNA primers in the No RT reaction (Fig. 5E) confirmed the absence of DNA in the RNA preparation. Mrhl RNA is expressed in testis, liver, kidney, and spleen but not in all tissues (26). We then checked whether mrhl RNA interacts with p68 in these mouse tissues. We carried out immunoprecipitation of p68 from the nuclear extract of these tissues, isolated the RNA, and probed for the 2.4-kb mrhl RNA by RT-PCR. As can be seen from Fig. 5F, mrhl RNA does interact with p68 in all these tissues. These experiments convincingly showed the interaction of mrhl RNA with p68, both in vitro and in vivo.
Fig 5.
p68 is an interacting partner of mrhl RNA. (A) Northwestern blot of total proteins from mouse total testicular cells and Gc1-Spg cells probed with full-length in vitro transcribed mrhl transcript. (B) Nuclear lysates from Gc1-Spg cells probed with anti-p68 antibody after RNA affinity pulldown using in vitro biotinylated mrhl RNA. (C) Western blot with anti-p68 antibody after in vivo mrhl RNA pulldown in Gc1-Spg cells transfected with plasmid containing mrhl fused with S1 aptamer. Mock is the pulldown experiment carried out in Gc1-Spg cells transfected with plasmid containing antisense mrhl fused with S1 aptamer. (D) Nuclear extract of Gc1-Spg cells immunoprecipitated (IP) with p68 antibody or preimmune IgG (negative control) and then immunoblotted (IB) with p68 antibody to check the specificity of pulldown. (E) Total RNA extracted from IP material was subjected to RT-PCR with the primers specific to mrhl (full-length and internal primers), SRA (positive control), and GAPDH (negative control). (F) mrhl RNA and p68 interaction in different mouse tissues demonstrated by immuno-pulldown of p68 from nuclear extracts of the tissues and subsequently scored for 2.4-kb mrhl RNA by RT-PCR. Images shown in panels A to G are from a representative experiment. Experiments were done in duplicate and repeated with two independent biological replicates.
Table 1.
Mrhl RNA interacting proteins identified through mass spectrometry
| Protein | Molecular mass | Identified peptides |
|---|---|---|
| Lim domain-containing protein | 120 kDa | K.FGE FCHGCSLLMT GPAMVAGEFK |
| K.RYTVVGNP YWMAPEMLNG K | ||
| K.RPPVEKATT K | ||
| DDX5 | 68 kDa | DR.GFGAPRFGGSKAGPLSG.K |
| R.GVEICIATPGK | ||
| Beta tubulin | 50 kDa | K.FWEVISDEHGIDPTGTYHGDSDLQLDR.I |
| R.ISVYYNEATGGK.Y | ||
| K.YVPR.A | ||
| R.AILVDLEPGTMDSVR.S | ||
| R.SGPFGQIFRPDNFVFGQSGAGNNWAK.G | ||
| K.GHYTEGAELVDSVLDVVR.K | ||
| K.IREEYPDR.I | ||
| R.IMNTFSVVPSPK.V | ||
| R.FPGQLNADLR.K | ||
| Dusp11 | 42 kDa | R.VFSGY SSAK.K |
| K.S FEKHLAPEEC FSPLDLFN.K | ||
| R.YLIDV EGMRPDDAIE LFS.R | ||
| R.GFED STHMMEPVFT AT.K |
mrhl RNA participates in nuclear retention of p68.
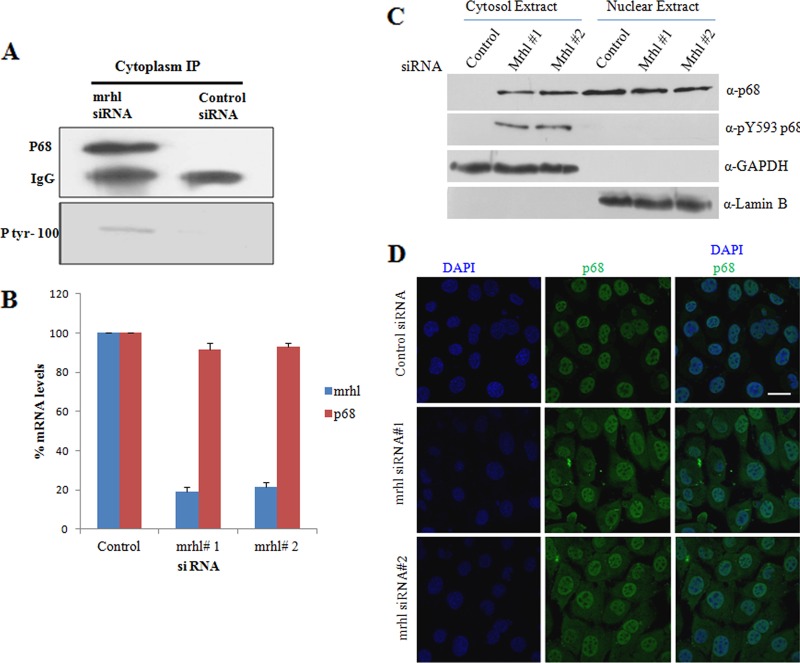
A previous report from Yang et al. (43) had shown a direct correlation between tyrosine-phosphorylated p68 and beta-catenin activation in human colon cancer cells. We were curious to know whether p68 is a part of activation of the Wnt signaling pathway that we had seen in our experiments following downregulation of mrhl RNA. We examined the subcellular distribution and tyrosine phosphorylation status of p68 in mrhl RNA-downregulated cells and control cells. Gc1-Spg cells were treated with a pool of 4 mrhl siRNAs and a control siRNA pool, and 48 h after transfection the cells were fractionated to yield nuclear and cytoplasmic fractions. p68 was immunoprecipitated from the cytoplasmic fraction, and the tyrosine phosphorylation was checked with phosphotyrosine antibody. Figure 6A shows p68 to be tyrosine phosphorylated and present in the cytoplasm of mrhl RNA-silenced cells but not in control cells. Yang et al. (43) reported the phosphorylation of cytoplasmic translocated p68 to occur at Tyr 593. Based on sequence context, Tyr 593 of human p68 is analogous to mouse Tyr 626 residue. We then repeated the cellular fractionation in treatment with mrhl siRNA1 and mrhl siRNA2. The downregulation of mrhl RNA expression was checked by qRT-PCR (Fig. 6B). The nuclear and cytoplasmic lysates of the experiment and the control were subjected to Western analysis with anti-p68 and phospho-Y593 p68 antibodies. As seen from Fig. 6C, p68 was present in the cytoplasmic fraction of mrhl RNA-downregulated cells compared to mock-transfected cells. The cytoplasmic p68 was illuminated with phospho-Y593 p68 antibody. GAPDH and lamin served as controls for cytosol and nuclear fractions, respectively. We further carried out immunofluorescence experiments in Gc1-Spg cells transfected with mrhl siRNA pool and stained with p68 antibody. The cytoplasmic staining of p68 in addition to the nuclear staining in mrhl RNA-silenced cells but not in the control is shown in Fig. 6D. Thus, these experiments conclusively showed that p68 translocates to cytoplasm and cytoplasmic p68 in its tyrosine-phosphorylated form and triggers translocation of beta-catenin in to the nucleus.
Fig 6.
mrhl RNA participates in nuclear retention of p68. (A) Cytoplasmic p68 under mrhl RNA downregulation is phosphorylated at tyrosine which was probed with phospho-Tyr antibody. Top panel, Western blotting of p68 IP in the mrhl siRNA-transfected cytoplasm and control cytoplasm; bottom panel, p68 immunoprecipitated protein subjected to Western analysis using phosphotyrosine antibodies in mrhl siRNA-treated cells and control cells. (B) Real-time PCR analysis for mrhl RNA and p68 expression upon mrhl RNA downregulation using mrhl siRNAs. Data represent three independent experiments done in triplicate. Error bars represent standard deviations. P < 0.001 by t test. (C) Western blot analysis using p68 and phosphor Y593 p68 antibodies in nuclear and cytoplasmic fractions of Gc1-Spg cells transfected with two independent mrhl siRNA pools or control siRNA. (D) Cytoplasmic localization of p68 in mrhl siRNA-treated cells as probed by immunofluorescence using anti-p68 antibody. Nuclei are stained with DAPI. Scale bar, 30 μm. Panels are representative images of two (A) or three (C and D) independent biological replicates.
p68 is required for beta-catenin nuclear translocation upon mrhl RNA downregulation.
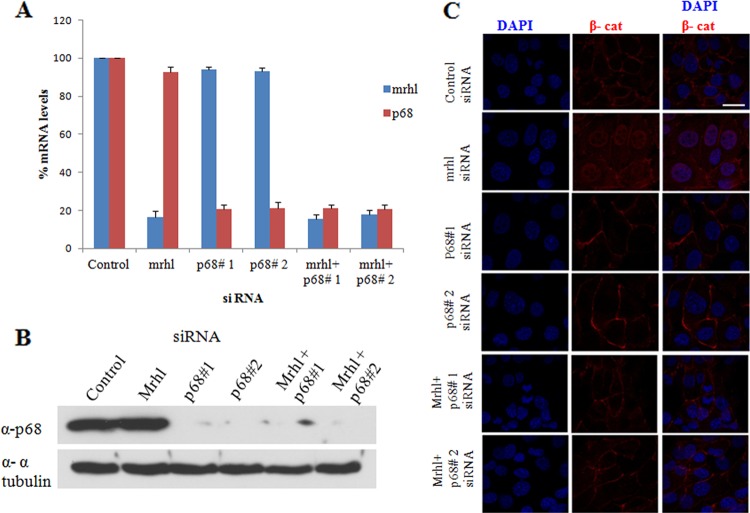
Next, we proceeded to investigate whether p68 is essential for the nuclear translocation of beta-catenin upon mrhl RNA downregulation. We resorted to silence the expression of mrhl RNA as well as p68. The downregulation of mrhl RNA and p68 RNA expression was probed by qRT-PCR (Fig. 7A), and the downregulation of p68 protein expression was checked by Western blotting (Fig. 7B) after 48 h of siRNA transfection. Beta-catenin localization was probed by immunofluorescence (Fig. 7C). On p68 silencing by two different siRNA pools, beta-catenin was membrane localized similarly to that seen in control cells. On silencing mrhl RNA, beta-catenin was found to be membranous as well as nucleus localized, which was consistent with our earlier observations. However, on silencing p68 along with mrhl RNA we found that the nuclear translocation of beta-catenin was largely blocked. This convincingly showed that beta-catenin nuclear translocation observed upon mrhl RNA downregulation is dependent on p68.
Fig 7.
p68 is required for beta-catenin nuclear translocation upon mrhl RNA downregulation. (A) Real-time PCR analysis showing relative levels of p68 RNA and mrhl RNA on treatment with the indicated siRNAs. Error bars represent standard deviations. P < 0.001 by t test. (B) Western blot showing downregulation of p68 protein expression. Alpha tubulin was used as a loading control. (C) Immunostaining in Gc1-Spg cells using anti-beta-catenin antibody on treatment with the indicated siRNAs. Scale bar, 25 μm. Images shown in panels A to C are from a representative experiment. Experiments were done in duplicate and repeated with two independent biological replicates.
Wnt3a ligand downregulates mrhl RNA.
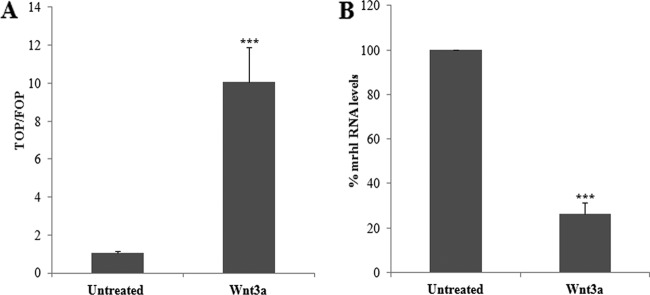
After establishing the negative regulatory role of mrhl RNA in Wnt signaling, it was important to know whether mrhl RNA is regulated upon Wnt signaling activation. For this, Gc1-Spg cells were treated with Wnt3a ligand for 12 h. Activation of Wnt signaling was assessed by standard TOP/FOP luciferase reporter assay, which showed about a 10-fold increase in TOP/FOP ratio upon Wnt3a treatment (Fig. 8A). mrhl RNA levels in the untreated and treated samples were examined by qRT-PCR, which showed about a 4-fold or 75% decrease in mrhl RNA expression upon Wnt3a treatment (Fig. 8B). We carried out the same experiment with Wnt5a ligand, which actually inhibits canonical wnt signaling (44), and found that mrhl RNA expression is not perturbed (data not shown). This experiment shows that the expression of mrhl RNA is responsive to Wnt3a ligand.
Fig 8.
Wnt3a ligand downregulates mrhl RNA in Gc1-Spg cells. (A) Luciferase reporter assay showing increased TOP/FOP ratio upon treatment of Gc1-Spg cells for 12 h with Wnt3a ligand. (B) qRT-PCR analysis showing about 75% reduction in mrhl RNA levels upon Wnt3a treatment. Error bars represent standard deviations. ***, P < 0.0001. Images are from a representative experiment. Experiments were done in triplicate and repeated with three biological replicates.
DISCUSSION
Noncoding RNAs have been shown to play multiple roles in many cellular processes. We have recently identified and characterized a novel noncoding RNA gene in the mouse genome (13, 26). In our present study, through a series of experiments, we have conclusively shown the activation of a canonical Wnt signaling pathway upon mrhl RNA downregulation. It is interesting to note that TCF4, a key transcription factor in canonical Wnt signaling, is also upregulated under mrhl RNA gene silencing. We speculate a feed-forward loop in the wnt activation process that in turn activates TCF4 upon mrhl RNA downregulation. Additionally, we have also observed downregulation of extracellular negative modulators like Dkk3 and Sfrp1, suggesting a close relationship between Wnt activation and the expression of negative modulators. We also point out here that beta-catenin is recruited with TCF4 at the promoter sites of Dkk3, Tsc22d1, Elk3, Sox8, and Sox12; this has not been reported earlier in the literature. It is interesting to note that the two genes Prickle 1 and Rac1, which are part of the noncanonical pathway (16, 45), are differentially perturbed upon mrhl RNA silencing, the significance of which cannot be commented upon at present. However, a recent report demonstrates that Rac1, a GTPase regulating cytoskeletal function, positively regulates nuclear localization of beta-catenin in the canonical Wnt signaling pathway (42). We find beta-catenin nuclear localization upon mrhl RNA downregulation in spite of about a 3-fold reduction in Rac1 mRNA levels (Fig. 3A). Although we have not measured Rac1 protein levels under these conditions, it is quite possible that the reduced levels of Rac1 protein in the activated (GTP-bound) form may be sufficient for beta-catenin nuclear translocation.
An important question that arises at this juncture is the mechanism of wnt activation under an mrhl RNA-downregulated condition. An important clue toward answering this question came from the Northwestern blot analysis and the mass spectrometric data of the RNA pulldown experiment. Among the four identified partners, Ddx5/p68 is a known RNA binding protein whose cytoplasmic translocation in its tyrosine-phosphorylated form is shown to stabilize cytoplasmic beta-catenin during PDGF-mediated epithelial-mesenchymal transition in colon cancer cells (43). In the present study, we observe tyrosine-phosphorylated p68 in the cytoplasm. In addition to p68, we have also identified Dusp11, a dual-specificity phosphatase, as another interacting protein of mrhl RNA (Table 1). It is likely that Dusp11 is a part of the p68-RNA complex regulating the dynamics of nucleocytoplasmic shuttling of p68. The role of mrhl RNA in this complex may be to serve as a scaffold for p68 protein binding and keep protein in a dephosphorylated state, thereby retaining it in the nucleus. Recently, long noncoding RNA Malat1 has been shown to regulate phosphorylation of SR proteins and in turn regulate alternative splicing (39). p68 is also shown to interact with another noncoding RNA, SRA, which is shown to have a coactivating role along with p68 in regulating gene expression at the chromatin level (5). Although we have not addressed the domain of mrhl RNA which interacts with p68 in this study, a preliminary exercise shows that a domain with a particular stem-loop secondary structure of SRA is also seen in mrhl noncoding RNA. Indeed, in our present study, we have observed that in addition to perturbation of expression of genes involved in the Wnt signaling pathway, many other genes belonging to other cellular processes are differentially regulated upon mrhl RNA silencing. It remains to be seen whether the p68-mrhl RNA complex also has an independent role, similar to that of the p68-SRA complex, in regulating chromatin-mediated gene expression.
In addition to Wnt pathway genes, many other genes were perturbed upon mrhl RNA downregulation. Perturbation of these non-Wnt pathway genes upon mrhl RNA silencing can be explained in part by perturbation of transcription factors and coactivators, which in turn can regulate many genes. Also, when we examined the perturbed gene list from the microarray data, we found that not all the Wnt target genes reported in the literature are perturbed under mrhl-downregulated conditions. Further, among the perturbed Wnt target genes, beta-catenin occupies the promoters of only a subset and not all of the genes (Fig. 4C). These observations can be explained by the fact that target gene regulation by the beta-catenin/TCF4 complex can be context and cell type specific and regulated by many other protein factors, chromatin remodelers, coactivators, and corepressors (25). We have also scanned a region of about 1 Mb upstream and downstream of the mrhl RNA gene and found that this region harbors genes like Gpt2, Itfg1, Dnaja2, and Vps35 (upstream) and Siah1a, Abcc12, Lonp2, and N4bp1 (downstream). Expression of these genes was not perturbed under mrhl RNA downregulation, indicating that the perturbation of gene expression is trans in nature with respect to the mrhl RNA gene locus. We also observe in the present study that Lim domain-containing protein, a transcription factor, is one of the interacting proteins of mrhl RNA (Table 1). To address these additional mechanisms of perturbation in gene expression, we have now begun to map the chromatin occupancy sites of mrhl RNA.
All the experiments reported in the present communication were carried out in a Gc1-Spg cell line derived from spermatogonial cells. Interestingly, we have come across a recent report by Yeh et al. (44) showing that Wnt signaling is suppressed in spermatogonial stem cells as well as spermatogonia but is activated in differentiating spermatocytes. Thus, it is tempting to speculate that mrhl RNA may function as one of the negative regulators of Wnt signaling in spermatogonial cells. We have also observed in the present study that mrhl RNA interacts with p68 in three other mouse tissues, namely, liver, kidney, and spleen, which had been shown earlier to express mrhl RNA (26). The functional significance of this association remains to be seen, and it would be most interesting to see whether mrhl RNA also acts as a negative regulator of Wnt signaling in these tissues.
ACKNOWLEDGMENTS
We acknowledge B. S. Suma of the confocal facility and G. Rupa of the mass spectrometry facility. We thank Jayashree Ladha and G. Anitha for assistance in microarray image analysis.
M.R.S.R. thanks the Department of Science and Technology for the J.C. Bose fellowship. This work was financially supported by the Department of Biotechnology, Government of India.
Footnotes
Published ahead of print 4 June 2012
REFERENCES
- 1. Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13:807–818 [DOI] [PubMed] [Google Scholar]
- 2. Aravin AA, Hannon GJ, Brennecke J. 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318:761–764 [DOI] [PubMed] [Google Scholar]
- 3. Barker N, et al. 2001. The chromatin remodelling factor Brg-1 interacts with β-catenin to promote target gene activation. EMBO J. 20:4935–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brembeck FH, Rosario M, Birchmeier W. 2006. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr. Opin. Genet. Dev. 16:51–59 [DOI] [PubMed] [Google Scholar]
- 5. Caretti G, Lei PE, Sartorelli V. 2007. The DEAD-box p68/p72 proteins and the noncoding RNA steroid receptor activator SRA: eclectic regulators of disparate biological functions. Cell Cycle 6:1172–1176 [DOI] [PubMed] [Google Scholar]
- 6. Clemson CM, et al. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33:717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cordin O, Banroques J, Tanner KN, Linder P. 2006. The DEAD-box protein family of RNA helicases. Gene 367:17–37 [DOI] [PubMed] [Google Scholar]
- 8. Dennis GJ, et al. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4:P3. [PubMed] [Google Scholar]
- 9. Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447:799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farazi TA, Juranek SA, Tuschl T. 2008. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 135:1201–1214 [DOI] [PubMed] [Google Scholar]
- 12. Fire A, et al. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811 [DOI] [PubMed] [Google Scholar]
- 13. Ganesan G, Rao SM. 2008. A novel noncoding RNA processed by Drosha is restricted to nucleus in mouse. RNA 14:1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gilbert SL, Pehrson JR, Sharp PA. 2000. XIST RNA associates with specific regions of the inactive X chromatin. J. Biol. Chem. 275:36491–36494 [DOI] [PubMed] [Google Scholar]
- 15. Gordon MD, Nusse R. 2006. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J. Biol. Chem. 281:22429–22433 [DOI] [PubMed] [Google Scholar]
- 16. Habas R, Dawid IB, He X. 2003. Coactivation of Rac and Rho by Wnt/Frizzled signaling is required for vertebrate gastrulation. Genes Dev. 17:295–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu Z, Mellor J, Wu J, DeLisi C. 2004. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics 5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reference deleted.
- 19. Kormish JD, Sinner D, Zorn MA. 2010. Interactions between SOX factors and Wnt/beta-catenin signaling in development and disease. Dev. Dyn. 239:56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Langfelder P, Horvarth S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li S, Weidenfeld J, Morrisey E. 2004. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol. Cell. Biol. 24:809–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu C, et al. 2002. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108:837–847 [DOI] [PubMed] [Google Scholar]
- 23. Logan YC, Nusse R. 2004. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20:781–810 [DOI] [PubMed] [Google Scholar]
- 24. Mohammad F, et al. 2008. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol. Cell. Biol. 28:3713–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mosimann C, Hausmann G, Basler K. 2009. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat. Rev. Mol. Cell Biol. 10:276–286 [DOI] [PubMed] [Google Scholar]
- 26. Nishant KT, Ravishankar H, Rao MRS. 2004. Characterization of a mouse recombination hot spot locus encoding a novel non-protein-coding RNA. Mol. Cell. Biol. 24:5620–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nusse R. 2005. Wnt signaling in disease and in development. Cell Res. 15:28–32 [DOI] [PubMed] [Google Scholar]
- 28. Oldham MC, et al. 2008. Functional organization of the transcriptome in human brain. Nat. Neurosci. 11:1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pandey RR, et al. 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32:232–246 [DOI] [PubMed] [Google Scholar]
- 30. Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH. 2002. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 298:1620–1623 [DOI] [PubMed] [Google Scholar]
- 31. Prasanth KV, et al. 2005. Regulating gene expression through RNA nuclear retention. Cell 123:249–263 [DOI] [PubMed] [Google Scholar]
- 32. Rasmussen TP, Mastrangelo MA, Eden A, Pehrson JR, Jaenisch R. 2000. Dynamic relocalization of histone MacroH2A1 from centrosomes to inactive X chromosomes during X inactivation. J. Cell Biol. 150:1189–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robb GB, Brown KM, Khurana J, Rana TM. 2005. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 12:133–137 [DOI] [PubMed] [Google Scholar]
- 34. Saldanha AJ. 2004. Java Tree view—extensible visualization of microarray data. Bioinformatics 20:3246–3248 [DOI] [PubMed] [Google Scholar]
- 35. Saris CG, et al. 2009. Weighted gene co-expression network analysis of the peripheral blood from amyotrophic lateral sclerosis patients. BMC Genomics 10:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Silverman E, Gilbert EG, Lin JR. 2003. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene 312:1–16 [DOI] [PubMed] [Google Scholar]
- 37. Sleutels F, Zwart R, Barlow DP. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 14:68–73 [DOI] [PubMed] [Google Scholar]
- 38. Srisawat C, Engelke DR. 2001. Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA 7:632–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tripathi V, et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39:925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Umlauf D, et al. 2004. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 36:1296–1300 [DOI] [PubMed] [Google Scholar]
- 41. Wang X, Song X, Glass CK, Rosenfeld MG. 2011. The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb. Perspect. Biol. 3:a003756 doi:10.1101/cshperspect.a003756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu X, et al. 2008. Rac1 activation controls nuclear localization of β-catenin during canonical Wnt signaling. Cell 133:340–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang L, Lin C, Liu ZR. 2006. P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta catenin. Cell 127:139–155 [DOI] [PubMed] [Google Scholar]
- 44. Yeh JR, Zhang X, Nagano MC. 2011. Wnt5a is a cell-extrinsic factor that supports self-renewal of mouse spermatogonial stem cells. J. Cell Sci. 124:2357–2366 [DOI] [PubMed] [Google Scholar]
- 45. Zallen JA. 2007. Planar polarity and tissue morphogenesis. Cell 129:1051–1063 [DOI] [PubMed] [Google Scholar]
- 46. Zhang B, Horvath S. 2005. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4:Article 17. doi:10.2202/1544-6115.1128 [DOI] [PubMed] [Google Scholar]