Abstract
Some cancers utilize the recombination-dependent process of alternative lengthening of telomeres (ALT) to maintain long heterogeneous telomeres. Here, we studied the recombinational telomere elongation (RTE) of the Kluyveromyces lactis stn1-M1 mutant. We found that the total amount of the abundant telomeric DNA in stn1-M1 cells is subject to rapid variation and that it is likely to be primarily extrachromosomal. Rad50 and Rad51, known to be required for different RTE pathways in Saccharomyces cerevisiae, were not essential for the production of either long telomeres or telomeric circles in stn1-M1 cells. Circles of DNA containing telomeric repeats (t-circles) either present at the point of establishment of long telomeres or introduced later into stn1-M1 cells each led to the formation of long tandem arrays of the t-circle's sequence, which were incorporated at multiple telomeres. These tandem arrays were extraordinarily unstable and showed evidence of repeated rounds of concerted amplification. Our results suggest that the maintenance of telomeres in the stn1-M1 mutant involves extreme turnover of telomeric sequences from processes including both large deletions and the copying of t-circles.
INTRODUCTION
The ability of human cancer cells to grow indefinitely requires maintenance of their telomeres (76). In most human cancers, this is achieved by upregulating telomerase activity (43). However, 5 to 10% of human tumor cells maintain their telomeres through a telomerase-independent mechanism termed alternative lengthening of telomeres (ALT) (for a review, see reference 14). Features of ALT cells include their highly elongated and extremely heterogeneous telomeres (36, 39, 56, 66) and the presence of subcellular structures called ALT-associated PML (promyelocytic leukemia) bodies (APB) (87), which contain telomeric DNA, as well as telomere-associated proteins and many other proteins involved in DNA repair, replication, and recombination (36, 40, 41, 69, 87). Several lines of evidence suggest that telomere maintenance in ALT cells depends on recombination. First, a DNA tag sequence inserted in the telomere was shown to be copied onto other telomeres in ALT cells, but not in telomerase-positive cells (22). Second, there are highly elevated levels of postreplicative telomere recombination, including sister chromatid exchanges, occurring in ALT cells (1, 6, 47). Third, ALT cells contain abundant extrachromosomal telomeric circles (t-circles), which are thought to form from intratelomeric recombination (11, 12, 24, 85, 88, 89). T-circles, with their C-rich strand at least partly single stranded, have emerged as a particularly reliable indicator of the ALT state (35). Despite these advances, the mechanism of recombination-dependent telomere maintenance in ALT cells, as well as the full extent of telomeric recombination in these cancers, remains poorly understood.
The mechanism of recombinational telomere elongation (RTE) has been extensively studied using telomerase deletion mutants in the budding yeasts Saccharomyces cerevisiae and Kluyveromyces lactis. Such mutants display gradual telomere shortening and growth senescence, followed by the RAD52-dependent emergence of better-growing postsenescence survivors that have elongated telomeres (48, 50, 52). Two types of RTE (termed I and II) have been found in S. cerevisiae. Type I survivors are characterized by amplification of subtelomeric Y′ elements and short telomeric tracts, while type II survivors are characterized by lengthened tracts of telomeric repeats (79). The two survivor types differ not only in the end structures they contain, but also in the genes responsible for their formation (17, 20, 44, 82).
Only type II RTE has been found to occur normally in K. lactis telomerase deletion mutants (50). The telomeric tracts present in K. lactis postsenescence survivors are longer than those seen in senescent cells but typically not more than hundreds of base pairs in length. Once lengthened by RTE, these telomeres are generally resistant to further recombination until gradual sequence loss again reduces them to very short lengths. Studies of these mutants suggested that type II RTE involves a roll-and-spread mechanism. According to this model, a tiny t-circle is copied by a rolling-circle mechanism to generate one long telomere, the sequence of which is then spread to all other telomeres by break-induced replication (BIR)-like events (33, 59, 60, 81). This model was originally suggested by the fact that postsenescence survivors generated in cells with two kinds of telomeric repeats often emerge with a repeating pattern common among most or all telomeres. Transformation of DNA circles containing telomeric repeats and a marker gene into a K. lactis telomerase deletion mutant led to addition of long tandem arrays of the transformed sequence onto telomeric ends. Mixing experiments performed with two t-circle species indicated that these arrays form from a single transformed molecule (60). T-circles as small as 100 nucleotides (nt) have been shown to promote telomere elongation in vivo in K. lactis (59). Even smaller DNA circles (including t-circles) have been shown to be suitable substrates for rolling-circle synthesis in vitro (25, 34, 46). T-circles from ∼100 bp to >30 kb have been shown to be abundant in certain yeast mutants with abnormally long telomeres (3, 33). Other important evidence for the roll-and-spread model came from the demonstration that the sequence of lengthened telomeres in postsenescence survivors originates from a single telomere source (81).
Work in recent years has expanded our views of the circumstances under which RTE can occur and the forms it can take. K. lactis mutants with very weak telomerase activity can use recombination to maintain very short telomeres or to occasionally generate survivor-like cells with all telomeres lengthened, the latter likely via a roll-and-spread mechanism (2). Certain mutant telomeric repeats, including those defective in binding the double-stranded telomere binding protein Rap1, lead to type IIR “runaway” RTE, which is characterized by a more ALT-like state with much longer and more heterogeneous telomeres than are seen in ter1-Δ mutants (4, 81). Similarly, rapid formation of long telomeric tracts by RTE occurring without gradual growth senescence was observed in certain telomere-capping mutants in S. cerevisiae (27, 30, 64). RTE, in at least some cases associated with an abundance of t-circles, is also thought to be responsible for the maintenance of telomeres at the ends of linear mitochondrial DNA in some species of ciliates and yeasts (55, 63, 80).
The initial description of type IIR RTE was done with the K. lactis stn1-M1 mutant (38), which contains an amino acid substitution in a component of the RPA-like Cdc13-Stn1-Ten1 (CST) complex that binds the 3′ single-stranded telomeric overhang and protects it from being degraded and engaging in recombination (for a review, see reference 70). Stn1 also contributes to the regulation of telomerase addition and is thought to help bring about second-strand synthesis at telomeres via its ability to interact with Polα/primase (29, 71). Compared to a K. lactis telomerase deletion mutant, the stn1-M1 mutant is distinctly more similar to human ALT cells. Both show the continuous presence of very long and heterogeneous telomeres generated from homologous recombination, and both produce abundant t-circles (3, 12, 85). Also, both lack growth senescence and survivor formation and appear instead to have chronic but slight growth defects (36, 38, 72). Finally, the presence of telomerase activity does not suppress the phenotype of either stn1-M1 or most ALT cells (36, 38). These facts are consistent with the phenotypes of both stn1-M1 mutants and ALT cells being due to continuous telomere-capping defects that are not repairable by telomere lengthening. The stn1-M1 mutant is therefore an excellent potential model to help understand ALT. In this report, we present data that demonstrate that t-circles are potent generators of lengthened telomeres in stn1-M1 cells and that an exceptionally high level of telomere instability occurs in stn1-M1 cells.
MATERIALS AND METHODS
Strains, plasmids, and circles.
All K. lactis strains used are derivative of the wild-type (WT) 7B520 (ura3-1 his2-2 trp1) strain (86). K. lactis stn1-M1 and stn1-M1 ter1-Δ strains were described previously (38). Circle N, which comprises 11.5 wild-type telomeric repeats and a 1.2-kb URA3 gene, was described previously (59). The plasmid pCXJ3 derivative circle A was constructed by inserting 11.5 K. lactis telomeric repeats that were obtained as an XhoI fragment from pAK25 (53) into an SalI site of pCXJ3 with the URA3 gene transcribed in the same direction as the telomeric repeats. The plasmid pSTN1 was constructed in two steps. First, a 3.4-kb fragment containing the 1.3-kb open reading frame (ORF) of the STN1 gene and 1.6-kb upstream and 0.5-kb downstream sequences was obtained by PCR (forward primer, 5′-ACGAGCTCTGGCAACCCACTTGTGACTA-3′; reverse primer, 5′-ACCTCGAGTGCTCAGCCAATTTCTGTTG-3′) using the genomic DNA of the WT 7B520 strain as the template. Second, the PCR fragment, which contains flanking SacI and XhoI sites, was inserted into the SacI and XhoI sites in the polylinker of pKL313(HIS3) (73) to generate pSTN1.
The stn1-M1 rad51 strain was generated by crossing the stn1-M1 strain with the rad51 strain (SAY516) (42), followed by random spore analysis and confirmation by restriction fragment length polymorphism (RFLP) analysis. The stn1-M1 ter1-Δ rad50, stn1-M1 rad50, and stn1-M1 rad50 rad51 strains were constructed as follows. The ∼3.4-kb rad50::KANMX disruption cassette was first amplified from the genomic DNA of SAY557 (42) by PCR (forward primer, 5′-AATTTGTGAGTCGGAGGACACT-3′; reverse primer, 5′-GTATTGGACATGATGGTGAGCTATT-3′) and transformed into stn1-M1 ter1-Δ cells complemented by pSTN1-TER1, stn1-M1 cells complemented by pSTN1, and stn1-M1 rad51 cells complemented by pSTN1, respectively. The replacement of the native RAD50 allele by the rad50::KANMX disruption cassette occurred through homologous recombination. The deletion of the RAD50 gene was confirmed by RFLP analysis and the loss of hybridization to a probe specific to the open reading frame of the RAD50 gene. The clones with the disrupted RAD50 allele were plated on medium containing 5-fluoroorotic acid (5-FOA) to select for the loss of pSTN1-TER1 and pSTN1 to generate the stn1-M1 ter1-Δ rad50::KANMX, the stn1-M1 rad50::KANMX, and stn1-M1 rad50::KANMX rad51 clones, respectively.
K. lactis transformation was performed by electroporation as described for S. cerevisiae (7), except for scaling down by 1,000-fold. Passaging of cells was carried out by serial streaking of single colonies on rich medium (yeast extract-peptone-dextrose [YPD] plates) at 30°C. Strains were streaked every 3 days down to single cells that grew into colonies. Each streak was estimated to be 20 to 25 cell divisions. Approximately 30 ng of circle N was used per transformation, and the average number of yeast cells used per transformation was ∼6 × 107.
The TER1/ter1-Δ STN1/stn1-M1 heterozygote was generated by mating the stn1-M1 ter1-Δ and GG1958 (STN1 TER1 ade2) strains (10). Diploid cells were sporulated and analyzed by random spore analysis as described previously (10). The stn1-M1 TER1 and stn1-M1 ter1-Δ clones were distinguished based on TER1 restriction fragment length polymorphisms seen in a Southern blot.
Southern hybridization and quantitation of URA3 copy number and telomere signals.
K. lactis genomic-DNA preparations were performed in two ways. The first way yielded high-quality genomic DNA and is based on the method of Philippsen et al. (68), except for two respects: (i) the starting cultures were 1.5 ml instead of 30 ml, and the reagents used in the protocol were scaled down proportionally; (ii) after step 11, the DNA pellet is dissolved completely in 100 μl of 10 mM Tris-HCl, 1 mM EDTA, followed by adding 1/3 volume of 5 M ammonium acetate, pH 4.8, to precipitate proteins on ice for 20 min; steps 9 to 11 were repeated, and the protocol was followed to the end. The second, more rapid DNA preparation method was used for screening large numbers of yeast clones and is based on the protocol of Hoffman (37), except that instead of starting with liquid overnight cultures, the yeast cells were collected by scooping them from the surfaces of solid media and suspended in 0.5 ml water prior to beginning the procedure. This rapid method was used only for uncut DNA samples isolated for Southern blotting (see Fig. 6B and C).
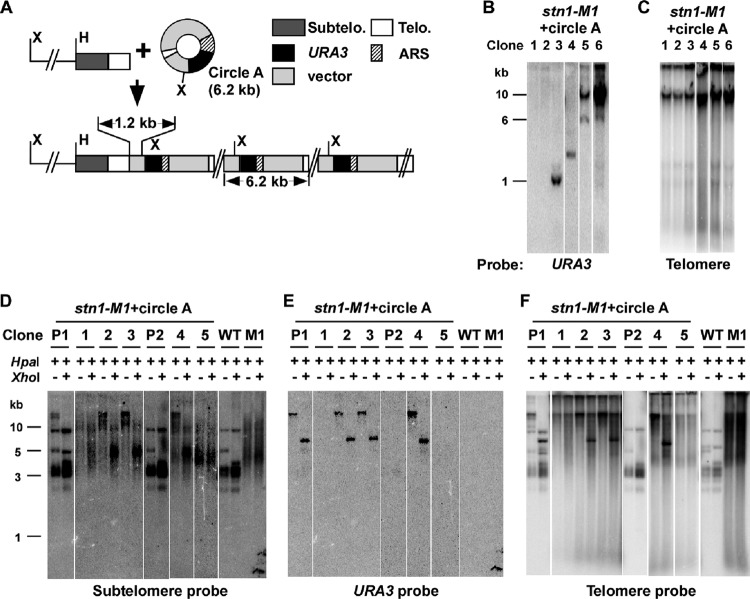
Fig 6.
A replicating t-circle present at the establishment of the stn1-M1 mutant state forms long tandem arrays at telomeres. (A) Diagram of circle A, a 6.2-kb replicating plasmid containing telomeric (Telo.) repeats, as well as the tandem array structure expected if the circle A sequence is added to telomere ends. Components of circle A, including ∼11.5 telomeric repeats and an ARS, are shown. The positions of HpaI (H) and XhoI (X) sites are indicated. The 1.2 kb of vector sequence from circle A between the basal telomeric repeats and the first XhoI site is indicated. Subtelo., subtelomeric. (B) Southern blot, hybridized with a URA3 probe, of undigested DNA from stn1-M1 clones derived from circle A transformants that had lost complementation by pSTN1. Clones exhibiting each of three types of outcomes are shown: loss of plasmid (lanes 1 and 2), retention of extrachromosomal plasmids with telomeric repeats deleted (lanes 3 and 4), and integration of plasmid into chromosomal DNA (lanes 5 and 6). (C) Same filter as in panel B after stripping and rehybridization with a telomere probe. (D) Southern blot, hybridized with a subtelomeric probe, of HpaI- and HpaI-plus-XhoI-digested DNA from stn1-M1 (pSTN1) circle A transformants (precursor clones P1 and P2) and stn1-M1 clones derived from circle A transformants that had lost pSTN1. Clones 1 to 3 are derived from P1, and clones 4 and 5 are derived from P2. Note that the clone designations do not match those in panels B and C. Untransformed wild-type STN1 (WT) and stn1-M1 control strains are also shown. Also note that one HpaI telomeric fragment in wild-type cells has an XhoI site in subtelomeric DNA. (E) Same filter as in panel D after stripping and rehybridization with a URA3 probe. (F) Same filter as in panel E after rehybridization and reprobing with a telomeric probe.
Yeast genomic DNA (cut or uncut) was run on 0.8% or 3% agarose gels and then transferred onto a Hybond N+ membrane. All hybridizations were carried out in Na2HPO4 and SDS as described previously (19). The telomeric probe used was Klac 1-25 (5′-ACGGATTTGATTAGGTATGTGGTGT-3′) (51). The URA3-HX probe was composed of a combination of three oligonucleotides (5′-CTTTCCAATTTTTTTTTTTTCGT-3′, 5′-CGTCATTATAGAAATCATTACG-3′, and 5′-TACGACCGAGATTCCC-3′), which are described in more detail below. Both the telomeric probe and the URA3-HX probe were end labeled with [γ-32P]ATP, with the temperature of hybridization and washing between 45 and 50°C. The subtelomeric probe was generated from pKL11-B [an insert of an ∼1-kb telomeric EcoRI-SmaI fragment into pBluescript SK(−)], which was digested with XbaI and ligated back together to excise all the telomeric sequence and was then digested by EcoRI and XbaI to generate an ∼600-bp subtelomeric fragment for a probe. The URA3 (from S. cerevisiae) and URA3 (from S. cerevisiae)-plus-RAD52 (from K. lactis) probes were described previously (53). The RAD50 gene probe was an ∼5.9-kb purified PCR product that contained a 3.9-kb ORF of the RAD50 gene from K. lactis genomic DNA (forward primer, 5′-AATTTGTGAGTCGGAGGACACT-3′; reverse primer, 5′-GTATTGGACATGATGGTGAGCTATT-3′). The RAD51 gene probe was an ∼2.4-kb purified PCR product that contained a 1.1-kb ORF of the RAD51 gene from K. lactis genomic DNA (forward primer, 5′-ACGCGGCCGCCTATCGCTGTTTA-3′; reverse primer, 5′-ACCTCGACGGACATGCGAGGCTGAT-3′). Subtelomeric, URA3, URA3+RAD52, RAD50, and RAD51 probes were prepared using an NEBlot Kit (NEB). The temperature of hybridization and washing for these probes was 65°C. The membranes were autoradiographed and visualized using a Molecular Dynamics Storm PhosphorImager. The method for the determination of the URA3 copy number was described previously (53). The quantitation of the relative level of internal telomeric blocks (see Fig. 4C and D) was performed by quantitating the signal from each small telomeric band and dividing it by the number of telomeric repeats in each block.
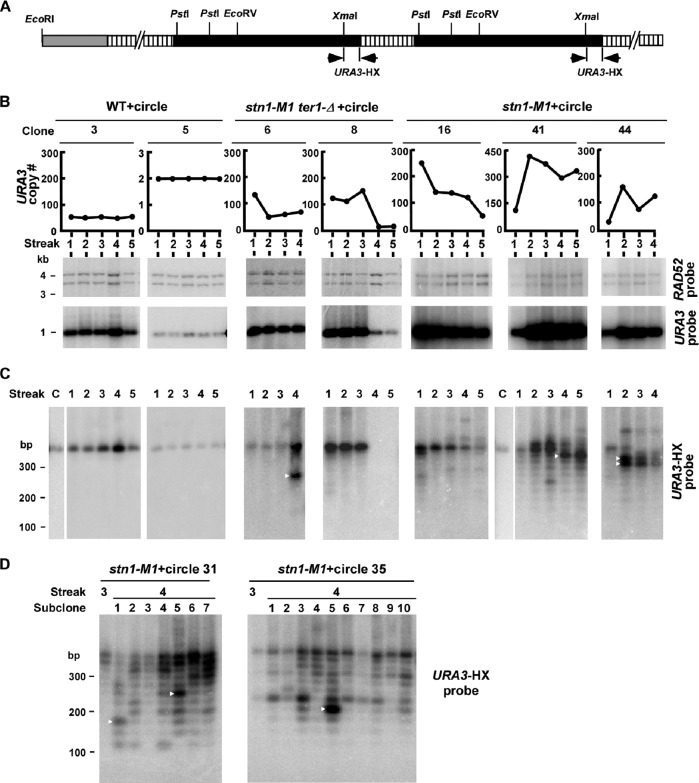
Fig 4.
Rapid changes occur in URA3 telomeric units from circle N-derived tandem arrays. (A) Diagram of a telomere with the circle N-derived tandem arrays of URA3 telomeric units (two units of which are shown) in transformed stn1-M1 and stn1-M1 ter1-Δ cells. The white boxes represent telomeric repeats, and the black boxes represent URA3. The positions of XmaI, PstI, EcoRI, and EcoRV sites are indicated. The locations of the URA3-HX probes used in panels C and D are also indicated. (B) Southern blotting and URA3 quantitation data from two independent lineages of both the wild type (clones 3 and 5) and stn1-M1 ter1-Δ (clones 6 and 8), as well as three independent lineages of stn1-M1 strains (clones 16, 41, and 44), all of which were transformed with circle N. Cells were serially passaged for 4 to 5 streaks on YPD plates, as indicated. The middle and bottom rows show Southern blots of XmaI-plus-PstI-digested DNA hybridized to the single-copy RAD52 gene (which is present in two pieces) and the URA3 gene. Only the largest of the XmaI-plus-PstI URA3 fragments is shown. The graphs show the copy numbers of URA3 at the streaks indicated, calculated from the Southern data. (C) XmaI-plus-PstI-digested DNA from the same samples as in panel B separated by 3% agarose gel electrophoresis and hybridized with the URA3-HX probe. The order of samples is identical to that in panel B except for the addition of lanes (labeled “C”) showing circle N digested with the same enzymes. The sample from streak 4 of stn1-M1 ter1-Δ clone 6 was overloaded relative to streaks 1 to 3 of the same clone. (D) Southern blotting of XmaI-plus-PstI-digested DNAs from multiple subclones from the 4th streak of each of two lineages of stn1-M1 strains transformed with circle N. The DNAs are shown separated on a 3% agarose gel and hybridized with the URA3-HX probe. Samples from the 3rd streak are also shown.
To measure the relative telomeric signal of stn1-M1 and stn1-M1 ter1-Δ lineages, liquid cultures of cell samples were divided into three subsamples of equal volume that underwent DNA preparation procedures independently. The EcoRI-digested DNA samples from each subsample were loaded at different positions (left, middle, or right) on the gels to control the variation of uneven blotting from different positions on the gels. Because of the potential for gene duplication from chromosome rearrangements in the mutants, we used signal levels from both RAD50 and RAD51 as internal single-copy controls. The ratios between RAD50 and RAD51 signals were between 0.8 and 1.3 in all the samples we analyzed, consistent with the relative dosage of the two genes remaining unchanged. The telomeric signals of the wild-type, stn1-M1, and stn1-M1 ter1-Δ strains were estimated by PhosphorImager analysis.
Pulsed-field gel electrophoresis.
Intact chromosome blocks were prepared as previously described (75). Contour-clamped homogeneous electric field–pulsed-field gel electrophoresis (CHEF-PFGE) was carried out in a CHEF Mapper XA Chiller system (Bio-Rad). Chromosomes were separated in 0.8% certified megabase agarose (Bio-Rad) in 1× TBE (90 mM Tris-borate, pH 8, 2 mM EDTA) at 16°C, using an alternating electric field angle of 120° at 2.7-V cm−1 electrode distance with a linear ramp time of field switch of 120 to 720 s for 72 h.
RESULTS
The amount of telomeric DNA in stn1-M1 cells varies during serial passaging.
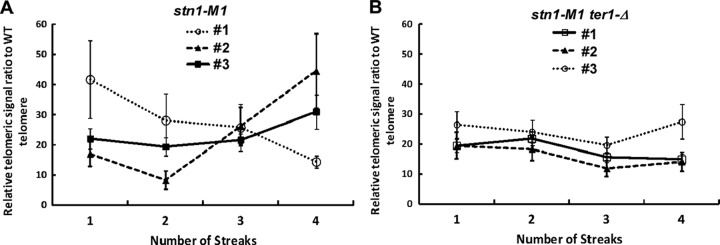
The pattern of long and heterogeneous telomeres seen in Southern blots of the stn1-M1 mutant remain generally similar in cell populations followed over extended passaging (38). This relatively steady state of telomeric DNA was suggested to be produced by high rates of telomeric recombination both extending and deleting telomeric sequences. To further examine the state of telomeric DNA in stn1-M1 cells, we serially restreaked six independent lineages of stn1-M1 mutants and four independent lineages of stn1-M1 mutants that had telomerase RNA gene deletions (ter1-Δ) and measured their relative telomeric DNA amounts at each of four consecutive streaks. Each overnight cell culture sample was divided into three parts from which genomic DNA was independently isolated. The average telomeric signal in each sample was measured relative to that of a wild-type control. The results for three representative lineages of each mutant are shown in Fig. 1. Total telomeric DNA signals in stn1-M1 and stn1-M1 ter1-Δ mutants among the 10 sets of samples studied varied from ∼8 to ∼46 times that of the wild-type strain and averaged ∼26 times greater in the stn1-M1 cells and ∼20 times greater in the stn1-M1 ter1-Δ cells, a difference that was statistically significant (P = 0.03). Given that the lengths of the 12 telomeres in a wild-type cell average ∼500 bp, the total range of telomeric DNA in the stn1-M1 and stn1-M1 ter1-Δ mutants was measured as 42 to 258 kb, consistent with the previous estimation (38). Our results showed that fluctuations in the total amount of telomeric DNA up to 2- to 3-fold could occur during a brief period of passaging of stn1-M1 cells (e.g., stn1-M1 clone 2 [Fig. 1A]). Thus, stn1-M1 cell net gains or losses of up to ∼100,000 bp of telomeric DNA commonly occur in the 20 to 25 cell divisions of a single streak of growth. Interestingly, the variation in the amount of telomeric DNA was significantly higher in stn1-M1 TER1 strains than in stn1-M1 ter1-Δ strains (P = 0.02).
Fig 1.
The amount of telomeric DNA in stn1-M1 and stn1-M1 ter1-Δ strains varies during passaging. Cells from three lineages each of the stn1-M1 (A) and stn1-M1 ter1-Δ (B) strains were passaged for 4 streaks on YPD plates. The total telomeric signal was measured on Southern blots from samples at each point. The samples were standardized using signal from two single-copy gene controls. The telomeric signal of the mutants is indicated as a ratio relative to the telomeric signal of a wild-type cell culture control. The error bars represent standard errors; n = 6 at each data point.
RAD50 and RAD51 are not essential for the stn1-M1 phenotype.
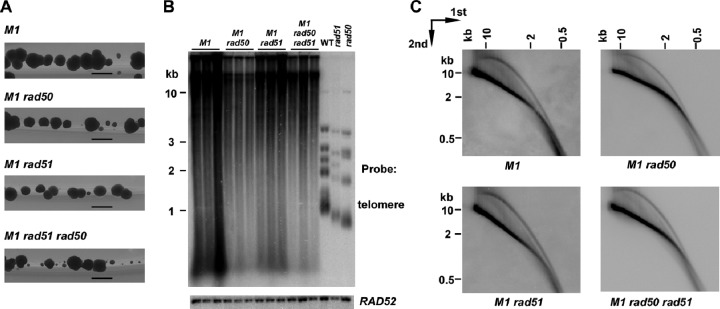
In S. cerevisiae telomerase deletion mutants, type I RTE requires the RAD51-dependent pathway of homologous recombination, while type II RTE requires the RAD50-dependent pathway (17, 44). Both pathways require RAD52, which is also essential for viability of K. lactis stn1-M1 cells, even in the presence of telomerase (38). To test if either gene is involved in telomere maintenance in the stn1-M1 mutant, we generated multiple independent stn1-M1 mutants that were disrupted for one or both of the RAD50 and RAD51 genes. The stn1-M1 rad51 mutants were generated by mating and sporulation, while the stn1-M1 rad50 and stn1-M1 rad50 rad51 mutants were generated by direct disruption of RAD50 in cells carrying a second copy of STN1 in a plasmid, followed by selection for loss of that plasmid (see Materials and Methods). Interestingly, we found that mutants of each type were viable. However, the stn1-M1 rad50 rad51 triple mutants initially grew particularly poorly on the 5-FOA plates where they were isolated (after selection for loss of the URA3-carrying pSTN1 plasmid) (data not shown). The growth phenotype of the triple mutants improved somewhat with subsequent streaks on rich medium, but colonies showed considerably more heterogeneity in size than the stn1-M1 rad50 and stn1-M1 rad51 mutants (Fig. 2A). These results indicated that neither RAD50 nor RAD51 was essential to viability but that each played some role in stn1-M1 cell growth, perhaps particularly at the initial establishment of the mutant state, when telomeres were presumably shortest.
Fig 2.
RAD50 and RAD51 are not essential for the viability of stn1-M1 cells. (A) Greater growth heterogeneity of stn1-M1 rad50 rad51 triple mutants. Colonies of stn1-M1 (M1), stn1-M1 rad50 (M1 rad50), stn1-M1 rad51 (M1 rad51), and stn1-M1 rad50 rad51 (M1 rad50 rad51) cells isolated from medium containing 5-FOA are shown 2 days after being streaked onto YPD medium. The photographs were taken with backlighting. Bars, 1 mm. (B) Southern blot, hybridized with a telomere probe (top) and a RAD52 probe (bottom), of EcoRI-digested DNAs from three independent clones of stn1-M1 (M1), stn1-M1 rad50 (M1 rad50), stn1-M1 rad51 (M1 rad51), and stn1-M1 rad50 rad51 (M1 rad50 rad51) strains, as well as WT, rad51 (SAY516), and rad50 (SAY557) strains. (C) Southern blot, hybridized with a telomere probe of EcoRI-digested DNA from stn1-M1 (M1), stn1-M1 rad50 (M1 rad50), stn1-M1 rad51 (M1 rad51), and stn1-M1 rad50 rad51 (M1 rad50 rad51) strains separated by two-dimensional agarose gel electrophoresis.
Examination of telomeres in the mutants using Southern blots of EcoRI-digested DNA (Fig. 2B and data not shown) showed that each clone of each type of mutant examined retained the characteristic smeared and highly elongated telomeric signal seen in the stn1-M1 single-mutant controls. This indicated that neither RAD50 nor RAD51 was essential for the long-telomere phenotype of the stn1-M1 mutant. The stn1-M1 rad50 and stn1-M1 rad50 rad51 mutants displayed less total telomeric signal (relative to a single-gene control) on average than stn1-M1 single mutants (Fig. 2B and data not shown). However, in other experiments, we found that stn1-M1 rad50 ter1-Δ cells constructed via a different parental source of the stn1-M1 mutation had telomeric signal levels similar to those of the stn1-M1 single mutant (data not shown). The reason for this difference is not known. Conceivably, telomerase could promote greater rates of telomeric deletions or interfere with the residual RTE in these cells. Any inhibitory effect of telomerase on net telomere lengthening by recombination would have to be greater than the direct telomeric-repeat synthesis provided by telomerase. Another possibility is that one or more differences in the genetic backgrounds other than the absence of telomerase are somehow responsible. In any event, the abundant telomeric DNA in stn1-M1 rad50 ter1-Δ cells demonstrated that, as expected, telomerase was not required for the formation of long telomeres in the absence of RAD50.
We next tested whether t-circle formation was affected by the rad50 or rad51 mutation. DNA from stn1-M1 rad50, stn1-M1 rad51, and stn1-M1 rad50 rad51 mutants, as well as the stn1-M1 single-mutant control, were analyzed by two-dimensional gel electrophoresis and Southern blotting. The results from this analysis (Fig. 2C) showed that all mutants retained the arc of telomeric signal shown previously to be composed of t-circles (3). Although we did not attempt to address whether there might be modest quantitative differences in t-circle levels between the mutants, these results indicated that t-circles can be efficiently generated in the absence of both RAD50 and RAD51 in stn1-M1 cells.
Long tandem arrays are generated in telomeres of stn1-M1 cells from a DNA circle containing telomeric repeats and the URA3 gene.
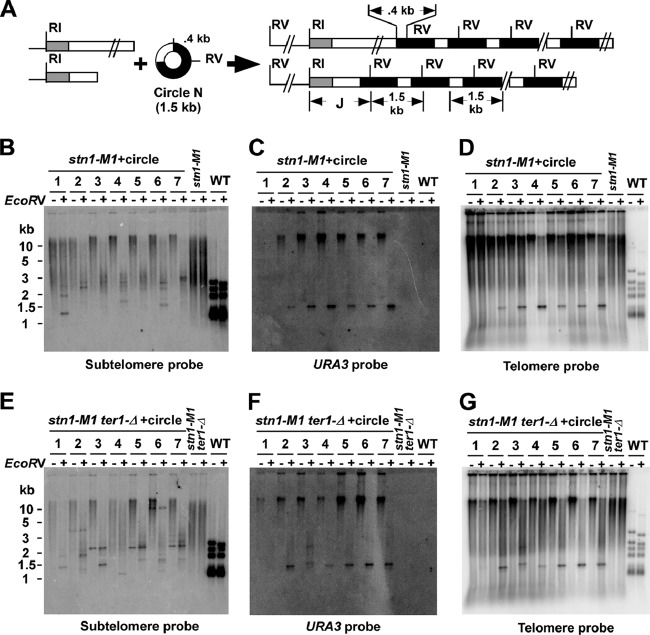
A prediction from the roll-and-spread model for recombinational telomere elongation (60) is that an exogenously transformed DNA circle containing telomeric repeats could be copied into telomeres as a long tandem array. Such an outcome is routinely observed in telomerase deletion mutants and even wild-type K. lactis (59, 60). To test this in the stn1-M1 mutant, we transformed the 1.5-kb circle N (59), which is composed of a fragment containing URA3 and 11.5 K. lactis telomeric repeats, into stn1-M1 and stn1-M1 ter1-Δ mutants (Fig. 3A). For simplicity, we refer to any circle containing telomeric repeats as a t-circle. We analyzed 47 stn1-M1 transformants and 55 stn1-M1 ter1-Δ transformants that grew on plates lacking uracil by Southern blotting and hybridization to determine their telomere structures. The terminal EcoRI fragments were detected using a subtelomeric probe that hybridizes to telomere-adjacent sequences at 11 of the 12 chromosome ends (Fig. 3B and E). While untransformed cells displayed a smear of subtelomeric signal in EcoRI digests running from <2 to >10 kb, most transformants in both mutants displayed a greater percentage of the subtelomeric signals running at >10 kb. This observation indicated that the average length of telomeres in transformants is greater than that in untransformed cells. In 34 of the 47 stn1-M1 transformants and a lower percentage (20 of the 55; P = 0.006 in an unpaired t test) of stn1-M1 ter1-Δ transformants, most subtelomeric signal was cleaved to shorter lengths (much of it 2 to 4 kb) by EcoRV, which has one cleavage site in the circle N sequence (Fig. 3B and E). In sharp contrast, subtelomeric signals from wild-type and untransformed stn1-M1 and stn1-M1 ter1-Δ cells were barely cleaved by EcoRV (Fig. 3B and E). The cleavage that did occur likely came from the single EcoRI telomeric fragment containing a subtelomeric EcoRV site. These data suggest that the sequence of circle N had been commonly incorporated into multiple, if not most, telomeres in stn1-M1 and stn1-M1 ter1-Δ transformants. As expected from this interpretation, URA3 and telomere probes hybridized to the same elongated subtelomeric EcoRI fragments (Fig. 3C, D, F, and G). In contrast, EcoRV cleaved the bulk of URA3 signal and a substantial part of telomeric signal to a band of ∼1.5 kb that was often visible in ethidium bromide staining of total yeast DNA in the gels used for blotting (Fig. 3C, D, F, and G and data not shown). These results are consistent with transformants commonly carrying a large number of copies of the circle N sequence integrated as tandem arrays at multiple telomeres. The relatively weak signal strength of the ∼1.5-kb band in the EcoRI-plus-EcoRV digests (relative to the >10-kb band in EcoRI digests) in Fig. 3C and F was not seen in equivalent experiments with other transformants (data not shown). It is therefore likely due to uneven blotting efficiencies in different parts of the particular gels shown.
Fig 3.
Long tandem arrays form at telomeres in stn1-M1 and stn1-M1 ter1-Δ circle N transformants. (A) Diagram of the 1.5-kb URA3 telomere circle (circle N) used for transformation and the structure of tandem arrays expected to be formed at telomeres by copying the sequence of circle N onto telomeric ends. The gray boxes represent subtelomeric sequence used as a probe in panels B and E; the white boxes indicate blocks of telomeric repeats, and the black boxes represent URA3. Subtelomeric EcoRI sites are ∼1 to 3.5 kb from telomere ends in untransformed K. lactis cells. The positions of EcoRI sites (RI) and EcoRV sites (RV) are indicated. J indicates subtelomere-telomere junction fragments. (B) Southern blot, hybridized with a subtelomeric probe, of EcoRI-digested (−) and EcoRI-plus-EcoRV-digested (+) DNAs from seven stn1-M1 strains transformed with circle N. Untransformed stn1-M1 and STN1 wild-type (WT) controls are also shown. (C) Same filter as in panel B after stripping and rehybridization with a URA3 probe. Note that the unequal amounts of URA3 signals between EcoRI and EcoRI-plus-EcoRV digests were not reproducible and are likely due to the uneven transfer of DNA during the Southern blotting. (D) Same filter as in panel B after stripping and rehybridization with a telomeric probe. (E) Southern blot, hybridized with a subtelomeric probe, of EcoRI- and EcoRI-plus-EcoRV-digested DNAs from seven stn1-M1 ter1-Δ strains transformed with circle N, along with untransformed stn1-M1 ter1-Δ and STN1 WT strains. (F) Same filter as in panel E after stripping and rehybridization with a URA3 probe. (G) Same filter as in panel E after stripping and rehybridization with a telomeric probe.
Slight variations in the sizes of the ∼1.5-kb band were observed in a few transformants (e.g., clone 3 of stn1-M1 ter1-Δ [Fig. 3F and G]). These likely were due to changes in the number of telomeric repeats present in each block of repeats derived from circle N, a phenomenon seen previously with t-circles transformed into short telomere mutants and thought to be due to recombination that altered the size of the t-circle before its sequence was copied onto telomeres (59). The hybridization of the telomeric probe to the stn1-M1 and stn1-M1 ter1-Δ transformants also showed substantial telomeric signals at very long lengths even after cleavage by EcoRV. This result suggests that some telomeric tracts much longer than the 11.5 repeats supplied by circle N are likely still present in the transformants.
Cleavage of DNA from circle N transformants with EcoRV is expected to remove all but one partial copy of circle N sequence from telomeres. In clonal isolates such as those examined, this was expected to produce one or more sharp bands of precise sizes that corresponded to junction fragments (Fig. 3A) containing subtelomeric DNA, some number of telomeric repeats, and ∼400 bp of URA3. Some such sharp bands, typically ranging from 1.2 to 3 kb, were observed with a subtelomeric probe in many transformants (e.g., clones 2 and 7 [Fig. 3B]). However, much of the subtelomeric signal that was shortened by EcoRV cleavage remained smeared in appearance, generally at sizes greater than those of the sharp bands. This indicated that these fragments, despite not being terminal in location, were nonetheless often heterogeneous in length. We conclude that the number of telomeric repeats that separate subtelomeric DNA from the closest integrated copy of circle N is not fully stable and often becomes heterogeneous in cell populations, presumably due to frequent ongoing recombination events.
An interesting observation with the stn1-M1 ter1-Δ transformants is the appearance of sharp bands that hybridize with the subtelomeric probe in some EcoRI-digested samples that are not affected by EcoRV digestion (e.g., clones 2, 3, 5, 6, and 7) (Fig. 3E). These EcoRV-resistant sharp bands occur in at least nine stn1-M1 ter1-Δ transformants but in only one stn1-M1 transformant. As these bands are not detected with URA3 or telomeric probes, they are likely to be independent of the sequence derived from circle N and may represent fusions between telomeric ends that have lost all, or almost all, of their telomeric repeats. Similar bands were also seen previously during passaging of stn1-M1 cells (38). The greater frequency of the sharp bands in stn1-M1 cells lacking telomerase might indicate that telomerase in stn1-M1 cells can lengthen short telomeres and render them resistant to nonhomologous end joining (NHEJ). Telomerase has previously been demonstrated to remain active in stn1-M1 cells (38).
The copy number of the 1.5-kb URA3 telomere units derived from circle N that were present in the transformants was estimated using a STN1 TER1 strain with a single integrated copy of URA3 as a control (53). For 10 stn1-M1 ter1-Δ transformants that were measured, the copy number of the URA3 telomere insert was estimated to be 3 to 135, with a mean of 43, and for 14 stn1-M1 transformants measured, the copy number of the URA3 telomere insert was estimated to be 6 to 247, with a mean of 99. The difference in mean copy numbers between the two mutants was close to significance (P = 0.06 in an unpaired t test). The highest copy number we observed is the equivalent of ∼4% of the total genomic DNA. The remaining transformants (e.g., clone 1 in Fig. 3B to D) did not have a detectable 1.5-kb fragment that hybridized to telomeric and URA3 probes in EcoRI-plus-EcoRV digests and presumably did not have precisely sized tandem copies of the URA3 telomere unit derived from circle N. Among 6 such examples of stn1-M1 transformants and 14 examples of stn1-M1 ter1-Δ transformants that were examined, the copy number of URA3 was always below 5 per cell (data not shown). The URA3 sequences in most or all of these clones are likely to be incorporated into telomeres, given that EcoRV produces at least one sharp band in the subtelomeric hybridization in at least 6 of the 13 stn1-M1 transformants without URA3 arrays and at least 17 of the 25 stn1-M1 ter1-Δ transformants without URA3 arrays (data not shown). However, tandem arrays of URA3 telomere units derived from circle N may not be formed, because EcoRV did not produce a detectable 1.5-kb fragment that hybridized to telomeric and URA3 probes. In these clones, circle N may integrate as single copies or form tandem arrays that are very short or are largely deleted after they form. The relatively short bands produced by EcoRV cleavage in these nonarray clones (e.g., clone 1 in Fig. 3B) (data not shown) suggest that at least some URA3 sequences reside close to subtelomeric sequence in these transformants.
URA3 telomere units are subject to very rapid changes in copy number during serial passaging.
The heterogeneously sized telomeric fragments and high rate of subtelomeric recombination of stn1-M1 suggested that telomeres in the mutant are very unstable and undergo high levels of recombination (38). The tandem URA3 telomere arrays in circle N transformants provided an excellent opportunity to further investigate this instability. Thirteen independent lineages of stn1-M1 circle N transformants and 10 independent lineages of stn1-M1 ter1-Δ circle N transformants were serially restreaked 4 or 5 times on rich-medium (YPD) plates and analyzed for their telomeric structure by Southern blotting. The copy number of URA3 genes at each streak of these lineages was estimated using a STN1 TER1 strain with a single integrated copy of URA3 as a control. The results from several of these clones are shown in Fig. 4B, and the copy number data for all the clones are shown in Table S1 in the supplemental material. In three control clones of wild-type strains transformed with circle N (two of which are shown in Fig. 4B), the copy number of URA3 genes remained essentially stable among all the streaks examined. None showed more than a 20% change in measured copy number from one streak to the next or from the first streak to the last. Whether the slight differences detected were due to actual copy number changes or to inaccuracies in measurement was not investigated. In contrast, the copy number of URA3 genes in all 23 of the stn1-M1 and stn1-M1 ter1-Δ transformants examined showed increases or decreases of >20% in at least one streak relative to the previous streak, and most showed much greater changes (Fig. 4B; see Table S1). Some lineages (e.g., clone 8 of stn1-M1 ter1-Δ and clone 16 of stn1-M1) (Fig. 4B) lost most or all of their initial URA3 copies during passaging, while others (e.g., clone 41 of stn1-M1) (Fig. 4B) showed pronounced increases in URA3 copy numbers. These results clearly show that the URA3 telomere arrays in the stn1-M1 mutants were highly dynamic, with either rapid deletion or amplification events often occurring.
Our data also suggested that the absence of telomerase in stn1-M1 mutants altered the extent or nature of the instability. Five of 10 stn1-M1 ter1-Δ clones showed at least a 2-fold reduction in URA3 copy number over the growth course studied, while only 2 of 13 stn1-M1 TER1 clones did (see Table S1 in the supplemental material). Furthermore, whereas only one of the 11 stn1-M1 TER1 clones with an initial URA3 copy number between 20 and 200 exhibited a drop to 10 or fewer copies at any point of the growth course, four out of five stn1-M1 ter1-Δ clones starting in the same copy number range did. The latter difference was statistically significant (P = 0.006; Fisher exact test).
One stn1-M1 TER1 clone (with a starting URA3 copy number of 6) and three stn1-M1 ter1-Δ clones (with starting URA3 copy numbers of 135, 11, and 45) were shown to lose all detectable URA3 copies by the fifth streak (see Table S1 in the supplemental material). To further study the potential for the loss of all copies of URA3 from the telomeres of these transformants, we patched cells from single colonies of stn1-M1 and stn1-M1 ter1-Δ transformants onto 5-FOA medium, which selects for Ura− cells. Thirteen of the 20 stn1-M1 transformants and 8 of the 13 stn1-M1 ter1-Δ transformants were found to produce 5-FOA-resistant colonies. One or two 5-FOAr clones from each transformant were examined by Southern blotting and hybridization to a URA3 probe (see Fig. S1A in the supplemental material) (data not shown). All 37 of these 5-FOA derivatives of stn1-M1 transformants (which initially contained 35 to 136 copies of URA3) and of stn1-M1 ter1-Δ transformants (which initially contained 8 to 152 copies of URA3) were confirmed to no longer contain any detectable copies of URA3 (see Fig. S1A). As with the circle N transformants examined (Fig. 3), the transformants producing 5-FOA-resistant colonies had URA3 genes at most of their telomeres, as judged by their sensitivity to EcoRV digestion (see Fig. S1B). Most transformants that did not produce 5-FOA-resistant colonies contained >200 copies of URA3. These results demonstrate that even copy numbers of the URA3 telomere units of >100 that are scattered among multiple telomeres can be completely lost from cells over a period of no more than ∼25 cell divisions.
Rapid variation in the lengths of telomeric repeat blocks within URA3 telomere arrays.
The circle N used to transform cells contains a block of ∼11.5 telomeric repeats. By studying the size of this block of telomeric repeats in the circle N transformants of stn1-M1 and stn1-M1 ter1-Δ mutants it is possible to determine whether any changes in its size took place after it entered stn1-M1 cells. DNA samples from 12 lineages of stn1-M1 transformants and six lineages of stn1-M1 ter1-Δ transformants described above were digested by XmaI and PstI, which excise the telomeric blocks and an adjacent small section of the URA3 gene from the bulk of the flanking URA3 sequences (Fig. 4A). A Southern blot of these samples was then probed with a combination of three labeled oligonucleotides specific to the small section of the URA3 gene still attached to the telomeric blocks (URA3-HX, indicated by arrows in Fig. 4A). In the two lineages of wild-type circle N transformants examined, a band of the same size as that in circle N was generated and remained the same size in the subsequent streaks (WT clones 3 and 5 [Fig. 4C]). However, all lineages of 12 stn1-M1 transformants and six stn1-M1 ter1-Δ transformants that were examined displayed different behavior. In lineages such as clone 8 of stn1-M1 ter1-Δ and clone 16 of stn1-M1, where URA3 copy numbers decreased greatly during passaging, the signal of telomeric bands decreased correspondingly. More strikingly, while most lineages of the stn1-M1 and stn1-M1 ter1-Δ transformants contained primarily a single band of the same size as that in circle N at the first streak, all of these transformants displayed multiple bands in one or more later streaks (Fig. 4C and data not shown). These extra bands were generally smaller than the original band, and in some cases (e.g., stn1-M1 clone 41), the bands were clearly in a ladder that appeared to vary by ∼25-bp steps. These results indicated that the telomeric blocks in the URA3 telomere tandem arrays were often deleted through loss of unit numbers of telomeric repeats.
In a few instances, a large percentage of the telomeric blocks changed in size from one streak to the next. For example, in stn1-M1 TER1 clone 41, the most intense band changed to a size apparently one repeat shorter from streak 3 to streak 4 (Fig. 4C). Using PhosphorImager analysis, the band in streak 4, indicated by the white arrowhead, was estimated at 177 copies (60% of 295 copies), whereas the same-size band one streak earlier was estimated as being only 39 copies (11% of 359 copies). Another example of this is stn1-M1 TER1 clone 44, where most of the signal from telomeric blocks changed from the original ∼300-bp fragment to bands of ∼225 bp and ∼250 bp between streak 1 and streak 2, with the URA3 copy number increased from ∼30 to ∼170. A third example is stn1-M1 ter1-Δ clone 6, where most of the signal from telomeric blocks changed to a much smaller fragment between streaks 3 and 4, with the URA3 copy number changing from ∼80 to ∼90 (Fig. 4C). These results indicate that a high percentage of the URA3 telomere units derived from circle N, even up to hundreds of copies, could be deleted and replaced by a large number of new copies of a unit with a different number of telomeric repeats in no more than 20 to 25 cell divisions.
To further study the change in the telomeric blocks in URA3 telomere arrays, we examined seven independent subclones from the 3rd streak of stn1-M1 clone 31 and 10 independent subclones from the 3rd streak of stn1-M1 clone 35 in the same way (Fig. 4D). Although subclones from the same transformant resembled one another in general, they still exhibited clear differences in banding patterns. For example, subclones 1 and 5 of stn1-M1 clone 31 and subclone 5 of stn1-M1 clone 35 showed prominent smaller bands that had been preferentially amplified from streak 3 to streak 4 (indicated by white arrowheads in Fig. 4D). Some other subclones contained bands corresponding to fragments with numbers of telomeric repeats from 1 to 11. The intense band in subclone 5 of stn1-M1 clone 35 (indicated by a white arrowhead in Fig. 4D) was estimated to be 65% of the total URA3 telomere units despite fragments of this size being faint or undetectable in other subclones. The sudden increase in the abundance of a particular faint band here and in the data shown in Fig. 4C suggests that a concerted mechanism for amplifying one particular URA3 telomere unit can occur during the maintenance of the long-telomere state of stn1-M1 cells. Whether these newly amplified novel-size bands reside together in tandem arrays or on a common subset of telomeres is not known.
Most of the telomeric signal from stn1-M1 cells does not migrate into a pulsed-field gel.
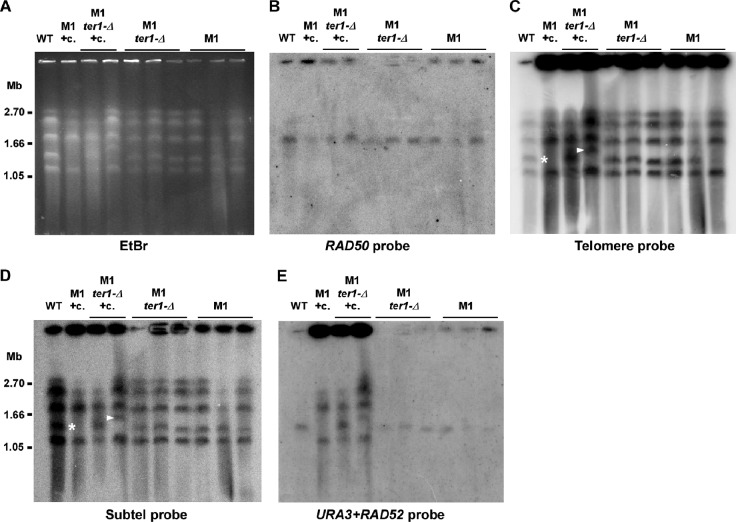
We next examined stn1-M1 mutants and their transformants with URA3 telomeric arrays using pulsed-field gel electrophoresis. Based upon the ethidium bromide-stained image of the gel (Fig. 5A) and upon hybridization to a single-copy RAD50 gene probe (Fig. 5B), chromosomes from both the wild-type and the various mutant strains were able to enter the gel with roughly similar efficiencies. Strikingly, however, 68 to 77% of signal to a telomeric probe of the stn1-M1 mutants was trapped in the wells compared to only 30% in the wild-type control (Fig. 5C). A similar but more limited retention of telomeric signal in the wells of standard gels has been seen for restriction enzyme-digested DNA from stn1-M1 cells (38) (Fig. 3D). Additionally, 32 to 69% of signal from the subtelomeric probe of stn1-M1 mutants was in the well compared to just 19% in the wild type (Fig. 5D). Why a single sample, the leftmost stn1-M1 ter1-Δ clone, behaved differently is not clear. Finally, a probe made from both S. cerevisiae URA3 and K. lactis RAD52 showed weak signal in wells and hybridization to a single chromosome in the wild type and in the stn1-M1 mutants that had not been transformed with circle N, a result consistent with detecting just the single-copy RAD52 gene (Fig. 5E). However, the same probe hybridized to most or all chromosomes in circle N-transformed strains, consistent with URA3 being present at most telomeres, and 70 to 85% of the total signal from this probe remained in the wells (Fig. 5E). Overall, these data show that sizable fractions of the telomeric repeats and the URA3 telomere units in transformed mutants, as well as a significant fraction of subtelomeric sequence from stn1-M1 and stn1-M1 ter1-Δ cells, were in structures that were both detached from chromosomes in our samples and present as structures that could not migrate into a pulsed-field gel. Such results conceivably could arise from very high rates of telomeric recombination, creating complex tangled molecules that often become detached from chromosomes in vivo or in vitro.
Fig 5.
Most telomeric signal of stn1-M1 and stn1-M1 ter1-Δ cells cannot enter a pulsed-field gel. (A) Ethidium bromide (EtBr)-stained pulsed-field gel showing chromosomes from wild-type K. lactis (WT), one stn1-M1 clone transformed by circle N (+c), two stn1-M1 ter1-Δ clones transformed by circle N (+c), and three each of untransformed stn1-M1 and stn1-M1 ter1-Δ clones. (B) Southern blot showing hybridization, using a RAD50 probe, of the DNA transferred from the agarose gel in panel A. (C) Same filter as in panel B after stripping and rehybridization with a telomeric probe. (D) Same filter as in panel B after stripping and rehybridization with a subtelomeric probe. (E) Same filter as in panel B after stripping and rehybridization with a probe composed of both URA3 and RAD52 gene sequences.
We also noticed that a chromosomal band in at least two of the stn1-M1 mutant samples either had shifted in size (marked with white arrowheads in Fig. 5C and D) or was missing (marked with asterisks in Fig. 5C and D). As the cells used were all haploid, it is likely that the latter event also involved a shift to a new size rather than chromosome loss. Interestingly, altered chromosomes corresponded to chromosome B, the only chromosome in the 7B520 strain background that contains a telomere lacking an R element, a sequence located immediately next to telomeres with homology to all other R elements (23, 61). Because the shared sequence of the R elements provides a backup means to repair deleted telomeres via homologous recombination (53), a telomere lacking an R element might be expected to be more prone to terminal deletions and rearrangements occurring as a consequence of telomere dysfunction.
A replicating t-circle present at the point of establishment of the stn1-M1 long-telomere state is readily incorporated at telomeres as long tandem arrays.
To test whether stn1-M1 cells could use t-circles to elongate telomeres without selection, we transformed plasmid pCXJ3 (18), which contains a K. lactis autonomously replicating sequence (ARS) and the URA3 gene, as well as circle A, a pCXJ3 derivative containing 11.5 telomeric repeats (Fig. 6A), into stn1-M1 cells complemented with a plasmid (pSTN1) containing the wild-type STN1 gene. We found that both pCXJ3 and circle A could be maintained extrachromosomally in these cells, which is indicated by the facts that the URA3 marker was genetically unstable and the URA3 probe could hybridize to some species of DNA running ahead of uncut genomic DNA in Southern blots (data not shown). We then streaked the plasmid-transformed cells on YPD plates and screened for the colonies losing pSTN1, which showed the rough colony phenotype of stn1-M1. Thirty-eight independent stn1-M1 clones were generated from the pCXJ3 transformants, and 163 were recovered from circle A transformants. Thirty-two of the 38 clones derived from pCXJ3 transformants and 130 of the 163 clones derived from circle A transformants were completely Ura− when tested on plates lacking uracil and showed no URA3 signal in Southern blots (Fig. 6B and data not shown), consistent with their having lost their respective plasmids after being placed on nonselective medium. For the 33 Ura+ stn1-M1 clones derived from circle A transformants, 10, including clones 3 and 4 in Fig. 6B, showed URA3 signal in bands that ran ahead of the bulk genomic DNA, which ran at the limit of mobility in the gel. The bands in these clones migrated at variable positions and did not hybridize with a telomere probe, suggesting that circle A remained in the clones but had suffered deletion of all telomeric repeats. These deletions appear to be dependent upon the stn1-M1 mutant state, as circle A molecules grown in multiple clones of wild-type cells did not show such deletions (data not shown). All 23 remaining Ura+ stn1-M1 clones derived from circle A transformants, including clones 5 and 6 (Fig. 6B), showed URA3 signal, often intense, at the limit of mobility in the gel, suggesting that the URA3 sequence from circle A was now incorporated into chromosomes in multiple copies. In contrast, none of the 6 Ura+ stn1-M1 mutants derived from pCXJ3 transformants showed varied sizes or had URA3 signal at the limit of mobility in the gel (data not shown), which indicates that integration of circle A into genomic DNA depends on the presence of telomeric DNA in the plasmid. Although we cannot rule out the possibility that the presence of an ARS influences the frequency of integration of circle A into genomic DNA, the results from circle N clearly show that an ARS is not required either for integration into genomic DNA or for formation of tandem arrays.
To examine the structure of telomeres in the 23 Ura+ stn1-M1 clones with integrated circle A sequences, we digested the DNA from the clones with HpaI and HpaI plus XhoI and probed with subtelomeric, URA3, and telomeric probes (Fig. 6D to F). HpaI does not cleave circle A and releases most telomeric ends in STN1 cells as 3- to 4-kb fragments (see the WT, P1, and P2 samples in Fig. 6D). XhoI cleaves once in circle A and cleaves only 1 of the 12 HpaI telomeric fragments in STN1 cells. Untransformed stn1-M1 (M1 in Fig. 6D to F) showed no URA3 signal and little or no sign of shortening of the long smeared telomeric and subtelomeric signals by XhoI. Ura− stn1-M1 clones derived from circle A transformants, including clones 1 and 5 (Fig. 6D to F), behaved similarly, consistent with the telomeres in these cells containing no sequence from circle A. In contrast, in the 23 Ura+ stn1-M1 clones with integrated URA3 sequence, including clones 2 to 4 (Fig. 6D to F), much of the subtelomeric signal was cleaved to substantially shorter lengths by XhoI (Fig. 6D). These data indicate that the sequence of circle A is present at multiple, if not most, telomeres in these clones. The URA3 signal in HpaI plus XhoI digests of these clones was present almost entirely in a band of ∼6 kb (Fig. 6E) that also hybridized intensely to a telomeric probe (Fig. 6F). These results are consistent with the circle A sequence in these 23 clones being integrated as tandem arrays at telomeres. The copy number of URA3 was measured in 11 of the clones and was estimated to range from 3 to 137, with a mean of 30 (data not shown). Our results indicate that in the absence of selection, DNA circles containing telomeric repeats can be incorporated very efficiently as tandem arrays into the telomeres of newly forming stn1-M1 mutants.
DISCUSSION
Telomere maintenance in the stn1-M1 mutant involves frequent and large telomeric deletions.
Increased recombination in and near telomeres is a common problem associated with defects in telomere capping (4, 31, 32, 51, 65). The K. lactis stn1-M1 mutant, which maintains very long telomeres through recombination, was previously shown to have elevated levels of single-stranded telomeric DNA, high levels of t-circles, and a greatly increased rate of subtelomeric recombination (3, 38). An important conclusion from our results here showed that the telomeres of stn1-M1 cells were subject to extreme levels of instability. One type of evidence for this was that the copy number of URA3 telomere units fluctuated dramatically, sometimes showing increases or decreases of more than 100 copies (totaling >150,000 bp) within a single streak (∼20 to 25 cell divisions) in stn1-M1 mutant cells, but not in wild-type cells. Moreover, even the subtelomere-adjacent block of telomeric repeats next to the URA3 telomere arrays exhibited sufficiently high levels of instability, such as not generally appearing as uniformly sized bands in Southern blots. We also showed that stn1-M1 cells could readily lose all of their >100 URA3 telomere units despite their being initially present at multiple telomeres, which argues that deletions affecting most or all telomeres are extremely common in this mutant. This conclusion is consistent with the past observations of an extremely high rate of loss of a subtelomeric URA3 gene in stn1-M1 (38). It is also consistent with the deep turnover of telomeric repeats reported in a K. lactis telomerase deletion mutant that had telomeres composed of repeats with a Rap1 binding deficiency, which undergoes a type IIR RTE similar to that of stn1-M1 (4, 38).
Another sign of extreme telomere instability in the stn1-M1 mutant was the fact that much of its telomeric DNA and some of its subtelomeric DNA were unable to enter pulsed-field gels. This occurred in spite of the fact that the bulk of the chromosomes as a whole, as judged by hybridizations to internal genes, did enter the gels. DNA structures previously shown not to enter pulsed-field gels include large nicked circles (8), DNA enriched with single-stranded tails or gaps (58), and branched replication and recombination intermediates (62, 67). Although t-circles are common in stn1-M1 cells (3), they do not appear to be abundant enough to account for the extent of retention observed. We suggest that most telomeric DNA of stn1-M1 cells exists as branched recombination intermediates that are unable to enter pulsed-field gels. The separation of telomeric DNA from bulk chromosomal DNA indicates that most telomeric DNA in stn1-M1 cells is either extrachromosomal at any given time or exists in fragile structures that become separated from chromosomes during the formation of the cell lysates used for pulsed-field gel analysis. Support for the former possibility comes from the phenotype of complemented stn1-M1 cells. While most long-telomere mutants that are complemented exhibit a very gradual return to normal telomere length due to replicative shortening, complemented stn1-M1 cells exhibit an essentially immediate return of telomeres to near-normal size (38). This is consistent with stn1-M1 telomeres experiencing abundant cleavage events. Extrachromosomal telomeric DNA in a variety of forms has been found to exist in human ALT cells, and material designated telomeric complex (t-complex), thought to be highly branched recombination intermediates, is known to be unable to enter gels (15, 57). Another example of long telomeric fragments that are unable to enter pulsed-field gels is found in fission yeast pot1-Δ rqh1-hd mutants (54, 78).
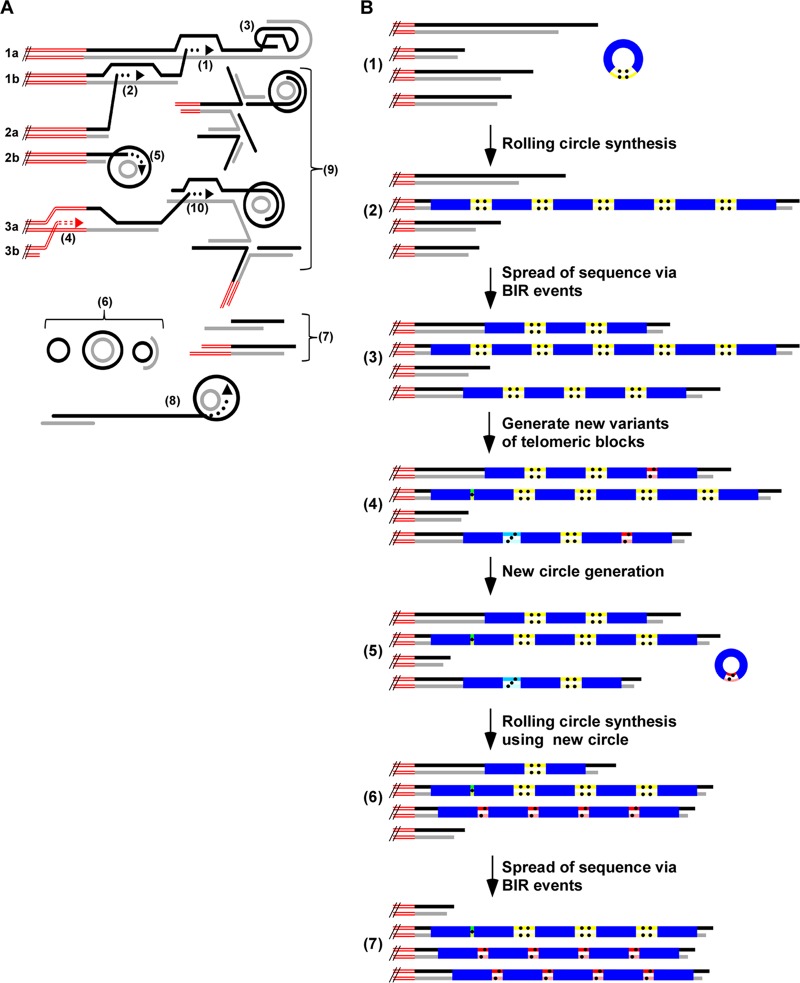
Conceivably, recombinationally entangled chromosomal ends may be difficult or impossible for cells to effectively resolve and would pose a barrier to proper chromosome segregation. Therefore, cell division in stn1-M1 cells may require cleavage or breakage events that regularly remove tangled t-complex from chromosome ends, much as removing matted hair can require a haircut. This could represent a major mechanism by which large deletions occur at multiple telomeres and URA3 telomere arrays in stn1-M1 cells. A model summarizing possible telomeric recombination structures and products is shown in Fig. 7A.
Fig 7.
Models for telomeric structure and maintenance in stn1-M1 cells. (A) Three pairs of sister chromatid ends (1a and b, 2a and b, and 3a and b), as well as structures containing telomeric repeats that are likely to be present in stn1-M1 cells, are illustrated. These structures include strand invasion between sister chromatids (1) and between nonsister chromatids (2); intratelomeric strand invasion to form a t-loop (3); subtelomeric strand invasion (4); strand invasion of a telomere end into a t-circle and rolling-circle copying (5); single- and double-stranded free t-circles (6); extrachromosomal linear telomeric fragments (7), some also containing subtelomeric sequence; extrachromosomal telomeric fragments elongated by rolling-circle synthesis from copying a t-circle (8); extrachromosomal t-complex involving base pairing between multiple molecules (9); and strand invasion of a telomere into t-complex (10). The double red lines represent subtelomeric sequences, and the black and gray lines represent G-rich and C-rich telomeric strands. (B) Model for telomere maintenance in stn1-M1 cells. (1) After a URA3 telomere circle N is transformed into stn1-M1 cells, it can be primed by a 3′ telomere end and used as a template for rolling-circle replication. The double red lines represent subtelomeric sequences, the black and gray lines represent G-rich and C-rich telomeric sequences, and the blue boxes represent the S. cerevisiae URA3 gene from circle N. The block of double yellow lines with four dots represents the block of 11.5 telomeric repeats in circle N. (2) Rolling-circle synthesis using the transformed circle N as the template produces a long tandem array of the sequence of the circle N at the end of a telomere. (3) With selection to maintain URA3, BIR events spread the sequence of the tandem array to multiple other telomeres. (4) With further cell divisions, ongoing high levels of telomeric recombination sometimes alter the number of repeats in the blocks between copies of URA3 in the tandem arrays (indicated by different colored boxes with different numbers of dots). (5 and 6) A URA3 telomere circle with a variant block of telomeric repeats is produced by recombination (5) and used as a template for another round of rolling-circle amplification (6). (7) The resulting newly generated tandem array is sometimes spread to one or more other telomere ends.
Another possible mechanism for large deletions is telomeric rapid deletion (TRD), a deletion process known to act on long telomeres, including those present in otherwise normal S. cerevisiae and K. lactis cells (5, 49, 85). TRD is proposed to involve the cleavage of a telomeric loop (t-loop)-like intermediate after strand invasion of a single-stranded telomeric 3′ end into a more internal double-stranded part of the same telomere. It may also be an important mechanism for producing t-circles (33, 83, 85). In support of these ideas, t-loop-like structures have been observed to be enriched in a K. lactis mutant with long dysfunctional telomeres and abundant t-circles (13). It is also important to note that cleavage of an intramolecular t-loop and cleavage of intermolecular entangled recombination intermediates might be mechanistically similar or identical.
The molecular basis of the stn1-M1 telomere-capping defect remains unclear. The amino acid substitution (I79K) present in the mutant resides in a conserved putative oligonucleotide/oligosaccharide binding domain suggested to be involved in binding the 3′ overhang of the telomere and in its interaction with Ten1 (26, 38, 77). A failure of Stn1 to bind well to the telomeric overhang might allow an overhang to become bound by recombination proteins. It would also be predicted to interfere with the ability of Polα/primase (which physically interacts with Stn1) to render the overhang double stranded at the end of DNA replication (29, 71).
The structure of tandem URA3 telomere arrays is consistent with a rolling-circle mechanism being involved in their generation.
Our results here demonstrate that stn1-M1 cells are highly able to incorporate and amplify the sequence of t-circles at their telomeres. As seen previously in ter1-Δ and, to a more limited extent, even wild-type cells (60), URA3 telomere circles transformed into stn1-M1 cells lead to long tandem arrays of the circle's sequence present at most chromosome ends. Notably, while the blocks of telomeric repeats between the URA3 genes within the tandem arrays eventually became highly heterogeneous in size (Fig. 4), immediately after their formation, they were mostly uniform, matching the size of the repeat block in the transformed DNA. This result is inconsistent with independent integration of multiple t-circle molecules but is consistent with the arrays being derived from a roll-and-spread mechanism, as indicated in steps 1 to 3 in Fig. 7B.
Another important finding in our work here was the repeated observation of instances where the predominant form of the URA3 telomere unit was rapidly and substantially replaced by a new form containing a different number of telomeric repeats. This argues strongly that the maintenance of telomeres in stn1-M1 cells includes a mechanism that results in concerted amplification of particular small telomeric regions. We propose that these sequence replacement events are the result of new rolling-circle copying events that occurred on URA3 telomere circles generated endogenously in the mutant cells (steps 5 to 7 in Fig. 7B). In favor of this, t-circles of a broad range of sizes are common in stn1-M1 cells (3). Additionally, ter1-Δ and stn1-M1 cells with tandem URA3 telomere arrays exhibit ladders of extrachromosomal URA3 telomere DNAs in gels (reference 60 and data not shown). This strongly suggests that URA3 telomere circles can be endogenously generated in stn1-M1 mutants.
A single secondary rolling-circle copying event by itself would probably be insufficient to amplify a novel form of the URA3 telomere unit to the extent we sometimes observed. We suggest that the greatest concerted amplifications are due to instances where stochastic BIR events happen to copy the first amplified array onto several other telomeres. As these would be occurring in the presence of other URA3 telomere arrays, as well as in the absence of any selective pressure, the extent of spreading to other telomeres would generally be expected to be less than that seen with the original circle N transformations. Because we cannot detect events where existing copies of the predominant form of the URA3 telomere unit are replaced with freshly amplified copies of the same unit, it is likely that our results considerably underestimate the occurrence of concerted array amplifications/replacements.
In three of the concerted amplification events we observed (streak 4 of stn1-M1 clone 41 [Fig. 4C] and subclone 1 of clone 31 and subclone 5 of clone 4 [Fig. 4D]), a single novel-size fragment of telomeric repeats was markedly amplified. If derived from a t-circle, these amplifications must either have copied a circle composed of a single URA3 telomere unit or copied a circle with >1 URA3 telomere unit where the telomeric blocks in each unit were the same size. In a fourth example of concerted amplification (streak 2 of stn1-M1 clone 44 [Fig. 4C]), two novel-size telomeric-repeat fragments were amplified to similar extents. This result might suggest either that a circle composed of two URA3 telomere units, each with different numbers of telomeric repeats, was used as a template for rolling-circle amplification or, alternatively, that two different-size circles were used as templates.
How variant URA3 telomere units with differing numbers of telomeric repeats arise is not known. The process presumably involves recombination events in which blocks of repeats between URA3 sequences are broken and modified prior to reforming. The significance of the tendency of the blocks to lose repeats is also unclear. It could reflect some bias in the mechanism that alters them, or it could be due to a selective advantage imparted by the shorter blocks. Conceivably, arrays with smaller blocks of telomeric repeats might be poorer targets for recombination events that cause breakages and deletions.
Evidence from ter1-Δ postsenescence survivors indicates that telomere-copying events can initiate either within telomeric sequences or within subtelomeric sequences (which share sequence among 11 of 12 K. lactis telomeres) (53, 81). This is almost certainly true in stn1-M1 cells as well, given the very high rate of BIR-like events affecting subtelomeric regions that have been reported there (38). The number of chromosome ends that acquire URA3 telomere tandem arrays after transformation with circle N is undoubtedly influenced by the selection for URA3. Because the arrays are highly prone to deletion, having them at multiple telomeres increases the likelihood that they will not be completely lost from cells. Alternatively, the spreading of URA3 telomere units from circles could come from multiple rounds of rolling-circle copying events using the same or different t-circles.
Although the bulk of our data in this report concern the behavior of URA3 telomere arrays, there is excellent reason to believe that these data are informative about the behavior of purely telomeric-repeat arrays in stn1-M1 cells. Our results showing that net changes of many tens of thousands of base pairs in the total amount of telomeric DNA can occur in the period of a single streak (20 to 25 cell divisions) clearly demonstrate that there is a very high level of instability in telomeric-repeat arrays in stn1-M1. The greater average length of telomeres containing URA3 telomere arrays compared to those of untransformed stn1-M1 cells (Fig. 3B, subtelomeric probe) might actually be an indication that URA3 telomere arrays are more stable than tracts of purely telomeric repeats. Conceivably, the more complex sequence of URA3 compared to the 25-bp repeats of the telomere might act to slow homology searches during recombination.
Extreme rates of telomere deletions such as occur in stn1-M1 cells would require equally large numbers of sequence additions to telomeric ends in order to maintain a more or less steady state of total telomeric DNA in the cell. As type IIR RTE is believed to allow unrestrained recombination to occur at telomeres of all lengths, there is no reason to believe that this should not be possible. It could occur not only via BIR events that copy other telomeres, but also through extrachromosomal telomeric DNA becoming reintegrated into chromosomal telomeres via recombination. Telomerase probably also contributes to telomere lengthening in those stn1-M1 cells that contain it in active form.
Establishment and maintenance of type II and type IIR RTE.
The roll-and-spread model was originally proposed to explain the generation of lengthened telomeres in senescing ter1-Δ mutants that previously contained only extremely short telomeres (60). The type II RTE in these cells, though it also involves elongation of telomeric-repeat tracts, is very different from that of the type IIR RTE of stn1-M1 cells. Because the telomere uncapping that drives it is due to short telomere length (less than ∼100 bp), type II RTE can essentially shut down once telomeres are even moderately elongated. Only after gradual telomere shortening again reduces telomere size to below a critical length will recombination again be induced. If senescing ter1-Δ cells contain even one telomere that is relatively long, the sequence of that telomere will be copied and spread to all other telomeres, presumably avoiding the need for copying a t-circle (81). As telomere lengths in ter1-Δ postsenescence survivors are generally more heterogeneous than those of senescent cells, this may imply that continued maintenance of lengthened telomeres by type II RTE (after the initial production of survivors) might occur with less use of copying of t-circles than at the initial establishment of postsenescence survivors.
Similarly, the role of t-circles might differ between the establishment and the maintenance phases of the type IIR RTE that occurs in stn1-M1 cells. At establishment (i.e., the moment a stn1-M1 mutant is first generated), there would be no long telomeres present. Rolling-circle synthesis copying a t-circle, therefore, might be an especially effective mechanism under such circumstances to make long tracts of telomeric DNA. Among our results here, only the experiments with circle A involved the study of the establishment stage of the stn1-M1 phenotype. Strikingly, we found that any newly established stn1-M1 mutant that retained the full circle A sequence did so by having it integrated as tandem arrays present at most telomeres. This integration occurred in spite of the absence of selection for the URA3 marker carried by circle A. These data demonstrate that t-circles present at the time of establishment of the stn1-M1 phenotype can be used with considerable efficiency as templates to elongate telomeres. Whether this use occurred during or slightly after establishment cannot be determined.
Previous work suggested that both the establishment and maintenance of the stn1-M1 mutant required the RAD52 gene (38). However, here, we showed that RAD50 and RAD51, genes required for type II and type I RTE in S. cerevisiae, respectively, were not absolutely required for cell viability, for the maintenance of very long telomeres, or for t-circle formation in stn1-M1 cells. Both genes likely do contribute to at least the establishment stage of the stn1-M1 mutant, as indicated by the especially poor growth of stn1-M1 rad50 rad51 triple mutants immediately after their formation. The viability of stn1-M1 rad50 rad51 mutants is perhaps not completely unexpected, as the formation of postsenescence survivors in K. lactis ter1-Δ mutants by type II RTE has recently been shown to also not be completely eliminated in a rad50 rad51 background (E. Basenko and M. J. McEachern, unpublished data). One possibility to help account for these observations may be that RAD59 is able to promote RTE even in the absence of RAD50 and RAD51. Absence of RAD59 has been shown to slow the growth of stn1-M1 mutants (38), suggesting that it is individually more important to the type IIR RTE of stn1-M1 cells than either RAD50 or RAD51. Recent work in S. cerevisiae has shown that telomerase deletion mutants of that yeast can remain viable even in the absence of RAD52 if they are generated in cells that already have long telomeres (28). It is conceivable that a similar process might also contribute to telomere maintenance in stn1-M1 rad50 rad51 mutants.
The formation of t-circles could also be envisioned to occur via multiple mechanisms, including pathways independent of RAD50 and RAD51. Extrachromosomal linear telomere fragments produced by telomeric deletions might become circularized through NHEJ or annealing of 3′ overhangs. More than one pathway for t-circle formation appears to exist in human cells. XRCC3 is required for t-circle formation in a TRFΔB mutant of human fibroblasts (85) but is not required for the t-circles formed in a TRFΔB mutant in Werner syndrome cells (45).
The effect of telomerase on stn1-M1 cells and URA3 telomere tandem arrays.
The initial characterization of stn1-M1 showed that telomerase is active in mutant cells but that its presence did not grossly affect growth or telomere phenotypes (38). However, here, we found evidence suggesting that the presence of telomerase does have some effects on stn1-M1 mutant cells. First, stn1-M1 TER1 circle N transformants were more likely to acquire longer telomeres with higher average copy numbers of URA3 from the integrated circle N than stn1-M1 ter1-Δ transformants. Second, even when comparing clones with a similar initial URA3 copy number, stn1-M1 TER1 transformants were less likely to exhibit net loss of URA3 telomere units during the following serial restreaking than were stn1-M1 ter1-Δ transformants. Other data argue that the presence of telomerase influences stn1-M1 cells even in the absence of URA3 telomere arrays. Both the average amounts of telomeric DNA and variation in the amount of telomeric signal during serial passaging are significantly higher in stn1-M1 TER1 strains than in stn1-M1 ter1-Δ strains. Past work also demonstrated that stn1-M1 spores produced through meiosis have poor viability (38). Through random spore analysis of a TER1/ter1 STN1/stn1-M1 heterozygote, we have shown that stn1-M1 ter1-Δ spores are recovered significantly less often than stn1-M1 TER1 spores (data not shown). This result indicates that the absence of telomerase further exacerbates stn1-M1 spore viability problems. Our work here also showed more bands likely to be telomere-telomere fusions in stn1-M1 ter1-Δ cells than in stn1-M1 TER1 cells.
Exactly how telomerase alters the behavior of stn1-M1 cells is not clear. Conceivably, it could provide some protection against degradation and recombination by simply binding to telomeric ends. Some data in S. cerevisiae suggest that telomerase plays a role in protecting telomeres whose capping structures are already defective (84). Perhaps more likely is that telomerase affects stn1-M1 cells through its ability to add new repeats to telomere ends. Telomerase is known to be active in stn1-M1 mutants (38). The sizable increase in the average amount of telomeric DNA in stn1-M1 cells relative to stn1-M1 ter1-Δ cells (∼6 times the amount in normal cells) suggests that its level of activity is greater than that seen in wild-type K. lactis cells. This would not be surprising, as examples of type IIR RTE in K. lactis that are caused by mutant telomeric repeats have been shown to deregulate sequence addition by both telomerase and recombination (4). Telomerase might be especially useful for lengthening of very short telomeric-repeat tracts. These could come from nearly complete deletions of telomeres or, in cells with URA3 telomere arrays, from breaks that occur in the short telomeric-repeat blocks within the arrays. Experiments with tagged telomeric repeats suggest that stn1-M1 cells undergo telomeric deletions to sizes shorter than wild type prior to the formation of long telomeres during establishment of the stn1-M1 state (J. Xu and M. J. McEachern, submitted for publication). This might help explain why telomerase appears to be helpful for viability of stn1-M1 spores. Very short telomeres are known to be relatively poor substrates for elongation by recombination (16). A modest improvement in telomere protection mediated directly or indirectly by telomerase might help reduce the likelihood of fusions between chromosome ends. It might also allow telomeres to be somewhat more stable in length, perhaps explaining the greater variability in telomere length we observed in stn-M1 cells than in stn1-M1 ter1-Δ cells.
Relationship of type IIR RTE to the telomere maintenance of ALT cancers.
The type IIR RTE of stn1-M1 and certain other mutant yeast cells is postulated to differ from the type II RTE of K. lactis ter1-Δ mutants in the basic nature of the telomere recombination that takes place. With ter1-Δ mutants, recombination is induced at telomeres that drop below ∼100 to 150 bp in length (81) and is shut down once telomeres become elongated to above that size. In type IIR RTE, however, telomeric recombination is induced by defects in telomere components that apparently cannot be shut down by telomere lengthening. This runaway telomere elongation is apparently balanced by sequence loss from extremely frequent deletion events, such as those characterized in this work.
The ALT that occurs in some human cancers shares a number of characteristics with the type IIR RTE of stn1-M1 cells that are not present in the type II RTE of K. lactis ter1-Δ cells. They include extreme telomere lengths; abundant extrachromosomal telomeric DNA, including t-circles; continuous telomere instability; absence of gross fluctuations in growth rates (senescence and survivor formation); and resistance to the presence of telomerase (9, 12, 85, 87). These similarities, combined with an apparent inability of ALT to arise from the mere absence of telomerase, may suggest that the ALT phenotype requires alterations in telomere components to increase the susceptibility of telomeres to homologous recombination. Evidence suggests that telomeres of ALT cells, like those of stn1-M1 cells, are subject to high rates of recombination on a continuous basis (47, 66, 74, 85). In one study, for example, multiple ALT strains, including some expressing telomerase, had telomeric exchanges detectable by chromosome orientation-fluorescence in situ hybridization (CO-FISH) in most metaphases (47). Other evidence is consistent with large telomeric deletions also being frequent in ALT cells and other mammalian cells with dysfunctional telomeres (47, 74, 85).
It is almost certain that telomere defects that allow type IIR RTE in yeast and human ALT cells do not represent complete losses of telomere function. If they did, such cells should experience high rates of telomere-telomere fusions from nonhomologous end joining. This raises the question of whether the high levels of telomeric recombination in the stn1-M1 mutant represent the maximum possible level or whether more extreme, possibly even lethal, levels might be possible with other telomere defects. Evidence from K. lactis telomerase deletion mutants with altered telomeric-repeat sequences indicates that type IIR RTE can occur with different levels of severity (4, 81). With ALT cancers, it might be predicted that selection would favor telomere defects that permit enough telomeric recombination to maintain telomeres but not so much that cell growth is overly compromised.
Our results here also suggest that copying of t-circles is likely to be at least part of the mechanism by which ALT cells maintain elongated telomeres. Consistent with this idea, depletion of NBS1, a component of the mammalian MRN complex, has been shown to reduce or eliminate t-circle production and lead to shortened telomeres in ALT cells, but not in telomerase-positive cells (21, 90). These results are consistent with t-circles playing an important, though perhaps not essential, role in telomere maintenance from ALT. We would also predict from our results that the relative extent to which sequence derived from a single t-circle can become amplified will be less in ALT cells than in K. lactis. The much greater number of telomeres in humans than in K. lactis would limit the spread of sequence from one telomere to a most other telomeres. Further studies are needed to better understand the mechanistic details of telomere maintenance in ALT cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jessica Kissinger and Wenyuan Xiao for technical support for the pulsed-field gel electrophoresis experiments. We also thank Evelina Basenko and Elizabeth Lucht for critical reading of the manuscript.
This work was supported by a grant to M.J.M. from the National Institutes of Health (GM 61645).
Footnotes
Published ahead of print 29 May 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Bailey SM, Brenneman MA, Goodwin EH. 2004. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 32:3743–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Basenko E, Topcu Z, McEachern MJ. 2011. Recombination can either help maintain very short telomeres or generate longer telomeres in yeast cells with weak telomerase activity. Eukaryot. Cell 10:1131–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Basenko EY, Cesare AJ, Iyer S, Griffith JD, McEachern MJ. 2010. Telomeric circles are abundant in the stn1-M1 mutant that maintains its telomeres through recombination. Nucleic Acids Res. 38:182–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bechard LH, Butuner BD, Peterson GJ, Topcu Z, McEachern MJ. 2009. Mutant telomeric repeats in yeast can disrupt the negative regulation of recombination-mediated telomere maintenance and create an alternative lengthening of telomeres-like phenotype. Mol. Cell. Biol. 29:626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bechard LH, Jamieson N, McEachern MJ. 2011. Recombination can cause telomere elongations as well as truncations deep within telomeres in wild-type Kluyveromyces lactis cells. Eukaryot. Cell 10:226–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bechter OE, Zou Y, Walker W, Wright WE, Shay JW. 2004. Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res. 64:3444–3451. [DOI] [PubMed] [Google Scholar]
- 7. Becker DM, Lundblad V. 1997. Introduction of DNA into yeast cells, p 13.7.5–13.7.7, Curr. Protoc. Mol. Biol., 4th ed, vol III John Wiley & Sons, Inc. Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 8. Beverley SM. 1988. Characterization of the ‘unusual’ mobility of large circular DNAs in pulsed field-gradient electrophoresis. Nucleic Acids Res. 16:925–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3:1271–1274 [DOI] [PubMed] [Google Scholar]
- 10. Carter SD, Iyer S, Xu J, McEachern MJ, Astrom SU. 2007. The role of nonhomologous end-joining components in telomere metabolism in Kluyveromyces lactis. Genetics 175:1035–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cerone MA, Autexier C, Londono-Vallejo JA, Bacchetti S. 2005. A human cell line that maintains telomeres in the absence of telomerase and of key markers of ALT. Oncogene 24:7893–7901 [DOI] [PubMed] [Google Scholar]
- 12. Cesare AJ, Griffith JD. 2004. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 24:9948–9957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cesare AJ, Groff-Vindman C, Compton S, McEachern MJ, Griffith JD. 2008. Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain. Mol. Cell. Biol. 28:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cesare AJ, Reddel RR. 2010. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 11:319–330 [DOI] [PubMed] [Google Scholar]
- 15. Cesare AJ, Reddel RR. 2008. Telomere uncapping and alternative lengthening of telomeres. Mech. Ageing Dev. 129:99–108 [DOI] [PubMed] [Google Scholar]
- 16. Chang M, Dittmar JC, Rothstein R. 2011. Long telomeres are preferentially extended during recombination-mediated telomere maintenance. Nat. Struct. Mol. Biol. 18:451–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen Q, Ijpma A, Greider CW. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen XJ. 1996. Low- and high-copy-number shuttle vectors for replication in the budding yeast Kluyveromyces lactis. Gene 172:131–136 [DOI] [PubMed] [Google Scholar]
- 19. Church GM, Gilbert W. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. U. S. A. 81:1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cohen H, Sinclair DA. 2001. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl. Acad. Sci. U. S. A. 98:3174–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Compton SA, Choi JH, Cesare AJ, Ozgur S, Griffith JD. 2007. Xrcc3 and Nbs1 are required for the production of extrachromosomal telomeric circles in human alternative lengthening of telomere cells. Cancer Res. 67:1513–1519 [DOI] [PubMed] [Google Scholar]
- 22. Dunham MA, Neumann AA, Fasching CL, Reddel RR. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26:447–450 [DOI] [PubMed] [Google Scholar]
- 23. Fairhead C, Dujon B. 2006. Structure of Kluyveromyces lactis subtelomeres: duplications and gene content. FEMS Yeast Res. 6:428–441 [DOI] [PubMed] [Google Scholar]
- 24. Fasching CL, Bower K, Reddel RR. 2005. Telomerase-independent telomere length maintenance in the absence of alternative lengthening of telomeres-associated promyelocytic leukemia bodies. Cancer Res. 65:2722–2729 [DOI] [PubMed] [Google Scholar]
- 25. Fire A, Xu SQ. 1995. Rolling replication of short DNA circles. Proc. Natl. Acad. Sci. U. S. A. 92:4641–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. 2007. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 14:208–214 [DOI] [PubMed] [Google Scholar]
- 27. Grandin N, Charbonneau M. 2003. The Rad51 pathway of telomerase-independent maintenance of telomeres can amplify TG1-3 sequences in yku and cdc13 mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 23:3721–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grandin N, Charbonneau M. 2009. Telomerase- and Rad52-independent immortalization of budding yeast by an inherited-long-telomere pathway of telomeric repeat amplification. Mol. Cell. Biol. 29:965–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grandin N, Damon C, Charbonneau M. 2000. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol. Cell. Biol. 20:8397–8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grandin N, Damon C, Charbonneau M. 2001. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J. 20:6127–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grandin N, Damon C, Charbonneau M. 2001. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20:1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grandin N, Reed SI, Charbonneau M. 1997. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 11:512–527 [DOI] [PubMed] [Google Scholar]
- 33. Groff-Vindman C, Cesare AJ, Natarajan S, Griffith JD, McEachern MJ. 2005. Recombination at long mutant telomeres produces tiny single- and double-stranded telomeric circles. Mol. Cell. Biol. 25:4406–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hartig JS, Kool ET. 2004. Small circular DNAs for synthesis of the human telomere repeat: varied sizes, structures and telomere-encoding activities. Nucleic Acids Res. 32:e152 doi:10.1093/nar/gnh149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Henson JD, et al. 2009. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 27:1181–1185 [DOI] [PubMed] [Google Scholar]
- 36. Henson JD, Neumann AA, Yeager TR, Reddel RR. 2002. Alternative lengthening of telomeres in mammalian cells. Oncogene 21:598–610. [DOI] [PubMed] [Google Scholar]
- 37. Hoffman CS. 1997. Preparation of yeast DNA, p 13.11.1–13.11.4, Curr. Protoc. Mol. Biol. John Wiley & Sons, Inc., Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 38. Iyer S, Chadha AD, McEachern MJ. 2005. A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol. Cell. Biol. 25:8064–8073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jeyapalan JN, Mendez-Bermudez A, Zaffaroni N, Dubrova YE, Royle NJ. 2008. Evidence for alternative lengthening of telomeres in liposarcomas in the absence of ALT-associated PML bodies. Int. J. Cancer 122:2414–2421 [DOI] [PubMed] [Google Scholar]
- 40. Jiang WQ, et al. 2005. Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol. Cell. Biol. 25:2708–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jiang WQ, Zhong ZH, Henson JD, Reddel RR. 2007. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene 26:4635–4647 [DOI] [PubMed] [Google Scholar]
- 42. Kegel A, Martinez P, Carter SD, Astrom SU. 2006. Genome wide distribution of illegitimate recombination events in Kluyveromyces lactis. Nucleic Acids Res. 34:1633–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim NW, et al. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266:2011–2015 [DOI] [PubMed] [Google Scholar]
- 44. Le S, Moore JK, Haber JE, Greider CW. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li B, Jog SP, Reddy S, Comai L. 2008. WRN controls formation of extrachromosomal telomeric circles and is required for TRF2DeltaB-mediated telomere shortening. Mol. Cell. Biol. 28:1892–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lindstrom UM, et al. 2002. Artificial human telomeres from DNA nanocircle templates. Proc. Natl. Acad. Sci. U. S. A. 99:15953–15958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Londono-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S, Reddel RR. 2004. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 64:2324–2327 [DOI] [PubMed] [Google Scholar]
- 48. Lundblad V, Blackburn EH. 1993. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell 73:347–360 [DOI] [PubMed] [Google Scholar]
- 49. Lustig AJ. 2003. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat. Rev. Genet. 4:916–923 [DOI] [PubMed] [Google Scholar]
- 50. McEachern MJ, Blackburn EH. 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10:1822–1834 [DOI] [PubMed] [Google Scholar]
- 51. McEachern MJ, Blackburn EH. 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376:403–409 [DOI] [PubMed] [Google Scholar]
- 52. McEachern MJ, Haber JE. 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 75:111–135 [DOI] [PubMed] [Google Scholar]
- 53. McEachern MJ, Iyer S. 2001. Short telomeres in yeast are highly recombinogenic. Mol. Cell 7:695–704 [DOI] [PubMed] [Google Scholar]
- 54. Miller KM, Cooper JP. 2003. The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Mol. Cell 11:303–313 [DOI] [PubMed] [Google Scholar]
- 55. Morin GB, Cech TR. 1986. The telomeres of the linear mitochondrial DNA of Tetrahymena thermophila consist of 53 bp tandem repeats. Cell 46:873–883 [DOI] [PubMed] [Google Scholar]
- 56. Murnane JP, Sabatier L, Marder BA, Morgan WF. 1994. Telomere dynamics in an immortal human cell line. EMBO J. 13:4953–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nabetani A, Ishikawa F. 2009. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol. Cell. Biol. 29:703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nakayama K, Kusano K, Irino N, Nakayama H. 1994. Thymine starvation-induced structural changes in Escherichia coli DNA. Detection by pulsed field gel electrophoresis and evidence for involvement of homologous recombination. J. Mol. Biol. 243:611–620 [DOI] [PubMed] [Google Scholar]
- 59. Natarajan S, Groff-Vindman C, McEachern MJ. 2003. Factors influencing the recombinational expansion and spread of telomeric tandem arrays in Kluyveromyces lactis. Eukaryot. Cell 2:1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Natarajan S, McEachern MJ. 2002. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 22:4512–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nickles K, McEachern MJ. 2004. Characterization of Kluyveromyces lactis subtelomeric sequences including a distal element with strong purine/ pyrimidine strand bias. Yeast 21:813–830 [DOI] [PubMed] [Google Scholar]
- 62. Nishitani H, Nurse P. 1995. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell 83:397–405 [DOI] [PubMed] [Google Scholar]
- 63. Nosek J, Rycovska A, Makhov AM, Griffith JD, Tomaska L. 2005. Amplification of telomeric arrays via rolling-circle mechanism. J. Biol. Chem. 280:10840–10845 [DOI] [PubMed] [Google Scholar]
- 64. Nugent CI, et al. 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8:657–660 [DOI] [PubMed] [Google Scholar]
- 65. Nugent CI, Hughes TR, Lue NF, Lundblad V. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274:249–252 [DOI] [PubMed] [Google Scholar]
- 66. Perrem K, Colgin LM, Neumann AA, Yeager TR, Reddel RR. 2001. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol. Cell. Biol. 21:3862–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Petit MA, Mesas JM, Noirot P, Morel-Deville F, Ehrlich SD. 1992. Induction of DNA amplification in the Bacillus subtilis chromosome. EMBO J. 11:1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Philippsen P, Stotz A, Scherf C. 1991. DNA of Saccharomyces cerevisiae. Methods Enzymol. 194:169–182 [DOI] [PubMed] [Google Scholar]
- 69. Potts PR, Yu H. 2007. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 14:581–590 [DOI] [PubMed] [Google Scholar]
- 70. Price CM, et al. 2010. Evolution of CST function in telomere maintenance. Cell Cycle 9:3157–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D. 2008. Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 27:2328–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rogan EM, et al. 1995. Alterations in p53 and p16INK4 expression and telomere length during spontaneous immortalization of Li-Fraumeni syndrome fibroblasts. Mol. Cell. Biol. 15:4745–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Roy J, Fulton TB, Blackburn EH. 1998. Specific telomerase RNA residues distant from the template are essential for telomerase function. Genes Dev. 12:3286–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sabatier L, Ricoul M, Pottier G, Murnane JP. 2005. The loss of a single telomere can result in instability of multiple chromosomes in a human tumor cell line. Mol. Cancer Res. 3:139–150. [DOI] [PubMed] [Google Scholar]
- 75. Sambrook J, Russell DW. 2001. Preparation of DNA for pulsed-field gel electrophoresis: isolation of intact DNA from yeast, p 5.65–5.67 In Sambrook J, Russell DW. (ed), Molecular cloning: a laboratory manual, 3rd ed, vol 1 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 76. Stewart SA, Weinberg RA. 2006. Telomeres: cancer to human aging. Annu. Rev. Cell Dev. Biol. 22:531–557 [DOI] [PubMed] [Google Scholar]
- 77. Sun J, et al. 2009. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 23:2900–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Takahashi K, et al. 2011. Fission yeast Pot1 and RecQ helicase are required for efficient chromosome segregation. Mol. Cell. Biol. 31:495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Teng SC, Zakian VA. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083–8093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tomaska L, Nosek J, Makhov AM, Pastorakova A, Griffith JD. 2000. Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance. Nucleic Acids Res. 28:4479–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Topcu Z, Nickles K, Davis C, McEachern MJ. 2005. Abrupt disruption of capping and a single source for recombinationally elongated telomeres in Kluyveromyces lactis. Proc. Natl. Acad. Sci. U. S. A. 102:3348–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tsai YL, Tseng SF, Chang SH, Lin CC, Teng SC. 2002. Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination. Mol. Cell. Biol. 22:5679–5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. van Steensel B, Smogorzewska A, de Lange T. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401–413 [DOI] [PubMed] [Google Scholar]
- 84. Vega LR, et al. 2007. Sensitivity of yeast strains with long G-tails to levels of telomere-bound telomerase. PLoS Genet. 3:e105 doi:10.1371/journal.pgen.0030105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang RC, Smogorzewska A, de Lange T. 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119:355–368 [DOI] [PubMed] [Google Scholar]
- 86. Wray LV, Jr, Witte MM, Dickson RC, Riley MI. 1987. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yeager TR, et al. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59:4175–4179 [PubMed] [Google Scholar]
- 88. Zellinger B, Akimcheva S, Puizina J, Schirato M, Riha K. 2007. Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol. Cell 27:163–169 [DOI] [PubMed] [Google Scholar]
- 89. Zeng S, et al. 2009. Telomere recombination requires the MUS81 endonuclease. Nat. Cell Biol. 11:616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhong ZH, et al. 2007. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J. Biol. Chem. 282:29314–29322 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.