Abstract
Chromosome segregation is powered by the kinetochore, a large macromolecular structure assembled on centromeric chromatin. Attachment of sister chromatids to microtubules is mediated by the highly conserved tripartite KMN (acronym for KNL-1–Mis12–Ndc80) kinetochore network. In the fission yeast Schizosaccharomyces pombe, the equivalent complex is called NMS (Ndc80-MIND-Spc7). Here, we show that not all components of the NMS complex had been identified previously. A 10th NMS component exists, the essential Sos7 protein, which is a genetic and physical interaction partner of Spc7. The analysis of sos7 kinetochore-null mutant yeast strains demonstrated that Sos7 is central to NMS function. In particular, Sos7 is required for kinetochore targeting of Spc7 as well as components of the MIND complex. sos7 mutant strains show severe chromosome missegregation phenotypes and have compromised microtubule-kinetochore interactions. Sos7 is the founding member of a functionally conserved fungal kinetochore family not present in the point centromere carrying Saccharomycotina clusters, suggesting that the new Sos7 family might be a signature motif of fungi with regional centromeres.
INTRODUCTION
The faithful transmission of eukaryotic chromosomes to the progeny is a complex process that requires coordinated protein interactions at the kinetochore-microtubule interface. The erosion of chromosome segregation fidelity leading to the loss/gain of chromosomes is of great medical relevance, as aneuploidy has been implicated to have a causative role in a number of diseases (1, 38, 68). The proper partitioning of sister chromatids in mitotis is mediated via direct contact of spindle microtubules with components of the outer kinetochore. Kinetochores are macromolecular structures assembled on the chromosomal centromere region. Apart from providing mechanical attachment sites, these organelles generate the spindle checkpoint signaling that ascertains proper sister chromatid biorientation before anaphase onset (44). Although kinetochores are very complex structures and the number of kinetochore proteins identified is still increasing, the direct interaction with spindle fibers is mediated largely by a highly conserved protein network called the KMN complex in higher eucaryotes and the NMS complex in the fission yeast Schizosaccharomyces pombe (2, 37, 47). The KMN-NMS network is composed of 3 subcomplexes: the Ndc80 complex and the MIND-Mis12 complex, each comprising 4 components, and a protein called KNL-1 in Caenorhabditis elegans, Blinkin-hSpc105 in vertebrates, Spc105 in Saccharomyces cerevisiae and Drosophila, and Spc7 in S. pombe (59, 67, 74). In addition, budding yeast Spc105 forms a subcomplex with the Kre28 protein, Blinkin binds metazoan Zwint via its C-terminal end, and KNL-1 interacts with KBP-5 (3, 29, 30, 46–48). The mitotic functions of Kre28 and KBP-5 have yet to be analyzed, whereas the Zwint protein is required for kinetochore residency of the mitotic checkpoint relevant RZZ (Rod-Zw10-Zwilch) complex (10, 65, 70). It has been suggested that Kre28 and Zwint are orthologs, although sequence similarities are very limited (48). In this work, we have identified the missing interaction partner of the S. pombe Spc7 protein.
Two synergistic microtubule-binding activities exist for the KMN-NMS network, one mediated by the Ndc80 complex via the Ndc80-Nuf2 homodimer and the other by the Spc7–Spc105–KNL-1–Blinkin family, which associates with microtubules via their N-terminal ends (2, 26, 48, 71, 73, 75). Kinetochore-microtubule interactions are regulated by Aurora B-mediated phosphorylation on several KMN-NMS targets, including the Spc7–Spc105–KNL-1–Blinkin family (4, 5, 32, 72). As the Spc7–Spc105–KNL-1–Blinkin family is required for kinetochore targeting of protein phosphatase 1 and spindle checkpoint components, it can be seen as a platform for proteins regulating and modulating the spindle checkpoint response (29, 30, 32, 35, 40, 48, 57).
Depletion of Spc7–Spc105–KNL-1–Blinkin proteins by RNA interference (RNAi) or expression of conditional lethal alleles demonstrates the essential function of this protein family in maintaining genome stability as spindle microtubule-kinetochore interactions are severely impaired or absent under such conditions (6, 27, 30, 46, 55) (22). The main phenotype of S. pombe mitotic cells expressing temperature-sensitive, kinetochore-null alleles of spc7 at the restrictive temperature is a failure to segregate the highly condensed chromatin, although spindle elongation had occurred (26). The constitutively kinetochore-localized Spc7 protein is needed for kinetochore targeting of MIND components and provides a link between the S. pombe NMS and the Sim4-Mal2-Mis6 kinetochore complexes while kinetochore linkage of the Ndc80 complex is independent of Spc7 (26). In this work, we have extended our analysis of Spc7 function and have isolated a new interaction partner, the Sos7 protein. This protein is an essential component of the NMS complex and the founding member of a new family of fungal kinetochore proteins.
MATERIALS AND METHODS
Strains and media.
All yeast strains used are shown in Table 1. Synthetic lethality of double mutant strains was scored by analyzing at least 12 tetrads of the relevant cross. S. pombe and Schizosaccharomyces japonicus strains were grown on MM minimal medium (42); the S. cerevisiae strain was grown according to the manufacturer's instructions (GAL4 two-hybrid system 3; BD Biosciences Clontech, Palo Alto, CA). The supplements used were as described in reference 26.
Table 1.
Yeast strains used in this study
| Name | Genotype | Source(s)a |
|---|---|---|
| UFY1596 | h− sos7-GFP/Kanr ade6-M210 leu1-32 his3Δ ura4-D18 | This study |
| UFY2154 | h− sos7-GFP/Kanr sad1-mCherry/Kanr leu1-32 ura4-D18 | This study |
| UFY2240 | h− sos7-GFP/Kanr mis6-mCherry/Kanr leu1-32 his3-D1 ade6− | This study |
| UFY1914 | h+ sos7-178/his3+ cnt1(NcoI):arg3+ ade6-M210 ura4-D18 arg3-D4 | This study |
| UFY1916 | h− sos7-Δ7/his3+ cnt1(NcoI):arg3+ ade6-M210 arg3-D4 | This study |
| UFY1918 | h+ sos7-+6/his3+ cnt1(NcoI):arg3+ arg3-D4 | This study |
| UFY1836 | h+ sos7-+6/his3+ otr1L(dg1a/HindIII):ura4+ ade6-M210 leu1-32 ura4− | This study |
| UFY1838 | h− sos7-178/his3+ otr1L(dg1a/HindIII):ura4+ ade6-M210 leu1-32 ura4− | This study |
| UFY1864 | h+ sos7-Δ7/his3+ otr1L(dg1a/HindIII):ura4+ ade6-M210 leu1-32 ura4− | This study |
| UFY1943 | h− sos7-Δ7/his3+ his3-D1 ade6− leu1-32 ura4-D18 | This study |
| UFY1945 | h− sos7-+6/his3+ his3-D1 ade6− leu1-32 ura4-D18 | This study |
| UFY1947 | h− sos7-178/his3+ his3-D1 ade6− leu1-32 ura4-D18 | This study |
| UFY1866 | h+ sos7-Δ7/his3+ fta2-GFP/Kanr ade6− leu1-32 ura4− | This study |
| UFY1921 | h+ sos7-178/his3+ LacI-GFP/his7+ LacO-repeat/lys1+ leu1− ura4− ade6-M210 | This study |
| UFY1924 | h90 sos7-+6/his3+ LacI-GFP/his7+ LacO-repeat/lys1+ leu1− ura4− ade6-M210 | This study |
| UFY1701 | h90 sos7-GFP/Kanr spc7-23/his3+ leu1-32 his3-D1 ura4-D18 ade6− | This study |
| UFY1872 | h90 spc7-GFP/Kanr sos7-Δ7/his3+ ade6-M210 leu1-32 ura4− | This study |
| UFY1847 | h+ sos7-cMyc/Kanr ade6− leu1-32 his3-D1 ura4-D18 | This study |
| UFY1734 | h− sos7-GFP/Kanr nuf2-1/ura4+ ade6-M210 leu1-32 his3-D1 ura4-D18 | This study |
| UFY1746 | h− sos7-GFP/Kanr mis12-537 leu1− ura4− his3-D1 | This study |
| UFY1987 | h+ sos7-Δ7/his3+ mis12-GFP/LEU2 leu1-32 | This study |
| UFY2002 | h− sos7-178/his3+ nuf2-GFP/ura4+ ade6− leu1-32 ura4-D18 | This study |
| UFY1937 | h+ mis12-537 nuf2-GFP/ura4+ ura4− leu1− | This study |
| UFY1796 | h− nuf2-1/ura4+ mis12-GFP/LEU2 | This study |
| UFY1979 | h− sos7-178/his3+ sim4-GFP/Kanr ade6-M210 leu1-32 his3-D1 ura4− | This study |
| UFY1981 | h− sos7-Δ7/his3+ sim4-GFP/Kanr ade6-M210 leu1-32 his3-D1 ura4− | This study |
| UFY1983 | h− sos7-+6/his3+ sim4-GFP/Kanr ade6-M210 leu1-32 his3-D1 ura4− | This study |
| UFY1732 | h+ sos7-GFP/Kanr sim4-193 ade6-M210 leu1-32 his3-D1 ura4-D18 arg3-D4 | This study |
| UFY1736 | h− sos7-GFP/Kanr fta2-291/his3+ ade6-M210 leu1-32 his3− ura4-D18 | This study |
| UFY1738 | h+ sos7-GFP/Kanr mal2-1 ade6-M210 leu1-32 his3-D1 ura4-D18 | This study |
| UFY1740 | h− sos7-GFP/Kanr mis15-68 ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1742 | h− sos7-GFP/Kanr mis17-362 ade6-M210 leu1-32 his3-D1 ura4-D18 | This study |
| UFY1745 | h+ sos7-GFP/Kanr mis6-302 leu1− ura4− ade6− | This study |
| UFY1844 | h− sos7-178/his3+ fta2-GFP/Kanr ade6− leu1-32 ura4− | This study |
| UFY1028 | h+ spc7-23/his3+ his3-D1 ade6-M216 leu1-32 ura4-D18 | U. Fleig |
| UFY1029 | h+ spc7-24/his3+ his3-D1 ade6-M216 leu1-32 ura4-D18 | U. Fleig |
| UFY1027 | h+ spc7-30/his3+ his3-D1 ade6-M216 leu1-32 ura4-D18 | U. Fleig |
| UFY210 | h+ spc7-GFP/Kanr ade6-M210 ura4-D6 Ch16[ade6-M216] | U. Fleig |
| UFY617 | h− spc7-HA/Kanr ade6-M210 leu1-32 ura4-D6 Ch16[ade6-M216] | U. Fleig |
| UFY852 | h− mal2-1 leu1-32 ade6-M210 ura4-D18 | U. Fleig |
| UFY1048 | h− fta2-291/his3+ his3− leu1-32 ura4-D18 ade6-M210 | U. Fleig |
| KG425 | h− ade6-M210 leu1-32 his3Δ ura4-D18 | K. Gould |
| KG554 | h− ade6-M216 leu1-32 his3Δ ura4-D18 | K. Gould |
| h+ LacI-GFP/his7+ LacO-repeat/lys1+ leu1− ura4− | M. Yanagida | |
| h− mis12-537 leu1-32 | M. Yanagida | |
| h− mis6-302 leu1-32 | M. Yanagida | |
| h− mis15-68 | M. Yanagida | |
| h− mis17-362 | M. Yanagida | |
| FY648 | h+ swi6Δ::his1+ otr1R(SphI)::ura4+ ura4-DS/E leu1-32 ade6-M210 | R. Allshire |
| FY4540 | h− sim4-193 cnt1(NcoI):arg3+ cnt3(NcoI):ade6+ otr2(HindIII):ura4+ tel1L:his3+ ade6-M210 leu1-32 ura4-D18 arg3-D4 his3-D1 | R. Allshire |
| FY5231 | h+ sim4-193 arg3-D4 ade6-M210 his3-D1 ura4-D18 leu1-32 | R. Allshire |
| ANF251-9A | h+ nuf2-1/ura4+ ura4-D18 | Y. Hiraoka |
| MY6760 | h− sad1-mCherry/Kanr leu1 | YGRC; M. Yanagida |
| SK130 | h− mis6-mCherry/Kanr leu1-32::SV40-GFP-atb2+[LEU2] ade6-M216 bub1::bub1-ΔGLEBS | YGRC; S. Hauf |
YGRC, Yeast Genetic Resource Center.
Identification of sos7+, generation of sos7 temperature-sensitive (sos7ts) alleles, and DNA methods.
A saturating multicopy extragenic suppressor analysis of the spc7-23 mutant phenotype was carried out at 32.5°C as described previously (25). In this screen, 102 of 370,000 transformants were able to grow at the restrictive temperature. These suppressors were put into 6 complementation groups. The largest complementation group consisted of the spc7+ gene, which was identified 66 times. The second largest group consisted of an uncharacterized open reading frame (ORF) with the systematic name SPAPB17E12.06. This ORF was isolated 11 times in the screen. The other suppressors were isolated at least 2 times.
The sos7+ cDNA sequence was isolated from an S. pombe cDNA bank (43). The sos7-null allele (Δsos7) was generated by replacing one of the 1,079-bp sos7+ ORFs with his3+ in diploid strain KG425xKG554. Tetrad analysis of 75 heterozygous Δsos7/sos7+ diploids revealed that only the 2 his− spores/tetrad grew. sos7ts variants replaced the endogenous sos7+ in strain KG425: transformants unable to grow at 36°C were backcrossed twice and sequenced as described previously (25). sos7 and spc7 plasmid-expressed variants were generated via PCR and cloning of the resulting DNA fragments into pJR2 vectors with the nmt1+ promoter (41). mis12+, mis13+, nnf1+, nuf2+, and spc25+ ORFs were obtained via PCR and cloned into pJR2 vectors. sos7 and spc7 variants were expressed in S. cerevisiae using GAL4 two-hybrid system 3 vectors (BD Biosciences Clontech, Palo Alto, CA).
Antibodies and immunoprecipitation.
Sos7 and Spc7 specific rabbit polyclonal antibodies were generated by using the entire 6×His-tagged peptide of Sos7 or the C-terminal 6×His-tagged peptide (amino acids [aa] 821 to 1364) of Spc7 (Eurogentec, Belgium). The dilution for Western blot analysis was 1:200. Protein extracts were prepared as described previously (27). Immunoprecipitations were carried out using the μMACS cMyc-, green fluorescent protein (GFP)-, or hemagglutinin (HA)-tagged-protein isolation kits (Miltenyi Biotec, GmbH, Bergisch Gladbach, Germany). For Western blots, we used the following first antibodies: anti-HA antibody (monoclonal mouse; Roche Diagnostics, Alameda, CA), anti-cMyc antibody (monoclonal mouse; Roche Diagnostics), anti-GFP antibody (polyclonal rabbit; Invitrogen), γ-tubulin antibody (monoclonal mouse GTU-88; Sigma-Aldrich, St. Louis, MO), and our anti-Sos7 antibody and anti-Spc7 antibody. The secondary antibodies used were anti-rabbit IgI (Fc)–alkaline phosphatase conjugate (Promega, Madison, WI) or anti-mouse IgG(H+L)–alkaline phosphatase conjugate (Promega), which were detected as described previously (53). Sos7-GFP bands on Western blots were quantified using ImageJ 1.44 (National Institutes of Health) and normalization to the γ-tubulin loading control. Chromatin immunoprecipitation (ChIP) was carried out as described previously (27). The quantification of the centromere-specific PCR products was done as described previously (52), using Scion Image software (NIH).
Microscopy.
Images of fixed cells were made with a Zeiss Axiovert 200 fluorescence microscope (Carl Zeiss) with a charge-coupled-device (CCD) camera (Orca-ER; Hamamatsu, Herrsching, Germany) and Openlab imaging software (PerkinElmer) or with a Zeiss spinning-disc confocal microscope with an AxioCam MRm camera and AxioVision software. Image editing was done with Adobe Photoshop CS2 (Adobe Systems Software Ireland Limited, Dublin, Ireland). Immunofluorescence microscopy was carried out as described previously (20, 61). For immunofluorescence, we used the following first antibodies: for tubulin, monoclonal antitubulin antibody TAT1, and for GFP fusion proteins, rabbit anti-GFP antibody (Invitrogen, Carlsbad, CA) and anti-Sos7 and anti-Spc7 antibodies. The last two antibodies were used at a dilution of 1:5 and were affinity purified with NHS-activated Sepharose 4 Fast Flow (GE Healthcare, NJ). The secondary antibodies used were fluorescein isothiocyanate-conjugated goat anti-mouse antibodies and Cy3-conjugated sheep anti-rabbit antibodies (Sigma-Aldrich) (see Fig. 1C and D, 2A, 3C and D, and 6B and C; see also Fig. S3A, S3B, and S4B in the supplemental material) and Alexa Fluor 488–goat anti-mouse IgG(H+L) and Alexa Fluor 594–goat anti-rabbit IgG(H+L) (Invitrogen) (see Fig. 3F and G; see also Fig. S4A and S4D). DNA was stained with 4,6-diamidino-2-phenylindole (DAPI).
Fig 1.
Extra sos7+ suppresses the phenotypes of spc7 mutant strains. (A) Serial dilution patch tests (104 to 101 cells) of the indicated spc7ts transformants grown under plasmid-selective conditions at the indicated temperatures for 5 to 6 days. “Vector” indicates plasmid without an insert; sos7+ denotes the presence of this ORF plus its wild-type promoter on plasmid pUR19. The right panel shows amino acid changes present in the Spc7 variants. (B) Overexpression of sos7+ suppresses the chromosome missegregation and spindle defects of the spc7-23 strain grown at the restrictive temperature. Diagrammatic representation of the chromatin distribution (left) and spindle morphology (right) of mitotic spc7-23 cells (n = 100). wt, wild-type phenotype. (C) (Left) A protein extract prepared from a strain expressing endogenous Spc7-GFP was used for immunoprecipitation using anti-GFP (α-GFP) antibody, followed by Western blotting with the same anti-GFP (middle panel) or the polyclonal anti-Spc7 antibody (right panel). (Right) Photomicrographs of fixed wild-type cells incubated with the anti-Spc7 antibody and DAPI. Bar, 5 μm. (D) Kinetochore localization of Spc7-23 at 34°C is dependent on sos7+ overexpression. Photomicrographs of fixed spc7-23 cells overexpressing sos7+ incubated with the anti-Spc7 antibody and DAPI. Bar, 5 μm.
Fig 2.
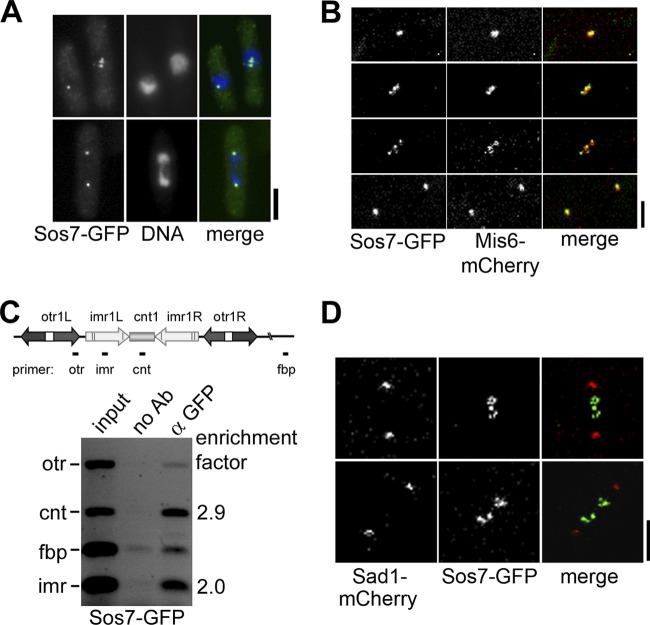
Sos7 is a new component of the kinetochore. (A) Localization of endogenous Sos7-GFP in fixed interphase, metaphase, and anaphase cells incubated with the anti-GFP antibody and DAPI. Bar, 5 μm. (B) Subcellular localization of Sos7-GFP in a Mis6-mCherry-expressing strain. Shown is a representative cell (n = 5) that goes through mitosis (time intervals from top to bottom: 13.5, 5, and 7.5 min). Bar, 2 μm. (C) Sos7 associates with the central domain of cen1. Cells expressing Sos7-GFP were fixed and processed for ChIP by using anti-GFP antibodies. Multiplex PCR analysis of the chromatin in immunoprecipitates shows an enrichment of the imr and cnt regions of cen1. (D) Live-cell images of 2 representative metaphase cells (n = 6) expressing Sad1-mCherry and Sos7-GFP. The top cell is seen in Movie S1 in the supplemental material.
Fig 3.
Sos7 mutant strains have severe mitotic defects. (A) Serial dilution patch tests (104 to 101 cells) of 3 sos7ts mutant strains. The indicated strains were grown at the various temperatures for 3 to 4 days. (B) Amino acid changes found in the Sos7 mutant proteins. (C) Kinetochore localization of Sos7-Δ7 at 25°C and after 6 h at 32°C. Fixed cells were stained with our anti-Sos7 antibody and the TAT-1 antitubulin antibody. Bar, 5 μm. (D) Photomicrographs of sos7ts cells incubated at 36°C for 6 h (a, sos7-+6; b and c, sos7-178; d and e, sos7-Δ7). Fixed cells were stained with antitubulin antibody and DAPI. Shown are all mutant phenotypes (unequally separated chromatin [cells a and b], segregated chromatin with lagging chromosomes [cell c], smeared chromatin [cell d], and nonseparated chromatin [cell e]) on an elongated spindle. (E) Quantification of the phenotypes described for panel D. The number of cells analyzed per strain and temperature was 100. (F) Photomicrographs of a mitotic sos7-Δ7 cell expressing endogenous Fta2-GFP. Cells were incubated at 36°C, fixed, and stained with antitubulin and anti-GFP antibodies. Bar, 2 μm. The number of mitotic Fta2-GFP signals analyzed was 327. (G) (Left) cen1-gfp sos7-178 cells incubated at 36°C for 6 h. Fixed cells were incubated with anti-GFP antibody, DAPI, and antitubulin antibody. Bar, 5 μm. (Right) Quantification of GFP signal distribution in wild-type and sos7ts strains at 25°C and 36°C. The number of cells analyzed per strain and temperature was 100.
Fig 6.
Sos7 is required for kinetochore targeting of MIND components. (A) Serial dilution patch tests (104 to 101 cells) of mis12-537 and nuf2-1 transformants grown under plasmid-selective conditions at the indicated temperatures for 5 days. sos7+, plasmid expressing sos7+. (B) Sos7-GFP localization in fixed mis12-537 and nuf2-1 cells incubated at the permissive (25°C) or restrictive (34°C) temperature. Diagrams above the photomicrographs show the positions of the GFP signal. Bar, 5 μm. (C) Kinetochore localization of Mis12-GFP (left panel) and Nuf2-GFP (right panel) in sos7ts strains. sos7ts cells expressing one of the above-mentioned tagged kinetochore proteins were grown at the indicated temperatures before fixation and stained with DAPI, anti-GFP, and TAT-1 anti-tubulin antibody. Bars, 5 μm. (D) Serial dilution patch tests of sos7-Δ7 cells transformed with plasmids expressing the indicated kinetochore proteins. Transformants were grown under plasmid-selective conditions at 25°C or 30°C for 5 to 6 days. (E) Summary of the interactions observed for the Ndc80-MIND-Spc7/Sos7 complex. An asterisk indicates that the sos7-Δ7 mutant phenotype is rescued only by extra Mis12 or Mis13, not by extra Nnf1 or Mis14.
For live-cell imaging, cells were pregrown in minimal medium at 30°C and slides were prepared by mounting cells on agarose pads as described previously (69). Images were obtained using a Zeiss spinning-disc confocal microscope equipped with an AxioCam MRm camera. The images shown are maximum-intensity projections of 15 z slices with a distance of 0.5 μm. The time interval between images was 30 s, and the exposure times were 600 ms for GFP (488 nm) and for mCherry (561 nm).
Signal intensity of GFP fluorescence in live-cell images was determined with ImageJ 1.44 (NIH). Signals were measured in a 10- by 10-pixel square, and the fluorescent background was subtracted. Fluorescent background was determined by measuring the fluorescence of a nearby nuclear area of the same size.
RESULTS
Identification of sos7+ as a suppressor of the spc7 temperature-sensitive phenotype.
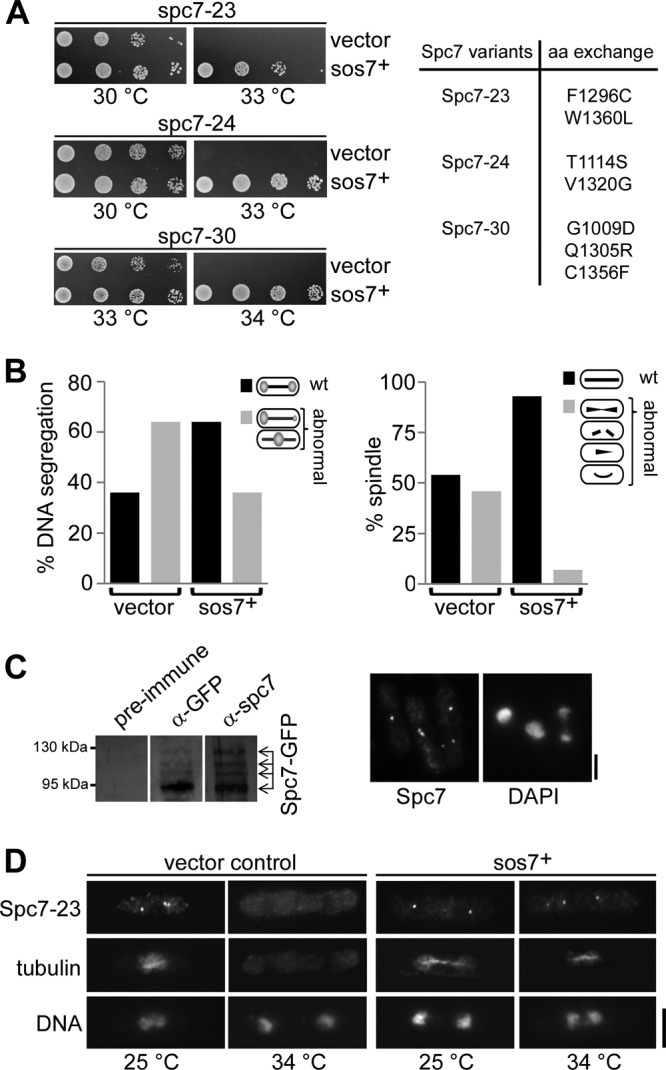
A saturating multicopy extragenic suppressor screen of the spc7-23 nongrowth phenotype at high temperatures identified an uncharacterized ORF with the systematic name SPAPB17E12.06. This ORF had been classified previously incorrectly as a pseudogene (S. pombe gene DB; Sanger Centre). The very recent reannotation of SPAPB17E12.06 is in accordance with our sequence analysis of this ORF isolated from an S. pombe cDNA library (56) (see Fig. S1 in the supplemental material). We named this ORF sos7+ (suppressor of spc7). The sos7+ ORF codes for 264 amino acids and thus for a 30.5-kDa protein, which is highly conserved (see below). Plasmid-borne overexpression of sos7+ rescued the temperature-sensitive phenotype of all spc7 mutant strains (Fig. 1A) by reducing the chromosome missegregation and spindle defects of such strains (Fig. 1B).
The temperature-sensitive Spc7-23 protein is present in the cell at the nonpermissive temperature but is not kinetochore associated (26). We therefore analyzed whether extra sos7+ rescued the temperature-sensitive phenotype of spc7 mutant strains by assisting kinetochore targeting of the mutant Spc7 protein. For this, an antibody against the C-terminal part of the Spc7 protein was generated (Fig. 1C). Immunofluorescence analysis revealed that overexpression of sos7+ resulted in kinetochore targeting of Spc7-23 at the nonpermissive temperature (Fig. 1D).
Sos7 is an essential constitutive component of the kinetochore.
The analysis of the subcellular localization of an endogenously expressed Sos7-GFP fusion protein identified Sos7 as a constitutive component of the S. pombe kinetochore. First, interphase cells showed a single fluorescent signal, while mitotic had 2 or more signals (Fig. 2A). Second, live-cell imaging of a Mis6-mCherry Sos7-GFP strain revealed colocalization of the two tagged proteins. Mis6 is part of the Sim4-Mal2-Mis6 kinetochore complex (Fig. 2B) (17, 37). Third, we determined association of Sos7-GFP with the centromere DNA using ChIP followed by multiplex PCR analysis with primers that amplified the otr, imr, and cnt centromere I regions and an unrelated euchromatic control region, fbp (50) (Fig. 2C, diagram). The Sos7 ChIP showed centromere enrichment of only the cnt and imr sequences, which were enriched 2.9- and 2-fold relative to the fbp DNA in the Sos7 ChIP and to the input control, respectively (Fig. 2C; see also Materials and Methods).
The C-terminal part of Sos7 shows homology to the S. cerevisiae spindle pole body (SPB) protein Bbp1 (35% identity in a 115-amino-acid-long region of Sos7) (49, 63). In fission yeast interphase and postmetaphase cells, the kinetochores are clustered at the SPB and thus the metaphase stage is used to discriminate between SPB and kinetochore localization. Using live-cell imaging, we determined Sos7-GFP localization in a strain expressing the SPB component Sad1-mCherry (19). Sos7-GFP did not colocalize with Sad1-mCherry in metaphase cells, indicating that Sos7 is not a component of the SPB (Fig. 2D; see also Movie S1 in the supplemental material).
As transcriptional silencing of centromeric chromatin is dependent on specific kinetochore proteins, we tested whether marker genes present within the centromere DNA were still repressed in sos7ts mutant strains (see below) (52). We found that Sos7 was not required for transcriptional silencing of the centromere region (see Fig. S2A in the supplemental material).
To determine if Sos7 was essential, we replaced one copy of the sos7+ gene with the his3+ marker in a diploid S. pombe strain where both endogenous his3+ alleles had been deleted. Tetrad analysis of 75 tetrads of this strain revealed that only the 2 his− spores in a tetrad grew (see Fig. S2B in the supplemental material).
Sos7 is essential for chromosome segregation and bipolar sister chromatid association.
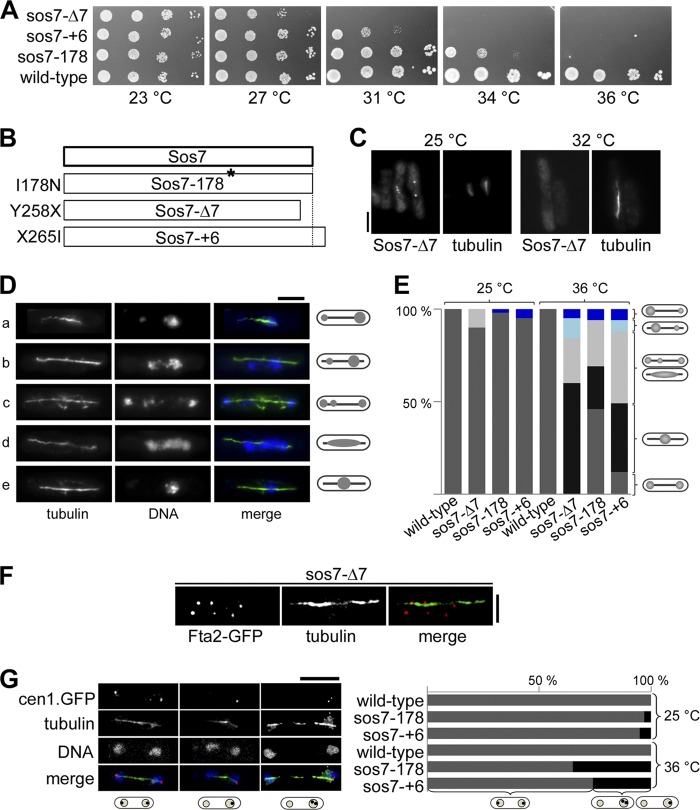
To elucidate the role of Sos7 at the kinetochore, we constructed sos7ts alleles. Three mutants were used for further analysis: those encoded by sos7-178, which carries a single base pair change that results in a single amino acid change at position 178 (isoleucine to asparagine) of the Sos7 protein; sos7-Δ7, which has a base pair substitution at position 1058 of the sos7 gene that results in a premature stop codon and a shorter Sos7 variant of 257 amino acids; and sos7-+6, where the presence of an extra A behind position 1076 leads to a frameshift mutation and a Sos7 variant with the 6 additional amino acids IMRRYR at the very C-terminal end (Fig. 3A and B; see also Fig. S1B in the supplemental material). The structure prediction program GORIV indicated that an extra α-helix might be present in the very C-terminal end of the Sos7-+6 protein (15).
The sos7 mutants grew similarly to the wild type at low temperatures, except that the sos7-Δ7 cells divided with an increased mean cell length of 25% (wild type, 13.6 μm; sos7-Δ7, 17.1 μm), indicating a cell cycle delay. Tentatively, we place this cell cycle delay at the G2-to-M transition, as 53% of the asynchronously growing sos7-Δ7 cells had duplicated SPBs, as shown by fluorescence signals of the SPB component Pcp1-GFP (8, 13).
Incubation of the sos7ts strains at temperatures above 31 to 34°C resulted in severe growth defects (Fig. 3A). To analyze whether the mutant Sos7 proteins were still kinetochore localized at the restrictive temperature, we generated and tested a polyclonal antibody against the entire Sos7 protein (see Fig. S3A in the supplemental material). Using this antibody, we found that the Sos7-Δ7 and Sos7-178 proteins were present at the kinetochore only at the permissive temperature while Sos7-+6 was also kinetochore localized at the restrictive temperature (Fig. 3C; see also Fig. S3B in the supplemental material). Thus, the mutant phenotypes of the sos7-Δ7 and sos7-178 strains can be explained by loss of kinetochore association.
Immunofluorescence analysis of sos7ts mutant cells revealed that at the permissive temperature, 5% of sos7-+6 and 2% of sos7-178 mitotic cells had unequal chromatin distributions, while 10% of sos7-Δ7 mitotic cells showed lagging chromosomes, possibly indicating merotelic attachment (Fig. 3E). Growth of these strains at the restrictive temperature led to a strong increase in the number of cells with chromosome segregation defects (Fig. 3D and E). The following abnormal phenotypes were scored for all three sos7 mutant strains: (i) unequal separation of the chromatin on an elongated spindle, (ii) unequally segregated chromatin with lagging chromosomes or chromatin smeared along the spindle, and (iii) nonseparated highly condensed chromatin on an elongated spindle. The sos7-178 strain showed the mildest mutant phenotype, as 46% of mitotic cells appeared to segregate their chromatin equally (Fig. 3E). In contrast, no wild-type mitosis was observed for sos7-Δ7 cells incubated at the restrictive temperature: 60% of such mitotic cells had an elongated spindle with no separation of the chromatin, 24% showed chromatin smeared along an anaphase spindle, and only 16% of the mitotic cells were able to segregate their chromatin, albeit unequally (Fig. 3E).
We analyzed whether microtubule-kinetochore interactions were compromised in sos7ts mutant strains by scoring the number of Fta2-GFP signals associated with an anaphase length-like spindle in sos7-Δ7 cells (Fig. 3F). Kinetochore targeting of the Sim4-Mal2-Mis6 complex component Fta2 is not affected by the presence of mutant Sos7 (see Fig. S4A in the supplemental material). Only 42.2% of Fta2-GFP signals were spindle associated (Fig. 3F). Thus, microtubule-kinetochore interactions were severely compromised in sos7 mutant cells (Fig. 3F). To assay bipolar chromosome orientation in sos7ts mutants, the subcellular localization of cen1-GFP was determined (45). At the permissive temperature, 3% of sos7-178 mitotic cells and 5% of sos7-+6 mitotic cells showed cosegregation of cen1-GFP sister centromeres, and these numbers increased to 35% and 24%, respectively, at the restrictive temperature (Fig. 3G).
Genetic and physical interactions of Sos7 and Spc7 place these proteins into an NMS subcomplex.
As kinetochore targeting of mutant Spc7 at the restrictive temperature was dependent on the presence of extra Sos7, we tested whether Sos7-GFP was localized correctly in an spc7-23 strain. The amounts of Sos7-GFP protein present in wild-type and spc7-23 cells were similar. However, in both strains, Sos7 levels were higher when cells were grown at 25°C than when they were grown at 33°C (Fig. 4A, left panel) (at 33°C, the Sos7 protein amount is approximately 60% of the amount present at 25°C). Live-cell imaging of Sos7-GFP in interphase cells incubated at 25°C revealed that kinetochore-localized Sos7-GFP was reduced in spc7-23 cells compared to the level in wild-type cells (Fig. 4A, middle and right panels) (approximately 50% of the wild-type signal). Incubation at the restrictive temperature for 6 h showed that kinetochore-localized Sos7-GFP was absent or severely reduced in spc7-23 interphase cells (Fig. 4A, middle and right panels) (approximately 13% of the wild-type signal). We conclude that kinetochore targeting of Sos7 depends on Spc7.
Fig 4.
Sos7 and Spc7 localization interdependencies. (A) (Left panel) Western blot analysis of Sos7-GFP immunoprecipitates from equal amounts of protein extracts from wild-type (wt) and spc7-23 cells grown at the indicated temperature. γ-Tubulin, loading control. (Middle panel) Live-cell images of Sos7-GFP in wt and spc7-23 cells grown at 25°C or incubated at 33°C for 6 h. (Right panel) Quantification of these fluorescence signals. The number of cells analyzed per strain and temperature was 30. (B) (Top) Serial dilution patch tests (104 to 101 cells) of the indicated strains grown at 18°C for 8 days. (Bottom) Live-cell images of Spc7-GFP in wt and sos7-Δ7 cells grown at 18°C. Bar, 5 μm. (Right panel) Quantification of these fluorescence signals. The number of cells analyzed per strain was 30. (C) Serial dilution patch tests (104 to 101 cells) of sos7ts cells transformed with plasmids overexpressing spc7+. Transformants were grown under plasmid-selective conditions at permissive (25°C or 30°C) or restrictive (28 to 34°C) temperatures for 5 to 6 days.
We next tested Spc7-GFP cellular localization in sos7-Δ7 cells by use of live-cell imaging. The spc7-gfp sos7-Δ7 strain is viable at 18°C but already shows a growth defect (Fig. 4B). A comparison of Spc7-GFP signals in wild-type and sos7-Δ7 cells analyzed at 18°C showed that the GFP signal in sos7-Δ7 cells was reduced strongly (Fig. 4B) (the signal in mutants cells is approximately 27% of the wild-type signal). At higher temperatures, no Spc7-GFP signal was detected in sos7-Δ7 cells.
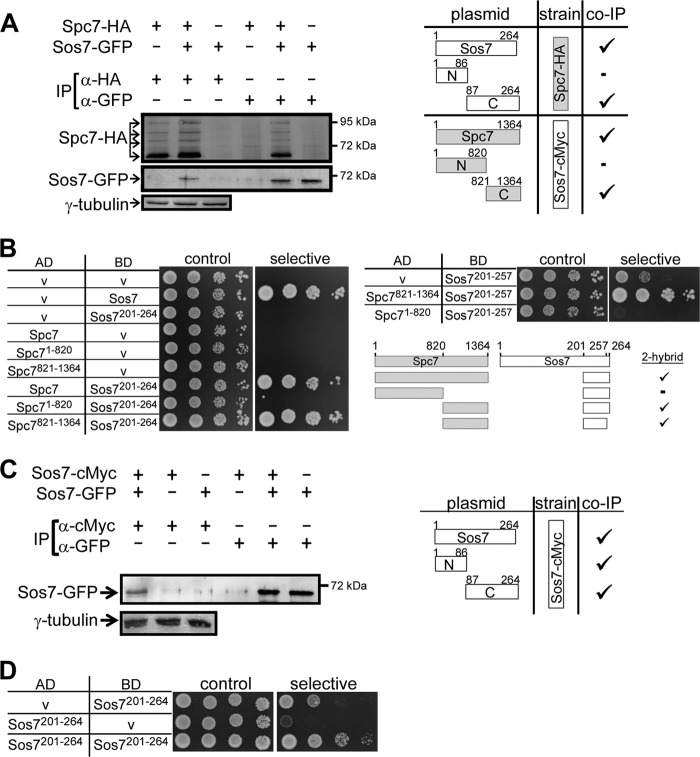
Our data suggest a tight interaction between Sos7 and Spc7, which is supported by the finding that overexpression of Spc7 rescued the temperature-sensitive nongrowth phenotype of all sos7ts mutants (Fig. 4C). To determine how these 2 proteins interacted with each other, we constructed strains where either sos7+ or spc7+ was tagged endogenously with cMyc or HA, respectively, and transformed these strains with plasmids that expressed gfp-tagged Sos7 or Spc7 variants. Coimmunoprecipitation followed by Western blot analysis revealed that Spc7 and Sos7 interacted with each other via their C-terminal ends (Fig. 5A; see also Fig. S5 in the supplemental material). In our coimmunoprecipitation assays, the endogenous wild-type nontagged version of the plasmid-expressed GFP-tagged protein is present. The endogenous protein will titrate some of the binding partners of the GFP-tagged version. We therefore tested the interaction between Sos7 and Spc7 also via a yeast 2-hybrid analysis: Sos7 and Spc7 interacted with each other, and this interaction between them occurred via the C-terminal 544 amino acids of Spc7 and a 57-amino-acid-long region in the C terminus of Sos7 (Fig. 5B).
Fig 5.
Interaction between Spc7 and Sos7 proteins. (A) Coimmunoprecipitation of tagged Spc7 and Sos7 proteins. Protein extracts of a Spc7-HA strain expressing plasmid-encoded Sos7-GFP variants or a vector control and of a Sos7-cMyc strain expressing plasmid-encoded Spc7-GFP variants or a vector control were used for immunoprecipitation (IP) using anti-HA and anti-GFP or anti-cMyc and anti-GFP antibodies, respectively. The immunoprecipitates were analyzed by Western blot analysis using the relevant antibodies. An example of such an immunoprecipitation is shown for the Spc7-HA strain transformed with a plasmid expressing full-length Sos7-GFP. γ-Tubulin, loading control. (Right panel) Summary of the interaction of various Spc7 and Sos7 variants.  , coimmunoprecipitation; −, no coimmunoprecipitation. Raw data are shown in Fig. S5 in the supplemental material. (B) Spc7 and Sos7 interact via their C-terminal ends. Yeast 2-hybrid analysis using the indicated fusion constructs. Transformants were grown on synthetic defined medium plates with (control) or without (selective) histidine and adenine for 3 days at 30°C. “AD” and “BD” denote the Gal4 activating and binding domains, respectively. (C) Coimmunoprecipitation of differently tagged Sos7 proteins. Protein extracts of a Sos7-cMyc strain expressing plasmid-encoded Sos7-GFP variants were used for immunoprecipitation using anti-cMyc antibodies. The immunoprecipitates were analyzed by Western blot analysis using an anti-GFP antibody. (Right panel) Summary of the interaction of Sos7 variants.
, coimmunoprecipitation; −, no coimmunoprecipitation. Raw data are shown in Fig. S5 in the supplemental material. (B) Spc7 and Sos7 interact via their C-terminal ends. Yeast 2-hybrid analysis using the indicated fusion constructs. Transformants were grown on synthetic defined medium plates with (control) or without (selective) histidine and adenine for 3 days at 30°C. “AD” and “BD” denote the Gal4 activating and binding domains, respectively. (C) Coimmunoprecipitation of differently tagged Sos7 proteins. Protein extracts of a Sos7-cMyc strain expressing plasmid-encoded Sos7-GFP variants were used for immunoprecipitation using anti-cMyc antibodies. The immunoprecipitates were analyzed by Western blot analysis using an anti-GFP antibody. (Right panel) Summary of the interaction of Sos7 variants.  , coimmunoprecipitation. Raw data are shown in Fig. S6 in the supplemental material. (D) Yeast 2-hybrid analysis showing Sos7201-264 self-interaction. Transformants were grown on SD plates with (control) or without (selective) histidine for 4 days. V, vector control; AD and BD, activating and binding domain plasmids, respectively.
, coimmunoprecipitation. Raw data are shown in Fig. S6 in the supplemental material. (D) Yeast 2-hybrid analysis showing Sos7201-264 self-interaction. Transformants were grown on SD plates with (control) or without (selective) histidine for 4 days. V, vector control; AD and BD, activating and binding domain plasmids, respectively.
We then tested whether Sos7 interacted with itself. An S. pombe strain expressing endogenous Sos7-cMyc was transformed with plasmids expressing full-length Sos7-GFP, Sos71-86–GFP (amino acids 1 to 86 of the Sos7 protein fused to GFP), or Sos787-264–GFP. Immunoprecipitation with an anti-cMyc antibody precipitated all three Sos7-GFP variants, albeit in small amounts (Fig. 5C; see also Fig. S6 in the supplemental material). The C-terminal interaction domain was mapped to a region of 64 amino acids with the help of the yeast 2-hybrid analysis (Fig. 5D). The N-terminal interaction domain was not mapped, as the Sos71-200 region shows autoactivation (unpublished observations).
Sos7 is required for kinetochore targeting of MIND but not of Ndc80 complex components.
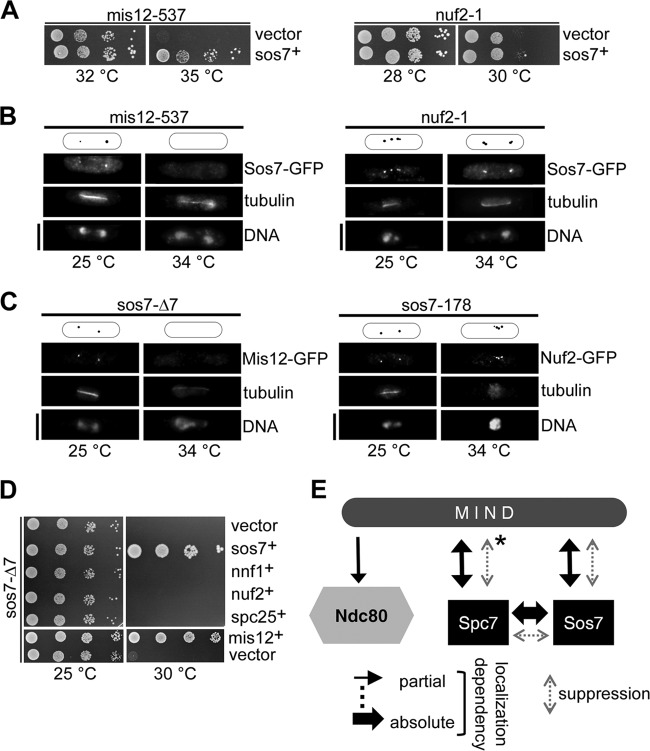
Spc7 is needed for kinetochore targeting of the MIND complex (and vice versa) but not of the Ndc80 complex (26). Given the tight interaction between Spc7 and Sos7, we next tested whether similar interdependencies existed between MIND components and Sos7. Indeed, plasmid-borne overexpression of sos7+ rescued the temperature-sensitive phenotype of the mutant MIND component mis12-537 but not that of the Ndc80 component nuf2-1 (Fig. 6A). In addition, kinetochore targeting of Sos7-GFP depended on a functional MIND complex, as the Sos7-GFP signal was reduced or absent in the mis12-537 strain grown at the permissive or restrictive temperature, respectively (Fig. 6B). Sos7-GFP kinetochore association was only slightly affected in the nuf2-1 strain, implying that a functional Ndc80 complex was not essential for correct Sos7 localization (Fig. 6B). The reverse scenario gave complementing results: the Mis12-GFP protein was severely reduced or absent from the kinetochore in sos7 mutant strains incubated at the restrictive temperature, as shown exemplarily for sos7-Δ7 (Fig. 6C). In contrast, Nuf2-GFP targeting was only slightly affected in the sos7 mutant strains (Fig. 6C). We next tested whether overexpression of components of the MIND and Ndc80 complexes could rescue the temperature-sensitive phenotype of the sos7 mutants. Overexpression of any of the components of the MIND complex, i.e., Mis12, Mis13, Mis14, or Nnf1, rescued the nongrowth phenotype of sos7-178 at the restrictive temperature. Extra Mis12 or Mis13 rescued sos7-Δ7 cells, while Mis14 and Nnf1 did not (Fig. 6D). Overexpression of the Ndc80 complex components Nuf2 and Spc25 did not suppress the temperature sensitivity of any sos7 mutants (Fig. 6D).
In summary, our data show that Sos7 and MIND components require each other for correct kinetochore targeting (Fig. 6E). Sos7, MIND components, and Spc7 can localize at the kinetochore independently of the Ndc80 complex, while kinetochore association of the Ndc80 component Nuf2 is influenced by MIND but is only moderately affected in a sos7ts strain (26, 58) (Fig. 6E; see also Fig. S4B in the supplemental material).
The Sim4-Mal2-Mis6 complex associates at the kinetochore independently of Sos7.
The 13-component Sim4-Mal2-Mis6 kinetochore complex was isolated separately from the Ndc80-MIND-Spc7 complex, although interactions exist between these two complexes (25–27, 37). We analyzed whether overexpression of sos7+ rescued the nongrowth phenotype of the mis6-302, mis15-68, mis17-362, sim4-193, fta2-291, and mal2-1 Sim4-Mal2-Mis6 complex mutants and found that this was not the case (unpublished observations) (12, 21, 25, 52, 66). Sos7-GFP was localized correctly in all of these Sim4-Mal2-Mis6 complex mutant strains grown at the restrictive temperatures, although kinetochore association was reduced moderately in the mal2-1 and fta2-291 strains (see Fig. S4C in the supplemental material). Furthermore, kinetochore targeting of Sim4-Mal2-Mis6 complex components was unaffected in sos7 mutants (see Fig. S4A and D). Thus, we conclude that Sos7 is not essential for kinetochore association of Sim4-Mal2-Mis6 complex components and vice versa.
Sos7 defines a highly conserved fungal kinetochore protein family.
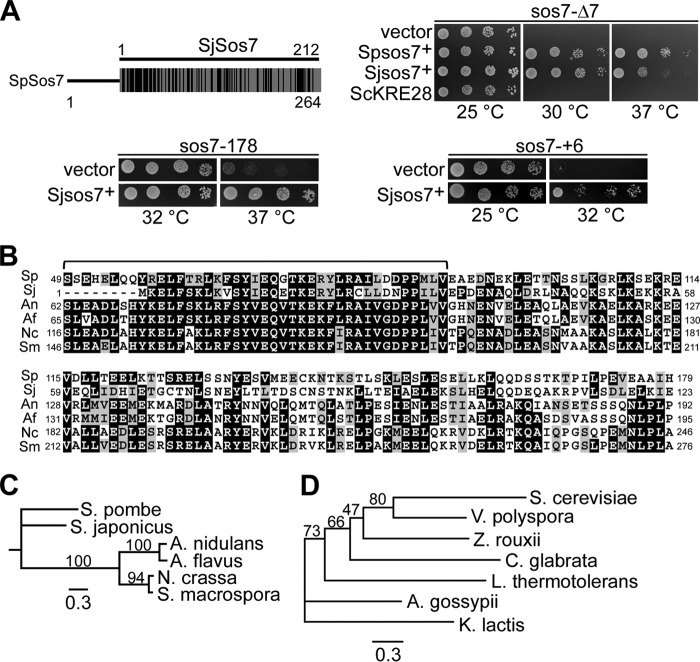
BLAST database searches revealed that Sos7 belongs to a highly conserved protein family present in fungi. The best match is a hypothetical protein encoded by the Schizosaccharomyces japonicus ORF SJAG_03327 (here called SjSos7), which has 36% identical and in total 60% identical and similar amino acids (Fig. 7A). To test whether these proteins were functional homologs, we analyzed whether expression of SjSos7 could rescue the temperature sensitivity of sos7 mutants. Plasmid-borne overexpression of this S. japonicus gene rescued the temperature-sensitive phenotype of all sos7ts mutant strains, although not as efficiently as S. pombe sos7+ (Fig. 7A).
Fig 7.
Sos7 belongs to a conserved protein family. (A) Diagrammatic representation of the comparison of Sos7 proteins from S. pombe and S. japonicus. Black bars indicate regions of identity and similarity. Clockwise from top right: serial dilution patch tests (104 to 101 cells) of sos7-Δ7 transformants expressing a vector control, S. pombe Sos7 (Spsos7+), S. japonicus Sos7 (Sjsos7+), or S. cerevisiae Kre28 (ScKRE28); sos7-+6 cells transformed with vector control or a plasmid with Sjsos7+; and sos7-178 cells transformed with vector control or a plasmid with Sjsos7+. (B) Amino acid sequence comparison of Sos7-like proteins from S. pombe, S. japonicus (XP_002174481), Aspergillus nidulans (CBF76747), Aspergillus flavus (XP_002383989), Neurospora crassa (CAE76380), and Sordaria macrospora (XP_003347116). Black boxes, ≥50% identity; gray boxes, ≥50% similarity (http://www.ch.embnet.org/software/BOX_form.html). A bracket indicates a highly conserved sequence motif. (C) Phylogenetic tree of Sos7 homologues shown in panel B. (D) Phylogenetic tree of Kre28 and V. polyspora homolog EDO18951, Z. rouxii CAR29059, A. gossypii AAS54100, C. glabrata XP_446867, L. thermotolerans CAR23808, and K. lactis XP_452234. The trees in panels C and D were engendered by PhyML 3.0 based on the maximum-likelihood method using the Jones Taylor Thornton substitution matrix (18). Bootstrap values (100 replicates) are shown. The scale bar indicates the length of the branch.
The Sos7 protein family is widely distributed among members of the phylum Ascomycota (Fig. 7B and C) and is also found in fungi belonging to the Basidiomycota, such as Cryptococcus neoformans. Sos7 family members have a highly conserved signature motif and smaller blocs of conserved sequence (Fig. 7B). However, Sos7-like proteins appear to be absent from one of the three Saccharomycotina clusters, namely, the “Saccharomyces complex” (33, 62).
Instead, these yeasts have a putative S. cerevisiae Kre28 homolog (Fig. 7D). The Kre28 kinetochore component is the interaction partner of Spc105, the Spc7 ortholog in S. cerevisiae (27, 46, 48). Sos7 and Kre28 show little sequence conservation, and plasmid-borne overexpression of Kre28 could not rescue sos7 mutant phenotypes (Fig. 7A). Furthermore, Kre28-GFP expressed from an S. pombe/S. cerevisiae shuttle vector was kinetochore targeted in S. cerevisiae but not in S. pombe wild-type or sos7 mutant strains (unpublished observations) (24). We conclude that although the Sos7 and Kre28 families are interaction partners of members of the Spc7–Spc105–KNL-1–Blinkin family in fungi, they do not appear to be functional homologs.
DISCUSSION
The Sos7 family appears to be a characteristic of fungi with regional centromeres.
In this paper, we have identified a highly conserved protein family and have characterized the founding member, the S. pombe Sos7 protein, which is an essential component of the kinetochore. Sos7-like proteins, which have a conserved signature motif, were found in a large number of fungal species. As the Sos7 ortholog from S. japonicus can rescue the mutant phenotypes of S. pombe sos7 mutant strains, we conclude that the Sos7 family consists of functionally homologous proteins. Interestingly, Sos7 family members seem to be absent in yeasts belonging to the Saccharomyces complex (33, 62). Instead, these organisms have proteins homologous to the S. cerevisiae Spc105 interaction partner Kre28. Kre28-like proteins are present in Vanderwaltozyma polyspora (EDO18951; e value, 3e−45), Zygosaccharmyces rouxii (CAR29059; e value, 1e−29), Ashbya gossypii (AAS54100; e value, 1e−28), Candida glabrata (XP_446867; e value, 3e−24), Lachancea thermotolerans (CAR23808; e value, 1e−22), and Kluyveromyces lactis (XP_452234; e value, 4e−08). In all of these organisms, point centromeres have been identified (7, 16, 23, 28, 54, 64). These findings raise the exciting possibility that fungi with point centromeres harbor a Kre28 family member while those with regional centromeres, such as S. pombe and S. japonicus, have a Sos7 ortholog as the Spc7-Spc105 interaction partner (11, 56). The centromere requirements of all other fungi with a putative Sos7 ortholog have not been analyzed yet. However, Candida albicans, which has a centromere whose size is between that of the short point centromeres and that of the regional centromeres flanked by heterochromatin, has a putative, albeit less well conserved Sos7 homolog (e value, 3e−6) (60).
Sos7 is the missing component of the S. pombe NMS complex.
The conserved NMS complex in fission yeast had been defined to consist of the four-subunit Ndc80 complex, the four-subunit MIND complex, and the single Spc7 protein (37, 47). As the Sos7-encoding ORF had been annotated incorrectly as a pseudogene until very recently, it was not identified in these biochemical approaches involving mass spectrometry. Our analysis shows that Sos7 represents the missing interaction partner of the Spc7–Spc105–KNL-1–Blinkin family in S. pombe.
Extra Sos7 rescued the nongrowth phenotype of all mutant spc7ts strains at the restrictive temperature, probably by assisting the kinetochore relocalization of the mutant Spc7 variants, as shown for spc7-23 (26). This rescue of the mutant spc7ts phenotypes did not require the presence of the Spc7 interaction partner Mal3, which is one of the Spc7 binding partner at the spindle-microtubule interface (27). Sos7 is a bona fide constitutive kinetochore protein that associates with the central domain of the centromere. However, Sos7 is not involved in the transcriptional silencing of this region, which is also a characteristic of the Spc7 protein (26, 27).
Our data demonstrate a tight interaction between Spc7 and Sos7: (i) Sos7 and Spc7 proteins coimmunoprecipitate in vivo; (ii) Sos7 and Spc7 proteins interact via their C-terminal regions, as shown by yeast 2-hybrid analysis; (iii) kinetochore localizations of Spc7 and Sos7 proteins were dependent on each other; (iv) overexpression of one protein rescued the phenotype of the mutant version of the other protein in a dose-dependent manner; and (v) sos7-Δ7 spc7ts double mutants were inviable, suggesting that cells harboring a mutant allele of one gene require the wild-type copy of the other gene for survival.
Hierarchical assembly of the NMS complex.
To determine the interdependencies of Sos7 with other components of the NMS complex, we scored reciprocal suppression and kinetochore localization dependencies. The emerging picture shows that Sos7 also interacts genetically with the MIND complex. The kinetochore targeting of Sos7 and that of the MIND component Mis12 were dependent on each other, and sos7ts mutants were suppressed by overexpression of MIND components. Thus, Sos7 establishes tight interactions with MIND and Spc7. This is in contrast to what has been described for Zwint, which might possibly represent the Sos7 metazoan ortholog. In vitro studies indicate that Zwint shows tight contacts only with KNL-1–Blinkin, not with the MIND complex (51). Analysis of the interactions of Sos7 with Ndc80 components suggests that these subunits of the NMS complex function mainly independently of each other, which is also the case for Spc7 and the Ndc80 subunit (26). Overall, our data imply that within the NMS complex, Spc7, Sos7, and the MIND complex show a tight functional interaction and Sos7 is a central component of this entity. Furthermore, kinetochore recruitment of the Ndc80 complex requires the MIND complex in S. pombe, similar to what has been shown for a number of other organisms (3, 31, 36, 39, 51).
Function of Sos7 in the cell cycle.
Our analysis showed that Sos7 is essential for proper segregation of sister chromatids, as all sos7ts mutants analyzed had severe mitotic defects. sos7-Δ7 cells showed the most severe phenotype: at the restrictive temperature, no wild-type mitosis was observed. Sixty percent of sos7-Δ7 mitotic cells were unable to separate their chromatin, although spindle elongation occurred, while the rest showed unequal or smeared chromatin in late anaphase cells. These phenotypes were caused by faulty microtubule-kinetochore interactions and are very similar to spc7ts mutant strains in frequency and severity of phenotype (26). This suggests that the phenotypes observed are caused at least in part by the loss of Spc7 kinetochore targeting in sos7ts cells. Apart from its requirement for the integrity of the NMS kinetochore complex, we speculate that Sos7 might also have a role in connecting the kinetochore with the SPB. First, the C-terminal part of Sos7 has homology to the C terminus of the S. cerevisiae Bbp1 protein, which is localized at the central plaque periphery of the SPB via its C-terminal domain (49, 63). Second, fission yeast kinetochores are clustered at the SPB during interphase, and thus, only a single, SPB-associated kinetochore signal is observed (14). We find that at the permissive temperature, 27% of sos7-Δ7 interphase cells have more than one kinetochore signal and these extra signals do not colocalize with the spindle pole body component Pcp1 (unpublished observations). This suggests that kinetochore clustering at the spindle pole body might be defective in sos7 mutant cells.
The Spc7–Spc105–KNL-1–Blinkin family interacts with distinct protein families.
Members of the Spc7–Spc105–KNL-1–Blinkin family have distinct interaction partners: Kre28 copurified with budding yeast Spc105, Zwint interacts with human Blinkin, KBP-5 coimmunoprecipitated with C. elegans KNL-1, and Sos7 is the interaction partner of the fission yeast Spc7 protein (3, 30, 46, 48). These four kinetochore components are members of different protein families which share little sequence homology. Expression of S. cerevisiae Kre28 or human Zwint protein in sos7 mutant strains could not compensate the mutant phenotypes of such strains, and neither Kre28-GFP nor Zwint-GFP could be kinetochore targeted in S. pombe (unpublished observations). However, these families might share a binding motif/structure, as Zwint and Kre28 both interacted with a Sos7 variant in a yeast 2-hybrid assay (unpublished observations).
Are the mitotic functions of the Sos7, Kre28, Zwint, and KBP-5 families similar? As shown for Sos7, Kre28 is also a constitutive and essential kinetochore component, but mutant alleles of KRE28 have not been described, and Kre28 function has been analyzed only in the context of spc105 mutants (46, 48). In contrast to our data concerning the hierarchy of the S. pombe NMS complex, the Spc105-Kre28 subcomplex appears to represent a third linker complex at the microtubule interface, as components of the MIND and Ndc80 subcomplexes do not require Spc105 for kinetochore targeting (48).
Zwint shows Blinkin-dependent kinetochore association which is mediated by an evolutionarily conserved region in Blinkin that is also present in Spc7 (amino acids 998 to 1157) (27, 29). Whether Zwint influences kinetochore localization of Blinkin and/or MIND components has not been determined, as kinetochore hierarchy in relation to Zwint has been analyzed predominantly for the mitotic checkpoint-relevant RZZ complex (10, 65, 70). However, as the mitotic phenotypes of Zwint RNAi-treated cells are similar to those seen when the RZZ component ZW10 is lost, and Bub1, which requires Blinkin for kinetochore targeting, is still kinetochore localized in Zwint minus cells, it is unlikely that Zwint plays a role in kinetochore targeting of Blinkin (30, 70). A requirement for Zwint in kinetochore targeting of the Ndc80 complex component Ndc80 has been shown for the Xenopus system (9). However, in human cells, Ndc80-Hec1 is a kinetochore localized in the absence of functional Zwint, as we have shown for S. pombe Sos7 and the Ndc80 complex member Nuf2 (34).
Lastly, the KNL-1 interaction partner KBP-5, which appears at the kinetochore in a time frame comparable to that of other C. elegans KMN components, has no essential mitotic function (3). Thus, the available experimental data suggest that the central role of Sos7 in the KMN-NMS hierarchy is not shared by Zwint and KBP-5. An extensive comparison of the mitotic roles of these 4 kinetochore protein families presupposes further analysis, especially with regard to Kre28 function.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eva Walla for excellent technical assistance, Johannes Hegemann for helpful discussion, Shelley Sazer for enlightening comments on the manuscript, and Robin Allshire, Kathleen Gould, Keith Gull, Silke Hauf, Yasushi Hiraoka, Hironori Niki, Takashi Toda, Mitsuhiro Yanagida, and the Yeast Genetic Resource Centre, Osaka, Japan, for reagents.
V.J. was supported by the Gründerstiftung zur Förderung von Forschung und wissenschaftlichem Nachwuchs an der Heinrich-Heine-Universität Düsseldorf.
Published ahead of print 18 June 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Baker DJ, et al. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36:744–749 [DOI] [PubMed] [Google Scholar]
- 2. Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127:983–997 [DOI] [PubMed] [Google Scholar]
- 3. Cheeseman IM, et al. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ciferri C, et al. 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133:427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. DeLuca JG, et al. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127:969–982 [DOI] [PubMed] [Google Scholar]
- 6. Desai A, et al. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dietrich FS, et al. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304–307 [DOI] [PubMed] [Google Scholar]
- 8. Ding R, West RR, Morphew DM, Oakley BR, McIntosh JR. 1997. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell 8:1461–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emanuele MJ, McCleland ML, Satinover DL, Stukenberg PT. 2005. Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol. Biol. Cell 16:4882–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Famulski JK, Vos L, Sun X, Chan G. 2008. Stable hZW10 kinetochore residency, mediated by hZwint-1 interaction, is essential for the mitotic checkpoint. J. Cell Biol. 180:507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fishel B, Amstutz H, Baum M, Carbon J, Clarke L. 1988. Structural organization and functional analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Mol. Cell. Biol. 8:754–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fleig U, Sen-Gupta M, Hegemann JH. 1996. Fission yeast mal2+ is required for chromosome segregation. Mol. Cell. Biol. 16:6169–6177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flory MR, Morphew M, Joseph JD, Means AR, Davis TN. 2002. Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ. 13:47–58 [PubMed] [Google Scholar]
- 14. Funabiki H, Hagan I, Uzawa S, Yanagida M. 1993. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121:961–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garnier J, Gibrat JF, Robson B. 1996. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266:540–553 [DOI] [PubMed] [Google Scholar]
- 16. Gordon JL, Byrne KP, Wolfe KH. 2011. Mechanisms of chromosome number evolution in yeast. PLoS Genet. 7:e1002190 doi:10.1371/journal.pgen.1002190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goshima G, Saitoh S, Yanagida M. 1999. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 13:1664–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 19. Hagan I, Yanagida M. 1995. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129:1033–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hagan IM, Hyams JS. 1988. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 89:343–357 [DOI] [PubMed] [Google Scholar]
- 21. Hayashi T, et al. 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118:715–729 [DOI] [PubMed] [Google Scholar]
- 22. Hayette S, et al. 2000. AF15q14, a novel partner gene fused to the MLL gene in an acute myeloid leukaemia with a t(11;15)(q23;q14). Oncogene 19:4446–4450 [DOI] [PubMed] [Google Scholar]
- 23. Heus JJ, Zonneveld BJ, de Steensma HY, van den Berg JA. 1993. The consensus sequence of Kluyveromyces lactis centromeres shows homology to functional centromeric DNA from Saccharomyces cerevisiae. Mol. Gen. Genet. 236:355–362 [DOI] [PubMed] [Google Scholar]
- 24. Jakopec V, Walla E, Fleig U. 2011. Versatile use of Schizosaccharomyces pombe plasmids in Saccharomyces cerevisiae. FEMS Yeast Res. 11:653–655 [DOI] [PubMed] [Google Scholar]
- 25. Kerres A, et al. 2006. Fta2, an essential fission yeast kinetochore component, interacts closely with the conserved mal2 protein. Mol. Biol. Cell 17:4167–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kerres A, Jakopec V, Fleig U. 2007. The conserved Spc7 protein is required for spindle integrity and links kinetochore complexes in fission yeast. Mol. Biol. Cell 18:2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kerres A, et al. 2004. The fission yeast kinetochore component Spc7 associates with the EB1 family member Mal3 and is required for kinetochore-spindle association. Mol. Biol. Cell 15:5255–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kitada K, Yamaguchi E, Hamada K, Arisawa M. 1997. Structural analysis of a Candida glabrata centromere and its functional homology to the Saccharomyces cerevisiae centromere. Curr. Genet. 31:122–127 [DOI] [PubMed] [Google Scholar]
- 29. Kiyomitsu T, Murakami H, Yanagida M. 2011. Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol. Cell. Biol. 31:998–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kiyomitsu T, Obuse C, Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell 13:663–676 [DOI] [PubMed] [Google Scholar]
- 31. Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A. 2006. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 173:9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koch A, Krug K, Pengelley S, Macek B, Hauf S. 2011. Mitotic substrates of the kinase aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast. Sci. Signal. 4:rs6 doi:10.1126/scisignal.2001588 [DOI] [PubMed] [Google Scholar]
- 33. Kurtzman CP, Robnett CJ. 2003. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res. 3:417–432 [DOI] [PubMed] [Google Scholar]
- 34. Lin YT, Chen Y, Wu G, Lee WH. 2006. Hec1 sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful chromosome segregation and spindle checkpoint control. Oncogene 25:6901–6914 [DOI] [PubMed] [Google Scholar]
- 35. Liu D, et al. 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188:809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu ST, Rattner JB, Jablonski SA, Yen TJ. 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175:41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu X, McLeod I, Anderson S, Yates JR, III, He X. 2005. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 24:2919–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lott IT, Head E. 2005. Alzheimer disease and Down syndrome: factors in pathogenesis. Neurobiol. Aging 26:383–389 [DOI] [PubMed] [Google Scholar]
- 39. Maskell DP, Hu XW, Singleton MR. 2010. Molecular architecture and assembly of the yeast kinetochore MIND complex. J. Cell Biol. 190:823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meadows JC, et al. 2011. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev. Cell 20:739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moreno MB, Duran A, Ribas JC. 2000. A family of multifunctional thiamine-repressible expression vectors for fission yeast. Yeast 16:861–872 [DOI] [PubMed] [Google Scholar]
- 42. Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 [DOI] [PubMed] [Google Scholar]
- 43. Moreno S, Nurse P. 1994. Regulation of progression through the G1 phase of the cell cycle by the rum1+ gene. Nature 367:236–242 [DOI] [PubMed] [Google Scholar]
- 44. Musacchio A, Salmon ED. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 [DOI] [PubMed] [Google Scholar]
- 45. Nabeshima K, et al. 1998. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell 9:3211–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nekrasov VS, Smith MA, Peak-Chew S, Kilmartin JV. 2003. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14:4931–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Obuse C, et al. 2004. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 6:1135–1141 [DOI] [PubMed] [Google Scholar]
- 48. Pagliuca C, Draviam VM, Marco E, Sorger PK, De Wulf P. 2009. Roles for the conserved spc105p/kre28p complex in kinetochore-microtubule binding and the spindle assembly checkpoint. PLoS One 4:e7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Park CJ, et al. 2004. Requirement for Bbp1p in the proper mitotic functions of Cdc5p in Saccharomyces cerevisiae. Mol. Biol. Cell 15:1711–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Partridge JF, Borgstrom B, Allshire RC. 2000. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 14:783–791 [PMC free article] [PubMed] [Google Scholar]
- 51. Petrovic A, et al. 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190:835–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pidoux AL, Richardson W, Allshire RC. 2003. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell Biol. 161:295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pohlmann J, Fleig U. 2010. Asp1, a conserved 1/3 inositol polyphosphate kinase, regulates the dimorphic switch in Schizosaccharomyces pombe. Mol. Cell. Biol. 30:4535–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pribylova L, Straub ML, Sychrova H, de Montigny J. 2007. Characterisation of Zygosaccharomyces rouxii centromeres and construction of first Z. rouxii centromeric vectors. Chromosome Res. 15:439–445 [DOI] [PubMed] [Google Scholar]
- 55. Przewloka MR, et al. 2007. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS One 2:e478 doi:10.1371/journal.pone.0000478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rhind N, et al. 2011. Comparative functional genomics of the fission yeasts. Science 332:930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rosenberg JS, Cross FR, Funabiki H. 2011. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol. 21:942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Saitoh S, Ishii K, Kobayashi Y, Takahashi K. 2005. Spindle checkpoint signaling requires the mis6 kinetochore subcomplex, which interacts with mad2 and mitotic spindles. Mol. Biol. Cell 16:3666–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Santaguida S, Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sanyal K, Baum M, Carbon J. 2004. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc. Natl. Acad. Sci. U. S. A. 101:11374–11379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sato M, Vardy L, Angel Garcia M, Koonrugsa N, Toda T. 2004. Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol. Biol. Cell 15:1609–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Scannell DR, Butler G, Wolfe KH. 2007. Yeast genome evolution—the origin of the species. Yeast 24:929–942 [DOI] [PubMed] [Google Scholar]
- 63. Schramm C, Elliott S, Shevchenko A, Schiebel E. 2000. The Bbp1p-Mps2p complex connects the SPB to the nuclear envelope and is essential for SPB duplication. EMBO J. 19:421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Souciet JL, et al. 2009. Comparative genomics of protoploid Saccharomycetaceae. Genome Res. 19:1696–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Starr DA, et al. 2000. HZwint-1, a novel human kinetochore component that interacts with HZW10. J. Cell Sci. 113(Pt 11):1939–1950 [DOI] [PubMed] [Google Scholar]
- 66. Takahashi K, Yamada H, Yanagida M. 1994. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell 5:1145–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tanaka TU. 2010. Kinetochore-microtubule interactions: steps towards bi-orientation. EMBO J. 29:4070–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Thompson SL, Compton DA. 2011. Chromosomes and cancer cells. Chromosome Res. 19:433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tran PT, Paoletti A, Chang F. 2004. Imaging green fluorescent protein fusions in living fission yeast cells. Methods 33:220–225 [DOI] [PubMed] [Google Scholar]
- 70. Wang H, et al. 2004. Human Zwint-1 specifies localization of Zeste White 10 to kinetochores and is essential for mitotic checkpoint signaling. J. Biol. Chem. 279:54590–54598 [DOI] [PubMed] [Google Scholar]
- 71. Wei RR, Al-Bassam J, Harrison SC. 2007. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 14:54–59 [DOI] [PubMed] [Google Scholar]
- 72. Welburn JP, et al. 2009. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell 16:374–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Welburn JP, et al. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 38:383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Westermann S, Drubin DG, Barnes G. 2007. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 76:563–591 [DOI] [PubMed] [Google Scholar]
- 75. Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. 2008. Orientation and structure of the Ndc80 complex on the microtubule lattice. J. Cell Biol. 182:1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.