Abstract
The Drosophila melanogaster Myb-MuvB/dREAM complex (MMB/dREAM) participates in both the activation and repression of developmentally regulated genes and origins of DNA replication. Mutants in MMB subunits exhibit diverse phenotypes, including lethality, eye defects, reduced fecundity, and sterility. Here, we used P-element excision to generate mutations in lin-52, which encodes the smallest subunit of the MMB/dREAM complex. lin-52 is required for viability, as null mutants die prior to pupariation. The generation of somatic and germ line mutant clones indicates that lin-52 is required for adult eye development and for early embryogenesis via maternal effects. Interestingly, the maternal-effect embryonic lethality, larval lethality, and adult eye defects could be suppressed by mutations in other subunits of the MMB/dREAM complex. These results suggest that a partial MMB/dREAM complex is responsible for the lethality and eye defects of lin-52 mutants. Furthermore, these findings support a model in which the Lin-52 and Myb proteins counteract the repressive activities of the other members of the MMB/dREAM complex at specific genomic loci in a developmentally controlled manner.
INTRODUCTION
The differentiation of unique cell types during metazoan development requires the establishment and maintenance of specific gene expression profiles. Central to this process are site-specific DNA-binding factors that regulate the transcriptional patterns that establish and maintain cellular identity. The tissue- and lineage-specific transcription factors involved in these processes are critical for the maintenance of cell identity and ultimately for the execution of metazoan development.
The Drosophila melanogaster Myb-MuvB complex (MMB), also known as the Rb, E2F, and Myb-associated protein (dREAM) complex, is a large (∼700 kDa), highly conserved, nine-subunit assembly that functions both in transcriptional activation and repression (4, 17, 18). The MMB/dREAM complex contains at least three sequence-specific DNA-binding components: Myb, the E2F2-DP heterodimer, and Mip120. The other subunits of the complex (Mip130, Mip40, Caf1/p55, Rbf1 or Rbf2, and Lin-52) are not known DNA-binding proteins but may contribute specific protein interaction surfaces for targeting the complex to chromatin. For example, a Myb mutant protein lacking its DNA-binding domain nevertheless localizes properly to chromosomes and can regulate gene expression (1, 35).
The testis-specific meiotic arrest complex (tMAC) is a paralogous assembly of proteins that contains two MMB/dREAM subunits, CAF1/p55 and Mip40, in addition to paralogs of Mip130 (Aly), Mip120 (Tomb), and Lin-52 (Wuc) (3, 7, 16). tMAC appears to regulate the transcription of genes involved in spermatogenesis (3, 37). Mutations of tMAC subunits prevent the completion of spermatogenesis and cause male sterility (3, 7, 16, 37).
Human cells contain complexes similar to MMB/dREAM, with an apparent preference for proteins encoded by specific members of multigene families. The human DREAM/LINC complexes include the homologs of Mip130 (Lin-9), Mip120 (Lin-54), p55/CAF1 (RBBP4/Lin-35), Mip40 (Lin-37), and Lin-52 (the MuvB core), together with either E2F4-DP and RBL2/p130 or with B-Myb and RBL1/p107 (21, 25, 27). Thus far, these complexes have been shown to regulate genes involved in cell cycle control in mammalian cells.
All of the MMB/dREAM subunits, with the exception of Myb, are present in a homologous complex in the nematode Caenorhabditis elegans (19). The genes encoding these proteins were identified in a screen for mutants that result in ectopic vulval development (5, 8). These studies identified two classes of loss-of-function mutants that redundantly suppress Ras pathway signaling, resulting in a synthetic multivulva (synMuv) phenotype. The combination of any class A synMuv mutant with any class B synMuv mutant causes a multivulva phenotype. Some of the proteins encoded by class B synMuv genes are present in a complex called DRM that is similar to MMB/dREAM (14). However, C. elegans appears to have lost its animal-specific Myb-related gene during the course of evolution (19).
Drosophila lin-52 is a homolog of C. elegans lin-52, a gene that functions to oppose let-60 (Ras) signaling in a lin-35 (Rb)-dependent manner (33). The 19.5-kDa Drosophila Lin-52 protein was previously identified as a stoichiometric subunit of the MMB/dREAM complex (18). Homologs of Lin-52 were subsequently identified in the human DREAM/LINC and nematode DRM complexes. Lin-52 lacks domains of known function and has no known enzymatic activity, but it is required for both activation and repression of genes regulated by the MMB/dREAM complex in Drosophila Kc cells in culture (10).
Drosophila Myb is a transcriptional activator whose steady-state levels in both adult flies and tissue culture cells is dependent on incorporation into the MMB/dREAM complex (2, 15, 24, 35). Drosophila myb null mutants die as late third-instar larvae (22). Cells in myb mutant animals exhibit aberrant mitotic spindle formation and defects in chromosome condensation (9, 22, 23). Previously, we showed that loss-of-function mutants of mip130, mip120, or mip40 could rescue the lethal phenotype of myb null mutants (2, 3). These results suggested that the lethality of myb mutants was dependent on the activity of Mip130, Mip120, and Mip40 in the context of a partially formed MMB/dREAM complex. Consistent with these observations, the failure to activate transcription of the Polo gene in the absence of Myb can be partially rescued by also removing Mip130 or E2f2 (35). Furthermore, the chromosome condensation defects that occur in the absence of Myb can be rescued by also removing Mip130. We thus hypothesized that Myb is required to override an intrinsic transcriptional repression by the other members of the MMB/dREAM complex in specific developmental contexts (10, 35). We have now used a combination of genetic and molecular techniques to demonstrate that Lin-52 can also act in opposition to the other members of the MMB/dREAM complex. However, the absence of Lin-52 causes different biological phenotypes than the absence of Myb. These genetic data imply that Lin-52 and Myb have distinct and key roles in activation of gene expression.
MATERIALS AND METHODS
Drosophila genetics and mosaic analysis.
All mutants, except for lin-5293, have been previously described (2, 3, 22). The lin-5293 mutant was generated by imprecise P-element excision following mobilization of P{EP}EP404 by the delta 2-3 transposase (13). Meiotic recombination was used to produce X chromosomes containing single, double, and triple mutants of lin-5293, mip1301-36, and/or mybMH107 together with the FRT19A recognition site for the FLP recombinase. Unless otherwise indicated, all flies were raised at 25°C. Unless otherwise indicated, all fly stocks were obtained from the Bloomington Drosophila Stock Center.
To generate standard mosaics by FLP-mediated mitotic recombination (11), female flies carrying the mutant of interest on an FRT19A-containing X chromosome and an FM7 balancer chromosome were crossed to male flies containing a P{hs-FLP}122 P{neoFRT}19A X chromosome. F1 larvae, pupae, or adults then were heat shocked for 1 h once or twice a day for 1 day or 2 or 3 consecutive days at 37°C.
To generate eyeless-GAL4 UAS-FLP mosaics with an eye-specific cell death transgene GMR-hid (EGUF/hid) bearing eyes composed solely of homozygous mutant cells (31), male flies of the genotype w1118 lin-5293 P{neoFRT}19A/Y P{lin-52+}/CyO were crossed to female flies of the genotype P{GMR-hid}SS1 y1 w* P{neoFRT}19A l (1)**/FM7; P{GAL4-ey.H}SS5 P{UAS-FLP1.D}JD2.
To generate female germ line mosaics (6), female flies carrying the mutant of interest on an FRT19A-containing X chromosome and an FM7 balancer chromosome were crossed to male flies of the genotype P{ovoD1-18}P4.1 P{hsp70-flp}1 y1 w1118 sn3 P{neoFRT}19A/Y. A dominant female-sterile mutation (ovoD) is used in this technique to eliminate heterozygous and homozygous wild-type cells for the gene of interest from the female germ line. Second-instar larvae were heat shocked at 37°C for 2 h on two consecutive days.
Hatch rate analysis.
Virgin females of the indicated genotype were collected and mated with w1118 males for 2 days. Zero- to twelve-hour-old embryos were collected from these flies and incubated for more than 24 h. The hatch rate was determined by dividing the number of hatched embryos by the total number of embryos deposited. The genotypes of the female flies used in hatch rate analyses were the following: (i) ovoD hs-FLP FRT19A/lin-5293 FRT19A; P{lin-52+}, (ii) ovoD hs-FLP FRT19A/mip1301-36 FRT19A, (iii) ovoD hs-FLP FRT19A/mip1301-36 lin-5293 FRT19A, (iv) ovoD, hs-FLP FRT19A/mybMH107 FRT19A, (v) ovoD hs-FLP FRT19A/lin-5293 mybMH107 FRT19A, (vi) mip40EY16520, (vii) ovoD hs-FLP, FRT19A/lin-5293 FRT19A; mip40EY16520/+; and (viii) ovoD hs-FLP FRT19A/lin-5293 FRT19A; mip12067A9/+.
Immunostaining.
Embryos were dechorionated in 100% bleach and fixed in a mixture of heptane and methanol (1:1) containing 10 mM EGTA for 5 min. The fixed embryos were treated with methanol to remove vitilline membranes and rehydrated with phosphate-buffered saline (PBS) containing 0.1% Triton X-100. Embryos were incubated with primary antibodies overnight at 4°C, washed, and incubated for 2 h with secondary antibodies and RNase and then washed again. The resulting embryos were mounted in Vectashield mounting medium (Vector Laboratories) with propidium iodide (PI) to stain DNA.
Ovaries were hand dissected and fixed in a mixture of 4% formaldehyde in PBS plus heptane for 15 min. After rinsing three times with PBS containing 0.1% Triton X-100, the fixed ovaries were incubated in PBS containing 0.6% Triton X-100 for an hour to remove lipid and increase antibody accessibility. Treated ovaries were incubated with primary and secondary antibodies as described above.
Imaginal discs, larval brains, and salivary glands were dissected from third-instar larvae and fixed with 4% formaldehyde in PBS for 15 min. The fixed tissues were washed and incubated in primary and secondary antibodies as described above.
The following primary antibodies were used: mouse anti-beta-tubulin (1:200; E7; Developmental Studies Hybridoma Bank), rabbit anticentrosomin (1:1,000; a gift from T. Kaufman, Indiana University), chicken anti-green fluorescent protein (anti-GFP) (1:3,000; 13970; Abcam), rabbit anti-Mip40 (1:600), rabbit anti-Mip120 (1:600), rabbit anti-Caf1/P55 (1:2,000), rabbit anti-E2f2 (1:500), rabbit anti-Mip130 (1:500), and rabbit anti-Lin-52 (1:300). The following secondary antibodies were used: Alexa Fluor 488–goat anti-mouse (1:500; Invitrogen), Alexa Fluor 488–goat anti-chicken (1:500; Invitrogen), and Alexa Fluor 633–goat anti-rabbit (1:500; Invitrogen) antibodies. DNA was visualized by staining with TOPRO-3 (1:1,000; Invitrogen) or propidium iodide (mounting medium from Vectashield).
Microscopy.
Immunofluorescence microscopy was performed as previously described using a Nikon PCM 2000 laser scanning confocal microscope (4, 35). Scanning electron microscopy and light microscopy of toluidine-stained or phase-contrast-visualized thick sections of adult Drosophila eyes were performed as previously described (28). Images were processed using Adobe Photoshop.
Molecular biology.
A transgene encoding the wild-type Lin-52 protein under the control of the lin-52 promoter was constructed by inserting a 1.6-kb fragment of a PCR-amplified genomic DNA fragment using the primers TTGCGGCCGCAGCGAATGAAAAACCGAAAGAAAGTG and TTGCGGCCGCCCTGCCTATTTATCTTTG into the NotI site of the pW8 P-element transformation vector plasmid (restriction enzyme sites indicated in bold). A transgene encoding GFP fused to the C terminus of the full-length Lin-52 protein was constructed by inserting the following three segments into the pCasper4 P-element transformation vector plasmid. (i) The region from the upstream neighboring gene CG15771 through the promoter and open reading frame of lin-52 was amplified by PCR with the primers GGCGGTACCGGAGCAGCGAATGAAAAA and TATGGATCCTCTTGGCTGCCGCTTAGGAG, and the resulting DNA fragment was digested with KpnI and BamHI. (ii) The coding sequence for GFP was amplified by PCR with the primers ACAGGATCCATGGTGAGCAAGGGCGAGGA and ATTCTAGATTACTTGTACAGCTCGTCCATG, and the resulting fragment was digested with BamHI and XbaI. (iii) The region from the end of the lin-52 open reading frame to the downstream neighboring gene CG15772 was amplified by PCR with the primers CCTCTAGACTGGATGATCCGATCCCTAA and AATCTCGAGCGTGTAGGACCCAAAATGCT, and the resulting fragment was digested with XbaI and XhoI.
Transgenes encoding N-terminally Flag-tagged wild-type Lin-52 utilized cDNA expressed under the control of the Drosophila Ubi-63E promoter (pSTE plasmid containing the Ubi-63E promoter, simian virus 40 [SV-40] polyadenylation sequence, and mini-white marker). These FLAG-lin-52 transgenes encoded an N-terminal FLAG sequence (DYKDDDDK) prior to the first methionine of Lin-52. PCR primers used for this construct were the following: 5′ FLAG-Lin-52, CTCGAGGCCTTCCACCATGGATTACAAAGACGATGACGATAAGATGAGCAACGCAGAGAC; and 3′ Lin-52, TCGAGGCCTTCATGGCTGCCGCTTAGGAGCGCG (initiation and termination codons of lin-52 are underlined).
FLAG–Lin-52 immunoprecipitation from Drosophila embryo extracts.
FLAG–Lin-52 complexes were purified from lin-5293/lin-5293; FLAG–Lin-52/FLAG–Lin-52 embryos that had been grown for 0 to 12 h. The embryos were processed as previously described (18). Dialyzed nuclear extracts (20 mM HEPES, pH 7.7, 0.3 M KCl, 0.5 mM EDTA, 0.01% NP-40, 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], and 5 mM 2-mercaptoethanol) were mixed with M2 anti-FLAG resin (Sigma). After incubation at 4°C, M2 beads were extensively washed with 0.3 M KCl buffer supplemented with 50 μg/ml ethidium bromide. The M2 beads were equilibrated in digestion buffer (20 mM HEPES, pH 7.7, 50 mM KCl, 2 mM CaCl2) and incubated with micrococcal nuclease (Roche) (1 U/μl) for 1 h at 4°C. The M2 beads were washed with 0.3 M KCl buffer followed by elution with 0.4 mg/ml FLAG peptide.
Gel filtration of FLAG–Lin-52-associated proteins.
FLAG–Lin-52-associated proteins from the M2 eluate were fractionated by gel filtration chromatography. Eluate from M2 agarose beads was loaded onto a Superdex 200 PC 3.2/30 gel filtration column (Amersham) on a Smart fast performance liquid chromatography (FPLC) system. Immunoblot analyses were conducted on Superdex 200 fractions (50 μl).
RNA interference (RNAi) of MMB/dREAM subunits in Kc cells.
Double-stranded RNA synthesis, RNA transfection, and immunoblotting for Mip130, Mip120, Myb, E2F2, and Lin-52 were performed as previously described (2).
RESULTS
Larval lethality of a lin-52 null mutant can be rescued by a mip40 null mutant.
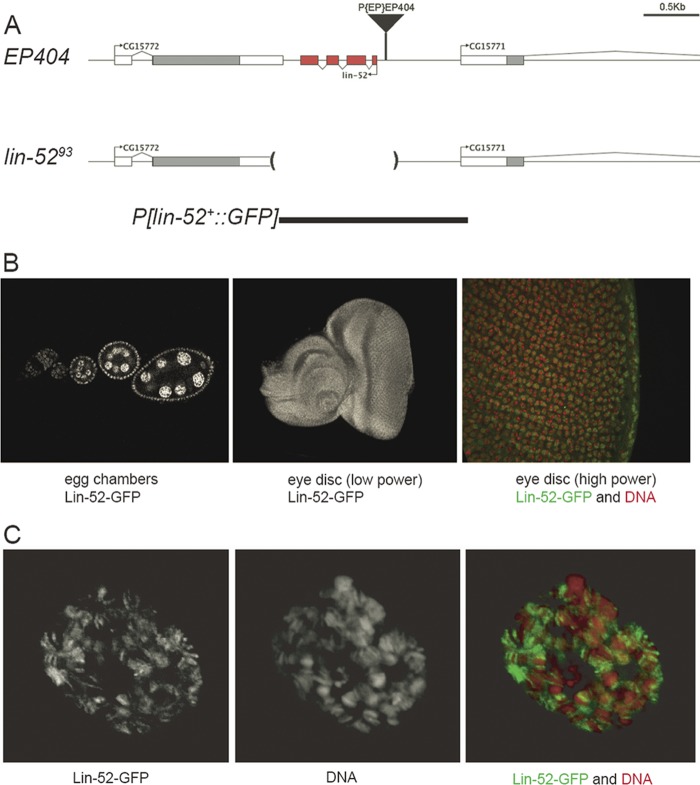
To address the role of lin-52 during development, we generated a deletion mutant by imprecise excision of P{EP}EP404, a P-element transposon inserted 108 bp upstream of the lin-52 coding region on the X chromosome (Fig. 1A). Southern blot analysis of genomic DNA from X-linked lethal mutants that resulted from P-element mobilization led to the identification of lin-5293. DNA sequencing of this mutant revealed that excision of EP404 had resulted in a 1.3-kb deletion that included the entire lin-52 gene (Fig. 1A). The late larval lethality of this mutant could be rescued by a P-element transgene containing a 1.6-kb genomic DNA fragment that included all four exons of lin-52 and the adjacent intergenic regions (Fig. 1A). A variant of this transgene encoding a GFP-Lin-52 fusion protein was also able to rescue mutant flies to adult viability.
Fig 1.
Generation of a lin-52 null mutant and lin-52+ transgenes. (A) Mobilization of a P element resulted in a 1,302-bp deletion that removed the entire lin-52 gene. A wild-type, 1.6-kb genomic DNA fragment was used to create P elements capable of rescuing the lin-5293 null mutant, one of which encoded a Lin-52–GFP fusion protein. (B) A Lin-52–GFP fusion protein produced under the control of a transgenic lin-52 promoter was present in the nuclei of ovarian germ line and somatic cells (left), in the nuclei of all cells of a larval eye imaginal disc (center), and in the euchromatic but not the heterochromatic regions of the nuclei of a larval eye imaginal disc (Lin-52–GFP in green, DNA in red) (right). (C) The Lin-52–GFP fusion protein was also present in polytene salivary gland nuclei and predominantly decorated the decondensed euchromatic regions of the chromosomal arms.
Analysis of third-instar larval tissues from flies bearing the GFP:lin-52+ transgene revealed that Lin-52 protein is normally present within the nuclei of ovarian germ line and somatic cells, of diploid tissues, including the imaginal discs and brain, and of polytene tissues, including the salivary glands (Fig. 1B and C). An analysis of the giant polytene chromosomes of larval salivary gland nuclei revealed that the Lin-52 protein predominantly decorated decondensed euchromatic regions of the chromosomal arms but not the more highly condensed heterochromatic regions (Fig. 1C). A similar euchromatic localization was seen in diploid cells within the imaginal eye discs (Fig. 1B).
Immunoblot analysis of Lin-52 levels in lin-5293 mutant animals during the course of development revealed that the maternally deposited stores of Lin-52 were depleted during larval development (Fig. 2C and D). These results are consistent with a model in which late larval lethality of the lin-5293 null mutant is due to a requirement for zygotically encoded Lin-52 once maternally deposited supplies have been exhausted.
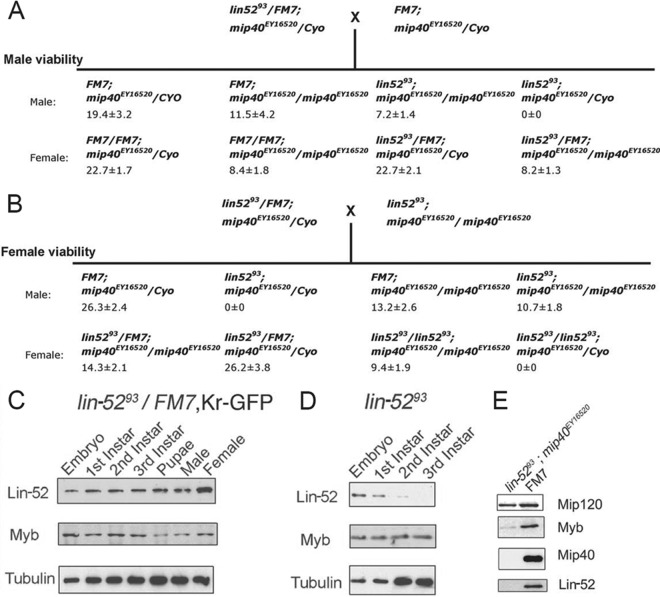
Fig 2.
lin-52 mutant larval lethality can be rescued by loss of mip40. (A and B) Two hundred to four hundred fifty progeny per indicated cross were counted. The values listed are the percent viability averages with standard deviations from two independent experiments. mip40EY16520 is a viable null mutation. FM7 and CyO are balancer chromosomes. FM7 is homozygous, viable, and female sterile. CyO is homozygous lethal. (C and D) Immunoblots for Lin-52 and Myb in lin-5293/FM7, Kr-GFP female and lin-5293/Y male embryos and larvae. Tubulin was used as a loading control. (E) Immunoblot of Mip120, Myb, Mip40, and Lin-52 protein in lin-52, mip40 double mutant adult female flies (lin-5293 mip40EY16520) or FM7 control flies.
We had previously shown that a Drosophila myb null mutation causes late larval lethality (22). Surprisingly, we then found that mutations in other subunits of the MMB/dREAM complex (mip40, mip120, and mip130) could suppress myb mutant lethality (2, 3). We therefore tested whether mutations in the same MMB/dREAM subunits could suppress lin-52 lethality. We were able to recover lin-52 mip40 double mutants at a frequency very similar to that of lin-52/+ mip40 animals (7.2 versus 8.2% for males in Fig. 2A; 9.4 versus 14.3% for females in Fig. 2B). In contrast, the presence of one or two copies of the wild-type mip40 gene resulted in uniform lethality of lin-52 mutant males and females (Fig. 2A and B). Immunoblotting of the lin-52 mip40 double mutants confirmed that both Lin-52 and Mip40 proteins were absent (Fig. 2E).
Lin-52 is exclusively found in the MMB/dREAM complex.
We previously found that Lin-52 was required for MMB-dependent transcriptional activation and repression via RNAi depletion in cultured Drosophila Kc cells (10). Furthermore, genome-wide analysis by chromatin immunoprecipitation (ChIP) indicated an excellent overlap between Lin-52 and several other MMB subunits, suggesting that chromatin-associated Lin-52 is exclusively found within MMB. Based on these data, we hypothesized that Lin-52 functions solely within the context of the MMB.
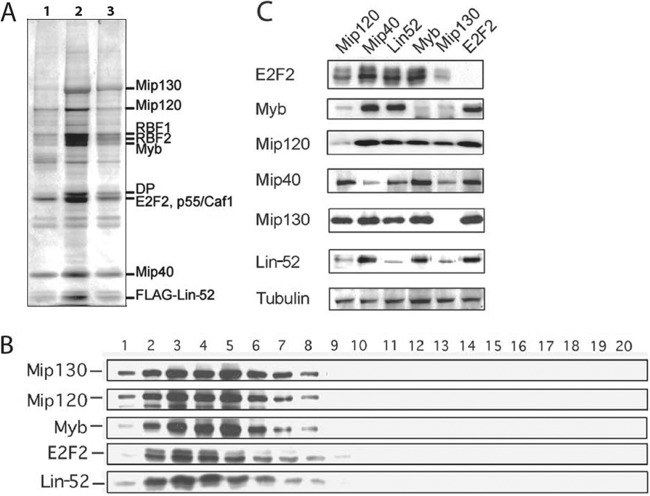
To determine which multiprotein complexes contain Lin-52, we first constructed a transgenic line of Drosophila that expressed a FLAG-tagged Lin-52 protein under the control of the ubiquitously expressed Ubi-63E promoter. Importantly, this transgene was capable of rescuing the lethality of the lin-5293 null mutant. We prepared a nuclear extract from lin-5293; FLAG–lin-52+ Drosophila embryos grown for 0 to 12 h. We then performed immunoprecipitation using anti-FLAG resin to recover FLAG–Lin-52 and coimmunoprecipitating proteins. Silver-stain analysis indicated that MMB/dREAM complex subunits represented the majority of proteins in the M2 eluate (Fig. 3A). To determine if FLAG–Lin-52 also is present in a non-MMB/dREAM complex or free from other MMB proteins, the affinity-purified proteins were size fractionated by gel filtration chromatography. Immunoblot analysis of the eluted fractions revealed that FLAG–Lin-52 cofractionated with other MMB/dREAM complex subunits (Fig. 3B).
Fig 3.
Lin-52 is present exclusively in the MMB/dREAM complex and interacts with the complex via its C terminus. (A) Silver-stained SDS-PAGE of eluate from M2 (anti-FLAG IgG) beads after incubation in extract of lin-5293/lin-5293; FLAG–Lin-52/FLAG–Lin-52 embryos grown for 0 to 12 h. The MMB/dREAM subunits were identified by immunoblotting (data not shown). Only the peak fractions are shown. (B) Gel filtration of FLAG–Lin-52-associated proteins. Eluate shown in panel A was fractionated on a Smart Superdex 200 gel filtration column. Sequential fractions were immunoblotted for the proteins indicated at the left. (C) RNA interference was performed in Drosophila Kc cells using double-stranded RNAs targeted against the gene indicated above each lane. After 3 days, cells were harvested and immunoblot analysis was performed with the antibodies indicated on the left.
We and others have previously found that the steady-state protein levels of some MMB/dREAM subunits are dependent upon the presence of other subunits (2, 10, 17). These results lead us to speculate that some MMB/dREAM subunits form a core complex that is necessary for the stability of other subunits. To determine if Lin-52 is necessary for the stability of MMB, we used RNA interference (RNAi) in Drosophila Kc cells to reduce the levels of specific subunits (Fig. 3C). During 3 days of RNAi treatment, we found no major effect on growth rates of the cells. We therefore performed immunoblots on cell pellets after a 3-day incubation with double-stranded RNA to determine the level of various MMB/dREAM subunits. We found that depletion of Lin-52 did not markedly affect the steady-state levels of other MMB/dREAM subunits in Kc cells, except for a moderate decrease in Mip40 levels. However, Lin-52 protein levels did decrease upon depletion of Mip120 and Mip130.
We also performed genetic mosaic analysis in vivo to determine the effect of long-term depletion of Lin-52 upon the levels of the MMB complex (Fig. 4). Immunostaining of relatively large lin-52 mutant clones of ovarian follicle cells revealed a diminution in the levels of other components of the complex, particularly Mip40, E2F2 to a lesser degree, and Mip130 to an even lesser degree. However, none of these proteins were completely absent in the absence of Lin-52. This effect was cell autonomous, since the levels of these proteins were unaltered in the adjacent heterozygous (lin-52+/−) tissue and homozygous wild-type (lin-52+/+) twin spots. The levels of p55/CAF1 and Mip120 were unaffected in the absence of Lin-52. The effects observed in mosaic analysis may be more dramatic because cells were homozygous for a lin-52 null mutant, whereas the RNAi experiments utilized a knockdown rather than a knockout of gene function. Taken together, the results from the biochemical fractionation, the RNAi experiments in cell culture, and mosaic analysis in vivo suggest that Lin-52 associates exclusively with other MMB/dREAM subunits in Drosophila. Additionally, these data suggest that all lin-5293 mutant phenotypes are likely related to MMB/dREAM function. The genetic interactions with other MMB genes presented in subsequent experiments described in the following sections of this paper are also readily explained by this hypothesis.
Fig 4.
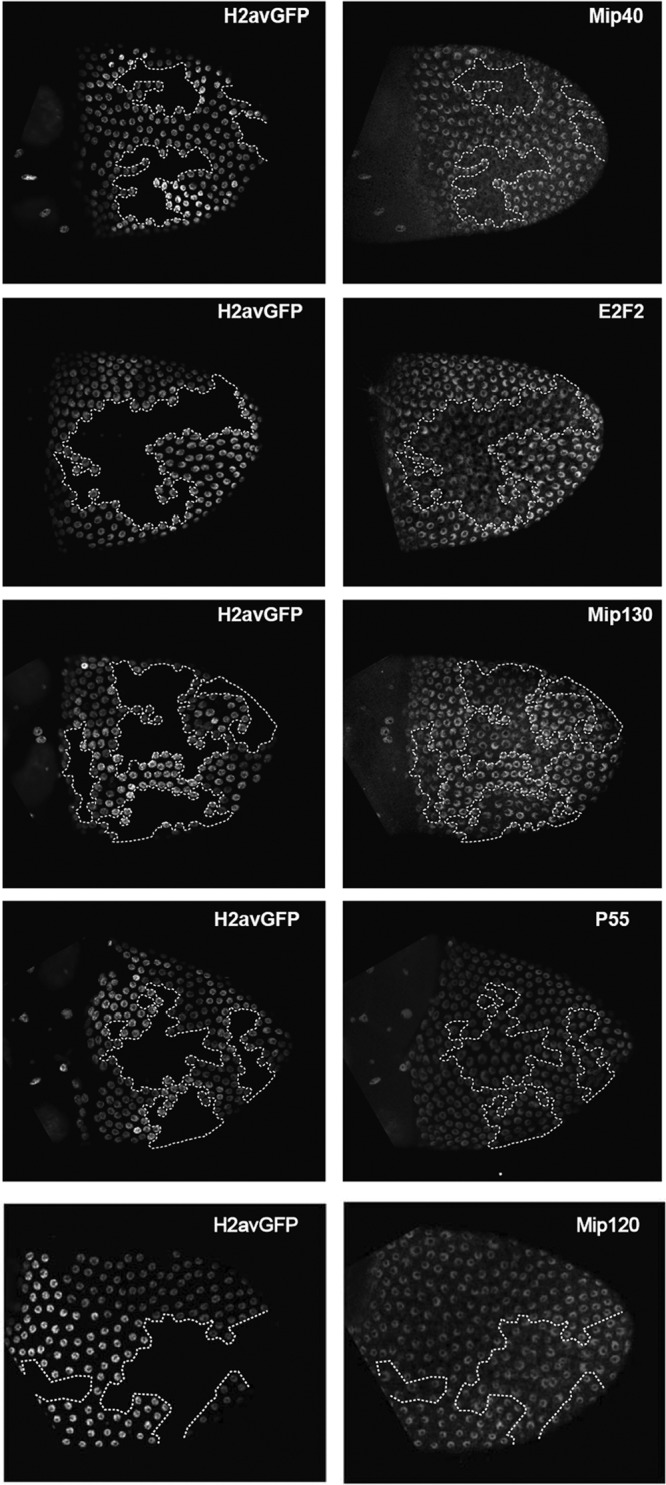
Absence of Lin-52 affects the levels of other MMB/dREAM components in vivo. Heat shock-FLP was used to induce FRT-mediated mitotic recombination during the development of heterozygous lin-52+/lin-5293 ovarian follicle cells. H2avGFP marks cells bearing the wild-type lin-52+ allele. The absence of H2avGFP indicates cells that are homozygous for the lin-5293 null allele. Doubly bright H2avGFP-positive cells represent twin spots bearing two copies of the wild-type lin-52+ allele. Ovaries were immunostained with the antibodies indicated on the right of each row. Dotted lines indicate the boundaries of homozygous lin-5293 mutant clones.
Adult eye defects caused by a lin-52 null mutant can be rescued by a mip130 null mutant.
The MMB/dREAM complex regulates genes involved in cell cycle regulation and in development (10, 17, 35). Previous studies have implicated different MMB/dREAM complex subunits in eye development in Drosophila. However, the absence of the E2F2, RBF1, RBF2, Mip130, Mip40, or Myb protein causes little or no adult eye phenotype (3, 23, 30). We therefore wished to test whether Lin-52 is required for eye development. To this end, we employed the EGUF/hid system to generate homozygous mutant eyes in an otherwise heterozygous animal (31). This method utilizes mitotic recombination caused by an eye-specific FLP recombinase, an FLP recombination target (FRT)-containing chromosome arm with the mutant allele of interest, and an FRT-containing chromosome arm with the wild-type allele, an eye-specific cell-lethal transgene (GMR-hid), and a recessive cell-lethal mutation. The result is adult eyes composed exclusively of cells derived from homozygous mutant clones. We observed a rough-eye phenotype in the absence of lin-52 that included small rhabdomeres and irregular ommatidia (Fig. 5). Importantly, a transgene expressing the wild-type allele (lin-52+) could rescue this rough-eye phenotype.
Fig 5.
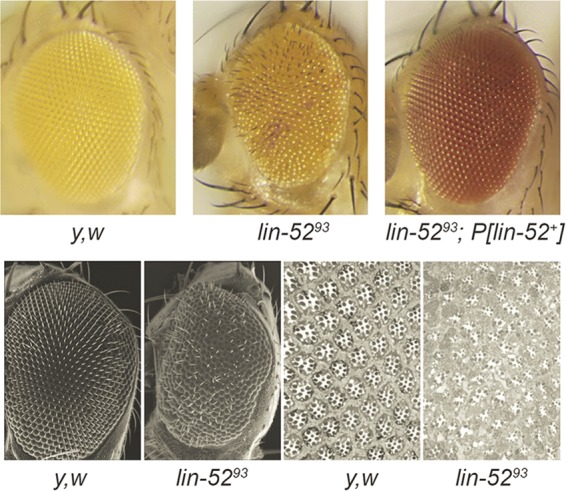
lin-52 is required for normal adult eye development. The EGUF/hid method was used to generate eyes composed completely of homozygous lin-5293 mutant cells (top row, center). The resulting rough-eye phenotype was rescued by the presence of a lin-52+ transgene (top row, right). No abnormalities were observed when the EGUF method was used with a y w control (top row, left). The different eye colors were due to the dosage of mini-white+ contributed by different transgenes. Scanning electron microscopy (bottom row, two left panels) and toluidine-stained sections (bottom row, two right panels) were used to further characterize the lin-5293 mutant eye defects.
To test whether the lin-52 mutant eye phenotype is cell autonomous, we performed standard mosaic analysis using hs-FLP to create patches of homozygous mutant cells adjacent to viable heterozygous and homozygous wild-type cells (11) (Fig. 6). For these studies, we used the wild-type white+ gene as a dominant marker to indicate the presence of the wild-type lin-52+ allele (26). We were able to identify small mutant clones by examination under a dissecting microscope (not shown). The mosaic eyes were imbedded in plastic, sectioned, and examined by phase-contrast microscopy. Homozygous mutant lin-52 clones could be readily identified by the absence of pigment granules of both the photoreceptor cells (black dots near rhabdomeres) and the surrounding pigment cells. The pigment granules are produced in the presence of the wild-type white+ gene. The mutant ommatidia were disorganized, with rhabdomeres that were distended and/or absent. However, the adjacent lin-52+ cells appeared normal in both structure and organization. These results demonstrate that unlike several other genes encoding members of the MMB/dREAM complex, lin-52 is required for normal eye development.
Fig 6.
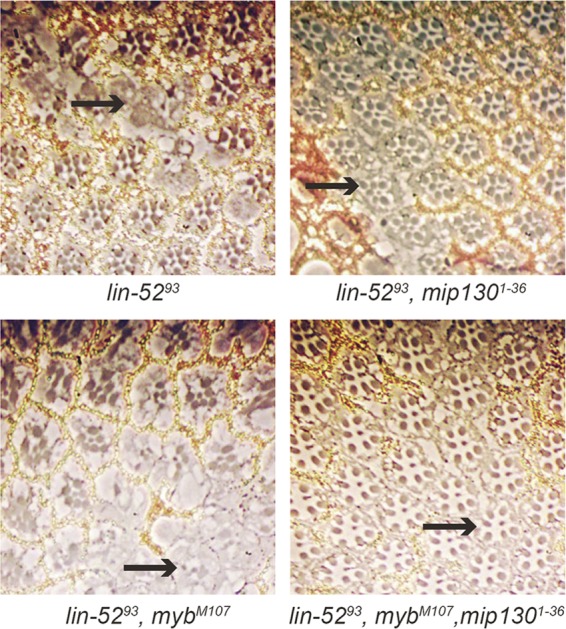
Adult eye defects caused by a lin-52 null mutant can be rescued by a mip130 null mutant. Mitotic recombination was induced via heat shock-FLP to produce mosaic eyes containing mutant clones that were homozygous for the indicated mutant combinations. Thick sections of adult eyes were then prepared and visualized by phase-contrast microscopy. The wild-type white+ gene is required for the formation of the pigment granules in both the photoreceptor and pigment cells. The pigment granules of the photoreceptor cells are marked by two black dots near the rhabdomeres. The pigment granules are absent (arrows) from homozygous mutant pigment cells and rhabdomeres.
Since the larval lethality of a lin-52 null mutant could be suppressed by the loss of mip40 (Fig. 2A and B), we wished to determine whether the adult eye phenotype that occurred in the absence of lin-52 could be modified by mutations in other MMB/dREAM complex subunits. The absence of either mip130 or myb does not cause an adult eye phenotype (2, 23). Standard mosaic analysis with hs-FLP as described above was used to generate clones mutant for both lin-52 and mip130, as evidenced by the absence of pigment (Fig. 6). Remarkably, these doubly mutant lin-52 mip130 clones appeared to be completely normal. In contrast, clones mutant for both lin-52 and myb appeared similar to clones mutant for lin-52 alone. However, triply mutant lin-52 myb mip130 clones once again appeared to be completely normal. These results indicate that the eye defects caused by the absence of lin-52 can be rescued by the loss of mip130 but not by the loss of myb.
Maternal lin-52 is required for early embryonic development.
Lin-52 protein was detected in both germ line cells (nurse cells and oocytes) and somatic cells (follicle cells) of the ovary (Fig. 1B). We therefore wished to test whether Lin-52 plays a role in oogenesis. Germ line cells and somatic cells that were homozygous for a lin-52 null mutation were generated with the FLP/FRT method using H2avGFP as a marker (see Fig. S1 in the supplemental material). Removal of lin-52 from the female germ line or somatic cells affected neither the morphology of those cells nor the progression of oogenesis.
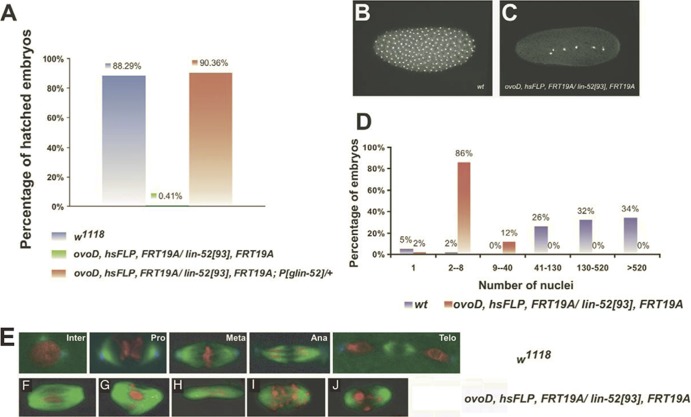
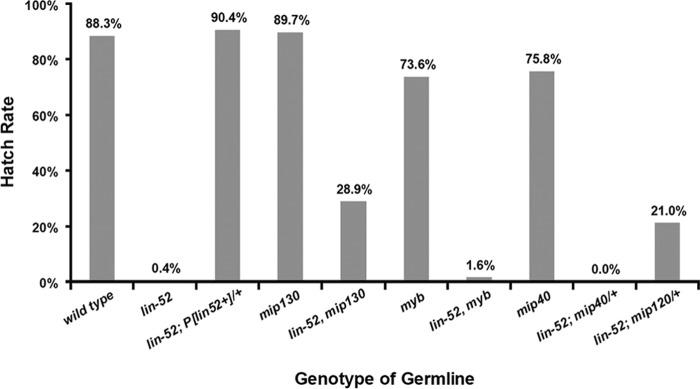
To ask if maternally contributed Lin-52 is essential for subsequent embryogenesis, an FLP/FRT-based dominant female-sterile technique was used to generate germ line clones that are derived exclusively from homozygous lin-5293 mutant cells. ovoD, a dominant mutation that causes female germ cells to arrest during early oogenesis, was used to eliminate both heterozygous mutant (lin-5293/+) and homozygous wild-type germ line cells (+/+). Therefore, all eggs laid by the heterozygous females arise from homozygous lin-52 mutant germ line clones. The heterozygous females producing lin-52 mutant germ line clones were then crossed to w1118 males to ensure that half of the progeny would carry one copy of a zygotically expressed wild-type lin-52+ gene, which is present on the X chromosome. Remarkably, less than 1% of the embryos arising from a lin-52 mutant female germ line developed into larvae, whereas nearly 90% of embryos arising from a lin-52 wild-type female germ line hatched (Fig. 7A). This result implies that the maternal contribution of lin-52 is essential for normal embryogenesis. Importantly, the maternal-effect lethal phenotype caused by the absence of lin-52 in the female germ line could be completely rescued by a lin-52+ transgene.
Fig 7.
Maternal lin-52 is required for early embryonic development. The dominant female-sterile method was used to generate female germ line clones that were homozygous for the lin-5293 null mutant in the absence or presence of a lin-52+ transgene. w1118 females were used as a control. Wild-type zygotic gene expression was provided by mating these females to w1118 males. (A) Hatch rate analysis of embryos derived from females of the indicated genotype. Depletion of lin-52 in the female germ line results in a maternal-effect lethal phenotype that can be rescued by a transgene carrying a lin-52 genomic rescue construct. (B) Embryos (60 to 90 min after egg laying) were stained with propidium iodide (PI) to detect nuclei. Eggs derived from lin-5293 females (C) exhibit a reduced number of nuclei compared to eggs from wild-type mothers (B). (D) Quantitation of the number of nuclei in lin-5293 or wild-type eggs. (E) Wild-type embryos (0 to 30 min after egg laying) showing different stages of mitosis. (F to J) Embryos derived from the lin-5293 mutant germ line (30 to 60 min after egg laying) displaying various mitotic defects. (F to H) Embryos were stained for DNA (PI, red), microtubules (alpha-tubulin, green), and a centrosome component (Cnn, blue). One or both centrosomes are often missing from mitotic spindles. (I and J) Embryos were stained for DNA (PI, red) and microtubules (alpha-tubulin, green) only. Additional defects include a broad spindle with no centrosomes (F), a branched spindle with no centrosomes (G), or an elongated spindle with no centrosomes (H). Abnormal chromosome distribution was often observed in mutant embryos (I and J).
Careful examination of the embryos produced in the absence of maternal lin-52 revealed a very early mitotic arrest (Fig. 7B, C, and D). Following fertilization, Drosophila embryos undergo 13 rounds of rapid, synchronous mitotic divisions without cytokinesis that are driven by maternally deposited products. These nuclear division cycles contain only S and M phases without any intervening gap phases. The vast majority (86%) of lin-52-deficient embryos collected for 30 min and then aged for 1 h arrested with only two to eight nuclei. In contrast, similarly aged wild-type embryos had continued to progress to contain 41 to 130 nuclei (26%), 130 to 520 nuclei (32%), or greater than 520 nuclei (34%). Immunostaining with anti-tubulin antibodies and counterstaining for DNA revealed the presence of various mitotic abnormalities in the lin-52-deficient embryos (Fig. 7E to J). In many cases the mitotic spindles and the metaphase plate failed to form properly, with condensed chromosomes often distributed diffusely between the spindle poles. These results are reminiscent of the mitotic defects observed in myb mutant somatic cells (9, 22, 23, 35). Mitotic defects have also been observed in Drosophila cell lines treated with RNAi against myb or mip130 (10, 12, 29). However, a similar analysis of germ line clones lacking mip130, mip40, or myb revealed relatively normal embryonic hatch rates (Fig. 8). These results imply that maternally deposited products of mip130, mip40, and myb are not essential for embryonic development. Because mip120 mutant females are sterile, it was not possible to perform hatch rate analysis (3). Since the larval lethality of a lin-52 null mutant could be suppressed by the loss of mip40 (Fig. 2A and B), and since the adult eye defects caused by a lin-52 null mutant could be suppressed by the loss of mip130 (Fig. 6), we wished to test whether the maternal-effect lethal phenotype of a lin-52 null mutant could be rescued by mutation of other members of the MMB/dREAM complex. Because lin-52, mip130, and myb are all located on the X chromosome, it was possible to create single chromosomes with pairs of mutants and an FRT19A FLP recognition site. Analyses of double-mutant germ line clones revealed that the absence of mip130 could partially rescue the maternal-effect lethal phenotype of the lin-52 mutant (Fig. 8). In contrast, the absence of myb did not modify the maternal-effect lethal phenotype of the lin-52 mutant.
Fig 8.
Maternal-effect lethal phenotype of a lin-52 mutant can be rescued by loss of mip130 or mip120. The dominant female-sterile method was used to generate homozygous lin-5293 mutant germ line clones in the presence or absence of a wild-type lin-52+ transgene or in the presence or absence of mutants of other genes encoding components of the MMB/dREAM complex. All mutants were homozygous unless the presence of a wild-type allele is indicated by a plus. Wild-type zygotic gene expression was provided by mating these females to w1118 males. Hatch rates were calculated as described in the legend to Fig. 7. The genotypes of the female flies used for hatch rate analyses are listed in Materials and Methods.
Because mip40 and mip120 are both located on the second chromosome, it was not possible to create double mutant combinations with lin-52 on a single chromosome. Therefore, we tested whether halving the maternal dose of either mip40 or mip120 could rescue the maternal-effect lethal phenotype of lin-52 mutant (Fig. 8). Strikingly, heterozygous deletion of mip120 was nearly as effective as the complete absence of mip130 in partially rescuing the maternal-effect lethal phenotype of the lin-52 mutant. In contrast, heterozygous deletion of mip40 did not modify the maternal-effect lethal phenotype of the lin-52 mutant. Taken together, these results imply that lin-52 is required in the female germ line for early embryogenesis. Furthermore, Lin-52 acts together with other members of the MMB/dREAM complex, including Mip130 and Mip120 in the female germ line and/or early embryo.
DISCUSSION
The primary objective of this study was to explore the function of Lin-52, the smallest subunit of the MMB/dREAM complex, which we had previously defined by biochemical purification (18). We generated a lin-52 null mutant and identified a regulatory interplay between different subunits of the MMB/dREAM complex. We showed that Lin-52 functions, in part, to inhibit the repressive activities of the other MMB/dREAM subunits, including Mip130, Mip120, and Mip40. Several phenotypes observed in lin-52 mutants (larval lethality, adult eye defects, and maternal-effect embryonic lethality) were suppressed by mutations in these core subunits of the MMB/dREAM complex. We had previously shown similar genetic interactions between myb mutants and the same core MMB/dREAM complex subunits (2, 3). However, lin-52 is essential for some biological processes that do not require myb, including embryonic development and adult eye development. Consistent with these observations in vivo, previous studies have shown that in cell culture the number of genes regulated by Lin-52 alone far exceeds the number of genes regulated by Myb alone (P < 0.001) or by both Lin-52 and Myb together (P < 0.001 by two-proportion z test) (see Fig. S2 in the supplemental material).
We hypothesize that in some contexts, Myb inhibits the repressive activities of Mip130, Mip120, and Mip40 and thereby aids in the activation of genes required for normal cell cycle progression and organismal development. This model, originally based on evidence from unexpected genetic interactions, was supported by subsequent RNAi studies in cultured cells and by gene expression studies in vivo (10, 17, 18, 35). Gene expression analysis after RNAi depletion in Drosophila Kc cells revealed a combinatorial requirement of MMB/dREAM subunits for either gene activation or repression. These studies showed that two-thirds of the genes repressed by MMB/dREAM did not require Myb protein, whereas most genes activated by MMB/dREAM required Myb protein (10).
The gene networks regulated by MMB/dREAM can be largely categorized into three main classes of repressed genes, namely, Myb-, E2F2-, or Mip120/Mip130-dependent repression, and a single major class of Myb-activated genes. Lin-52 is required for 76% of Myb-activated genes (for comparison, 56% of genes were dependent on Mip40), supporting a model in which Lin-52 plays an important role in gene activation. Our thinking is that regulation is hierarchical, such that specific loci may be repressed and then activated (or vice versa). The members of the MMB complex would help to establish one level of regulation at a primary stage of determination, while other cis-acting factors would determine the normal basal and induced levels of expression at another level of transcriptional control. The mechanistic basis of this process remains to be elucidated. However, it is clear that repression of many genes requires E2F2 and associated Rbf proteins and often the recruitment of L(3)MBT. Activation at this first level may require Myb, and as suggested here, Lin-52 may be key as an activator of transcription early in embryogenesis and in the developing eye.
Our finding that Drosophila lin-52 appears to counteract the activities of other MMB/dREAM complex subunits may extend to paralogous genes. The Drosophila gene wake-up-call (wuc) was recently identified as a lin-52 paralog that is exclusively expressed in testes (7). Wuc protein directly interacts with Aly, a Mip130 paralog that is also a subunit of the testis meiotic arrest complex (tMAC) (3, 37). Three subunits of tMAC are testis-specific paralogs of MMB/dREAM subunits: Aly (Mip130), Tomb (Mip120), and Wuc (Lin-52). The other two subunits of tMAC are also present in MMB/dREAM: Mip40 and p55/Caf1. The loss of tMAC function results in a decrease in testis-specific gene expression and meiotic arrest in spermatocytes (36). Interestingly, wuc mutants could partially rescue expression of several putative tMAC target genes in aly mutant testes (7). These data suggest that wuc opposes aly function in a manner analogous to the opposition of lin-52 and mip130 shown here.
Recent work in mammalian systems has demonstrated an important function for Lin-52 homologs in the DREAM complex. Phosphorylation of Lin-52 by the DYRK1A kinase was required for full assembly of the human DREAM complex (20, 34). These results indicate that Lin-52 plays an important role in mediating protein-protein interactions among DREAM complex subunits.
Unlike Drosophila, C. elegans lacking lin-52 is viable. However, the mutant worms are sterile (33). Genome-wide ChIP and gene expression analyses indicate that DRM directly represses 119 genes, primarily those involved in reproduction and development (32). Expression of these genes in specific developmental programs or tissues presumably requires the inhibition of the repressive DRM complex. C. elegans appears to have lost an animal-type myb gene during the course of evolution (19). It is not currently known if the DRM complex contains another transcription factor that may function to inhibit the repressive activities of the DRM complex (14). In the absence of a dedicated activator subunit, Lin-52 may serve in both the activation and repression of DRM-regulated genes. Perhaps phosphorylation by a DYRK1A-like kinase or some other posttranslational modification alters Lin-52 function in order to inactivate DRM activity. Additional studies will be required to determine whether such posttranslational modifications of Drosophila Lin-52 are required to regulate MMB/dREAM complex stability and function.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants R37CA030490 (to M.R.B.) and R01CA128836 (to J.S.L.).
We thank Mike Simon for helpful advice about mosaic analysis of adult eyes, Laura Andrejka for preparation of antibodies, and Lajja Mani for maintaining fly stocks.
Footnotes
Published ahead of print 11 June 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Andrejka L, et al. 2011. Animal-specific C-terminal domain links myeloblastosis oncoprotein (Myb) to an ancient repressor complex. Proc. Natl. Acad. Sci. U. S. A. 108:17438–17443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beall EL, Bell M, Georlette D, Botchan MR. 2004. Dm-myb mutant lethality in Drosophila is dependent upon mip130: positive and negative regulation of DNA replication. Genes Dev. 18:1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beall EL, et al. 2007. Discovery of tMAC: a Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B. Genes Dev. 21:904–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beall EL, et al. 2002. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420:833–837 [DOI] [PubMed] [Google Scholar]
- 5. Ceol CJ, Stegmeier F, Harrison MM, Horvitz HR. 2006. Identification and classification of genes that act antagonistically to let-60 Ras signaling in Caenorhabditis elegans vulval development. Genetics 173:709–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chou TB, Perrimon N. 1992. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics 131:643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doggett K, Jiang J, Aleti G, White-Cooper H. 2011. Wake-up-call, a lin-52 paralogue, and Always early, a lin-9 homologue physically interact, but have opposing functions in regulating testis-specific gene expression. Dev. Biol. 355:381–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferguson EL, Horvitz HR. 1989. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics 123:109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fung SM, Ramsay G, Katzen AL. 2002. Mutations in Drosophila myb lead to centrosome amplification and genomic instability. Development 129:347–359 [DOI] [PubMed] [Google Scholar]
- 10. Georlette D, et al. 2007. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes Dev. 21:2880–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Golic KG. 1991. Site-specific recombination between homologous chromosomes in Drosophila. Science 252:958–961 [DOI] [PubMed] [Google Scholar]
- 12. Goshima G, et al. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316:417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greenspan RJ. 1997. Fly pushing: the theory and practice of Drosophila genetics. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 14. Harrison MM, Ceol CJ, Lu X, Horvitz HR. 2006. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc. Natl. Acad. Sci. U. S. A. 103:16782–16787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hou DX, Akimaru H, Ishii S. 1997. Trans-activation by the Drosophila myb gene product requires a Drosophila homologue of CBP. FEBS Lett. 413:60–64 [DOI] [PubMed] [Google Scholar]
- 16. Jiang J, Benson E, Bausek N, Doggett K, White-Cooper H. 2007. Tombola, a tesmin/TSO1-family protein, regulates transcriptional activation in the Drosophila male germline and physically interacts with always early. Development 134:1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Korenjak M, et al. 2004. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119:181–193 [DOI] [PubMed] [Google Scholar]
- 18. Lewis PW, et al. 2004. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 18:2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lipsick JS. 2004. synMuv verite–Myb comes into focus. Genes Dev. 18:2837–2844 [DOI] [PubMed] [Google Scholar]
- 20. Litovchick L, Florens LA, Swanson SK, Washburn MP, DeCaprio JA. 2011. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 25:801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Litovchick L, et al. 2007. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell 26:539–551 [DOI] [PubMed] [Google Scholar]
- 22. Manak JR, Mitiku N, Lipsick JS. 2002. Mutation of the Drosophila homologue of the Myb protooncogene causes genomic instability. Proc. Natl. Acad. Sci. U. S. A. 99:7438–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manak JR, Wen H, Tran V, Andrejka L, Lipsick JS. 2007. Loss of Drosophila Myb interrupts the progression of chromosome condensation. Nat. Cell Biol. 9:581–587 [DOI] [PubMed] [Google Scholar]
- 24. Okada M, Akimaru H, Hou DX, Takahashi T, Ishii S. 2002. Myb controls G(2)/M progression by inducing cyclin B expression in the Drosophila eye imaginal disc. EMBO J. 21:675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pilkinton M, Sandoval R, Colamonici OR. 2007. Mammalian Mip/LIN-9 interacts with either the p107, p130/E2F4 repressor complex or B-Myb in a cell cycle-phase-dependent context distinct from the Drosophila dREAM complex. Oncogene 26:7535–7543 [DOI] [PubMed] [Google Scholar]
- 26. Ready DF, Hanson TE, Benzer S. 1976. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 53:217–240 [DOI] [PubMed] [Google Scholar]
- 27. Schmit F, et al. 2007. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle 6:1903–1913 [DOI] [PubMed] [Google Scholar]
- 28. Simon MA, Bowtell DDL, Dodson GS, Laverty TR, Rubin GM. 1991. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell 67:701–716 [DOI] [PubMed] [Google Scholar]
- 29. Somma MP, et al. 2008. Identification of Drosophila mitotic genes by combining co-expression analysis and RNA interference. PLoS Genet. 4:e1000126 doi:10.1371/journal.pgen.1000126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stevaux O, et al. 2002. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J. 21:4927–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stowers RS, Schwarz TL. 1999. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152:1631–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tabuchi TM, et al. 2011. Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex. PLoS Genet. 7:e1002074 doi:10.1371/journal.pgen.1002074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thomas JH, Ceol CJ, Schwartz HT, Horvitz HR. 2003. New genes that interact with lin-35 Rb to negatively regulate the let-60 ras pathway in Caenorhabditis elegans. Genetics 164:135–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tschop K, et al. 2011. A kinase shRNA screen links LATS2 and the pRB tumor suppressor. Genes Dev. 25:814–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wen H, Andrejka L, Ashton J, Karess R, Lipsick JS. 2008. Epigenetic regulation of gene expression by Drosophila Myb and E2F2-RBF via the Myb-MuvB/dREAM complex. Genes Dev. 22:601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. White-Cooper H. 2010. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction 139:11–21 [DOI] [PubMed] [Google Scholar]
- 37. White-Cooper H, Leroy D, MacQueen A, Fuller MT. 2000. Transcription of meiotic cell cycle and terminal differentiation genes depends on a conserved chromatin associated protein, whose nuclear localisation is regulated. Development 127:5463–5473 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.