Abstract
Sphingomyelin synthase (SMS) catalyzes the formation of sphingomyelin, a major component of the plasma membrane and lipid rafts. To investigate the role of SMS in cell signaling and migration induced by binding of the chemokine CXCL12 to CXCR4, we used mouse embryonic fibroblasts deficient in SMS1 and/or SMS2 and examined the effects of SMS deficiency on cell migration. SMS deficiency promoted cell migration through a CXCL12/CXCR4-dependent signaling pathway involving extracellular signal-regulated kinase (ERK) activation. In addition, SMS1/SMS2 double-knockout cells had heightened sensitivity to CXCL12, which was significantly suppressed upon transfection with the SMS1 or SMS2 gene or when they were treated with exogenous sphingomyelin but not when they were treated with the SMS substrate ceramide. Notably, SMS deficiency facilitated relocalization of CXCR4 to lipid rafts, which form platforms for the regulation and transduction of receptor-mediated signaling. Furthermore, we found that SMS deficiency potentiated CXCR4 dimerization, which is required for signal transduction. This dimerization was significantly repressed by sphingomyelin treatment. Collectively, our data indicate that SMS-derived sphingomyelin lowers responsiveness to CXCL12, thereby reducing migration induced by this chemokine. Our findings provide the first direct evidence for an involvement of SMS-generated sphingomyelin in the regulation of cell migration.
INTRODUCTION
Sphingomyelin synthase (SMS) is an enzyme involved in sphingomyelin (SM) biosynthesis that transfers the phosphorylcholine moiety from phosphatidylcholine onto the primary hydroxyl of ceramide, producing sphingomyelin and diacylglycerol (20, 61). There are two isoforms of mammalian SMS (SMS1 and SMS2), both of which are predicted to have six transmembrane domains with an active site. The regulation of SMS activity has been proposed to determine cellular levels of ceramide, diacylglycerol, and sphingomyelin (13, 20, 54, 55, 61, 64). Ceramide is a bioactive lipid that plays a role in cell death, proliferation, and differentiation (17, 42), whereas diacylglycerol activates protein kinase C and promotes cell survival and proliferation (16). Sphingomyelin is a major component of the plasma membrane and lipid rafts, and we very recently uncovered that SMS1-generated sphingomyelin plays an important role in transferrin trafficking (48). However, the role of SMS in cellular function still remains poorly understood.
Lipid rafts are membrane microdomains where glycosphingolipids, such as GM1 and sphingomyelin, are enriched and held together mainly by hydrophobic interactions. Lipid rafts are biochemically characterized by resistance to cold detergent lysis (8). They have been proposed to function as platforms, participating in the sorting of receptors, such as G protein-coupled receptors (GPCRs) and tyrosine kinase-coupled receptors, and in the regulation of receptor-mediated signal transduction (33, 51).
GPCRs mediate cell migration toward a concentration gradient of the cognate chemokine ligand (32). The chemokine CXCL12 binds and signals through a limited number of GPCRs, including CXCR4 and CXCR7 (5, 19). Signaling through the CXCL12 (SDF1)-CXCR4 pathway is essential for homing of hematopoietic stem cells to the bone marrow and for the survival of vascular endothelial cells. It is also involved in the migration and metastasis of tumor cells (9, 36, 37, 53). CXCR4 forms a complex with CCR2, CCR5, or CXCR7 (21, 41, 49). CXCL12 treatment induces the formation of CXCR4 homodimers, thereby promoting cell migration (4, 56). The formation of homodimers can be inhibited by cholesterol depletion, which disrupts lipid rafts (58). Because CXCR4 is partially incorporated into lipid rafts after stimulation with CXCL12, lipid rafts have been proposed to play a key role in CXCL12/CXCR4 signaling (39).
In this paper, we examined the roles of SMS and sphingomyelin in the regulation of cell migration. We employed mouse embryonic fibroblasts (MEFs) from SMS knockout (KO) mice to assess the effects of SMS and sphingomyelin deficiency on cell migration mediated by the CXCL12/CXCR4 pathway. Furthermore, we examined how SMS and sphingomyelin affect CXCR4 activation in these cells.
MATERIALS AND METHODS
Antibodies and reagents.
AMD3100 octahydrochloride hydrate (sc-252367), fusin small interfering RNA (siRNA; sc-35422), and antibodies specific to extracellular signal-regulated kinase 2 (ERK2; C-14), actin (I-19; sc-1616) and caveolin-1 (N-20; sc-894) were from Santa Cruz Biotechnology. Anti-active ERK polyclonal antibody (V8031) was from Promega. Anti-CXCR4 polyclonal (ab2074) and anti-alpha 1 sodium potassium ATPase monoclonal (ab7671) antibodies were from Abcam (United Kingdom). Anti-maltose binding protein (anti-MBP; 05-912) antibody was from Upstate. Anti-flotillin-1 monoclonal antibody (610820) was from BD Transduction Laboratories. Allophycocyanin (APC)-conjugated anti-CXCR4 antibody (247506) was from R&D Systems. Alexa Fluor 488-conjugated goat anti-mouse IgM (A-21042) and anti-rabbit IgG(H+L) (A-11008) antibodies were from Invitrogen. Peroxidase-conjugated donkey anti-mouse IgG(H+L) and rabbit IgG(H+L) antibodies were from Jackson ImmunoResearch. Nuclei were visualized using 4′,6′-diamidino-2-phenylindole (DAPI; Invitrogen). Lysenin with MBP was kindly provided by T. Kobayashi (Riken, Japan). C6-ceramide (1900) and C6-NBD-ceramide (1841; 6{N-[(7-nitrobenzo-2-oxa-1,3-diazol-4-yl)amino]caproylsphingosine}) were from Matreya. C6-sphingomyelin (860582) was from Avanti Polar Lipids. Recombinant mouse CXCL12 (460-SD) and 125I-CXCL12 (NEX346) were from R&D Systems and from PerkinElmer Life and Analytical Sciences, respectively. Recombinant murine epidermal growth factor (EGF; 315-09) and recombinant murine platelet-derived growth factor (PDGF)-BB (315-18) were from Peprotech. U-0126 (70970) was from Cayman Chemical.
Plasmid construction.
Human CXCR4 cDNA was obtained from HEK cell cDNA. pCAGGS-CFP (where CFP is cyan fluorescent protein) and pCAGGS-Venus vectors were kindly provided by M. Matsuda and E. Kiyokawa (Kyoto University, Japan). CFP- and Venus-C-terminally tagged CXCR4 cDNAs were cloned into pCAGGS vectors (35).
Cell culture and transfection.
MEFs were isolated from mice in which the SMS1 and/or the SMS2 gene was disrupted (34, 62). These SMS KO MEFs were immortalized by transfecting the simian virus 40 (SV40) large T antigen. Wild-type and SMS KO MEFs were cultured in RPMI 1640 medium (Sigma-Aldrich) containing 10% fetal bovine serum at 34°C in 5% CO2. Cells were transfected with plasmid constructs or siRNAs using Lipofectamine 2000 (Invitrogen), and after incubation for 24 to 48 h, experiments were performed as described below.
SMS activity.
For the in vitro SMS activity assay, cells were homogenized in ice-cold buffer (20 mM Tris-HCl, pH 7.4, 2 mM EDTA, 10 mM EGTA, 1 mM phenylmethylsulfonyl fluoride, 2.5 μg/ml leupeptin, and 2.5 μg/ml aprotinin), and 70 μg of total protein was mixed with the reaction solution (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 20 μM C6-NBD-ceramide, 120 μM phosphatidylcholine) and incubated for 90 min at 37°C. Lipids were extracted using the Bligh and Dyer method (7), applied onto thin-layer chromatography (TLC) plates, and developed with a solvent consisting of chloroform–methanol–12 mM MgCl2 (65:25:4, vol/vol/vol). The fluorescent lipids were visualized using LAS-1000 plus (Fujifilm, Japan) and quantified using Multi Gauge 3.1 (Fujifilm). For the in vivo SMS activity assay, cells were incubated with 4 μM C6-NBD-ceramide for 60 min at 37°C and washed twice with ice-cold phosphate-buffered saline (PBS). Lipids were extracted and quantified as for the in vitro SMS activity assay.
HPLC-MS/MS analysis.
Measurement of sphingomyelin in lipid extracts was performed using high-performance liquid chromatography–tandem mass spectrometry (HPLC-MS/MS) (34). The quantity of both sphingomyelin species (d18:1, 16:0 and d18:1, 24:1) were determined.
Electrophoresis and Western blotting.
Cells were homogenized in SDS sample buffer (40 mM Tris-HCl, pH 6.8, 50 mM dithiothreitol, 1% SDS, 4.5% glycerol, and bromophenol blue), and the cell lysates were subjected to SDS-PAGE. Proteins were transferred onto a nitrocellulose membrane (0.45-μm pore size; Bio-Rad) using a transfer buffer (40 mM glycine, 48 mM Tris base). Membranes were blocked with 5% skim milk dissolved in PBS and incubated with primary antibody overnight at 4°C. After four washes with PBS containing 0.02% Tween 20, membranes were incubated with secondary antibody conjugated to peroxidase, washed four times with PBS, and developed with enhanced chemiluminescence (ECL) Western blot detection reagents (Amersham, United Kingdom) or with SuperSignal West Dura extended duration substrate (Thermo Fisher Scientific).
Quantitative real-time PCR.
A 5-μg sample of total RNA, isolated using an RNeasy minikit (Qiagen, Germany), was used in reverse transcription reactions using a cDNA synthesis kit (TaKaRa Shuzo, Japan). Real-time PCR was performed using an Applied Biosystems 7300 real-time PCR system, according to the standard TaqMan PCR kit protocol.
Transwell migration assay.
The chemotaxis assay was performed using a modified transwell migration assay (8-μm pore size; BD Falcon). The lower chamber was filled with 700 μl of chemoattractant in serum-free culture medium. After cells were harvested with trypsin-EDTA, 2 × 105 cells were resuspended in 250 μl of serum-free culture medium and transferred to the upper chamber. The cells were allowed to migrate for 6 to 8 h at 34°C. Nonmigrating cells on the upper surface of the membrane were scraped off, and migratory cells attached to the lower surface were stained with DAPI and counted.
Two-dimensional chemotaxis assay.
Cell migration was also examined using a μ-Slide chemotaxis assay (Ibidi, Germany) and was performed according to the manufacturer's instructions. Briefly, a suspension of MEFs (7 μl of 3 × 106 cells/ml) was seeded into the center of a μ-Slide chamber and incubated for 3 h. After cells were washed, they were stimulated with a medium containing 2.5 nM CXCL12. Cell movement was observed every 30 min over a period of 12 h by live-cell imaging in a cell observation chamber mounted to a Biozero fluorescence microscope (Keyence, Japan) with a Plan Apo 20× (0.75-numerical-aperture [NA]) dry lens. Images were acquired using the Biozero application software (Keyence, Japan). Cell tracking analysis was performed on the images using the manual tracking plug-in (Fabrice Cordelieres, France) and the chemotaxis and migration tool (Ibidi) for ImageJ, version 1.43u (National Institutes of Health), as described in the μ-Slide chemotaxis protocol.
Fractionation by discontinuous OptiPrep step gradient.
Membrane fractionation was performed using an OptiPrep gradient by Axis-Shield (Axis-Shield, Norway). Briefly, cells were washed once with PBS to remove the culture medium, harvested with a scraper, and resuspended in homogenization buffer (0.25 M sucrose, 20 mM KCl, 1 mM EDTA, 20 mM HEPES-KOH, pH 7.6, protease inhibitor cocktail). Cells were lysed by passage 20 times through a fine syringe needle. The homogenates were centrifuged at 1,000 × g for 5 min to obtain a postnuclear supernatant. The cell lysates were loaded onto an OptiPrep gradient consisting of 5, 10, 15, and 20% (wt/vol) iodixanol solutions. Centrifugation was performed on a Beckman SW 41Ti rotor at 100,000 × g for 16 h at 4°C. Twenty fractions were collected from the top of each centrifuge tube. Each fraction was analyzed by SDS-PAGE and Western blotting. Sphingomyelin in fraction 2 (the sodium potassium ATPase-enriched plasma membrane fraction) was measured by HPLC-MS/MS as described above.
Detergent-based fractionation.
Cells were harvested and lysed with buffer (1% Brij 58, 10 mM Tris buffer, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 mM Na3VO4, and protease inhibitor cocktail). The lysates were centrifuged for 5 min at 1,000 × g. The supernatant was adjusted to 40% sucrose and placed into the bottom of an OptiPrep gradient consisting of 5 and 30% (wt/vol) iodixanol solutions. The gradients were centrifuged at 100,000 × g in a Beckman SW41 Ti rotor for 16 h at 4°C. Fractions (1 ml/fraction) were collected and used for HPLC-MS/MS analysis and Western blotting.
Immunofluorescence.
Cells on coverslips were fixed with 3.7% paraformaldehyde-PBS for 15 min. When permeabilization was required, cells were treated with 0.2% Triton X-100 for 4 min. After samples were washed four times with PBS, coverslips were blocked with 1% bovine serum albumin (BSA)-PBS for 15 min and incubated with the primary antibody for 1 h. For lysenin staining, the coverslips were incubated with MBP-lysenin for 1 h before the addition of the primary antibody. After cells were washed four times with PBS, they were incubated with Alexa Fluor-conjugated secondary antibody for 30 min. Subsequently, cells were washed four times with PBS and mounted on a microscope slide with Fluoromount (Diagnostic BioSystems) to preserve fluorescence. Specimens were observed with a Leica TCS SP2 confocal microscope using a HCX PL APO 63×/1.32 NA oil Ph3 lens. Images were acquired using Leica Confocal Software (LCS).
FRET sensitized emission.
After cells were cotransfected with CXCR4-CFP and CXCR4-Venus vectors for 24 h, they were imaged with a Leica TCS SP2 confocal microscope equipped with a thermally regulated stage that allowed us to obtain time-lapse images at 37°C. Image processing was controlled with a fluorescence resonance energy transfer (FRET) sensitized-emission application (LCS 2.611537) following the manufacturer's instructions. After background subtraction, the FRET/CFP ratio was determined using the software, and subsequently the FRET/CFP ratios of the plasma membrane regions in these images were analyzed with ImageJ, version 1.43u, software.
Flow cytometry analysis.
Cells were cultured in 10-cm dishes to 30% confluence in normal culture medium and harvested with 1.37 mM EDTA. Cells were then washed with PBS and incubated with APC-conjugated anti-CXCR4 for 30 min at 4°C. After cells were washed with PBS, the fluorescence intensity was determined by flow cytometry using a MoFlo XDP Cell Sorter (Beckman Coulter).
Ligand-receptor binding assay.
CXCR4-transfected MEFs (1 × 105 cells) were resuspended and incubated in 50 μl of binding buffer (0.5% BSA in PBS) with [125I]CXCL12 for 30 min on ice. After cells were washed five times with binding buffer, they were lysed with the buffer (0.1 M NaOH, 0.1% Triton X-100), and cell-associated CXCL12 was measured by counting radioactivity (1470 Wizard; Perkin Elmer).
RESULTS
Biochemical characterization of immortalized MEFs from SMS knockout mice.
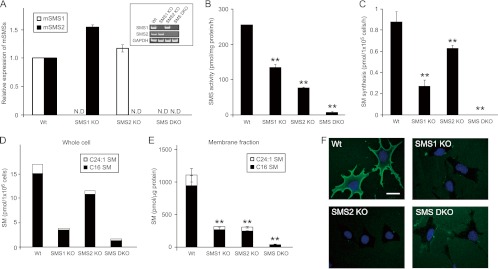
To examine the role of SMS in cell migration, we employed SMS-deficient MEFs. As shown in Fig. 1A, SMS mRNA was undetectable in the SMS KO MEFs. Cellular sphingomyelin content was determined by HPLC-MS/MS. Loss of the SMS gene led to a deficiency of sphingomyelin (Fig. 1D) that paralleled the decrease in SMS activity in vitro (Fig. 1B) and in vivo (Fig. 1C). Sphingomyelin is a major sphingolipid of lipid microdomains in the plasma membrane. We measured sphingomyelin content in the lipids extracted from the sodium/potassium ATPase-enriched plasma membrane fraction. Plasma membrane sphingomyelin was significantly reduced by SMS1 or SMS2 KO, and cells with the SMS1/SMS2 double-KO (SMS DKO) exhibited a 96.4% decrease in plasma membrane sphingomyelin (Fig. 1E). Cell surface sphingomyelin was also analyzed by staining cells with lysenin (60), which specifically binds to sphingomyelin. Cell surface lysenin staining was decreased in SMS1 KO, SMS2 KO, and SMS DKO MEFs compared to levels in wild-type MEFs (Fig. 1F). Thus, SMS deficiency leads to a reduction in sphingomyelin biosynthesis in MEFs.
Fig 1.
Biochemical characterization of SMS knockout MEFs. (A) SMS1 and SMS2 mRNA expression levels in SMS knockout MEFs. mRNA expression was measured by RT-PCR (inset, GAPDH was used as an equal loading marker) and real-time PCR. The real-time PCR was performed for both the target gene and β-actin. Expression was calculated after values were normalized against β-actin in each sample and is presented as relative mRNA expression. Values are means ± standard deviations (n = 3). (B) In vitro SMS activity of SMS knockout MEFs. The lysates from 2.3 × 103 cells (70 μg protein) were added to the reaction mix and incubated at 37°C for 90 min. The extracted lipids were quantified using a Fluoro-Image Analyzer. Values are means ± standard deviations (n = 3). **, P < 0.01. (C) In vivo SMS activity in SMS knockout MEFs. Cells were incubated with 4 μM C6-NBD-ceramide for 60 min at 37°C, and then lipids were extracted and quantified using a Fluoro-Image Analyzer. Values are means ± standard deviations (n = 3). **, P < 0.01. (D) Sphingomyelin level in SMS knockout MEFs. Cells (3 × 105 cells) were harvested and dissolved. The lipids were extracted, and both sphingomyelin species (d18:1, 16:0 and d18:1, 24:1) were analyzed by HPLC-MS/MS to determine quantity. (E) The amount of sphingomyelin in the plasma membrane fraction. The sodium/potassium ATPase-enriched fraction was obtained from the MEF lysate by discontinuous OptiPrep step gradient, and the lipids were extracted from a sample containing 100 ng of protein and analyzed by HPLC-MS/MS. Values are means ± standard deviations (n = 3). **, P < 0.01. (F) MBP-lysenin staining of the plasma membrane in MEFs. MBP-lysenin staining was performed to detect plasma membrane sphingomyelin. Scale bar, 10 μm. Wt, wild type; ND, not detected.
SMS regulates CXCL12/CXCR4-mediated cell migration.
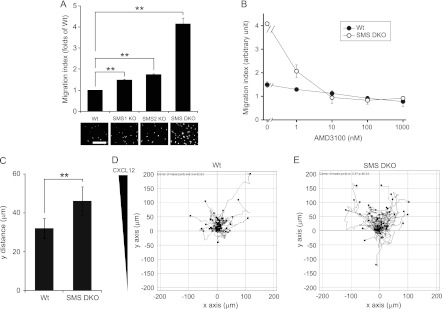
To assess whether SMS is involved in regulating cell migration, we employed a transwell chamber assay (Fig. 2A and B) and a μ-Slide chemotaxis assay (Fig. 2C to E). CXCL12 treatment stimulated migration of SMS1 KO, SMS2 KO, and SMS DKO MEFs to a significantly greater extent than wild-type MEFs, with the last of these displaying the highest motility (Fig. 2A). Similar results were obtained with 10% fetal bovine serum (Table 1). CXCL12 is known to bind to CXCR4 and CXCR7 (5, 10). To determine the target receptors of CXCL12, the transwell migration assay was performed in the presence of the CXCR4 antagonist, AMD3100. CXCL12-induced cell migration was inhibited by AMD3100 in a dose-dependent manner, and 10 nM AMD3100 completely inhibited cell migration (Fig. 2B). These results suggest that CXCR4 specifically mediates cell migration induced by CXCL12 in this assay.
Fig 2.
SMS deficiency promotes CXCL12-induced cell migration. (A) MEF migration was measured using a transwell migration assay. CXCL12 (0.25 nM) was added to the lower chamber of the vessel. Cells were added to the upper chamber and incubated for 6 h at 34°C. Migrating cells that had moved to the far side of the upper chamber were stained with DAPI and counted. The number of cells migrating in response to the ligand was divided by that of unstimulated cells for each genotype. The migration index represents the relative value of each genotype to the wild type. Values are means ± standard deviations (n = 3). **, P < 0.01. Scale bar, 200 μm. (B) Cell migration was measured using a transwell migration assay. AMD3100 at the indicated concentration was added to both chambers of the vessel, and 2.5 nM CXCL12 was added to the lower chamber. Cells were then stimulated for 6 h with CXCL12. The migration index was defined as migration relative to that of cells not stimulated with CXCL12 (which was set at 1). Values are means ± standard deviations (n = 3). (C to E) MEF migration was also measured with a two-dimensional chemotaxis assay using a μ-Slide chemotaxis assay. MEFs were seeded onto the μ-Slide, stimulated with 2.5 nM CXCL12, and then monitored by microscopy every 30 min for 12 h. The migration distance along the y pathway was determined and defined as the y distance (C). Values are means ± standard deviations (n = 50). **, P < 0.01. Track plots of wild type (D) and SMS double-knockout MEFs (E) were also analyzed and are shown in the graphs. The starting point of each individual cell is located in the center of the diagram.
Table 1.
Chemotaxis of MEFs induced by fetal bovine serum and growth factors
| Cell type | Migration with:a |
||
|---|---|---|---|
| 10% FBS (P < 0.05) | 50 ng/ml EGF (P > 0.05) | 50 ng/ml PDGF (P > 0.05) | |
| Wild type | 1 | 1 | 1 |
| SMS1 KO | 4.19 ± 0.14 | 1.02 ± 0.07 | 0.91 ± 0.33 |
| SMS2 KO | 5.81 ± 0.08 | 1.01 ± 0.22 | 0.79 ± 0.11 |
| SMS DKO | 8.48 ± 2.43 | 0.98 ± 0.35 | 1.35 ± 0.15 |
MEF migration was stimulated by 10% FBS (fetal bovine serum), 50 ng/ml EGF, and 50 ng/ml PDGF and measured using a transwell migration assay. Values are means ± standard deviations and are relative to wild-type migration, which was set at 1. P values in parentheses were calculated from comparison between wild-type MEFs and SMS KO MEFs based on original data (Dunn's multiple comparison test). P values with Kruskal-Wallis tests were as follows: 10% FBS, P < 0.05; 50 ng/ml EGF, P > 0.05; and 50 ng/ml PDGF, P > 0.05 (n = 3).
The CXCL12/CXCR4 pathway is known to potently induce chemotaxis. We further tested the effects of SMS deficiency on CXCL12/CXCR4 pathway-dependent chemotaxis using the μ-Slide assay. Similar to the transwell migration assay, SMS-deficient cells (SMS DKO cells) displayed significant enhancement of chemotaxis compared to wild-type cells (Fig. 2C). Figure 2D and E show the track plots of 50 cells for each genotype (wild type and SMS DKO). These results indicate that SMS deficiency enhances chemotaxis via the CXCL12/CXCR4 pathway.
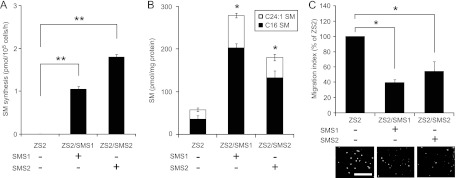
We further examined whether introducing the SMS gene into SMS DKO MEFs could block CXCL12/CXCR4-dependent cell migration. SMS DKO MEFs were transfected with either the mouse SMS1 or SMS2 gene to establish the stably expressing cell lines named ZS2/SMS1 and ZS2/SMS2, respectively. Gain of SMS gene function led to an increase in SMS activity (Fig. 3A), thereby elevating sphingomyelin levels (Fig. 3B). As shown in Fig. 3C, overexpression of either SMS1 or SMS2 significantly suppressed migration of SMS DKO MEFs (Fig. 3C). These results suggest that SMS activity has an inhibitory effect on cell migration and chemotaxis mediated by CXCL12/CXCR4 signaling.
Fig 3.
Overexpression of the SMS1 or SMS2 gene suppresses CXCL12-induced cell migration. (A) In vivo SMS activity in ZS2 MEFs. SMS double-knockout MEFs stably expressing either empty vector (ZS2 cells), mouse SMS1 (ZS2/SMS1 cells) or mouse SMS2 (ZS2/SMS2 cells) were generated. After being cultured for 24 h at 34°C, 1 × 106 cells were incubated with 4 μM C6-NBD-ceramide for 60 min at 37°C, and the lipids were extracted. Quantitation of the lipids was performed using a Fluoro-Image Analyzer. Values are means ± standard deviations (n = 3). **, P < 0.01. (B) Sphingomyelin level in ZS2 MEFs. Cells (3 × 105 cells) were harvested and dissolved. The lipids were extracted, and both sphingomyelin species (d18:1, 16:0 and d18:1, 24:1) were analyzed by HPLC-MS/MS to determine the quantity. Values are means ± standard deviations (n = 3). *, P < 0.05. (C) Cell migration was measured using a transwell migration assay. CXCL12 (0.25 nM) was added to the lower chamber of the vessel. Cells were added to the upper chamber and incubated for 6 h at 34°C. The migration index is represented as relative migration with that of ZS2 set at 100%. Values are means ± standard deviations (n = 3). *, P < 0.05 (Dunn's multiple comparison test using original data) and P < 0.05 (Kruskal-Wallis test).
Exogenous sphingomyelin inhibits CXCL12-induced cell migration.
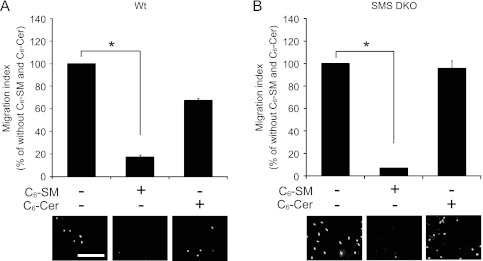
SMS catalyzes the synthesis of sphingomyelin from ceramide and phosphatidylcholine. Consequently, its inactivation should decrease sphingomyelin content while increasing ceramide levels. To identify lipids that influence cell migration, the effects of exogenous sphingolipids on CXCL12/CXCR4-induced cell migration were examined. Initially, we assessed the impact of various bioactive lipids on cell viability. Treatment of MEFs with 6 μM C6-sphingomyelin or C6-ceramide for 8 h had no effect on cell viability (data not shown). Notably, C6-sphingomyelin treatment significantly inhibited CXCL12-induced cell migration in both wild-type (Fig. 4A) and SMS DKO MEFs (Fig. 4B). In contrast, C6-ceramide treatment had no effect on cell migration in SMS DKO MEFs although it slightly increased migration of wild-type MEFs (Fig. 4). As wild-type MEFs, but not SMS DKO MEFs, have the ability to convert ceramide into sphingomyelin (Fig. 1C), it is likely that a portion of the C6-ceramide (which does not directly influence migration) incorporated into wild-type MEFs was metabolized to C6-sphingomyelin (which inhibits cell migration). These results suggest that sphingomyelin suppresses migration toward chemoattractants.
Fig 4.
Exogenous sphingomyelin, but not ceramide, inhibits CXCL12-induced cell migration. Cell migration was measured using a transwell migration assay. Wild-type (A) and SMS double-knockout (B) MEFs were incubated for 30 min with or without 6 μM C6-sphingomyelin (C6-SM) or 6 μM C6-ceramide (C6-Cer), which was added to both chambers of the vessel used to measure cell migration. They were then stimulated for 8 h with 0.25 nM CXCL12. Cell motility is represented as a migration index, defined as migration relative to that of cells not treated with C6-sphingomyelin or C6-ceramide (which was set at 100%). Values are means ± standard deviations (n = 3). *, P < 0.05 (Dunn's multiple comparison test using original data) and P < 0.05 (Kruskal-Wallis test).
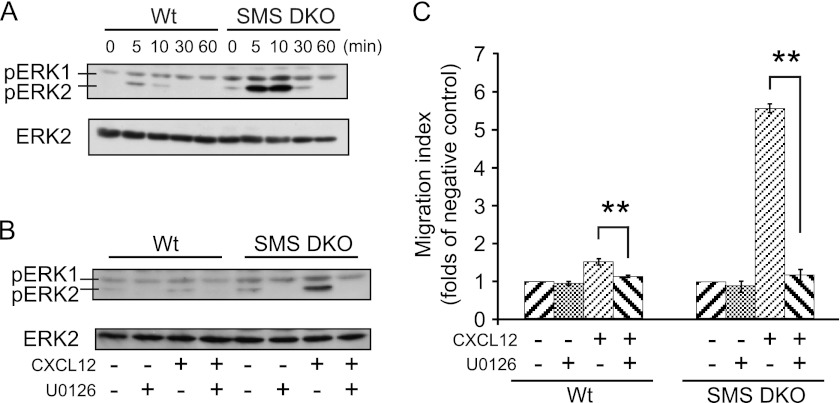
SMS deficiency promotes ERK phosphorylation/activation in CXCL12-stimulated cells.
To assess the effects of SMS deficiency on the ERK1 and ERK2 (ERK1/2) signaling pathway, which regulates cell migration (9, 36, 37, 53), we investigated ERK1/2 phosphorylation/activation. ERK1/2 phosphorylation was induced by CXCL12 treatment in wild-type MEFs, and phosphorylation was stimulated to a significantly greater extent in SMS DKO MEFs than in wild-type MEFs (Fig. 5A). Interestingly, ERK1/2 activation by the CXCL12/CXCR4 pathway was sustained for a longer period (up to 30 min) than in the wild type (10 min). The upregulation of ERK1/2 phosphorylation in SMS DKO MEFs was blocked by C6-sphingomyelin (data not shown). We also showed that treatment with the ERK kinase (MEK) inhibitor U0126 strongly inhibited CXCL12-induced ERK phosphorylation (Fig. 5B) and cell migration (Fig. 5C), confirming ERK-dependent cell migration in both wild-type and SMS DKO MEFs.
Fig 5.
SMS deficiency increases CXCL12-induced cell migration through MEK/ERK activation. (A) Cells were starved for 3 h and then stimulated with 1.25 nM CXCL12 for the indicated periods of time. Cell lysates (1 × 106 cells) were analyzed by Western blotting to detect phospho-ERK1/2 (pERK1 and pERK2). Similar results were obtained in three independent experiments. (B) Cells were starved for 3 h and then stimulated with 1.25 nM CXCL12 for 10 min in the presence or absence of 10 μM U0126. Cell lysates (1 × 106 cells) were analyzed by Western blotting to detect phospho-ERK1/2. Similar results were obtained in three independent experiments. (C) Cells were incubated for 30 min with 10 μM U0126, which was added to the upper chamber of the vessel used to measure cell migration. They were then stimulated for 8 h with 1.25 nM CXCL12. The migration of the cells was measured as a migration index. Values are means ± standard deviations (n = 3). **, P < 0.01.
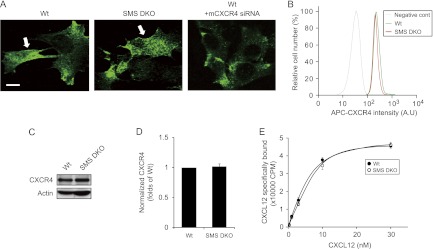
SMS deficiency does not affect CXCR4 expression, the binding affinity of CXCL12 for CXCR4, or receptor internalization.
Plasma membrane sphingomyelin was significantly decreased in SMS DKO MEFs (Fig. 1E and F). Therefore, we investigated if SMS-derived sphingomyelin affects the plasma membrane localization of CXCR4. CXCR4 was found on the plasma membrane and the cytoplasm in both SMS DKO and wild-type MEF lines as shown by immunocytochemistry using the CXCR4 antibody, whose specificity was confirmed by siRNA-mediated knockdown of CXCR4 expression in wild-type MEFs (Fig. 6A). Wild-type and SMS DKO MEFs displayed no difference in CXCR4 localization (Fig. 6A). Moreover, cell surface and whole-cell expression of CXCR4, as determined by flow cytometry (Fig. 6B) and Western blotting (Fig. 6C and D), were not altered in these MEF lines.
Fig 6.
SMS deficiency has no effect on CXCR4 expression and its binding affinity to CXCL12. (A) CXCR4 localization in MEFs. Cells were fixed with 3.7% paraformaldehyde and processed for confocal microscopy to detect CXCR4. The arrows indicate its membrane localization. Scale bar, 20 μm. (B) The amount of cell surface CXCR4 in MEFs. Cells were harvested with EDTA solution, incubated with APC-conjugated CXCR4 antibody, and analyzed by flow cytometry. (C) The total amount of CXCR4 in MEFs. Whole-cell lysates were processed for Western blotting using CXCR4 antibody. Equivalent amounts of protein were loaded onto each well (see actin staining). (D) The density of CXCR4 bands shown in panel C was calculated, normalized to that of the wild-type MEFs, and represented as normalized CXCR4. Values are means ± standard deviations (n = 6). (E) CXCR4-transfected MEFs were incubated with 0 to 30 nM [125I]CXCL12 for 30 min on ice. Cell-associated CXCL12 binding was measured by counting radioactivity. Specific binding was determined by subtracting the nonspecific binding from the total binding. Nonspecific binding was determined by adding a 100-fold excess of unlabeled CXCL12. AU, arbitrary units.
To assess the effects of SMS-derived sphingomyelin on the CXCL12-CXCR4 interaction, we examined the affinity for CXCL12 binding using a ligand-receptor binding assay. The dissociation constant was 5.1 nM in both MEF lines (Fig. 6E), ruling out significant effects of SMS deficiency on the molecular interaction between CXCL12 and CXCR4. These results suggest that SMS-derived sphingomyelin targets downstream of CXCL12/CXCR4 binding.
Ligand binding to many plasma membrane receptors leads to internalization and receptor desensitization, a key process that prevents potentially detrimental sustained receptor signaling. Lagane et al. reported that a mutant CXCR4 lacking the β-arrestin binding site, which prevents internalization, was constitutively expressed on the cell surface, enhancing CXCL12-induced cell migration and ERK phosphorylation (26). To assess the effects of SMS deficiency on CXCR4 internalization, we performed time-lapse visualization of CXCR4-Venus for a period of 60 min following CXCL12 treatment. The cytoplasmic CXCR4 fluorescence intensity did not change in either wild-type or SMS DKO MEFs for a period of 10 min following CXCL12 treatment. At the 30- and 60-min time points, the intensity levels increased similarly in both MEF lines (data not shown), showing that SMS deficiency had no significant effect on CXCR4 internalization.
SMS deficiency facilitates CXCR4 accumulation into lipid rafts.
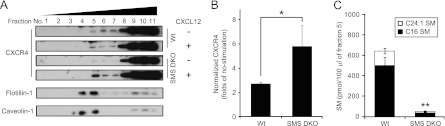
It has been reported that CXCR4 accumulates into lipid rafts (39), which are mainly composed of cholesterol, sphingomyelin, and glycosphingolipids (2). To determine whether SMS-derived sphingomyelin affects CXCR4 relocalization to lipid rafts, we fractionated lipid rafts and determined protein levels of CXCR4. First, we confirmed that the density of the expected CXCR4 protein band detected by immunoblotting was significantly diminished by treatment with siRNA specific for mouse CXCR4 (data not shown). In both MEF lines, CXCL12 treatment promoted the relocalization of CXCR4 into flotillin-1- and caveolin-1-enriched detergent resistant membrane (DRM) fractions (Fig. 7A, fraction 5). Notably, SMS DKO MEFs, which had significantly reduced sphingomyelin content in the DRM fraction (Fig. 7C), displayed elevated levels of CXCR4 in the DRM fraction compared to wild-type MEFs (Fig. 7A and B). These results suggest that SMS-derived sphingomyelin prevents CXCR4 from relocalizing to lipid rafts.
Fig 7.
SMS deficiency increases CXCL12-induced accumulation of CXCR4 in DRM. (A) Cells were stimulated with or without 2.5 nM CXCL12 for 2 min at 34°C and extracted with lysis buffer containing 1% Brij 58. Equivalent amounts of proteins were fractionated in OptiPrep discontinuous gradients (5% and 30%). Fractions were collected from the top to the bottom of the gradient and analyzed by Western blotting with antibodies to CXCR4 and to the lipid raft markers flotillin-1 and caveolin-1. Similar results were obtained in three independent experiments. (B) The density of CXCR4 bands was calculated and divided by the flotillin-1 density and then normalized to that of nonstimulated cells. Values are means ± standard deviations (n = 3). *, P < 0.05. (C) Each lipid raft fraction was extracted to compare sphingomyelin in lipid rafts. Lipids were extracted, and both sphingomyelin species (d18:1, 16:0 and d18:1, 24:1) were analyzed by HPLC-MS/MS to determine the quantity. Values are means ± standard deviations (n = 4). **, P < 0.01.
SMS-derived sphingomyelin inhibits CXCR4 dimerization on the plasma membrane.
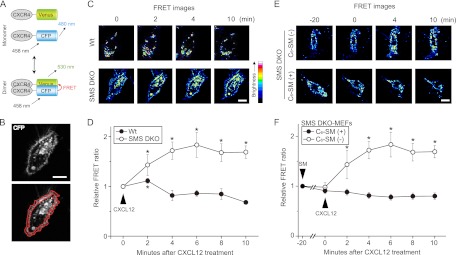
It has been reported that lipid raft-dependent dimerization of CXCR4 is required for CXCL12-induced actin polymerization and cell migration (58) To examine the effects of SMS-derived sphingomyelin on CXCR4 dimerization, MEFs coexpressing CXCR4-CFP and CXCR4-Venus were stimulated with CXCL12, and then FRET sensitized emission and acceptor photobleaching analysis for plasma membrane regions were performed. At first, FRET sensitized emission using CFP-tagged K-ras (a plasma membrane localizing protein) (40) and CXCR4-Venus was performed, establishing the negative control for FRET analysis (data not shown). In MEFs coexpressing CXCR4-CFP and CXCR4-Venus, FRET intensity was normalized to CFP intensity, yielding a relative FRET ratio. Upon CXCL12 stimulation, the relative FRET ratio increased at the plasma membrane (Fig. 8B, lower panel) within 2 min in both MEF lines (Fig. 8C and D). Subsequently, at 4 min, the relative FRET ratio decreased in wild-type MEFs. In contrast, the relative FRET ratio at 4 min in SMS DKO MEFs was higher than at the zero time point (Fig. 8C and D), and these cells displayed filopodium formation (data not shown) and ERK1/2 phosphorylation (Fig. 5A). Similar results were obtained in an acceptor photobleaching experiment (data not shown). These results show that SMS deficiency promotes CXCR4 dimerization in response to CXCL12.
Fig 8.
Increase in CXCL12-induced CXCR4 dimerization in the plasma membrane by SMS deficiency and its inhibition by exogenous sphingomyelin. (A) Schematic representation of FRET to monitor CXCR4 dimerization. CXCR4 was tagged with CFP or Venus at the C terminus to yield CXCR4-CFP and CXCR4-Venus, respectively. FRET occurs when the excited-state energy of CFP is transferred to Venus when they are close physical proximity to each other, occurring, for example, when CXCR4 undergoes dimerization. (B) The fluorescence image of a cell overexpressing CXCR4-CFP and a diagram of the plasma membrane region are shown. The region enclosed by two red lines (lower panel) was defined as the plasma membrane region. Scale bar, 20 μm. (C) Time-lapse series of FRET ratio images of MEFs after addition of CXCL12. Cells were transfected with vectors containing CXCR4-Venus and CXCR4-CFP and then used for FRET experiments. After the addition of 6.25 nM CXCL12, a time-lapse series of FRET images was obtained at 1-min intervals, and these are displayed with their respective times. Scale bar, 20 μm. (E) After SMS double-knockout MEFs were treated with 6 μM exogenous C6-sphingomyelin (C6-SM) for 20 min, they were stimulated with CXCL12. A time-lapse series of FRET images was obtained at 1-min intervals and are displayed with their respective times, with CXCL12-stimulation at 0 min. Scale bar, 20 μm. (D and F) The FRET/CFP ratio intensity (the FRET intensity divided by CFP intensity in the same area and at the same time point) of the plasma membrane region. Based on the FRET images, the FRET/CFP ratio intensity of the plasma membrane was calculated. The relative FRET ratio is represented as a relative intensity, with the FRET/CFP ratio intensity at 0 min (D) and −20 min (F) set at 1. Values are means ± standard deviations (n = 6). *, P < 0.05.
To obtain additional evidence for the involvement of sphingomyelin in CXCR4 dimerization, SMS DKO MEFs were treated with C6-sphingomyelin for 20 min and then stimulated with CXCL12. FRET analysis showed that treatment of SMS DKO MEFs with C6-sphingomyelin inhibited the elevation in the relative FRET ratio following CXCL12 stimulation (Fig. 8E and F). These results suggest that sphingomyelin synthesized by SMSs prevents CXCR4 dimerization at the plasma membrane.
DISCUSSION
The findings from this study reveal, for the first time, that SMS-derived sphingomyelin regulates signaling by the CXCL12/CXCR4 pathway. Sphingomyelin in lipid rafts prevents CXCR4 dimerization and incorporation into lipid rafts, decreasing responsiveness to CXCL12. These novel findings should help further our understanding of the role of SMS-derived sphingomyelin in a myriad of cellular responses.
SMS activity modulates a number of different biological functions, including cell growth (54, 61) and apoptosis (46, 47). However, the biochemical characteristics of SMS are not yet fully understood. SMS DKO MEFs have little in vitro or in vivo SMS activity, and the remaining SMS activity in SMS1 and SMS2 KO MEFs is derived from SMS2 and SMS1, respectively. SMS1 KO reduced in vitro SMS activity by half (Fig. 1B), while in vivo SMS activity (Fig. 1C) and sphingomyelin content in whole cells (Fig. 1D) were further decreased by about two-thirds compared to levels in wild-type cells. These results suggest that, in contrast to SMS1, SMS2 is constitutively suppressed in cells, consistent with our recent observations using a human SMS overexpression system (48). Interestingly, in vivo SMS activity (Fig. 1C) was not correlated with sphingomyelin levels in the plasma membrane fractions (Fig. 1E) in either SMS1 or SMS2 KO MEFs. This may be due to compartmentalization of these SMS isoforms to distinct suborganelles. However, the molecular mechanisms underlying SMS regulation remain obscure.
SMS activity is physiologically tightly regulated. For example, upregulation of SMS activity is associated with cell proliferation induced by hepatic regeneration (1), SV40 transformation (31), and basic fibroblast growth factor stimulation (44). In contrast, certain stress stimuli, such as tumor necrosis factor (24), Fas ligand (24), ionizing radiation (24), staurosporine (59), UVB irradiation (12) and photodynamic therapy (15), decrease sphingomyelin synthesis. Therefore, knowledge of the biological roles of SMS1, SMS2, and sphingomyelin is crucial to our understanding of numerous biological phenomena.
Sphingomyelin, but not ceramide, had an inhibitory effect on cell migration via the CXCL12/CXCR4 pathway (Fig. 4). Wang et al. reported that sphingomyelin hydrolysis by neutral sphingomyelinase (nSMase) leads to the formation of ceramide, which in turn promotes cell polarization and motility (57). In contrast, Heakal and Kester demonstrated that treatment with exogenous nanoliposomal short-chain ceramide inhibits breast cancer cell migration (18). These results seem to be contradictory. However, the results of Wang et al. can also be interpreted to suggest that reduced sphingomyelin levels promote cell migration. Similarly, the results of Heakal and Kester can be interpreted to suggest that elevated ceramide levels increase sphingomyelin production by SMS, which inhibits migration (although sphingomyelin levels were not measured in their paper).
Recent reports indicate that sphingolipid metabolism modulates cell migration. Downregulation of glycosphingolipids by a glucosylceramide synthase inhibitor prevents CXCR4-dependent cell migration (30). Diacylglycerol, another SMS product, activates protein kinase C isozymes and consequently promotes cell migration through the ERK signaling pathway (16). In this study, however, cell migration via the CXCL12/CXCR4 pathway was not enhanced in C6-ceramide-treated SMS DKO MEFs (Fig. 4) or in SMS1/SMS2-overexpressing cells (Fig. 3), in which cellular levels of C6-glucosylceramide (formed by glucosylceramide synthase) or diacylglycerol were, respectively, likely increased. These results appear to rule out significant roles for ceramide and diacylglycerol in cell migration. Therefore, sphingomyelin is likely the key lipid regulating cell migration in our cellular model.
In our experiments, unique effects of SMS1 and SMS2 on cell migration were not found (Fig. 2 and 3), most probably because membrane sphingomyelin deficiency was similar in both SMS1 and SMS2 KO MEFs (Fig. 1F). The specific roles of SMS1 and SMS2 in sphingomyelin production and cell migration remain to be clarified.
The results of this study significantly extend our understanding of lipid raft biology. Cholesterol and sphingomyelin are abundant in lipid rafts. Cholesterol contributes to the stability of lipid rafts, and its depletion by methyl-β-cyclodextrin dissociates several proteins from rafts and inactivates signaling cascades (50, 52). In contrast, the role of sphingomyelin in lipid rafts is largely unknown. Li et al. demonstrated that sphingomyelin deficiency by SMS knockdown had no effect on cholesterol in lipid rafts (27). Our results also show that the compartmentalization of specific proteins, including flotillin-1 and caveolin-1, in the lipid raft fraction was intact in sphingomyelin-deficient cells (Fig. 7A). Importantly, sphingomyelin deficiency increased responsiveness to signaling by the CXCL12/CXCR4 pathway, indicating that, in addition to cholesterol, sphingomyelin in lipid rafts modulates the cellular response to cell motility cues.
We previously proposed that lipid rafts play a role in generating T-cell antigen receptor signaling in Jurkat T cells (23). Our present study indicates that sphingomyelin in lipid rafts modulates signal transduction by interacting with cell surface receptors. Our results also provide insight into the regulation of GPCR dimerization. Ligand-induced dimerization of receptor tyrosine kinases, such as the EGF and PDGF receptors, triggers autophosphorylation and receptor activation (11). Similarly, homodimerization has been described for numerous GPCRs, including the β2-adrenergic receptor (3), the dopamine receptor (38), the δ- and κ-opioid receptors (43), the M3 muscarinic receptor (65), metabotropic glutamate receptor subtype 5 (mGluR5) (45), neuropeptide Y (NPY) receptors (14), CXCR4 (21, 22), and CCR5 (22). The sphingosine-1-phosphate receptor family has also been reported to form homo- and heterodimers (25). In this study, we demonstrated that the migration of SMS-deficient MEFs increased in response to specific GPCR ligands, including CXCL12 (Fig. 2) and sphingosine-1-phosphate (data not shown). In contrast, SMS deficiency had no effect on EGF- or PDGF-stimulated cell migration (Table 1). Thus, our results suggest that sphingomyelin may be a selective modulator of GPCR signaling, modulating the accumulation and dimerization of these receptors in lipid rafts.
Our findings also provide the foundation for a novel pharmacological approach for targeting CXCR4 in cancer metastasis. CXCR4 has been detected in diverse malignancies and appears to play an important role in numerous cancers (6). There are a number of different strategies for targeting CXCR4 for cancer therapy. Targeting CXCL12/CXCR4 with antibodies, antagonists, and siRNA has been shown to inhibit cancer cell growth and migration (28, 29, 63). Our results, demonstrating that sphingomyelin inhibits CXCL12-induced cell migration suggest that treatments which increase SMS activity and sphingomyelin levels may hold promise for cancer therapy.
ACKNOWLEDGMENTS
We thank E. Kiyokawa (Kanazawa Medical University, Japan) and S. Shimozono (RIKEN, Japan) for critical discussion and for reading the manuscript.
This study was supported in part by the Sapporo Biocluster Bio-S, the Knowledge Cluster Initiative of the Ministry of Education, Culture, Sports, Science and Technology (Japan), the Japan Society for the Promotion of Science (Grant-in-Aid for Young Scientists 21890144 and 23790366 to K.K.), and the Sumitomo Foundation (to K.K.).
Footnotes
Published ahead of print 11 June 2012
REFERENCES
- 1. Albi E, Cataldi S, Rossi G, Magni MV. 2003. A possible role of cholesterol-sphingomyelin/phosphatidylcholine in nuclear matrix during rat liver regeneration. J. Hepatol. 38:623–628 [DOI] [PubMed] [Google Scholar]
- 2. Anderson RG, Jacobson K. 2002. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science 296:1821–1825 [DOI] [PubMed] [Google Scholar]
- 3. Angers S, et al. 2000. Detection of beta 2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET). Proc. Natl. Acad. Sci. U. S. A. 97:3684–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Babcock GJ, Farzan M, Sodroski J. 2003. Ligand-independent dimerization of CXCR4, a principal HIV-1 coreceptor. J. Biol. Chem. 278:3378–3385 [DOI] [PubMed] [Google Scholar]
- 5. Balabanian K, et al. 2005. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J. Biol. Chem. 280:35760–35766 [DOI] [PubMed] [Google Scholar]
- 6. Balkwill F. 2004. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin. Cancer Biol. 14:171–179 [DOI] [PubMed] [Google Scholar]
- 7. Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 8. Brown D. 2002. Structure and function of membrane rafts. Int. J. Med. Microbiol. 291:433–437 [DOI] [PubMed] [Google Scholar]
- 9. Burger JA, Kipps TJ. 2006. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood 107:1761–1767 [DOI] [PubMed] [Google Scholar]
- 10. Burns JM, et al. 2006. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J. Exp. Med. 203:2201–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cadena DL, Gill GN. 1992. Receptor tyrosine kinases. FASEB J. 6:2332–2337 [DOI] [PubMed] [Google Scholar]
- 12. Charruyer A, et al. 2008. Decreased ceramide transport protein (CERT) function alters sphingomyelin production following UVB irradiation. J. Biol. Chem. 283:16682–16692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding T, et al. 2008. SMS overexpression and knockdown: impact on cellular sphingomyelin and diacylglycerol metabolism, and cell apoptosis. J. Lipid Res. 49:376–385 [DOI] [PubMed] [Google Scholar]
- 14. Dinger MC, Bader JE, Kobor AD, Kretzschmar AK, Beck-Sickinger AG. 2003. Homodimerization of neuropeptide y receptors investigated by fluorescence resonance energy transfer in living cells. J. Biol. Chem. 278:10562–10571 [DOI] [PubMed] [Google Scholar]
- 15. Dolgachev V, et al. 2004. De novo ceramide accumulation due to inhibition of its conversion to complex sphingolipids in apoptotic photosensitized cells. J. Biol. Chem. 279:23238–23249 [DOI] [PubMed] [Google Scholar]
- 16. Griner EM, Kazanietz MG. 2007. Protein kinase C and other diacylglycerol effectors in cancer. Nat. Rev. Cancer. 7:281–294 [DOI] [PubMed] [Google Scholar]
- 17. Hannun YA, Obeid LM. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9:139–150 [DOI] [PubMed] [Google Scholar]
- 18. Heakal Y, Kester M. 2009. Nanoliposomal short-chain ceramide inhibits agonist-dependent translocation of neurotensin receptor 1 to structured membrane microdomains in breast cancer cells. Mol. Cancer Res. 7:724–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Horuk R. 2001. Chemokine receptors. Cytokine Growth Factor Rev. 12:313–335 [DOI] [PubMed] [Google Scholar]
- 20. Huitema K, van den Dikkenberg J, Brouwers JF, Holthuis JC. 2004. Identification of a family of animal sphingomyelin synthases. EMBO J. 23:33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Isik N, Hereld D, Jin T. 2008. Fluorescence resonance energy transfer imaging reveals that chemokine-binding modulates heterodimers of CXCR4 and CCR5 receptors. PLoS One 3:e3424 doi:10.1371/journal.pone.0003424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Issafras H, et al. 2002. Constitutive agonist-independent CCR5 oligomerization and antibody-mediated clustering occurring at physiological levels of receptors. J. Biol. Chem. 277:34666–34673 [DOI] [PubMed] [Google Scholar]
- 23. Jin ZX, et al. 2008. Impaired TCR signaling through dysfunction of lipid rafts in sphingomyelin synthase 1 (SMS1)-knockdown T cells. Int. Immunol. 20:1427–1437 [DOI] [PubMed] [Google Scholar]
- 24. Kolesnick RN, Haimovitz-Friedman A, Fuks Z. 1994. The sphingomyelin signal transduction pathway mediates apoptosis for tumor necrosis factor, Fas, and ionizing radiation. Biochem. Cell Biol. 72:471–474 [DOI] [PubMed] [Google Scholar]
- 25. Kostenis E. 2004. Novel clusters of receptors for sphingosine-1-phosphate, sphingosylphosphorylcholine, and (lyso)-phosphatidic acid: new receptors for “old” ligands. J. Cell Biochem. 92:923–936 [DOI] [PubMed] [Google Scholar]
- 26. Lagane B, et al. 2008. CXCR4 dimerization and beta-arrestin-mediated signaling account for the enhanced chemotaxis to CXCL12 in WHIM syndrome. Blood 112:34–44 [DOI] [PubMed] [Google Scholar]
- 27. Li Z, et al. 2007. Inhibition of sphingomyelin synthase (SMS) affects intracellular sphingomyelin accumulation and plasma membrane lipid organization. Biochim. Biophys. Acta 1771:1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liang Z, et al. 2004. Inhibition of breast cancer metastasis by selective synthetic polypeptide against CXCR4. Cancer Res. 64:4302–4308 [DOI] [PubMed] [Google Scholar]
- 29. Liang Z, et al. 2005. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 65:967–971 [PMC free article] [PubMed] [Google Scholar]
- 30. Limatola C, et al. 2007. Evidence for a role of glycosphingolipids in CXCR4-dependent cell migration. FEBS Lett. 581:2641–2646 [DOI] [PubMed] [Google Scholar]
- 31. Luberto C, Hannun YA. 1998. Sphingomyelin synthase, a potential regulator of intracellular levels of ceramide and diacylglycerol during SV40 transformation. Does sphingomyelin synthase account for the putative phosphatidylcholine-specific phospholipase C? J. Biol. Chem. 273:14550–14559 [DOI] [PubMed] [Google Scholar]
- 32. Luster AD. 1998. Chemokines—chemotactic cytokines that mediate inflammation. N. Engl. J. Med. 338:436–445 [DOI] [PubMed] [Google Scholar]
- 33. Manes S, Ana Lacalle R, Gomez-Mouton C, Martinez AC. 2003. From rafts to crafts: membrane asymmetry in moving cells. Trends Immunol. 24:320–326 [DOI] [PubMed] [Google Scholar]
- 34. Mitsutake S, et al. 2011. Dynamic modification of sphingomyelin in lipid microdomains controls development of obesity, Fatty liver, and type 2 diabetes. J. Biol. Chem. 286:28544–28555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mochizuki N, et al. 2001. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature 411:1065–1068 [DOI] [PubMed] [Google Scholar]
- 36. Muller A, et al. 2001. Involvement of chemokine receptors in breast cancer metastasis. Nature 410:50–56 [DOI] [PubMed] [Google Scholar]
- 37. Nagasawa T, et al. 1996. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature 382:635–638 [DOI] [PubMed] [Google Scholar]
- 38. Ng GY, et al. 1996. Dopamine D2 receptor dimers and receptor-blocking peptides. Biochem. Biophys. Res. Commun. 227:200–204 [DOI] [PubMed] [Google Scholar]
- 39. Nguyen DH, Taub D. 2002. CXCR4 function requires membrane cholesterol: implications for HIV infection. J. Immunol. 168:4121–4126 [DOI] [PubMed] [Google Scholar]
- 40. Parton RG, Hancock JF. 2004. Lipid rafts and plasma membrane microorganization: insights from Ras. Trends Cell Biol. 14:141–147 [DOI] [PubMed] [Google Scholar]
- 41. Percherancier Y, et al. 2005. Bioluminescence resonance energy transfer reveals ligand-induced conformational changes in CXCR4 homo- and heterodimers. J. Biol. Chem. 280:9895–9903 [DOI] [PubMed] [Google Scholar]
- 42. Pettus BJ, Chalfant CE, Hannun YA. 2002. Ceramide in apoptosis: an overview and current perspectives. Biochim. Biophys. Acta 1585:114–125 [DOI] [PubMed] [Google Scholar]
- 43. Ramsay D, Kellett E, McVey M, Rees S, Milligan G. 2002. Homo- and hetero-oligomeric interactions between G-protein-coupled receptors in living cells monitored by two variants of bioluminescence resonance energy transfer (BRET): hetero-oligomers between receptor subtypes form more efficiently than between less closely related sequences. Biochem. J. 365:429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Riboni L, Viani P, Bassi R, Giussani P, Tettamanti G. 2001. Basic fibroblast growth factor-induced proliferation of primary astrocytes. evidence for the involvement of sphingomyelin biosynthesis. J. Biol. Chem. 276:12797–12804 [DOI] [PubMed] [Google Scholar]
- 45. Romano C, Yang WL, O'Malley KL. 1996. Metabotropic glutamate receptor 5 is a disulfide-linked dimer. J. Biol. Chem. 271:28612–28616 [DOI] [PubMed] [Google Scholar]
- 46. Separovic D, et al. 2007. Sphingomyelin synthase 1 suppresses ceramide production and apoptosis post-photodamage. Biochem. Biophys. Res. Commun. 358:196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Separovic D, et al. 2008. Suppression of sphingomyelin synthase 1 by small interference RNA is associated with enhanced ceramide production and apoptosis after photodamage. Exp. Cell Res. 314:1860–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shakor AB, et al. 2011. Sphingomyelin synthase 1-generated sphingomyelin plays an important role in transferrin trafficking and cell proliferation. J. Biol. Chem. 286:36053–36062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sierro F, et al. 2007. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc. Natl. Acad. Sci. U. S. A. 104:14759–14764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Silvius JR. 2003. Role of cholesterol in lipid raft formation: lessons from lipid model systems. Biochim. Biophys. Acta 1610:174–183 [DOI] [PubMed] [Google Scholar]
- 51. Simons K, Ikonen E. 1997. Functional rafts in cell membranes. Nature 387:569–572 [DOI] [PubMed] [Google Scholar]
- 52. Simons K, Toomre D. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1:31–39 [DOI] [PubMed] [Google Scholar]
- 53. Tachibana K, et al. 1998. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature 393:591–594 [DOI] [PubMed] [Google Scholar]
- 54. Tafesse FG, et al. 2007. Both sphingomyelin synthases SMS1 and SMS2 are required for sphingomyelin homeostasis and growth in human HeLa cells. J. Biol. Chem. 282:17537–17547 [DOI] [PubMed] [Google Scholar]
- 55. Tafesse FG, Ternes P, Holthuis JC. 2006. The multigenic sphingomyelin synthase family. J. Biol. Chem. 281:29421–29425 [DOI] [PubMed] [Google Scholar]
- 56. Vila-Coro AJ, et al. 1999. The chemokine SDF-1α triggers CXCR4 receptor dimerization and activates the JAK/STAT pathway. FASEB J. 13:1699–1710 [PubMed] [Google Scholar]
- 57. Wang G, Krishnamurthy K, Chiang YW, Dasgupta S, Bieberich E. 2008. Regulation of neural progenitor cell motility by ceramide and potential implications for mouse brain development. J. Neurochem. 106:718–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang J, He L, Combs CA, Roderiquez G, Norcross MA. 2006. Dimerization of CXCR4 in living malignant cells: control of cell migration by a synthetic peptide that reduces homologous CXCR4 interactions. Mol. Cancer Ther. 5:2474–2483 [DOI] [PubMed] [Google Scholar]
- 59. Wiesner DA, Dawson G. 1996. Staurosporine induces programmed cell death in embryonic neurons and activation of the ceramide pathway. J. Neurochem. 66:1418–1425 [DOI] [PubMed] [Google Scholar]
- 60. Yamaji-Hasegawa A, et al. 2003. Oligomerization and pore formation of a sphingomyelin-specific toxin, lysenin. J. Biol. Chem. 278:22762–22770 [DOI] [PubMed] [Google Scholar]
- 61. Yamaoka S, Miyaji M, Kitano T, Umehara H, Okazaki T. 2004. Expression cloning of a human cDNA restoring sphingomyelin synthesis and cell growth in sphingomyelin synthase-defective lymphoid cells. J. Biol. Chem. 279:18688–18693 [DOI] [PubMed] [Google Scholar]
- 62. Yano M, et al. 2011. Mitochondrial dysfunction and increased reactive oxygen species impair insulin secretion in sphingomyelin synthase 1-null mice. J. Biol. Chem. 286:3992–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yasumoto K, et al. 2006. Role of the CXCL12/CXCR4 axis in peritoneal carcinomatosis of gastric cancer. Cancer Res. 66:2181–2187 [DOI] [PubMed] [Google Scholar]
- 64. Yeang C, et al. 2008. The domain responsible for sphingomyelin synthase (SMS) activity. Biochim. Biophys. Acta 1781:610–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zeng FY, Wess J. 1999. Identification and molecular characterization of m3 muscarinic receptor dimers. J. Biol. Chem. 274:19487–19497 [DOI] [PubMed] [Google Scholar]