Abstract
Enterotoxigenic Escherichia coli (ETEC) is an important cause of diarrhea, mainly in developing countries. Although there are 25 different ETEC adhesins described in strains affecting humans, between 15% and 50% of the clinical isolates from different geographical regions are negative for these adhesins, suggesting that additional unidentified adhesion determinants might be present. Here, we report the discovery of Coli Surface Antigen 23 (CS23), a novel adhesin expressed by an ETEC serogroup O4 strain (ETEC 1766a), which was negative for the previously known ETEC adhesins, albeit it has the ability to adhere to Caco-2 cells. CS23 is encoded by an 8.8-kb locus which contains 9 open reading frames (ORFs), 7 of them sharing significant identity with genes required for assembly of K88-related fimbriae. This gene locus, named aal (adhesion-associated locus), is required for the adhesion ability of ETEC 1766a and was able to confer this adhesive phenotype to a nonadherent E. coli HB101 strain. The CS23 major structural subunit, AalE, shares limited identity with known pilin proteins, and it is more closely related to the CS13 pilin protein CshE, carried by human ETEC strains. Our data indicate that CS23 is a new member of the diverse adhesin repertoire used by ETEC strains.
INTRODUCTION
Enterotoxigenic Escherichia coli (ETEC) strains are a frequent cause of diarrhea worldwide. ETEC causes a cholera-like watery diarrhea in developing countries, mostly in children less than 5 years of age (4). ETEC is also the main etiologic agent of traveler's diarrhea, affecting mostly adults traveling to regions of endemicity (8). Currently, there is no effective vaccine to prevent ETEC-associated diarrhea, although some formulations based on attenuated strains or virulence factors have conferred some protection in volunteer studies (32, 33). ETEC is defined as an E. coli strain able to produce at least one of two types of enterotoxins, the heat-labile toxin (LT) and/or the heat-stable toxin (ST) (23), responsible for the movement of electrolytes and water from the intestinal cells to the intestinal lumen, resulting in watery diarrhea (12). To colonize the intestine, ETEC strains carry a diverse repertoire of adhesion factors (19). The classical ETEC adhesins are the colonization factors (CFs) or coli surface antigens (CS), proteinaceous organelles generally composed of one or two repeating subunits arranged in different conformations to produce fimbrial, fibrillar, or afimbrial structures (19). There are 22 different CF variants currently described (including colonization factor antigen I [CFA/I]), and the majority are usually designated “CS” followed by a number that indicates their placement in an order arranged according to the date of discovery (35). Furthermore, three nonclassical adhesins, Tia, TibA, and EtpA, not considered in the CS nomenclature, have been discovered in ETEC prototype strain H10407 (4).
Studies have been carried out attempting to identify adhesins more frequently found in ETEC isolates, with the ultimate goal of including them in vaccine formulations conferring broad immune protection (32). Epidemiological studies from different geographical regions have identified CFA/I, CS1, CS2, CS3, CS4, CS5, CS6, CS7, CS14, CS17, and CS21 as the more frequent variants present in ETEC strains (23). EtpA has also been frequently detected, although the distribution of nonclassical adhesins has been less thoroughly evaluated. As for most bacterial pathogens, one ETEC strain can encode more than one adhesin, ranging from two to four variants (35).Interestingly, a percentage of clinical isolates remain negative for all current identified adhesin variants. According to reports published within the last 10 years detecting at least 18 different ETEC CFs in four separate countries, the percentages of CF-negative isolates ranged from 44% in Guinea-Bissau (31) to 19.5% in Bangladesh (25), 35.5% in Bolivia (25), and 51% in Peru (24). In a previous study in Chile, which also included the detection of three nonclassical adhesin genes beside 19 different CF genes, 15% of strains were negative for all the adhesin variants (6). These results strongly suggest that uncharacterized adhesins are expressed by clinical ETEC strains of human origin.
Here, we report the identification of a novel adhesion factor in ETEC strain 1766a, which was negative for all previously described ETEC adhesin genes, despite displaying the capacity to attach to intestinal epithelial cells. We combined two strategies previously used to identify adhesins in ETEC and other pathogenic E. coli strains, allowing the identification of an 8.8-kb locus, composed of 9 genes, encoding proteins predicted to participate in the assembly of an adhesin associated with the attachment of this ETEC strain to epithelial cells.
MATERIALS AND METHODS
Bacterial strains and vectors.
E. coli strains and DNA vectors used in this study are described in Table 1. ETEC 1766a was isolated from a child suffering from diarrhea in a periurban community in Santiago, Chile (17). Strains were commonly grown in Luria-Bertani broth (LB) or Dulbecco's modified Eagle medium (DMEM) at 37°C or 20°C to obtain heat-extracted proteins. Antibiotics (100 μg/ml ampicillin or 50 μg/ml kanamycin) were added when it was necessary.
Table 1.
Plasmids and strains used in this study
| Vector or strain | Description | Reference or source |
|---|---|---|
| Vectors | ||
| pHC79 | Cosmid vector | 14 |
| pKD4 | Template plasmid for allelic replacement | 5 |
| pKD46 | Plasmid carrying genes encoding lambda red recombinase system | 5 |
| pG12 | pHC79::aal adherence+ | This work |
| pTn3 | G12::EZ-Tn5<KAN-2> adherence+ | This work |
| pTn6 | G12::EZ-Tn5<KAN-2> adherence+ | This work |
| pTn11 | aalB::EZ-Tn5<KAN-2> adherence− | This work |
| Strains | ||
| ETEC 1766a | O4 ETEC LT+STh+ | 6 |
| E. coli HB101 | Nonadherent laboratory strain (upE44, hsdS20(rB− mB−), recA13, ara-14, proA2, lacY1, galK2, rpsL20, xyl-5, mtl-1, leuB6, thi-1) | 2 |
| ETEC H10407 | Prototype O78 ETEC LT+ STh+ STp+a CFA/I+ Tia+ TibA+ EtpA+ | 3 |
| ETEC 1766a ATE | ETEC 1766a ΔaalA ΔaalB | This work |
| ETEC 1766 UJ1 | ETEC 1766aΔaalB::kanR | This work |
STp, ST porcine.
Cell cultures.
Intestinal epithelial Caco-2 cells, derived from human colonic carcinoma, were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a humidified atmosphere of 5% CO2. In order to propagate cells, they were seeded in 56.7-cm2 dishes and grown to midconfluence, detached with 0.25% trypsin-EDTA, and split at a ratio of 1:4. For adhesion assays, cells were seeded in 6-well plates and left to reach confluence. Medium replacements were performed every 2 days.
Adhesion assays.
Attachment ability was evaluated on Caco-2 epithelial cells. Strains were grown overnight in LB broth at 37°C without shaking and then diluted 1:100 in DMEM. Confluent Caco-2 cultures on 6-well plates (∼1.75 × 106 cells per well) were infected with 1.75 × 107 CFU for 30 min at 37°C and 5% CO2. When necessary, 1% d-mannose was included in the culture medium. Nonadherent bacteria (planktonic) were removed by repeated washings with phosphate-buffered saline (PBS), and attached bacteria were recovered by lysing the cell layer with 0.1% Triton X-100. The bacterial suspension was serially diluted and seeded in LB agar. Counts were performed after 16 h, and the adhesion level was expressed as the percentage of adhered bacteria with respect to the total population (i.e., planktonic and adhered bacteria). To evaluate the participation of the AalE protein, adhesion assay was performed in the presence of the preimmune or the anti-AalE serum. Bacterial suspensions were incubated for 20 min at room temperature with a 1:10 dilution of sera and then added over Caco-2 cells. Alternatively, Caco-2 cells were grown in glass coverslips and then infected to visualize the bacterial adhesion pattern. These cell layers were fixed with 70% methanol for 5 min and stained with Giemsa solution for 40 min at room temperature. Coverslips were mounted over glass slides and analyzed by light microscopy.
EM.
A 3-ml volume of overnight bacterial cultures grown at 37°C in DMEM was concentrated 10-fold, applied to Formvar-carbon-coated copper grids, negatively stained with 2% phosphotungstic acid (pH 6.8) adjusted with 1 N KOH, and examined under a Philips 201 electron microscope (EM) at 60 kV.
Heat-extracted proteins.
Overnight bacterial cultures (10 ml) in DMEM, incubated at 37°C without shaking, were centrifuged at 3,000 × g for 10 min, and the pellet was washed in PBS and centrifuged again. Bacteria were suspended in 100 μl of PBS and heated at 60°C for 30 min. Suspensions were centrifuged again at 3,000 × g for 10 min, and heat-extracted proteins were recovered in the supernatant. Protein quantitation was carried out by the Bradford assay. Approximately 800 ng of proteins was mixed with 5× Laemmli buffer, boiled for 5 min, separated using sodium dodecyl sulfate–15% polyacrylamide gel electrophoresis (SDS-15% PAGE) gels, and stained with Coomassie brilliant blue. To identify proteins by matrix-assisted laser desorption ionization–tandem time of flight (MALDI-TOF/TOF), bands were selected, cut, and sent to the Mass Spectrometry Core at the University of Texas Medical Branch (Galveston, TX). To determine the N-terminal sequence, approximately 120 μg of heat extracts was subjected to SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane. Proteins were stained with Ponceau S, and the selected band was cut and sent to the Edman Sequencing & Protein Analysis Core Facility at the University of Maryland School of Medicine (Baltimore, MD).
Polyclonal sera and immunoblot detection.
Rat polyclonal serum against AalE protein and preimmune serum were obtained from Rockland Immunochemicals (Boyertown, PA) using gel slices containing the 28-kDa AalE band. Two rats were injected with approximately 240 μg each, and the serum providing the best immunoreactivity, over the background observed with preimmune serum, was selected. For immunoblot detection, 200 ng of heat extracts was subjected to SDS-PAGE and transferred to a nitrocellulose membrane. Nonspecific protein binding was blocked with 5% nonfat milk for 12 h at 4°C, and the membrane was incubated with 1:1,000 rat anti-AalE serum, or preimmune serum at the same dilution, for 1 h at room temperature. To detect immunoreactive bands, the membrane was incubated with 1:1,000 horseradish peroxidase-linked anti-rat IgG for 1 h at room temperature and its enzymatic activity was revealed with tetramethylbenzidine (Novex; Invitrogen).
Immunogold staining.
Strains were grown in LB broth until the optical density at 600 nm (OD600) reached approximately 0.6 and concentrated 100-fold by centrifugation. A drop of suspended bacteria was placed on Formvar-carbon-coated nickel grids (200 mesh) and allowed to adhere. Grids were incubated with the anti-AalE serum diluted 1:100 for 1 h and, after 5 washes in PBS–1% BSA–1% Tween 20, incubated with 10-nm-diameter-gold-particle-labeled donkey anti-rat IgG diluted 1:100 for 1 h. Grids were washed again, negatively stained as was indicated above, and analyzed at the Laboratory of Electron Microscopy, Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile.
Cosmid library.
Genomic DNA (gDNA) (20 μg) was partially digested with Sau3AI (2 U) for 10 s at room temperature. The enzyme was inactivated by heating at 65°C for 15 min, and products were separated by pulsed-field agarose gel electrophoresis (low-melting-point 1% agarose) at 6 V/cm for 16 h at 14°C with a ramped switch time of 4 s to 40 s. Nonstained gel pieces containing products between 20 and 40 kb were cut (using stained molecular weight ladder markers as a reference), melted, and digested with agarase. Nondigested agarose was pelleted by centrifugation at 16,000 × g for 10 min and discarded. Oligosaccharides were removed from the supernatant by dialysis in a nitrocellulose membrane, and DNA was concentrated in a speed vacuum centrifuge. In parallel, 5 μg of pHC79 cosmid vector was linearized with BamHI and calf intestinal phosphatase. Lineal gDNA (4 μg; 20 to 40 kb) was incubated with 500 ng of lineal vector in the presence of 30 U of T4 DNA ligase for 16 h at 16°C. Recombinant cosmids were packaged in lambda heads, and the whole mixture was used to transduce E. coli HB101. Transductants were selected in LB supplemented with ampicillin (100 μg/ml).
Selective screening of adherent recombinant cosmid clones.
To select cosmid clones with cell-attachment ability, 1,000 clones were pooled in groups of 50 and adhesion assays were performed as described above. Each group was used to infect one well of a six-well plate for 3 h. Cell lysates were serially diluted in PBS, and, for each group, colonies recovered from the most diluted suspensions were pooled and used to perform another adhesion assay following the same procedure. Colonies recovered after the second cycle were selected, and adherence ability was individually evaluated in 30-min adhesion assays.
Identification of functional sequences within constructions.
To identify sequences directing the adherence ability of the recombinant cosmid clone, EZ-Tn5 <KAN-2> transposons (Epicentre, Illumina, Inc., Madison) were used according to instructions provided by the manufacturer. Random in vitro transposition was carried out in the isolated recombinant cosmid, and the mix was used to electroporate E. coli HB101. Kanamycin- and ampicillin-resistant clones were selected (in order to rule out insertions disrupting the bla gene) and evaluated for adherence ability as described above. The insertion point within the cosmid was determined by direct sequencing from both transposon ends and identified by performing comparisons with databases using blastn (BLAST).
Allelic replacement.
The usher-like sequence disrupted by transposon insertion in clone Tn11 was inactivated in the ETEC 1766a genome by allelic replacement (5). Briefly, ETEC 1766a was transformed with pKD46 plasmid encoding the lambda red recombinase system and subsequently with a DNA fragment containing the kanamycin resistance gene (kanR) flanked by 40-nucleotide ends identical to the target sequence in the ETEC 1766a genome (primers PUJ-kanF and PUJ-kanR listed in Table 2). Kanamycin-resistant colonies were evaluated by PCR in order to prove that replacement had taken place. A primer that recognizes kanR (kanR-R) and another one that recognizes the aalA gene were used (Table 2).
Table 2.
Oligonucleotides used in this study
| Primer | Sequence (5′ to 3′) | Protocol(s) | Source |
|---|---|---|---|
| cshA-F | CTGTCATGTGGTGGTGGATG | Sequencing/PCR/RT-PCR | This work |
| cshA-R | CATCCACCACCACATGACAG | Sequencing | This work |
| cshB-F | GTTATCTGGTTGACCTGTCC | Sequencing/PCR/RT-PCR | This work |
| cshB-R | GGACAGGTCAACCAGATAAC | Sequencing | This work |
| cshB2-F | CGAAGGATGAATGGAATGCC | Sequencing | This work |
| cshB2-R | GGCATTCCATTCATCCTTCG | Sequencing/PCR/RT-PCR | This work |
| cshC-F | GCAGACCAGACCCGCTATAT | Sequencing/PCR/RT-PCR | This work |
| cshC-R | ATATAGCGGGTCTGGTCTGC | Sequencing | This work |
| cshD-F | CCCTGATGGTTGACGGAAAG | Sequencing | This work |
| cshD-R | CTTTCCGTCAACCATCAGGG | Sequencing/PCR/RT-PCR | This work |
| faeI-F | GTGAGTCATTTCAGTGATG | Sequencing | This work |
| faeI-R | CATCACTGAAATGACTCAC | Sequencing | This work |
| PUJ-kanF | GTCACGTATCAGTAACGGAGAAAAGGATTGTGTGTGCAGGGTGTAGGCTGGAGCTGCTTC | Allelic replacement | This work |
| PUJ-kanR | TGTACCGGTAACGCCCGGTCATCGATGCGGGGTAATAACTCATATGAATATCCTCCTTAG | Allelic replacement | This work |
| kanR-R | GCCATGATGGATACTTTCT | Allelic replacement | This work |
| aalR-F | CGGGTGTCTTAATGGAAAAT | PCR/RT-PCR | This work |
| aalR-R | CCTTTCCTGCTCCCCATAAT | PCR/RT-PCR | This work |
| aalA-R | CGTGACCACAAAGGTCAGTG | PCR/RT-PCR | This work |
| aalC-R | TCACCACATTTCCCTTCACC | PCR/RT-PCR | This work |
| aalD-F | GCACTCAGTATTCAGTCAGC | PCR/RT-PCR | This work |
| aalE-F | CTGCTATGGCGTGGACTGTA | PCR/RT-PCR | This work |
| aalE-R | AGGTGAATAGGGGGTTCTCG | PCR/RT-PCR | This work |
| aalF-F | CGCCACATGCAGATATTCTT | PCR/RT-PCR | This work |
| aalF-R | AAGCCCTGGTTCATCAGAAG | PCR/RT-PCR | This work |
| aalG-F | AAAAACCACCCTGACCACAC | PCR/RT-PCR | This work |
| aalG-R | GTGCCGTTCAGACGTAATTC | PCR/RT-PCR | This work |
| aalH-F | CGGATGAGTACACCAGATGC | PCR/RT-PCR | This work |
| aalH-R | GCTGCCATTCTCCTGTGAC | PCR/RT-PCR | This work |
| T7/KAN2 FP-1 | ACCTACAACAAAGCTCTCATCAACC | Sequencing | Epicentre |
| T7/KAN2 RP-1 | GCAATGTAACATCAGAGATTTTGAG | Sequencing | Epicentre |
| T7 | TAATACGACTCACTATAGGG | Sequencing | Universal primer |
Locus sequencing.
Regions flanking a transposon insertion site in the Tn11 cosmid were sequenced. Three E. coli fimbrial loci (accession numbers 315063705, X03675, and 209915368) which contained the previously known usher genes chsB and faeD were aligned using ClustalW2, and PCR primers were designed to amplify conserved regions in an attempt to span the entire length of the insertion by obtaining amplification products of ≤1,500 bp (for sequencing from both ends; see Table 2). Primers were tested in PCRs using ETEC 1766a genomic DNA and purified recombinant cosmid from G12 (pG12) as templates, and sequences were further determined at Macrogen (Rockville, MD) starting from pG12. Fragments were assembled and analyzed to find similar sequences in databases, using BLAST, and to predict open reading frames (ORFs) by using the ORF Finder server. The sequence of the fimbrial locus was further checked and completed by sequencing the whole genome of ETEC 1766a. In this case, 5 μg of purified genomic DNA was sheared to 300 bp (average length), processed via the standard Illumina paired-end protocol, and sequenced on an Illumina Hiseq2000 analyzer at the University of Maryland School of Medicine (Baltimore, MD). Reads were assembled de novo using the Short Oligonucleotide Analysis Package (SOAPdenovo, version 1.3), and contigs that included sequences previously determined in pG12 were identified. PCRs using primers which recognize every ORF within the locus were performed to check the genetic organization, and direct-sequencing reactions were carried out to accurately determine the unknown junctions between contigs when required.
CF phylogenetic tree.
Amino acid sequences of mature CF structural subunits or chaperone proteins were aligned using ClustalW2. N-terminal signal sequences for export to the periplasm, described in GenBank records or predicted by bioinformatics analysis using Signal-IP software, were removed. A distance-based phylogenetic tree was constructed by the neighbor-joining method using Mega 5.05 software with bootstrap confidence tests (1,000 replicates).
RT-PCR.
In order to detect expression of aal genes, RNA was obtained with an SV total RNA isolation system (Promega) and treated with RNase-free DNase I (Fermentas). Reverse transcription was carried out starting from 1 μg of RNA by the use of RevertAid reverse transcriptase (M-MulV RT; Fermentas) and random hexamers as primers. PCRs were performed to detect each gene within aal, or junctions between genes, using cDNA as the template and primers listed in Table 2.
Statistical analysis.
Three independent adhesion assays were performed, and data were subjected to analysis of variance (ANOVA) and Student's t tests. Differences were considered statistically significant when P < 0.05.
Nucleotide sequence accession number.
The GenBank accession number for the aal nucleotide sequence is JQ434477.
RESULTS
ETEC 1766a has the capacity to adhere to Caco-2 cells.
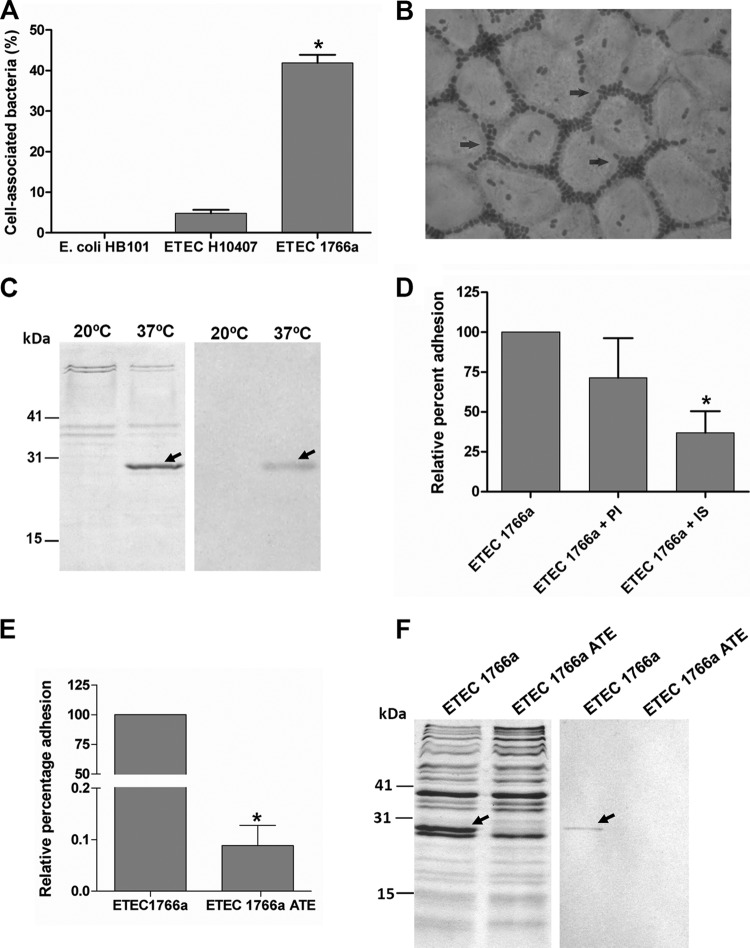
Strain ETEC 1766a was obtained from a child suffering from watery diarrhea, and it was characterized in a previous study (6). It belongs to the O4 serogroup; it carries the human ST (STh)- and LT-encoding genes and was negative for the currently known ETEC adhesin genes (encoding CFA/I, CS1 to CS8, CS12 to CS15, CS17 to CS22, Tia, TibA, and EtpA). Despite all this, ETEC 1766a was able to adhere to Caco-2 cells, showing an adhesion level significantly higher than that of nonadherent laboratory strain E. coli HB101 and almost 8-fold higher than that of the prototype ETEC H10407 strain (Fig. 1A). d-Mannose did not reduce this level (99% ± 13.9% relative to the percentage obtained without d-mannose), suggesting that the observed adhesion capacity was not mediated by type I fimbriae. Microscopic examination of Caco-2 cells infected with ETEC 1766a showed that bacteria adhere mainly to the periphery of the cell (Fig. 1B).
Fig 1.
Adherence of ETEC 1766a to Caco-2 cells and finding of AalE as an adhesion determinant. (A) Adhesion level of E. coli HB101, ETEC H10407, and ETEC 1766a expressed as the percentage of cell-associated bacteria in relation to the total bacteria present after a 30-min infection (mean ± standard error of the mean [SEM]). *, percentage significantly higher than E. coli HB101 and ETEC H10407 percentages (P < 0.001). (B) Giemsa staining of Caco-2 cell layers infected with ETEC 1766a. Arrows indicate the presence of bacteria at the cell borders. Magnification, ×1,000. (C) Heat-extracted proteins from ETEC 1766a cultured at 20°C and 37°C (left) and immunoblot analysis using polyclonal serum obtained after immunization of a rat with the 28-kDa-band protein (right). In both cases, the presence of the protein band is indicated by arrows. (D) Relative levels of adherence of ETEC 1766a in the presence of preimmune serum (PI) or immune serum (IS) against the 28-kDa protein whose results are shown in panel C (mean ± SEM). *, percentage significantly lower than the percentage of adherence seen in the absence of serum (P < 0.05). (E) Relative adherence of spontaneously attenuated ETEC 1766a (ETEC 1766a ATE) compared to the wild type (mean ± SEM). *, percentage significantly lower than wild-type strain percentage (P < 0.001). (F) Absence of AalE in ETEC 1766a ATE heat extracts determined by Coomassie blue staining (left) and immunoblot analysis (right).
ETEC 1766a carries a 28-kDa putative adhesin.
Protein heat extraction is a procedure that has been used to identify some CFs in ETEC strains and other adhesins in pathogenic E. coli strains under different environmental conditions (21, 34). We used this approach to obtain proteins from ETEC 1766a cultured at 20°C and 37°C, which were then separated by SDS-PAGE. After Coomassie blue staining, a 28-kDa band was clearly visible when proteins were extracted from cultures incubated at 37°C but not at 20°C (Fig. 1C) (10). A polyclonal serum prepared against the excised protein recognized the same band in immunoblot assays (Fig. 1C) and was able to significantly block ETEC 1766a adhesion to intestinal epithelial cells (Fig. 1D). In contrast, when preimmune serum was used, no significant reduction in the adhesion level was observed (Fig. 1D). These results suggest that the 28-kDa protein acts as an adhesin.
ETEC 1766a carries fibrillar structures on its surface, which was evident upon electron microscopic analysis, suggesting that the appendages could be part of its adhesion repertoire (Fig. 2A). By immunogold-labeling electron microscopy, we observed that the polyclonal serum recognized antigens in nonpermeabilized bacteria, suggesting that the 28-kDa adhesin is located at the bacterial surface (Fig. 2C and E). However, the labeling pattern did not seem to match with the fibers observed when negative staining was performed (Fig. 2A). Instead, it suggested that the adhesin was arranged as an afimbrial structure. To identity this protein, the 28-kDa band was excised and analyzed by MALDI-TOF/TOF but no significant matches were found in the protein databases. Therefore, N-terminal sequencing was performed and 9 amino acids (TVNGDGGSV) that did not display identity with any peptide sequence stored in the databases were identified, suggesting that the 28-kDa adhesin was a previously uncharacterized protein. We designated this novel protein AalE (protein encoded by an adhesion-associated locus of ETEC), and we propose the inclusion of this adhesin in the current CS nomenclature as CS23.
Fig 2.
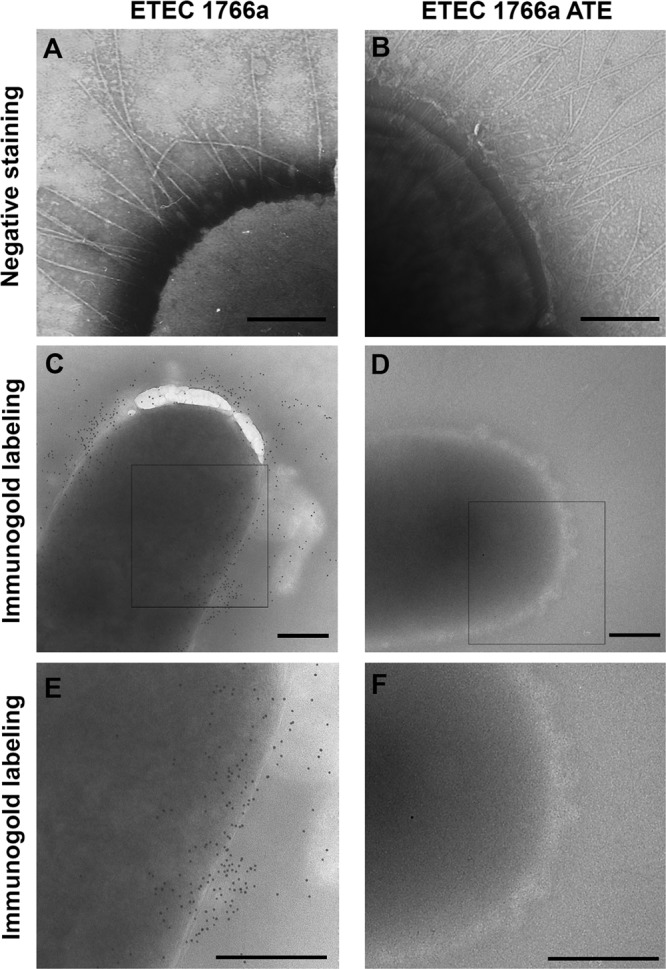
Transmission electron microscopy (TEM) analysis of ETEC 1766a and its spontaneously attenuated variant ETEC 1766a ATE. (A and B) Negatively stained bacteria. Magnification, ×47,000. (C and D) Immunogold labeling using anti-AalE polyclonal serum. Magnification, ×22,000. (E and F) Digital zoom to squares shown in panels C and D. Bars, 0.3 μm.
A spontaneous mutation affected the adhesion ability of ETEC 1766a.
We observed that some colonies recovered from the ETEC 1766a master stock stored at room temperature in an agar slant showed a significant reduction in adhesion properties compared to the fully virulent wild-type strain. Surprisingly, the AalE protein was not detected in heat extracts of these isolates, although the rest of the electrophoretic protein profile seemed to be identical to that obtained with the wild-type strain (Fig. 1F). Further, the AalE protein was not detected on the bacterial surface after immunogold labeling, even though fibrillar structures were clearly observed (Fig. 2B, D, and F). These observations further supported the role for AalE in adhesion of ETEC 1766a and indicated that this protein is not part of the fimbrial structures observed on the surface of this strain.
AalE is produced by a cosmid clone with adherence ability.
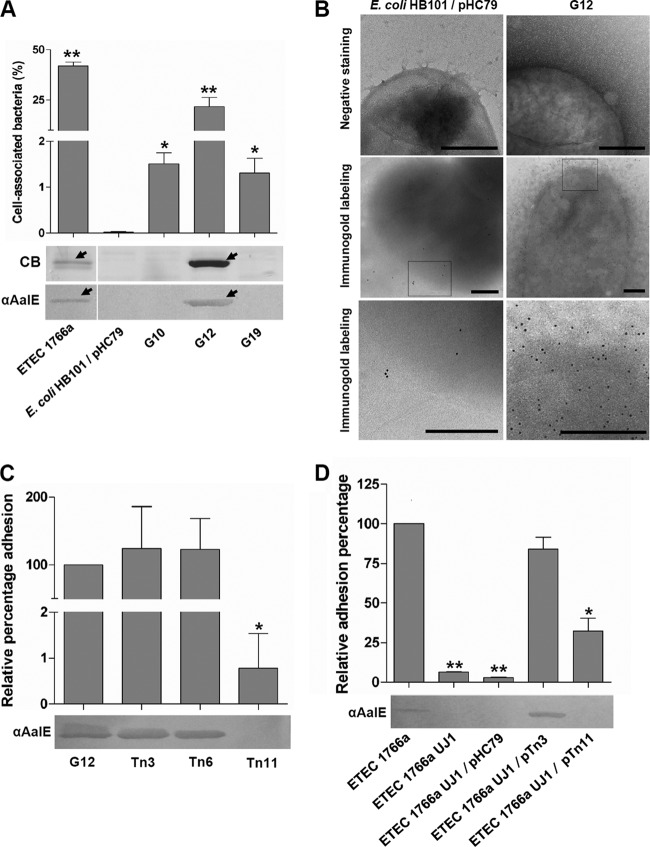
To identify the genomic sequences encoding the ETEC 1766a AalE adhesin, we constructed a cosmid library containing ETEC DNA fragments in the E. coli HB101 background, using pHC79 as vector. A total of 2,160 ampicillin-resistant clones were obtained, and 1,000 of them were screened in adhesion assays. Three clones, named G10, G12, and G19, were selected. All of them showed an adherence level significantly higher than that of E. coli HB101 carrying the empty cosmid (Fig. 3A).
Fig 3.
Clones selected from the cosmid library and finding sequences responsible for adherence ability. (A) Adherence ability of selected clones (mean ± SEM) and presence of AalE in heat extracts detected by Coomassie blue staining (CB) and immunoblot analysis. *, percentage significantly higher than that seen with E. coli HB101 carrying the empty pHC79 cosmid (P < 0.05); **, percentage significantly higher than those seen with G10 and G19 (P < 0.001). (B) Analysis of bacterial surface by TEM of negatively stained (upper row) host strain and clone G12. Detection of AalE by immunogold labeling (middle row, ×22,000 magnification; bottom row, digital zoom of squares in middle row). Bars, 0.3 μm. (C) Relative levels of adhesion of mutants generated by random transposon insertion in pG12 (mean ± SEM) and detection of AalE in heat extracts by immunoblot analysis. *, percentage significantly lower than G12 percentage (P < 0.001). (D) Relative levels of adhesion of mutants generated by allelic replacement of the usher-like sequence in ETEC 1766a (ETEC 1766a UJ1) and further effect of transformation with empty pHC79 cosmid or G12-derived recombinant cosmids (mean ± SEM). AalE in heat extracts was detected by immunoblot analysis. *, percentage significantly lower than wild-type ETEC 1766a percentage (P < 0.05). **, P < 0.001.
To determine whether the putative adhesin AalE was produced by any of these clones, heat extracts were obtained and separated by SDS-PAGE. The 28-kDa band was clearly observed in Coomassie-stained extracts from the G12 cosmid clone, and its presence was confirmed by immunoblot analysis (Fig. 3A). AalE was not detected in heat extracts from E. coli with strain HB101 only or HB101 carrying empty pHC79 as well as other two adherent cosmid clones (Fig. 3A). No fibrillar structures were visible at the G12 surface (Fig. 3B). Production of AalE by G12 was also evidenced by immunogold-labeling EM, and the distribution of gold particles suggested once again its arrangement in afimbrial structures (Fig. 3B). No fibrillar structures were evident in the host strain carrying the empty pHC79 cosmid, and just a few gold particles, reflecting background staining, were observed by immunogold labeling (Fig. 3B).
A putative usher gene is required for both AalE extracellular localization and adherence to Caco-2 cells.
To identify the adhesion determinant located within pG12, we performed random transposon mutagenesis using the EZ-Tn5 <KAN-2> transposon. Twenty kanamycin-/ampicillin-resistant clones were selected and analyzed by heat-protein extraction and by their adherence properties. The AalE protein was not detected in heat extracts from one clone, named Tn11 (Fig. 3C), and this observation was consistent with the significant reduced adherence level compared to G12 (Fig. 3C). AalE was detected in heat extracts from two other G12-derived transposon insertion clones (Tn3 and Tn6) which did not decrease their adherence capacity. The DNA sequence disrupted by transposon insertion in the Tn11 cosmid (pTn11) was identified by direct outward sequencing from both transposon ends. A fragment of 1,588 bp was obtained by merging the two sequences, and blastn analysis indicated that it shares identity with cshB and faeD genes, encoding usher proteins involved in assembly of K88-related fimbriae. The prototype members of the K88 or F4 fimbriae were first identified in ETEC strains that infect pigs (16). Amino acid sequences derived from both genes did not contain the peptide sequence found in the AalE N terminus (TVNGDGGSV), suggesting that the identified novel putative usher protein is involved in the export of AalE to the bacterial surface.
The putative usher gene identified by the transposition protocol was mutated in ETEC 1766a by allelic replacement. As predicted, the AalE protein was not detected in heat extracts obtained from this mutant and its adhesion ability was significantly reduced compared to that of the wild-type strain (Fig. 3D). The presence of AalE in heat extracts and wild-type adherence levels were reestablished by transforming the mutant strain with pTn3 (cosmid containing the transposon inserted in an irrelevant site) but not with pTn11. Therefore, these data, together with those previously obtained, indicated that the adhesion abilities of ETEC 1766a and G12 were conferred by a genomic segment carrying a locus encoding a surface structure that contains the AalE protein and is assembled by the usher/chaperone pathway (20).
AalE is encoded by a K88-like locus.
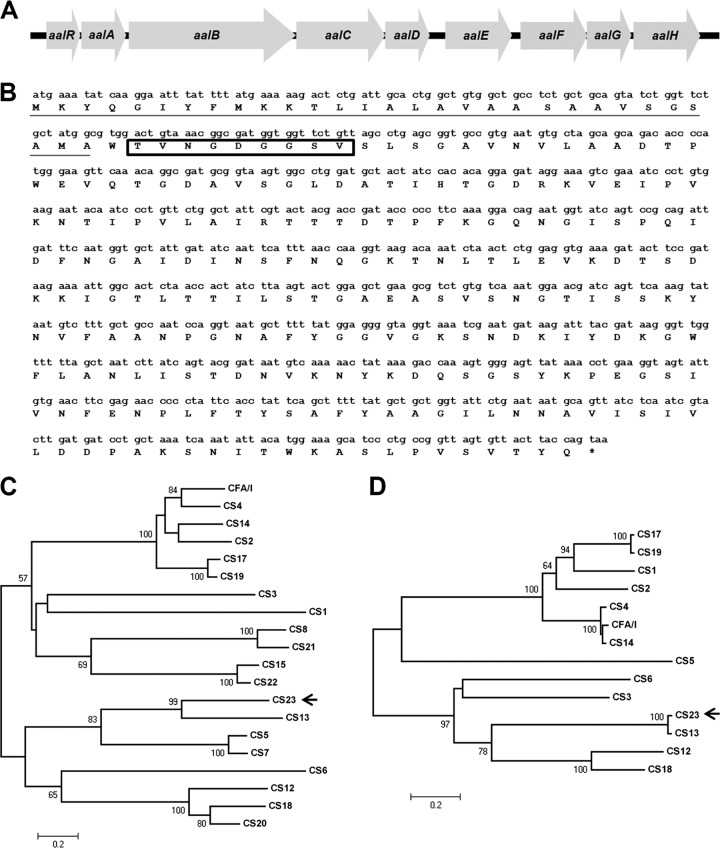
Typically, structures assembled by the usher/chaperone pathway are encoded by genetic clusters that include one or more ORFs for structural subunits which constitute the structure at the bacterial surface, one ORF for the usher protein, which forms a pore inserted at the external membrane and allows export of structural subunits, and one ORF for the chaperone, a periplasmic protein which assists in the export to the extracellular space (22). Therefore, the identification of a putative usher gene as a functional determinant for the adhesion phenotype suggested the presence of additional genes that could fulfill the above-mentioned functions. Thus, the DNA sequence flanking the putative usher gene was determined. PCRs performed with primers directed against conserved regions of three different previously known K88-like clusters and, further, performance of partial sequencing of the ETEC 1766a genome by the use of Illumina-based technology allowed identification of an 8.8-kb K88-related locus that carries 9 ORFs encoding 6 putative structural components, a transcriptional regulator, a chaperone, and the usher proteins (Table 3). Deduced amino acid sequences from 7 of the 9 ORFs share significant (>90%) identity with proteins involved in the assembly of K88-related fimbriae (Table 3). As expected, the predicted protein product from one ORF included the TVNGDGGSV peptide sequence identified in AalE (Fig. 4B). The deduced full-length AalE amino acid sequence shared discrete identity with the sequence of a putative fimbrial structural subunit of the plant pathogen Erwinia amylovora (44%; E value 8e-47, accession number YP_003529604) and with the sequence of CshE, the major structural subunit of CS13, whose adhesin activity has not been demonstrated (13). Among proteins of proven function, AalE shared low identity with FaeG, the major structural subunit and adhesin of the K88 fimbriae (37). In order to follow the previously proposed nomenclature, we named this locus aal and its genes aalR, aalA, aalB, aalC, aalD, aalE, aalF, aalG, and aalH, where aalB is the usher-encoding gene first identified in the G12 recombinant cosmid (Table 3 and Fig. 4A). Deduced amino acid sequences of AalA, AalB, AalC, AalD, AalF, AalG, and AalH proteins were shown to share ≥90% identity with Fae proteins, which participate in the biogenesis of the K88 fimbria (Table 3). The dissimilarity between the aal locus and the previously known K88-related loci lies in the ORFs encoding the putative transcriptional regulator (aalR) and the major structural subunit (aalE) (Table 3). It is commonly observed that clusters encoding usher/chaperone-assembling adhesins contain genes encoding transcriptional regulators. In this case, AalR shares homology with the PapB transcriptional regulator, controlling expression of genes involved in synthesis of the P pili carried by uropathogenic E. coli strains (36).
Table 3.
ORFs predicted within aal locus and similarity of their products to proteins of known functions in databanks
| Gene | Gene length (nt)/protein length (aa)a | Predicted protein domain | Similar protein/organism | Function assigned to similar protein | Identity (%)b | E value |
|---|---|---|---|---|---|---|
| aalR | 318/105 | PapB superfamily (pfam0333) | PapB/Escherichia coli W | Transcriptional regulator | 57/94 (61) | 1e-22 |
| aalA | 537/178 | FimA (COG3539) | FaeC/Escherichia coli | Tip structural subunit | 154/172 (90) | 5e-88 |
| aalB | 2319/772 | Fimbrial usher protein (pfam 00577) | FaeD/Escherichia coli B185 | Usher protein | 756/772 (98) | 0.0 |
| aalC | 876/291 | Fimbrial chaperone PefD (PRK15211) | FaeE/Escherichia coli SE11 | Fimbrial chaperone | 232/237 (98) | 5e-134 |
| aalD | 492/163 | No domains predicted | FaeF/Escherichia coli SE11 | Minor fimbrial structural subunit | 154/163 (93) | 7e-83 |
| aalE | 798/265 | K88 superfamily (pfam 02432) | FaeG Fimbrial protein/Escherichia coli UMNK88 | Major fimbrial structural subunit | 84/303 (28) | 1e-10 |
| aalF | 798/265 | K88 superfamily (pfam 02432) | FaeH/Escherichia coli UMNK88 | Minor fimbrial structural subunit | 236/265 (89) | 4e-173 |
| aalG | 534/177 | K88 superfamily (pfam 02432) | FaeI/Escherichia coli B185 | Minor fimbrial structural subunit | 174/177 (98) | 5e-123 |
| aalH | 741/246 | No domains predicted | FaeJ/Escherichia coli B185 | Minor fimbrial structural subunit | 238/246 (97) | 1e-173 |
nt, nucleotides; aa, amino acids.
Identity, number of amino acids showing identity/total number of amino acids (no gaps between sequences).
Fig 4.
Genetic organization of the aal locus and sequence coding for the major structural subunit AalE. (A) Organization of aal locus showing the nine predicted ORFs. (B) Nucleotide sequence of aalE and translated amino acid sequence. The predicted signal sequence for secretion is underlined, and the peptide sequence identified by its N-terminal sequence is shown inside the rectangle. (C) Dendrogram derived from the alignment of AalE (CS23) with 19 known human CF major structural subunits. Sequences corresponded to CfaB (CFA/I fimbria, GenBank accession number AAC41415), CsoA (CS1, CBL93542), CotA (CS2, CAA87761), CstA (CS3, CAA34820), CsaB (CS4, AAK97135), CsfA (CS5, CAA11820), CssA (CS6, AAC45093), CsvA (CS7, AAK09045), CofA (CS8, BAA07174), CswA (CS12, AAK09047), CshE (CS13, CAA50789), CsuA1 (CS14, CAA66122), NfaA (CS15, CAA45906), CsbA (CS17, AAS89777), FotA (CS18, AAB41914), CsdA (CS19, AAQ19775), CsnA (CS20, AAL31639), LngA (CS21, AAC33154), and CseA (CS22, AAD30557). The arrow indicates the branch that includes the CS23 and CS13 major structural subunits. Only bootstrap values higher than 50% are shown above or under the nodes. (D) Dendrogram derived from the alignment of AalC (CS23) with 13 known human CF chaperone proteins. Sequences corresponded to CfaA (CFA/I chaperone, GenBank accession number AAC41414), CsoB (CS1, CAA44672), CotB (CS2, CAA87760), an unnamed protein (CS3, CAA34815), CsaA (CS4, AAK97134), CsfB (CS5, CAD90920), CssC (CS6, AAC45095), CswB (CS12, AAK09048), CshC (CS13, CAA50787), CsuB (CS14, AAQ20104), CsbB (CS17, AAS89776), FotB (CS18, AAO73847), and CsdB (CS19, AAQ19774). The arrow indicates the branch that includes CS23 and CS13. Only bootstrap values higher than 50% are shown above or under the nodes.
The TVNGDGGSV peptide identified by N-terminal sequencing, which did not match with any peptide in databases, was found within the AalE protein starting at residue 32 (Fig. 4B). As expected for a structural subunit, the presence of a signal peptide for secretion was predicted and the most probable proteolytic cleavage would take place between residues 30 and 31, leaving a tryptophan residue (W) as the first amino acid in the mature protein. AalE clustered with the CS13 major structural subunit CshE (sharing 40% identity) in a phylogenetic tree built from the alignment of the amino acid sequences of mature AalE and those of the other 19 known mature major structural subunits of the CFs carried by human ETECs (Fig. 4C). In addition, close relationships such as those previously described between subunits CS8 and CS21 (9), CS15 and CS22 (21), and CS17 and CS19 (11) were observed. Association between CS23 and CS13 was also evident by constructing a phylogenetic tree from an alignment of 13 available chaperone amino acid sequences involved in assembly of ETEC CFs (data not shown).
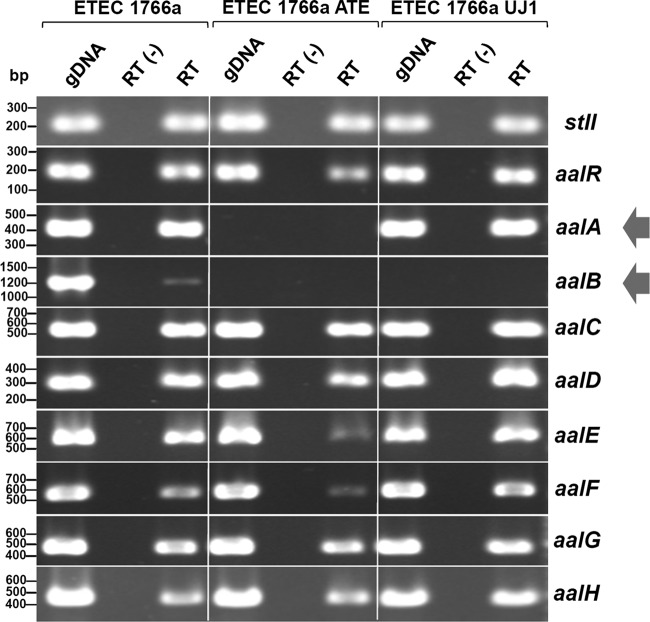
Transcripts generated from the 9 aal genes were detected by RT-PCR in the wild-type ETEC 1766a strain, whereas, in the spontaneously attenuated strain (ETEC 1766a ATE), amplification products were obtained for all the genes, except for aalA and aalB (Fig. 5). No product was obtained for ETEC 1766a ATE when PCR was performed to amplify the fragment between aalR and aalC or the fragment between aalR and aalH. In contrast, the fragment between aalC and aalH was amplified, suggesting that the loss of adherence ability in ETEC 1766a ATE was due to a replacement of the original aalA-aalB segment by the insertion of a DNA sequence of unknown size. As expected, only aalB (encoding the usher protein) transcript was not detected in the mutant strain generated by allelic replacement (ETEC 1766a UJ1), confirming that there was no polar effect after insertion of the kanR gene (Fig. 5).
Fig 5.
Presence and expression of aal genes in ETEC 1766a and their derived mutant strains. PCR was carried out using purified genomic DNA (gDNA) obtained from wild-type ETEC 1766a, the spontaneously attenuated strain (ETEC 1766a ATE), and the mutant generated by allelic replacement (ETEC 1766a UJ1). In parallel, RT-PCR was performed starting from cDNA synthetized in the absence [RT(-)] or presence (RT) of reverse transcriptase. Products were subjected to electrophoresis in agarose gels and stained with ethidium bromide. Arrows at the right indicate genes, or their transcripts, not detected in mutant strains.
To determine whether the CS23 adhesin was carried by other ETEC strains, detection of aalE gene was performed by PCR in 102 previously characterized ETEC strains obtained in Chile, including different serogroups with diverse virulence repertoires (6). For all the isolates, no product was obtained, suggesting that, at least in this geographic region, the presence of the CS23 aalE gene is not frequent.
DISCUSSION
Although there are more than 20 different adhesins described in ETEC strains, there are still isolates obtained from patients suffering from diarrhea which are negative during detection of all the different adhesin variants (35). In a similar way, adhesion has been established as a crucial step within the ETEC infectious cycle (7); therefore, it is presumed that every strain should carry adhesins in order to cause illness. Furthermore, it is known that some ETEC classical and nonclassical adhesins stimulate a humoral immune response during infections, making them potential antigens for vaccine development. Therefore, identification of adhesins carried by clinically relevant ETEC strains lacking the previously identified adhesins and further detection of new variants are pending tasks to obtain a complete picture of the distribution of these potentially immunogenic proteins.
Here, we report the identification of the aal locus, which encodes CS23 as an adherence determinant of ETEC 1766a. This strain obtained from a child suffering from diarrhea was negative for 19 CF genes and for nonclassical tia, tibA, and etpA genes. The aal locus is composed of nine genes, and, according to their predicted functions, they are able to direct the assembly of a functional adhesin at the bacterial surface. This structure would be mainly constituted by the major structural subunit AalE, which determines the difference between CS23 and the previously described K88-related fimbriae. The AalA, AalD, AalF, AalG, and AalH proteins share significant identity with the K88-structural Fae proteins and are predicted to be part of the CS23 structure as minor subunits. Immunogold-labeling EM performed with antibodies directed against AalE indicated that it is located on the bacterial surface, and it suggested that CS23 is an afimbrial adhesin. The K88-related adhesins, also known as the κ family within the usher-chaperone-assembled structural group, are fibers 2 to 5 mm in diameter, except for that encoded by the locus of diffuse adherence (lda) gene carried by atypical enteropathogenic E. coli (EPEC) strains, which is an afimbrial adhesin (20, 27). The minor structural subunits and proteins involved in assembly of K88, Lda, and CS23 share a high level of similarity, so it is not surprising that CS23 might be arranged as an afimbrial structure. Even though the AalE immunogold-labeling electron micrographs support this observation, the results are not conclusive and further analysis might be required. In fact, the reduction of ETEC 1766a adhesion capacity in the presence of anti-AalE serum was milder than that observed after the usher gene inactivation. The level was significantly lower than the adhesion capacity in the absence of antibodies but not significantly lower than the adhesion capacity in the presence of preimmune serum. This indicates that, probably, the polyclonal AalE serum is useful to detect the denatured protein by immunoblot analysis but only recognizes some immunogenic regions within the assembled CS23 structure.
Genetic relationships between human and porcine ETEC fimbriae as found for CS23 and K88 have been previously reported. Loci encoding CS12, CS18, and CS20, in strains causing diarrhea in humans, share similarity with the 987P locus carried by strains that infect pigs (18). These studies have suggested that some pathogenic ETEC strains evolved from a common ancestor. In the case of CS23 and K88, the divergence would have arisen from mutations in the major structural subunit gene which could account for the host specificity.
Interestingly, after comparison of the AalE amino acid sequence with those found in databases, the similarity was found with putative fimbrial structural subunits, displaying K88-family-type domains, produced by the plant pathogens Erwinia amylovora and Erwinia pyrifoliae. The Erwinia genus is part of the Enterobacteriaceae family and includes bacteria affecting mainly fruit trees and potatoes (15, 29). In addition to their putative role in human infection, their presence in plant pathogens suggests that K88-like fimbriae can mediate adherence to plant tissues. In a similar manner, pili, flagella, and the type III secretion system, normally required for adherence of enterohemorrhagic E. coli O157:H7 to animal tissues, have been associated with colonization of spinach leaves (26). To our knowledge, there are no reports showing similar results for ETEC structures, but considering that ETEC strains have been detected in vegetables (28), it is possible that their adhesins are also important to bind plant surfaces.
CS6, CS10, CS15, and CS22 have been previously described as afimbrial CFs, of which CS6 is one of the most frequently carried by ETEC strains causing diarrhea (19). Although it does not seem to possess a fimbrial morphology, CS23 is more closely related to the CS13 fimbria, which is also a K88-related structure. The presence of CS13 is not frequent within ETEC strains causing diarrhea in humans (23), and the absence of the aalE gene in Chilean strains suggests a similar scenario for CS23. If widely distributed ETEC adhesins are to be considered in a strategy for vaccine development, CS23 would most likely not be a suitable candidate compared with others that are more commonly identified in clinical isolates. Other CFs such as CS14, CS17, and CS18 were not frequently found in Chilean ETEC strains (6); however, they have been commonly detected in Guinea-Bissau (31), Bolivia (25), and Peru (24). Therefore, detection of CS23 could be performed in ETEC strains from other geographical regions in order to determine its actual global distribution.
A spontaneous attenuation causing reduction in the adherence ability of ETEC 1766a was noticed during the course of the experiments reported here. A similar event involving loss of an ST-encoding gene in an ETEC strain resulting from the deletion of DNA fragments from the plasmid which carried the gene has been previously described (30). In the case of ETEC 1766a, a mutation within the region that included aalA and aalB was the cause of the decrease in the adherence. The presence and expression of the remaining aal genes were detected, suggesting that the mutated fragment was essential to assemble a functional CS23 at the bacterial surface. This observation was further proved by allelic replacement of aalB. Results presented here suggest that the fragment containing aalA and aalB was replaced by a longer fragment in the attenuated ETEC 1766a strain, thus explaining the failure of the attempt to amplify the segment between aalR and aalC. It is well known that virulence-related traits are encoded within mobile elements and that these kinds of events have been attributed to recombinases encoded by transposons, insertion sequences, or phages, even when the specific stimuli that trigger these phenomena remain largely unidentified (1).
Rigid fibers were observed on the surface of ETEC 1766a, although they did not seem to be the main determinant of its adhesion capacity because they were also produced by the spontaneously attenuated strain, which was poorly adherent to Caco-2 cells. These fibers may be acting as adhesins in vivo, and other in vitro experimental models must be used to evaluate its functionality. Indeed, their identity remains to be determined. The presence of additional ETEC 1766a adhesion determinants was suggested by selection of G10 and G19 clones from the cosmid library. These clones carry sequences which conferred adhesion ability to E. coli HB101 at a lower level than that showed by G12. Further, the DNA restriction pattern of the purified recombinant cosmids was different. This suggested that the acquired adherence phenotype is determined by a factor distinct from the one encoded by the aal locus. Further investigation should allow identification of the fibrillar structures and/or additional ETEC 1766a adhesins. In conclusion, we have identified CS23 as a novel adhesin of this human ETEC strain.
ACKNOWLEDGMENTS
We thank Ann Mari Svennerholm and Halvor Sommerfelt for their suggestions and Miguel O'Ryan for critical reading of the manuscript. We thank Andrew Quest and Tulio Nuñez for providing us the cell line and Gabriela Mercado and Rosalba Lagos for providing us the cosmid vector.
The doctoral thesis of F.D.C. was supported by MECESUP higher education program project UCH407 and Doctoral Thesis Support Fellowship 24091103 (CONICYT, Ministerio de Educación, Gobierno de Chile). The work in the R.V. laboratory was supported by FONDECYT 1110260 and grant ID 38874, “Diarrheal disease in infants and young children in developing countries,” from the Bill and Melinda Gates Foundation. The work in the A.G.T. laboratory was supported by NIH/NIAID grant 5R01AI079154.
Footnotes
Published ahead of print 29 May 2012
REFERENCES
- 1. Ahmed N, Dobrindt U, Hacker J, Hasnain SE. 2008. Genomic fluidity and pathogenic bacteria: applications in diagnostics epidemiology and intervention. Nat. Rev. Microbiol. 6:387–394 [DOI] [PubMed] [Google Scholar]
- 2. Boyer HW, Roulland-Dussoix D. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459–472 [DOI] [PubMed] [Google Scholar]
- 3. Crossman LC, et al. 2010. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J. Bacteriol. 192:5822–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Croxen MA, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8:26–38 [DOI] [PubMed] [Google Scholar]
- 5. Datsenko KA, Wanner BL. 2000. One step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Del Canto F, et al. 2011. Distribution of classical and nonclassical virulence genes in enterotoxigenic Escherichia coli isolated from Chilean children and tDNA screening for putative insertion sites for genomic islands. J. Clin. Microbiol. 49:3198–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dorsey FC, Fischer JF, Fleckenstein JF. 2006. Direct delivery of heat-labile enterotoxin by enterotoxigenic Escherichia coli. Cell. Microbiol. 8:1516–1527 [DOI] [PubMed] [Google Scholar]
- 8. Gascón J. 2006. Epidemiology, etiology and pathophysiology of traveler's diarrhea. Digestion 73:102–108 [DOI] [PubMed] [Google Scholar]
- 9. Gómez-Duarte OG, et al. 1999. Identification of lngA, the structural gene of longus type IV pilus of enterotoxigenic Escherichia coli. Microbiology 145:1809–1816 [DOI] [PubMed] [Google Scholar]
- 10. Göransson M, Forsman K, Uhlin BE. 1989. Regulatory genes in the thermoregulation of Escherichia coli pili gene transcription. Genes Dev. 3:123–130 [DOI] [PubMed] [Google Scholar]
- 11. Grewal HMS, et al. 1997. A new putative fimbrial colonization factor, CS19, of human enterotoxigenic Escherichia coli. Infect. Immun. 65:507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guttman JA, Finlay BB. 2008. Subcellular alterations that lead to diarrhea during bacterial pathogenesis. Trends Microbiol. 16:535–542 [DOI] [PubMed] [Google Scholar]
- 13. Heuzenroeder MW, Elliot TR, Thomas CJ, Halter R, Manning PA. 1990. A new fimbrial type (PCFO9) on enterotoxigenic Escherichia coli O9:H−LT+ isolated from a case of infant diarrhea in central Australia. FEMS Microbiol. Lett. 54:55–60 [DOI] [PubMed] [Google Scholar]
- 14. Hohn B, Collins J. 1980. A small cosmid for efficient cloning of large DNA fragments. Gene 11:291–298 [DOI] [PubMed] [Google Scholar]
- 15. Kim WS, Gardan L, Rhim SL, Geider K. 1999. Erwinia pyrifoliae sp. nov., a novel pathogen that affects Asian pear trees (Pyrus pyrifolia Nakai). Int. J. Syst. Bacteriol. 49:899–906 [DOI] [PubMed] [Google Scholar]
- 16. Korea CG, Ghigo JM, Beloin C. 2011. The sweet connection: solving the riddle of multiple sugar-binding fimbrial adhesins in Escherichia coli. Bioessays 33:300–311 [DOI] [PubMed] [Google Scholar]
- 17. Levine MM, et al. 1993. Epidemiologic studies of Escherichia coli diarrheal infections in low socioeconomical level peri-urban community in Santiago, Chile. Am. J. Epidemiol. 138:849–869 [DOI] [PubMed] [Google Scholar]
- 18. Nada RA, et al. 2011. Discovery and phylogenetic analysis of novel members of class b enterotoxigenic Escherichia coli adhesive fimbriae. J. Clin. Microbiol. 49:1403–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nuccio SP, Bäumler AJ. 2007. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71:551–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pichel M, Binsztein N, Viboud G. 2000. CS22, a novel human enterotoxigenic Escherichia coli adhesin, is related to CS15. Infect. Immun. 68:3280–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pizarro-Cerdá J, Cossart P. 2006. Bacterial adhesion and entry into host cells. Cell 124:715–727 [DOI] [PubMed] [Google Scholar]
- 23. Qadri F, Svennerholm AM, Faruque ASG, Sack RB. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment and prevention. Clin. Microbiol. Rev. 18:465–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rivera FP, et al. 2010. Genotypic and phenotypic characterization of enterotoxigenic Escherichia coli strains isolated from Peruvian children. J. Clin. Microbiol. 48:3198–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rodas C, et al. 2009. Development of multiplex PCR assays for detection of enterotoxigenic Escherichia coli colonization factors and toxins. J. Clin. Microbiol. 47:1218–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saldaña Z, Sánchez E, Xicohntencatl-Cortes J, Puente JL, Girón J. 2011. Surface structures involved in plant stomata and leaf colonization by Shiga-toxigenic Escherichia coli O157:H7. Front. Microbiol. 2:119 doi:10.3389/fmicb.2011.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Scaletsky ICA, Michalski J, Torres AG, Dulger MV, Kaper JB. 2005. Identification and characterization of the locus for diffuse adherence, which encodes a novel afimbrial adhesin found in atypical enteropathogenic Escherichia coli. Infect. Immun. 73:4753–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Singh G, Vajpayee P, Ram S, Shanker R. 2010. Environmental reservoirs for enterotoxigenic Escherichia coli in south Asian gangetic riverine system. Environ. Sci. Technol. 44:6475–6480 [DOI] [PubMed] [Google Scholar]
- 29. Smits TH, et al. 2010. Complete genome sequence of the fire blight pathogen Erwinia amylovora CFBP 1430 and comparison to other Erwinia spp. Mol. Plant Microbe Interact. 23:384–393 [DOI] [PubMed] [Google Scholar]
- 30. Sommerfelt H, et al. 1989. Mechanism of spontaneous loss of heat-stable toxin (STa) production in enterotoxigenic Escherichia coli. APMIS 97:436–440 [DOI] [PubMed] [Google Scholar]
- 31. Steinsland H, et al. 2002. Enterotoxigenic Escherichia coli infections and diarrhea in a cohort of young children in Guinea-Bissau. J. Infect. Dis. 186:1740–1747 [DOI] [PubMed] [Google Scholar]
- 32. Svennerholm AM, Tobias J. 2008. Vaccines against enterotoxigenic Escherichia coli. Expert Rev. Vaccines 7:795–804 [DOI] [PubMed] [Google Scholar]
- 33. Torres AG. 2008. Intestinal pathogenic Escherichia coli, p 1013–1029 In Barrett A, Stanberry L. (ed), Vaccines for biodefense and emerging and neglected diseases. Academic Press, St. Louis, MO [Google Scholar]
- 34. Torres AG, et al. 2007. Bile salts induce expression of the afimbrial LDA adhesin of atypical enteropathogenic Escherichia coli. Cell. Microbiol. 9:1039–1049 [DOI] [PubMed] [Google Scholar]
- 35. Turner SM, Scott-Tucker A, Cooper LM, Henderson IR. 2006. Weapons of mass destruction: virulence factors of the global killer enterotoxigenic Escherichia coli. FEMS Microbiol. Lett. 263:10–20 [DOI] [PubMed] [Google Scholar]
- 36. Xia Y, Forsman K, Jass J, Uhlinm BE. 1998. Oligomeric interaction of the PapB transcriptional regulator with the upstream activating region of pili adhesin gene promoters in Escherichia coli. Mol. Microbiol. 30:513–523 [DOI] [PubMed] [Google Scholar]
- 37. Zhang W, Fang Y, Francis D. 2009. Characterization of the binding specificity of K88ac and K88ad fimbriae of enterotoxigenic Escherichia coli by constructing K88ac/K88ad chimeric FaeG major subunits. Infect. Immun. 77:699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]