Abstract
Urease represents a critical virulence factor for some bacterial species through its alkalizing effect, which helps neutralize the acidic microenvironment of the pathogen. In addition, urease serves as a nitrogen source provider for bacterial growth. Pathogenic mycobacteria express a functional urease, but its role during infection has yet to be characterized. In this study, we constructed a urease-deficient Mycobacterium tuberculosis strain and confirmed the alkalizing effect of the urease activity within the mycobacterium-containing vacuole in resting macrophages but not in the more acidic phagolysosomal compartment of activated macrophages. However, the urease-mediated alkalizing effect did not confer any growth advantage on M. tuberculosis in macrophages, as evidenced by comparable growth profiles for the mutant, wild-type (WT), and complemented strains. In contrast, the urease-deficient mutant exhibited impaired in vitro growth compared to the WT and complemented strains when urea was the sole source of nitrogen. Substantial amounts of ammonia were produced by the WT and complemented strains, but not with the urease-deficient mutant, which represents the actual nitrogen source for mycobacterial growth. However, the urease-deficient mutant displayed parental colonization profiles in the lungs, spleen, and liver in mice. Together, our data demonstrate a role for the urease activity in M. tuberculosis nitrogen metabolism that could be crucial for the pathogen's survival in nutrient-limited microenvironments where urea is the sole nitrogen source. Our work supports the notion that M. tuberculosis virulence correlates with its unique metabolic versatility and ability to utilize virtually any carbon and nitrogen sources available in its environment.
INTRODUCTION
Mycobacterium tuberculosis, the etiological agent of tuberculosis (TB), establishes infection in adult humans primarily in the lungs. The pathogenesis of M. tuberculosis is generally and simplistically represented as a bimodal distribution between active and latent TB based on the presence or absence of clinical symptoms, respectively. However, at the bacterial cell level, it has been increasingly recognized that within the same infected individual, a wide continuous spectrum of physiological states exists between actively replicating and dormant bacilli, dictated by the host microenvironments encountered by the pathogen (2). Currently, the WHO estimates that one-third of the world's population is latently infected with TB, with 9.27 million new cases and 1.76 million deaths annually (45).
It is generally agreed that M. tuberculosis relies on its exceptional ability to replicate and/or persist within the changing and adverse microenvironments encountered within its human host. Previous studies have proposed various mechanisms for M. tuberculosis survival and persistence within the macrophage, suggestive of its extraordinary adaptability as an intracellular human pathogen. Several mycobacterial factors have been implicated in the phagosome maturation arrest in resting macrophages early in the infection, hence allowing effective intracellular bacterial multiplication. Examples include cell wall components such as lipoarabinomannan (LAM) (10), trehalose dimycolate (TDM) (15), and sulfolipids (12), but also proteins, including the serine-threonine kinase PknG (41), SapM phosphatase (31), or the rv3778c gene product (27). Upon onset of host adaptive immunity, however, mycobacteria translocate into a bactericidal phagolysosome, which is characterized by an acidic (pH 5.5), highly nitro-oxidative, nutrient-limiting, and possibly hypoxic microenvironment (17, 32). Mycobacterial genes involved in detoxification of reactive nitrogen intermediates and reactive oxygen intermediates (RNI/ROI), in protein and DNA repair, and in protection have been identified with proposed mechanisms to defend against the host stresses encountered within the phagolysosomal compartment (7). Moreover, a (limited) number of genes have been identified to be specifically involved in M. tuberculosis survival within activated macrophages. rv3671c, a gene encoding a membrane-associated serine protease, was recently found to be largely responsible for the ability of M. tuberculosis to resist acidity and maintain intrabacterial pH under in vitro acidic conditions and in activated macrophages (40). Previous work has also shown that the absence of a key enzyme of the glyoxylate shunt, isocitrate lyase (ICL), leads to an attenuated phenotype in gamma interferon (IFN-γ)-activated macrophages and in mice (24), suggesting a critical role for the central metabolism in intracellular persistence, in particular the ability to metabolize host-derived lipids.
Ureases have been described as a virulence factor in several pathogenic bacteria. Specifically, the alkalizing effect of ammonia produced upon ureolysis has been proposed to contribute to their pathogenesis. It has been evidently shown that the presence of urease activity does neutralize the acidic environment encountered by Helicobacter pylori and Yersinia enterocolitica, thereby promoting colonization and survival of these pathogens within the gastric mucosa (3, 19, 47). The M. tuberculosis urease has been functionally characterized previously (4), but its actual physiological role and contribution during infection have not been determined. Several studies have illustrated that the mycobacterial (Mycobacterium bovis BCG) urease contributes to alkalize the mycobacterium-containing vacuole in macrophages (13, 23, 34) and it was therefore proposed that the mycobacterial urease activity helps promote a more favorable environment for the intracellular persistence of bacilli. However, a urease-deficient M. bovis BCG mutant was not impaired in its ability to survive within THP-1 human macrophages and in mice (30). The role of the M. tuberculosis urease activity has not been reported so far.
The ability of the mycobacterial (BCG) urease to alkalize the microenvironment has been exploited to develop a novel BCG-based vaccine candidate consisting of a urease knockout BCG mutant expressing the Listeria monocytogenes listeriolysin (13). In the absence of the mycobacterial urease activity, BCG maintained a mild acidic pH within the bacillus-containing vacuole, providing an ideal pH environment for lysteriolysin to perforate the vacuole membrane. Lysis of the phagosome then promoted mycobacterial antigen translocation into the cytoplasm and apoptosis of infected macrophages, which resulted in stronger immune responses that provided better protection upon M. tuberculosis challenge. Similarly, immunization of mice and guinea pigs with a urease-deficient BCG strain expressing perfringolysin O and overexpressing key immunodominant antigens resulted in enhanced immune responses (37). Both the urease KO listeriolysin (VMP1002)- and perfringolysin O (AERAS-422)-expressing BCG vaccine candidates are currently undergoing phase I clinical trial.
Furthermore, the alkalizing effect produced by M. bovis BCG urease activity was also demonstrated to attenuate major histocompatibility complex (MHC) class II molecule expression in infected THP-1 human macrophages (34), which consequently led to the inhibition of CD4+ T-cell activation (23), thereby suggesting that the mycobacterial urease has a probable role in suppressing host immune responses.
In addition to their ability to raise the pH in their microenvironment, most ureolytic bacteria are also capable of assimilating urea as a nitrogen source in anabolic processes, such as amino acid biosynthesis, for bacterial growth (Fig. 1) (20, 43). While carbon metabolism has emerged as a major determinant of M. tuberculosis pathogenicity, nitrogen metabolism has received little attention. Previous microarray and gene deletion studies have concluded that M. tuberculosis preferentially utilizes carbon-based sources for in vivo persistence and sustainability (5, 9, 14, 33, 38). These carbon-based sources include fatty acids and cholesterol (25, 46) and lipids (28). Conversely, mycobacteria can utilize amino acids, in particular asparagine, glutamate, and aspartate, and are able to assimilate inorganic sources such as ammonium salts for in vitro replication (29). The M. tuberculosis urease activity and its gene expression were found to increase upon nitrogen deprivation (4), thus suggesting that urea may be a potential source of nitrogen for M. tuberculosis and other pathogenic mycobacteria. However, the role of the mycobacterial urease in nitrogen metabolism has not been reported.
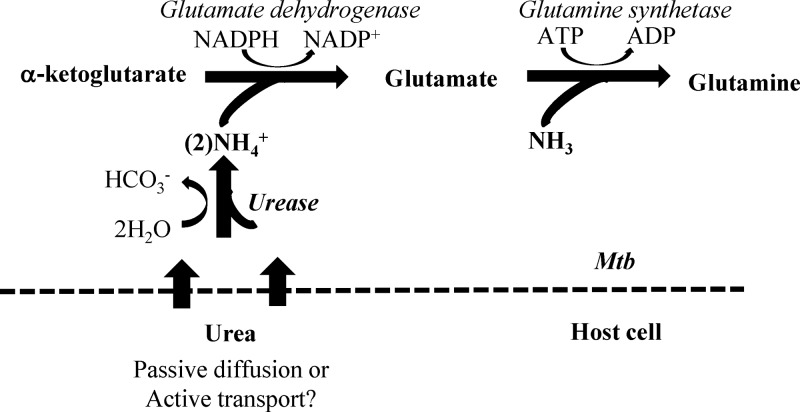
Fig 1.
Glutamate biosynthesis in Mycobacterium tuberculosis (Mtb) and urea hydrolysis. The proposed assimilation route of urea by M. tuberculosis for the biosynthesis of glutamate is shown (http://www.kegg.com).
In this study, we investigated the role of the urease activity in M. tuberculosis infection and nitrogen metabolism. We constructed a urease-deficient M. tuberculosis strain and confirmed the alkalizing effect of the M. tuberculosis urease activity in macrophages, although it did not confer any growth advantage to the pathogen. In contrast, we demonstrated that the M. tuberculosis urease activity plays a role in the bacterium central metabolism by providing a source of nitrogen when urea is readily available as a substrate.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. tuberculosis CDC1551 wild-type, derived mutant, and complemented strains were grown in Middlebrook 7H9 medium (Difco) supplemented with 10% ADS [50 g bovine fraction V albumin, 20 g d-(+)-glucose, 8.1 g sodium chloride per liter], 0.05% Tween 80, and 0.5% glycerol or on Middlebrook 7H11 agar containing oleic acid-albumin-dextrose-catalase (OADC; Becton Dickinson), 0.5% glycerol, and cycloheximide (100 μg/ml). When appropriate, hygromycin, kanamycin, and gentamicin were added at 80, 20, and 10 μg/ml, respectively.
To study the source of nitrogen, mycobacteria were grown in minimal medium which consisted of 2.5 g Na2HPO4, 1 g KH2PO4, 1 mg pyridoxine, 0.5 mg biotin, 15 mg ferric ammonium citrate, 40 g MgSO4, 0.5 mg CaCl2, 0.6 mg ZnSO4, 0.6 mg CuSO4, 0.8 g NaCl, and 0.5 g tyloxapol per liter of medium. Glutamate, urea (Sigma), or ammonium sulfate (Sigma) was added to the minimal medium when appropriate and at various concentrations. M. tuberculosis strains were grown in Middlebrook 7H9 medium to mid-log phase. The bacterial cells were washed in minimal medium. Subsequently, 10 ml minimal medium supplemented with 0.1% fatty-acid free bovine serum albumin (BSA; Sigma A8806), 0.2% glycerol, and the desired nitrogen source was inoculated with the washed bacterial culture at an initial optical density at 600 nm (OD600) of 0.05 ± 0.003 in a T25 flask (Nunc NunclonΔ flasks with filter caps). The flasks were incubated at 37°C for up to 10 days. The OD600 was measured every 2 days. Growth was scored as positive when the OD600 readout was 0.05 or higher.
ΔureC and complemented-strain construction.
A urease-negative mutant strain was generated in the M. tuberculosis CDC1551 background by homologous recombination using suicide plasmid backbone pYUB854, as previously described (1). Briefly, fragments of around 0.9 kb flanking the ureC open reading frame (ORF) were amplified from M. tuberculosis CDC1551 wild-type genomic DNA using the primers UreC5FP (TTACTAGTAGGTGCTCGGCCGTGACGAT), UreC5RP (TTAAGCTTGGTGGACGGCCCGACGAC), UreC3FP (TTTCTAGAGTCGTGCTCAAAGGTGGGGCG), and UreC3RP (TTCTTAAGTGTAGACGAGGTGCAAGGCGT) for 5′- and 3′-end PCR, respectively, with restriction sites underlined for cloning purposes. The PCR products were cloned into TOPO vector (Invitrogen), sequenced, and subcloned into the pYUB854 plasmid using the corresponding sites flanking the hygromycin resistance gene, hyg. A PacI-restricted fragment containing the selection genes lacZ and sacB was obtained from pGOAL17 (26) and cloned into the pYUB854-PCR5′-3′ construct to obtain the final delivery vector pYUB854-ureC. M. tuberculosis CDC1551 bacteria were electroporated with 1 μg of the UV-irradiated plasmid solutions as described previously (35). Hygromycin-resistant (Hygr) white colonies were selected, and deletion at the correct locus was verified by PCR using 2 sets of primers overlapping the deleted region (set 1, TGCGCCTCAATCATCCGGAGG [forward] and CACGAGCAGACCTCACTAGC [reverse]; set 2, ACTGCTTGTCCGATATCTGAT [forward] and TCTGCTCGCACAGTTCGGACA[reverse]) and Southern blot analysis (see method below). To generate an unmarked ΔureC mutant, the hygromycin resistance gene was removed by electroporating a plasmid encoding χδ-resolvase (tnpR) (1). The gentamicin-resistant clones were first selected after incubation at 31°C for the resolvase activity and selected again on 7H11 containing 2% sucrose after incubation at 39°C. Complementation of the unmarked ΔureC mutant was performed by amplifying the ureABC region from M. tuberculosis CDC1551 wild-type genomic DNA using primers UreAFP (TTTCTAGAACAACGCTTCGTGGACTTGG) and UreCRP (TTAAGCTTAGAACAGGAAATACCGTTGTG). The 3.2-kb PCR fragment was sequenced and cloned into integrative plasmid pMV306 (36). Upon electroporation of the ΔureC mutant, Kanr colonies were obtained and successfully complemented clones were confirmed by phenotypic growth in minimal medium with 3.5 mM urea.
Southern blot analysis.
Chromosomal DNA (1 μg) prepared from each M. tuberculosis strain was digested with SalI for 4 h and subjected to 0.8% agarose gel electrophoresis. The agarose gel containing the digested DNA was chemically treated and transferred onto a nitrocellulose membrane (Millipore) according to Roche's digoxigenin (DIG) application manual. The membrane was UV fixed for 1 min and equilibrated with 10 ml preheated DIG Easy Hyb solution (Roche) at 65°C for 20 min, with gentle agitation. A DIG-labeled probe was amplified using the PCR DIG probe synthesis kit (Roche) according to the manufacturer's instructions and using the following primers: CGCACCGTCGCAGAGTTGAT (forward) and GTATCCGGTCGCCGGTGGTA (reverse). For hybridization, about 5 to 25 ng/ml heat-denatured DIG-labeled DNA probe in DIG Easy Hyb solution was incubated with the membrane overnight at 65°C. Detection was performed using alkaline phosphatase (AP)-conjugated anti-DIG antibody (Roche) at a dilution of 1:5,000. The membrane was developed using nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP)-AP substrate (Chemicon).
RNA extraction and real-time PCR.
Mid-log-phase cultures of the M. tuberculosis strains were incubated with RNAprotect Bacteria reagent (Qiagen) for RNA stabilization. The pelleted bacteria were then resuspended in 100 μl Tris-EDTA (TE) containing 20 mg/ml lysozyme and incubated at room temperature for 20 min. RNA extraction was then performed using RNeasy Minikit (Qiagen) according to the manufacturer's instructions. Purified RNA was treated using the RNase free-DNase set (Qiagen) to remove contaminant DNA. Reverse transcription was performed on 10 ng bacterial RNA using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed in a 96 well-plate with each well containing 2 μl cDNA mix, 0.5 μl forward (F) and reverse (R) primers (0.5 μM final), and 25 μl SYBR green supermix with ROX (Bio-Rad) to a final volume of 50 μl. The primers used were ureC-F (GTCACCGAAGACCGGTGTGG), ureC-R (GGTTGCCGCTGATGATTTCGG), ureF-F (CCCTGGAAGCGTTCCTGAAAC), ureF-R (GTTCCTCCCAGCCGGAATCG), sigA-F (GCGACCAAAGCAAGCACGGC), and sigA-R (GTCGTAGTGTCCTGGGGTGC). Samples were run in triplicate. Real-time PCR amplification was conducted with the ABI Prism 7500 sequence detector (Applied Biosystems) over 40 cycles and with an annealing temperature of 61°C. Expression of each target gene was based on relative quantification using the comparative critical threshold (CT) value method. Relative quantification of a specific gene was evaluated in each reaction by normalization to the CT value obtained for the endogenous control gene, sigA. Control reactions without cDNA were used as negative controls.
Urease activity assay.
Urease activity in the M. tuberculosis WT, mutant, and complemented strains was determined using a BD BBL Taxo differentiation disc urea kit (Becton Dickinson, Inc., Sparks, MD), as previously described (16). Briefly, the strains were grown to mid-log phase and concentrated to 1 × 109 CFU/ml in Middlebrook 7H9 medium. The bacterial culture was incubated with the urea disc at 37°C. Colorimetric changes were monitored after 1 day and up to 10 days after urea disc introduction. A change of color of the suspension to red was scored as urease positive.
Ammonia detection assay.
The bacterial cultures were filtered using a 0.22-μm filter (Millipore) in order to obtain a cell-free spent culture medium. The ammonia (NH3) content in the spent culture medium was measured using an ammonia assay kit (Sigma; AA0100) by following the manufacturer's instructions. Briefly, 20 μl sample (undiluted and 10-fold dilution in minimal medium) was added to 200 μl ammonia assay reagent in a flat-bottom 96-well tissue culture plate and the mixture was incubated for 30 min before initial absorbance reading at 340 nm (OD340). l-Glutamate dehydrogenase (10 μl of a 10-fold dilution in DNase/RNase-free distilled water) was added to each well, and the reaction mixture was incubated for 5 min before absorbance was read at 340 nm. NH3 concentrations were calculated based on the formula given in the manufacturer's protocol.
Anaerobic shift-down assay.
The anaerobic shift-down assay was performed on the M. tuberculosis strains as previously described (39). Briefly, aerated precultures in Dubos complete medium were harvested at mid-log phase, washed twice in phosphate-buffered saline (PBS) supplemented with 0.05% Tween 80 (PBS-T), and resuspended in Dubos complete medium at a final OD600 of 0.1. Where indicated, the culture medium was supplemented with urea at a final concentration of 3.5 mM. The bacterial suspension was then distributed in 24-well tissue culture plates (1 ml/well). Methylene blue (1.5 μg/ml) was added as an indicator of oxygen depletion in control wells. The plates were incubated in airtight anaerobic jars (bioMérieux) with Anaerogen gas packs (Oxoid) to generate anaerobic atmospheric conditions. Atmospheric oxygen depletion was indicated by the anaerobic indicator strip (BD Diagnostics). Hypoxia was typically achieved within 24 h after incubation under anaerobic conditions, as evidenced by the complete decolorization of the oxygen sensor methylene blue. Survival was monitored by CFU count for up to 10 days after methylene decolorization. Each experimental sample consisted of triplicate wells.
Macrophage infection assay.
Marrow cells flushed from femurs of BALB/c mice were differentiated into macrophages over 7 days in Dulbecco modified Eagle medium (DMEM) (Difco) containing 0.58 g/liter l-glutamine, 1 mM sodium pyruvate, 10% fetal bovine serum (FBS), 10 mM HEPES, 100 U/ml penicillin-streptomycin, and 10 ng/ml recombinant macrophage colony-stimulating factor (M-CSF) (R&D Systems). For activated macrophages, 10% horse serum (Difco) was supplemented in the medium and activated with 100 U/ml recombinant mouse IFN-γ (Chemicon) and 10 ng/ml lipopolysaccharide (LPS) for 48 h prior to infection. Monolayers (5 × 104 cells/well) in 24-well plates (Nunc) were incubated with mycobacteria at a multiplicity of infection (MOI) of 2 for 45 min in incomplete culture medium (DMEM containing 0.58 g/liter l-glutamine, 1 mM sodium pyruvate, 10 mM HEPES, and 10 ng/ml recombinant M-CSF), after which the monolayers were washed and complete culture medium (DMEM containing 0.58 g/liter l-glutamine, 1 mM sodium pyruvate, 10 mM HEPES, 10% FBS, and 10 ng/ml recombinant M-CSF) was added to each well. At various time points, the monolayers were washed with PBS and lysed with 0.1% Triton X-100 (Sigma) to release the intracellular bacteria. The cell lysates were serially diluted in 7H9 medium and plated on 7H11 agar. The number of CFU was determined after incubation at 37°C for 16 days.
Phagosomal pH assay.
Assessment of the pH of M. tuberculosis phagosomes was performed according to previous literature (44). Briefly, M. tuberculosis bacteria were labeled with 40 μM pHrodo succinimidyl ester (SS; Invitrogen), which emits red fluorescence in an acidic environment, in PBS supplemented with 0.05% Tween 80 (pH 9) at 37°C for 1 h. The bacteria were then washed thrice with 7H9 to remove the excess dye. Resting or activated mouse bone marrow-derived macrophages (5 × 105/well) were then infected with the labeled M. tuberculosis strains at an MOI of 10 for 2 h. As a control, concanamycin C (200 nM), a potent inhibitor of the V-ATPase proton transport, was added to the infected macrophages. Noninternalized bacteria were then washed away, and the macrophages were further incubated for 2 h. The cells were trypsinized and fixed with 4% paraformaldehyde. The fixed cells were then analyzed by fluorescence-activated cell sorting (FACS) on BD LSR Fortessa and FlowJo 7.6.5 software.
Mouse infection.
All animal procedures were approved by the ethics committee of the Scientific Institute for Public Health-Veterinary and Agrochemical Research Centre (IPH-VAR) under protocol number 100219-01. B6D2/F1 mice (Janvier, Le Genest, France) were inoculated under anesthesia with M. tuberculosis WT and ΔureC strains via the intratracheal route at 104 CFU per mouse. Four animals per group were sacrificed. The lungs, spleen, and liver were aseptically harvested and homogenized in PBS with the antibiotic mixture PANTA (bioMérieux) by mechanical disruption using a homogenizer. The organ homogenates were appropriately diluted in PBS and plated onto 7H11 agar plates. CFU were enumerated after 3 to 4 weeks of incubation at 37°C.
Statistics.
Statistical significance was assessed by the Student t test, and two-tailed P values of less than 0.05 were considered statistically significant.
RESULTS
Construction of an unmarked M. tuberculosis ΔureC mutant and its complemented counterpart.
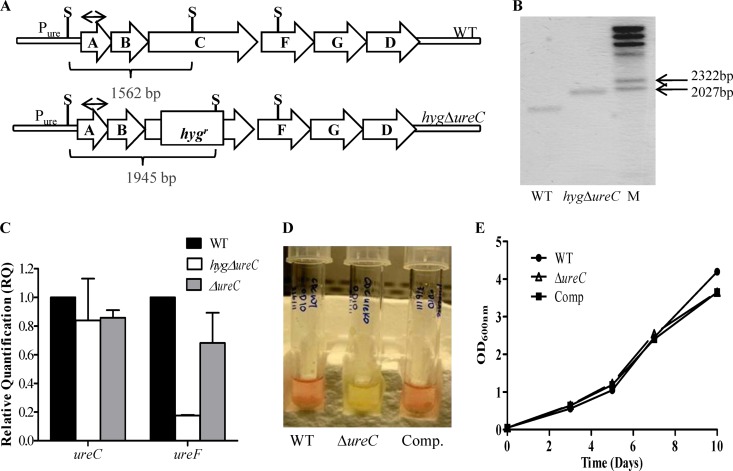
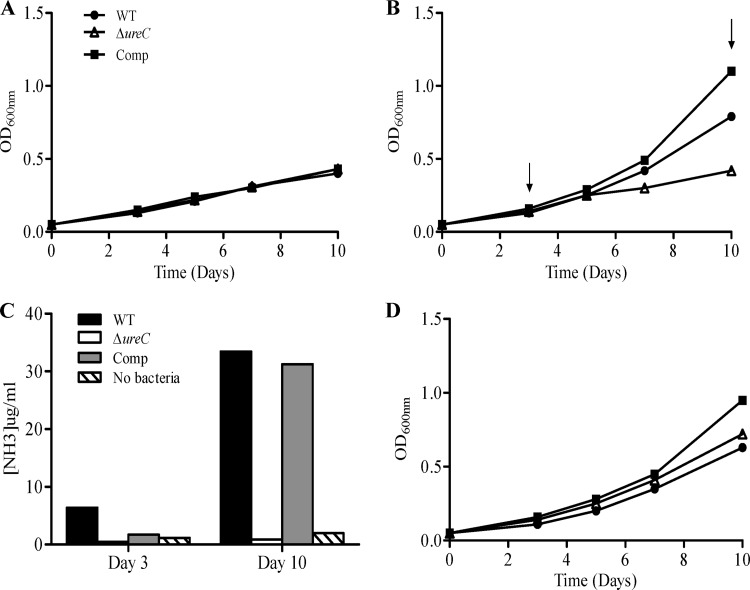
A urease-negative (ΔureC) mutant was constructed in the M. tuberculosis CDC1551 background. The ureC open reading frame (ORF), which encodes one of the three structural subunits of the holoenzyme (Fig. 2A), was deleted via double homologous recombination, leading to integration of a hygromycin resistance (hygr) cassette as described previously (1), which was further confirmed by Southern blotting (Fig. 2B). As the ureC ORF is part of an operon (Fig. 2A), the insertion of the hygr cassette had exerted a polar effect on the downstream genes of the operon as evidenced by their lower transcriptional activity, determined by real-time PCR analysis (Fig. 2C). Since such a polar effect would have precluded complementation studies, the hygr gene cassette was thus removed as previously described (1) to obtain the unmarked ΔureC mutant, in which the wild-type (WT) transcriptional level of the downstream genes in the operon was restored (Fig. 2C). The ΔureC mutant was then complemented whereby the ureABC portion of the urease operon expressed under the control of its original promoter was introduced in the ΔureC mutant using the promoterless integrative plasmid pMV306 (36). A colorimetric assay was performed to assess the urease activity in the ΔureC mutant, WT, and complemented strains as described elsewhere (16). While both cultures of the WT and complemented strains turned pink upon incubation with a urea disc, the culture of the ΔureC mutant remained yellow, indicating the absence of urease activity in the mutant strain (Fig. 2D). Furthermore, no growth defect was observed for the ΔureC mutant when grown aerobically in a standard culture medium (7H9) (Fig. 2E), indicating that the loss of the M. tuberculosis urease activity does not affect the general in vitro bacterial fitness.
Fig 2.
Construction of the M. tuberculosis ΔureC mutant. (A) Schematic organization of M. tuberculosis urease locus (ureABCFGD). The ΔureC mutant was obtained by introducing a hyg cassette into the ureC ORF (hygΔ ureC). Removal of the hyg cassette led to an unmarked ΔureC mutant. Restriction sites (RE) and Southern blot probe are indicated (S, SalI; probe represented by double-headed arrow). (B) Southern blot analysis. M, DIG-labeled molecular ladder. (C) Transcriptional activity of ureC and ureF ORFs in the hygΔ ureC and ΔureC mutants as determined by real-time PCR analysis. (D) Urease activity in WT, ΔureC, and complemented (Comp) strains determined by a colorimetric biochemical assay. (E) In vitro growth kinetics in 7H9 medium of the WT, ΔureC, and Comp strains.
M. tuberculosis urease contributes to the alkalization of the phagosomal pH but not to the persistence of bacilli in macrophages.
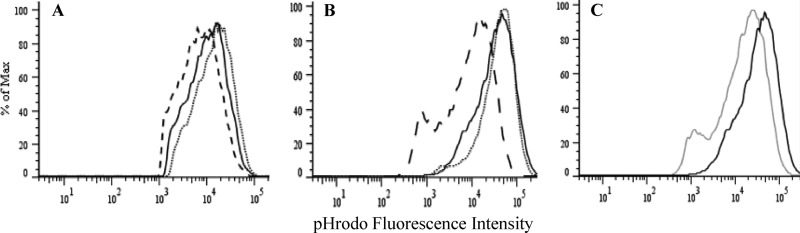
Earlier studies have suggested that the mycobacterial (BCG) urease may be involved in the intracellular persistence of the bacilli through alkalization of the pH within the mycobacterium-containing vacuole in macrophages (13, 23, 34). To examine this hypothesis in M. tuberculosis, resting or activated (with IFN-γ plus LPS) primary mouse bone-marrow derived macrophages (BMMΦ) were infected with WT, ΔureC, or complemented mycobacteria labeled with pHrodo, a pH-sensitive fluorescent dye which increases its fluorescence intensity with acidity (44). FACS analysis of the infected macrophages revealed a slight but significant shift for the ΔureC-infected resting BMMΦ compared to that observed with the WT strain, indicating a more acidic phagosomal environment for the mutant bacteria (Fig. 3A). This shift was not observed in activated infected BMMΦ, in which the mean fluorescence intensities for the ΔureC- and WT-containing vacuoles were comparable (Fig. 3B). This observation thus suggested that the mycobacterial urease activity in the WT strain is not sufficient to alkalize significantly the more acidic phagolysosomal environment within activated macrophages, unlike concanamycin C, a potent vacuolar-type H+-ATPase inhibitor (Fig. 3C). Interestingly, lower fluorescence intensities were clearly measured for the complemented strain than for the WT strain in both resting and activated macrophages, indicating a less acidic microenvironment for the complemented mycobacteria. This may indicate a greater urease activity in the complemented strain than in the WT strain. Integration of the ureABC fragment at a different genetic locus in the complemented strain may have resulted in its upregulation.
Fig 3.
Phagosomal pH measurement of infected macrophages. Shown are overlaid FACS histograms of pHrodo fluorescence intensities of resting (A) and activated (with IFN-γ plus LPS) (B) murine bone marrow-derived macrophages (BMMΦ) infected with pHrodo-labeled M. tuberculosis WT (solid line), ΔureC (dotted line), or Comp (dashed line) strains and activated BMMΦ infected with pHrodo-labeled WT with (gray line) or without (black line) 200 nM concanamycin (C). Results are representative of two independent experiments.
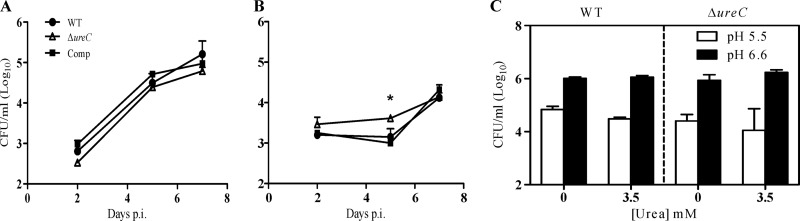
To correlate the alkalizing effect of the urease activity to the intracellular persistence of the bacilli, resting and activated BMMΦ were infected with the ΔureC mutant, WT, and complemented strains and their infection profiles were monitored over time. Similar infection profiles were observed in resting macrophages for all the strains (Fig. 4A). Interestingly, significantly higher CFU counts were obtained with the ΔureC mutant at day 5 postinfection in activated macrophages (Fig. 4B). One could speculate that the M. tuberculosis urease activity leads to accumulation of toxic product(s), although such toxicity was not seen in resting macrophages.
Fig 4.
Infection profile of M. tuberculosis ΔureC mutant in macrophages. Resting (A) and activated (with IFN-γ plus LPS) (B) murine bone marrow-derived macrophages (BMMΦ) were infected with M. tuberculosis WT, ΔureC, and complemented (Comp) strains at a multiplicity of infection (MOI) of 2. The infected cells were lysed and their bacterial CFU were assessed at 2, 5, and 7 days postinfection (p.i.). (C) Viability of WT and ΔureC mutant under anaerobic conditions at neutral (6.6) or mildly acidic (5.5) pH, in the presence or absence of 3.5 mM urea at day 10. Results are expressed as the means ± standard deviations of triplicates. Results are representative of two independent experiments. *, P = 0.0164.
In addition, the survival ability of the WT and ΔureC strains was assessed in a hypoxic acidic in vitro model, which is believed to partially mimic the microenvironment encountered by the bacilli within the phagosome of macrophages (39). Here, the culture medium was supplemented with 3.5 mM urea in order to test whether the alkalizing effect of the urease activity could improve mycobacterial survival. However, comparable bacterial counts between the WT and ΔureC strains were obtained under all the culture conditions tested, i.e., pH 6.6 or 5.5 and in the presence or absence of urea (Fig. 4C).
Together, these data thus indicate that the alkalizing effect of the urease activity does not provide M. tuberculosis with a growth advantage in macrophages or in an acidic hypoxic in vitro environment.
Urea utilization by M. tuberculosis occurs exclusively via the urease-dependent pathway.
In addition to their ability to neutralize the acidic environment for survival and colonization, urease-expressing human-pathogenic bacteria such as Bacillus cereus and Actinomyces naeslundii are able to utilize urea as a source of nitrogen during infection (20, 21). Furthermore, the M. tuberculosis urease activity was reported to be upregulated under nitrogen starvation conditions (4). We therefore hypothesized that the M. tuberculosis urease activity may be involved in the bacterium's central metabolism, by providing a source of nitrogen through the hydrolysis of urea and production of ammonia (Fig. 1). To test this hypothesis, the in vitro growth kinetics of the WT, the ΔureC mutant, and its complemented counterpart were examined in a minimal medium containing glycerol as the sole carbon source and urea as the sole nitrogen source. As expected, the absence of any source of nitrogen prevented the WT, ΔureC, and complemented strains from growing substantially (Fig. 5A), whereas the presence of 3.5 mM urea allowed exponential replication of the WT and complemented strains but not the ΔureC mutant (Fig. 5B). To correlate the utilization of urea by the strains with their respective growth kinetics, the amount of ammonia (NH3) produced in the culture medium was measured. Increasing levels of NH3 were detected over time in the culture medium inoculated with the WT and complemented strains (Fig. 5C), whereas no significant production of NH3 was detected in the culture medium inoculated with the ΔureC mutant, even at day 3 postinoculation, when the bacterial growth of the mutant is comparable to that of the WT and complemented strains (Fig. 5B).
Fig 5.
Utilization of urea as nitrogen source by M. tuberculosis. WT, ΔureC, and complemented (Comp) strains were grown in minimal media containing 0.2% glycerol only (A) or supplemented with 3.5 mM (B) or 71.4 mM (D) urea. The OD600s of the cultures were monitored over a period of 10 days. Arrows indicate time points where the amount of NH3 present in the culture media was measured (C). Results are representative of two independent experiments.
Interestingly, a higher exogenous urea concentration (71.4 mM) did not lead to enhanced growth of the WT or complemented strain compared to that observed with 3.5 mM urea, but it allowed the ΔureC mutant to replicate as efficiently as the WT strain (Fig. 5D). However, substantial levels of ammonia were detected in noninoculated culture medium supplemented with 71.4 mM urea, suggesting the occurrence of spontaneous hydrolysis of urea, thereby allowing the ΔureC mutant to grow (Table 1).
Table 1.
Spontaneous hydrolysis of ureaa
| Urea concn in defined liquid media (mM) | NH3 concn (mg/ml) |
|---|---|
| 0 | 1.72 |
| 3.5 | 1.97 |
| 71.4 | 5.34 |
Levels of NH3 in fresh minimal medium without urea, or supplemented with 3.5 or 71.4 mM urea, not inoculated with bacteria, were measured after 10 days of incubation. Results are representative of two independent experiments.
Together, these data indicated that the defect in urease activity impairs mycobacterial growth when urea is the sole source of nitrogen and that the by-product of ureolysis, ammonia, is the likely source of nitrogen for its growth.
Ammonia is the actual nitrogen source utilized for mycobacterial replication.
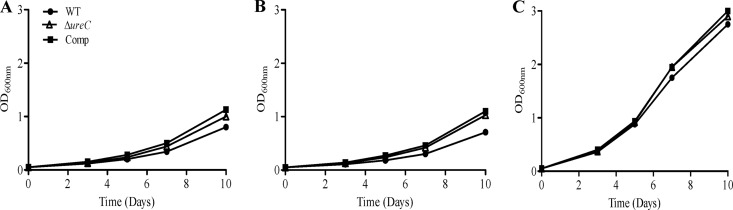
To confirm that the growth impairment observed with the ΔureC mutant is due to the inability to provide a source of ammonia, we assessed its growth profile in the presence of ammonium sulfate (NH4SO4). Consistently, the ΔureC mutant was able to grow as efficiently as the WT and complemented strains in the presence of 1 or 10 mM ammonium sulfate (Fig. 6A and B). Notably, the growth rates obtained with the WT and complemented strains in the presence of 1 or 10 mM NH4SO4 were comparable to that obtained when grown in 3.5 or 71.4 mM urea (Fig. 5B and D), supporting the idea that ammonia generated from urea hydrolysis through the urease activity is the actual source of nitrogen when urea is present in the culture medium. Furthermore, addition of l-glutamic acid, an essential amino acid that is synthesized from ammonia (Fig. 1), led to significantly greater growth rates for all the strains, indicating that the conversion of ammonia into l-glutamic acid is likely a limiting step in M. tuberculosis (Fig. 6C).
Fig 6.
Utilization of other nitrogen sources by M. tuberculosis. WT, ΔureC, and complemented (Comp) strains were grown in minimal media containing 0.2% glycerol supplemented with 1 mM (A) or 10 mM (B) NH4SO4 or 0.424 g/liter glutamic acid. OD600s of the cultures were monitored over a period of 10 days. Results are representative of two independent experiments.
Role of the mycobacterial urease activity in the mouse model of TB.
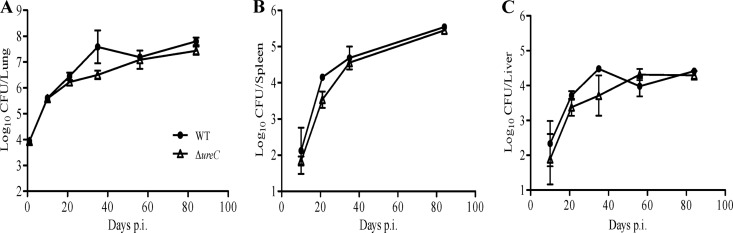
To assess the role and importance of the mycobacterial urease activity during in vivo TB infection, adult immunocompetent mice were infected via the intratracheal route with the WT and ΔureC strains and their infection profiles were monitored by determining the bacterial loads in their lungs, spleen, and liver at various time points postinfection. Comparable infection profiles in these three organs were obtained for both strains (Fig. 7). Therefore, these data indicated that the mycobacterial urease activity is not required for effective replication and persistence of M. tuberculosis in the mouse lungs, liver, and spleen.
Fig 7.
Survival of M. tuberculosis ΔureC mutant in the mouse model of tuberculosis. Bacterial loads were quantified in lungs (A), spleen (B), and liver (C) from mice infected with WT and ΔureC strains, as indicated, via the intratracheal route. Results are expressed as the means ± standard deviations for 4 mice per time point. Results are representative of two independent experiments.
DISCUSSION
In this study, we addressed the physiological role of M. tuberculosis urease activity. A urease-deficient mutant was constructed from the M. tuberculosis CDC1551 WT strain by deleting the ureC ORF, which encodes one of the structural subunits of the urease holoenzyme. The unmarked in-frame ureC deletion did not affect the transcriptional activity of the downstream genes in the urease operon, and complementation was achieved by expressing ureABC under the control of its original promoter.
Our results indicated that an absence of the urease activity did not impair the general in vitro bacterial fitness of M. tuberculosis. Using pHrodo-labeled mycobacteria, we demonstrated the alkalizing effect of the M. tuberculosis urease activity in resting macrophages, in which mycobacteria reside in a neutral phagosomal vacuole, consistent with previous studies performed with BCG (13, 23, 34), the surrogate model organism for M. tuberculosis. However, the M. tuberculosis urease activity failed to significantly alkalize the more acidic microenvironment of the phagolysosomal compartment in activated macrophages. This observation thus suggested that the alkalizing effect provided by the mycobacterial urease activity is somewhat modest. In addition, we showed that the urease activity and its alkalizing effect do not confer on the pathogen a greater ability to replicate or survive within macrophages, as evidenced by comparable infection profiles between the WT and urease-deficient mutant.
Understanding the metabolic needs of M. tuberculosis during infection is fundamental for identifying potential novel drug targets (29). Over the years, in vitro metabolism-related studies have demonstrated that M. tuberculosis is a flexible bacterium that is able to metabolize a wide variety of sources for growth (6). Here, we showed that M. tuberculosis is able to assimilate urea for growth and that this ability is urease dependent. When there is a deletion for urease activity, M. tuberculosis is greatly impaired in its growth ability when urea is provided as the sole source of nitrogen. We also showed that ammonia arising from ureolysis is the actual nitrogen source utilized by M. tuberculosis for its in vitro growth. Therefore, we propose that M. tuberculosis assimilates urea via its urease activity and that this process generates ammonia, which is one of the key precursors for the biosynthesis of essential amino acids such as glutamate.
Whereas no difference in growth profile was observed for the WT and urease-deficient mutant in macrophages ex vivo in culture medium containing a variety of carbon and nitrogen sources, the mycobacterial urease activity may still be important for intracellular growth and replication under nutrient-limited conditions. It has been reported that arginine hydrolytic enzyme arginase 1 was considerably induced in primary mouse macrophages upon mycobacterial infection (8). As urea is one of the products of arginase catalysis (22), M. tuberculosis could presumably use urea as a nitrogen source for production of compounds required for survival within the mammalian cells.
The apparent dispensability of the M. tuberculosis urease activity in the mouse model is further supported by the fact that upregulation of the mycobacterial urease operon or nitrogen metabolism-related genes has never been reported from microarrays of M. tuberculosis-infected activated macrophages and mouse models of tuberculosis, in contrast to genes involved in carbon-based metabolism (33, 38). It is possible that other readily available nitrogen sources within the host cell, such as ammonia from the metabolism of nitrogenous compounds, and nitrate (39), could bypass the need for M. tuberculosis to metabolize urea. It has indeed become very clear that functional redundancy and compensatory mechanisms are hallmarks of M. tuberculosis virulence, as illustrated by a recent study on M. tuberculosis fumarate reductase activity (42). In that study, the authors reported on the role of fumarate reductase and succinate dehydrogenase in energy metabolism and succinate secretion, but gene deletion did not lead to an attenuation phenotype in the Wayne model or in the mouse model. Thus, the absence of in vivo phenotype observed with a urease-deficient M. tuberculosis mutant likely reflects the metabolic versatility which provides M. tuberculosis with the ability to adapt to virtually any microenvironment encountered in its host, in which carbon and nitrogen sources may vary qualitatively and quantitatively.
Alternatively, the mycobacterial urease activity may be important for bacillus persistence or replication at specific sites of infection within the host. Indeed, the occurrence of extrapulmonary TB cases and the ability to infect virtually any organs and tissues imply the capability of M. tuberculosis to adapt extensively within its host and likely rely on its ability to utilize various sources of energy. Consequently, it is conceivable that the ability to utilize urea as a source of nitrogen could be critical at specific sites of infection where other sources of nitrogen are limited. For instance, during intestinal tuberculosis infection, urea-containing body fluids such as saliva (11) and tissue exudates (18), which coat the mucosal surfaces of the gastrointestinal tract, could provide a constant source of energy for the bacilli to maintain bacterial replication at the site of infection, hence promoting disease.
In conclusion, whereas we show that the alkalizing effect of the M. tuberculosis urease during macrophage infection does not appear to be involved in the pathogen's survival and persistence, we demonstrate a role for the ureolytic activity in nitrogen metabolism which could be critical for the pathogen's survival under conditions where urea represents the sole source of nitrogen. While it remains a challenge to address the actual metabolic needs of M. tuberculosis during in vivo infection, our findings hereby further support the metabolic versatility of this pathogen and contribute to fill the existing gaps in the knowledge of M. tuberculosis metabolic networks.
ACKNOWLEDGMENTS
We gratefully thank Pablo Bifani from the Novartis Institute for Tropical Diseases in Singapore for helpful discussions during the course of the study. We also thank Paul Edward Hutchinson, Lew Fei Chuin, and Tan Kar Wai for their assistance in FACS analysis.
This work was supported by the Biomedical Research Council of Singapore (Individual Research Grant BMRC 05/1/21/19/395 allocated to S.A.) and the Singapore-MIT Alliance for Research and Technology (pilot study grant allocated to S.A.).
Footnotes
Published ahead of print 29 May 2012
REFERENCES
- 1. Bardarov S, et al. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–3017 [DOI] [PubMed] [Google Scholar]
- 2. Barry CE, III, et al. 2009. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 7:845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burne RA, Chen YY. 2000. Bacterial ureases in infectious diseases. Microbes Infect. 2:533–542 [DOI] [PubMed] [Google Scholar]
- 4. Clemens DL, Lee BY, Horwitz MA. 1995. Purification, characterization, and genetic analysis of Mycobacterium tuberculosis urease, a potentially critical determinant of host-pathogen interaction. J. Bacteriol. 177:5644–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cole ST, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 6. Cook GM, et al. 2009. Physiology of mycobacteria. Adv. Microb. Physiol. 55:81–182, 318–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ehrt S, Schnappinger D. 2009. Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell. Microbiol. 11:1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. El Kasmi KC, et al. 2008. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat. Immunol. 9:1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fontán P, Aris V, Ghanny S, Soteropoulos P, Smith I. 2008. Global transcriptional profile of Mycobacterium tuberculosis during THP-1 human macrophage infection. Infect. Immun. 76:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fratti RA, Chua J, Vergne I, Deretic V. 2003. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc. Natl. Acad. Sci. U. S. A. 100:5437–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Golub LM, Borden SM, Kleinberg I. 1971. Urea content of gingival crevicular fluid and its relation to periodontal diseases in humans. J. Periodontal Res. 6:243–251 [DOI] [PubMed] [Google Scholar]
- 12. Goren MB, D'Arcy Hart P, Young MR, Armstrong JA. 1976. Prevention of phagosome-lysosome fusion in cultured macrophages by sulfatides of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 73:2510–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grode L, et al. 2005. Increased vaccine efficacy against tuberculosis of recombinant Mycobacterium bovis bacille Calmette-Guerin mutants that secrete listeriolysin. J. Clin. Invest. 115:2472–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hampshire T, et al. 2004. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis (Edinb.) 84:228–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Indrigo J, Hunter RL, Jr, Actor JK. 2003. Cord factor trehalose 6,6′-dimycolate (TDM) mediates trafficking events during mycobacterial infection of murine macrophages. Microbiology 149:2049–2059 [DOI] [PubMed] [Google Scholar]
- 16. Jassal MS, et al. 2010. 13[C]-urea breath test as a novel point-of-care biomarker for tuberculosis treatment and diagnosis. PLoS One 5:e12451 doi:10.1371/journal.pone.0012451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. MacMicking JD, Taylor GA, McKinney JD. 2003. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science 302:654–659 [DOI] [PubMed] [Google Scholar]
- 18. McLean RJ, Nickel JC, Cheng KJ, Costerton JW. 1988. The ecology and pathogenicity of urease-producing bacteria in the urinary tract. Crit. Rev. Microbiol. 16:37–79 [DOI] [PubMed] [Google Scholar]
- 19. Mobley HL, Island MD, Hausinger RP. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mols M, Abee T. 2008. Role of ureolytic activity in Bacillus cereus nitrogen metabolism and acid survival. Appl. Environ. Microbiol. 74:2370–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morou-Bermudez E, Burne RA. 1999. Genetic and physiologic characterization of urease of Actinomyces naeslundii. Infect. Immun. 67:504–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morris SM., Jr 2002. Regulation of enzymes of the urea cycle and arginine metabolism. Annu. Rev. Nutr. 22:87–105 [DOI] [PubMed] [Google Scholar]
- 23. Mukai T, Maeda Y, Tamura T, Miyamoto Y, Makino M. 2008. CD4+ T-cell activation by antigen-presenting cells infected with urease-deficient recombinant Mycobacterium bovis bacillus Calmette-Guerin. FEMS Immunol. Med. Microbiol. 53:96–106 [DOI] [PubMed] [Google Scholar]
- 24. Muñoz-Elías EJ, McKinney JD. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11:638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pandey AK, Sassetti CM. 2008. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U. S. A. 105:4376–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parish T, Stoker NG. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146(Pt 8):1969–1975 [DOI] [PubMed] [Google Scholar]
- 27. Pethe K, et al. 2004. Isolation of Mycobacterium tuberculosis mutants defective in the arrest of phagosome maturation. Proc. Natl. Acad. Sci. U. S. A. 101:13642–13647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peyron P, et al. 2008. Foamy macrophages from tuberculous patients' granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 4:e1000204 doi:10.1371/journal.ppat.1000204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ratledge C. 1976. The physiology of the mycobacteria. Adv. Microb. Physiol. 13:115–244 [DOI] [PubMed] [Google Scholar]
- 30. Reyrat JM, Lopez-Ramirez G, Ofredo C, Gicquel B, Winter N. 1996. Urease activity does not contribute dramatically to persistence of Mycobacterium bovis bacillus Calmette-Guerin. Infect. Immun. 64:3934–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saleh MT, Belisle JT. 2000. Secretion of an acid phosphatase (SapM) by Mycobacterium tuberculosis that is similar to eukaryotic acid phosphatases. J. Bacteriol. 182:6850–6853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schaible UE, Sturgill-Koszycki S, Schlesinger PH, Russell DG. 1998. Cytokine activation leads to acidification and increases maturation of Mycobacterium avium-containing phagosomes in murine macrophages. J. Immunol. 160:1290–1296 [PubMed] [Google Scholar]
- 33. Schnappinger D, et al. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sendide K, Deghmane AE, Reyrat JM, Talal A, Hmama Z. 2004. Mycobacterium bovis BCG urease attenuates major histocompatibility complex class II trafficking to the macrophage cell surface. Infect. Immun. 72:4200–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spratt JM, Britton WJ, Triccas JA. 2010. In vivo persistence and protective efficacy of the bacille Calmette Guerin vaccine overexpressing the HspX latency antigen. Bioeng. Bugs 1:61–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stover CK, et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460 [DOI] [PubMed] [Google Scholar]
- 37. Sun R, et al. 2009. Novel recombinant BCG expressing perfringolysin O and the over-expression of key immunodominant antigens; pre-clinical characterization, safety and protection against challenge with Mycobacterium tuberculosis. Vaccine 27:4412–4423 [DOI] [PubMed] [Google Scholar]
- 38. Talaat AM, Lyons R, Howard ST, Johnston SA. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. U. S. A. 101:4602–4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tan MP, et al. 2010. Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS One 5:e13356 doi:10.1371/journal.pone.0013356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vandal OH, Pierini LM, Schnappinger D, Nathan CF, Ehrt S. 2008. A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat. Med. 14:849–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walburger A, et al. 2004. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science 304:1800–1804 [DOI] [PubMed] [Google Scholar]
- 42. Watanabe S, et al. 2011. Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog. 7:e1002287 doi:10.1371/journal.ppat.1002287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Williams CL, Preston T, Hossack M, Slater C, McColl KE. 1996. Helicobacter pylori utilises urea for amino acid synthesis. FEMS Immunol. Med. Microbiol. 13:87–94 [DOI] [PubMed] [Google Scholar]
- 44. Wong D, Bach H, Sun J, Hmama Z, Av-Gay Y. 2011. Mycobacterium tuberculosis protein tyrosine phosphatase (PtpA) excludes host vacuolar-H+-ATPase to inhibit phagosome acidification. Proc. Natl. Acad. Sci. U. S. A. 108:19371–19376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. World Health Organization 2011. Global tuberculosis control report 2011. World Health Organization, Geneva, Switzerland: http://www.who.int/topics/tuberculosis/en/ [Google Scholar]
- 46. Yang X, Nesbitt NM, Dubnau E, Smith I, Sampson NS. 2009. Cholesterol metabolism increases the metabolic pool of propionate in Mycobacterium tuberculosis. Biochemistry 48:3819–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Young GM, Amid D, Miller VL. 1996. A bifunctional urease enhances survival of pathogenic Yersinia enterocolitica and Morganella morganii at low pH. J. Bacteriol. 178:6487–6495 [DOI] [PMC free article] [PubMed] [Google Scholar]