Abstract
Previously, we proposed a two-stage model for an in vitro neural correlate of eyeblink classical conditioning involving the initial synaptic incorporation of glutamate receptor A1 (GluA1)-containing α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid type receptors (AMPARs) followed by delivery of GluA4-containing AMPARs that support acquisition of conditioned responses. To test specific elements of our model for conditioning, selective knockdown of GluA4 AMPAR subunits was used using small-interfering RNAs (siRNAs). Recently, we sequenced and characterized the GluA4 subunit and its splice variants from pond turtles, Trachemys scripta elegans (tGluA4). Analysis of the relative abundance of mRNA expression by real-time RT-PCR showed that the flip/flop variants of tGluA4, tGluA4c, and a novel truncated variant tGluA4trc1 are major isoforms in the turtle brain. Here, transfection of in vitro brain stem preparations with anti-tGluA4 siRNA suppressed conditioning, tGluA4 mRNA and protein expression, and synaptic delivery of tGluA4-containing AMPARs but not tGluA1 subunits. Significantly, transfection of abducens motor neurons by nerve injections of tGluA4 flop rescue plasmid prior to anti-tGluA4 siRNA application restored conditioning and synaptic incorporation of tGluA4-containing AMPARs. In contrast, treatment with rescue plasmids for tGluA4 flip or tGluA4trc1 failed to rescue conditioning. Finally, treatment with a siRNA directed against GluA1 subunits inhibited conditioning and synaptic delivery of tGluA1-containing AMPARs and importantly, those containing tGluA4. These data strongly support our two-stage model of conditioning and our hypothesis that synaptic incorporation of tGluA4-containing AMPARs underlies the acquisition of in vitro classical conditioning. Furthermore, they suggest that tGluA4 flop may have a critical role in conditioning mechanisms compared with the other tGluA4 splice variants.
Keywords: GluA1, GluA4 siRNA, turtles, Trachemys scripta elegans, truncated, alternative splicing
ionotropic glutamate receptors of the α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid type (AMPARs) are heteromeric complexes assembled from four subunits—glutamate receptors A1–4 (GluA1–4)—each of which is expressed as alternatively spliced isoforms (Santos et al. 2009). Alternative splicing in the extracellular region of the third transmembrane domain near the C-terminal end yields variants termed flip and flop for each subunit. The alternative splicing results in a difference of only a few amino acids but differentially affects the kinetics of receptor function (Penn and Greger 2009). This region is not involved directly in ligand binding but modulates receptor desensitization and channel conductance. The expression of different GluA variants can be modulated by development, stress, stimulation parameters, and disease states. In spite of numerous studies characterizing AMPAR alternative splicing, the role of different GluA variants in synaptic plasticity and learning is not well understood. To address this question, our previous work characterized the sequence structure of the turtle GluA4 subunit, Trachemys scripta elegans (tGluA4), and identified 10 splice variants in the turtle brain (Sabirzhanov and Keifer 2011). Analysis of the expression pattern of tGluA4 mRNA and its alternatively spliced isoforms showed that tGluA4 flip and flop were the most abundantly expressed variants in the adult turtle brain, followed by tGluA4c flip and flop and a novel variant that is truncated at the C-terminal end, tGluA4trc1. Here, we were interested in determining whether these alternatively spliced isoforms of tGluA4 were differentially regulated in an in vitro model of classical conditioning (Keifer and Zheng 2010).
It is widely accepted that trafficking of AMPARs mediates rapid synaptic modifications underlying long-term potentiation (LTP), long-term depression, and learning (Derkach et al. 2007; Keifer and Zheng 2010). Previously, we developed an in vitro brain stem preparation from the turtle, which generates a neural analog of eyeblink classical conditioning in which to study the cellular and molecular mechanisms of this form of learning [see Keifer et al. (1995), Keifer and Houk (2011), and Keifer and Zheng (2010) for reviews]. Brain tissue from turtles is remarkably resistant to hypoxia, thereby allowing preservation of neural circuits in a dish for extensive periods, which can be subjected to pharmacological agents or incubation procedures. In place of using a tone or airpuff for behaving animals, stimulation of the auditory nerve [the “tone” conditioned stimulus (CS)] is paired with the trigeminal nerve [the “airpuff” unconditioned stimulus (US)], which results in the gradual acquisition of burst discharge in the abducens nerve (controlling blinking in this species) in response to the CS that is representative of a neural correlate or “fictive” blink conditioned response (CR). Our previous studies of in vitro eyeblink classical conditioning suggested that acquisition of CRs is initiated by synaptic insertion of AMPARs containing tGluA1 subunits followed by the synthesis and synaptic incorporation of tGluA4 subunits (Keifer and Houk 2011; Keifer and Zheng 2010; Mokin et al. 2007; Zheng and Keifer 2009). Since the sequence and domain structure of tGluA4 and its splice variants have now been characterized (Sabirzhanov and Keifer 2011), we developed a small-interfering RNA (siRNA) to selectively suppress expression of tGluA4 AMPAR subunits to directly test our two-stage model of AMPAR trafficking during in vitro classical conditioning. The results show that the acquisition of CRs corresponds with the expression and synaptic delivery of tGluA4 subunits and that this is inhibited by the anti-tGluA4 siRNA. Interestingly, conditioning suppressed by the siRNA can be rescued by a tGluA4 flop rescue plasmid but not a rescue plasmid to the flip variant. Finally, a siRNA directed against GluA1 inhibits conditioning as well as synaptic delivery of tGluA1- and tGluA4-containing AMPARs. These data support our model that synaptic incorporation of tGluA1 subunits is required to unsilence auditory synapses to allow for the later delivery of tGluA4-containing AMPARs required for acquisition of CRs.
MATERIALS AND METHODS
Conditioning procedures.
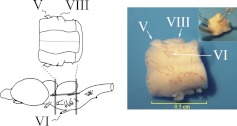
Freshwater pond turtles, T. scripta elegans, purchased from commercial suppliers, were anesthetized by hypothermia until torpid and decapitated. All experiments involving the use of animals were performed in accordance with the guidelines of the National Institutes of Health, and protocols were approved by the University of South Dakota Institutional Animal Care and Use Committee. The brain stem was transected at the levels of the trochlear and glossopharyngeal nerves, and the cerebellum was removed as described previously (Anderson and Keifer 1999). The preparation consists of only the pons with intact cranial nerves V through VIII, as shown in Fig. 1. The brain stem was bathed continuously (2–4 ml/min) with physiological saline containing (in mM): 100 NaCl, 6 KCl, 40 NaHCO3, 2.6 CaCl2, 1.6 MgCl2, and 20 glucose, which was oxygenated with 95% O2/5% CO2 and maintained at room temperature (22–24°C) at pH 7.6. Suction electrodes were used for stimulation and recording of cranial nerves (Fig. 1). The US was a twofold threshold single shock applied to the trigeminal nerve, and the CS was a 100-Hz, 1-s train stimulus applied to the ipsilateral auditory nerve, which was below threshold amplitude required to produce activity in the abducens nerve. Neural responses were recorded from the ipsilateral abducens nerve that innervates the extraocular muscles controlling movements of the eye, nictitating membrane, and eyelid. The CS–US interval was 20 ms, which was defined as the time between the CS offset and the onset of the US. The intertrial interval between the paired stimuli was 30 s. A pairing session was composed of 50 CS–US presentations, followed by a 30-min rest period, during which no stimuli were delivered. CRs were defined as abducens nerve activity that occurred during the CS and exceeded an amplitude of double the baseline recording level. All treatment groups received paired CS–US stimulation except for pseudoconditioned controls, which received the same number of CS and US exposures that were explicitly unpaired using a CS–US interval randomly selected between 300 ms and 25 s.
Fig. 1.
Illustration showing the in vitro brain stem preparation used in the present study. The preparation consists of only the pons with intact cranial nerves V through VIII. The cerebellum is removed. The drawing shows the pons dorsal-side up, viewing the 4th ventricle. The photograph shows a typical preparation ventral-side up, in which the abducens (VI) nerve is accessible for recording of reflex responses by a suction electrode (inset: anterior is downward). Stimulation of the trigeminal (V) and auditory nerves (posterior VIII) is also achieved with suction electrodes. Since turtles lack muscles of facial expression, nerve VII does not contribute to blinking.
Design of siRNAs and application procedures.
To target the tGluA4 AMPAR gene, dicer-substrate duplex RNAs consisting of 27-mer with two base 3′ overhangs (siRNA) were designed by RNA interference (RNAi) design software (Integrated DNA Technologies, Coralville, IA). First, dicer-substrate duplex RNA targeting the tGluA4 target gene was designed to bind to all 10 isoforms of tGluA4 (GenBank Accession Numbers: HM209319, HM209320, HM209321, HM209322, GU079942, GU079943, GU079944, GU079945, GU079946, and GU079947) (Sabirzhanov and Keifer 2011) at site 895–921 bp. This site is within the leucine/isoleucine/valine binding protein-like encoding region of tGluA4 mRNA, which is common to all isoforms. The sense sequence was 5′-GUACACAUCUGCUCUGACCUACGAT-3′, and the antisense sequence was 5′-AUCGUAGGUCAGAGCAGAUGUGUACUU-3′. The designed duplex RNAs were checked by the Basic Local Alignment Search Tool (BLAST) database (http://blast.ncbi.nlm.nih.gov/) to test for unique targets. Preliminary experiments showed that the tGluA4 siRNA had the most effective suppression of tGluA4 mRNA or protein expression in cell cultures at a concentration of 100 nM. For a negative control (NC), we used the Silencer NC #7 siRNA (Ambion, Austin, TX). This siRNA was designed to have no significant homology to any known gene sequences from human, mouse, or rat species and was used in a previous study (Keifer et al. 2009). The GluA1 siRNA was obtained from commercial suppliers (sc-35485, Santa Cruz Biotechnology, Santa Cruz, CA). Our preliminary experiments using this siRNA showed the most effective inhibition occurred after incubation for 6–12 h at 100 nM. Therefore, turtle brain stem preparations were incubated with 100 nM anti-tGluA4 siRNA or Silencer NC #7 mixed in Lipofectamine RNAiMAX (Invitrogen, Life Technologies, Carlsbad, CA; 2.5 μl/ml) and oxygenated physiological saline at room temperature for 25 h or anti-GluA1 siRNA for 12 h. Preparations were washed for 1 h in saline, followed by conditioning for two pairing sessions (C2). After conditioning, they were frozen in liquid nitrogen for later analysis by real-time RT-PCR or Western blotting or fixed in cold 0.5% paraformaldehyde for immunocytochemistry.
tGluA4 siRNA-resistant constructs (rescue plasmids) and application.
Rescue plasmids were designed for major tGluA4 isoforms found in the adult turtle brain. The sequence corresponding to full-length tGluA4 flip, tGluA4 flop, or tGluA4trc1 was inserted into a pCIG expression vector between the XhoI and HindIII sites. The rescue plasmids for tGluA4 flip and flop duplex RNA were made by introducing four single nucleotide mutations into the target region of the anti-tGluA4 siRNA using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA; Table 1). Rescue plasmids for tGluA4trc1 contained three single nucleotide mutations (Table 1). The plasmids will express mRNA for only the tGluA4 variant of interest because of sequence differences among them. For introduction of plasmids into brain stem preparations, a 0.135-pM concentration (1 μl total volume) of rescue plasmid was pressure microinjected into the cut end of the abducens nerve to allow for axonal transport, and preparations were incubated in oxygenated physiological saline for 8 h. This procedure allowed for transfection of abducens motor neurons that convey the CR and unconditioned blink response (UR) (Keifer et al. 2009). After incubation, 100 nM anti-tGluA4 siRNA mixed in Lipofectamine RNAiMAX (Invitrogen, Life Technologies; 2.5 μm/ml) was added to the bath, and preparations were incubated for an additional 17 h, followed by washing and conditioning for two pairing sessions as described above. After the physiological experiments, tissue was frozen in liquid nitrogen and analyzed for expression of tGluA4 and its variants by real-time RT-PCR or detected by immunocytochemistry.
Table 1.
Mutations in the tGluA4 siRNA target region for the rescue plasmids (mutations are underlined)
| Rescue plasmids for tGluA4 flip and flop dsRNA | |
| Target region of siRNA | AAGTACACATCTGCTCTGACCTACGAT |
| mutation | AAGTATACATCCGCTCTCACGTACGAT |
| Rescue plasmids for tGluA4trc1 dsRNA | |
| Target region of siRNA | GGGTCTGTCTGTCTATTGAAATACTAA |
| mutation | GGGTCGGTCTGTCGATTGGAATACTAA |
Plasmids for glutamate receptor A4 subunit and its splice variants from pond turtles, Trachemys scripta elegans, (tGluA4) flip and flop contained 4 nucleotide mutations into the target region of the small-interfering RNA (siRNA), whereas plasmids for a novel, truncated variant of tGluA4 at the C-terminus (tGluA4trc1) contained 3 nucleotide mutations. dsRNA, double-stranded RNA.
Cell culture and transfection.
COS-1 cells were maintained in DMEM, supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C and 5% CO2 atmosphere. Sequences corresponding to full-length tGluA4 flip or flop or the tGluA4trc1 variants were inserted into a pcDNA3.1/V5-HisTOPO TA expression vector. The resulting recombinant proteins contained a V5 epitope and polyhistidine region at the C-terminus. Cells at 70% confluence were transfected with 15 μg plasmid, which was diluted in 100 μl Opti-MEM I reduced serum medium. Transfection of cells was performed using Lipofectamine LTX reagent (Invitrogen, Life Technologies; 25 μl), incubated for 30 min at room temperature.
Real-time RT-PCR.
Total RNA was isolated from turtle brain stems or cell cultures using TRI Reagent (Molecular Research Center, Cincinnati, OH). Brain stem samples were obtained by dissecting a thick slab of tissue from the pons containing the abducens nuclei, only on the stimulated side. Real-time RT-PCR was performed using 50 ng total RNA/reaction. RNA was combined with primer/probe sets and TaqMan Gold RT-PCR Master Mix, which contains MultiScribe RT and RNase inhibitor. Gene-specific primers and probes were created for T. scripta elegans to total tGluA4 and its variants tGluA4 flip, tGluA4 flop, tGluA4c flip, tGluA4c flop, tGluA4trc1, and turtle actin, using Primer Express Software (Applied Biosystems, Life Technologies, Carlsbad, CA), and these are shown in Table 2 (Sabirzhanov and Keifer 2011). Target specificity of each primer and probe set was confirmed by performing real-time RT-PCR reactions with clones of identified tGluA4 variants. Each primer/probe set was specific only for its target tGluA4 variant. Efficiency of reactions for each set of primers was near 100% (Sabirzhanov and Keifer 2011) and was measured using the comparative threshold (CT) slope method. The CT values were plotted vs. the log of the dilution, and a linear regression was performed. Efficiency = (10−1/slope − 1) × 100% (Pfaffl 2001). Real-time assays were run on an ABI PRISM 7000 analyzer (Applied Biosystems, Life Technologies). The RT-PCR profile consisted of one cycle at 48°C for 30 min and 95°C for 10 min, followed by 55 cycles at 95°C for 15 s and 60°C for 1 min. All reactions were performed twice. Samples were confirmed to be free of DNA contamination by performing reactions without RT. Total RNA from six animals was used for each experimental group. Real-time RT-PCR data were normalized to turtle actin, which was unchanged by the conditioning procedures, and analyzed by the 2-ΔCT method (Livak and Schmittgen 2001) by an investigator blinded to the identity of the experimental groups.
Table 2.
Primers and probes used for real-time RT-PCR
| Target | Forward primer | Reverse primer | MGB probe |
|---|---|---|---|
| tGluA4 | Total tGluA4 forward CTATTATGGAAAAAGCGGGACAAA | Total tGluA4 reverse CATTGGCTCCTCCATGCATA | Total tGluA4 ACATCGTTGCAAACCTGGGATTCAAAGATAT |
| tGluA4 flip | tGluA4/tGluA4c flip forward GGTGAATGTGGAGCCAAGGACT | tGluA4 flip/flop reverse TTCAGAAAAGGTCAACTTCATTCGCTT | tGluA4/tGluA4c flip/flop CTTCTACATTCTGGTTGGAGGCTTGGGCT |
| tGluA4 flop | tGluA4/tGluA4c flop forward GCAGCGGGGGAGGTGAC | tGluA4 flip/flop reverse TTCAGAAAAGGTCAACTTCATTCGCTT | tGluA4/tGluA4c flip/flop CTTCTACATTCTGGTTGGAGGCTTGGGCT |
| tGluA4c flip | tGluA4/tGluA4c flip forward GGTGAATGTGGAGCCAAGGACT | tGluA4c flip/flop reverse GAGGAAGTTGGATTAAAAGTCTGTGC | tGluA4/tGluA4c flip/flop CTTCTACATTCTGGTTGGAGGCTTGGGCT |
| tGluA4c flop | tGluA4/tGluA4c flop forward GCAGCGGGGGAGGTGAC | tGluA4c flip/flop reverse GAGGAAGTTGGATTAAAAGTCTGTGC | tGluA4/tGluA4c flip/flop CTTCTACATTCTGGTTGGAGGCTTGGGCT |
| tGluA4trc1 | tGluA4trc1 forward GGAAATGTTCAGTTTGATCACTATGG | tGluA4trc1 reverse TCAATAGACAGACAGACCCTTTCAGATA | tGluA4trc1 probe TCGCAGAGTCAACTACACAATGGATG |
| Actin | Actin forward AGGGAAATCGTGCGTGACAT | Actin reverse GCGGCAGTGGCCATCTC | Actin probe AAGCTGTGCTATGTTGC |
MGB, minor groove binder.
Western blot analysis.
Immediately after the physiological experiments, brain stem samples were obtained by dissecting a thick slab of tissue from the pons containing the abducens nuclei on the stimulated side, frozen in liquid nitrogen, and stored at −70°C. Tissue was homogenized in lysis buffer (20 mM Tris, pH 8.0, 1 mM EDTA, 1% Nonidet P-40, 0.15 M NaCl, 10 mM Na4P2O7, and 5% glycine) with a protease (Roche, Mannheim, Germany) and phosphatase inhibitor cocktail (Sigma, St. Louis, MO), rotated at 4°C for 2 h, and centrifuged at 14,000 g for 20 min at 4°C, and the supernatants were aliquoted and stored at −70°C. Protein sample concentrates were solubilized in 2× SDS/β-mercaptoethanol and boiled for 5 min before separation by 8% SDS-PAGE. After electrophoresis, samples were transferred to polyvinylidene difluoride membranes and blocked with 5% nonfat dry milk in Tris-buffered saline/0.1% Tween-20 for 1 h at room temperature. Membranes were probed with the following primary antibodies overnight at 4°C: GluA1 (1:750; Millipore, Billerica, MA), GluA4 (1:1,000; Millipore), or GluA2/3 (1:1,000; Millipore). The specificity of the antibodies was confirmed by Western blot. They were then incubated with horseradish peroxidase-conjugated secondary antibodies (1:10,000) for 1 h at room temperature. Loading controls were performed using a primary antibody to actin (1:1,000), followed by fluorescently tagged secondary antibody (1:5,000). Proteins were detected by the ECL Plus chemiluminescence system (Amersham Pharmacia, GE Healthcare Biosciences, Piscataway, NJ) or the Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE).
Glutamate receptor localization, confocal imaging, and data analysis.
Immediately after the physiological experiments, brain stems were immersion fixed in cold 0.5% paraformaldehyde. Tissue sections were cut at 30 μm and incubated in primary antibody overnight at 4°C with gentle shaking. The primary antibodies used were a goat polyclonal, which recognizes the GluA4 subunit of AMPARs (1:100; Santa Cruz Biotechnology; Cat. #7614), a rabbit polyclonal, which recognizes GluA1 (1:200; Millipore; Cat. #1504), a rabbit polyclonal, which recognizes GluA2/3 (1:500; Millipore; Cat. #1506), and a mouse monoclonal, which recognizes synaptophysin (Syn; 1:1,000; Sigma; Cat. #5768). After the primary antibodies, sections were rinsed and incubated with secondary antibodies for 2 h, using a concentration of 1:100 for GluA4 and GluA1 or 1:200 for synaptophysin. The secondary antibodies were a Cy3-conjugated rabbit anti-goat IgG for GluA4, a DyLight 648-conjugated goat anti-rabbit IgG for GluA1, and a DyLight 488-conjugated goat anti-mouse IgG for synaptophysin (Jackson ImmunoResearch Laboratories, West Grove, PA), which were used to visualize the primary antibodies. After incubation in the secondary antibodies, sections were rinsed, mounted on slides, and coverslipped. Images of labeled neurons (∼25 cells/preparation) in the principal or accessory abducens motor nuclei were obtained using an Olympus FluoView 1000 laser-scanning confocal microscope. Tissue samples were scanned using a 60 × 1.4 numerical aperture oil-immersion objective with triple excitation with a 488-nm argon laser, a 543-nm HeNe laser, and a 635-nm diode laser. Examples of unprocessed confocal images (except that the contrast was increased) of abducens motor neurons from a conditioned preparation and one treated by the tGluA4 siRNA are shown in Fig. 2. Quantification of punctate staining of at least twofold greater intensity above background was performed using stereological procedures (Mokin and Keifer 2006) with MetaMorph software (Universal Imaging, Downington, PA). The software separates the image into individual color files (Fig. 2) where they are thresholded and puncta selected for analysis. Images of two consecutive optical sections were taken. Protein puncta were counted in one optical section (sample section) if they were not present in the optical section immediately below the sample section (look-up section) and if they were within the inclusion boundaries of the unbiased counting frame. Colocalized staining, indicating the presence of glutamate receptor subunits at synaptic sites, was determined when red, blue, and green puncta were immediately adjacent to one another or if they were overlapping. Data were analyzed with StatView software using a one-way ANOVA, followed by post hoc analysis using a Fisher's test, and treatment groups were compared with pseudoconditioning for two sessions (Ps2). Values are presented as means ± SE.
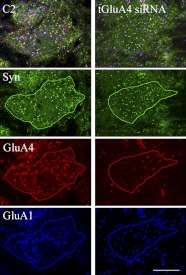
Fig. 2.
Confocal images of abducens motor neurons after normal conditioning for 2 pairing sessions (C2) or conditioning following treatment with the glutamate receptor A4 (GluA4) subunit small-interfering RNA (tGluA4 siRNA). Images are unprocessed except that the contrast was increased for the illustration. After drawing the outline of the cell of interest by the investigator, the software breaks the original image into its individual color channels, revealing punctate staining for synaptophysin (Syn; green), GluA4 (red), or GluA1 (blue) for quantitative analysis. Original scale bar = 10 μm.
RESULTS
Specificity of siRNAs and rescue plasmids in cell cultures and brain stems.
The GluA4 AMPAR subunit in turtle brain has 10 identified, alternatively spliced isoforms, including the major variants tGluA4 flip and flop, tGluA4c flip and flop, and tGluA4trc1 (Sabirzhanov and Keifer 2011). tGluA4 flip/flop together are the most abundantly expressed variants (55% of total tGluA4), followed by tGluA4c flip/flop (26% of the total) and tGluA4trc1 (15%). The tGluA4 siRNA used here was designed to bind to a common sequence shared by all of the variants and resulted in suppressed expression of the major isoforms. As shown in Fig. 3, COS-1 cells were transfected with tGluA4 flip, tGluA4 flop, or tGluA4trc1. Real-time RT-PCR showed that the levels of mRNA for all three variants were reduced significantly when cells were treated with the anti-tGluA4 siRNA compared with mRNA expression levels in untreated cells (Fig. 3; n = 3/group; tGluA4 flip, P < 0.0001; tGluA4 flop, P < 0.0001; tGluA4trc1, P < 0.0001, siRNA vs. control). Rescue of the RNAi effect by expression of a siRNA-resistant form of the targeted mRNA (rescue plasmids) is a powerful tool for determining the specificity of siRNA-directed inhibition of gene expression, and therefore, rescue experiments were performed. Because it is not possible to distinguish the flip and flop isoforms of tGluA4 by Western blot, we used real-time RT-PCR analysis of mRNA levels to verify the specificity of our rescue plasmids (Fig. 3). Compared with samples treated by the tGluA4 siRNA, cell cultures transfected with tGluA4 flip, tGluA4 flop, or tGluA4trc1 rescue plasmids containing nucleotide mutations in the target region of the siRNA showed substantial and selective resistance to the inhibitory effect of the tGluA4 siRNA. For example, cells transfected with tGluA4 flip and the anti-tGluA4 siRNA, followed by treatment with the rescue plasmid for tGluA4 flip, showed significantly elevated expression of tGluA4 flip mRNA (Fig. 3; P < 0.0001, rescue vs. siRNA) but not for tGluA4 flop mRNA (P = 0.46). Alternatively, cells transfected with tGluA4 flop and siRNA, followed by treatment with the rescue plasmid to tGluA4 flop, resulted in increased mRNA expression only for the flop variant (P = 0.0002). Likewise, the tGluA4trc1 rescue plasmid had no effect on expression of tGluA4 flip or flop but resulted in increased mRNA only for the truncated isoform (P < 0.0001). Similar data for brain stem preparations can also be seen in Fig. 7, where mRNA expression for all of the major tGluA4 isoforms was suppressed by the anti-tGluA4 siRNA, whereas there was selective rescue of the flip, flop, and trc1 isoforms by application of the rescue plasmids. These experiments confirmed that mRNA expression of the major tGluA4 isoforms was suppressed by treatment with the tGluA4 siRNA and could be reversed selectively by treatment with the rescue plasmids.
Fig. 3.
Real-time RT-PCR analysis of tGluA4 mRNA expression in cell cultures treated with anti-tGluA4 siRNA and rescue plasmids. Cells transfected specifically with constructs for tGluA4 flip or flop and truncated variant of tGluA4 (tGluA4trc1) were examined. Control cells were transfected with expression constructs alone. *Significant differences from control; #significant differences from siRNA and rescue treatment groups. P values are given in the text.
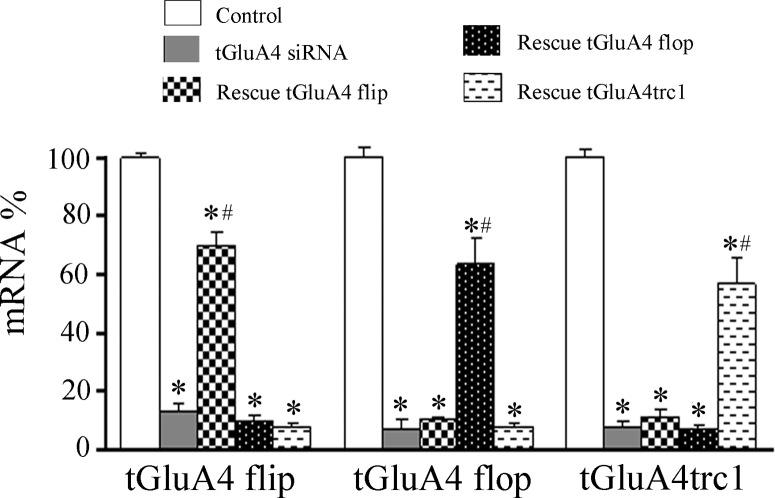
Fig. 7.
Expression of the major mRNA splice variants of tGluA4 after conditioning or treatment with siRNA followed by conditioning or application of rescue plasmid and anti-tGluA4 siRNA followed by conditioning. Total tGluA4 expression (splice variants combined) after pseudoconditioning was set at 100% for comparison with the other groups. *Significant differences from Ps2.
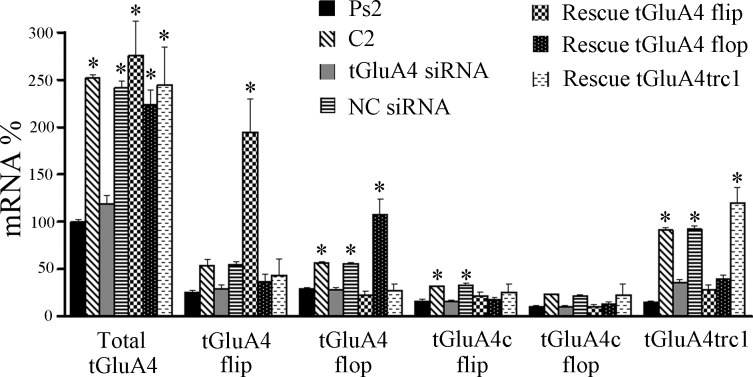
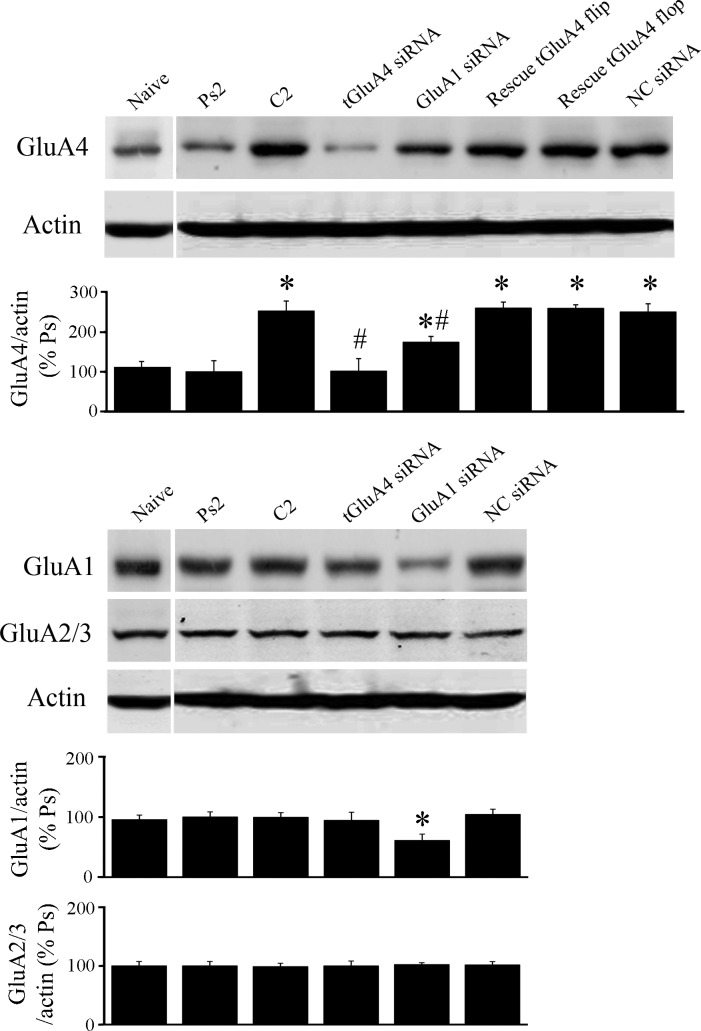
Analysis of protein expression in brain stems (n = 3 preparations/group), which underwent the conditioning procedure under different treatment conditions, was used to further assess the specificity of our RNAi reagents. Under basal or naïve conditions, there is normally a relatively low level of GluA4 expression that is not altered significantly after two sessions of pseudoconditioning (Fig. 4; P = 0.19, Ps2 vs. naive). However, blots of GluA4 showed a significant increase in protein expression after two pairing sessions of conditioning compared with pseudoconditioning (Fig. 4; P = 0.0003, C2 vs. Ps2), as has been observed previously (Mokin et al. 2007). Treatment with the anti-tGluA4 siRNA, followed by the conditioning procedure, resulted in greatly reduced GluA4 levels, which were similar to pseudoconditioning (P = 0.96, tGluA4 siRNA vs. Ps2; P = 0.0003 vs. C2), demonstrating the effectiveness of this siRNA on protein suppression. Preparations treated with the GluA1 siRNA showed a level of GluA4 protein expression that was elevated with respect to pseudoconditioning but was not as high as that observed for normal conditioning (P = 0.03, GluA1 siRNA vs. Ps2; P = 0.02 vs. C2). We have hypothesized that the expression of GluA4 is dependent on the first step of GluA1 AMPAR synaptic incorporation, which should be inhibited by the anti-GluA1 siRNA. However, it can be seen from Fig. 4 that the GluA1 siRNA did not completely eliminate GluA1 protein expression. We surmise that a small number of GluA1 AMPARs were incorporated into synapses during the anti-GluA1 siRNA/conditioning procedure and resulted in low levels of GluA4 synthesis but not enough to generate CRs. Preparations treated by coapplication of the tGluA4 flip or flop rescue plasmids with the anti-tGluA4 siRNA showed high levels of GluA4 (P = 0.0002 for both vs. Ps2), as did the NC group (P = 0.0003) which was treated with a nonspecific siRNA followed by conditioning. Importantly, analysis of the same preparations showed that the tGluA4 siRNA minimally affected expression of GluA1 compared with pseudoconditioning (P = 0.70), whereas the siRNA directed against GluA1 resulted in a significant reduction in GluA1 protein (Fig. 4; P = 0.02, GluA1 siRNA vs. Ps2). The relative amounts of GluA2/3 protein remained unchanged following the different treatments (F5,12 = 0.05, P = 0.99). These data indicate that the tGluA4 siRNA has its primary inhibitory effect on the GluA4 subunit and not the other AMPAR subunits. However, whereas the GluA1 siRNA suppressed expression of that subunit, it also reduced GluA4, likely through indirect effects on GluA1 trafficking, and had no effect on GluA2/3.
Fig. 4.
α-Amino-3-hydroxy-5-methylisoxazole-4-propionic acid type receptor (AMPAR) subunit protein expression in brain stem preparations from the different treatment groups. Some preparations were either given no stimulation (Naïve) or underwent pseudoconditioning (Ps2) or conditioning (C2) without further treatment. Other preparations were incubated in siRNA alone, followed by conditioning [tGluA4 siRNA, GluA1 siRNA, negative control (NC) siRNA]. Finally, preparations were injected with rescue plasmids into the abducens nerve for transfection of abducens motor neurons, followed by anti-tGluA4 siRNA treatment and C2 (rescue tGluA4 flip; rescue tGluA4 flop). Actin-loading controls are also shown. *Significant differences from Ps2; #significant differences from C2.
Conditioning is inhibited by tGluA4 siRNA and selectively rescued by tGluA4 flop plasmid.
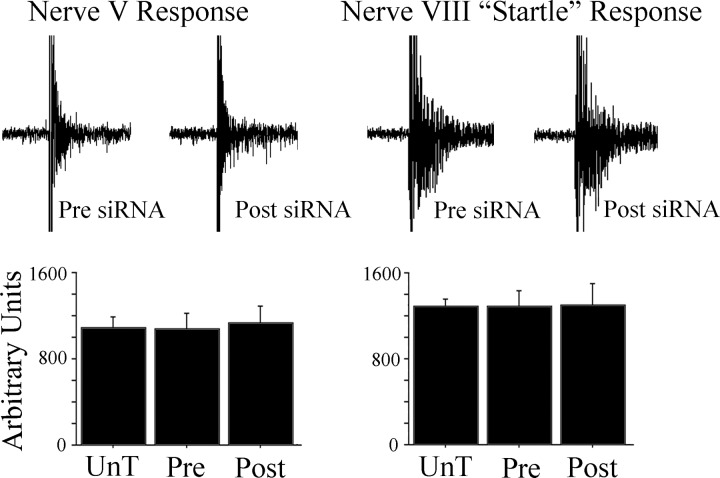
Prior to the conditioning experiments, the responsiveness of cranial nerve reflexes was used to assess whether basal synaptic transmission was adversely affected by the anti-tGluA4 siRNA. The neural correlates of the abducens UR produced by stimulation of the trigeminal nerve and the “startle” response evoked by strong stimulation of the auditory nerve were measured using area of the total response (Fig. 5). There were no significant differences in nerve V URs among untreated preparations (UnT) and the anti-tGluA4 siRNA treatment group for recordings taken either before (Pre) or after (Post) incubation in the siRNA (Fig. 5; n = 5 preparations/group; F2,12 = 0.1, P = 0.96). Similarly, neural correlates of the startle response generated to stimulation of the auditory nerve were not different Pre or Post anti-tGluA4 siRNA treatment or compared with group UnT (Fig. 5; F2,12 = 0.01, P = 0.99). Therefore, the general responsiveness of abducens motor neurons to stimulation of either the trigeminal or the auditory nerves was not affected by the siRNA treatment. This may be due to the lack of effect on tGluA2/3-containing AMPARs, which are likely to mediate, at least in part, these responses.
Fig. 5.
Physiological responsiveness of the nerve V unconditioned blink response (UR) and the nerve VIII “startle” responses is unaffected by treatment with the anti-tGluA4 siRNA. Comparison of data from untreated (UnT) preparations, those before treatment (Pre) with the GluA4 siRNA, and those after treatment (Post) shows no significant differences among the groups for either evoked reflexes.
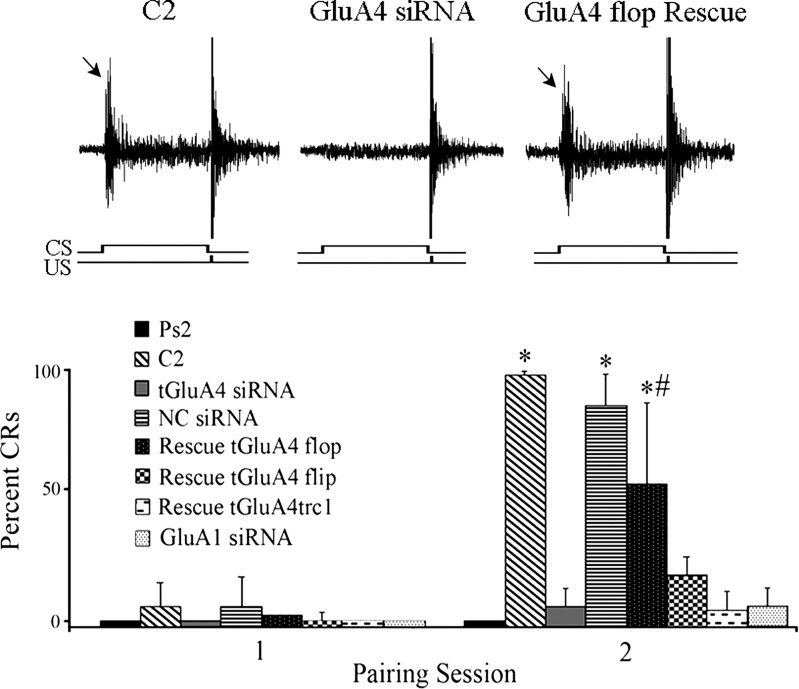
Application of the conditioning stimuli to brain stem preparations typically results in generation of abducens nerve CRs by the second pairing session. Here, the mean percent of CRs for all of the experimental groups, which were subjected to real-time RT-PCR analysis, is shown in Fig. 6, and the corresponding pattern of tGluA4 splice variant mRNA expression is presented in Fig. 7. Paired training in untreated preparations resulted in an average of 100% CRs in the second pairing session, whereas pseudoconditioning using unpaired stimuli resulted in 0% CRs (Fig. 6; n = 6 preparations/group; P < 0.0001, C2 vs. Ps2). A representative recording of a CR generated in response to normal conditioning is shown in Fig. 6. Conditioning resulted in a significant increase in the total amount of tGluA4 mRNA compared with pseudoconditioning (Fig. 7; n = 6 preparations/group; P < 0.0001), specifically for tGluA4 flop (P = 0.008) but not flip (P = 0.20), tGluA4c flip (P = 0.004) but not tGluA4c flop (P = 0.052), and tGluA4trc1 (P < 0.0001). These results confirm many of our previous studies showing that conditioning is associated with enhanced synthesis of tGluA4 AMPAR subunits. Transfection of brain stems with the anti-tGluA4 siRNA was performed, followed by conditioning, which significantly inhibited the generation of CRs to a mean of 6% (Fig. 6; P < 0.0001 vs. C2). As can be seen in the physiological record, a CR is not generated, but the UR appears robust. Correspondingly, all of the tGluA4 splice variant mRNA levels examined were reduced to nonsignificant values (Fig. 7; F1,10 = 4.2, P = 0.07 vs. Ps2). These findings verify that all of the major tGluA4 variants are suppressed by the siRNA used here. Application of the NC siRNA to preparations prior to the conditioning procedure, in contrast, failed to have an inhibitory effect and resulted in 87% CRs by the second pairing session (Fig. 6). This group also showed high levels of expression for total tGluA4 and most of the splice variants, consistent with our findings for normal conditioning (Fig. 7). Significantly, preparations injected with the rescue plasmid to tGluA4 flop into the abducens nerve and incubation in the anti-tGluA4 siRNA followed by training showed restored but fewer CRs to a level of 52% (Fig. 6; P < 0.0001 vs. Ps2). In these preparations, the amount of total tGluA4 mRNA was reinstated to significant levels above pseudoconditioning (Fig. 7; P = 0.0003 vs. Ps2), and expression of tGluA4 flop was approximately twofold greater compared with normal conditioning (P < 0.0001), whereas there was no effect on flip (P = 0.49) or the other variants. Treatment with the tGluA4 flip rescue plasmid failed to result in substantial conditioning, with only 18% CRs (Fig. 6; P = 0.09 vs. Ps2), although it was dramatically overexpressed (Fig. 7; P < 0.0001). Similar results were obtained with the rescue plasmid to the truncated version of tGluA4, tGluA4trc1, which averaged 4% CRs (Fig. 6; P = 0.70 vs. Ps2), although levels of tGluA4trc1 mRNA were elevated (Fig. 7; P < 0.0001). These data support the conclusion that the expression of tGluA4 AMPAR subunits is critical for the acquisition of in vitro classical conditioning and also indicate that tGluA4 flop specifically has an important role in conditioning mechanisms compared with the other tGluA4 splice variants. Surprisingly, the novel, truncated AMPAR subunit tGluA4trc1 also appears to be regulated in conditioning, although its rescue alone fails to result in CRs.
Fig. 6.
Acquisition of conditioning is inhibited by anti-tGluA4 or anti-GluA1 siRNA treatment and rescued by tGluA4 flop rescue plasmid. Percent of conditioned response (CR) acquisition after 1 and 2 pairing sessions for the different treatment groups is shown. Treated preparations were incubated in the siRNA alone followed by conditioning or pretreated with rescue plasmids followed by tGluA4 siRNA and the conditioning procedure. Physiological traces from some of these groups show representative recordings of CRs (arrows) and URs during paired stimulation. *Significant differences from Ps2; #significant differences from C2. CS, conditioned stimulus; US, unconditioned stimulus.
Synaptic delivery of tGluA4 is inhibited by siRNA and restored by tGluA4 flop rescue plasmid.
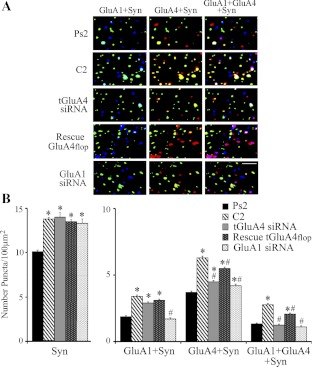
Immunocytochemistry and confocal imaging were performed to assess the synaptic localization of AMPARs in conditions in which expression of tGluA4 was manipulated during the training procedures. These data are summarized in Fig. 8 (n = 5 preparations/group). Our previous studies have shown that tGluA1- and tGluA4-containing AMPARs are delivered to synaptic sites during normal conditioning. tGluA1 subunits are delivered to synapses early in conditioning, followed by incorporation of tGluA4 subunits and withdrawal of tGluA1 (Mokin et al. 2007). Assessment of AMPAR synaptic localization after two pairing sessions here supports these findings. We used the vesicle-associated protein synaptophysin as a marker of synapses. Interestingly, synaptophysin has been implicated in regulating endocytosis, which controls synaptic vesicle availability during distinct states of neuronal activity (Kwon and Chapman 2011). After conditioning, in which there was a mean of 99% CRs, there was a significant increase in colocalization of tGluA1- or tGluA4-containing AMPARs with synaptophysin compared with pseudoconditioning (mean of 0% CRs), indicating that AMPARs containing these subunits were delivered to synapses (Fig. 8B; tGluA1 + Syn: F4,619 = 72.5, P < 0.0001; tGluA4 + Syn: F4,619 = 96.3, P < 0.0001, comparison of C2 vs. Ps2). Moreover, there was a significant increase in triple-label colocalization of tGluA1, tGluA4, and synaptophysin at this same time period (F4,618 = 74.5, P < 0.0001, C2 vs. Ps2), presumably when tGluA4 is being inserted and tGluA1 is withdrawn from synapses. These findings are illustrated in the confocal images shown in Fig. 8A. Another group of preparations was pretreated with the anti-tGluA4 siRNA and then presented with the conditioning stimuli for two pairing sessions in normal saline. This group had an average of 4% CRs and showed dramatically reduced levels of colocalization of tGluA4 with synaptophysin compared with normal conditioning (Fig. 8B; P < 0.0001, tGluA4 siRNA vs. C2). Synaptic localization of tGluA1 remained elevated significantly after anti-tGluA4 siRNA treatment compared with pseudoconditioning (Fig. 8B; P < 0.0001, tGluA4 siRNA vs. Ps2), whereas synaptic colocalization of tGluA1 with tGluA4 was reduced to pseudoconditioned values (P = 0.44, tGluA4 siRNA vs. Ps2). These findings suggest that tGluA4-containing AMPARs were not delivered to synapses in significant enough quantities to induce conditioning but that delivery of tGluA1 subunits was intact. Finally, the abducens nerve of brain stem preparations was injected with rescue plasmid to tGluA4 flop, followed by incubation in the anti-tGluA4 siRNA, and subjected to the training protocol. AMPAR localization data from these preparations, in which there was a mean of 60% CRs, showed that synaptic incorporation of tGluA4 recovered to a level near normal conditioning (Fig. 8B; P < 0.0001, rescue vs. Ps2), as did colocalization of tGluA1 with tGluA4 at synaptic sites (P < 0.0001, rescue vs. Ps2). It can also be seen from the analysis of punctate staining that there is a uniform and significant increase in the levels of synaptophysin in the experimental groups that received paired stimulation (Fig. 8B; P < 0.0001, all groups vs. Ps2; F3,495 = 1.7, P = 0.16, comparison of all groups excluding Ps2). This finding has been observed consistently in our studies (Zheng and Keifer 2008, 2009). Interestingly, the enhanced synaptophysin induced by pairing is not affected by application of the siRNAs or the rescue plasmids that act on AMPARs postsynaptically, suggesting that it reflects presynaptic mechanisms operating independently of those controlling AMPAR trafficking. This observation will be discussed further below. Although interesting, this finding indicates that the observed changes in AMPAR subunit colocalization among the different experimental groups are not due to alterations in synaptophysin staining. Finally, we assessed whether knockdown of tGluA4 had any effect on trafficking of tGluA2/3-containing AMPARs, even though their synthesis was not affected by treatment (Fig. 4). Colocalization of tGluA2/3 AMPAR subunits with synaptophysin showed no significant changes from pseudoconditioning after either conditioning or conditioning performed during treatment with the tGluA4 siRNA but instead maintained a stable synaptic presence throughout the procedures (Fig. 9; F2,147 = 0.7, P = 0.49, comparison of Ps2, C2, and tGluA4 siRNA groups). Together, these data strongly support our hypothesis that synaptic incorporation of tGluA4-containing AMPARs in abducens motor neurons underlies the acquisition of in vitro classical conditioning. Synaptic delivery of tGluA1 AMPARs alone does not support CRs. Moreover, the tGluA4 flop splice variant appears to have a critical role in conditioning.
Fig. 8.
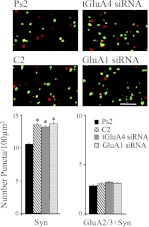
Synaptic localization of tGluA4- or tGluA1-containing AMPARs after conditioning or treatment with siRNA and rescue plasmids. A: representative confocal images of abducens motor neurons from the different treatment groups showing punctate staining for synaptophysin (green), GluA1 AMPAR subunits (blue), and GluA4 (red). Double colocalization of GluA1 with synaptophysin (GluA1 + Syn) is cyan, GluA4 with synaptophysin (GluA4 + Syn) is yellow, and triple colocalization of GluA1 and GluA4 with synaptophysin (GluA1 + GluA4 + Syn) is white. Original scale bar = 2 μm. B: quantitative analysis of synaptophysin punctate staining and the colocalization of AMPAR subunits with synaptophysin for the different treatment groups. *Significant differences from Ps2; #significant differences from C2.
Fig. 9.
Synaptic localization of tGluA2/3-containing AMPARs after conditioning or treatment with the siRNAs. Levels of synaptophysin punctate staining increased significantly in all of the groups that received paired stimulation compared with Ps2 (P < 0.0001). Conditioning in normal medium or after treatment with either the tGluA4 or GluA1 siRNA did not alter the colocalization of these AMPARs with synaptophysin. Representative images of abducens motor neurons are also shown (synaptophysin, green; tGluA2/3, red; colocalization, yellow). *Significant differences from Ps2; scale bar = 2 μm.
Conditioning and synaptic delivery of AMPARs following GluA1 siRNA treatment.
We have hypothesized that delivery of tGluA1-containing AMPARs is required to unsilence auditory synapses and activate N-methyl-d-aspartate receptors (NMDARs) to allow synthesis and delivery of tGluA4 required for acquisition of CRs (Keifer and Zheng 2010). Delivery of tGluA1 does not, by itself, initiate conditioning. According to this proposal, inhibition of tGluA1 would be expected to suppress synaptic incorporation of both tGluA1- and tGluA4-containing AMPARs, as well as CR acquisition. This prediction was confirmed here. A commercially available siRNA to mammalian GluA1 was found to be effective in suppressing expression of this subunit in turtles (Fig. 4). When tested during the conditioning procedure, the average number of CRs recorded from this group was only 6% (Fig. 6). Colocalization analysis with synaptophysin showed that synaptic delivery of AMPARs containing tGluA1 was inhibited significantly (Fig. 8B; P < 0.0001, GluA1 siRNA vs. C2), as was tGluA4 (P < 0.0001, GluA1 siRNA vs. C2) compared with normal conditioning. Correspondingly, colocalization of tGluA1 and tGluA4 subunits at synaptic sites was reduced to pseudoconditioned values (Fig. 8B). Again, trafficking of tGluA2/3 subunits was examined for any compensatory activity after conditioning during the GluA1 siRNA treatment. Colocalization of tGluA2/3 with synaptophysin showed no differences compared with the pseudoconditioned, conditioned, or tGluA4 siRNA-treated groups (Fig. 9; P = 0.76). These data suggest that suppression of synaptic delivery of tGluA1 inhibits delivery of tGluA4-containing AMPARs and CR acquisition.
DISCUSSION
Function of tGluA4 splice variants in classical conditioning.
A number of studies have characterized the effect of alternative splicing and post-transcriptional RNA editing on AMPAR channel properties (Pei et al. 2009; Ravindranathan et al. 2000). For example, mutations in the GluA4 C-terminus, in particular, alter its trafficking properties (Coleman et al. 2006), and GluA4 knockout mice exhibit slower channel kinetics in auditory brain stem nuclei (Yang et al. 2011). However, the relative function of AMPAR splice variants in synaptic plasticity and learning is largely unknown. Recently, we identified 10 alternative splice variants of the GluA4 AMPAR subunit in the turtle brain, in which expression of tGluA4 flip/flop, tGluA4c flip/flop, and a novel, truncated variant tGluA4trc1 are major isoforms (Sabirzhanov and Keifer 2011). Here, we observed that the mRNA levels of most of these major isoforms were increased significantly with conditioning, including tGluA4 flop, tGluA4c flip, and tGluA4trc1 (an increase in tGluA4 flip and tGluA4c flop was observed, but these were not statistically significant). The functional importance of some of these isoforms in conditioning was determined by combining application of an anti-tGluA4 siRNA with rescue plasmids selective for specific splice variants. Knockdown of tGluA4 using our siRNA suppressed the conditioning-induced increase in expression of the major tGluA4 isoforms and inhibited conditioning. Surprisingly, only the rescue plasmid to tGluA4 flop resulted in significant recovery of conditioning, suggesting an important functional role for this isoform. Supporting the possible importance of tGluA4 flop in AMPAR trafficking and learning, Coleman et al. (2006) found that GluA4 flop becomes readily detectable on the membrane surface of cultured cells when coexpressed with stargazin, a transmembrane AMPAR regulatory protein (TARP) (Chen et al. 2000; Tomita et al. 2005). Moreover, coassembly of GluA4 flop with flip isoforms increased the surface expression of flop (Coleman et al. 2006), suggesting that flip/flop heteromeric GluA4 may be more efficiently trafficked and favored after induction of plasticity states. Studies show that stargazin plays a critical role in promoting AMPAR clustering and surface expression at excitatory synapses through its interaction with postsynaptic density protein 95 (Bats et al. 2007; Chen et al. 2000; Tomita et al. 2005). Stargazin has also been shown to increase the single channel conductance and channel burst duration of GluA4-mediated currents (Tomita et al. 2005). In the case of in vitro classical conditioning, there was a twofold overexpression of GluA4 flop during the rescue experiments and reinstatement of normal amounts of GluA4, resulting in 52% conditioning, but not higher levels closer to normal. Likewise, a threefold overexpression of GluA4 flip failed to rescue conditioning entirely. Overexpression of these isoforms may result in greater numbers of homomeric receptors. However, assembly of heteromeric flip/flop AMPARs appears to predominate in native cells and results in increased surface expression of flop (Brorson et al. 2004; Coleman et al. 2006). We have not yet determined whether stargazin functions as a TARP in our model system, which might serve to facilitate synaptic delivery of GluA4 flop, thereby restoring some level of conditioning. Based on the findings of others, it seems likely that rescue of both the flip and flop tGluA4 isoforms together would enhance the recovery of conditioning over flop or flip rescue alone.
We previously identified a novel, truncated tGluA4 splice variant, tGluA4trc1, which is comprised of only an ∼400-amino acid N-terminal domain (NTD) (Sabirzhanov and Keifer 2011). Surprisingly, the tGluA4trc1 isoform is expressed as protein in the turtle brain stem in relatively high amounts (15%) and is also characterized in human, mouse, and rat. The extracellular NTD forms dimers, possibly appearing as an ∼84-kD band in Western blots of brain stem preparations (Sabirzhanov and Keifer 2011), and is involved in AMPAR subunit-specific recognition and assembly (Kuusinen et al. 1999; Leuschner and Hoch 1999). Here, we show that the expression of tGluA4trc1 is regulated during conditioning; however, rescue of tGluA4trc1 alone fails to restore learning. Studies by others (Sia et al. 2007) using cell cultures showed that transfection with a GluA4 AMPAR subunit construct lacking the NTD resulted in diffuse immunocytochemical staining and lack of protein clustering. In contrast, transfection with a GluA4 construct containing the NTD showed clusters that were recruited to sites of synaptic contact, suggesting that the NTD alone is sufficient for synaptic delivery. These data suggest that tGluA4trc1 may contribute to the synaptic recruitment of AMPARs during conditioning. Interestingly, we have found that tGluA4trc1 coimmunoprecipitates with full-length tGluA4 in transfected cells (unpublished data). Although tGluA4trc1 does not contain a ligand-binding domain and can be considered physiologically silent, it may assemble with full-length subunits and aid in chaperoning them to synapses during conditioning. However, rescue and subsequent overexpression of this truncated subunit alone after anti-tGluA4 siRNA treatment would be expected to lead to assembly of mainly nonfunctional receptors and loss of conditioning, as was observed here. For comparison, overexpression of tGluA4trc1 by application of the rescue plasmid alone without the tGluA4 siRNA resulted in high levels of CR acquisition due to the availability of normal AMPARs. A truncated version of the GluA1 subunit has also been described previously, which is most similar to our tGluA4s (Gomes et al. 2008). Although it generates nonfunctional homomeric receptors, coexpression with full-length GluA1 gives rise to receptors that produce whole-cell currents. There is currently only limited understanding of the function of the truncated isoforms of AMPAR subunits.
Enhanced levels of synaptophysin are not affected by the siRNAs.
Previously, we showed that presynaptic structural modifications of bouton growth and enrichment with synaptophysin protein accompanied conditioning with a similar time course as the postsynaptic trafficking of AMPARs (Li and Keifer 2012; Li et al. 2011). These coordinate pre- and postsynaptic alterations require brain-derived neurotrophic factor (BDNF), and bouton growth specifically uses the Eph/ephrin trans-synaptic signaling system. In some of our studies (Zheng and Keifer 2008, 2009), in response to experimental manipulation, we observed a disassociation in these coordinated responses to conditioning in which AMPAR trafficking was inhibited, whereas elevated levels of synaptophysin remained unaffected. For example, bath application of an antagonist to calcium calmodulin-dependent protein kinase II inhibited synaptic delivery of GluR4-containing AMPARs and conditioning but not the increased synaptophysin levels (Zheng and Keifer 2009). Here, similar findings were observed with the siRNAs, which specifically targeted AMPARs. Such findings likely reflect independent mechanisms underlying pre- and postsynaptic modification, as signal transduction mechanisms not dependent on AMPAR trafficking would be unaffected by the treatment. Similar dissociation of BDNF induced coordinated pre- and postsynaptic modification suggesting independent mechanisms have also been shown in cultured hippocampal neurons derived from mutant mice (Alder et al. 2005). These observations will be useful in determining signaling pathways specific for pre- and postsynaptic plasticity mechanisms.
Two-stage model of AMPAR trafficking during classical conditioning.
We have proposed a two-stage model of AMPAR trafficking during in vitro classical conditioning, which underlies the acquisition of abducens CRs (Keifer and Houk 2011; Keifer and Zheng 2010; Zheng and Keifer 2009). In this model, the first stage involves synaptic incorporation of tGluA1-containing AMPARs, which act to unsilence auditory nerve synapses by activating NMDARs, allowing postsynaptic entry of calcium. This step is rapid and does not involve protein synthesis of tGluA1 but translocation of existing receptors to synaptic sites (Mokin et al. 2007). This initiates a second stage of NMDAR-induced synthesis and synaptic incorporation of tGluA4 AMPAR subunits, which are hypothesized to replace tGluA1 and generate the CRs. The present study presents several findings that are key to supporting this model. First, treatment with the GluA1 siRNA resulted in a reduction in not only tGluA1 subunits and colocalization with synaptophysin but also significantly reduced tGluA4 protein and synaptic localization, as well as inhibition of conditioning. These data support our hypothesis that the initial synaptic delivery of tGluA1-containing AMPARs is required for the later delivery of AMPARs containing tGluA4 subunits. Second, selective suppression of tGluA4 expression by treatment with an anti-tGluA4 siRNA attenuated tGluA4 synaptic localization and conditioning, whereas synaptic incorporation of tGluA1 was unaffected. Furthermore, coapplication of the anti-tGluA4 siRNA with a tGluA4 flop rescue plasmid restored tGluA4 synaptic delivery and CR acquisition. These findings support our conclusion that synaptic delivery of tGluA4 AMPAR subunits specifically underlies acquisition of CRs. Therefore, tGluA1 synaptic incorporation is an early event in conditioning—followed by synaptic incorporation of tGluA4 subunits—which is dependent on completion of the first stage of AMPAR trafficking. To further support our model, assessment of the synthesis and trafficking of tGluA2/3 showed that there are no significant compensatory changes of either process in response to the various treatments affecting tGluA4 or tGluA1 subunits [see also, Mokin et al. (2007)]. Therefore, as was proposed some time ago (Malinow and Malenka 2002), it appears that tGluA2/3-containing AMPARs are constitutively cycled into and out of synapses, and that process is not altered, at least as determined here, during synaptic plasticity. Multistage AMPAR trafficking has been proposed for other models of learning, and their similarities and differences have been detailed elsewhere (Keifer and Zheng 2010). It is worth pointing out that this form of learning requires both GluA1 and GluA4 subunits that are targeted to synapses rather than GluA1 alone, which appears to occur in hippocampal LTP (Derkach et al. 2007), a conclusion that may require further analysis. Both subunits are well represented in the cranial nerve nuclei (Keifer and Carr 2000; Petralia and Wenthold 1992), and GluA4 subunits, particularly the flop isoform, specifically contribute to rapidly decaying, fast synaptic currents necessary for high-fidelity auditory processing (Mosbacher et al. 1994; Ravindranathan et al. 2000). Since the auditory nerve initiates conditioning in this model system, it is not surprising that tGluA4 AMPAR subunits are recruited to synapses during training.
GRANTS
Support for this work was provided by National Institute of Neurological Disorders and Stroke (Grant NS051187) and National Center for Research Resources (Grant P20 RR015567) and is designated as Centers of Biomedical Research Excellence (COBRE) to J. Keifer.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: Z.Z., B.S., and J.K. conception and design of research; Z.Z. and B.S. performed experiments; Z.Z. and B.S. analyzed data; Z.Z., B.S., and J.K. interpreted results of experiments; Z.Z. and J.K. prepared figures; Z.Z., B.S., and J.K. drafted manuscript; J.K. edited and revised manuscript; Z.Z., B.S., and J.K. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Frances Day for assistance with the confocal microscopy.
REFERENCES
- Alder J, Thakker-Varla S, Crozler RA, Shaheen A, Plummer MR, Black IB. Early presynaptic and late postsynaptic components contribute independently to brain-derived neurotrophic factor-induced synaptic plasticity. J Neurosci 25: 3080–3085, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CW, Keifer J. Properties of conditioned abducens nerve responses in a highly reduced in vitro brainstem preparation from the turtle. J Neurophysiol 81: 1242–1250, 1999 [DOI] [PubMed] [Google Scholar]
- Bats C, Groc L, Choquet D. The interaction between stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron 53: 719–734, 2007 [DOI] [PubMed] [Google Scholar]
- Brorson JR, Li D, Suzuki T. Selective expression of heteromeric AMPA receptors driven by flip-flop differences. J Neurosci 24: 3461–3470, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 408: 936–943, 2000 [DOI] [PubMed] [Google Scholar]
- Coleman SK, Moykkynen T, Cai C, von Ossowski L, Kuismanen E, Korpi ER, Keinanen K. Isoform-specific early trafficking of AMPA receptor flip and flop variants. J Neurosci 26: 11220–11229, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkach VA, Oh MC, Guire ES, Soderling TR. Regulatory mechanisms of AMPA receptors in synaptic plasticity. Nat Rev Neurosci 8: 101–113, 2007 [DOI] [PubMed] [Google Scholar]
- Gomes AR, Ferreira JS, Paternain AV, Lerma J, Duarte CB, Carvalho AL. Characterization of alternatively spliced isoforms of AMPA receptor subunits encoding truncated receptors. Mol Cell Neurosci 37: 323–334, 2008 [DOI] [PubMed] [Google Scholar]
- Keifer J, Armstrong KE, Houk JC. In vitro classical conditioning of abducens nerve discharge in turtles. J Neurosci 15: 5036–5048, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keifer J, Carr MT. Immunocytochemical localization of glutamate receptor subunits in the brain stem and cerebellum of the turtle Chrysemys picta. J Comp Neurol 427: 455–468, 2000 [DOI] [PubMed] [Google Scholar]
- Keifer J, Houk JC. Modeling signal transduction in classical conditioning with network motifs. Front Mol Neurosci 4: 1–8, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keifer J, Sabirzhanov BE, Zheng Z, Li W, Clark TG. Cleavage of proBDNF to BDNF by a tolloid-like metalloproteinase is required for acquisition of in vitro eyeblink classical conditioning. J Neurosci 29: 14956–14964, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keifer J, Zheng Z. AMPA receptor trafficking and learning. Eur J Neurosci 32: 269–277, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusinen A, Abele R, Madden DR, Keinanen K. Oligomerization and ligand-binding properties of the ectodomain of the alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunit GluRD. J Biol Chem 274: 28937–28943, 1999 [DOI] [PubMed] [Google Scholar]
- Kwon SE, Chapman ER. Synaptophysin regulates the kinetics of synaptic vesicle endocytosis in central neurons. Neuron 70: 847–854, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner WD, Hoch W. Subtype-specific assembly of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunits is mediated by their N-terminal domains. J Biol Chem 274: 16907–16916, 1999 [DOI] [PubMed] [Google Scholar]
- Li W, Keifer J. Rapid enrichment of presynapatic protein in boutons undergoing classical conditioning is mediated by brain-derived neurotrophic factor. Neuroscience 203: 50–58, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zheng Z, Keifer J. Transsynaptic EphB/ephrin-B signaling regulates growth of presynaptic boutons required for classical conditioning. J Neurosci 31: 8441–8449, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the −2ddCT method. Methods 25: 402–408, 2001 [DOI] [PubMed] [Google Scholar]
- Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci 25: 103–126, 2002 [DOI] [PubMed] [Google Scholar]
- Mokin M, Keifer J. Quantitative analysis of immunofluorescent punctate staining of synaptically localized proteins using confocal microscopy and stereology. J Neurosci Methods 157: 218–224, 2006 [DOI] [PubMed] [Google Scholar]
- Mokin M, Zheng Z, Keifer J. Conversion of silent synapses into the active pool by selective GluR1–3 and GluR4 AMPAR trafficking during in vitro classical conditioning. J Neurophysiol 98: 1278–1286, 2007 [DOI] [PubMed] [Google Scholar]
- Mosbacher J, Schoepfer R, Monyer H, Burnashev N, Seeburg PH, Ruppersberg JP. A molecular determinant for submillisecond desensitization in glutamate receptors. Science 266: 1059–1062, 1994 [DOI] [PubMed] [Google Scholar]
- Pei W, Huang Z, Wang C, Han Y, Park JS, Niu L. Flip and flop: a molecular determinant for AMPA receptor channel opening. Biochemistry 48: 3767–3777, 2009 [DOI] [PubMed] [Google Scholar]
- Penn AC, Gregor IH. Sculpting AMPA receptor formation and function by alternative RNA processing. RNA Biol 6: 517–521, 2009 [DOI] [PubMed] [Google Scholar]
- Petralia RS, Wenthold RJ. Light and electron immunocytochemical localization of AMPA-selective glutamate receptors in the rat brain. J Comp Neurol 318: 329–354, 1992 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: 2002–2007, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranathan A, Donevan SD, Sugden SG, Greig A, Rao MS, Parks TN. Contrasting molecular composition and channel properties of AMPA receptors on chick auditory and brainstem motor neurons. J Physiol 523: 667–684, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabirzhanov B, Keifer J. Cloning and characterization of glutamate receptor subunit 4 (GluA4) and its alternatively spliced isoforms in turtle brain. J Mol Neurosci 44: 159–172, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SD, Carvalho AL, Caldeira MV, Duarte CB. Regulation of AMPA receptors and synaptic plasticity. Neuroscience 158: 105–125, 2009 [DOI] [PubMed] [Google Scholar]
- Sia GM, Beique JC, Rumbaugh G, Cho R, Worley PF, Huganir RL. Interaction of the N-terminal domain of the AMPA receptor GluR4 subunit with the neuronal pentraxin NP1 mediates GluR4 synaptic recruitment. Neuron 55: 87–102, 2007 [DOI] [PubMed] [Google Scholar]
- Tomita S, Adesnik H, Sekiguchi M, Zhang W, Wada K, Howe JR, Nicoll RA, Bredt DS. Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature 435: 1052–1058, 2005 [DOI] [PubMed] [Google Scholar]
- Yang YM, Aitoubah J, Lauer AM, Nuriya M, Takamiya K, Jia Z, May BM, Huganir RI, Wang LY. GluA4 is indispensable for driving fast neurotransmission across a high-fidelity central synapse. J Physiol 589: 4209–4227, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Keifer J. PKA has a critical role in synaptic delivery of GluR1- and GluR4-containing AMPARs during initial stages of acquisition of in vitro classical conditioning. J Neurophysiol 101: 2539–2549, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Keifer J. Protein kinase C-dependent and independent signaling pathways regulate synaptic GluR1 and GluR4 AMPAR subunits during in vitro classical conditioning. Neuroscience 156: 872–884, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]