Abstract
Background and Objectives
Cystic fibrosis (CF) is an autosomal recessive genetic disease. Infections in these patients are mostly caused by three bacteria: Staphylococcus aureus, Haemophilus influenza and particularly Pseudomonas aeruginosa. Carbapenems including antibiotics are used to combat infections with Pseudomonas aeruginosa. In recent years, carbapenems resistant strains of P. aeruginosa isolated from clinical specimens are being reported. Decrease in drug penetration and production of metalobeta lactamase (MBLS) have been proposed as mechanisms of resistance.
Materials and Methods
In this descriptive study, the population under investigation was 27 patients suffering from CF in Alzahra hospital of Isfahan. Clinical specimens were taken by deep swabbing from throat and data from every patient was recorded in a questionnaire. The specimens were cultured and isolated organisms were identified as P. aeruginosa using standard tests. Kirby-Bauer disk diffusion method was used to determine the bacterial drug resistance pattern. Strains of P. aeruginosa were checked for production of MBLS using disk impregnated with IPM-EDTA and PCR targeting of bla VIM.
Results
Among the 27 patients, 7 (26%) had P. aeruginosa infection. In total, 11 P. aeruginosa isolates were taken. All isolates were susceptible to imipenem, ticarcillin, ciprofloxacin and piperacillin. The lowest scale of susceptibility belonged to ceftazidime (72.2%) followed by tobramycin (45.4%). None of the strains were positive for the bla VIM gene.
Conclusion
Isolates of P. aeruginosa from CF patients in Isfahan were susceptible to antibiotics during the study period.
Keywords: Pseudomonas aeruginosa, Cystic fibrosis, VIM
INTRODUCTION
Cystic fibrosis is an autosomal recessive genetic disease (1). The abnormal characteristic of this disease is the movement of water and ions through the epithelial cells that leads to formation of a dense mucosa and decrease in mucosal clearance in the lungs (2). The disease also causes pancreatic and reproductive organ failure (3, 4). Defect in the cftr gene which is located on chromosome number 7 is the main cause of CF (2, 5). In patients suffering from CF, because of the failure in chlorine transition in the upper membrane of respiratory epithelial cells, chlorine is engulfed in these cells and will entail severe absorption of sodium and water through the respiratory duct that will cause the mucosa in the channels to lose moisture and become sticky which finally lead to obstruction in the respiratory channels (1). The existence of dense discharges in these patients will limit the movement of the respiratory cilia. As a result, the air channels will not be refined (6, 7). Bacterial colonization in respiratory duct will lead to the enfeebling of lung function (2). Most infections in these patients are mostly caused by P. aeruginosa but also by Staphylococcus aureus, and Haemophilus influenza (2, 8). The outbreak of P. aeruginosa infection among patients with CF varies in the world. The rate of infection with this organism among CF patients was reported as 80% in Belgium and Canada (8, 9). Occurrence of cross infections are common (10). The colonization of this bacterium in the lungs of patient with CF following an initial infection leads to destruction of tissue and decrease in respiratory function (11). Antibiotics are the main ways to control the infection (12, 13). However, P. aeruginosa is resistant to most antibiotics and this phenomenon has created problems in treatment of patients (14, 15). The carbapenems including imipenem and meropenem are being used to treat infections caused by P. aeruginosa (16). Resistance to carbapenems by P. aeruginosa strains has been reported in recent years (17). Decrease in drug penetration and creation of effective betalactamase on carbapenems such as metlo beta lactamase (MBLs) was proposed as reasons for such resistance (17, 18).
Classification of MBLs into three sub groups of B1, B2 and B3 is based on molecular structure. The B1 group includes, GIM, VIM, SPM, and IMP (19, 20). Their producing gene is situated on integrons and can integrate into plasmids or chromosomes. Accordingly, bacteria posessing these genes can transform the genes to other pseudomonas and even other Gram negative rods. So far, no inhibitor has been developed for these enzymes. The recognition of these enzymes and the scale of their outbreak in clinical samples are important to prevent the spread of infection. In this study we investigate the presence of MBLs among P. aeruginosa isolated from patients with CF using both phenotypic and PCR assays.
MATERIALS AND METHODS
In this descriptive study, the population under study consisted of patients suffering from CF (n = 27) in Alzahra hospital of Isfahan. The sputa specimens were taken from these patients using deep swabbing of throat. For every patient a questionnaire (containing age, sex, and history of antibiotic use data) was prepared. The specimens were immediately transferred to blood agar and Mac Conkey agar. The media were incubated for 18 -24 hours at 37°C and then P. aeruginosa bacteria were detected through biochemical tests and were kept at -20°C until used (20).
Antimicrobial susceptibility testing
The isolated organisms were subjected to the Kirby-Bauer disk diffusion method (CLSI) using disks containing imipenem, ceftazidime (CAZ), piperacillin (PIP), ciprofloxacin (CIP), ticarcillin (TIC) and tobramycin. P. aeruginosa ATCC 27853 was used as control.
Pure colonies of P. aeruginosa were inoculated to tube containing TSB broth (Trypticase Soy Broth) so that the turbidity of the media reached 0.5 MacFarland. Then using sterile swab, this bacterial suspension was cultured in Muller Hinton agar and the disk IMP-EDTA was placed in agar area.
PCR experiments for detection of MBLs gene
DNA was extracted by boiling method (22). The reaction was prepared in final volume of 25 µl containing 2.5 µl, MgCl, 2 mM, mixture of oligonucleotides (0.2 mM), 0.25 mM of each primers (F- GTTTGGTGCCATATCGCAAC and R-ATTGCGCAGCACCAGGATAG) flanking a 382 bp of bla VIM gene, 100ng of template DNA, and 2U of Taq polymerase enzyme.
The thermocycler was programmed as follows: Initial denaturation at 94°C for 5 minutes, and 30 cycles of 94° C for one minute, 56.5° C for 30 seconds, 72°C for 20 seconds with the final extension step at 72° C for 5 minutes.
P. aeruginosa containing the bla VIM gene (Pasteur Institue of Iran) was used as positive control. The PCR product was electrophoresed on 1.5% agarose gel and following ethidium bromide staining was viewed under UV light.
SPSS 10 software was used for statistical analysis of data.
RESULTS
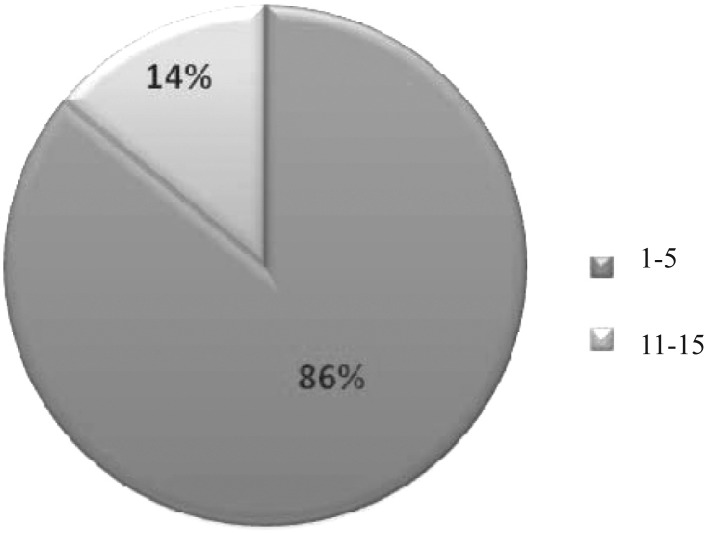
Of 27 CF patients, 18 (66.6%) were male and 9 (33.4%) were female. Distribution of age groups is shown in Fig. 1. The most prevalent age group belongs to patients 1 to 5 years old and the least were in age group 16 to 25. From the total of 27 patients under study, only 2 (7.4%) had never been hospitalized from birth to the time of sampling and were negative for P. aeruginosa in culture. They were female and belonged to the age group 6 to 15. Previous hospitalizations were noticed twice in 11 and three times in 14 of patients. 13 patients underwent respiratory problems at the time of sampling. One patient (4%) had digestive problems and 13 patients (48%) had no notable problem at the time of sampling. From 27 patients, 7 (26%) were infected with Pseudomonas aeruginosa. Totally, 11 P. aeruginosa isolates were taken (2 patients yielded 3 isolates of P. aeruginosa). From 7 patients infected with P. aeruginosa, one patient (14.3%) was female and 6 (85.7%) were male. Patients were in age group 1 to 15; however, they were mostly in the 1 to 5 years of age group (86%) and one patient was in age group 6 to 15. No significant relation was found between the frequency of hospitalization and P. aeruginosa infection.
Fig. 1.
The age groups of cystic fibrosis patients infected with Pseudomonas aeruginosa
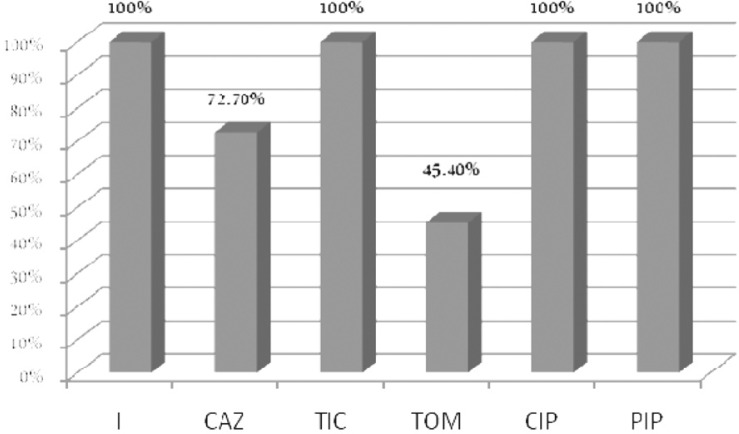
None of the isolates showed resistance to imipenem. The highest rate of resistance belonged to tobramycin (45.4%). Results of susceptibility testing are shown in Fig. 2. None of the clinical isolates was positive for the bla VIM gene.
Fig. 2.
Susceptibility of P. aeruginosa isolated from cystic fibrosis patients to antibacterial agents
I: imipenem, CAZ: ceftazidime, TIC: Ticarcillin, TOM: Tobramycin, CIP: Ciprofloxacin, PIP: Piperacillin
DISCUSSION
Bacterial resistance to carbapenems is a clinical concern (20, 23). There is more concern about P. aeruginosa which is an opportunistic pathogen in patients with immune deficiency (9, 23). Katarin et al. isolated 92 isolates (76.6%) from a total of 120 patients infected with CF in 1996 (24). In Ireland, the prevalence has been reported as 56.2% (25). In Germany, the prevalence of the bacteria in patients having CF has been reported as 50% in 2008 (26). In a 2003 study conducted in Iran, of 64 patients suffering from CF, 21 (32.8%) were infected with this bacterium (27). There are reasons for differences in prevalence of infection with this organism. Hospitalization and duration of infection with P. aeruginosa can be a reason since the bacteria can transfer from one infected patient to another (10). Family awareness and educational level of mothers of sick children has a significant role with the spread of infection with P. aeruginosa in CF cases.
In this research, all isolates were susceptible to imipenem, ticarcillin, ciprofloxacin and piperacillin. The lowest rate of susceptibility belonged to tobramycin (45.4%) followed by ceftazidime (72.2%). Eftekhar et al. did not detect any strain resistance to imipenem in P. aeruginosa isolated from patients having CF in Iran in 2003 and reported the rates of susceptibility to ciprofloxacin, ceftazidime, tobramycin, piperacillin and ticarcillin as 7.5, 85.9, 85.7, 81 and 76% respectively (27). It appears that the scale of antibiotic susceptibility of P. aeruginosa recovered from people suffering from CF has almost been similar in Tehran and Isfahan. Patients 1-5 years old made the dominant group in both studies. Since the studied group of children had not been bedridden, this may be a reason why multi drug resistant strains of P. aeruginosa could rarely be isolated from them. In a study conducted in Portugal, P. aeruginosa producing MBLs were reported from a 14 year old patient with CF (28). Isolates of P. aeruginosa producing bla VIM has already been reported in Iran (Shahcheraghi et al 2010). Because of increase in resistance of P. aeruginosa to carbapenems in Iran, it was assumed that we can see the resistance strains of P. aeruginosa isolated from patients with CF. However, this hypothesis was not true. It seems that age of patients, hospital stay and contact with CF cases might be risk factors to acquire carbapenems resistant strains of P. aeruginosa. All our patients were ambulant and none of them was bedridden at the time of sampling. Most of them (86%) were children from 1 to 5 years old and had less chance of hospitalization and thus the possibility of getting resistance genes in strains isolated from them was far from imagination. It seems that carbapenems remain as effective drug against P. aeruginosa isolated from children with CF in Iran.
ACKNOWLEDGEMENT
We thank Dr. Bahram Nasr Esfahani, Dr. Sorushnia, Ms. Gilda Amini and the staff of the Microbiology Departments of Isfahan University of Medical Sciences.
REFERENCES
- 1.Arancibia F, Bauer TT, Ewig S, Mensa J, Gonzalez J, Niederman MS, et al. Community-acquired pneumonia due to Gram-negative bacteria and P. aeruginosa . Arch Intern Med. 2002;162:1849–1858. doi: 10.1001/archinte.162.16.1849. [DOI] [PubMed] [Google Scholar]
- 2.Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: Mucoid Pseudomonas aeruginosa and Burkholderia cepacia . Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Institute of Health. Learning about cystic fibrosis. 2009. http://www.genome.gov/10001213.
- 4.Chee LC, Durie P. Genotype and phenotype in cystic fibrosis. Hospit Pract. 1997;15:115–142. doi: 10.1080/21548331.1997.11443512. [DOI] [PubMed] [Google Scholar]
- 5.Gilligan PH. Microbiology of airway disease in patient with cystic fibrosis. Clin Micobiol Rev. 2002;4:35–51. doi: 10.1128/cmr.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-beta-lactamases: the quiet before the storm? 2005;18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerem E. The role of Pseudomonas aeruginosa in the pathogenesis of lung disease in cystic fibrosis: more questions than answers. Pediatr Pulmonol. 1997;16(Suppl):265–266. doi: 10.1002/ppul.19502308137. [DOI] [PubMed] [Google Scholar]
- 8.West SE, Zeng L, Lee BL, Kosorok MR, Laxova A, Rock MJ, Splaingard MJ, Farrell PM. Respiratory infections with Pseudomonas aeruginosa in children with cystic fibrosis: early detection by serology and assessment of risk factors. JAMA. 2002;287:2958–2967. doi: 10.1001/jama.287.22.2958. [DOI] [PubMed] [Google Scholar]
- 9.Spilker T, Coenye T, Vandamme P, LiPuma JJ. PCR-based assay for differentiation of Pseudomonas aeruginosafrom other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004;42:2074–2079. doi: 10.1128/JCM.42.5.2074-2079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ojeniyi B, Frederiksen B, Hoiby N. Pseudomonas aeruginosacross-infection among patients with cystic fibrosis during a winter camp. Pediatr Pulmonol. 2000;29:177–181. doi: 10.1002/(sici)1099-0496(200003)29:3<177::aid-ppul4>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 11.Gillspie SH. Second edition. 2006. Principles and Practice of Clinical Bacteriology. Wiley Online. [Google Scholar]
- 12.Webb AK. The treatment of pulmonary infection in cystic fibrosis. Scand J Infect Dis. 1995;96(Suppl):24–27. [PubMed] [Google Scholar]
- 13.Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev. 2010;23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mouton JW. Combination therapy as a tool to prevent emergence of bacterial resistance. Infection. 1999;27(Suppl 2):S24–28. doi: 10.1007/BF02561666. [DOI] [PubMed] [Google Scholar]
- 15.Mouton JW, den Hollander JG, Horrevorts AM. Emergence of antibiotic resistance amongst Pseudomonas aeruginosaisolates from patients with cystic fibrosis. J Antimicrob Chemother. 1993;31:919–926. doi: 10.1093/jac/31.6.919. [DOI] [PubMed] [Google Scholar]
- 16.Sonnesyn SW, Gerding DN. Antimicrobials for the treatment of respiratory infections. In: Niedeman MS, Sarosi GA, Glassroth J, editors. Respiratory infections- A Scientifics Basis for Management. Philadelphia: Saunders; 1994. pp. 511–537. [Google Scholar]
- 17.Lee K, Lim YS, Yong D, Yum JH, Chong Y. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamaseproducing isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2003;41:4623–4629. doi: 10.1128/JCM.41.10.4623-4629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varaiya A, Kulkarni N, Kulkarni M, Bhalekar P, Dogra J. Incidence of metallo beta lactamase producing Pseudomonas aeruginosa in ICU patients. Indian J Med Res. 2008;127:398–402. [PubMed] [Google Scholar]
- 19.Oh EJ, Lee S, Park YJ, Park JJ, Park K, Kim SI, et al. Prevalence of metallo-betalactamase among Pseudomonas aeruginosa and Acinetobacter baumannii in a Korean university hospital and comparison of screening methods for detecting metallo-betalactamase. J Microbiol Methods. 2003;54:411–418. doi: 10.1016/s0167-7012(03)00090-3. [DOI] [PubMed] [Google Scholar]
- 20.Pitout JD, Gregson DB, Poirel L, McClure JA, Le P, Church DL. Detection of Pseudomonas aeruginosa producing metallo-beta-lactamases in a large centralized laboratory. J Clin Microbiol. 2005;43:3129–3135. doi: 10.1128/JCM.43.7.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mouton JW, den Hollander JG, Horrevorts AM. Emergence of antibiotic resistance amongst Pseudomonas aeruginosa isolates from patients with cystic fibrosis. J Antimicrob Chemother. 1993;31:919–926. doi: 10.1093/jac/31.6.919. [DOI] [PubMed] [Google Scholar]
- 22.Hall BG, Barlow M. Revised Ambler classification of {beta}-lactamases. J Antimicrob Chemother. 2005;55:1050–1051. doi: 10.1093/jac/dki130. [DOI] [PubMed] [Google Scholar]
- 23.Cheng K, Smyth RL, Gvan JR, Doherty C, Winstannley C, Denning N, et al. Spread of beta-lactam resistant Pseudomonas aeruginosain a Cystic Fibrosis clinic. Lancet. 1996;348:639–642. doi: 10.1016/S0140-6736(96)05169-0. [DOI] [PubMed] [Google Scholar]
- 24.Xu J, Moore JE, Murphy PG, Millar BC, Elborn JS. Early detection of Pseudomonas aeruginosa--comparison of conventional versus molecular (PCR) detection directly from adult patients with cystic fibrosis (CF) Ann Clin Microbiol Antimicrob. 2004;3:21. doi: 10.1186/1476-0711-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valenza G, Tappe D, Turnwald D, Frosch M, König C, Hebestreit H, et al. Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J Cyst Fibros. 2008;7:123–127. doi: 10.1016/j.jcf.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Eftekhar F, Rostamizadeh F, Khodadad A, Henry D, Speert DP. Isolation and genetic fingerprinting of Pseudomonas aeruginosafrom Iranian patients with cystic fibrosis using RAPD-PCR. Iran J Biotech. 2003;1:95–100. [Google Scholar]
- 27.Cardoso O, Alves AF, Leitão R. Metallo-beta-lactamase VIM-2 in Pseudomonas aeruginosa isolates from a cystic fibrosis patient. Int J Antimicrob Agents. 2008;31:375–9. doi: 10.1016/j.ijantimicag.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Chee LC, Durie. P. Genotype and phenotype in cystic fibrosis. Hospit Pract. 1997;15:115–142. doi: 10.1080/21548331.1997.11443512. [DOI] [PubMed] [Google Scholar]