Summary
Background
An interlocked transcriptional-translational feedback loop (TTFL) is thought to generate the mammalian circadian clockwork in both the central pacemaker residing in the hypothalamic suprachiasmatic nuclei and in peripheral tissues. The core circadian genes, including Period1 and Period2 (Per1 and Per2), Cryptochrome1 and Cryptochrome2 (Cry1 and Cry2), Bmal1, and Clock are indispensable components of this biological clockwork. The cycling of the PER and CRY clock proteins has been thought to be necessary to keep the mammalian clock ticking.
Results
We provide a novel cell-permeant protein approach for manipulating cryptochrome protein levels to evaluate the current transcription and translation feedback model of the circadian clockwork. Cell-permeant cryptochrome proteins appear to be functional on the basis of several criteria, including the abilities to (1) rescue circadian properties in Cry1−/−Cry2−/− mouse fibroblasts, (2) act as transcriptional repressors, and (3) phase shift the circadian oscillator in Rat-1 fibroblasts. By using cell-permeant cryptochrome proteins, we demonstrate that cycling of CRY1, CRY2, and BMAL1 is not necessary for circadian-clock function in fibroblasts.
Conclusions
These results are not supportive of the current version of the transcription and translation feed-back-loop model of the mammalian clock mechanism, in which cycling of the essential clock proteins CRY1 and CRY2 is thought to be necessary.
Introduction
Circadian (daily) rhythms regulate gene-expression patterns, cellular activities, and behavioral phenomena [1]. In mammals, a circadian oscillator in the suprachiasmatic nuclei (SCN) of the hypothalamus appears to be a central pacemaker that coordinates circadian oscillators in peripheral tissues and cells, including liver, lung, kidney, muscle, cornea, and fibroblasts [2–7]. A network of evidence supports a model for the mammalian clockwork that proposes autoregulatory transcriptional and translational feedback loops (TTFLs) of key “clock proteins” that are rhythmically abundant and that interact with one another to feed back upon the transcriptional activity of their respective genes [8–11]. In mammals, positive transcriptional “drive” is provided by a heterodimer of the bHLH-PAS factors, BMAL1 and CLOCK (and/or NPAS2 [12]), and this heterodimer activates transcription at E box enhancers [13]. Negative feedback in the system is primarily accomplished by Period (especially PER1 and PER2) and Cryptochrome (CRY1 and CRY2) gene products that translocate to the nucleus and repress the transcription of their own genes [14–23]. Although an important role for posttranslational modification of key mammalian clock proteins is recognized [24–26], the rhythmic expression of mRNAs and proteins encoded by key clock genes has led to the understandable conclusion that these rhythmic expression patterns are essential [8, 9, 14–16, 19, 20, 27–29]. However, this is obviously not necessarily true; an essential gene product might have a rhythmic expression pattern that is not inevitably functional. For example, in plants, LhcII transcript levels and translational rates exhibit daily or circadian oscillations, but these rhythmic patterns are not converted into downstream patterns of LHCII protein abundance [30, 31].
The TTFL model can be used to predict whether specific components need to oscillate [8–11, 32, 33]. Cycling of the positive components (especially CLOCK and BMAL1) appears to be unnecessary for circadian precession [34–36], and consequently, testing the TTFL’s prediction of rhythmically expressed factors has focused upon the negative limb components PER1/2 and CRY1/2. Consistent with the TTFL model, constitutive overexpression of mCry1 mRNA in mammalian cells has been reported to enhance damping [37]. In addition to constitutive-overexpression studies, however, an important support for the TTFL model in other organisms has been perturbation analyses where experimental expression of clock proteins as pulses or steps was used to determine whether the oscillator is reset in a phase-dependent fashion. For example, in the cases of Drosophila, Neurospora, and cyanobacteria, inducible promoters that rapidly turn on or off were used to drive expression of clock genes, thereby eliciting phase setting or phase shifting [38–41]. This approach has not yet been reported in mammals for pulse or step perturbations, presumably because of the lack of an inducible promoter system in which clock-protein expression can be turned on or off rapidly [42, 43].
A better methodology for perturbation analyses is needed to provide definitive tests of the predictions of the TTFL model in mammalian cells. An alternative approach that has not yet been applied to the study of circadian rhythms is to fuse a protein transduction domain (PTD) to a clock protein and transduce it directly across the cell membrane. This novel method would allow the rapid introduction of a clock protein into cells without the complications of transcriptional or posttranscriptional regulation. If the turnover rate of the modified clock protein is sufficiently high, this technology would permit the administration of pulses of the clock protein as well. In other applications, PTDs have been used to import functional peptides and proteins into multiple cell types so as to probe signal transduction pathways, to introduce recombinases, to alter cell-division kinetics, and to act for other purposes [44–49]. The wide range of cell types and the ease of translocation across the plasma membrane overcome the limitations of other, more invasive, methods of introducing proteins directly into cells such as microinjection or membrane-permeabilizing reagents [44, 49, 50]. By using this technology to test predictions of the circadian TTFL model in mammalian fibroblasts, we found that cell-permeant CRY proteins (CP-CRYs) (1) can rescue the arhythmia of Cry1−/−Cry2−/− mammalian fibroblasts, (2) can phase shift the fibroblast oscillator, (3) can upregulate BMAL1 expression levels, and (4) can demonstrate that cycling of CRY1, CRY2, and BMAL1 proteins is not necessary for clock function in mammalian fibroblasts. Therefore, in contrast to the current TTFL model of the mammalian clockwork, our results question whether rhythmicity in the abundances of mCRY1 and mCRY2 is essential for operation of the mammalian circadian oscillator.
Results
Translocation of Cell-Permeable Mouse CRY1 and CRY2 into Mammalian Cells
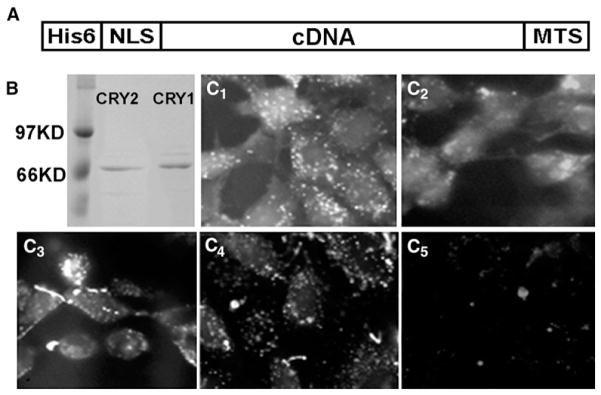
Cell-permeant-protein technology has used PTDs that are composed of either (1) positively charged residues such as the TAT peptide [44] or a series of arginine residues [45] or (2) hydrophobic residues [46–48].We chose the latter type of PTD for making cell-permeant mouse CRY1 and CRY2 (CP-CRY1 and CP-CRY2), as shown in Figure 1A. Recombinant CP-CRY1 and CP-CRY2 fusion proteins have a C-terminal hydrophobic sequence, AAVLLPVLLAAP from Kaposi FGF-4 (called the MTS for “membrane-transduction sequence”), which can mediate uptake of proteins directly through membranes [46], and a N-terminal NLS (nuclear localization signal) from simian virus 40 large T antigen to ensure the translocation of the protein into the nucleus (and the NLS has additionally been found in some cases to aid the translocation across the plasma membrane as well [47]). A His6 tag was included for protein purification. The CP-CRYs can be expressed as soluble proteins in E. coli and purified (Figure 1B). For testing the uptake of CP-proteins in cultured cells, fluorescein-labeled CP-proteins were incubated with Rat-1 fibroblasts and HEK293 cells. CP-CRYs, CP-mutCRY1, and CP-CRE are taken up into Rat-1 fibroblasts (Figure 1C) and into HEK293 cells (data not shown). There appears to be some aggregation of CP-CRY1, CP-mutCRY1, and CP-CRE in the cells that might have cytoplasmic loci; these might be analogous to the cytoplasmic loci found for PER and TIM in Drosophila [51], whereas CP-CRY2 appears to be more homogeneously distributed. In all cases, however, there appears to be nonaggregated CP proteins throughout the cytoplasm and nuclei. As has been found for CP proteins in other studies, the transduction efficiency of CP-CRYs into fibroblasts is approximately 95%. Uptake of extracellular CP-CRYs can also be detected by immunoblotting of treated cells with CRY1 and CRY2 polyclonal IgG antibodies. When CP-CRY1 or CP-CRY2 is present continuously in the medium, the intracellular levels of CP-CRY1 or CP-CRY2 are present at high levels that do not fluctuate with a circadian period (Figure S1 in the Supplemental Data available online).
Figure 1. Uptake of Cell-Permeant Cryptochrome Proteins into Rat-1 Fibroblasts.
(A) Schematic structure of the recombinant CP proteins.
(B) Affinity-purified CP-CRY1 and CP-CRY2 proteins expressed in E. coli.
(C) Rat-1 cells were incubated with 50 nM of fluorescein-labeled CP-CRY1 (C1), CP-CRY2 (C2), CP-mutCRY1 (C3), CP-CRE (C4), or FITC (no protein) (C5). After a 30 min incubation, cells were treated with proteinase K to remove any fluorescein-labeled protein that nonspecifically adhered to the surface of the cells.
Repression of BMAL1/CLOCK-Mediated Transcription by CP-CRYs
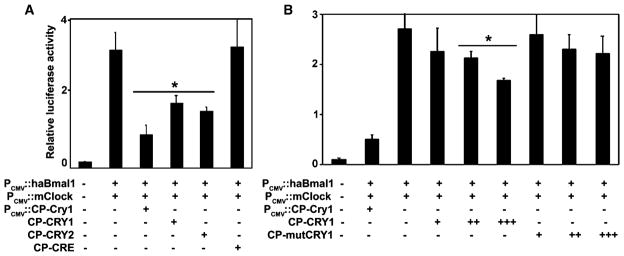
A standard assay for CRY activity is the inhibition of BMAL1/CLOCK-dependent transactivation of E box-containing promoters [19, 52]. In transient-transfection assays, cotransfected hamster Bmal1 (haBmal1) and mouse Clock (mClock) can activate the E box-containing PK2 promoter (PPK2; see [53]), and this activation can be significantly repressed by cotransfected CP-mCry1 that is intracellularly expressed under the control of PCMV (Figures 2A and 2B). It also can be suppressed by the extracellular addition of CP-CRYs (Figures 2A and 2B). Repression by either intracellularly expressed CP-CRY1 or by extracellularly applied CP-CRY1 was dose dependent (Figure 2B and Figure S2). We used a cell-permeant version of Cre Recombinase (CP-CRE) that has previously been shown to translocate into cells and confer intracellular loxP-specific recombination activity [47] as a control protein for our studies. CP-CRE uses the same MTS and NLS as our CP proteins, and it translocates into fibroblasts (Figure 1C4), but it does not repress PPK2::Fluc activity when it is added to the extracellular medium (Figure 2A). As another control, a mutant CP-CRY1 (CP-mutCRY1) was constructed, on the basis of a study showing that mutation of aspartate residue 387 of mouse CRY1 to alanine (D387A) results in a reduced (but not eliminated) inhibition of E box-containing promoters [54]. In transient-transfection assays, both intracellularly expressed and extracellularly applied CP-mutCRY1 showed significantly less inhibition than CP-CRY1 at low dosages. As found in the original publication [54], however, the D387A mutant is able to inhibit E box promoters at high dosage (Figure 2B and Figure S2). The data in Figure 2 show the suppressive action on PPK2::Fluc activity, but essentially identical results were obtained for the repression of PmPer1::Fluc activity by CP-CRY (data not shown).
Figure 2. Repression of BMAL1/CLOCK-Mediated Transcription by CP-CRY1 and CP-CRY2.
HEK293 cells were transfected with haBmal1, mouse Clock, and an E box-containing promoter PPK2-driven luciferase reporter (PPK2::luc).
(A) Intracellularly expressed CRY (PCMV::Cry1) and extracellularly applied CP-CRYs on PPK2 activity were evaluated with a luciferase assay of PK2 promoter activity. The concentrations of CP-CRY1, CP-CRY2, and CP-CRE proteins were 250 nM.
(B) Both transfected PCMV::Cry1 and extracellularly applied CP-CRY can inhibit PPK2 activity. Extracellularly applied CP-CRY1 and CP-mutCRY1 both inhibit PPK2 activity in a dose-dependent fashion, but CP-mutCRY1 is less effective than CP-CRY1. Protein concentrations are as follows: +, 5 nM; ++, 50 nM; and +++, 250 nM. The plotted values are mean±SD of three replicates. Asterisks indicate significant differences from haBmal1/mClock controls at the p < 0.01 level (Student’s t test).
CP-CRY1 and CP-CRY2 Can Rescue Circadian Properties in mCry1−/−mCry2−/− Mouse Fibroblasts
In fibroblasts and SCN slices derived from mice in which the mCry1 and mCry2 genes have been knocked out (mCry1−/−mCry2−/−), mPer1 and mPer2 RNA levels are arrhythmic and expressed at middle to high values compared with wild-type mouse fibroblasts and SCN slices [5, 18]. In fibroblasts derived from mCry1−/−mCry2−/− double-knockout mice [55], we found that PER1 levels did not oscillate and were higher than those in fibroblasts derived from wild-type mice (Figures 3A and 3B). After treatment with CP-CRY1 + CP-CRY2, the PER1 level in knockout fibroblasts was reduced from the high levels that are diagnostic of mCry1−/−mCry2−/− fibroblasts to levels similar to that of the wild-type fibroblasts (Figures 3A and 3B). This effect is probably mediated by the suppressive activity of cryptochromes on the BMAL1/CLOCK complex [19].
Figure 3. Circadian Properties Can Be Rescued by CP-CRYs in Cry1−/−Cry2−/− Knockout Mice Fibroblasts.
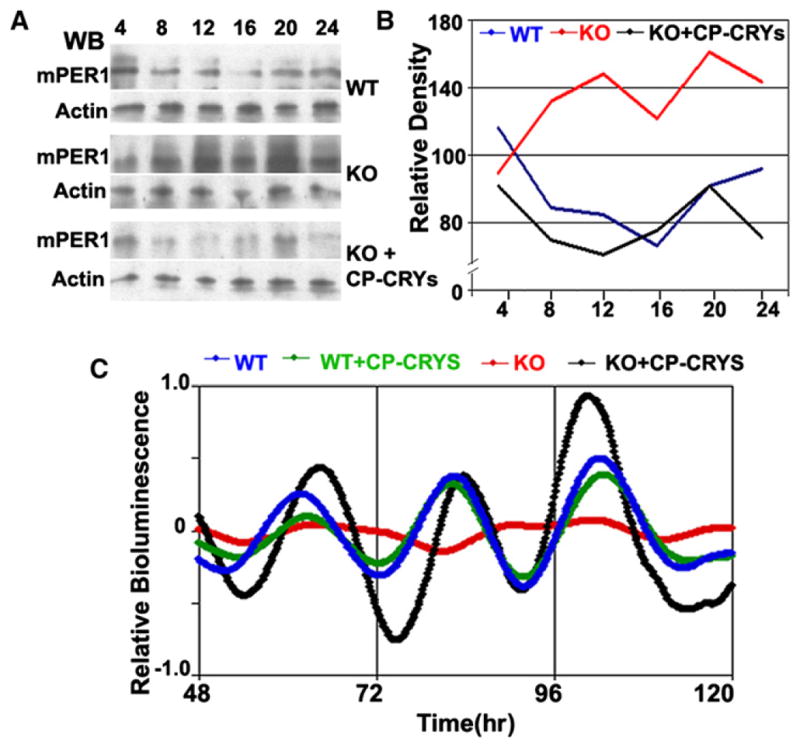
(A) mPER1 expression level is high and arrhythmic in Cry1−/−Cry2−/− mouse fibroblasts. Treatment with CP-CRY1 and CPCRY2 reduces mPER1 expression levels to wild-type levels.
(B) Quantification of results shown in (A). Protein levels of mPER1 were normalized to the actin loading control.
(C) “Rescue” of circadian rhythmicity in Cry1−/−Cry2−/− knockout mice fibroblasts. The PBmal1::Fluc luminescence reporter was introduced to the fibroblasts by nucleofection. After CP-CRY1 and CP-CRY2 treatment, the PBmal1::Fluc expression level is elevated and rhythmicity is recovered. In the wild-type mouse fibroblasts, rhythms are sustained in both the untreated and the CPCRY1- + CP-CRY2-treated cell lines. Data were detrended with LumiCycle software. Colors represent the following: wild-type (blue), wild-type + CP-CRYs (green), knockout (red), and knockout + CP-CRYs (black).
CP-CRY1 and CP-CRY2 can also rescue the rhythmicity of mCry1−/−mCry2−/− fibroblasts. We transfected wild-type and mCry1−/−mCry2−/− fibroblasts with a PBmal1::Fluc luminescence reporter plasmid by nucleofection. Wild-type mouse fibroblasts express a circadian rhythm of luminescence, whereas mCry1−/−mCry2−/− fibroblasts are arrhythmic (Figure 3C). However, mCry1−/−mCry2−/− fibroblasts treated with CP-CRY1 + CP-CRY2 recovered rhythmicity. The data depicted in Figure 3C are detrended data because the luminescence rhythms of these mouse fibroblasts—even of the wild-type fibroblasts—are consistently less robust than that of the Rat-1 fibroblasts shown below. These data indicate that CP-CRY1 and CP-CRY2 are functionally active in vivo.
CP-CRY1 and CP-CRY2 Induce BMAL1 but Not CLOCK Expression
Coordination between the positive (BMAL1 and CLOCK/NPAS2) and negative (PER1/2 and CRY1/2) limbs of the mammalian TTFL is facilitated by the orphan nuclear receptor REV-ERBα—the BMAL1/CLOCK heterodimer activates REV-ERBα, which then represses further Bmal1 transcription [34, 56]. CRY proteins repress the activity of BMAL1/CLOCK, thereby repressing REV-ERBα expression [34]. Therefore, because CRY proteins repress the repressor of BMAL1 (i.e., REV-ERBα), CRY proteins activate Bmal1 transcription [57]. Moreover, CRY1 binds to the C terminus of BMAL1 and protects BMAL1 from degradation [36]. Therefore, CP-CRY proteins could enhance BMAL1 levels both by transcriptional activation of Pbmal1 activity and by inhibition of BMAL1 degradation. On the basis of these predictions, we confirmed the functional activity of CP-CRY1 and CP-CRY2. With conditions in which control endogenous levels of BMAL1 are undetectable by immunoblotting (“PBS” in Figure 4A), CP-CRE does not elevate BMAL1, but both CP-CRY1 and CP-CRY2 dramatically upregulate BMAL1 protein levels within 30 min after treatment with the CP proteins (Figures 4A and 4B). This upregulation is at least 10- to 20-fold and is constitutive (Figure S3). This effect is likely to be mediated (at least partially) by induction of PBmal1 because PBmal1 activity is stimulated acutely by treatment with CP-CRY proteins in both Rat-1 and mCry1−/−mCry2−/− fibroblasts (Figure S4). Pulse treatments of CP-CRY1 or CP-CRY2 for 3 hr or 6 hr upregulate BMAL1 for only the time of exposure to these proteins. After wash out of CP-CRY1, the upregulated BMAL1 disappears within 1 hr (Figure 4B). As was true for the transient-transfection assay of PPK2 activity (Figure 2), CP-mutCRY1 (D387A) is less active than CP-CRY1 in the induction of BMAL1; i.e., a low concentration (5 nM) of CP-mutCRY1 is less effective, whereas higher concentrations of CP-mutCRY1 (25 and 100 nM) appear to be equivalent to those of CP-CRY1 (Figure 4C). Finally, CP-CRYs do not nonspecifically induce all proteins in Rat-1 fibroblasts (or even all core clock proteins) because neither CP-CRY1 nor CP-CRY2 had a significant effect on CLOCK protein levels (Figure S5). The data of Figure 4A show clearly that the intracellular activity of CP-CRY is not rhythmic, as judged by the activation of BMAL1 levels. Apparently, extracellular CP-CRY is continuously entering the cells, where it is inactivated or degraded relatively rapidly as judged by the rapid decline in BMAL1 levels when extracellular CP-CRY is removed (Figure 4B).
Figure 4. Impact of CP-CRY1 and CP-CRY2 Treatment upon BMAL1-Protein Abundance in Rat-1 Fibroblasts as Measured by Immunoblotting.
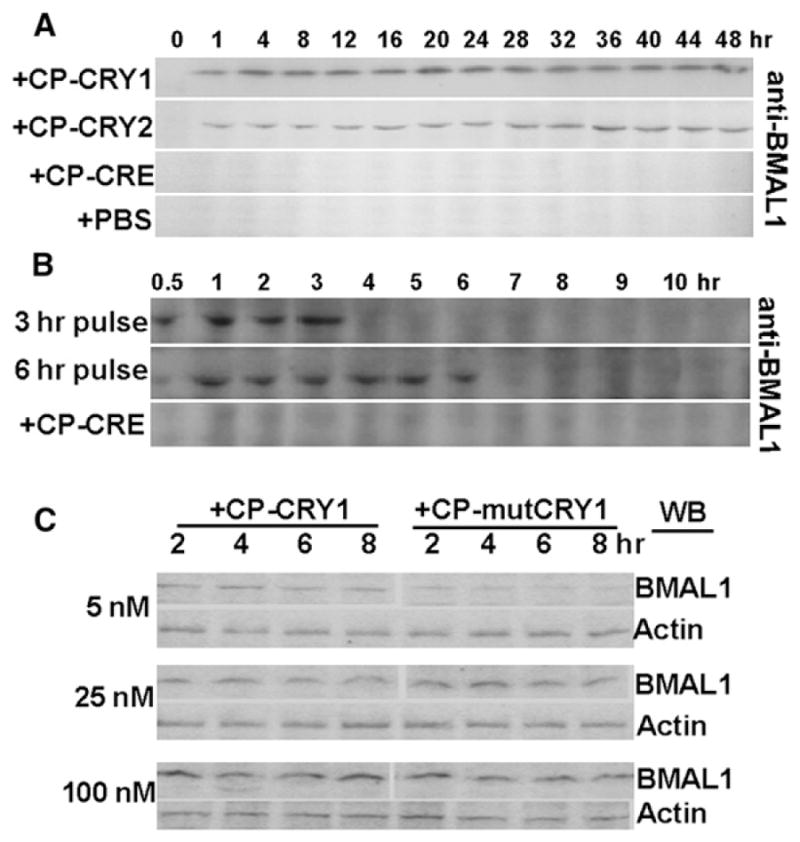
(A) BMAL1-protein expression in response to continuous treatment of CP-CRY1, CP-CRY2, or CP-CRE. PBS is the phosphate-buffered saline control.
(B) BMAL1 expression in response to 3 hr or 6 hr pulse treatments of CP-CRY1.
(C) Comparison of the BMAL1 expression level of Rat-1 cells treated with different concentrations of CP-mCRY1 or CP-mutCRY1 as compared with the actin control.
Phase Shift of the Rhythm in Rat-1 Fibroblasts by CP-CRYs
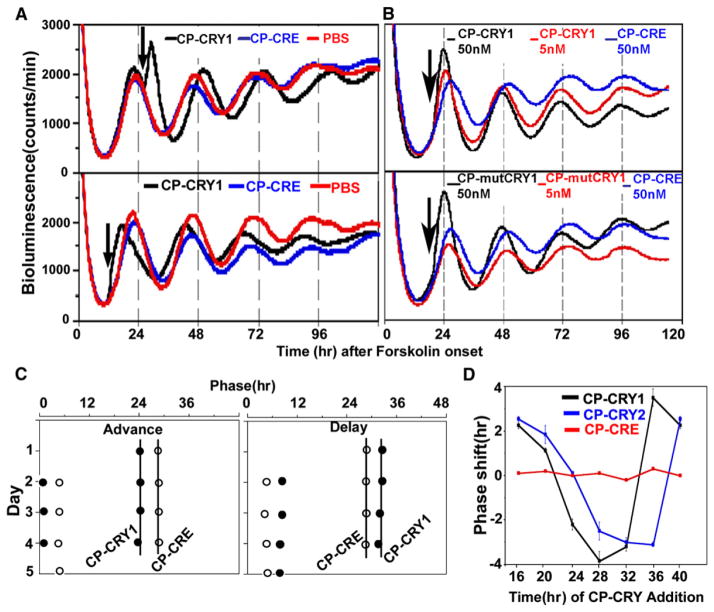
As judged by the upregulation of BMAL1 levels (Figure 4) and PBmal1 activity (Figure S4), the CP-CRY proteins are functionally active soon after they are administered in the extracellular medium. This makes them ideal for perturbation experiments. We used Rat-1 fibroblasts that have been stably transfected with a PPer2::Fluc reporter because they exhibit robust circadian oscillations of luminescence for several days after synchronization by 10 μM forskolin. The addition of either CP-CRY1 or CP-CRY2 (“step” treatments, i.e., continuous treatments) can phase shift the PPer2::Fluc reporter rhythm in a phase-dependent manner, whereas CP-CRE and PBS have no effect. Step treatments of CP-CRY1 or CP-CRY2 can elicit phase shifts up to 3 hr (either advances or delays, depending upon the phase of the treatment onset; Figure 5). At low concentrations (5 nM), CP-mutCRY1 has less (or no) phase-resetting activity effect on the Rat-1 rhythms (Figure 5B), and this finding is consistent with its reduced efficacy in the transient-transfection assay (Figure 2B). When the data for CP-CRY1 and CP-CRY2 are plotted as phase-response curves (PRCs), the resetting effects are clearly phase dependent (Figure 5D) and stable (Figure 5C). The phase position of PRCs for CP-CRY1 versus CP-CRY2 is similar, consistent with the similar phasing of CRY1 and CRY2 expression [28], but the PRC for CP-CRY2 may be slightly delayed relative to that of CP-CRY1 (Figure 5D). The phase resetting and BMAL1 activation we observe with CP-CRY1 and CP-CRY2 is likely to be due to a CRY-specific action and not due to a nonspecific increase of membrane permeability. This was demonstrated by the observation that none of the CP proteins stimulated a Ca2+ influx into fibroblasts under conditions in which ATP does elicit a Ca2+ flux (Figure S6).
Figure 5. Phase Shifts Induced by Step Treatments of CP-CRY1 and CP-mCRY2.
Rhythms in Rat-1 cells with stably transfected PPer2::Fluc reporters were initiated by a 2 hr forskolin pulse (10 μM).
(A) CP-CRY1 (50 nM) was added to the assay medium at the phases indicated by arrows. Addition of CP-CRE (50 nM) or PBS were controls.
(B) CP-CRY1 is more effective than CP-mutCRY1 as a phase-shifting agent at a 5 nM concentration.
(C) Regression analyses of CP-CRY1-induced phase shifts. The left panel shows phase advance, and the right panel shows phase delay; open symbols are CP-CRE-treated controls, and filled symbols are CP-CRY1-treated samples.
(D) PRCs to step treatments of CP-CRY1 and CP-CRY2 on Rat-1 fibroblasts. Black and blue symbols are CP-CRY1 and CP-CRY2 treatments, respectively. Red symbols are CP-CRE treatments. Error bars represent ± SD.
There were two surprises in the data shown in Figure 5. First, the continuous treatments with CP-CRY1 or CP-CRY2 caused phase resetting but did not cause a period change (Figure 5C). Second, there was no acute repressive effect on PPer2 activity at the time of CP-CRY addition as would have been predicted by the data derived from the transient-transfection assay (Figure 2 and [19]). In fact, at most phases when CP-CRY1 or CP-CRY2 was added, the acute effect on PPer2 activity was either stimulatory (as shown in Figure 5A) or had no effect. (Also note the acute stimulation of PBmal1 in Figure S4.) This discrepancy will be addressed in the Discussion.
CRY1, CRY2, and BMAL1 Cycling Is Not Required for the Circadian Oscillations of Rat-1 Fibroblasts
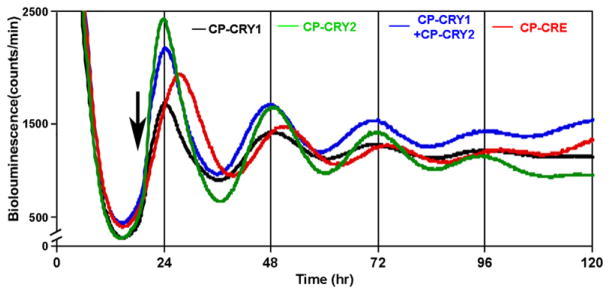
Ueda and coworkers [37] have reported that intracellular overexpression of mCry1 from the PCMV promoter inhibits rhythmicity in transiently transfected Rat-1 fibroblasts. CP-CRY1 and CP-CRY2 afford an alternative approach to address the issue of CRY suppression of rhythmicity. As shown in Figure 4A, the addition of either CP-CRY1 or CP-CRY2 results in a 10× to 20× BMAL1 overexpression that persists as long as the CP protein is present in the extracellular medium. Although the BMAL1 level is not clamped at an absolutely constant level, it is clamped at a very high level and its fluctuations are not circadian. Nevertheless, Figure 6 shows that treatment with either CP-CRY1 or CP-CRY2 or both does not suppress the circadian clock, as monitored by rhythmicity of the PPer2::Fluc reporter in Rat-1 cells. Table 1 summarizes the data from four separate experiments that show the addition of CP-CRY1 or CP-CRY2, or both, does not significantly enhance the damping rate of the rhythm.
Figure 6. Administration of CP-CRY Proteins Does Not Suppress Circadian Rhythmicity of the PPer2::Fluc Reporter in Rat-1 Cells.
CP-proteins were added at the time indicated by the arrow (concentration = 50 nM for each protein).
Table 1.
Damping Rate of Luminescence Rhythms under Different Continuous Treatments
| Treatment | Damping Rate (d)
|
|||
|---|---|---|---|---|
| Exp. 1 | Exp. 2 | Exp. 3 | Exp. 4 | |
| CP-CRY1 | 2.09 ± 0.26 | 1.70 ± 0.08 | 2.06 ± 0.01 | 1.54 ± 0.04 |
| CP-CRY2 | 1.89 ± 0.21 | 1.64 ± 0.12 | 2.08 ± 0.08 | 1.59 ± 0.03 |
| CP-CRY1 + CP-CRY2 | 1.96 ± 0.12 | — | — | 1.65 ± 0.03 |
| CP-CRE | 2.01 ± 0.14 | 1.73 ± 0.08 | 2.04 ± 0.04 | 1.63 ± 0.19 |
| PBS | — | 1.73 ± 0.09 | 2.03 ± 0.06 | 1.67 ± 0.12 |
Damping rates were determined by LUMICYCLE data-analysis software (Actimetrics). The damping rate (d) is the number of days (as 24 hr increments) required for the amplitude of the rhythm to decrease to 1/e (~36.8 %) of the starting value [6]. Values in the table are mean damping rate ±SD for each treatment (n = 4 for experiments 1–3, n = 3 for experiment 4). A two-factor ANOVA analysis revealed that there are not significant differences among the five treatments in any of the four experiments (p value of 0.27), but among the four experiments there are differences in the average damping rate that might be attributable to differences in the condition of the fibroblasts in the various experiments.
Discussion
Manipulating Intracellular Clock-Protein Concentrations with Cell-Permeant Proteins
Direct manipulation of clock-protein abundances in Neurospora, Drosophila, and cyanobacteria has been accomplished by use of inducible promoters to drive expression of clock genes to elicit phase setting or phase shifting [38–41]. On the other hand, rapid transitions of the abundance of clock proteins have not been reported for mammalian studies involving either inducible promoter systems or transfected DNA plasmids [37, 58–60]. For example, a study that applied a doxycycline- responsive promoter system to drive mPER2 expression in mammalian fibroblasts used the system only for constitutive on or off expression rather than as pulses or steps [58]. Moreover, even when mPer2 mRNA transcription is constitutively driven in mammalian cells, posttranscriptional processes confer rhythmicity on the protein’s abundance [59, 60], as had already been reported for Drosophila PER [61]. Another approach has been to transiently transfect mammalian cells with a construct driving clock-gene expression from a constitutive promoter; an investigation using that strategy reported that constant expression of mCry1 abolished circadian rhythmicity [37]. We have taken a different approach toward manipulating the intracellular levels of clock proteins. Our cell-permeantprotein strategy enabled the perturbation of CRY levels without the complications of transcriptional or translational regulation. Moreover, the intracellular activity of CP-clock proteins can be rapidly turned on or off; in the case of our studies, we have used the induction of BMAL1 protein as a gauge of the intracellular activity of the extracellularly applied CP-CRYs (Figure 4A). BMAL1 levels changed rapidly in response to the addition or removal of extracellular CP-CRYs (Figure 4B). This rapid response demonstrates the usefulness of using CP-proteins for perturbation analyses of circadian clocks.
We found that step treatments reset phase and that a PRC composed of both advance and delay shifts can be obtained from these stimuli. These step treatments of CP-CRYs do not cause a significant change in period, nor do they suppress the amplitude of the rhythm (see next section). Surprisingly, even though CP-CRYs enhance PBmal1 activity as expected (Figure S4; see [34, 57]), CP-CRYs do not acutely repress PmPer2 activity in cells in which the reporter’s luminescence is continuously monitored, whereas in transiently transfected HEK293 cells, CP-CRYs repressed BMAL1/CLOCK activation of the E box-containing promoters PPK2 (Figure 2) and PmPer1 (not shown). These differences in the acute repression of PPer activity may stem from the fact that BMAL1 and CLOCK are usually overexpressed from plasmids in the standard transient transfection assay (as shown in Figure 2 and Figure S2), and therefore the stoichiometries of transcriptional factors are different from the case of our stably transfected Rat-1 reporter cells that have only chromosomal copies of Bmal1 and Clock.
Does the Clock Require Cyclic Expression of Clock Proteins?
The current model of the mammalian clockwork is based on a TTFL model that posits central clock proteins being transcribed and translated rhythmically in an “autoregulatory” negative-feedback loop. A crucial prediction of the model is that there will be at least one essential clock protein whose oscillation is necessary. The TTFL model has been mathematically modeled with realistic values for state variables and parameters assigned to the known components of this clockwork [32, 33]. At the current time, essential components of the mammalian clockwork include PER1/2, CRY1/2, BMAL1, and CLOCK/NPAS2. For which of these clock proteins can it be shown that rhythmic abundance is indispensable? Rhythmicity of the positive elements CLOCK and BMAL1 does not appear to be necessary [34–36]. CLOCK is not rhythmically expressed in Rat-1 fibroblasts (Figure 4D), although it might exhibit low-amplitude oscillations in some mammalian tissues as inferred from the low-amplitude mRNA-abundance rhythms seen in some peripheral tissues [62]. BMAL1 is rhythmically expressed, but several studies have shown that constitutive expression of BMAL1 does not interfere with normal clock operation in animals [34, 35] or in cell cultures [36]. We also find by using our cell-permeant- clock-protein approach that the constitutive overexpression of BMAL1 evoked by treatment with CP-CRY1 or CP-CRY2, or both, does not significantly affect the circadian oscillator in Rat-1 cells (Figures 4A and 6).
On the other hand, rhythmicity of the levels of the essential negative elements PER1/2 and CRY1/2 has been thought to be indispensable. Ueda and coworkers [37] report that the overexpression of mCry1 from a transfected plasmid abolishes rhythmicity. By using cell-permeant CRY proteins, we obtained a different result. CP-CRY1 or CP-CRY2 treatment, or both, causes a large and constitutive increase of BMAL1 levels in Rat-1 fibroblasts (Figure 4), indicating continued intracellular activity of the cell-permeant CRY proteins. However, this treatment does not abolish (or even significantly dampen) the oscillator as judged by the rhythm of PPer2 activity (Figure 6 and Table 1). Our results are not consistent with the conclusions of Ueda and coworkers. However, our methods are different from their methods; by using cell-permeant clock proteins, we directly perturb the cellular levels of CRY proteins, whereas the intracellular overexpression methodology of Ueda and coworkers implicates additional layers of transcriptional and posttranscriptional regulation [37]. Moreover, by assaying the activation of BMAL1 by CP-CRYs, we could ascertain the relative dose of active cryptochrome, whereas in the study of Ueda and coworkers, there was no way of knowing how much the CRY levels have been elevated.
Another important methodological distinction that might explain the discrepancy between our results and those of Ueda and coworkers [37] is that different PPer2 elements were used in the luminescence reporters: Our PPer2 region was larger (−1670 to +53; see [7]), whereas their PPer2 region was considerably smaller (−219 to +76; see [37]). Therefore, because our PPer2:: Fluc reporter includes more potential regulatory sites, it is probable that our results reflect a broader spectrum of regulatory inputs to PPer2 activity. Finally, it might be argued that the cell-permeant recombinant proteins used in this study lack CRY’s usual cofactors [21, 22] and therefore behave differently from endogenously produced CRY that might contain a full complement of cofactors. However, recombinant CP-CRY represses PPK2 activity in the transcriptional assay (Figure 2), indicating that it is functionally active; moreover, spectral measurements of CP-CRY confirm that it has incorporated the cofactors FAD and pterin (Figure S7).
Broader Implications
Do our results topple the TTFL model? Certainly not. Rhythmic abundance(s) of the essential clock proteins PER1/2 may still be required for a functional clockwork. A number of studies support the necessity of PER1 or PER2 rhythms, or both, in animals [63, 64] or in cell cultures [58–60]. Our results indicate that rhythmicity of the essential clock proteins CRY1/2 is not necessary and focus attention upon PER1/2 as the bastion upon which rhythmic transcription and translation could be founded. Nevertheless, it might be prudent to consider whether it is time for a major tune-up of the TTFL model for the mammalian clockwork. Such a re-evaluation has occurred in the case of the circadian-clock mechanism in cyanobacteria, where the same kind of evidence that supports a TTFL in the mammalian clockwork was previously used to support a TTFL mechanism in cyanobacteria [39, 41, 65]. Recent studies have shown that a TTFL mechanism is not necessary in cyanobacteria under some conditions (e.g., DD; see [65]). Moreover, three cyanobacterial clock proteins purified and mixed in a test tube can reconstitute a circadian oscillation in vitro [66, 67]. This result unequivocally demonstrates that a posttranslational circadian oscillator can operate under some conditions without a negative-feedback transcriptional and translational loop, and this insight could be relevant to the case of mammalian clocks.
Is it possible that—by analogy with the cyanobacterial system—there is a core mammalian clockwork that is purely posttranslational? This possibility cannot be ignored, and it is possible that our results with CP-CRY proteins intimate that underlying posttranslational clock. An intriguing final speculation upon the discrepancy between our results and those of Ueda and coworkers [37] is that the intracellular expression of CRY1 in their study has the effect of continuously flooding the cells with newly synthesized protein that is not posttranslationally modified, whereas our CP-CRYs have already incorporated the FAD/pterin cofactors (Figure S7) and may be otherwise posttranslationally modified. Perhaps if a posttranslational oscillator is the core mammalian clockwork, the introduction of post-translationally modified CRY (as with CP-CRYs) may perturb the core oscillator less than the new synthesis of unmodified CRY proteins. It may be in the fullness of time that this era of re-evaluation will lead to minor alterations of our understanding of the mammalian clockwork and that the TTFL model will emerge triumphant. On the other hand, this time of re-evaluation may lead to a very different comprehension of the mammalian clock mechanism.
Experimental Procedures
Expression and Purification of Cell-Permeant CRY1 and CRY2 Proteins
NLS-mCry1-MTS and NLS-mCry2-MTS were constructed by amplification of mouse Cry1 (NM_007771) and mouse Cry2 (NM_009963) sequences with primers A and B for mCry1 and primers C and D for mCry2 (see below) (templates were a gift from Dr. Steven Reppert). The PCR products were cloned into the expression vector pET28a+ (Novagen). The resulting constructs were used to express NLS-mCRY1-MTS (=CP-mCRY1) and NLS-mCRY2-MTS (=CP-mCRY2) fusion proteins that were purified by metal affinity chromatography as described in the Supplemental Data. As a control, a mutant NLS-CRY1-MTS (CP-mutCRY1 with D387A) was constructed on the basis of the sequence described in [54]. For the transient-transfection assay of PPK2 activity, the following constructs were made in pcDNA3.1: pPCMV::NLS-mCry1-MTS, pPCMV::NLS-mCry2-MTS, and pPCMV::NLS-mutCry1-MTS.
Primer A: 5′-GACACATATGCCCAAGAAGAAGAGAAAGATGGG GGTGAACGCC GTGCAC-3′
Primer B: 5′-CGTCTCTCGAGTTACGGTGCGGCAAGAAHAACA GGGAGAAGAAC GGCTGCGTTACTGCTCTGCCGCTGG-3′
Primer C: 5′-GACACATATGCCCAAGAAGAAGAGAAAGATGGC GGCGGCTGCTG TGGTG-3′
Primer D: 5′-GTCTCTCGAGTTACGGTGCGGCAAGAAGAACAG GGAGAAGAAC GGCTGCGGAGTCCTTGCTTGCTGGCT-3′
Uptake of CP Proteins
The intracellular presence of CP-CRY proteins in Rat-1 fibroblasts and HEK293 cells was demonstrated by fluorescence microscopy. Purified CP proteins were labeled with fluorescein isothiocyanate (FITC) with a FITC-labeling kit according to the manufacturer’s instructions (Promega). After extensive dialysis against PBS for removal of free FITC, 25 nM concentrations of labeled CP-CRY1, CP-CRY2, CP-mutCRY1, and CP-CRE were added to cultured HEK293 cells and Rat-1 fibroblasts for 30 min (“CP-CRE” = CP-Cre recombinase; see [47, 48]). Then, cells were washed with PBS and treated with proteinase K (5 μg/μl) for 10 min at 37°C. Proteinase K is a broad spectrum protease that was used for eliminating nonspecific adherence of proteins to the cell surface, thereby distinguishing between CP proteins translocated into cells from those sticking to the outside of the cell. Cells were then washed three times with PBS and observed with a fluorescence microscope (Axioscope; Zeiss) with a CCD camera (Princeton Instruments).
Transient-Transfection Assay
HEK293 cells were grown at 37°C in DMEM (11965-092, GIBCO/Invitrogen) supplemented with 10% FBS, 50 units/ml penicillin, and 50 μg/ml streptomycin. Approximately 2 × 104 cells were seeded in each well in a 96-well plate 1 day before transfection. When the cells reached 90%–95% confluency, they were transfected with Lipofectamine 2000 (Invitrogen) with various combinations of the following constructs (all in pcDNA3.1): 100 ng pPCMV::mClock, 100 ng pPCMV::haBmal1, 0.5 ng pPCMV::Rluc, and/or 100 ng pPPK2::Fluc. Twenty-four hours after transfection, 250 nM CP-CRY1, 250 nM CP-CRY2, 250 nM CP-mutCRY1, and 250 nM CP-CRE were added to the medium and incubated for 4 hr. Then, the cells were lysed with passive lysis buffer and firefly luciferase (Fluc), and Renilla (Rluc) luciferase activities were assayed with the Dual-Luciferase Assay Kit (Promega).
“Rescue” of Circadian Properties in mCry1−/−mCry2−/− Mouse Fibroblasts
Wild-type and Cry1−/−Cry2−/− double knockout mouse fibroblasts (provided by Dr. Aziz Sancar) were cultured at 37°C in DMEM (11965-092, GIBCO/Invitrogen) supplemented with 10% FBS, 50 units/ml penicillin, and 50 μg/ml streptomycin. For testing the effect of CP-CRY treatment on PER1 levels, 1 × 106 mouse fibroblast cells were seeded in 35 mm dishes 2 days before treatment with CP proteins. Then, both wild-type and knockout fibroblasts were treated with forskolin (10 μM) for 2 hr to synchronize the cells in the population (this incubation included CP proteins for the samples to be treated). After this synchronization treatment, the cells were washed with DMEM and resuspended in DMEM without or with 0.1 μM CP-CRY1 + 0.1 μM CP-CRY2. Every 4 hr thereafter, cells were washed with cold PBS twice and lysed. Proteins in the lysates were separated by SDS-PAGE (in 7.5% acrylamide gels) and then transferred to nitrocellulose membranes for immunoblotting. Anti-mPER1 polyclonal IgG (#516045, Calbiochem) was used for detection of the PER1 levels.
For “rescue” of rhythmicity in Cry1−/−Cry2−/− double-knockout mouse fibroblasts, 1 × 106 cells were seeded in 35 mm dishes 2 days before transfection. After reaching 90% confluency, cells were transfected by “nucleofection” electroporation with 2 μg of the PBmal1::Fluc reporter plasmid in accordance with the manufacturer’s protocol (NHDF Nucleofector Kit # VPD-1001, Amaxa Biosystems). After electroporation, cells were grown at 37°C in DMEM + 10% FBS for 1 day. Then, wild-type fibroblasts were treated with forskolin (10 μM) for 2 hr. Knockout fibroblasts were treated with forskolin (10 μM) + 0.1 μMCP-CRY1 + 0.1 μMCP-CRY2 for 2 hr. After this treatment, the medium was replaced with DMEM + 10% FBS including luciferin (0.1 mM) and, in the case of the “rescue” samples, also including 0.1 μM CP-CRY1 and 0.1 μM CP-CRY2. The dishes were sealed and luminescence was monitored in an automated luminescence recording apparatus designed for circadian experiments (the “LumiCycle,” Actimetrics) at 36°C.
Immunoblotting
Rat-1 cells stably transfected with the PPer2::Fluc reporter construct [7] were cultured and synchronized with forskolin as above. Eighteen hours after forskolin synchronization, 50 nM CP-CRY1, 50 nM CP-CRY2, 50 nM CP-CRE, 50 nM CP-mutCRY1, and PBS were added to cells. Every 4 hr after the treatment onset, cells were washed twice with ice-cold PBS and lysed in 100 μl lysis buffer (lysis buffer: 50mMTris-HCl [pH 7.5], 150mMNaCl, and 1%NP40). Lysate proteins were separated by SDS-PAGE (in 10% gels) and then transferred to nitrocellulose membranes for immunoblotting. Primary antibodies were rat anti-mCRY1 monoclonal IgG (from Dr. Aziz Sancar), rabbit anti-mCRY2 IgG (Alpha Diagnostic International), rabbit anti-BMAL1 polyclonal IgG (Oncogene catalog #PC539, lot# D13920-1; see Figure S8 for its characterization), and rabbit anti-CLOCK polyclonal IgG (#233170, Calbiochem). The secondary antibodies were AP-conjugated anti-rabbit IgG (#S3731, Promega) and AP-conjugated anti-rat IgG (Promega, #S3831).
Phase Shift of Fibroblast Oscillations Elicited by CP-CRY Proteins
These experiments used the Rat-1 line that has been stably transfected with a PPer2::Fluc reporter construct and that shows a robust circadian rhythm of luciferase activity [7]. The cells were cultured in DMEM (11965-092, GIBCO/Invitrogen) supplemented with 5% FBS, 50 units/ml penicillin, and 50 μg/ml streptomycin in a 5% CO2 incubator at 37°C. Approximately 5 × 105 cells were seeded in a 35 mm dish at least 5 days before the experiment. Three days after the cells reached 100% confluency, the cells were treated with 10 μM forskolin (Sigma) for 2 hr to synchronize the oscillators among the cells in the population. At the end of the forskolin treatment, the medium was replaced with assay medium (DMEM without phenol red, supplemented with bicarbonate [350 mg/L], 5% FBS, 10 mM HEPES [pH 7.2], antibiotics [25 units/ml penicillin and 25 μg/ml streptomycin], and 0.1 mM luciferin [Promega]). Culture dishes were sealed with a 40 mm microscope glass cover and high-vacuum grease to prevent the evaporation of culture medium. The luminescence rhythm was monitored in the LumiCycle (Actimetrics). For the “step” treatments, CP-CRY1 or CP-CRY2, as well as 50 nM of the control proteins CP-CRE and CP-mutCRY1, were added to the cell samples at different phases and left continuously with the cells thereafter while the luminescence patterns were recorded for 5 days or more. Regression analyses to determine period and phase of the luminescence rhythms were performed with the Chrono II program (courtesy of Dr. Till Roenneberg). Rate of damping was assessed with the software included with the Lumicycle (Actimetrics).
Supplementary Material
Acknowledgments
We appreciate the feedback and assistance with initial studies of Guo Huang and Dr. Shin Yamazaki, Dr. Steven Dowdy, Dr. Yao-Zhong Lin, Dr. Vincent Cassone, Dr. Stein Servick, and Dr. Masayuki Matsushita. We are grateful to Dr. Steven Reppert and Dr. Charles Weitz of their generous gifts of plasmids expressing mCry1, mCry2, mClock, and hBmal1, to Dr. H. Earl Ruley for CP-CRE, and to Dr. Dao-Qi Zhang for his assistance with the fluorescence microscopy shown in Figure 1. We are indebted to Dr. Aziz Sancar for mCRY1/mCRY2 antibodies, the mCry1−/−mCry2−/− knockout mouse fibroblasts, and for his expert advice and gracious support concerning this project, including insightful comments on our manuscript. We thank Dr. Till Roenneberg for the analysis program Chrono II. This work has been supported by the Air Force Office of Sponsored Research (grant no. AFOSR F49620-01-1-0448), and the National Institute of Mental Health (grants no. R21 MH65910 and R01 MH43836).
Footnotes
Additional Experimental Procedures and eight figures are available at http://www.current-biology.com/cgi/content/full/17/13/1091/DC1/.
References
- 1.Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology: Biological Timekeeping. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- 2.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 3.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 5.Yagita K, Tamanini F, van Der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 6.Izumo M, Johnson CH, Yamazaki S. Circadian gene expression in mammalian fibroblasts revealed by real-time luminescence reporting: Temperature compensation and damping. Proc Natl Acad Sci USA. 2003;100:16089–16094. doi: 10.1073/pnas.2536313100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izumo M, Sato TR, Straume M, Johnson CH. Quantitative analyses of circadian gene expression in mammalian cell cultures. PLoS Comput Biol. 2006;10:e136. doi: 10.1371/journal.pcbi.0020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 9.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 10.Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 11.Lowrey PL, Takahashi JS. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10:543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 14.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 15.Sun ZS, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee CC. Rigui [mper], a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 16.Zylka M, Shearman L, Weaver D, Reppert S. Three period homologs in mammals: Differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside the brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 17.Van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, Wit J, Verkerk A, Eker APM, Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 18.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, et al. Differential regulation of mammalian Period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin A, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 20.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 21.Van der Schalie E, Green CB. Cryptochromes. Curr Biol. 2005;15:R785. doi: 10.1016/j.cub.2005.09.030. [DOI] [PubMed] [Google Scholar]
- 22.Partch CL, Sancar A. Cryptochromes and circadian photoreception in animals. Methods Enzymol. 2005;393:726– 745. doi: 10.1016/S0076-6879(05)93038-3. [DOI] [PubMed] [Google Scholar]
- 23.Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 25.Partch CL, Shields KF, Thompson CL, Selby CP, Sancar A. Posttranslational regulation of mammalian circadian clock by cryptochrome and protein phosphatase 5. Proc Natl Acad Sci USA. 2006;103:10467–10472. doi: 10.1073/pnas.0604138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A. Differential effects of PER2 phosphorylation: Molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hastings MH, Field MD, Maywood ES, Weaver DR, Reppert SM. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: New insights into a core clock mechanism. J Neurosci. 1999;19:RC11. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyamoto Y, Sancar A. Circadian regulation of cryptochrome genes in the mouse. Brain Res Mol Brain Res. 1999;71:238–243. doi: 10.1016/s0169-328x(99)00192-8. [DOI] [PubMed] [Google Scholar]
- 29.Miyamoto Y, Sancar A. Vitamin B2-based bluelight photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in mammals. Proc Natl Acad Sci USA. 1998;95:6097–6102. doi: 10.1073/pnas.95.11.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piechulla B, Gruissem W. Diurnal mRNA fluctuations of nuclear and plastid genes in developing tomato fruits. EMBO J. 1987;6:3593–3599. doi: 10.1002/j.1460-2075.1987.tb02690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riesselmann S, Piechulla B. Diurnal and circadian light-harvesting complex and quinone B-binding protein synthesis in leaves of tomato (Lycopersicon esculentum) Plant Physiol. 1992;100:1840–1845. doi: 10.1104/pp.100.4.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leloup JC, Goldbeter A. Modeling the mammalian circadian clock: Sensitivity analysis and multiplicity of oscillatory mechanisms. J Theor Biol. 2004;230:541–562. doi: 10.1016/j.jtbi.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 33.Forger DB, Peskin CS. Stochastic simulation of the mammalian circadian clock. Proc Natl Acad Sci USA. 2005;102:321–324. doi: 10.1073/pnas.0408465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 35.von Gall C, Noton E, Lee C, Weaver DR. Light does not degrade the constitutively expressed BMAL1 protein in the mouse suprachiasmatic nucleus. Eur J Neurosci. 2003;18:125–133. doi: 10.1046/j.1460-9568.2003.02735.x. [DOI] [PubMed] [Google Scholar]
- 36.Kiyohara YB, Tagao S, Tamanini F, Morita A, Sugisawa Y, Yasuda M, Yamanaka I, Ueda HR, van der Horst GTJ, Kondo T, et al. The BMAL1 C terminus regulates the circadian transcription feedback loop. Proc Natl Acad Sci USA. 2006;103:10074–10079. doi: 10.1073/pnas.0601416103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 38.Aronson B, Johnson K, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: Autoregulation in the clock gene frequency. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 39.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 40.Edery I, Rutila JE, Rosbash M. Phase shifting of the circadian clock by induction of the Drosophila period protein. Science. 1994;263:237–240. doi: 10.1126/science.8284676. [DOI] [PubMed] [Google Scholar]
- 41.Xu Y, Mori T, Johnson CH. Circadian clock-protein expression in cyanobacteria: Rhythms and phase-setting. EMBO J. 2000;19:3349–3357. doi: 10.1093/emboj/19.13.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furth PA, Onge LS, Boger H, Gruss P, Gossen M, Kistner A, Bujard H, Hennighausen H. Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter. Proc Natl Acad Sci USA. 1994;91:9302–9306. doi: 10.1073/pnas.91.20.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lübber H, Bujard H. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci USA. 1996;93:10933–10938. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: Delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 45.Matsushita M, Tomizawa K, Moriwaki A, Li ST, Terada H, Matsui H. A high-efficiency protein transduction system demonstrating the role of PKA in long-lasting long-term potentiation. J Neurosci. 2001;21:6000–6007. doi: 10.1523/JNEUROSCI.21-16-06000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rojas M, Donahue JP, Tan Z, Lin YZ. Genetic engineering of proteins with cell membrane permeability. Nat Bio-technol. 1998;16:370–375. doi: 10.1038/nbt0498-370. [DOI] [PubMed] [Google Scholar]
- 47.Jo D, Nashabi A, Doxsee C, Lin Q, Unutmaz D, Chen J, Ruley HE. Epigenetic regulation of gene structure and function with a cell-permeable Cre recombinase. Nat Biotechnol. 2001;19:929–933. doi: 10.1038/nbt1001-929. [DOI] [PubMed] [Google Scholar]
- 48.Peitz M, Pfannkuche K, Rajewsky K, Edenhofer F. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: A tool for efficient genetic engineering of mammalian genomes. Proc Natl Acad Sci USA. 2002;99:4489–4494. doi: 10.1073/pnas.032068699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsushita M, Matsui H. Protein transduction technology. J Mol Med. 2005;83:324–328. doi: 10.1007/s00109-004-0633-1. [DOI] [PubMed] [Google Scholar]
- 50.Morris MC, Depollier J, Mery J, Heiz F, Divita G. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat Biotechnol. 2001;19:1173–1176. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- 51.Meyer P, Saez L, Young MW. PER-TIM interactions in living Drosophila cells: An interval timer for the circadian clock. Science. 2006;311:226–229. doi: 10.1126/science.1118126. [DOI] [PubMed] [Google Scholar]
- 52.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 53.Cheng MY, Bullock CM, Li C, Lee AG, Bermak JC, Belluzzi J, Weaver DR, Leslie FM, Zhou Q. Prokineticin 2 transmits the behavioral circadian rhythm of the superachiasmatic nucleus. Nature. 2002;417:405–410. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- 54.Froy O, Chang DC, Reppert SM. Redox potential: Differential roles in dCRY and mCRY1 functions. Curr Biol. 2002;12:147–152. doi: 10.1016/s0960-9822(01)00656-x. [DOI] [PubMed] [Google Scholar]
- 55.Gauger MA, Sancar A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005;65:6828–6834. doi: 10.1158/0008-5472.CAN-05-1119. [DOI] [PubMed] [Google Scholar]
- 56.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 57.Yu W, Nomura M, Ikeda M. Interactivating feed-back loops within the mammalian clock: BMAL1 is negatively autoregulated and upregulated by CRY1, CRY2, and PER2. Biochem Biophys Res Commun. 2002;290:933–941. doi: 10.1006/bbrc.2001.6300. [DOI] [PubMed] [Google Scholar]
- 58.Yamamoto Y, Yagita K, Okamura H. Role of cyclic mPer2 expression in the mammalian cellular clock. Mol Cell Biol. 2005;25:1912–1921. doi: 10.1128/MCB.25.5.1912-1921.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishii K, Yamanaka I, Yasuda M, Kiyohara YB, Kitayama Y, Kondo T, Yagita K. Rhythmic post-transcriptional regulation of the circadian clock protein mPER2 in mammalian cells: A real-time analysis. Neurosci Lett. 2006;401:44–48. doi: 10.1016/j.neulet.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 60.Fujimoto Y, Yagita K, Okamura H. Does mPER2 protein oscillate without its coding mRNA cycling?: Post-transcriptional regulation by cell clock. Genes Cells. 2006;11:525–530. doi: 10.1111/j.1365-2443.2006.00960.x. [DOI] [PubMed] [Google Scholar]
- 61.Yang Z, Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- 62.Yamamoto T, Nakahata Y, Soma H, Akashi M, Mamine T, Takumi T. Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol Biol. 2004;5:18. doi: 10.1186/1471-2199-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 64.Numano R, Yamazaki S, Umeda N, Samura T, Sujino M, Takahashi R, Ueda M, Mori A, Yamada K, Sakaki Y, et al. Constitutive expression of the Period1 gene impairs behavioral and molecular circadian rhythms. Proc Natl Acad Sci USA. 2006;103:3716–3721. doi: 10.1073/pnas.0600060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 66.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 67.Mori T, Williams DR, Byrne MO, Qin X, Mchaourab HS, Egli M, Stewart PL, Johnson CH. Elucidating the ticking of an in vitro circadian clockwork. PLoS Biol. 2007;5:e93. doi: 10.1371/journal.pbio.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.