Abstract
Introduction
Since the first application of antibiotics to treat bacterial infections, the development and spread of resistance has been a persistent threat. An ever-evolving pipeline of next-generation therapeutics is required for modern medicine to remain one step ahead of pathogens.
Areas covered in this review
This review describes recent efforts to develop drugs that interrupt the assimilation of iron by bacteria, a process that is vital to cellular homeostasis and is not currently targeted by antibiotics used in the clinic. We cover the mechanisms through which bacteria acquire iron for their environment and detail efforts to intervene in these processes with small molecule inhibitors that target key steps in these pathways, with a special emphasis on recent advances published during the 2010–2012 period.
Expert Opinion
For decades, the routes used by bacteria to assimilate iron from host and environmental settings have been the subject of intense study. While numerous investigations have identified inhibitors of these pathways, many have stopped short of translating the in vitro results to in vivo proof of concept experiments. Extension of preliminary findings in this manner to validate the clinical potential of iron assimilation pathways for therapeutic development will significantly increase the impact of the field.
1. Introduction
The marketing of Prontosil, the world’s first small molecule anti-infective with proven clinical efficacy, by Bayer in the 1930’s dawned a new era in anti-infective research and ignited the Golden Age of antibiotic discovery & development. The three decades spanning 1940 to 1970 saw the approval of 270 antibacterial drugs derived from 11 classes of structurally distinct classes [1]. Further modification of these drugs satiated the clinic for the next 20 years, where iterative improvements were made to provide new therapeutics with broader spectrum of activity, reduced toxicity, and enhanced potency in organisms that had developed resistance.
Despite the industry’s best efforts to continue to modify the core structures into new compounds to remain ahead of pathogens’ evolution of resistance, the clinical and agricultural misuse has resulted in the development of “Super Bugs” that are insensitive to first-line therapeutics. It is alarming that outbreaks of extremely drug resistant strains showing tolerance to “drugs of last resort” are occurring and may spread to create a pandemic situation [2–4]. This has been compounded by the divestment in the field, where it is estimated that only five major corporations have retained active antibacterial programs [5]. The burden of resolving this clearly unmet medical need is placed largely on the shoulders of academic, government, and nonprofit researchers (as well as small- to medium-size private enterprises). Remarkable efforts have been made to draw attention to the dearth of the antibiotic pipeline, as well as calls for Government intervention through the implementation of policy changes that incentivize work in the field and reduce regulatory hurdles [4–8].
Iron is an essential mineral for all forms of life, playing key structural and chemical roles in protein cofactors, most notably in heme and Fe-S clusters [9]. The vital nature of this nutrient in cellular homeostasis is confounded by the astounding insolubility of the ferric ion under biological conditions (i.e. 10−18 M at neutral pH) and this situation is exacerbated for pathogens during the establishment of infection due to sequestration of free Fe(III) by serum proteins and the liver [10, 11].
As the molecular details of iron retrieval pathways have been unveiled, it has been noted that genetic deletion mutants are unviable in an iron limited setting and, in the case of human pathogens, virulence is either ablated or markedly reduced [10, 11]. Herein, we review the mechanisms by which bacteria acquire iron from the host in an infection-setting and survey the literature for recent efforts to leverage the essential nature of these pathways to develop antibacterial agents.
2. Pathways of Bacterial iron assimilation
The relative scarcity of soluble iron in the environment has led to the evolution of specific and efficient pathways for its bioaccumulation. However, the proclivity of this transition metal to generate reactive oxygen species through Fenton chemistry necessitates a stringent control of iron atoms once they are inside the cell. The careful orchestration of these acquisition and storage pathways in bacteria are governed by the ferric uptake regulator (fur) that controls global changes bacterial metabolism in response to iron limitation [12, 13]. When iron is plentiful, it is bound by Fur protein. The holo-protein in turn binds to Fur box sequences and represses the transcription of iron scavenge and storage pathway genes. In addition to directly repressed genes, Fur contributes to global changes in bacterial physiology to help the organism deal with iron-limiting conditions through the regulation of sigma factors. Under control of these transcriptional promoters are homologs of iron-containing proteins (e.g. superoxide dismutase) that utilize different reaction centers/chemistries to accomplish the same goal, as well as flagellum components and genes involved in biofilm formation. These latter characteristics indicate that iron starvation triggers immobilized cultures to switch from sessile to planktonic states, where the organisms are more sensitive to antibiotics. These findings accentuate the role of fur in pathogenesis and drug resistant phenotypes [14, 15]. In an infection, bacteria utilize three general iron acquisition pathways that are summarized in Fig. 1. These routes are outlined in the following sections.
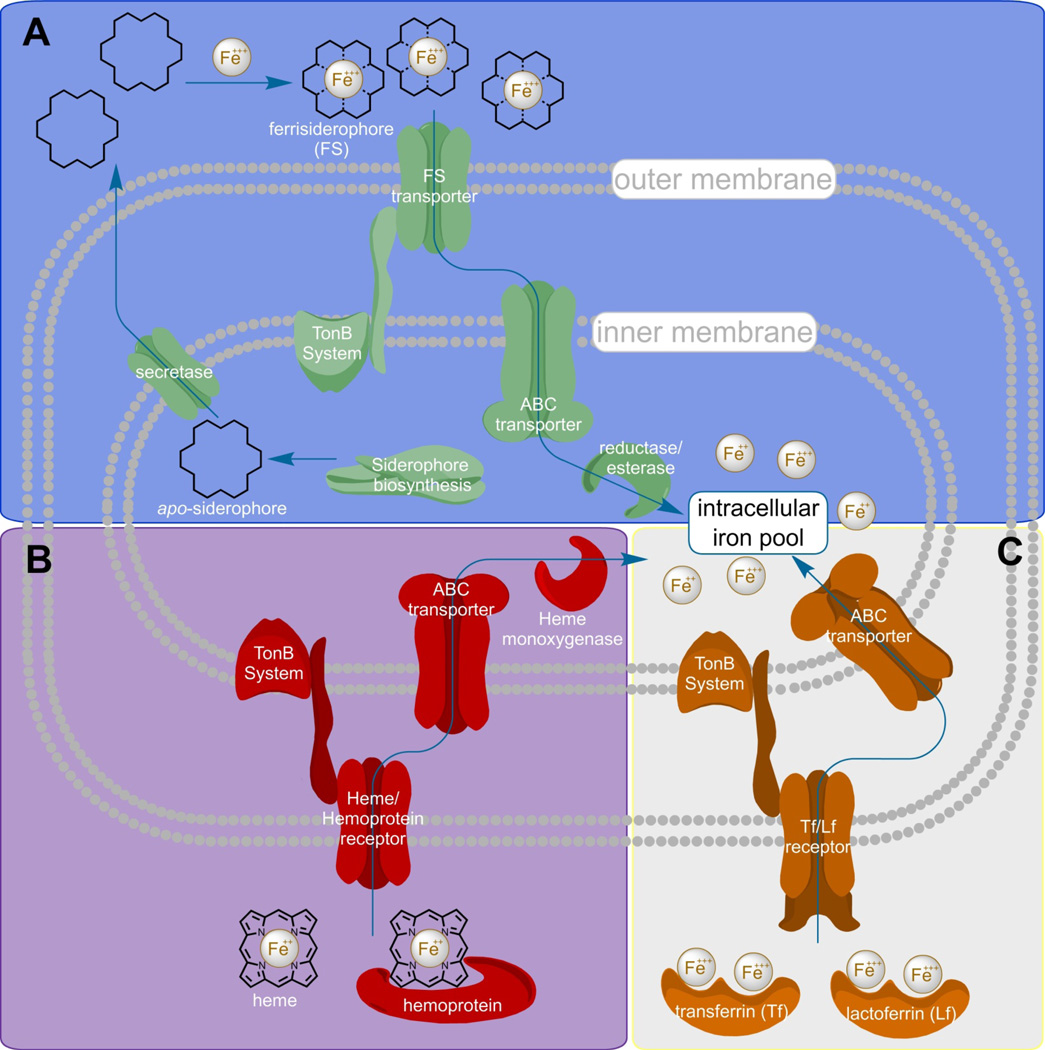
Figure 1. Gram negative bacterium pathways to iron acquisition during infection.
The three general pathways used by Gram negative bacteria to procure iron from the host are diagrammed. A) After production of apo-siderophores by Siderophore biosynthesis machinery, the chelators are transported to the periplasmic space by a specific secretase. After being pumped into the extracellular environment, apo-siderophores strip iron from host proteins, and shuttle it back to the bacterium. Ferrisiderophore transporters bind the ferrisiderophore and translocate it to the periplasm through interaction with the TonB system. Ferrisiderophores are then shuttled to an ABC-transporter that pumps the Fe(III)-loaded molecule into the cytoplasm. Once inside the cell, the iron complex is dissociated by reductases/esterases to liberate the iron atom where it joins the intracellular iron pool. B) Heme and hemoprotein assimilation system initiate by the binding of heme or hemoproteins to specific receptors. In either case, only heme is translocated across the outer membrane though interaction of the receptors with the TonB energizing system. Heme is then trafficked to an ABC transporter embedded in the inner membrane where it is actively pumped into the cell. Finally, heme monooxygenase utilizes the chemistry of the ferrous iron center to perform a ring-opening oxidation of the porphyrin ring that releases iron to the intracellular iron pool. C) Accessing iron from host sequestration systems begins by binding of transferrin or lactoferrin to receptors on the outer membrane that are capable of removing Fe(III) from the host protein. These receptors translocate iron atoms to the periplasm where they are shuttled to ABC-transporters that pump the ferric ion into the cytosol where it joins the intracellular pool.
2.1 Siderophores as a primary mechanism for iron acquisition
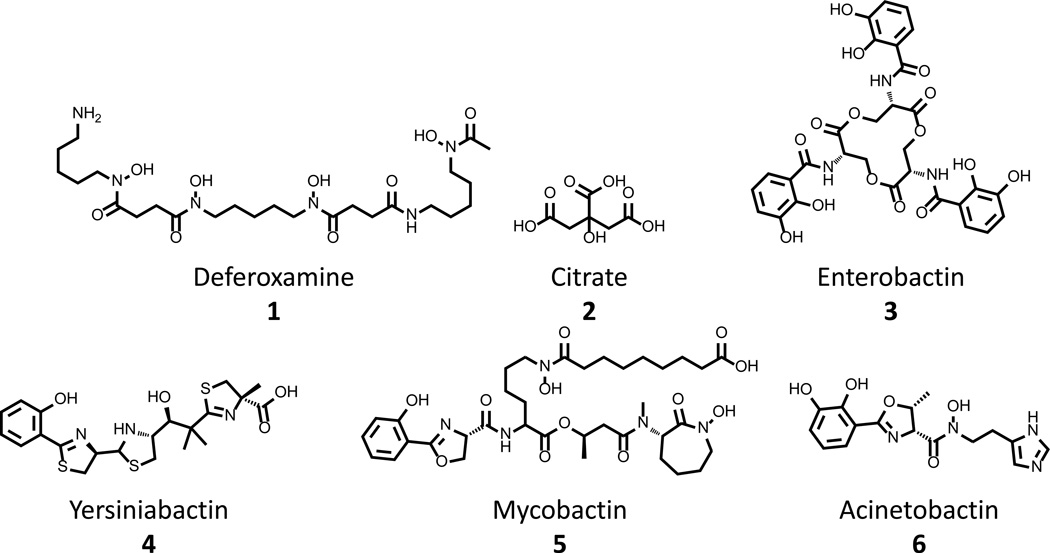
In an effort to scavenge iron from both natural environmental and infection settings, bacteria have developed pathways to synthesize, secrete, and retrieve small molecule chelators that display unprecedented affinity for ferric ions. These compounds display dissociation constants for iron that range from 10−20 to 10−52 M [16], interactions so strong that they are capable of driving the dissolution of insoluble salts in the environment or the stripping of Fe(III) from host sequestering proteins [9]. These siderophores fall into three general groupings based on chemical functionality: hydroxamates, alpha-hydroxyacids, and catecholates. Prototypical compounds that represent these classes of compounds are deferoxamine 1, citrate 2, and enterobactin 3, respectively (Fig. 2). Additionally, hybrid compounds, such as yersiniabactin 4, mycobactin 5, and acinetobactin 6 (Fig. 2) exist that employ ligands from more than one class to form a tight Fe(III) complex. The mechanisms through which siderophores are used to scavenge iron are similar irrespective of chemical functionality or bacterial cell wall structure: cell wall-associated receptor proteins of Gram positive organisms generally substitute in the pathways for outer membrane proteins of Gram negative organisms. As such, details of the system used by E. coli, a model Gram negative bacterium, to acquire iron through the production, secretion and retrieval of enterobactin 3 are discussed below to exemplify the processes of these pathways, and a general landscape is presented in Figure 1A.
Figure 2. Select Representative structures of siderophores used by human pathogens.
Deferoxamine 1, citrate 2, and enterobactin 3 are representative structures of the hydoxamate, α-hydroxyacid and catecholate classes of siderophores. Yersiniabactin 4, mycobactin 5, and acinetobactin 6 demonstrate the diverse structures of siderophores that mix functionalities from multiple classes in order to bind iron.
Enterobactin 3 is biosynthesized by a series of enzymes (siderophore biosynthesis, Fig. 1A) encoded by the ent operon. This pathway begins with the production of 2,3-dihydroxybenzoate (DHB) from chorismate, a primary metabolite utilized in the synthesis of aromatic amino acids. DHB is both secreted into the culture medium (where it acts as a low-affinity ligand for ferric ions) and is further elaborated by a nonribosomomal peptide synthetase-like pathway into the triscatecholate enterobactin 3 (apo-siderophore, Fig. 1A).
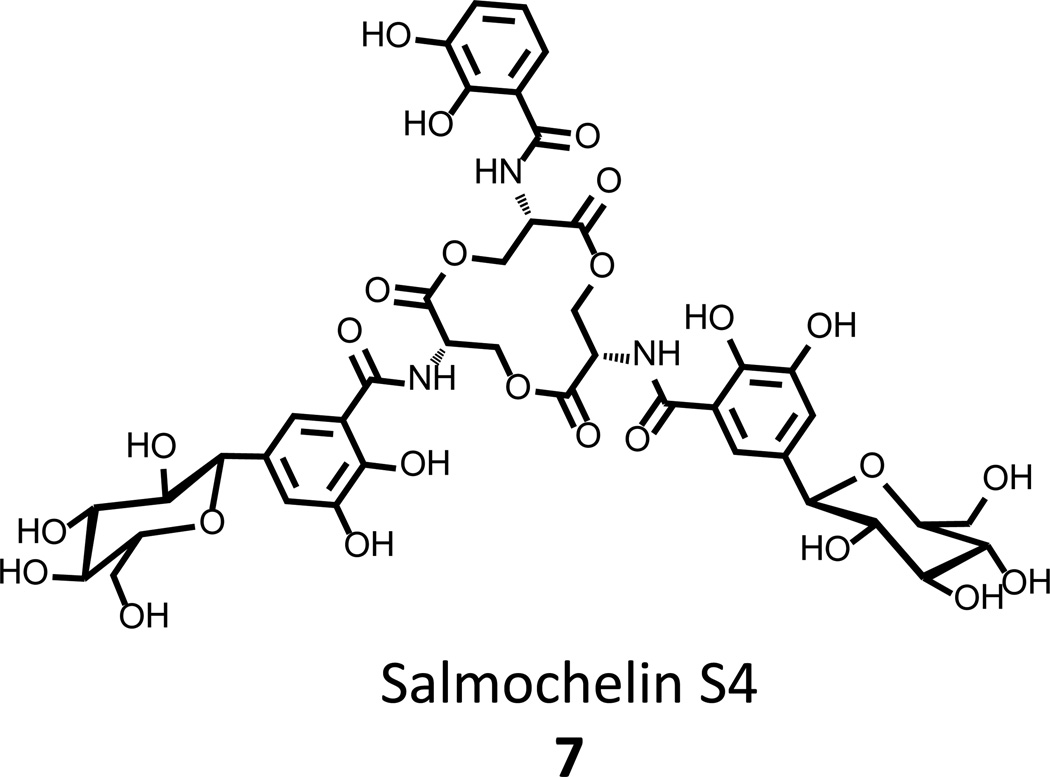
Efficient secretion of 3 requires the EntS protein (secretase, Fig. 1A) that translocates 3 into the periplasm where it is then pumped out to the extracellular environment [17]. It has also been found that, prior to secretion, 3 can be further modified into a derivative siderophore, salmochelin S4 7 (Fig. 3), by the addition of 1 or 2 C-linked glucosyl units [18–20]. This further elaboration has no deleterious effects on the ability of the triscatecholate to bind iron, but provides an advantage to the bacterium with respect to the host’s innate immune defenses (vide infra) [21].
Figure 3. Salmochelin S4, a C-linked glucosyl derivative of enterobactin that evades the innate immune system.
After binding ferric ion, ferrisiderophores are retrieved from the environment by ferrisiderophore receptors on the outer membrane that translocate the chelate into the periplasmic space by interaction with the TonB system that derives energy from the protonmotive force at inner membrane. At this point, periplasmic ferrisiderophore binding proteins shuttle the molecules to specific ABC-transporters, which hydrolyse ATP to transport the iron-bearing cargo into the bacterial cytosol [22, 23].
Releasing ferric iron from receptors that possess such high affinity for their ligand is a daunting task and is typically accomplished through the exploitation of iron’s labile chemistry through the reduction of Fe(III) to Fe(II) by ferrisiderophore reductases to weaken the affinity of the system. However, in the case of enterobactin 3, the affinity is so high that dismantling of the molecule is required for efficient release, and a ferri-enterobactin esterase is employed to cleave the macrocyclic backbone and liberate the nutrient [20, 24, 25].
2.2 Heme acquisition and utilization
In the contexts of a bacterial infection, the most abundant source of iron in the body is the heme cofactor bound to hemoglobin in red blood cells. Thus, it is no surprise that human pathogens possess the capacity to acquire iron from this source and a typical assimilation pathway is outlined in Figure 1B. Both Gram positive and Gram negative bacteria secrete a variety of exotoxins, rhamnolipids, and surfactants to lyse red blood cells and capture either the released hemoglobin or heme (made accessible by the action of secreted proteases) through the expression of soluble and membrane-bound receptors [26]. After binding heme is stripped from hemoproteins and translocated to the periplasm in a TonB dependent mechanism, and shuttled to ABC transporters that pump heme into to the cytosol. Fe(II) is then liberated from the porphyrin by a ring-opening oxidation that is accomplished heme monooxygenase enzymes, to yield various biliverdins and carbon monoxide as co-products.
2.3 Host sequestration of iron
Iron levels in the human body are tightly controlled at an estimated concentration of 10−24M; this both limits the damage caused by its inherent reactivity, as well as represents an inborn strategy to thwart microbial growth. Circulating iron is tightly bound by transferrin, where the holo-protein is efficiently captured by hepatocytes, stripped of its cargo, and the apo-protein shuttled back into the bloodstream [27, 28]. During times of iron sufficiency, excess atoms are stored in hepatocytes as insoluble salts within the subunits of ferritin and the overall homeostasis is regulated by hepcidin, a 25-residue peptide hormone [29].
The high affinity of transferrin for Fe(III) (Kd = 10−23 M) necessitates the production of siderophore scavengers by microbes to outcompete the host proteins for the nutrient. Some bacteria overcome this by secreting acidic compounds into their microenvironment, a response that lowers environmental pH and has the effect of lowering the Kd of transferrin for iron when colonizing a host and makes Fe(III) more soluble during growth in the open environment [9]. To combat this, activation of the innate immune system induces the production of lactoferrin, a defense protein with an even greater affinity for iron that is stable to reductions in pH. This further reduces the availability of the nutrient [30–32].
The strong pressure applied by host iron-sequestration mechanisms has guided the evolution of receptors for host sources, namely heme and hemoproteins (e.g. hemoglobin) (vide supra section 2.2) as well as direct receptors for transferrin and lactoferrin [33] and is outlined in Fig. 1C. Similar to hemoprotein scavenging mechanisms, these receptors interact with the TonB transport system in Gram negative bacteria to strip the cargo and translocate Fe(III) into the intermembrane space, where it is shuttled to an ABC transporter that actively pumps the ion into the cytosol. While transferrin binding proteins have been identified in Gram positive pathogens, the mechanisms of utilization are less well understood [34, 35].
In an additional line of defense, the host immune system has also developed approaches to combat the bacterial mechanisms to acquire iron, namely their siderophore assimilation pathways. Numerous human proteins are known to bind to ferrisiderophores and limit their availability in the bloodstream [36]. Among these are human serum albumin, that exhibits a Kd of 10−5 M for ferrienterobactin [37], but this interceptor is not potent enough to sequester the transporter from the bacterial cell. Recently, the neutrophil-gelatinase-associated lipocalin (NGAL) was serendipitously found to bind ferrienterobactin with an impressive affinity (Kd of 0.4 nM) [38–40]. NGAL is secreted by mononuclear cells in response to activation of the lipolysaccharide-recognizing Toll-like receptor 4 and is a specific response of the innate immune system toward invasion of Gram negative organisms.
While the siderophore binding function of NGAL appears to be efficient at dispelling and restricting systemic infection by normal Gram negative flora of the gut, it has recently been found that the iro gene cluster, contained within pathogenicity-associated islands or on plasmids, encodes machinery to circumvent this [41]. The iro locus encodes numerous proteins that are responsible for the modification of enterobactin 3 into salmochelin S4 7. While the glucosyl appendages added by the Iro enzymes do not affect the triscatecholate’s high affinity for iron (vide supra), they reduce the Kd that NGAL diplays for the ferri-siderophore equivalent of 7 by >3 orders of magnitude [42]. By providing bacterial species with a siderophore system that is no longer bound by host proteins, the iro locus enables organisms conventionally restricted to niche environments to flourish in other settings and cause disease, and best exemplified by the virulent nature of estraintestinal and hemorrhagic E. coli in recent years [36, 43, 44].
3 Targeting iron assimilation pathways
The necessity of iron for growth and the requirement of functional assimilation pathways for host colonization make these attractive targets for the rational development of targeted therapeutics. In this section, we discuss approaches that have been investigated as a means to inhibit or exploit the essential nature of these pathways of bacterial virulence to provide compounds that may prove suitable for development into new antibacterial agents.
3.1 Chelation therapy
The most superficial approach to control bacterial growth through perturbation of iron assimilation pathways is to directly compete with the bacterium for the nutrient. Under normal conditions, this is accomplished systemically through the binding of free Fe(III) by transferrin and locally at the site of infection by the inducible secretion of lactoferrin by neutrophils.
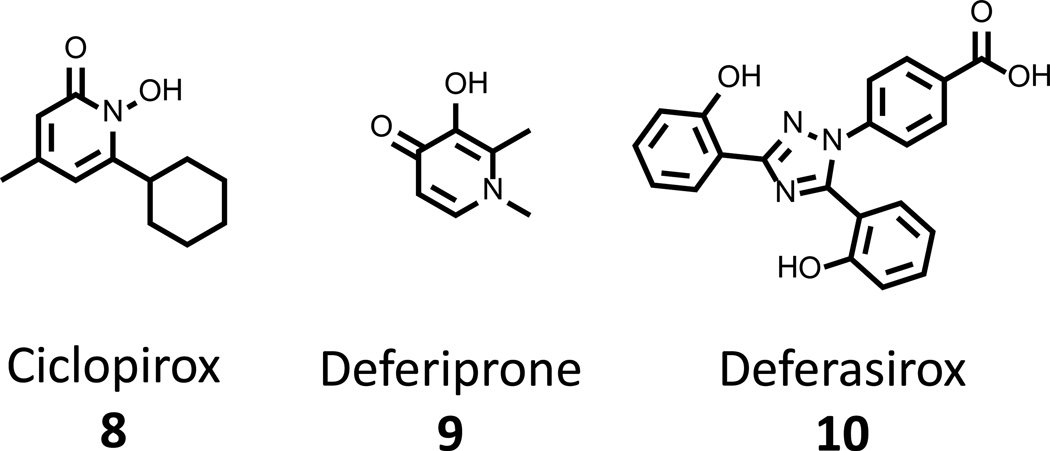
Given the high affinity and selectivity of siderophores, modern medicine has employed natural siderophores such as deferoxamine 1(Fig. 2), a siderophore from Streptomyces pilosus, as well as synthetic chemicals ciclopirox 8, deferiprone 9, and deferasirox 10 (Figure 4), to treat iron overload in thalassemia patients [45, 46]. Agents 1 and 8 – 10 have proven to be well tolerated in the clinic [47] and their low toxicity has led to the testing of their antimicrobial activity against a number of human pathogens, including Plasmodium, Pseudomonas and Staphylococcal species [47–49]. While these compounds do not possess impressive minimum growth inhibitory concentration (MIC) values in broth dilution assays (MIC for 1 of >400 µg/mL against S. aureus) [49], they have been effective in a coadministration setting, potentiating the activity of known antibiotics up to 50-fold in some strains. However, at least in the case of 1, a marked growth enhancement can be observed in some instances, arising from the agent’s natural origins. This is because in addition to expressing receptors for their own siderophores, bacteria also express surface proteins that enable the assimilation of xenosiderophores from other species and thus exploit the efforts of their cohabitants in the battle to acquire the nutrient. It is through such mechanisms that the paradoxical effects of 1 are expected to function. Additionally, these xenosiderophore pathways are the root cause of increased morbidity and mortality of thalassemia patients undergoing chelation therapy with 1, where an acute susceptibility to specific organisms (i.e. Yersinia eterocolitica and fungal pathogens) has been identified [50].
Figure 4. Chelation therapy agents with reported antibacterial activity.
Ciclopirox 8, deferiprone 9 and deferasirox 10 are three FDA-approved chelation therapy agents that have been investigated for their antifungal activities.
Given this liability, synthetic chelators have been the subject of considerable development efforts. Ciclopirox 8 and deferiprone 9 (Fig. 4) typify approved chelation therapeutics, and both have been found to possess potent antibacterial and antifungal activities in vitro. In studies of ciclopirox 8, it was found that this compound possesses modest in vitro activity against Aspergillus fumigates, a fungal pathogen, and that it synergizes with clinically useful antifungals [51].
Deferiprone 9 has seen a much more thorough investigation, but those efforts have centered around fungal infections. While not a bacterial indication, the results described below are relevant to the discussion of repurposing chelation therapy agents for antimicrobial applications, and warrant discussion. Efforts have focused the antifungal attributes of 9 against Rhizopus oryzae, the major cause of mucormycosis: a fungal colonization prominently afflicting uncontrolled diabetes mellitus patients with ketoacidosis. In these tests, 9 was shown to be a potent inhibitor of fungal growth in vitro, with a MIC of 3.2 µg/mL [52]. These results were further extended to a proof of concept study in a mucormycosis model using diabetic mice with ketoacidosis. Test groups treated with 100 mg/kg deferiprone 9 were completely protected against R. oryzae challenge, a result similar to that observed for a control group that received liposomal amphotericin B, the current standard of treatment for the disease [52]. These findings have not been further pursued, to the best of our knowledge.
Finally, deferasirox 10, a new chelation therapeutic agent, was approved for use by the FDA in 2005 for indications of chronic iron overload. In vitro testing of this new drug with Mucorales and Rhizopus spp. revealed MIC potencies of 3–6 µg/mL [53]. R. oryzae challenges of diabetic-ketoacidotic mice demonstrated that administration of 10 at 10 mg/kg twice daily displayed effectiveness in resolving the disease state similar to a treatment with liposomal amphotericin B [53]. Additionally, these studies evaluated a combined treatment of 10 with liposomal amphotericin B and revealed a synergetic activity for the cotreatment relative to monotreatment controls of 10 or liposomal amphotericin B. The promising results of these animal experiments stimulated further testing in single patient and small group human trials, where patients with recalcitrant mucormycosis were cured after coadministration of 10 with amphotericin B [54, 55]. However, these studies were not properly controlled: further experiments have called the initial results into question this year when a double-blind placebo-controlled Phase II clinical trial evaluating efficacy of this cotherapy regimen was not able to demonstrate significant benefit in the test group, and in fact showed a marked increase in mortality (NCT00419770, www.clinicaltrials.gov) [56]. While the limited patient population in this study unintentionally biased the test group with a higher burden of coexisting conditions, these findings are a cause for alarm, and careful evaluation of the data will be required before the regimen is evaluated further.
3.2 Inhibitors of siderophore biosynthesis/assimilation
The biosynthesis of siderophores has also received much attention. Work along these lines has primarily been focused on the catecholate class of siderophores, a narrowing that has arisen from the deep understanding of nonribosomal peptide synthetase pathways and the extension of this knowledge toward the rational development of therapies to treat human disease. Collectively, studies involving the inhibition of siderophore production have focused on the biosynthetic routes to enterobactin 3, yersiniabactin 4, mycobactin 5, and acinetobactin 6, which represent virulence factors of E. coli, Yersinia pestis, M. tuberculosis, and Acinetobacter baumannii, respectively. All of these compounds (Fig. 2) possess a salicyl- or DHB-chelating units and are assembled by multienzyme complexes that polymerize the aryl- and amino-acid monomers into the respective products. During polymerization, amino acid sidechains may be further elaborated through oxidation and/or cyclisation to provide Lewis acid functionalities to actively participate in Fe (III) coordination.
In considering enzymatic activities to target for inhibition, the processes involving the biosynthesis and incorporation of the salicyl- and DHB- fragments have been a primary focus as such functionalities are absent from human metabolism and thus provide an opportunity for a high therapeutic potential in the absence of polypharmacology.
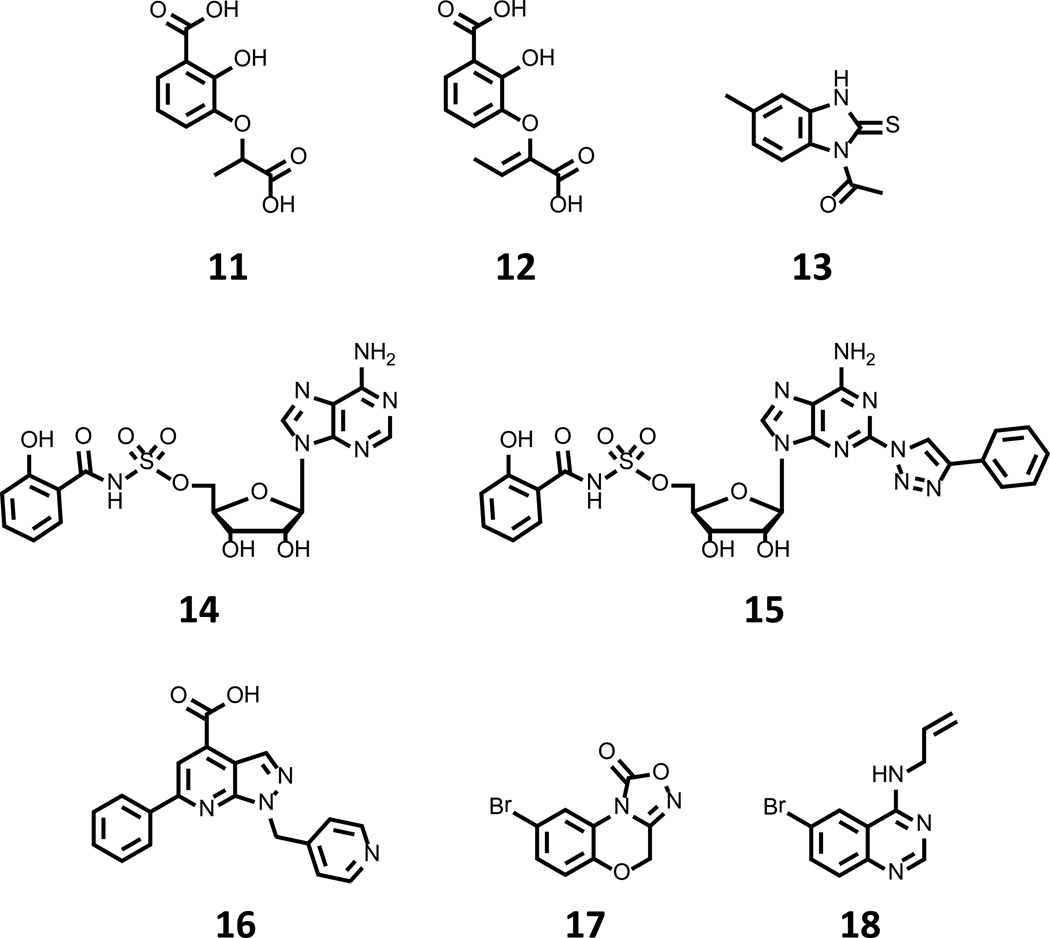
Efforts to inhibit the production aryl acid moieties have targeted salicylate synthase, the first committed step in the production of siderophores such as mycobactin 5 and yersiniabactin 4. Top compounds arising from these studies are shown in Fig. 5. Payne et al. first described work in this arena with a study of Irp9, the salicylate synthase of Y. pestis where benzoate analogs of chorismate were investigated as a means to rationally derive an inhibitor of the enzyme [57]. Iterative studies led to the synthesis of 2,3-dihydroxybenzoate ethers that possessed the greatest potency in the series, and provided 11 (Fig. 5) as a top lead with a Ki of 11 µM. A follow up study has since investigated similar compounds with the M. tuberculosis salicyl synthase MtbI and again identified this 2,3-substitution of the phenyl ring to possess the greatest potency [58]. While 11 exhibited a markedly reduced inhibitory behavior with the tuberculosis enzyme, subtle elaboration of the alkyl ether provided a series of compounds with Ki values again in the 10 µM range, with the best in class 12 (Fig. 5) displaying Ki value of 13 µM.
Figure 5. Compounds targeting siderophore biosynthetic pathways.
Compounds 11–13 have been identified to possess modest in vitro inhibitory activity with salicylate synthase enzymes of Y. pestis and M. tuberculosis. 5’-O-[N-salicyl-O-sulfamoyl]-adenosine 14 is a transition state analog of the reaction intermediate generated by the salicyl adenyl transferase enzyme of siderophore biosynthetic pathways, and has been shown to possess modest anti-mycobacterial activity. 15 is an optimized lead originating from medicinal chemistry optimization of 14. High throughput screening against BasE identified 16 as a potent inhibitor of A. baumannii salicyl adenyl transferase. Compounds 17 and 18 are members of LOPAC1280 that inhibit P. aeruginosa PvdQ with modest affinity.
Additionally, Vasan et al. have recently described the initiation of a program to identify inhibitor of MtbI via a target-based high throughput screening approach [59]. The authors screened 104,802 compounds and identified benzimidazole-2-thiones as novel series of reversible, noncompetitive inhibitors of MtbI with observable structure-activity relationship. Thiourea 13 (Fig. 5) was identified as a top candidate, with an IC50 of 7.6 µM in the biochemical assay, on par with the potencies observed for 12. While these reports have identified modest biochemical inhibitors of salicyl synthase, the compounds’ effects on bacterial growth, siderophore production, and/or viability in an iron-restricted setting have not been disclosed, and such results are eagerly awaited.
With regard to efforts targeting siderophore assembly, the activation of aryl acid functionalities has also received much attention. The activation of salicyl- and DHB- units is accomplished by adenylation enzymes that catalyse two half-reactions: first, the formation of the aryl adenylate, and then subsequent decomposition to provide an enzyme-linked aryl thioester. Given the similarity of this chemistry to that possessed by aminoacyl-tRNA synthetases, the first rationally designed inhibitors were a logical extension of non-hydrolysable transition state mimics for this enzyme class, specifically the 5’-O-[N-acyl-sulfamoyl]-adenosines [60]. The first compound developed for this purpose, 5’-O-[N-salicyl-sulfamoyl]-adenosine 14 (Fig. 5) was found to inhibit the salicyl-adenylating enzymes of Y. pestis, M. tuberculosis, and P. aeruginosa enzymes [61] with such a high affinity that delicate characterization was required to derive the apparent inhibitor constant, which was found to be in the single digit nM range. This value is approximately five orders of magnitude lower than the Km values exhibited for either of the substrates. Furthermore, this compound was characterized in cultures of Y. pestis and M. tuberculosis, where it was found to stifle siderophore biosynthesis and to also be a modest inhibitor of pathogen growth under iron limiting conditions in vitro [61].
Extensive SAR studies of 14 have focused on substitutions of the nucleobase, glycosyl, sulfamoyl and aryl acid portions of the molecule [62–64]. This work has provided 15 (Fig. 5) as an optimized structure that possesses Kiapp value of 27 pM for MtbA and inhibits the growth of M. tuberculosis H37RV by 99% at a concentration of 49 nM. These remarkable in vitro potencies, along with the suggestive results for target engagement inside the cell put forth by Ferreras et al. [61], provide a strong case for further evaluation of the compound class in a rodent model of tuberculosis. Toward this direction, the final disclosure on the subject is the preparation of a perdeuterospecies for pharmacokinetic studies [65], but subsequent work by the Aldrich group (vide infra) suggests a poor outcome from these studies [66].
Continued work in this vein has targeted BasE, the adenylation enzyme initiating the synthesis of acinetobactin 6 by A. baumannii, a Gram negative pathogen and considerable source of drug resistant nosocomial infection. A recent HTS campaign targeting this enzyme identified numerous chemotypes, with the most potent hit, 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid 16 (Fig. 5), exhibiting a KD of 78 nM [66]. Structural characterization of BasE in complex with 16 has demonstrated an unanticipated binding mode for the inhibitor with the phenyl ring positioned in the channel of the enzyme through which the acceptor nucleophile of the second half reaction would normally access the AMP-intermediate, which leaves the adenine binding pocket nearly unoccupied with the carboxylate pointing toward the region occupied by ribose in the AMP structure.
BasE is classified as adenylation domain of a nonribosomal peptide synthetase-enzyme, and is a member of the acyl/aryl CoA synthetase/adenylation domain of nonribosomal peptide synthetase/firefly luciferase (ANL) superfamily [67], and thus shows structural and functional similarities to firefly luciferase (Fluc). Given this homology, the potency of 16 in the biochemical assay relative to other hits, and the presence of a carboxylate functionality, it is an enticing conjecture to consider the possibility of the enzyme catalyzing the formation of a bisubstate adduct in the presence of 16 and ATP. Just such a mechanism has been observed for PCT124, a compound disputed to target nonsense codon suppression in Fluc cell-based assays as it was additionally identified as a potent Fluc inhibitor [68]. Later evidence showed that PTC124 exerted its extraordinary inhibitory potency against Fluc through the formation of an AMP adduct upon the catalytic action of luciferase [69]. If this mechanism were operational here, it could explain the unprecedented potency and peculiar binding mode of 16. Furthermore, blockage of the nucleophile access channel could stand to potentiate inhibition of the pathway by preventing breakdown of bisubstrate adduct. Reports describing the interrogation of 16 through medicinal chemistry efforts are awaited.
As a final entry in this section, Gulick et al. have recently detailed the biochemical activity and pilot screening of PvdQ, an N-terminal nucleophile hydrolase involved in the maturation of pyoverdine, the quinoline siderophore of P. aeruginosa [70]. This study assigned enzyme function to both PvdL and PvdQ, and identified an unprecedented and transient myristoylation event whose purpose is still poorly understood but essential for efficient growth of the organism in iron-restricted media. This new understanding of enzyme function allowed the development of an HTS-compatible assay for PvdQ that was used to profile the LOPAC1280 collection of bioactive molecules; as a result, compounds 17 and 18 (Fig. 5) were identified as modest inhibitors of the enzyme. Cocrystal structures for both 17 and 18 with PvdQ were also solved, and these data may allow for the further elaboration of either compound into more potent inhibitors of the enzyme that may prove useful to probe PvdQ as a target for antibacterial development.
3.3 Siderophore conjugates
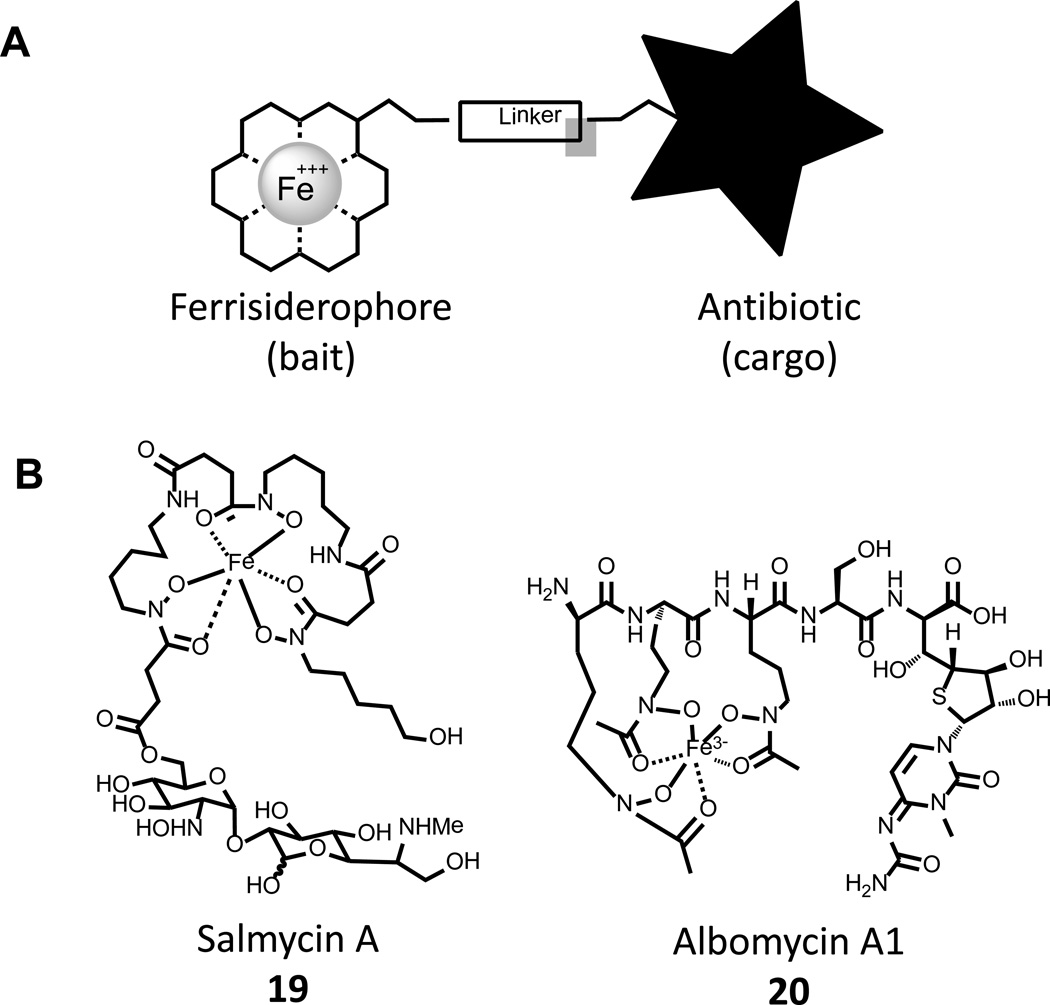
The competition invoked by the use of siderophores places a significant amount of selective pressure on bacterial cells of different species and a marked advantage is gained by organisms expressing receptors for xenosiderophores. As such, some organisms have developed the capacity to produce dual-function chemical warfare agents to combat the co-opting of their siderophores by coupling them to toxic moieties. A schematic of such compounds is outlined in Fig. 6A, where the siderophore is used as bait to deliver toxic cargo into the periplasm or cytosol of competing strains; akin to a “trojan horse”. This approach has particular attractiveness in the case of Gram negative pathogens, where the permeability of the outer membrane poses a significant barrier [71].
Figure 6. Sideromycins- “Trojan Horse” antibiotics.
A) Sideromycins are bifunctional compounds that use a siderophore as bait to provide for the active transport of antibiotic molecules into bacterial cells. B) Natural sideromycins Salmycin A 19 and Albomycin A1 20.
The term sideromycin is used to describe compounds of this class that are produced in Nature, and two such compounds are salmycin 19 and albomycin A1 20 (Fig. 6B). A detailed study of 19 and 20 found that both display truly remarkable MIC values of 8 – 10 ng/mL in Y. enterocolitica [72], and it is noted for comparison that ampicillin exhibits a MIC of 100 ng/mL in this strain. Testing of 19 in vivo found limited effectiveness in a mouse model of S. aureus septicemia, where it reduced bacterial titer 30–80 fold 72 h post infection; the compound displayed poor pharmacokinetic characteristics and was not pursued further [72]. In contrast, similar studies of 20 demonstrated a remarkable in vivo effectiveness where a single 1 mg/kg dose 6 h post infection reduced Y. enterocolitica titer by 3–4 log units, a result similar to those observed for the gentamicin control [73]. Even more impressive results were observed in models of S. pneumonia infection, where complete resolution of infection was observed at 7 days following a routine administration of 20 at doses as low as 1 mg/kg [73]. However, the frequency with which resistant mutants evolved was very high and likely due to the simple downregulation of siderophore receptors from the cell surface. While these findings are considered excellent proof of concept, 20 has not been pursued further, and no clinical trials have been initiated, possibly due to the combination of limited spectrum of activity, modest pharmacokinetic parameters, and high resistance rates [72, 73].
There has also been marked activity in the development of synthetic molecules conjoining siderophores with existing antibacterials, and their detailed description is beyond the scope of this review; instead, we will focus only on very recent advances. Interested readers are directed to [74] and [75] for timely summaries of the accomplishments of the field. Very recent and noteworthy siderophore-antibiotic conjugates are shown in Fig. 7.
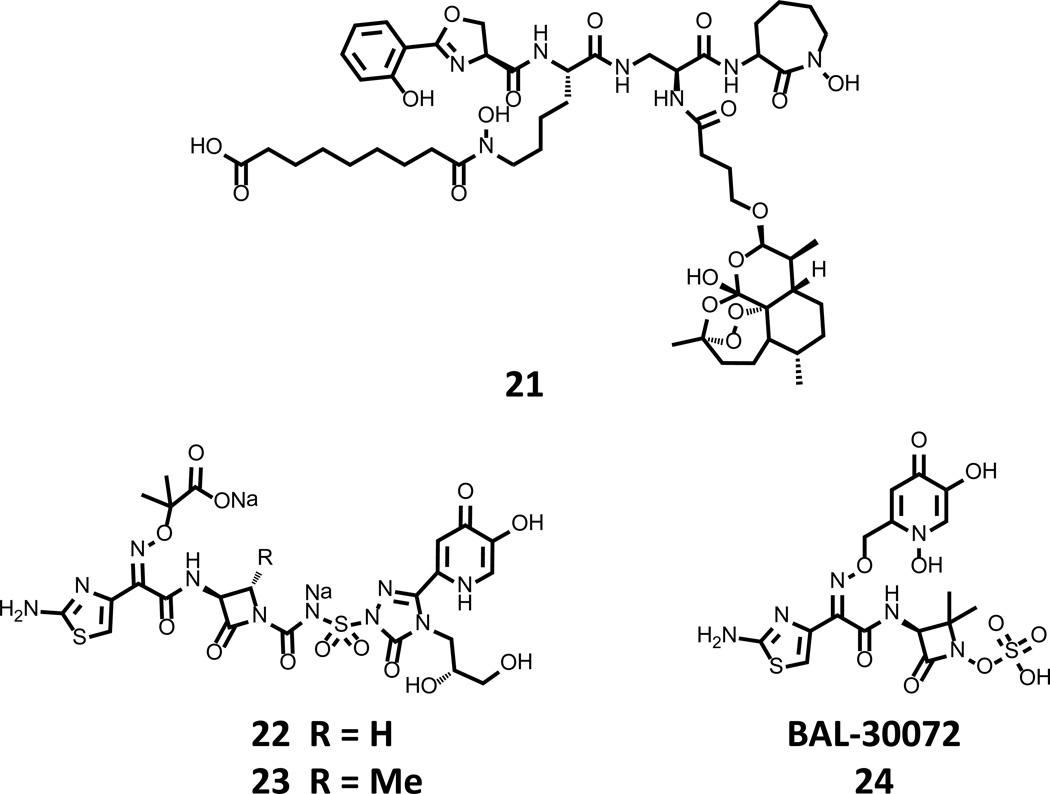
Figure 7. Recently disclosed synthetic siderophore-antibiotic conjugates.
Compound 21, contains artemisinin functionality that is linked to mycobactin to afford for uptake by mycobacteria, as well as its proximal localization to a Fenton chemistry potentiator that activates the toxicity of the endoperoxide. Compounds 22 and 23 were recently described as leads by Pfizer, and are potent inhibitors of fluoroquin-resistant P. aeruginosa. BAL-30072 24 has successfully completed Phase I clinical trials and is cleared to start Phase II evaluations for effectiveness to treat multidrug resistant Gram negative infections.
The Miller lab has been very active in this field and recently reported compound 21 (Fig. 7) to test the ability to rationally design a new antimycobacterial. 21 combines a reactive-oxygen species generating warhead (artemisinin) and the mycobactin siderophore core to target the cargo to the mycobacterial cell [76]. It is noteworthy that artemisinin itself, a source structure for several antimalarials, is not toxic to Mycobacteria spp. Putatively, 21 chelates Fe (III) in bacterial culture medium, where it is then taken up by the bacterial cell. However, cellular penetration of 21 requires further activation by a ferrisiderophore reductase before it becomes a Fenton chemistry potentiator (ferrous iron) and renders artimisinin toxic to the cell. Testing of 21 in vitro confirmed the activation hypothesis and revealed a remarkable antimycobacterial activity < 2 µg/mL for numerous strains of the M. tuberculosis, including multidrug and extremely drug resistant strains. Additionally impressive was the anti-Plasmodium activity of 21 that displayed a 4 ng/mL IC50 in P. falciparum cultures, similar to natural artemisinin. Taken together, the Miller lab has created a truly remarkable compound that displays potent activity against two of the primary causes of morbidity worldwide.
Pfizer recently described an extensive reevaluation of prior work by Upjohn [77]. The work centers around the development of MC-1 22 (Fig. 7) that links a monocyclic β-lactam (monocarbam) to a hydroxypryidinone chelating functionality. This heterocycle is sufficient to co-opt outer membrane influx pathways and deliver the toxic cargo to the bacterial periplasm. Detailed evaluation of the biological data for MC-1 22 identified high plasma protein binding to be the source of reduced in vivo efficacy, and a systematic interrogation of the structure produced 23 as an optimized lead with excellent in vitro activity against major Gram negative pathogens (0.25 – 2 µg/mL MIC values) and a protective dose with 50% efficacy (PD50) in a murine respiratory tract infection model with P. aeruginosa of 1.9 mg/kg. Clinical advancement of 22, 23 or related derivatives has not yet been reported.
Another siderophore-β-lactam conjugate, BAL-30072 24 (Fig. 7), was recently disclosed by Basilea Pharmaceutica. 24 is a hybrid compound containing an hydroxypyridinone bait tethered to a metallo-β-lactamase resistant sulfolactam core [78–82]. A thorough in vitro evaluation of 24 against Gram negative pathogens revealed MIC90 values in the single digit micromolar range for Actinobacter spp., E. coli, and P. aeruginosa, among many others [81]. Furthermore, these results were extended to a detailed in vitro and in vivo evaluation of 24 with six A. baumannii strains [78]. Statistically significant resolution of the bacterial infection in their rat model was observed for five of the six strains and mirrored the in vitro results, a significant feat for any new compound. While complications with the unoptimized in vivo model were noted, the results were satisfactory enough to allow for the initiation of Phase I clinical trials. Press releases from Basilea Pharmaceutica indicate that 24 was well tolerated, and Phase II trials evaluating efficacy in multidrug-resistant Gram-negative infections are planned to begin in 2013 [83].
3.4 Heme degradation
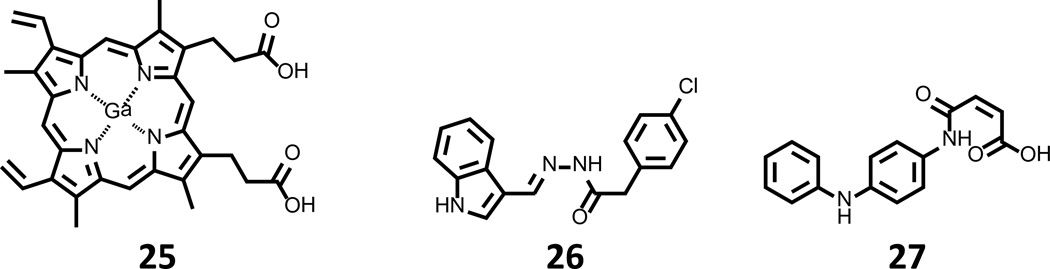
The acquisition of heme is a major source of iron for blood-borne pathogens. While the mechanisms by which major agents of septicemia acquire heme have been and continue to be actively studied, few efforts have been put forth to interrogate the druggability of heme assimilation pathways. Top compounds arising from these limited campaigns are shown in Fig. 8. Heme assimilation was first targeted with small molecules by Stojiljkovic et al. using gallium-loaded protoporphyrin IX 25 (Fig. 8). While initial studies attributed the antibacterial activity of 25 to the release of the toxic transition-metal cargo [84, 85], it was later discovered that 25 is incapable of being oxidized by heme monooxygenase [86] and may in fact exhibit its action through inhibition of this enzyme. Aside from work on 25, only a single additional effort investigating the druggability of heme assimilation has been reported, where compounds 26 and 27 (Fig. 8) were identified as new inhibitors of P. aeruginosa and Nesseria heme monoxygenase by virtual screening [87]. In addition to profiling these compounds with the bacterial enzymes, 26 and 27 were further characterized in a whole cell α-biliverdin production assay using E. coli strains that ectopically express the P. aeruginosa and Neisseria enzymes. In this evaluation, 26 and 27 were capable of blocking the oxidation of heme at concentrations of 250 µM. Further, testing against P. aeruginosa MPAO1 under iron-restricted conditions with heme as the sole source of iron demonstrated significant growth inhibition, a result that could be overcome by supplementation with ferric pyoverdine. Taken together, these results indicate that the inhibition of heme monooxygenase halts bacterial growth in conditions where heme is the sole iron source for P. aeruginosa, as the case would likely be in a clinical setting. While the authors acknowledge that 26 and 27 may not be suitable for further development, the proof of concept demonstrated in this study warrants further pursuit of higher potency inhibitors of this enzyme, and we anxiously await the results of follow up studies.
Figure 8. Inhibitors of heme assimilation.
Gallium protoporphyrin IX 25 has been described as a possible inhibitor of heme monooxygenase, while 26 and 27 inhibit the formation of α-bilirubin in cell-based assays for heme monooxygenase.
4.0 Expert opinion
Despite significant advances in the development of inhibitors that target steps in iron assimilation pathways, further in vivo testing to validate targets and accomplish proof of concept studies with preliminary chemotypes will be essential to the success of these programs. Many of the compounds detailed above were characterized with limited in vitro biological testing to recapitulate a phenotype of sensitization to iron-restricted growth conditions. While this provides sufficient proof of involvement in iron assimilation pathways, it does not speak to the clinical relevance of the targets or pathways. Microorganisms possess numerous mechanisms through which to access iron from their host during infection. The role of this redundancy, and its capacity to provide inborn mechanisms of resistance through the upregulation of compensatory pathways, has not been evaluated. Further testing of the compounds described herein under conditions more similar to those encountered during infection, such as culture in whole blood to better predict performance in an animal model, as well as more frequent extension of work to such in vivo models, would increase the impact and translational relevance of this work.
With regard to resistance mechanisms, a recent report highlights the role of pathway redundancy: virulence was unaffected after disruption of the enterobactin and salmochelin pathways in extraintestinal pathogenic E. coli using a chicken model of sepsis [88]. It was further shown that infectivity was sustained by the production of aerobactin, a hydroxamate siderophore purported to sufficiently to supply iron in vivo. This is in contrast to results observed in mouse, where the salmochelin production locus iroA was required for infectivity [42]. These disparities may represent differing capacities of the bacterial strains and/or alternatively suggest that successful targeting of iron assimilation may be restricted to organisms possessing singular acquisition pathways or the development of cotherapies. Nonetheless, the findings necessitate further study of iron assimilation and inhibitors thereof in the most disease-relevant models to ascertain the true therapeutic potential of these pathways.
The end of the Golden era of antibiotic discovery has left us in a dangerous situation where our pipeline of reserve and next-generation therapeutics may soon run dry. To stay ahead of the selective pressures placed upon pathogens through the continued use of antibiotics, the paradigm followed for the last 60 years needs to be reconsidered: Broad spectrum targets have been, by and large, exploited. The prokaryotic portion of the evolutionary tree is remarkably diverse. It has been suggested that humans have more in common with paramecia that do Gram positive and Gram negative organisms [89]. This distance makes the frequency of highly identical molecular targets vanishingly small. Indeed, the majority of current therapeutics act through limited mechanisms: inhibition of the bacterial ribosome, cell wall assembly intermediates (i.e. lipid II) or the cell wall biogenic machinery itself (e.g. penicillin binding proteins). Industry has exerted tremendous effort to develop compounds that act highly conserved molecular processes and targets [90], but the future of the anti-infectives field may very well focus on therapeutics with limited coverage that interfere with disease- and strain-specific pathways and targets [90, 91]. While this adjustment in scope would not provide broad-spectrum drugs, such compounds would address specifically unmet medical needs. Efforts to target pathways used by bacteria to acquire iron, where a remarkable amount of diversity is possessed, fall into this category [92]. Furthermore, as we begin to better understand the human microbiome, limited spectral coverage can in principle, be advantageous. Specific therapeutics are anticipated perturb the ecology of the microbiome less than traditional antibiotics and may thereby limit annihilation of the normal gastrointestinal flora and consequently reduce incidences of opportunistic secondary infections (e.g. Clostridium difficile).
With this realization, our approaches to policy regarding anti-infective drug approval for needs an update. As the era of personalized medicine dawns, infectious disease associations have begun to call for the regulatory field to develop a Limited Population Antibiotic Drug status for indications with low patient populations and high need for novel intervention. Such a characterization would provide for similar benefits as drugs for rare diseases that are awarded orphan drug status under the Orphan Drug Act of 1983, a designation which could markedly reduce the time and cost of obtaining regulatory approval for much needed drugs, and thereby improve the human condition though the more rapid release of new therapeutic regiments to the general public. This case has recently become a cause championed by the Infectious Disease Society of America [4–8], and we await to see the impact that their efforts are able to have in terms of policy augmentation. For a further discussion of regulatory and policy shifts that may accelerate the progression of antimicrobial therapeutics to the clinic, the readership is directed to [7] and [8].
Highlights.
In an infection setting, bacteria assimilate iron from the host environment by (i) secreting siderophores to strip iron from host proteins, (ii) binding, internalizing and degrading heme, and (iii) expressing receptors for host proteins that strip ferric ion from transferrin and lactoferrin.
Chelation therapy agents are being investigated for their potential to be repurposed into adjuvant therapies to treat fungal infections.
Studies of siderophore biosynthesis enzymes have provided inhibitors with nanomolar affinities, and preliminary studies show that inhibition of these enzymes quell bacterial growth in iron-restricted media.
Investigations targeting heme monooxygenase have identified compounds that inhibit α-bilirubin production in bacterial cell assays and limit bacterial growth when heme is the only source of iron.
Synthetic siderophore-β-lactam conjugates have shown effectiveness in proof of concept rodent models of Gram negative bacterial infection, and Phase II clinical trials of one such compound are set to begin in 2013.
Evolving trends in antibacterial development and calls for regulatory-related policy changes are discussed.
List of abbreviations
- ABC
ATP binding cassette
- DHB
2,3-dihydroxybenzoate
- IC50
concentration that elicits 50% inhibition
- MIC
minimum inhibitory concentration
- MIC90
minimum concentration that inhibits 90% of growth
- NGAL
neutrophil-gelatinase-associated lipocalin
- PD50
dose that protects 50% of the challenged animals
- SAR
structure-activity relationship
References
- 1.Power E. Impact of antibiotic restrictions: the pharmaceutical perspective. Clinical Microbiology and Infection. 2006 Aug;12:25–34. doi: 10.1111/j.1469-0691.2006.01528.x. [DOI] [PubMed] [Google Scholar]
- 2.Norrby R, Powell M. The bacterial challenge: time to react. Stockholm: European Centre for Disease Prevention and Control; 2009. [Google Scholar]
- 3.Choffnes ER, Relman DA, Mack A. Antibiotic Resistance: Implications for global health and novel intervention strategies. Washington, D.C.: National Academies Press; 2010. [PubMed] [Google Scholar]
- 4.America IDSo. Bad bugs, no drugs: as antibiotic discovery stagnates… a public health crisis brews. Alexandria, VA: Infectious Disease Society of America; 2004. [Google Scholar]
- 5. Gilbert DN, Guidos RJ, Boucher HW, et al. The 10 × '20 Initiative: Pursuing a Global Commitment to Develop 10 New Antibacterial Drugs by 2020. Clinical Infectious Diseases. 2010 Apr 15;50(8):1081–1083. doi: 10.1086/652237. Description of the Infectious Disease Society of America's intiative to bring 10 new antibacterial compounds to the clinic by 2020.
- 6.Boucher HW. Challenges in Anti-Infective Development in the Era of Bad Bugs, No Drugs: A Regulatory Perspective Using the Example of Bloodstream Infection as an Indication. Clinical Infectious Diseases. 2010 Jan 1;50:S4–S9. doi: 10.1086/647937. [DOI] [PubMed] [Google Scholar]
- 7.Boucher HW, Talbot GH, Bradley JS, et al. Bad Bugs, No Drugs: No ESKAPE! An Update from the Infectious Diseases Society of America. Clinical Infectious Diseases. 2009 Jan;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 8.Guidos RJ, Spellberg B, Blaser M, et al. Combating Antimicrobial Resistance: Policy Recommendations to Save Lives. Clinical Infectious Diseases. 2011 May 1;52:S397–S428. doi: 10.1093/cid/cir153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neilands JB. Iron absorption and transport in microorganisms. Annu Rev Nutr. 1981;1:27–46. doi: 10.1146/annurev.nu.01.070181.000331. [DOI] [PubMed] [Google Scholar]
- 10.Ratledge C. Iron metabolism and infection. Food Nutr Bull. 2007 Dec;28(4 Suppl):S515–S523. doi: 10.1177/15648265070284S405. [DOI] [PubMed] [Google Scholar]
- 11.Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 12.Crosa JH. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997 Sep;61(3):319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999 Oct;181(20):6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosa biofilm formation. Proceedings of the National Academy of Sciences of the United States of America. 2005 Aug 2;102(31):11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock V, Dahl M, Klemm P. Abolition of Biofilm Formation in Urinary Tract Escherichia coli and Klebsiella Isolates by Metal Interference through Competition for Fur. Applied and Environmental Microbiology. 2010 Jun;76(12):3836–3841. doi: 10.1128/AEM.00241-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollack JR, Neilands JB. Enterobactin, an iron transport compound from Salmonella typhimurium. Biochem Biophys Res Commun. 1970 Mar 12;38(5):989–992. doi: 10.1016/0006-291x(70)90819-3. [DOI] [PubMed] [Google Scholar]
- 17.Furrer JL, Sanders DN, Hook-Barnard IG, et al. Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol Microbiol. 2002 Jun;44(5):1225–1234. doi: 10.1046/j.1365-2958.2002.02885.x. [DOI] [PubMed] [Google Scholar]
- 18.Bister B, Bischoff D, Nicholson GJ, et al. The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica. Biometals. 2004 Aug;17(4):471–481. doi: 10.1023/b:biom.0000029432.69418.6a. [DOI] [PubMed] [Google Scholar]
- 19.Fischbach MA, Lin HN, Liu DR, et al. In vitro characterization of IroB, a pathogen-associated C-glycosyltransferase. Proceedings of the National Academy of Sciences of the United States of America. 2005 Jan 18;102(3):571–576. doi: 10.1073/pnas.0408463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin H, Fiscchbach MA, Liu DR, et al. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. Journal of the American Chemical Society. 2005 Aug 10;127(31):11075–11084. doi: 10.1021/ja0522027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crouch M-LV, Castor M, Karlinsey JE, et al. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2008 Mar;67(5):971–983. doi: 10.1111/j.1365-2958.2007.06089.x. [DOI] [PubMed] [Google Scholar]
- 22.Wiener MC, Horanyi PS. How hydrophobic molecules traverse the outer membranes of Gram-negative bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2011 Jul;108(27):10929–10930. doi: 10.1073/pnas.1106927108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caza M, Lepine F, Milot S, et al. Specific roles of the iroBCDEN genes in virulence of an avian pathogenic Eschetichia coli O78 strain and in production of salmochelins. Infection and Immunity. 2008 Aug;76(8):3539–3549. doi: 10.1128/IAI.00455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abergel RJ, Zawadzka AM, Hoette TM, et al. Enzymatic Hydrolysis of Trilactone Siderophores: Where Chiral Recognition Occurs in Enterobactin and Bacillibactin Iron Transport. Journal of the American Chemical Society. 2009 Sep 9;131(35):12682–12692. doi: 10.1021/ja903051q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen NA, Lin H, Wei R, et al. Structural characterization of enterobactin hydrolase IroE. Biochemistry. 2006 Aug 29;45(34):10184–10190. doi: 10.1021/bi060950i. [DOI] [PubMed] [Google Scholar]
- 26.Wilks A, Burkhard KA. Heme and virulence: how bacterial pathogens regulate, transport and utilize heme. Nat Prod Rep. 2007 Jun;24(3):511–522. doi: 10.1039/b604193k. [DOI] [PubMed] [Google Scholar]
- 27.Gkouvatsos K, Papanikolaou G, Pantopoulos K. Regulation of iron transport and the role of transferrin. Biochimica Et Biophysica Acta-General Subjects. 2012 Mar;1820(3):188–202. doi: 10.1016/j.bbagen.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 28.Siddique A, Kowdley KV. Review article: the iron overload syndromes. Alimentary Pharmacology & Therapeutics. 2012 Apr;35(8):876–893. doi: 10.1111/j.1365-2036.2012.05051.x. [DOI] [PubMed] [Google Scholar]
- 29.Ganz T. Hepcidin, a key regulator of iron metabolism and mediator of anemia of inflammation. Blood. 2003 Aug 1;102(3):783–788. doi: 10.1182/blood-2003-03-0672. [DOI] [PubMed] [Google Scholar]
- 30.Abril Garcia-Montoya I, Siqueiros Cendon T, Arevalo-Gallegos S, et al. Lactoferrin a multiple bioactive protein: An overview. Biochimica Et Biophysica Acta-General Subjects. 2012 Mar;1820(3):226–236. doi: 10.1016/j.bbagen.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crowley RC, Leigh JA, Ward PN, et al. Differential Protein Expression in Streptococcus uberis under Planktonic and Biofilm Growth Conditions. Applied and Environmental Microbiology. 2011 Jan;77(1):382–384. doi: 10.1128/AEM.01099-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamiya H, Ehara T, Matsumoto T. Inhibitory effects of lactoferrin on biofilm formation in clinical isolates of Pseudomonas aeruginosa. Journal of Infection and Chemotherapy. 2012 Feb;18(1):47–52. doi: 10.1007/s10156-011-0287-1. [DOI] [PubMed] [Google Scholar]
- 33.Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 34.Modun B, Williams P. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect Immun. 1999 Mar;67(3):1086–1092. doi: 10.1128/iai.67.3.1086-1092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor JM, Heinrichs DE. Transferrin binding in Staphylococcus aureus: involvement of a cell wall-anchored protein. Mol Microbiol. 2002 Mar;43(6):1603–1614. doi: 10.1046/j.1365-2958.2002.02850.x. [DOI] [PubMed] [Google Scholar]
- 36.Fischbach MA, Lin H, Liu DR, et al. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol. 2006 Mar;2(3):132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 37.Konopka K, Neilands JB. Effect of serum albumin on the siderophore-mediated utilization of transferrin iron. Biochemistry. 1984;23(10):2122–2127. doi: 10.1021/bi00305a003. 1984. [DOI] [PubMed] [Google Scholar]
- 38.Smith KD. Iron metabolism at the host pathogen interface: lipocalin 2 and the pathogen-associated iroA gene cluster. Int J Biochem Cell Biol. 2007;39(10):1776–1780. doi: 10.1016/j.biocel.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borregaard N, Cowland JB. Neutrophil gelatinase-associated lipocalin, a siderophore-binding eukaryotic protein. Biometals. 2006 Apr;19(2):211–215. doi: 10.1007/s10534-005-3251-7. [DOI] [PubMed] [Google Scholar]
- 40.Goetz DH, Holmes MA, Borregaard N, et al. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Molecular Cell. 2002 Nov;10(5):1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 41.Sorsa LJ, Dufke S, Heesemann J, et al. Characterization of an iroBCDEN gene cluster on a transmissible plasmid of uropathogenic Escherichia coli: Evidence for horizontal transfer of a chromosomal virulence factor. Infection and Immunity. 2003 Jun;71(6):3285–3293. doi: 10.1128/IAI.71.6.3285-3293.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fischbach MA, Lin H, Zhou L, et al. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc Natl Acad Sci U S A. 2006 Oct 31;103(44):16502–16507. doi: 10.1073/pnas.0604636103. The authors evaluate the role of the iroA gene cluster in the virulence of extraintestinal pathogenic E. coli, and demonstrate their findings in a mouse model of infection.
- 43.Johnson JR, Russo TA. Uropathogenic Escherichia coli as agents of diverse non-urinary tract extraintestinal infections. Journal of Infectious Diseases. 2002 Sep 15;186(6):859–864. doi: 10.1086/342490. [DOI] [PubMed] [Google Scholar]
- 44.Johnson JR, Russo TA. Extraintestinal pathogenic Escherichia coli: "The other bad E coli". Journal of Laboratory and Clinical Medicine. 2002 Mar;139(3):155–162. doi: 10.1067/mlc.2002.121550. [DOI] [PubMed] [Google Scholar]
- 45.Kovacevic Z, Kalinowski DS, Lovejoy DB, et al. The medicinal chemistry of novel iron chelators for the treatment of cancer. Curr Top Med Chem. 2011;11(5):483–499. doi: 10.2174/156802611794785190. [DOI] [PubMed] [Google Scholar]
- 46.Rund D, Rachmilewitz E. Beta-thalassemia. N Engl J Med. 2005 Sep 15;353(11):1135–1146. doi: 10.1056/NEJMra050436. [DOI] [PubMed] [Google Scholar]
- 47.Nick H, Acklin P, Lattmann R, et al. Development of tridentate iron chelators: From desferrithiocin to ICL670. Current Medicinal Chemistry. 2003 Jun;10(12):1065–1076. doi: 10.2174/0929867033457610. [DOI] [PubMed] [Google Scholar]
- 48.Brock JH, Liceaga J, Kontoghiorghes GJ. The effect of synthetic iron chelators on bacterial growth in human serum. FEMS Microbiol Immunol. 1988 Jan;1(1):55–60. doi: 10.1111/j.1574-6968.1988.tb02490.x. [DOI] [PubMed] [Google Scholar]
- 49.Hartzen SH, Frimodtmoller N, Thomsen VF. The antibacterial activity of a siderophore. 2. The influence of deferoxamine alone and combined with ascorbic acid on the activity of antibiotics against Staphyloccocus aureus. Apmis. 1991 Oct;99(10):879–886. [PubMed] [Google Scholar]
- 50.Zurlo MG, Destefano P, Borgnapignatti C, et al. Survival and causes of death in thalassemia major. Lancet. 1989 Jul;2(8653):27–30. doi: 10.1016/s0140-6736(89)90264-x. [DOI] [PubMed] [Google Scholar]
- 51.Zarember KA, Cruz AR, Huang C-Y, et al. Antifungal Activities of Natural and Synthetic Iron Chelators Alone and in Combination with Azole and Polyene Antibiotics against Aspergillus fumigatus. Antimicrob Agents Chemother. 2009 Jun;53(6):2654–2656. doi: 10.1128/AAC.01547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ibrahim AS, Edwards JE, Fu Y, et al. Deferiprone iron chelation as a novel therapy for experimental mucormycosis. Journal of Antimicrobial Chemotherapy. 2006 Nov;58(5):1070–1073. doi: 10.1093/jac/dkl350. [DOI] [PubMed] [Google Scholar]
- 53.Ibrahim AS, Gebermariam T, Fu Y, et al. The iron chelator deferasirox protects mice from mucormycosis through iron starvation. Journal of Clinical Investigation. 2007 Sep;117(9):2649–2657. doi: 10.1172/JCI32338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spellberg B, Andes D, Perez M, et al. Safety and Outcomes of Open-Label Deferasirox Iron Chelation Therapy for Mucormycosis. Antimicrob Agents Chemother. 2009 Jul;53(7):3122–3125. doi: 10.1128/AAC.00361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reed C, Ibrahim A, Edwards JE, et al. Deferasirox, an iron-chelating agent, as salvage therapy for rhinocerebral mucormycosis. Antimicrob Agents Chemother. 2006 Nov;50(11):3968–3969. doi: 10.1128/AAC.01065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Spellberg B, Ibrahim AS, Chin-Hong PV, et al. The Deferasirox-AmBisome Therapy for Mucormycosis (DEFEAT Mucor) study: a randomized, double-blinded, placebo-controlled trial. Journal of Antimicrobial Chemotherapy. 2012 Mar;67(3):715–722. doi: 10.1093/jac/dkr375. A timely report on the Phase II clinical trial of deferasirox for efficacy in human cases of mucormycosis.
- 57.Kerbarh O, Bulloch EM, Payne RJ, et al. Mechanistic and inhibition studies of chorismate-utilizing enzymes. Biochem Soc Trans. 2005 Aug;33(Pt 4):763–766. doi: 10.1042/BST0330763. [DOI] [PubMed] [Google Scholar]
- 58.Manos-Turvey A, Bulloch EM, Rutledge PJ, et al. Inhibition studies of Mycobacterium tuberculosis salicylate synthase (MbtI) Chemmedchem. 2010 Jul 5;5(7):1067–1079. doi: 10.1002/cmdc.201000137. [DOI] [PubMed] [Google Scholar]
- 59.Vasan M, Neres J, Williams J, et al. Inhibitors of the Salicylate Synthase (MbtI) from Mycobacterium tuberculosis Discovered by High-Throughput Screening. Chemmedchem. 2010 Dec;5(12):2079–2087. doi: 10.1002/cmdc.201000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finking R, Neumuller A, Solsbacher J, et al. Aminoacyl adenylate substrate analogues for the inhibition of adenylation domains of nonribosomal peptide synthetases. Chembiochem. 2003 Sep;4(9):903–906. doi: 10.1002/cbic.200300666. [DOI] [PubMed] [Google Scholar]
- 61. Ferreras JA, Ryu JS, Di Lello F, et al. Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis. Nat Chem Biol. 2005 Jun;1(1):29–32. doi: 10.1038/nchembio706. The authors present the first work demonstrating the inhibition of aryl acid adenylating enzymes involved in siderophore biosynthesis and their utility as targets for therapeutic development.
- 62.Duckworth BP, Nelson KM, Aldrich CC. Adenylating Enzymes in Mycobacterium tuberculosis as Drug Targets. Curr Top Med Chem. 2012 Apr;12(7):766–796. doi: 10.2174/156802612799984571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gupte A, Boshoff HI, Wilson DJ, et al. Inhibition of Siderophore Biosynthesis by 2-Triazole Substituted Analogues of 5'-O- N-(Salicyl)sulfamoyl adenosine: Antibacterial Nucleosides Effective against Mycobacterium tuberculosis. Journal of Medicinal Chemistry. 2008 Dec;51(23):7495–7507. doi: 10.1021/jm8008037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qiao CH, Gupte A, Boshoff HI, et al. 5 '-O- (N-Acyl)sulfamoyl adenosines as antitubercular agents that inhibit MbtA: An adenylation enzyme required for siderophore biosynthesis of the mycobactins. Journal of Medicinal Chemistry. 2007 Nov;50(24):6080–6094. doi: 10.1021/jm070905o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gupte A, Subramanian M, Remmel RP, et al. Synthesis of deuterium-labelled 5 '-O- N-(Salicyl)sulfamoyl adenosine (Sal-AMS-d(4)) as an internal standard for quantitation of Sal-AMS. Journal of Labelled Compounds & Radiopharmaceuticals. 2008 Jan-Feb;51(1–2):118–122. doi: 10.1002/jlcr.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drake EJ, Duckworth BP, Neres J, et al. Biochemical and Structural Characterization of Bisubstrate Inhibitors of BasE, the Self-Standing Nonribosomal Peptide Synthetase Adenylate-Forming Enzyme of Acinetobactin Synthesis. Biochemistry. 2010 Nov;49(43):9292–9305. doi: 10.1021/bi101226n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gulick AM. Conformational Dynamics in the Acyl-CoA Synthetases, Adenylation Domains of Non-ribosomal Peptide Synthetases, and Firefly Luciferase. Acs Chemical Biology. 2009 Oct;4(10):811–827. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Auld DS, Thorne N, Maguire WF, et al. Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression. Proceedings of the National Academy of Sciences of the United States of America. 2009 Mar 3;106(9):3585–3590. doi: 10.1073/pnas.0813345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Auld DS, Lovell S, Thorne N, et al. Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124. Proceedings of the National Academy of Sciences of the United States of America. 2010 Mar 16;107(11):4878–4883. doi: 10.1073/pnas.0909141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drake EJ, Gulick AM. Structural Characterization and High-Throughput Screening of Inhibitors of PvdQ, an NTN Hydrolase Involved in Pyoverdine Synthesis. Acs Chemical Biology. 2011 Nov;6(11):1277–1286. doi: 10.1021/cb2002973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hancock REW. The bacterial outer membrane as a drug barrier. Trends in Microbiology. 1997 Jan;5(1):37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 72.Braun V, Pramanik A, Gwinner T, et al. Sideromycins: tools and antibiotics. Biometals. 2009 Feb;22(1):3–13. doi: 10.1007/s10534-008-9199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pramanik A, Stroeher UH, Krejci J, et al. Albomycin is an effective antibiotic, as exemplified with Yersinia enterocolitica and Streptococcus pneumoniae. International Journal of Medical Microbiology. 2007 Oct;297(6):459–469. doi: 10.1016/j.ijmm.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 74.Miller MJ, Zhu H, Xu Y, et al. Utilization of microbial iron assimilation processes for the development of new antibiotics and inspiration for the design of new anticancer agents. Biometals. 2009 Feb;22(1):61–75. doi: 10.1007/s10534-008-9185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ji C, Juarez-Hernandez RE, Miller MJ. Exploiting bacterial iron acquisition: siderophore conjugates. Future Med Chem. 2012 Mar;4(3):297–313. doi: 10.4155/fmc.11.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miller MJ, Walz AJ, Zhu H, et al. Design, Synthesis, and Study of a Mycobactin-Artemisinin Conjugate That Has Selective and Potent Activity against Tuberculosis and Malaria. Journal of the American Chemical Society. 2011 Feb 23;133(7):2076–2079. doi: 10.1021/ja109665t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Flanagan ME, Brickner SJ, Lall M, et al. Preparation, Gram-Negative Antibacterial Activity, and Hydrolytic Stability of Novel Siderophore-Conjugated Monocarbam Diols. Acs Medicinal Chemistry Letters. 2011 May;2(5):385–390. doi: 10.1021/ml200012f. A thorough report detailing the medicinal chemistry optimization of a β-lactam/siderophore conjugate for efficacy in a mouse model of P. aeruginosa infection.
- 78.Higgins PG, Stefanik D, Page MGP, et al. In vitro activity of the siderophore monosulfactam BAL30072 against meropenem-non-susceptible Acinetobacter baumannii. The Journal of antimicrobial chemotherapy. 2012 May;67(5):1167–1169. doi: 10.1093/jac/dks009. 2012. [DOI] [PubMed] [Google Scholar]
- 79.Mima T, Kvitko BH, Rholl DA, et al. In vitro activity of BAL30072 against Burkholderia pseudomallei. International Journal of Antimicrobial Agents. 2011 Aug;38(2):157–159. doi: 10.1016/j.ijantimicag.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mushtaq S, Warner M, Livermore D. Activity of the siderophore monobactam BAL30072 against multiresistant non-fermenters. Journal of Antimicrobial Chemotherapy. 2010 Feb;65(2):266–270. doi: 10.1093/jac/dkp425. [DOI] [PubMed] [Google Scholar]
- 81.Page MG, Dantier C, Desarbre E. In vitro properties of BAL30072, a novel siderophore sulfactam with activity against multiresistant gram-negative bacilli. Antimicrob Agents Chemother. 2010 Jun;54(6):2291–2302. doi: 10.1128/AAC.01525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Page MGP, Desarbre E, Gebhardt K, et al. Synthesis and antimicrobial activity of the novel siderophore sulfactam BAL30072. International Journal of Infectious Diseases. 2011 Jul;15:S17–S17. [Google Scholar]
- 83.Pharmaceutica B. Basilea announces conclusion of BAL30072 phase I multiple ascending dose study. Basel, Switzerland: 2011. [Google Scholar]
- 84.Stojiljkovic I, Evavold BD, Kumar V. Antimicrobial properties of porphyrins. Expert Opinion on Investigational Drugs. 2001 Feb;10(2):309–320. doi: 10.1517/13543784.10.2.309. [DOI] [PubMed] [Google Scholar]
- 85.Stojiljkovic I, Kumar V, Srinivasan N. Non-iron metalloporphyrins: potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol Microbiol. 1999 Jan;31(2):429–442. doi: 10.1046/j.1365-2958.1999.01175.x. [DOI] [PubMed] [Google Scholar]
- 86.Reniere ML, Torres VJ, Skaar EP. Intracellular metalloporphyrin metabolism in Staphylococcus aureus. Biometals. 2007 Jun;20(3–4):333–345. doi: 10.1007/s10534-006-9032-0. [DOI] [PubMed] [Google Scholar]
- 87.Furci LM, Lopes P, Eakanunkul S, et al. Inhibition of the bacterial heme oxygenases from Pseudomonas aeruginosa and Neisseria meningitidis: novel antimicrobial targets. J Med Chem. 2007 Aug 9;50(16):3804–3813. doi: 10.1021/jm0700969. [DOI] [PubMed] [Google Scholar]
- 88. Caza M, Lepine F, Dozois CM. Secretion, but not overall synthesis, of catecholate siderophores contributes to virulence of extraintestinal pathogenic Escherichia coli. Mol Microbiol. 2011 Apr;80(1):266–282. doi: 10.1111/j.1365-2958.2011.07570.x. The authors evaluate the role of siderophore secretion in extraintestinal pathogenic E. coli using a chicken sepsis model. Their findings the null effect that ablation of catecholate siderophore production had on infectivity.
- 89.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997 May 2;276(5313):734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 90. Payne DJ, Gwynn MN, Holmes DJ, et al. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nature Reviews Drug Discovery. 2007 Jan;6(1):29–40. doi: 10.1038/nrd2201. An honest summary of the antibiotic discovery efforts at GlaxoSmithKline, including difficulties encountered and failures.
- 91.Broetz-Oesterhelt H, Sass P. Postgenomic strategies in antibacterial drug discovery. Future Microbiology. 2010 Oct;5(10):1553–1579. doi: 10.2217/fmb.10.119. [DOI] [PubMed] [Google Scholar]
- 92.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007 Sep;71(3):413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]