Abstract
Clostridium difficile is an important, emerging nosocomial pathogen. The transition from harmless colonization to disease is typically preceded by antimicrobial therapy, which alters the balance of the intestinal flora, enabling C. difficile to proliferate in the colon. One of the most perplexing aspects of the C. difficile infectious cycle is its ability to survive antimicrobial therapy and transition from inert colonization to active infection. Toxin-antitoxin (TA) systems have been implicated in facilitating persistence after antibiotic treatment. We identified only one TA system in C. difficile strain 630 (epidemic type X), designated MazE-cd and MazF-cd, a counterpart of the well-characterized Escherichia coli MazEF TA system. This E. coli MazF toxin cleaves mRNA at ACA sequences, leading to global mRNA degradation, growth arrest, and death. Likewise, MazF-cd expression in E. coli or Clostridium perfringens resulted in growth arrest. Primer extension analysis revealed that MazF-cd cleaved RNA at the five-base consensus sequence UACAU, suggesting that the mRNAs susceptible to cleavage comprise a subset of total mRNAs. In agreement, we observed differential cleavage of several mRNAs by MazF-cd in vivo, revealing a direct correlation between the number of cleavage recognition sites within a given transcript and its susceptibility to degradation by MazF-cd. Interestingly, upon detailed statistical analyses of the C. difficile transcriptome, the major C. difficile virulence factor toxin B (TcdB) and CwpV, a cell wall protein involved in aggregation, were predicted to be significantly resistant to MazF-cd cleavage.
INTRODUCTION
Clostridium difficile is a Gram-positive, spore-forming anaerobe that has recently emerged as an important nosocomial pathogen. The normal colonic flora usually serves to repress infection by this pathogen. However, in patients receiving antimicrobial therapy, C. difficile populations are able to increase as the normal flora decreases, typically resulting in infection (C. difficile infection, or CDI) that manifests clinically as severe diarrhea (8, 43). One of the most challenging aspects of C. difficile-associated disease is the high incidence of recurrent infections, due either to reinfection by spore ingestion or relapse resulting from the germination of spores that remained in the intestinal lumen after the inadequate treatment of the primary infection (37). The incidence and severity of nosocomial CDI have been increasing in the past decade, and treatment is difficult and expensive. The emergence of the highly virulent BI/NAP1/027 (toxinotype III) strain accounts for some of this increase in severity and contributes to rising mortality rates. Progressively more cases of community-acquired (versus nosocomial) disease are being documented, and the number of cases in those who were considered to be at low risk of acquiring infections (generally healthy individuals who were not in hospitals or taking antibiotics) is also rising (43).
The virulence of this pathogen is attributed to two major exotoxins, toxin A and toxin B, and the binary toxin CDT (Clostridium difficile transferase). Contrasting reports on the pathological importance of the toxins have recently been published. One group found that toxin B alone was essential for C. difficile virulence (30), while another found that both A+B− and A−B+ strains can cause disease in a hamster model (28). Both toxin A and B proteins are monoglucosyltransferases that cause the disruption of tight junctions and the actin cytoskeleton, resulting in the wide-scale destruction of the intestinal epithelium. The cytotoxic and inflammatory pathological features characteristic of the disease are attributed to the physical effects of the toxins. The role of CDT in disease is not well understood but has recently been shown to increase microtubule formation to presumably facilitate bacterial adherence (47). Surface layer proteins involved in the adherence of the pathogen to the intestinal mucosa are also considered to be important for C. difficile pathogenesis. Surface layer protein A, SlpA, was recently demonstrated to be altered in certain hypervirulent strains, facilitating tighter C. difficile adherence (43). Finally, the ability to form spores is a major phenomenon associated with C. difficile virulence. The spores are highly resistant to desiccation, chemical shock, and extreme temperatures and are the most important mode of dissemination.
Recent reports suggest a link between pathogenesis and toxin-antitoxin (TA) systems (40, 54, 55). TA systems comprise a stable toxin and an unstable antitoxin. TA toxins, in contrast to exotoxins, are intracellular and only affect an essential process, such as translation or replication within the producing cell (11, 16, 34). The antitoxin is able to sequester the effects of the toxin through the formation of a stable complex. When physiological conditions favor the degradation of the antitoxin by proteases, the toxin is free to act on the cell.
The MazE-MazF TA system has been studied in detail in several bacteria. Escherichia coli MazF is a sequence-specific endoribonuclease that cleaves single-stranded mRNA at ACA sequences (53); the MazE antitoxin inhibits the cleavage of mRNA by MazF. MazF facilitates bacterially programmed cell death in E. coli and Myxococcus xanthus which is mechanistically distinct from that in eukaryotic cells (3, 26, 33). Although MazF toxins in two pathogens (Mycobacterium tuberculosis and methicillin-resistant Staphylococcus aureus [MRSA]) are also sequence-specific endoribonucleases, they have 5-nucleotide (nt) recognition sequences which are proposed to selectively target distinct mRNA transcripts for degradation (54, 55).
Although CDIs cause an estimated 15,000 to 20,000 deaths per year in the United States (43) and C. difficile treatment results in the expenditure of billions of health care dollars annually, our understanding of the spectrum of molecular mechanisms (apart from its exotoxins) that contribute to its pathogenicity, virulence, and ability to survive antibiotic treatment are not known. To this end, we studied the characteristics of the MazEF-cd TA system in C. difficile. As in Mycobacterium tuberculosis and Staphylococcus aureus, the MazF-cd toxin appears to selectively target a subset of transcripts for degradation by the recognition of a 5-nt consensus sequence followed by cleavage. Interestingly, this UACAU consensus sequence was significantly underrepresented in mRNAs for toxin B, the cell wall protein CwpV, and two proteases (Lon and ClpC) which may play a role in MazE-cd antitoxin degradation.
MATERIALS AND METHODS
Strains, plasmids, and reagents.
The E. coli strains BL21(DE3) [F− ompT hsdSβ(rβ-mβ) dcm gal(DE3) tonA] (Novagen) and BW25113 (lacIq rrnBT14 Δlac-ZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) were used for all protein expression and toxicity studies. E. coli K-12 Mach1 T1 cells [ΔrecA1398 endA1 tonA Φ80ΔlacM15 ΔlacX74 hsdR(rk+ mk+); Invitrogen] were used for all cloning experiments. CD3461 (mazF-cd) and CD3462 (mazE-cd) genes were PCR amplified from C. difficile 630 genomic DNA (ATCC BAA-1382D-5). mazF-cd was cloned into pBAD33 (17) with NdeI/HindIII ends to create pBAD33-mazF-cd. mazE-cd was cloned into pINIII using NdeI/BamHI ends to create pINIII-mazE-cd. mazF-cd and mazE-cd were also cloned into pColdTF-FT using NdeI/HindIII or NdeI/BamHI, respectively, to create pColdTF-FT-mazF-cd and pColdTF-FT-mazE-cd for expression. pColdTF-FT is a derivative of pColdTF (TaKaRa Bio) and was created by inserting an in-frame flag epitope tag before the NdeI sequence of the multiple cloning site; this plasmid was a generous gift from Jared Sharp. Cultured E. coli was grown in M9 minimal media supplemented with either 0.2% glucose or 0.21% glycerol at 37°C unless otherwise noted. The working concentrations of ampicillin and chloramphenicol were 100 and 34 μg/ml, respectively. The DNA sequences of PCR fragments used for cloning were confirmed by automated DNA sequence analysis. All oligonucleotides used in this study are listed in Table 1.
Table 1.
Oligonucleotides used in this work
| Primer name | Sequence |
|---|---|
| B4 | 5′-GGTCCGTCCCACCGAAGAAC-3′ |
| C | 5′-GACACGAACGTTTTACGAAG-3′ |
| D2 | 5′-TCTCTATTTATCTGACCGCG-3′ |
| E1 | 5′-TGCATTGCCTTAACAATAAG-3′ |
| G | 5′-GAGCCGTTGCCTGATTAATG-3′ |
| H | 5′-AGCGTCAACGCTTATGATGG-3′ |
| J1 | 5′-GACGGCCATCTAACTTGATG-3′ |
| K | 5′-GCGGAGCGCCTGGCGCC-3′ |
| 1351 | 5′-GATCGATTGATCATTCAGG-3′ |
| ompF | 5′-CCCACAGCAACGGTGTCGTC-3′ |
| tufA-1 | 5′-CTTCCATTTCAACCAGTTCC-3′ |
| tufA-2 | 5′-CCGGCATTACCATCTCTACG-3′ |
| NWO1845 | 5′-GCAACCTAGCTTCTCTAATATAGTAAGATTAGTTTTATAATC-3′ |
| NWO1760 | 5′-CGCGGATCCATGGAAATATGTAGTTCTAATAGCATAAGAAATATGG-3′ |
| NWO1848 | 5′-GGTTTATAAATTAGGATTTTTAAATTACAAGAGCTTTGAATATTTAGGC-3′ |
| NWO1849 | 5′-GTTCCTGTCATTCCTTCGTTTAACTTAATAGATAATTCAC-3′ |
| NWO1861 | 5′-CTTGGCTCTGATTTCTTCGCTTGCTCTGTTTTAGACG-3′ |
| NWO1862 | 5′-CGTTGTTTTTTAACCAACATAAGTGAGTGTTATTTCCG-3′ |
Transformation and expression in C. perfringens.
The pCM14 plasmid was created from a pCM7 backbone, a derivative of pCM3 (GenBank accession no. AB562893). pCM3 contains origins of replication for E. coli and C. perfringens, the cat (chloramphenicol acetyltransferase) gene conferring high-level chloramphenicol resistance, and a multiple cloning site. pCM7 was digested with PacI and NheI and ligated to the rrnB terminator containing compatible ends. The bgaR-bgaL promoter fragment was inserted into the AvrII and NdeI sites. The NdeI site of the bgaR gene was removed by site-directed mutagenesis. Truncated genes (such as bla and cop) were removed by mutagenesis. A similar expression plasmid, using the same promoter and terminator, was recently published (19). pCM14-mazF-cd or pCM14 (see Fig. S1 in the supplemental material) was transformed into C. perfringens strain 13 as previously described (2). Bacteria were cultured anaerobically in modified R & S (maltose) medium containing 10 μg/ml chloramphenicol at 37°C, and growth was monitored by optical density. Modified R & S (maltose) medium is based on R & S medium (14, 42). Induction was performed by the addition of lactose to 0.5%.
Preparation of recombinant MazE-cd and MazF-cd.
pColdTF-FT-mazF-cd and pColdTF-FT-mazE-cd BL21(DE3) transformants were used to inoculate 1 liter of M9 liquid medium and grown to an optical density at 600 nm (OD600) of 0.3 to 0.4. The culture was transferred to a 15°C water bath and incubated for 30 min before being induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and expressed for 18 h. Cells were subsequently disrupted by sonication, and the protein extracts were applied to nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) to purify the proteins as recommended by Qiagen. Elutions were pooled and subjected to thrombin cleavage to excise trigger factor (TF)-(His)6 from the target protein. Recombinant proteins were further purified over an anti-FLAG resin to remove TF from the flag-tagged MazF-cd. All elution fractions were visualized by 17.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue.
Determination of MazF-cd-FT activity in vitro.
MS2 RNA (1.6 μg) was added to a reaction mixture containing either MazF-cd-FT or MazE-cd-FT in 10 mM Tris-HCl, pH 7.8, for 15 min at 37°C. Increasing ratios of toxin and antitoxin were preincubated for 30 min at 4°C, followed by 5 min at room temperature before being added to MS2 RNA. The reaction products were then separated on a 1% agarose-morpholinepropanesulfonic acid (MOPS)-formaldehyde gel and visualized by ethidium bromide staining.
MS2 primer extension analysis.
The MazF-cd MS2 cleavage assays were carried out as previously described (56). Full-length MS2 RNA was partially digested with or without purified toxin protein MazF-cd-FT and with or without purified CspA(His)6 protein at 37°C for 15 min. The digestion reaction mixture (10-μl total volume) consisted of 0.8 μg of MS2 RNA substrate, 1.25 μM MazF-cd-FT, 32 μg of CspA(His)6, and 0.5 μl of RNase inhibitor (Roche) in 10 mM Tris-HCl (pH 7.8). Primer extensions were carried out at 47°C for 1 h in a 20-μl reaction mixture. The reactions were stopped by the addition of 12 μl of sequence loading buffer (95% formamide, 20 mM EDTA, 0.05% bromphenol blue, and 0.05% xylene cyanol EF). The samples were incubated at 90°C for 5 min prior to electrophoresis on a 6% polyacrylamide, 6 M urea gel. Primers were 5′-end labeled with [32P]ATP using T4 polynucleotide kinase. An IPTG-inducible pColdI-cspA plasmid was generously provided by Yoshi Yamaguchi to generate CspA(His)6 pure protein that was purified over Ni-NTA resin (Qiagen).
Analysis of steady-state mRNA levels.
Total RNA was extracted using a hot phenol method. The radiolabeled DNA fragments used for Northern analysis were derived from PCR products comprising open reading frames (ORFs) of the E. coli genes lpp (major outer membrane lipoprotein) and ompF (outermembrane porin protein F). The tufA (EF-Tu)-specific fragment was developed using primers that partially overlapped the fusA upstream gene and ended just before the beginning of the tufA ORF, allowing the detection of tufA and not tufB (note that this fragment hybridizes to two transcripts, one containing tufA and fusA and the other containing tufA, fusA rpdG, and rpsL).
In vivo primer extension.
Primer extension analysis was carried out as previously described (39). Primers used were ompF, tufA-1, and tufA-2.
Statistical analysis of UACAU frequency in the C. difficile transcripts.
We retrieved the genomic sequence of Clostridium difficile 630 from NCBI RefSeq (accession no. NC_009089), as well as plasmid pCD630 (accession no. NC_008226), and extracted all ORFs from the records. We first calculated the nucleotide composition of each ORF. The probability, p, of the cleavage motif UACAU appearing anywhere in the ORF is p = (percentage of U)2 × (percentage of A)2 × (percentage of C). Let L be the length of the ORF. The expected number, E, of the motifs in the ORF is E = p(L − 4). Let K be the actual number of the motifs in the ORF. The probability, P, of having K or fewer motifs in the ORF is
An ORF with a very small P value suggests that it has evolved to eliminate the motif from its sequence.
Target mRNA expression, purification, and reverse transcriptase PCR (RT-PCR).
C. perfringens strain 13 DNA was prepared by standard phenol-chloroform-isoamyl alcohol extraction followed by ethanol precipitation. C. perfringens CPE0292 and CPE1662 target genes were PCR amplified from C. perfringens strain 13 DNA. CPE0292 was cloned into pET21c using BamHI/HindIII ends to make pET21c-CPE1662. CPE1662 was cloned into pET21c using NdeI/BamHI ends to make pET21c-CPE0292. Target genes were expressed in E. coli BW25113 cells for 30 min prior to the induction of MazF-cd. Total RNA was extracted with Tri-Reagent, followed by ethanol precipitation. Residual DNA contamination was removed using the Turbo DNA-free kit (Ambion). The reverse transcription of the isolated RNA was performed using the Superscript III first-strand synthesis supermix kit (Invitrogen) following the manufacturer's protocol. cDNA was synthesized using 0.75 μg of total RNA and either NWO1845 to detect a single cut region of CPE0292, NWO1849 to detect a region with three cut sites in CPE0292, or NWO1862 to detect a region with no cuts within CPE1662. The PCR included a single melting step at 94°C for 45 s, cooling to either 51.9°C for NWO1845 and NWO1849 or 54.8°C for NWO1862, and then a 40-s extension at 72°C for 28 cycles. The following primer pairs were used to detect each region: NWO1845 and NWO1760, NWO1849 and NWO1848, and NWO1862 and NWO1861. All products were stained with ethidium bromide and visualized on a 1.5% agarose gel.
RESULTS
A single MazEF TA system in the genome of a pathogenic C. difficile strain.
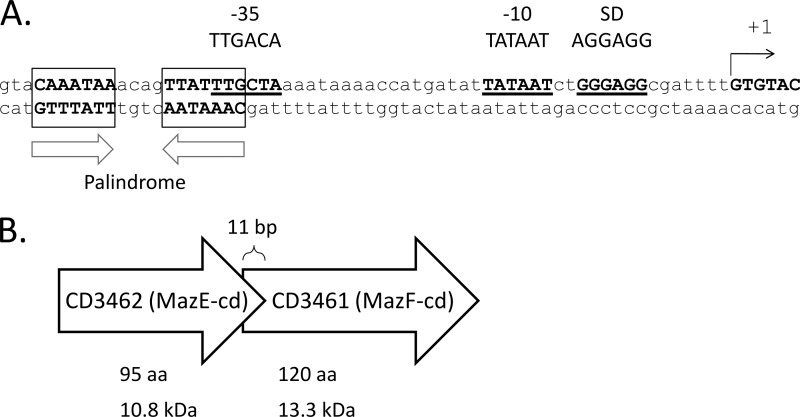
Any factor that alters the colonic flora increases the risk of C. difficile infection. Antibiotic treatment, especially broad spectrum, is the predominant risk factor for colonization that leads to C. difficile proliferation and disease (5, 38). The physiological and metabolic changes in C. difficile that underlie infection during or after antibiotic stress are not known. Since some TA systems have been linked to the establishment of the persistent state (25, 27, 50) and the expression of TA toxins in E. coli appears to facilitate cell survival during stress (16, 34), we initiated studies aimed at determining if TA systems contribute to C. difficile colonization and/or infection. We analyzed the genome of the C. difficile 630 (epidemic type X) strain for the presence of genes encoding orthologs of the following cognate antitoxin-toxin pairs using the Blastp algorithm: MazEF, ChpBIK, YefM-YoeB, DinJ-YafQ, RelBE, HipBA, HigBA, VapBC, YafNO, HicAB, PrlF-YhaV, and MqsR-YgiT (16, 23, 31, 34, 46, 51). Genes encoding candidate toxin and antitoxin orthologs were then analyzed for characteristic features of TA modules. Only one apparent TA system was identified whose genes encoded proteins with size and sequence similarity to E. coli MazE antitoxin (13% identity, 24% similarity; 10.8 kDa) and MazF toxin (21% identity, 36% similarity; 13.3 kDa); they were designated MazE-cd and MazF-cd, respectively. Characteristic genomic features of TA modules that were also exhibited by mazEF-cd are summarized in Fig. 1. The antitoxin and toxin genes were adjacent, the antitoxin gene preceded the toxin gene, the open reading frames overlapped (11 bp in this case), and a palindrome in the promoter region (used for module autoregulation) was identified next to the predicted −10 and −35 promoter sequences.
Fig 1.
Features of the MazEF-cd TA module. (A) The palindrome is boxed, with arrows below indicating directions. It overlaps the predicted sigma70 promoter region, designated −10 and −35, with consensus sequences indicated. The predicted Shine-Dalgarno sequence is present 8 bases upstream of the MazE-cd start. (B) Illustration of MazEF-cd gene orientation demonstrating the 11-bp overlap of MazF-cd by MazE-cd. Amino acid length and molecular mass are shown below. Relative lengths of arrows are not drawn to scale.
MazF-cd is toxic in E. coli and C. perfringens.
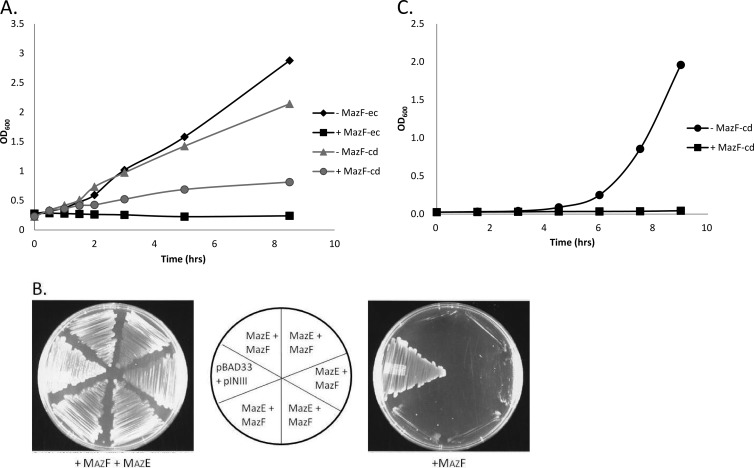
To date, all E. coli MazF counterparts exhibit the same general enzymatic activity, i.e., they function as single-stranded, sequence-specific endoribonucleases (sometimes referred to as mRNA interferases) that target mRNA for cleavage. We expressed mazF-cd in E. coli K-12 host cells using the arabinose-inducible vector pBAD33. As with E. coli MazF, the expression of MazF-cd led to cell growth arrest (i.e., stasis) (Fig. 2). However, the kinetics of arrest were markedly slower for MazF-cd. While growth arrest occurs within 15 min of induction for E. coli MazF (1), the expression of MazF-cd began to significantly slow growth after 2 h and took approximately 8 h to reach a state of growth arrest (Fig. 2A). The longer lag in E. coli cells may be due to lower levels of MazF-cd, since many C. difficile genes are difficult to express in E. coli. We next used a functional assay to demonstrate that MazE-cd acts as the cognate antitoxin to MazF-cd in vivo. MazF-cd toxicity was rescued by the coexpression of both the toxin and antitoxin gene from separate inducible plasmids (Fig. 2B). The molecular biological tools for clostridia are very limited; an inducible vector enabling the tight on-off expression of genes (especially critical when expressing toxic genes, as in our case) is not yet available. Alternatively, we created a lactose-inducible plasmid for tightly regulated, inducible toxin expression in C. perfringens. Interestingly, the induction of MazF-cd in C. perfringens resulted in an immediate effect, precluding any increase in the optical density of the toxin-expressing cells (Fig. 2C).
Fig 2.
Expression of MazF-cd inhibits growth and MazE-cd prevents toxicity. (A) Growth curve of E. coli BW25113 cells harboring the pBAD33-mazF-cd or pBAD33-mazF-ec plasmid either induced (with arabinose) or uninduced (without arabinose). Optical density was monitored beginning at time zero, the point of induction. One representative experiment (of three replicates) is shown. (B) pBAD33-mazF-cd and pINIII-mazE-cd constructs were cotransformed into BW25113 cells and streaked onto M9 minimal medium plates containing either 0.015% arabinose (+MazF) or 0.015% arabinose plus 1 mM IPTG (+MazF +MazE). The coinduction of both toxin and antitoxin permits normal growth in these cells (left); however, toxin-only expression prevents cell growth (right). The center diagram identifies the strains in each sector of the left and right plates. Five of the six sectors are individual transformants of the BW25113 strain containing both pBAD33-mazF-cd and pINIII-mazE-cd plasmids, while the sixth was a control BW25113 strain that contained empty pBAD33 and pINIII plasmids. (C) Growth curve of C. perfringens strain 13 cells harboring pCM14-mazF-cd plasmid either induced (with lactose) or uninduced (without lactose) at time zero. Optical density was monitored immediately after the addition of lactose.
MazF-cd expression leads to mRNA degradation of selected transcripts in vivo.
Since all characterized MazF toxins act by cleaving mRNA, we first sought to confirm that MazF-cd also targets mRNA. We performed Northern analysis to assess steady-state levels of three E. coli mRNAs before and after MazF-cd induction: lpp, ompF, and tufA (Fig. 3). Curiously, MazF-cd expression led to the cleavage of two of the three transcripts (ompF and tufA); lpp mRNA was not cleaved. The rate of ompF and tufA cleavage was slower and degradation was incomplete relative to that of E. coli MazF. For example, the lpp, secE, and ompA transcripts were completely degraded by E. coli MazF (with no detectable degradation intermediates) within 20 min of toxin induction (53). In contrast, MazF-cd cleavage resulted in the accumulation of one or more degradation intermediates. These results suggest that MazF-cd may have lower enzymatic efficiency than its E. coli counterpart, or that it recognizes a more complex RNA consensus sequence.
Fig 3.
Expression of MazF-cd in E. coli decreases steady-state levels of ompF and tufA mRNAs but not lpp in vivo. RNA was extracted at the time points indicated from BW25113 cells expressing MazF-cd, followed by Northern analysis. The cleavage of ompF and tufA was detected, and cleavage products are denoted by arrows. No additional products were detected below the cropped images. No cleavage products were detected for lpp. A minimal level of cleavage occurred at time zero due to the leaky expression of pBAD33-mazF-cd before induction.
MazF-cd cleaves mRNA at UACAU sequences.
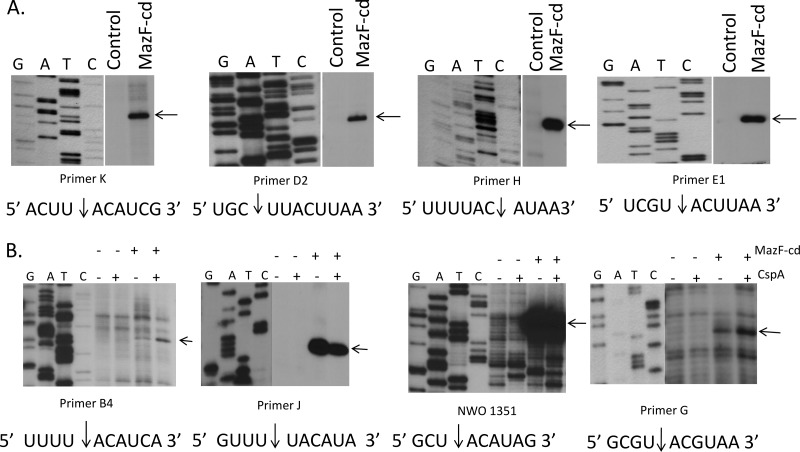
We first tested whether the slower growth inhibition and weaker RNA degradation were consequences of MazF-cd cleavage of mRNA at a longer recognition sequence relative to the short ACA consensus for E. coli MazF. To this end, we used an in vitro cleavage assay developed in the Inouye laboratory that utilizes a long, single-stranded RNA substrate (bacteriophage MS2 RNA) which enabled the determination of the recognition sequences for two M. tuberculosis MazF toxins, MazF-mt3 and MazF-mt7 (55). MS2 RNA is relatively large (∼3,500 nt) and commercially available, and it comprises a nearly equal base content (26% G, 23% A, 26% C, 25% U). A long test substrate increases the odds for the identification of target sequences but also tends to form extensive secondary structures (∼70% of the nucleotides in MS2 are estimated to be double stranded). Therefore, the major cold shock protein/RNA chaperone CspA is added at saturating levels to prevent secondary structure formation (22). The site specificity for RNA cleavage by MazF-cd was determined after performing primer extensions with oligonucleotides that enabled us to determine toxin cleavage sites along the entire 3.5-kb MS2 RNA.
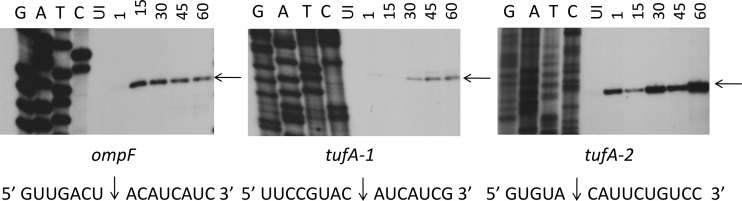
We prepared recombinant MazF-cd as a fusion to trigger factor (TF) to facilitate robust expression without any toxicity; we used the same approach for the production of recombinant MazE-cd. The TF tag was proteolytically removed after the purification of each recombinant protein to preclude functional perturbation. As a pilot experiment, we incubated MS2 RNA with the purified, recombinant MazF-cd alone or with increasing molar ratios of MazE-cd (Fig. 4). MazF-cd cleaved MS2 RNA into several discrete products (Fig. 4, lane 4). As expected, the MazE-cd antitoxin alone did not cleave RNA (Fig. 4, lane 3). Increasing molar ratios of the antitoxin were preincubated with the toxin, and no cleavage was observed with the 2:1 and 3:1 antitoxin-to-toxin ratios (Fig. 4, lanes 5 to 7). We then performed primer extensions and identified eight cleavage sites (Fig. 5) with an apparent consensus sequence of UACAU (five of the eight were UACAU and three varied at position 4, UACUU or UACGU). In vivo primer extension experiments (i.e., from RNA prepared from E. coli cells after MazF-cd induction) were next performed to assess cleavage site specificity within the two transcripts (ompF and tufA) that were degraded by MazF-cd in Fig. 3. Primers were designed for nearly complete coverage of the coding regions of each transcript (excluding the 3′ ends corresponding to regions near or annealing to the distal primers), ensuring that each of the UACAU preliminary consensus sequences, as well as variants at position 4, were detected (Fig. 6). Interestingly, we observed cleavage only at each UACAU sequence in both transcripts, the predominant cleavage site detected in vitro. The 1,185-nt tufA coding region contains no UACGU sequences but has one UACUU and two UACAU sequences; MazF-cd only cleaved at the two UACAU sequences in tufA. Similarly, the 1,089-nt ompF coding region was cleaved only at the single UACAU sequence, even though it contained two UACUU sequences. We also did not observe cleavage at either single UACCU sequence (the alternate substitution at position 4 was not detected with MS2 RNA) represented in tufA and ompF. Finally, lpp mRNA is devoid of UACAU, UACUU, UACGU, and UACCU sequences, in agreement with our Northern analyses in which no cleavage was detected (Fig. 3). Therefore, under physiological conditions, MazF-cd exhibited strict specificity for UACAU sequences; as with E. coli MazF, the cleavage recognition sequence was invariant. Regarding the position of cleavage within the 5-nt consensus, the predominant site (6 of 11) mapped between the U and A at positions 1 and 2 (U↓ACAU). Variation in cleavage position within the consensus sequence is not unusual, as E. coli MazF also exhibits cleavage before or after the first A of its short ACA consensus sequence (A↓CA or ↓ACA [52]). There is also an inherent margin of error of 1 to 2 nt on either side of the actual cleavage site when cleaved RNA product lengths are assessed according to their alignment with the adjacent DNA sequencing ladder. In summary, our data demonstrate that MazF-cd specifically cleaves mRNA at UACAU consensus sequences in vivo (Table 2).
Fig 4.
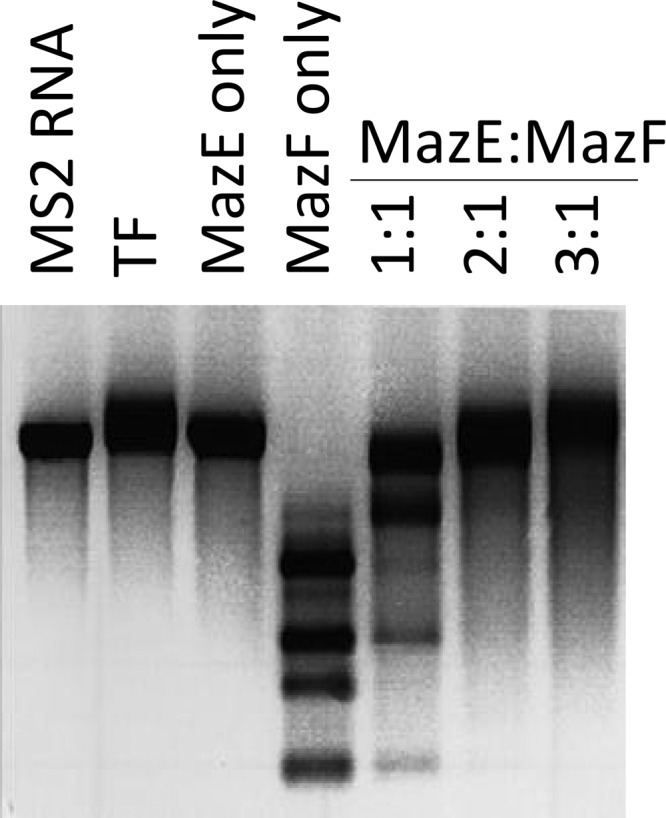
Recombinant MazF-cd cleaves MS2 mRNA, and MazE-cd prevents cleavage of MS2 mRNA. Trigger factor (TF) was used as a control for the purification of recombinant MazE-cd and MazF-cd. Products were visualized by the addition of ethidium bromide prior to electrophoresis.
Fig 5.
In vitro primer extension using an MS2 RNA template suggests a MazF-cd cleavage consensus sequence. Purified MazF-cd was first incubated with MS2 target mRNA to allow cleavage. Radiolabeled primers were annealed to the cleaved mRNA, and a reverse transcription reaction was initiated. Reverse transcriptase is able to convert the target mRNA into cDNA until it reaches a point of mRNA cleavage. The products of the transcriptional runoff are resolved on a high-resolution polyacrylamide gel, and the exact position of RNA cleavage by MazF-cd was accurately deduced by the comparison of the mobility of the primer extension product relative to the mobility of bands comprising the sequencing reactions on the MS2 RNA using the same primer and reverse transcriptase. (A) Four cleavage sites were detected when purified MazF-cd was incubated with MS2 RNA (without the addition of CspA) before performing the extension reaction. Control lanes contain MS2 RNA only, yielding a full-length extension product distant from the MazF-cd cleavage site. (B) Four additional cleavage sites were detected when purified MazF-cd was incubated with MS2 RNA and CspA before performing the extension reaction. MS2 RNA was added to all four primer extension reaction lanes. −MazF-cd/−CspA denotes that the extension reaction was performed on MS2 RNA only, −MazF-cd/+CspA denotes that CspA and MS2 RNA were incubated before the extension reaction was performed. Full-length MS2 RNA extension products were cropped from the images shown. Cleavage sites/products are designated by arrows, and the sequences flanking the MazF-cd cleavage site are shown below. DNA sequencing ladders using the same primer were loaded in the adjacent lanes.
Fig 6.
Primer extension of ompF and tufA pinpoints the MazF-cd cleavage site as UACAU. The RNA used for Northern analysis in Fig. 3 was subjected to primer extension analysis; DNA sequencing ladders using the same primer were loaded in the adjacent lanes.
Table 2.
In vitro and in vivo primer extension cleavage sites
| Cleavage site and primer name | Sequencea |
|---|---|
| In vitro | |
| G | 5′ CGCGU↓ACGUAAAG 3′ |
| K | 5′ GACUU↓ACAUCGAA 3′ |
| D2 | 5′ GC↓UUACUUAAGGG 3′ |
| H | 5′ UUUUAC↓AUAAAAC 3′ |
| E1 | 5′ UCGU↓ACUUAAAUA 3′ |
| B4 | 5′ UUUU↓ACAUCAAGA 3′ |
| 1351 | 5′ UCGCU↓ACAUAGCG 3′ |
| J | 5′ UUU↓UACAUAAACG 3′ |
| In vivo | |
| ompF | 5′ UGACU↓ACAUCAUC 3′ |
| tufA-1 | 5′ UCCGUAC↓AUCAUC 3′ |
| tufA-2 | 5′ AGUGUA↓CAUUCUG 3′ |
| Consensus | UACAU |
Boldface letters indicate conserved sequences adjacent to detected cut sites.
MazF-cd shares its pentad cleavage consensus sequence with counterparts in S. aureus and Bacillus subtilis.
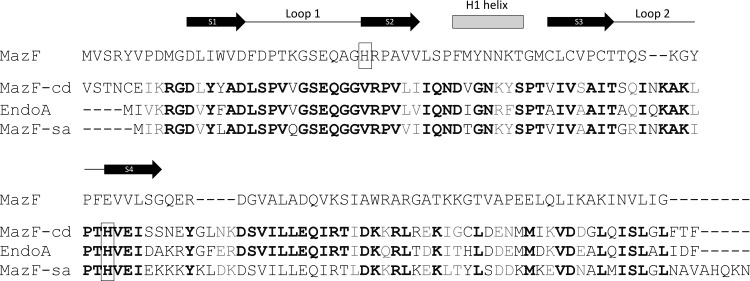
Interestingly, MazF orthologs derived from three distinct eubacteria, B. subtilis (35, 36), S. aureus (13, 54), and C. difficile, each cleave mRNA at UACAU. The single transcript tested with MazF from B. subtilis (also known as EndoA) generated the cleavage pattern UAUAAU↓ACAUA, containing the UACAU consensus (underlined) (36). Subsequent cleavage analyses for B. subtilis MazF using multiple templates revealed a clear minimal consensus of UACAU (35). The conservation in consensus recognition sequences among these three toxins provides a platform that may aid in the identification of amino acids that contribute to sequence specificity. The alignment of these three MazF toxins revealed substantial shared sequence identity that contrasts with marginal similarity to E. coli MazF (Fig. 7). Curiously, although nuclear magnetic resonance studies by Li et al. implicate E. coli MazF H28 in substrate recognition (29), which is consistent with its solvent exposure in the X-ray crystal structure of the MazE-MazF complex (24), there are no histidines at or near this position in the three UACAU-cleaving MazF toxins (Fig. 7). In other MazF orthologs without a histidine at position 28, there is one in proximity that is instead proposed to contribute to substrate recognition (e.g., H17 of Kid and H23 of ChpBK) (29). However, the three UACAU-cleaving MazF toxins have an absolutely conserved histidine (H63 in MazF-cd) that is predicted as the first residue of β-sheet S4 immediately following loop 2 (Fig. 7). It is possible that the position of the histidine influences substrate specificity, as proposed by Li et al. (29). However, they also proposed that the relatively hydrophobic site formed by the binding site created in the MazF dimer (when the S3-S4 loop 2 of one MazF monomer interacts with the H1 helix of the other) could also directly interact with the hydrophobic moieties of the RNA substrate (29). This would also be consistent with the sequence relationships shown in our alignment, since residues comprising these two regions are conserved among the three UACAU-cleaving MazF toxins but distinct from E. coli MazF.
Fig 7.
MazF-cd exhibits significant sequence similarity to counterparts in S. aureus and B. subtilis. EndoA and MazF-sa were compared to each other and to E. coli MazF. Identical amino acids are highlighted in boldface, while conserved substitutions (+1 or greater on the BLOSUM62 alignment score matrix) are highlighted in gray. Histidines believed to be involved in sequence recognition are boxed. Only the secondary structure motifs relevant to this work are illustrated above the respective amino acids of E. coli MazF. Black arrows, β-sheets; gray rectangle, α-helix; black line, unstructured loops.
MazF-cd cleavage sites are significantly underrepresented in toxin B and the cell wall surface protein CwpV.
C. difficile strain 630 contains 3,776 predicted open reading frames (ORFs) (48). We analyzed the overall nucleotide content of the ORFs, determined the probability of the occurrence of the UACAU consensus of each ORF (Table 3, Expected), and compared it to the actual occurrence (Table 3, Actual) (described in Materials and Methods). The top 20 proteins whose mRNAs contain more UACAU motifs than predicted (with P values close to 0) are listed in Table S1 in the supplemental material. These proteins are predicted to have shorter half-lives under conditions where the MazF-cd toxin is activated in vivo. None of these proteins have an obvious role in pathogenesis, nor was there any apparent bias for proteins involved in one or more cellular processes. However, the analysis of transcripts with fewer UACAU motifs than predicted (Table 3) revealed a protein with a prominent role in C. difficile virulence (toxin B) and the aggregation-promoting cell surface protein CwpV with sequence similarity to known C. difficile adhesins SlpA and Cwp66 (9, 41). Toxin B appears to be an important determinant of C. difficile virulence (28, 30). Interestingly, toxin A mRNA contains 12 predicted cleavage sites, suggesting a differential rate of cleavage between these two important virulence factors. The precise role of CwpV is not yet clear; however, it constitutes a significant proportion of cell surface proteins, it promotes autoaggregation that may influence the formation of biofilm-like aggregates in infected mice (41), and its expression is phase variable (9). Also of note, the protease Lon as well as ClpC (the ATPase component of the Clp proteolytic complex) contain no UACAU motifs (Table 3). This implicates these proteases in the regulation of MazF-cd activation, since the degradation of MazE-cd would result in excess toxin. In fact, Lon and Clp family members have been frequently linked with antitoxin degradation in E. coli (16).
Table 3.
Top mRNA transcripts less susceptible to cleavage by MazF-cd
| Locus | Length (nt) | Expected | Actual | Probability | Gene | Synonym | Product |
|---|---|---|---|---|---|---|---|
| CD0423 | 8,724 | 11.52 | 2 | 0.000779381 | Putative helicase | ||
| CD1862 | 9,036 | 12.45 | 4 | 0.005514112 | Putative conjugative transposon DNA recombination protein | ||
| CD0660 | 7,101 | 12.23 | 4 | 0.006431016 | tcdB | toxB | Toxin B |
| CD0514 | 4,869 | 6.78 | 1 | 0.008796151 | cwpV | Cell surface protein (aggregation promoting factor) (41) | |
| CD0204 | 2,235 | 4.24 | 0 | 0.014369553 | Putative signaling protein | ||
| CD1856 | 2,430 | 3.76 | 0 | 0.023300809 | Putative conjugal transfer protein | ||
| CD1657 | 2,475 | 3.70 | 0 | 0.024543046 | Putative bifunctional glycine dehydrogenase/aminomethyl transferase protein | ||
| CD1120 | 2,373 | 3.62 | 0 | 0.026575548 | dhaB1 | Glycerol dehydratase | |
| CD1366 | 2,454 | 3.55 | 0 | 0.028631025 | Hypothetical protein | ||
| CD0313 | 2,388 | 3.48 | 0 | 0.03059477 | Putative heavy metal-transporting ATPase | ||
| CD0006 | 2,427 | 3.47 | 0 | 0.031170373 | gyrA | DNA gyrase subunit A | |
| CD0026 | 2,448 | 3.41 | 0 | 0.032943761 | clpC | mecB | ATP-dependent Clp protease |
| CD1769 | 4,254 | 6.85 | 2 | 0.033039303 | Hypothetical protein | ||
| CD3301 | 2,370 | 3.40 | 0 | 0.033136288 | lon | ATP-dependent protease La | |
| CD1105 | 4,032 | 3.38 | 0 | 0.034062837 | Putative DNA primase | ||
| CD0418 | 2,394 | 3.37 | 0 | 0.034245223 | Putative conjugal transfer protein | ||
| CD1403 | 2,190 | 3.35 | 0 | 0.035123302 | Putative nonribosomal peptide synthetase | ||
| CD3246 | 2,220 | 3.27 | 0 | 0.037784489 | Putative surface protein group II intron reverse | ||
| CD0506 | 1,830 | 3.21 | 0 | 0.04006084 | Transcriptase/maturase | ||
| CD1380 | 2,505 | 4.97 | 1 | 0.041450174 | Putative transporter |
Finally, the UACAU motif is not represented in transcripts for 1,303 C. difficile proteins, comprising approximately one-third of all C. difficile transcripts. We investigated the statistical significance of this finding. First, the average length of the ORFs that do not have UACAU is 631 nt, and the average length of ORFs having at least one UACAU is almost twice that, 1,108 nt. Therefore, the absence of UACAU in the 1,303 C. difficile ORFs is due to the shorter relative ORF length of this subset. Second, we found that essentially the same number of ORFs (1,305) lacked an AUCUA test motif with the same base composition as but different order from our real motif. Therefore, the absence of UACAU in a third of the C. difficile ORFs is not unexpected given the base composition and length. However, unlike the ACA-cleaving E. coli MazF, MazF-cd is predicted to target selected transcripts because it has a longer recognition sequence.
mRNA transcript stability upon MazF-cd induction is dependent on the number of UACAU sequences.
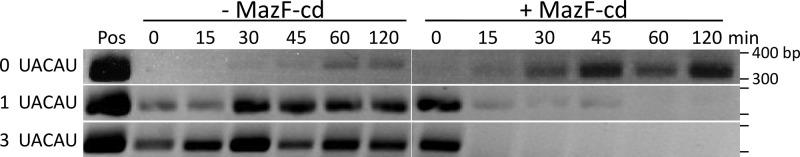
To establish if there was a relationship between the number of toxin cleavage sites and transcript stability, we induced the expression of target mRNAs in E. coli and estimated their steady-state levels after the subsequent induction of MazF-cd using reverse transcriptase PCR (RT-PCR). Two target transcripts were selected for analysis: CPE0292, a carbohydrate kinase family gene, and CPE1662, a Ppx/GppA phosphatase family gene. We chose C. perfringens genes for targets based on a similar bioinformatic analysis of C. perfringens transcripts for MazF-cd susceptibility to that previously described. These two genes were chosen for their differing numbers of cuts within equal lengths (both coding regions are approximately 1,500 nt). Each C. perfringens gene was cloned into a pET expression vector, and its expression was induced in E. coli for 30 min, followed by the induction of MazF-cd for 15, 30, 45, 60, or 120 min. We used gene-specific primers to examine two 300-nt regions of CPE0292, one with a single UACAU and the other with three UACAU sequences (Fig. 8). The relative stability of these regions was estimated by RT-PCR of total RNA harvested. The region containing a single UACAU site was detectable up to 45 min after MazF-cd induction. In contrast, the region with three UACAU sites was completely degraded within 15 min of MazF-cd induction. As a control, we monitored a 300-base region of CPE1662 mRNA with no cleavage sites. No degradation of this control mRNA was observed; instead, the steady-state level increased with time (as expected, since the target gene was continually expressed). Curiously, we detected significantly lower steady-state levels of CPE1662 mRNA in the uninduced controls than in the MazF-cd-induced samples. We hypothesize that the transcription of this gene is naturally low (many C. perfringens genes do not express well in E. coli). However, upon the induction of MazF-cd, this transcript may be more abundant, since other cellular mechanisms that control its transcription or stability are perturbed in the presence of MazF-cd. For example, an RNase that normally regulates CPE1662 levels may be preferentially disabled due to the cleavage of its transcript by MazF-cd.
Fig 8.
Degradation of mRNA is directly related to cleavage site number. Target mRNAs were induced in E. coli for 30 min prior to MazF-cd induction for the times shown. Total RNA was extracted from cells harvested at each time point, and mRNA stability was monitored by RT-PCR. Data are representative of at least two independent experiments. A discussion of the low mRNA levels in the uninduced CPE1662 relative to the induced lanes is presented in Results.
Consistent with the results shown in Fig. 8, the stability of three endogenous E. coli transcripts (lpp, ompF, and tufA) assessed by Northern analysis of total RNA harvested from MazF-cd-induced cells (Fig. 3) was also dependent on the number of UACAU sequences. lpp transcripts contained no cleavage consensus sites and were stable throughout the induction period. ompF transcripts (containing a single cleavage site) were less stable; a very low level of mRNA is detectable at 60 min postinduction. tufA, a transcript with two cleavage sites, was completely undetectable 15 min after MazF-cd induction. Therefore, our results demonstrate that there is a direct correlation between the number of cleavage recognition sites within a given transcript and its susceptibility to degradation by MazF-cd.
DISCUSSION
The pathogenic C. difficile 630 strain contains only one recognizable TA system, MazEF-cd. The body of data derived from the study of chromosomal TA systems in E. coli suggests that they represent an evolutionary adaptation designed to survive relatively short pulses of stress by reversibly controlling growth rate. TA toxin-mediated growth arrest is intended to be transient in E. coli; cells eventually die when stress is prolonged (4, 11, 44). Thus, MazF-mediated cell death is often referred to as bacterial programmed cell death, although the molecular features are markedly distinct from those of mammalian cells (3, 26). E. coli MazF toxin cleaves mRNA at a short and abundant sequence (ACA). In the case of MazF-cd, the recognition sequence is more complex, yet the effect on growth appears to be analogous when the toxin is expressed in an E. coli host. Physiological triggers of toxin activation leading to growth arrest for the array of TA toxins are largely uncharacterized with the exception of E. coli MazF, which has been shown to be activated by thymine starvation, oxidative stress, high temperatures, DNA damage, and the antibiotics chloramphenicol, rifampin, and spectinomycin (20, 44, 45). In addition, gene profiling studies of ampicillin-treated E. coli cells revealed an increase in MazEF transcripts with time of exposure, suggesting toxin activation (25).
E. coli TA systems have also been linked to the establishment of the persistent state upon antibiotic exposure. The HipA TA toxin was discovered as a mutant allele that increases the recovery of persisters at a 1,000-fold higher level than wild-type cells (32). The ΔhipBA strain exhibits a 10- to 100-fold decrease in persisters (25), while HipA overexpression causes a 10- to 10,000-fold increase in persisters (12, 27, 50). Similarly, RelE or MazF overexpression increases the level of surviving persisters 10- to 10,000-fold (25) or 100- to 10,000-fold (50), respectively. The deletion of the gene encoding the YafQ TA toxin reduces cell survival in E. coli biofilms treated with bactericidal antibiotics; the overproduction of YafQ in E. coli makes biofilm cells more resistant to antibiotics (18).
Although the physiological triggers and role of MazF-cd are still unclear, extensive studies on E. coli TA systems provide a framework to hypothesize possible roles for MazF-cd in its natural host. First, since the expression of MazF-cd in E. coli and C. perfringens resulted in growth arrest, and since this state has been linked to survival during relatively short periods of stress, MazF-cd may be important in (i) aiding survival during colonization when the inciting antibiotic is still present and/or (ii) facilitating persistence that leads to recurrent infections.
Two key features of TA systems underlie toxin activation: coregulation at the transcriptional level and antitoxin instability. More specifically, toxin and antitoxin genes are adjacent and coregulated in their own operon. Toxin activation is the result of a dynamic process that exploits the short half-life of the intrinsically unfolded cognate antitoxin (6, 7, 15) that is consequently readily susceptible to degradation by cellular proteases. Therefore, an increase in transcript levels of toxin alone or both antitoxin and toxin are hallmarks of physiological toxin activation, because each scenario results in a net increase in toxin levels.
We mined the data sets from two published C. difficile microarray studies to determine if they supported a role for MazF-cd in persistence and/or if any apparent toxin activation could be documented under the conditions used. In the first study, Fairweather and colleagues tested the effects of clindamycin or the β-lactam amoxicillin (at subinhibitory concentrations to preclude cell death) on global gene expression (10). They also analyzed the gene expression profile of C. difficile cells exposed to a subinhibitory concentration of metronidazole, the antibiotic traditionally enlisted as first-line therapy for CDI. The effects of other stress conditions on global gene expression were also analyzed, allowing the comparison of the classes of genes whose transcript levels increase upon exposure to each type of stress (10). In this data set, both toxin and antitoxin mRNA levels increased by 25% or more with clindamycin and amoxicillin exposure (Table 4). However, only the MazE-cd antitoxin mRNA levels increased in metronidazole-treated C. difficile cells. In contrast to the antibiotic stresses imparted, there was no significant increase (i.e., relative levels were below the 25% increase threshold) in MazEF-cd upon heat, acid, alkali, and aerobic shock (Table 4). The significance of these data is unclear, because (i) the increase in steady-state toxin and antitoxin transcript levels was marginal (perhaps due to the subinhibitory concentration of antibiotic used), (ii) the antitoxin/toxin ratio (1.3 to 1.4) favored the antitoxin with each of the three antibiotics, and (iii) RNA was also prepared from mid-logarithmic-phase C. difficile cells, conditions where the transcription of the pathogenicity locus (PaLoc; comprising genes for toxins A and B, a putative holin, and two regulatory elements) is repressed by CodY. Consequently, the inherently low level of toxin B mRNA prevented us from seeing any direct effect of MazF-cd on its transcript levels in vivo. The levels of the CwpV transcripts were essentially unchanged with all three antibiotics. In the second study, C. difficile expression profiles were compared before and 30, 60, and 120 min after the infection of colorectal epithelial Caco-2 cells (21). There was no statistically significant trend for MazE-cd or MazF-cd gene expression; however, the upregulation of CodY was again observed, precluding toxin B expression. Therefore, neither study used conditions that supported a notable trend regarding TA toxin activation.
Table 4.
mazE-cd and mazF-cd transcripts are upregulated under antibiotic stressa
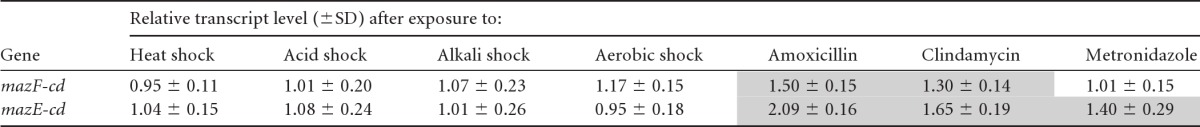
Shaded values are transcripts upregulated by 25% or more and are adapted from data reported by Emerson et al. (10). Antibiotic concentrations used were 1 mg/ml amoxicillin, 50 mg/ml clindamycin, and 0.15 mg/ml metronidazole. Values shown are averages from duplicate microarray experiments.
A battery of primer extension experiments led to the identification of a clear consensus sequence for MazF-cd cleavage, UACAU. Curiously, the MazF counterparts in B. subtilis and S. aureus possess the same recognition sequence (13, 54). This longer recognition is predicted to result in more selective mRNA degradation than that which occurs with ACA-cleaving E. coli MazF. In fact, in Fig. 8 we demonstrated that the number of UACAU sequences dictates mRNA stability in the presence of MazF-cd. mRNAs with no UACAU sequences were resistant to MazF-cd degradation; as the number of UACAU sequences increased, so did the relative rate of transcript degradation. These proof-of-principle experiments enabled us to reliably apply bioinformatics approaches to predict which C. difficile transcripts would be preferentially protected or degraded upon toxin activation. Nearly one-third of the C. difficile ORFs (encoding proteins possessing a spectrum of functions) did not contain an UACAU sequence and are consequently protected from MazF cleavage, including the CodY global repressor. We did not detect a common theme among the functions of proteins whose transcripts have more UACAU motifs than expected (see Table S1 in the supplemental material), nor did any protein stand out as having an important role in pathogenesis. In fact, the majority of hits (17/20) were annotated as hypothetical proteins or putative counterparts of known bacterial proteins.
However, some of the top statistical hits for the protein candidates predicted to have transcripts that are resistant or less susceptible to MazF-cd cleavage have clear roles in C. difficile pathogenesis (Table 3). Toxin B, whose gene in the C. difficile PaLoc is a major determinant of a toxigenic C. difficile strain, is third on the list. Although the toxin B transcript contains four cleavage sites, its relative degradation rate is predicted to be much slower than that of toxin A (which contains 12). This difference is very provocative and may represent a novel posttranscriptional method for the regulation of toxin A and B protein production. The cell wall protein CwpV is fourth on the list (41). Fairweather and colleagues have demonstrated that CwpV is important for agglutination and hypothesize that CwpV is important for biofilm-like formation or adhesion (41). Expression of CwpV is phase variable, and only 0.1 to 10% of C. difficile cells express this protein on the cell surface under standard laboratory conditions (9, 41). Phase variation in this case is controlled by a novel DNA inversion event mediated by the site-specific recombinase RecV and inverted repeats upstream of the gene encoding CwpV, such that transcription is off in one orientation and on in the other (9, 41). The most widely accepted theory for the enlistment of phase variation by pathogens is to evade the host immune system (49). Curiously, the first and second hits in Table 3 are annotated as a putative helicase and a putative DNA recombination protein, respectively. Although a recombinase that facilitates CwpV phase variation has been identified (9), these first two hits may have a role in controlling other, as-yet unknown genes, that may also be turned on and off through phase variation and play important roles in C. difficile cell survival or virulence.
Future gene profiling experiments using cells in late-exponential/early stationary phase under nutrient-limiting conditions or from an expanded panel of antibiotics at higher than subinhibitory concentrations should provide useful insights into whether the MazEF-cd TA module is upregulated by selected antibiotics and/or other triggers. If so, we should be able to better understand the physiological role of MazF-cd and assess whether the statistical predictions regarding transcript stability upon toxin activation are supported in vivo.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ling Zhu for providing reagents and assistance with MS2 primer extension analysis and Masayori Inouye for helpful discussions and expert advice.
This work was supported in part by National Institutes of Health T32 training grant AI07403, Virus-Host Interactions in Eukaryotic Cells, from the NIAID (to F.P.R. and J.M.H., awarded to G. Brewer).
Footnotes
Published ahead of print 27 April 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Aizenman E, Engelberg-Kulka H, Glaser G. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. U. S. A. 93:6059–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allen SP, Blaschek HP. 1990. Factors involved in the electroporation-induced transformation of Clostridium perfringens. FEMS Microbiol. Lett. 58:217–220 [DOI] [PubMed] [Google Scholar]
- 3. Amitai S, Kolodkin-Gal I, Hananya-Meltabashi M, Sacher A, Engelberg-Kulka H. 2009. Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins.” PLoS Genet. 5:e1000390 doi:10.1371/journal.pgen.1000390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amitai S, Yassin Y, Engelberg-Kulka H. 2004. MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 186:8295–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bignardi GE. 1998. Risk factors for Clostridium difficile infection. J. Hosp. Infect. 40:1–15 [DOI] [PubMed] [Google Scholar]
- 6. Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R. 2005. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 30:672–679 [DOI] [PubMed] [Google Scholar]
- 7. De Jonge N, et al. 2009. Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol. Cell 35:154–163 [DOI] [PubMed] [Google Scholar]
- 8. Dodson AP, Borriello SP. 1996. Clostridium difficile infection of the gut. J. Clin. Pathol. 49:529–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emerson JE, et al. 2009. A novel genetic switch controls phase variable expression of CwpV, a Clostridium difficile cell wall protein. Mol. Microbiol. 74:541–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Emerson JE, Stabler RA, Wren BW, Fairweather NF. 2008. Microarray analysis of the transcriptional responses of Clostridium difficile to environmental and antibiotic stress. J. Med. Microbiol. 57:757–764 [DOI] [PubMed] [Google Scholar]
- 11. Engelberg-Kulka H, Glaser G. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43–70 [DOI] [PubMed] [Google Scholar]
- 12. Falla TJ, Chopra I. 1998. Joint tolerance to beta-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA. Antimicrob. Agents Chemother. 42:3282–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fu Z, Donegan NP, Memmi G, Cheung AL. 2007. Characterization of MazFSa, an endoribonuclease from Staphylococcus aureus. J. Bacteriol. 189:8871–8879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fuchs AR, Bonde GJ. 1957. The nutritional requirements of Clostridium perfringens. J. Gen. Microbiol. 16:317–329 [DOI] [PubMed] [Google Scholar]
- 15. Garcia-Pino A, et al. 2010. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell 142:101–111 [DOI] [PubMed] [Google Scholar]
- 16. Gerdes K, Christensen SK, Lobner-Olesen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371–382 [DOI] [PubMed] [Google Scholar]
- 17. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harrison JJ, et al. 2009. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agents Chemother. 53:2253–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hartman AH, Liu H, Melville SB. 2011. Construction and characterization of a lactose-inducible promoter system for controlled gene expression in Clostridium perfringens. Appl. Environ. Microbiol. 77:471–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hazan R, Sat B, Engelberg-Kulka H. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186:3663–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Janvilisri T, Scaria J, Chang YF. 2010. Transcriptional profiling of Clostridium difficile and Caco-2 cells during infection. J. Infect. Dis. 202:282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang W, Hou Y, Inouye M. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272:196–202 [DOI] [PubMed] [Google Scholar]
- 23. Jorgensen MG, Pandey DP, Jaskolska M, Gerdes K. 2009. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 191:1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamada K, Hanaoka F, Burley SK. 2003. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol. Cell 11:875–884 [DOI] [PubMed] [Google Scholar]
- 25. Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kolodkin-Gal I, Hazan R, Gaathon A, Carmeli S, Engelberg-Kulka H. 2007. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 318:652–655 [DOI] [PubMed] [Google Scholar]
- 27. Korch SB, Hill TM. 2006. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J. Bacteriol. 188:3826–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuehne SA, et al. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713 [DOI] [PubMed] [Google Scholar]
- 29. Li GY, et al. 2006. Characterization of dual substrate binding sites in the homodimeric structure of Escherichia coli mRNA interferase MazF. J. Mol. Biol. 357:139–150 [DOI] [PubMed] [Google Scholar]
- 30. Lyras D, et al. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Makarova KS, Grishin NV, Koonin EV. 2006. The HicAB cassette, a putative novel, RNA-targeting toxin-antitoxin system in archaea and bacteria. Bioinformatics 22:2581–2584 [DOI] [PubMed] [Google Scholar]
- 32. Moyed HS, Bertrand KP. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155:768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nariya H, Inouye M. 2008. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 132:55–66 [DOI] [PubMed] [Google Scholar]
- 34. Pandey DP, Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park JH, Yamaguchi Y, Inouye M. 2011. Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett. 585:2526–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pellegrini O, Mathy N, Gogos A, Shapiro L, Condon C. 2005. The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Mol. Microbiol. 56:1139–1148 [DOI] [PubMed] [Google Scholar]
- 37. Poutanen SM, Simor AE. 2004. Clostridium difficile-associated diarrhea in adults. CMAJ 171:51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Privitera G, et al. 1991. Prospective study of Clostridium difficile intestinal colonization and disease following single-dose antibiotic prophylaxis in surgery. Antimicrob. Agents Chemother. 35:208–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Prysak MH, et al. 2009. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol. Microbiol. 71:1071–1087 [DOI] [PubMed] [Google Scholar]
- 40. Ramage HR, Connolly LE, Cox JS. 2009. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 5:e1000767 doi:10.1371/journal.pgen.1000767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reynolds CB, Emerson JE, de la Riva L, Fagan RP, Fairweather NF. 2011. The Clostridium difficile cell wall protein CwpV is antigenically variable between strains, but exhibits conserved aggregation-promoting function. PLoS Pathog. 7:e1002024 doi:10.1371/journal.ppat.1002024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Riha WE, Jr, Solberg M. 1971. Chemically defined medium for the growth of Clostridium perfringens. Appl. Microbiol. 22:738–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 [DOI] [PubMed] [Google Scholar]
- 44. Sat B, et al. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 183:2041–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sat B, Reches M, Engelberg-Kulka H. 2003. The Escherichia coli mazEF suicide module mediates thymineless death. J. Bacteriol. 185:1803–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schmidt O, et al. 2007. prlF and yhaV encode a new toxin-antitoxin system in Escherichia coli. J. Mol. Biol. 372:894–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schwan C, et al. 2009. Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog. 5:e1000626 doi:10.1371/journal.ppat.1000626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sebaihia M, et al. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779–786 [DOI] [PubMed] [Google Scholar]
- 49. van der Woude MW, Baumler AJ. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vazquez-Laslop N, Lee H, Neyfakh AA. 2006. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J. Bacteriol. 188:3494–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yamaguchi Y, Park JH, Inouye M. 2009. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J. Biol. Chem. 284:28746–28753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y, Zhang J, Hara H, Kato I, Inouye M. 2005. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 280:3143–3150 [DOI] [PubMed] [Google Scholar]
- 53. Zhang Y, et al. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12:913–923 [DOI] [PubMed] [Google Scholar]
- 54. Zhu L, et al. 2009. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J. Bacteriol. 191:3248–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhu L, et al. 2008. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 69:559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhu L, et al. 2006. Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 281:18638–18643 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.